Abstract
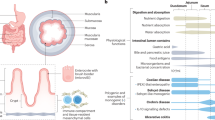
The intestinal epithelium undergoes fast turnover, and the villus length in the small intestine gradually decreases from the duodenum to the ileum. However, the underlying mechanisms remain poorly understood. In this study, we investigate the regulatory mechanism underlying the regional disparity of villus length. A progressive strengthening of BMP signaling from the duodenum to the jejunum and ileum establishes a signaling gradient, resulting in differences in the rates of cell proliferation and apoptosis. We show that BMP signaling regulates the survival of the small intestine epithelial cells by inhibiting integrin expression and thereby inducing cell apoptosis. Combined with mathematical modeling, our data reveal that BMP signaling provides positional cues and antagonizes Wnt signaling to control villus growth, while Wnt signaling promotes BMP signaling to counteract excessive proliferation, thus maintaining villus length. Our findings provide insights into the signaling dynamics governing epithelial turnover and villus length in the small intestine.
Similar content being viewed by others
Introduction
The small intestine is a crucial organ for food digestion, nutrient absorption, and pathogen defense in the body. To carry out these functions, the intestinal epithelium, which is composed of single-layer cells, undergoes a rapid renewal process, with a turnover cycle of 3–5 days in most adult mammals1,2,3,4. The number of cells in the intestinal epithelium, reflected in villus length, is associated with nutrient absorption. Elongated villi are associated with obesity5, while abnormal cell death can lead to villus atrophy and impaired nutrient absorption in certain pathological conditions6. Different segments of the small intestine exhibit distinct nutrient absorption preferences7. Interestingly, villi length is gradually shortened from proximal to distal small intestine, forming a “Villus Length Gradient”, enabling the fulfillment of varying nutritional requirements8. Additionally, the epithelial cells of the villi can be regenerated and repaired to form the initial structure when injured9. Therefore, the intestinal epithelium serves as an excellent model for studying cell turnover and the processes involved in cell population size control.
Morphogens control embryonic development and organ formation by providing positional information through the reaction–diffusion mechanism to determine cell fate10,11,12. In the small intestine, the balance between the proliferation of intestinal stem cells and the apoptosis of differentiated cells regulates both epithelial cell number and villus length13,14. Key morphogen signaling, notably Wnt and BMP, plays crucial roles in this process by influencing both proliferation and differentiation of intestinal stem cells14,15,16,17,18,19,20,21. Both Wnt and BMP signaling exhibit a gradient activity along the villus-crypt axis: high Wnt activity is at the base of the crypt and low activity is present in the villus, while BMP activity shows the opposite distribution9,22. BMP signaling establishes a gradient through autocrine and paracrine mechanisms, which are antagonized by Grem1 and Grem2 secreted by the mesenchymal cells around the crypt21. BMP ligands bind to BMPR1a (ALK3) or BMPR1B (ALK6) and BMPR223, then induce phosphorylation of Smad1/5/8, which form a complex with Smad4 and activate target genes in the nucleus. Concurrently, high levels of R-spondin around the bottom of the crypt potentiate Wnt/β-catenin signaling and play a critical role in maintaining the stemness and proliferation of intestinal stem cells24,25. However, how signaling pathways interplay to govern the villus length of the intestinal epithelium remains elusive.
Turnover of intestinal epithelial cells is mediated by apoptosis triggered by cell exfoliation, a process known as anoikis6,26. Various factors such as TGF-β, TNF-α, and fructose regulate cell survival and shedding5,27,28,29, but the mechanisms by which intestinal epithelial cells perceive their relative position in the villus, initiate apoptosis, and maintain a stable turnover rate remain unclear. Our study reveals distinct apoptosis and proliferation rates of epithelial cells in the proximal and distal segments of the small intestine, with BMP signaling establishing a gradient difference from proximal to distal regions. Through the integration of mathematical models, we discover that the intensity of BMP signaling is closely associated with cell position and regulates apoptosis through the integrin family in a concentration-dependent manner. We find that excessive proliferation of stem cells does not necessarily lead to changes in villi length, whereas loss of BMP signaling leads to longer villi. The interaction between BMP and Wnt signaling also contributes to the steady-state turnover of the epithelial cells, thereby maintaining the villus length in the small intestine. Our findings shed light on the homeostatic length control of villi in the small intestine, offering valuable insights into treating intestinal absorption disorders.
Results
Distinct proliferation and apoptosis patterns along the villus axis in different small intestine segments
The small intestinal epithelial cells undergo gradual differentiation to become mature functional cells along the crypt-villus axis and eventually undergo apoptosis when reaching the villus tips20,30. In agreement with this, we observed that TUNEL (TdT-mediated dUTP Nick-End Labeling) signal intensity was gradually increased from the base to the apex of villi (Fig. 1a). We also found that the distal small intestine exhibited higher TUNEL signal at the base of villi compared to the proximal small intestine (Fig. 1a). To determine whether the villus length difference between the proximal and distal small intestines results from variations in proliferation or apoptosis (cell survival), we first stained cleaved caspase-3 to assess cell death rate. The results revealed that most cells underwent apoptosis when reaching the villus tip, as indicated by cleaved caspase-3 signals that were predominantly localized at the tips of both proximal and distal small intestines (Fig. 1b). To quantify apoptotic cells, we employed whole-mount staining and normalized the cleaved caspase-3-positive cells to the total cell count in each villus. As shown in Fig. 1c, apoptosis was more active in the distal small intestine.
a Cell death in the proximal and distal small intestinal segments of wild-type mice revealed by TUNEL assay. TUNEL signal intensity was quantified along the villus epithelium in ten bins per segment, with error bars representing the standard deviation (SD), visualized as shaded regions in the plots. N = 3 mice/group. Scale bars, 200 µm. b Cleaved-caspase 3 staining on paraffin-embedded section in the proximal and distal small intestine. N = 3 mice/group. Scale bars, 200 µm. c Cleaved-caspase 3 staining in whole mount proximal and distal small intestine. Normalized cell death probability was calculated by dividing apoptotic cells (cleaved-caspase 3+) by total epithelial cells in each villus. Apoptotic cells (cleaved-caspase 3+) were counted from the top view, and total epithelial cells were counted from the side view of agarose sections. The average total count per villus was used for each intestinal segment. N = 3 mice/group. Scale bars, lateral view on the left: 50 µm, top view on the right: 200 µm. d Lineage tracing of Lgr5+ cells in the proximal and distal small intestine of Lgr5CreERT2; Rosa26loxp-stop-loxp-ZsGreen mice on day 1 and day 4. Measurement of length from the villus top end to the ZsGreen front on day 4, indicated by the distance between two arrowheads in one villus, is shown on the left panel. Arrowheads indicate two positions: the first marks the true villus tip, and the second marks the region where a cluster of migrating ZsGreen+ cells is located. Each individual data point represents one villus measurement. N = 3 mice/group. Scale bars, 200 µm (Created in BioRender. Liu (2025) https://BioRender.com/do3glyu). e Proliferation dynamics revealed by EdU/BrdU dual labeling. Representative images show EdU (green) and BrdU (red) labeling, with nuclei counterstained by DAPI (blue). High magnification images show EdU+ (green) and BrdU+ (red) cells in the epithelium. EdU+, BrdU− cells (arrows) indicate cells exiting S-phase. The quantification was presented in the right panel. N = 4 mice. Scale bar: 50 µm. The data were analyzed by an unpaired t-test with Welch-correction (two-sided) (c–e). Data represent mean ± SD.
To determine cell survival time in different regions of the small intestine, we conducted lineage tracing experiments using tamoxifen-induced Lgr5CreERT2; Rosa26loxp-stop-loxp-ZsGreen mice. One day after tamoxifen treatment, stem cells were labeled green at the bottom of crypts as green (Fig. 1d). At day 4, we observed that the topmost ZsGreen+ cells in the villi of the distal segment were located close to the villus tip. In contrast, ZsGreen+ cells in the villi of the proximal intestine were situated further away from the tip, suggesting that the cells derived from Lgr5-ZsGreen+ cells reach the villus tips faster in the distal intestine compared to those in the proximal intestine.
Moreover, we performed a dual EdU/BrdU pulse-chase experiment to compare the proliferation rates of the proximal and distal small intestines and quantified the mean duration of synthesis-phase (Ts) by following the report of Harris et al. 31. The results showed that the proximal intestine has a shorter Ts, indicating faster proliferation compared to the distal region (Fig. 1e). These results together indicate that regional differences in the dynamics of proliferation and apoptosis correlate with variations in villus lengths along the small intestine, with the proximal segment exhibiting faster migration and delayed apoptosis initiation compared to the distal segment.
Distinct BMP signaling activity in different segments of the small intestine
We then explored whether distinct gene expression programs were associated with different regions of the small intestine. We analyzed Wnt, TGF-β, and BMP signaling pathways in epithelial cells. Wnt signals, indicated by Axin2-GFP and β-catenin intensity, were quantified only in crypts, as no detectable signals were found in villi. The intensities and gradients of these signals showed similar trends from the duodenum to the ileum (Fig. 2a and Supplementary Fig. 1a). TGF-β signaling, as shown by phosphorylated Smad3 levels, exhibited a slight increase in the ileum (Supplementary Fig. 1b). BMP signaling, measured by phosphorylated Smad1/5 levels, BRE-tdTomato signals, and BMP target gene Id1, increased gradually from the proximal to distal segments (Fig. 2b–e). Similar BMP signaling patterns were detected in both rats and monkeys (Supplementary Fig. 1c), suggesting evolutionary conservation of BMP signaling between proximal and distal segments across different species.
a Ki67 immunofluorescence staining in Axin2-mGFP knock-in mice across small intestine segments, showing grayscale and fluorescence intensity. Axin2 intensity was quantified from stem to TA cells within crypts, with error bars representing the standard deviation (SD), visualized as shaded regions in the plots. N = 3 mice/group. Scale bars, 50 μm. b p-Smad1/5 immunofluorescence staining showing BMP signaling intensity along the villus axis. Data is presented by absolute position (left) and relative position (right) for comparison with error bars representing the standard deviation (SD), visualized as shaded regions in the plots. N = 5 mice. Scale bars, 500 μm (left); 100 μm (zoomed-in images on the right). c Immunoblotting of p-Smad1/5, total Smad1, and β-actin from the villus region of the proximal and distal small intestine. N = 3 mice/group. d tdTomato signals in 4xBRE-tdTomato knock-in mice, with magnified views in insets. N = 3 mice/group. Scale bars, 100 μm. e Id1 expression detected by RNAscope, showing punctate signals of Id1 mRNA. Quantification is shown with mean values and standard error of the mean (SEM). N = 3 mice. Scale bar, 100 μm. f Expression of BMP-suppressed enterocyte genes in the proximal and distal small intestine. N = 3 mice/group. g Both BMP ligand and inhibitor undergo generation (\(\varnothing\)B, \(\varnothing\)I), diffusion (DB, DI), and degradation (decB, decI). The competitive binding of the inhibitor to the ligand impedes the interaction between the BMP ligand and receptor (R and decBR) (Created in BioRender. Liu (2025) https://BioRender.com/7ob2gem). h Spatial distribution of BMP ligand, receptor, and inhibitor from villus apex to stroma, with BMP distribution in the proximal and distal small intestine with the model solutions (smooth lines). i Scatter plots comparing BMP and inhibitor diffusion rates, with model solutions fitting p-Smad1/5 distribution. j Grem1 protein distribution in the stromal cells and crypt region of the proximal small intestine, quantified in both regions. N = 3 mice/group. Scale bar: 50 µm. k BMP signaling intensity variation from proximal to distal regions (Created in BioRender. Liu (2025) https://BioRender.com/x1duy74). The data were analyzed by an unpaired t-test with Welch-correction (two-sided) (j). Data represent mean ± SD.
By comparing gene expression profiles in different segments of the small intestine in control and Bmpr1a knockout mice, we observed distinct patterns of gene regulation mediated by BMP signaling. Specifically, genes repressed by BMP signaling were enriched in the proximal small intestine (Fig. 2f). For instance, Fabp1, which has been reported to be a marker for the proximal segment32, was upregulated in Bmpr1a knockout intestine (Fig. 2f). Dhcr24, which showed higher expression in the proximal region compared to the distal region, was found to be suppressed by BMP signaling (Supplementary Fig. 1d). These results suggest that BMP signaling exhibits a gradient from the proximal to distal small intestine.
To explore the factors contributing to differences in BMP signaling along the small intestine, we developed a mathematical model using reaction–diffusion equations to simulate the interactions of BMP ligands, their receptors, and inhibitors (Fig. 2g). The simulation focused on the one-dimensional stroma-crypt-villus axis. We tested varying node densities (41, 64, 128, 256, and 512 nodes) and observed minimal effects on the simulations, indicating that the model’s behavior was relatively insensitive to simulation nodes within this range (Supplementary Fig. 2a). In our reaction–diffusion model, we initially considered mechanisms for BMP gradient formation, including BMP mRNA transcription, BMP ligand diffusion, and a counter-gradient regulated by Grem1. However, BMP ligand mRNA, such as Bmp2 and Bmp4, was higher in the proximal region (Supplementary Fig. 2b). We found that the models, including BMP mRNA transcription (Supplementary Fig. 2c, model 1) and combining BMP ligand diffusion (Supplementary Fig. 2c, model 2), could not fully explain the observed higher p-Smad1/5 expression in the distal region. Additionally, without incorporating quantified Bmpr1a receptor expression, the model failed to adequately capture BMP activity at the villus tip (Supplementary Fig. 2c, model 3; Supplementary Fig. 2d). Therefore, we developed a model (Supplementary Fig. 2c, model 4), which incorporated both inhibitors and receptors, providing a more accurate simulation of BMP gradient formation (Fig. 2h). We performed 100,000 computational simulations in our reaction–diffusion model, testing 11 parameters related to BMP signaling, including ligand/inhibitor diffusion, production, and decay rates (Supplementary Fig. 2e). By setting a normalized root mean squared deviation (NRMSD) threshold of 0.085 or lower, we identified optimal parameter sets that aligned the model’s predictions with observed p-Smad1/5 data (Fig. 2h and Supplementary Data 1). Notably, a majority of these solutions revealed a higher diffusion rate of the inhibitor in the proximal segment compared to the distal segment (Fig. 2i). The sensitivity analysis revealed that variations in BMP inhibitor parameters had a greater impact than BMP ligands, highlighting the crucial role of BMP inhibitors in gradient regulation (Supplementary Fig. 2f). To determine whether the observed variations in the BMP gradient between the proximal and distal segments could be attributed to differences in inhibitor diffusion, we specifically varied the inhibitor diffusion rate, while keeping all other parameters constant, to isolate its contribution to the observed proximal-distal differences in the BMP gradient (Supplementary Fig. 2g). This finding suggests that in order to establish a BMP signal gradient in the proximal segment, inhibitors expressed in the stroma cells need to diffuse over a greater distance. In contrast, the remaining biophysical parameters, including inhibitor production, did not exhibit distinguishable patterns (Supplementary Fig. 2h). To further validate our hypothesis from mathematical modeling, we conducted smFISH analysis, which revealed that Grem1 mRNA is predominantly localized to stroma cells subjacent to the crypts (Supplementary Fig. 2i). Additionally, Grem1 protein is primarily found in stromal cells, with diffusion extending into the crypt region of the proximal intestine. The elevated expression of Grem1 in the proximal region also accounts for the comparatively lower BMP signaling in this segment (Fig. 2j). Taken together, these results suggest that the observed variations in BMP signaling among distinct small intestinal segments may be attributable to differential expression and diffusion of the inhibitors (Fig. 2k).
BMP signaling regulates villus length
Considering the gradient difference of BMP signaling between the proximal and distal segments of the small intestine, we examined the involvement of BMP signaling in the regulation of villus length using genetically engineered mice. Villin-creERT2-driven conditional knockout (cKO) of Bmpr1a led to body weight loss in the early time, which gradually recovered over time (Supplementary Fig. 3a). Bmpr1a cKO and Smad4 cKO mice exhibited increased villus length and cell number, accompanied by enhanced stem cell proliferation shown by Ki67+ cells and EdU/BrdU tracing (Fig. 3a, b and Supplementary Fig. 3b–d). In contrast, Grem1 cKO mice showed reduced villus cell number and a less organized nuclear arrangement, alongside diminished stem cell proliferation (Fig. 3c and Supplementary Fig. 3e). Notably, even after six weeks of Bmpr1a cKO, the overall villus length was longer than in the control (Fig. 3d). In addition, Bmpr1a cKO increased the number of absorptive epithelial cells as shown by Apoa1-mCherry (Supplementary Fig. 3f). Consistently, the genes related to nutrient absorption were upregulated in both the proximal and distal segments of the Bmpr1a cKO small intestine compared to control groups (Supplementary Fig. 3g, h). These data together indicate that BMP signaling plays a crucial role in regulating villus length and absorptive function.
a Immunofluorescence staining of Ki67 in control and VillinCreERT2; Bmpr1aflox/flox (Bmpr1a cKO) mice at day 12 post-tamoxifen injection (dpi). Quantification of villus length and cell number from the proximal to the distal small intestine and Ki67+ cells in the jejunum. N = 3 mice/group. Scale bars, 200 µm. b Immunofluorescence staining of Ki67 in control and VillinCreERT2; Smad4flox/flox (Smad4 cKO) mice at 12 dpi. Quantification of villus length and cell number from the proximal to the distal small intestine, and Ki67+ cells in the jejunum. N = 3 mice/group. Scale bars, 200 µm. c Immunofluorescence staining of Ki67 in control and CaggCreERT2; Grem1flox/flox (Grem1 cKO) mice at 12 dpi. Quantification of villus length and cell number from the proximal to the distal small intestine, and Ki67+ cells in the jejunum. N = 3 mice/group. Scale bars, 200 µm. d Immunofluorescence staining of Ki67 and E-cadherin in control and Bmpr1a cKO mice at 42 dpi. Quantification of villus length from the proximal to the distal small intestine. N = 3 mice/group. Scale bars, 500 µm. In (a–c) the white arrows indicate the apex of the villus, and the original blue color of DAPI staining in microscopy images has been adjusted to green for better visualization. This adjustment applies uniformly across the entire image. Quantification of villus length and cell number was analyzed by two-way ANOVA with Tukey’s multiple comparison test; Quantification of Ki67+ cells was analyzed by unpaired t-test (two-sided). Data represent mean ± SD.
BMP signaling promotes anoikis by suppressing integrin expression
As both cell proliferation and death can contribute to the turnover rate of the intestinal epithelium, we examined cell death with the TUNEL assay. Interestingly, TUNEL staining and apoptosis genes were diminished in the Bmpr1a cKO small intestine (Fig. 4a and Supplementary Fig. 4a), confirming the important role of BMP signaling in cell death control17. To investigate the underlying mechanisms of villus growth and epithelial cell turnover, we developed a two-stage mathematical model combining (i) progenitor self-renewal in crypts and (ii) differentiation into terminal cells in the villus. The model captured the behavior of progenitors and differentiated cells, including cell proliferation, migration, and apoptosis. In the mathematical model, BMP signaling inhibited stem cell proliferation probabilities (\({p}_{0}\)) using the Hill equation. BMP modulated apoptosis (\({d}_{1}\)) through either a threshold mechanism or the integral mechanism involving cumulative integration of BMP concentration over time33,34. We first focused on testing the integral mechanism, where apoptosis is modulated by the cumulative effect of BMP concentration over time (Fig. 4b, and see Results section for model 5). In this model, we incorporated differential Grem1 expression and its diffusion rates across the proximal and distal small intestine, based on both the reaction–diffusion model (Fig. 2h) and experimental data (Fig. 2j). Using the results predicted in Fig. 2h–j as input, we treated Grem1 activity as a spatially variable factor to more accurately simulate how regional variations in Grem1 influence BMP gradients (Fig. 4c) and villus growth (Fig. 4d). This refinement allows the model to predict differing steady-state villus lengths between the proximal and distal segments, with the villus length in the proximal segment reaching approximately twice that of the distal segment (Fig. 4d). This indicates that differential BMP signaling can generate varying villus lengths in silico. The villi in the distal segment accumulated a similar amount of BMP (t1–t3) as the proximal villi (t1–17) but reached this level more rapidly (Fig. 4e). This observation implies that the time required for BMP accumulation correlates with the time needed to achieve steady-state villus length in both the distal and proximal segments (Fig. 4d). Given that our mathematical model indicating that cell death rate depends on BMP concentration integration, we hypothesize that the accelerated apoptosis observed in the distal villi is due to the more rapid accumulation of BMP in the distal small intestine. The caspase-3 activity assay in intestinal organoids confirmed that prolonged BMP exposure induced an increased time-dependent apoptosis (Supplementary Fig. 4b). The analysis of p-Smad1/5 and TUNEL signals along the villi confirmed that accelerated apoptosis in the distal villi was due to more rapid BMP accumulation, with a corresponding earlier onset and higher intensity of apoptosis in the different regions along the villus (Fig. 4f).
a TUNEL assay (control and Bmpr1a cKO mice) with grayscale enlargements. TUNEL+ cells quantified across intestinal regions (Villus lengths-scaled) with error bars representing the standard deviation (SD), visualized as shaded regions in the plots. N = 3 mice/group. Scale bars, 200 µm. b Two-stage cell lineage model: progenitor cell proliferation and differentiation leading to cell death (d_1) controlled by BMP activity (Created in BioRender. Liu (2025) https://BioRender.com/jyzljkj). c Computational modeling shows that BMP signaling progressively amplifies during villus elongation (t1–t8). Distal segments, which exhibit lower expression and diffusion rate of Grem1, develop earlier and stronger BMP peaks compared to proximal regions. d Simulated villus growth (proximal/distal) with progenitor cell proportions. The dashed line marks the crypt-villus boundary. e Model prediction of BMP signaling activity in the apical cells of the villi upon reaching a state of equilibrium in villus growth. f p-Smad1/5 and TUNEL intensity profiles along villus (proximal P6 vs distal D3 positions). P6, 6th position from a base in the proximal segment. D3, 3rd position from base in the distal segment. g Integrin gene heatmap (control vs cKO). N = 3 mice/group. h Immunofluorescence staining and quantification of integrin α6 with error bars representing the standard deviation (SD), visualized as shaded regions in the plots. N = 3 mice/group. Scale bars, 200 µm. i Organoid immunoblots (48 h treatments: E [EGF], N [Noggin], R [R-spondin], B [BMP 6.7/20 ng/mL]). N = 3 cultures. N = 3 independent organoid cultures. j Ki67 staining and villus length quantification in control and Pyrintegrin-injected mice. N = 3 mice/group. Scale bars, 200 µm. The white arrows indicated the apex of the villus. k Villus growth prediction under the threshold mechanism, modifying the differentiated cell removal rate (d_1) using a Hill equation. l Villus length modeling under different conditions (WT, constant d_1, γ_B = 0) of the model shown in Fig. 4b. m EdU+ cells 24 h after injection. N = 3 mice/group. Scale bars, 200 µm. The data were analyzed by two-way ANOVA with Tukey’s multiple comparison test (i) and unpaired t-test with Welch-correction (two-sided) (m). Data represent mean ± SD.
Anoikis, the integrin-mediated apoptosis, is important for the steady-state maintenance of the intestinal epithelium35. However, the mechanisms controlling integrin expression are not well understood. We discovered that integrin mRNA levels were upregulated in Bmpr1a cKO intestinal epithelium (Fig. 4g and Supplementary Fig. 4c), and that BMP suppressed their expression (Supplementary Fig. 4d). In control mice, Itga6 expression decreased from the base to the apex of the villi, but in Bmpr1a cKO mice, elevated Itga6 expression persisted (Fig. 4h). We further found that Smad1 and Smad4 bound to the promoters of the Itgb4 and Itga6 genes, indicating direct regulation of integrin expression by BMP signaling (Supplementary Fig. 4e and Supplementary Data 2)17,36. BMP inhibited the expression of Itgb4 and Itga6 in a concentration-dependent manner (Fig. 4i). This inhibition was accompanied by increased levels of cleaved caspase-3 and decreased levels of Bcl2, verifying the prediction of the mathematical model that BMP promotes apoptosis in the intestinal epithelium in a concentration-dependent manner (Fig. 4b–f). Supporting this, the ablation of integrin enhanced cleaved caspase-3 levels (Supplementary Fig. 4f). Consistently, BMP had no effect on organoid budding number and survival rates in Itgb4 KO organoids (Supplementary Fig. 4g). Interestingly, Pyrintegrin, an integrin agonist that enhances cell-extracellular matrix adhesion-mediated integrin signaling37, upregulated Itga6 and increased villus length in vivo (Fig. 4j and Supplementary Fig. 4h). Next, we tested the threshold mechanism (Supplementary Fig. 5, Model 6) using a mathematical model. As high BMP level inhibits villus elongation, the model predicted that the distal small intestine exhibited a very short villus length due to the high BMP signaling at the villus base (Fig. 4k). Therefore, the threshold mechanism would make it difficult for the distal villi to grow, which contradicts the in vivo observations. Taken together, these results suggest the integral mechanism by which BMP signaling induces anoikis by suppressing integrin expression.
BMP signaling exerts dual effects in the intestinal epithelium: inhibiting cell proliferation while promoting apoptosis17,20. However, distinguishing their different impacts on the intestinal epithelium using mouse models poses a challenge. To address this problem, we utilized mathematical modeling by altering a single variable in the role of BMP signaling. Initially, we presumed that a constant cell death rate would be unaffected by variations of BMP concentration to minimize the impact of BMP gradient differences on cell apoptosis. This resulted in a reduced discrepancy in villus length between the proximal and distal small intestine in the model (Fig. 4l, i and ii). Subsequently, we disabled the role of BMP in stem cell proliferation. Simulations showed that inhibition of stem cell proliferation had a minimal effect on villus length in the proximal small intestine, where BMP levels were low. However, in the distal small intestine, where BMP-regulated stem cell proliferation was disabled, villus length significantly increased, reducing the difference in villus length between the proximal and distal regions (Fig. 4l, iii). Similar proliferating cells in the proximal and distal segments of the Bmpr1a cKO intestine validated the mathematical model (Fig. 4l). Therefore, BMP regulates villus length by modulating both apoptosis and proliferation.
BMP signaling provides positional cues and generates negative feedback on Wnt signaling
To track the process of villus growth, mice were injected with poly(I:C), which induces apoptosis in villus epithelial cells and selectively removes villi in the duodenum and ileum38. Cell proliferation was active and extended into the villus region at 48 hpi (hours post-injection) (Fig. 5a). At 72 hpi, the proliferative zone was reduced to the crypt region. Since both BMP and Wnt signaling play important roles in the regulation of proliferation and death of intestinal epithelial cells, we aimed to determine whether they are involved in the rapid cell proliferation in the crypts during early villus growth and the subsequent deceleration as villi approach steady-state length. Bmpr1a deletion led to elevated levels of the Wnt-responsive protein Axin2 in both the stem cell and transit-amplifying zones (Fig. 5b). Long-term BMP treatment resulted in the suppression of Wnt signaling targets such as Sox9 and Cd44, along with a decrease in β-catenin activity in the nucleus in intestinal organoids (Fig. 5c and Supplementary Fig. 6a). To investigate the negative feedback of BMP signaling on the Wnt pathway during villus growth, we employed a mathematical model (Fig. 5d and Supplementary Fig. 5, model 7). In the early stages of villus growth, low levels of BMP signaling in the crypts did not hinder Wnt activity, allowing stem cell proliferation (Fig.5e, t1–t4). As the villus elongated, BMP signaling was intensified, leading to suppression of Wnt signaling and villus growth deceleration (Fig.5e, t5–t6). BMP antagonists secreted from the stromal cells bind BMP at the base of the villi, preventing the interaction with its receptors and establishing a stable boundary beneath the crypts39,40. Mathematical modeling showed that BMP signaling intensity was correlated with cell spatial arrangement during villus growth (Fig. 5e, right). This suggests that BMP signaling carries positional information, guiding cell proliferation during villus growth. Consistent with the model, following the poly(I:C) injection, the BMP signaling indicator p-Smad1/5 was detected only on the villus tips at 48 hpi and increased, extending to the whole villus region at 72 hpi (Fig. 5f). Additionally, Grem1 expression was initially elevated during villus regeneration but diminished as the villus elongated (Supplementary Fig. 6b). However, p-Smad3 levels were detected at all the time points (Fig. 5f), indicating that TGF-β signaling remained consistently high during villus regeneration. These dynamic changes suggest that BMP signaling intensity adjusts with villus length, reflecting cellular positioning. To assess villus regeneration, we injected mice with Poly(I:C). While control mice showed limited villus regeneration in the duodenum and ileum after 48 h, Bmpr1a cKO mice exhibited rapid villus growth, reaching similar villus lengths in both the duodenum and ileum (Fig. 5g). Taken together, these results indicate that BMP signaling dynamically modulates its intensity in response to cellular spatial positioning, and by inhibiting Wnt signaling, BMP signaling regulates the rate of cell proliferation to coordinates the regeneration and elongation of villi.
a Immunofluorescence staining of Ki67 at 0 h, 24 h, 48 h, and 72 h after poly(I:C) injection (hpi). N = 3 mice/group. Scale bars, 100 µm. The expression regions of Ki67 were indicated by arrowheads (Created in BioRender. Liu (2025) https://BioRender.com/mycsfrf). b GFP fluorescence imaging of Axin2-mGFP in control and Bmpr1a cKO mice in the proximal segment (left). The images showed fluorescence intensity of Axin2-mGFP, highlighting the Wnt-responsive protein expression (middle). Quantification of Axin2 intensity was performed from the stem cell zone to the transit-amplifying (TA) zone within crypts for both Bmpr1a cKO and control mice (right). N = 3 mice/group. Scale bars, 50 µm. c RT-qPCR of Sox9, Cd44, Id1, Id2, and Bmpr1a in control and Bmpr1a cKO organoids cultured in ENR and ERB medium for 24 h. E, EGF, 50 ng/mL; N, Noggin, 100 ng/mL; R, R-spondin, 500 ng/mL; B, BMP, 20 ng/mL. N = 3 independent experiments. d A schematic diagram of a two-stage cell lineage model regulated by Wnt and BMP signaling. Wnt signaling promotes proliferation of progenitor cells (\({p}_{0}\)), while BMP signaling suppresses cell proliferation and Wnt activity (Created in BioRender. Liu (2025) https://BioRender.com/2tio9zc). e Mathematical model of villus growth with corresponding spatial distribution of Wnt and BMP activity in the distal small intestine. t1 through t6 are characterized by identical temporal intervals. The colormap axis represents the proportion of progenitor cells ranging from 0 to 1 along the crypt-villus axis. The dashed line demarcates the boundary between the crypt and the villus. f Immunofluorescence staining of p-Smad1/5 and p-Smad3 at 0 h, 24 h, 48 h, and 72 h after poly(I:C) injection. N = 3 mice/group. Scale bars, 100 µm. g Immunofluorescence staining of Ki67 from the duodenum to the ileum of control and Bmpr1a cKO mice at 48 h after poly(I:C) injection. The white arrows indicate the corresponding position of the duodenum and the ileum. N = 3 mice/group. Scale bars, 500 μm. The original blue color of DAPI staining in microscopy images has been adjusted to green for better visualization (a, g). This adjustment applies uniformly across the entire image. The data were analyzed by an unpaired t-test with Welch-correction (two-sided) (b). Data represent mean ± SD.
Positive feedback of Wnt signaling on BMP signaling contributes to the villus length control
We also investigated the impact of Wnt signaling on villus length using Apc heterozygous knockout mice. Apc heterozygous knockout led to higher cell proliferation, reduced villus length in the proximal region, but had no effect on villus cell number (Fig. 6a). EdU lineage tracing revealed that faster migration of EdU+ cells to the apex of the villus and shorter survival time for intestinal epithelial cells in Apc heterozygous mice (Supplementary Fig. 7a), indicating that the activation of Wnt signaling results in an accelerated cell turnover. When mice were injected with adenovirus overexpressing R-spondin1, they exhibited elongated crypts and excessive cell proliferation (Supplementary Fig. 7b, c), as early reported41. Interestingly, R-spondin1 overexpression also increased cell number in villi, but had no effect on villus length in most parts of the small intestine, even shorter villi were observed in the most proximal small intestine. These results indicate that Wnt-induced excessive cell proliferation does not lead to an increase in villus length.
a Immunofluorescence staining of Ki67 in VillinCreERT2; Apcflox/+mice at 16 dpi. Quantification of the villus length and epithelial cell number from the proximal to the distal small intestine and Ki67+ cells in the jejunum. N = 3 mice/group. Scale bars, 200 µm. The white arrows indicate the apex or the base of the villus. b RT-qPCR of Axin2, Ki67, Bmp2, Bmpr1a, Pmp22, and Slc34a2 in intestinal organoids derived from control and Apc heterozygous (hez) mice. N = 3 independent experiments. c RT-qPCR of Axin2, Ki67, Id1, and Id2 in intestinal organoids cultured in E and ER medium. E, EGF; R, R-spondin,. N = 3 independent experiments. d Immunofluorescence staining and quantification of p-Smad1/5 in the proximal region of the small intestine between control and Apc hez mice, with error bars representing the standard deviation (SD), visualized as shaded regions in the plots. N = 3 mice/group. Scale bars, 200 µm. e A schematic diagram of a two-stage cell lineage model regulated by Wnt and BMP signaling, modeled using the Hill equation. Wnt signaling exhibits a feedback regulation on BMP signaling. Conversely, BMP signaling inhibits Wnt signaling (Created in BioRender. Liu (2025) https://BioRender.com/2tio9zc). f The mathematical model shows BMP activity, cell removal rate, and villus length under three conditions: (i) normal; (ii) Wnt activation without BMP regulation (γ_1 = 0); (iii) Wnt activation with BMP regulation (γ_1). The colormap represents progenitor cell proportion along the crypt-villus axis, with a dashed line marking the crypt-villus boundary. g Immunofluorescence staining of Ki67 in the proximal small intestine of the indicated mice. Mice were injected with tamoxifen for five days and then sacrificed 2 days later. N = 3 mice/group. Scale bars, 200 µm. The white arrows indicate the apex of the villus. For a, villus length and cell number were analyzed by two-way ANOVA with Tukey’s multiple comparison test; Ki67+ cells were analyzed by unpaired t-test with Welch-correction (two-sided). b, c Analyzed by unpaired t-test with Welch-correction (two-sided); g was analyzed by one-way ANOVA with Tukey’s multiple comparison test. Data represent mean ± SD.
Given the significant role of BMP signaling in regulating villus length, we investigated whether Wnt signaling has any impact on BMP activity. Notably, the Wnt signaling target Axin2 and BMP signaling genes (Bmp2, Bmpr1a, Pmp22, and Slc34a2) were upregulated in Apc heterozygous mice (Fig. 6b). In organoids cultured in the ER medium containing EGF and R-spondin1, Axin2 and the BMP target genes Id1 and Id2 were upregulated compared to those in the E medium containing EGF only (Fig. 6c). Furthermore, p-Smad1/5 was detected in the lower half of the villus epithelium in Apc heterozygous mice (Fig. 6d). These data suggest that elevated Wnt signaling promotes BMP signaling activity. To further explore the effect of Wnt signaling on BMP signaling, we developed a mathematical model using the Hill equation to add Wnt-BMP positive feedback regulation based on experimental findings (Fig. 6a–d) and confirmed its impact on villus growth (Fig. 6e and Supplementary Fig. 5, model 8). This positive feedback mechanism enhanced BMP signaling and apoptosis, therefore maintaining villus length (Fig. 6f, ii, iii) compared to the control group (Fig. 6f, i). Consistent with this, in Apc heterozygous mice, TUNEL signals were detected starting from the base of the villi (Supplementary Fig. 7d), indicating that cell apoptosis was initiated at an earlier stage of differentiation upon excessive Wnt activation. Consistently, proliferation and pro-apoptosis genes were upregulated in Apcmin/+ intestinal epithelial cells (Supplementary Fig. 7e).
According to the model, high Wnt signaling activity alone should lead to longer villi due to increased proliferation (Fig. 6f, (ii)). However, the positive feedback between Wnt and BMP signaling counteracts this by inducing cell death, thereby maintaining stable cell numbers and villus length (Fig. 6f, (iii)). To experimentally validate this, we generated Apc and Bmpr1a double conditional knockout (DKO) mice. Both Bmpr1a cKO and DKO mice exhibited increased cell number and villus length, whereas Apc heterozygous mice did not (Fig. 6g). These results indicate that while Wnt signaling promotes cell proliferation, it also upregulates BMP signaling, which induces cell apoptosis. This balance ensures that increased cell proliferation does not lead to excessive villus elongation. Blocking BMP signaling alone is sufficient to increase villus length, highlighting the essential role of BMP signaling in villus length maintenance.
Discussion
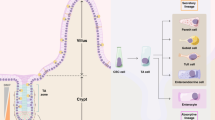
Previous analyses of small intestinal segments have primarily focused on comparative studies of differential genes and cell types using single-cell and spatial transcriptomic data32,42,43. However, a notable difference among these segments is the variable length of villi. The mechanisms underlying the gradient differences in villus length along the small intestine remain unexplored. By integrating experimental data and mathematical modeling, we established a framework for understanding the regulatory mechanism governing villus length. We observed differential BMP signaling activities in different regions of the small intestine, which control villus length along the proximal-distal axis. Our results also revealed the crucial role of the interplay between BMP and Wnt signaling in regulating epithelial turnover (Fig. 7).
BMP signaling exhibits a spatial gradient from the proximal duodenum to the distal ileum. As BMP signaling increases from the proximal to distal segment, Wnt signaling activity remains relatively constant. BMP signaling inhibits cell proliferation, promotes epithelial cell apoptosis by inhibiting integrin expression, and suppresses Wnt signaling. This gradient of BMP signaling correlates with the gradual reduction in villus length from the proximal to distal small intestine. Along the crypt-villus axis, increased BMP signaling coupled with its negative feedback on Wnt signaling attenuates cell proliferation as villi reach a certain length. Furthermore, the positive feedback of Wnt signaling on BMP signaling prevents excessive proliferation to maintain stable villus length (Created in BioRender. Liu (2025) https://BioRender.com/u0fu7hg).
Gradually reduced villus length from the proximal to distal small intestine is associated with the turnover rate of epithelial cells28. Cell turnover involves a finely tuned balance between cell proliferation and apoptosis, which are constrained in different zones along the crypt-villus axis: cell proliferation occurs in the crypts while apoptosis takes place in the villus tip1. The turnover time of small intestinal epithelial cells has been investigated using isotope labeling44 and Lgr5-LacZ tracing methods1. However, the turnover differences along the small intestine from the proximal to distal segments have not been examined. We observed faster proliferation rates in the proximal region and higher apoptotic activity in the distal small intestine. These differences are in agreement with the change in villus length along the small intestine, with long villi in the proximal segments and short villi in the distal segments.
Intestinal segment transplantation showed that villus length adapts to the surrounding environment: transplantation of a segment of proximal small intestine to a distal location resulted in shorter villi, whereas transplantation of a distal segment to a proximal location led to longer villi45. This suggests that villus length is dictated by the local environmental factors at different intestinal segments. This raises the question of whether distinct molecules are expressed in different intestinal segments or whether some signaling pathways exhibit differential activities in these regions. Our study supports the latter, demonstrating that BMP signaling is differentially activated, generating a gradient that controls villus length along the proximal-distal axis of the small intestine.
The formation and maintenance of organ structure rely on intricate signaling networks that govern cell fate decisions46. BMP signaling regulates cell differentiation, apoptosis, and proliferation, which could be concentration-dependent47. Previous studies show that cells interpret their spatial positions by sensing and responding to the varying BMP concentrations in their microenvironment33,48. BMP inhibitors play a crucial role in establishing the gradient of BMP signals14,49. Consistently, a high Gremlin1 level at the base of the crypts establishes a robust BMP boundary, where cells respond to its spatial gradient along the villus-crypt axis to regulate cell death.
Anoikis, a major type of cell death that differentiates intestinal epithelial cells, plays a critical role in maintaining intestinal epithelial homeostasis6. Integrin proteins prevent anoikis by anchoring cells to the extracellular matrix50. Integrin α6β4 subunits are exclusively expressed in epithelial cells and suppress anoikis in human differentiated intestinal cells35. We demonstrated that BMP signaling downregulates the expression of integrins, including α6β4. Consistent with the role of apoptosis in villus length determination, we found that activation of integrin signaling promoted the elongation of small intestinal villi. As epithelial cells migrate along the crypt-villus axis towards the apical tips of villi, they encounter prolonged BMP signaling activation, a result of integration of signal concentration over time rather than simple concentration-dependent linear effects, leading to induction of apoptosis. Such phenomena are also observed in the studies of morphogen transduction51,52.
Organ homeostasis critically relies on complex feedback loops integrating cell proliferation and apoptosis, essential for maintaining tissue integrity and functionality in various physiological and pathological scenarios53,54,55. The interplay between Wnt and BMP signaling plays an important role in the maintenance of intestinal epithelial homeostasis. BMP is known to antagonize Wnt signaling in controlling stem cell proliferation16. Here, we observed that BMP generates negative feedback on Wnt signaling, enabling information transmission from the villus to the crypt region to regulate the cell number of the intestinal epithelium. We also found that Wnt signaling upregulates the expression of BMP ligands and receptors. This reciprocal regulation forms a feedback loop wherein Wnt signaling activation strengthens BMP activity, contributing to the maintenance of intestinal villus length. Specifically, Wnt signaling activation promotes cell proliferation, and meanwhile, increases BMP signaling, which limits cell number by enhancing anoikis to regulate villus length. It will be interesting to study the regulatory mechanisms that orchestrate the adaptation of villus length by the interplay between epithelial and mesenchymal cell signaling pathways, and in response to environmental changes56,57.
Our current mathematical model focuses on the proliferation and apoptosis of epithelial cells and their combined effect on villus length. However, the process of villus formation also involves complex interactions with other cell types, such as mesenchymal and immune cells58. Future modeling improvements will need to incorporate these cell types and their signaling pathways to better reflect the complexities of villus structure and enhance the model’s predictive capacity.
Methods
Mice
Bmpr1aflox/flox mice were kindly provided by Dr. Yuji Mishina59, and villincreERT2 mice were graciously shared by Dr. Sylvie Robine, Institute Curie-CNRS60. Smad4flox/flox mice originated from Dr. Xiao Yang61. CaggcreERT2 and Grem1flox/flox mice were purchased from Saiye Biotech Limited Company (Guangzhou, China). Apcflox/flox mice were acquired from Jackson Laboratory. Apc min/+ mice were kindly shared by Dr. Wei Wu. Lgr5CreERT2 and Rosa26loxp-stop-loxp-ZsGreen mice from Dr. Xiao-Dong Wang (National Institute of Biological Sciences, Beijing). Rosa26loxp-stop-loxp-Cas9-GFP mice were provided by Dr. Jianwei Wang (Tsinghua University, Beijing). Apoa1-mCherry mice were commercially generated by Gem Pharmatech (Nanjing). Axin2-mGFP mice were kindly provided by Dr. Yi Zeng62. All mice were systematically backcrossed into the C57BL/6 genetic background for a minimum of 10 generations. Mice of both genders, aged between 2 and 4 months, were included in this study. The determination of sample size was not predicated on statistical methods. Typically, a minimum of three mice per genotype were utilized in each experimental assay. No exclusions were made from the study cohort, and randomization of the animals was not employed. Tamoxifen (Sigma) was solubilized in corn oil to attain a concentration of 20 mg/mL, with an administration of 75 mg/kg injected into the mice over a span of five days. Pyrintegrin (Selleckchem, #E0462) was dissolved in DMSO to prepare a stock solution. A volume of 15 μL of the stock solution (66 μg/μL) was added to 120 μL of PEG300 and mixed until clear. Subsequently, 15 μL of Tween 80 was added and mixed until clear. Then, 150 μL of water was added. Each mouse received a daily injection of 100 μL for five consecutive days, followed by sacrifice on the sixth day. For the administration of EdU, a dosage of 42.5 mg/kg was injected at 1 h and 24 h prior to sacrifice. In the context of adenovirus injections, an adenovirus carrying the full-length human R-spondin1 was generated and purified through cesium chloride gradient centrifugation. A dose of 5 × 108 pfu of the virus was intravenously injected into each mouse via tail vein injection. Mice injected with adenovirus expressing R-spondin1 were sacrificed 5 days later. One milligram/mL poly(I:C) HMW (InvivoGen, tlrl-pic) was made, and 20 mg/kg was injected. All procedures involving animals adhered to the pertinent guidelines and were conducted with the approval of the Institutional Animal Care and Use Committee of Guangzhou Laboratory (GZLAB-AUCP-2023-01-A01).
Rat and monkey
Eight-week-old rats were acquired from SJA Laboratory Animal Co., Ltd. The small intestine of cynomolgus monkey (Macaca fascicularis) were obtained from Yunnan Key Laboratory of Primate Biomedical Research, and the experimental protocols were approved in advance by the Institutional Animal Care and Use Committee of Yunnan Key Laboratory of Primate Biomedical Research.
Generation of 4xBRE-tdTomato knock-in mice
DKO-AG-haESCs were obtained by optimizing AG-haESCs63, which can obtain mice with the genotype of interest more efficiently64. Plasmid pX330-mCherry-Rosa26-SG was constructed by inserting the sgRNA sequence of the Rosa26 gene trap range into the pX330-mCherry plasmid (Addgene, Plasmid# 98750) containing the sgRNA scaffold and Cas9 DNA sequence. To obtain the donor plasmid pT-4xBRE-miniP-tdTomato-PEST, the sequence containing pT-4xBRE-miniP-tdTomato-PEST and the homologous arm was ligated into pMD19T-Vector. pX330-mCherry-Rosa26-SG and pT-4xBRE-miniP-tdTomato-PEST were transfected to DKO-AG-haESCs by lipo2000 (Invitrogen, Cat#11668019). Cells were then obtained by sorting the haploid cells by FACS (Fluorescence Activated Cell Sorting) and genotype identification by DNA sequencing. The cells were injected into mature oocytes, and semi-cloned mice were obtained by embryo transfer into the uterus of pseudo-pregnant female mice. Then Rosa26-4xBRE-miniP-tdTomato-PEST mice were obtained after mating and genotype identification.
Isolation of intestinal crypts and organoid culture
Intestinal crypts were isolated and cultured following established methodologies65. Briefly, the murine intestine was longitudinally sectioned and subjected to thorough washing with cold PBS. Subsequent removal of villi was performed with precision, and intestine fragments of 5–10 mm were immersed in 2 mM EDTA dissolved in PBS. The suspension was incubated at 4 °C for 30 min, vigorously agitated within cold PBS, and then passed through a 70 μm cell strainer (BD Biosciences). The crypt fraction was enriched via centrifugation (3 min at 300–400 g), and then mixed with Matrigel (BD Biosciences) and seeded onto 24-well plates. Upon solidification of the Matrigel, crypt culture medium designated as ENR (Advanced DMEM/F12 supplemented with EGF (50 ng/mL, Novoprotein, C09), Noggin (100 ng/mL, OrganRagen, 807-NOG), and R-spondin1 (500 ng/mL, OrganRagen, 861-RS1), as well as Penicillin/Streptomycin, GlutaMAX-I, N2, B27, and N-acetylcysteine (Sigma-Aldrich)) was meticulously introduced and replenished at three-day intervals. For passaging, organoids embedded within Matrigel were suspended within 1 mL of cold PBS, pelleted via centrifugation (3 min at 300–400 g), and then re-embedded in fresh Matrigel prior to seeding onto a plate and subsequent medium supplementation. After two generations, organoids were subsequently maintained in EN (withdraw Noggin from ENR), ENR, or ERB (ER with BMP4 (20 ng/mL, R&D, 314-BP-050)) medium for a duration of 24 or 48 h. Five hundred nanometers 4-OH-tamoxifen (4-OHT, Sigma, H7904) or ethanol was used for 48 h to induce Bmpr1a knockout in organoids.
Lentivirus production and Itgb4 knock-out in organoids
HEK293FT were cultured in a 6-cm dish with Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen, 31053028) supplemented with 10% fetal bovine serum (FBS, Hyclone, SH20074.03), 1% penicillin/streptomycin (Gibco, 15070-063), 0.1 mM MEM Non-Essential Amino Acids (NEAA, Gibco, 11150-050), 2 mmol/L L-glutamine (Gibco, 25030-081), and 1 mM Sodium Pyruvate (Gibco, 11360-070) under the selection with Geneticin (500 μg/mL, G418). The cells were transfected with 4 μg of the plasmids LV04-gRNA (Sigma Aldrich) targeting mouse Itgb4 (sgRNA1+PAM: GAGCTGTACCGAGTGCATCCGG; sgRNA2 + PAM: TTGTCCACCCGGATGCAC TCGG), 6 μg Pax2.0, and 2 μg pMD2.G, using Polyetherimide (PEI, Polysciences, PT-101-01N), and cultured with G418-free medium. Three days post-transfection, the supernatant was passed through a 0.45-μm filter, added Lenti-concentin virus precipitation solution (5X, ExCell Bio, EMB810A), and refrigerated at 4 °C overnight. After centrifugation at 1500 g for 30 min, the pellet was re-suspended in 250 μL of infection medium (the ENR medium containing 6.67 μM blebbistatin, 2.5 μM CHIR-99021, 10% Wnt3a conditional medium, and 10 μg/mL polybrene (Macgene, MC032)).
Organoid infection was performed as previously described in ref. 66. Briefly, organoids derived from the crypts of Rosa26loxp-stop-loxp-Cas9-GFP mice were cultured with the ENR culture medium. After 2.5 μM CHIR-99021 and 10 mM nicotinamide treatment for 2 days, the organoids were digested and re-suspended with infection medium containing virus. Then, infected cells were placed on solidified Matrigel and incubated overnight at 37 °C. Two days after infection, change the medium with ENR plus 2 μg/mL puromycin. Once the organoids grew and passed normally under the puromycin selection, the CAS9 protein was induced with 4-OHT. Immunoblotting was performed on day 4 after 4-OHT treatment.
TUNEL staining
The mouse intestine was rinsed with ice-cold PBS, and fixed with 4% paraformaldehyde at 4 °C overnight. Then the tissues were dehydrated and embedded in paraffin wax. Freshly cryopreserved intestinal samples were arranged in a “Swiss-roll” configuration and embedded within paraffin wax. Embedded tissues were sectioned with a thickness of 5 μm. After deparaffinization, the sections were subjected to TUNEL staining in accordance with the manufacturer’s guidelines (Roche), and counter-stained with DAPI.
TUNEL signal intensities were quantified by calculating the mean intensity along the villus axis. Each villus was subdivided into ten equally sized bins along its length (from base to tip) to assess regional variations in signal intensity. The decision to use ten segments was based on optimizing the resolution of intensity gradients along the villus while maintaining sufficient statistical power for comparison. For each segment, the mean TUNEL intensity was calculated, excluding non-positive values. These results, along with standard deviations, are presented to highlight the trends observed in control and Bmpr1a cKO samples. The central line represents the mean, and the shadow regions depict the standard deviation (SD) for each bin. Additionally, for the analysis of proximal and distal small intestine regions, we carefully selected representative sections from both ends of the intestine for comparison. Each region was delineated based on anatomical landmarks, ensuring consistent sampling. Improved image presentation and analysis results are now provided to enhance clarity.
Immunofluorescence and histological staining
For immunohistochemical (IHC) analysis, sections of formalin-fixed and paraffin-embedded intestine were deparaffinized with isopropanol and alcohol solutions. Subsequently, antigen retrieval was performed, and endogenous peroxidase activity was quenched with H2O2. The sections were subjected to a blocking procedure involving 3% serum for 30 min and incubated with primary antibodies overnight at 4 °C. The primary antibodies were: rabbit anti-Ki67 (Abcam, ab15580, used at a dilution of 1:100), goat anti-Gremlin1 (R&D, AF956, used at a dilution of 1:50), and rabbit cleaved caspase-3 (CST, 9664S, 1:300). Then horseradish peroxidase-conjugated anti-rabbit or anti-goat antibody (Invitrogen, diluted at 1:200) was added for 2 h. Finally, DAB substrate (Dako) was added. For immunofluorescence (IF) analysis, similar antigen retrieval and blocking procedures were performed, and sections were incubated overnight at 4 °C with the following primary antibodies: p-Smad1/5 (CST, 9516 s, 1:100), p-Smad3 (CST, 9520s, 1:100), β-catenin (Sigma, C7207, 1:100), Gremlin1 (Abmart, PK12814S,1:100), and Itga6 (HUABI, ET1602-1, 1:100), and rabbit cleaved caspase-3 (CST, 9664S,1:300).
Tyramide signal amplification (TSA)
A Fluorescence kit based on TSA technology (Huilanbio BiologicalTechnology, Shanghai, China) was employed to detect p-Smad1/5, p-Smad3, β-catenin, and Itga6 (HUABI, ET1602-1, 1:100). The intestinal tissues were fixed in 10% formalin at room temperature for 2 h, followed by overnight immersion in 30% sucrose solution. Subsequently, the tissues were embedded for cryo-sections. For antigen retrieval, sections were exposed to a pH 6.0 citric acid buffer in a microwave-enabled retrieval chamber and boiled at 100 °C for 10 min. Following natural cooling, the sections were washed with PBS (pH 7.4) three times for 5 min. To inhibit endogenous peroxidase activity, sections were incubated with a 3% hydrogen peroxide solution at room temperature for 15 min in a light-protected environment. This was followed by three additional 5-min washes with PBS. After drying, a hydrophobic barrier was outlined around the tissue using a super-pap pen. A solution of 3% Bovine Serum Albumin (BSA) in PBS was applied within the outlined area, covering the tissue uniformly. The tissue was blocked with BSA for 30 min at room temperature. The primary antibody was then applied after removing the blocking solution. The sections were placed horizontally in a humid chamber to prevent light exposure and incubated overnight at 4 °C for optimal antibody binding. Subsequently, the sections were washed for 5-min, three times with PBS (pH 7.4) using a decolorization shaker. After a brief drying period, the tissue was incubated with a horseradish peroxidase (HRP)-conjugated secondary antibody for 50 min at room temperature in the absence of light and washed three times with PBS. Fluorescence labeling was achieved by treating the sections with TYR570 fluorescent dye for 10–15 min, followed by PBS washing for visualization.
EdU and BrdU labeling
To assess proliferation rates and cell cycle dynamics in the proximal and distal small intestine, we performed a dual EdU/BrdU pulse-chase experiment as previously described in ref. 31. BrdU (10 mg/mL) and EdU (5 mg/mL) were prepared in PBS. Mice were intraperitoneally injected with EdU (50 mg/kg) at time-point 0, followed by BrdU (50 mg/kg) 1 h later. At 1.5 h post-injection, mice were euthanized by cervical dislocation, and the gravid uterus was removed and placed in ice-cold PBS for 3 min.
To assess epithelial turnover and cell migration, EdU and BrdU labeling were performed as follows: mice were injected with EdU (50 mg/kg) 48 h prior to euthanasia to label cells in the S-phase at this time point. Twenty-four hours later, BrdU (50 mg/kg) was injected to label cells in the S-phase at the second time point. After the second injection, the mice were euthanized 24 h later, and intestinal tissues were collected for further processing.
Calculation of synthesis phase duration
The synthesis phase duration was calculated based on EdU and BrdU labeling in epithelial cells of the crypt region. Cells were classified into two groups based on their EdU and BrdU reactivity: EdU+; BrdU+: the cells that remained in S-phase during the EdU and BrdU injection interval; EdU+BrdU-: the cells that exited S-phase during the EdU to BrdU injection interval. The synthesis phase duration (Ts) was determined using the following formula:
This ratio reflects the proportion of cells that remained in S-phase during the injection interval relative to those that exited S-phase, with shorter Ts indicating faster cell cycle progression in the crypt epithelium.
BrdU immunostaining
Tissue sections underwent antigen retrieval with citrate buffer (pH 6.0) in a microwave for 15 min, followed by cooling and washing with PBS. Sections were blocked with 3% BSA and 0.1% Triton-X100 in PBS for 1–2 h at room temperature, then incubated overnight with APC-conjugated anti-BrdU antibody (1:200, BD Biosciences, Lot No. 0041156). After washing, secondary antibody incubation was performed for 1 h at room temperature.
EdU labeling
EdU incorporation was detected using the BeyoClick™ EdU-555 Kit (Beyotime, Cat No. C0075S, Lot No. A018240930) according to the manufacturer’s instructions. Sections were incubated with the EdU reagent for 30 min at room temperature, washed with PBS, and mounted with DAPI-containing medium. Fluorescent images were captured using a fluorescence microscope.
To quantify EdU+ cells in the small intestine, EdU-labeled cells were manually counted along the villus axis in both proximal and distal regions of the small intestine from control and mutant mice. Specifically, EdU+ cells were counted along the villus axis between two designated points: the crypt base (near the stem cell region) and the point where the highest accumulation of EdU+ cells was observed. The number of EdU+ cells within this defined region was recorded for each crypt-villus unit. The total number of EdU+ cells within each crypt-villus unit was quantified, and data were averaged across biological replicates.
smFISH staining and quantitative image analysis of mRNA expression
Bmp2 mRNA (ACD Bio, 406661-C1), Bmp4 mRNA (ACD Bio, 401301-C2), Id1 mRNA (ACD Bio, 312221-C3), Grem1 mRNA (ACD Bio, 314741), Dhcr24 mRNA (ACD Bio, 491361-C2) were detected with RNAscope by following the protocol (ACD Bio, 323100). After hybridization according to the manufacturer’s protocols, tissue sections were washed for 5 min in PBS containing 0.1% Tween-20, blocked for 1 h at room temperature in PBS containing 5% FBS. All the steps were applied.
For quantifying smFISH mRNA expression, we utilized a systematic image segmentation approach based on the log filter method to identify and count mRNA puncta in small intestinal tissue sections from three mice. The filtered images were normalized to a 0–1 scale, and a threshold value of 0.1 was applied to create a binary mask. The binary image was then processed to identify connected components using bwlabel in MATLAB, where each labeled region was considered a distinct mRNA punctum. The number of puncta in each region was recorded as the quantification metric. For Grem1 mRNA quantification, to ensure comparable data, mRNA puncta were quantified within a defined tissue area of 625 µm² per region, with each region carefully chosen to represent both proximal and distal sections of the small intestine. For each mouse, mRNA counts were obtained from ten corresponding proximal and distal regions, ensuring consistent tissue area coverage. The selected regions were of uniform size to minimize variability between samples. The same area was used consistently across all samples to allow for accurate comparisons between proximal and distal regions within the same mouse, as well as across different mice. To assess the significance of differences in mRNA abundance between proximal and distal regions, a paired t-test was performed on the normalized puncta counts for each mouse, with statistical significance set at p < 0.05. Bmp2, Bmp4, and Id1 mRNA expression exhibit clustering of mRNA puncta. Therefore, the method for quantifying mRNA expression was consistent with the approach used to quantify the gradient of p-Smad1/5 signals in villi, both relying on the measurement of fluorescence intensity.
Villus length measurement
To assess villus length, each individual small intestine was first aligned in accordance with its initial point (duodenum) and concluding point (ileum). The aligned intestines were subsequently partitioned into six equidistant segments to ensure consistent sampling across the entire length. These villi were chosen based on their proximity to the corresponding division points, ensuring that they were representative of the segment’s characteristics. To minimize measurement bias, only fully intact villi, with no visible damage or distortions, were included. For each selected villus, the length was measured from the base to the tip. The measured values from positions 1 through 6 were then used to calculate the mean villus length for the entire small intestine, providing a comprehensive assessment of villus morphology across different intestinal regions
Quantitative RT-PCR
Total RNA extraction was carried out using the TRIZOL reagent (Catalog No: 15596026, Thermo Fisher Scientific). Subsequently, 1 μg of total RNA was subject to reverse transcription into cDNA using the HiScript II Q Select RT SuperMix (Vazyme). The resultant cDNA was amplified via real-time PCR employing the ChamQ SYBR Color qPCR Master Mix (Vazyme). The determination of relative mRNA expression levels was effected utilizing the 2−ΔΔCq method, with normalization against GAPDH or β-actin mRNA. The statistical analysis was performed with the Student’s t-test. The specific primer sequences employed for quantitative PCR are shown in Supplementary Table 1.
Chromatin immunoprecipitation PCR (ChIP-PCR)
ChIP assay was performed in Lgr5+ ISCs sorted from cultured organoids. One million cells were obtained through FACS and crosslinked with 1% formaldehyde for 8 min at room temperature. ChIP-PCR was performed by following the manufacturer’s instructions (iDeal ChIP-seq kit, Diagenode), and the lysates were incubated with Rabbit Control IgG antibody (Abclonal, AC005), anti-Smad467 or anti-Smad1 (CST, 6944, 1:2000) antibodies. Real-time PCR was performed (LightCycler 480 system, Roche). All the primers used were shown in Supplementary Table 2.
Cytoplasmic/nuclear fractionation
Wash cells twice with PBS and add 250 μL of lysis buffer (10 mM Hepes-NaOH, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.5 mM β-mercaptoethanol) supplemented with protease inhibitor mixture and phosphatase inhibitors. Scrape cells, vortex, and incubate on ice for 15–20 min. Add 4–5 μL of 10% NP-40 to the lysate, vortex, and incubate on ice for 2 min. Centrifuge at 16,000 g for 10–15 min. The resulting supernatant contains cytoplasmic proteins, and the pellet is reserved for nuclear protein extraction. Wash the pellet with ice-cold PBS twice (200 μL each). Add 60-80 μL of nuclei lysis buffer (10 mM Tris-HCl, pH 7.6, 420 mM NaCl, 0.5% Nonidet P-40, 1 mM DTT, 1 mM PMSF, 2 mM MgCl2, protease and phosphatase inhibitors) to the pellet. Disperse the pellet with pipette tips and incubate on ice for 20 min. Centrifuge at 16,000 × g for 10–15 min. The resulting supernatant contains nuclear proteins. Adjust the NaCl concentration to 150 mM using lower salt buffer (10 mM Tris-HCl, pH 7.6, 1 mM DTT, 1 mM PMSF, 2 mM MgCl2,& protease and phosphatase inhibitors).
Immunoblotting
Protein lysates were prepared from intestinal epithelium, the villus region, or organoids, as described previously in ref. 68. Antibodies are as follows: rabbit anti-integrin β4 (Abcam, ab182120, utilized at a dilution of 1:1000), rabbit anti-integrin alpha 6 (HUABI, ET1602-1, 1:1000), rabbit p-Smad1/5 (CST, 9516s,1:1000), rabbit Smad1(CST, 6944T, 1:1000), rabbit anti-Bcl2 (CST, 3498s, 1:1000), mouse anti-BAX (Santa, sc7480,1:1000), rabbit anti-cleaved caspase 3 (CST, 9664L, 1:1000), rabbit anti-total caspase 3 (Proteintech, 19677-1-AP, 1:1000), rabbit anti active β-catenin (CST, 8814, 1:1000), mouse anti-GAPDH (Ruikang, RM2002,1:10000), rabbit anti-β-actin (CST, 4970S,1:5000), and mouse anti-Tubulin (Proteintech, 66031-1-Ig, 1:20000), rabbit anti-Histone H3(CST, 4499T, 1:5000). Original immunoblots are shown in Supplementary Fig. 8.
GreenNuc™ caspase-3 activity detection in small intestinal organoids and CC3+ cells quantification in vivo
For GreenNuc™ caspase-3 activity detection in small intestinal organoids, organoids are harvested using 1× PBS, centrifuged, and resuspended in 500 µL PBS. After pipetting to separate buds, the mixture is centrifuged again, and Matrigel is added before plating in a 96-well plate. Each well receives 2 µL of Matrigel, incubated at 37 °C for 10 min, followed by the addition of 50 µL ENR medium. Organoids are cultured for 3-4 days until budding, after which BMP (20 ng/mL) + ER are applied to five wells, and ENR to five control wells.
At each time point (0 h, 4 h, 8 h, and 24 h), a 96-well plate is collected, and 50 µl of 3 µM GreenNuc™ Caspase-3 Substrate (1 mM) is diluted in 1:300 PBS and added to each well. After 30 min of incubation at room temperature, images are captured. The GreenNuc™ Caspase-3 Substrate (1 mM) used in this protocol is available under catalog number C1168M (Beyotime, Lot No. Z905241007).
CC3+ cells were manually counted from the top view of whole mount villus images, and the total DAPI+ cell count was manually determined from the side view of agarose sections for each intestinal segment. The number of CC3+ cells per villus was divided by the average total number of DAPI+ cells in the same villus to calculate the CC3+ cell density. The points in the statistical graph represent the ratio of CC3+ cells to DAPI+ cells for each villus.
RNA-seq analysis
cDNA libraries were generated using the Ovation RNA-Seq System V2 kit (NuGEN). Sequencing was performed on the Illumina platform, employing a paired-end 150-base strategy, under the supervision of Novogene Bioinformatics Technology Co., Ltd (Beijing, China). The obtained sequencing reads were aligned to the mouse genome reference (mm10) using STAR (Spliced Transcripts Alignment to a Reference) with default parameters. Differential expression analysis was conducted using DESeq2 software (version 1.28.1). To assess functional enrichment, Gene Ontology (GO) enrichment analysis was performed using ClusterProfiler software (version 3.16.1).
Quantification of ZsGreen+ cell migration
ZsGreen+ cells were quantified by measuring the distance from the true villus tip to the leading edge of the ZsGreen+ cell cluster. The villus tip was visually identified, and the front of the migrating ZsGreen+ cells was defined as the leading edge of the cell cluster. These two positions were marked in images, and the distance between them was measured. Distances were calculated for both the proximal and distal small intestine. These measurements were manually performed based on the whole mount images, and the distances between these defined points were used for subsequent statistical analysis.
Quantification of Axin2-GFP signals along the crypt
After performing frozen tissue sectioning and staining, different segments of the intestine (duodenum, jejunum, and ileum) were selected for analysis. To assess the gradient of Axin2-GFP expression along the crypt, from stem cells to transit-amplifying (TA) cells, the following steps were carried out: Crypt boundaries were manually delineated using Fiji’s Segmented Line tool, starting from the crypt base (stem cell region) to the crypt-villus junction. Axin2-GFP signal intensities were quantified along the absolute physical length of the crypt, enabling the analysis of spatial differences in signal intensity along the proximal-distal axis of the small intestine. Positive Axin2-GFP signals were identified by applying a threshold intensity level above the background noise, ensuring consistent identification of positive signals across all images. Final data were averaged across biological replicates and presented as relative protein signal profiles.
Quantification of p-Smad1/5 signaling along the villus axis
To quantify p-Smad1/5 signaling, images were acquired using a VS200 microscope (Olympus) and processed using custom MATLAB scripts. Representative villi were selected based on their integrity and length to ensure consistent analysis. Each villus was manually oriented vertically and independently sorted to eliminate interference from adjacent structures. Signals from the epithelial region were specifically analyzed, excluding any contribution from mesenchymal cells to ensure accuracy.
To compare p-Smad1/5 profiles across villi of varying lengths, the p-Smad1/5 signal was quantified along the physical length of the villus, allowing for analysis of spatial differences in signal intensity across the proximal-distal axis of the small intestine. The MATLAB pipeline incorporated interpolative methods to standardize signal measurements across villi with varying physical lengths. Specifically, villi were interpolated to the average length of the relative intestinal segment using a linear interpolation algorithm within each region (i.e., duodenum, jejunum, or ileum). This allowed uniform signal intensity profiles, enabling direct comparisons across different samples. For each image, p-Smad1/5 intensities were summed across interpolated rows, generating a robust signal intensity matrix for further analysis. Final data were averaged across biological replicates and plotted as absolute and relative p-Smad1/5 profiles.
Quantification of Grem1 protein signals
To analyze Grem1 signals in the stromal cell regions, 30 regions of interest (ROIs), each measuring 625 µm², were selected from three mice, from both the proximal and distal areas. For each selected ROI, the average signal intensity for Grem1 was quantified. The average Grem1 signal intensity within each ROI was calculated, and the differences in signal intensity between the proximal and distal regions were compared.
Mathematical modeling
Reaction–diffusion model
The mathematical formulations were applied to the entire length of the proximal (600 μm) and distal (335 μm) small intestine, spanning from the villus to stroma cells. The model elucidates the interaction between BMP signaling, inhibitory factors, and receptors, whereby upon expression of signaling molecules (\({\phi }_{B}\), \({\phi }_{I}\), and R); BMP signaling and inhibitory factors can undergo diffusion (DB, DI, and DBI), degradation (decB, decI, and decBR); \({k}_{{onI}}\) and \({k}_{{offI}}\) are the reaction rates for the binding and dissociation of BMP and its inhibitor, and \({k}_{1}\) and \({k}_{2}\) are the reaction rates for the binding and dissociation of BMP and its receptor. In our reaction–diffusion model, we explored several potential mechanisms underlying BMP gradient formation, including: (1) A transcription gradient of BMP ligand mRNA that directly influences BMP protein distribution (Supplementary Fig. 2c, model 1);
(2) Source-sink diffusion, wherein BMP ligands diffuse across the tissue (Supplementary Fig. 2c, model 2);
(3) A counter-gradient mechanism, involving the diffusion of Gremlin1 across the embryonic tissue to regulate BMP activity without BMP receptor (Supplementary Fig. 2c, model 3)
or with BMP receptor (Supplementary Fig. 2c, model 4
Spatial distribution data aided in determining the production regions of Bmp2b, Bmpr1a, and Inhibitor30. Discretization into 41 equidistant nodes was applied across the entire crypt-villus structure, and the finite difference method was employed for the resolution of the second-order spatial derivative. Temporal evolution of the simulation was executed utilizing the adaptive solver ode15s in MATLAB, employing a relative tolerance threshold of 1e-9. The model was simulated over a developmental interval of 2 h, allowing for comprehensive molecule interactions and achievement of an equilibrium state. Evaluation of model performance was facilitated by computing the NRMSD, comparing the simulated outcomes with the p-Smad1/5 distribution along both the proximal and distal segments of the small intestine. Model simulations exhibited a favorable agreement with experimental observations. All parameters were varied, encompassing all biologically feasible values (Supplementary Fig. 2e).
A computational model for villus growth coupled with cell lineage and signaling
We considered a two-stage cell lineage where progenitor cells undergo self-proliferation in the crypt region and differentiate into terminally differentiated (TD) cells. The model incorporates the regulatory roles of BMP, Wnt, and BMP inhibitor, whose concentrations are denoted by [B], [W], and [I], respectively. The crypt-villus axis is characterized by a one-dimensional space with the cell density distributions of progenitor cells \({C}_{0}\) and the cell density distributions of TD cells \({C}_{1}\), in which \(x=0\) represents the crypt base and \(x={x}_{\max }\) is the position of villus top tracking the growth of villus69,70. At the initial time point (\(t=0\)), the progenitor cell density \({C}_{0}\) is assumed to be concentrated in the crypt region and the TD cell density \({C}_{1}=0\) throughout the crypt-villus axis. The density of progenitor cells \({C}_{0}\) and the density of TD cells \({C}_{1}\) are governed by.
where \({p}_{0}\left(x,\left[B\right],\left[W\right]\right)\) is the proliferation probability of progenitor cells, which is spatially dependent and regulated by BMP and Wnt signaling20, \({v}_{0}\) is the reciprocal of the cell cycle length multiplied by ln 2, \({d}_{1}\) is the removal rate for TD cells, corresponding to the anoikis of TD cells, and \(V\) is the velocity of cell migration, which is related to the self-proliferation of progenitor cells in the crypt region and the removal of TD cell in the villus region, maintaining the uniform cell density along the crypt-villus axis i.e., \({C}_{0}+{C}_{1}=1\) for \(x\in [0,{x}_{\max })\).
in which \(\sigma \left({C}_{0},{C}_{1}\right)\) is a cell density constraint defined as Eq. (4) to penalize the overcrowding of cells along the crypt-villus axis.
The velocity of cell migration at the crypt base is designated as zero \(V(0,t)=0\), and the dynamics of the crypt-villus length is determined by the velocity at the villus top,
The proliferation probability of progenitor cells is spatially dependent and regulated by Wnt and BMP20. To maintain a homeostatic population of progenitor cells, the proliferation probability \({p}_{0}\) is assumed to be approximately 1 in the crypt region and approximately 0 in the villus region, resulting in differentiation from progenitor cells into TD cells after the progenitor cells migrate out of the crypt region. The proliferation probability \({p}_{0}\) is defined as
where \(\frac{1}{2}\left(1-\tanh \left(\frac{x-w}{\epsilon }\right)\right)\) represents the spatial range of progenitor cell proliferation in the absence of regulatory effects of BMP and Wnt signaling. The parameter \(w\) dictates the baseline crypt progenitor zone \(x\in \left(0,w\right)\), which ensures the crypt retains a finite size. \(\epsilon\) is a small value ensuring that \({p}_{0}\) diminishes sharply from 1 to 0 for \(x > w\). The regulatory effects of BMP and Wnt signaling influence the spatial range of progenitor cell proliferation, represented by Hill functions \({\phi }_{p0}\left(\left[B\right],\left[W\right]\right)\) defined in Eqs. (20), (27), and (32).
In the investigation of the regulation of BMP signaling on villus length, only the inhibition of BMP signaling on the proliferation probability and promotion of BMP signaling on the apoptosis rate were under consideration (Fig. 4b k). BMP and BMP inhibitor are produced by TD cells and stroma cells, respectively. The binding of the inhibitor to BMP removes BMP from regulation and generates \(\left[{BI}\right]\). The dynamics of BMP and BMP inhibitor are described by a reaction–diffusion system.
Each species of molecule assumes individual diffusion rates \({D}_{B}\), \({D}_{I}\), \({D}_{{BI}}\). The production of BMP is assumed to be jointly determined by the regulations and the density of TD cells. The production of BMP inhibitor in the stroma region is represented by a production term \({\phi }_{I}(x)={\bar{\phi }}_{I}\) for \(x\in (-{w}_{s},0)\) and \({\phi }_{I}(x)=0\) for \(x > 0\), in which \({w}_{s}\) indicates the size of the stroma region adjacent to the crypt base. \({k}_{{\mbox{on}}}\) and \({k}_{{\mbox{off}}}\) are the reaction rates for the binding and dissociation of BMP and its inhibitor. The degradation of molecules is proportional to the concentrations with decay rates \({d}_{B}\), \({d}_{I}\), \({d}_{{BI}}\), respectively. The regulation of BMP signaling on progenitor cell proliferation is represented by \({\phi }_{p0}\left(\left[B\right]\right)\), where \({\gamma }_{B}\) is the reciprocal of half maximal concentrations of BMP inhibiting progenitor cell proliferation, \(n\) is the Hill coefficient. All Hill coefficients \(n\) in the model are assigned to 3.
Regarding the BMP-induced apoptosis, we propose two different models based on two hypotheses. The first is the integral mechanism (Fig. 4b and Supplementary Fig. 5, model 5), which posits that cell fate decisions are influenced by the cumulative effect of BMP signaling over time. The enhancement of TD cell anoikis by BMP signaling is described as a removal rate,
that is proportional to the total amount of BMP \({\left[B\right]}_{{\mbox{total}}}\), calculated by integrating the concentration of BMP molecule \(\left[B\right]\) over the crypt-villus axis. The second is the threshold mechanism (Fig. 4k and Supplementary Fig. 5, model 6), which assumes that the intensity of BMP signaling determines cell fate, with a specific threshold of signaling intensity triggering apoptosis. The enhancement of TD cell anoikis by BMP signaling is calculated by the Hill equation of the concentration of BMP molecule \(\left[B\right]\) over the crypt-villus axis.
In the investigation of the regulation of Wnt-BMP signaling on villus length, the activation of Wnt signaling on progenitor cell proliferation and the regulation of BMP by Wnt were introduced to the model (Fig. 5d and Supplementary Fig. 5, model 7). The production of Wnt is self-activating and a downregulated by BMP. The dynamics of BMP, Wnt, and BMP inhibitor are described by a reaction–diffusion system.
The regulation of BMP signaling on the proliferation probability \({p}_{0}\) reduces the spatial range of progenitor cell proliferation, while the regulation of Wnt signaling counterbalances the effect of BMP signaling. \({D}_{W}\) and \({d}_{W}\) are the diffusion rates and decay rates of Wnt, where \({\gamma }_{W}\), \({\gamma }_{B}\), \({\gamma }_{1}\), \({\gamma }_{2}\), denote the reciprocal of the half maximal concentrations. \({\bar{\phi }}_{B}\), \({\bar{\phi }}_{W}\) are the production rates of BMP and Wnt.
To further study the impact of the positive feedback of Wnt to BMP on the maintenance of villus length homeostasis, the enhancement of Wnt on BMP production is introduced (Fig. 6e and Supplementary Fig. 5: model 8). The dynamics of BMP, Wnt, and BMP inhibitor are described by a reaction–diffusion system.
The system of Eqs. (11−13) and Eqs. (17–32) were solved numerically using the Fourier spectral method for the spatial approximation and a semi-implicit scheme for the temporal discretization, and implemented on MATLAB. The parameters involved in the model are listed in Supplementary Table 3.
Statistical analysis
Each experiment was performed more than 3 times, each with at least three biological replicates. In instances where the assumption of normal distribution was tenable, the statistical comparison of disparities between two groups was performed through either Student’s t-test or two-way analysis of variance (ANOVA), as denoted in the figure legends. The statistical evaluations were executed employing the GraphPad Prism 8 software package. For methodologies encompassing immunoblotting, immunohistochemistry, and immunofluorescence, illustrative images of a representative nature are showcased.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The RNA-seq data generated in this study have been made publicly accessible via the Gene Expression Omnibus (GEO) database under the accession code GSE247044. Source data are provided with this paper.
Code availability
Original Code for reaction–diffusion modeling of BMP gradient formation has been deposited to GitHub: https://github.com/XuWangYGClab/PDE-modeling_crypt_villus_BMP. https://doi.org/10.5281/zenodo.15429803. Original Code for the villus growth has been deposited to GitHub: https://github.com/XuWangYGClab/computational-model-for-villus-growth-coupled-with-cell-lineage-and-signaling. https://doi.org/10.5281/zenodo.15429760.
References
Barker, N. et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007 (2007).
Sato, T. & Clevers, H. Growing self-organizing mini-guts from a single intestinal stem cell: mechanism and applications. Science 340, 1190–1194 (2013).
Reddien, P. W. The purpose and ubiquity of turnover. Cell 187, 2657–2681 (2024).
Gehart, H. & Clevers, H. Tales from the crypt: new insights into intestinal stem cells. Nat. Rev. Gastroenterol. Hepatol. 16, 19–34 (2019).
Taylor, S. R. et al. Dietary fructose improves intestinal cell survival and nutrient absorption. Nature 597, 263–267 (2021).
Patankar, J. V. & Becker, C. Cell death in the gut epithelium and implications for chronic inflammation. Nat. Rev. Gastroenterol. Hepatol. 17, 543–556 (2020).
Montoro-Huguet, M. A., Belloc, B. & Dominguez-Cajal, M. Small and large intestine (I): malabsorption of nutrients. Nutrients 13, 1254 (2021).
Kiela, P. R. & Ghishan, F. K. Physiology of intestinal absorption and secretion. Best. Pr. Res. Clin. Gastroenterol. 30, 145–159 (2016).
Hageman, J. H. et al. Intestinal regeneration: regulation by the microenvironment. Dev. Cell 54, 435–446 (2020).
Stapornwongkul, K. S. & Vincent, J. P. Generation of extracellular morphogen gradients: the case for diffusion. Nat. Rev. Genet 22, 393–411 (2021).
Wang, Y. F., Liu, C. & Xu, P. F. Deciphering and reconstitution of positional information in the human brain development. Cell Regen. 10, 29 (2021).
Tuazon, F. B., Wang, X., Andrade, J. L., Umulis, D. & Mullins, M. C. Proteolytic restriction of chordin range underlies BMP gradient formation. Cell Rep. 32, 108039 (2020).
Beumer, J. & Clevers, H. Cell fate specification and differentiation in the adult mammalian intestine. Nat. Rev. Mol. Cell Biol. 22, 39–53 (2021).
Zhang, M., Liu, Y. & Chen, Y. G. Generation of 3D human gastrointestinal organoids: principle and applications. Cell Regen. 9, 6 (2020).
Farin, H. F. et al. Visualization of a short-range Wnt gradient in the intestinal stem-cell niche. Nature 530, 340–343 (2016).
He, X. C. et al. BMP signaling inhibits intestinal stem cell self-renewal through suppression of Wnt-beta-catenin signaling. Nat. Genet. 36, 1117–1121 (2004).
Qi, Z. et al. BMP restricts stemness of intestinal Lgr5(+) stem cells by directly suppressing their signature genes. Nat. Commun. 8, 13824 (2017).
Wang, Y. et al. Stromal BMP signaling regulates mucin production in the large intestine via interleukin-1/17. Sci. Adv. 9, eadi1827 (2023).
Beumer, J. et al. Enteroendocrine cells switch hormone expression along the crypt-to-villus BMP signalling gradient. Nat. Cell Biol. 20, 909–916 (2018).
Beumer, J. et al. BMP gradient along the intestinal villus axis controls zonated enterocyte and goblet cell states. Cell Rep. 38, 110438 (2022).
Zhou, W., Yan, K. & Xi, Q. BMP signaling in cancer stemness and differentiation. Cell Regen. 12, 37 (2023).
Brugger, M. D. & Basler, K. The diverse nature of intestinal fibroblasts in development, homeostasis, and disease. Trends Cell Biol. 33, 834–849 (2023).
Beck, S. E. et al. Bone morphogenetic protein signaling and growth suppression in colon cancer. Am. J. Physiol. Gastrointest. Liver Physiol. 291, G135–G145 (2006).
de Lau, W. et al. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature 476, 293–297 (2011).
Nusse, R. & Clevers, H. Wnt/beta-catenin signaling, disease, and emerging therapeutic modalities. Cell 169, 985–999 (2017).
Frisch, S. M. & Screaton, R. A. Anoikis mechanisms. Curr. Opin. Cell Biol. 13, 555–562 (2001).
Dunker, N., Schmitt, K., Schuster, N. & Krieglstein, K. The role of transforming growth factor beta-2, beta-3 in mediating apoptosis in the murine intestinal mucosa. Gastroenterology 122, 1364–1375 (2002).
Parker, A. et al. Elevated apoptosis impairs epithelial cell turnover and shortens villi in TNF-driven intestinal inflammation. Cell Death Dis. 10, 108 (2019).
Liu, S., Ren, J., Hu, Y., Zhou, F. & Zhang, L. TGFbeta family signaling in human stem cell self-renewal and differentiation. Cell Regen. 13, 26 (2024).
Moor, A. E. et al. Spatial reconstruction of single enterocytes uncovers broad zonation along the intestinal villus axis. Cell 175, 1156–1167 e1115 (2018).
Harris, L., Zalucki, O. & Piper, M. BrdU/EdU dual labeling to determine the cell-cycle dynamics of defined cellular subpopulations. J. Mol. Histol. 49, 229–234 (2018).
Zwick, R. K. et al. Epithelial zonation along the mouse and human small intestine defines five discrete metabolic domains. Nat. Cell Biol. 26, 250–262 (2024).
Teague, S. et al. Time-integrated BMP signaling determines fate in a stem cell model for early human development. Nat. Commun. 15, 1471 (2024).
Wartlick, O. et al. Dynamics of Dpp signaling and proliferation control. Science 331, 1154–1159 (2011).
Beauséjour, M. et al. Suppression of anoikis in human intestinal epithelial cells: differentiation state-selective roles of α2β1, α3β1, α5β1, and α6β4 integrins. BMC Cell Biol. 14, 53 (2013).
Chen, L. et al. A reinforcing HNF4-SMAD4 feed-forward module stabilizes enterocyte identity. Nat. Genet. 51, 777–785 (2019).
Lee, H. W. et al. A podocyte-based automated screening assay identifies protective small molecules. J. Am. Soc. Nephrol. 26, 2741–2752 (2015).
Ohara, T. E., Colonna, M. & Stappenbeck, T. S. Adaptive differentiation promotes intestinal villus recovery. Dev. Cell 57, 166–179.e166 (2022).
Lander, A. D., Nie, Q. & Wan, F. Y. Do morphogen gradients arise by diffusion?. Dev. Cell 2, 785–796 (2002).
Barrio, L. & Milan, M. Boundary Dpp promotes growth of medial and lateral regions of the Drosophila wing. Elife. https://doi.org/10.7554/eLife.22013 (2017).
Kim, K. A. et al. Mitogenic influence of human R-spondin1 on the intestinal epithelium. Science 309, 1256–1259 (2005).
Hickey, J. W. et al. Organization of the human intestine at single-cell resolution. Nature 619, 572–584 (2023).
Wang, Y. et al. Single-cell transcriptome analysis reveals differential nutrient absorption functions in human intestine. J Exp Med. https://doi.org/10.1084/jem.20191130 (2020).
Cheng, H. & Leblond, C. P. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. I. Columnar cell. Am. J. Anat. 141, 461–479 (1974).
Altmann, G. G. & Leblond, C. P. Factors influencing villus size in the small intestine of adult rats as revealed by transposition of intestinal segments. Am. J. Anat. 127, 15–36 (1970).
Dong, J. et al. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell 130, 1120–1133 (2007).
Reddi, A. H. Role of morphogenetic proteins in skeletal tissue engineering and regeneration. Nat. Biotechnol. 16, 247–252 (1998).
Madamanchi, A., Mullins, M. C. & Umulis, D. M. Diversity and robustness of bone morphogenetic protein pattern formation. Development. https://doi.org/10.1242/dev.192344 (2021).
Plikus, M. V., Widelitz, R. B., Maxson, R. & Chuong, C. M. Analyses of regenerative wave patterns in adult hair follicle populations reveal macro-environmental regulation of stem cell activity. Int. J. Dev. Biol. 53, 857–868 (2009).
Vachon, P. H. Integrin signaling, cell survival, and anoikis: distinctions, differences, and differentiation. J. Signal Transduct. 2011, 738137 (2011).
Liberali, P. & Schier, A. F. The evolution of developmental biology through conceptual and technological revolutions. Cell. https://doi.org/10.1016/j.cell.2024.05.053 (2024).
Briscoe, J. & Small, S. Morphogen rules: design principles of gradient-mediated embryo patterning. Development 142, 3996–4009 (2015).
McCarthy, N. et al. Distinct mesenchymal cell populations generate the essential intestinal BMP signaling gradient. Cell Stem Cell 26, 391–402.e395 (2020).
Yao, X., Shen, H., Peng, Q. & Yu, J. TP53/miR-129/MDM2/4/TP53 feedback loop modulates cell proliferation and apoptosis in retinoblastoma. Cell Cycle 20, 603–615 (2021).
Shields, A. et al. Toll-9 interacts with Toll-1 to mediate a feedback loop during apoptosis-induced proliferation in Drosophila. Cell Rep. 39, 110817 (2022).
Shyer, A. E., Huycke, T. R., Lee, C., Mahadevan, L. & Tabin, C. J. Bending gradients: how the intestinal stem cell gets its home. Cell 161, 569–580 (2015).
Huycke, T. R. et al. Patterning and folding of intestinal villi by active mesenchymal dewetting. Cell 187, 3072–3089 e3020 (2024).
Zhu, G., Hu, J. & Xi, R. The cellular niche for intestinal stem cells: a team effort. Cell Regen. 10, 1 (2021).
Mishina, Y., Hanks, M. C., Miura, S., Tallquist, M. D. & Behringer, R. R. Generation of Bmpr/Alk3 conditional knockout mice. Genesis 32, 69–72 (2002).
Ireland, H. et al. Inducible cre-mediated control of gene expression in the murine gastrointestinal tract: effect of loss of β-catenin. Gastroenterology 126, 1236–1246 (2004).
Yang, X., Li, C., Herrera, P. L. & Deng, C. X. Generation of Smad4/Dpc4 conditional knockout mice. Genesis 32, 80–81 (2002).
Wang, J. et al. Endothelial Wnts control mammary epithelial patterning via fibroblast signaling. Cell Rep. 34, 108897 (2021).
Yang, H. et al. Generation of genetically modified mice by oocyte injection of androgenetic haploid embryonic stem cells. Cell 149, 605–617 (2012).
Zhong, C. et al. CRISPR-Cas9-mediated genetic screening in mice with haploid embryonic stem cells carrying a guide RNA library. Cell Stem Cell 17, 221–232 (2015).
Zhao, B. et al. The non-muscle-myosin-II heavy chain Myh9 mediates colitis-induced epithelium injury by restricting Lgr5+ stem cells. Nat. Commun. 6, 7166 (2015).
Liu, Y. et al. Segregation of the stemness program from the proliferation program in intestinal stem cells. Stem Cell Rep. 18, 1196–1210 (2023).
Fei, T. et al. Genome-wide mapping of SMAD target genes reveals the role of BMP signaling in embryonic stem cell fate determination. Genome Res. 20, 36–44 (2010).
Wang, S. et al. Non-muscle myosin heavy chain 9 maintains intestinal homeostasis by preventing epithelium necroptosis and colitis adenoma formation. Stem Cell Rep. 16, 1290–1301 (2021).
Chou, C. S. et al. Spatial dynamics of multistage cell lineages in tissue stratification. Biophys. J. 99, 3145–3154 (2010).
Zhang, L., Lander, A. D. & Nie, Q. A reaction–diffusion mechanism influences cell lineage progression as a basis for formation, regeneration, and stability of intestinal crypts. BMC Syst. Biol. 6, 93 (2012).
Acknowledgements
We thank Drs. Yuji Mishina, Sylvie Robine, Xiao Yang, Xiaodong Wang, Jianwei Wang, Yi Zeng for mice, and Dr. Haiyong Zhao for the 4xBRE-tdTomato construct. We also thank Dr. Naihe Jing and Huidong Liu for their insightful comments. This work was supported by grants from the National Natural Science Foundation of China (32588201, 32200637, and 92354306), the National Key Research and Development Program of China (2024YFA1307401 and 2023YFA1800603), and the Natural Science Foundation of Jiangxi Province (20224ACB209001), the Shenzhen Medical Research Fund (B2302022), and the Beijing Science and Technology Plan (Z231100007223006).
Author information
Authors and Affiliations
Contributions
Conceptualization: X.W., S.L., Y.L., and Y.-G.C. Methodology: X.W., S.L., Y.L., X.K., L.Z., and Y.-G.C. Software: X.W., X.K., and L.Z. Investigation: X.W., S.L., Y.L., X.K., J.C., P.Y., L.Z., Z.Z., M.H., L.L., M.Z., Y.W., W.J., J.L., L.Z., and Y.-G.C. Formal analysis: X.W., S.L., Y.L., X.K., J.C., P.Y., L.Z., Z.Z., M.H., L.L., M.Z., Y.W., L.Z., and Y.-G.C. Writing—original draft: X.W. and Y.-G.C. Writing—review and editing: X.W., S.L., Y.L., X.K., L.Z., and Y.-G.C. Supervision: Y.-G.C. Project administration: Y.-G.C. Funding acquisition: X.W., Y.L., and Y.-G.C.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Linheng Li and the other anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wang, X., Li, S., Liu, Y. et al. Positional BMP signaling orchestrates villus length in the small intestine. Nat Commun 16, 5461 (2025). https://doi.org/10.1038/s41467-025-60643-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-60643-2