Abstract
Oncogene-induced replicative stress (RS) drives tumor progression by disrupting genome stability, primarily through transcription-replication conflicts (TRCs), which promote R-loop accumulation and trigger the DNA damage response (DDR). In this study, we investigate the role of chromatin regulators in exacerbating TRCs and R-loop accumulation in cancer. We find that in breast cancer patients, the simultaneous upregulation of MYC and the H2A.Z-specific chaperone ANP32E correlates with increased genomic instability. Genome-wide analyses reveal that ANP32E-driven H2A.Z turnover alters RNA polymerase II processivity, leading to the accumulation of long R-loops at TRC sites. Furthermore, we show that ANP32E overexpression enhances TRC formation and activates an ATR-dependent DDR, predisposing cancer cells to R-loop-mediated genomic fragility. By exploiting the vulnerability of ANP32E-expressing cancer cells to ATR inhibitors, we find that tumors relied on this DDR pathway, whose inhibition halts their pro-metastatic capacity. These findings identify ANP32E as a key driver of TRC-induced genomic instability, indicating ATR inhibition as a potential therapeutic strategy for ANP32E-overexpressing tumors.
Similar content being viewed by others
Introduction
Triple-negative breast cancer (TNBC) is the most aggressive breast cancer (BC) subtype due to its high recurrence rates, metastatic potential, and heterogeneity1. One determinant of the molecular variability within tumors is genomic instability, which enhances the adaptability and survival of pro-metastatic cancer cells in hostile microenvironments. Genomic instability manifests in various forms, including increased frequency of point mutations, copy number alterations (CNA), or chromosomal instability (CIN), characterized by structural and numerical chromosomal changes over time2. While the underlying molecular mechanisms remain under investigation, it has been established that oncogene-induced replicative stress (RS)3 plays a central role in tumor progression. Among oncogenes, MYC is a major contributor to RS across various cancers and is the most frequently amplified gene in TNBC4,5,6. Certain genomic regions known as common fragile sites (CFSs), are susceptible to RS, making them hotspots for genomic instability7.
Although eukaryotic cells have evolved mechanisms to spatially and temporally separate transcription and replication, RS may inevitably lead to transcription-replication conflicts (TRCs). These collisions between transcription and replication machinery frequently cause DNA damage and genomic lesions8,9. If unresolved or improperly managed by the DNA damage response (DDR), TRCs can result in double-strand breaks (DSBs), further exacerbating genome instability10. Accordingly, cancer cells harboring frequent TRCs rely on DNA repair pathways to withstand these genomic challenges, thus providing an opportunity for therapeutic intervention11. Cancer patients with DDR-addicted tumors would therefore benefit from selective DDR inhibition.
DNA damage resulting from TRC is further worsened by R-loops, which are co-transcriptional DNA:RNA hybrids12. R-loops can exacerbate TRCs by stalling RNA polymerase II (RNApol II) along the gene body or impeding replication fork progression12,13. Recent studies have highlighted a complex interplay between R-loops and many chromatin factors to control RNApol II density and processivity14,15. Different histone chaperones like FACT, INO80, and PSIP1, as well as helicases like DH9, have been implicated in R-loop homeostasis associated TRCs16,17,18,19,20. Indeed, R-loops play a role in regulating RNApol II promoter-proximal pause-release21, a key checkpoint in transcriptional elongation. Nucleosome composition and positioning represent the first barrier controlling RNApol II escape and processivity along the gene body22. Specific chromatin factors, including remodelers and histone chaperones, participate in different steps of nucleosome assembly and recycling23. For instance, H2A.X and H2A.Z variants of histone H2A are exchanged by specific chaperones at sites of DNA damage and in the proximity of the transcription start site (TSS), respectively24. H2A.Z is enriched at the +1 nucleosome of active promoters and at enhancers, correlating with paused RNApol II25,26,27. Compared to other histone variants, H2A.Z weakens nucleosome stability, facilitating transcriptional elongation. Its turnover is modulated by the histone chaperone ANP32E28,29, playing a critical role in modulating RNApol II pause-release30,31. Notably, emerging evidence suggests distinct functions for H2A.Z at promoters and gene bodies, underscoring the complexity of its regulatory function26.
Based on these insights, we hypothesized that altering chromatin players could amplify oncogene-induced RS, increasing TRC and R-loop formation, thereby promoting genomic instability. In the present work, we found that ANP32E is the most frequently amplified and overexpressed histone chaperone in TNBC patients characterized by MYC deregulation. ANP32E is encoded on the q21 arm of chromosome 1, which is a frequently amplified genomic region in BC patients, often detected as an early initiating event characterizing non-cancer lesions and in situ carcinomas32,33,34. While 1q21 amplification has been linked to BC recurrence35, no certain connection with specific oncogenes was reported36,37. ANP32E has been identified as a prognostic marker in TNBC, where its overexpression correlates with reduced disease-free survival and increased cancer growth in mouse xenograft models38. However, the molecular mechanisms by which ANP32E elicits tumor progression remains unexplored. One of ANP32E’s best-characterized roles is its cooperation with the NuA4/P400/TIP60 complex to regulate H2A.Z turnover. Among its function, P400 is rapidly recruited to DSB to initially deposit H2A.Z followed by ANP32E-mediated eviction, which contributes to chromatin remodeling and proper DNA repair39,40,41. Therefore, dysregulation of the chromatin regulatory network in cancer cells may not only influence oncogene activation but also affect DDR, contributing to genome instability.
In this study, we demonstrate that ANP32E overexpression exacerbates oncogene-induced RS by promoting TRC formation, R-loop accumulation, and genomic instability. Furthermore, we explore the therapeutic potential of targeting ANP32E-driven vulnerabilities in TNBC.
Results
ANP32E expression correlates with MYC and genome instability in TNBC
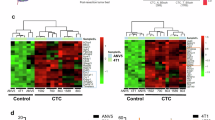
To identify histone chaperones that drive genomic instability in TNBC undergoing MYC-induced RS, we analyzed public datasets of BC patients. Given the positive correlation between MYC expression and its genomic amplification in Basal BC (mostly TNBC) (Supplementary Fig. 1a)42, we screened for histone-binding proteins upregulated in MYC-amplified Basal BC patients, when compared to the other-BC subtypes. Among chromatin binding factors, the histone chaperone ANP32E emerged as the most significantly upregulated, contrasting with other genome-maintenance-related chaperones, whose expression remained constant (Fig. 1a and Supplementary Data 1). Importantly, we observed that ANP32E is among the most frequently amplified genes in Basal BC patients (Supplementary Fig. 1b) and its expression correlates with increased copy number alterations (CNA), indicating potential clinical relevance (Fig. 1b). To explore whether MYC and ANP32E deregulation jointly contribute to genomic instability, we stratified BC patients based on MYC and ANP32E overexpression and corresponding copy number gains (hereafter referred to as MYC-ANP32E). We found that patients with concomitant MYC and ANP32E upregulation exhibited a significant increase in the fraction of the altered genome (FGA) (Fig. 1c). Importantly, this increase was not observed when ANP32E is co-upregulated with other RS-inducing oncogenes like BCL2, MDM2, and CCND1 (Supplementary Fig. 1c), reinforcing the specificity of the MYC-ANP32E axis in the context of BC genomic instability (Supplementary Fig. 1c)43. Moreover, MYC and ANP32E co-amplification was more frequent in Basal BC with respect to the other-BC subtypes (Supplementary Fig. 1d). Basal BCs are the most affected by genome instability in comparison with the other-BC subtypes, as confirmed by higher mutation counts, FGA, and microsatellite instability (MSI) (Supplementary Fig. 1e–g)44. To further validate the correlation between MYC-ANP32E upregulation and genome instability, we leveraged data from the Metabric study, which clustered patients based on cancer driver CNA affecting gene expression45,46. Interestingly, we observed that Cluster 9 and Cluster 10, resembling genomically unstable ER+ and Basal TNBC patients respectively, were characterized by the highest frequency of MYC-ANP32E co-upregulation (Fig. 1d and Supplementary Fig. 1h). This trend was particularly evident when comparing Cluster 10 with the other clusters of TNBC patients (Cluster 4ER−), which is more genomically stable and characterized by low frequency of MYC-ANP32E co-upregulation (Fig. 1d and Supplementary Fig. 1h). Although ANP32E overexpression has been reported as a negative prognostic factor in TNBC38, our analysis revealed that MYC-ANP32E co-upregulation further worsens TNBC prognosis, surpassing the adverse effects of individual MYC or ANP32E upregulation (Fig. 1e and Supplementary Fig. 1i). Considering that TNBC mortality is linked to metastasis, we analyzed an additional dataset segregating patients based on metastatic status (Metastatic Breast Cancer Project 2021). We found a higher frequency of MYC-ANP32E co-upregulation in TNBC metastatic patients, suggesting that it may provide a selective advantage to pro-metastatic cancer cells (Fig. 1f).
a Volcano plot of differentially expressed genes between MYC-CNA patients with Basal-BC (n = 61) or Other-BC subtypes (n = 92), based on TCGA data. Wald test p values (Null-hp: log2FC = 0) shown. Dashed lines indicate significance thresholds: log2FC > ±1.5 and −log10 p value >5. b Density plot showing the frequency of Basal BC patients with genomic CNA, stratified by ANP32E expression quartiles (mRNA z-scores, log RNA-Seq V2 RSEM). Statistical significance: unpaired two-tailed t-test, n = 171, exact p values shown in plots (TCGA data). c Tukey Boxplot (25th–75th percentile and median) of FGA genome in BC patients grouped by MYC and ANP32E gain/expression. Double positivity: 75° expression for both genes + copy number gain. Two-tailed Wilcoxon test, exact p values, and n are shown in the plot (TCGA data). d Stacked barplot of MYC/ANP32E upregulation in BC patients, stratified by Metabric IntClust, sorted by genomic instability. e Kaplan–Meier survival plot for Basal BC patients with MYC/ANP32E upregulation (TCGA data). Double positivity as in (c). Log-rank test p values indicate statistical significance. f Barplot of TNBC patients with MYC/ANP32E co-amplification, grouped by metastasis status (data from Metastatic BC project 2021). g Normalized rank sums of somatic CNA initiation times in 22 BC patients (ref. 34). Zoom-ins of chr1q/chr8q highlight ANP32E/MYC positions. Dashed line: 95th percentile significance threshold. h Barplot of DDR pathway genes correlating with ANP32E expression. Significance threshold from random gene set analysis. Pearson correlation score ±0.85 threshold.
To determine whether ANP32E contributes to the observed augmented genome instability, we investigated the evolution of CNA in BC patients34. Timeline analysis highlighted that CNA of chr1q encompassing ANP32E represents an early event, preceding MYC amplification (Fig. 1g). Furthermore, among the genes belonging to the chr1q, ANP32E showed the strongest association with genomic instability (Supplementary Fig. 2a), correlating with MYC upregulation (Supplementary Fig. 2a). Clinical relevance of these findings was corroborated by a multivariate analysis, which identified ANP32E upregulation as a specific predictor of poor survival in basal BC (Supplementary Fig. 2c).
Genomic instability often accumulates in cancers defective in DDR pathways47,48,49 and basal BCs commonly harbor BRCA1/2 and homologous recombination repair (HRR) deficiencies50,51. However, ANP32E overexpression-driven genomic instability was independent of BRCA1/2 or HRR mutations (Supplementary Fig. 2d, e). We, therefore, looked at the correlation between ANP32E expression and the transcriptional regulation of specific DDR pathway genes in Basal BC patients, using FOXA1 and MYC expression as negative and positive controls, respectively. We found that several DDR pathway genes show an upregulation correlating with ANP32E expression (Fig. 1h left and Supplementary Fig. 2f). This correlation was not observed with MYC or FOXA1 as reference genes, the latter being inversely associated with hormone receptor-negative BCs52. On the contrary, we found no DDR pathway that is downregulated in accordance with ANP32E expression (Fig. 1h right and Supplementary Fig. 2f). Importantly, we found that the Fanconi Anemia (FA) pathway, involved in resolving RS-derived genome instability, exhibited the strongest correlation with ANP32E deregulation (Fig. 1h left). Collectively, these findings indicate that in BC patients, upregulation of ANP32E and MYC correlated with an increase in genome instability and tumor progression, with ANP32E amplification representing an early oncogenic event.
ANP32E elicits R-loop-mediated TRCs and DNA damage
Despite its relevance in cancer, the mechanism by which ANP32E contributes to tumor aggressiveness and genome instability remains unclear. At first, we investigated the functional relationship between MYC and ANP32E and found that MYC binds to the ANP32E promoter in multiple cancer cell lines (https://maayanlab.cloud/Harmonizome/gene_set/MYC/ENCODE+Transcription+Factor+Targets). In addition, the induction of MYC expression in immortalized human mammary epithelial cells (IMEC)48 led to a concomitant upregulation of ANP32E and RS factors Chk1, pChk1, RPA32, and pRPA32 (Supplementary Fig. 3a, b)53,54. Deregulated transcription can lead to collisions between the transcriptional and replicative machineries, giving rise to TRCs and DNA damage. Since ANP32E regulates the turnover of the histone variant H2A.Z, which contributes to transcription regulation55, we investigated its role in TRC formation in BC. Using transformed IMECs (tIMECs) stably expressing MYC and mutant PI3KCA53, we constitutively overexpressed ANP32E to match its TNBC expression levels, generating tIMEC-A cells (Supplementary Fig. 3c, d). Since TRCs associate with R-loops accumulation12,56, we monitored their abundance in tIMEC and tIMEC-A cells. As a control, we also generated tIMEC-A cells that stably overexpressed RNaseH1 (thereafter named tIMEC-A-H1), which specifically degrades DNA:RNA hybrids (Supplementary Fig. 3e). ANP32E overexpression significantly increased R-loop formation, which was rescued by both exogenous treatment with RNaseH1 and its endogenous overexpression (Fig. 2a and Supplementary Fig. 3f). This result was further confirmed by proximity ligation assays (PLA) showing R-loop accumulation at RNA polymerase II (RNApol II) elongation sites and in proximity to ANP32E, supporting a direct involvement of ANP32E in R-loops dynamics57 (Fig. 2b, d and Supplementary Fig. 3h). In addition, TRCs frequency, measured as the collision between elongating RNApol II and PCNA, was elevated in tIMEC-A cells and reversed by RNaseH1 (Fig. 2c, d). Importantly, the increase of TRCs in tIMEC-A and its dependency on R-loops was also confirmed by co-immunoprecipitation of pS2RNApol II and PCNA (Supplementary Fig. 3i).
a Dot blot showing R-loop abundance in tIMEC, tIMEC-A, and tIMEC-A-H1 cells ± RNaseH1. dsDNA used as internal normalizer. Quantification from three biological replicates (mean ± s.e.m.), unpaired two-tailed t-test p values shown. b Representative PLA images showing proximity between ANP32E and R-loops. Scale bar = 10 µm. c PLA images of pS2RNApol II-PCNA proximity as TRCs marker. Scale bar = 10 µm. d Quantification of PLA foci/nucleus from (b, c): tIMEC n = 106/104, tIMEC-A n = 85/84, tIMEC-A-H1 n = 72/75. Unpaired two-tailed t-test p values are shown. e Top: FANCD2 immunofluorescence in EdU+ cells (EdU channel excluded for clarity). Bottom: quantification of FANCD2 foci/nucleus in EdU+ (green violins) and EdU– (gray violins) cells; unpaired two-tailed t-test p values shown. Scale bar = 10 µm. f Alkaline comet assay ± VE822 (1.25 µM, 24 h). Top: representative images, comet quantification (OpenComet)94, and DNA intensity heatmaps (CometScore)95. Bottom: violin plots of comet tail length; 150–300 cells/condition, three biological replicates. Unpaired two-tailed t-test was used for p value calculation. g Left: representative binucleated (BN) cells with micronuclei (MNi) after Cytochalasin B + Mitomycin C treatment. Right: %BN cells with MNi ± Mitomycin C (0.03 µg/ml) or VE822 (0.027, 0.08, 0.25 µM) treatment. ≥3000 cells/condition on three biological replicates. The barplot reports mean ± s.d. with two-way ANOVA adjusted p values. h Immunostaining of BN cells with MNi that are positive or negative for centromere staining (CREST) after VE822. Phalloidin marks cytoplasm; arrows indicate MNi. Scale bar = 10 µm. i Barplot showing the percentage of BN cells with DSB-derived (acentric) MNi ± Mitomycin C or VE822 treatment. Mean ± s.e.m. from three replicates; 25–60 BN cells/condition were quantified per replicate.
Given the observed ANP32E-dependent increase of R-loops and TRCs, we investigated its impact on DNA double-strand break (DSB) accumulation. By immunofluorescence analysis, we found that S-phase (EdU+) tIMEC-A showed an increased number of ubiquitinated FANCD2 foci, an ATR-dependent DDR factor that is recruited to chromatin by binding R-loops in proximity of stalled replication forks (Fig. 2e)58,59. Next, we performed a Comet assay in the presence or absence of two DNA damage-stimulating drugs: Etoposide (ETP)60 and VE822, an ATR inhibitor (ATRi) that impedes ATR-mediated DDR activation61. We observed that upon ETP treatment, ANP32E overexpression induced a R-loop-dependent increase in DSBs that was reversed by RNaseH1 upregulation (Supplementary Fig. 4a). ATR inhibition (VE822) further exacerbated DSB accumulation in tIMEC-A cells, highlighting their dependency on DDR activation (Fig. 2f). To test the severity of the accumulated DSBs, we measured the formation of Micronuclei (MNi), small nuclear-like structures containing massively damaged DNA. Despite the absence of a significant difference in untreated cells, VE822 treatment led to a significant increase in MNi formation in tIMEC-A (Fig. 2g and Supplementary Fig. 4b). However, R-loops resolution by RNaseH1 overexpression did not rescue the accumulation of MNi. To gain further insights, we tested whether R-loops are specifically implicated in clastogenic MNi subclass, that derive from unresolved DSBs62. Thus, we measured the fraction of MNi containing clastogenic chromosomes, characterized by the absence of centromeres (CREST-negative). We found that clastogenic MNi were more abundant in cells overexpressing ANP32E and their increase elicited by VE822 treatment was both ANP32E and R-loop-dependent (Fig. 2h, i and Supplementary Fig. 4b, d). To exclude that the described mechanism is restricted to the employed cellular model, we selected MDA-MB-231 and SUM-159-PT as two alternative TNBC cell lines that express either high or low levels of ANP32E, respectively, in the presence of upregulated MYC (Supplementary Fig. 5a, b). By either silencing or overexpressing ANP32E in such alternative models (named MB-231sh and SUM-159-A, respectively), we confirmed that ANP32E overexpression correlates with R-loop accumulation, TRCs, and DNA DSBs (Supplementary Fig. 5c–k). In addition, we excluded that the observed differences were caused by ANP32E-mediated alterations of S-phase or R-loop prone genes (Supplementary Fig. 5l, m). In summary, we found that ANP32E upregulation in TNBC is associated with increased TRCs and DSBs, which predispose cells to genomic fragility via a mechanism involving R-loops. This phenotype resulted being worsened upon ATR inhibition, highlighting a putative vulnerability of BC cells with such a biological background.
ANP32E stimulates ATR-dependent DDR and alteration of transcription and replication dynamics
The augmented genome instability observed upon ATR inhibition in TNBC cells with ANP32E overexpression suggested a sustained activation of ATR-mediated DDR in conjunction with high ANP32E levels. We therefore performed immunofluorescence analysis of two key ATR-dependent DDR players, p(S33)RPA32 and 53BP163,64,65,66,67 in S-phase cells (EdU+), when TRCs are most likely to occur. We observed a pronounced punctate distribution of these DDR markers in EdU+ tIMEC-A cells, indicating DDR activation at discrete genomic loci that was rescued by R-loops degradation. As expected, ATR inhibition prevented their activation (Fig. 3a–d). Of importance, cell cycle analysis revealed no substantial differences among the cell lines, with a mild accumulation of cells in G1/S upon ATRi treatment (Supplementary Fig. 6a). Furthermore, we found that the number of foci-positive cells for both DDR markers increased in tIMEC-A cells, indicating a more widespread DDR response across the cell population (Fig. 3e). Interestingly, ATR inhibition by VE822 led to a unique phenotype, where nuclei in tIMEC-A cells were fully positive for pRPA32 staining, indicative of excessive accumulation of DNA damage68 preceding cell death (Fig. 3a, f). While 53BP1 foci were not entirely abolished, they were larger in size in ATR-inhibited cells, suggesting persistence of unresolved damage from previous cell divisions (Supplementary Fig. 6b)69,70. These findings were further validated in MDA-MB-231 and SUM-159-PT cell lines, where silencing or overexpression of ANP32E led to corresponding decreases and increases in pRPA32 and 53BP1 recruitment, respectively (Supplementary Fig. 6c–f).
a Representative p(S33)RPA32 immunofluorescence images in EdU+ cells (S-phase), untreated (left) or treated with VE822 (0.25 µM, 6 h, right). Merge shows DAPI (blue), EdU (green), and pRPA32 (red); EdU channel excluded in zoom-ins for clarity. Scale bar = 10 µm. b Quantification of pRPA32 foci/nucleus in EdU+ cells ± VE822 treatment. Unpaired two-tailed t-test p values are shown. c Representative 53BP1 immunofluorescence in EdU+ cells, untreated or VE822-treated as in (a). Merged DAPI, EdU, and 53BP1 channels shown; EdU excluded in zoom-ins. Scale bar = 10 µm. d Quantification of 53BP1 foci/nucleus in EdU+ cells ± VE822. Unpaired two-tailed t-test p values reported. e Barplots of % EdU+ cells with ≥ 1 foci for pRPA32 (left) or 53BP1 (right) under untreated conditions. Mean ± s.d. from four biological replicates; unpaired two-tailed t-test p values shown. f Barplot of % cells with pan-nuclear pRPA32 staining after VE822 (0.25 µM, 6 h) treatment. Mean ± s.d. from four replicates; p values from unpaired two-tailed t-test. g Representative images of ongoing (top) and multiorigin (bottom) replication forks. Scale bar = 10 µm. Schematic of CldU–IdU sequential labeling is shown below. h Tukey boxplots of replication fork speed (top) and inter-origin distance in multiorigin fibers (bottom) from a representative experiment. Total fibers: tIMEC = 339, tIMEC-A = 335, tIMEC-A-H1 = 248; multiorigin fibers: tIMEC = 90, tIMEC-A = 68, tIMEC-A-H1 = 43. Unpaired two-tailed t-test p values reported.
An alternative explanation for the differential activation of DDR markers is the possibility that ANP32E directly participates in recruiting DDR factors to damaged sites. To test this, UV laser microirradiation was used to induce DNA damage independent of TRCs, and recruitment of pRPA32 and 53BP1 was assessed in cells with either ANP32E knockdown or overexpression. Importantly, we observed that the recruitment of both DDR factors was functional and comparable regardless of ANP32E levels, indicating that ANP32E modulation does not interfere with DDR factor recruitment (Supplementary Fig. 7a–c).
DNA repair is a time-costly biological process that can negatively impact transcription and replication dynamics71,72. We therefore asked whether the increased frequency of TRCs and sustained DDR activation observed in ANP32E-overexpressing cells might interfere with transcription and replication kinetics. To test this, we performed EU labeling of nascent transcripts following transcription elongation synchronization with DRB treatment73. We measured reduced levels of nascent RNA in tIMEC-A cells at the steady state, with the difference becoming more pronounced after DRB treatment and RNApol II release. This suggested that transcriptional dynamics were slower in tIMEC-A cells (Supplementary Fig. 7d, e). By performing a DNA fiber assay to measure replication fork speed74 we observed a slower progression of replication forks and an increased frequency of multiple origin fibers in tIMEC-A, with origins firing closer together. The slowed replication dynamics were rescued by R-loops degradation via RNaseH1 overexpression, suggesting that R-loops interfere with fork progression (Fig. 3g, h, and Supplementary Fig. 7f). The presence of fork stalling or barriers was further supported by increased asymmetry in bidirectional replication forks, indicating that the decreased speed of replication did not involve fork arrest mediated by checkpoint pathway activation (Supplementary Fig. 7g, h). In summary, ANP32E overexpression resulted in sustained activation of ATR-dependent DDR in actively replicating cells through a mechanism involving R-loops formation and altering transcription and replication dynamics.
ANP32E overexpression leads to stalling of the elongating RNApol II
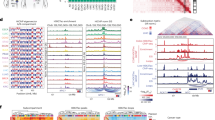
To determine the molecular mechanisms by which ANP32E elicits TRCs, we investigated whether its overexpression enhanced the turnover of histone variant H2A.Z across the genome. By performing CUT&RUN, we found that ANP32E overexpression increased its chromatin binding at genomic sites enriched for H2A.Z and P400, a core component of the P400/TIP60 complex that interacts with ANP32E24 (Supplementary Fig. 8, 10a and Supplementary Table 1). Of note, the overexpression of ANP32E reduced the levels of H2A.Z at open chromatin regions, resulting in a gain of acetylated H2A.Z (Supplementary Fig. 10b, c), in line with previous findings75,76. Notably, both ANP32E and its partner P400 exhibited decreased chromatin binding at these sites, consistent with the recent finding that ANP32E evicts H2A.Z to form stable ANP32E-H2A.Z dimers that are retained in the nucleoplasm (Supplementary Fig. 10c)28,77. We next employed CUT&RUN data for initiating (pS5) and elongating (pS2) RNApol II to evaluate ANP32E effect on RNApol II promoter-proximal pausing and its stalling along the gene body. We observed a decreased abundance of initiating RNApol II in the proximity of the TSS of ANP32E-overexpressing cells, possibly reflecting the less stable barrier posed by H2A.Z eviction at the +1 nucleosomes and the increased escape from transcription initiation (Fig. 4a and Supplementary Figs. 8, 9)78,79. This shift in RNApol II positioning was confirmed by the redistribution of paused pS5 RNApol II, which moved downstream from the TSS in ANP32E-overexpressing cells, while it remained near the TSS in control cells (Fig. 4b). Furthermore, by mapping the distance of pS2RNApol II from the TSS, we observed that while in tIMEC it spread more uniformly along the gene body, in tIMEC-A it was enriched towards promoter-proximal regions (Fig. 4a, b). This suggests that elongating RNApol II encounters stalling events within the gene body in ANP32E-overexpressing cells. Both observations were confirmed by ChIP-qPCR on selected regions (Supplementary Fig. 10d). To further explore this finding, we calculated the Pausing Index (PI) and Stalling Index (SI), a measure of RNApol II promoter-proximal pausing and stalling along the gene body, respectively (Supplementary Fig. 10e). Quantitative evaluation of PI and SI revealed that, while initiating RNApol II showed a lower PI in ANP32E-overexpressing cells, the SI was significantly increased for the elongating RNApol II in tIMEC-A cells. This indicated that ANP32E overexpression is associated with more frequent stalling of RNApol II during transcription elongation. Interestingly, R-loops degradation rescued this effect (Fig. 4a, b and Supplementary Fig. 10e). Further studies are needed to determine whether R-loops degradation restores RNApol II processivity or it contributes to resolve premature termination of stalled RNApol II, reducing transcriptional stress15,80.
a Cumulative CUT&RUN signal plots for pS5- and pS2RNApol II at ATAC-seq promoter clusters 1–5 (10,821 accessible sites), sorted by pS5RNApol II signal. Regions were scaled on gene body (TSS–TES) ± 3 kb. b Heatmaps of pS5 (pausing) and pS2 (stalling) peak density, plotted as distance from the nearest TSS (–0.5 to +10 kb). Dashed lines indicate 50%, mean, and 75% of peak distributions for pS5- and pS2RNApol II samples. c Cumulative plot of nucleosome occupancy scores in tIMEC at high (Q4) pS5RNApol II accessible regions; number of regions shown on each plot. d Heatmap of H2A.Z peak density at the highest (Q4) pS2RNApol II stalling regions, shown relative to nearest TSS (–0.5 to +10 kb); 50%, mean, and 75% values connected by dashed lines across cell lines. e Cumulative CUT&RUN signal plots for immunoprecipitated factors at RNApol II-sorted ATAC-seq promoter clusters 1–5 (10,821 sites), centered on TSS ± 500 bp. f Cumulative CUT&RUN signal plots for immunoprecipitated factors at pS2RNApol II stalling regions, grouped by stalling index quartiles (n = 2827 regions per quartile). Signal shown from −3 kb to +6 kb relative to the stalling start site (SSS).
We hypothesized that the misregulated processivity of RNApol II directly depends on the altered H2A.Z turnover due to its effects on nucleosome stability. Therefore, we employed ATAC-seq data to calculate the nucleosome occupancy at sites enriched for initiating RNApol II and harboring H2A.Z (Supplementary Fig. 10f). In ANP32E-overexpressing cells, nucleosome occupancy was reduced downstream of the TSS and within the gene bodies, which was consistent with the enhanced removal of H2A.Z (Fig. 4c, d). Of note, this difference was lost when we analyzed TSS characterized by a low abundance of initiating RNApol II (Supplementary Fig. 10g). By analyzing H2A.Z peaks we found a different distribution downstream the TSS among the cell lines, consistently with the enhanced removal of H2A.Z occurring in ANP32E-overexpressing cells (Fig. 4d). Given the partial recovery of H2A.Z distribution upon RNaseH1 overexpression, we compared peaks of chromatin-bound ANP32E, finding that 85% of the target genes were preserved between tIMEC-A and tIMEC-A-H1 cells (Supplementary Fig. 10h). This result excluded the possibility that the observed phenotype was driven by an altered binding of ANP32E due to R-loops degradation. Overall, these findings suggest that ANP32E overexpression leads to transcriptional stress by altering H2A.Z turnover and nucleosome stability, resulting in the stalling of elongating RNApol II during transcription elongation.
ANP32E-dependent accumulation of DDR factors at RNApol II stalling sites
We hypothesized that the increased pausing of the RNAPol II may raise the frequency of TRC downstream of the TSS. Therefore, by using the initiating RNApol II as a reference of transcriptionally active genes, we looked at the binding of the TRC-related DNA damage factors pRPA32, γH2A.X, and TOP2A, a topoisomerase involved in the resolution of DNA supercoiling occurring in between the colliding transcriptional and replicative machinery71 (Fig. 4e, Supplementary Fig. 8, and Supplementary Table 1). We observed that actively transcribed regions in ANP32E-overexpressing cells showed an increased recruitment of pRPA32 downstream of the TSS, with a milder accumulation of γH2A.X in the surrounding genomic regions (Fig. 4e). Notably, this pattern was partially rescued by RNaseH1-mediated R-loop resolution, suggesting that ANP32E overexpression may enhance the loading of TRC-related DNA damage factors (Fig. 4e). To clarify whether the TRC-associated DDR factors are deposited in proximity of RNApol II stalling sites, we clustered the genomic regions based on the SI quartiles of elongating RNApol II and analyzed their relative accumulation. Interestingly, we observed the shift between pS5- and pS2RNApol II from the start of the stalling site (SSS) with an intensity that correlates with higher SIs of elongating RNApol II (Supplementary Fig. 10i). This pattern was mirrored by a gradual increase of H2A.Z abundance in the surrounding of stalling sites, which was counteracted by the chaperone activity of ANP32E (Fig. 4f). Moreover, pRPA32 and, to a lesser extent, TOP2A accumulated at stalling sites in ANP32E-overexpressing cells, while γH2A.X was more broadly distributed but primarily localized at or downstream of the SSS. Considering that an additional function of H2A.Z is to keep chromatin in an open state at sites of DSBs where its acetylation is necessary for the initial stages of DDR39,77,81, we monitored the relative distribution of acH2A.Z (Fig. 4f). We found that regions with the highest SI accumulated acH2A.Z in an ANP32E-dependent manner, thus recapitulating the observed pattern of the TRC-associated DDR factors. We excluded that the observed differences depended on the alteration of chromatin accessibility resulting from H2A.Z turnover since ATAC-seq signal increased proportionally with the magnitude of stalling but independently from ANP32E status (Supplementary Fig. 10j). Additionally, the direct correlation between the altered H2A.Z turnover and the recruitment of DDR factors was strengthened by observing an increase in the deposition of pRPA32, TOP2A and acH2A.Z proportionally to the magnitude of H2A.Z eviction at pS2RNApol II SSS in tIMEC-A compared to tIMEC cells (Supplementary Fig. 11a, b). Overall, our data indicate that the increased turnover of H2A.Z led to an augmented stalling of the elongating complexes along the gene body, which was recovered upon R-loops degradation. Stalling sites with higher pS2RNApol II accumulation also show higher recruitment of DNA damage markers and retention of acH2A.Z, indicative of active DDR.
H2A.Z altered turnover primes for the formation of long R-loops
We next investigated whether R-loops accumulated at the ANP32E-associated damage sites by performing DRIP-seq. We found that ANP32E-overexpressing cells showed a higher number of R-loop peaks (Fig. 5a and Supplementary Fig. 12a). Of note, the increased abundance of ANP32E-driven R-loops was rescued by the RNaseH1 overexpression (Fig. 5b, c). The efficacy of RNaseH1 was further validated by its acute expression in tIMEC-A, which further reduced R-loops abundance (Supplementary Figs. 8, 12b, c). Of importance, we found that R-loops length was increased in tIMEC-A, with respect to both tIMEC and tIMEC-A-H1 (Fig.5d and Supplementary Fig. 12c). These findings were further confirmed by performing CUT&RUN of R-loops (Supplementary Figs. 8, 12a–f). Interestingly, R-loop length increased at genomic sites showing H2A.Z-loss in ANP32E-overexpressing cells (Fig. 5e and Supplementary Fig. 12g). We observed a similar behavior at sites of reduced accessibility in tIMEC-A (Fig. 5f and Supplementary Fig. 12h). Of importance, the expression of RNaseH1 re-established a shortening of R-loops without affecting their relative abundance (Fig. 5e, f and Supplementary Fig. 12g, h).
a DRIP-seq peak counts per cell line representing R-loops abundance. Individual replicate values are shown as mean ± s.d.; mean indicated above each sample. b Cumulative DRIP-seq signal at RNaseH1-sensitive tIMEC-A R-loop peaks (n = 47,940), aligned at peak start and shown from –500 to +800 bp. c Pie chart showing the percentage of ANP32E-induced R-loops that are RNaseH1-sensitive or -resistant. d Tukey boxplots (25th to 75th percentile and median) of R-loop peak lengths (bp) per cell line. Peaks present in three biological replicates: tIMEC = 34,125; tIMEC-A = 82,494; tIMEC-A-H1 = 90,009. P values from a two-tailed Wilcoxon test. e Density plot of R-loop lengths overlapping H2A.Z-loss regions in tIMEC vs tIMEC-A. Dashed line marks the 75th percentile; shaded for tIMEC-A. f Density plot of R-loop lengths intersecting regions of chromatin accessibility loss in tIMEC vs tIMEC-A. Dashed 75th percentile marked as in (e). g Pie charts showing number of R-loop-associated genes with upregulated (UP), downregulated (DOWN), or unchanged (NO) expression in tIMEC vs tIMEC-A (based on RNA-seq). h Cumulative CUT&RUN signal for H2A.Z at TSS (top) and gene body (bottom) of genes stratified by expression (High, Medium, Low) from RNA-seq. i Tukey Boxplot (25th to 75th percentile and median) of gene expression values (log2TPM) in R-loop-forming genes in tIMEC-A vs other cell lines. n: tIMEC = 4092, tIMEC-A = 5387, tIMEC-A-H1 = 6055; four replicates/conditions. P values from a two-tailed unpaired t-test. j Barplot showing mean ± s.d. log2FC of R-loop length between tIMEC and tIMEC-A, grouped by gene expression pattern retrieved from four biological replicates: Low (n = 2293/4271), Mid (n = 2963/5773), High (n = 2364/4456). Two-tailed Wilcoxon test p values reported. k Left: Tukey boxplot Boxplot (25th to 75th percentile and median) of differential pausing index (PI) sites (log2FC tIMEC-A vs tIMEC), representative data from three biological replicates. Horizontal lines define Faster, Equivalent, or Slower pS5RNApol II release in tIMEC-A vs tIMEC. Right: Barplot of H2A.Z FRiP (fraction of reads in peak) values at regions with faster pS5RNApol II release in tIMEC-A (n = 12,660).
To verify whether the enhanced R-loop formation in tIMEC-A was not solely a consequence of alterations in the transcriptome, we determined differential gene expression profiles by RNA-seq. This analysis revealed that about 500 genes had altered expression levels in tIMEC-A with respect to tIMEC cells, while less significant variation was observed upon RNaseH1 overexpression (Supplementary Fig. 13a, b and Supplementary Data 2). Of note, most of the genes harboring R-loops did not exhibit a significant change in gene expression (Fig. 5g and Supplementary Fig. 13c), thus excluding a significant contribution of global transcriptional alteration to the enhanced formation of R-loops in ANP32E-overexpressing cells. These findings strengthened the hypothesis that H2A.Z turnover is responsible for accumulating long R-loops. Considering that the distribution of H2A.Z occupancy along the gene is proportional to the transcriptional output60, we further analyzed the relationship between H2A.Z status and gene expression31 (Fig. 5h and Supplementary Fig. 13d). Low-expressed genes exhibited reduced H2A.Z levels at the promoter, but relatively high levels along the gene body, consistent with previous findings31. This trend was preserved across cell lines at promoters, but in tIMEC-A cells, we observed preferential depletion of H2A.Z at the gene body of low-expressed genes (Fig. 5h). Of importance, R-loop-forming genes in tIMEC-A cells were, on average, less expressed (Fig. 5i). Moreover, genes with low expression levels displayed a more pronounced variation in R-loop length between tIMEC and tIMEC-A cells (Fig. 5j and Supplementary Fig. 13e). In addition, regions where H2A.Z is lost in tIMEC-A cells showed both longer R-loops and a faster release of the initiating RNApol II (Fig. 5e, k and, Supplementary Fig. 13f). These results suggest that longer R-loops may be a consequence of faster release of initiating RNAPol II, due to the destabilization of the chromatin barrier by H2A.Z removal, particularly in low-expressed genes. Notably, changes in global chromatin accessibility were less impactful in regions with active pS5RNApol II (Supplementary Fig. 13g).
ANP32E overexpression induces R-loop dependent TRCs affecting genome stability
The formation of R-loops can lead to the exposure of the ssDNA strand opposite to the DNA:RNA hybrid. If not promptly resolved, this state may lead to DNA breaks and eventually to genomic fragility15. We hypothesized that the increased DDR activity in ANP32E-overexpressing cells is linked to DNA damage occurring specifically at R-loop sites. To validate this observation at the genome-wide level, we intersected the genomic regions harboring R-loops with those characterized by DNA damage. Notably, we observed a significant overlap between R-loops and the DDR factors pRPA32 and γH2A.X, as well as TOP2A, in ANP32E-overexpressing cells with respect to tIMEC, confirming that DNA damage occurs predominantly at R-loop sites (Fig. 6a). This finding is consistent with previous evidence showing that pRPA32 is recruited at the opposite DNA strand at TRCs sites71,82. By plotting the DRIP-seq signal at genomic regions with increasing pS2RNApol II stalling, we noticed higher and more widespread R-loop occupancy in tIMEC-A cells, which was resolved upon expression of RNaseH1 (Fig. 6b). Given this correlation and the substantial overlap of R-loops with pRPA32 (Fig. 6a), we defined R-loop-dependent TRC sites as genomic regions harboring both R-loops and pRPA3282 (thereafter named R-TRCs). Strikingly, we observed a very defined deposition of pRPA32 at the extremities of R-TRCs characterized by long R-loops (Fig. 6c). Moreover, the occupancy of elongating RNApol II matched the location of conflict sites, indicating that we detected the transcriptional side of the TRC (Supplementary Fig. 14a). Interestingly, γH2A.X accumulated in between the two conflict bubbles, indicating the occurrence of DSBs (Supplementary Fig. 14a). We further examined the frequency of R-loops at R-TRCs and found that, in ANP32E-overexpressing cells, R-loops were more frequent at both conflict and non-conflict sites compared to tIMEC cells. However, their fold change enrichment was sensibly higher at R-TRCs (Supplementary Fig. 14b). Genes affected by R-TRCs were more abundant in ANP32E-overexpressing cells, while genes with non-TRC R-loops or R-loop-independent DNA damage (pRPA32-only) were equivalently shared between cellular conditions (Fig. 6d and Supplementary Fig. 14c). Furthermore, in tIMEC-A cells R-loops associated with R-TRCs were longer than those not involved in ATR-dependent DDR activation, confirming that both the higher frequency and length of R-loops correlate with DNA damage (Fig. 6e).
a Overlap of genes showing both R-loops and DDR markers from DRIP-seq and CUT&RUN data, respectively. Statistical significance of intersections assessed via one-tailed hypergeometric test (Null-hp P[X ≤ x]). Raw gene numbers and overlap percentages are shown. b Cumulative plot of R-loop distribution relative to pS2RNApol II stalling start sites (SSS), grouped into quartiles (Q1–Q4, increasing stalling). A −0.5 to +0.8 Kb window centered on SSS is displayed. Red shading highlights R-loop coverage in tIMEC-A for each stalling quartile. c Heatmaps of pRPA32 and R-loops at regions where both signals co-occur (n = 7486), sorted by decreasing size. The same region order is used across panels. Dashed lines mark region ends. Schematic of proposed TRC model aligns with heatmap features observed in tIMEC-A. d Barplot of the percentage of genes exhibiting R-TRCs (pRPA32 + R-loops) in each cell line. Raw gene counts are indicated above the bars. e Tukey boxplot (25th to 75th percentile and median) of R-loop length (bp) at TRC regions (pRPA32 + R-loops; tIMEC = 376, tIMEC-A = 5527, tIMEC-A-H1 = 5211) vs. non-TRC regions (R-loops only; tIMEC = 33,626, tIMEC-A = 76,270, tIMEC-A-H1 = 83,901). R-loop peaks are shared by three biological replicates intersected with a representative replicate for pRPA32. Two-tailed unpaired t-test p values are shown. f Gene ontology enrichment for R-TRC genes. Terms and −log10(p value) from Fisher's exact test are shown. Colors indicate the reference database from which the terms were obtained. g Distribution of common fragile sites (CFSs) and their chromosomal locations (blue), alongside alteration frequencies in basal BC patients (black lines). h Density plot showing proximity of TRC regions (red) to nearest CFS breakpoint. R-loop-only (green), pRPA32-only (blue), and three sets of matched random regions (dashed lines) serve as controls.
Since ANP32E-overexpressing BC tumors correlated with an augmented aggressiveness and worst prognosis38, our next question was how R-TRCs enhance cancer cell fitness. To address this, we first performed a gene ontology analysis to determine whether the affected genes play a role in BC. Strikingly, we found that several affected genes belong to pathways related to BC-associated processes and tumor progression, including deregulated proliferation and migration (Fig. 6f). Interestingly, about 80% of TRC genes in ANP32E-overexpressing cells are targets of ANP32E, reinforcing its involvement in TRC formation (Supplementary Fig. 14d). We next examined whether R-TRCs contribute to genome instability by analyzing common fragile sites (CFS), which are implicated in early cancer development. We assessed the proximity between high-fidelity R-TRC sites in our cell model and the nearest CFS breakpoint (Fig. 6g)83. Notably, we found that TRC sites show higher proximity with CFSs with respect to regions with non-TRC R-loops and R-loop-independent sites of DNA damage (pRPA32-only) (Fig. 6h). The robustness of this finding was further validated by comparing the datasets against randomly generated regions (Fig. 6h). To further establish the link between R-loop-dependent TRCs and clinically relevant genomic instability, we stratified basal BC patients from TCGA based on FA pathway upregulation. Notably, the FA pathway was upregulated in patients with ANP32E amplification (Fig. 1d) and is involved in resolving R-loop-associated DNA damage59. Consistently, TRC sites were enriched within ~10 kb of the closest copy number variant (CNV) breakpoint, particularly in patients with an active FA pathway (Supplementary Fig. 14e). To explore whether genomic instability in these regions contributes to metastasis, we analyzed public datasets and found that genomic alterations in actively expressed TRC-associated genes near CFS breakpoints were more frequent in TNBC metastatic patients (Supplementary Fig. 14f). Together, these results indicate that R-loops-dependent TRCs occur at genes involved in BC-related pathways in proximity to CFSs, potentially driving genome instability.
ANP32E effects through genic and cis-regulator elements
R-loops can influence both genic regions and cis-regulatory elements21. By analyzing DRIP-seq peaks annotation in our cellular models, we observed that while R-loops at both TRC and non-TRC sites were predominantly intragenic, they were also present in intergenic and non-coding regions (Supplementary Fig. 15a). Comparing the annotation abundance of the two R-loops categories, we observed that despite there were no substantial differences in the overall distribution across functional elements, TRC R-loops were more abundant and sensibly longer at promoters/TTS of ANP32E-overexpressing cells, (Supplementary Fig. 15a, b). In contrast, non-TRC R-loops were enriched in intergenic and intronic regions and were generally shorter (Supplementary Fig. 15a, b), indicating that R-TRC-associated genome instability primarily affects intragenic regions. Focusing on cis-regulatory elements, we found that TRC R-loops in ANP32E-overexpressing cells were strongly enriched at tRNA and other RNA elements, with increased length compared to non-TRC R-loops of the same functional category (Supplementary Fig. 15c, d)21. Non-TRC R-loops, however, exhibited a more uniform distribution across cell lines (Supplementary Fig. 15c, d). This suggests that ANP32E overexpression influences transcription and replication dynamics not only by altering promoter-proximal chromatin but also through cis-regulatory elements. Since H2A.Z is enriched at active enhancers84,85, we hypothesized that ANP32E overexpression may deplete enhancer-bound H2A.Z and affect enhancer RNAs (eRNAs). Therefore, we employed ATAC-seq data analysis to retrieve putative active enhancers defined as chromatin-accessible distal elements (>1 kb from the nearest TSS) and identified eRNAs using our total-RNA-seq data86. By plotting the CUT&RUN signal of H2A.Z, ANP32E, and P400 at eRNAs, we observed a comparable behavior to that observed at TSS (Supplementary Figs. 10c, 15e). Specifically, ANP32E-overexpressing cells showed a strong decrease in H2A.Z signal, with a concomitant reduction in chromatin-bound ANP32E and P400. Furthermore, we observed a reduction in initiating RNApol II, consistent with prior studies showing that H2A.Z depletion impairs RNApol II recruitment at enhancers, resulting in a reduced eRNAs synthesis84 (Supplementary Fig. 15f, g). Accordingly, pRPA32 and γH2A.X signals increased in tIMEC-A cells (Supplementary Fig. 15f), suggesting that ANP32E overexpression depletes H2A.Z at both intragenic regions and active enhancers, leading to DNA damage activation.
ANP32E overexpression triggers R-TRCs and vulnerability to ATRi in vivo
Our results indicate that ANP32E overexpression promotes TRC formation and DNA damage. To assess the pathogenic relevance of these findings in vivo, we orthotopically injected tIMEC and tIMEC-A cells in immunocompromised mice and analyzed the formed tumors to determine the frequency of TRCs by PLA. Tumors derived from tIMEC-A cells displayed a significant enrichment in TRC foci, along with increased ATR-dependent DNA damage, as indicated by elevated pRPA32 foci (Fig. 7a–c). These findings suggest that ATR inhibition may be a valuable therapeutic strategy for ANP32E-overexpressing tumors. To test this hypothesis, we evaluated the cytotoxicity of the ATR inhibitor VE822 in vitro. ANP32E overexpression in TNBC models, increased sensitivity to ATR inhibition (Fig. 7d, Supplementary Fig. 16a, and Supplementary Table 2), while its silencing improved cell growth and metabolic activity upon VE822 exposure (Supplementary Fig. 16b, c). Additionally, R-loops degradation by RNaseH1 overexpression in tIMEC-A cells partially rescued VE822-induced cytotoxicity, supporting the role of R-loops in ATR-dependent DNA damage (Fig. 7d). This synthetic lethal interaction was confirmed using an alternative ATR inhibitor (Ceralasertib) and the ATR-independent DNA-damaging agent ETP (Supplementary Fig. 16d, e). Caspase activation assay further confirmed ATRi efficacy on tIMEC-A cells, which displayed a significantly higher percentage of apoptotic cells compared to tIMEC and tIMEC-A-H1 (Supplementary Fig. 16f).
a Representative PLA images showing TRCs between pS2RNApol II and PCNA in mouse xenografts derived from tIMEC and tIMEC-A cells. Three zoom-ins of single cells are shown per condition. Scale bar = 10 µm. b Representative immunofluorescence images of pRPA32 (red) and human-mitochondria (green) in xenografts from tIMEC and tIMEC-A. Three zoom-ins per condition are shown. c Violin plots showing the number of PLA (left) or pRPA32 (right) foci per nucleus across two xenografts per condition, corresponding to (a, b). Unpaired two-tailed t-test p values are indicated. d Dose-response curves of tIMEC, tIMEC-A, and tIMEC-A-H1 cells treated with VE822 for 48 h. EC50 indicated by vertical dashed lines. Data were reported as Mean ± SD of three technical replicates. e Experimental scheme: luciferase-tagged tIMEC or tIMEC-A cells were orthotopically injected into mice. Once tumors were palpable, VE822 (4x/week) or vehicle was administered. After 4 weeks, tumors and metastases were analyzed. f Dot plot representing tumor growth curves (mean ± s.e.m.) for tIMEC and tIMEC-A xenografts with or without VE822 treatment, monitored over 21 days. Two-way ANOVA p values are shown. n: tIMEC vehicle = 11, VE822 = 9; tIMEC-A vehicle = 10, VE822 = 11 (pooled from two independent experiments). g Barplot showing mean ± s.e.m. photon flux per metastasis in vehicle- or VE822-treated mice with tIMEC or tIMEC-A tumors. n: tIMEC vehicle = 4, VE822 = 3; tIMEC-A vehicle = 5, VE822 = 4. Two-tailed t-test p values indicated. Representative lung metastases are shown below. h Mean cell number per metastasis (±SD) in tIMEC or tIMEC-A tumors ± VE822. n: tIMEC vehicle = 8, VE822 = 3; tIMEC-A vehicle = 27, VE822 = 43. Two-tailed unpaired t-test p values are shown. i Left: Barplot of cell number per metastasis (mean ± s.e.m.) categorized into DTCs, micro-, and macrometastases. Two-tailed Wilcoxon t-test p values are shown. Right: Representative IHC images (human-mitochondria) identifying metastasis types. Scale bar = 100 µm; zoom-in = 15 µm. Cell counts used to classify each category are provided.
To assess ATRi efficacy in vivo, VE822 was administered to mice harboring tIMEC- and tIMEC-A-derived xenografts and compared with vehicle-treated controls (Fig. 7e). ANP32E overexpression led to the formation of larger tumors, which were specifically responsive to ATR inhibition (Fig. 7f). Of importance, tIMEC-A-derived xenografts displayed a more aggressive phenotype characterized by poorly differentiated cellular elements with a high nucleus-to-cytoplasm ratio and high proliferation index, representing a hallmark of poor prognosis in BC patients (Supplementary Fig. 16g, h). In contrast, tIMEC-derived xenografts displayed well-differentiated areas with squamous epithelial aggregates and keratinization. Treatment with VE822 induced a more differentiated phenotype in tIMEC-A tumor xenografts, along with a reduction in the proliferation index (Supplementary Fig. 16g, h). Concomitantly to the formation of primary tumors, mice developed metastatic lesions, whose abundance was enriched in mice injected with ANP32E-overexpressing cells, yet this effect was strongly mitigated by ATR inhibition upon VE822 treatment (Fig. 7g)38. To better characterize the effect of ANP32E overexpression and ATRi on metastasis formation, we quantified the size of metastatic lesions. On average, metastases derived from ANP32E-overexpressed tumors were larger, and VE822 treatment strongly reduced their size (Fig. 7h). Specifically, ANP32E overexpression led to a higher frequency of micro- and macrometastases at the expense of disseminated tumor cells (DTCs) (Fig. 7i, nr in figure legend). Importantly, VE822 treatment significantly impaired the metastatic burden, resulting in the formation of scattered DTCs that did not evolve into macrometastases, at least at the time of observation (Fig. 7i). In summary, ANP32E overexpression is associated with increased TRCs and ATR-dependent DDR activation in vivo, uncovering a vulnerability to ATR inhibitor, which reduced the primary tumor volume and metastatic potential of BC cells.
Discussion
The molecular processes leading to genomic instability in cancer cells are diverse, but replication stress (RS) has emerged as a common factor driving its occurrence2. In the present work, we identified the overexpression of the histone chaperone ANP32E as a factor that exacerbates RS in the presence of MYC upregulation, revealing a dependency of cancer cells on DNA repair pathways, representing a potential therapeutic target. We showed that ANP32E deregulation leads to an increased H2A.Z turnover, resulting in a higher frequency of RNApol II stalling, R-loop dependent TRCs, and genome instability. While other chaperones like FACT were shown to be implicated in R-loop-dependent TRCs homeostasis16,17,18,19,20, ANP32E is the first described chaperone that worsen TRCs effect instead of resolving them. From a molecular perspective, ANP32E differs from FACT in its specificity toward H2A.Z. While FACT targets H2A/H2B and promotes both their removal and restoration, ANP32E promotes H2A.Z removal followed by P400-mediated reintroduction of acetylated histones to repress transcription. Therefore, this uncontrolled turnover of H2A.Z can lead to unwanted changes in chromatin composition and compaction, exacerbating the harmful effects of TRCs and R-loops accumulation.
Genomic instability in cancer is often linked to deficiencies in DNA repair genes, such as germline mutations in BRCA1/287. However, in BC patients with MYC-ANP32E deregulation, we did not observe a predominant deficiency in DNA damage response (DDR) pathways. Instead, we found upregulation of several DDR pathways, particularly the Fanconi anemia (FA) pathway, which plays a critical role in resolving R-loop-derived DNA damage in response to ATR signaling58,59. Concordantly, by recapitulating the deregulation of ANP32E in a basal BC model, we observed that its overexpression correlates with increased R-loops formation and ubiquitinated FANCD2, which is a known effector of the active FA cascade. In line with previous reports, we observed that concomitantly to R-loops increase there was an augmented frequency of TRCs, that most probably were Head-ON conflicts, as only frontal collisions between the transcriptional and replicative machineries can be detected by RNApol II-PCNA PLA88. Importantly, Head-ON collisions are the most harmful for DNA integrity and are frequently associated with R-loops formation and ATR signaling activation12. Indeed, downstream to ANP32E overexpression also led to heightened activation of the ATR-dependent DNA damage markers pRPA32 and 53BP1, which are indicative of DNA lesions. These findings highlight the RS experienced by ANP32E-overexpressing cells and their dependence on ATR-mediated repair to maintain genomic stability. As a proof of concept, treatment with an ATR inhibitor (VE822) triggered genomic instability in these cells, as reflected by the increase in DSBs and micronuclei, the occurrence of DNA damage catastrophes, and the accumulation of 53BP1 bodies68,70.
To unveil the possible molecular mechanism by which ANP32E contributes to genome instability, we explored the downstream events of its action on H2A.Z turnover, which resulted being differentially regulated at cis-regulatory elements (promoters and enhancers) and gene bodies of high- and low-expressed genes31. Although we could not distinguish between the direct and indirect effects of ANP32E overexpression, H2A.Z removal at promoter-proximal regions, correlated with RNApol II stalling, R-loop accumulation, and pRPA32 marking of DNA damage. In this biological context, we also determined a spurious firing of replication origins, possibly contributing to the increased frequency of TRCs. Notably, R-loop-dependent TRCs may further exacerbate the activation of non-canonical replication origins, hindering fork restart at stalled sites13,15. Of note, H2A.Z is also enriched at replication origins, regulating their firing and replication timing89, indicating that ANP32E overexpression may disrupt the regulation of replication origins, contributing to the observed instability. Therefore, despite we cannot exclude the contribution of other factors to the observed phenotype, we propose a molecular mechanism in which ANP32E-mediated eviction of H2A.Z from promoter-proximal regions leads to a faster release of paused RNApol II26, resulting in an accumulation of elongating complexes and their stalling along the gene body. This process is accompanied by an increased recruitment of DNA damage factors and long R-loops formation. Additionally, ANP32E preferentially evicts H2A.Z from low-expressed gene bodies, which further contributes to R-loop accumulation.
Based on previous studies, we mapped R-loop-dependent TRCs as genomic regions harboring both R-loops and pRPA32 marks82. This allowed us to visualize the transcriptional conflicts across the genome. Given the relative distribution of R-loops, pRPA32, γH2A.X, and elongating RNApol II, we suggest that the conflicts involved at least two transcriptional bubbles, with frequent occurrence of DNA DSBs in between. Importantly, we found that the genomic regions that are affected by R-loops-dependent TRCs involve genes that give rise to long transcripts8,13. This result suggests a peculiarity of ANP32E action that specifically affects late-replicating, long transcripts, being responsible for the onset of CFS at these specific genomic sites. Importantly, TRCs were also enriched in genes of BC-associated pathways, primarily involved in cell migration and proliferation. According to the hypothesis that unresolved TRCs give rise to DSBs and ultimately to genome instability, we found that R-loops-dependent TRC formed consequently to ANP32E overexpression were significantly enriched in proximity of known CFSs. The biological relevance of this finding is further supported by recent results obtained from patients of breast adenocarcinoma relating APOBEC3-dependent kataegis in R-loop regions with similar proximity to structural variant breakpoints90.
Increasing the relevance of the molecular mechanism that links ANP32E overexpression to genome instability, we confirmed in vivo that ANP32E stimulates the recruitment of pRPA32 and the occurrence of TRCs. Moreover, inhibition of ATR with VE822 strongly reduced the formation of primary tumors and metastatic burden. This result is of particular importance since it gives a rationale for proposing ANP32E expression status alongside MYC amplification as a diagnostic marker to identify patients that would benefit from ATRi-based therapies. Since the available clinical trials suggest that DDR inhibitor drugs, including ATRi, are more effective in combinational therapies, it would be worth evaluating possible drug combinations to maximize treatment success91,92,93. Additionally, extending ATRi treatment to other cancers with elevated ANP32E expression, either alone or with RS-inducing oncogenes, warrants further investigation.
In summary, this study proposes a molecular mechanism through which ANP32E overexpression exacerbates RS by inducing epigenetic changes that lead to the accumulation of R-loop-dependent TRC and genomic instability, which can be exploited for the rational administration of ATRi drugs to increase therapeutic success.
Methods
Cell culture
All cell lines were cultured at 37 °C and 5% CO2. IMEC-TetON-MYC, tIMEC, tIMEC-A, and tIMEC-A-H1 cells were maintained in DMEM-F12 (Gibco, 11320-084) supplemented with human EGF (DBA, AF-100-15-1mg), Insulin (Merck, I6634-250MG), hydrocortisone (Voden, 74144), and bovine pituitary extract (Gibco, 13028-014) and 1x PenStrep (Gibco, 15070-063). SUM-159 cell line was cultured in Ham’s-F12 (Gibco, 11765054) supplemented with insulin 5 μg/ml (Merck, I6634-250MG), hydrocortisone 1 μg/ml (Voden, 74144), and fetal bovine serum 5% (Gibco 10270106) and 1x PenStrep (Gibco, 15070-063). MDA-MB-231 cell line was cultured in DMEM (Gibco 11960044) supplemented with fetal bovine serum 10% (Gibco 10270106), 1x glutamine (Gibco 25030024), and 1x PenStrep (Gibco, 15070-063). Myc induction in IMEC-TetON-MYC was stimulated with 2 h Doxycycline treatment.
Generation of stable cell lines and transient transfection
3xFLAG-ANP32E cDNA was cloned in a pGK plasmid backbone (Addgene, 169744) to transduce tIMEC and SUM-159-PT cells and generate the tIMEC-A and SUM-159-A cell lines. SUM-159-A cell population was next seeded at a single-cell level for clonal selection of FLAG-ANP32E positive cells, which were screened by IF and WB. Three independent clones were next employed for the experiment, named SUM-159-Acl1,cl2,cl3, respectively. tIMEC-A-H1 cell line was generated by stable nucleofection of ppyCAG_RNaseH1_WT plasmid (Addgene, 111906)21 using the P1 Primary Cell 4D-Nucleofector X kit (Lonza, V4XP-1032) and selected by keeping them in culture under hygromycin (Applichem, A5347) 500 ng/ml selection for at least 2 weeks. MB-231sh1 and MB-231sh2 cell lines are two different cell populations, generated by transduction of MDA-MB-231 (named in short MB-231) with lentiviral vectors carrying pLKO.1 plasmid (Addgene, 8453) cloned with shANP32E-808 (sh sequence: GGATTTGATCAGGAGGATAAT). Transient transfection of ppyCAG_RNaseH1_WT plasmid was performed 24 h after cell seeding using Lipofectamine 3000 (Thermo Fisher Scientific, L3000008), and cells were analyzed starting from 10 h after transfection.
Dot blot assay
Genomic DNA was isolated from tIMEC, tIMEC-ANP32E, and tIMEC-ANP32E-H1 starting from an 80% confluent six-well plate. Cells were pelleted and washed in PBS 1x (Gibco, 10010023). Next the cell pellet was resuspended in lysis buffer (Tris-HCl pH 7.2 10 mM, EDTA 10 mM, NaCl 150 mM, SDS 0.4%) + Proteinase-K 4 mg/ml (Roche, 3115887001) and incubated for 2 h at 55 °C. Samples were next precipitated with 1V cold isopropanol, centrifuged for 2 min at 4 °C max speed, and washed with ice-cold EtOH 70%. After centrifugation the pellet was dried at RT and resuspended in ddMilliQ water by incubation 1–2 h at 42 °C. Dot blot samples were prepared from 1 µg gDNA with or without RNaseH1 2500 U treatment (NEB, M0297) O/N 37 °C. Samples were denatured in denaturation buffer (NaOH 400 mM, EDTA 10 mM in ddMilliQ water) for 5 min at 95 °C and then kept on ice to avoid reannealing. Denaturation was stopped by addition of 1V Ammonium Acetate 2 M. After equilibration and washing of a nitrocellulose membrane 0.2 µm (GE Healthcare, 10600001) in SSC 10x buffer (NaCl 3 M, Sodium citrate 0.3 M in ddMilliQ water, pH 7.0) the prepared samples were spotted on the membrane using a Dot Blot apparatus (Bio-Rad, 170-6545) and UV-crosslinked for 3 min at 1200 J/m2. Next, the membrane was blocked with PBS-milk 5% and incubated with primary antibodies s9.6 1:500 (Millipore, MABE1095) and dsDNA 1:3000 (Abcam, ab273137) O/N 4 °c. After washing, species-specific HRP secondary antibody (Sigma-Aldrich, AP160P) 1:3000 was applied, and the membrane was imaged on a ChemiDoc apparatus (Bio-Rad).
Proximity ligation assay (PLA)
Cells were seeded on gelatin 0.1% (Sigma-Aldrich, G1393) coated glass coverslips in a 24-well plate. After 24 h, cells were fixed with ice-cold MeOH for 10 min at −20 °C, blocked and permeabilized in Blocking Solution (Goat serum 5%, BSA 5%, Triton 0.5% in PBS 1x) for 1 h at RT. PLA was performed using Duolink in situ detection kit (Sigma-Aldrich, DUO92008) with Duolink PLUS anti-rabbit (Sigma-Aldrich, DUO92002) and MINUS anti-mouse probes (Sigma-Aldrich, DUO92004) according to the manufacturer's instructions with 10 min washes. The following primary antibodies diluted 1:250 were incubated for 2 h at RT: anti-pS2RNApol II (Abcam, ab5095) + anti-PCNA (Merck, WH0005111M2), anti-pS2RNApol II (Abcam, ab5095) + anti-R-loop (Millipore, MABE1095), anti-ANP32E (LB-Bio, LS-C344600-400) + anti-R-loop (Millipore, MABE1095). DAPI 1:1000 (Invitrogen, D1306) diluted in PBS 1x was applied for 30 min at RT for nuclei staining. Images were acquired at a Nikon Ti2 confocal microscope equipped with a 60x oil-immersion ocular objective. PLA foci quantification was performed using a custom-made ImageJ macro.
Western Blot
Whole-cell lysates were extracted using RIPA lysis buffer (Tris-HCl pH 8.0 10 mM, EDTA 1 mM, NaCl 140 mM, SDS 0.1%) supplemented with protease inhibitor cocktail 1x (Roche, 11873580001), phosphatase inhibitors 1x (Sigma-Aldrich, P5726 and P0044), PMSF 1 mM (Thermo Fisher, 36978), Na3VO4 1 mM (Sigma-Aldrich, S6508), okadaic acid 1x (Sigma-Aldrich, 459616). Whole-cell lysates were then subjected to sonication with a Bioruptor sonicator for ten cycles of 30 sec on/ 30 s off, followed by two centrifugation steps at max speed for 10 min each. Protein concentration was measured from supernatant using a BCA assay kit (Thermo Fisher, 23250). Depending on the target protein, 15–35 μg of the lysate were run on SDS-PAGE gels and transferred using a iBlot2 apparatus (Invitrogen) on nitrocellulose membrane (Invitrogen, IB23001/2), then blocked in either PBST-milk 5% (anti-ANP32E Abcam ab5993, anti-FLAG Sigma-Aldrich F1804, anti-V5 Cell-Signaling 13202; anti-H3 Cell-Signaling 13202 4499, anti-β-actin Sigma-Aldrich A5441) or PBST-BSA 5% (anti-H2A.Z Abcam ab4174, anti-pS2RNApol II abcam, ab5095, pS5 RNApol II Active Motif, 102660). Primary antibodies were incubated at 4 °C O/N. Membranes were imaged with ECL substrate (Cytiva, RPN2232) on a Bio-Rad Chemidoc imager.
Alkaline comet assay
Comet assay was performed using Comet Assay Kit (Abcam, ab238544) according to the manufacturer's instructions. Briefly, 150,000 cells were seeded in six-well plates, grown O/N, and when indicated, treated with either ETP 2.5 µM (MedChemExpress, HY-13629) or VE822 1.25 µM for tIMEC, 2.5 μM for MDA-MB-231, 4.5 μM for SUM-159 (MedChemExpress, HY-13902) for 24 h. Cells were scraped, embedded in low-melting agarose at a 1/10 ratio (v/v), and distributed on a glass microscope slide. Cells were lysed by incubating the slides for 45 min in lysis buffer (NaCl, EDTA, DMSO, Comet lysis solution 10x provided with the kit, pH 10.0). After incubation in an alkaline solution (NaOH, EDTA) for 30 min at 4 °C, cells were subjected to alkaline electrophoresis for 45 min at 300 mA. Slides were washed in ddH2O and ice-cold 70% EtOH before letting the agarose dry at RT. Once dried, the slides were incubated for 15 min at RT with Vista Green dye. Comets were acquired with a FITC filter of a Leica DMIL LED epifluorescence microscope equipped with a Leica DFC 450C camera. About 150–300 cells for every condition were acquired and quantified using both the OpenComet94 plugin for ImageJ and CometScore software95.
Micronuclei (MN) assay and centromere detection in MN
For the MN assay, 7000 cells/well were seeded in 96-well plates. After 24 h, 0.03 µg/ml Mitomycin C (MitoC, Sigma-Aldrich M5353), increasing concentrations of VE822 as indicated in figure legends, or fresh medium (NT condition) were incubated at 37 °C for 6 h. Next, Cytochalasin B (Sigma-Aldrich, C6762) 0.6 µg/ml was added and incubated for 30 h at 37 °C to block cytokinesis. Cells were then washed with PBS 1x and fixed in 4% PFA for 10 min at RT. Nuclei and MNi were stained using Hoechst solution (Invitrogen, H3570) 1:2000 in PBS 1x upon 30 min incubation at RT. After washing with PBS 1x, plates were acquired using an Operetta imager with a 20x objective (PerkinElmer). Three technical replicates per condition were acquired for a total of 1500–3000 BN cells per condition. The experiment was performed in three biological replicates. MNi in BN cells were quantified by adapting the parameters of the Operetta MN quantification analysis function.
For the detection of centromeres in MNi of BN cells, 30,000 cells were seeded on gelatin 0.1% coated coverslips in 24-well plates, grown ON at 37 °C, and treated as described above. After fixation, cells were blocked and permeabilized in Blocking solution (BSA 1%, Goat serum 5%, Triton X-100 0.5%, PBS 1x) for 1 h at RT. Centromeres were stained using CREST serum (Antibodies Inc, 15‐234) diluted 1:250 in PBS-BSA 3% and incubated for 2 h at RT. Cells were washed 3x in PBS 1x, then incubated for 1 h at RT with a solution containing: secondary antibody Alexa anti-human 647 1:1000 Invitrogen, A21445, Phalloidin 555 (Abcam, ab176756) 1:2000, and Hoechst 1:2000 in PBS 1x. Images were acquired with a Leica SP8 confocal microscope using a 63x objective and 2x zoom. Phalloidin staining was used to identify MNi that are in the cytoplasm of BN cells, which were divided after MitoC or VE822 treatment. An in-house ImageJ macro was used to measure the CREST signal in MNi.
RNA extraction and RT-qPCR
Total RNA was extracted from 70% confluent cells using TRIzol (Invitrogen, 15596026). Residual DNA was digested with DNaseI treatment (Roche, 11284932001). Expression analysis of S-phase related and R-loops prone genes CDK2, CCNE1, RPL13A, and TFPT genes was performed through RT-qPCR using the SuperScript™ III Platinum™ SYBR™ Green One-Step qPCR Kit w/ROX (Thermo Fisher, 11746-500). The used primers are listed in Supplementary Table 4. RT-qPCR results were analyzed through the ΔCt method, using GAPDH as the housekeeping gene.
Cell cycle analysis by FACS
Cells were seeded in six-well plates, grown O/N, and treated with VE822 0.25 µM for 6 h when indicated. For the evaluation of cell cycle arrest cells were fixed in the presence of 70% ethanol for 15 min at 4 °C, after washing 1 μl of FxCycle™ Far Red Stain (Invitrogen, F10348) and 5 μl DNase-free RNase A [10 mg/ml] (Thermo Scientific, EN0531) were added and incubated for 30 min at RT. For every sample, 20k events were acquired on a FACS Canto (BD) instrument and analyzed through FlowJo™ software.
Laser microirradiation
About 3500 cells were seeded on gelatin-coated 96-well plates with optical bottom (Costar, Corning # CLS3614) and grown O/N. Before laser microirradiation, cells were pre-sensitized using 15 min incubation with 10 µg/ml Hoechst at 37 °C. Live cells were detected using a 560 nm laser to avoid UV-crosslinking. Next, they were microirradiated using the Leica SP8 FRAP module with a 405 nm laser, 80% laser power, two line averages, two iterations, and a dwell time of 15.39 µs/pxl. Within 1 h from the irradiation, cells were pre-extracted with Triton X-100 0.25 % in PBS for 5 min on ice, MeOH fixed for 15 min at −20 °C, incubated again in pre-extraction solution for 5 min on ice, and next subjected to pRPA32 and 53BP1 IF as described above. The recruitment of DDR factors to the microirradiated site was quantified using the Damage Analyzer ImageJ plugin (https://bitbucket.org/daniel_garneau/microirradiation_analysis/src/master/README.md).
Immunofluorescence and EdU staining for nascent DNA
For IF staining, 35,000 cells were seeded on gelatin 0.1% coated coverslips in 24-well plates and grown ON at 37 °C. When indicated, cells were treated with VE822 0.25 µM for 6 h. EdU treatment was performed using the Click-IT EdU Imaging Kits (Invitrogen, C10337) by treating cells for 1 h with EdU 7.5 µM. Next coverslips were washed in PBS 1x and cells fixed in cold MeOH for 10 min at −20 °C. After PBS-BSA 3% washes, fixed cells were blocked and permeabilized in Blocking solution (BSA 1%, Goat serum 5%, Triton X-100 0.5%, PBS 1x) for 1 h at RT. EdU detection was allowed by click-it reaction of Alexa-Fluor 488 as described in the Click-IT EdU Imaging Kits (Invitrogen, C10337). Primary antibodies against FANCD2 1:100 (NovusBio, NB100-182SS), pRPA32 1:1000 (Bethyl, A300-246A-8), and 53BP1 1:100 (Millipore, MAB3802) were diluted in blocking solution and incubated at RT for 2 h. Next, coverslips were washed in PBS 1x before incubation with Alexa-Fluor-647 specie-specific secondary antibodies (Thermo Fisher, A32728 and A-21245) and either Hoechst 1:2000 for EdU labeled cells or DAPI 1:1000 for simple IF. For simple IF, all washing steps were performed in PBS 1x and primary antibody directly incubated after cells blocking and permeabilization. For R-loops IF and V5 tagged RNaseH1, cells were incubated with pre-extraction buffer (PBS, Triton X-100 0.25 %) for 5 min on ice before and after MeOH fixation for 10 min at −20 °C. Next, fixation was quenched with 10 min incubation at RT with glycine 0.1 M, coverslips washed 2x with PBS-Tween-20 0.5%, and incubated for 1 h at RT with s9.6 and anti-V5 primary antibodies 1:250 (Millipore, cat. MABE1095 and Cell Signaling, cat. 13202). Secondary antibody and DAPI diluted 1:1000 in PBS 1x were incubated for 1 h at RT before mounting coverslips. Images were acquired using either a Leica SP8 or Nikon Ti2 confocal microscopes. Imaging quantification was performed with custom-made ImageJ macros.
EU labeling and DRB treatment
Cells were seeded on gelatin 0.1% coated coverslips in 24-well plates and grown ON at 37 °C. Cells were treated with 100 µM DRB (Sigma-Aldrich, D1916) for 3 h to inhibit transcription elongation, next medium containing DRB was washed out to allow for synchronized release of transcription. Nascent DNA was labeled through EU incorporation (Click-iT® RNA Imaging Kits, Invitrogen, cat. C10329) for 1 h starting from either: 1 h before end of DRB treatment, together with DRB release or 2 h after DRB release. Next, samples were fixed for EU staining in 4% PFA for 10 min RT, permeabilized in PBS-Triton 0.3%, and stained according to the manufacturer's instructions. Nuclei staining was performed with DAPI 1:1000 incubated for 30 min at RT. Images were acquired using a Leica SP8 confocal microscope equipped with a 63x oil-immersion ocular objective. Image quantification was performed with a custom-made ImageJ macro.
DNA fiber assay
DNA fiber assay was adapted from ref. 96, briefly 200,000 cells were seeded in six-well plates and grown ON at 37 °C in order to reach about 50% confluency. CldU 25 µM (Sigma-Aldrich, C6891) was added to the medium and incubated for precisely 20 min at 37 °C. Next, IdU 250 µM (Sigma-Aldrich, I7125) was added without intermediate washing and incubated for precisely 20 min at 37 °C. Cells were washed twice in ice-cold PBS 1x, trypsinized, pelleted by centrifugation at 200×g for 5 min RT, and resuspended in ice-cold PBS 1x at a density of about 400,000 cells/ml. About 2 µl of the cell suspension were pipetted at the top a positively charged microscope slide (Epredia, J1800AMNZ), air dried for max 5 min and lysed for 2 min in 7 µl of Spreading Buffer (Tris-HCl pH 7.4 260 mM, EDTA 40 mM, SDS 0.5 % w/v). Slides were next tilted to an angle of 25°−40° until the cells' droplet reached the bottom edge of the slide. Slides were air dried and fixed in 1 ml methanol:acetic acid 3:1 for 10 min RT, washed twice in ddH2O, rinsed, and incubated in HCl 2.5 M for 1 h at RT. After PBS 1x washes to remove the excess of HCl, the slides were blocked in PBS-BSA 1 + 0.001% Tween-20 for 1 h at RT and incubated with a mix of rat monoclonal anti-BrdU antibody (1:500, Abcam Ab6325) and mouse monoclonal anti-BrdU antibody (1:500, BD Bioscience 347580) for 1 h at RT. After 3x washes in PBS 1x the slides were re-fixed in PFA 4% for 10 min RT, washed again in PBS 1x and blocking solution, and incubated for 2 h at RT with secondary antibody mix of Alexa-Fluor 647 anti-rat (Invitrogen, A-21247) and Alexa-Fluor 488 anti-mouse (Invitrogen, 11001) 1:500 in blocking solution. Lastly, the slides were washed in PBS 1x, dried to remove the excess liquid, and mounted using aqueous mounting medium (Histo-Line, PMT030). Images were acquired using a Nikon-AX confocal microscope with a 60x objective and 2048 × 2048 pxl resolution. Three biological replicates were performed, and for each one, about 300 fibers were analyzed using DNA Stranding software97.
ATAC-seq samples and library preparation
ATAC-seq samples were prepared according to the Omni-ATAC protocol98. Three biological replicates were performed for tIMEC, tIMEC-ANP32E, and tIMEC-ANP32E-H1 cells. For each sample, 100,000 cells were collected and centrifuged at 500 rcf 4 °C for 5 min. Cells were then resuspended in 50 μL of cold ATAC-Resuspension buffer (RSB; Tris-HCl pH 7.4, 10 mM NaCl, 10 mM MgCl2, and 3 mM, distilled water) containing 0.1% NP40 and 0.1% Tween-20 and 0.01% Digitonin (Promega, G9441) and incubated for 3 min to allow cells permeabilization. Nuclei were next isolated by washing cells with 1 mL cold ATAC-RSB buffer containing 0.1% Tween-20, and centrifuged at 500 rcf 4 °C for 10 min. Isolated nuclei were resuspended in 50 μL of Transposition mix (Illumina Tagment DNA Enzyme 100 nM, TD-Buffer 2X (kit from Illumina, 20034197), PBS 0.33%, 0.01% Digitonin, 0.1% Tween-20, distilled water) and incubated at 37 °C for 30 min in a thermomixer with 1000 rpm mixing, in order to allow the transposition reaction. The transposed samples were purified using Zymo DNA Clean and Concentrator-5 Kit (Zymo Research, D4013) and PCR-amplified using customized primers (Supplementary Table 4) designed following Illumina Adapter Sequences manual (Illumina, Document #1000000002694 v16) instructions. PCR-enriched libraries were, then, purified using AMPure XP Beads (Beckmann Coulter, A63881). Libraries quality and size distribution was analyzed at 2100 Bioanalyzer Instrument (Agilent, Model G2939BA) using Agilent High Sensitivity DNA Kit (Agilent, 5067-4626). Samples were sequenced using an Illumina NovaSeq6000 instrument.
CUT&RUN samples and library preparation
CUT&RUN protocol was adapted from ref. 99: cells were seeded and grown for 24 h at 37 °C in order to reach 70–80% confluency, and 250,000 cells per sample were collected afterward. For TOP2A CUT&RUN, before collection, cells were treated with Etoposide 5 μM for 1 h at 37 °C to improve the TOP2A signal100. Cells were next centrifuged 3 min at 600×g, washed three times in 1.4 mL Wash Buffer (HEPES pH 7.5 20 mM, NaCl 150 mM, Spermidine 0.5 mM, cOmplete™ EDTA-free Protease Inhibitor Cocktail 100X (Merck, 11873580001), and distilled water) and resuspended in 1 mL Wash buffer. Meanwhile, 150 μl Concavalin-A slurry (Polyscience, 86057-3) per sample were activated by washing the bead slurry three times in 1.5 mL Binding Buffer (HEPES pH 7.5 20 mM, KCl 10 mM, CaCl2 1 mM, MnCl2 1 mM). Activated beads were added to the cells while gentle vortexing and left rotating (25 rpm) for 10 min at RT. Afterward, cells bound to beads were divided into aliquots, one for each primary antibody to be used (Supplementary Table 5). The supernatant was removed by placing samples on a magnetic stand, the beads were resuspended in 100 μL of Antibody buffer (Wash buffer, 0.025% Digitonin (Merck, 300410), EDTA 2 mM), and 1 μg of the antibody was added. Tubes were left rotating ON at 4 °C. Samples were washed twice in 1 mL Dig-Wash buffer (Wash buffer, 0.025% Digitonin) and resuspended in 150 μL pAG-MNase 700 ng/ml diluted in Dig-Wash Buffer while gentle vortexing; tubes were left rotating (25 rpm) for 1 h at 4 °C. Samples were washed, resuspended in 100 μL Dig-wash Buffer and put on ice, next 2 μL of CaCl2 100 mM was added to allow chromatin digestion reaction. After 30 min of incubation 100 μL of 2X STOP buffer (NaCl 340 mM, EDTA 20 mM, EGTA 4 mM, 5% Digitonin, 100 μg/mL RNase A (Merck, R6513), 50 μg/mL Glycogen (Thermo Scientific, R0561), and distilled water) were added and incubated 30 min at 37 °C. The supernatant containing digested chromatin was transferred to clean 1.5 mL DNA LoBind® tubes. Samples were phenol/chloroform extracted and quantified using a Qubit™ 4 Fluorometer (Invitrogen, Q33238) using the Qubit™ 1X dsDNA High Sensitivity kit (Invitrogen, Q33265). 3 ng of DNA were used to perform library preparation with NEBNext® Ultra™ II DNA Library Prep Kit for Illumina® (E7645), following the modified protocol from ref. 101 specific for small-fragment libraries. Libraries quality and size distribution was analyzed at 2100 Bioanalyzer Instrument (Agilent, Model G2939BA) using Agilent High Sensitivity DNA Kit (Agilent, 5067-4626). Samples were sequenced using an Illumina NovaSeq6000 instrument.
ChIP-qPCR and Co-IP
Cells were crosslinked in 1% Formaldehyde (Sigma-Aldrich, 8187081000) for 10 min RT at a density of 3 mln cell/ml, quenched for 5 min at RT with Glycine 0.125 µM and sonicated for 17 min with Covaris M220 (peak power 75 W, duty factor 10, cycles per burst 250) in lysis buffer (50 mM Tris-HCl pH 8.0, 10 mM EDTA, 0.5% SDS, and protease inhibitor cocktail). Sheared chromatin was pelleted at 20,000×g for 10 min at 4 °C, and the supernatant diluted into Buffer A (10 mM Tris-HCl pH 7.5, 1 mM EDTA, 0.5 mM EGTA, 1% Triton X-100, 0.1% SDS, 0.1% Na-deoxycholate, and 140 mM NaCl). ProteinG-Dynabeads (Pierce™ Thermo Fisher, 10004D) were functionalized with primary antibodies anti-pS2RNApol II (Abcam, ab5095), pS5 RNApol II (Active Motif, 102660), or rabbit IgG (Millipore, PP64B). Sheared chromatin was next incubated O/N with functionalized beads at 4 °C upon rotation, conserving 5% of the Input. Beads were washed four times in Buffer A and once in Buffer C (10 mM Tris-HCl pH 8.0 and 10 mM EDTA). Samples were decrosslinked and eluted by incubation at 95 °C for 15 min in 4x Loading Buffer (Thermo Scientific, B0007) for Co-IP and then subjected to WB. For ChIP-qPCR samples were incubated for 30 min at 37 °C with Elution buffer (10 mM Tris-HCl pH 8.0, 5 mM EDTA, 300 mM NaCl, and 0.5% SDS) and RNase A (Thermo Fisher, 12091039), following 1 h incubation at 55 °C with Proteinase-K (Thermo Fisher, EO0492) and O/N decrosslinking at 65 °C. Decrosslinked DNA was purified using the MinElute kit (Qiagen, 28004) and subjected to RT-qPCR (SensiFAST™ SYBR® No-ROX Kit, Bioline BIO-98020) using primers listed in Supplementary Table 3.
Pausing index analysis
Pausing index (PI) and Stalling index (SI) were determined by the ratio of read density at the promoter to the read density at the corresponding gene body102; therefore, the PI for pS5- and SI for pS2-RNApol II were determined to evaluate the differential presence of paused/stalled elongating enzyme along the gene body of Pol II-occupied genes. The PI/SI analysis was performed, giving pS5- and pS2RNApol II BigWig files to the getPausingIndices() function of BRGenomics (Version v1.8.0; DeBerardine M., 2022; https://bioconductor.org/packages/release/bioc/html/BRGenomics.html) R package (4.1.2.2 R software). PI/SI were calculated at common pS5- and pS2RNApol II annotated genes, respectively, longer than 1 kb, in all three cell lines, defining the promoter region as −50/+300 bp interval from the TSS and the gene body as +301 bp from the TSS and 1 kb downstream the TES.
Nucleosome occupancy calculation
The NucleoATAC tool was employed for nucleosome mapping analysis starting from ATAC-seq data103. The analysis was performed on biological replicates of merged reads, with genomic regions set as ±500 bp windows from line-specific accessible TSS. Plots were produced using deepTools (4.1.2.3 DeepTools) computeMatrix and plotProfile utilities, using as input nucleoatac_signal.smooth.bedgraph.gz or occ.bedgraph.gz files converted to BigWiG.
DNA:RNA immunoprecipitation and sequencing (DRIP-seq)
DRIP-seq samples were prepared based on the protocol described in Sanz et al. 2019104. Briefly, genomic DNA was extracted in native conditions and digested by restriction enzyme cocktail fragmentation (BsrGI-HF NEB cat. R3575, EcoRI-HF NEB cat. R3101, HindIII-HF NEB cat. R31045, SspI-HF Neb cat. R3132, and XbaI NEB cat. R01455). Immunoprecipitation was performed using 8 µg of fragmented DNA incubated for 14 h at 16 °C with 20 µg of S9.6 antibody (Millipore, cat. MABE1095). Next, the complexes were captured on ProteinA/G agarose beads (Thermo Fisher Scientific R0561), washed, eluted, and purified by phenol/chloroform extraction. qPCR of control regions was performed to check the quality of the material. Libraries for sequencing were prepared by using NEB-Next end repair module (NEB, cat. E6050), followed by Klenow fragment exo- A-tailing (NEB, cat. M0212), adapters ligation with Quick Ligation kit (NEB, cat. M2200) and amplification with Phusion Flash HF PCR master mix (Thermo Fisher Scientific, cat. F548S). All purification and size selection steps were performed using AMPure XP Beads. Libraries were sequenced on an Illumina NovaSeq6000 platform with SR 100 bp to obtain 40–60M reads per sample.
RNA-seq
Total RNA for RNA-seq experiments was extracted from 75% confluent cells. Then, 1 million cells were trypsinized, pelleted, and resuspended in 1 ml TRIzol (Invitrogen, 15596018). Total RNA was extracted by phenol/chloroform and purified through the RNA Clean and Concentrator-25 kit (Zymo Research, R1018), following the manufacturer’s instructions. RNA integrity was checked through Agilent Bioanalyzer on Agilent RNA 6000 Pico Chips (Agilent Technologies, 5067-1513). 450 ng of total RNA were next subjected to library preparation using the Universal Plus Total RNA-seq kit (TECAN, 9156-A01) according to the manufacturer's instructions. Libraries were purified using AMPure XP Beads and fragment distribution checked through Agilent DNA High Sensitivity Chips (Agilent Technologies, 5067-4626). Samples were sequenced on an Illumina NovaSeq6000 platform with SR 100 bp to obtain 40–60M reads per sample. The experiment was done in four independent biological replicates.
Data analysis
Raw FASTQ reads of CUT&RUN, ATAC-seq, and DRIP-seq were trimmed using the Trimmomatic tool (https://chipster.csc.fi/manual/trimmomatic.html) to remove adapter contamination and aligned to the primary assembly of the human reference genome version GRCh38 using Bowtie2 (https://bowtie-bio.sourceforge.net/bowtie2/manual.shtml) with –very-sensitive and –dovetail options for CUT&RUN data. CUT&RUN bam files were downsampled using the BEDOPS bedextract utility (https://bedops.readthedocs.io/en/latest/index.html) to obtain the same reads number for every sample and assess unbiased comparison. ATAC-seq and DRIP-seq were instead normalized on the fraction of reads in peaks and library size, respectively, by using bedtools (https://bedtools.readthedocs.io/en/latest/). To account only for the specific signal, DRIP-seq, the ratio over RNaseH1 exogenous treatment was performed from bigwig files, and only the regions with positive enrichment over noise were kept. Peak calling was performed with MACS2 in the case of CUT&RUN and DRIP-seq data (https://github.com/jsh58/MACS) by using -broadpeak option and a p value of 0.00001 and 0.0001, respectively, and in the case of ATAC-seq with Genrich tool (https://github.com/jsh58/Genrich) using a p value of 0.001, AUC 100 and maximum peaks distance of 50 bp, PCR duplicates and ChrM corresponding reads were removed. ATAC-seq differential analysis was performed using the DiffBind R package (http://bioconductor.org/packages/release/bioc/html/DiffBind.html). The peaks annotation was performed with Homer software annotatePeaks.pl function (http://homer.ucsd.edu/homer/) and the frequency of each annotation was normalized according to their overall genomic abundance in bp retrieved from UCSC Table Browser (https://genome.ucsc.edu/cgi-bin/hgTables). The deepTools module CountReadsPerBin was used to calculate the fraction of reads in peaks (FRiP) by sampling the genome into 10,000 positions of size 1 bp (https://deeptools.readthedocs.io/en/develop/source/deeptools.html).
RNA-seq reads were aligned to the human reference genome with STAR, generating gene counts and removing PCR duplicates. Bam files were normalized according to the library size using bedtools. Transcripts expression levels were quantified as transcripts per million by mean of Salmon (https://salmon.readthedocs.io/en/latest/salmon.html#using-salmon). PATCHED chromosomes and mtDNA were removed from the analysis, and differentially expressed genes were identified using the Bioconductor package DESeq2 and “apeglm” for LFC shrinkage (https://bioconductor.org/packages/devel/bioc/manuals/DESeq2/man/DESeq2.pdf). GO terms enrichment analysis was performed using EnrichR (https://cran.r-project.org/web/packages/enrichR/vignettes/enrichR.html). eRNAs were determined from total RNA-seq data by using the featureCount function of DESeq2 applied to ATAC-seq accessible regions farer than 1 kb from TSSs86.
All further statistical analysis was performed using R software.
Cytotoxicity curves and EC50 calculation
To evaluate the cytotoxicity of VE822, Ceralasertib (MedChemExpress, HY19323) and Etoposide, cells were plated in 96-well plates, grown O/N at 37 °C, and treated with serial dilutions of the indicated compound concentrations. Cell numbers were counted before treatment and 24 or 48 h after treatment using Incucyte® Live-Cell Analysis System (Sartorius) that automatically counts cells based on dedicated analysis adapted to specific cell lines morphology. EC50 values were calculated based on the dose-response curve fitting with a dedicated analysis available from Incucyte 2021C software. The dose-response curve was generated using the ratio of cell counts per image, normalized to the counts at time 0 h. Five images per each well were acquired, and each condition was prepared at least in technical duplicate.
Caspase assay
To evaluate early apoptosis in cells treated with VE822, CellEvent Caspase-3/7 Green ReadyProbes™ Reagent (Invitrogen, R37111) was used. In brief, cells were plated in 96-well plates, grown O/N at 37 °C, and treated with different dilutions of VE822 together with the CellEvent™ Caspase-3/7 Green reagent consisting of a DEVD peptide. Cell growth was monitored for 48 h after treatment using Incucyte® Live-Cell Analysis System (Sartorius) equipped with a FITC filter allowing the detection of cleaved caspase-3/7, leading to the cleavage of DEVD peptide and the release of green fluorescence. The percentage of apoptotic cells was calculated using the Incucyte® Live-Cell Analysis System 2021C software.
Cell titer blue
To evaluate the cytotoxicity of VE822, cells were plated in 96-well plates at least in technical duplicate for each condition to be tested, grown O/N at 37 °C, and treated with serial dilutions of the indicated compound concentrations. At the end of treatment, cells were incubated O/N with CellTiterBlue reagent (Promega, cat. G8080). The day after, 48 h after treatment, fluorescence was acquired on a Varioskan plate reader (Thermo Scientific). Fluorescence values were converted into a percentage of viable cells based on the non-treated control. Technical replicates were used to calculate the mean and SD. Each experiment was performed at least in biological triplicate.
In vivo tumor analysis
About 200,000 tIMEC or tIMEC-A cells, transduced with a lentiviral vector encoding for luciferase in 1:6 matrigel (BD Biosciences) were orthotopically injected in 5-week-old NSG mice (Charles River Laboratories). Once tumor xenografts were palpable (0.2 × 0.2 cm), mice were randomized (n = 6) and treated with vehicle (10% D-a-tocopherol polyethylene glycol 1000 succinate in PBS, Sigma-Aldrich) or VE822 (60 mg/kg) administered by oral gavage every other day for 4 weeks. Tumor volume was measured two times a week with an electronic caliper using the formula π/6 × (smaller diameter)2 × larger diameter. At the end of the treatments, bioluminescent signals were measured by using the IVIS Lumina III system (PerkinElmer) and, after mice sacrifice, primary tumors and lungs were collected for immunochemical analyses. All studies on mice were conducted in strict accordance with the institutional guidelines for animal research and approved by the Italian Ministry of Health; Department of Public Health, Animal Health, Nutrition, and Food Safety in accordance to the law on animal experimentation (D.Lgs. 26/2014), Italian Ministry of Health authorization (IACUC 373/2015-PR).
FFPE tissue immunofluorescence and PLA
For immunofluorescence and PLA experiments 5-μm-thick FFPE sections of primary tumors were subjected to antigen retrieval by performing sections rehydration with successive incubations in xylene, ethanol 96%, ethanol 70%, ddH2O followed by 20-min incubation in sodium citrate solution pH 6.0 10 mM or Tris-EDTA pH 9.0, respectively both supplemented with Tween-20 0.05% at 90 °C. Next sections were blocked in AB solution (Goat serum 3%, Triton 0.3% in PBS) for 1 h RT and incubated overnight with primary antibodies against pRPA32 1:1000 (Bethyl, A300-246A-8), anti-human-mitochondria (Abcam, ab92824) 1:1000 or anti-pS2RNApol II (Abcam, ab5095) + anti-PCNA (Merck, WH0005111M2) 1:400. Samples were washed in PBS-Triton 0.3% and either incubated with specie-specific secondary antibodies Alexa-Fluor-647 and Alexa-Fluor-488 or subjected to PLA using Duolink in situ detection kit (Sigma-Aldrich, DUO92008) with Duolink PLUS anti-rabbit (Sigma-Aldrich, DUO92002) and MINUS anti-mouse probes (Sigma-Aldrich, DUO92004) according to manufacturer instruction.
FFEPE tissue immunohistochemistry
Immunohistochemical analyses were performed on 5-μm-thick FFPE sections of primary tumors and lung metastases as previously described53. Sections were exposed to ki-67 (D3B5, Cell Signalling Technology) or mitochondria (113-1, Abcam) antibodies. The number of lung metastatic cells was assessed by counting human-mitochondria-positive cells with an optical microscope. For Hematoxylin/Eosin, tissues were stained with hematoxylin for 2 min and subsequently with eosin for 1 min.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Public BC patients data were retrieved from cBioPortal (https://www.cbioportal.org) using the brca_tcga_pan_can_atlas_2018 dataset (Breast Invasive Carcinoma TCGA PanCancer Atlas) and Breast Cancer METABRIC45,46 for Breast Cancer Specific analysis and the Metastatic Breast Cancer Project 2021 dataset (www.mbcproject.org, Provisional, December 2021), for metastatic BC patients analysis. Chromatin player genes were retrieved from GO terms: GO:0042393 and GO:0140713. The list of DDR pathway genes used for expression correlation analysis with ANP32E, MYC, and FOXA1 was retrieved from ref. 49. MYC transcription factor validated targets from ENCODE were retrieved from Harmonizome database (https://maayanlab.cloud/Harmonizome/gene_set/MYC/ENCODE+Transcription+Factor+Targets). ANP32E mRNA expression data in BC cell lines were retrieved from the public dataset 23Q2 available from the DepMap portal (https://depmap.org/portal/). All sequencing data produced in the present manuscript have been deposited in NCBI’s Gene Expression Omnibus with the GEO Series accession number GSE250562. Source data are provided with this paper.
References
Kumar, P. & Aggarwal, R. An overview of triple-negative breast cancer. Arch. Gynecol. Obstet. 293, 247–269 (2016).
Negrini, S., Gorgoulis, V. G. & Halazonetis, T. D. Genomic instability — an evolving hallmark of cancer. Nat. Rev. Mol. Cell Biol. 11, 220–228 (2010).
Woo, R. A. & Poon, R. Y. C. Activated oncogenes promote and cooperate with chromosomal instability for neoplastic transformation. Genes Dev. 18, 1317–1330 (2004).
Duffy, M. J., O’Grady, S., Tang, M. & Crown, J. MYC as a target for cancer treatment. Cancer Treat. Rev. 94, 102154 (2021).
AlSultan, D. et al. The novel low molecular weight MYC antagonist MYCMI-6 inhibits proliferation and induces apoptosis in breast cancer cells. Invest. New Drugs 39, 587–594 (2021).
Xu, J., Chen, Y. & Olopade, O. I. MYC and breast cancer. Genes Cancer 1, 629–640 (2010).
Kumar, R. et al. HumCFS: a database of fragile sites in human chromosomes. BMC Genomics 19, 985 (2019).
Le Tallec, B. et al. Updating the mechanisms of common fragile site instability: how to reconcile the different views? Cell. Mol. Life Sci. 71, 4489–4494 (2014).
Stork, C. T. et al. Co-transcriptional R-loops are the main cause of estrogen-induced DNA damage. eLife 5, e17548 (2016).
Bayona-Feliu, A. et al. The chromatin network helps prevent cancer-associated mutagenesis at transcription-replication conflicts. Nat. Commun. 14, 6890 (2023).
Topatana, W. et al. Advances in synthetic lethality for cancer therapy: cellular mechanism and clinical translation. J. Hematol. Oncol. 13, 118 (2020).
Hamperl, S., Bocek, M. J., Saldivar, J. C., Swigut, T. & Cimprich, K. A. Transcription-replication conflict orientation modulates R-loop levels and activates distinct DNA damage responses. Cell 170, 774–786.e19 (2017).
Hamperl, S. & Cimprich, K. A. Conflict resolution in the genome: how transcription and replication make it work. Cell 167, 1455–1467 (2016).
Sanz, L. A. et al. Prevalent, dynamic, and conserved R-loop structures associate with specific epigenomic signatures in mammals. Mol. Cell 63, 167–178 (2016).
Brickner, J. R., Garzon, J. L. & Cimprich, K. A. Walking a tightrope: the complex balancing act of R-loops in genome stability. Mol. Cell 82, 2267–2297 (2022).
Prendergast, L. et al. Resolution of R-loops by INO80 promotes DNA replication and maintains cancer cell proliferation and viability. Nat. Commun. 11, 4534 (2020).
Cristini, A., Groh, M., Kristiansen, M. S. & Gromak, N. RNA/DNA hybrid interactome identifies DXH9 as a molecular player in transcriptional termination and R-loop-associated DNA damage. Cell Rep. 23, 1891–1905 (2018).
Jayakumar, S. et al. PSIP1/LEDGF reduces R-loops at transcription sites to maintain genome integrity. Nat. Commun. 15, 361 (2024).
Yang, J. et al. The histone chaperone FACT contributes to DNA replication-coupled nucleosome assembly. Cell Rep. 14, 1128–1141 (2016).
Herrera-Moyano, E., Mergui, X., García-Rubio, M. L., Barroso, S. & Aguilera, A. The yeast and human FACT chromatin-reorganizing complexes solve R-loop-mediated transcription-replication conflicts. Genes Dev. 28, 735–748 (2014).
Chen, L. et al. R-ChIP using inactive RNase H reveals dynamic coupling of R-loops with transcriptional pausing at gene promoters. Mol. Cell 68, 745–757.e5 (2017).
Eigenhuis, K. N., Somsen, H. B. & van den Berg, D. L. C. Transcription pause and escape in neurodevelopmental disorders. Front. Neurosci. 16, 846272 (2022).
Oberbeckmann, E. et al. Ruler elements in chromatin remodelers set nucleosome array spacing and phasing. Nat. Commun. 12, 3232 (2021).
Burgess, R. J. & Zhang, Z. Histone chaperones in nucleosome assembly and human disease. Nat. Struct. Mol. Biol. 20, 14–22 (2013).
Rudnizky, S. et al. H2A.Z controls the stability and mobility of nucleosomes to regulate expression of the LH genes. Nat. Commun. 7, 12958 (2016).
Day, D. S. et al (2016). Comprehensive analysis of promoter-proximal RNA polymerase II pausing across mammalian cell types. Genome Biol. https://doi.org/10.1186/s13059-016-0984-2 (2016).
Piquet, S. et al. The histone chaperone FACT coordinates H2A.X-dependent signaling and repair of DNA damage. Mol. Cell 72, 888–901.e7 (2018).
Mao, Z. et al. Anp32e, a higher eukaryotic histone chaperone directs preferential recognition for H2A.Z. Cell Res. 24, 389–399 (2014).
Obri, A. et al. ANP32E is a histone chaperone that removes H2A.Z from chromatin. Nature 505, 648–653 (2014).
Colino-Sanguino, Y., Clark, S. J. & Valdes-Mora, F. The H2A.Z-nucleosome code in mammals: emerging functions. Trends Genet. 38, 273–289 (2022).
Vardabasso, C. et al. Histone variant H2A.Z.2 mediates proliferation and drug sensitivity of malignant melanoma. Mol. Cell 59, 75–88 (2015).
Nishimura, T. et al. Evolutionary histories of breast cancer and related clones. Nature 620, 607–614 (2023).
Kaplan, H. G. et al. Multi-omic profiling of simultaneous ductal carcinoma in situ and invasive breast cancer. Breast Cancer Res. Treat. 205, 451–464 (2024).
Wang, Z. et al. Evolving copy number gains promote tumor expansion and bolster mutational diversification. Nat. Commun. 15, 2025 (2024).
Goh, J. Y. et al. Chromosome 1q21.3 amplification is a trackable biomarker and actionable target for breast cancer recurrence. Nat. Med. 23, 1319–1330 (2017).
Orsetti, B. et al. Genetic profiling of chromosome 1 in breast cancer: mapping of regions of gains and losses and identification of candidate genes on 1q. Br. J. Cancer 95, 1439–1447 (2006).
Ibragimova, M. K., Tsyganov, M. M., Pevzner, A. M. & Litviakov, N. V. Transcriptome of breast tumors with different amplification status of the long arm of chromosome 8. Anticancer Res. 41, 187–195 (2021).
Xiong, Z. et al. ANP32E induces tumorigenesis of triple-negative breast cancer cells by upregulating E2F1. Mol. Oncol. 12, 896–912 (2018).
Xu, Y. et al. Histone H2A.Z controls a critical chromatin remodeling step required for DNA double-strand break repair. Mol. Cell 48, 723–733 (2012).
Idrissou, M. et al. TIP60/P400/H4K12ac plays a role as a heterochromatin back-up skeleton in breast cancer. Cancer Genomics Proteom. 17, 687–694 (2020).
Gursoy-Yuzugullu, O., Ayrapetov, M. K. & Price, B. D. Histone chaperone Anp32e removes H2A.Z from DNA double-strand breaks and promotes nucleosome reorganization and DNA repair. Proc. Natl Acad. Sci. USA 112, 7507–7512 (2015).
Prat, A. et al. Clinical implications of the intrinsic molecular subtypes of breast cancer. Breast 24, S26–S35 (2015).
Kotsantis, P., Petermann, E. & Boulton, S. J. Mechanisms of oncogene-induced replication stress: jigsaw falling into place. Cancer Discov. 8, 537–555 (2018).
Pikor, L., Thu, K., Vucic, E. & Lam, W. The detection and implication of genome instability in cancer. Cancer Metastasis Rev. 32, 341–352 (2013).
Pereira, B. et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat. Commun. 7, 11479 (2016).
Dawson, S.-J., Rueda, O. M., Aparicio, S. & Caldas, C. A new genome-driven integrated classification of breast cancer and its implications. EMBO J. 32, 617–628 (2013).
Schär, P. Spontaneous DNA damage, genome instability, and cancer—when DNA replication escapes control. Cell 104, 329–332 (2001).
Jeggo, P. A., Pearl, L. H. & Carr, A. M. DNA repair, genome stability and cancer: a historical perspective. Nat. Rev. Cancer 16, 35–42 (2016).
Wang, Z. et al. Comutations in DNA damage response pathways serve as potential biomarkers for immune checkpoint blockade. Cancer Res. 78, 6486–6496 (2018).
Park, W. K. et al. Long-term oncologic outcomes of unselected triple-negative breast cancer patients according to BRCA1/2 mutations. npj Precis. Onc. 8, 1–7 (2024).
Yndestad, S. et al. Homologous recombination deficiency across subtypes of primary breast cancer. JCO Precis. Oncol. 7, e2300338 (2023).
Ruff, G. L., Murphy, K. E., Smith, Z. R., Vertino, P. M. & Murphy, P. J. Subtype-independent ANP32E reduction during breast cancer progression in accordance with chromatin relaxation. BMC Cancer 21, 1342 (2021).
Poli, V. et al. MYC-driven epigenetic reprogramming favors the onset of tumorigenesis by inducing a stem cell-like state. Nat. Commun. 9, 1024 (2018).
Saxena, S. & Zou, L. Hallmarks of DNA replication stress. Mol. Cell 82, 2298–2314 (2022).
Li, B., Carey, M. & Workman, J. L. The role of chromatin during transcription. Cell 128, 707–719 (2007).
Stoy, H. et al. Direct visualization of transcription-replication conflicts reveals post-replicative DNA:RNA hybrids. Nat. Struct. Mol. Biol. 30, 348–359 (2023).
Mosler, T. et al. R-loop proximity proteomics identifies a role of DDX41 in transcription-associated genomic instability. Nat. Commun. 12, 7314 (2021).
Okamoto, Y. et al. Replication stress induces accumulation of FANCD2 at central region of large fragile genes. Nucleic Acids Res. 46, 2932–2944 (2018).
Okamoto, Y. et al. FANCD2 protects genome stability by recruiting RNA processing enzymes to resolve R-loops during mild replication stress. FEBS J. 286, 139–150 (2019).
Montecucco, A., Zanetta, F. & Biamonti, G. Molecular mechanisms of etoposide. EXCLI J. 14, 95–108 (2015).
Charrier, J.-D. et al. Discovery of potent and selective inhibitors of ataxia telangiectasia mutated and Rad3 related (ATR) protein kinase as potential anticancer agents. J. Med. Chem. 54, 2320–2330 (2011).
Terradas, M., Martín, M. & Genescà, A. Impaired nuclear functions in micronuclei results in genome instability and chromothripsis. Arch. Toxicol. 90, 2657–2667 (2016).
Maréchal, A. & Zou, L. RPA-coated single-stranded DNA as a platform for post-translational modifications in the DNA damage response. Cell Res. 25, 9–23 (2015).
Olson, E., Nievera, C. J., Klimovich, V., Fanning, E. & Wu, X. RPA2 is a direct downstream target for ATR to regulate the S-phase checkpoint. J. Biol. Chem. 281, 39517–39533 (2006).
Vassin, V. M., Anantha, R. W., Sokolova, E., Kanner, S. & Borowiec, J. A. Human RPA phosphorylation by ATR stimulates DNA synthesis and prevents ssDNA accumulation during DNA-replication stress. J. Cell Sci. 122, 4070–4080 (2009).
Bigot, N. et al. Phosphorylation-mediated interactions with TOPBP1 couple 53BP1 and 9-1-1 to control the G1 DNA damage checkpoint. eLife 8, e44353 (2019).
Her, J., Ray, C., Altshuler, J., Zheng, H. & Bunting, S. F. 53BP1 mediates ATR-Chk1 signaling and protects replication forks under conditions of replication stress. Mol. Cell Biol. 38, e00472-17 (2018).
Toledo, L. I. et al. ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell 155, 1088–1103 (2013).
Harrigan, J. A. et al. Replication stress induces 53BP1-containing OPT domains in G1 cells. J. Cell Biol. 193, 97–108 (2011).
Lukas, C. et al. 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat. Cell Biol. 13, 243–253 (2011).
Tsai, S. et al. ARID1A regulates R-loop associated DNA replication stress. PLoS Genet. 17, e1009238 (2021).
Heilbrun, E. E., Merav, M., Parnas, A. & Adar, S. The hardwired transcriptional response to DNA damage. Curr. Opin. Syst. Biol. 19, 1–7 (2020).
Debès, C. et al. Ageing-associated changes in transcriptional elongation influence longevity. Nature 616, 814–821 (2023).
Chodosh, L. A., Fire, A., Samuels, M. & Sharp, P. A. 5,6-Dichloro-1-β-D-ribofuranosylbenzimidazole inhibits transcription elongation by RNA polymerase II in vitro. J. Biol. Chem. 264, 2250–2257 (1989).
Murphy, K. E., Meng, F. W., Makowski, C. E. & Murphy, P. J. Genome-wide chromatin accessibility is restricted by ANP32E. Nat. Commun. 11, 5063 (2020).
Valdés-Mora, F. et al. Acetylation of H2A.Z is a key epigenetic modification associated with gene deregulation and epigenetic remodeling in cancer. Genome Res. 22, 307–321 (2012).
Gursoy-Yuzugullu, O., Ayrapetov, M. K. & Price, B. D. Histone chaperone Anp32e removes H2A.Z from DNA double-strand breaks and promotes nucleosome reorganization and DNA repair. Proc. Natl Acad. Sci. USA https://doi.org/10.1073/pnas.1504868112 (2015).
Weber, C. M., Ramachandran, S. & Henikoff, S. Nucleosomes are context-specific, H2A.Z-modulated barriers to RNA polymerase. Mol. Cell 53, 819–830 (2014).
Chen, Z. et al. High-resolution and high-accuracy topographic and transcriptional maps of the nucleosome barrier. Elife 8, e48281 (2019).
Mérida-Cerro, J. A., Maraver-Cárdenas, P., Rondón, A. G. & Aguilera, A. Rat1 promotes premature transcription termination at R-loops. Nucleic Acids Res. 52, 3623–3635 (2024).
Hayakawa, K. et al. H2A O-GlcNAcylation at serine 40 functions genomic protection in association with acetylated H2AZ or γH2AX. Epigenetics Chromatin 10, 51 (2017).
Promonet, A. et al. Topoisomerase 1 prevents replication stress at R-loop-enriched transcription termination sites. Nat. Commun. 11, 3940 (2020).
Li, S. & Wu, X. Common fragile sites: protection and repair. Cell Biosci. 10, 29 (2020).
Brunelle, M. et al. The histone variant H2A.Z is an important regulator of enhancer activity. Nucleic Acids Res. 43, 9742–9756 (2015).
Giaimo, B. D., Ferrante, F., Herchenröther, A., Hake, S. B. & Borggrefe, T. The histone variant H2A.Z in gene regulation. Epigenetics Chromatin 12, 37 (2019).
Ahmed, I. et al. eRNA profiling uncovers the enhancer landscape of oesophageal adenocarcinoma and reveals new deregulated pathways. eLife 12, e80840 (2023).
Sporikova, Z., Koudelakova, V., Trojanec, R. & Hajduch, M. Genetic markers in triple-negative breast cancer. Clin. Breast Cancer 18, e841–e850 (2018).
Bhowmick, R., Mehta, K. P. M., Lerdrup, M. & Cortez, D. Integrator facilitates RNAPII removal to prevent transcription-replication collisions and genome instability. Mol. Cell 83, 2357–2366.e8 (2023).
Long, H. et al. H2A.Z facilitates licensing and activation of early replication origins. Nature 577, 576–581 (2020).
McCann, J. L. et al. APOBEC3B regulates R-loops and promotes transcription-associated mutagenesis in cancer. Nat. Genet. 55, 1721–1734 (2023).
Abel, M. L. et al. Targeting replication stress and chemotherapy resistance with a combination of sacituzumab govitecan and berzosertib: a phase I clinical trial. Clin. Cancer Res. 29, 3603–3611 (2023).
Takahashi, N. et al. Berzosertib plus topotecan vs topotecan alone in patients with relapsed small cell lung cancer: a randomized clinical trial. JAMA Oncol. https://doi.org/10.1001/jamaoncol.2023.4025 (2023).
Villaruz, L. C. et al. NCI 9938: Phase I clinical trial of ATR inhibitor berzosertib (M6620, VX-970) in combination with irinotecan in patients with advanced solid tumors. J. Clin. Oncol. 40, 3012–3012 (2022).
Gyori, B. M., Venkatachalam, G., Thiagarajan, P. S., Hsu, D. & Clement, M.-V. OpenComet: an automated tool for comet assay image analysis. Redox Biol. 2, 457–465 (2014).
RexHoover.com. CometScore http://rexhoover.com/index.php?id=cometscore (2017).
Dhar, S., Datta, A., Banerjee, T. & Brosh, R. M. Single-molecule DNA fiber analyses to characterize replication fork dynamics in living cells. Methods Mol. Biol. 1999, 307–318 (2019).
Li, L. et al. Automatic DNA replication tract measurement to assess replication and repair dynamics at the single-molecule level. Bioinformatics 38, 4395–4402 (2022).
Corces, M. R. et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods 14, 959–962 (2017).
Janssens, D. & Henikoff, S. CUT&RUN: targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nat. Protoc.13, 1006–1019 (2019).
Dellino, G. I. et al. Release of paused RNA polymerase II at specific loci favors DNA double-strand-break formation and promotes cancer translocations. Nat. Genet. 51, 1011–1023 (2019).
Liu, N. Library Prep for CUT&RUN with NEBNext® UltraTM II DNA Library Prep Kit for Illumina® (E7645). (2019).
Adelman, K. & Lis, J. T. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat. Rev. Genet. 13, 720–731 (2012).
Schep, A. N. et al. Structured nucleosome fingerprints enable high-resolution mapping of chromatin architecture within regulatory regions. Genome Res. 25, 1757–1770 (2015).
Sanz, L. A. & Chédin, F. High-resolution, strand-specific R-loop mapping via S9.6-based DNA-RNA immunoprecipitation and high-throughput sequencing. Nat. Protoc. 14, 1734–1755 (2019).
Acknowledgements
We thank Maria Luce Negri and Dr. Sven Beyes from the Zippo laboratory, for helpful discussion and technical support. We thank Dr. Veronica De Sanctis, Dr. Roberto Bertorelli, and Dr. Paolo Cavallerio of the next-generation sequencing (NGS) core facility for their support, technical contribution, and sample sequencing. The cell technology (CTF) and high throughput screening (HTS) facilities at the CIBIO Department are also gratefully acknowledged. We thank Prof. Luca Fava, Prof. Alessandro Provenzani, and Prof. Matthias Altmeyer for critical comments and suggestions. This project has been realized with the financial contribution from Associazione Italiana per la Ricerca sul Cancro (AIRC), project code 25373 (to S.L.). Funding from AIRC under IG 2018 - ID. 21492 project, - PNRR M4C2I1.3-HEAL ITALIA project PE 00000019 CUP B73C22001310006 from Italian Ministry for University - PRIN 2022 prot. 20223NY37M supported MT work. European Union- NextGenerationEU- Fondi MUR D.M. 737/2021- Project title: “Sviluppo di terapie innovative nel trattamento del carcinoma alla mammella” supported AT work. European Union- NextGenerationEU- Fondi MUR D.M. 737/2021- Project title: “Ruolo di MERTK nell’interazione tra cellule tumorali e il microambiente nella progressione del tumore alla mammella” supported MG work. Work in the Zippo group was supported by grants from the AIRC foundation (IG 2019-22911 and IG 2024-30745), European Union under the Horizon 2020 Framework Program H2020 Future and Emerging Technologies (801336; - PROCHIP), and Worldwide Cancer Research (25-0268).
Author information
Authors and Affiliations
Contributions
S.L., V.P., and A.Z. conceived the study, designed the experiments, and interpreted the data. S.L. and L.F. performed chromatin, expression, and R-loops profiling experiments and computational analyses. S.L., V.P., M.B., F.B., L.F., and A.F. performed the cellular and molecular biology studies, participating in data analyses. A.T. and M.G. performed the in vivo experiments and IHC analyses. M.T. designed the in vivo experiments and interpreted the data. S.L., Y.C., and G.D’A. performed computational analyses of cancer patients’ datasets under the supervision of A.Z. and F.D. A.Z. supervised the work, S.L. and A.Z. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Madapura Pradeepa and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Lago, S., Poli, V., Fol, L. et al. ANP32E drives vulnerability to ATR inhibitors by inducing R-loops-dependent transcription replication conflicts in triple negative breast cancer. Nat Commun 16, 4602 (2025). https://doi.org/10.1038/s41467-025-59804-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-59804-0