Abstract
Lipopolysaccharide (LPS) and lipoprotein, two essential components of the outer membrane (OM) in Gram-negative bacteria, play critical roles in bacterial physiology and pathogenicity. LPS translocation to the OM is mediated by LptDE, yet how lipoproteins sort to the cell surface remains elusive. Here, we identify candidate lipoproteins that may be transported to the cell surface via LptDE. Notably, we determine the crystal structures of LptDE from Pseudomonas aeruginosa and its complex with an endogenous Escherichia coli lipoprotein LptM. The paLptDE-LptM structure demonstrates that LptM may translocate to the OM via LptDE, in a manner similar to LPS transport. The β-barrel domain serves as a passage for the proteinaceous moiety while its acyl chains are transported outside. Our finding has been corroborated by results from native mass spectrometry, immunofluorescence, and photocrosslinking assays, revealing a potential surface exposed lipoproteins (SLPs) transport mechanism through which lipoproteins are loaded into LptA by LolCDE prior to assembly of the LptB2FGCADE complex. These observations provide initial evidence of functional overlap between the Lpt and Lol pathways, potentially broadening current perspectives on lipoprotein sorting.
Similar content being viewed by others
Introduction
Gram-negative bacteria feature a double-membraned cell envelope, of which the outer membrane (OM) serves as a selective permeability barrier. It shields the cells against a wide variety of harmful chemicals while allowing efficient exchange of nutrients and wastes in and out of the cells1,2,3. Structurally, the OM is an asymmetric lipid bilayer with lipopolysaccharides (LPS) enriched in its outer leaflet and phospholipids in its inner leaflet. Additionally, the OM harbors two types of proteins, namely integral outer membrane proteins (OMPs) and lipoproteins. The OMPs are almost exclusively β-barrel structured and effectuate essential functions including OM biogenesis, secretion and efflux2,4,5 (Supplementary Fig. 1A). The lipoproteins are attached to the membrane via its N-terminal lipid tails. Although the globular domains of most OM lipoproteins reside within the periplasmic space, a subset can be detected by probes outside of the cell and are accordingly termed “surface exposed lipoproteins” (SLPs). Apart from certain capsular polysaccharides, LPS and SLPs are the two major lipid-containing structural elements of the OM that are exposed to the extracellular milieu (Supplementary Fig. 1A, B), where they can be recognized by Toll-like receptors (TLRs) 4 and 2, respectively, on host cells, eliciting an inflammatory response6,7,8.
Synthesized in the cytoplasm and processed in the inner membrane (IM), both LPS and SLPs are amphipathic molecules. In Gram-negative bacteria, trafficking of these molecules across the aqueous periplasm to the OM is carried out by two distinct pathways, namely, the lipopolysaccharide transport (Lpt) and the localization of lipoprotein (Lol) pathways (Supplementary Fig. 1A). The Lpt pathway comprises seven essential Lpt proteins (LptA-G)9,10. Among them the heteromeric ATP-binding cassette (ABC) transporter LptB2FG extracts LPS from the outer leaflet of the IM, and transfers it to the membrane-bound protein LptC. The periplasmic protein LptA connects the LptB2FGC complex with the LptDE complex in the OM9,10,11,12,13,14,15,16,17,18,19. Release of the LPS from LptDE is facilitated via lateral opening at the first and last β strands of the LptD β-barrel12,20,21,22. Efficient transport of LPS to the cell surface thus requires assembly of a transenvelope complex that consists of all seven Lpt proteins and utilizes the energy derived from the ABC transporter LptB2FG to drive the unidirectional LPS export10,23,24.
In contrast, lipoprotein sorting to the OM via the classical Lol pathway does not involve the assembly of a transenvelope complex. Rather, a periplasmic protein LolA ferries nascent lipoproteins, extracted from the IM by the ABC transporter LolCDE, across the periplasmic space to the OM-attached lipoprotein LolB. LolB receives the lipoproteins from LolA and inserts them to the inner leaflet of the OM25,26,27,28 (Supplementary Fig. 1A). Despite the current state of knowledge that lipoproteins are retained on the periplasmic sides of either the OM or the IM, a plethora of lipoproteins has been found to be surface-exposed in Gram-negative bacteria26,28,29,30,31,32. However, how these SLPs reach the cell surface remains enigmatic and posts new questions in understanding lipoprotein transport mechanisms33.
Yang et al. reported that LptM (formerly YifL), a lipoprotein conserved in Enterobacteriaceae, stably and functionally interacts with LptDE in Escherichia coli and is involved in the oxidative maturation of LptD34. Here, we determine the crystal structures of LptDE from Pseudomonas aeruginosa (paLptDE) and its complex with an endogenous Escherichia coli (E. coli) lipoprotein LptM. By combining structural, biochemical, and mass spectrometric approaches, we identify multiple lipoprotein candidates that may be transported to the cell surface via LptDE. This finding aligns with the work of He et al., who reported that the LptD homolog BB0838 may function as a surface lipoproteins flippase in the LPS-deficient Borrelia burgdorferi35. Our findings suggest a potential mechanism in which LptA of the Lpt pathway may mediate transfer of some SLPs (notably LptM) from LolCDE to LptDE, potentially facilitating their cell surface exposure.
Results
Identification of a subset of lipoproteins that interact with the LptDE complex
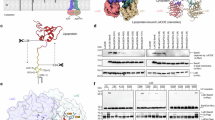
During purification of LptDE proteins, we noticed that certain endogenous proteins invariably co-purify with LptDE complexes from both Pseudomonas aeruginosa (paLptDE) and Shigella flexneri (sfLptDE) (Fig. 1A, B). Proteomic analysis using mass spectrometry identified the co-purified proteins as predominantly LptM and Lpp (Braun’s lipoprotein), two of the most abundant lipoproteins in E. coli29,34,36. Further analysis yielded seven additional lipoproteins that co-purify with LptDE. These proteins, present in relatively low abundance in E. coli, were detected via concentration of a low-yield paLptDE sample to ca. 20 mg ml−1 (Fig. 1B). Therefore, our initial analysis led to an inventory of 9 lipoproteins-namely LptM, Lpp, YedD, YbjP, SlyB, YajG, DolP, Slp and Blc-that are enlisted as candidates for interaction partners of LptDE (Supplementary Table 1).
A SDS-PAGE gel showing that endogenous lipoproteins LptM and/or Lpp were co-purified with affinity-purified LptDEHis proteins. Endogenous LptM migrated to a similar position to the recombinant LptMHis on the SDS-PAGE gel (rightmost lane). Data are representative of three independent experiments. B SDS-PAGE analysis of the purified paLptDEHis protein showing the co-purified endogenous lipoproteins (LptM, Lpp, YedD, YbjP, YajG, SlyB, DolP, Slp and Blc). The lipoproteins were identified by mass spectrometry analysis. The paLptDEHis sample was concentrated to ~20 mg ml−1 and 2 μl of sample was loaded. Data are representative of three independent experiments. C Immunoblotting using anti-Strep tag II antibody showing that the affinity-purified sfLptDEHis pulled down all nine lipoproteins identified in (B). The bottom panel shows the over-expression of each lipoprotein in the cell by using membrane fractions solubilized with 1% LDAO. Each lipoprotein including BamE with a C-terminal Strep tag II was co-expressed with sfLptDEHis in E. coli SF100 strain in the assays. Note that sfLptDEHis only pulled down a small amount of Blc due to the low expression levels of Blc. Data are representative of three independent experiments. D Native mass spectrometry analysis of the purified sfLptDEHis protein. The spectrum shows the presence of three protein complexes, sfLptDEHis, sfLptDEHis-LptM and sfLptDEHis-YedD, in the sample. Note: The 17+ charge state of sfLptDEHis-YedD and the 15+ charge state of sfLptDEHis overlap in the MS spectrum. E Native mass spectrometry analysis of the purified paLptDEHis protein. The spectrum indicates the presence of six different protein complexes in the sample, paLptDΔ12EHis, paLptDΔ12EHis-LptM, paLptDΔ23EHis, paLptDΔ23EHis-LptM, paLptDΔ23EHis-YbjP and paLptDΔ23EHis-SlyB. paLptDΔ12 and paLptDΔ23 stand for the N-terminal 12 and 23 residues of the mature paLptD were degraded, respectively. Species assignments in this spectrum are based on a minimum of three consecutive charge states to ensure reliability. The molecular masses of the mature lipoproteins are listed in Supplementary Table 1, and theoretical and experimentally determined masses of LptDE-lipoprotein complexes are listed in Supplementary Table 2. Source data are provided as a Source Data file.
We next sought to confirm the interactions between LptDE and the identified lipoproteins. For this, we co-expressed all of the 9 identified lipoproteins together with sfLptDE. The six-histidine-tagged sfLptDEHis was used as a bait protein captured all of the nine lipoproteins that have a Strep-tag II fused to their C-terminus (Fig. 1C, top panel). It is evident from our results that LptDE indeed forms physical interactions, with the nine lipoproteins established by proteomics. However, not all lipoproteins are tightly associated with LptDE.
To further confirm the interaction and to investigate the binding specificity of the identified lipoproteins to LptDE, we subjected the purified sfLptDE and paLptDE samples to native MS analysis. Native MS spectra clearly showed that two of the lipoproteins, LptM and YedD formed stable complexes with sfLptDE (Fig. 1D), whereas paLptDE exhibited tight association with LptM, SlyB and YbjP, three of the earlier MS-identified lipoproteins (Fig. 1E). These observations reinforced the notion that LptDE selectively binds a subset of lipoproteins.
Overall structure of the paLptDE-LptM complex
Confirmation of the LptDE:lipoprotein interaction comes from crystallographic studies. We obtained an atomic structure of paLptDE-LptM and uncovered the interaction details. PaLptD bears a low sequence identity (<25%) with the two LptD homologs with full-length structures to date21,22,37, containing an extra 120-residue loop named insertion22 at the N-terminus (Fig. 2A and Supplementary Fig. 2). After extensive crystallization trials, crystals obtained from the purified paLptDE sample diffracted to 3.0-Å resolution. The structure was determined using molecular replacement with the structure of a jellyroll-truncated paLptDE as search model22 and was refined to Rwork/Rfree = 0.23/0.26 (Supplementary Table 3). Throughout model building, we noticed a continuous, strong positive difference Fourier electron density that could not be accounted for by paLptDE itself (Fig. 2B). The ribbon-shaped electron density is lodged at the interface between the β-jellyroll and the β-barrel domains of paLptD. The electron density in the paLptD β-barrel lumen displays characteristic polypeptide features with discernible side chains, whereas the trifurcated branches extending into the membrane boundary are reminiscent of three acyl chains of a lipoprotein (Fig. 2B). We therefore conclude that the extra density corresponds to a molecule of LptM in association with paLptDE. To further confirm that the additional density in the paLptDE structure corresponds to the lipoprotein LptM, we purified paLptDE protein from the lptM-depleted E coli SF100 strain, and determined the structure at a resolution of 3.3 Å (Supplementary Table 3). In the newly obtained paLptDE structure, the additional density was absent, resulting in the final apo-paLptDE structure (Fig. 2B). Moreover, SDS-PAGE analysis of the dissolved crystals confirmed the presence of LptM in the crystals of paLptDE-LptM (Supplementary Fig. 3A). Moreover, in the final paLptDE-LptM structure, the N-terminal 81 residues of the mature paLptD and the C-terminal 35 residues of LptM (a total of 48 residues) are disordered. We attribute this to intrinsic flexibility as well as partial protein degradation in the aforementioned regions. The latter has been confirmed by native mass spectrometry analysis (Fig. 1E and Supplementary Table 2). Overall, our paLptDE-LptM structure resembles a previous β-jellyroll-truncated paLptDE structure22 with a root-mean-square deviation (RMSD) of 2.0 Å over 740 aligned Cα atoms. Additionally, we also observed a long α-helix at the C-terminus of paLptE (Fig. 2B), a feature that is consistent with the NMR structure of the isolated paLptE but missing in the previously-reported truncated paLptDE structure38.
A Schematic structures of paLptD (green), paLptE (magenta) and LptM (orange). The two conserved inter-domain disulfide bonds (yellow) and the jellyroll-barrel connecting loop (residues P316-P328, red) in paLptD are labeled and highlighted, respectively. The signal peptides of paLptD (residues 1-33), paLptE (residues 1–19) and LptM (residues 1–19) are labeled. B Crystal structure of the paLptDE-LptM (left) and apo-paLptDE (right) complex. The paLptD (green) and paLptE (magenta) are shown in cartoon. Unbiased Fo − Fc difference Fourier electron density (blue mesh, contoured at 2.0 σ) calculated before modeling LptM molecule. C Zoomed-in view of the atomic model of LptM superimposed with the electron density. The LptM (stick mode) was placed in the electron densities, showing that the electron density fragment in the paLptD barrel displays polypeptide features with bulky side chains (left top insert). The jellyroll-barrel connecting loop and the two pairs of disulfide bonds are highlighted in red and magenta, respectively. Numbering of the LptM residues in the structure follows the nomenclature of bacterial lipoproteins (left bottom insert). D Close-up view of the LptM-paLptDE interactions. Residues of paLptD (F281, I313, Y314, F325, I892, L894 and L897, in green) that interact with the three acyl chains (R1, R2 and R3) are shown in stick mode (left panel); residues of paLptD (Q322, Y357, E371, R373, I900 and V901, in green) that interact with the proteinaceous part of LptM are also shown in stick mode and labeled (right panel). LptM is shown in stick mode (brown). R1, R2 and R3 stand for three acyl chains of LptM.
Importantly, our structure reveals that three hydrophobic acyl chains (R1, R2 and R3) of LptM are positioned in a hydrophobic environment surrounded by the last V-shaped β strand of the β-jellyroll and the exterior surface of the paLptD β-barrel. This observation suggests that the hydrophobic acyl chains of LptM directly access the OM upon dissociation from the paLptD β-jellyroll (Fig. 2C). In contrast, the proteinaceous moiety of the mature LptM (residues Gly21-Tyr27) interacts with residues lining the hydrophilic lumen of the LptD β-barrel (Fig. 2D, right panel). The structural arrangement indicates that the proteinaceous moiety of LptM is positioned to tuck into the hydrophilic lumen of the LptD β-barrel. Remarkably, LptM is enclosed in a region that is covalently sealed by the jellyroll-barrel connecting loop at one side and two conserved inter-domain disulfide bonds at the opposite side (Fig. 2C, left panel and Supplementary Fig. 3B and Supplementary Movie 1). Crystallographic data suggest that release of LptM from paLptD β-barrel to the OM requires disruption of the non-covalent interface between β1 and β26 of the paLptD β barrel21.
LptDE-bound lipoproteins can be surface-exposed
The paLptDE-LptM structure implies that lipoprotein LptM, like LPS, may be eventually transported to the cell surface through LptDE (Fig. 2B). To test whether LptM and the other LptDE-bound lipoproteins are indeed surface-exposed, we carried out immunofluorescence assays. For this, with the exception of RcsF, which carries an N-terminal Flag tag, the other lipoproteins are tagged with a C-terminal His tag for detection. As shown in Fig. 3A, E. coli cells that harbored each of LptDE-bound lipoprotein-expressing plasmids (except Blc) and RcsF (as a positive control) displayed red fluorescence circles, suggesting that the C-termini of these eight lipoproteins are surface-exposed. By contrast, E. coli cells transformed with a plasmid that expressed BamB, a lipoprotein of the β-barrel-assembly machinery (BAM) complex that attaches to the inner leaflet of the OM39,40, displayed red fluorescence only when the cell membranes were permeabilized (Fig. 3A).
A Immunofluorescence assays showing that LptM, Lpp, YedD, YbjP, YajG, Slp, DolP and SlyB are surface-exposed. DAPI and Alexa Fluor 546-conjugated secondary antibody are blue- and red- fluorescencing, respectively. BamB was selected as a negative control. Membrane permeabilization was carried out by treatment with 0.5% Triton X-100 and 5 mM EDTA. Data are representative of three independent experiments. B Dot blot assays (both using intact cells and cell lysates) showing that the identified LptDE-bound lipoproteins (LptM, Lpp, YedD, YbjP, YajG, Slp, DolP and SlyB) are surface-exposed at least for certain percentage of the expressed lipoproteins in the cells. Note that only a small amount of Blc was detected in the lysate, consistent with the low expression levels shown in Fig. 1C. BamB was selected as a negative control, and RcsF was selected as a positive control. With the exception of RcsF, which carries an N-terminal Flag tag, the other lipoproteins are tagged with a C-terminal His tag for visualization or detection. Data are representative of three independent experiments.
Dot blot analyzes corroborated these findings, showing consistent surface localization patterns across all tested LptDE-associated lipoproteins except Blc (Fig. 3B). The limited detection of Blc in both assays aligns with its low expression levels observed in pulldown experiments (Fig. 1C), suggesting that its surface presence remains below detectable thresholds under these experimental conditions. Notably, while complete translocation efficiency wasn’t demonstrated for all molecules, the consistent detection of accessible surface-localized species across multiple independent lipoproteins confirms functional LptDE-mediated transport capability. These results collectively suggest that LptDE may facilitate cell surface localization for some SLPs (predominantly LptM).
LptM is extracted from the IM by LolCDE rather LptB2FG
Next, we ask how lipoprotein LptM is transferred to LptDE. As both matured lipoproteins and LPS anchor to the outer leaflet of the IM via their acyl chains prior to trafficking, two potential routes are possible for lipoproteins sorting to the cell surface. First, LptM might be promiscuously extracted by LptB2FG from the IM and then transported to the cell surface via the Lpt pathway, given that both lipoproteins and LPS share hydrophobic acyl chains and hydrophilic components (Supplementary Fig. 1B). The second possibility is that LptM, like conventional lipoproteins, is extracted by LolCDE in the Lol pathway but is delivered to LptA, the periplasmic component that bridges the periplasmic space between LptB2FGC and LptDE in the Lpt pathway. To clarify the LptM sorting route, we first probed whether LptM passes through the β-jellyroll domain of LptD. To this end, a photocrosslinkable unnatural amino acid, pBpa, was introduced at either I230 (paLptD P328) or Y112 (paLptD L210), two LPS crosslinkable sites in Shigella flexneri LptD41,42 (Supplementary Fig. 4A). Photocrosslinking was performed when either sfLptDI230pBpaE or sfLptDY112pBpaE was co-expressed with LptM23. We observed LptM cross-linked to LptD at each site (Fig. 4A), suggesting that LptM was delivered to the jellyroll-barrel interface as was observed in the structure of paLptDE-LptM complex through the β-jellyroll domain of LptD.
A In vivo photocrosslinking showing that LptM cross-linked to pBpa-containing LptD (I230pBpa or Y112pBpa) when LptM and sfLptDE were co-expressed in E. coli. LptM contained a C-terminal Strep tag II for immunoblotting detection. B–D Reconstitution of LptM membrane-to-membrane transport. Residues G21 of LptC and Y320 of LptG are located in the LPS-binding cavity of LptB2FGC, but neither LptC (G21pBpa) nor LptG (Y320pBpa) cross-linked to LptM (B). Residues R212 and R223 of LptF are located in the β-jellyroll of LptF, but neither LptF (R212pBpa) nor LptF (R223pBpa) cross-linked to LptM (C). Residues T47 and F78 of LptC are located in the β-jellyroll of LptC. Neither LptC (T47pBpa) nor LptC (F78pBpa) cross-linked to LptM (D). Cartoons show experimental designs of the reconstituted system. LptM can be inserted into nanodisc in either orientation, but only the productive orientation is shown for simplicity. The red arrow denotes LPS transport direction in LptB2FGC. None of these selected sites cross-linked to LptM, with or without ATP, but all of these selected sites in LptB2FGC cross-linked to LPS upon being substituted with pBpa (Supplementary Fig. 6A-C). E LptM cross-linked to LolC in LolCDE upon UV radiation. Residues L256 and V260 of LolC are located in the lipoprotein-binding cavity of LolCDE. The LptM×LolC crosslinks decreased with the increase of incubation time. F LolA cross-linked LptM upon being released from LolCDE in an ATP-dependent manner. Residue W70 of LolA was substituted with pBpa in the assays. Cartoons show experimental designs of the reconstituted system. The red star denotes the position of pBpa in LolA. G In vitro LPS or LptM membrane-to-membrane transport assays showing that LPS was efficiently transported from LptB2FGC-A to LptDY112pBpaE in an ATP-dependent manner (middle), but LptM was not transported from LptB2FGC-A to LptDY112pBpaE (right). Cartoons show experimental designs of the reconstituted system. Data in Fig. 4 are representative of three independent experiments. Source data are provided as a Source Data file.
To investigate whether LptM is extracted by LptB2FGC from the IM, we sought to reconstitute LptM transport using purified protein components. First, to monitor whether LptM enters into the LPS-binding cavity of LptB2FGC, two LPS-photocrosslinkable sites (LptCG21 and LptGY320) in the LPS-binding cavity of E. coli LptB2FGC were substituted with pBpa (Supplementary Fig. 4B, left). The purified LptB2FGCG21pBpa or LptB2FGY320pBpaC complexes were reconstituted in nanodiscs together with either LPS or LptM (Supplementary Fig. 5). We observed that LPS cross-linked to LptC and LptG (Supplementary Fig. 6A). However, LptM did not cross-link to LptC or LptG under the same conditions (Fig. 4B). Furthermore, LPS×LptC (or LPS×LptF) crosslinks (Supplementary Fig. 6B, C) were detected at each site in the β-jellyroll bridge of E. coli LptB2FGC (LptFR212, LptFR223, LptCT47 and LptCF78)23 that was substituted with pBpa (Supplementary Fig. 4B, right) in an ATP-dependent manner, but none of these sites cross-linked to LptM (Fig. 4C, D). These observations are consistent with previous reports that LptB2FGC is responsible for extracting and transporting LPS, thus arguing against LptM entering the LPS-binding pocket of LptB2FGC and passing through β-jellyroll domains of LptF and LptC.
We next explored whether LptM is extracted by the ABC transporter LolCDE in the Lol pathway. To test this, we also carried out photocrosslinking assays using purified protein components. Based on the LolCDE structures43,44,45, we purified E. coli LolCDE with pBpa substituted at either L256 or V260 in LolC, two sites in the lipoprotein-binding cavity of LolCDE (Supplementary Fig. 4C). We detected LptM×LolCL256pBpa and LptM×LolCV260pBpa crosslinks upon UV radiation in the absence of ATP (Fig. 4E), suggesting that LptM entered the lipoprotein-binding pocket of LolCDE. To test whether LolCDE delivers LptM to LolA, we purified E. coli LolA that contains pBpa at W70 (Supplementary Fig. 4D) and incubated LolAW70 with nanodisc-embedded LolCDE and LptM (Supplementary Fig. 5). We observed LptM×LolA crosslinks only in the presence of ATP (Fig. 4F). Our combined results suggest that LptM, like other lipoproteins, is extracted from the IM by LolCDE, followed by delivery to LolA. Our dot blot assays also reveal (Fig. 3B) that some LptM molecules remain attached to the inner leaflet of the OM.
To further confirm that LptM is not transferred to LptDE from LptB2FGC, we performed membrane-to-membrane lipoprotein transfer assays using purified protein components. For this, the nanodisc-embedded LptDI230pBpaE (a site in LptD that cross-links to both LPS and LptM) was incubated with either nanodisc-embedded LptB2FGC-A+LptM or LptB2FGC-A + LPS. LPS strongly cross-linked to LptD in the presence of ATP, but we did not observe any crosslinks between LptM and LptD (Fig. 4G). We therefore concluded that it is not likely that LptB2FGC extracts LptM from the IM. Rather, a crosstalk between the Lpt and the Lol pathways for LptM surface localization must exist, which possibly involves LolCDE.
LptA crosstalks with the Lol pathway in LptM surface localization
To investigate potential mechanisms for LptM delivery to LptDE and explore interactions between the Lpt and Lol pathways, we first tested whether LptM could associate with sfLptDE in vivo under LolA-depleted conditions. We co-expressed sfLptDE and LptM in the lolA-depleted MG3324 (NR754 Δlpp ΔrcsB zii::Tn10 cpxA24 ΔlolA) E. coli strain46. SDS-PAGE analysis of the affinity-purified sample showed that LptM formed a stable complex with sfLptDE (Fig. 5A). While this result does not exclude the possibility of LolA’s involvement in LptM localization, it prompted further exploration of candidate pathways for periplasmic shuttling.
A SDS-PAGE (left) and western blotting (middle and right) analysis of the affinity-purified LptDEHis when both LptDEHis and LptMStrep were co-expressed in the lolA-depleted MG3324 E. coli strain. LptM was copurified with LptDEHis in the absence of LolA, indicating that LolA is not required for LptM transport to LptDE. B In vivo photocrosslinking assays showing that LptM cross-linked to pBpA-containing LptA (F95pBpA or I36pBpA) when LptM and LptA were co-expressed in E. coli. C In vitro LptM transport assays showing that pBpa-containing LptA (F95pBpa) cross-linked to LptM that was reconstituted into nanodisc together with LolCDE in a time- and ATP- dependent manner. The red star denotes the location of F95pBpa in the LptA β-jellyroll. Cartoons show experimental designs of the lipoprotein transfer assays. D The Hook motif of LolC in LolCDE is required for LptM transfer from LolCDE to both LolA and LptA. Data in Fig. 5 are representative of three independent experiments. Source data are provided as a Source Data file.
Given LptA’s unique role as a central hub in the Lpt pathway-interacting with both LptDE and LptB2FGC-we hypothesized that LptA might serve as an intermediate chaperone linking LolCDE-released lipoproteins to LptDE. To test this, we first co-expressed LptM with LptA that contains pBpa at F95 or I36 in E. coli (Supplementary Fig. 4E). We observed that LptM cross-linked to LptA upon UV radiation (Fig. 5B). By contrast, LolB and NlpA, two lipoproteins that respectively anchor to the inner leaflet of the OM and outer leaflet of the IM did not cross-link to LptA in the same scenario (Supplementary Fig. 6D). This directly demonstrates that LptA interaction is specific to some SLPs like LptM and not a nonspecific effect. To carry out in vitro lipoprotein transfer assays, nanodisc-embedded LolCDE-LptM was incubated with LptAF95pBpa. Upon UV radiation, we observed that LptA cross-linked to LptM in an ATP-dependent manner (Fig. 5C). However, when LolCDE was replaced by a variant which lacks the Hook motif (residues P167-P179) in LolC (Supplementary Fig. 4C), LptAF95pBpa did not cross-link LptM (Fig. 5D)47. The results reveal that LptA interacts with LolCDE in a manner analogous to the Lol pathway, likely recognizing cargo via the same Hook motif-mediated mechanism. These findings, together with our earlier lipoprotein transfer assays results, consistently suggest that LptA may crosstalk to the Lol pathway by shuttling LptM across the periplasm and presenting it to LptDE.
LptM translocates across the OM in an Lpt-dependent manner
Our earlier studies establish that the LptB2FGC complex is not responsible for shuttling LptM across the periplasm, but we inquired whether LptB2FGC takes part in LptM transport to the cell surface. First, we reconstituted membrane-to-membrane delivery of LPS by incorporating purified inner and outer membrane transport complexes into separate nanodisc. We found that the purified LptA protein is prone to form oligomers, hampering its binding with LptB2FGC or LptDE with high affinity. In order to increase LPS transport efficiency in vitro, we constructed an LptC-LptA fusion protein that formed a stable complex with LptB2FG. After mixing the nanodisc-embedded LptDY112pBpaE protein complexes with nanodisc-embedded LptB2FGC-A + LPS, we observed that LPS cross-linked to LptD in an ATP-dependent manner (Supplementary Fig. 7). By contrast, in the absence of LptA, we did not observe any LPS×LptD crosslinks (Supplementary Fig. 7). The results demonstrate the in vitro reconstitution of Lpt pathway in which all seven Lpt proteins and ATP are required for LPS transport23. Next, we investigated whether the reconstituted Lpt pathway drives the export of LptDE retained LptM across the OM. We then prepared three different types of nanodisc-embedded protein complex samples (LptB2FGC-A, LptB2FGC-A+LptM and LptB2FGC-A + LPS) and incubated them with the nanodisc-embedded LptDI230pBpaE-LptM complex in the presence ATP over a time course. Upon exposure to ultraviolet (UV) radiation, we observed LptM crosslinked to LptD for all three samples. Interestingly, we found that in the presence of ATP and LPS, the ratio of crosslinking decreased with increasing incubation time (Fig. 6A). The results suggest that both LPS and ATP hydrolysis by LptB2FG are required for LptM OM translocation, and the stream of LPS molecules push LptM towards its destination. Our paLptDE-LptM structure reveals that LptM is positioned in a region that is covalently sealed on one side by the jellyroll-barrel connecting loop and on the opposite side by two conserved inter-domain disulfide bonds. Given the positioning of lipoprotein LptM in the structure, once released from LptDE, the only feasible destination for unbound LptM is translocation across the OM to the cell surface. These findings provide critical indirect evidence supporting the hypothesis that LptM translocates across the OM in an Lpt-dependent manner. The constant levels of LptD×LptM crosslinks for the nanodisc-embedded LptB2FGC-A+LptM sample are consistent with our earlier conclusion that LptB2FGC alone cannot transfer LptM to LptDE (Fig. 6A).
A In vitro membrane-to-membrane transport assays showing that LptM translocates across the OM in an ATP- and LPS- dependent manner. Four different types of nanodiscs were prepared for the assays: LptB2FGC-A, LptB2FGC-A+LptM, LptB2FGC-A + LPS and LptDI230pBpaE-LptMStrep. The LptD×LptM crosslinks decreased in an ATP- and time- manner only when nanodisc-embedded LptDI230pBpaE-LptMStrep was incubated with the nanodisc containing both LptB2FGC-A and LPS. Cartoons show experimental designs of the in vitro LPS transport assays. Data are representative of three independent experiments. B The proposed model of SLP transport to the cell surface. The red dash line indicates that LptA crosstalks to the Lol pathway during OM biogenesis. Source data are provided as a Source Data file.
Taken together, our research and that of Yang et al. complement and validate each other. Both we and Yang et al. observed the interaction between LptDE and LptM. Yang et al. demonstrated that LptM promotes the oxidative maturation of LptD, highlighting the biological significance of the LptDE-LptM interaction. Our paLptDE-LptM structure further validates the interaction between LptDE and LptM, offering a more detailed and precise depiction of their interaction sites. Yang et al. showed that the LptM-LptD interaction does not affect the LptD-LptA interaction; in contrast, it activates the LPS translocon. We demonstrated that the flow of LPS molecules then drives LptM toward its destination, with both LPS and ATP hydrolysis by LptB2FGC being required. Based on the positioning of lipoprotein LptM in the structure, LptM is inevitably localized to the cell surface after completing its function. Moreover, our findings suggest a potential pathway for the transfer of LptM to LptDE. Based on our results, we propose that LptM is matured on the inner membrane, transported via LolCDE to LptA, and then transferred to LptDE.
Discussion
In this study, we identified candidate lipoproteins that may be transported to the cell surface via LptDE. Our paLptDE-LptM structure suggested an alternative mechanism for how a subset of lipoproteins can be transported to the outer leaflet of the OM. First, the paLptDE-LptM structure provids a snapshot of an intermediate state of lipoprotein transport to the cell surface, suggesting that the hydrophobic acyl chains and hydrophilic proteinaceous moiety pass through the OM via the exterior and interior of the LptD β-barrel, respectively. Second, the LptM-binding site in paLptDE could imply that the destination of LptM is potentially at the outer leaflet of the OM. This highlights a critical role of the two pairs of conserved interdomain disulfide bonds within LptD in preventing its cargo, LPS or some SLPs, from mis-localization in the inner leaflet of the OM. We further show that LptA, a periplasmic component of the Lpt pathway, can shuttle lipoproteins through the periplasmic space after they are extracted from the inner membrane by LolCDE, thus suggesting a possible crosstalk between the Lol and Lpt pathways during OM biogenesis. Based on our findings, we propose a previously unidentified sorting route for lipoprotein surface presentation (Fig. 6B). OM lipoproteins including SLPs, are first extracted from the IM by LolCDE. Both periplasmic chaperones LolA and LptA can accept lipoproteins upon their release from LolCDE. The LolA-bound lipoproteins are targeted to the inner leaflet of the OM via the classical Lol pathway. Alternatively, LptA-bound lipoproteins are incorporated into the Lpt pathway by bridging LptB2FGC with LptDE, and are thereby transported to the cell surface. Lipoprotein surface translocation relies on energy provided by ATP binding and hydrolysis, catalyzed both by LolCDE, to load LptA, and by LptB2FGC in the presence of LPS, to push SLPs through to their cell surface destination24,46. However, our in vitro and in vivo crosslinking experiments only provided preliminary evidence suggesting potential crosstalk between the Lol and Lpt pathways. To further validate this model, additional experiments including bacterial genetic analyzes are warranted in future.
In contrast to a number of previously-reported surface-exposed lipoproteins that require either a dedicated translocon48,49,50 or the BAM complex51,52 for surface translocation, the SLPs identified here share the same OM translocon (LptDE) and employ the periplasmic chaperone, LptA. These SLPs all lack large protein domains that can potentially provide energy to drive OM translocation via folding, in the energy-deficient OM milieu53,54,55. In this regard, it is not surprising that these SLPs utilize the trans-periplasmic scaffold of the Lpt pathway, in which the cytoplasmic ATP serves as energy source driving the SLPs through the hydrophobic channel to the cell surface. Our model that a particular size of lipoprotein, such that it approximates the shape and volume of an LPS molecule, could determine the range of SLPs selected by LptA also predicts that smaller lipoproteins might be recognized with lower affinity. It is intriguing that many other lipoproteins in the same size range, and expressed under similar growth conditions, like OsmB, are not SLPs. Clearly, the selection of SLPs by LptA is not restricted to acyl chain recognition like for LolA. There must be some structural determinant in the proteinaceous domain of SLPs that enables them to be distributed to the inner leaflet of the outer membrane in a LolCDE and LolAB-dependent manner, while a proportion is simultaneously distributed to the cell surface in a LolCDE and Lpt-dependent manner. However, structure and/or sequence features that dictate surface exposure of a specific lipoprotein await further clarification.
To validate the heterologous LptDE-LptM interaction in our paLptDE-LptM structure, we predicted paLptDE-LppL (LptM homolog in Pseudomonas aeruginosa) and ecLptDE-LptM complexes using AlphaFold Server. Both models showed high similarity to our paLptDE-LptM structure (Cα RMSD 0.71 Å and 2.15 Å), with the N-terminal loops of LppL and LptM adopting analogous binding geometries at the β-jellyroll-barrel interface (Supplementary Fig. 8A, B). The predicted palmitoyl chain orientations in LppL/LptM precisely matched the spatial arrangement of the R2 acyl chain observed in our paLptDE-LptM structure. Similarly, N-terminal loop of LptM in Yang et al.‘s predicted ecLptDE-LptM adopts analogous binding geometries at the β-jellyroll-barrel interface (Supplementary Fig. 8C)34. In conclusion, these structural comparisons indicate that the heterologous LptDE-LptM interaction might faithfully recapitulate the homologous interaction.
Comparative analysis of paLptDE-LptM and apo-paLptDE structures revealed significant weakening of β1-β26 strand interactions in the LptD β-barrel, particularly diminished hydrogen bonding between S329-Q891 and S334-D885 (Supplementary Fig. 9A). This supports Yang et al.‘s hypothesis that LptM facilitates lateral gate opening through β-sheet destabilization, consistent with their MD simulations based on AlphaFold2-predicted LptDE models34. However, our crystallographic structures reveal critical discrepancies: (i) Distinct hydrogen polar interaction disruption sites compared to computational predictions, and (ii) The C134-C270 distance exhibited a slight contraction rather than expansion upon LptM binding, and (iii) Minimal structural perturbations at β-jellyroll-barrel interface upon LptM binding (Supplementary Fig. 9A, B), contrasting with the substantial conformational changes proposed in their simulations.
To explore the potential substrate diversity of LptDE beyond LptM, we employed AlphaFold Server to predict eight lipoproteins identified in our proteomic analysis (Supplementary Table 1). Remarkably, six predicted complexes (paLptDE-Lpp, paLptDE-YedD, paLptDE-YbjP, paLptDE-SlyB, paLptDE-YajG and paLptDE-Blc) display interaction pattern similar to LptM: (i) Palmitoyl chains anchoring at the LptD β-jellyroll-barrel hydrophobic interface, and (ii) the N terminal proteinaceous loops of lipoproteins extend from the jellyroll-barrel connecting loop into the β-barrel lumen of LptD (Supplementary Fig. 10). These predictions, corroborated by functional experimental data, suggest that LptDE might have a broader substrate specificity rather than exclusive LptM recognition, implying a generalized transport mechanism for structurally diverse SLPs. He et al. reported that knockdown of LptD homolog BB0838 in the LPS-deficient Borrelia burgdorferi disrupted the translocation of surface lipoproteins through the spirochetal outer membrane35. This suggests that the BB0838 may function as a surface lipoprotein flippase, and supports our findings that SLPs are transported to the cell surface via LptDE. We recognize that while these findings suggest a broader substrate specificity for LptDE, additional experimental validation is required to confirm this. Further research should focus on other SLPs and bacterial genetic analyzes to validate the wider applicability of this transport mechanism.
Methods
Protein expression, purification and crystallization
We cloned lptDE genes from two different species into vector pBAD22 under control of the arabinose promoter. The plasmids contained the genes for lptD and lptE in this order, each with a separate translation-initiation site. The full-length lptD and lptE genes were amplified from Pseudomonas aeruginosa PAO1-LAC (47085D-5) and Shigella flexneri (700030D) genomic DNAs (ATCC) by polymerase chain reactions (PCR). All the generated plasmids (pBAD22-palptDE and pBAD22-sflptDE) contained a hexahistidine tag at the C-terminus of LptE that was introduced in the PCR primer to facilitate subsequent affinity purification. The plasmid pBAD22-lptDE was then transformed into the protease-deficient E. coli strain SF100 [KS272∆ (ompT-entF)] (ATCC) for co-expression. Protein expression was induced by the addition of 0.5% L-arabinose for 12 hrs at 26 °C when the O.D.600 of the culture reached about 1.0. Cells were harvested by centrifugation at 4500 g for 30 min at 4 °C. Cell pellets were resuspended in 1×PBS (pH 7.4), lysed by a single passage through a French Press (JN-3000 PLUS, China) at 16,000 psi, and centrifuged at 39,000 g for 45 min at 4 °C to collect the total cell membranes. The total membranes were solubilized with a buffer containing 1×PBS (pH 7.4) and 0.5% N-lauroylsarcosine sodium (Sigma-Aldrich) for 1 hr at 4 °C. The outer membranes were isolated by centrifugation at 39,000 g for 1 hr at 4 °C and were further solubilized for 1 hr with buffer A [20 mM Tris-HCl (pH 8.0), 150 mM NaCl, 20 mM imidazole and 1% N,N-Dimethyldodecylamine N-oxide (LDAO)]. The supernatant was collected after centrifugation at 39,000 g for 1 hr at 4 °C and incubated with pre-equilibrated Ni-NTA agarose beads for 2 hrs at 4 °C. Protein-bound Ni-NTA agarose beads were rinsed with buffer B [20 mM Tris-HCl (pH 8.0), 150 mM NaCl, 30 mM imidazole and 0.2% LDAO], and detergent exchange was performed with buffer C [20 mM Tris-HCl (pH 8.0), 150 mM NaCl, and 1% tetraethylene glycol monooctyl ether (C8E4)]. The LptDE complex was eluted from the Ni-NTA agarose beads using buffer D [20 mM Tris-HCl (pH 8.0), 150 mM NaCl, 200 mM imidazole and 1% C8E4]. The eluted LptDE complex was subsequently applied to a Resource-Q column (GE Healthcare), and followed by a SuperdexTM 200 10/300 size exclusion column (GE Healthcare) that was pre-equilibrated with 20 mM Tris-HCl (pH 8.0), 150 mM NaCl and 0.6% C8E4. The purified paLptDE and sfLptDE samples were shown to contain lipoprotein LptM as revealed by 15% SDS-PAGE analysis. Normally, we obtained ~1.5 mg protein for paLptDE complex from about 80 liters of Terrific Broth (TB) culture, which is a 10-fold lower than the yield of sfLptDE.
Crystallization was conducted at 16 °C using the hanging drop vapor diffusion method, mixing 1 μl each of paLptDE-LptM (15 mgml−1) and reservoir solutions at a ratio of 1:1. Initial crystallization condition was found using a broad screening. After optimization, the best crystals were obtained in a buffer containing 175 mM citric acid/Bis-Tris (pH 5.0), 17% (v/v) PEG3350 and 3% methanol (as additive). The paLptDE-LptM crystals appeared overnight and grew to their final size ~100 × 120 × 60 μm in about one week. They belong to space group P21 and the best crystals diffracted to 3.0 Å at synchrotron. The apo-paLptDE protein was purified from the lptM-depleted E. coli SF100 strain using a similar protocol as described for paLptDE-LptM. The apo-paLptDE crystals were obtained in a condition that contained 2% tacsimate (pH5.0), 16% PEG3350 and 100 mM sodium citrate tribasic dehydrate at 16 °C and the best crystals diffracted to 3.3 Å. The crystals belong to space group P212121 with two apo-paLptDE complexes in one asymmetric unit. All the crystals were flash frozen in liquid nitrogen by the addition of 20% glycerol into the reservoir solutions for data collection.
Structure determination and refinement
All diffraction data were collected at Shanghai Synchrotron Radiation Facility (SSRF, Shanghai, China). X-ray data were processed with HKL200056. For both paLptDE-LptM and paLptDE, an initial molecular replacement solution was found using Phaser with the jellyroll-truncated paLptDE (PDB code: 5IVA) as search model57.The electron densities for the jellyroll domains of paLptD were resolved after molecular replacement57, followed by several rounds of manual building for the jellyroll domain of paLptD with Coot58. β-barrel lumen exhibited polypeptide features with discernible bulky side chains, which allowed us to manually build LptM into the structure. Further SDS-PAGE analysis using dissolved crystals confirmed the presence of LptM in the crystals. Refinements of the paLptDE-LptM and apo-paLptDE datasets were performed using Phenix and CCP4 refmac559. PaLptDE-LptM and paLptDE structures were refined to 3.0-Å and 3.3-Å resolution with Rwork/Rfree = 0.23/0.26 and Rwork/Rfree = 0.22/0.26, respectively. The poorly defined residues (matured proteins) in the best-refined structures of paLptDE-LptM (paLptD: 1-81; LptM: 12-46) and apo-paLptDE (paLptD: 1-70) presumably indicate conformational flexibility and partial degradation of the paLptD N-terminus. All structure figures were rendered using PyMOL60. The detailed refinement statistics for both paLptDE-LptM and paLptDE structures are listed in Supplementary Table 3.
Construction of the lptM-depleted E. coli SF100 strain
To delete the lptM gene in E. coli SF100 strain, the linear fragments were amplified by PCR using primers P1 (5’-TCCTGCGATG ATAGAAAGCA GAAAGCGATG AACTTTACAG GCAATCCATA GTGTAGGCTGGAGCTGCTTC-3’) and P2 (5’-TTCGAGAACT GCATCATTTA CTCCAATCAC GCGGGTACAG AAACTGACTT CATATGAATATCCTCCTTAG-3’) with 50-nt extensions that are homologous to regions adjacent to the lptM gene using the plasmid pKD3 as template. PCR fragments were then transformed by electroporation into the E. coli SF100 strain that carries the pKD46 plasmid to obtain lptM::Cm. Mutants were colony-purified once non-selectively at 37 °C to eliminate pKD46. The Cm cassette was then removed by introducing the helper plasmid pCP20 into the mutant lptM::Cm and colony-purified once non-selectively at 43 °C. The lptM-depleted E. coli SF100 strain was used for over-expressing paLptDE proteins to obtain apo-paLptDE crystals.
Liquid chromatography-tandem mass spectrometry
The gel bands containing the protein samples were decolorized, reduced with DTT, and alkylated with iodoacetamide, followed by overnight digestion with trypsin. The resulting peptides were then extracted using 60% acetonitrile. The peptide mixture obtained after digestion was analyzed using a liquid chromatography-linear ion trap-orbitrap (nanoLC-LTQ-Orbitrap XL, Thermo, San Jose, CA) mass spectrometer. The chromatographic column was a self-packed C18 reverse-phase column with an inner diameter of 75 μm and a length of 15 cm, filled with 3 μm ReproSil-Pur C18-AQ (Dr. Maisch GmbH, Ammerbuch) packing material. The sample loading column had an inner diameter of 150 μm and a length of 3 cm, filled with 5 μm ReproSil-Pur C18-AQ (Dr. Maisch GmbH, Ammerbuch). The mobile phase consisted of A: 0.5% FA/H2O and B: 0.5% FA/ACN, with a flow rate of 300 nL min−1. A 90-minute gradient was employed. Data analysis was performed using Proteome Discoverer (version 1.4.0.288, Thermo Fischer Scientific). MS2 spectra were searched using the SEQUEST engine against the E. coli_NCBI20170519.fasta databases. Search parameters included trypsin digestion with two missed cleavage sites, precursor ion mass tolerance of less than 20 ppm, and fragment ion mass tolerance of less than 0.6 Da. Carbamidomethylation of cysteine was set as a fixed modification, and oxidation of methionine was set as a variable modification. The peptide spectral matches (PSM) were filtered using the Percolator algorithm with a q-value of less than 1% (1% FDR). The identified peptides were grouped into proteins based on the principle of parsimony.
Native mass spectrometry
Prior to mass spectrometry (MS) analysis, the purified paLptDE or sfLptDE protein samples were buffer exchanged into 150 mM ammonium acetate (pH 8.0) containing 0.5% C8E4 (v/v) on a Superdex 200 gel filtration column (GE Healthcare). LptDE samples were subsequently introduced via gold-coated nanospray capillaries prepared in house and MS spectra were recorded on a Synapt G2-Si instrument (Waters, Manchester, UK) modified for high mass detection. A partial denaturation experiment was carried out by adding dilute acetic acid to the LptDE samples to a final concentration of 0.05% immediately prior to MS sample introduction. Optimized instrument parameters include capillary voltage 1.8 kV, sampling cone 150 V, sampling offset 100 V, trap collision energy 150 V, transfer collision energy 35 V, trap gas flow 5.0 mlmin−1. MS data were analyzed using MassLynx version 4.1 (Waters, Manchester, UK).
Pulldown and immunoblotting assays
To confirm the interactions between sfLptDE and the nine identified lipoproteins (and lipoprotein BamE as negative control), plasmids contained the genes for sflptD, sflptEHis and a lipoprotein with a C-terminal Strep tag II in this order, each with a separate translation-initiation site, were cloned in the pBAD22 vector. The constructed plasmids were individually transformed into E. coli SF100 cells for co-expression. Protein expression and purification followed a similar procedure as described for paLptDE, but only 1 liter of LB culture was prepared for each construct. All the cell lysates were prepared from the total membranes solubilized with 1% LDAO. Each of the purified sfLptDEHis proteins and its cell lysate was subjected to SDS-PAGE (15%) analysis.
After electrophoresis, the proteins were transferred to a PVDF membrane and blocked using TBST buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.05% Tween-20) that contained 5% skim milk for 1 hr. The PVDF membrane was then incubated with anti-Strep tag II mouse monoclonal antibody (1:3500) (Beijing Xuheyuan Biotech) at room temperature for 1 hr and subsequently washed with TBST buffer twice and further incubated with anti-mouse IgG (H + L)-HRP (1:3500) (LabLead) at room temperature for 1 hr. PVDF membranes were exposed using enhanced chemiluminescence reagents (EasySee Western Blot Kit, Trans™).
Immunofluorescence assays
To carry out immunofluorescence assays, each of the identified lipoproteins (with a C-terminal His tag), BamB (as a negative control, with a C-terminal His tag) and RcsF (as a positive control, with an N-terminal Flag tag) were subsequently cloned into the pET22b vector and transformed into E. coli SF100 cells. For each sample, 1 ml E. coli cell culture (O.D.600 = 1.0) was loaded into a 1.5-ml Eppendorf tube. After removing the supernatant by centrifugation, each cell pellet was washed twice with PBS. The cell pellet for each sample was then resuspended with 1 ml PBS, and 200 µl suspension for each sample was transferred into the glass bottom cell culture dish (NEST, ø15 mm) that was pretreated with 0.01% poly-L-lysine. Following incubation for 5 min, the redundant suspension was discarded. The bacteria attached to the dish were fixed by addition of 200 µl 4% (w/v) paraformaldehyde in PBS for 2 hrs, followed by wash twice with 1 ml PBS for each sample.
The fixed bacteria were then blocked in 10% normal goat serum in PBS for 30 min. After being blocked, the samples were incubated with primary antibody (mouse anti-His 1:100, CMCTAG or anti-Flag, 1:100, F1005, LabLead) for 2 hrs, and then washed with PBS three times, 5 min each. The samples were then stained with Alexa Fluor 546-conjugated secondary antibody (goat anti-mouse, 1:200, Invitrogen) for 1 hr. After being washed for three times with PBS, they were mounted in Moviol Mounting Media with 4’,6’-diamidino-2-phenylindole (DAPI, blue-fluorescing), a membrane-penetrating fluorescent dye that binds strongly to adenine-thymine rich regions in DNA. For E coli cells that harbored the pET22b-bamB plasmid, an addition step was performed by treating with 0.5% Triton X-100 and 5 mM EDTA for 30 min to punch the membrane before proceeding to block with 10% normal goat serum in PBS.
The bacteria images were acquired on the Delta Vision OMX V3 image system (GE Healthcare) with a × 100/1.40 NA oil-immersion objective lens (Olympus UPlanSApo) and a camera (Evolve 512×512, Photometrics). Images were processed and analyzed using Image J software (NIH). The experiments were performed in duplicate and repeated three times, and the results are representative of replicates.
Dot blot assays
Dot blot assays were carried out similarly as described in a previous study61. Briefly, E. coli SF100 cells that carried either identified lipoproteins, BamB (as a negative control) or RcsF (as a positive control)-expressing plasmid (pET22b-lipoproteinHis, pET22b-bamBHis or pET22b-FlagrcsF) were grown to OD600 = 1.0. 1 ml of cell culture was withdrawn for each sample, and was washed twice with PBS (10 mM Na2HPO4, 1.8 mM KH2PO4, 2.7 mM KCl, 137 mM NaCl, pH 7.4). Cell lysates were prepared by sonication of the cell suspension in PBS supplemented with 10 mM EDTA. Three microliters of cell suspension (whole cell) or cell lysate were spotted on an Immobilon-PSQ transfer membrane (Merck Millipore Ltd.) and air dried. Membranes were blocked with 1% (w/v) skim milk in PBS for 30 min at room temperature and probed with anti-His and anti-Flag mouse monoclonal antibody (TansGen Biotech) for 1 hr at room temperature. The membranes were washed three times for 5 min with PBS and probed with goat anti-mouse IgG (H + L) HRP-conjugated secondary antibody (1:3500) (LabLead) for 1 hr at room temperature. Membranes were exposed using enhanced chemiluminescence reagents (EasySee Western Blot Kit, Trans™). The experiments were performed in triplicate and repeated at least three times, and the results are representative of replicates.
Nanodisc preparation
E. coli Total Lipid Extract (Avanti Lipids) was solubilized in chloroform, dried under nitrogen to form a thin lipid film. The lipid film was hydrated and resuspended at a concentration of 25 mM Total Lipid in 250 mM sodium cholate. Each membrane protein complex (LptDE, LptDE-LptM, LptB2FGC, LptB2FGC-A or LolCDE), MSP1D1 membrane scaffold protein62, and Total Lipid were mixed at a molar ratio of 1:2.4:80 in a buffer containing 15 mM sodium cholate and incubated for 30 min at 4 °C. Detergents were removed by incubation with 0.8 mg ml−1 Bio-Beads SM2 (Bio-Rad) overnight at 4 °C. Nanodisc-embedded complexes were further purified using a Superose6 increase 10/300GL column (GE Healthcare) in a buffer containing 20 mM Tris-HCl (pH 8.0) and 150 mM NaCl. Peak fractions were combined and concentrated to ~0.6 mg ml−1.
In vivo photocrosslinking
To test whether LptM cross-links to LptDE in E. coli, expression plasmids pBAD22-lptDE and pET28a-lptMStrep were constructed. pBAD22-lptDE plasmids contain an amber (TAG) codon at either LptDI230 or LptDY112 for incorporation of pBpa. E. coli BL21 (DE3) cells were transformed with three plasmids pSup-BpaRS-6TRN22, pBAD22-lptDE and pET28a-lptMStrep simultaneously for protein expression. The transformed E. coli cells were grown at 37 °C in LB in the dark, supplemented with ampicillin sodium (30 μg ml−1), kanamycin (15 μg ml−1) and chloramphenicol (15 μgml−1). When the culture reached O.D.600 = 1.0, pBpa was added into LB at a final concentration of 0.5 mM. After 1 h, protein expression was induced by the addition of 0.05 mM IPTG and 0.05% L-arabinose at 18 °C for 4 hrs. 200 μl culture aliqouts withdrawn were used either directly or exposed to UV light (365 nm, 100 W; Thermo Fisher Scientific) for 10 min. Proteins were separated on a 15% SDS-PAGE gel, and crosslinks were detected with Strep tag II monclonal antibody (Thermo Fisher Scientific).
To investigate whether LptA cross-links to LptM, LolB or NlpA, expression plasmids pQlinkN-lptAHis, pQlinkN-lptAHis-lptMStrep, pET28a-lolBStrep and pET28a-nlpAStrep were constructed. The pQlinkN-lptAHis and pQlinkN-lptAHis-lptMStrep plasmids contain an amber (TAG) codon at either LptAF95 or LptAI36 for incorporation of pBpa. E. coli BL21 (DE3) cells were transformed with two plasmids including pSup-BpaRS-6TRN22 and pQlinkN-lptAHis-lptMStrep plasmids, or three plasmids including pSup-BpaRS-6TRN22, pQlinkN-lptAHis and pET28a-lolBStrep or pET28a-nlpAStrep. The transformed E. coli cells were grown at 37 °C in the dark. When the cultures reached O.D.600 = 1.0, pBpa was added to LB at a final concentration of 0.5 mM. After 1 hr, protein expression was induced by the addition of 0.05 mM IPTG at 18 °C for 4 hrs. Photocrosslinks were detected in a similar way as described above.
In vitro photocrosslinking assays
To express and purify LptM proteins for nanodisc reconstitution, the E. coli lptM gene (along with the C-terminal Strep tag II coding sequence) was cloned into vector pET28a. The generated pET28a-lptMStrep plasmid was transformed into E. coli BL21 (DE3) strain. The pET28a-LptMStrep- harboring E. coli cells were grown at 37 °C in LB with 35 μgml−1 kanamycin. Protein expression was induced by addition of 0.1 mM IPTG at 25 °C for 12 hrs when the optical density of the culture reached 1.0 at 600 nm. LptM was purified from the membrane fractions by using 1.0% DDM. After the detergent-solubilized supernatants were incubated with pre-equilibrated Strep-tactin beads for 1 hr, LptMStrep protein was eluted with wash buffer containing 2.5 mM d-Desthiobiotin (Sigma).
LptM(or LPS), pBpa-containing LptB2FGC proteins (or pBpa-containing LolCDE proteins), MSP1D1 and Total Lipid were mixed at a molar ratio of 1:1:2.4:80 and incubated for 30 min at 4 °C. Detergents were removed by incubation with 0.8 mg ml−1 Bio-Beads SM2 (Bio-Rad) overnight at 4 °C. Nanodisc-embedded complexes were further purified using a Superose6 increase 10/300GL column (GE Healthcare) in a buffer containing 20 mM Tris-HCl (pH 8.0) and 150 mM NaCl. Peak fractions were combined and concentrated to ~0.6 mg ml−1. The reconstituted nanodisc samples were incubated with or without 2 mM ATP (and 2 mM MgCl2) at room temperature for various lengths of time (0–10 min), followed by UV radiation for 10 min when needed.
To carry out LptM transfer assays, nanodisc-embedded LolCDE-LptM complexes were mixed with either LolA(W70pBpa) or LptA(F95pBpa) at a molar ratio of 1:1 at room temperature. After addition of 2 mM ATP (and 2 mM MgCl2), the mixture was incubated for 10 min at room temperature, followed by UV radiation for 10 min when needed. LPS/LptM membrane-to-membrane transport was performed using a similar procedure but using nanodisc that contained different membrane protein complexes.
For western blot detection of crosslinks, the reaction mixtures were first separated by 15% SDS–PAGE gel, and transferred to PVDF membranes (Bio-Rad). Following blocking for 1 hr with TBST buffer [20 mM Tris-HCl (pH 8.0), 150 mM NaCl, 0.1% Tween-20] containing 8% skim milk, the PVDF membrane was incubated with either Strep tag II antibody (1:3000 dilution, AP1013a, YTHX, China) or anti-E. coli LPS antibody (1:3000 dilution, ab35654, Abcam) at room temperature for 1 hr. After washing with TBST buffer three times, the PVDF membrane was incubated with goat anti-rabbit or goat anti-mouse horseradish peroxidase (HRP)-conjugated secondary antibody (1:5000 dilution, LabLead, China) at room temperature for 1 hr. LptM or LPS crosslinks were visualized using an enhanced chemiluminescence detection kit (Applygen, China).
AlphaFold server structure prediction
Models of paLptDE-LppL, ecLptDE-LptM, paLptDE-SLPs (Lpp, YedD, YbjP, SlyB, SolP, Slp, YajG and Blc) were predicted in AlphaFold Server, which has additional capabilities, especially the ability to predict the structures of complexes containing multiple non-protein molecules63. The structure of the paLptDE-LppL complex was predicted using the residues 34-924 of Q9I5U2 for LptD, residues 20-207 of Q9HX32 for LptE, and residues 21-46 of P17323 for LppL (a homolog of LptM in Pseudomonas aeruginosa). The structure of the ecLptDE-LptM complex was predicted using the residues 25-784 of P31554 for LptD, residues 19–193 of P0ADC1 for LptE, and residues 20-67 of P0ADN6 for LptM. The structures of the paLptDE-SLPs (Lpp, YedD, YbjP, SlyB, SolP, Slp, YajG and Blc) complexes were predicted using the same paLptDE sequences mentioned above, along with the 20 amino acids following the N-terminal cysteine in each SLPs. Notably, the cysteine at the N-terminus of each SLPs is palmitoylated when predicted. All predicted models are available for download at https://figshare.com/s/0cfec19bc5da80ee484c.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The structural model and the diffraction data of the apo-paLptDE and paLptDE-LptM crystal structures are deposited in the Protein Data Bank with the accession codes 8H1S and 8H1R, respectively. All predicted models are available for download at https://figshare.com/s/0cfec19bc5da80ee484c. The MS data and original immunofluorescence images are available for download at https://figshare.com/s/3321115d11373a6b67d7. Source data are provided with this paper.
Change history
22 May 2025
In the version of the article initially published, Long Miao's second affiliation was incorrectly listed as affiliation 3 and has now been amended to affiliation 7 (Key Laboratory of RNA Biology, CAS Center for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing, 100101, China). An incorrect list names was originally published in the Peer review information. The affiliation and peer reviewers list are amended in the HTML and PDF versions of the article.
References
Nikaido, H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev.: MMBR 67, 593–656 (2003).
Konovalova, A., Kahne, D. E. & Silhavy, T. J. Outer membrane biogenesis. Annu. Rev. Microbiol. 71, 539–556 (2017).
May, K. L. & Silhavy, T. J. Making a membrane on the other side of the wall. Biochimica et. Biophysica Acta 1862, 1386–1393 (2017).
Chng, S. S., Ruiz, N., Chimalakonda, G., Silhavy, T. J. & Kahne, D. Characterization of the two-protein complex in Escherichia coli responsible for lipopolysaccharide assembly at the outer membrane. Proc. Natl Acad. Sci. USA 107, 5363–5368 (2010).
Bos, M. P., Robert, V. & Tommassen, J. Biogenesis of the gram-negative bacterial outer membrane. Annu. Rev. Microbiol. 61, 191–214 (2007).
Aliprantis, A. O. et al. Cell activation and apoptosis by bacterial lipoproteins through toll-like receptor-2. Science 285, 736–739 (1999).
Vijay, K. Toll-like receptors in immunity and inflammatory diseases: past, present, and future. Int. Immunopharmacol. 59, 391–412 (2018).
Poltorak, A. et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science 282, 2085–2088 (1998).
Sperandeo, P. et al. Functional analysis of the protein machinery required for transport of lipopolysaccharide to the outer membrane of Escherichia coli. J. Bacteriol. 190, 4460–4469 (2008).
Okuda, S., Sherman, D. J., Silhavy, T. J., Ruiz, N. & Kahne, D. Lipopolysaccharide transport and assembly at the outer membrane: the PEZ model. Nat. Rev. Microbiol. 14, 337–345 (2016).
Sperandeo, P. et al. Characterization of lptA and lptB, two essential genes implicated in lipopolysaccharide transport to the outer membrane of Escherichia coli. J. Bacteriol. 189, 244–253 (2007).
Ruiz, N., Gronenberg, L. S., Kahne, D. & Silhavy, T. J. Identification of two inner-membrane proteins required for the transport of lipopolysaccharide to the outer membrane of Escherichia coli. Proc. Natl Acad. Sci. USA 105, 5537–5542 (2008).
Sherman, D. J. et al. Decoupling catalytic activity from biological function of the ATPase that powers lipopolysaccharide transport. Proc. Natl Acad. Sci. USA 111, 4982–4987 (2014).
Tran, A. X., Dong, C. & Whitfield, C. Structure and functional analysis of LptC, a conserved membrane protein involved in the lipopolysaccharide export pathway in Escherichia coli. J. Biol. Chem. 285, 33529–33539 (2010).
Luo, Q. et al. Structural basis for lipopolysaccharide extraction by ABC transporter LptB2FG. Nat. Struct. Mol. Biol. 24, 469–474 (2017).
Dong, H., Zhang, Z., Tang, X., Paterson, N. G. & Dong, C. Structural and functional insights into the lipopolysaccharide ABC transporter LptB2FG. Nat. Commun. 8, 222 (2017).
Li, Y., Orlando, B. J. & Liao, M. Structural basis of lipopolysaccharide extraction by the LptB2FGC complex. Nature 567, 486–490 (2019).
Owens, T. W. et al. Structural basis of unidirectional export of lipopolysaccharide to the cell surface. Nature 567, 550–553 (2019).
Tang, X. et al. Cryo-EM structures of lipopolysaccharide transporter LptB2FGC in lipopolysaccharide or AMP-PNP-bound states reveal its transport mechanism. Nat. Commun. 10, 4175 (2019).
Dong, H. et al. Structural basis for outer membrane lipopolysaccharide insertion. Nature 511, 52–56 (2014).
Qiao, S., Luo, Q., Zhao, Y., Zhang, X. C. & Huang, Y. Structural basis for lipopolysaccharide insertion in the bacterial outer membrane. Nature 511, 108–111 (2014).
Botos, I. et al. Structural and functional characterization of the LPS transporter LptDE from gram-negative pathogens. Structure 24, 965–976 (2016).
Sherman, D. J. et al. Lipopolysaccharide is transported to the cell surface by a membrane-to-membrane protein bridge. Science 359, 798–801 (2018).
Okuda, S., Freinkman, E. & Kahne, D. Cytoplasmic ATP hydrolysis powers transport of lipopolysaccharide across the periplasm in E. coli. Science 338, 1214–1217 (2012).
Okuda, S. & Tokuda, H. Lipoprotein sorting in bacteria. Annu. Rev. Microbiol. 65, 239–259 (2011).
Konovalova, A. & Silhavy, T. J. Outer membrane lipoprotein biogenesis: lol is not the end. Philos. Trans. R. Soc. Lond. Ser. B, Biol. Sci. 370, 26370942 (2015).
Narita, S. I. & Tokuda, H. Bacterial lipoproteins; biogenesis, sorting and quality control. Biochimica et. biophysica acta 1862, 1414–1423 (2017).
Wilson, M. M. & Bernstein, H. D. Surface-exposed lipoproteins: an emerging secretion phenomenon in gram-negative bacteria. Trends Microbiol. 24, 198–208 (2016).
Cowles, C. E., Li, Y., Semmelhack, M. F., Cristea, I. M. & Silhavy, T. J. The free and bound forms of Lpp occupy distinct subcellular locations in Escherichia coli. Mol. Microbiol. 79, 1168–1181 (2011).
Sukupolvi, S. & O’Connor, C. D. TraT lipoprotein, a plasmid-specified mediator of interactions between gram-negative bacteria and their environment. Microbiological Rev. 54, 331–341 (1990).
Grabowicz, M. Lipoprotein transport: greasing the machines of outer membrane biogenesis: re‐examining lipoprotein transport mechanisms among diverse gram‐negative bacteria while exploring new discoveries and questions. Bioessays 40, 1700187 (2018).
Hooda, Y., Lai, C. C. & Moraes, T. F. Identification of a large family of slam-dependent surface lipoproteins in gram-negative bacteria. Front. Cell. Infect. Microbiol. 7, 207 (2017).
Cole, G. B., Bateman, T. J. & Moraes, T. F. The surface lipoproteins of gram-negative bacteria: protectors and foragers in harsh environments. J. Biol. Chem. 296, 100147 (2021).
Yang, Y. et al. LptM promotes oxidative maturation of the lipopolysaccharide translocon by substrate binding mimicry. Nat. Commun. 14, 6368 (2023).
He, H. et al. A Borrelia burgdorferi LptD homolog is required for flipping of surface lipoproteins through the spirochetal outer membrane. Mol. Microbiol. 119, 752–767 (2023).
Braun, V. & Bosch, V. Sequence of the murein-lipoprotein and the attachment site of the lipid. Eur. J. Biochem. 28, 51–69 (1972).
Botte, M. et al. Cryo-EM structures of a LptDE transporter in complex with Pro-macrobodies offer insight into lipopolysaccharide translocation. Nat. Commun. 13, 1–10 (2022).
Moehle, K. et al. Solution structure and dynamics of LptE from Pseudomonas aeruginosa. Biochemistry 55, 2936–2943 (2016).
Han, L. et al. Structure of the BAM complex and its implications for biogenesis of outer-membrane proteins. Nat. Struct. Mol. Biol. 23, 192–196 (2016).
Wu, T. et al. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell 121, 235–245 (2005).
Li, X., Gu, Y., Dong, H., Wang, W. & Dong, C. Trapped lipopolysaccharide and LptD intermediates reveal lipopolysaccharide translocation steps across the Escherichia coli outer membrane. Sci. Rep. 5, 11883 (2015).
Ryu, Y. & Schultz, P. G. Efficient incorporation of unnatural amino acids into proteins in Escherichia coli. Nat. Methods 3, 263–265 (2006).
Sharma, S. et al. Mechanism of LolCDE as a molecular extruder of bacterial triacylated lipoproteins. Nat. Commun. 12, 1–11 (2021).
Tang, X. et al. Structural basis for bacterial lipoprotein relocation by the transporter LolCDE. Nat. Struct. Mol. Biol. 28, 347–355 (2021).
Bei, W. et al. Cryo-EM structures of LolCDE reveal the molecular mechanism of bacterial lipoprotein sorting in Escherichia coli. PLoS Biol. 20, e3001823 (2022).
Grabowicz, M. & Silhavy, T. J. Redefining the essential trafficking pathway for outer membrane lipoproteins. Proc. Natl Acad. Sci. USA 114, 4769–4774 (2017).
Kaplan, E., Greene, N. P., Crow, A. & Koronakis, V. Insights into bacterial lipoprotein trafficking from a structure of LolA bound to the LolC periplasmic domain. Proc. Natl Acad. Sci. 115, E7389–E7397 (2018).
Hooda, Y. et al. Slam is an outer membrane protein that is required for the surface display of lipidated virulence factors in Neisseria. Nat. Microbiol. 1, 16009 (2016).
Hooda, Y. & Moraes, T. F. Translocation of lipoproteins to the surface of gram negative bacteria. Curr. Opin. Struct. Biol. 51, 73–79 (2018).
Hooda, Y., Shin, H. E., Bateman, T. J. & Moraes, T. F. Neisserial surface lipoproteins: structure, function and biogenesis. Pathog. Dis. 75, 28158534 (2017).
Konovalova, A., Perlman, D. H., Cowles, C. E. & Silhavy, T. J. Transmembrane domain of surface-exposed outer membrane lipoprotein RcsF is threaded through the lumen of β-barrel proteins. Proc. Natl Acad. Sci. 111, E4350–E4358 (2014).
Oomen, C. J. et al. Structure of the translocator domain of a bacterial autotransporter. EMBO J. 23, 1257–1266 (2004).
Dong, C. et al. Wza the translocon for E. coli capsular polysaccharides defines a new class of membrane protein. Nature 444, 226–229 (2006).
Goyal, P. et al. Structural and mechanistic insights into the bacterial amyloid secretion channel CsgG. Nature 516, 250–253 (2014).
Cao, B. et al. Structure of the nonameric bacterial amyloid secretion channel. Proc. Natl Acad. Sci. USA 111, E5439–E5444 (2014).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 276, 307–326 (1997).
Adams, P. D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D. 58, 1948–1954 (2002).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr D. 60, 2126–2132 (2004).
Bailey, S. The Ccp4 Suite - programs for protein crystallography. Acta Crystallogr D. 50, 760–763 (1994).
DeLano, W. L. PyMOL molecular viewer: updates and refinements. Abstr. Pap. Am. Chem. Soc. 238 (2009).
Konovalova, A., Perlman, D. H., Cowles, C. E. & Silhavy, T. J. Transmembrane domain of surface-exposed outer membrane lipoprotein RcsF is threaded through the lumen of beta-barrel proteins. Proc. Natl Acad. Sci. USA 111, E4350–E4358 (2014).
Ritchie, T. et al. Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol. 464, 211–231 (2009).
Abramson, J. et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 630, 493–500 (2024).
Acknowledgements
The authors would like to thank Dr. Chérine Bechara at University of Oxford for preliminary native mass spectrometry analysis of the paLptDE protein sample; Dr. Marcin Grabowicz at Emory University School of Medicine provided the lolA-depleted MG3324 (NR754 Δlpp ΔrcsB zii::Tn10 cpxA24 ΔlolA) E. coli strain; Xing Jia and Shuoguo Li from Center for Biological Imaging (CBI), Institute of Biophysics, Chinese Academy of Sciences for their help of taking and analyzing SIM images; Xiang Ding and Lili Niu from the Mass Spectrometry Core Facility of the Institute of Biophysics for mass spectrometry identification. The diffraction data were collected at beamlines BL19U and BL17U of the Shanghai Synchrotron Radiation Facility (SSRF, China). This work was supported by grants from National Key Research and Development Project of the Ministry of Science and Technology, China (2021YFF0700201·to YH.), Special·Fund for Strategic Pilot Technology·of Chinese Academy of Sciences (XDB37020201 to·Y.H.) and the National Science Foundation (MCB-1727508 and MCB-1810695 to W.I.).
Author information
Authors and Affiliations
Contributions
Y.H. conceived and supervised the project. Q.L., C.W., S.Q., S.Y., W.B., L.C., S.K., J.Z. Y.Z. and K.W. performed the experiments. C.W., S.Q., Q.L., J.Z., and Y.H. collected diffraction data, built the model, and refined the structure; K.W. and M.Z. performed the native mass spectrometry analysis; L.C. and L.M. carried out the immunofluorescence assays; S.K. and W.I. contributed to the structure analysis; F.W., X.L. and M.H. provided materials. X.L., J.L., Q.L., M.Z., W.B. and Y.H. wrote the manuscript with the input from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Paola Sperandeo, Christopher Cassidy, Bert van den Berg, Shan Feng and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Luo, Q., Wang, C., Qiao, S. et al. Surface lipoprotein sorting by crosstalk between Lpt and Lol pathways in gram-negative bacteria. Nat Commun 16, 4357 (2025). https://doi.org/10.1038/s41467-025-59660-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-59660-y