Abstract
Prions are infectious agents that initiate transmissible spongiform encephalopathies, causing devastating neuronal destruction in Creutzfeldt-Jakob and Kuru disease. Rapid cell death depends on presence of the endogenous prion protein PrPC, but its mechanistic contribution to pathogenesis is unclear. Here we investigate the molecular role of PrPC, reactive oxygen species and lipid metabolism in ferroptosis susceptibility, a regulated cell death process characterized by lipid peroxidation. We discover that elevated expression of the cellular prion PrPC creates a relaxed oxidative milieu that favors accumulation of unsaturated long-chain phospholipids responsible for ferroptotic death. This condition is sustained by the luminal protein glutathione peroxidase 8, which detoxifies reactive species produced by protein misfolding. Consequently, both PrPC and infectious Creutzfeldt-Jakob disease (CJD) prions trigger ferroptotic markers and sensitization. This lethality is further enhanced by RAC3, a small GTPase. Depletion of RAC3 is observed solely in pathologically afflicted cortices in CJD patients, revealing a synergistic modulation of lipids and reactive species that drives ferroptosis susceptibility. Together, the results show that PrPC initially suppresses oxidative stress, attenuates cellular defenses, and establishes a systemic vulnerability to the ferroptotic cascade. These results provide insight into the mechanism underlying regulation of ferroptosis in prion diseases and highlight potential therapeutic targets for diseases involving dysregulated cell death processes.
Similar content being viewed by others
Introduction
Prion diseases such as Creutzfeldt-Jakob disease (CJD) in humans and scrapie in sheep are characterized by the conversion of normal cellular prion protein PrPC into a misfolded, beta-sheet-rich isoform (PrPSc). PrPSc is thought to promote conversion of PrPC into the misfolded pathological form that then propagates in the brain and aggregates into the major component of scrapie-associated fibrils1. Although the precise mechanism by which prions kill neurons has not been established, it is thought to involve the abnormal accumulation of aggregated PrPSc2,3,4. The misfolded prion protein can disrupt cellular function, damage cellular membranes, and trigger intracellular signaling pathways that ultimately result in neuronal death5,6. Crucially, neurotoxicity requires expression of the native endogenous prion protein, demonstrating its key role in the process6. Yet, pinpointing the cause of this toxicity has been difficult, resulting in a complex cell death pattern that has so far remained elusive in mechanistic investigations.
Mesenchymalization is a transitional process that leads to increased sensitivity to ferroptosis7, a form of regulated cell death that involves iron-dependent lipid peroxidation. A central player in ferroptosis, glutathione peroxidase 4 (GPX4), resists this process via glutathione-mediated detoxification of lipid peroxides. Mesenchymalization elevates proteins involved in lipid metabolism and limits antioxidant defense proteins, which can increase susceptibility. These changes can increase vulnerability to ferroptosis-inducing agents, such as erastin and RSL3. Notably, expression of the prion protein is intricately linked to increased mesenchymal markers and metastatic susceptibility in patients8,9. Metastatic transformation is associated with increased membrane fluidity, typically achieved via enrichment of membrane polyunsaturated fatty acid (PUFA)-containing phospholipids. The peroxidation of these lipids, which serve as the primary substrate for ferroptotic oxidative stress (reactive oxygen species, ROS), is a key biochemical process directly linked to prion toxicity10.
In this study, we investigated the consequences of elevated expression of the major prion protein (PrPC) to uncover a molecular basis for cell death in prion-induced diseases. Although cells with elevated PrPC were lost with extended culture, short-term conditional expression limited cellular oxidative stress via a closely related glutathione peroxidase in the endoplasmic reticulum, GPX8. This decline of ROS led to a new composition of membrane lipids in which ferroptosis-susceptible PUFA-glycerophospholipids comprised the major component. As a result, PrPC-expressing cells are significantly more sensitive to ferroptotic ROS. A bona fide CJD model showed that the pathological prion PrPSc sensitizes to ferroptosis in organoids, while PRNP-knockout increased resistance. We moreover identified the small-GTPase RAC3 as a key contributor to prion-induced ferroptosis susceptibility due to its impact on membrane ROS, PUFA levels and drive toward mesenchymalization. Cerebral cortex and synaptic junctions from CJD patient brains show a dramatic loss of RAC3 positivity, indicating that the accumulation of PrPSc and RAC3 together provokes cellular loss. Together, these results uncover a mechanistic basis for ferroptotic cell death susceptibility in prion-related diseases.
Results
PrPC inhibits endoplasmic ROS
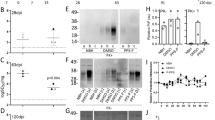
Earlier reports indicated the prion protein can act as a surface ferrireductase to enable iron uptake and increase oxidative stress11,12,13,14,15, which could potentiate ferroptosis. We sought to substantiate these observations in ferroptosis-sensitive HT-1080 fibrosarcoma cells ectopically expressing full-length PrPC (PrPC OE). We measured intracellular labile iron using calcein acetoxymethyl but observed no changes relative to vector-containing controls, whereas ferric ammonium citrate or deferoxamine treatment decreased and increased fluorescence signal, respectively. Ferritin heavy and light chain protein levels and FerroOrange were also unchanged in PrPC OE, suggesting that iron balance is unaffected by elevating PrPC (Fig. 1A, Supplementary Fig. 1A). An evolutionary and informatics approach also failed to identify a basis for ferrireductase homology in the major prion protein (Supplementary Fig. 1B, C). Finally, we specifically measured iron concentration using a nondestructive approach with Mössbauer spectroscopy. Mössbauer spectra for control and PrPC OE samples have same Doublet1 with equal Mössbauer parameters (Supplementary File 1), which correspond best to the Fe3+ in ferritin (and/or hemosiderin) and have no significant difference in relative transmissions. Thus, iron is mainly found as Fe³⁺, and its content was not changed in PrPC OE samples compared to control. The same Fe³⁺ content in both samples confirms that any significant conversion of Fe³⁺ to Fe²⁺ was not observed in PrPC OE cells.
A Cellular free iron content in empty vector control (control OE) and PrPC OE HT-1080 cells measured by 5 nM calcein or 1 μM FerroOrange staining. Wild-type HT-1080 treated with 500 μM ferric ammonium citrate (FAC) and 100 μM deferoxamine (DFO) compared to DMSO as the positive and negative control, respectively, a typical FACS histogram is depicted. Insets show ferritin heavy chain (FTH, 21kD), ferritin light chain (FTL, 20kD) and PrPC levels by Western blot with normalization by beta-actin (ACTB, 42kD). B Relative PRNP mRNA expression levels change up to 21 days post-infection with a lentiviral expression vector-containing PrPC (OE) (encoded by PRNP) and a separate cistron for puromycin resistance, in 1 μg/mL puromycin-containing media. Total RNA was normalized to empty vector control (C). C Cytosolic ROS levels detected by 2,7-Dichlorodihydrofluorescein diacetate (DCFH-DA) using flow cytometry in indicated HT-1080 cells at 7 dpi (days post-infection). A representative flow cytometry histogram of three independent repetitions is depicted (left panel). Boxplot shows (right panel) fluorescence intensity in control and PrPC OE cells after 0.1 μM RSL3 for 0 h and 2 h, with mean, max and min indicated. D Pearson correlation analysis of GPX8 mRNA expression with PRNP mRNA expression (R = 0.621; two-tailed P < 0.0001; 95% confidence interval 0.587 to 0.651) of 18,575 genes determined for 1393 individual cell lines. GAPDH is included as a reference gene. TPM, transcripts per million. E Subcellular colocalization of PrPC together with the endoplasmic reticulum-specific marker concanavalin A (10 μg/mL). Pictures shown are representative results of at least three independent repetitions performed with similar outcomes (scale bar = 20 µm). F Kinetic analysis of ER ROS using Hyper-3 ER fluorescent reporter (emission ratio 488/405) upon PrPC expression (PRNPtet +dox). Samples were measured by flow cytometry for 1 min to determine baseline intensity before supplementation with DTT (∆ 5 mM) or H2O2 (▲ 50 mM). -dox/+dox are samples treated with vehicle or doxycycline, respectively. DTT, dithiothreitol. G Glutathione peroxidase (GPX) activity in PrPC OE compared to empty vector control showing in bar graph. Insets show PrPC and GPX8 levels by Western in PrPC OE and control cells. H Fluorescence intensity of Hyper-3 ER in HT-1080 cells expressing GPX8 compared to empty vector control cells. Average fluorescence intensity values (emission ratio 488/405) were determined by flow cytometry. Insets show GPX8 level by Western. A, C FACS histograms of at least three independent experiments are depicted. Boxplot (C) is shown with whiskers min to max. Relative mRNA expression (B) is shown as mean ± SEM of n = 3 technical replicates. Boxplot data (C) is plotted as whiskers min to max, showing all points of n = 6 each replicates of duplicate repetitions of the experiment with similar results. Significance was determined by two-tailed t-test (C). Activity data (G) is plotted as representative mean ± SEM of n = 4 biological replicates for independent experiments. Intensity (H) was calculated versus control, shown as mean ± SEM of n = 3 biological replicates. P-values of two-tailed t-test (B, G, H) or ANOVA multiple comparisons with Tukey post-test are shown for comparisons. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Source data are provided as a Source Data file.
We observed that cells constitutively expressing PrPC lose expression over time, despite selection with a dicistronic puromycin resistance gene, while PRNP (the gene encoding PrPc) knockout cells grow faster (Fig. 1B, Supplementary Fig. 1D). The prion protein has been implicated in cellular stress processes16, thus we suspected PrPC OE cells were lost due to increased oxidative stress. Surprisingly, testing dihydrodichlorofluorescein diacetate (DCFH-DA) revealed instead a significant decrease in cytosolic reactive species that rose to the same levels as control cells upon short treatment with the ferroptosis inducer 1S,3R-RSL3 (Fig. 1C), consistent with earlier reports showing that the prion protein protects against oxidative stress17,18. We speculated that co-regulated proteins could mitigate ROS in response to PrPc expression. A linear regression analysis of 18,575 genes co-expressed with PRNP unexpectedly revealed GPX8, a paralog of GPX4, to have the highest degree of correlation of all genes (Pearson = 0.621, P (two-tailed) <0.0001; 1393 lines, Cancer Cell Line Encyclopedia; Fig. 1D). As an endoplasmic reticulum resident protein, GPX8 detoxifies H2O2 generated during disulfide isomerase mediated protein (re-)folding19,20,21,22 and is tightly associated with mesenchymalization processes23. Analysis of the top 100 genes associated with PRNP expression by Pearson correlation additionally revealed genes involved in epithelial-mesenchymal transition as the highest associated category (Supplementary Fig. 1E).
We observed primary localization to the ER in cells overexpressing PrPC (Fig. 1E), suggesting the ROS environment in the ER could be affected24. The ER redox state is intricately connected to the maintenance of protein-folding equilibrium25. To avoid the aforementioned long-term culture effects, we generated an HT-1080 cell line conditional for PrPC expression (PRNPtet; Supplementary Fig. 1F) and tested whether PrPC influenced luminal H2O2 levels using an endoplasmic-specific H2O2 sensor, Hyper3-ER26. After 3 days of doxycycline treatment, we observed a striking loss in basal fluorescence signal and in H2O2-supplemented PRNPtet cells, whereas dithiothreitol uniformly led to lower levels in both treated and untreated cells (Fig. 1F). Due to its association with PRNP, we tested next if ectopic PrPC also triggered GPX8 accumulation and observed a corresponding increase in protein and glutathione peroxidase activity, measured by increased reduced glutathione consumption (Fig. 1G). Increased GPX8 expression was additionally found to be sufficient to inhibit luminal H2O2 (Fig. 1H) and Hyper-ER fluorescence, while knockdown of GPX8 increased luminal H2O2 (Supplementary Fig. 2A). We next explored if GPX8 directly affects PrPC. Using PrPC cell surface staining, we observed that GPX8 co-transfection strongly depletes surface PrPC (Supplementary Fig. 2B). Thus, we investigated if total PrPC protein was decreased and saw that GPX8 potently diminishes cellular PrPC signal. This effect was reversed with the addition of the proteosome inhibitor MG132, indicating that GPX8 steers PrPC proteosomal degradation. Taken together, these results indicate that PrPC acts to depress cellular and specifically ER H2O2, and this redox balance is controlled by GPX8, which facilitates PrPC degradation.
PrPC expression induces a ferroptosis-sensitive phenotype
To investigate loss of PrPC-overexpressing cells over time, we tested cell death inhibitors against apoptosis, necroptosis and ferroptosis using resazurin as a viability indicator. We infected cells with PrPC-containing lentivirus for 3 days and selected for puromycin resistance for 24 h, counted equal numbers of cells, and treated with inhibitors for 3 days. Only Ferrostatin-1 (Fer-1) and alpha-tocopherol (aToc) significantly protected cell viability, suggesting that ferroptosis susceptibility in PrPC OE cells drives their continual loss (Fig. 2A). Quantitative PCR also showed significantly higher PRNP expression in Fer-1 and aToc-treated cultures, while a live/dead Calcein-AM/PI stain confirmed cell loss due to lack of viability (Supplementary Fig. 2C, D). Thus, we reasoned that diminished oxidative stress in PrPC OE cells may relax antioxidative defenses, rendering them more susceptible to strong oxidative bursts like ferroptosis.
A Cell survival of HT-1080 PrPC OE (OE) and empty vector control (C) cells treated with cell death inhibitors. Cells were infected for 3 days, selected with puromycin for 24 h, then cultured with inhibitors 10 μM α-tocopherol (aToc) or 2 µM ferrostatin-1 as a ferroptosis inhibitor, 10 μM z-VAD-FMK (zVAD) as an apoptosis inhibitor, and 10 μM necrostatin-1 (Nec-1) as a necroptosis inhibitor, respectively, for 3 days. B (Left) Dose-response curves illustrating sensitivity of PrPC-expressing HT-1080 cells (PrPC OE) compared to empty vector control cells (control OE) to IKE treatment over 16 h. (Right) Survival of PrPC OE cells compared to control against ferroptosis inducer 0.3 μM IKE (16 h). (Lower panels) Sensitivity of GPX8 knockout (KO) cells with (+dox) and without (-dox) PrPC OE. Inset shows GPX8 protein levels by Western and total protein loading compared to knockout control cells. Viability data are plotted as representative mean ± SD of n = 3 technical replicates for independent experiments repeated at least three times with similar outcomes. C PrPC OE effect on lipid peroxidation induced by 0.1 μM RSL3 induction in HT-1080 OE and control cells measured by BODIPY 581/591 C11 (BODIPY-C11). A typical FACS histogram of n = 3 technical replicates of three independent repetitions is depicted. The bar graph shown on the right illustrates the BODIPY-C11 median intensity ± SEM. D Comparative analysis of lipid levels in PrPC OE HT-1080 cells compared to control cells, highlighting fatty acid acyl chain saturation in normalized lipidomics data. Normalization was performed by glucose peak area for each sample and each feature. XCMS data analysis and tentative annotation (of 617 known lipids) were according to the Bruker UPLC-Q-TOF protocol in positive mode, detecting 10,521 total features. Significant lipids have -log10 P > 1.30. Viability data are representative means of n = 9 (A) biological or n = 3 (B) technical replicates for experiments repeated independently at least three times. P-values of two-way ANOVA multiple comparisons with Tukey post-test are shown (A–C). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Lipid statistics (D) were determined by P-values (P < 0.05, n = 5, two-tailed Welch’s t-test). n = 5 PrPC OE and n = 5 control samples. Source data are provided as a Source Data file.
To test this, we treated PrPC OE cells with a specific inhibitor of system xC-, imidazole ketone erastin (IKE), which blocks glutathione uptake (Fig. 2B). Consistent sensitization was observed at all tested concentrations but was rescued by the addition of aToc, demonstrating specificity for ferroptosis. A simultaneous knockout of GPX8 together with PrPC expression in PRNPtet cells revealed increased sensitivity to IKE, supporting the role of GPX8 in maintaining the redox environment inside these cells. Lipid peroxidation is an essential hallmark of ferroptosis. Application of RSL3 for 4 h resulted in a potent shift in lipid peroxidation in PrPC OE cells, as detected by the fluorescent membrane peroxidation sensor Bopidy-C11 (Fig. 2C). A substantial portion of PrPC OE cells die very rapidly from this treatment (Supplementary Fig. 2E) and may only transiently/weakly show Bodipy-positivity. Finally, testing of a panel of anticancer agents moreover showed no discernible nonspecific sensitivity to cell death (Supplementary Fig. 2F). Together, these results demonstrate a specific sensitivity to ferroptosis in PrPC OE cells.
To further investigate why lower levels of basal oxidative stress lead to ferroptosis sensitivity, we conducted a global lipid mass spectrometry analysis of PrPC OE compared to control cells. According to lipids determined by total ion chromatogram, significant fold changes were observed for highly unsaturated long-chain containing glycerophospholipids, the primary substrates of lipid peroxidation during ferroptosis (Fig. 2D). Further accumulation of saturated and monounsaturated phospholipids indicated a high level of membrane plasticity that suggested global lipid remodeling. We found that expression of PRNP correlates with ELOVL1 and FADS3, which respectively confirm the elongation and desaturation of fatty acid precursors (Supplementary Fig. 2G). Interestingly, ATP2A3, which increases calcium concentrations in the ER driving sensitive PUFA-phospholipid formation27, was found to be strongly inversely correlated, suggesting additional PUFA-lipids in PrPC OE are being compensated, to a degree.
Specifically, early ferroptosis lipoxygenase targets of arachidonate and adrenate-containing phospholipids28 PC(O-38:4) and PI(14:1/20:4) were found to be elevated by 1.91 and 1.96 fold, respectively. Long-chain PUFA-PLs containing two diacyl-PUFA chains PC(22:6/22:6, 1.89 fold) and ether lipids can selectively fuel the spread of lipid peroxidation in a ferroptosis-sensitive environment29,30,31,32. Thus, we surmise that elevated PrPC levels initially limit oxidative stress, allowing for a higher abundance of oxidation-sensitive PUFA-containing phospholipids. However, some cells may exceed this ROS threshold, thus accounting for the protective effect of Fer-1 in untreated PrPC cells (Fig. 2A).
Ferroptosis in bona fide CJD model
Ferroptosis is challenging to detect retrospectively, but several contemporaneous markers are available. One highly specific marker of ferroptosis, fatty acid binding protein five (FABP5), a marker used for retrospective ferroptosis detection in humans33, was found significantly upregulated in PrPC OE cells (Fig. 3A–C). Other proposed markers cyclooxygenase-2 (PTSG2), transferrin receptor (TFRC; by RNA but not Western), heme oxygenase 1 (HMOX1) and the lipid degradation product 4-Hydroxynonenal (4-HNE) were found to be significantly elevated in PrPC OE cells (Fig. 3B), signifying the background presence of ferroptosis in cultured cells upon prion overexpression.
A FABP5 changes in PrPC OE and empty vector control cells by immunostaining and Western with densitometry. Pictures shown are representative results of three independent repetitions performed in triplicate with similar outcomes (scale bar = 20 µm). B Relative expression of ferroptosis markers in PrPC OE (OE) and control (C) cells detected by qPCR and Western. Lower blots display total protein loading. C Comparison of FABP5 staining intensity in iPS-derived brain organoids infected with normal brain homogenates (NBH) and sCJD prion homogenates (169 days post-infection, dpi). Pictures shown are representative results of at least three independent repetitions performed with similar outcomes of minimum 10 organoids (scale bar = 200 µm (left panels) and 50 µm (right panels)). D Western blotting and densitometry comparing FABP5 protein levels in the brain tissue of mice inoculated with normal brain homogenate (NBH; control) or RML scrapie brain homogenate and sacrificed at 80, 108, and 160 days post inoculation (dpi). E Western blotting and densitometry comparing FABP5 protein levels in the brain tissue of people who died from sporadic CJD with tissue from patients who died of a non-brain-related condition. F Lactate dehydrogenase (LDH) release level of brain organoids treated with increasing RSL3 concentrations and aToc rescue for ferroptosis specificity. G Normalized survival of brain organoids infected with sCJD (MM1) prion homogenates for 90 days compared to NBH controls against an RSL3 treatment at indicated concentrations (scale bar = 200 µm). Cell death was measured by LDH release activity (48 h, relative to DMSO). H Normalized survival of PRNP KO compared to NBH controls treated with RSL3 and measured by LDH release (48 h, relative to DMSO). Western data (A, B) is shown as mean ± SD of n = 3 technical replicates. Relative mRNA expression (B) is shown as mean ± SEM of n = 3 technical replicates. LDH release data and Western data (E) are plotted as representative mean ± SEM of n = 3 (D, F) or n = 6 (E, G, H) biological replicates for independent experiments. Significance was determined by two-tailed t-test (A, B, F), two-tailed Welch’s t-test (E) and two-way ANOVA comparisons, Tukey post-test (D, G, H). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Source data are provided as a Source Data file.
Next, we sought to examine if ferroptosis susceptibility is exclusive to cells expressing the native PrPC protein or also present in a bona fide infectious prion model. For this, we chose an established iPS-derived organoid method34,35,36, inoculated with homogenates from sporadic Creutzfeldt-Jakob disease (sCJD)-infected individuals or normal brain homogenates (NBH), then cultured for 169 days. The pathogenic sCJD prion is detected in the former homogenates as observed by the RT-QuIC assay37, while HT-1080 PrPC OE are classified as non-infectious/non-seeding even in the presence of RSL3 (Supplementary Fig. 3A, B). In contrast to control organoids, sCJD-treated organoids showed pronounced FABP5 staining that was concentrated in areas of evident cell death as partially indicated by empty or “ghost” cellular structures devoid of organelles (Fig. 3C, Supplementary Fig. 4A). This snapshot indicated that the ferroptosis marker FABP5 is expressed in higher levels in cells from sCJD-treated organoids that may potentially die by ferroptosis. 4-HNE detection revealed a significant increase in lipid peroxide products in sCJD organoids at 56 dpi, but only mildly increased at 169 dpi, possibly due to loss of these unstable products prior to the time of measurement (Supplementary Fig. 4B), while FABP5 staining may persist after cells have died.
Therefore, to further investigate whether FABP5 increases according to the progression of prion disease, we compared mice intracranially mock inoculated with normal brain homogenates (NBH) or RML scrapie prions for protein levels at 80- (mid-incubation period), 108- (early symptom onset), and 160- (terminal disease onset) days post-infection. In isolated brains from these animals at 80 dpi no significant difference was observed, however, at 108 and 160 dpi a striking increase in FABP5 in RML mice was observed (Fig. 3D).
To further correlate the emergence of ferroptosis marker FABP5 in mice to CJD in humans, we examined sCJD brain homogenates and found a significant increase in cumulative FABP5 (Fig. 3E). Thus, the increase of the FABP5 ferroptosis marker is conserved from HT-1080 PrPC OE to RML mice to CJD patients.
We then sought to examine the direct effect of infectious prions on ferroptosis in vitro. We tested iPS-derived organoids by treatment with RSL3, collected medium fractions and measured cell death by lactate dehydrogenase (LDH) release. In this assay, RSL3 treatment was rescued with alpha-tocopherol, demonstrating ferroptosis specificity up to 80 nM (Fig. 3F). We infected organoids with sCJD homogenates, incubated these for 110 days (a time point where the earliest metabolic changes were observed)35 and subsequently exposed them to 40 or 80 nM RSL3 for 48 h. The results showed a remarkable susceptibility compared to control NBH cultures (Fig. 3G). In contrast, genetic ablation of PRNP in iPSC-derived organoids showed a significant resistance to ferroptosis induction by RSL3 (Fig. 3H). To exclude the possibility of CJD-treated organoids being nonspecifically susceptible to lethal stimuli, we treated NBH and CJD samples with staurosporine but observed no changes in sensitivity (Supplementary Fig. 4C). The specific susceptibility to ferroptosis was further supported by increased FABP5 marker deposition in wild-type organoids treated with 40 nM RSL3 compared to PRNP KO organoids, particularly in densely neuronal regions as demarcated by MAP2 expression (Supplementary Fig. 5). Finally, to test if neural cells are directly susceptible to ferroptosis, we expressed human PrPC or knocked out Prnp in glutamate-sensitive neuronal HT22 cells. We observed the same effect as in HT-1080: HT22 PrPC OE significantly sensitizes cells to RSL3, while Prnp KO has no effect on viability (Supplementary Fig. 6A). Together, these results show that both PrPC and infectious prions drive ferroptosis sensitivity.
Synergistic vulnerability with RAC3
In HT-1080 PrPC OE, sCJD-infected organoids and HT22 cells, we observed a fraction of cells that were highly responsive to ferroptosis induction. This suggested that a more complex mechanism or local milieu could influence cellular vulnerability. We therefore chose to identify factors that sensitize to PrPC by conducting a whole-genome CRISPR-activation38 screen in doxycycline-inducible PRNPtet cells. The inducible system enables transduction with the sgRNA library prior to PrPC overexpression and ensures that control cells have the same complement of guides before ferroptosis induction.
We infected cells with the guide library 3 days prior to doxycycline treatment, split the culture, then supplemented media with a low dosage of ferroptosis inducer IKE (500 nM) for 16 h to induce synthetic lethality (Fig. 4A). Next, we isolated surviving cell genomic DNA for PCR amplification and guide sequencing, and compared frequencies in the control population versus PrPC-overexpressing cells, in which depleted guides are expected to correlate to genes promoting PrPC-sensitized ferroptosis.
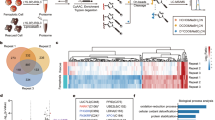
A Schematic of a whole-genome CRISPR-activation depletion screen. HT-1080 conditionally expressing PrPC (PRNPtet) were transduced with a synergistic activation mediator pooled human library and dCas9-VP64 lentiviruses. PrPC expression was induced for 3 days with doxycycline or vehicle, followed by treatment with 500 nM IKE, resulting in a viability loss of 5–10%. Normalized guide frequencies amplified from viable cells in sensitized PrPC cells were scored against (-)doxycycline control cells to identify enriched or depleted guides. B Fishtail plot of prion-facilitated ferroptosis by differential CRISPRa guide representation. Effect size is determined by the average PRNPtet (+)doxycycline / (−)doxycycline ratio. Depleted guides (Effect size <1) are positive regulators of PrPC-facilitated ferroptosis. C Inverse relationship of RAC3 and PRNP in PrPC OE or RAC3 OE, respectively, by qPCR and Western blot. Relative mRNA expression is shown as mean ± SD of n = 3 technical replicates. D Gene effect characterized by principal component analysis. CRISPR gene effect (individual gene knockout effect on viability and growth, scaled for whole-genome libraries) in 1095 cell lines is displayed in two dimensions. The distance between two genes in the scatter plot can be interpreted to have similar or inverse effects on cellular processes across individual cell lines, relative to all other cell lines. Significance was determined by two-tailed t-test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Source data are provided as a Source Data file.
A clear pattern emerged with significantly depleted guides in the surviving PRNPdoxi population corresponding to RAC3 (p = 0.0007, two-tailed t-test) and its partner RHOC (p = 0.010) small GTP-ases39, while RHO-interactors40 EXOC3 (p = 0.0267) and EPHB4 (p = 0.0678) were enriched (Fig. 4B). A highly depleted hit, glycosyltransferase ABO (p = 0.001), was recently implicated in Covid pathogenesis via its glycosylation activity. In contrast, SNX7, a factor proposed to assist autophagosome lipid degradation and the transferrin receptor41, was significantly enriched (p = 0.0015). Given the evidence of Rho-GTPase involvement, we focused on the most depleted gene, RAC3, a member of the RAS family of small GTPases.
Similar to GPX8, high Rho/RAC family expression is strongly associated with cancer mesenchymalization, metastasis, and stemness42,43,44. Ferroptosis sensitivity has been strongly linked to a high mesenchymal state7. Thus, if RAC3 facilitates PrPC-induced ferroptosis, we hypothesized it would be hazardous for cells to simultaneously express both genes. Indeed, we observed a significant decrease in RAC3 RNA and protein in PrPC OE cells, while ectopic expression of RAC3 also diminished PrPC expression (Fig. 4C). In light of this relationship, we investigated their nearest gene neighborhood by CRISPR gene effect analysis (Fig. 4D). Profiling of 17,931 genes and their survival scores in 1095 whole-genome screens by principal component analysis revealed a striking 2D proximity of RAC3 and GPX8, strongly suggesting a shared role in cellular processes and paralleling the observed expression correlation of GPX8 and PRNP (Fig. 1D).
RAC3 expression sensitizes cells to PrPC-induced ferroptosis
Ferroptosis inducers like RLS3 were identified by inducing cell death in KRAS-expressing lines. We tested whether RAC3 increases ferroptosis vulnerability. We tested the inhibitor EHop-016, which inhibits RAC activity and RAC-directed membrane effects via guanine-exchange activity inhibition. Stimulation of ferroptosis with IKE in HT-1080 cells was partially blocked via co-treatment with EHop-016 compared to vehicle, while elevated RAC3 expression, in contrast, sensitized to ferroptosis induction (Fig. 5A, B). We then sought to test whether combined overexpression of PRNP and RAC3 potentiated ferroptosis sensitivity, but were unable to maintain such clones.
A Survival of HT-1080 cells treated with RAC-inhibitor EHop-016 (2 μM) and ferroptosis inducer (IKE) rescued by 10 μM α-tocopherol (aToc). B Viability bar graph of RAC3 expressing cells (RAC3 OE) compared to empty vector control with ferroptosis inducer 1.25 μM IKE and 10 μM αToc as a ferroptosis inhibitor. A Western indicates increased expression of RAC3 protein. C Cell survival of PrPC OE, RAC3 OE, and RAC3+PrPc co-expressing cells was detected in freshly infected cell. Cells were treated subsequently with either 2 μM Ferrostatin-1 or DMSO for 3 days. D A flow cytometry histogram depicts the effect of RAC3 OE on induced ROS level via 0.3 μM RSL3 treatment in HT-1080 cells for 3 h measured by DCFH-DA after the indicated time. E Principal component analysis of all metabolomic and lipidomic mass spectrometry features for RAC3 OE cells and cells treated with IKE, compared to control cells. (PC, principal component). F Bar graph illustrates differential expression of 210 annotated phospholipids and lysophospholipids with varying total numbers of double bonds in RAC3 OE cells compared to control cells with an empty vector. Data are representative means ± SD of n = 5 technical replicates. G Differential expression of 210 annotated phospholipids and lysophospholipids with varying total numbers of double bonds in IKE-treated cells compared to vehicle-treated controls. Data are representative means ± SD of n = 5 technical replicates. H Brightfield images of control, RAC3 OE, TGFbeta-1 treated, and RAC3+PrPc OE co-infected cells indicating various levels of roundness (scale bar = 20 µm). I Aspect ratio depicting the effects of TGFbeta-1 treatment, RAC3 OE cells, and RAC3+PrPc co-expressing HT-1080 cells (mean shape, length/width ratio). J Relative expression of epithelial and mesenchymal markers in RAC3 OE and TGFbeta-1 treated cells (3 days) compared to empty vector or DMSO-treated control detected by qPCR. Relative mRNA expression is shown as mean ± SEM of n = 3 technical replicates. Viability data (A–C) are representative means ± SEM of n = 3 biological replicates for experiments repeated independently at least three times. The aspect ratio (I) are representative means of n = 12 replicates measured by high content microscopy. Significance was determined by two-way ANOVA multiple comparisons with Tukey post-test (A–C, I) or two-tailed t-test (J). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Source data are provided as a Source Data file.
We therefore compared HT-1080 cells freshly infected with either a PrPC or RAC3-containing construct and an empty overexpression vector to doubly infected PrPC/RAC3 cells (Fig. 5C). In all instances, treatment with Ferrostatin-1 significantly rescued viability. We also tested in parental cells whether RAC3 knockdown abrogated cell death and observed a significant increase in survival (Supplementary Fig. 6B).
RAC GTPases can activate NADPH oxidases to produce ROS45. We tested cytosolic ROS levels in RAC3 OE with DCFH-DA. While unstimulated cells show no difference from parental controls, stimulation with RSL3 for 3 h revealed a sharp increase in oxidized signal marker (40.8% vs 22.2.%, Fig. 5D), demonstrating potent ROS induction. This revealed a contrasting phenotype to PrPC OE cells, which suppress cytosolic ROS prior to RSL3 treatment (Fig. 1C). A comprehensive metabolite analysis revealed a clear separation of RAC3 OE and control cells by principal component analysis (Fig. 5E), indicating that a biochemical restructuring has already occurred following increased expression. Interestingly, RAC3 OE cells were separated on the second component axis to a degree comparable to IKE treatment of control cells, suggesting they affect many metabolites similarly. However, treatment of control cells with IKE increased PC 1 differences, while treatment of RAC3 OE only triggered minimal changes. These differences may be due to the long (14 day) duration of RAC3 overexpression vs short (16 h) IKE treatment. These results suggest that RAC3 OE induces a partial metabolic signature, similar to IKE-treated cells. If this hypothesis were valid, metabolites, such as lipids, would be similarly affected in susceptible cells.
To test this hypothesis, we analyzed total ion chromatograms of 210 annotated PUFA and ferroptosis-resistant MUFA-containing phospholipids. We observed a correspondent distribution in RAC3 OE and IKE-treated cells, suggesting that RAC3-functions similarly to deplete ferroptosis targets of PUFA-glyceryophospholipids (Fig. 5F, G). If these lipids were degraded, replenishment would likely require de novo lipid synthesis or scavenging. We observed correspondent expression of RAC3 and fatty acid synthase (FASN) by linear regression analysis (Pearson R = 0.465), suggesting cellular restocking of lipids occurs partially via de novo lipogenesis (Supplementary Fig. 6C).
One notable feature of HT-1080 RAC3 OE cells is a clear loss of adhesion along with cellular rounding, a phenotype typically associated with mesenchymalization (Fig. 5H). We quantified this rounding using length-versus-width parameters and compared it to treatment with soluble TGFbeta-1 protein, which induces epithelial-to-mesenchymal transition. While RAC3 OE resulted in roundness comparable to TGFbeta-1, co-expression with PRNP potentiated this phenotype significantly (Fig. 5H, I). Further confirmation of partial mesenchymalization was observed in RAC3 OE by increased transcription of ZEB1, N-cadherin, Vimentin, and Snail1, while E-cadherin, a marker for epithelium, was decreased. The observed effect was as potent or stronger than the addition of TGFbeta-1 (Fig. 5J, Supplementary Fig. 6D, E). PRNP expression is moreover tightly linked to the TGF-induced gene TGF1I1 expression as well as Hippo pathway members responsible for communicating TGF signals (Supplementary Fig. 7A). TGF and associated metastatic processes are strongly linked to poor survival; thus, it is also critically noted that high PRNP levels also predict poor survival in Kaplan–Meier analyses of colon and liver carcinomas as well as glioblastoma (Supplementary Fig. 7B). Taken together, RAC3 and PrPC synergize to induce a mesenchymal phenotype which ultimately sensitizes cells to ferroptosis.
Loss of RAC3 expression in CJD pathology
Since both PrPC and pathogenic prion can sensitize to ferroptosis, we reasoned that rapidly progressing neurodegeneration and mortality (mean: 4–5 months) in most sporadic CJD patients may produce similar evidence46. If this were the case, RAC3 would be expected to decline with the severity of the disease. We analyzed RAC3 protein in infected RML mouse brain homogenates and observed a distinctive and significant loss of RAC3 protein in these samples over time (Fig. 6A). This loss of RAC3 was mirrored in end-stage CJD brains in humans, indicating conservation of this mechanism (Fig. 6B), together with a slight but concurrent increase in GPX8 protein (Supplementary Fig. 8A). We thus investigated the spatial localization of RAC3 in CJD brains. Immunohistochemistry with antibodies against RAC3 revealed widespread loss of signal in the frontal neocortex of sCJD autopsy cases compared to control autopsy cases and in sCJD-treated organoids (Fig. 6C, Supplementary Fig. 8B, C). In the frontal cortex of control cases RAC3 is expressed in the neuropil predominantly showing a fine punctate distribution pattern which suggests a mainly synapse-associated localization, while cell bodies do not contain RAC3 (Fig. 6C). In all analyzed sCJD cases (Table 1) RAC3 expression is dramatically reduced: the synaptic pattern is only faintly recognizable indicating not only a decreased synaptic RAC3 expression but also a loss of synaptic structures.
A Western blotting and densitometry comparing RAC3 protein levels in the brain tissue of mice inoculated with normal brain homogenate (NBH; control) or RML scrapie brain homogenate and sacrificed at 80, 108, and 160 days post inoculation (dpi). Total protein is shown in the lower blot. Data are representative means ± SEM of n = 3 (NBH) or n = 4 (RML) biological replicates. B Western blotting and densitometry comparing RAC3 protein levels in the brain tissue of people who died from sporadic CJD with tissue from patients who died of a non-brain-related condition. Total protein is shown in the lower blot. Data are representative means ± SEM of n = 3 (NBH) or n = 6 (sCJD) biological replicates. C (Left panels) Immunohistochemical detection of prion protein deposits in the cortex of the superior frontal gyrus of Creutzfeldt-Jakob disease (CJD) case (left column) and control case (upper panels, scale bar = 20 µm). A higher magnification shows a punctate synaptic distribution pattern of prion protein deposits typical for both sporadic MM/MV1 CJD cases, whereas no deposits are detectable in the control case (lower panels; scale bar = 200 µm). (Right panels) Immunohistochemical detection of RAC3 in the superior frontal gyrus in a control case (left column) and a case with Creutzfeldt-Jakob disease (CJD, right column). RAC3 is strongly expressed in the neocortical neuropil of the control case (upper left; scale bar = 2 mm), showing a punctate synaptic distribution pattern at higher magnification without expression in cell bodies (lower left; scale bar = 20 µm). In the CJD case, total immunohistochemical signal for RAC3 is strongly reduced in the overview image (upper right; scale bar = 2 mm) compared to the control case. The synaptic distribution pattern is diffuse and unrecognizable at higher magnification (lower right; scale bar = 20 µm). The baseline characteristics of the 6 enrolled patients (3 neuropathological cases and 3 controls) are listed in Table 1. D A model of prion-induced ferroptosis sensitivity. PrPC expression lowers ER ROS levels via increased GPX8 activity, leading to a relaxed oxidative environment that favors the accumulation of ferroptosis-sensitive PUFA-PL. Oxidative stress facilitated by RAC3 simultaneously depletes these lipids and triggers mesenchymalization, sensitizing to ferroptosis. PrPSc is speculated to contribute to ferroptosis by increased lipid peroxidation, consistent with observations in infected mice, or by cellular sensing of misfolded PrP, resulting in GPX8 upregulation and sensitivity in the presence of RAC3. Significance was determined two-tailed t-test (A, B), P-values = 0.0135 (A) and 0.0003 (B), respectively. Source data are provided as a Source Data file.
Discussion
Prion infection was previously shown to increase lipid peroxidation and sensitize to oxidative stress47, but viability loss has been underinvestigated due to the absence of other characterized cell death mechanisms at the time of discovery. Here, we revisited the mechanism of prion-induced cell death sensitivity in the context of ferroptosis-sensitive cells and infectious prion organoids to uncover key hallmarks of ferroptosis in the form of elevated biomarkers, sensitizing lipids and peroxidation, a ROS imbalance, and a strong linkage to mesenchymalization processes. A disparity in the interpretation of the role of ROS in prion pathology has been noted in several studies48,49. Importantly, these discrepancies likely result from different cellular systems and treatment durations, as ferroptosis-sensitized cells will eventually disappear from culture and therefore are not subjected to further investigation, while cells that develop resistance to ferroptosis are likely to be maintained and studied. For this reason, we focused on acutely expressing prion cultures, specifically those using an inducible system.
We found that ectopic expression of the native prion PrPC drives expression of the ER-resident protein GPX8, whose primary role is to detoxify H2O2 resulting from protein (re)folding. This increase in glutathione peroxidase activity results in a low-ROS state that permits accumulation of ferroptosis-sensitive lipids such as the arachidonic acid-containing glycerophospholipid, rendering cells sensitive to a strong oxidative “pulse” and lipid peroxidation. Although in the first instance it is counterintuitive that cells with lower ROS burden are more susceptible to ferroptosis, this concept is supported by the protection of PrPC-OE cells with long-term treatment of Fer-1. It is expected that cells experience a transient increase in ROS during cell metabolism or division, and that cells with lower ROS levels readily succumb to these stresses via ignition of the lipid peroxidation cascade and are therefore protected by Fer-1. Moreover, cells with higher tonic ROS levels must have established an antioxidant system or a more saturated lipid repertoire to cope with these higher levels.
We observed that GPX8 facilitates PrPC degradation. One interpretation of this might be that the favorable oxidation conditions in the ER allow for efficient proteosome targeting of PrPC. This would be consistent with the body of literature showing PrPC conformational changes that block degradation50,51. Notably, GPX8 expression is tightly linked to mesenchymal processes. This may be due to the fact that highly motile cells require flexible membranes comprised of PUFA-containing phospholipids. These lipids are, in turn, sensitive to membrane ROS, which thus requires protection via the glutathione peroxidase system. This would account for the PrPC/GPX8 mesenchymalization axis and a dampened oxidative state in the ER, likely leading to an overall lowering of antioxidant systems in the cytosol, which render the membrane sensitive to strong oxidative pulses delivered by ferroptosis inducers, or by exposure to infectious prion particles (Fig. 6D)52.
Native PrPC protein is an absolute requirement for prion propagation and disease progression, as shown by the absence of degeneration in Prnp-ablated species53. However, the role of PrPC in neuronal toxicity is much less defined. PrPC levels have been shown to decrease over the course of prion disease54. Like RAC3, this may be explained by the selective loss of cells or structures containing high levels of PrPC. Alternatively, if the PrPC levels are dropping due to consumption as the substrate for conversion into prions and deposition of these within the brain, disease subtypes or strains that convert and deposit fastest may demonstrate slower pathogenic decline, due to less functional PrPC remaining to prime cells for ferroptosis.
Cells displaying a mesenchymal phenotype are exquisitely sensitive to ferroptotic ROS7. A high representation of Hippo genes in the YAP1/TAZ pathway was also associated with high PrPC levels, consistent with a role in mesenchymalization and cancer stem cells found in patient cohorts8 (Supplementary Fig. 6B). Focal adhesion genes strongly dovetail with PRNP expression, further underscoring a change in cellular phenotype. Under nonpathological conditions, this phenotype may persist indefinitely. However, insult due to intrinsic phenomena such as inflammation or extrinsic oxidative stress may be sufficient to cause a chain reaction, ultimately resulting in ferroptosis in these sensitized cells. Interestingly, expression of vimentin is highly associated with dendritic damage in Alzheimer disease55, suggesting that mesenchymalization may be a common misfolding- or damage-induced response in both Alzheimer and CJD neurons.
RAC is reported to drive epithelial-to-mesenchymal transition in a ROS-induced feedback loop via activation of the NADPH oxidase (NOX) complex56. NOXes are responsible for the production of hydrogen peroxide and ROS at the membrane where ferroptosis is initiated57,58. Particularly interesting are the differences in cytosolic ROS measured by DCFH-DA in PrPC and RAC3 overexpressing cells. While PrPC OE appears to depress cytosolic ROS levels in response to RSL3, RAC3 reverses this phenotype. Hence, the favorable environment for PUFA-phospholipids becomes toxic when RAC3 is present. In this respect, it is striking that RAC3 expression is depleted in sCJD-infected organoids, RML mice, and CJD patient brains. However, both the organoids and the patients may be classified as “end-stage,” after the majority of cell death has taken place. This would imply that cells with higher levels of RAC3 have already succumbed to prion-induced cell death, and that remaining cells express lower RAC3. Interestingly, RAC3 has been found to protect against ferroptosis in ES cells59; thus, the context of RAC3 and particularly the proportion in the active GTP-bound state or inactive GDP-bound are likely critical in determining its function and localization. Synaptic loss is another key feature of CJD. Thus, it is tempting to speculate that punctate RAC3 immunopositivity is directly involved in synaptic degradation linked to cell death.
Early studies of neuronal death in prion diseases focused on apoptosis as the prominent mechanism60; however, other methods have since been implicated5. These include necroptosis61 and autophagy62. Although both autophagy and necroptosis pathways exhibit some cross-talk with mesenchymalization and ferroptosis63,64,65, the latter has not been investigated as a mechanism of prion-induced cell death. In addition to our current data, previous studies also indicate the involvement of ferroptosis. For example, the early increase in lipid peroxidation seen in mice accompanying misfolded prion accumulation10 and analysis of lipid profiles in terminal CJD brain tissue showing a decrease in fatty acid oxidizability66 support a shift toward neuronal vulnerability to ferroptosis during disease. Even earlier investigations hinted at the possibility of mesenchymal transition, with transgenic PRNP OE mice demonstrating heightened susceptibility to oxidative stress insults and increased glutathione activity67. The mechanism by which PrPC influences mesenchymalization is not fully understood; however, its expression is increased during EMT more than tenfold to enhance glycosylation of the NCAM1 adhesion molecule68. Furthermore, PrPC has recently been linked with a mesenchymal phenotype and poor prognosis in colorectal cancer69. How PrPSc specifically contributes to ferroptosis is difficult to decipher, but possibilities include direct effects on lipid peroxidation47, retrograde transport70 and cellular detection of misfolding that contributes to GPX8 activity increase71,72, or an early compensatory increase in PrPC73 that drives these processes. Certainly, the presence of RAC3 is detrimental, yet it remains an open question whether these processes are directly linked or if the cumulative effect on lipid peroxidation is sufficient to drive lethality.
In conclusion, this study provides insights into the complex mechanisms underlying prion-induced cell death sensitivity. We show that the native PrPC is responsible for maintaining a low-ROS state through GPX8 activity. The state is favorable for the accumulation of PUFA-phospholipids; however, the state is highly susceptible to ferroptotic ROS, driving sensitivity. Additionally, the results highlight the impact of RAC3, which promotes ROS and disrupts the favorable cellular environment for PUFA-phospholipids in PrPC OE cells. The loss of RAC3-positive structures in RML mice and Creutzfeldt-Jakob patients strongly supports its involvement in prion toxicity. Understanding these processes contributes to our fundamental understanding of prion biology to help guide future therapeutic approaches.
Methods
Ethics approval
This retrospective study involving human participants was in accordance with the ethical standards of the institutional and national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. Data analyses were performed on paraffin-embedded tissue samples from archived material at the Edinger Institute. Use of CJD human pathological material was endorsed by the local ethical committee (Goethe-University Medical School/UCT Frankfurt, Ref. no. 19–431). Postmortem analyses on the material were conducted with informed consent from families. Organoids were approved by: Prion protein function in redox homeostasis and associated failure in prion disease, 17-NIAID-00212, 8/3/2017. The human-induced pluripotent stem cells and brain homogenates used in this study were de-identified before being provided to the researchers at the NIH. Thus, the NIH Office of Human Subjects Research Protections (OHSRP) has determined these samples to be exempt from IRB review as per approved project; Prion protein function in redox homeostasis and associated failure in prion disease, 17-NIAID-00212, 8/3/2017.
All animal experiments were approved by the RML Animal Care and Use Committee under protocols 2019-043 and 2022-045. All mice were housed at the Rocky Mountain Laboratories (RML) in an AAALAC-accredited facility in compliance with guidelines provided by the Guide for the Care and Use of Laboratory Animals (Institute for Laboratory Animal Research Council) with Dark/light cycle: 12 h/12 h, ambient temperature: 70 ± 2 F, humidity: approximately 40%. All experiments were approved by the RML Animal Care and Use Committee under protocols 2019-043 and 2022-045.
Cell lines and culture conditions
Cell lines used in this study: HT-1080 (ATCC #CCL-121), HEK-293T (ATCC #CRL-3216), HT22 (Sigma #SCC129). Each cell line was grown under recommended conditions DMEM (Thermo Fisher Scientific) supplemented with 10% fetal bovine serum. Additionally, the growth medium for all cell lines was enriched with 1% penicillin-streptomycin (Thermo Fisher Scientific) and 1% L-glutamine (Thermo Fisher Scientific). HT-1080 was maintained with 1% NEAA (non-essential amino acids, Thermo Fisher Scientific).
PrPC expression and immunostaining
3 × 103 PrPC OE and control cells were seeded in each well of a 96-well plate overnight, washed two times with PBS and fixed with 4% PFA for 10 min. Cells were blocked in 5% BSA Fraction V, 10% FBS, 0.3% Triton-X in PBS and incubated in primary antibody (Supplementary Reagents Table) overnight at 4 °C, followed by two times PBS wash and secondary antibody. Images were taken on a confocal microscope in individual channels and merged in Adobe Photoshop.
Cell viability assays
2 × 103 cells were plated in 96-well plates and exposed to IKE (See Supplementary Reagents Table) for an overnight incubation, following the specific conditions outlined in the legends. Subsequently, resazurin (Sigma) was introduced to the wells to a final concentration of 50 µM. After an 8-h incubation period, the fluorescence readings were recorded using an Envision 2104 Multilabel plate reader (PerkinElmer) with excitation at 540 nm and emission at 590 nm. For each condition, a minimum of three wells were subject to measurement and averaged; in general, all viability is presented as percentage relative to the respective control as mean ± SEM.
Generation of overexpression and suppression cell lines
Human cDNA for PRNP and RAC3 containing was amplified from HT-1080 cDNA and cloned into lentiviral expression vector pLVTHM (Addgene #12247) or pLV (Addgene #39481) with IRES puromycin. A human RAC3 and three GPX8 knockdown guides (See Supplementary Reagents Table) were cloned independently into pLV hU6-sgRNA hUbC-dCas9-KRAB-T2a-Neo (modified from Addgene #71236). The viruses were packaged using a pantropic lentiviral 2nd-generation system. Control cells were infected with pLVTHM empty or lentiCRISPRv2 empty vectors, respectively. Following 3 days of infection, overexpressing cells were selected by corresponding drug resistance markers. Knockdown cells were collected on specified days after infection. Knockout vectors for GPX8 and murine Prnp were designed and cloned into lentiCRISPRv2 (Addgene #52961), transiently transfected 16 h into recipient cells, selected with 1 μg/mL puromycin, then cultivated until assay timepoints.
Western blot
For each experimental condition, approximately 5 × 105 cells were seeded overnight in a 6-well plate. Protein samples were collected using 100 µL of RIPA buffer and subjected to sonication and concentration determination by BCA. Subsequently, the samples were separated on a 15% SDS-PAGE gel and transferred onto PVDF membranes. Membranes were blocked with 5% skim milk in TBS-T for 1 h at room temperature, then incubated overnight at 4 °C with the primary antibody. Afterward, the membranes were washed three times with TBS-T and incubated with HRP-coupled secondary antibodies for 1 h at room temperature. Following three additional washes with TBS-T, the signal detection was carried out using ECL (Bio-Rad) according to the manufacturer’s instructions.
For human brain homogenate blotting, 15 µl of ~2% (w/v) brain homogenate was boiled in 1x Bolt LDS sample buffer (Invitrogen) containing 6% (v/v) BME for 5 min, loaded into 10-well Bolt 4–12% Bis-Tris gels (Invitrogen), and resolved at a constant 200 V in 1x MES buffer (Invitrogen) for 35 min. Proteins were transferred to a PVDF membrane (Millipore) using the iBlot 2 transfer system (Invitrogen), which was then blocked in SuperBlock Blocking Buffer (Invitrogen) for 10 min at room temperature. RAC3 (ABCAM; AB124943; 1:1000), FABP5 (Proteintech; Cat#: 12348-1-AP; 1:2000) and GPX8 (Genetex; GTX125992; 1:1000) primary antibodies, followed by anti-rabbit HRP conjugated secondary antibody (Abcam; 1:5000) were detected by ECL SuperSignal West Atto Ultimate Sensitivity Chemiluminescent Substrate (Invitrogen) and imaged with a iBright imaging system (Invitrogen). Total protein was labeled with a Ponceau stain solution (0.1% w/v Ponceaus and 5% v/v acetic acid) and imaged by the iBright imaging system. ImageJ 1.52n was used to measure the densitometry density of the bands as well as the total protein. The levels of test protein were normalized to the total protein, and the average levels between CJG and control groups were compared by Welch’s unpaired t-test (Two-tailed with 95% confidence interval).
GPX activity detection
The GPX activity assay (ELA-E-BC-K096-S) was carried out using instructions provided by the manufacturer.
Cytosolic ROS detection
Cells were seeded in a 6-well plate, and the following day, the medium was replaced with 2 mL of medium containing RSL3 at a concentration of 100 nM for 2 h. 2,7-Dichlorodihydrofluorescein diacetate (DCFH-DA) from Biomol (Cay85155-50) was added to the wells at a final concentration of 25 µM to detect cytosolic ROS. The cells were then incubated for an additional 30 min. After removing the medium, the wells were rinsed twice with 2 mL of and 200 µL of Accutase from Sigma (A6964) was added to each well. The detached cells were resuspended in 800 µL of PBS per well and analyzed using an Attune acoustic flow cytometer from Applied Biosystems. Histogram intensities were collected from the BL-1 channel excited by a 488 nm laser. The median fluorescence intensity of each well was determined and normalized to the DMSO-treated control cells using FlowJo 10 software.
Lipid peroxidation detection
Flow cytometry was employed to detect lipid peroxides. In each well of a 6-well plate, 1 × 105 cells were seeded. The following day, the medium was replaced with 2 mL of medium containing the ferroptosis inducer RSL3 at a concentration of 100 nM for 4 h. Afterward, BODIPY 581/591 C11 (BODIPY-C11, Thermo Fisher Scientific) was added to the wells at a final concentration of 2 µM to detect lipid peroxidation. The cells were then incubated for an additional 30 min. Following the removal of the medium, the wells were rinsed twice with 2 mL of PBS. To detach the cells, 200 µL of Accutase (Sigma) (A6964) was added to each well. Detached cells were subsequently resuspended in 800 µL of PBS per well and analyzed using an Attune acoustic flow cytometer (Applied Biosystems). Histogram intensities were collected from the BL-1 channel excited by a 488 nm laser. The median fluorescence intensity of each well was determined and normalized to the DMSO-treated control cells using FlowJo 10 software.
Ferroptosis assay
A total of 4 × 104 cells were seeded in a 6-well plate for infection. Following a 3-day infection period, cells were selected using puromycin overnight for 16 h. Afterward, inhibitors were added to the respective wells, and a 3-day co-culture with the inhibitors was carried out. Following trypsinization and counting, an empirically determined volume of control cells containing 2 × 103 cells was plated in 96-well plates, with the same volume of each experimental condition being added to each well. Resazurin (Sigma) was introduced to the wells (final concentration 50 µM) for 8 h. Fluorescence readings were taken using an Envision 2104 Multilabel plate reader from PerkinElmer (excitation, 540 nm; emission, 590 nm). For each experiment, a minimum of three wells were measured for each experimental condition. The viability data were presented as a percentage relative to the control and expressed as a mean ± SEM.
High content assay
For ferroptosis biomarker detection, 5 × 103 cells for each experimental condition were seeded in 96-well plates and allowed to incubate overnight. Immunofluorescence intensity assays were conducted using the Operetta imaging system from PerkinElmer. During the assay, DAPI staining was utilized to identify primary objects, and a Cy3-labeled secondary antibody from Dianova was employed for intensity measurements. The secondary antibody was diluted at a ratio of 1:500. Antibodies used were listed in the Supplementary Reagents Table.
For cell shape, cells were stained with FITC-conjugated phalloidin and imaged in Thermo Scientific CX-7. The parameter “ObjectShapeLWRCh1” was used to measure cell shape based on the ratio of length to width using object-aligned boundary boxes.
Organoid generation, culture and infection
The undirected organoids used for the current study were differentiated from ACS-1023 (ATCC) or RAH019A74 induced pluripotent stem cells, maintained in long-term culture and infected with 0.1% (tissue wet weight/volume) sCJD prions (ACS-1023 with MV1 and RAH019A with MM1) from postmortem brain homogenate as described previously35. Organoids were infected at 5 months post beginning differentiation and harvested when substantially infected at 179 dpi or treated with RSL3 at 90 days post-infection; the disease timeframe associated with the beginning of metabolic changes35. PRNP KO organoids were differentiated from RAH019A cells that had been engineered with a frameshift mutation to disrupt the PRNP open reading frame as described by Groveman and Williams75. Four-five-month-old RAH019A WT and PRNP KO organoids were used for RSL3 treatments. Media lactate dehydrogenase (LDH) release assays were carried out using the Cytotoxicity Detection Kit Plus (LDH) from Roche as per the manufacturer’s instructions. For histological analysis, organoids were fixed in neutral buffered formalin for approximately 24 h before embedding in paraffin. Two- or five-micron sections were cut for staining as described below.
RT-QuIC
Cells were cultured − or + doxycycline for 3 days and then seeded at 130,000 cells/well in 6-well culture plates. The next day, cells were harvested by trypsinization and washed with culture medium. Three replicate wells were pooled together to make 4 final replicates of each condition. The pooled cells were centrifuged for 5 min 1500 RPM. The media was aspirated, and the pellets were snap frozen in liquid nitrogen and then stored at −80 °C. Naïve or 22 L infected N2a cells were used as negative and positive controls, respectively.
For RT-QuIC, the cell pellets were resuspended in 250 µL of 0.1% SDS in PBS and briefly sonicated. Lysates were centrifuged at 2000×g for 2 min, and the supernatants were recovered and diluted 1:10 or 1:100 in 0.1% (w/v) SDS in PBS with 1x N2. RT-QuIC was performed as previously described with some modifications76. Black 384-well assay plates with a clear bottoms (Nunc) were pre-loaded with 49 µL of RT-QuIC reaction mix containing final concentrations of 10 mM sodium phosphate (pH 7.4), 300 mM NaCl, 10 μM thioflavin T (ThT), 1 mM ethylenediaminetetraacetic acid tetrasodium salt (EDTA), and 0.1 mg/mL hamster recombinant PrP 90–231 (purified as described in ref. 77). Eight replicate reaction wells were seeded with 1 μL of diluted lysate for each sample, also contributing 0.002% SDS to the final reaction mix. Plates were sealed (Nalgene Nunc International sealer) and then incubated in a BMG FLUOstar Omega plate reader at 50 °C for 50 h with cycles of 60 s of shaking (700 rpm, double-orbital) and 60 s of rest throughout the incubation. ThT fluorescence measurements (excitation, 450 ± 10 nm; emission, 480 ± 10 nm [bottom read]) were taken every 45 min. Wells that showed an increase in ThT fluorescence >10% of the maximum baseline-subtracted ThT fluorescence value on the experimental plate were considered positive reactions. A sample was considered positive for prion seeding activity if >25% of the replicate reaction wells had positive responses.
Lipidomics
See schematic overview (Supplementary Fig. 9).
RAC3 overexpressing lipidomics study
A total number of samples was fifteen in three groups. There were five technical replicates for each group (5 samples of the parental HT-1080 cells (control), 5 samples of IKE-treated samples, 5 samples of the RAC3 OE cells). Lipid extraction was performed using the following procedure. Initially, cell pellets were collected in Eppendorf Safe-Lock tubes and treated with 400 µL of methanol cooled on dry ice. The tubes were then vortexed to ensure thorough mixing. Subsequently, the pellets were transferred to MACHEREY-NAGEL GmbH & Co. KG bead tubes (MN Bead Tubes Type A). Any remaining pellet residue was washed with an additional 400 µL of methanol cooled on dry ice, and the mixture was vortexed for 30 s before being transferred to the bead tubes. The pellets were then homogenized in the bead tubes using a homogenizer (Precellys, Bertin Technologies) at 4 °C using the soft mode. After homogenization, the bead tubes were centrifuged at 19,000×g for 15 min at 4 °C. Following centrifugation, 400 µL of the supernatant was transferred to a vial. Finally, the supernatant was injected into an LC–MS/MS system, which consisted of a liquid chromatography system (Exion LC series UHPLC, Sciex) and a time-of-flight mass spectrometer (X500QTOF, Sciex). A reverse-phase chromatography column (Cortecs UPLC C18 column, 150 mm × 2.1 mm ID, 1.6 µm, Waters Corporation) was used for lipid separation. The eluents A and B were composed of 60% CH3CN, 40% H2O, 1% HCOOH, 1% HCOONH4 (10 mM) and 90% 2-Propanol, 10% CH3CN, 1% HCOOH, 1% HCOONH4 (10 mM), respectively. A gradient elution method was employed for lipid analysis with a total duration of 25 min and a flow rate of 0.25 mL/min. The gradient program followed the schedule below: 0 min, 68.0% A; 1.50 min, 68.0% A; 21 min, 3.0% A; 25 min, 3.0% A.
Mass spectrometry parameters were set in the electrospray ionization positive mode, with a TOF start mass of 100 Da and a TOF stop mass of 1700 Da. The MS experiment was conducted in Information-dependent acquisition mode (Data-dependent acquisition mode). Raw data was converted into mzXML format using MSConverter (ProteoWizard, Palo Alto, CA) in peak picking mode to obtain centroid data. Feature detection, processing, alignment, and identification were performed using MZmine software78. For tentative annotation of lipids, the MoNA library was utilized. Lipid differential expression and lipid characteristics were analyzed using LipidSig79. PCA analysis was conducted in Metaboanalyst online, based on MZmine data processing80. Ellipses show 95% confidence region.
PrPC overexpressing lipidomics study
A total number of samples was ten in two groups. There were five technical replicates for each group (5 samples of parental HT-1080 cells (control) and 5 samples of PrPC overexpressing (PrPC OE)). Lipid profiling of PrPC-overexpressing (PrPC OE) cells and control HT-1080 cells was performed using a Bruker maXis II quadrupole time-of-flight (Q-TOF) mass spectrometer (Bruker Daltonik GmbH, Bremen, Germany) coupled to a Waters ACQUITY UPLC system. Lipid separation was achieved using a 29-min gradient elution on a UPLC column. The gradient program was as follows: 0–1.5 min, 68% solvent A; 21–25 min, 3% solvent A; 29 min, 68% solvent A. The injection volume was 5 μL. Mass spectrometric analysis was conducted in positive ion mode using electrospray ionization (ESI). The mass range scanned was from 100 to 2000 Da in data-dependent acquisition (DDA) mode. Raw data files were converted to mzXML format after recalibration in enhanced quadratic mode using the Bruker Compass Data Analysis Software (Bruker Daltonik GmbH, Bremen, Germany). Feature detection, peak processing, alignment, and tentative lipid annotation were performed using the XCMS online service81 with the “UPLC / Bruker Q-TOF pos” data analysis protocol.
Immunohistochemistry on human brain tissue and organoids
Tissue samples of the anterior superior or medial frontal gyrus were cut out of autopsy brains fixed in 4% neutral phosphate-buffered formaldehyde, put into 100% formic acid for 1 h at room temperature (RT), washed under running tap water for 30 min, fixed again for 24–48 h in neutral phosphate-buffered formaldehyde at RT and paraffinized.
Immunohistochemistry for prion protein including counterstaining with hematoxylin was performed on 5 µm paraffin sections in a Bench Mark Ultra (Roche) according to the instructions of the manufacturer using a mouse monoclonal antibody (clone L42; RIDA, catalogue-no. R8005) diluted 1:300 and a diaminobenzidine/peroxidase based detection system (optiView; Roche). For RAC3, Abcam AB129062 was used at 1:300. Sections were pretreated with proteinase (Roche) for 8 min at RT and boiled in citrate buffer (buffer CC1, Roche) for 32 min.
Images of prion protein and RAC3 stains were taken with an Olympus BX23 camera at an Olympus BX45 microscope using cellsSens Entry software v3.2.
Organoids were immersed in 3.7% neutral buffered formalin for ~ 24 h prior to standard embedding in paraffin. Two or five-micron sections were cut. Organoid histochemistry staining for FABP5 and RAC3 was carried out as above. MAP2 (1:500; Abcam) and GFAP (1:2500, Dako) were detected using ChromoMap DAB (Roche/Ventana #NC1859896) and photographed using Aperio Imagescope software.
Real-time PCR
1 × 106 cells per sample were seeded, and total RNA was isolated using the TRIzol RNA extraction protocol. Subsequently, 2 μg of total RNA was reverse transcribed using the AMV Reverse Transcriptase Kit (NEB, M0277S). Quantitative PCR reactions were carried out on a LightCycler480 (Roche) instrument using Power SYBR Green Master Mix (Lager, 5000989). Differences in mRNA levels compared to control were calculated using the ΔΔCp method, with RPL27 or TBP as housekeeper gene. The qPCR primer sequences are listed in the Supplementary Reagents Table.
Iron and H2O2 determination
The cytosolic LIP was determined as described82. For each measurement, 4 × 105 cells were loaded with 5 nM calcein-AM (Molecular Probes) in PBS at 37 °C for 15 min. After washing non-internalized calcein, the cells were transferred to a 1.5 mL ep tube, and the basal calcein fluorescence was recorded (excitation, 488 nm; emission, 517 nm). HyperER endoplasmic reticulum hydrogen peroxide measurement was performed as described83. 500 µM Ferric ammonium citrate (FAC) or 100 µM Deferoxamine (DFO) were treated overnight as the positive and negative control.
FerroOrange. For each FerroOrange measurement, 4 × 105 cells were loaded with 1uM FerroOrange in non FBS medium at 37 °C for 20 min. After washing two times with PBS, the cells were transferred to 1.5 mL ep tube, and the fluorescence was recorded (excitation, 532 nm; emission, 572 nm). 500 µM Ferric ammonium citrate (FAC) or 100 µM Deferoxamine (DFO) overnight treated condition were measured as the positive and negative control. For Mössbauer measurements, see Supplementary File 1.
Informatic analyses
All gene expression correlations and gene effect correlations are derived from Cancer Cell Line Encyclopedia data downloaded 22Q1 and 23Q2 from Depmap databases, respectively. Pearson’s expression correlation and P-values were calculated for display using Python libraries pandas, scipy, statsmodels, sklearn and matplotlib. All scripts are available upon request.
CRISPR screen
Human Synergistic Activation Mediator (SAM) library (#1000000057, Addgene) was infected together with lenti dCAS-VP64_Blast (#61425, Addgene) and MS2-P65-HSF1_Hygro (#61426, Addgene) into PRNPtet cells and selected with respective antibiotics. After 3 days selection, PrPC was induced for 3 days with doxycycline, or vehicle for controls, followed by treatment with 300 nM IKE, resulting in viability loss of 5–10%. Normalized guide frequencies amplified from viable cells in the sensitized PrPC cells were scored against to (-)doxycycline control cells to identify enriched or depleted guides using ENCoRE84.
Animal studies
Prion inoculation and brain homogenate preparation for western blotting analysis.
Six-week-old wild-type C57BL10 mice were intracerebrally inoculated with 30 µL of 1% (w/v) brain homogenate from RML prion strains, which were prepared from stocks previously determined to have a 50% infective dose (ID50) of 2.4 × 104 RML. Uninfected control mice were ‘mock’ inoculated with uninfected or normal brain homogenates (NBH). Mice were monitored twice weekly prior to clinical onset and every 1–3 days throughout the clinical phase. Mice were euthanized at 80, 108, 160 dpi, and brains were collected and sliced into 300 µm thick coronal sections. Those slices harboring the hippocampus, thalamus, hypothalamus, and amygdala (areas of known disease pathology) were homogenized in 1 × PBS into 1% w/v brain homogenate for western blot analysis.
Proteins were denatured by boiling in 1 × Bolt LDS sample buffer (Invitrogen) containing 10% Beta-mercaptoethanol for 5 min at 100 °C and resolved into various sizes in 4–12% Bis-Tris gels (Invitrogen). Transfer, blotting and quantification were done as for the human brain samples. The levels of test protein were normalized to the total protein, and the average levels between uninfected and infected groups were compared by Welch’s unpaired t-test (Two-tailed with 95% confidence interval).
Sex and gender
Sporadic CJD qualifies as a rare disease with 1 to 2 new cases per million inhabitants per year. sCJD has a 1:1 male-to-female ratio, and thus sex does not appear to be a factor determining susceptibility or progression of the disease. Thus, sex is not considered in the study design, and the pathological material was assigned to gender according to a licensed pathologist during the autopsy. Anonymized reporting of individual sexes does not influence outcome or predictability in the study.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data presented in the manuscript are available in the Supplementary files. Repository accessions and links RAC3 overexpressing and imidazole ketone erastin (IKE) Lipidomics: MassIVE MSV000097840 and PrPC overexpressing Lipidomics: MassIVE MSV000097841. Source data are provided with this paper.
References
Prusiner, S. B. Novel proteinaceous infectious particles cause scrapie. Science 216, 136–144 (1982).
Collinge, J. Prion diseases of humans and animals: their causes and molecular basis. Annu. Rev. Neurosci. 24, 519–550 (2001).
Ugalde, C. L., Finkelstein, D. I., Lawson, V. A. & Hill, A. F. Pathogenic mechanisms of prion protein, amyloid-β and α-synuclein misfolding: the prion concept and neurotoxicity of protein oligomers. J. Neurochem. 139, 162–180 (2016).
Chang, S. C. et al. Propagation of PrPSc in mice reveals impact of aggregate composition on prion disease pathogenesis. Commun. Biol. 6, 1162 (2023).
Saá, P., Harris, D. A. & Cervenakova, L. Mechanisms of prion-induced neurodegeneration. Expert Rev. Mol. Med. 18, e5 (2016).
Mercer, R. C. C. & Harris, D. A. Mechanisms of prion-induced toxicity. Cell Tissue Res. 392, 81–96 (2023).
Viswanathan, V. S. et al. Dependency of a therapy-resistant state of cancer cells on a lipid peroxidase pathway. Nature 547, 453–457 (2017).
Le Corre, D. et al. The cellular prion protein controls the mesenchymal-like molecular subtype and predicts disease outcome in colorectal cancer. EBioMedicine 46, 94–104 (2019).
Li, H. et al. Pro-prion, as a membrane adaptor protein for E3 ligase c-Cbl, facilitates the ubiquitination of IGF-1R, promoting melanoma metastasis. Cell Rep. 41, 111834 (2022).
Brazier, M. W. et al. Correlative studies support lipid peroxidation is linked to PrPres propagation as an early primary pathogenic event in prion disease. Brain Res. Bull. 68, 346–354 (2006).
Kim, N. H. et al. Increased ferric iron content and iron-induced oxidative stress in the brains of scrapie-infected mice. Brain Res. 884, 98–103 (2000).
Singh, A. et al. Prion protein regulates iron transport by functioning as a ferrireductase. J. Alzheimers Dis. 35, 541–552 (2013).
Fernaeus, S. & Land, T. Increased iron-induced oxidative stress and toxicity in scrapie-infected neuroblastoma cells. Neurosci. Lett. 382, 217–220 (2005).
Bogdan, A. R., Miyazawa, M., Hashimoto, K. & Tsuji, Y. Regulators of iron homeostasis: new players in metabolism, cell death, and disease. Trends Biochem. Sci. 41, 274–286 (2016).
Tripathi, A. K. et al. Prion protein functions as a ferrireductase partner for ZIP14 and DMT1. Free Radic. Biol. Med. 84, 322–330 (2015).
Castle, A. R. & Gill, A. C. Physiological functions of the cellular prion protein. Front. Mol. Biosci. 4, 19 (2017).
Rachidi, W. et al. Expression of prion protein increases cellular copper binding and antioxidant enzyme activities but not copper delivery. J. Biol. Chem. 278, 9064–9072 (2003).
Zeng, F., Watt, N. T., Walmsley, A. R. & Hooper, N. M. Tethering the N-terminus of the prion protein compromises the cellular response to oxidative stress. J. Neurochem. 84, 480–490 (2003).
Gross, E. et al. Generating disulfides enzymatically: reaction products and electron acceptors of the endoplasmic reticulum thiol oxidase Ero1p. Proc. Natl. Acad. Sci. USA 103, 299–304 (2006).
Tu, B. P. & Weissman, J. S. The FAD- and O2-dependent reaction cycle of Ero1-mediated oxidative protein folding in the endoplasmic reticulum. Mol. Cell 10, 983–994 (2002).
Ramming, T., Hansen, H. G., Nagata, K., Ellgaard, L. & Appenzeller-Herzog, C. GPx8 peroxidase prevents leakage of H2O2 from the endoplasmic reticulum. Free Radic. Biol. Med. 70, 106–116 (2014).
Nguyen, V. D. et al. Two endoplasmic reticulum PDI peroxidases increase the efficiency of the use of peroxide during disulfide bond formation. J. Mol. Biol. 406, 503–515 (2011).
Khatib, A. et al. The glutathione peroxidase 8 (GPX8)/IL-6/STAT3 axis is essential in maintaining an aggressive breast cancer phenotype. Proc. Natl. Acad. Sci. USA 117, 21420–21431 (2020).
Rane, N. S., Yonkovich, J. L. & Hegde, R. S. Protection from cytosolic prion protein toxicity by modulation of protein translocation. EMBO J. 23, 4550–4559 (2004).
Malhotra, J. D. & Kaufman, R. J. Endoplasmic reticulum stress and oxidative stress: a vicious cycle or a double-edged sword? Antioxid. Redox Signal 9, 2277–2293 (2007).
Malinouski, M., Zhou, Y., Belousov, V. V., Hatfield, D. L. & Gladyshev, V. N. Hydrogen peroxide probes directed to different cellular compartments. PLoS ONE 6, e14564 (2011).
Xin, S. et al. MS4A15 drives ferroptosis resistance through calcium-restricted lipid remodeling. Cell Death Differ. 29, 670–686 (2021).
Kagan, V. E. et al. Oxidized arachidonic and adrenic PEs navigate cells to ferroptosis. Nat. Chem. Biol. 13, 81–90 (2017).
Wiernicki, B. et al. Excessive phospholipid peroxidation distinguishes ferroptosis from other cell death modes including pyroptosis. Cell Death Dis. 11, 922 (2020).
Zou, Y. et al. Plasticity of ether lipids promotes ferroptosis susceptibility and evasion. Nature 585, 603–608 (2020).
Qiu, B. et al. Phospholipids with two polyunsaturated fatty acyl tails promote ferroptosis. Cell 187, 1177–1190.e1118 (2024).
Kraft, V. A. N. et al. GTP cyclohydrolase 1/tetrahydrobiopterin counteract ferroptosis through lipid remodeling. ACS Cent. Sci. 6, 41–53 (2020).
Peng, H. et al. Fatty acid-binding protein 5 is a functional biomarker and indicator of ferroptosis in cerebral hypoxia. Cell Death Dis. 15, 286 (2024).
Lancaster, M. A. & Knoblich, J. A. Generation of cerebral organoids from human pluripotent stem cells. Nat. Protoc. 9, 2329–2340 (2014).
Groveman, B. R. et al. Sporadic Creutzfeldt-Jakob disease prion infection of human cerebral organoids. Acta Neuropathol. Commun. 7, 90 (2019).
Groveman, B. R. et al. Sporadic Creutzfeldt-Jakob disease infected human cerebral organoids retain the original human brain subtype features following transmission to humanized transgenic mice. Acta Neuropathol. Commun. 11, 28 (2023).
Green, A. J. E. RT-QuIC: a new test for sporadic CJD. Pract. Neurol. 19, 49–55 (2019).
Konermann, S. et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517, 583–588 (2015).
Wan, C. et al. Panorama of ancient metazoan macromolecular complexes. Nature 525, 339–344 (2015).
Kovalski, J. R. et al. The functional proximal proteome of oncogenic Ras includes mTORC2. Mol. Cell 73, 830–844.e812 (2019).
Shatz, O. & Elazar, Z. ATG9 raises the BAR for PI4P in autophagy. J. Cell Biol. 218, 1432–1433 (2019).
Machado, M. S. et al. The inflammatory cytokine TNF contributes with RAC3-induced malignant transformation. EXCLI J. 17, 1030–1042 (2018).
Panelo, L. C. et al. High RAC3 expression levels are required for induction and maintaining of cancer cell stemness. Oncotarget 9, 5848–5860 (2018).
Clark, E. A., Golub, T. R., Lander, E. S. & Hynes, R. O. Genomic analysis of metastasis reveals an essential role for RhoC. Nature 406, 532–535 (2000).
Hordijk, P. L. Regulation of NADPH oxidases. Circ. Res. 98, 453–462 (2006).
Sitammagari, K. K. & Masood, W. Creutzfeldt Jakob disease in StatPearls. (StatPearls, 2023).
Milhavet, O. et al. Prion infection impairs the cellular response to oxidative stress. Proc. Natl. Acad. Sci. USA 97, 13937–13942 (2000).
Westergard, L., Christensen, H. M. & Harris, D. A. The cellular prion protein (PrP(C)): its physiological function and role in disease. Biochim. Biophys. Acta 1772, 629–644 (2007).
Singh, N., Singh, A., Das, D. & Mohan, M. L. Redox control of prion and disease pathogenesis. Antioxid. Redox Signal 12, 1271–1294 (2010).
Andre, R. & Tabrizi, S. J. Misfolded PrP and a novel mechanism of proteasome inhibition. Prion 6, 32–36 (2012).
Yedidia, Y., Horonchik, L., Tzaban, S., Yanai, A. & Taraboulos, A. Proteasomes and ubiquitin are involved in the turnover of the wild-type prion protein. EMBO J. 20, 5383–5391 (2001).
Haigh, C. L. et al. Acute exposure to prion infection induces transient oxidative stress progressing to be cumulatively deleterious with chronic propagation in vitro. Free Radic. Biol. Med. 51, 594–608 (2011).
Büeler, H. et al. Mice devoid of PrP are resistant to scrapie. Cell 73, 1339–1347 (1993).
Mays, C. E. et al. Prion disease tempo determined by host-dependent substrate reduction. J. Clin. Invest. 124, 847–858 (2014).
Levin, E. C. et al. Neuronal expression of vimentin in the Alzheimer’s disease brain may be part of a generalized dendritic damage-response mechanism. Brain Res. 1298, 194–207 (2009).
Radisky, D. C. et al. Rac1b and reactive oxygen species mediate MMP-3-induced EMT and genomic instability. Nature 436, 123–127 (2005).
Acevedo, A. & González-Billault, C. Crosstalk between Rac1-mediated actin regulation and ROS production. Free Radic. Biol. Med. 116, 101–113 (2018).
Hordijk, P. L. Regulation of NADPH oxidases: the role of Rac proteins. Circ. Res. 98, 453–462 (2006).
Chen, H. et al. SETD8 inhibits apoptosis and ferroptosis of Ewing’s sarcoma through YBX1/RAC3 axis. Cell Death Dis. 15, 494 (2024).
Giese, A. & Kretzschmar, H. A. Prion-induced neuronal damage—the mechanisms of neuronal destruction in the subacute spongiform encephalopathies. Curr. Top. Microbiol. Immunol. 253, 203–217 (2001).
Ma, Y. et al. Stimulations of the culture medium of activated microglia and TNF-α on a scrapie-infected cell line decrease the cell viability and induce marked necroptosis that also occurs in the brains from the patients of human prion diseases. ACS Chem. Neurosci. 10, 1273–1283 (2019).
Liberski, P. P., Sikorska, B., Bratosiewicz-Wasik, J., Gajdusek, D. C. & Brown, P. Neuronal cell death in transmissible spongiform encephalopathies (prion diseases) revisited: from apoptosis to autophagy. Int. J. Biochem. Cell Biol. 36, 2473–2490 (2004).
Zhou, Y., Liao, J., Mei, Z., Liu, X. & Ge, J. Insight into crosstalk between ferroptosis and necroptosis: novel therapeutics in ischemic stroke. Oxid. Med. Cell Longev. 2021, 9991001 (2021).
Gugnoni, M., Sancisi, V., Manzotti, G., Gandolfi, G. & Ciarrocchi, A. Autophagy and epithelial–mesenchymal transition: an intricate interplay in cancer. Cell Death Dis. 7, e2520 (2016).
Gao, M. et al. Ferroptosis is an autophagic cell death process. Cell Res. 26, 1021–1032 (2016).
Pamplona, R. et al. Increased oxidation, glycoxidation, and lipoxidation of brain proteins in prion disease. Free Radic. Biol. Med. 45, 1159–1166 (2008).
Brown, D. R. & Besinger, A. Prion protein expression and superoxide dismutase activity. Biochem. J. 334, 423–429 (1998).
Mehrabian, M. et al. The prion protein controls polysialylation of neural cell adhesion molecule 1 during cellular morphogenesis. PLoS ONE 10, e0133741 (2015).
Mouillet-Richard, S. et al. Wnt, glucocorticoid and cellular prion protein cooperate to drive a mesenchymal phenotype with poor prognosis in colon cancer. J. Transl. Med. 22, 337 (2024).
Béranger, F., Mangé, A., Goud, B. & Lehmann, S. Stimulation of PrPC retrograde transport toward the endoplasmic reticulum increases accumulation of PrPSc in prion-infected cells. J. Biol. Chem. 277, 38972–38977 (2002).
Hetz, C. et al. The disulfide isomerase Grp58 is a protective factor against prion neurotoxicity. J. Neurosci. 25, 2793–2802 (2005).
Iuchi, K., Takai, T. & Hisatomi, H. Cell death via lipid peroxidation and protein aggregation diseases. Biology 10, 399 (2021).
Orsi, A., Fioriti, L., Chiesa, R. & Sitia, R. Conditions of endoplasmic reticulum stress favor the accumulation of cytosolic prion protein. J. Biol. Chem. 281, 30431–30438 (2006).
Foliaki, S. T. et al. Pathogenic prion protein isoforms are not present in cerebral organoids generated from asymptomatic donors carrying the E200K mutation associated with familial prion disease. Pathogens 9, 482 (2020).
Groveman, B. R. et al. Lack of transmission of chronic wasting disease prions to human cerebral organoids. Emerg. Infect. Dis. 30, 1193–1202 (2024).
Williams, K. et al. Neural cell engraftment therapy for sporadic Creutzfeldt-Jakob disease restores neuroelectrophysiological parameters in a cerebral organoid model. Stem Cell Res. Ther. 14, 348 (2023).
Orrú, C. D. et al. Factors that improve RT-QuIC detection of prion seeding activity. Viruses 8, 140 (2016).
Schmid, R. et al. Integrative analysis of multimodal mass spectrometry data in MZmine 3. Nat. Biotechnol. 41, 447–449 (2023).
Lin, W.-J. et al. LipidSig: a web-based tool for lipidomic data analysis. Nucleic Acids Res. 49, W336–W345 (2021).
Pang, Z. et al. Using MetaboAnalyst 5.0 for LC-HRMS spectra processing, multi-omics integration and covariate adjustment of global metabolomics data. Nat. Protoc. 17, 1735–1761 (2022).
Tautenhahn, R., Patti, G. J., Rinehart, D. & Siuzdak, G. XCMS Online: a web-based platform to process untargeted metabolomic data. Anal. Chem. 84, 5035–5039 (2012).
Epsztejn, S., Kakhlon, O., Glickstein, H., Breuer, W. & Cabantchik, Z. I. Fluorescence analysis of the labile iron pool of mammalian cells. Anal. Biochem. 248, 31–40 (1997).
Zhang, Y. et al. Targeting the functional interplay between endoplasmic reticulum oxidoreductin-1α and protein disulfide isomerase suppresses the progression of cervical cancer. EBioMedicine 41, 408–419 (2019).
Trümbach, D. et al. ENCoRE: an efficient software for CRISPR screens identifies new players in extrinsic apoptosis. BMC Genomics 18, 1–13 (2017).
Acknowledgements
We thank Maika Dunst for expert technical assistance with tissue processing and immunohistochemistry. Human sCJD brain samples were a kind gift from Prof Gianluigi Zanusso (University of Verona) and include 3 each of two sCJD subtypes (MM1 and MV2). We also acknowledge the help of the Innovation Platform for Academicians of Hainan Province. This project was supported by the German Research Foundation (DFG, Project 501860452) to J.S., T.A., and S.M. and DFG (511521600) to J.S., the EnABLE initiative by the state of Hessen to S.M., and Helmholtz Center Munich to J.S., Genetics and Cell Engineering Group. This work was also supported by the Specific Research Fund of the Innovation Platform for Academicians of Hainan Province (Grant Nos. YSPTZX2022011, YSPTZX2025011) to X.J. and J.S.; and the Undergraduates Training Program for Innovation and Entrepreneurship of Hainan Province (X.J.). This work was also funded in part (C.H., B.G., K.W., S.F., B.R., T.T.) by the intramural research program of the National Institutes of Health (NIAID).
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
J.S. conceived the study, supervised the research, and wrote the manuscript together with C.H. and T.A. H.P. and S.P. performed all genetic modifications and cellular in vitro assays, CRISPR screening, interpretation and statistics. M.Q., C.S. and S.F. performed Western and RT-PCR analyses. X.J., C.C. and X.Z. performed informatics analyses. B.G. and K.W. performed all organoid and RT-QuiC assays. K.K. performed Mössbauer analyses. B.V. and C.M. performed the mass spectrometry and lipidomics analysis. B.R. and S.F. performed animal time course infections and analyzed the tissues. T.A. and S.M. collected patient material, performed histology and immunostainings, and evaluated results for the manuscript. T.T. performed IHC analysis on certain human tissue and organoids. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s Note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Peng, H., Pfeiffer, S., Varynskyi, B. et al. Prion-induced ferroptosis is facilitated by RAC3. Nat Commun 16, 5385 (2025). https://doi.org/10.1038/s41467-025-60793-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-60793-3