Abstract
Single-cell sequencing provides transcriptomic profiling at single-cell resolution, uncovering cellular heterogeneity with unprecedented precision. Yet, current single cell data analysis suffers from the inherent data noises, batch effects, and sparsity, highlighting the requirement of a unified model to represent cellular states. To circumvent this problem, many recent efforts focus on training single-cell foundation models based on large datasets. However, current human foundation models are still limited by the sizes of training data and model parameters. Here, we have collected a diverse dataset of 100 million human cells, on which we train a single-cell foundation model (CellFM) containing 800 million parameters. To balance efficiency and performance, the model is trained through a modified RetNet framework on the MindSpore. Extensive experiments have shown that CellFM outperforms existing models in cell annotation, perturbation prediction, gene function prediction, and gene-gene relationship capturing.
Similar content being viewed by others
Introduction
Single-cell RNA sequencing (scRNA-seq) technologies have revolutionized molecular biology by enabling the measurement of transcriptome profiles with unparalleled scale and precision1,2. As single-cell technologies advance, the rapid accumulation of extensive datasets has posed significant analytical challenges3,4, primarily due to the data’s inherent noise, sparsity, and batch effects. Despite the development of numerous single-cell-specific tools5,6,7 to address these challenges, their performance often falls short when applied to new datasets and struggles to scale with the growing data size. More importantly, these tools fail to fully leverage the rich information embedded in large atlas datasets, underscoring the need for novel computational strategies.
To address this challenge, several single-cell foundation models have been developed to analyze single-cell data8. Drawing inspiration from the remarkable success of large language models (LLMs) in natural language processing (NLP)9, and aiming to reduce training costs, researchers have begun exploring the fine-tuning of these LLMs using relatively small amounts of single-cell data. For example, Cell2Sentence10 converts gene expression profiles of individual cells into sequences of gene names ordered by expression levels and uses these sequences to fine-tune the GPT-2 model. Similarly, GenePT11 utilizes GPT-3.5 to generate gene embeddings based on gene descriptions and metadata. While these approaches have improved by fine-tuning the GPT models, they still fall short of fully harnessing the rich gene expression data in large single-cell datasets, highlighting the need for more comprehensive strategies.
To create single-cell foundation models from scratch, three types of single-cell foundation models, including ordering, value categorization, and Value projection were proposed. We first introduce gene-ranking-based models, such as iSEEEK12, which was the first model trained on over 10 million cells by predicting gene rank. Similarly, tGPT13 learns gene embeddings by autoregressively modeling gene ranks relative to their neighbors, processing sequences of genes ordered by expression levels, and predicting the next gene’s rank based on prior context. Trained on 22.3 million single-cell transcriptomes from humans and mice, tGPT demonstrated superior performance across multiple datasets. Geneformer14 predicts gene positions within the cellular context to derive rank-based gene embeddings. With training on a dataset of 30 million single-cell transcriptomes spanning diverse human tissues, Geneformer has achieved outstanding predictive performance.
Recently, value categorization strategies were applied to leverage gene counts. By representing each gene with an embedding and binning its RNA counts, these models convert the continuous gene expression data into categorical values, enabling the use of methods designed for categorical data. For example, scBert15 bins gene expression values into discrete “buckets," transforming the continuous task of predicting gene expression into a classification problem. Trained on millions of human cells, scBert has shown improved performance across various datasets. Similarly, scGPT16 also segments gene expression values but enhances the process with an attention mask mechanism for autoregressive prediction. Using a self-supervised approach, scGPT optimizes both cell and gene representations. Trained on over 33 million human cells, scGPT excels in various single-cell tasks. The Universal Cell Embedding (UCE)17 captures molecular diversity across species by integrating genetic data using protein language models. It uses self-supervised learning to predict gene expression by masking a subset of genes, refining predictions with binary cross-entropy loss. With over 650 million parameters, UCE is trained on more than 36 million cells, offering insights into gene expression across diverse cellular contexts.
To further predict precise gene values, the value projection-based single-cell models were proposed. In this strategy, the gene expression vector xi is expressed as the sum of two components: a projection of the gene expression vector and a positional or gene embedding. Unlike ordering and value categorization methods, the key advantage of value projection is that it preserves the full resolution of the data. For example, scFoundation18 directly predicts raw gene expression values using a mask autoencoder (MAE). Trained on a large dataset of around 50 million human cells with ~0.1 billion parameters, scFoundation demonstrates decent performance in single-cell analysis. Similarly, GeneCompass19 incorporates four types of biological prior knowledge to enhance the understanding of gene regulatory mechanisms during gene expression prediction. Trained on about 50 million human and 50 million mouse cells, GeneCompass has around 100 million parameters. To integrate metadata information, scELMo20 leverages LLMs like GPT-3.5 to generate metadata descriptions and embeddings, combining them with raw data in both zero-shot and fine-tuning frameworks to address a variety of tasks.
Despite the endeavor, the potential of foundation models trained exclusively on 100 million human cells has yet to be fully explored. The limited availability of sufficient single-species training data, such as for human cells, hindered the development of large-scale, single-species models. Existing single-species models are typically trained on around 50 million cells, resulting in fewer than 100 million parameters. One reason for this limitation is the difficulty in collecting single-cell datasets, which are often stored in various formats (e.g., FASTQ, h5ad, Seurat objects, 10x Genomics) and dispersed across different repositories to accommodate diverse data processing and analysis needs.
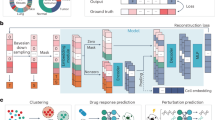
Here, we collect a lot of single-cell datasets from public databases and then make these data cleansing and standardization of unified formats, resulting in compiling a dataset of approximately 100 million human cells sequenced through various technologies. These datasets are twice as large as those used in the current largest single-species model, providing a rich foundation for training a larger model21,22. We proposed a robust single-cell foundation model CellFM (Fig. 1) with an impressive 800 million parameters, marking an eightfold increase in model parameters over the current largest single-species model. To enhance the training of CellFM’s extensive parameters and to handle its substantial dataset, we have integrated ERetNet, a Transformer architecture variant with linear complexity. ERetNet’s design ensures a balance between efficiency and performance, serving as the backbone of our model. CellFM is categorized as a value-projection-based single-cell foundation model, as it aims to recover the vector embeddings of masked genes derived from their linear projections based on gene expression values. CellFM is developed using the MindSpore AI framework from Huawei and is trained on four Huawei Altas800 servers, each equipped with eight Ascend910 NPUs. Our comprehensive experiments have shown that CellFM outperforms existing models across diverse applications, such as cell annotation, perturbation prediction, and gene function prediction.
a CellFM consists of the Embedding module, the ERetNet module, and the LoRA module. The expression of the cell is first fed into the Embedding module to obtain the initial token values of each gene. The embedded gene tokens of the cell are then input into the ERetNet module to learn the gene-to-gene relationships and gene embeddings. Finally, the Low-Rank Adaptation (LoRA) is implemented to minimize the number of training parameters of CellFM. b Each ERetNet Layer block integrates Multi-Head Attention (MHA), the Simple Gated Linear Unit (SGLU), and Layer Normalization (LN). c The collecting workflow and constituents of the training dataset employed within CellFM. d The pre-trained CellFM model is adaptable for a multitude of single-cell downstream analyzes including cell type annotation, perturbation prediction, gene network inference, and gene function prediction.
Results
Overview of cellFM
Single-cell sequencing technology is crucial for revealing the detailed landscape of cellular diversity and function at the single-cell resolution. With the development of single-cell sequencing technologies, a vast array of datasets has been amassed, laying a robust groundwork for the training of single-cell foundation models. However, these datasets are available across various public repositories, including the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO)23, the European Nucleotide Archive (ENA)24, the Genome Sequence Archive (GSA)25,26, and the ImmPort27. We have meticulously curated single-cell data from these esteemed public databases (Fig. 1a). These datasets are stored in multiple formats, such as FASTQ data, expression matrices, or Seurat/Scanpy objects. We first process raw FASTQ data into the gene expression matrix through primary analysis software provided by manufacturers. Subsequently, all acquired expression matrices were processed using a standardized data analysis workflow facilitated by the SynEcoSys® single-cell database (Singleron Biotechnologies)28. This process involved three key steps including quality control for filtering cells and genes, gene name standardization according to HUGO Gene Nomenclature Committee (HGNC) guidelines, and converting the data to a unified sparse matrix format for subsequent analysis. Our efforts have successfully aggregated 19,914 samples, totaling 102,304,686 human cells from different organs and single-cell sequencing technologies. We have provided a dataset summary (Supplementary Fig. S1) and shared a detailed list of the sources for the training datasets used in CellFM (Supplementary_Metadata_information.xlsx). Concretely, 46.3 million cells were derived from normal donors, the other cells were from diseased donors, such as 7.1 million cells from Viral infections donors and 3.5 million cells from lung cancer donors. Most datasets were sequenced by 10x genomics \({3}^{{\prime} }\) containing 66.7 million cells. Among the curated dataset, approximately 70 million cells had annotated cell types. The training dataset includes a diverse range of cell types, such as T cells (19.2 million), mononuclear phagocytes (7.01 million), neurons (6.29 million), and fibroblasts (3 million).
Building on our comprehensive collection of human cell data, we introduce CellFM, an efficient foundation model endowed with 800 million parameters, designed to streamline the analysis of single-cell data (Fig. 1b). The model’s core is comprised of an embedding module, a series of stacked ERetNet Layers, and the low-rank adaptive module (LoRA) mechanism. CellFM begins by converting scalar gene expression data into rich, high-dimensional embedding features through its embedding module. These gene embeddings are then fed into L ERetNet Layers, which are adept at capturing the nuanced relationships among genes based on their expression profiles. Each ERetNet Layer is composed of several key components: the Gated Multi-head Attention (MHA) unit, the Simple Gated Linear Unit (SGLU), and Layer Normalization (LN). Collectively, these elements empower the ERetNet Layer to achieve training parallelism, cost-effective inference, and superior performance (Fig. 1c). Furthermore, CellFM integrates the LoRA module to reduce the number of trainable parameters during the fine-tuning phase when adapting the model to new datasets. Once pre-trained, CellFM can be applied to multiple single-cell downstream applications, such as gene function prediction, cell type annotation, perturbation effect prediction, and gene network analysis (Fig. 1d).
CellFM improves the accuracy of gene function prediction
Gene function prediction is a cornerstone for deciphering the roles and properties of genes under diverse conditions14. With the human genome comprising ~20,000 protein-encoding genes29, and a significant portion lacking functional annotations, the accurate prediction of gene functions is imperative for a deeper understanding of their roles within biological systems. Here, we evaluated the performance of CellFM for identifying gene function through three distinct gene categories including Dosage sensitivity (referred to as T1), Bivalent methylation status versus non-methylated (T2), and Bivalent methylation versus Lys4-only methylation (T3). These categorizations represent binary classification challenges, where model predictions are assessed against actual gene function labels. Since the limited number of genes in these three tasks, typically fewer than 1000, fine-tuning existing single-cell foundation models presents a challenge.
To make a fair comparison, we adopted a zero-shot learning strategy for each model on the gene function prediction task. As shown in Fig. 2a, our model demonstrated remarkable results, achieving the best performance on three tasks. CellFM surpassed existing methods, with a 5.68% and 5.86% increase over the top two competing models UCE and scGPT in terms of average accuracy values, respectively. A similar trend was observed for Macro-F1 scores (Fig. 2b). The superior performance of our model was further substantiated by visualization results generated by Uniform Manifold Approximation and Projection (UMAP) using the gene embeddings from each pre-trained model. Our model’s ability to distinctly categorize dosage-sensitive from non-sensitive genes was evident in Fig. 2c and Supplementary Fig. S2. However, scGPT and Geneformer exhibited an overlap in the embedding space that could undermine gene function prediction accuracy. In summary, our findings underscore the model’s proficiency in accurately predicting gene functions utilizing a zero-shot approach, showcasing its efficacy without the need for extensive model fine-tuning.
a accuracy (ACC) scores and (b) Macro-F1 values for CellFM along with other competing single-cell foundation models on the binary classification data. c The visualization results of CellFM, scGPT, and Geneformer are plotted by the Uniform Manifold Approximation and Projection (UMAP) using the gene embedding generated through each model. d The Area Under the Precision-Recall Curve (AUPR) values for multiple gene functions predicted by CellFM and other competing single-cell foundation models on Gene Ontology (GO) data, where each gene is annotated with numerous functions. MF Molecular Function, CC Cellular Component, and BP Biological Process. Source data are provided as a Source Data file.
To evaluate the ability of CellFM on multi-class classification, We further performed the gene function prediction using the data from the Gene Ontology (GO). This dataset includes three major categories: biological process (BP), cellular component (CC), and molecular function (MF). Detailed information about the GO dataset can be found in the referenced study30. Given the complexity of predicting all gene functions (BP: 1578, CC: 253, MF: 299), we focused our evaluation on the top 10 most frequent functions within each category. This approach ensures a realistic yet manageable benchmark for model comparison. To guarantee fairness across methods, we intersected the gene sets from each foundational single-cell model and maintained consistent training, validation, and test sets. As shown in Fig. 2d, CellFM demonstrated superior average performance, outperforming the top two models GeneCompass and UCE by 1.6% and 1.94% in average AUPR, respectively. Other models such as scFoundation and Geneformer also delivered competitive results with scGPT achieving an AUPR of 71.3%. We didn’t compare with scELMo since scELMo employed large language models (LLMs) like GPT-3.5 as generators to generate embeddings from metadata descriptions in the training phase, which has included gene function information.
CellFM enables predicting perturbation responses
Recent advancements in sequencing and gene editing have enabled large-scale experimental perturbation simulations to study changes in gene expression and cellular behavior. These simulations are essential for understanding cellular responses to various stimuli and are increasingly applied to investigate drug effects, disease mechanisms, and therapeutic interventions. However, the vast combinatorial space of potential gene perturbations quickly exceeds the practical limits of experimental feasibility. To overcome this, single-cell foundation models adopt perturbation modeling, leveraging knowledge from known experimental perturbations to predict responses to unknown perturbations. By utilizing self-attention mechanisms over the gene dimension, these models can capture intricate gene interactions and accurately predict gene expression responses in unseen perturbations. The predictive power of perturbation modeling becomes especially valuable in AI-driven drug discovery, where it is used to forecast how existing drugs or genes will affect cellular processes, identify new drug targets, and explore drug repurposing opportunities. Additionally, these models offer deep insights into cellular heterogeneity, crucial for developing personalized medicine strategies.
To assess CellFM’s proficiency in predicting perturbation responses, we utilized two Perturb-seq datasets: (1) the Adamson dataset31, encompassing 87 single-gene perturbations with roughly 100 cells per perturbation and at least 7000 control cells; and (2) the Norman dataset32, which includes 131 dual-gene and 105 single-gene perturbations. As depicted in Fig. 3a, we employed the Pearson correlation metric on the top 20 differentially expressed genes (De) to evaluate each model, where Δ denotes the degree of gene expression alteration post-perturbation relative to the control state. We evaluated all single-cell foundation models on the perturbation task by combining them with the classic perturbation model GEARS, as suggested by the study scFoundation. GEARS is a computational tool specifically designed for predicting single and multi-gene perturbations based on scRNA-seq datasets. GEARS operates by integrating a gene-gene interaction network as prior knowledge, which allows it to leverage existing biological information to improve prediction accuracy. GEARS has demonstrated state-of-the-art performance in gene perturbation predictions, making it a leading choice in the field. Concretely, we replaced GEARS’ gene embeddings with those derived from CellFM (Fig. 3a). As shown in Fig. 3b-c, our model consistently outperformed all competing single-cell foundation models, achieving improvements of 1% and 1.45% in average PCC and MSE compared to the second-ranked model scFoundation, respectively. Additionally, CellFM consistently surpassed GEARS with 4.75% and 7% improvement regarding average PCC and MSE values, respectively. As shown in Supplementary Fig. S4, CellFM consistently outperformed all other single-cell foundation models, as well as the non-foundational model GEARS, when measured by the R2 metric. CellFM achieved an R2value that was 1.3% higher than the second-best scGPT in terms of average R2. The visual results for two specific perturbation cases from the Adamson dataset in Fig. 3d have further shown that CellFM could accurately predict the direction of perturbation.
a A diagram showcasing the perturbation prediction model leveraging cell-specific gene embeddings derived from CellFM. b The mean square error (MSE) between predicted and actual post-gene expressions for the top differentially expressed (DE) genes in a zero-shot setting. c Comparison of CellFM with other single-cell foundation models and the perturbation prediction method GERAS. Pearson correlation coefficients between predicted and actual gene expression changes are reported for the top differentially expressed (DE) genes in a zero-shot setting. d Analysis of gene expression changes following perturbations of AHR+ctrl (n = 464 cells) and BPGM+ZBTB1 (n = 280 cells). The plots compare predicted versus actual expression changes for the top 20 differentially expressed genes. Box plot elements represent: center line, median (50th percentile); box limits, upper (75th) and lower (25th) quartiles; whiskers, 1.5 × interquartile range (IQR) from the box; points beyond whiskers are considered outliers. The horizontal dashed line indicates the null effect baseline (0 change). Minimum and maximum values are represented by the whisker endpoints, with all percentiles calculated from the empirical distribution of expression changes. e A graphical representation of potential perturbation combinations across a 20-gene space, differentiated by experiment type (train, valid, test, unseen). Predicted perturbations are indicated by square boxes, with the actual source perturbation marked by a cross. The boxes are colored as follows: dark purple for test data, light purple for validation, medium purple for training, and gray for unseen. f The accuracy of each model in predicting the correct source of perturbation among the top 10 predictions for test cases in a fine-tuning setting. Source data are provided as a Source Data file.
The Norman dataset targeted 105 genes with 236 perturbations and represented just 5% of the expansive 5565 possible gene combinations, highlighting the vast unexplored perturbation space. Consequently, we harnessed CellFM to extend the scope of perturbations virtually and graphically represent the anticipated average response for each gene combination. Concretely, we trained CellFM on the existing knockouts (KOs) from the Norman dataset and extrapolated to other perturbations. CellFM was trained on the original Norman dataset, which covers 236 perturbations targeting 105 genes. We then used the fine-tuned model to predict and expand the responses to untested perturbation combinations in silico. These predictions were visualized as the response for each perturbation combination. We excluded all perturbed genes and plotted the UMAP graph. As shown in Supplementary Fig. S5, the clusters exhibited overlap in certain regions while remaining distinct in others, consistent with the expectation that several perturbations either have no effect or produce similar effects. The genes depicted in each cluster represent the “dominant gene" within the perturbation combinations. The results have demonstrated a strong association between the clusters and their respective dominant genes. For example, the cluster associated with the SET gene indicates that the data points in this cluster correspond to combined perturbations involving SET and another gene (e.g., SET+CLDN6, SET+MIDN).
In addition to the gene perturbation, we further validate the performance of CellFM on drug-perturbed data. Similarly, we also combined CellFM with a non-single foundation model CellOT33, which was a classic model for drug perturbation prediction. Concretely, we integrated CellFM and CellOT by replacing the input cell representation of CellOT with the cell representation from CellFM (Supplementary Fig. S9a), as CellOT is specifically tailored for drug perturbation and does not require gene embeddings. In this setting, we compared CellFM with CellOT, scGEN, and Identity. As indicated in Figure S9b, CellFM outperformed CellOT in perturbation prediction on four drugs, achieving improvements of 66.6% and 2.2% in average l2 and PCC, respectively.
Reverse perturbation prediction in silico using CellFM
Beyond forecasting the outcomes of gene perturbations, the accurate prediction of CRISPR target genes that prompt cellular recovery from disease states is equally significant. Here, we conducted “in silico reverse perturbation prediction” following the study scGPT16, utilizing the Norman dataset. Concretely, we followed scGPT, selecting 20 perturbation genes from the Norman dataset to construct perturbation cases for fine-tuning and testing. This combinatorial space consists of 210 one-gene or two-gene perturbation combinations. The subset was selected to maximize the representation of ground truth perturbation data across both training and testing cases, using a random train-test split. Since scGPT did not specify a particular seed for splitting, we used the default seed. The resulting dataset contained 47 (22%) known perturbations, including 33 training cases, 3 validation cases, and 11 test cases, with the remaining perturbation cases left as unseen. Both scGPT and CellFM were evaluated on this newly split dataset, as we were unable to replicate the exact splits used in the original scGPT study.
CellFM demonstrated remarkable success, accurately predicting the perturbations that would yield the observed cellular outcomes. For example, it accurately identified the combination of CNN1 and ETS2, as well as the pairing of ETS2 and IGDCC3, as the top predictions for a specific test case (Fig. 3e). A similar accuracy trend was observed for the perturbations involving the combinations of IGDCC3 and MAPK1, and CEBPE and CNN1 genes (Supplementary Fig. S6). CellFM and scGPT achieved similar performance in relevant perturbations (purple bars in Supplementary Fig. S8). However, CellFM achieved an average of correctly identified perturbations in 81.8% of the top 10 predictions (Fig. 3f), which was 18.1% higher than scGPT. As the number of top predictions considered increased, both CellFM and scGPT demonstrated enhanced performance. However, GEARs did not exhibit a comparable level of improvement in accuracy. The performance of GEARS was consistent with the findings reported in the scGPT, where the hit rates for top 1 to top 8 predictions also remained constant. This behavior is likely due to the limitations of GEARS as a smaller model, which struggles to identify perturbation combinations effectively. To further assess the robustness of CellFM, we performed additional evaluations by varying the random seeds during the dataset splitting process to generate the new training and test cases. As shown in Supplementary Fig. S7b, CellFM outperformed scGPT and GEARS in terms of average hit rate accuracy. Specifically, CellFM achieved an average of 36.3% and 54.5% correctly identified perturbations in the top 3 and top 5 predictions, which were 18.1% and 18.2% higher than scGPT, respectively. To further examine whether the performance of CellFM was affected by random seeds during model fine-tuning, we re-evaluated CellFM, scGPT, and GEARS under different random seeds. The results demonstrated that CellFM consistently maintained comparable top 1 prediction performance, outperforming scGPT by 9.1% (Supplementary Fig. S7a). For the reverse perturbation prediction, we maintained the original comparison for CellFM and only evaluated it against scGPT and GEARS since the newly added single-cell foundation models neither provided code nor published corresponding results in their original studies (Supplementary Fig. S3) and reimplementing these models would have required substantial programming effort and time.
The current study is limited to gene perturbation without considering drug molecules. Thus, our method doesn’t perform reverse perturbation prediction in silico on the drug perturbation datasets such as sciPlex, since drug perturbation datasets require additional information specific to the drug molecules themselves. In the future, we will expand CellFM’s capabilities to support drug perturbation datasets, which will involve adapting the model architecture to incorporate drug-specific molecular information.
Cell type annotation with CellFM
Cell type annotation is a cornerstone of single-cell data analysis, essential for uncovering the cellular heterogeneity within biological samples. To evaluate CellFM’s competency in cell type annotation, we conducted an exhaustive benchmark against several recent single-cell foundation models. We have also included two baseline methods—SVM and scmap—which were also suggested simultaneously in the benchmark studies34,35. The cell annotation benchmarks used in our study, including intra-dataset and inter-dataset evaluations, were based on the latest benchmarking framework, scEval8, designed to evaluate single-cell foundation models on cell annotation tasks. Following prior studies such as scGPT and scEval, we utilized randomized train-test splits for intra-dataset evaluation to assess model performance under consistent experimental conditions. While intra-datasets were a part of our evaluation, we also performed inter-dataset testing, which better reflects real-world scenarios involving large batch effects. For inter-datasets, we partitioned the data by batch or patient ID, iteratively using each batch as the test set while training on the remaining batches. For hPancreas, we directly adopted the train-test settings established in scGPT, ensuring training and testing data came from different batches. This cross-batch validation ensures a thorough and fair assessment of model robustness across batches. Importantly, the cell annotation datasets used in our study were not included during the pre-training of CellFM. For the zero-shot cell type annotation task, all foundation models, including ours, were fully frozen during training. Classifiers (e.g., MLP or CNN+MLP) were then trained on embeddings extracted from these frozen models using labeled training data and the cross-entropy loss function. These trained classifiers were subsequently used to predict cell types on test datasets. We used the default classifiers implemented in each single-cell foundation model. For models like UCE, which lack a predefined classifier, we implemented a multi-layer perceptron (MLP) approach, in line with standard foundation model practices. For the CellFM, we followed it with the MLP classifier for the classification task.
Based on the findings in the “Scaling of data and model size” section (Supplementary Note 4), we used CellFM with 80 million model parameters (CellFM-80M) for cell annotation and batch effect correction tasks. Our initial evaluation involved eight intra-datasets. Following the methodology established by scGPT16, we segmented each intra-dataset, allocating 70% for training and the remainder for testing. As illustrated in Fig. 4a, CellFM(80M) excelled baselines in terms of ACC across intra-datasets (we simply refer to CellFM(80M) as CellFM). The average ACC for CellFM was 92.91%, surpassing the second-ranked single-cell foundation model scFoundation by 2.02%. CellFM were also 1.97%, 26.86%, and 10.2% higher than those of SVM, scmap, and scBERT, respectively. A similar trend could be found when measured by the Macro-F1 values (Fig. 4b). To substantiate the superior outcomes of CellFM, we present a case study of the immune dataset. The predictions were visualized in Fig. 4c, d and Supplementary Fig. S10, 11, CellFM achieved high precision for most cell types. To evaluate the efficacy of our model in the context of batch effects, we assessed its performance on 7 paired cross-batch datasets. In each scenario, a distinct batch of data was designated as the test set, with the remaining data constituting the training set. As depicted in Fig. 4e, f and Supplementary Fig. S12, our model continued to outperform its competitors and was 2.3% higher than the second-ranked single-cell foundation model scFoundation. CellFM outperformed SVM, scmap, and scBERT on inter-dataset cell annotation tasks, with average accuracy improvements of 2%, 21.1%, and 3.4%, respectively. We further present a visualization case study of the hPancreas dataset in Fig. 4g, h and Supplementary Fig. S10-11. CellFM showed a high precision of most cell types. We also evaluated the embedding quality of all single-cell foundation models on inter-datasets using the scIB metric scores. As shown in Supplementary Fig. S15, CellFM outperformed competing models in AvgBio scores, aligning with its enhanced cell classification performance. Additionally, we incorporated the scIB metric scores to evaluate the impact of MLP layers in CellFM and found minimal performance variation when adjusting layers from one to three. Supplementary Fig. S16 shows a positive correlation between classification accuracy and AvgBio scores, but both metrics exhibited only minor changes across layer configurations.
Heatmaps illustrating (a) classification accuracy and (b) Macro-F1 scores of each model across intra-datasets. Red indicates lower performance, and blue represents the highest performance. c The river plot of CellFM illustrates the predicted cell types and their relationships to the actual cell types on the Immune dataset. d The river plot of scGPT illustrates the predicted cell types and their relationships to the actual cell types on the Immune dataset. Heatmaps show (e) classification accuracy and (f) Macro-F1 scores of each model across inter-datasets, with each value representing the average accuracy calculated from five independent runs using different random seeds. Red indicates lower performance, and blue represents the highest performance. g The river plot of CellFM illustrates the predicted cell types and their relationships to the actual cell types on the hPancreas dataset. h The river of scGPT plot illustrates the predicted cell types and their relationships to the actual cell types on the hPancreas dataset. Source data are provided as a Source Data file.
To further assess CellFM’s capability to distinguish subtypes such as exhausted and activated CD8+ T cells, we evaluated it on the basal cell carcinoma (BCC) dataset (GSE123813) and the liver hepatocellular carcinoma (LIHC) dataset (GSE140228), both obtained from a database article36 due to their availability in h5ad format and detailed cell type annotations. As shown in Supplementary Figs. S13 and S14, CellFM consistently outperformed other models across all cell types, achieving an average accuracy 2.3% higher than the second-best model, UCE. Notably, CellFM demonstrated exceptional performance in distinguishing exhausted and activated CD8+ T cells, surpassing UCE by an average of 6.5%. On the BCC_GSE123813 dataset, CellFM achieved accuracy scores of 77% and 74% for exhausted and activated CD8+ T cells, respectively, outperforming UCE by 6% and 7%. A similar trend was observed on the LIHC_GSE140228 dataset, further confirming CellFM’s robustness in identifying these cell states.
We made CellFM, scGPT, GeneCompass, and Geneformer to fine-tune. As shown in Supplementary Fig. S17, the performance of fine-tuned scGPT aligns closely with the results reported in its original article and the benchmark study scEval8. For instance, fine-tuned scGPT achieved 92.2% cell type annotation accuracy on the human pancreas dataset, comparable to its original study and that reported in scEval. Similarly, fine-tuned Geneformer reached 85.3% accuracy on the same dataset, consistent with the performance reported in scEval, although its original paper did not provide results for cell type classification. The fine-tuned GeneCompass achieved comparable results with the fine-tuned CellFM. CellFM(800M) obtained low performance on the intra- and inter-datasets when evaluated through zero-shot. However, across all inter-datasets, CellFM (800M) demonstrated an average fine-tuning accuracy that was 12.8% and 15.92% higher than scGPT and Geneformer, respectively. This performance indicated the potential power of the larger model. To evaluate the performance of CellFM affected by LoRA during the fine-tuning phase, we conducted the ablation experiments on the cell type annotation task using the inter-datasets. As shown in Supplementary Fig. S18, the performance of CellFM showed minimal changes when LoRA was applied compared to when it was not, across the inter-datasets. However, using LoRA reduced the time required for fine-tuning. Based on these findings, we recommend applying LoRA during the fine-tuning of CellFM to enhance efficiency without sacrificing performance.
Since Long Non-Coding RNAs (lncRNAs) were included in CellFM’s training data, we evaluated its ability to identify cell-type-specific lncRNAs using attention scores. By analyzing CLS-gene attention, we selected the top 100 genes per cell type, identifying critical lncRNAs for classification. Trained on PBMC data, CellFM highlighted HOTAIRM1 as a top gene for CD14+ Monocytes. This myeloid-specific lncRNA regulates monocyte differentiation via miR-3960 and HOXA1, with silencing reducing CD14 and monocyte marker expression. Including HOTAIRM1 in scRNA-seq annotation improves CD14+ Mono identification accuracy, demonstrating lncRNAs’ value in cell type classification37.
To evaluate whether alternative normalization methods might further improve CellFM’s performance, we tested scTransform38, which corrects the variance-mean bias. Due to computational constraints, we performed these experiments using a smaller CellFM model with 80 million parameters instead of the original 800 million. As shown in Supplementary Fig. S21(a), the performance with scTransform was slightly lower than with log1p normalization used in CellFM. These results suggested that while scTransform addresses variance-mean bias explicitly, it does not yield substantial improvements in CellFM’s performance for the cell-type annotation tasks evaluated.
To evaluate the efficiency of the modified ERetNet model used in CellFM, we further evaluated two key modifications to RetNet: replacing the traditional feedforward network with a gated bilinear network, and substituting the pre-layer LayerNorm with the DeepNorm layer normalization technique. We also benchmarked it against the classic Transformer model. All ablation experiments were conducted using a newly trained CellFM model with 80 million parameters, with a focus on the cell type annotation task, due to limitations in time and computational resources. As shown in Supplementary Fig. S21(b), removing the Simple Gated Linear Unit and DeepNorm resulted in decreases of 0.8% and 0.9%, respectively. Additionally, the removal of the L_cls loss led to a slight drop (0.4%) in performance. When compared to the classic Transformer, CellFM demonstrated a 1.2% improvement. In conclusion, these modifications collectively contributed to the robust performance of CellFM, as evidenced by the benchmarking results. Additionally, the implementation of Gated Multi-head Attention (MHA) in CellFM improved the computational complexity from \(O({l}_{\max }^{2}d)\) to \(O({l}_{\max }{d}^{2}/h)\), where d was set to 1536 and the number of attention heads (h) was set to 48. Consequently, the actual computational complexity of CellFM is \(O\left(2048\times \frac{153{6}^{2}}{48}\right),\) which is smaller than \(O\left(204{8}^{2}\times 1536\right).\) The formula derivation can be found in Supplementary Note 1.
To further evaluate the performance of CellFM in integrating datasets with batch effects, we conducted a comparison among three single-cell foundation models: scELMo, scGPT, and UCE. We included scGPT, scELMo, and UCE in the comparison, as their original studies reported batch correction capabilities. The deep learning framework used for CellFM, MindSpore, does not support the Gradient Reversal Layer (GRL) technique. Similarly, other single-cell foundation models with batch effect correction functionalities also lack GRL implementation. To ensure a fair comparison, we re-evaluated scGPT by removing the GRL loss while retaining its other loss functions. We evaluated CellFM across multiple datasets, including PBMC 10k, the human brain cell atlas, and two versions of Tabula Sapiens. As demonstrated in Supplementary Fig. S19, CellFM achieved the highest average AvgBio scores on these datasets, outperforming the second-best method, UCE, by 2.1%.
Deciphering gene relationships with CellFM
The intricate interplay among target genes within a Gene Regulatory Network (GRN) is pivotal for orchestrating key biological processes. Here, we examined the ability of CellFM to encode these gene relationships through its gene embeddings and attention maps. To evaluate the gene relationships efficiently captured by the pre-trained CellFM, we fine-tuned CellFM using 32484 immune cells from the Immune data and about 200000 non-immune cells from the human brain data. As shown in Fig. 5a and Supplementary Fig. S22, we present three gene relationship graphs: Fig. 5(a) shows the pre-trained CellFM, Figure S22(b) displays CellFM trained on immune cells, and Figure S22(c) illustrates CellFM trained on non-immune cells. The results show that the relationships among genes IL-2, IL-3, and IL-4, observed in the pre-trained CellFM, were preserved when CellFM was fine-tuned on immune cells. However, these relationships were absent when CellFM was trained solely on non-immune cells. Previous studies have shown that IL-2, IL-3, and IL-4 are involved in the JAK/STAT pathway. IL-2, secreted primarily by Th1 cells, promotes immune activation, while IL-4, secreted by Th2 cells, facilitates anti-inflammatory signaling. Together, they regulate immune responses by mediating cell proliferation, differentiation, and immune homeostasis. Additionally, IL-3 stimulates the STAT5 pathway, which regulates cell proliferation, differentiation, and anti-apoptotic signaling39,40. In summary, CellFM effectively preserved biologically relevant immune gene relationships. On the other hand, we observed that the gene relationship structures between IL1RAP and IL1R1 were only present in the CellFM trained on immune cells. Previous studies have shown that IL1RAP and IL1R1 form a functional receptor complex that mediates IL-1β signaling. This interaction plays a critical role in neutrophilic inflammation, exacerbating airway inflammation and contributing to worsened pulmonary obstruction.
a A gene cluster comprising IL2, IL3, and IL4 was identified through the cosine similarity of gene embeddings generated by the pre-trained CellFM (zero-shot). b This analysis compares the number of enriched pathways derived from gene programs extracted by CellFM (zero-shot) and a coexpression network within an immune-related human dataset, across various Leiden clustering resolutions. c This workflow outlines the process of identifying the most influenced genes through attention maps, where attention scores from perturbed cell states are sequentially ranked to select the most impacted genes. d The heatmap displays the connectivity changes in the network of the top 20 genes most influenced by the gene SPI1 in a fine-tuning setting. The color gradient represents the correlation strength between SPI1 and its perturbed genes, ranging from blue (weakest) to purple (strongest positive). e The network graph represents the top 20 genes, with ChIP-seq predicted targets validated in the ChIP-Atlas database, highlighted in light blue. f The heatmap displays KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways enriched for the top 20 SPI1-impacted genes, identified through one-sided hypergeometric tests with Benjamini-Hochberg correction for multiple comparisons.
To further verify the identified gene programs, we followed scGPT to perform Leiden clustering gene programs on the gene similarity graph construed by K-Nearest Neighbors (KNN) and extracted gene programs from gene clusters that consisted of five or more genes. Subsequently, we conducted a comprehensive pathway enrichment analysis based on the gene programs using the Kyoto Encyclopedia of Genes and Genomes (KEEG). As illustrated in Fig. 5b, we juxtaposed the results yielded by CellFM with those obtained from co-expression network analysis. CellFM consistently revealed a significantly higher number of enriched pathways across all clustering resolutions, except for the resolution at 10. To further validate the efficiency of the pathways identified by CellFM, we conducted a comparative analysis of the pathways between CellFM and the co-expression network at a resolution of 40. As shown in Supplementary Table S1–S3, both methodologies identified 25 common pathways. CellFM uniquely identified an additional 59 pathways, 7 of which were pertinent to immune system processes. Conversely, the co-expression network uniquely identified 32 pathways, of which only 2 were associated with immune functions. These comprehensive findings underscore CellFM’s superior capacity to capture subtle and intricate gene-gene interactions, thereby enabling the elucidation of specific biological mechanisms within a broader cellular context.
CellFM efficiently identified genes most affected by perturbations
In this section, we analyzed the perturbed genes in the perturbation experiments and their most significantly affected genes through the attention map (Fig. 5c). Concretely, we modeled the effects of the perturbing gene by providing the model with the control cell expression profiles (non-perturbed) and explicitly indicating the gene as the perturbed gene. The model then predicted how the gene perturbation would affect the expression of other genes. Using the attention mechanism, we identified the 20 genes most influenced by perturbing the gene.
We have provided nine cases in Supplementary Fig. S23. Across nine case genes, among the top 20 genes most influenced by CellFM for each perturbation gene, an average of 18 were found in the ChIP-Atlas database41. The results showed that CellFM can correctly identify influenced genes through attention scores. Additionally, we further conducted the pathway analysis on case perturbation genes JUN and SPI1 to show that CellFM captured distinct pathway-activation patterns through the genes most influenced by perturbation genes. As illustrated in Fig. 5d, in the Adamson dataset, CellFM identified the top 20 genes most influenced by gene SPI1. Most genes were confirmed to be associated with SPI1, as validated by the ChIP-Atlas database (Fig. 5e). We noted that SPI1 in the column has connectivity, while the SPI1 in the row doesn’t. The reason may be caused by the attention mechanism used in CellFM (Supplementary Note 3). Most gene pairs in the heatmap of Fig. 5d have values near zero due to our normalization strategy. We ranked all genes in the perturbed dataset from Adamson based on their attention scores and recalculated the connection scores by dividing these ranked scores by the total number of genes. Additionally, the partial relationships between CCNB2, CCNA2, TOP2A, MKI67, KPNA2, and CENPF in Fig. 5d weren’t captured likely because CellFM focused on capturing relationships with SPI1. When not designating perturbation targets in CellFM (i.e., during regular gene recovery tasks), we could observe relations between these genes as Supplementary Fig. S24. In addition, based on gene embeddings derived from CellFM, the cosine similarity scores of these genes were significantly higher compared to those of other genes, indicating that CellFM can learn strong intrinsic relationships among them.
To further directly demonstrate the relationships between SPI1 and the SPI1 perturbed genes, we obtained the SPI1 ChIP-seq BigWig files (SRX2770855) from the public ChIP-Atlas database (https://chip-atlas.org/peak_browser) and visualized them using the Integrative Genomics Viewer (IGV). To select the top 5 genes with the strongest SPI1 binding, we identified those with the highest MACS2 scores, indicating the most significant SPI1 binding at their genomic loci. These genes were prioritized based on peak intensity and the extent of overlap with key genomic regions. The IGV snapshots allowed us to visually confirm the presence and strength of SPI1 binding at these loci, providing direct evidence of SPI1’s regulatory impact on these genes. As shown in Supplementary Fig. S25, the results clearly show the binding peaks of SPI1 at the genomic loci of the target genes, indicating potential regulatory interactions.
To construct a comprehensive gene-transcription factor (TF) interaction network for the genes influenced by SPI1, we utilized the TRRUST database42 (http://www.grnpedia.org/trrust) to identify the top 300 genes most influenced by SPI1. As shown in Supplementary Fig. S26, the network diagram illustrates the visualization of these interactions, where nodes represent genes or transcription factors and edges denote the interactions between them. Red nodes represent transcription factors, while blue nodes represent genes. Notably, genes such as MYC emerged as hub genes, interacting with multiple key TFs, including CTNNB1, JUNB, and CCNB143,44,45. This network analysis provides valuable insights into the regulatory mechanisms underlying SPI1-mediated gene expression, highlighting potential transcription factor interactions that may drive downstream cellular processes.
Furthermore, CellFM was able to capture distinct pathway-activation patterns (Fig. 5f) through the genes most influenced by SPI146,47,48. For pathway enrichment analysis, we included all genes from the KEGG pathway database as the background gene set to provide a broader biological context. This approach was intentional to highlight the pathways enriched by the top 20 genes most influenced by SPI1 perturbation. The enriched pathways presented in Fig. 5f emphasize those in which SPI1 and its perturbed genes may exert a coordinated influence on important biological processes. For instance, the Human T-cell leukemia virus 1 infection (HTLV-1) contained SPI1, CCNB2, and CCNA2, holding particular significance. First, the SPI1 gene plays a critical role in leukemia stem cell (LSC) self-renewal49. In HTLV-1 infection, SPI1 activation may promote the expansion of leukemia stem cells, enhancing their self-renewal capacity and accelerating leukemia progression. Second, the CCNA2 gene primarily regulates cell cycle transitions. HTLV-1-encoded oncoproteins Tax and HBZ disrupt CCNA2 expression, causing cell cycle dysregulation and increasing cell immortalization potential50. In HAM/TSP patients, observed downregulation of CCNA2 may be due to IRF-1 suppression, which could prevent transformation into ATLL. For the other pathway Acute myeloid leukemia (AML) enriched SPI1 and CCNA2 holding particular significance. First, SPI1 is crucial for normal blood cell differentiation, and its dysregulation in AML leads to uncontrolled cell growth and blocked differentiation, promoting leukemia development46. In AML, CCNA2 is abnormally overexpressed in certain subtypes, particularly in therapy-related AML (t-AML)51. This overexpression, along with other cell cycle regulation genes like CCNE2 and CDC2, is linked to poor prognosis and is often associated with chromosomal deletions of 5 and 7, which correlate with lower survival rates.
We also showed the case perturbed gene JUN (Supplementary Fig. S27) and obtained the ChIP-seq BigWig files (SRX10976151) for the JUN transcription factor. The ChIP-seq data for JUN were also obtained from publicly available datasets and visualized using IGV. Representative snapshots of JUN binding sites across various genomic regions are provided in the Supplementary Fig. S28. These visualizations highlight significant ChIP-seq peaks, offering direct evidence of JUN’s binding to specific genomic loci. Additionally, the pathway Focal adhesion in Supplementary Fig. S27c identified key genes such as JUN, CCND3, and CTNNB1, all of which play essential roles in cell adhesion and proliferation. Concretely, JUN encodes c-Jun, a component of the AP-1 complex that regulates gene expression and proliferation. It is closely linked to focal adhesion pathways, particularly through FAK (Focal Adhesion Kinase), which mediates cell attachment to the extracellular matrix (ECM) and influences migration and invasion52. CCND3, a regulator of the G1/S phase transition, is connected to FAK signaling, linking cell adhesion to cell cycle progression53. Additionally, CTNNB1 (β-catenin) through the Wnt/β-catenin pathway promotes cell adhesion by interacting with cadherins and modulating FAK activity. This coordinated regulation of JUN, CCND3, and CTNNB1 suggests their collaborative role in driving cancer development and progression through cell adhesion and proliferation pathways54.
Discussion
To aid efficient analysis of the single-cell data and harness the wealth of knowledge contained within single-cell atlas datasets, we have introduced a state-of-the-art foundation model known as CellFM. This model was pre-trained on our meticulously curated datasets, encompassing about 100 million human cells. These datasets empower CellFM to generate an expansive set of 800 million model parameters, marking an eightfold increase over the parameters present in the current single-cell models trained on a single species. To augment the training efficiency of CellFM, we have adopted the ERetNet architecture as its core. This network represents an advancement over the traditional RetNet framework, offering enhanced parallel training capabilities and cost-effective inference. These features collectively contribute to CellFM’s exceptional performance. Moreover, CellFM incorporates a Low-Rank Adaptive module designed to minimize parameter count during fine-tuning, thereby optimizing the model for specific tasks without compromising its generalizability. Through a series of comprehensive experiments, CellFM has demonstrated its effectiveness across a range of single-cell tasks including cell type annotation, prediction of responses to perturbations, gene network analysis, and gene function prediction.
To satisfy the model’s training on large-scale datasets, we have chosen a variant of the RetNet architecture as CellFM’s foundation, diverging from the Transformers used in other single-cell foundation models. The RetNet architecture facilitates parallel, recurrent, and chunkwise processing, which we have refined by integrating the SGLU module, amplifying training efficiency. Additionally, we have embedded the Low-Rank Adaptive (LoRA) strategy within CellFM, optimizing its training on new datasets with similar characteristics. The combination of this efficient training architecture and the comprehensive datasets forms the basis for developing the current largest CellFM model, equipped with 800 million parameters. CellFM is developed using the MindSpore AI framework from Huawei and is trained on four Huawei Altas800 servers, each equipped with eight Ascend910 NPUs. Our rigorous experiments demonstrated the model’s adaptability and potency in multiple single-cell downstream tasks. In the spirit of research collaboration, we are dedicated to sharing our progress by making the CellFM codes and the pre-trained model publicly available. This initiative aims to provide researchers with a unified framework that streamlines the adoption of pre-trained models for their distinct research goals. While large human datasets have also been used to train multi-species models like UCE and GeneCompass, the number of human cells in these models did not exceed 50 million. In contrast, our model was trained on ~100 million human cells. Moreover, our model’s parameter size is eight times larger than GeneCompass and 1.23 times larger than UCE. As demonstrated in Figs. 2–4, our model consistently outperformed GeneCompass and UCE in tasks such as cell type annotation, gene function prediction, and cell perturbation.
Despite the advances in CellFM, several limitations remain to be explored. Firstly, the attention map in CellFM was limited in capturing gene relationships related to static or global biological knowledge. In the future, we will explore new explainability techniques to overcome this challenge. Furthermore, the current model is limited by the absence of multi-species data, which restricts its potential for broader biological contexts and cross-species comparisons. Finally, the model’s construction did not leverage existing biological prior knowledge, which could affect its depth and accuracy in interpreting biological phenomena.
Methods
Data collection
All training data utilized in this study were sourced from reputable public databases. Specifically, from April 2021 to August 2023, we identified datasets leveraging keywords like “single-cell RNA sequencing,” “single-cell transcriptome,” and “single-cell RNA.” These keywords were used to search through databases such as NCBI GEO23, ENA24, GSA25,26, ImmPort27, and others. In our selection process, we carefully curated the datasets, retaining only those human single-cell datasets that were relevant to our study. These datasets were encountered in multiple formats, including FASTQ data, expression matrices, and Seurat/Scanpy objects. Our initial step involved transforming the raw FASTQ data into expression matrices using primary analysis software supplied by the manufacturers. Following this, all obtained and transformed expression matrices underwent pre-processing through a standardized workflow provided by the SynEcoSys® single-cell database from Singleron Biotechnologies28. This workflow included several critical steps: (1) Quality control involved filtering cells based on a minimum gene count threshold of 200 genes per cell; (2) Gene name standardization was conducted by the HUGO Gene Nomenclature Committee (HGNC) guidelines, ensuring that gene aliases in each dataset were converted to their respective HGNC-approved gene symbols. This step guaranteed the uniqueness and consistency of gene names across all datasets. (3) Finally, the expression matrices for each sample were converted into a unified sparse matrix format, preparing them for subsequent model training.
CellFM architecture
The CellFM model comprises three core components, including the embedding module, the ERetNet module, and the LoRA module (Fig. 1). The embedding module in CellFM maps one-dimensional scalar values of gene expression to high-dimensional embedding features for model training, enabling the representation of gene expressions in a high-dimensional space. CellFM then applies the ERetNet module to learn the relationships among genes based on the gene expression information. In parallel, CellFM uses the LoRA module to help train CellFM more efficiently by reducing the number of parameters when adjusting model weights with new data.
The embedding module
To efficiently train CellFM, we have set an upper limit on the number of genes it inputs, defined by the threshold \({l}_{\max }=2048\). For each cell, if the number of expressed genes exceeds \({l}_{\max }\), we randomly select \({l}_{\max }\) genes with high expression values. Conversely, if a cell has fewer expressed genes than \({l}_{\max }\), we pad the gene IDs and set the padded values as zero because the model architecture has a fixed length \({l}_{\max }\) for parallel computing. These padded values won’t participate in the calculations during CellFM’s training and thus don’t influence our models. We then apply a Multilayer Perceptron (MLP) to map the scalar expression values of genes to embedding vectors necessary for the ERetNet module as follows:
where \({W}_{0}\in {{\mathbb{R}}}^{1\times b}\), \({W}_{1}\in {{\mathbb{R}}}^{b\times b}\), and \({W}_{2}\in {{\mathbb{R}}}^{b\times d}\) are learnable parameter matrices. The coefficient α is a learnable residual coefficient. The hyperparameters b and d are set to b = 256 and d = 1536, respectively. The term \({X}_{0}\in {{\mathbb{R}}}^{{l}_{\max }\times 1}\) represents the initial input cell pre-processed by the aforementioned workflow.
As performed in previous LLMs, we randomly mask the expressions of 20% of the genes (denoted as M) and then recover them based on the non-masked genes. During gene masking, the 20% of genes masked during the pre-training task were exclusively selected from non-padded genes, ensuring that CellFM focused on reconstructing meaningful gene expression patterns using the remaining relevant gene information. This design allows the model to learn effective compression and meaningful representations without interference from padding values. Specifically, for the M masked gene expressions, we replace the gene expressions of the cell with a learnable weight vector \({X}_{M}\in {{\mathbb{R}}}^{1\times d}\) initialized to zero. Consequently, the feature Xtmp can be obtained as follows:
where ⊙ is the element-wise product, and \(\overrightarrow{M}\in {\{0,1\}}^{{l}_{\max }}\) is the mask vector indicating the position of masked genes with value 0.
To learn the specific characteristic of each gene, we initialize a learnable embedding matrix \({E}^{G}\in {{\mathbb{R}}}^{24079\times d}\). The term 24079 represents the number of genes ID and the d = 1536 means the dimension of vector embeddings initialized based on each unique gene ID. We then integrate the gene expression and gene ID embeddings as follows:
Furthermore, we incorporate an additional learnable weight Xcls ∈ R1×d, which is appended to the gene expression embeddings. This weight facilitates the learning of cell-level features by aggregating gene information in the following manner:
where the ∥ symbol denotes the concatenation of two vectors.
The ERetNet module
CellFM learns gene embeddings and relationships from gene expression through the ERetNet module, a variant of the RetNet55. RetNet is an efficient, high-performance variant of the Transformer architecture. To better adapt large-scale single-cell datasets, we have modified the RetNet module in two ways: First, we’ve replaced the traditional feedforward network in RetNet with a gated bilinear network, which has led to improved model performance and a smoother training process. Second, we’ve refined the model’s training stability and performance by substituting the pre-layer LayerNorm in RetNet with the DeepNorm layer normalization technique56. Collectively, these modifications have resulted in the ERetNet module, which includes a Gated Multi-head Attention (MHA), a Gated Linear Unit (SGLU), and layer normalization (LN), all contributing to a more stable and effective gene expression analysis model.
Gated multi-head attention (MHA)
The Gated Multi-head Attention (MHA) block is used to learn dependencies between genes, which is a variant of the Retention mechanism in the RetNet. To address the computational inefficiency of the exponential attention operations (\({{\mathcal{O}}}({l}_{\max }^{2}d)\)) implemented in RetNet, we adopt the method proposed by Shen et al.57. By first computing the keys (K) and values (V), followed by the queries (Q), this approach achieves a linear complexity of \({{\mathcal{O}}}\left(\frac{{l}_{\max }{d}^{2}}{h}\right)\), significantly reducing overhead. Furthermore, we scaled Q, K, and V following the study57 to ensure the half-precision training as follows:
where \({l}_{cell}={l}_{\max }+1\) denotes the number of expressed genes within each cell, \(\overrightarrow{M}\) denotes the mask vector.
To enhance the CellFM’s representational power, we use h = d/dhead attention heads in each ERetNet layer, where dhead = 32 is the head dimension. Each head consists of three parameter matrices WQ, WK, \({W}_{V}\in {{\mathbb{R}}}^{{d}_{head}\times {d}_{head}}\). In addition, we add a swish gate58 to increase the non-linearity of ERetNet layers. Formally, given input Xexpr, we define the MHA layer as follows:
where WG, \({W}_{O}\in {{\mathbb{R}}}^{d\times d}\) are learnable parameters, and GroupNorm normalizes59 the output of each head, following SubLN proposed in the study60.
Simple Gated Linear Unit (SGLU)
To improve model performance and a smoother training process, we’ve replaced the traditional feedforward network in RetNet with a gated bilinear network. The gated unit GLU introduces a multiplicative gating mechanism that explicitly indicates the model’s memory of each feature dimension, thereby smoothing the training process and facilitating better integration between channels. Considering that the gating mechanism inherently introduces nonlinear relationships, to further accelerate the model’s computation, this work, referring to literature61, adopts the SGLU, which is based on the GLU formula62 but omits the Swish activation function:
where ⊙ is the element-wise product.
Layer normalization (LN)
The Transformer architecture in large single-cell models typically uses post-norm normalization after residual connections to enhance model depth and convergence. However, this can cause a gradient explosion as the model size grows. To counteract this, a pre-norm strategy is applied in RetNet for a stabilized training process, albeit with a potential performance trade-off. To address this gap, CellFM employs the new post-norm normalization method DeepNorm56. DeepNorm reduces the contribution ratio of each network block to the output, thereby reducing the amount of gradient that needs to be updated and ensuring the stability of training.
where LN( ⋅ ) is LayerNorm and λ is a hyperparameter.
Low-rank adaptation (LoRA) module
Large models typically comprise hundreds of millions of parameters, resulting in considerable time consumption for full model training. To alleviate the burden of training on various datasets, we employ the Low-Rank Adaptation (LoRA) algorithm63. LoRA operates under the assumption that updates to pre-trained weights during fine-tuning can be decomposed by low rank. Hence, for a pre-trained weight matrix W0 ∈ Rn×k, we utilize low-rank decomposition to constrain the weight increment ∇ W:
where B ∈ Rd×r, A ∈ Rr×k, and the rank \(r < < \min (d,k)\).
During the forward computation, both W0 and matrices A and B are used in calculations with the input X; however, during the backward propagation, the W0 parameter is frozen and does not undergo gradient updates, while only A and B are updated. It can be observed that in regular training, the training parameter count for W0 is n × k, whereas, with LoRA-based training, the training parameter count for W0 is the sum of the parameters of A and B, which is (n + k) × r. Since the dimension r is significantly smaller than n and k, the number of training parameters for the weights is greatly reduced, leading to a substantial decrease in computational overhead.
In the ERetNet architecture, the MHA has five weight matrices: WQ, WK, WV, WG, and WO, and the SGLU gating unit has three weight matrices: Wu, Wv, and Wo. We consider the dimensions of these 8 matrices to be d × d. In this experiment, we limit the application of LoRA to only the ERetNet encoder part and freeze all model parameters for updates except for the weights of the LayerNorm layer.
Loss functions
Mean squared error (MSE)
In CellFM, we focus on minimizing the Mean Squared Error (MSE) as the primary metric because it effectively measures the discrepancy between the predicted and actual gene vector embeddings for masked genes. MSE is particularly suitable in this context as it penalizes larger errors more heavily, making it crucial for accurately recovering gene representations. Additionally, MSE has been widely adopted in similar tasks involving gene expression prediction and representation learning. For example, scFoundation18 and GeneCompass19 employ MSE to optimize gene expression prediction in high-dimensional spaces, demonstrating its effectiveness in promoting precise modeling. Specifically, we employ a fully connected MLP followed by the ERetNet module to estimate the expression value for M genes. The optimization of this objective involves utilizing the MSE loss at the masked positions, denoted as M mask. The MSE works as follows:
∣Mmask∣ denotes the count of ones in the mask gene vector Mmask for each cell. \({X}_{i}^{(L)}\) signifies the features derived from the ERetNet for gene i at the L layer.
To further enhance CellFM’s learning capabilities and channel aggregation, the feature corresponding to token cls is also leveraged, represented as \({X}_{\,{\mbox{cls}}\,}^{(L)}\in {{\mathbb{R}}}^{1\times d}\). This feature passes through an additional network designed to predict expression values. Specifically, a learnable parameter matrix \({W}_{{{\rm{cls}}}}\in {{\mathbb{R}}}^{d\times d}\) and an activation function are employed. After mapping \({X}_{\,{\mbox{cls}}\,}^{(L)}\), the result is multiplied with the embeddings of the genes to be predicted in the vocabulary EG to compute inner products, yielding another set of predicted values. These are then compared with the actual values to calculate the mean squared error loss.
The @ symbol represents matrix multiplication. σ is an activation function Sigmoid. \({E}_{i}^{G}\) denotes the embedding of gene i in the vocabulary EG. Finally, the total loss functions of CellFM can be obtained as follows:
Baseline single-cell foundation models
We have incorporated several recent single-cell foundation models into our benchmarking, including scFoundation, GeneCompass, UCE, scELMo, scBERT, scGPT, and Geneformer. These models represent different approaches to single-cell analysis, and we have categorized them based on their methodological focus. Specifically, models including CellFM, scFoundation, scELMo, and GeneCompass fall under the value projection category, while UCE, scGPT, and scBERT belong to the value categorization category, and Geneformer is placed in the ordering category.
Implementation details
CellFM consists of 40 stacked ERetNet blocks, with each block having 48 attention heads. During pre-training, we used ~100 million cells for training CellFM. The model was optimized using the Adam optimizer with a starting learning rate of 1e-7 and trained for a total of 2 epochs. The total batch size was 128, distributed equally across 4 Huawei Altas800 servers, each equipped with 8 Ascend910 NPUs. The MindSpore AI development framework powered the automatic data parallel training.
The decision to train the CellFM model for two epochs was informed by standard practices in large-scale model training9, where rapid convergence is typically observed within the initial epochs. To validate this convergence of CellFM, we conducted the experiment using the 80-million-parameter version of CellFM on all training datasets. The results confirmed the same pattern: the loss dropped sharply-from 8 to below 1-during the first epoch, with only minimal changes in the second epoch (in Supplementary Fig. S29). This behavior reflects the typical convergence dynamics of large-scale models and supports our choice to limit training to two epochs to balance efficiency and performance.
Pre-processing
The gene symbols across all raw count gene expression matrices were standardized using the reference mapping provided by the HUGO Gene Nomenclature Committee. This process included both human protein-coding genes and common mitochondrial genes, resulting in a comprehensive gene set G consisting of 24,078 genes. Ultimately, normalization and a log1p transformation were employed across all gene expression matrices to alleviate skewness in the data. For the cell type annotation task, we excluded cell types with fewer than 10 cells in the training data, as well as those present in the query data but absent in the reference data.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The datasets used in this study are publicly accessible through the cited publications, with detailed links provided in Supplementary Table S4. Due to the large cell population and computational time and resource limitations, the human brain CellAtlas dataset, originally comprising over 3 million cells, was downsampled to 126,339 cells using a sampling rate of 3.75%, with cells randomly selected by tissue type. Similarly, for the Tabula Sapiens dataset, both versions (V1 and V2) were downsampled by tissue type at a rate of 25%, resulting in 111,013 and 172,999 cells, respectively, to ensure computational efficiency while maintaining biological representativeness. Source data are provided with this paper. The data used in this study have been uploaded to Zenodo and are freely available at: https://doi.org/10.5281/zenodo.1513866564.
Code availability
All codes used in this study can be available at: https://github.com/biomed-AI/CellFM. The codes used in this study have been uploaded to Zenodo and are freely available at: https://doi.org/10.5281/zenodo.1515590065.
Change history
02 February 2026
In the version of this article initially published, a Supplementary Data file and Source Data files for Figs. 3, 4 and Supplementary Figs. 4, 7–9, 12–21 were missing and are now included in the HTML and PDF versions of the article.
References
Han, X. et al. Construction of a human cell landscape at single-cell level. Nature 581, 303–309 (2020).
Lotfollahi, M., Hao, Y., Theis, F. J. & Satija, R. The future of rapid and automated single-cell data analysis using reference mapping. Cell 187, 2343–2358 (2024).
Xu, C. et al. Automatic cell-type harmonization and integration across human cell atlas datasets. Cell 186, 5876–5891 (2023).
Lu, Y. et al. A single-cell atlas of the multicellular ecosystem of primary and metastatic hepatocellular carcinoma. Nat. Commun. 13, 4594 (2022).
Wolf, F. A., Angerer, P. & Theis, F. J. Scanpy: large-scale single-cell gene expression data analysis. Genome Biol. 19, 1–5 (2018).
Hao, Y. et al. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat. Biotechnol. 42, 293–304 (2023).
Zeng, Y., Wei, Z., Pan, Z., Lu, Y. & Yang, Y. A robust and scalable graph neural network for accurate single-cell classification. Brief. Bioinforma. 23, bbab570 (2022).
Liu, T., Li, K., Wang, Y., Li, H. & Zhao, H. Evaluating the utilities of large language models in single-cell data analysis. Preprint at bioRxiv https://doi.org/10.1101/2023.09.08.555192 (2023).
Touvron, H. et al. Llama: open and efficient foundation language models. Preprint at arXiv https://doi.org/10.48550/arXiv.2302.13971.09685 (2023).
Levine, D. et al. Cell2sentence: teaching large language models the language of biology. Preprint at bioRxiv https://doi.org/10.1101/2023.09.11.557287 (2023).
Chen, Y. T. & Zou, J. Genept: a simple but hard-to-beat foundation model for genes and cells built from ChatGPT. Preprint at bioRxiv https://doi.org/10.1101/2023.10.16.562533 (2023).
Shen, H. et al. A universal approach for integrating super large-scale single-cell transcriptomes by exploring gene rankings. Brief. Bioinforma. 23, bbab573 (2022).
Shen, H. et al. Generative pretraining from large-scale transcriptomes for single-cell deciphering. Iscience 26, 106536 (2023).
Theodoris, C. V. et al. Transfer learning enables predictions in network biology. Nature 618, 616–624 (2023).
Yang, F. et al. scbert as a large-scale pretrained deep language model for cell type annotation of single-cell RNA-seq data. Nat. Mach. Intell. 4, 852–866 (2022).
Cui, H. et al. scgpt: toward building a foundation model for single-cell multi-omics using generative AI. Nat. Methods 21, 1470–1480 (2024).
Rosen, Y. et al. Universal cell embeddings: a foundation model for cell biology. bioRxiv Preprint at https://doi.org/10.1101/2023.11.28.568918 (2023).
Hao, M. et al. Large-scale foundation model on single-cell transcriptomics. Nat. methods 21, 1481–1491 (2024).
Yang, X. et al. Genecompass: deciphering universal gene regulatory mechanisms with a knowledge-informed cross-species foundation model. Cell Res. 34, 830–845 (2024).
Liu, T., Chen, T., Zheng, W., Luo, X. & Zhao, H. Scelmo: embeddings from language models are good learners for single-cell data analysis. Preprint at bioRxiv https://doi.org/10.1101/2023.12.07.569910 (2023).
Hoffmann, J. et al. Training compute-optimal large language models. arXiv preprint https://doi.org/10.48550/arXiv.2203.15556 (2022).
Kaplan, J. et al. Scaling laws for neural language models. arXiv Preprint https://doi.org/10.48550/arXiv.2001.08361 (2020).
Barrett, T. et al. Ncbi geo: archive for functional genomics data sets-"update. Nucleic Acids Res. 41, D991–D995 (2012).
Cochrane, G. et al. Priorities for nucleotide trace, sequence and annotation data capture at the ensembl trace archive and the EMBL nucleotide sequence database. Nucleic Acids Res. 36, D5–D12 (2007).
Chen, T. et al. The genome sequence archive family: toward explosive data growth and diverse data types. Genomics, Proteom. Bioinforma. 19, 578–583 (2021).
Bai, X. et al. Database resources of the national genomics data center, china national center for bioinformation in 2024. Nucleic Acids Res. 52, D18–D32 (2024).
Bhattacharya, S. et al. Immport, toward repurposing of open access immunological assay data for translational and clinical research. Sci. data 5, 1–9 (2018).
Zhang, Y. et al. Synecosys: a multifunctional platform of large-scale single-cell omics data analysis. Preprint at bioRxiv https://doi.org/10.1101/2023.02.14.528566 (2023).
Salzberg, S. L. Open questions: how many genes do we have? BMC Biol. 16, 94 (2018).
Yuan, Q., Xie, J., Xie, J., Zhao, H. & Yang, Y. Fast and accurate protein function prediction from sequence through pretrained language model and homology-based label diffusion. Brief. Bioinforma. 24, bbad117 (2023).
Adamson, B. et al. A multiplexed single-cell crispr screening platform enables systematic dissection of the unfolded protein response. Cell 167, 1867–1882 (2016).
Norman, T. M. et al. Exploring genetic interaction manifolds constructed from rich single-cell phenotypes. Science 365, 786–793 (2019).
Bunne, C. et al. Learning single-cell perturbation responses using neural optimal transport. Nat. methods 20, 1759–1768 (2023).
Ma, W., Su, K. & Wu, H. Evaluation of some aspects in supervised cell type identification for single-cell RNA-seq: classifier, feature selection, and reference construction. Genome Biol. 22, 1–23 (2021).
Abdelaal, T. et al. A comparison of automatic cell identification methods for single-cell RNA sequencing data. Genome Biol. 20, 1–19 (2019).
Han, Y. et al. Tisch2: expanded datasets and new tools for single-cell transcriptome analyses of the tumor microenvironment. Nucleic Acids Res. 51, D1425–D1431 (2023).
Xin, J. et al. Downregulation of long noncoding RNA hotairm1 promotes monocyte/dendritic cell differentiation through competitively binding to endogenous mir-3960. Onco Targets Ther. 10, 1307–1315 (2017).
Hafemeister, C. & Satija, R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 20, 296 (2019).
Lu, Q. et al. Effect of penthorum chinense pursh compound on AFB1-induced immune imbalance via jak/stat signaling pathway in spleen of broiler chicken. Vet. Sci. 10, 521 (2023).
Gündogdu, M. S. et al. The haematopoietic gtpase rhoh modulates IL3 signalling through regulation of stat activity and IL3 receptor expression. Mol. cancer 9, 1–13 (2010).
Zou, Z., Ohta, T., Miura, F. & Oki, S. Chip-atlas 2021 update: a data-mining suite for exploring epigenomic landscapes by fully integrating chip-seq, atac-seq and bisulfite-seq data. Nucleic Acids Res. 50, W175–W182 (2022).
Han, H. et al. Trrust v2: an expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic acids Res. 46, D380–D386 (2018).
Liu, X. et al. β-catenin overexpression in malignant glioma and its role in proliferation and apoptosis in glioblastma cells. Med. Oncol. 28, 608–614 (2011).
Acosta, J. C. et al. Myc inhibits p27-induced erythroid differentiation of leukemia cells by repressing erythroid master genes without reversing p27-mediated cell cycle arrest. Mol. Cell Biol. 24, 7286–95 (2008).
Yin, X.-Y. et al. Inverse regulation of cyclin b1 by c-myc and p53 and induction of tetraploidy by cyclin b1 overexpression. Cancer Res. 61, 6487–6493 (2001).
Wang, X., Jin, P., Zhang, Y. & Wang, K. Circspi1 acts as an oncogene in acute myeloid leukemia through antagonizing spi1 and interacting with micrornas. Cell death Dis. 12, 297 (2021).
Zhu, H. et al. T-all leukemia stem cell’stemness’ is epigenetically controlled by the master regulator spi1. Elife 7, e38314 (2018).
Lawrence, A.-L. E. et al. Salmonella enterica serovar typhimurium Spi-1 and Spi-2 shape the global transcriptional landscape in a human intestinal organoid model system. MBio 12, 10–1128 (2021).
Hegde, S., Hankey, P. & Paulson, R. F. Self-renewal of leukemia stem cells in friend virus-induced erythroleukemia requires proviral insertional activation of Spi1 and hedgehog signaling but not mutation of p53. Stem Cells 30, 121–130 (2012).
Saffari, M. et al. Coevolution of htlv-1-hbz, tax, and proviral load with host irf-1 and ccna-2 in ham/tsp patients. Infect., Genet. Evolution 103, 105337 (2022).
Qian, Z. et al. Cytogenetic and genetic pathways in therapy-related acute myeloid leukemia. Chem.-Biol. Interact. 184, 50–57 (2010).
Wang, Y., Ma, J., Chen, L., Xie, X.-L. & Jiang, H. Inhibition of focal adhesion kinase on hepatic stellate-cell adhesion and migration. Am. J. Med. Sci. 353, 41–48 (2017).
Yamamoto, D. et al. FAK overexpression upregulates cyclin D3 and enhances cell proliferation via the pkc and pi3-kinase-akt pathways. Cell. Signal. 15, 575–583 (2003).
Francalanci, P. et al. Focal adhesion kinase (fak) over-expression and prognostic implication in pediatric hepatocellular carcinoma. Int. J. Mol. Sci. 21, 5795 (2020).
Sun, Y. et al. Retentive network: a successor to transformer for large language models. Preprint at https://doi.org/10.48550/arXiv.2307.08621 (2023).
Wang, H. et al. Deepnet: scaling transformers to 1000 layers. IEEE Transactions on Pattern Analysis and Machine Intelligence (IEEE, 2024).
Shen, Z., Zhang, M., Zhao, H., Yi, S. & Li, H. Efficient Attention: Attention With Linear Complexities. In Proceedings of the IEEE/CVF Winter Conference On Applications Of Computer Vision, 3531–3539 (2021).
Ramachandran, P., Zoph, B. & Le, Q. V. Swish: a self-gated activation function. arXiv Prepr. arXiv:1710. 05941 7, 5 (2017).
Wu, Y. & He, K. Group normalization. In Proceedings of the European conference on computer vision (ECCV), 3–19 (2018).
Shoeybi, M. et al. Megatron-lm: training multi-billion parameter language models using model parallelism. Preprint at arXiv https://doi.org/10.48550/arXiv.1909.08053 (2019).
Qin, Z. et al. Transnormerllm: a faster and better large language model with improved transnormer. arXiv 2307.14995 (2023).
Shazeer, N. et al. Glu variants improve transformer. Preprint at arXiv https://doi.org/10.48550/arXiv.2002.05202 (2020).
Hu, E. J. et al. Lora: low-rank adaptation of large language models. ICLR 1.2 (2022).
Zeng, Y. et al. CellFM: a large-scale foundation model pre-trained on transcriptomics of 100 million human cells. Zenodo https://doi.org/10.5281/zenodo.15138665 (2025).
Zeng, Y. et al. CellFM: a large-scale foundation model pre-trained on transcriptomics of 100 million human cells. Zenodo https://doi.org/10.5281/zenodo.15155900 (2025).
Acknowledgements
This study has been supported by the National Key R&D Program of China (2023YFF1204900 to Z.H.), the Fundamental Research Funds for the Central Universities (2024IAIS-QN020 to Z.Y.), the National Natural Science Foundation of China (T2394502 to Y.Y. and 62402071 to Z.Y.), the Postdoctoral Fellowship Program of CPSF (GZC20233321 to Z.Y.), the China Postdoctoral Science Foundation (2024M763866 to Z.Y.), and the National Science Foundation of Jiangsu Province (BK20230278 to J.F.).
Author information
Authors and Affiliations
Contributions
Y.Z., W.Y. and Y.Y. conceived and supervised the project. Y.Z., Z.W., J.X., N.S.,Y.S., C.Z. and W.L. contributed to the algorithm implementation. Y.Z. and Y.Y.wrote the manuscript. Y.Z., S.Y., J.Z., N.F., H.Zhang, Y.L., H.Zhao, J.F. and Y.Y.were involved in the discussion and proofreading.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Yang Xu, who co-reviewed with Min Dai, George Gavriilidis and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zeng, Y., Xie, J., Shangguan, N. et al. CellFM: a large-scale foundation model pre-trained on transcriptomics of 100 million human cells. Nat Commun 16, 4679 (2025). https://doi.org/10.1038/s41467-025-59926-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-59926-5
This article is cited by
-
Harnessing artificial intelligence to advance CRISPR-based genome editing technologies
Nature Reviews Genetics (2026)
-
Toward informed batch correction for single-cell transcriptome integration
Nature Computational Science (2026)
-
Single-cell foundation models: bringing artificial intelligence into cell biology
Experimental & Molecular Medicine (2025)
-
Insights, opportunities, and challenges provided by large cell atlases
Genome Biology (2025)