Abstract
The mechanisms of the pectoral girdle transformation at the origin of terrestrial locomotion in vertebrates remain an outstanding problem. The loss of intramembranous bones and the enlargement of endochondral bones resulted in the disarticulation of the pectoral girdle from the skull and the formation of the neck during the fish-to-tetrapod transition. Despite the functional implications of this skeletal shift in the emergence of terrestrial vertebrates, the underlying genetic-developmental alterations have remained enigmatic. Here, we show that in zebrafish pectoral girdle mesodermal cells expressing gli3, a transcription factor gene in the Hedgehog signaling pathway, differentiate into both intramembranous and endochondral bones. Intriguingly, Gli and Hedgehog compound knockout fish exhibited an unexpected combination of actinopterygian fish and stem-tetrapod pectoral girdle characteristics. These ontogenetic and anatomical data suggest that a trade-off between the two distinct ossification pathways is a deeply embedded developmental program in bony fishes and that tuning of this trade-off can generate novel pectoral girdle akin to those of stem-tetrapods at the dawn of vertebrate terrestrialization.
Similar content being viewed by others
Introduction
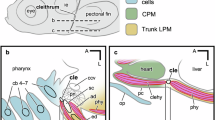
Across the fish-to-tetrapod transition, the amount of intramembranous (direct ossification of mesenchyme) bones decreased and that of endochondral bones (ossification replacing a cartilage template) increased throughout the vertebrate body, facilitating terrestrial feeding, locomotion, and breathing1,2,3,4. The pectoral, or shoulder, girdle is a dramatic example of this intramembranous-to-endochondral skeletal shift. In bony ray-finned (actinopterygian) and lobe-finned (sarcopterygian) fishes, the pectoral girdle consists of a relatively small endochondral component (scapula and coracoid bones) and a large series of intramembranous bones (i.e., cleithrum, clavicle, and supracleithrum) that link the scapula and coracoid to the skull5 (Fig. 1a). As vertebrates transitioned onto land in the Late Devonian, the scapula and coracoid expanded, serving as robust attachment sites of forelimb musculature for terrestrial locomotion6,7,8. Concomitantly, a reduction of the intramembranous bones disconnected the pectoral girdle from the skull and triggered the evolution of a functional neck, providing greater mobility to the head9. Despite the profound functional implications of this intramembranous-to-endochondral shift in the pectoral girdle of early tetrapods, the underlying genetic and developmental changes of this evolutionary transformation remain unexplored.
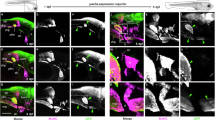
a The adult zebrafish pectoral girdle bones. The intramembranous bones and endochondral bones are highlighted by green and orange, respectively. The intramembranous series is connected to the posterior skull via the post-temporal bone (pt). The pectoral fin (pf) articulates on the scapula (sca). cr, coracoid; cl, cleithrum; me, mesocoracoid; scl, supracleithrum. b, c Digitally sectioned immunofluorescence of EGFP with membrane (CellMask) and nucleus (DAPI) staining of CNE14:egfp embryos at 32 hpf (b) and 48 hpf (c). Bright EGFP expression is observed in the mesenchymal cells at the prospective pectoral fin region at 32 hpf. As the pectoral fin grows, EGFP-positive cells are localized in the fin mesenchyme, including the proximal fin, at 48 hpf. These images were 3D-digital sections obtained by a confocal microscope and at the same scale. n = 12. d A stereotype fluorescent microscope image of CNE14:egfp transgenic embryo at 48 hpf. The transgenic embryos exhibit bright EGFP expression in the pectoral fin. n = 49. e, f HCR of CNE14:egfp pectoral fin region with probes complementary to col2a1a for chondrocytes (scapulocoracoid “sc”, magenta), sp7 for osteoblasts (“cl”, white), and egfp (gli3-positive cells, green) with DAPI staining (nuclei, blue) at 72 hpf. n = 19 (See Supplementary Tables 5–7). f The enlarged image of the proximal pectoral fin marked by the rectangle in e. Egfp-positive cells express col2a1a in the scapulocoracoid and sp7 in the cleithrum, indicating that gli3-positive cells differentiate into proliferating chondrocytes and osteoblasts. n = 23. g–j Live fluorescent imaging of the pectoral fin in CNE14:egfp; sp7:mCherry (g and h) or CNE14:egfp; col2a1aBAC:mCherry (i, j) embryo from 32 to 74 hpf. The shown images are representative 3D projections at 35 hpf (g), 54 hpf (h), 56 hpf (i), and 74 hpf (j). Images in g-jwere generated by Imaris (Method). The prospective scapulocoracoid and cleithrum cells reside inside or proximal to the pectoral fin (g, i). These cells migrate proximally and form pectoral girdle bones as development proceeds (h, j). Cell migration trajectories are dragon tails created by Imaris. The unit of the time bars is hours. n = 3. The images in (g–j) are at the same scale. EGFP-positive cells contribute to the endochondral disc (ed), the scapulocoracoid (sc), and the cleithrum (cl). Scale bars are 20 μm (b), 300 μm (d), 50 μm (e, i), and 500 μm (j). A anterior, P posterior, D distal, and Pro proximal, c chondrocytes, cl cleithrum, ed endochondral disc, ff fin fold, o osteoblast, pf pectoral fin, sc scapulocoracoid, and y yolk.
It was long hypothesized that these two bone types arose from distinct cell populations. Intramembranous bones were proposed to develop from neural crest cells, whereas endochondral bones were thought to develop from mesodermal cells10. However, recent studies have challenged this paradigm and provided evidence that the two distinct ossification modes could share common developmental origins in tetrapods11,12,13. For example, in chicken and mouse embryos, the clavicle develops via intramembranous ossification from the lateral plate mesoderm (LPM) cells and cardiopharyngeal mesoderm (CPM)14,15,16,17,18. Additionally, the scapula, which is endoskeletal, originates as an admixture of the LPM and the paraxial mesoderm (PAM) cells in mice, chicken, axolotls, and, tentatively, in turtles19,20,21,22,23,24. A previous study in mice suggested that neural crest cells also contribute to the muscle attachment site of the scapula25. Because it was assumed that the cleithrum of actinopterygian fishes has a neural crest origin, it was hypothesized that the neural crest contribution to the scapula is a remnant of the lost cleithrum—“the cleithrum’s ghost25.” Neural crest contribution to the scapula, however, has not been detected in other studies in tetrapods18,25,26, and, thus, is still under debate. Intriguingly, a recent study discovered that the zebrafish (Danio rerio) cleithrum develops from four distinct embryonic cell populations (neural crest, CPM, LPM, and PAM), while the scapula develops solely from LPM27. This knowledge illuminates how the compositional changes in the embryonic origins contributed to the shift from intramembranous to endochondral bones in the pectoral girdle during the fish-to-tetrapod transition. Yet taxonomically broad investigations of pectoral girdle embryonic origins in other vertebrates with paired fins (chondrichthyan, actinopterygian, and sarcopterygian fishes except for tetrapods) would answer how tetrapod intramembranous and endochondral bones obtained common developmental origins.
Zebrafish belong to Ostariophysan clade, which is one of the largest groups in actinopterygian fishes and includes 28% of extant teleost fishes28. Zebrafish are a prominent fish model system for dissecting the molecular mechanisms underlying skeletal development due to its superb amiability to genetic approaches. D.rerio also represent a powerful model for analyzing pectoral girdle evolution in early tetrapods; zebrafish exhibit numerous evolutionarily conserved anatomical features of their girdle that are seen in basal sarcopterygian fishes and stem-tetrapods, including the cleithrum, supracleithrum, coracoid, scapula, mesocoracoid, and supracleithrum except for anocleithrum8,29,30. Some ossification processes of teleost and basal actinopterygian fishes, however, differ from that of tetrapods, for example, many bones conventionally called endochondral bones, including pectoral fins and girdle, use perichondral ossification during early embryonic development in teleosts31,32,33,34. Also, miniaturization of teleosts fishes altered ossification modes, bone structures, and morphology35,36,37,38. Despite these ontogenetic and structural difference, the genes indispensable for osteoblast and chondrocyte differentiation are tightly conserved between actinopterygians and tetrapods39. These findings make zebrafish a compelling model to scrutinize the ossification molecular mechanisms in ray-finned fishes and to illuminate the water-to-land transition.
Limited knowledge about the genetic pathways responsible for pectoral girdle formation of vertebrates with paired fins represents a major obstacle to discovering the molecular mechanisms responsible for the evolution of the terrestrial pectoral girdle over the course of the fish-to-tetrapod transition. In mice, the Pbx genes, which encode TALE homeoprotein transcription factors, play pivotal roles in endochondral acromion and scapular blade formation, inducing the expression of various transcription factor genes, such as Alx1, Tbx15, Gli3, and Pax140,41,42,43. Additionally, Tbx5-/- mice exhibit a loss of the entire scapula44. In contrast to the accumulated knowledge of the genetic mechanisms required for mammalian and avian pectoral girdle formation, the knowledge of genes involved in fish pectoral girdle formation is limited to only a few studies45,46,47,48.
Gli gene products belong to the Kruppel family of zinc finger protein family and show a profound evolutionarily conservation across invertebrates and vertebrates49,50. Mechanistically, Gli proteins are components of the Hedgehog (Hh) - Gli signaling pathway, where they function as a transcriptional activator upon the binding of Hh ligands to Patched (Ptch) membrane receptors or as a transcriptional repressors without Hh ligands51. Gli proteins are indispensable for animal body development and homeostasis. Coding sequence mutations or large genome deletions at the human GLI3 locus, one of the GLI family genes cause congenital skeletal diseases, including Greig cephalopolysyndactyly characterized by macrocephaly, preaxial polydactyly, intellectual disability, and other severe phenotypes52. GLI3-related Pallister-Hall syndromes (PHS) is another congenital disease paired with hypothalamic hamartoma and mesoaxial polydactyly53. Yet the evolutionary conservation of Gli3 functions during the ontogeny of the aquatic fish body remains largely undetermined.
Previous studies showed that Gli3 plays a key regulatory role in shoulder girdle formation as well as in the patterning of distal appendages in tetrapods54,55,56, making gli3 a compelling candidate gene for understanding the development of the pectoral girdle in fishes. In limb development, Gli3 is expressed in the anterior limb bud as compared to the posteriorly localized Sonic hedgehog (Shh) expression and, thus, is mostly processed to the repressor form without Hh ligands55. Gli3 deletion in mice causes a wide scapula blade and polydactyly, demonstrating that the GLI3 repressor restricts scapula width and digit number41,55,57,58. Deletion of gli3 in the teleost fish medaka further supports the notion that it has an evolutionary conserved function in repressing fin ray and endochondral bone number in the pectoral fin59. Moreover, the previous studies demonstrated that shha (one of the shh genes created in the teleost-specific whole genome duplication (TGD60)) is indispensable for the formation of the cleithrum and the scapulocoracoid, a primordium of the scapula and coracoid, in the zebrafish and medaka pectoral girdles61,62. However, further investigation of Hh-Gli signaling is crucial to elucidate the genetic mechanisms of intramembranous and endochondral bone specification and development in vertebrates with paired fins.
In this study, we investigated the functions of Hh-Gli signaling in zebrafish pectoral girdle development with an emphasis on the balancing mechanism of intramembranous and endochondral ossification. In addition, by conducting the unbiased genomic screening, we identified the downstream target genes of the Hh-Gli signaling, including activin A receptor type 1-like (acrv1l). These newly obtained results suggest that changes in the Hh-Gli signaling or its associated genes might have contributed to the transformation of the pectoral girdle during the water-to-land transition.
Results
Contribution of gli3-positive cells to intramembranous and endochondral pectoral girdle bones
To identify the embryonic origins of the two types of ossification in the pectoral girdle of vertebrates with paired fins, we genetically mapped the contribution of gli3-positive cells, which are of lateral plate mesoderm origin in tetrapods58,63,64,65, to pectoral girdle bones using the zebrafish transgenic system. We conducted an evolutionary sequence comparison of conserved noncoding element 14 (CNE14), a gli3 enhancer for pectoral appendage expression58 among the ray-finned zebrafish and spotted gar, the chondrichthyan elephant shark, and tetrapod Western clawed frog, mouse, and human (Supplementary Fig. 1). CNE14 is highly conserved among these species, except for zebrafish, which showed a lower conservation compared to other species. Next, we cloned both zebrafish and elephant shark CNE14 upstream of a basal promoter and EGFP and tested their activities by injecting them into fertilized zebrafish eggs (see Methods section). Intriguingly, while zebrafish CNE14 did not show EGFP expression in the pectoral fin, elephant shark CNE14 showed conspicuously fin-localized EGFP fluorescence from 32 h post-fertilization (hpf) and beyond (Fig. 1b–d, and Supplementary Fig. 1). Chondrichthyans have not undergone the TGD, and we therefore regard the conspicuous fin-localized activity produced by the elephant shark enhancer as most likely due to a retention of an evolutionarily conserved cis-regulatory sequence and activity that would have been plesiomorphic to gnathostomes66. Next, we established elephant shark CNE14:egfp transgenic zebrafish (hereafter “CNE14:egfp”). Using multiplexed fluorescent in situ hybridization chain reaction (HCR), we confirmed that CNE14:egfp fluorescence marks the endogenous gli3-positive mesenchymal cells in the entire endochondral disc and, therefore, is a practical reporter of gli3-positive cells (Supplementary Fig. 1).
To determine whether gli3-positive cells contribute to the two distinct types of ossification, we conducted HCR with probes complementary to egfp (gli3-positive cells) and sp7 (osteoblast marker67) or col2a1a (proliferative chondrocyte marker68) using CNE14:egfp transgenic embryos. We found that EGFP-positive cells express sp7 in the cleithrum and col2a1a in the scapulocoracoid at 72 hpf (Fig. 1e, f). To further validate whether gli3-positive cells differentiate into osteoblasts or chondrocytes in the pectoral girdle, we performed fluorescent live-cell imaging of CNE14:egfp; sp7:mCherry69 and CNE14:egfp; col2a1aBAC:mCherry-NTR70 fish using a confocal microscope with a temperature and humidity control from 32 to 74 hpf. This live imaging and subsequent 3D cell migration analysis showed that EGFP-positive cells inside and peripheral to the proximal pectoral fin at 32 hpf migrate to proximal direction and start express mCherry driven by sp7 enhancer in the cleithrum at 55 hpf or mCherry driven by the col2a1a BAC in the scapulocoracoid at 63 hpf (Fig. 1g–j). Taken together, these results demonstrate that gli3-positive cells inside and peripheral to the early fin bud migrate and differentiate to osteoblasts in the cleithrum or chondrocytes in the scapulocoracoid in zebrafish embryos.
Morphological phenotype of gli3 knockout zebrafish
To test the function of Gli3 in zebrafish ossification, we generated stable gli3 homozygous knockout zebrafish mutants bearing a frame-shift mutation in exon5 (gli314ins/14ins) using CRISPR/Cas9 (Supplementary Fig. 2 and Supplementary Table 1). At 7 and 30 days post fertilization (dpf), gross observation indicated that gli314ins/14ins fish possess the normal body morphology, including the eye, yolk, median fin fold, and cloaca (Supplementary Fig. 3). We also conducted acid-free skeletal staining of 13 and 30 dpf juveniles and did not find significant difference in ossification of cranial, trunk, and tail skeletons, including the opercular, parashenoid, vertebrae, and hypural bones between wildtype and gli314ins/14ins fish. The dorsal extention of the cleithrum, however, is slightly shorter than that of wildtype in some gli314ins/14ins juveniles (n = 2/8, Supplementary Fig. 3). Survival of gli314ins/14ins fish to adulthood allowed us to determine the functional roles of the Hh-Gli signaling pathway in adult body structures (Supplementary Fig. 4). Gross observation indicated that some adult gli314ins/14ins fish possess an external notch posterior to the jaw bones (Supplementary Fig. 4, 28/159). We then scrutinized alterations of skeletal morphology throughout the body of adult gli314ins/14ins fish using bone and cartilage staining followed by dissections (Methods, Supplementary Figs. 4–6). Compared to wildtype fish, gli314ins/14ins fish showed excessively mineralized skull bones, including of the parietal and frontal bones, and ectopic ossification in the eye lens at 3 months-old (Supplementary Fig. 4, n = 5/7). The excessive mineralization phenotype matches GLI3-related human congenital diseases, such as Greig cephalopolysyndactyly and Pallister-Hall syndromes52,71. Moreover, gli314ins/14ins zebrafish exhibited an abnormally expanded telencephalon at three months old (Supplementary Fig. 7, Supplementary Table 3). Intriguingly, we did not find any increase of fin ray or endochondral radial numbers in the pectoral fin as previously observed in gli3 knockout medaka fish (Supplementary Table 4)72. The lack of ossification phenotype in gli314ins/14ins pectoral fin is consistent with another independent study that generated gli3 knockout zebrafish with a different genetic mutation73.
Gli3 determines the balance of osteoblast and chondrocyte differentiation
Next, to analyze the function of Gli3 in the intramembranous and endochondral ossification of the pectoral girdle, we conducted HCR with probes complementary to sp7 and col2a1a using gli314ins/14ins embryos. The quantification of sp7-positive cells showed that fewer osteoblasts are present in the cleithrum at the level of the scapulocoracoid in gli314ins/14ins mutants compared to wildtype embryos at 72 hpf (Fig. 2a and b, Supplementary Table 5–7, p = 0.00018). We also compared sp7:egfp expression in the cleithrum of wildtype and gli314ins/14ins embryos. EGFP expression is weaker in the ventral part of the cleithrum in gli314ins/14ins embryos than wildtype (Supplementary Fig. 8). Gli3 functions as a transcriptional activator or repressor depending on the presence of Hh28. Therefore, osteoblast reduction could arise from a loss of either or both functions. To differentiate between these possibilities, we treated wildtype zebrafish embryos with BMS-833923, a Smoothened antagonist (i.e., Hedgehog inhibitor74) that decreases the amount of Gli activator from 32 hpf to 72 hpf. In BMS-833923-treated embryos, we observed fewer osteoblasts in the cleithrum at the level of the scapulocoracoid than wildtype embryos in a comparable manner to gli314ins/14ins embryos at the equivalent total body length stage (wildtype embryos at 68 hpf and BMS-833923-treated embryos at 72 hpf embryos, p = 0.0028) (Supplementary Fig. 9, Supplementary Table 5–7). This demonstrates the functional significance of the Gli activator in osteoblast differentiation in the cleithrum.
a–c HCR of col2a1a (magenta) and sp7 (green) with DAPI staining (cyan). 3D stack lateral visualization of confocal scanning of gli3+/+ (a), gli314ins/14ins (b), and gli2b17ins/17ins; gli314ins/14ins embryos (c). The osteoblasts around the cleithrum at the level of the scapulocoracoid are reduced in gli314ins/14ins and gli2b17ins/17ins; gli314ins/14ins embryos (b, c). n = 6 for each genotype. The scales are the same from (a–c). d, e Dissected pectoral fins of gli3+/+ (d) and gli314ins/14ins (e) embryos at 72 hpf after HCR with the same probes used in (a–c). The scapulocoracoid is larger in gli314ins/14ins embryos (e) than in gli3+/+ embryos (d). The scale is the same between (d, e). f The comparison of the osteoblast (sp7-positive) and chondrocyte (col2a1a-positive) numbers in the cleithrum and the scapulocoracoid, respectively, between gli3+/+ and gli314ins/14ins embryos (n = 18 for gli3+/+ and n = 16 for gli314ins/14ins embryos). **; p = 0.0000178 by a two-tailed student t-test. Scale bars are 300 μm in (a) and 50 μm in (d). cl cleithrum, sc scapulocoracoid.
To genetically corroborate that Gli functions as an activator in the zebrafish pectoral girdle, we decreased the amount of Gli activator by introducing a stable frameshift mutation into the coding sequence of gli2b, encoding another Gli family zinc finger protein that mainly functions as a transcriptional activator50, in gli314ins/14ins embryos using CRISPR/Cas9 (Supplementary Fig. 2, Supplementary Table 1). Consistent with the phenotype of BMS-833923-treated embryos, gli2b17ins/17ins; gli314ins/14ins double knockout embryos have fewer osteoblasts in the cleithrum at the position where the scapulocoracoid attaches than gli314ins/14ins embryos at 72 hpf (Fig. 2c, Supplementary Tables 5, 6. average number; 23.6 cells in gli314ins/14ins vs 16.77 cells in gli2b17ins/17ins; gli314ins/14ins embryos). However, an ANOVA test did not detect a statistically significant difference between these two genotypes (p = 0.12, Supplementary Table 7). Intriguingly, we also observed that the number of col2a1a-positive proliferative chondrocytes in the scapulocoracoid of gli3 14ins/14ins embryos increased compared to wildtype embryos (Fig. 2f, g and Supplementary Fig. 10, Supplementary Table 7, p = 0.0000178). Albeit the difference of averaged chondrocyte numbers between in wildtype and gli2b17ins/17ins; gli314ins/14ins embryos (165.67 vs 193.69, Supplementary Table 5), we did not detect a statistically significant difference between them (p = 0.06). These results suggest that the Gli3 activator form, and possibly Gli2b with a relatively weak contribution, induces osteoblast differentiation in the cleithrum at the expense of chondrocytes in the scapulocoracoid.
Regulation of BMP signaling via activin A receptor type 1-like expression by Gli3
To elucidate the Gli3 pathway that determines osteoblast and chondrocyte differentiation, we identified Gil3 target genes in an unbiased way by combining RNA-sequencing (RNA-seq) and Assay for Transposase-Accessible Chromatin using sequencing (ATAC-seq) in wildtype and gli314ins/14ins embryos (Fig. 3a). We separately isolated EGFP-positive cells from CNE14:egfp; gli3+/+ and CNE14:egfp; gli314ins/14ins embryos by Fluorescence Activated Cell Sorting (FACS) and conducted RNA-seq at 55hpf, when both the cleithrum and the scapulocoracoid are at their early developmental stages (Fig. 3a). A comparative analysis of transcriptome profiles between EGFP-positive cells in gli3+/+ and gli314ins/14ins embryos identified 351 significantly upregulated genes and 232 downregulated genes (p < 0.05) (Fig. 3b, Supplementary Data 1). This gene list includes klf3 and zic3, which were previously identified as direct GLI3 target genes in mouse limb buds75, supporting the efficacy of our method. Next, we conducted ATAC-seq with EGFP-positive cells sorted from CNE14:egfp; gli3+/+ embryos at 55 hpf, identified accessible chromatin regions (ACRs) in a genome-wide manner, and annotated these ACRs to genes depending on the proximity to transcription start sites (Supplementary Data 2). The gene with one of the lowest p-values from RNA-seq and an ACR promoter is activin A receptor type 1-like (acvr1l), which encodes a BMP receptor type 176 (Fig. 3c). Given the necessity of BMP signaling in osteoblast and chondrocyte differentiation and its involvement in human skeletal diseases77,78,79,80, zebrafish acvr1l is a compelling Gli3 target for pectoral girdle development. An analysis of previously published Hi-C data indicates that the acvr1l promoter domain is in a ~ 90 kbp topologically associating domain (TAD), suggesting its cis-regulatory elements lie in this region81 (Fig. 3d). Intriguingly, our analysis of the Activin receptor 1 gene family history indicated that the Acvr1l gene is evolutionarily conserved in all extant jawed fish lineages yet got lost multiple times independently in the major extant tetrapod lineages (Supplementary Fig. 11). The notable exception are crocodilians that retained Acvr1l. We therefore propose that the functional importance of the Acvr1l gene gradually faded in tetrapods, concomitantly with the intramembranous-to-endochondral skeletal shift of the pectoral girdle starting during the fish-to-tetrapod transition (Supplementary Fig. 11, Supplementary Table 8, and Supplementary Note 1).
a The experimental scheme of the unbiased high-throughput genomic screening (see Methods). From CNE14:egfp; gli3+/+ and CNE14:egfp; gli314ins/14ins embryos, EGFP-positive cells were separately isolated by FACS. The sorted cells were subjected to RNA-seq and ATAC-seq. Three biological replicates were conducted for RNA-sequencing and two were for ATAC-seq. b The heat map of top 2000 differentially expressed genes in CNE14:egfp; gli3+/+ and CNE14:egfp; gli314ins/14ins embryos. Color code: red indicates high and blue indicates low enrichment. c The volcano plot of the RNA-seq result. The genes with p < 0.5 and >two-fold expression change are coded by red. The genes on the left side (Log2 fold change <0) are down-regulated genes in gli314ins/14ins embryos while the genes on the right side (Log2 fold change > 0) are up-regulated genes in the mutant embryos. Acvr11 is one of the prominent Gli3 candidate genes. The statistical analysis was conducted by EdgeR program. d The genome browser visualization of zebrafish RNA-seq (wildtype and gli314ins/14ins samples), ATAC-seq, and HiC results, and skate RNA-seq and ATAC-seq results at the acvr1l locus. In zebrafish, the transcript level is lower at the exons of acvr1l in gli314ins/14ins embryos than gli3+/+ embryos. Zebrafish ATAC-seq and HiC showed that the promoter region of acvr1l is ACR and in a TAD. Zebrafish HiC result was produced from the previously published data81. Skate RNA-seq and ATAC-seq data were generated from the previous paper118. e, f The expression pattern of acvr1l in gli3+/+ and gli314ins/14ins embryos. Expression in the pectoral fin is decreased in gli314ins/14ins embryos compared to gli3+/+ embryos. n = 24. The scale is the same in (e, f). g–i Immunofluorescence of phosphorylated-Smad 1/5/8. The strong signal is observed in cells surrounding the cleithrum of wildtype embryos (g, arrows), which decreases to gli314ins/14ins (h) and to gli2b17ins/17ins; gli314ins/14ins embryos (i). n = 6 for wildtype, 9 for gli314ins/14ins, and 5 for gli2b17ins/17ins; gli314ins/14ins embryos. j a bar graph of phosphorylated Smad1/5/8 staining level in gli3+/+, gli314ins/14ins and gli2b17ins/17ins embryos. The fluorescence intensity in the cleithrum was quantified and standardized by the area size in the cleithrum (“Method”). All raw data points were plotted. Raw data are in Supplementary Table 9. The data were analyzed and compared among samples by two-tailed student’s t-test. * indicates p < 0.05. p-value between gli3+/+ and gli314ins/14ins is 0.049 and between gli3+/+ (WT) and gli314ins/14ins is 0.025. While gli314ins/14ins and gli2b17ins/17ins; gli314ins/14ins embryos exhibit statistically significant reduction of the PSmad1/5/8 signal from wildtype embryos, gli314ins/14ins and gli2b17ins/17ins; gli314ins/14ins embryos do not show statistically significant change (p = 0.28). The unit of vertical axis is arbitrary unit. n = 6, 9, and 5 biologically independent samples for wildtype embryos, gli314ins/14ins and gli2b17ins/17ins; gli314ins/14ins embryos, respectively. Error bars show standard deviations. k-l) LDN193189-treated pectoral fins stained by HCR with sp7 and col2a1a probes and DAPI staining. Sp7 expression was diminished by 10 μM of the inhibitor treatment. Arrows indicate sp7 expression, and arrowheads show its weak or no expression. sc scapula. n = 23 for 0 and 10 μM. The scale bar in (e, g) are 50 μm. g–i, k, l are the same scale.
Subsequent whole-mount RNA in situ hybridization corroborated the regulation of acvr1l by Gli3; acvr1l transcripts are enriched in the endochondral disc of the wildtype pectoral fin but are decreased in gli314ins/14ins fin at 55 hpf (Fig. 3e, f). Moreover, we found acvr1l expression in the pectoral fin is a shared feature with chondrichthyans (little skate, Leucoraja erinacea) and non-teleost actinopterygians (spotted gar, Lepisosteus oculatus) (Supplementary Fig. 12). In accordance with the reduction of acvr1l expression in gli314ins/14ins zebrafish embryos, we also found a significant decrease in staining levels of phosphorylated Smad1/5/8 (the active form of Smad1/5/8 in BMP signaling) in osteoblasts surrounding the cleithrum bone from wildtype to gli314ins/14ins embryos (p = 0.02), and possibly to gli2b17ins/17ins; gli314ins/14ins embryos but not at a statistically significant level (p = 0.28) (Fig. 3g–i, Supplementary Table 9). This may reflect a relatively weak function of Gli2b compared to Gli3 in pectoral girdle development. Finally, to test the function of Acvr1l in the differentiation of osteoblasts and chondrocytes in shoulder girdle formation, we treated wildtype zebrafish embryos with an Activin receptor type 1 inhibitor LDN19318982 from 32 to 72 hpf. Compared to the proper expression of sp7 and col2a1a in the control pectoral girdle, sp7 expression was diminished in LDN193189-treated embryos (Fig. 3k, l). Therefore, the differentiation of osteoblasts requires BMP signaling via Acvr11 in the fish pectoral girdle.
Hedgehog-Gli mutant zebrafish recapitulate evolutionary trajectories of intramembranous and endochondral bones along the water-to-land axis
The pectoral girdle in non-teleost actinopterygians consists of a series of intramembranous (supracleithrum, cleithrum, and clavicle) and endochondral bones (scapulocoracoid and mesocoracoid)83. Teleosts, including zebrafish, share these fundamental components, except for the separation of the scapula and coracoid and the loss of the clavicles84 (Fig. 4a). Gross examination and skeletal staining showed that in approximately 20 % of gli314ins/14ins zebrafish either the left or right pectoral fin shifted dorsally due to the dorsoventrally shortened cleithrum (28/159 fish, Supplementary Fig. 4). Intriguingly, subsequent µCT scanning and three-dimensional morphometric analysis revealed mixed characteristics of actinopterygian and stem-tetrapodomorph pectoral girdle morphologies in these severely affected girdles; the dorsal blade of the cleithrum and the posterior branchial lamina, which posteriorly supports opercular chamber movement in fish, were markedly reduced (Fig. 4a, d, j, Supplementary Fig. 13 and Supplementary Data 3). Moreover, the supracleithrum was absent (Fig. 4a, d, Supplementary Fig. 4), resembling the condition of stem-tetrapods such as Tulerpeton7,85. The cleithrum on the opposite side in these severely affected fish and on both sides in the remaining 80% of glis314ins/14ins zebrafish was dorsoventrally shorter compared to those of wildtype individuals (Supplementary Fig. 14, Supplementary Data 3, n = 23/23). In addition to the reduction of the intramembranous series, the scapula of gli314ins/14ins zebrafish exhibited the characteristics of stem-tetrapod traits; an enlarged supraglenoid buttress and the anteromedially extended flange for the glenoid (Fig. 4b, e). The glenoid was ovoid and concave in gli314ins/14ins fish, and it is posterolaterally opened compared to the flat glenoid in wildtype fish (Fig. 4b, c, e, f; n = 3/6).
a–c, d–f, g–i Volumetric renderings of the skeleton, which was segmented manually from µCT scans of three-months old adult fish. a–c wildtype, d–f) gli314ins/14ins, g–i gli314ins/14ins; shha30%KO fish. a, d, g the whole-body 3D reconstruction, (b, c, e, f, h, i) the enlarged images of the scapulocoracoid and mesocoracoid (b, e, h); lateral view, (c, f, i); posterior view). In contrast to the thin and long dorsal cleithrum with the postbranchial lamina in gli3+/+ fish (a), the dorsal cleithrum was reduced in either the left or right side of gli314ins/14ins fish (n = 28/159) (d) and both sides of gli314ins/14ins; shha30%KO fish (3/3) (g). In the scapula of gli3+/+ fish, the glenoid is anteroposteriorly wide and flat (b, c), but it is concave and posterolaterally opened, and delineates the proximal surface of basal radials of the pectoral fin in gli314ins/14ins and gli314ins/14ins; shha30%KO fish (e, f, h, i) (n = 5/6 for each genotype). The supraglenoid buttress is more developed in gli314ins/14ins and gli314ins/14ins; shha30%KO fish compared to wildtype fish (b, e, h). j Quantitative analysis of the cleithrum height, scapula width, mesocoracoid width, and glenoid depth. The length and width of each bone were measured and standardized by the dorsoventral length of the skull (see Methods). The unit is aribitrary unit [a.u.]. All raw measurements are summarized in Supplementary Data 3. While the cleithrum is dorsoventrally shorter in the severely affected pectoral girdles of gli314ins/14ins and gli314ins/14ins; shha30%KO fish than those of gli3+/+ fish, the scapula and mesocoracoid are larger in gli314ins/14ins and gli314ins/14ins; shha30%KO fish. The glenoid is more concave in gli314ins/14ins and gli314ins/14ins; shha30%KO fish than wildtype fish. ANNOVA test and following Tukey-Kramer Post Hoc test were conducted to detect statistically significant differences of bone size among wildtype and knockout fish. **; absolute mean value > Q critical value in Tukey-Kramer Post Hoc Test. 5.07 and 5.83 > 3.73 in the cleithrum, 5.10 and 6.16 > 3.73 in the scapula, and 6.07 > 3.88 in the mesocoracoid, and 6.05 > 3.88 in the glenoid). Maximum, minimum, median, and average values are shown in the box and whisker plots. Two or three biologically independent fish samples were analyzed in each genotype. Both left and right pectoral girdle measurements are plotted. See all replicates and measurements in Supplementary Data 3. c coracoid, cl cleithrum, g glenoid, ms mesocoracoid, pl postbranchial lamina, sb supraglenoid buttress, and scl supracleithrum. The scale bar indicates 2.5 mm.
The mesocoracoid is another endochondral bone that bridges the coracoid and the cleithrum and is present in actinopterygian fishes but not in sarcopterygian fishes or stem-tetrapods such as Tiktaalik or Acanthostega. In these stem-tetrapods, the supraglenoid buttress is the evolutionary counterpart of the mesocoracoid86. The mesocoracoid/supraglenoid buttress becomes stouter from fishes with paired fins to stem-tetrapods to serve as an attachment site for the dorsomedial musculature of the forelimb87. In gli314ins/14ins zebrafish, the mesocoracoid is laterally wider than in the wildtype (Fig. 4c, f, n = 4/5).
Additionally, to investigate whether the pectoral girdle phenotype is exacerbated by a further reduction of Gli activator, we injected gRNA complementary to zebrafish shha with Cas9 mRNA into gli314ins/14ins eggs, raised them for 3 months, and genotyped them by deep sequencing of shha (see Methods). Strikingly, gli314ins/14ins; shha30% fish (30% of the intact shha is detected) exhibited a dorsoventrally short cleithrum comparable to the severely affected cleithrum in gli314ins/14ins fish, but on both the left and right sides (Fig.4g, n = 3/3). The shape of the scapula in this compound knockout fish is also concave and posterolaterally opened (Fig.4h, i). gli314ins/14ins; shha30% fish also showed smaller ratio of cleithrum/scapulocoracoid volume than those of wildtype fish (Supplementary Fig. 15 and Supplementary Data 4). These results indicate that the Hedgehog-Gli signaling regulates not only the shoulder girdle bone morphology, but also the ratio of intramembranous and endochondral ossification in pectoral girdle development.
Discussion
The phenotype of gli3 knockout zebrafish is consistent with those described in tetrapod models, suggesting that the gene’s functions are evolutionarily conserved across bony fishes. We have shown that gli314ins/14ins zebrafish exhibited abnormal skeletogenesis in the skull, eye, and shoulder girdle. The dense mineralization of intramembranous skull bones is a coherent phenotype with human diseases, such as Greig cephalopolysyndactyly52. Moreover, expansion of the endochondral scapula in shha and gli3 compound knockout zebrafish is consistent with the scapula phenotype in Gli3 knockout mice41. These comparable ossification defects in zebrafish and mammals thus suggest that Gli3 functions are evolutionarily conserved in bone formation at least between actinopterygians and tetrapods. Importantly, previous studies indicate that a reduction of Hedgehog signaling severly affects cleithrum, scapula, and coracoid development in zebrafish88. Given these results, we propose that conserved Gli3 functions form the actinopterygian fish pectoral girdle, balancing intramembranous and endochondral ossification.
Polydactyly is a pervasive phenotype in Gli3-deficient mammals, and even gli3 knockout medaka show an increase of fin rays and endochondral radials72. The fin phenotype of gli314ins/14ins zebrafish, however, differs from the previously published medaka phenotype59; gli314ins/14ins zebrafish did not show the increase of fin rays and endochondral radials. The lack of a bone-increase phenotype in the pectoral fin of gli314ins/14ins zebrafish is consistent with results from a different zebrafish gli3 knockout allele produced in an independent study89 and, thus, is a validated phenotype from multiple studies. This phenotypic difference between zebrafish and medaka might reflect diverse functions of the Hh-Gli signaling in formation of exceptionally diverse pectoral fins and girdles in actinopterygian fishes, including various fin ray numbers, possibly due to differential retention of TGD-duplicated Hh-Gli pathways90. Alternatively, the short DNA oligo insertion into gli3 coding sequence by CRISPR/Cas9 might induce genetic compensation that increases similar gene transcription in zebrafish91. In addition to Gli3, Gli2 functions as both activator and repressor depending on Hedgehog ligand binding to its receptors92. Yet, even gli2b; gli3 double knockout zebrafish did not manifest the increase of fin rays and radials. Gli2a and Gli2b repressor forms may functionally compensate Gli3 repressor function to restrict the number of fin rays and radials along the anterior-posterior axis in zebrafish. The comprehensive elimination of all gli2 and gli3 family genes (i.e., gli2a, gli2b, and gli3) would likely provide “polydactyly” phenotype in zebrafish although the elimination of all Gli2 and Gli3 activator and repressor forms might not provide a conspicuous phenotype, as both activator and repressor forms are critical for distinct domains of paired appendage development in mice92. These potential functional compensations among Gli proteins might explain the absence of statistically significant differences of osteoblast and chondrocyte numbers in the pectoral girdle between gli3 and gli2b; gli3 knockout embryos.
The morphological transformation of the vertebrate pectoral girdle during the water-to-land transition has been extensively described87,93, yet the underlying ontogenetic changes remain uninvestigated. In the last few decades, zebrafish have emerged as a compelling model to dissect the genetic and molecular pathways underlying diverse arrays of skeletal phenotypes because zebrafish are amenable to various genetic manipulations, live imaging, and produce ample fertilized eggs for embryological experimental studies. Leveraging these zebrafish strengths, our current study illuminates the developmental mechanisms underlying the pectoral girdle evolution, which have been long-debated94. Despite the fact that the crown teleost ancestor underwent the TGD that led sub- and neo-functionalization of many gene duplicates60,95, our Gli3 knockout zebrafish show the evolutionarily conserved the pectoral girdle phenotype. Distinct from sarcopterygians, teleosts mostly use perichondral ossification during development, including pectoral fin and girdle formation31,96,97. Additionally, in teleost fishes, the scapulocoracoid is divided into two distinct elements, the scapula and coracoid. Despite these genomic, ontogenetic, and anatomical specializations, zebrafish Gli3 functions in developing bones, including the scapula, are substantially conserved with extant mammals. Moreover, we identified acvr1l expression in spotted gar and skate embryonic fin and girdle structures, in which the scapula and coracoid are unified in a single bone, further supporting the evolutionary conservation of Gli3 signaling in ancestral jawed vertebrate pectoral girdle development. Thus, our present study with basal and derived actinopterygians and chondrichthyans provides the first genetic and ontogenetic insights into the pectoral girdle transformation from water to land.
While zebrafish are a compelling model organism to illuminate the molecular mechanisms of skeletal development and evolution, they represent a miniaturized teleost species. Miniaturization causes structural reduction, simplification, and novelty98. Haemal arches, for example, develop via intramembranous ossification in many small teleost fishes, including zebrafish, but via endochondral ossification in larger teleosts99. The coracoid and scapula of Priocharax ariel, one of the miniaturized teleost fishes in South America, are not ossified even in adult fish in contrast to full ossification of the cleithrum36. Thus, a caveat of using minituarized teleost fish for understanding of the pectoral girdle evolution is potential changes in ossification patterns.
During the evolution of the miniaturized body, many endochondral bones in zebrafish shifted from endochondral to perichondral ossification, including the pectoral girdle. The zebrafish scapulocoracoid, the primordium of the scapula and coracoid, is a thin cartilage plate during early development84. The coracoid ossification initiates at the dorsal edge of the anterior extension of the scapulorocraoid and spreads to the posterior side via perichondral ossification100. The different ossification modes of endochondral bones between miniaturized teleosts and stem-tetrapods may hamper us from a direct comparison of pectoral girdle developmental mechanisms. In this study, we hypothesize that the final size and morphology of the scapula and coracoid simply reflect those of cartilage primordium common for perichondral and endochondral ossification. Accordingly, we discuss the molecular and ontogenetic mechanisms underlying changes in the ratio of endochondral and intramembranous bones during the water-to-land transition using zebrafish pectoral girdle as a proxy. However, one may argue that the final size and morphology of the scapula and coracoid might have been modified during the shift from endochondral to perichondral ossification, irrespective of cartilage primordium morphology. Despite such differences in ossification modes between miniaturized teleosts and stem-tetrapods, the molecular mechanisms that induce chondrocyte and osteoblast differentiation are tightly conserved101. The exceptional conservation of ossification genes could justify an evolutionary prevalence of the Gli3 signaling pathway that determines the balance of endochondral and intramembranous ossification. The pectoral girdle evolutionary mechanisms would be further scrutinized by studies with non-miniaturized actinopterygians such as spotted gar, an emerging model without a 3rd round of whole genome duplication and body miniaturization95.
The evolutionary trajectory of the intricate shoulder girdle development at the origin of stem-tetrapods has been uncharacterized. By taking advantage of the ontogenetic and genetic accessibility of zebrafish, we discovered that gli3-positive mesodermal cells give rise to both intramembranous and endochondral bones in the pectoral girdle (Fig. 5). Our results reveal that modulations in Hh-Gli signaling are plausible evolutionary mechanisms to assemble the tetrapodmorph pectoral girdle through mesodermal cell fate alternations, which have been also reported in the evolution of digits from fin rays11,12. Thus, changes in the fate of mesodermal cells appear to have occurred repeatedly in appendages across the fish-to-tetrapod transition and represent a major path for the evolutionary origin of novel traits.
From fish to tetrapods, endochondral bones (scapula and coracoid) expanded at the expense of the intramembranous bones (anocleithrum, cleithrum, and clavicles). In gli314ins/14ins zebrafish, the cleithrum becomes dorsoventrally short, reminiscent of the pectoral girdles of early tetrapods, such as Acanthostega, which were evolving to live on land. Gli3-positive cells migrate into the cleithrum and scapulocoracoid, and Gli3 regulates BMP signaling via acvr11 expression. In crown-group tetrapods, including mice and chickens, the prospective pectoral girdle cells reside in the proximal region of forelimb buds. Pbx1/2/3 family genes regulate the expression of alx, tbx15, and gli3 in the superior scapula blade (ssbl), inferior scapula blade (isbl), and spine/central scapula blade (sp/csbl)41,119. The cellular origins of the shoulder girdle bones, shown by light green and orange in Acanthostega and Eryops are estimations from histological characters. an anocleithrum, c coracoid, cla clavicle, cl cleithrum, g glenoid, ic interclavicle, ms mesocoracoid arch, pl postbranchial lamina, sc scapula, scc scapulocoracoid, scl supracleithrum, sgb supraglenoid buttress. The pectoral girdle of Mus was reproduced from the previous study with permission (ref. 41). Acanthostega shoulder girdle was reproduced from the previously published article (ref. 7). Lepisosteus and Eryops were reproduced from previous studies with permission (ref. 87 Copyright © 1924 Wiley-Liss, Inc.). Photos of the pectoral girdle of an adult Lepisosteus oculatus are in Supplementary Fig. 16.
The dorsoventrally short cleithrum, the loss of the supracleithrum, the posterolaterally oriented concave glenoid, and the robust mesocoracoid in gli3 single and gli3;shh compound knockout zebrafish conform to the shared features of the pectoral girdle bones in Devonian-era stem-tetrapods, such as Tiktaalik, Acanthostega, and Ichthoystega6,7,8 (Fig. 5). Moreover, the concave and posterolaterally-opened glenoid in gli314ins/14ins fish is similar to a prominent feature of the stem-tetrapods. These morphological features permit the rotation, flexion, extension, protraction, and retraction of the humerus6. The current work suggests that the Gli3-Acvr1l pathway and their functions in the pectoral girdle formation are evolutionarily conserved at least from basal to derived actinopterygian fishes. Genetic alterations in Hh-Gli or associated signaling pathways, such as the evolutionary reduction and eventual loss of tetrapod Acvr1l orthologs, might have released a deeply embedded developmental program to generate tetrapod morph pectoral girdle phenotypes in bony vertebrates.
Methods
Animal maintenance
All fish work was conducted according to standard protocols approved by the animal committees of Rutgers University (protocol number 201702646) and Michigan State University (PROTO202200367). Skate embryos (Leucoraja erinacea) were purchased from Marine Biological Laboratory (Woods Hole, MA), fixed by 4% PFA, and kept in methanol at −20 °C for whole-mount in situ hybridization.
Evolutionary comparisons of CNE14
The evolutionary conservation of gli3 CNE14 sequence was conducted in mVISTA102. For this sequence comparison, the following genome regions were downloaded from Ensembl and compared in mVISTA: human: GRCh38:7:41954886:42270163, mouse: GRCm39:13:15632485:15909946, tropical clawed frog: UCB_Xtro_10.0:6:57560778:57692896, elephant shark: Callorhinchus_milii-6_1_3_KI635914_1_1211641_1411175, spotted gar: LepOcu1_LG9_37116819_37285109, and zebrafish: GRCz11:24:11522406:11827369.
Establishment of CNE14:egfp transgenic fish
The region of the zebrafish and elephant shark genome homologous to human CNE1426 was amplified by PCR and subcloned pCR™8/GW/TOPO® (Invitrogen) (the primer sequences are in Supplementary Table 1). The cloned CNE14 fragment was transferred into pXIG-cfos-EGFP vector by Gateway™ LR Clonase™ II (Invitrogen)46. Each CNE14 / pXIG-cofs-EGFP vector was injected into over 250 one-cell stage zebrafish eggs with Tol2 mRNA as previously described47. EGFP fluorescence was observed at 48 hpf under a fluorescence stereomicroscope. For elephant shark CNE14, F0 embryos with EGFP expression were raised for three months, and the EGFP expression pattern was confirmed in F1 embryos obtained from outcrossing the F0 to wildtype fish under a stereotype fluorescent microscope. The embryos with EGFP expression in the pectoral fin were reared to adult stage and further crossed to wildtype fish to establish stable transgenic embryos, which were used for immunofluorescence, HCR in situ hybridization, and live fluorescent imaging.
Whole mount in situ hybridization
The 3’UTR or coding sequence of the zebrafish, skate, and spotted gar genes were amplified from zebrafish (48hpf), skate (stage 31), and spotted gar (stage 28) cDNA by PCR and cloned into pCR™II-TOPO® vector (Invitrogen) (PCR primer sequences are in Supplementary Table 1). The RNA probes were synthesized from the vectors by T7 or SP6 RNA polymerase for zebrafish and skate and from vector insert PCR amplificate for gar.
Chromogenic in situ hybridization for acvr1l in zebrafish and skate embryos was performed as previously described48. The spotted gar ISH method is based on Thisse & Thisse 2008103. In situ hybridization chain reaction (HCR) was performed as previously described49 using complementary probes to zebrafish sp7, col2a1a, egfp, gli3, which were all synthesized by Molecular Instruments (CA). To observe the HCR results at the single cell resolution, the pectoral fin and girdle complex were manually dissected out from the body with tweezers, mounted on slide glasses, and scanned by a confocal microscope (Zeiss LSM510 META). The number of chondrocytes in the scapulocoracoid (col2a1a+) and osteoblasts surrounding the cleithrum at the level of the scapulocoracoid (sp7+) were manually counted and statistically compared among different genotypes or embryos treated by different concentrations of the inhibitor by a Student’s t-test. For HCR of sp7, col2a1a and egfp, n = 23 (Fig. 1e, f). For the staining of sp7 and col2a1a in Fig. 2d–f, all raw data and replicates are shown in Supplementary Table 5.
For fluorescence in situ hybridization of gli3 mRNA, we followed the protocol previously published in Lauter et al. 104. Briefly, embryos were fixed at 48 hpf by 4% PFA. Next day, following DEPC-PBST (phosphate buffered saline, 0.1% Tween-20, pH 7.3) rinse, the embryos were treated by 2% hydrogen peroxide in methanol for 20 min. After rehydration, embryos were subjected to the previously published whole mount in situ hybridization protocol48. For fluorescent color development, two solutions were prepared: 500 μg/ml Fast Blue (Sigma F3378) or 500 μg/ml NAMP (Sigma N5000) in 0.1 M Tris-HCl pH 8.2, containing 50 mM MgCl2, 100 mM NaCl, and 0.1% Tween-20 were prepared. Before color development, we mixed these two solutions at 1:1 ratio and soaked embryos in the mixed solution until the color developed.
Live fluorescence cell imaging
CNE14:egfp transgenic fish were crossed to sp7:mCherry fish or col2a1aBAC:mCherry fish, and fertilized eggs were collected. Embryos with EGFP expression in the pectoral fin primordium were selected under a fluorescent stereotype microscope at 31 hpf and embedded in 1% low-melt agarose gel in a glass-bottom 3.5 cm dish50. The gel covering the anterior half of embryos was removed to allow normal growth of the embryos. E3 medium including tricaine at 0.003% was added to the dishes, and the dishes were kept inside an incubator chamber at 28 °C by a confocal microscope (Zeiss LSM510) from 32 to 74 hpf. Z-stack images (3 µm z-interval, ~30–40 slices for each embryo) were obtained at 30 minutes time intervals. The time lapse data were analyzed with Imaris (version 10.1) to manually track EGFP-positive cell movement and contribution to the cleithrum and scapulocoracoid. The replicate number is three for each transgenic fish imaging.
Establishment of gli3, gli2b;gli3, and gli3;shha compound knockout fish
Gli3 single knockout fish with frameshift mutations in exon5 and exon14 were generated by injection of gRNAs complementary to target sites and Cas9 mRNA into wildtype (*AB) fertilized eggs as previously reported12,105. The detailed information of gRNAs and identified frame shift mutations are summarized in Supplementary Fig. 1. Briefly, gRNA and Cas9 mRNA were injected into zebrafish one-cell stage eggs, and the injected eggs were reared to adult fish, which were subjected to T7 assay and sequencing to determine genetic mutations at the target loci. The fish with frame-shift mutations (F0) were out- crossed to wildtype fish to obtain heterozygous knockout fish (F1). To avoid any off-target effects of CRISPR/Cas9 gene deletions on the phenotypic analysis, we repeated outcrossing the heterozygous knockout fish to wildtype fish (*AB) and raising offspring twice, obtaining F4 fish for the analysis conducted in the paper. Then, gli3 ex514ins/+ fish (F4) were crossed with each other and gli3 ex514ins/14ins fish were established. The survival rate of gli3 ex514ins/14ins zebrafish is not significantly different from that of wildtype fish and the ratio of gli3 +/+, gli3 ex514ins/+ and gli3 ex514ins/14ins follow Mendelian inheritance ratio. To obtain gli2/gli3 compound knockout fish, gRNA complementary to a target site in gli2b exon1 and Cas9 mRNA was injected into one-cell stage gli3 ex514ins/14ins fertilized eggs. The injected eggs were reared to an adult stage and subjected to a T7 assay and sequencing to identify frameshift mutations. The fish with frameshift mutations were outcrossed to gli3 ex514ins/14ins fish and the collected eggs were raised for three months and genotyped for gli2b. Obtained gli2b17ins/+; gli3 ex514ins/14ins fish were crossed with each other and embryos were subjected to HCR in situ hybridization.
To obtain gli3/shha compound knockout fish, gRNA complementary to shha and Cas9 mRNA were injected into gli3 ex514ins/14ins fertilized eggs. The tail fins of 3 month old adult fish were cut and subjected to a T7 assay for the shha gene. The tail lysis of the mutant fish was further subjected to deep sequencing of shha target loci to determine frameshift mutation ratio among all mutations (Supplementary Table 1).
All gRNA sequence and genotyping primers are summarized in Supplementary Table 1. The frameshift mutations in all mutant fish are in Supplementary Fig. 1.
Genotyping of gli3 and gli2b knockout fish
Adult or embryonic tails were excised and lysed as previously described12. Using these tail lysis solutions as PCR templates, the gli3 and gli2 loci were amplified by PCR. The PCR primers used for PCR amplification are summarized in Supplementary Table 1. For gli3, a 14 bp difference between wildtype and mutant fish was confirmed on 3% agarose gels. BSAJ1 was added to gli2b PCR products and the treated PCR products were confirmed on 1% agarose gel (wildtype; 232 bp, gli2b17ins; 130 + 123 bp).
Live fluorescence imaging of sp7 expression in the cleithrum
The whole body EGFP expression patterns of sp7:egfp; gli3+/+, sp7:egfp; gli314ins/14ins, sp7:egfp; gli2b17ins/17ins; gli314ins/14ins embryos were photographed at 72 hpf under Leica M205. The representative images are shown in Supplementary Fig. 8. n = 8 for each genotype.
Chemical inhibitor treatment of zebrafish embryos
Zebrafish embryos were cultured in 40 ml of E3 medium (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4) containing Smoothened inhibitor (BMS-833923, Selleckchem) or Activin receptor type 1 inhibitor (LDN-193189, TOCRIS) in 10 cm plastic dishes from 32 hpf to 72 hpf. The concentration of BMS-833923 was 5 μM, and of LDN-193189 was 10 μM. At 72 hpf, these inhibitor-treated embryos were fixed by 4% PFA and subjected to immunofluorescence staining or HCR. All raw data, cell count results, and replicates are summarized in Supplementary Table 5.
FACS sorting of EGFP-positive cells from CNE14;egfp fish
Approximately 100,000 EGFP-positive cells were collected from CNE14:egfp; gli3+/+ and gli314ins/14ins fish by FACS as previously described51. Briefly, CNE14:egfp; gli3+/+ or gli314ins/14ins fish were crossed to wildtype fish or gli314ins/14ins fish, respectively, and fertilized eggs were collected. The eggs were raised to 55 hpf at 27.5 °C. Approximately 200 embryos from each genotype were separately dissociated into single cells. The yolk was removed by de-yolk buffer106 (55 mM NaCl, 1.8 mM KCl, 1.25 mM NaHCO3), and embryos were rinsed by PBS. Then, embryos were dissociated into single cells by 0.25% trypsin-EDTA and 4 mg/ml Collagenase with mechanical stress of pipetting at 30 °C. Trypsin was inactivated by adding DMEM-10%FBS medium, and dissociated cells were collected by centrifugation. The cells were resuspended in DMEM-10%FBS and strained by a 40 μm filter (Falcon). Purified single cell suspensions were subject to FACS at Rutgers Flow Cytometry Core Facility (http://rwjms1.rwjms.rutgers.edu/flow/).
RNA-sequencing
Total RNA was immediately extracted from 100,000 EGFP-positive cells isolated by FACS using Trizol (Invitrogen). Briefly, the cell suspension was mixed with 1 ml of Trizol by vigorous vortexing and kept for 5 min at room temperature. The mixed solution was centrifuged, and the supernatant was transferred to a new tube. Chloroform (0.2 ml) was added, vigorously vortexed, and centrifuged for 15 min. The supernatant was mixed with 0.5 ml of isopropanol, kept for 10 min at room temperature, and then centrifuged for 15 minutes. The precipitated RNA was washed by 70% ethanol and reconstituted in 30 μL water. The RNA samples were submitted to Novogene (CA, USA), converted to sequencing library, and sequenced.
ATAC-sequencing
ATAC-sequencing was performed according to the previously described protocol52. Approximately 100,000 EGFP cells isolated by FACS were rinsed in PBS and resuspended in lysis buffer (10 mM Tris-HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.1% NP40). Cells were kept on ice for 10 minutes and then centrifuged at 500 g for 10 min at 4 °C. The pellets were resuspended in the reaction buffer (25 μL TD Buffer (Illumina Cat #FC-121-1030), 2.5 μL Tn5 Transposase (Illumina Cat #FC-121-1030), and 22.5 μL H2O). Cells were kept at 37 °C for 30 minutes and then purified by Qiagen MinElute Kit. The DNA was eluted with 10ul of Elution buffer. Open chromatin regions flanked by the adapter sequence were amplified with 14 cycles by the thermal cycler with Illumina/Nextera i5 common adapter and i7 index adapters. The amplified sequencing library was submitted to BGI and sequencing was performed by the paired-end, 100 bp reading.
Sequencing data analysis
The quality of RNA-sequencing data (FASTQ format) was analyzed by FastQC107. Then, after the adapter sequencing primers were removed by Trimmomatic108, the data were mapped on zebrafish genome (GRcz10) by HISAT2109. The read numbers for genes were counted by HTSeq110, and differentially expressed genes were determined by EdgeR111 in R (R 4.2.2). Following differential gene expression analysis, a heatmap and volcano plot were generated using the “Heatplus” and “EnhancedVolcano” packages in R, respectively.
ATAC-Seq analysis was carried out through multiple programs in Linux. FastQC was used to get a raw quality read of the data, then low-quality reads and sequencing adapters were removed by Trimmomatic. HISAT2 mapped these reads on the zebrafish genome (GRCz10). PCR duplicates were removed with Picard (http://broadinstitute.github.io/picard/), and MACS2 was used to call peaks of open chromatin regions112. HOMER was used to identify Gli3 binding regions in ATAC-seq peaks and provide their gene name/specific chromosomal location113.
Phylogeny and synteny analysis
Activin receptor type 1 gene family members across vertebrate lineages were identified via Blast searches in the Ensembl and NCBI genome databases using sequences from lineage-representative species as queries (e.g., spotted gar for ray-finned fishes). Survey results and accession numbers are listed in Supplementary Table 8. Protein sequences were aligned using MAFFT v7.490114. A Maximum Likelihood phylogeny from lineage-representative species was generated with PhyML v3.3.20180621115 (LG model, 100 bootstrap replicates). Synteny conservation around activin receptor genes was analyzed with the Genomicus Browser version 04.02116.
CT scanning and quantitative analysis
To identify adult zebrafish bone phenotypes, three-month old fish (wildtype, gli3, and gli;Shh compound mutant lines, the total body length spanning from 2.7 to 3.3 cm) were fixed with 10% formalin overnight, stained by 0.5% phosphomolybdic acid for a week followed by water rinse several times, and micro-CT scanned at The University of Chicago (PaleoCT: https://luolab.uchicago.edu/paleoct/). Voxel size varied from 5.18 to 8.26 μm depending on the distance between the specimens and the beam directional tube. The pectoral girdle bones were manually segmented, reconstructed into 3D, and visualized using Amira 2019.4 (Fisher).
To quantify the size and shape of the pectoral girdle, twenty landmarks were used (see Supplementary Fig. 13). The dorsal, anterior, and posterior extremities of the cleithrum were defined as A, G, and C, respectively. The attachment position of the scapula to the cleithrum was defined as D. The length between the dorsal and posteroventral extremities (A-D) was defined as the cleithrum height. The anterior-posterior cleithrum length was measured by extending the horizontal line anteriorly from C (B-C). The anterior-posterior coracoid length was determined by the anterior and posterior extremities of the coracoid (H-I). Following, a perpendicular line to the H-I line was drawn to maximize its length (J-K) as a coracoid height. The scapula height was determined by the dorsal and ventral extremities of the scapula (D-L). To measure the scapula width, a perpendicular line to the D-L was drawn at its center position (E-F). From the posterior view, a straight line was drawn between the dorsal and ventral edges of the glenoid fossa (M and N). Then, a perpendicular line from the M-N line to the bottom of the glenoid fossa was drawn to maximize its length (O-P; the glenoid fossa depth). The mesocoracoid height was determined by the dorsal and ventral extremities of the mesocoracoid from the posterior view (R-Q). An orthogonal line to the R-Q line at its center position was drawn as the mesocoracoid width (S-T). Each length was measured in Amira and standardized by the length of the skull height. The raw measurements are in Supplementary Data 3. The volumes of the cleithrum, coracoid, and scapula of wildtype, gli314ins/14ins, gli314ins/14ins; shha30% fish were digitally measured in Amira and summarized in Supplementary Data 4. Replicates of CT analysis represent 3, 5, and 2 individuals for wildtype, gli3, and gli3;shh compound mutant fishes, respectively. The telencephalon length along the anterior-posteior axis was measured and standardized by the length from the most anterior tip of the head to the first vertebrate in wildtype and gli314ins/14ins fish (three replicates each).
Immunofluorescence staining and signal quantification
Immunofluorescence staining of EGFP and phosphorylated Smad 1/5/8 was conducted following the previously described protocol53. After EGFP or phosphorylated Smad 1/5/8 was stained, the plasma membrane and nucli were stained by CellMask (1/1000 dilution, Invitrogen) and DAPI (1/4000 dilution) in PBS containing 0.1% TritonX-100. EGFP antibody (Abcam #ab290) and phosphorylated Smad 1/5/8 antibody (Cell Signaling #13820) were used at 1/1000 and 1/100 dilution, respectively. For the detection of the first antibody, anti-mouse Alexa 488 (Invitrogen #A21206) was used at 1/1000 dilution. The stained pectoral fin and girdles were photographed by a confocal microscope (LSM510). After photographing of the staining samples, the signal intensity of phosphorylated Smad 1/5/8 was quantified using ImageJ. The cleithrum region is manually identified as Region Of Interest and the sinal intensity/are size was measured by ImageJ function.
Gross observation and whole-mount bone and cartilage staining
Gross morphology observation of adult gli314ins/14ins, gli2b17ins/17ins, gli2b17ins/17ins; gli314ins/14ins, and gli314ins/14ins; shha30% fish were conducted under Leica M205 steromicroscopy. The replicate number of each genotype is 159, 21, 9, and 3, respectively. The total body length of these fish were between 2.8 to 3.4 cm. The fish used for gross observation was subjected to whole mount skeletal staining. Whole-mount skeletal staining of adult zebrafish with Alizarin red and Alcian blue was conducted as follows. The specimens were anesthetized and fixed by 10% neutral-buffered formalin at room temperature overnight. After rinsing with water, their entrails were manually removed, and the remaining body was immersed in 70% ethanol. The soluion was replaced by 70% ethanol / 30% acetic acid with 0.02% of Alcian Blue 8GX (Sigma-Aldrich A5268), and the samples were incubated at room temperature overnight. The specimens were rinsed by a series of ethanol/water mix solutions (75%, 50%, 25%, and 0% ethanol) for an hour each. Then they were immersed in 30% saturated sodium borate for an hour. The specimens were treated by 0.25% trypsin for five hours and washed by 1% potassium hydroxide solution twice. The solution was replaced by 0.005% Alizarin red S (Sigma-Aldrich A5533) /1% potassium hydroxide, and incubated at room temperature overnight. The stained specimens were bleached in 25% glycerol/75% 0.1% potassium hydroxide/0.15% hyrdoxidase solution for five hours. The specimens were soaked through a series of 0.1% potassium hydroxide/glycerol solutions (75%, 50%, 25%, and 0% potassium hydroxide) and preserved in 100% glycerol solution. The replicate number of adult gli314ins/14ins, gli2b17ins/17ins, gli2b17ins/17ins; gli314ins/14ins, and gli314ins/14ins; shha30% fish used for skeletal staining were 7, 8, 5, and 3, respectively.
For juveniles at 13-30 dpf, to avoid acid demineralization of bones, we conducted acid-free skeletal staining as previously published117. Briefly, we prepared two stock solutions. One is 0.02% alcian blue, 60 mM (13 dpf) or 120 mM (30 dpf) MgCl2, and 70% ethanol, and the other is 0.5% alizarin red S powder dissolved in water. We mixed these Alcian blue and alizarine red solutions at 100:1 ratio. Then, juveniles fixed by 4% PFA for two hours at room temperature were stained in this solution overnight. Next day, the juveniles were bleached in 1.5% H2O2 and 1% KOH solution and moved to 50 % glycerol solution. The juvenile sizes for gross observation and skeletal staining are shown in the Supplementary Fig. legends. Replicate number for each genotype at each stage varies from 2 to 8. The number of obtained juveniles is summarized in Supplementary Table 2.
A quantification for the dorsal shift of the pectoral girdle
The dorsoventral body height at the pectoral position was measured with wildtype and gli314ins/14ins fish. The vertical length between the proximal end of the first fin ray and the ventral edge of the body was also measured and standardized by the dorsoventral body height.
Skeletal preparation of the spotted gar pectoral girdle bone
An adult, wild-caught spotted gar (Lepisosteus oculatus, total length was 61 cm) was euthanized with overdose of MS-222 (Tricaine) and then boiled for 10 min in water. The pectoral girdle complex was dissected out, and muscles and connective tissues were manually removed by tweezers. The cleaned pectoral girdle bones were photographed by Canon EOS R6 camera.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Raw sequence and processed BAM files of RNA-seq and ATAC-seq are available at the National Center for Biotechnology Information Sequence Read Archives (NCBI SRA), www.ncbi.nlm.nih.gov/sra (BioProject accession code no PRJNA767802). Source data are provided with this paper.
References
Clack, J. A. The fish-tetrapod transition: new fossils and interpretations. Evolut. Educ. Outreach 2, 213–223 (2009).
Hirasawa, T. & Kuratani, S. Evolution of the vertebrate skeleton: morphology, embryology, and development. Zool. Lett. 1, 2 (2015).
Wood, T. W. P. & Nakamura, T. Problems in fish-to-tetrapod transition: genetic expeditions into old specimens. Front. Cell Dev. Biol. 6, 70–70 (2018).
Goodrich, E. S. Studies on the structure & development of vertebrates. (MacMillan and Co., Limited, 1930).
Romer, A. S. The Vertebrate Body. p. 643 (W. B. Saunders Company, Philadelphia, 1949).
Shubin, N. H., Daeschler, E. B. & Jenkins, F. A. The pectoral fin of Tiktaalik roseae and the origin of the tetrapod limb. Nature 440, 764–771 (2006).
Coates, M. I. The Devonian tetrapod Acanthostega gunnari Jarvik: postcranial anatomy, basal tetrapod interrelationships and patterns of skeletal evolution. Trans. R. Soc. Edinb. Earth Sci. 87, 363–421 (1996).
Ahlberg, P. E., Clack, J. A. & Blom, H. The axial skeleton of the Devonian tetrapod Ichthyostega. Nature 437, 137–140 (2005).
Daeschler, E. B., Shubin, N. H. & Jenkins, F. A. A Devonian tetrapod-like fish and the evolution of the tetrapod body plan. Nature 440, 757–763 (2006).
Smith, M. M. & Hall, B. K. Development and evolutionary origins of vertebrate skeletogenic and odontogenic tissues. Biol. Rev. Camb. Philos. Soc. 65, 277–373 (1990).
Hawkins, M. B., Henke, K. & Harris, M. P. Latent developmental potential to form limb-like skeletal structures in zebrafish. Cell 184, https://doi.org/10.1016/j.cell.2021.01.003 (2021).
Nakamura, T., Gehrke, A. R., Lemberg, J., Szymaszek, J. & Shubin, N. H. Digits and fin rays share common developmental histories. Nature 8, 225–228 (2016).
Shimada, A. et al. Trunk exoskeleton in teleosts is mesodermal in origin. Nat. Commun. 4, 1639–1639 (2013).
Nagashima, H. et al. Developmental origin of the clavicle, and its implications for the evolution of the neck and the paired appendages in vertebrates. J. Anat. 229, 536–548 (2016).
McGonnell, I. M., McKay, I. J. & Graham, A. A population of caudally migrating cranial neural crest cells: functional and evolutionary implications. Dev. Biol. 236, 354–363 (2001).
Chevallier, A. Origines des ceintures scapulaires et pelviennes chez l’embryon d’oiseau. J. Exp. Morphol. Embryol. 42, 275–292 (1997).
Heude, E. et al. Unique morphogenetic signatures define mammalian neck muscles and associated connective tissues. Elife 7, https://doi.org/10.7554/eLife.40179 (2018).
Adachi, N., Bilio, M., Baldini, A. & Kelly, R. G. Cardiopharyngeal mesoderm origins of musculoskeletal and connective tissues in the mammalian pharynx. Development 147, https://doi.org/10.1242/dev.185256 (2020).
Huang, R., Zhi, Q., Patel, K., Wilting, J. & Christ, B. Dual origin and segmental organisation of the avian scapula. Development 127, 3789–3794 (2000).
Piekarski, N. & Olsson, L. A somitic contribution to the pectoral girdle in the axolotl revealed by long-term fate mapping. Evolut. Dev. 13, 47–57 (2011).
Durland, J. L., Sferlazzo, M., Logan, M. & Burke, A. C. Visualizing the lateral somitic frontier in the Prx1Cre transgenic mouse. J. Anat. 212, 590–602 (2008).
Valasek, P. et al. Somitic origin of the medial border of the mammalian scapula and its homology to the avian scapula blade. J. Anat. 216, 482–488 (2010).
Shearman, R. M., Tulenko, F. J. & Burke, A. C. 3D reconstructions of quail-chick chimeras provide a new fate map of the avian scapula. Dev. Biol. 355, 1–11 (2011).
Burke, A. C. The development and evolution of the turtle body plan: inferring intrinsic aspects of the evolutionary process from experimental embryology. Am. Zool. 31, 616–627 (1991).
Matsuoka, T. et al. Neural crest origins of the neck and shoulder. Nature 436, 347–355 (2005).
Epperlein, H.-H., Khattak, S., Knapp, D., Tanaka, E. M. & Malashichev, Y. B. Neural crest does not contribute to the neck and shoulder in the axolotl (Ambystoma mexicanum). PloS one 7, e52244–e52244 (2012).
Kuroda, S., Lalonde, R. L., Mansour, T. A., Mosimann, C. & Nakamura, T. Multiple embryonic sources converge to form the pectoral girdle skeleton in zebrafish. Nat. Commun. 15, 6313 (2024).
Chakrabarty, P. et al. Phylogenomic systematics of ostariophysan fishes: ultraconserved elements support the surprising non-monophyly of characiformes. Syst. Biol. 66, 881–895 (2017).
Andrews, S. M. & Westoll, T. S. IX.—The postcranial skeleton of Ensthenopteron foordi Whiteaves. Earth Environ. Sci. Trans. R. Soc. Edinb. 68, 391–489 (1970).
Mansuit, R. et al. Development and growth of the pectoral girdle and fin skeleton in the extant coelacanth Latimeria chalumnae. J. Anat. 236, 493–509 (2020).
Desvignes, T., Carey, A., Braasch, I., Enright, T. & Postlethwait, J. H. Skeletal development in the heterocercal caudal fin of spotted gar (lepisosteus oculatus) and other lepisosteiformes. Dev. Dyn. 247, 724–740 (2018).
Hall, B. K. Evolutionary Developmental Biology, Second edition, Chapter 16, 255–279. (Springer Netherlands, 1999).
Hall, B. K. Fins into limbs: evolution, development, and transformation, Part 2, 6, 79–92. (University of Chicago Press, 2007).
Witten, P. E. & Villwock, W. Growth requires bone resorption at particular skeletal elements in a teleost fish with acellular bone (Oreochromis niloticus, Teleostei: Cichlidae). J. Appl. Ichthyol. 13, 149–158 (1997).
Weitzman, S. H. & Fink, W. L. Relationships of the neon tetras, a group of South American freshwater fishes (Teleostei, Characidae), with comments on the phylogeny of New World characiforms. Bull. Mus. Comp. Zool. Harv. Coll. 150, 339–395 (1983).
Weitzman, S. H. & Vari, R. P. Two new species and a new genus of miniature characid fishes (Teleostei: Characiformes) from Northern South America. Proc. Biol. Soc. Wash. 100, 640–652 (1987).
Roberts, T. R. Danionella-Translucida, a new genus and species of cyprinid fish from Burma, one of the smallest living vertebrates. Environ. Biol. Fishes 16, 231–241 (1986).
Huysseune, A. The Laboratory Fish, Handbook of Experimental Animals, Chapter 18–Skeletal System, 307–317. (Academic Press, 2000).
Lleras-Forero, L., Winkler, C. & Schulte-Merker, S. Zebrafish and medaka as models for biomedical research of bone diseases. Dev. Biol. 457, 191–205 (2020).
Capellini, T. D. et al. Scapula development is governed by genetic interactions of Pbx1 with its family members and with Emx2 via their cooperative control of Alx1. Development 137, 2559–2569 (2010).
Kuijper, S. et al. Genetics of shoulder girdle formation: roles of Tbx15 and aristaless-like genes. Development 132, 1601–1610 (2005).
Aubin, J., Lemieux, M., Moreau, J., Lapointe, J. & Jeannotte, L. Cooperation of Hoxa5 and Pax1 genes during formation of the pectoral girdle. Dev. Biol. 244, 96–113 (2002).
Timmons, P. M., Wallin, J., Rigby, P. W. & Balling, R. Expression and function of Pax 1 during development of the pectoral girdle. Development 120, 2773–2785 (1994).
Rallis, C. et al. Tbx5 is required for forelimb bud formation and continued outgrowth. Development 130, 2741–2751 (2003).
Ahn, D.-g, Kourakis, M. J., Rohde, L. A., Silver, L. M. & Ho, R. K. T-box gene tbx5 is essential for formation of the pectoral limb bud. Nature 417, 754–758 (2002).
Heude, É., Shaikho, S. & Ekker, M. The dlx5a/dlx6a genes play essential roles in the early development of zebrafish median fin and pectoral structures. PLoS ONE 9, e98505–e98505 (2014).
Grandel, H. et al. Retinoic acid signalling in the zebrafish embryo is necessary during pre-segmentation stages to pattern the anterior-posterior axis of the CNS and to induce a pectoral fin bud. Development 129, 2851–2865 (2002).
Farmer, D. T., Patel, P., Choi, R., Liu, C. Y. & Crump, J. G. A comprehensive series of Irx cluster mutants reveals diverse roles in facial cartilage development. Development 148, dev197244 (2021).
Ingham, P. W. & McMahon, A. P. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 15, 3059–3087 (2001).
Hui, C.-c & Angers, S. Gli proteins in development and disease. Annu. Rev. Cell Dev. Biol. 27, 513–537 (2011).
Matissek, S. J. & Elsawa, S. F. GLI3: a mediator of genetic diseases, development and cancer. Cell Commun. Signal. 18, 1–20 (2020).
Vortkamp, A., Gessler, M. & Grzeschik, K. H. GLI3 zinc-finger gene interrupted by translocations in Greig syndrome families. Nature 352, 539–540 (1991).
Biesecker, L. G. in GeneReviews((R)) (eds M. P. Adam et al.) (1993).
Wang, B., Fallon, J. F. & Beachy, P. A. Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell 100, 423–434 (2000).
Litingtung, Y., Dahn, R. D., Li, Y., Fallon, J. F. & Chiang, C. Shh and Gli3 are dispensable for limb skeleton formation but regulate digit number and identity. Nature 418, 979–983 (2002).
Riddle, R. D., Johnson, R. L., Laufer, E. & Tabin, C. Sonic hedgehog mediates the polarizing activity of the ZPA. Cell 75, 1401–1416 (1993).
Sheth, R. et al. Hox genes regulate digit patterning by controlling the wavelength of a Turing-type mechanism. Science 338, 1476–1480 (2012).
Te Welscher, P. et al. Progression of vertebrate limb development through SHH-mediated counteraction of GLI3. Science 298, 827–830 (2002).
Letelier, J. et al. The Shh/Gli3 gene regulatory network precedes the origin of paired fins and reveals the deep homology between distal fins and digits. Proc. Natl. Acad. Sci. USA 118, https://doi.org/10.1073/pnas.2100575118 (2021).
Pasquier, J. et al. Evolution of gene expression after whole-genome duplication: new insights from the spotted gar genome. J. Exp. Zool. B Mol. Dev. Evol. 328, 709–721 (2017).
Neumann, C. J., Grandel, H., Gaffield, W., Schulte-Merker, S. & Nusslein-Volhard, C. Transient establishment of anteroposterior polarity in the zebrafish pectoral fin bud in the absence of sonic hedgehog activity. Development 126, 4817–4826 (1999).
Letelier, J. et al. A conserved Shh cis-regulatory module highlights a common developmental origin of unpaired and paired fins. Nat. Genet. 50, 504–509 (2018).
te Welscher, P., Fernandez-Teran, M., Ros, M. A. & Zeller, R. Mutual genetic antagonism involving GLI3 and dHAND prepatterns the vertebrate limb bud mesenchyme prior to SHH signaling. Genes Dev. 16, 421–426 (2002).
Buscher, D., Bosse, B., Heymer, J. & Ruther, U. Evidence for genetic control of Sonic hedgehog by Gli3 in mouse limb development. Mech. Dev. 62, 175–182 (1997).
Hui, C. C., Slusarski, D., Platt, K. A., Holmgren, R. & Joyner, A. L. Expression of three mouse homologs of the Drosophila segment polarity gene cubitus interruptus, Gli, Gli-2, and Gli-3, in ectoderm- and mesoderm-derived tissues suggests multiple roles during postimplantation development. Dev. Biol. 162, 402–413 (1994).
Venkatesh, B. et al. Elephant shark genome provides unique insights into gnathostome evolution. Nature 505, 174–179 (2014). 2014 505:7482.
Nakashima, K. et al. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell 108, 17–29 (2002).
Spranger, J., Winterpacht, A. & Zabel, B. The type II collagenopathies: a spectrum of chondrodysplasias. Eur. J. Pediatr. 153, 56–65 (1994).
Kague, E. et al. Osterix/Sp7 limits cranial bone initiation sites and is required for formation of sutures. Dev. Biol. 413, 160–172 (2016).
Askary, A. et al. Iroquois proteins promote skeletal joint formation by maintaining chondrocytes in an immature state. Dev. Cell 35, 358–365 (2015).
Naruse, I. et al. Birth defects caused by mutations in human GLI3 and mouse Gli3 genes. Congenit. Anom. 50, 1–7 (2010).
Letelier, J. et al. The Shh/Gli3 gene regulatory network precedes the origin of paired fins and reveals the deep homology between distal fins and digits. Proc Natl. Acad. Sci. USA 118, e2100575118 (2021).
Tanaka, Y. et al. K. Anterior-posterior constraint on Hedgehog signaling by hhip in teleost fin elaboration. Development 151, dev202526 (2024).
Almuraikhi, N. et al. Hedgehog signaling inhibition by smoothened antagonist BMS-833923 reduces osteoblast differentiation and ectopic bone formation of human skeletal (mesenchymal) stem cells. Stem Cells Int. 2019, https://doi.org/10.1155/2019/3435901 (2019).
Vokes, S. A., Ji, H., Wong, W. H. & McMahon, A. P. A genome-scale analysis of the cis-regulatory circuitry underlying sonic hedgehog-mediated patterning of the mammalian limb. Genes Dev. 22, 2651–2663 (2008).
Mullins, M. C. et al. Genes establishing dorsoventral pattern formation in the zebrafish embryo: the ventral specifying genes. Development 123, 81–93 (1996).
ten Dijke, P. et al. Identification of type I receptors for osteogenic protein-1 and bone morphogenetic protein-4. J. Biol. Chem. 269, 16985–16988 (1994).
Lavery, K., Swain, P., Falb, D. & Alaoui-Ismaili, M. H. BMP-2/4 and BMP-6/7 differentially utilize cell surface receptors to induce osteoblastic differentiation of human bone marrow-derived mesenchymal stem cells. J. Biol. Chem. 283, 20948–20958 (2008).
Fujii, M. et al. Roles of bone morphogenetic protein type I receptors and Smad proteins in osteoblast and chondroblast differentiation. Mol. Biol. Cell 10, 3801–3813 (1999).
Zhang, D. et al. ALK2 functions as a BMP type I receptor and induces Indian hedgehog in chondrocytes during skeletal development. J. Bone Min. Res 18, 1593–1604 (2003).
Franke, M. et al. CTCF knockout in zebrafish induces alterations in regulatory landscapes and developmental gene expression. Nat. Commun. 12, 5415 (2021).
Yu, P. B. et al. BMP type I receptor inhibition reduces heterotopic ossification. Nat. Med. 14, 1363–1369 (2008).
Grande, L. An empirical synthetic pattern study of gars (Lepisosteiformes) and closely related species, based mostly on skeletal anatomy: the resurrection of Holostei. (American Society of Ichthyologists and Herpetologists (ASIH), 2010).
Grandel, H. & Schulte-Merker, S. The development of the paired fins in the zebrafish (Danio rerio). Mech. Dev. 79, 99–120 (1998).
Lebedev, O. A. & Coates, M. I. The postcranial skeleton of the Devonian tetrapod Tulerpeton curtum Lebedev. Zool. J. Linn. Soc. 114, 307–348 (1995).
Johanson, Z., Joss, J. M. P. & Wood, D. The scapulocoracoid of the Queensland lungfish Neoceratodus forsteri (Dipnoi: Sarcopterygii): morphology, development and evolutionary implications for bony fishes (Osteichthyes). Zoology 107, 93–109 (2004).
Romer, A. S. Pectoral limb musculature and shouldergirdler structure in fish and tetrapods. Anat. Rec. 27, 119–143 (1924).
Grandel, H., Draper, B. W. & Schulte-Merker, S. Dackel acts in the ectoderm of the zebrafish pectoral fin bud to maintain AER signaling. Development 127, 4169–4178 (2000).
Tanaka, M. Alterations in anterior-posterior patterning and its accompanying changes along the proximal-distal axis during the fin-to-limb transition. genesis 56, e23053–e23053 (2018).
Furutani-Seiki, M. & Wittbrodt, J. Medaka and zebrafish, an evolutionary twin study. Mech. Dev. 121, 629–637 (2004).
El-Brolosy, M. A. et al. Genetic compensation triggered by mutant mRNA degradation. Nature 568, 193–197 (2019).
Bowers, M. et al. Limb anterior-posterior polarity integrates activator and repressor functions of GLI2 as well as GLI3. Dev. Biol. 370, 110–124 (2012).
Dial, K. P., Shubin, N. & Brainerd, E. L. (eds) Great transformations in vertebrate evolution. (University of Chicago Press, Chicago, 2015).
Westoll, T. S., Andrews, S. M., Miles, R. S. & Walker, A. D. Problems in vertebrate evolution: essays presented to Professor T.S. Westoll. (Linnean Society of London by Academic Press, 1977).
Braasch, I. et al. The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons. Nat. Genet. 48, 427–437 (2016).
Hall, B. K. Fins into limbs: evolution, development, and transformation. (University of Chicago Press, 2007).
Hall, B. K. Evolutionary developmental biology. (Springer Science & Business Media, 2012).
Hanken, J. & Wake, D. B. Miniaturization of body-size—organismal consequences and evolutionary significance. Annu. Rev. Ecol. Syst. 24, 501–519 (1993).
Le Pabic, P., Dranow, D. B., Hoyle, D. J. & Schilling, T. F. Zebrafish endochondral growth zones as they relate to human bone size, shape and disease. Front Endocrinol. (Lausanne) 13, 1060187 (2022).
Cubbage, C. C. & Mabee, P. M. Development of the cranium and paired fins in the zebrafish Danio rerio (Ostariophysi, Cyprinidae). J. Morphol. 229, 121–160 (1996).
Tonelli, F. et al. Zebrafish: a resourceful vertebrate model to investigate skeletal disorders. Front. Endocrinol. 11, 489 (2020).
Frazer, K. A., Pachter, L., Poliakov, A., Rubin, E. M. & Dubchak, I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 32, W273–W279 (2004).
Thisse, C. & Thisse, B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3, 59–69 (2007). 2008 3:1.
Lauter, G., Söll, I. & Hauptmann, G. Two-color fluorescent in situ hybridization in the embryonic zebrafish brain using differential detection systems. BMC Dev. Biol. 11, 43 (2011).
Nakayama, T. et al. Simple and efficient CRISPR/Cas9-mediated targeted mutagenesis in Xenopus tropicalis. Genes 51, 835–843 (2013).
McGarvey, A. C. et al. Single-cell-resolved dynamics of chromatin architecture delineate cell and regulatory states in zebrafish embryos. Cell Genom. 2, 100083 (2022).
Andrews, S. FastQC: A quality control tool for high throughput sequence data. (2011).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Goldstein, L. D. et al. Prediction and quantification of splice events from RNA-seq data. PLoS ONE 11, e0156132–e0156132 (2016).
Putri, G. H., Anders, S., Pyl, P. T., Pimanda, J. E. & Zanini, F. Analysing high-throughput sequencing data in Python with HTSeq 2.0. Bioinformatics 38, 2943–2945 (2022).
Kvam, V. M., Liu, P. & Yaqing, S. A comparison of statistical methods for detecting differentially expressed genes from RNA-seq data. Am. J. Bot. 99, 248–256 (2012).
Grytten, I. et al. Graph peak caller: calling chip-seq peaks on graph-based reference genomes. PLoS Comput. Biol. 15, e1006731–e1006731 (2019).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Guindon, S. et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 (2010).
Parey, E. et al. Genome structures resolve the early diversification of teleost fishes. Science 379, 572–575 (2023).
Walker, M. B. & Kimmel, C. B. A two-color acid-free cartilage and bone stain for zebrafish larvae. Biotech. Histochem. 82, 23–28 (2007).
Marletaz, F. et al. The little skate genome and the evolutionary emergence of wing-like fins. Nature 616, 495–503 (2023).
Young, M., Selleri, L. & Capellini, T. D. Genetics of scapula and pelvis development: an evolutionary perspective. Curr. Top. Dev. Biol. 132, 311–349 (2019).
Acknowledgements
We thank A. Gillis for kindly sharing elephant shark tissue. J. Lemberg provided technical assistance for µCT scanning and analysis. We thank N.H. Shubin for providing fish space, A.A. Abbasi for providing the PXIG vector, G. Senevirathne for sharing the HCR protocol, Y.J. Chang and Office of Advanced Research Computing Rutgers for computational assistance, J.J. Tena for visualization of high-throughput sequencing results, D. Remsen, S. Bennett, and staffs at the Marine Resource Center at MBL for skate embryos, J. Talbot for technical advice of live fluorescent imaging, G.Crump and C. Arata for sharing osteoblast and chondrocyte fluorescence transgenic fish, the Bayousphere Lab (A. Ferrara) at Nicholls State University for help with gar sampling, and Brett Racicot (Michigan State University) for gar husbandry. We thank M.B. Hawkins for providing critical comments to the manuscript. This work was performed with Honors College with Johnson & Johnson Women in STEM2D Life Sciences Summer Undergraduate Research Fellowship (to J.W.), The Paul Robeson Scholarship, the Enrico P. Veltri Scholarship for the Life Sciences, Duncan and Nancy MacMillan Award for Research Excellence (to T.W.), the Division of Life Sciences Summer Undergraduate Research Fellowship provided by Macmillan foundation (to A.S.), The INSPIRE program (IRACDA New Jersey/New York for Science Partnerships in Research & Education) (to D.N.), NSF EDGE grant 2029216 (to I.B.), the institutional support provided by the Rutgers University School of Arts and Sciences and the Human Genetics Institute of New Jersey, the National Science Foundation under Grant IOS 2210072, Marine Biological Laboratory Whitman Fellowship (2018), Society for Developmental Biology Innovation Grant (2020), and Busch Biomedical Grant (to T.N.).
Author information
Authors and Affiliations
Contributions
T.W., I.B., and T.N. designed research. K.F. and T.N. created genetically modified fish. J.W., A.S., and T.N. analyzed RNA-sequencing data, ATAC-sequencing data, and conducted chromogenic whole mount in situ hybridization and HCR. A.E., A.A., K.F., D.B., and H.C. genotyped transgenic and knockout fish. T.S. and T.N. conducted CT scan. T.W. created pectoral girdle bone 3D reconstructions from CT scan data. I.B. and O.F. conducted phylogenetic and synteny analyses and spotted gar in situ hybridization. D.G. conducted statistical analyses. S.A. conducted CT analysis of the telencephalon. D.N. conducted in situ hybridization of skate embryos. T.W., J.W., and T.N. analyzed the data. S.K., T.S., I.B., and T.N. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Sarah K McMenamin and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wei, J., Wood, T.W.P., Flaherty, K. et al. Distinct ossification trade-offs illuminate the shoulder girdle reconfiguration at the water-to-land transition. Nat Commun 16, 4983 (2025). https://doi.org/10.1038/s41467-025-60236-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-60236-z
This article is cited by
-
Ossicle inferior angle of the scapula: a palpable Albeit normal entity
Surgical and Radiologic Anatomy (2025)