Abstract
Therapeutic monoclonal antibodies can prevent severe disease in SARS-CoV-2 exposed individuals. However, currently circulating virus variants have evolved to gain significant resistance to nearly all neutralizing human immune system-derived therapeutic monoclonal antibodies that had previously been emergency-authorized for use in the clinic. Here, we describe the discovery of a panel of single-domain antibodies (VHHs) directed against the spike protein S2 subunit that broadly neutralize SARS-CoV-1 and −2 with unusually high potency. One of these VHHs tightly clamps the spike’s monomers at a highly conserved, quaternary epitope in the membrane proximal part of the trimeric Heptad Repeat 2 (HR2) coiled-coil, thereby locking the HR2 in its prefusion conformation. Low dose systemic administration of a VHH-human IgG1 Fc fusion prevented SARS-CoV-2 infection in two animal models. Pseudovirus escape selection experiments demonstrate that the very rare escape variants are rendered almost non-infectious. This VHH-based antibody with a highly potent mechanism of antiviral action forms the basis for a new class of pan-sarbecovirus neutralizing biologics, which are currently under development. In addition, the unique quaternary binding mode of the VHHs to the prefusion HR2 could be exploited for other class I fusion proteins.
Similar content being viewed by others
Introduction
The spike of SARS-CoV-2 is a major target of neutralizing antibodies. This class I fusion protein consists of a membrane distal S1 and a membrane proximal S2 subunit. The S1 subunit comprises the receptor-binding domain (RBD) which is the target of the majority of the SARS-CoV-2-neutralizing antibodies1. The RBD is also immunodominant and tolerates mutations that result in SARS-CoV-2 immune escape2. As a result of this antigenic evolution, nearly all of the currently emergency use approved monoclonal antibodies fail to neutralize the recently circulating SARS-CoV-2 variants, i.e. BA.4, BA.5, BQ.1.1, XBB, BA.2.86.1 and JN.13,4. The S2 subunit exerts membrane fusion, a process that involves major conformational changes in the spike, which ultimately result in the formation of a so-called six-helix bundle (6HB) between heptad repeat 1 (HR1) and heptad repeat 2 (HR2)5. The S2 subunit is more conserved than S1 and, therefore, considered an attractive target for the development of neutralizing antibodies with broad anti-sarbecovirus potential. Several monoclonal antibodies that recognize sites in the S2 subunit, such as the stem helix or the fusion peptide of SARS coronaviruses, have been described6,7,8,9,10. In general, however, S2-specific monoclonal antibodies have poor virus neutralizing activity. Single-domain antibodies (VHHs, also known as nanobodies) directed against the SARS-CoV-2 S2 subunit have also been reported, which, even as Fc-fusions, have very low SARS-CoV-2-neutralizing activity11,12. Here we report a family of VHHs that can potently neutralize SARS-CoV-1 and −2, including past and currently circulating SARS-CoV-2 variants, by binding to a highly conserved membrane proximal region in the HR2 coiled-coil. These VHHs uniquely bind to a quaternary epitope, locking the HR2 coiled-coil in its prefusion conformation, thereby blocking virus entry.
Results
Isolation of SARS-CoV-1 and −2 S2 subunit-specific VHHs
We generated a VHH phage display library from a llama that was immunized with recombinant spike proteins of SARS-CoV-1, MERS-CoV and SARS-CoV-2 (Wuhan sequence with prefusion-stabilizing K986P and V987P mutations and RRAR-GSAS mutations to inactivate the furin cleavage site, referred to as S-2P) and performed biopanning against immobilized SARS-CoV-2 S-2P in the presence of an excess of SARS-CoV-2 RBD in solution to isolate VHHs that target the spike protein outside of the RBD. After 3 (R3) or 4 (R4) rounds of bio-panning, phagemid clones were selected for small scale VHH production in E. coli, binding to coronavirus spike proteins, as well as recombinant RBD and S2 subunits, and neutralization of VSV pseudotyped with the SARS-CoV-2 spike (D614G) protein (Fig. 1a). VHHs that recognized the S2 subunit of SARS-CoV-2 and displayed virus neutralizing activity clustered into a single family (Fig. 1b, Supplementary Fig. 1A). Ten VHHs from this family were selected for purification and further analysis. The VHHs bound the S2 subunit of the SARS-CoV-2 spike (Wuhan) and the spikes of all SARS-CoV-2 variants that we tested, of SARS-CoV-1 but not of MERS-CoV (Fig. 1c, Supplementary Fig. 1B). The selected VHHs neutralized SARS-CoV-1 and a broad range of SARS-CoV-2 variant VSV pseudotypes with unexpectedly high potency, with IC50 values as low as 1.1 ng/mL (R3DC23) (Fig. 1d, Supplementary Fig. 1C–F). R3DC23 and R4DC6 also neutralized authentic SARS-CoV-2 viruses with remarkably strong potency (Fig. 1e). Bio-Layer Interferometry (BLI) analysis revealed that the most potent VHH, R3DC23, binds immobilized spike trimers with favorable kinetics, while the poor fitting of the association sensorgram using a 1:1 model suggested that the spike trimer is bound by multiple VHHs (Fig. 1f).
a Screen of E. coli periplasmic extracts (PE) of VHH clones isolated after 3 (R3) or 4 (R4) rounds of bio-panning on SARS-CoV-2 spike protein for binding to the indicated recombinant proteins (blue heat map) and for neutralization of VSV particles pseudotyped with SARS-CoV-2 spikes (red heat map). b Amino acid sequences alignment of S2 binding, neutralizing VHHs. Numbering according to Kabat. The boxes indicate CDR1, CDR2 and CDR3 as defined by IMGT. The amino acids indicated in red are part of the paratope. c Binding of VHHs to HEK293T cells expressing the spike protein of SARS-CoV-2 D614G, BA.1, BA.2, BA.5, BQ.1.1, MERS or transfected with empty vector (EV). S309 was used as a positive control and GBP, a GFP-binding VHH, as a negative control. d Neutralization of VSV pseudotyped with spike protein of the indicated SARS-CoV-2 variants and SARS-CoV-1 (SC-1) by R3DC23. The graph shows the mean (line) and individual (dots) IC50 values calculated from at least 2 independent neutralization assays. e Neutralization of authentic SARS-CoV-2 D614G and BA.1 virus by S2-binding VHHs. The graphs show the mean ± S.D. (N = 2) number of counted plaques for each VHH dilution. S309 was used as positive control. f Binding kinetics of R3DC23 to S-2P trimer immobilized on a BLI biosensor. *Due to the poor fitting for the association phase, the calculated KD, app and kon, app might not be accurate. Source data are provided as a source data file.
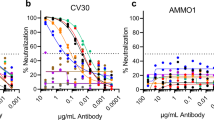
With clinical development in mind, we humanized the framework regions of R3DC23 and two related VHHs (R3C4 and R4DC20) and fused these VHHs to a human IgG1-Fc_YTE to create bivalency and extend the half-life in circulation (Supplementary Fig. 2A). These Fc-fusions neutralized SARS-CoV-2 D614G and BA.5 even more efficiently than their monovalent formats, with huR3DC23-Fc being the most potent (Fig. 2a, Supplementary Fig. 2B). Humanization of R3DC23 did not affect the neutralizing activity of huR3DC23-Fc and further lowered its already favorable hydrophobicity (Supplementary Fig. 2C, D). huR3DC23-Fc could neutralize VSV pseudotypes displaying the spike protein of SARS-CoV-2 BA.1, BA.2, BA.2.75, BA.5, BA.4.6, BQ.1.1, XBB, BA.2.86, EG.5.1, KP.3, KP.2.3, KP.1.1, LB.1 and XEC with IC50 values close to or below 1 ng/mL and potently neutralized authentic viruses (Fig. 2b, c, Supplementary Fig. 2E, F). To test if huR3DC23-Fc can protect in vivo, K18-hACE2 mice were treated with huR3DC23-Fc or control IgG1 antibody (palivizumab) prior to challenge with SARS-CoV-2. In contrast to palivizumab-treated K18-hACE2 mice, huR3DC23-Fc protected the challenged K18-hACE2 mice from body weight loss and death (Fig. 2d). In addition, huR3DC23-Fc treatment strongly controlled viral replication in challenged K18-hACE2 mice and reduced recruitment of monocytes to the lungs but enabled the presence of eosinophils, cell types which are considered to promote and control pulmonary disease, respectively (Fig. 2e, f, Supplementary Fig. 2G)13,14. We also fused humanized R3DC23 to an Fc containing the half-life-extending LS substitutions15. Similar to huR3DC23-Fc, huR3DC23-Fc_LS displayed low apparent hydrophobicity and potently neutralized authentic SARS-CoV-2 variants and pseudotyped VSV (Fig. 2g, Supplementary Table S3, Supplementary Fig. 2H, I). We evaluated the therapeutic potential of huR3DC23-Fc_LS, administered shortly after exposure, in the Syrian Golden hamster model. Hamsters were challenged with SARS-CoV-2 and, 4 h later, treated with either 10 or 2 mg/kg huR3DC23-Fc_LS, 10 mg/kg bebtelovimab (biosimilar; positive control), or 10 mg/kg palivizumab (negative control). At 4 days post infection, high levels of huR3DC23-Fc_LS were detected in the serum of all hamsters treated with 2 mg/kg and in 4 out of 6 hamsters treated with 10 mg/kg. No or very low levels of huR3DC23-Fc_LS were detected in the sera of two animals in the 10 mg/kg huR3DC23-Fc_LS treatment group (Supplementary Fig. 2J). This most likely results from unsuccessful injection, which has been reported by others16. Apart from these two hamsters, the lung virus loads, sampled on day 4 after challenge, were below the detection limit in the huR3DC23-Fc_LS treated hamsters whereas control treated animals had high lung virus loads (Fig. 2h, left panel). In accordance, a strong reduction in viral RNA was observed in the lungs of hamsters treated with either huR3DC23-Fc_LS or bebtelovimab (Fig. 2h, right panel). In addition, the body weight loss was significantly lower in hamsters treated with 10 mg/kg huR3DC23-Fc_LS (Supplementary Fig. 2K). These data show that Fc-fusion of the S2 targeting VHH can strongly restrict SARS-CoV-2 replication in vitro and in vivo.
a Neutralization of VSV pseudotyped with the spike protein of SARS-CoV-2 D614G by Fc-fusions of humanized R3DC23 and related VHHs (mean ± SEM (N = 3) normalized GFP MFI level of infection). b IC50 values (calculated from at least two independent neutralization assays) of neutralization of VSV pseudotyped with spikes of SARS-CoV-2 variants by huR3DC23-Fc. Numbers on top of the graphs: number of assays in which no neutralization was observed for the respective virus. c Neutralization of authentic SARS-CoV-2 D614G and BA.1 virus by huR3DC23-Fc and R3DC23 (mean ± S.D. (N = 4 for huR3DC23-Fc and R3DC23, N = 2 for S309 and N = 1 for palivizumab) number of counted plaques). d Prophylactic treatment with huR3DC23-Fc protects K18-hACE2 mice from lethal SARS-CoV-2 infection. Kaplan–Meier curve of animal survival portion of the indicated groups (left). Mean relative bodyweight of K18-hACE2 mice treated with huR3DC23-Fc (N = 5 ± SEM) or palivizumab (N = 5 ± SEM) and of WT mice treated with palivizumab (N = 5 ± SEM) (right). e, f Prophylactic treatment with huR3DC23-Fc controls viral replication and pulmonary inflammation at 5DPI. e Median and individual viral titer in the lungs of mice (N = 5 in all groups). f Mean ± SEM level (N = 5 in all groups) of the indicated lung-infiltrated leukocytes as percentage of living cells. (*P < 0.05, **P < 0.01). g In vitro neutralization (geometric mean IC50 values) of authentic SARS-CoV-2 variants by huR3DC23-Fc_LS (neutralization curves in Supplementary Fig. 2I). ND: not possible to determine IC50 value, NT: not tested. h Median TCID50/gram (left) and viral RNA copies/gram lung tissue (right) in Syrian golden hamsters (N = 6) after intraperitoneal administration of huR3DC23-Fc_LS 4 h post-infection with SARS-CoV-2. Two animals in the 10 mg/kg group were not exposed to huR3DC23-Fc_LS treatment and were excluded from statistical analysis (#) (Supplementary Fig. 2J). P values are shown in gray above graphs. Statistical tests are specified in the Methods section. Source data are provided as a source data file.
R3DC23 prevents spike-mediated membrane fusion
Enveloped viruses can be neutralized by preventing receptor binding and/or membrane fusion. VHH R3DC23 did not prevent the binding of ACE2-Fc to SARS-CoV-2 Spike-2P (Fig. 3a, Supplementary Fig. 3A). In addition, R3DC23, R3C4 and huR3DC23-Fc did not induce premature shedding of the S1 subunit, another way to interfere with receptor binding, in contrast to CB6 and VHH72-S56A (Fig. 3b and Supplementary Fig. 3B)17,18. R3DC23 or huR3DC23-Fc also failed to prevent ACE2-Fc binding to SARS-CoV-2 spike-expressing cells (Fig. 3c, Supplementary Fig. 3C). These results strongly suggest that our S2-specific VHHs prevent virus entry after attachment.
a Binding of hACE2-muFc to coated recombinant 2P-stabilized spike proteins in the presence of a dilution series of R3DC23, GBP (negative control), VHH72_S56A and CB6 (positive controls), measured in ELISA (mean (N = 2) OD450 signal). b Anti-S1 Western blot analysis of the growth medium (SN) and cell lysates (LYS) of HEK293T cells expressing the SARS-CoV-2 D614G spike protein (HEK S (D614G)) or not (HEK EV) incubated for 30 min with the indicated VHHs, CB6 (positive control), S309 and GBP (negative controls). Arrowheads at the right indicate cell-associated uncleaved spike and the S1 spike subunit. c Binding of hACE2-muFc to cell surface-expressed spike in the presence of R3DC23, VHH72_S56A and GBP (N = 2 samples). Lower dotted line: ACE2-muFc binding to cells that do not express spike, upper dotted line: ACE2-muFc binding to spike-expressing cells in the absence of antibody. d GFP expression of Vero E6 cells that were treated with PBS, GBP, palivizumab (negative controls), R3DC23 or huR3DC23-Fc 4 h after co-transfection of a GFP and a spike expression vector. Scale bar: 250 μm. e Syncytia formation by spike-expressing cells in the presence of R3DC23, GBP or VHH55 (negative control) with live cell imaging. Mean ± SD (N = 3 wells) GFP-positive area of wells is shown. f ACE2-independent spike-mediated syncytia formation, as measured by GFP reconstitution of split GFP. HEK293T cells, co-transfected with a spike and a GFP1-10 expression plasmid, were stimulated with CB6 (20 µg/mL) or PBS, followed by treatment with R3DC23, GBP or S309 and added to HEK293T cells that were transfected with a GFP11 expression plasmid in a 96 well plate. Median (line) and individual (dots) fluorescence intensity (N = 4 wells) is shown. g Syncytia formation of VeroE6/TMPRSS2 cells infected with GFP-expressing replication-competent VSV pseudotyped with SARS-CoV-2 spikes, in the presence of a dilution series of R3DC23, huR3DC23-Fc, GBP or S309. Mean (N = 2) GFP fluorescence of infected wells is shown, images: GFP fluorescence of the indicated samples at 40 h post-infection. Source data are provided as a source data file.
Upon co-transfection of the SARS-CoV-2 spike and GFP, Vero E6 cells form GFP-positive syncytia, the formation of which was completely prevented by R3DC23 added after transfection (Fig. 3d). This was confirmed by live cell imaging in which R3DC23 and huR3DC23-Fc prevented syncytia formation (Fig. 3e, Supplementary Fig. 3D). R3DC23 (and S30919) also blocked ACE2-independent fusion of spike-expressing HEK293T cells on which spike activation by ACE2 binding and subsequent cell-cell fusion was mimicked by a short preincubation with CB620 (Fig. 3f). Finally, addition of R3DC23 4 h after infection with replication-competent GFP-expressing VSV pseudotyped with SARS-CoV-2 spike strongly (IC50 = 9.2 ng/mL) prevented syncytium formation whereas moderate inhibition was observed with S309 (Fig. 3g, Supplementary Fig. 3E)21. We conclude that the S2-targeting VHHs described here can neutralize SARS-CoV-1 and -2 by preventing membrane fusion.
S2-binding VHHs bind to the membrane proximal region of heptad repeat 2
To understand the mechanism by which R3DC23 inhibits spike-mediated membrane fusion and to identify its likely binding region in the S2 subunit, we performed escape virus selection in which R3DC23 was added to VeroE6/TMPRSS2 cells after infection with replication-competent GFP-expressing VSV pseudotyped with SARS-CoV-2 spike (Supplementary Fig. 4A). Nine escape clones from wells in which R3DC23 did not prevent syncytium formation and spread were isolated and their spike coding sequence was determined. Each of these viral clones had acquired a single amino acid substitution at one out of four positions: N1192D (1 clone), L1197P (2 clones), L1200P (1 clone), Q1201R (4 clones), and Q1201K (1 clone), all located within the membrane proximal region of HR2 (Fig. 4a, b; Supplementary Fig. 4B). Escape VSV pseudotyped viruses with mutations L1197P, L1200P, Q1201R, or Q1201K were resistant to R3DC23 neutralization, whereas the N1192D mutant virus remained sensitive, especially on Vero E6 cells (Fig. 4c, d; Supplementary Fig. 4C, D). In line with this, R3DC23 retained full binding capacity to cell surface-expressed N1192D, whereas binding to spike muteins with any of the other escape selection mutations was lost (Fig. 4e).
a Complete SARS-CoV-2 spike trimer model (6VXX_1_1_1), red and blue sticks: amino acids at which substitutions were observed in the escape viruses; red and blue helices: region corresponding to the two peptides in the spike that were protected from HDX by R3DC23. Helices in black: transmembrane domain. b HR2 amino acid sequence of the indicated viruses. Dots: amino acids that are identical to the SARS-CoV-2 HR2. Red: positions at which substitutions were observed in the escape variants, blue: N-glycosylation site. The sequence in normal font: the part of the HR2 that contains the heptad repeat motif, gray italic: the flexible linker between the HR2 and transmembrane region, red and blue braces: two spike peptides that were protected by R3DC23 as revealed by HDX-MS. c, d Neutralization of replication-competent VSV pseudotyped with SARS-CoV-2 spike viral escape variants by R3DC23 on VeroE6/TMPRSS2 cells (c) and Vero E6 (d) cells. The graphs show the mean ± SEM (N = 4 replicates of the same dilution series) level of normalized GFP MFI. e Binding of R3DC23 to cells expressing the Wuhan (WT), N1192D, L1197P, L1200P, Q1201R, or Q1201K spike variants. f Kinetics of viral replication of replication-competent VSV-GFP reporter viruses pseudotyped with SARS-CoV-2 Wuhan (parental, WT) or the selected escape variants as measured by life cell imaging of infected Vero E6 cells. The graph shows the mean ± SEM (N = 5 wells) GFP+ area per well of infected cells at the indicated time points post-infection. g Left: the HR2 coiled-coil structure (PDB: 2FXP) as determined by NMR. Red: substitutions observed in the escape viruses, green: glycans conjugated at N1194 as modeled in 6VXX_1_1_1. Right: top view of part of the HR2 (K1191-K1205) coiled-coil, red: substitutions observed in the escape viruses, blue: N1194 on one protomer. The first GlcNac residues of the N-glycans to N1194 are shown in green69. h Binding of R3DC23, GBP (10 μg/mL), or S309 (1 μg/mL) to cells expressing SARS-CoV-2 spike protein variants. Source data are provided as a source data file.
The amino acids in the escape viruses that abolish R3DC23 binding are positioned between the viral membrane and a tetra-antennary N-glycan at position N1194 of the spike22. The HR2 sequences of SARS-CoV-1 and -2 are identical but differ in MERS and HKU1, explaining the specificity of the R3DC23-related VHHs (Fig. 4b). In the SARS-CoV-2 spike prefusion conformation, the membrane-proximal part of HR2 assembles into a coiled-coil of 3 parallel alpha helices that, upon transition to the postfusion conformation, initially disassemble and then bring the viral and host cell membrane in proximity by zippering up with the HR1 coiled-coil to form a stable 6HB23. This crucial function of the membrane proximal HR2 region in the fusion process may explain why the VSV pseudotyped R3DC23 escape viruses were severely attenuated (Fig. 4f, Supplementary Fig. 4E). In the reported NMR structure of the SARS-CoV-1 HR2 (PDB 2FXP24), the side chain of Q1183 (corresponding to Q1201 in SARS-CoV-2) is oriented outwards but both L1179 (L1197 in SARS-CoV-2) and L1182 (L1200 in SARS-CoV-2) are oriented towards the center of the coiled-coil (Fig. 4g). This suggests that R3DC23 either binds to the inner face of the HR2 coiled-coil, if these residues are directly involved in R3DC23 binding, or that proline substitutions, known to break alpha helices, at L1197 and L1200 disrupt the folding of the HR2 helix, and thus are indirectly involved in R3DC23 binding25. R3DC23, however, retained full binding to L1197A/F and L1200A/V spikes, indicating that the proline substitutions have an indirect effect on R3DC23, and as a result, that an intact HR2 helix is essential for binding (Fig. 4h, Supplementary Fig. 4F). The R3DC23 binding site was confirmed by hydrogen-deuterium exchange mass spectrometry (HDX-MS) (Supplementary Table S1) in which R3DC23 protected two adjacent peptides spanning residues 1187 through 1205 (Fig. 5a, b, Supplementary Fig. 5). One peptide neighboring the epitope region, originating from the trimerizing foldon domain, also shows protection, likely due to the proximity of this peptide to the epitope. No protection outside of this region was seen (Fig. 5b, Supplementary Fig. 5A). The efficient binding of R3DC23 to cells expressing merely the HR2 at their surface formally demonstrated that the HR2 is indeed the target of R3DC23 (Fig. 5c, Supplementary Fig. 6A).
a, b Identification of the R3DC23 binding region on recombinant spike protein by HDX-MS. The panels in A show the HDX-MS uptake plots of the two peptides with high degrees of protection from deuteration in the presence of R3DC23. The asterisk indicates the glycosylated Asn residue. b Difference HDX uptake plot (Woods Plot). Each line is a peptide and the residues spanned by the peptide are indicated by the positioning along the x-axis. The position along the y-axis is the difference in exchange between the apo condition and the R3DC23 bound condition (apo – R3DC23 bound) at an individual exchange time. The color of each line indicates the exchange time. c R3DC23 binding to cells that express the SARS-CoV-2 spike (left panel) or HR2 (right panel) at their surface. d R3DC23 does not recognize cells that express the SARS-CoV-2 6HB or 5HB at their surface. e R3DC23 binds to trimerized HR2 coiled-coil constructs. The graphs show binding of huR3DC23-Fc (left panel) or palivizumab (right panel) to various HR2 constructs, that are fused to His-SUMO (monomeric) or foldon (trimeric). f R3DC23 VHHs bind with a 3:1 stoichiometry to recombinant SARS-CoV-2 spike trimers (S-2P). The BLI sensorgram shows the immobilization of S-2P trimer on a biolayer interferometry biosensor followed by association with 20 nM R3DC23 monomer in solution. Raw data (no reference subtraction) of one of triplicate experiments is shown. The sensongrams of the two other runs are shown in Supplementary Fig. 6D. Source data are provided as a source data file.
In the spike postfusion conformation, HR2 helices are folded around the HR1 coiled-coil, forming the 6HB. Binding of R3DC23 to the HR2 as part of the 6HB was tested by the use of cell surface-expressed 6HB and 5HB, corresponding to the 6HB that lacks one HR2 helix. Although monomeric HR2 (SUMO-HR2) bound to cells expressing the 5HB, R3DC23 failed to bind these cells or to cells expressing the 6HB at their surface (Fig. 5d, Supplementary Fig. 6B). During the transition of the spike from its pre- to postfusion conformation, the HR2 coiled-coil unfolds. Therefore, we tested binding of R3DC23 to both trimeric and monomeric HR2 formats by ELISA. Although NMR has demonstrated that HR2 peptides spontaneously assemble into coiled-coils of 3 parallel alpha helices, we stabilized this trimeric conformation by fusing C-terminally truncated HR2 variants to a trimeric foldon24. HR2 monomers were obtained by fusing HR2 to SUMO via a 3C protease cleavage site (Supplementary Fig. 6C). Whereas efficient binding of huR3DC23-Fc was observed for protease-cleaved HR2 (to remove the His-SUMO tag to enable HR2 trimerization) and all three HR2-foldon fusions, truncated at different positions of the HR2 C-terminus, binding to monomeric HIS-SUMO-HR2 was very poor (Fig. 5e). These data suggest that the epitope of R3DC23 does not extend beyond L1203 and that R3DC23 has a strong preference for trimeric HR2 as present in the prefusion spike. BLI of R3DC23 binding to immobilized S-2P proteins under saturating conditions revealed a stoichiometry of 3.53 ± 0.133 (mean ± S.D. of n = 3) VHHs per spike trimer (Fig. 5f, Supplementary Fig. 6D). Collectively, these data indicate that R3DC23 binds the spike protein at the membrane proximal half of HR2, most likely at the outside of the prefusion trimeric HR2 coiled-coil with a 3:1 stoichiometry.
R3DC23 recognizes a quaternary epitope in HR2
We resolved the crystal structure of R3DC23 in complex with a peptide spanning the complete HR2 (H1159-K1211). The crystal asymmetric unit shows an HR2 coiled-coil trimer in complex with three R3DC23 molecules, each binding the interface between two HR2 peptides (Fig. 6). The R3DC23 binding site spans residues N1192 to Y1206 (Fig. 6a–c), where R3DC23 binds two adjacent HR2 helices, encompassing a 407 Å2 buried surface area, 8 H-bonds, 2 salt bridges and a calculated solvation free energy gain ΔiG for complex formation (i.e. hydrophobic contribution to binding) of −4.7 kcal/mol for helix (i), and a 461 Å2 buried surface area, 6 H-bonds, 1 salt bridge and a calculated ΔiG of −2.4 kcal/mol for helix (ii) (Fig. 6c, Supplementary Fig. 7A, B). CDR3 forms the dominant contact surface in the complex, where N100a and Y100b form an extensive H-bond network with N1194, Q1201, and E1202 in helix (ii) and S1196 in helix (i), Y96 goes in H-bond contact with helix (ii) main chain carbonyls, S98 goes in H-bond interaction with E1195 in helix (i), and V97 binds a hydrophobic patch formed by L1200 and L1203 in helix (i). CDR1 and CDR2 are involved, respectively, in hydrophobic interactions and two salt bridges (R52 – D1199) with helix (i) (Fig. 6c). The identification of Q1201 as a R3DC23 escape mutant agrees with its central role in the H-bond network with CDR3. L1197 and L1200, however, are not in direct contact with R3DC23, suggesting that substitutions by proline contribute indirectly to escape, likely as a result of a destabilization and/or conformational adjustment of the epitope. The conformation of the HR2 trimer in complex with R3DC23 closely matches that observed in the prefusion form of the S protein, where it corresponds to an approximately 3 nm high region between the membrane and the HR2 and linker regions covered by N-glycosylation (Fig. 6a). In the postfusion form, HR2 helices rearrange to bind the surface of a HR1 coiled-coil, resulting in partial unfolding of the HR2 C-terminal region and disruption of the R3DC23 binding site (Fig. 6d). Conversely, the interfacial binding of R3DC23 to the HR2 coiled-coil likely stabilizes the prefusion S protein.
a left: model of full-length prefusion S (6VSB_1_1_269) with superimposition of the R3DC23 – HR2 complex, all shown in molecular surface representation, with N-glycans in stick representation. R3DC23 VHHs are colored in orange, labeled S protein regions: cytoplasmic domain (red; CP), transmembrane domain (gray; TM), heptad repeat 2 (blue; HR2), S2 stem helix (deep pink; SH), heptad repeat 1 (lemon, HR1), central helix and connector domain (pink; CH-CD), the S1 regions encompassing the N-terminal domain (NTD) and receptor binding domain (RBD) (light blue). Right: model of the proteolytically processed postfusion S2 subunit (7E9T70), color coded as in prefusion spike. b Side and axial view (inset) of the R3DC23 – HR2 complex (sand and blue, respectively) superimposed with prefusion HR2 coiled-coil (light blue). In R3DC23, CDR1, 2, and 3 are colored magenta, yellow, and cyan, respectively. The HR2-binding epitope spanning D1192 – Y1206 is shown in stick representation. c Close-up of boxed region, encompassing a single VHH and two HR2 copies (i and ii) forming the adjoined binding epitope. Escape mutant positions are labeled in red. d Axial view of HR1-HR2 (lemon and blue) region of postfusion S protein, superimposed with R3DC23 in the HR2 complex (gray). e The amino acids involved in interactions between R3DC23 and two HR2 helices as observed in the crystal of the R3DC23-HR2 complex are indicated in red.
The importance of the amino acid residues that are directly involved in the interaction as revealed by X-ray crystallography was validated on cell surface-expressed spikes (Fig. 6e). All non-escape positions between N1194 and L1203 were substituted by at least one other amino acid while S309 binding was used to confirm similar expression levels of the spike mutants (Fig. 7a). Substitutions at N1194 or S1196, respectively, completely and strongly reduced binding of R3DC23(-Fc). Substitutions D1199N, E1202K and L1203G had a more modest effect whereas the E1195Q, I1198V, and G1204E substitutions had no obvious impact on binding (Fig. 7a, Supplementary Fig. 7C, D). VSV particles pseudotyped with these spike variants were generated. Whereas none of these mutations affected neutralization by S309, huR3DC23-Fc could not neutralize VSV particles pseudotyped with spike variant S1196F, which completely escaped binding, or the N1194E spike variant, for which binding was only partly abrogated (Fig. 7b, Supplementary Fig. 7E, F). Moderate loss of binding resulted in a more pronounced loss in neutralization, especially for the D1199N substitution. A clear loss in neutralization by huR3DC23-Fc also resulted in a clear loss in sensitivity for huR3DC23-Fc-mediated inhibition of membrane fusion (Fig. 7c, Supplementary Fig. 8). Overall there was a tendency for spike variants that totally escape huR3DC23-Fc-mediated inhibition of membrane fusion to have lower fusogenic activity, except for the Q1201K and Q1201R variants (Fig. 7c, Supplementary Fig. 8B). Similar to D1199N, VSV particles pseudotyped with the spike of BA2.75.2 (which contains D1199N) were less sensitive to huR3DC23-Fc-mediated neutralization than its parental BA.2.75 counterpart, despite the only limited difference in binding to cell-expressed spikes, at both pH 7.5 and pH 5.5 (Fig. 7d, Supplementary Fig. 8C–E). No other SARS-CoV-2 emerging variants have a fixed mutation in HR2 and the variance within the R3DC23 epitope remains very low (Supplementary Fig. 9). Among sarbecoviruses the R3DC23 epitope is highly conserved, including in Khosta-2, which can use hACE2 as a receptor26 (Supplementary Fig. 10). In line with this, R3DC23 efficiently bound to cells expressing Khosta-2 spike (Fig. 7e).
a Binding of R3DC23 to cells expressing the indicated spike mutants, measured by flow cytometry. The left graph shows the ratio of the MFI of R3DC23 binding at the indicated concentrations to transfected (GFP+) cells expressing spikes harboring the indicated point mutation and that of non-transfected (GFP-) cells. The right graph shows the mean EC50 of R3DC23 binding to cells expressing spikes harboring the indicated point mutations (n = 2). b Neutralization of VSV particles pseudotyped with SARS-CoV-2 spikes harboring the indicated point mutations. Mean neutralization IC50 (n = 2) is shown. c Fusogenicity of spike variants with point mutations introduced in the HR2 region, in the presence of palivizumab (negative control) and huR3DC23-Fc. Cells were treated with antibody (1 µg/mL), 2 h after co-transfection of a spike variant and a GFP expression vector, and syncytium formation was monitored with live-cell imaging. Shown in the graph is the mean ± SD (N = 3 samples) slope of the curve derived from the area of green objects at time points 24 h after transfection and 38 h after transfection as a measure for fusogenicity of the HR2 mutant. d Neutralization of VSV particles pseudotyped with BA.2.75 or BA2.75.2 spikes by huR3DC23-Fc, S309 (positive control) and palivizumab (negative control). Mean ± SD (N = 3 replicates of the same dilution series) of normalized GFP MFI is shown. e Binding to surface-expressed Khosta-2 spike by a dilution series of R3DC23 (lef graph) or huR3DC23-Fc, palivizumab or S309 at 10 µg/mL. The ratio of MFI of bound antibody to GFP+ cells expressing spikes or no spikes over that of non-transfected GFP- cells is shown. Source data are provided as a source data file.
R3DC23 inhibits fusion by preventing 6HB formation
Next to the conventional closed prefusion conformation, parts of the recombinant soluble prefusion-stabilized spike protein, including the HR2 coiled-coil, can adopt a conformation where the trimeric interface is exposed to solvent, referred to as an open-interface trimer27. Temperature incubation monitored by HDX-MS revealed that R3DC23 did not affect the transition of prefusion spike from a closed to an open trimer interface conformation, which is likely due to close proximity of the helices in the HR2 coiled-coil in the latter (Fig. 8a). Obstructing shedding of S1 can prevent membrane fusion, as exemplified by monoclonal antibody S30928. In contrast to S309, R3DC23 did not impair S1 shedding induced by the CB6 antibody (Fig. 8b)28,29. After S1 shedding and cleavage at the TMPRSS2 site, the three HR1 regions stretch out to form a trimeric HR1 coiled-coil, thereby anchoring the fusion peptides into the target cell membrane. This is the extended intermediate of the spike protein (Fig. 8c). We noticed that the HR1 coiled-coil on spike-expressing cells that were triggered to undergo S1 shedding, was recognized by SUMO-HR2 when added before the induction of shedding. This allowed us to assess whether R3DC23 can interfere with the transition of the spike from its prefusion to its extended intermediate conformation. huR3DC23-Fc pre-treatment of spike-expressing cells that were subsequently triggered with trypsin and VHH72_S56A-Fc to undergo shedding did, however, not prevent the binding of monomeric SUMO-HR2 to the extended HR1 coiled-coil (Fig. 8d and Supplementary Fig. 11A). Moreover, huR3DC23-Fc even promoted binding of SUMO-HR2 to trypsin-treated cells which might result from locking of spikes that have (spontaneously) shed S1 in the extended intermediate conformation. As mere binding of SUMO-HR2 to HR1 in the extended spike intermediate might also interfere with refolding to the postfusion conformation23, we investigated if huR3DC23-Fc could interfere with this refolding by applying huR3DC23-Fc before shedding and SUMO-HR2 after shedding. huR3DC23-Fc could indeed prevent the transition of triggered spikes to their postfusion conformation, as binding of SUMO-HR2 added after shedding was notably higher to cells treated with huR3DC23-Fc before shedding as compared to cells pre-treated with palivizumab or to huR3DC23-Fc pre-treated cells that were not stimulated to undergo shedding (Fig. 8e). The modest but significant binding of SUMO-HR2 to the latter cells can be explained by either spontaneous shedding or by limited binding of SUMO-HR2 to accessible R3DC23 of huR3DC23-Fc that engaged only one VHH in the spike binding (Supplementary Fig. 11B). By locking the HR2 coiled-coil, R3DC23 can prevent the formation of the postfusion 6HB either by interfering with the migration of the HR2 helices towards the HR1 coiled-coil or/and by directly interfering with the zippering of the HR2 helices with the HR1 coiled-coil. To test the latter possibility, we investigated if pretreatment of trimeric HR2-foldon with R3DC23 could prevent the binding of the former to cells exposing the spike HR1 coiled-coil. We used HR2(MERS)-foldon as a control that can associate with the HR1 coiled-coil of SARS-CoV-2 but is not recognized by R3DC23 (Supplementary Fig. 11C, D). In contrast to HR2(MERS)-foldon, binding of trimeric SARS-CoV-2 HR2-foldon to cells exposing HR1 was inhibited by pretreatment with R3DC23 (Fig. 8f). Collectively, these data demonstrate that R3DC23 does not hamper the refolding of spike to the extended intermediate after S1 shedding, but interferes with the subsequent formation of the 6HB.
a Transition of S-2P from its open-interface conformation to the prefusion conformation (left) or vice versa (right). The fraction prefusion S−2P as determined by hydrogen-exchange pulse-labeling is plotted as a function of incubation time prior to the deuterium pulse27. b Anti-S1 Western blot analysis of the growth medium (SN) and cell lysates (LYS) of HEK293T cells expressing the spike protein (SC2 D614G S) or not (HEK EV) incubated sequentially with the indicated VHHs/antibodies. The shown western blot is representative for two experiments. The right panel shows the mean ratio (N = 2) of S1 detected in the supernatant to S1 detected in the cell lysate. c Schematic overview of membrane fusion (top) and proposed steps where R3DC23 interferes with membrane fusion (bottom). RBD: green, S1: turquoise, FP: fusion peptide (brown), HR1: heptad repeat 1 (pink), CH: central helix (blue), CD: connector domain (orange), HR2: heptad repeat 2 (red), MPR: membrane proximal region (light gray), TM+cytosolic: transmembrane domain and cytosolic domain (dark gray). Created in BioRender. Saelens, X. (2025) https://BioRender.com/5i3waej. d Binding of SUMO-HR2 that was added to spike-expressing cells before stimulation with VHH72_S56A-Fc to induce S1 shedding in the absence/presence of huR3DC23-Fc (N = 8 samples pooled from three independent experiments, line indicates the mean). e Binding of SUMO-HR2 (left) and control SUMO-HR1 peptides (right) to spike-expressing cells that were on beforehand stimulated with VHH72_S56A-Fc to undergo S1 shedding in the absence or presence of huR3DC23-Fc (N = 9 samples pooled from three independent experiments, line indicates the mean) (*P < 0.05 and ****P < 0.0001). f Binding of respectively HR2(SARS-CoV-2)-foldon (left) and HR2(MERS)-foldon (right) pretreated with either R3DC23 or control nanobody (GBP) to cells displaying the extended intermediate of spikes that have shed S1 (N = 6 samples pooled from two independent experiments; in each condition, line indicates the mean) (*P < 0.05, **P < 0.005, ****P < 0.0001). P-values are shown in gray above graphs. Statistical tests are specified in the Methods section. Source data are provided as a source data file.
Discussion
The SARS-CoV-2 neutralizing activity evoked by licensed COVID-19 vaccines is primarily mediated by RBD-specific antibodies30. RBD-targeting monoclonal antibodies have been approved to treat or prevent COVID-1919,20,31,32,33. However, novel SARS-CoV-2 variants that escape from these RBD-targeting antibodies continuously emerge34. In contrast, the R3DC23 epitope is conserved among the SARS-CoV-2 variants of concern, variants of interest or variants under monitoring that have circulated up to date. Moreover, with few exceptions, the epitope is conserved among sarbecoviruses. In the fall of 2022 BA2.75.2 carrying a D1199N substitution, transiently circulated to revert back to D1199 in the successor CH.1.1 variant35. Like the D1199N point mutation, pseudotypes with the BA2.75.2 spike were less sensitive to R3DC23 neutralization. The low frequency and rapid replacement of BA2.75.2 by CH.1.1 might indicate that D1199N was rather a passenger mutation and has a likely cost in viral fitness35. Although the membrane proximal region of HR2 is relatively conserved between SARS-CoV-2, MERS, and HKU1, the region recognized by R3DC23 contains several differences. One of these is the Q1201K substitution that abrogated the binding of R3DC23 in escape variants. Moreover, in the MERS spike L1197 is replaced by Tyr at the corresponding position. As L1197 is oriented towards the center of the HR2 coiled-coil, it is likely that Tyr disturbs the HR2 coiled-coil formation.
The HR2 in recombinant S-2P spike that was used for immunization and bio-panning, and the HR2 on the virion contain a complex-type glycan at N1194 that is critical for infectivity36. Although the HR2-targeting VHHs can efficiently bind to their epitope that is delineated between the viral membrane and this N-glycan, this narrow site of approximately 3 nm might be hard to target by conventional antibodies22,37. Recently, a human monoclonal antibody that targets the membrane proximal region of HR2 was described that was isolated from a memory B cell from a SARS-CoV2 (ancestral) convalescent individual38. This monoclonal antibody, which likely binds a non-quaternary epitope in HR2, neutralizes SARS-CoV-2 variants with a much lower potency (IC50 = 4.8–35.4 µg/mL) while not exceeding a plateau of maximum 50% neutralization. In addition, natural infection with SARS-CoV-2 can elicit antibodies that can cross-react with a peptide that corresponds to the short flexible linker (K1205YEQYIKW1212) positioned between the HR2 and the transmembrane region of SARS-CoV-2 spikes39. Vaccination of mice with membrane-anchored S2 subunit induced antibodies recognizing this peptide40. Presumably, this antibody response is facilitated by the flexibility of the spike, which allows it to tilt along its axis to up to 90°, thereby exposing the membrane proximal region41. A few monoclonal antibodies targeting the N-terminal half of the SARS-CoV-1 HR2, above the N1194 glycosylation site, have been described. The humanized antibody hMab5.17, for example, can neutralize SARS-CoV-2 with very moderate potency (IC50 approximately 12 μg/mL) and reduce viral replication in Syrian hamsters42,43. Similarly, monoclonal antibody 1G10, obtained from mice vaccinated with a bacteria-produced HR2-GST fusion construct, can neutralize SARS-CoV-1 with an IC50 of 13 µg/mL44. In sharp contrast to these reported HR2 targeting antibodies, R3DC23 has very strong neutralizing activity. The highly potent fusion inhibition activity of R3DC23 can be explained by the quaternary nature of its epitope: each R3DC23 VHH binds two adjacent HR2 helices and each HR2 helix is bound by two VHHs. R3DC23 thus prevents the unraveling of the HR2 coiled-coil, a critical step that precedes the 6HB formation in which HR2 helices wrap around the central HR1 coiled-coil45. This hypothesis is in line with the observation that in contrast to R3DC23, hMab5.17 binds single HR2 strands and neutralizes SARS-CoV-2 far less efficiently43 (Supplementary Fig. 12). The flexibility of the spike at its “hip” (near the stem helix), “knee” (between the stem helix and HR2), and “ankle” (between the HR2 and the membrane proximal region) is important for multiple steps in the fusion process: (i) to engage multiple ACE2 proteins at the host cell surface, (ii) for efficient anchoring of the fusion peptide into the host cell membrane, and (iii) for the transition of the extended intermediate to the postfusion conformation of the spike (Fig. 8c)23,37,46.
Because R3DC23 neutralizes SARS-CoV-2 via a novel mode of action it was important to test if R3DC23-Fc fusions can also control viral replication in vivo. Using K18-hACE2 mice and the Syrian hamster model we demonstrated that prophylactic or therapeutic administration of humanized R3DC23 fused to Fc(YTE/LS) restricts replication of SARS-CoV-2 in the lungs of challenged animals. These results indicate that Fc fusions of R3DC23 either as monotherapy or as part of an antibody cocktail represent a valuable strategy to prevent or treat disease caused by SARS-CoV-2 and other sarbecoviruses. Moreover, this novel mode of action could be applied to other enveloped viruses that use a class 1 fusion protein with an accessible HR2 coiled-coil in the prefusion state such as the fusion protein of henipaviruses.
Methods
Cell lines
FreeStyle293F cells (Thermo Fisher Scientific) and HEK293-S cells (Thermo Fisher Scientific) were cultured in FreeStyle293 expression medium (Life Technologies), cultured at 37 °C with 8% CO2 while shaking at 130 rpm. HEK293T cells (ATCC) and Vero E6 cells (ATCC) were cultured at 37 °C in the presence of 5% CO2 in DMEM supplemented with 10% heat-inactivated FBS, 1% penicillin, 1% streptomycin, 2 mM L-glutamine, non-essential amino acids (Invitrogen) and 1 mM sodium pyruvate. ExpiCHO-S cells (GIBCO) were cultured at 37 °C with 8% CO2 while shaking at 130 rpm in ExpiCHO expression media (GIBCO). VeroE6/TMPRSS2 cells that stably express human TMPRSS2 (NIBIOHN, JCRB1819)47 were cultured in DMEM containing 10% FBS, Penicillin (100 unit/mL), Streptomycin (100 μg/mL), Geneticin (G418) (1 mg/mL). VeroE6/TMPRSS2 cells were seeded in medium without Geneticin for infectivity assays.
Expression and purification of recombinant SARS-CoV-2 spike
ExpiCHO cells were transfected with a plasmid encoding the SARS-CoV-2 spike ectodomain (residues 1-1208), with 2P (K986 and V987) and RRAR-GSAS (residues 682-685) mutations to stabilize the protein in the prefusion conformation48, a kind gift from Dr. Jason McLellan. Twelve days after transfection, the supernatant was collected, and the protein was purified from the supernatant with a 5 mL HisTrapTM High Performance column (Cytiva), which had been equilibrated with 20 mM NaH2PO4 pH 7.4, 0.5 M NaCl, and 20 mM imidazole. The protein was eluted with 20 mM NaH2PO4 pH 7.4, 0.5 M NaCl and 0.5 M imidazole. Peak fractions were desalted to PBS using a HiPrepTM 26/10 desalting column (Cytiva) and sterile filtered with a low protein binding 0.22 µm filter.
VHH phage display and panning
A llama that was previously immunized with recombinant prefusion stabilized SARS-CoV-1 and MERS spike protein was additionally immunized 3 times with recombinant SARS-CoV-2 spike protein (S-2P, Wuhan sequence) stabilized in its prefusion conformation49,50. Five days after the third immunization, peripheral blood lymphocytes were isolated from the llama and a VHH-displaying phagemid library was constructed. Phages that display SARS-CoV-2 spike specific VHHs were enriched from this phage library by 2 successive rounds of biopanning on 100 ng of HIS-tagged SARS-CoV-2 spike 2P protein49, that was immobilized in a well of a microtiter plate (type II, F96 Maxisorp, Nunc) via precoated anti-HIS antibodies in the presence of 10 μg/mL RBD-SD1-mouse IgG. Two additional rounds of biopanning were performed using anti-HIS captured spike proteins (R3C and R4C series) or using directly coated spike proteins (R3DC and R4DC series). Also these two series of additional rounds of biopanning were performed in the presence of 10 μg/mL RBD-SD1-mouse IgG. For each panning round an uncoated well was used as a negative control. The wells were then washed 5 times with phosphate-buffered saline (PBS) + 0.05% Tween 20 and blocked with 4% milk powder in PBS (Regilait) in the first panning round and Pierce protein free blocking buffer (Thermo Fisher Scientific), SEA blocking buffer (Thermo Fisher Scientific) and 1% BSA in PBS in the subsequent panning rounds. Non-specifically bound phages were removed by extensive washing with PBS + 0.05% Tween 20. The retained phages were eluted with TEA-solution (14% trimethylamine (Sigma) pH 10) and subsequently neutralized with 1M Tris-HCl pH 8.0. The collected phages were amplified in exponentially growing E. coli TG1 cells, infected with VCS M13 helper phages and subsequently purified using PEG 8000/NaCl precipitation for the next round of selection. Enrichment after each panning round was evaluated by infecting TG1 cells with 10-fold serial dilutions of the collected phages after which the bacteria were plated on LB agar plates with 100 μg/mL− ampicillin and 1% glucose.
Preparation of periplasmic extracts
After 3 or 4 panning rounds individual colonies of phage infected bacteria were randomly selected for further analysis. The selected colonies were inoculated in 2 mL of terrific broth (TB) medium with 100 μg/mL ampicillin in 24-well deep well plates. After 5 h incubation at 37 °C, isopropyl β-D-1-thiogalactopyranoside (IPTG) (1 mM) was added to induce VHH expression during overnight incubation at 37 °C. To prepare periplasmic extract, the bacterial cells were pelleted and resuspended in 250 μL TES buffer (0.2 M Tris-HCl pH 8.0, 0.5 mM EDTA, 0.5 M sucrose) and incubated at 4 °C for 30 min. Subsequently 350 μL water was added to induce an osmotic shock. After 1 h incubation at 4 °C followed by centrifugation, the periplasmic extract was collected.
Production of VHHs in E. coli and purification
For the production of VHH in E. coli, a pMECS vector containing the VHH of interest was transformed into WK6 cells (the non-suppressor E. coli strain) and plated on an LB plate containing ampicillin. The next day clones were picked and grown overnight in 2 mL LB containing 100 μg/mL ampicillin and 1% glucose at 37 °C while shaking at 200 rpm. One mL of this preculture was used to inoculate 25 mL of TB supplemented with 100 µg/mL ampicillin, 2 mM MgCl2 and 0.1% glucose and incubated at 37 °C with shaking (200–250 rpm) until an OD600 of 0.6–0.9 was reached. VHH expression was induced by addition of IPTG to a final concentration of 1 mM followed by overnight incubation at 28 °C while shaking at 200 rpm. The VHH-containing fraction was extracted from the periplasm and purified as described in Wrapp et al.50. In short, the VHHs were purified from the periplasmic extract using Ni Sepharose beads (GE Healthcare). After elution with 500 mM imidazole, the VHH-containing fractions were buffer-exchanged with PBS using a Vivaspin column (5 kDa cutoff, GE Healthcare). The purified VHHs were analyzed by SDS-PAGE and Coomassie staining and by intact mass spectrometry. For crystallography, R3DC23 was produced in a 1 L culture of E. coli WK6. The periplasmic extract was loaded on a 1 mL HisTrapTM High Performance column (Cytiva), which had been equilibrated with 20 mM NaH2PO4 pH 7.5, 0.5 M NaCl, and 20 mM imidazole. Bound VHH was eluted with 20 mM NaH2PO4, 20 mM NaCl and 0.5 M imidazole. The peak fractions were pooled and further purified by size exclusion chromatography, using a HiLoad 16/600 Superdex® 75 prep grade column (Sigma-Aldrich), in PBS.
Enzyme-linked immunosorbent assay
Wells of microtiter plates (type II, F96 Maxisorp, Nunc) were coated overnight at 4 °C with 100 ng of recombinant SARS-Cov-2 S-2P, SARS-CoV-2 S-6P protein, recombinant SARS-CoV-1 spike protein, recombinant MERS-CoV spike protein, recombinant HKU1 spike protein (all kindly provided by Dr. Jason McLellan), SARS-CoV-2 Omicron BA.1 S protein (ACROBiosystems), mouse Fc-tagged SARS-CoV-2 RBD (Sino Biological, 40592-V05H), the SARS-CoV-2 spike S2 subunit (ACROBiosystems, S2NC52H5) or BSA. The coated plates were blocked with 5% milk powder in PBS. Dilution series of the VHHs, VHH-Fcs or antibodies were added to the wells and plates were further incubated for 90 min at room temperature. After washing, binding of VHHs was detected by HRP-conjugated rabbit anti-camelid VHH antibodies (Genscript, catalog number A01861-200, 1/5000) or a mouse anti-HA antibody (clone 16B12, BioLegend, catalog number 901501, 1/2000), followed by anti-mouse IgG-HRP (Cytiva, NA931V, 1/2000). Binding of VHH-Fcs or conventional human monoclonal antibodies was detected by HRP-conjugated rabbit anti-human IgG (Sigma, catalog number A8792, 1/2000). After washing, 50 μL of TMB substrate (tetramethylbenzidine, BD OptEIA) was added to the plates and the reaction was stopped by addition of 50 μL of 1 M H2SO4. The absorbance at 450 nm was measured with an iMark Microplate Absorbance Reader (Bio Rad). Curve fitting was performed using nonlinear regression using Graphpad 8.0.
In the PE-ELISA shown in Fig. 1a, the blue-white heat map shows for each PE sample (10-fold diluted), the ratio of the ELISA OD450 signal for the indicated antigen over the ELISA OD450 signal of the corresponding PE sample for the control antigen (BSA), normalized to the signal for a well with TES buffer (used to prepare the PEs) (0%) and the highest OD450 ratio in the ELISA (100%). PEs prepared from E. coli cells that express an RBD binding VHH were used as control. Buffer used to prepare the PEs was used as negative control (TES).
Generation of replication-deficient VSV pseudotyped viruses
To generate replication-deficient VSV pseudotyped viruses, HEK293T cells, transfected with SARS-CoV-1 S or SARS-CoV-2 S expression vectors were inoculated with a replication-deficient VSV vector containing eGFP and firefly luciferase expression cassettes48,51. After 1 h incubation at 37 °C, the inoculum was removed, cells were washed with PBS and incubated in media supplemented with an anti-VSV G mAb (ATCC, catalog number CRL-2700, RRID:CVCL_G654; 1/1000) for 16 h. Pseudotyped particles were then harvested and clarified by centrifugation51.
Pseudovirus neutralization assay with periplasmic extracts
Pseudoviruses expressing the SARS-CoV-2 spike (D614G) were incubated for 30 min at 37 °C with a 1/100 dilution of periplasmic extract in Fluorobrite DMEM medium (Invitrogen), supplemented with 5% heat-inactivated FBS, 1% penicillin, 1% streptomycin, 2 mM L-glutamine, non-essential amino acids (Invitrogen) and 1 mM sodium pyruvate. The incubated pseudoviruses were subsequently added to subconfluent monolayers of Vero E6 or Vero E6/TMPRSS2 cells from which the original growth medium was removed. Sixteen hours later, the cells were lysed using passive lysis buffer (Promega). The transduction efficiency was quantified by measuring the GFP fluorescence in the prepared cell lysates using a Tecan infinite 200 pro plate reader. GFP fluorescence was normalized using the GFP fluorescence of non-infected cells and infected cells treated with PBS.
Flow cytometric analysis of VHH-binding to HEK293T cells expressing SARS-CoV spike proteins or fragments thereof
Binding of VHHs to spike proteins on the surface of mammalian cells was determined by flow cytometry using pcG1-expression plasmids containing the coding sequence of the ancestral SARS-CoV-2 spike protein from which the C-terminal 18 amino acids were deleted (AA1-1255) and in which the D614G substitution was introduced by QuickChange site-directed mutagenesis (Agilent) according to the manufacturer’s instructions. For analysis of VHH-binding to cells expressing the SARS-CoV-2 HR2 coiled coil, a gene fragment containing the SARS-CoV-2 signal peptide (MFVFLVLLPVVSSQ), fused via a GSG-linker to an HA-tag and fused via a GSG-linker to the SARS-CoV-2 P1162-K1255 sequence was ordered at IDT and cloned into a pcDNA3.4 expression vector via TOPO-cloning (pcDNATM 3.4 TOPOTM TA cloning kit, Thermo Fisher Scientific). Two days after transfecting HEK293T cells with a GFP expression plasmid in combination with either a spike expression plasmid or a control expression plasmid the cells were collected. All further procedures were performed on ice. The cells were washed once with PBS and blocked with 1% BSA. The cells were then stained with antibody or VHH dilution series for 90 min and subsequently washed 3 times with PBS containing 1% BSA. Binding of VHHs was detected using a mouse anti-HIS-tag antibody (Biorad, MCA1396, 1/2000) and an AF647 conjugated donkey anti-mouse IgG antibody (Invitrogen, catalog number A31571, 1/1000). Binding of VHH-Fcs or antibodies was detected using an AF633 donkey goat anti-human IgG antibody (Invitrogen, catalog number A21091, 1/1000) and dead cells were stained using Live/Dead stain (Invitrogen, 15560607, 1/1000). Following 3 washes with PBS containing 0.5% BSA, the cells were analyzed by flow cytometry using a BD LSRII flow cytometer (BD Biosciences). Binding was calculated as the ratio of the AF647 MFI of antibodies binding GFP+ cells over that of GFP- cells. The binding curves were fitted using nonlinear regression (Graphpad 9.10).
For cells transfected with 6HB and 5HB expression vectors, the expression of the 6HB and 5HB was confirmed by mouse anti-HA staining of the HA-tag that was fused to the N-terminus of these protein constructs, and for the 5HB also by binding of monomeric HIS-SUMO-HR2, which was detected with a mouse anti-HIS-tag antibody. Binding of HIS-SUMO-HR2 or HR2-foldon to spike-expressing cells expressing were detected using a mouse anti-HIS-tag antibody and a mouse anti-strep-tag antibody (Qiagen, catalog number 34850, 1/1000), respectively, in combination with an AF647-conjugated anti-mouse IgG antibody (Invitrogen, A31571, 1/1000). Binding was calculated as the ratio of the AF647 MFI of antibodies binding GFP+ cells over that of GFP- cells. An example of the gating strategy for data analysis is shown in Supplementary Fig. 13A.
Generation of spike protein expression vectors for the production of VSVdelG pseudovirus particles expressing spike proteins of SARS-CoV-2 variants
The pCG1 expression vector for the SARS-CoV-2 spike protein containing the D614G mutation was generated from the pCG1-SARS-2-Sdel18 vector by introducing the specific RBD mutation(s) via QuickChange mutagenesis using appropriate primers, according to the manufacturer’s instructions (Aligent). For the pCG1-SARS-2-Sdel18 expression vector for the omicron BA.1 variant, a codon-optimized spike protein nucleotide sequence containing the BA.1 mutations (A67V, Δ69-70, T95I, G142D, Δ143-145, N211I, Δ212, ins215EPE, G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H, N764K, D796Y, N856K, Q954H, N969K, L981F) and flanking BamHI and SalI restriction sites was ordered at GeneArt (Thermo Fischer Scientific) and cloned in the pCG1 vector as a BamHI/SalI fragment. For the pCG1-SARS-2-BA.2 Sdel18 expression vector, a codon-optimized spike protein nucleotide sequence containing the BA.2 mutations (T19I, ΔL14-P26, A27S, G142D, V213G, G339D, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, S477N, T478K, E484A, Q493R, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K) and flanking BamHI and SalI restriction sites was ordered at GeneArt (Thermo Fischer Scientific) and cloned in the pCG1 vector as a BamHI/SalI fragment. After sequencing, clones containing the correct spike coding sequence were prepared using the Qiagen plasmid kit. For the pCG1-SARS-2-BA.2.75 Sdel18 expression vector, a codon-optimized spike protein nucleotide sequence containing the BA.2.75 mutations (T19I, ΔL14-P26, A27S, G142D, S147E, W152R, F157L, I210V, V213G, G257S, G339D, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, G446S, S477N, N460K, T478K, E484A, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K) and flanking BamHI and SalI restriction sites was ordered at GeneArt (Thermo Fischer Scientific) and cloned in the pCG1 vector as a BamHI/SalI fragment. To generate expression plasmids for the BA.5 spike protein, mutations ΔLH96-V70, L452R, F486V and R493Q were introduced in the BA.2 spike expression construct by QuickChange mutagenesis according to the manufacturer’s instructions (Aligent). Likewise, to generate the BA.4.6 and BQ.1.1 spike expression vectors, mutations R346T and N658S and mutations R346T and K444T were respectively introduced in the BA.5 spike expression vector by QuickChange mutagenesis. The BF.7 spike expression vector was generated by introducing the R346T substitution in the BA.5 spike expression vector by QuickChange mutagenesis. After sequencing, plasmids from clones containing the correct spike coding sequence were prepared using the Qiagen plasmid kit. To generate the pCG1-SARS-2-XBB, a gBlock corresponding to the SARS-CoV-2 amino acid sequence V70-V510 containing the XBB mutations (V83A, G142D, ΔY144, H146Q, Q183E, V213E, D339H, R346T, L368I, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, V445P, G446S, S477N, T478K, E484A, F486S, F490S, Q498R, N501Y, Y505H) was ordered (IDT) and cloned into the pCG1-SARS-2 BA.2 vector via Gibson assembly according to the manufacturer’s instructions (New England Biolabs). To this end, the pCG1-SARS-2-BA.2 vector was amplified by PCR using appropriate primers (5’ GTGCCATTGGTGCCGGACACG 3’ and 5’ CACCAGCCTTACAGAGTGG 3’) and the Phusion High-Fidelity DNA polymerase (New England Biolabs). The XBB.1.5(-G252V) spike expression vector was generated by introducing the F496P substitutions in the XBB spike expression vector by QuickChange mutagenesis. The KP.3 spike expression vector was generated by introducing F456L, Q493E, V1104L in the JN.1 spike expression vector by Gibson assembly (using primers 5’ CCCAGATCATCACCACCGACAACACCTTCGTGTCTGGC 3’ and 5’ TAATCGTAGTTGCCGgaGTGTTTGGAGTCCAGCTTGTTG 3’). The KP.2.3 spike expression vector was generated by introducing Δ31, H146Q, R346T and E493Q (reversion to WT) in the KP.3 spike expression vector by Gibson assembly (using primers 5’ CAGACCCACATATGGCGTGGGCCACCAGCCTTACAGAG 3’ and 5’ TCACACACTGGCTGGACACCAGAGGCAGCAGCACCAGAAAC 3’. The KP.3.1.1 spike expression vector was generated from the KP.3 spike expression vector by introducing Δ31 with QuickChange mutagenesis. The LB.1 spike expression vector was generated from the JN.1 spike expression vector by introducing Δ31, Q183H, R346T and F456L by Gibson assembly (using primers 5’ TCCACCGAGATCTATCAGGCCGGCAACAAGCCCTGTAAG 3’ and 5’ TTCACACACTGGCTGGACACCAGAGGCAGCAGCACCAG 3’). The KP.1.1 spike expression vector was generated from the KP.3 spike expression vector by introducing R346T, E493Q (reversion to WT) and K1086R by Gibson assembly (using primers 5’ AGAGAAGGCGTGTTCGTGTCCAACGGCACCCATTGGTTC 3’). The XEC spike expression vector was generated from the KP.3 spike expression vector by introducing T22N and F59S by Gibson assembly (using primers 5’ ATGGCACCAAGAGATTCGACAACCCCGTGCTGCCCTTC 3’ and 5’ GGCAGCAGCACCAGAAACACGAACATGGTGGCGGATCCG 3’). The nucleotide sequence of all purified spike expression plasmids was verified by Sanger sequencing. The Khosta-2 spike expression vector was a kind gift from Dr. Michael Letko.
For the validation of the amino acids that are important for binding of R3DC23 as determined by X-ray crystallography, every position in HR2 between N1194 and L1203 that was not involved in the viral escape analysis was substituted to at least one amino acid that occurred at that position in the GISAID database by Quickchange mutagenesis. Expression levels of each of the mutated spikes were confirmed by staining with S309, an RBD-binding monoclonal antibody.
SARS-CoV pseudovirus neutralization assay
For the VSV pseudotype neutralization experiments, the pseudoviruses were incubated for 30 min at 37 °C with different dilutions of purified VHH or VHH-Fc fusions or with GFP-binding protein (GBP: a VHH specific for GFP). The incubated pseudoviruses were subsequently added to subconfluent monolayers of Vero E6 or VeroE6/TMPRSS2 cells. Sixteen hours later, the cells were lysed using passive lysis buffer (Promega). The transduction efficiency was quantified by measuring the GFP fluorescence in the prepared cell lysates using a Tecan infinite 200 pro plate reader. GFP fluorescence was normalized using either the GFP fluorescence of non-infected cells and infected cells treated with PBS or the lowest and highest GFP fluorescence value of each dilution series. The IC50 was calculated by non-linear regression curve fitting, log(inhibitor) vs. response (four parameters). Alternatively, dilution series of VHHs or antibodies were mixed with 100 PFU of GFP-expressing replication competent VSV virus particles pseudotyped with the SARS-CoV-2 spike protein derived from an early isolate. Of note, during propagation of this viral clone (S1-10a) on Vero E6 cells, the furin cleavage site was mutated and as such inactivated21. After 30 min incubation at 37 °C the virus-antibody mixtures were added to monolayers of Vero E6, VeroE6/TMPRSS2 or A549-hACE2 cells and allowed to infect and replicate for three days. In all neutralization assays using pseudotyped VSV viral particles, FluoroBrite DMEM medium (Invitrogen) supplemented with 5% heat-inactivated FBS, 100 unit/mL penicillin, 100 unit/mL streptomycin, 2 mM L-glutamine, non-essential amino acids (Invitrogen) and 1 mM sodium pyruvate was used to prepare the VHH- or antibody-virus mixtures. The mixtures were added to cells from which the original growth medium was removed. For all VSV pseudotype neutralization assays using huR3DC23-Fc_LS, huR3DC23-Fc_LS from stable cell pools was used.
Neutralization assays using authentic SARS-CoV-2 performed at VIB-UGent CMB
In vitro neutralization experiments with authentic SARS-CoV-2 viruses were performed in the Biosafety level 3 laboratory at VIB-UGent Center for Medical Biotechnology. The plaque reduction assays using authentic viruses were performed with SARS-CoV-2 D614G strain SARS-CoV-2/human/FRA/702/2020, obtained from the European Virus Archive (EVAG) and with an SARS-CoV-2 BA.1 virus that was obtained from KULeuven52 and grown at KULeuven on Vero E6 cells. Further propagation of the virus was performed on VeroE6/TMPRSS2 cells. Both viruses were titrated using a plaque assay in which monolayers of VeroE6/TMPRSS2 cells were infected with dilutions series prepared in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 2% fetal bovine serum (FBS) in duplicate for 2 vials of each virus. Two hours after infection, Avicel was added to a final concentration of 0.3% (w/v). Dose-dependent neutralization was assessed by mixing the constructs at different concentrations (5-fold serial dilutions) with 40 PFU of SARS-CoV-2, followed by incubation of the mixture at 37 °C for 1 h. The VHH-virus mixes were then added to VeroE6/TMPRSS2 cell monolayers in 24-well plates and incubated at 37 °C for 1 h. Subsequently, Avicel was added to a final concentration of 0.3% (w/v). After 2 days of incubation at 37 °C, the overlays were removed, and the cells were fixed with 3.7% paraformaldehyde (PFA) and stained with 0.5% crystal violet. Half-maximum neutralization titers (PRNT50) were defined as the VHH or VHH-Fc concentration that resulted in a plaque reduction of 50% across two independent plates.
Analysis of binding kinetics and stoichiometry by biolayer interferometry
The SARS-CoV-2 spike 2 P (S2-P) binding kinetics of R3DC23 monomer were assessed by biolayer interferometry on an Octet RED96 system (FortéBio). Streptavidin (SA) biosensors (Sartorius) were soaked in 1x kinetics buffer (10 mM HEPES pH 7.4, 150 mM NaCl, 0.1 mg/mL bovine serum albumin, 0.02% Tween-20 and 0.02% sodium azide) for 20 min before use. Trimeric S2-P-foldon-His-Strep (in-house HEK293S produced) was thawed from −20 °C and incubated at 37 °C for 24 h followed by 30 °C for 30 min, before immobilizing on the SA biosensors at 36.5 µg/mL (80 nM) for 450 s, to a signal of 1.5–1.7 nm. Association (120 s) and dissociation (480 s) of a two-fold dilution series starting from 20 nM monomeric R3DC23 VHH in 1x kinetics buffer were measured at 30 °C. Between analyses, biosensors were regenerated by three times 5 s exposure to regeneration buffer (10 mM glycine pH 3). The response in nm for stoichiometry analysis was determined from the raw data curves. Data for kinetics analysis were double reference-subtracted and aligned to each other in Octet Data Analysis software v9.0 (FortéBio). Association and dissociation of the full dilution series were fit in a global 1:1 model. In Fig. 1f, the blue lines are the sensorgrams for the indicated concentrations of R3DC23 and the black lines are the fitted curves. The poor fitting of the association sensorgram using a 1:1 model suggests that most likely the spike trimer is bound by multiple R3DC23 VHHs. If a ligand is captured to a biosensor in such a way it is fully available for analyte binding, stoichiometry of the interaction between ligand and analyte can be described upon saturation of the captured ligand as:
Generation of R3DC23-Fc(YTE)
A humanized (Q1D, Q5V, A13P, D15G, T19R, M63V, S73N, T84L, K83R and Q98L; according to Kabat numbering) version of R3DC23 was fused via a (G4S)2 linker to a human IgG1 Fc (EPKSCdel_YTE_K447del) and ordered synthetically at IDT as gBlocks. Upon arrival, gBlocks were solubilized in ultraclean water at a concentration of 20 ng/µL. gBlocks were A-tailed using the NEBNext-dA-tailing module (NEB), purified using CleanPCR magnetic beads (CleanNA) and inserted in pcDNA3.4-TOPO vector (Thermo Fisher Scientific). The ORF of positive clones was fully sequenced, and pDNA of selected clones was prepared using the NucleoBond Xtra Midi kit (Machery-Nagel).
Production of YTE variants of VHH-Fc in mammalian cells
HEK293S cells were transfected with VHH-Fc encoding plasmids using poly-ethylene imine (PEI). Briefly, suspension-adapted and serum-free HEK293S cells were seeded at 3 × 106 cells/mL in Freestyle-293 medium (Thermo Fisher Scientific). Next, 4.5 μg/mL of pcDNA3.4-VHH-Fc plasmid DNA was added to the cells and the cells were incubated on a shaking platform at 37 °C and 8% CO2, for 5 min. Next, 9 μg of PEI/mL was added to the cultures, and cells were further incubated for 5 h, after which an equal culture volume of Ex-Cell-293 (Sigma) was added to the cells. Transfected cells were incubated for 4 days, after which the cells were pelleted (10 min, 300 × g) and supernatants were filtered before further use.
Additionally, huR3DC23-Fc_YTE was expressed in ExpiCHO-STM cells (Thermo Fisher Scientific), according to the manufacturer’s protocol. Briefly, a 50 mL culture of 6 ×106 cells per mL, grown at 37 °C and 8% CO2, was transfected with 40 µg of pcDNA3.4-huR3DC23-Fc_YTE plasmid DNA using ExpiFectamineTM CHO reagent. One day after transfection, 300 µL of ExpiCHOTM enhancer and 8 mL ExpiCHOTM feed was added to the cells, and cultures were further incubated at 32 °C and 5% CO2. Cells were fed a second time on day 5 after transfection. Productions were collected as soon as cell viability dropped below 75%.
For purification of the VHH-Fc proteins, supernatants were loaded on a 5 mL MAbSelect SuRe column (Cytiva). Unbound proteins were removed by a wash step with McIlvaine buffer pH 7.2, and bound proteins were eluted using McIlvaine buffer pH 3.0. Immediately after elution, protein-containing fractions were neutralized with 30% (v/v) of a saturated Na3PO4 buffer. Next, these fractions were pooled, and loaded on a HiPrep Desalting column for buffer exchange to PBS pH 7.4.
Hydrophobic interaction chromatography (HIC) assay
Apparent hydrophobicity was assessed using a hydrophobic interaction chromatography (HIC) assay employing a Dionex ProPac HIC-10 column, 100 mm ×4.6 mm (Thermo Fisher Scientific, catalog nr. 063655), containing a stationary phase consisting of a mixed population of ethyl and amide functional groups bonded to silica. All separations were carried out on an Agilent 1100/1260 HPLC equipped with a UV/VIS detector. The column temperature was maintained at 25 °C throughout the run and the flow rate was 0.8 mL/min. The mobile phases used for HIC were 1.6 M ammonium sulfate and 50 mM phosphate pH 7.0 (buffer A), and 50 mM phosphate pH 7.0 (buffer B). Protein and calibrator samples were diluted 1:1 with buffer A and injected onto the column. Following a 5 min hold at 50% B, bound protein was eluted using a linear gradient from 50 to 100% B in 50 min followed by 5 min hold at 100% B. The column was washed with 100% B, followed by 50 mM ammonium acetate pH 5.0 and re-equilibration in 50% B for 10 min prior to the next sample. The separation was monitored by absorbance at 280 nm with a 16 nm bandwidth, without reference subtraction.
SARS-CoV-2 infection in K18-hACE2 Tg mice
K18-hACE2Tg/+ mice were obtained from The Jackson Laboratory (Strain #034860). Male and female K18-hACE2Tg/+ mice were housed under specific pathogen-free conditions before they were transferred to the Biosafety level 3 laboratory at VIB-UGent Center for Medical Biotechnology. In the BSL-3 laboratory, mice were housed with a 16 h light (6 A.M.–10 P.M.), 8 h dark (10 P.M.–6 A.M.) cycle, at room temperature (21 °C) and 45-65% humidity. Mice were used between 8 and 11 weeks of age. All experiments were approved by the animal ethics committee at Ghent University (EC2020-125) and were in accordance with Belgian animal protection law. Mice received 150 µg huR3DC23-Fc via an intraperitoneal injection in 200 µL. The irrelevant antibody palivizumab (anti-RSV monoclonal antibody) was used as a negative control. Twenty-four hours later, animals were anesthetized by isoflurane inhalation and 3*10^2 PFU of SARS-CoV-2 614 G (SARS-CoV-2/human/FRA/702/2020, obtained from the European Virus Archive) was administered by intratracheal instillation. Each treatment group contained 5 mice. Animals were monitored on a daily base by a blind observer who measured weight change and scored for humane endpoints (hunchback (1 point), piloerection (1 point), less movement upon opening cage (1 point), motionless upon touching (2 points), neurological symptoms (shaking, balance, 3 points), heavy breathing (3 points)). Mice that lost more than 25% of their initial bodyweight or reached a humane endpoint with a score of 5 points were euthanized.
In a separate experiment, mice were sacrificed with an overdose of pentobarbital on day five post infection to measure viral replication and inflammatory cell influx in the lung. The left lung lobe was collected in PBS and frozen for virological analysis. The right lung lobes were isolated, cut with scissors and then digested for 30 min in RPMI-1640 (Gibco, Thermo Fisher Scientific) containing 20 µg/mL liberase TM (Roche), 10 U/mL DNase I (Roche), and 5% of FCS (Bodinco) at 37 °C. Next, lungs were filtered through a 70 µm cell strainer. For quantification of the viral lung load, the left lung lobe was homogenized using a Precellys Tissue Homogenizer (Bertin) and 2 mL Precellys tubes. After centrifugation (1000 × g for 5 min), 5-fold dilution series of the supernatant were prepared in duplicate and added to a monolayer of VeroE6/TMPRSS2 cells in 24-well plates and incubated at 37 °C for 1 h. Subsequently, Avicel was added to a final concentration of 0.3% (w/v). After 2 days of incubation at 37 °C, the overlays were removed, the cells were fixed with 3.7% paraformaldehyde (PFA) and stained with 0.5% crystal violet. Per dilution series, the viral plaques were counted in at least 2 wells and used to calculate the number of PFU. Finally, for each mouse the PFU per g was calculated as the mean of 2 duplicates. For the quantification of the viral RNA, 150 µL of cleared lung homogenate supernatant was used to prepare RNA using the Macherey-Nagel NucleoSpin RNA Virus, Mini kit for viral RNA. cDNA was prepared using random hexamer primers (Roche Transcriptor First strand cDNA synthesis kit) and used to performing qPCR using the following Nucleocapsid specific primers and Roche probe: FW primer 5’-TTA CAA ACA TTG GCC GCA AA-3’, RV primer 5’-TTA CAA ACA TTG GCC GCA AA-3’ and probe 5’-FAM-ACA ATT TGC CCC CAG CGC TTC AG-BHQ1-3’.
The following statistical tests were used to test statistically significant differences between groups: Log-rank, Mantel-cox test for animal survival (shown in Fig. 2j, left panel), two-way ANOVA with Tukey’s multiple comparisons test for mean relative bodyweight (Fig. 2j, right panel), Kruskal-Wallis test with Dunn’s multiple comparisons test for lung viral titers (Fig. 2k), one-way ANOVA with Tukey’s multiple comparisons test when data are normally distributed (AMs, eosinophils, monocytes, T cells and B cells) or Kruskal-Wallis test with Dunn’s multiple comparisons test when data are not normally distributed (DCs, neutrophils) (Fig. 2l).
Flow cytometric analysis of the leukocyte influx in the lungs
Lung single cell suspensions were incubated with the eBioscience™ Fixable Viability Dye eFluor™ 506 (Thermo Fisher Scientific, 1/300) to identify dead cells. Fc Block 2.4.G2 (Bioceros, 1/800) was used to prevent aspecific antibody binding. Cell surface markers were stained for 30 min at 4 °C in the dark.
Lung single-cell suspensions were stained for flow cytometry using PerCP/Cy5.5-conjugated CD88 (clone 20/70; BioLegend, catalog number 135812, 1/200), eFluor450-conjugated Ly-6C (clone HK1.4; eBioscience, catalog number 48-5932-82, 1/500), BV605-conjugated CD11b (cline M1/70; BioLegend, catalog number 101237, 1/800), BV650-conjugated Ly-6G (clone 1A8; BD Biosciences, catalog number 740554, 1/400), BV711-conjugated CD64 (clone X54-5/7.1; BioLegend, catalog number 139311, 1/100), BV786-conjugated Siglec-F (clone E50-2440; BD Biosciences, catalog number 740956, 1/400), AF700-conjugated CD45 (clone 30-F11, BioLegend, catalog number 103127, 1/800), APC-eFluor780 conjugated anti-I-A/I-E (clone M5/114.15.2; Thermo Fisher Scientific, catalog number 47-5321-82, 1/1000), PE-Cy5-conjugated CD3e (clone 145-2c11; Thermo Fisher Scientific, catalog number 15-0031-81, 1/200), PE-Cy5-conjugated NK1.1 (clone PK136, BioLegend, catalog number 108713, 1/200), PE-Cy5-conjuaged CD19 (clone 1D3; Thermo Fisher Scientific, catalog number 15-0193-82, 1/400) and PE-Cy7-conjugated CD11c (clone N418; Thermo Fisher Scientific, catalog number 25-0114-82, 1/800).
After staining, lung cells were washed and fixation was performed using BD cytofix fixation buffer (BD Biosciences) for 20 min. Samples were washed and 123count eBeads™ Counting Beads (Thermo Fisher Scientific) were added to each sample to determine absolute cell numbers. Settings were calibrated using UltraComp eBead™ Compensation Beads (Thermo Fisher Scientific). Data were acquired and analyzed on a NovoCyte Quanteon flow cytometer (Agilent). All procedures were performed in BSL-3 conditions. The gating strategy for data analysis is shown in Supplementary Fig. 13B.
Production of huR3DC23-Fc_LS following transient transfection
The gene encoding huR3DC23-Fc_LS was codon optimized, synthesized, and cloned into the pXLG6 backbone vector at ATUM’s laboratories. Upon gene and codon optimization, the R3DC23 DNA sequence was inserted into pXLG6 expression vector and transfected in CHOExpress™ cells at a cell density of 4 ×106 cells/mL. TGE supernatant was harvested by centrifugation and clarified by filtration (0.2 µm) after 10 days when cell viability dropped below 90%. The protein was further purified by Protein A affinity chromatography.
Fed batch production of huR3DC23-Fc_LS from stable pool at 1 L scale
The gene encoding huR3DC23-Fc_LS was codon optimized, synthesized, and cloned into the pXLG6 backbone vector at ATUM’s laboratories. Upon expansion to a density of about 4 ×106 cells/mL, parental CHOExpress™ cells were co-transfected with the expression vector and the pXLG5 helper vector. The stable pool was generated under 50 mg/L puromycin selective pressure (applied daily) and further expanded. The stable pool research cell bank was banked at day 14 when cell viability reached 95%.
The RCB pool was then expanded for protein production at 1 L scale and cultured until day 12 (cell density 3.5 ×107 cells/mL, cell viability 96%). The supernatant was harvested by centrifugation and clarified by filtration (0.2 µm). The protein was further purified by Protein A using MabSelect SuRe LX resin. Consecutive washed were performed with 20 mM sodium phosphate and 110 mM NaCl at pH 7.2; 100 mM sodium acetate and 500 mM NaCl at pH 5.5; and 20 mM sodium phosphate at pH 7.2. The eluate in 100 mM sodium acetate pH 3.5 was neutralized to pH 7.0 by addition of 1 M Tris pH 11.0 (10%v/v). After filter sterilization (0.22 µm), the protein was aliquoted at 2 mg/mL.
Protein preparation for biophysical analysis of Fc-fusions
Preceding biophysical characterization, MAbSelect SuRe-purified protein samples were further purified via size-exclusion chromatography (SEC) on a 12 °C-cooled Superdex 200 column (Cytiva) equilibrated with either phosphate-buffered saline (PBS, Sigma-Aldrich) supplemented with 0.02% sodium azide to prevent microbial growth, or a sample buffer comprising 50 mM L-histidine and 150 mM L-arginine (Sigma-Aldrich), 0.02% polysorbate-20 and 0.02% sodium azide, set to pH 7.0 at 25 °C. After filter sterilization (0.22 µm), 1 mg/mL aliquots were snap-frozen in polypropylene tubes in liquid nitrogen and stored at −80 °C.
Sample composition
Purified VHH-Fc samples were characterized by analytical SEC to determine the molecular composition of each sample. After rapid thawing in a water bath at 25 °C, 10 min centrifugation at 16,000 × g and transfer of supernatant to fresh tubes, 5 µg was injected on an AdvanceBio SEC column, 4.6 ×300 mm (Agilent) with 2.7 µm porous particle size and 300 Å pore size, calibrated with PBS. The separation was monitored by absorbance at 280 nm with a 16 nm bandwidth, without reference subtraction. For additional quality control, proteins were separated on reducing 15% SDS-PAGE with Coomassie staining.
Acquisition of sotrovimab (without LS), cilgavimab, bebtelovimab biosimilars and palivizumab
Bebtelovimab biosimilar (PX-TA1750), cilgavimab biosimilar (PX-TA1033) and sotrovimab biosimilar (without LS) (PX-TA1637) were purchased from Proteogenix. Clinical grade palivizumab was obtained from the Ghent University hospital.
Neutralization assays using authentic SARS-CoV-2 to test the neutralizing activity of huR3DC23-Fc_LS
SARS-CoV-2 viruses belonging to different lineages (D614G, Delta, Omicron BA.1, Omicron BA.2 and Omicron BA.5) were isolated from nasopharyngeal swabs taken from patients/travelers between January 2020 and July 2022. More specifically, the following clinical isolates were used: SARS-CoV-2 Isolate BavPat1/2020/Germany (09 Feb 2020); Delta variant SARS-Related Coronavirus 2, Isolate hCoV-19/USA/MD-HP05647/2021; Omicron BA.1 variant SARS-CoV-2 hCoV-19/Netherlands/NH-RIVM-72291/2021; Omicron BA.2 variant Clinical isolate hCoV-19/Netherlands/VCB-20220303-1/2022; and Omicron BA.5 variant Clinical isolate hCov19/NL/VCB-20220714-2/2022. Dose-dependent neutralization of the test item (huR3DC23-Fc_LS), the positive controls (bebtelovimab biosimilar, cilgavimab biosimilar, sotrovimab biosimilar (without LS) and a negative control (isotype control) were assessed in an authentic virus neutralization assay. For all assays in which authentic D614G, Delta, BA.1, and BA.2 SARS-CoV-2 were tested, huR3DC23-Fc_LS produced from transiently transfected cells was used. For all assays in which SARS-CoV-2 BA.5 was tested, huR3DC23_Fc_LS produced from stable cell pools was used. For each variant, three independent runs were performed. Different system controls were included in the assay: cell only (medium only), virus only, and an internal positive control (human serum). Briefly, 5-fold or 7-fold serial dilutions of the test items and controls were incubated with a fixed amount of plaque-forming units (PFUs) of the virus for 1 h at room temperature. Afterwards, the Vero E6 cell monolayer was inoculated with virus-antibody mixtures for 1 h at 37 °C. In a next step, the inoculum was removed and cells were incubated at 37 °C with infection medium (up to 18–24 h post-infection). Afterwards, the SARS-CoV2 infected cells were fixed and immunostained with a SARS-CoV Nucleocapsid Antibody (Sino Biological, catalog number: 40143-MM05, clone #5), followed by HRP-conjugated Goat anti-Mouse IgG (H + L) Secondary Antibody (Invitrogen, catalog number A16072). Spots (infected cells) were counted using an Immunospot Image Analyser. For each test item, the compound concentration showing 50 % reduction in infection (IC50) was calculated based on the Zielinska method. The geometric mean values were calculated based on three independent runs. (ND) Not possible to determine IC50 value for sotrovimab (without LS) within the tested concentration range (0.128-10 000 ng/mL). (NT) Cilgavimab was not tested for Omicron BA.5. (*) The IC50 of cilgavimab for BA.1 is based on 2 biological replicates in which the highest concentration of cilgavimab (500 ng/mL) resulted in a more than 50 % reduction of viral replication but not in a third biological replicate. (Sotrovimab biosimilar) = sotrovimab biosimilar without LS mutation.
SARS-CoV-2 challenge model in Syrian golden hamsters
In brief, 9-to 10-weeks-old male Syrian golden hamsters (Mesocricetus auratus) weighing 89.8–132.3 g were obtained from Janvier (France). Housing conditions and experimental procedures were approved by the ethics committee in the Netherlands (study was registered under number 27700202114492-WP49). The hamsters were housed according to SOP VCX-P073 (Animal housing and welfare management) in elongated type 2 IVC group cages with one or two animals per cage under BSL-II conditions during acclimatisation and in elongated type 2 group cages under BSL-III conditions (isolators) during challenge. Six hamsters per group were infected intranasally with 100 TCID50/dose SARS-CoV-2 (Wuhan strain) in a total dose volume of 100 µL, divided equally over both nostrils. The test item huR3DC23-Fc_LS (2 and 10 mg/kg), palivizumab (10 mg/kg) and bebtelovimab (10 mg/kg) were administered by intraperitoneal injection 4 h after the SARS-CoV-2 challenge. The irrelevant antibody palivizumab (anti-RSV monoclonal antibody) was used as a negative control, while bebtelovimab was used as a positive control. The huR3DC23-Fc_LS used in the hamster study was produced from stable cells pools. Hamsters were monitored daily for behavior, appearance and body weight.
On day 4 post-infection, animals were sacrificed. At the time of necropsy, gross pathology was performed and abnormalities were recorded. Samples from the right lung lobes were collected and frozen for virological analysis. To determine virus titers, quadruplicate 10-fold serial dilutions were used in confluent layers of Vero E6 cells. To this end, serial dilutions of the samples (lung tissue homogenates) were incubated on Vero E6 monolayers for 1 h at 37 °C. Vero E6 monolayers were then washed and incubated for 5 or 6 days at 37 °C, after which plates were stained and scored based on cytopathic effect (CPE) by using the vitality marker WST8 (colorimetric readout). Viral titers (Log10 TCID50/g) were calculated using the Spearman-Karber method. For the viral titration, the lower limit of detection (LLOD) ranged between 1.1 and 1.3 log10 TCID50/g. To detect viral RNA, lung tissue and homogenates were used. RNA was isolated and Taqman PCR was performed. The number of copies (Log10 CP/g) in the different samples was calculated against a standard included in each run. For the viral RNA, the LLOD was 3.5 Log10 CP/g. In the graphs in Fig. 2h, the dotted horizontal lines indicate the LLOD.
Blood samples were collected prior to the start of the study on day -2 ( ~ 200 μl of blood was collected for serum preparation under isoflurane anesthesia) and on day 4 post-infection (p.i.), at time of necropsy for pharmacokinetic analysis. Blood samples for serum preparation were immediately transferred to appropriate tubes containing a clot activator. Serum was collected and stored frozen. To inactivate potential infectious material present and to allow the testing of the sera samples in a BSL-2 environment, day 4 post-infection sera was heat-treated at 56 °C for 30 min.
One-way ANOVA with Dunn’s multiple comparisons test was used to test statistical differences in TCID50/gram lung tissue and viral RNA copies/gram lung tissue between treatment groups in hamsters (Fig. 2n).
To assess whether there was a difference in hamster body weight (BW) at any time during the follow up of 4 days, we used the per hamster normalized BW. This was calculated as the BW at each of the 4 days day minus the BW 2 days before the start of the experiment. This normalized BW was the outcome variable in a linear mixed model, run in R53 using the nlme54,55 and splines packages. As predictor variables, we used the time (in days) and the treatment group (positive control, negative control, VHH-Fc low and VHH-Fc high dose groups). The time was modeled with a natural spline with 1 internal knot (df = 2) at day 2.5. An interaction between time and treatment group allowed for different courses of BW in the different treatment groups. Further, we allowed for a random intercept and a random slope for time in each hamster to model the within-hamster correlation between the time points. We obtained confidence intervals for pairwise differences in BW between the negative control group and all other treatment groups using the contrast56 and rms57 packages. The intervals were calculated with robust variance-covariance estimators and corrected for multiple testing with the multcomp58 package.
ELISA for detection of huR3DC23-Fc_LS in hamster sera
Pharmacokinetic analysis (PK) of the hamster serum samples was done using an ELISA based assay. Streptavidin-coated 96-wells microtiter plates were pre-blocked with superblock T20 (Thermo Fisher Scientific, catalog number 37516) at room temperature. Afterwards, the plates were washed 3 times with 200 µL of washing buffer (PBS with 0.05% Tween20). Biotinylated anti-VHH monoRab monoclonal antibody (Genscript, catalog number A01995-200) was captured at a concentration of 0.1 μg/mL (10 ng/well in 0.1 mL) onto the streptavidin plate for 2 h at room temperature with gentle shaking (400 rpm). Next, the supernatant was removed, and plates were washed three times with washing buffer. As a next step, 100 µL of 1.7-fold dilution series of standard calibrator (huR3DC23-Fc_LS), freshly prepared quality control samples (QCs) or hamster serum samples were added to the plates (according to a minimal required dilution of 10 or 50), which allowed huR3DC23-Fc_LS to bind to the captured anti-VHH antibody. After 1 h incubation with shaking at room temperature, plates were again washed three times with washing buffer. To detect bound huR3DC23-Fc_LS onto the plates, anti-VHH monoRab antibody conjugated to horse-radish peroxidase (Genscript, catalog number A01861-200) was added at a concentration of 0.2 µg/mL (0.1 mL per well) and incubated for 30 min at room temperature with shaking. Plates were washed three times with PBS buffer to remove the remaining Tween20. The final step was the addition of 100 µL of 1-Step Ultra TMB-ELISA Substrate solution (Thermo Fisher Scientific, catalog number 34029) into wells for 5 min to allow a colorimetric reaction. Following the color development, the reaction was stopped by adding 50 µL of Stop solution (Thermo Fisher Scientific, Catalog number N600). The colorimetric output was read on a Tecan Spark instrument with SparkControl software.
S1 shedding assay
Antibody or VHH was added at a final concentration of 10 µg/mL to 500 000 HEK293T cells that were previously transfected with a SARS-CoV-2 spike (D614G)-expressing plasmid, or an empty control vector. The antibody-cell mixture was incubated for 30 min at 37 °C and 5% CO2. After incubation, cells were pelleted by centrifugation, supernatant was transferred to a fresh tube and the cell pellet was lysed with 250 µL of RIPA lysis buffer (50 mM Tris-HCl pH 8.0, 100 mM NaCl, 1 mM EDTA, 1 mM EGTA, 0.1% SDS, 1% NP-40). Twenty µL samples of supernatant and lysate were separated on 8% SDS-PAGE gels, and electroblotted onto nitrocellulose membranes. Membranes were blocked with 4% milk, stained with rabbit anti-SARS-S1 antibody (1/1000, Sino Biological, 40591-T62) followed by anti-rabbit IgG-HRP (1/2000, Cytiva, NA934V) and developed using Pierce™ ECL Western Blotting Substrate (Thermo Fisher Scientific).
Fusion inhibition assay using spike-expressing Vero E6 cells
Vero E6 cells were transfected with a GFP expression vector together with either a control expression vector (no spike) or a SARS-CoV-2 spike expression vector using Fugene (Promega). Two hours after transfection, PBS, monoclonal antibodies, or VHHs were added to a final concentration of 10 μg/mL. Twenty-two or forty hours later, the cells were imaged using an Olympus fluorescence microscope using a 10x lens. Alternatively, monoclonal antibodies or VHHs were added to transfected cells at a final concentration of 1 µg/mL and GFP expression was monitored hourly over time with an Incucyte Zoom live cell analysis device (Sartorius) and analyzed with the provided Incucyte software. To derive fusogenicity of spike variants, the mean slope of the curve derived from the area of green objects at time points 24 h after transfection (maximum GFP signals in negative control cells, indicating maximal GFP expression) and 38 h after transfection (maximal size of syncytia, before cell death) was calculated.
Fusion inhibition assay using replication-competent GFP report VSV virus pseudotyped with Wuhan SARS-CoV spikes(del-18)
VeroE6/TMPRSS2 cells were infected with 40 PFU of SARS-CoV-2 spike GFP-expressing pseudotyped replication-competent VSV-GFP virus, generously provided by Dr. Florian Schmidt21. Two hours later the indicated monoclonal antibodies or VHHs were added. Non-infected cells were used as negative controls. Infected cells were incubated overnight and imaged with an Olympus fluorescence microscope using a 4x lens. GFP fluorescence was measured with a fluorimeter. Of note, different from the clone of replication-competent pseudotyped VSV particles used in the neutralization assays, the furin cleavage site of the virus used in the fusion assays was intact as confirmed by Sanger sequencing.
Fusion inhibition assay using split GFP
HEK293T cells in a 96 well plate were transfected with a GFP11 expression vector (Addgene, plasmid #68716), and HEK293T cells in a 6 well plate were cotransfected with a GFP1-10 expression vector (Addgene, plasmid #68715) and a surface-expressed SARS-CoV-2 spike expression vector, or an empty vector. One day after transfection, the HEK293T cells expressing GFP1-10 and the spike protein were detached with trypsin, incubated for 15 min at room temperature with CB6 (which destabilizes the spike by arresting the RBD in an up conformation and as such induces S1 shedding and subsequent cell-cell fusion was induced by CB6) or with a control antibody, and subsequently incubated with VHH or antibody at a final concentration of 10 µg/mL, for 30 min at room temperature. After incubations, the cells were added to the HEK293T cells that express GFP11. Cell-cell fusion was monitored by measuring the GFP fluorescence with a Tecan infinite 200 pro plate reader 16 h after co-incubation of the cells.
Viral escape selection
Monolayers of VeroE6/TMPRSS2 cells seeded in 96 well plates were infected with 200 PFU of GFP expressing replication-competent VSV virus particles pseudotyped with the SARS-CoV-2 spike protein containing an intact furin cleavage site, generously provided by Dr. Florian Schmidt21. Two hours after infection, 10 μg/mL of VHH R3DC23 was added to 59 wells, and as control 1 well was treated with PBS. Rapid syncytium formation and spread of the infection to neighboring cells was observed in the PBS treated well, whereas these events were blocked or strongly delayed in wells with R3DC23. From the growth medium of wells that displayed syncytia formation or viral replication in the presence of VHH R3DC23, nine single clones were isolated by limiting dilution on fresh VeroE6/TMPRSS2 cells seeded in 96 well plates. Growth medium of wells with a single PFU was used to propagate possible escape viruses on monolayers of VeroE6/TMPRSS2 cells seeded in 6 well plates in the presence of VHH R3DC23. From these infected cells, RNA was prepared using a nucleospin RNA virus kit (Macherey Nagel Bioanalysis) and used to generate cDNA using random hexamer primers. This cDNA was used to amplify the spike coding sequences by PCR. The resulting PCR fragments were purified and sequenced using Sanger sequencing. The obtained nucleotide sequences were analyzed and aligned to spike proteins of WT SARS-CoV-2 and clade 1, 2, and 3 sarbecoviruses using CLC Main Workbench 20.0.4. Each of these viral clones had acquired a single amino acid substitution at one out of four positions: N1192D (1 clone), L1197P (2 clones), L1200P (1 clone), Q1201R (4 clones), and Q1201K (1 clone). Mutations were visualized on a model of full-length glycosylated spike protein obtained from Charmm-gui.org (PDB: 6VXX_1_1_1 model) or the SARS-CoV-2 HR2 coiled-coil as determined by NMR (PDB: 2FXP) using Pymol.
Growth kinetics of viral escape variants
Vero E6 cells seeded in a 96 well-plate were infected with 50 PFU of GFP-expressing replication-competent VSV virus particles pseudotyped with the SARS-CoV-2 spike protein, that were obtained during escape selection. GFP-expression was monitored hourly with an Incucyte Zoom device and analyzed with accompanying software.
HDX-MS epitope mapping
SARS-CoV-2 S-2P trimer at 3.33 μM was incubated overnight at 37 °C. The protein was then diluted to 1.66 μM trimer in the presence or absence of 6.25 μM R3DC23 in 1x PBS (pH 7.4, Sigma-Aldrich P4417). To initiate exchange, the protein was diluted tenfold into 25 °C temperature-equilibrated deuterated buffer made by lyophilizing 1x PBS and resuspending in D2O (Sigma-Aldrich 151882). Samples were quenched at each time point (15 s, 3 min, 30 min, 3 h) by mixing 60 µL of the exchange reaction with 60 µL of ice-cold 2x quench buffer (3.6 M guanidinium chloride, 500 mM TCEP, 200 mM glycine pH 2.4). The quenched samples were incubated on ice for 1 min and then flash frozen in liquid nitrogen and stored at −80 °C until LC-MS. LC-MS and data analysis was conducted as previously described27. The glycosylated peptide has taken up more deuterium than the number of backbone exchangeable sites (11 sites) because the glycan can uptake and retain deuterium at amide sites similarly to the backbone as noted by Guttman et al.59. No bimodal spectra were noted outside of populations <5% in regions previously described in Costello et al.27 which are associated with the prefusion and open-interface trimer conformational change. One biological replicate (same VHH preparation, other S-2P preparation) was performed for R3DC23 bound to S-2P in the prefusion conformation.
To allow access to the HDX data of this study, the HDX data summary table (Supplementary Table 1) and the HDX data tables (included in the Source Data file) are included as per consensus guidelines60.
Generation of 5HB and 6HB constructs
For the 5HB construct, the following sequence was ordered synthetically at IDT as a gBlock: the transmembrane domain and part of the stalk of an influenza NA mutant that assembles in monomers instead of tetramers61, followed by HR1 (DNA encoding AA911-988), a SGGRGG linker, HR2 (DNA encoding AA1162-1206), a GGGGKGGSG linker, HR1, SGGRGG linker, HR2, GGGGKGGSG linker, HR1, GGGGS linker and finally an HA tag. This gBlock was cloned into the pcDNATM 3.4 TOPOTM vector with the pcDNATM 3.4 TOPOTM TA cloning kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. A secreted version of such a 5HB was shown to bind to HR2 peptides and to neutralize SARS-CoV-2 with µM-range potency (31). Similarly, we observed binding of monomeric SUMO-HR2 fusion proteins but not SUMO-HR1 fusion proteins to 5HB expressing cells, demonstrating correct folding of the 5HB (Supplementary Fig. S15). To create the 6HB construct, an extra HR2 was cloned into this construct, after the third HR1 repeat, with Gibson assembly. In brief, a gBlock encoding the last residues of HR1 (KVE), the SGGRGG linker and HR2 sequence was ordered synthetically at IDT. Both the HR2 gBlock and existing 5HB were amplified with PCR using primers that created homologous regions at the 3’ and 5’ ends. Finally, the insert and vector were joined using the NEBuilder® HiFi DNA assembly kit (New England Biolabs). The ORFs were sequence-verified with Sanger sequencing.
HR2 expression and purification
For structural biology purposes the HR2 protein was expressed in a bacterial expression system. Therefore, the synthetic gene encoding residues H1159-K1211 of the SARS-CoV-2 spike was cloned into a pFloat-SUMO vector, generating a His-tagged SUMO-HR2 fusion protein. The construct also contained a 3C protease cleavage site to remove the His-SUMO-tag. The pFloat-SUMO-HR2 plasmid was transformed in BL21(DE3) cells and plated on kanamycin (100 µg/mL) containing LB agar plates. A small LB culture, supplemented with 100 µg/mL kanamycin, was inoculated with a single colony of BL21(DE3)(pFloat-SUMO-HR2) and grown overnight at 37 °C. One L LB cultures were subsequently inoculated with 20 mL of this preculture and grown at 37 °C until OD600 reached 0.8. At this point protein expression was induced by adding 0.5 mM IPTG to the E. coli culture. Cells were incubated further overnight at 20 °C and subsequently harvested by centrifugation (Beckman rotor 8.1000, 5000 rpm (4752 g), 15 min, 4 °C). The pellet was resuspended in PBS, 500 mM NaCl, 10 mM imidazole, 5 mM ß-mercaptoethanol, 0.1 mg/mL 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride (AEBSF), 1 µg/mL leupeptine, 50 µg/mL DNaseI and 20 mM MgCl2. The cells were lysed using a French press (Constant Systems) at 20 kpsi and the cell debris was removed by centrifugation. The cell lysate was loaded on a Ni-sepharose FF HiLoad column (GE Healthcare), equilibrated in 20 mM Tris-HCl pH 7.5, 500 mM NaCl, 10 mM imidazole, 5 mM ß-mercaptoethanol. The bound proteins were eluted using a linear gradient to 500 mM imidazole. Fractions containing the His-SUMO-HR2 protein were pooled and dialyzed overnight to 20 mM Tris-HCl pH 7.5, 150 mM NaCl at 4 °C, followed by 2 h incubation with 3 C protease at room temperature. The cleaved sample was loaded again on a Ni-sepharose FF HiLoad column, equilibrated in the same buffer. The flow through, containing the HR2 protein, was concentrated and applied to a BioRad Enrich70 10/30 size exclusion column (SEC), equilibrated in 20 mM Tris-HCl pH 7.5, 150 mM NaCl. The HR2-containing SEC fractions were pooled.
Generation, production and purification of HR2-foldon constructs
For the HR2-foldon constructs, the following sequences were ordered synthetically at IDT as gBlocks: the SARS-CoV-2 signal peptide, followed by the HR2 sequence (in Wuhan numbering: for the SARS-CoV-2 HR2 (QELGKYEQ-GS-GYIPE): AA1159-1208, for the SARS-CoV-2 HR2 (QELGKY-IPE): AA1159-1206), for the SARS-CoV-2 HR2 (QEL-QEAQGYIPE): AA1159-1203), for the MERS-CoV HR2: AA1242-1291) linked by a GS linker to the foldon sequence, followed by a PreCission cleavage site, 8x His-tag and 2x StrepII tag. This gBlock was cloned into the pcDNATM 3.4 TOPOTM vector with the pcDNATM 3.4 TOPOTM TA cloning kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. HEK293T cells were transfected with the sequence verified construct for production. Five days after transfection, the supernatant of cells was collected and cleared from cell debris by centrifuging twice (10 min, 400 × g). HR2-foldon constructs were purified with Ni SepharoseTM High Performance beads (Cytiva), which were first equilibrated with 20 mM NaH2PO4 pH 7.5, 0.5 M NaCl, and 20 mM imidazole. Bound HR2-foldon was eluted with 20 mM NaH2PO4, 20 mM NaCl and 0.5 M imidazole. The eluted fraction was then buffer exchanged to PBS with 3 kDa cutoff centrifugal units (Amicon).
R3DC23-HR2 crystallization, X-ray data collection, processing, and structure determination
For crystallization, R3DC23 was added to HR2 in 1.2 times molar excess and concentrated to 19 mg/mL using Amicon Ultra 3 kDa cut off centrifugal filter devices. Crystallization screens were set up using the sitting drop vapor diffusion technique, mixing 0.1 µL of R3DC23-HR2 and 0.2 µL bottom solution. Crystals were grown from the Molecular Dimensions Proplex crystallization screen, in 0.1 M magnesium chloride hexahydrate, 0.1 M sodium citrate pH 5.0, and 15% PEG4000. For X-ray data collection, crystals were flash frozen in liquid nitrogen. X-ray data were collected on the i24 beamline at the Diamond Light Source synchrotron facility (Didcot, UK). X-ray data were processed using autoPROC+Staraniso62,63. The structure of the R3DC23-HR2 complex was solved using the automatic molecular replacement workflow in the CCP4 cloud64. The initial model was further build manually in Coot65 and refined using phenix.refine66 from the Phenix crystallographic software suite67. Data collection parameters, as well as processing and refinement statistics are shown in Supplementary Table S2. The electron density map of R3DC23 bound to two HR2 helices is presented in Supplementary Fig. 14.
SARS-CoV-2 spike sequence variant analysis
SARS-CoV-2 spike protein sequences were downloaded from GISAID (N = 17,063,827 sequences available on November 10th 2024). Only sequences originating from infected human hosts were retained. Very short spike sequences retrieved from incompletely sequences genomes were removed. In the next step, we removed spike protein sequences with a high frequency of undetermined amino acids (X), derived from poor sequencing results (Ns). X frequency threshold was defined as (IQR * 1.5) + Q3 = 5.0% X per protein, where IQR and Q3 are inter-quartile range and the third quartile, respectively. Further, proteins with premature stop codon(s) were excluded. The resulting 13,564,753 quality-controlled spike protein sequences were aligned using the Muscle algorithm in R version 4.1.1 package msa version 1.24.0 with 12 as gap opening and 3 as gap extension penalties and the BLOSUM80 substitution matrix.
To calculate sequence variation, amino acid positions present in the reference spike sequence annotated in NC_045512.2 (Wuhan-Hu-1 isolate, NCBI RefSeq) were extracted from multiple sequence alignment (MSA), disregarding insertions. Conservation matrixes were calculated using Biostrings version 2.60.0 for different subsets of the MSA: sequences derived from the Omicron variant (N = 7,305,876), sequences of Omicron sub-lineages (see figure for respective sequence counts) or sequences obtained before 31/10/2021 (prior to the onset of Omicron, N = 4,697,133). Variant frequencies were derived from conservation matrixes, while undetermined amino acid X frequencies were excluded. Minor variant frequencies were summed to obtain overall sequence variability and calculate conservation percentage as compared to the Wuhan reference.
Sequence variation was overlaid on the surface of spike protein structure model published by the Amaro lab (https://amarolab.ucsd.edu/files/covid19/PSF-PDB_spike_open_prot_glyc_memb_wat_ions_amarolab.tar.gz). R package Bios2cor 2.2.1 was used to calculate Shannon’s entropy score of each position in the MSA, except positions with more than 20 percent of gaps, which were excluded.
HDX-MS analysis of the transition kinetics of prefusion to open-interface spike trimer
3.33 µM SARS-CoV-2 S-2P trimer was incubated at 37 °C for 2 days. The protein was then diluted to 1.66 µM trimer in the presence or absence of 6.25 µM R3DC23 in 1× PBS (pH 7.4, Sigma-Aldrich P4417). The conversion from prefusion to open-interface trimer was initiated by incubating the protein at 4 °C. At 1 h, 3.5 h, 7.5 h, 25 h and 100 h, 6 µL of protein was removed from the incubation, diluted tenfold with deuterated 1× PBS (made as indicated in other HDX method sections), allowed to label for 60 seconds at 25 °C and then quenched with 60 µL of ice-cold 2× quench buffer (3.6 M guanidinium chloride, 500 mM TCEP, 200 mM glycine pH 2.4). The quenched samples were incubated on ice for 1 min and then flash frozen in liquid nitrogen and stored at −80 °C until LC-MS. At 100 h of incubation at 4 °C the final timepoint for measuring the prefusion to open-interface trimer conversion was taken. One experiment was performed. LC-MS and data analysis was conducted as previously described27.
HDX-MS analysis of the transition kinetics of open-interface trimer to prefusion spike
After 100 h of incubation at 4 °C to convert spike from predominantly prefusion to predominantly open-interface trimer, the same aliquot of protein (1.66 µM trimer in the presence or absence of 6.25 µM R3DC23) was moved from 4 °C to 37 °C to initiate the conformational change from open-interface trimer to prefusion. At 10 s, 1 min and 24 h, 6 µL of protein was removed from the incubation, diluted tenfold with deuterated 1× PBS (made as indicated in other HDX method sections), allowed to label for 60 sec at 25 °C and then quenched with 60 µL of ice-cold 2× quench buffer (3.6 M guanidinium chloride, 500 mM TCEP, 200 mM glycine pH 2.4). The quenched samples were incubated on ice for 1 min and then flash frozen in liquid nitrogen and stored at −80 °C until LC-MS. One experiment was performed. LC-MS and data analysis was conducted as previously described27.
Flow cytometric analysis of binding of HIS-SUMO-HR2 or HR2-foldon to cells displaying the extended spike intermediate after antibody-evoked S1 shedding
HEK293T cells were co-transfected with a GFP expression vector and a SARS-CoV-2 spike(del18) expression vector. Two days after transfection, the cells were detached using ice-cold trypsin and subsequently blocked with 1% BSA. Alternatively, when indicated the cells were detached with enzyme-free dissociation buffer and subsequently activated by trypsin. To refold the prefusion spike into the extended intermediates containing the HR1 coiled-coil, the cells were stimulated with 10 µg/mL VHH72_S56A-Fc for 10 min at 37 °C to trigger shedding of S1, or as control stimulated with BSA. This was performed after pre-incubating the cells with either 10 µg/mL huR3DC23-Fc or palivizumab control antibody on ice for 20 min. Refolding to the extended intermediate spike conformation containing the HR1 coiled-coil was monitored by staining with 10 µg/mL HIS-SUMO-HR2, or as control with HIS-SUMO-HR1. Binding of HIS-SUMO-HR2 and HIS-SUMO-HR1 was detected using a mouse anti-HIS antibody and an AF647-conjugated anti-mouse IgG antibody. Binding was calculated as the ratio AF647 MFI of GFP+ cells over that of GFP- cells. Alternatively, when indicated, stable display of the extended intermediate was obtained by pre-treating cells with huR3DC23-Fc (10 µg/mL) for 20 min on ice before inducing S1 shedding by treatment with 10 µg/mL VHH72_S56A-Fc for 10 min at 37 °C. After washing twice, the cells were stained with 5 µg/mL HR2(SARS-CoV-2)-foldon or HR2(MERS)-foldon preincubated with a 2-fold molar excess of either monomeric VHH R3DC23 or a control VHH (GBP). Binding of HR2-foldon was detected using a mouse anti-strep-tag antibody and AF647-conjugated anti-mouse IgG antibody. Binding was calculated as the ratio of the AF647 MFI of GFP+ cells over that of GFP- cells
The following tests were used to test statistical significance between groups: two-way ANOVA with Sidak’s multiple comparisons for binding of SUMO-HR2 to shedded spike when added before S1 shedding (Fig. 8d), one-way ANOVA with Dunn’s multiple comparisons test for binding of SUMO-HR2 (or SUMO-HR1) to the HR1 coiled coil when added after S1 shedding (Fig. 8e), two-way ANOVA with Sidak’s multiple comparisons test for binding of HR2-foldon to the HR1 coiled coil (Fig. 8f).
Prediction of protein structures with AlphaFold-Multimer
The trimer structures were predicted with AlphaFold-Multimer (v2.3.1)68. Here, a protein complex was predicted from a FASTA file with the sequence of interest three times. Multiple sequence alignments were computed with the default databases and default HMMER strategy. Each of the five AlphaFold-Multimer models were used to predict five structures, and of the 25 resulting structures the optimal prediction was selected based on the highest predicted template modeling (pTM) score. Finally, Amber relaxation is applied to the final structure.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The X-ray crystallographic data generated in this study have been deposited in the PDB database under accession code 9FCM. Other structures in this study are PDB 2FXP (structure of the SARS-CoV-1 HR2, determined by NMR) and 6VXX_1_1_1 (fully-glycosylated full-length spike model). All raw data generated in this study is available in the Source Data file. Source data are provided with this paper.
References
Wheatley, A. K. et al. Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain. Cell Rep. 37, 109822 (2021).
Piccoli, L. et al. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell, https://doi.org/10.1016/j.cell.2020.09.037 (2020).
Planas, D. et al. Resistance of Omicron subvariants BA.2.75.2, BA.4.6, and BQ.1.1 to neutralizing antibodies. Nat. Commun. 14, 824 (2023).
Focosi, D., Franchini, M., Casadevall, A. & Maggi, F. An update on the anti-spike monoclonal antibody pipeline for SARS-CoV-2. Clin. Microbiol. Infect. 30, 999–1006 (2024).
Dodero-Rojas, E., Onuchic, J. N. & Whitford, P. C. Sterically confined rearrangements of SARS-CoV-2 Spike protein control cell invasion. Elife 10, e70362 (2021).
Vanderheijden, N. et al. Functional analysis of human and feline coronavirus cross-reactive antibodies directed against the SARS-CoV-2 fusion peptide. Front. Immunol. 12, 790415 (2021).
Dacon, C. et al. Rare, convergent antibodies targeting the stem helix broadly neutralize diverse betacoronaviruses. Cell Host Microbe 31, 97–111.e12 (2023).
Pinto, D. et al. Broad betacoronavirus neutralization by a stem helix-specific human antibody. Science 373, 1109–1116 (2021).
Piepenbrink, M. S. et al. Potent universal beta-coronavirus therapeutic activity mediated by direct respiratory administration of a Spike S2 domain-specific human neutralizing monoclonal antibody. PLOS Pathog. 18, e1010691 (2022).
Andreano, E. et al. Extremely potent human monoclonal antibodies from COVID-19 convalescent patients. Cell, https://doi.org/10.1016/j.cell.2021.02.035 (2021).
Mast, F. D. et al. Highly synergistic combinations of nanobodies that target SARS-CoV-2 and are resistant to escape. Elife 10, e73027 (2021).
Rossotti, M. A. et al. Arsenal of nanobodies shows broad-spectrum neutralization against SARS-CoV-2 variants of concern in vitro and in vivo in hamster models. Commun. Biol. 5, 933 (2022).
Fumagalli, V. et al. Administration of aerosolized SARS-CoV-2 to K18-hACE2 mice uncouples respiratory infection from fatal neuroinvasion. Sci. Immunol. 7, eabl9929 (2022).
Yan, B., Yang, J., Xie, Y. & Tang, X. Relationship between blood eosinophil levels and COVID-19 mortality. World Allergy Organ. J. 14, 100521 (2021).
Zalevsky, J. et al. Enhanced antibody half-life improves in vivo activity. Nat. Biotechnol. 28, 157–159 (2010).
Starr, T. N. et al. SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape. Nature 597, 97–102 (2021).
Schepens, B. et al. An affinity-enhanced, broadly neutralizing heavy chain-only antibody protects against SARS-CoV-2 infection in animal models. Sci. Transl. Med. 13, eabi7826 (2021).
Walls, A. C. et al. Unexpected receptor functional mimicry elucidates activation of coronavirus fusion. Cell 176, 1026–1039.e15 (2019).
Pinto, D. et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature 583, 290–295 (2020).
Shi, R. et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature 584, 120–124 (2020).
Koenig, P.-A. et al. Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape. Science 371, eabe6230 (2021).
Casalino, L. et al. Beyond shielding: the roles of glycans in the SARS-CoV-2 spike protein. ACS Cent. Sci. 6, 1722–1734 (2020).
Marcink, T. C. et al. Intermediates in SARS-CoV-2 spike-mediated cell entry. Sci. Adv. 8, eabo3153 (2022).
Hakansson-McReynolds, S., Jiang, S., Rong, L. & Caffrey, M. Solution structure of the severe acute respiratory syndrome-coronavirus heptad repeat 2 domain in the prefusion state. J. Biol. Chem. 281, 11965–11971 (2006).
Richardson, J. S. & Richardson, D. C. Amino acid preferences for specific locations at the ends of alpha helices. Science 240, 1648–1652 (1988).
Seifert, S. N. et al. An ACE2-dependent Sarbecovirus in Russian bats is resistant to SARS-CoV-2 vaccines. PLoS Pathog. 18, e1010828 (2022).
Costello, S. M. et al. The SARS-CoV-2 spike reversibly samples an open-trimer conformation exposing novel epitopes. Nat. Struct. Mol. Biol. 29, 229–238 (2022).
Tortorici, M. A. et al. Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms. Science 370, 950–957 (2020).
Raghuvamsi, P. V. et al. SARS-CoV-2 S protein:ACE2 interaction reveals novel allosteric targets. eLife 10, e63646 (2021).
Bowen, J. E. et al. SARS-CoV-2 spike conformation determines plasma neutralizing activity elicited by a wide panel of human vaccines. Sci. Immunol. 7, eadf1421 (2022).
Jones, B. E. et al. The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates. Sci. Transl. Med. 13, eabf1906 (2021).
Westendorf, K. et al. LY-CoV1404 (bebtelovimab) potently neutralizes SARS-CoV-2 variants. Cell Rep. 39, 110812 (2022).
Kim, C. et al. A therapeutic neutralizing antibody targeting receptor binding domain of SARS-CoV-2 spike protein. Nat. Commun. 12, 288 (2021).
Cao, Y. et al. Imprinted SARS-CoV-2 humoral immunity induces convergent Omicron RBD evolution. Nature https://doi.org/10.1038/s41586-022-05644-7 (2022).
Qu, P. et al. Enhanced neutralization resistance of SARS-CoV-2 Omicron subvariants BQ.1, BQ.1.1, BA.4.6, BF.7, and BA.2.75.2. Cell Host Microbe 31, 9–17.e3 (2023).
Huang, H.-Y. et al. Vaccination with SARS-CoV-2 spike protein lacking glycan shields elicits enhanced protective responses in animal models. Sci. Transl. Med. 14, eabm0899 (2022).
Turoňová, B. et al. In situ structural analysis of SARS-CoV-2 spike reveals flexibility mediated by three hinges. Science 370, 203–208 (2020).
Planchais, C. et al. Broad sarbecovirus neutralization by combined memory B-cell antibodies to ancestral SARS-CoV-2. iScience 110354, https://doi.org/10.1016/j.isci.2024.110354 (2024).
Ng, K. W. et al. Preexisting and de novo humoral immunity to SARS-CoV-2 in humans. Science 370, 1339–1343 (2020).
Ng, K. W. et al. SARS-CoV-2 S2–targeted vaccination elicits broadly neutralizing antibodies. Sci. Transl. Med. 14, eabn3715 (2022).
Yao, H. et al. Molecular architecture of the SARS-CoV-2 Virus. Cell 183, 730–738.e13 (2020).
Lai, S.-C. et al. Characterization of neutralizing monoclonal antibodies recognizing a 15-residues epitope on the spike protein HR2 region of severe acute respiratory syndrome coronavirus (SARS-CoV). J. Biomed. Sci. 12, 711–727 (2005).
Wu, W.-L. et al. Monoclonal antibody targeting the conserved region of the SARS-CoV-2 spike protein to overcome viral variants. JCI Insight 7, e157597 (2022).
Lip, K.-M. et al. Monoclonal antibodies targeting the HR2 domain and the region immediately upstream of the HR2 of the S protein neutralize in vitro infection of severe acute respiratory syndrome coronavirus. J. Virol. 80, 941–950 (2006).
Shi, W. et al. Cryo-EM structure of SARS-CoV-2 postfusion spike in membrane. Nature 619, 403–409 (2023).
Song, Y. et al. In situ architecture and membrane fusion of SARS-CoV-2 Delta variant. Proc. Natl Acad. Sci. USA. 120, e2213332120 (2023).
Matsuyama, S. et al. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc. Natl Acad. Sci. USA. 117, 7001–7003 (2020).
Berger Rentsch, M. & Zimmer, G. A vesicular stomatitis virus replicon-based bioassay for the rapid and sensitive determination of multi-species type I interferon. PLoS ONE 6, e25858 (2011).
Wrapp, D. et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367, 1260–1263 (2020).
Wrapp, D. et al. Structural basis for potent neutralization of betacoronaviruses by single-domain camelid antibodies. Cell 181, 1004–1015.e15 (2020).
Hoffmann, M. et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181, 271–280.e8 (2020).
Planas, D. et al. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature 602, 671–675 (2022).
R Foundation for Statistical Computing, Vienna, Austria. R: a language and environment for statistical computing (R Foundation for Statistical Computing, 2024).
Pinheiro, Bates & R Core Team. nlme: linear and nonlinear mixed effects models (R Core Team, 2023).
Mixed-effects models in S and S-PLUS (Springer-Verlag, 2000). https://doi.org/10.1007/b98882.
Kuhn, M., Weston, S., Wing, J., Forester, J. & Thaler, T. contrast: A Collection of Contrast Methods. R package version 0.24.2, https://CRAN.R-project.org/package=contrast (2022).
Harrell, F. E. Jr. rms: Regression Modeling Strategies. R package version 6.8-0, https://CRAN.R-project.org/package=rms (2024).
Hothorn, T., Bretz, F. & Westfall, P. Simultaneous inference in general parametric models. Biometrical J. 50, 346–363 (2008).
Guttman, M., Scian, M. & Lee, K. K. Tracking hydrogen/deuterium exchange at glycan sites in glycoproteins by mass spectrometry. Anal. Chem. 83, 7492–7499 (2011).
Masson, G. R. et al. Recommendations for performing, interpreting and reporting hydrogen deuterium exchange mass spectrometry (HDX-MS) experiments. Nat. Methods 16, 595–602 (2019).
da Silva, D. V., Nordholm, J., Madjo, U., Pfeiffer, A. & Daniels, R. Assembly of subtype 1 influenza neuraminidase is driven by both the transmembrane and head domains. J. Biol. Chem. 288, 644–653 (2013).
Vonrhein, C. et al. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. D. Biol. Crystallogr. 67, 293–302 (2011).
Vonrhein, C. et al. Advances in automated data analysis and processing within autoPROC, combined with improved characterisation, mitigation and visualisation of the anisotropy of diffraction limits using STARANISO. Acta Crystallogr. Sect. A 74, a360–a360 (2018).
Krissinel, E. et al. CCP4 Cloud for structure determination and project management in macromolecular crystallography. Acta Cryst. D. 78, 1079–1089 (2022).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 (2004).
Afonine, P. V. et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D. Biol. Crystallogr. 68, 352–367 (2012).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 66, 213–221 (2010).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Woo, H. et al. Developing a fully glycosylated full-length SARS-CoV-2 spike protein model in a viral membrane. J. Phys. Chem. B 124, 7128–7137 (2020).
Tai, L. et al. Nanometer-resolution in situ structure of the SARS-CoV-2 postfusion spike protein. Proc. Natl Acad. Sci. USA 118, e2112703118 (2021).
Acknowledgements
We are grateful to G. Zimmer for providing reagents to generate VSV pseudotype particles; J. McLellan and D. Wrapp for providing SARS-CoV-2, SARS-CoV, MERS-CoV, and HKU1 spike and SARS-CoV-2 RBD proteins and expression plasmids for these proteins; We thank L. de Waal from Viroclinics for coordinating the hamster challenge study. We thank Leander Meuris for helping with statistical analysis of the hamster study body weight loss. We thank Piet Maes for providing a SARS-CoV-2 Omicron BA.1 isolate and Florian I Schmidt for providing replication competent VSV pseudotyped with SARS-CoV-2 spike. We thank the staff of the VIB Flow Core Ghent for providing access to flow cytometry equipment and the support with flow cytometry experiments and the staff of the VIB Bioimaging Core Ghent for outstanding support with microscopy imaging. We thank F. Gibson and T. Swift for support during writing. We are grateful to M. Letko and to K. Vermeire who provided a Khosta-2 spike expression vector and A549-hACE2 cells, respectively. We thank the VIB protein core for performing SEC-MALS analysis. We acknowledge all Global Initiative on Sharing All Influenza Data (GISAID) contributors for sharing the sequencing data. The VeroE6/TMPRSS2 cells were provided by the National Institute for Biological Standards and Control (NIBSC) Research Reagent Repository, UK, with thanks to Dr. Makoto Takeda. S.M. is a Chan Zuckerberg Biohub Investigator. This work was supported by a PhD fellowship from the Fund for Scientific Research Flanders (FWO) (S.D.C.), EOS joint programme of Fonds de la recherche scientifique—FNRS and Fonds wetenschapellijk onderzoek–Vlaanderen—FWO (G0H7518N EOS ID: 30981113 and G0H7322N EOS ID: 40007527) (X.S., B.S.), FWO Research project G0B1917N (X.S.), FWO Research project G0G4920N (X.S., N.C., J.N.), FWO Research project 3G0G4820 (J.N., B.N.L.), Belgian Federal Government support for the VirusBank Platform (J.N.), VIB Grand Challenges project IBCORI (X.S., N.C.), VIB Center for Medical Biotechnology institutional core funding (X.S., N.C.), European Union’s Horizon 2021 research and innovation programme (HORIZON-HLTH-2021-CORONA-01), grant agreement No 101045949 (V.B., N.C., X.S.). The BSL-3 facility at CMB was established with generous support from FWO Research Infrastructure grant, VIB, VIB-CMB and Ghent University institutional funds.
Author information
Authors and Affiliations
Contributions
Conceptualization: S.D.C., S.M., B.N.L., N.C., H.R., X.S., B.S.; Methodology: S.D.C., I.V.M., L.v.S., S.R.S., J.D., N.D., S.L., W.N., K.R., D.F., S.D., A.-S.D.S., J.C.Z.M., T.V., K.S., L.M., M.B., M.V., C.D.W., H.D., J.Z., P.V., G.H.G., C.L., V.B., M.R., R.A., J.N., H.R., B.S.; Investigation: S.D.C., I.V.M., L.v.S., S.R.S., J.D., N.D., S.L., W.N., D.F., T.V., C.D.W., J.Z., B.S.; Visualization: S.D.C., I.V.M., L.v.S., S.R.S., D.F., J.Z., V.B., M.R., H.R., B.S.; Funding acquisition: V.B., J.N., S.M., B.N.L., N.C., H.R., X.S., B.S.; Supervision: L.v.S., G.H.G., V.B., J.S., S.M., B.N.L., N.C., H.R., X.S., B.S.; Writing – original draft: S.D.C., I.V.M., S.R.S., X.S., B.S.; Writing – review and editing: S.D.C., S.R.S.,W.N., V.B., H.R., X.S., B.S.
Corresponding authors
Ethics declarations
Competing interests
S.D.C., B.S., X.S., L.v.S., W.N., K.R., N.C., G.H.G., H.R., I.V.M., V.B., and R.M. are named as inventors on patent application WO2023/222825 A1 “Sarbecovirus Spike S2 Subunit Binders”. N.C. and X.S. are scientific founders of ExeVir Bio BV and are in receipt of ExeVir Bio BV share options. T.V. and V.B. are and M.R. and C.L. were employed by ExeVir Bio BV and are in receipt of ExeVir Bio BV share options. All other authors declare that they have no competing interests.
Peer review
Peer review information
Nature Communications thanks Joseph G. Jardine, Lei Sun and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
De Cae, S., Van Molle, I., van Schie, L. et al. Ultrapotent SARS coronavirus-neutralizing single-domain antibodies that clamp the spike at its base. Nat Commun 16, 5040 (2025). https://doi.org/10.1038/s41467-025-60250-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-60250-1