Abstract
Glutamate, a crucial player in hepatic amino acid metabolism, has been relatively unexplored in immune cell activation. We show in a study with male mice that hepatic glutamate accumulates in vesicles of perivenous hepatocytes through vesicular glutamate transporter 3 (VGLUT3), regulated by the aryl hydrocarbon receptor upon chronic alcohol intake. Additional binge drinking triggers the exocytosis of glutamate by altering the intracellular Ca2+ level, stimulating metabotropic glutamate receptor 5 (mGluR5) and subsequent NADPH oxidase 2 (NOX2)-mediated ROS production in Kupffer cells (KCs). This interaction between hepatocytes and KCs is facilitated by pseudosynapse formation, arising from alcohol-induced ballooning of perivenous hepatocytes. Genetic or pharmacological interference of mGluR5 or NOX2 in KCs alleviates alcohol-related steatohepatitis (ASH). Analysis of patient samples confirmed some of the findings from mice, showing that plasma glutamate concentration and VGLUT3 levels correlate with ASH development. Conclusively, our findings highlight glutamate storage and release in mediating ASH, particularly through the pseudosynapse between hepatocytes and KCs.
Similar content being viewed by others
Introduction
Alcohol-related liver disease (ALD) has emerged as a major public health issue due to increasing global alcohol consumption1,2. While the majority of heavy drinkers develop alcohol-related fatty liver (AFL), approximately 20% of AFL patients progress into alcohol-related steatohepatitis (ASH) through mechanisms that remain incompletely understood. ASH is characterized by ballooned hepatocytes with fat droplets, Mallory body formation, and neutrophil infiltration3 and requires active medical intervention due to high short-term mortality and development of cirrhosis1,2. Thus, elucidating the mechanisms underlying the transition from AFL to ASH is of critical importance.
Kupffer cells (KCs), the liver’s resident macrophages, adhere to liver sinusoidal endothelial cells (LSECs) where they phagocytose gut-derived pathogens, and communicate with hepatocytes (HEPs) via intercellular mediators through the LSEC fenestrae and the space of Disse4,5. During ASH development, KCs not only respond to elevated portal levels of lipopolysaccharide by generating reactive oxygen species (ROS) through NADPH oxidases (NOXs), but also by secreting pro-inflammatory cytokines and chemokines, including interleukin (IL)−1β, tumor necrosis factor (TNF)-α, and C–C motif chemokine ligand 2 (CCL2) to activate pro-inflammatory immune cells including neutrophils, exacerbating hepatocyte injury and ASH progression4,6. Although C-X-C motif chemokine ligand 1 (CXCL1)-dependent infiltration of neutrophils has been well established as a hallmark of ASH7,8, little is known about how KCs recruit neutrophils during ASH progression.
Glutamate, a non-essential amino acid, plays an important role in hepatic amino acid metabolism9. Recently, glutamatergic signaling has been reported to be critical in the progression of liver diseases10,11. Specifically, increased hepatic glutamate activates metabotropic glutamate receptor 5 (mGluR5) in hepatic stellate cells (HSCs) and natural killer (NK) cells, promoting de novo lipogenesis of adjacent HEPs in AFL10 and inducing apoptosis of activated HSCs in liver fibrosis11, respectively. Other studies have suggested that hepatic mGluR5 blockade offers protection against acetaminophen-induced ROS production and high-fat diet-mediated hepatic steatosis in mice12,13. Additionally, glutamate-mediated cellular signaling has also been observed in other non-neuronal cells, including pancreatic islet cells14 and osteoclasts15. As an excitatory neurotransmitter, the storage, release, and uptake of glutamate are tightly regulated by vesicular glutamate transporters (VGLUTs)16 and excitatory amino acid transporters (EAATs)17. However, the regulation and role of glutamate by VGLUTs and EAATs in the liver have not been extensively investigated particularly during ASH development.
Thus, we aimed to investigate the mechanism of glutamate-mediated cellular interaction in ASH. We demonstrate that chronic alcohol consumption induces the accumulation of hepatic glutamate through VGLUT3, whose expression is upregulated by the aryl hydrocarbon receptor (AHR). We also elucidate the mechanism by which additional binge drinking triggers glutamate release, activating mGluR5 of KC in the vicinity and leading to NOX2-mediated ROS production and neutrophil recruitment. In addition, we show that alcohol-induced morphological changes in hepatocytes facilitate their glutamatergic interaction with neighboring KCs through the formation of a pseudosynapse. These findings provide clear insights into the glutamatergic signaling during ASH progression.
Results
Chronic-plus-binge ethanol drinking stimulates KC activation in ASH
To investigate the precise mechanism of ASH progression caused by alcohol consumption, various ethanol concentrations were exposed to either primary HEPs or mice. This study utilized three in vivo models: acute ethanol intake (4 g/Kg EtOH, Binge), 2-week ethanol consumption (4.5 % liquid EtOH diet for 2 weeks, 2w-EtOH), and 2-week ethanol feeding plus a binge (2w-EtOH + Binge), which resembles acute on chronic conditions similar to those seen in ASH patients18 (Fig. 1a). In vitro 6-h exposure of 20 to 100 mM EtOH concentrations to isolated HEPs did not change medium levels of alanine aminotransferase (ALT), aspartate aminotransferase (AST) and lactate dehydrogenase (LDH), whereas blood EtOH concentration (approximately 15 mM) by single binge intake significantly increased serum ALT levels in vivo (Fig. 1b and Supplementary Fig. 1a). Additionally, the absence of liver damage after 2w-EtOH feeding appears to be due to the lesser effect of the toll-like receptor 4 (TLR4) and lipopolysaccharide (LPS) signal, but 2w-EtOH + Binge resulted in significantly greater liver injury than a single binge (Fig. 1b). These data confirmed the synergistic effects of chronic low-dose ethanol consumption on single binge-induced liver injury18, hence suggesting that alcohol-induced liver injury may be affected by other cells such as neutrophils rather than ethanol itself19.
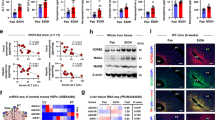
a Schematic representation of different ethanol (EtOH) treatments applied to WT mice and hepatocytes (HEPs). b Measurement of ALT in the cultured media of HEPs treated with EtOH for 6 h (left, n = 3/group). Serum ALT and EtOH levels from in vivo experiments (right, n = 6/group). c–g Analysis parameters in time-course tracing of acute on chronic EtOH-fed mice, comparing each timepoint after binge alcohol administration relative to the EtOH only (0 h). c Evaluation of serum ALT, AST, and TG in mice with a 2-week EtOH diet plus binge drinking (n = 4/group). d Representative staining of liver sections with H&E solutions and antibodies of Ki-67, MPO and CLEC4F. Scale bar, 50 μm. e Bar and linear graphs or flow cytometry panels showing frequencies of liver mononuclear cells (MNCs) and neutrophils in liver (n = 4/group). f Quantification of the number of CLEC4F+ Kupffer cells (KCs) per HEPs by immunostaining (n = 12/group), as well as their frequency in flow cytometry and counts per liver weight (n = 4/group). g mRNA expression of Cxcl1 and Ccl2 in isolated HEPs and Cxcl1 (n = 5/group), Il1b, Il6, Cybb, and Grm5 in isolated KCs (n = 4/group). Statistical comparisons were made using one-way ANOVA with Tukey’s multiple comparisons. Data are presented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001. Source data are provided as a Source Data file.
Besides hepatic E-selectin- or CXCL1-mediated neutrophil migration by binge drinking7,19, inflammatory liver injury after acute-on-chronic alcohol consumption may have been deteriorated by unknown factors. To clearly dissect this conundrum, we investigated the kinetics of liver injury, regeneration, and changes of inflammatory cells time-dependently in 2w-EtOH + Binge (Fig. 1a, c). After binge, serum levels of ALT and AST peaked at 6 h (Fig. 1c). However, the number of binucleated or Ki-67-positive HEPs was significantly increased at 3 h but slightly decreased at 6 h (Fig. 1d). Moreover, immunostaining and flow cytometry revealed that hepatic migration of MPO-positive neutrophils (Ly6G+CD11b+ cells) was significantly increased18 (Fig. 1d, e). Unexpectedly, CLEC4F+ cells in liver tissues and F4/80highCD11bintCLEC2+TIM4+ cells identified through flow cytometry were recognized as KCs (Fig. 1d and Supplementary Fig. 1b, c). These KCs gradually disappeared from the perivenous areas at 3 h following binge drinking (Fig. 1d, f). The frequencies of liver macrophages (referred to as F4/80intCD11bhigh cells) and eosinophils (SiglecF+CD11b+ cells) were reduced after binge drinking (Supplementary Fig. 1d). However, a single binge drinking without chronic alcohol consumption did not change the frequency and characteristics of CLEC4F+ KC (Supplementary Fig. 1e). In qRT-PCR analyses, in addition to HEPs, the gene expression of Cxcl1, along with inflammation-related genes (Il1b, Il6, and Cybb) and glutamate receptor gene (Grm5) in KCs, were sharply increased only at 1 h post-binge and then quickly decreased at 3 h (Fig. 1g). Among other chemokines involved in neutrophil migration, although the expression level of Cxcl2 was lower than that of Cxcl1, it showed a significant increase at 1 and 3 h in KCs, with a slight rise in HEPs at 3 h. Conversely, the expression levels of Cxcl3 and Cxcl5 were undetectable in HEPs and only weakly elevated in KCs (Supplementary Fig. 1f). However, these changes were not observed in vitro EtOH treatment to KC (Supplementary Fig. 1g, h). Collectively, these data suggest that activation of KCs is rapidly induced at 1 h after binge in chronic alcohol consumption, preceding their disappearance, which may subsequently promote hepatocyte damage and neutrophil migration. Additionally, the rapidly upregulation of mGluR5 expression in KCs implies elevated hepatic glutamate release immediately after binge.
Perivenous HEPs execute glutamate synthesis, uptake and storage after chronic alcohol consumption
Prompted by the up-regulation of Grm5 in KCs, we measured the change of glutamate level in blood and liver after 2w-EtOH feeding. Higher plasma glutamate level was reported in AFL patients than healthy subjects, with the highest level in ASH patients10. Similarly, glutamate concentration was slightly increased in both sera and livers of 2w-EtOH-fed mice compared to those of isocaloric diet (Pair)-fed mice. While hepatic glutamate level was notably decreased in mice fed with 2w-EtOH + Binge, serum glutamate was increased compared to that of 2w-EtOH (Fig. 2a), suggesting hepatic glutamate release after binge. To explore the source of hepatic glutamate, we first analyzed bulk RNA sequencing (RNA-seq) of whole liver tissues of Pair- and 2w-EtOH-fed mice and found several enriched gene sets related to glutamate transporters and transaminases in 2w-EtOH-fed mice (Fig. 2b). Of note, not only glutamate synthesis-related genes such as Aldh4a1 and Oat10, but also glutamate transporter-related genes including Slc17a8 (encoding VGLUT3) and Slc1a2 (encoding EAAT2) were significantly increased in 2w-EtOH group (Fig. 2b and Supplementary Fig. 2a, b). To clearly demonstrate hepatic zone-specific expression of these genes, we evaluated public single-cell RNA-seq (scRNA-seq) data of mouse liver20, and confirmed the level of glutamate-related proteins and mRNA using liver tissues and isolated primary HEPs from 2w-EtOH-fed mice (Fig. 2c–f). Intriguingly, several expressions of genes related to glutamate synthesis (e.g. Aldh4a1, Oat), vesicular (Slc17a8) and membrane-bound (Slc1a2) glutamate transporters were mainly expressed in the perivenous HEPs (Fig. 2c). Both protein and mRNA levels of ALDH4A1, OAT, VGLUT3, and EAAT2 remarkably increased and expanded their expression from perivenous to mid-zonal HEPs of EtOH-fed mice compared to Pair-fed mice (Fig. 2d–f and Supplementary Fig. 2c, d). These expressions were exactly co-localized with EtOH-mediated cytochrome P4502E1 (CYP2E1) in HEPs, suggesting the involvement of EtOH metabolism on the expression of Aldh4a1, Oat, Slc17a8, and Slc1a2.
a Glutamate concentrations in serum and liver from Pair-fed and EtOH-fed WT mice with/without binge drinking (n = 8/group). b RNA-sequencing analysis of the whole liver tissues from WT mice fed with isocaloric (Pair) or EtOH diet for 2 weeks (n = 3/group). Graphical representation and heatmaps about pathways of glutamate production and transportation. c Relative mRNA expression of glutamate transporters according to the liver zonation in single-cell RNA (scRNA)-sequencing of WT mouse HEPs (GSE84498). d Representative immunofluorescence staining of VGLUT3, EAAT2, OAT, and ALDH4A1 in liver sections merged with DAPI. Central vein (CV) and portal triad (PT). Scale bar, 50 μm. e Western blot analysis in isolated HEPs from WT mice. f Relative mRNA expression of Aldh4a1, Oat, Slc1a2, and Slc17a8 in the whole liver tissues from EtOH-fed mice and EtOH-treated HEPs compared to control (n = 4/group). g Immunostaining of VGLUT3 after liver tissue expansion by the epitope-preserving magnified analysis of proteome protocol. Scale bar, 100 μm. h Schematic protocol of isolating microvesicles (MVs) by stepwise centrifugation. i, j Representative Western blot analysis and glutamate concentrations in isolated MVs from Pair- or EtOH-fed WT mice (n = 6/group). Statistical comparisons were made using one-way ANOVA with Tukey’s multiple comparisons test (a) or two-tailed unpaired t-test (f, j). p < 0.05 was considered statistically significant. Data are presented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001. Source data are provided as a Source Data file.
VGLUT3 immunostaining showed increased expression upon EtOH exposure, but it was hard to clearly identify VGLUT3 vesicles (Supplementary Fig. 2c). Using a super-resolution imaging method by expanding liver tissues with the epitope-preserving magnified analysis of proteome protocol21,22, we observed a precise distribution of VGLUT3 vesicles (Fig. 2g). The results revealed that VGLUT3 is expressed in perivenous HEPs after EtOH consumption (Upper panel), and VGLUT3 is more densely clustered at the basolateral membrane (Lower panel). However, VGLUT3 vesicles completely disappeared after binge (Fig. 2g). Next, to clarify the level of glutamate in VGLUT3 vesicles16, we isolated microvesicles (MVs) from HEPs in stepwise centrifugation (Fig. 2h). First, we verified that only VGLUT3 was expressed in isolated MVs from HEPs compared to VGLUT1 and VGLUT2 levels in brain (Fig. 2i and Supplementary Fig. 2e, f), with significantly higher glutamate level in 2w-EtOH-fed MVs compared to Pair-fed MVs (Fig. 2j). These data suggest that chronic alcohol consumption induces the storage of glutamate in VGLUT3 vesicles at perivenous HEPs, which could be released by binge drinking.
NRF2 and AHR up-regulate VGLUT3 and EAAT2 expression in perivenous HEPs
Based on the co-localization of elevated VGLUT3 and EAAT2 levels, we assessed protein-protein interactions of L-glutamate transporter activity (GO: 0005313) through the STRING database. Increased Slc17a8 (VGLUT3) and Slc1a2 (EAAT2) showed direct interaction pathways (Fig. 3a). Moreover, the conserved promoter region of VGLUT3 in mouse strains comprises the putative binding sites for the AHR transcription factor23. Similarly, EAAT2 expression was strongly up-regulated by endogenous AHR agonist, 2-(1′H-indole-3′-carbonyl)-thiazole-4-carboxylic acid methyl ester (ITE) in mouse astrocytes24, indicating the role of AHR in the induction of VGLUT3 and EAAT2. Concordantly, when we re-evaluated AHR ligand-stimulated genes in mouse lung fibroblasts25, Slc1a2 was found to be one of the most highly induced genes (Fig. 3b). Additionally, our previous RNA-seq analysis also revealed that along with increased expression of Ahr and Nfe2l2, that are mutually regulated26 and inducible by alcohol intake10,27, representative AHR- and nuclear factor erythroid-2-related factor 2 (NRF2)-related genes were up-regulated (Fig. 3c and Supplementary Fig. 3a, b). Furthermore, NRF2- and AHR-related genes were highly enriched in perivenous HEPs (Fig. 3d), and the protein levels of NRF2 and AHR were significantly increased and co-stained with each other after chronic EtOH intakes at perivenous HEPs (Fig. 3e, f). These findings imply that VGLUT3 and EAAT2 expression may be regulated by NRF2 and AHR.
a Protein-protein interactions of L-glutamate transporter activity (GO:0005313) in bulk RNA-seq data of mouse livers through the STRING database. b Top 10 up-regulated genes stimulated by AHR agonists (TCDD, ITE) in mouse lung fibroblast, re-analyzed using public microarray data. c mRNA expression of NRF2 and AHR-related genes and AHR target genes from RNA-seq analysis of WT mice fed with Pair or EtOH diet for 2 weeks (n = 3/group). d Relative mRNA expression of AHR-related genes according to liver zonation in scRNA-seq of WT mouse liver (GSE84498). e Western blot of NRF2 and AHR in isolated HEPs. f Representative immunofluorescent staining of AHR and NRF2 in liver sections. Scale bar, 50 μm. g, h mRNA expression of Nfe2l2, Slc1a2, and Slc17a8 in freshly isolated HEPs treated with 20 mM EtOH and N-acetyl-L-cysteine (NAC) for 24 h (g, n = 4/group), along with their Western blot analysis of NRF2, VGLUT3, and EAAT2. i In situ closed liver perfusion performed with VEH, 100 mM EtOH, or 1 μM AHR agonist (ITE) containing media in WT mice (2 h). Relative mRNA (n = 6/group) and protein levels of Ahr (AHR), Nfe2l2 (NRF2), Slc17a8 (VGLUT3), and Slc1a2 (EAAT2) in isolated HEPs. j Representative immunofluorescence staining of VGLUT3 and EAAT2 in liver after in situ closed perfusion. Scale bar, 50 μm. Statistical comparisons were made using one-way ANOVA with Tukey’s multiple comparisons test. Data are presented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001. Source data are provided as a Source Data file.
To further demonstrate above findings, isolated HEPs were exposed to EtOH in the presence or absence of anti-oxidant N-acetyl-L-cysteine (NAC) to alleviate ethanol-induced oxidative stress, which acts as an activator of NRF2. In qRT-PCR and Western blot, EtOH treatment significantly increased mRNA and protein levels of NRF2, VGLUT3, and EAAT2, while NAC treatment reduced their expression levels dose-dependently (Fig. 3g, h). Next, after confirming the elevated levels of AHR ligands such as tryptophan (Trp), kynurenine (Kyn), and indole-3-acetic-acid (IAA) in sera of 2w-EtOH-fed mice (Supplementary Fig. 3c), AHR activation by ITE in isolated HEPs also increased the expressions of Nfe2l2, Ahr, Slc17a8, and Slc1a2 in dose- and time-dependent manners (Supplementary Fig. 3d, e). We further tested the effects of EtOH and AHR activation on the expression of VGLUT3 and EAAT2 in vivo using in situ liver perfusion system28. AHR activation by ITE in combination with EtOH synergistically up-regulated the mRNA and protein levels of NRF2, AHR, VGLUT3, and EAAT2 compared to isolated HEPs treated with EtOH alone (Fig. 3i and Supplementary Fig. 3f). In immunostaining, VGLUT3, EAAT2, NRF2, and AHR were synergistically increased at the perivenous HEPs by EtOH and ITE (Fig. 3j and Supplementary Fig. 3g). Furthermore, as pharmacologic inhibition of VGLUT3 significantly attenuated liver injury (Supplementary Fig. 4). These data indicate that upon EtOH exposure, the NRF2-AHR signaling pathway up-regulates the expression of VGLUT3 and EAAT2 in HEPs, and that VGLUT3 plays a crucial role in alcohol-related liver injury.
Glutamate released from ballooned hepatocytes triggers mGluR5-mediated ROS production in KCs
Given the significance of VGLUT3 in the above findings, we investigated the mechanisms underlying the processes of VGLUT3 exocytosis and glutamate release. The soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptor (SNARE) complex mediates calcium (Ca2+)-dependent exocytosis of glutamate from VGLUT3 vesicles in astrocytes29,30, while chronic alcohol consumption enhances mitochondrial matrix Ca2+ level and hormone-mediated Ca2+ signaling in rat HEPs31,32. Thus, we explored the changes in intracellular Ca2+, as well as SNARE complex, for the exocytosis of hepatic glutamate vesicles. In RNA-seq analysis, 2w-EtOH feeding increased the gene expressions of Ca2+ channels and transporters33,34 in plasma (NCX, VDCC, TRPC) and mitochondrial (MCU) membranes, whereas those in ER (SERCA, RyR, IP3R) showed no differences (Fig. 4a). In addition, elevated expression of MCU-related genes decreased after additional binge drinking (Supplementary Fig. 5a). Further, we investigated the change in hepatic Ca2+ level. Using the intravital liver imaging technique and Fluo-4 AM (fluorescent Ca2+ indicator)35, we observed that both the number and intensity of Fluo-4 AM+ HEPs were significantly increased in perivenous HEPs of 2w-EtOH-fed mice, while binge drinking immediately decreased intracellular Ca2+ level in HEPs within 15 min (Fig. 4b, Supplementary Fig. 5b and Supplementary Movie 1–3). However, there was no prominent change in Ca2+ level in HEPs of Pair-fed mice (Supplementary Fig. 5c and Supplementary Movie 4). In line with these results, the expression of SNARE complex assembly-related genes was increased in 2w-EtOH-fed mice, whereas its disassembly-related genes were up-regulated in 2w-ETOH + Binge (Fig. 4c). Accordingly, in vitro high-dose EtOH treatment significantly raised medium level of glutamate in HEPs isolated from EtOH-fed mice compared to those of Pair-fed mice (Fig. 4d). Together, these data suggest that the SNARE complex assembly and intracellular Ca2+ change release glutamate from VGLUT3+ vesicles from HEPs into the extracellular space upon binge EtOH exposure.
a A schematic figure of Ca2+ channels and transporters in HEPs. Heatmaps of genes in Pair and EtOH group (n = 3/group). b Representative intravital liver imaging stained with Fluo-4 AM and lectin. Statistical analysis of Fluo-4 AM-positive cells per image field (n = 4/group). Scale bar, 100 μm. c Heatmaps showing relative mRNA expression of SNARE complex-related genes analyzed by RNA-sequencing before and after binge drinking in EtOH-fed mice (n = 3/group). d Glutamate concentration in the supernatant after in vitro additional EtOH treatment of HEPs isolated from 2-week Pair-fed and EtOH-fed mice (n = 3/group). e Flow cytometry analyses of mGluR5 expression and ROS generation in isolated KCs. f ROS generation and gene expression of Grm5, Cybb, and Cxcl1 in KCs treated with mGluR5 selective agonist (CHPG) or monosodium glutamate (MSG) compared to vehicle (n = 4/group). g Size distribution analysis of HEPs isolated from Pair- or EtOH-fed mice. Bar graph shows the average diameter of HEPs in each group (n = 6/group). h Transmission electron microscopy in liver section fed with Pair or EtOH diet for 2 weeks. Bar graph indicates the average distance between adjacent hepatocytes (HEPs) and Kupffer cells (KCs), measured across multiple cell pairs per mouse (n = 12 HEP–KC pairs/group). Scale bar, 5 μm. i Co-culturing of KCs directly or indirectly with HEPs isolated from EtOH-fed mice, followed by treatment with 50 mM EtOH for 6 h. Relative mRNA expression of Cybb, and Il1b in isolated KCs (n = 6/group). LDH was measured in the supernatant (n = 4/group). Statistical comparisons were made using one-way ANOVA with Tukey’s multiple comparisons test (b, i), and multiple comparisons by two-stage step-up method of Benjamini, Krieger and Yekutieli. F (d), or two-tailed unpaired t-test (f–h). Data are presented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001. Source data are provided as a Source Data file.
Next, we questioned the possible effects of hepatic glutamate on mGluR5 in neighboring KCs (Supplementary Fig. 5d), and found that the mGluR5 expression was increased by 2w-EtOH feeding, and ROS production was significantly enhanced in KCs at 1 h after binge (Fig. 4e). In vitro stimulation of mGluR5 by CHPG or monosodium glutamate (MSG) increased ROS production, as well as the expression of Grm5, Cybb, and Cxcl1 in KCs (Fig. 4f and Supplementary Fig. 5e, f), suggesting hepatocyte injury by NADPH oxidase 2 (NOX2)-mediated oxidative stress and CXCL1-derived neutrophil migration. Then we asked how hepatic glutamate stimulates mGluR5 in KCs. Hepatocyte ballooning is a specific phenotype in ASH3, where the plasma membrane of hepatocyte is highly permeable to water due to intracellular amino acid accumulation (Na+-dependent amino acid transport, e.g. EAAT) or osmotic stress (Na+/K+ pump)36. Despite unchanged expression of Na+/K+ pump-related genes (Supplementary Fig. 5g), enhanced EAAT2 expression indicated high osmolarity in HEPs due to glutamate accumulation from EtOH consumption (Fig. 3h, j). In addition, alcohol-mediated alteration of the hepatocyte cytoskeleton promotes swelling37, which was confirmed in our gene expression profile. Expression of genes related to tubulin was decreased whereas that of filament (actin and keratin)-related genes was considerably up-regulated in EtOH-fed mouse liver (Supplementary Fig. 5g). When the volume of hepatocytes was measured in culture36, the diameter of isolated EtOH-fed HEPs was significantly increased compared to those of Pair-fed HEPs (Fig. 4g). Strikingly, electron microscopy revealed ballooning in perivenous HEPs including storage of fat droplets, dilated mitochondria, and loss of microvilli, inducing close contact of HEPs with neighboring KCs in EtOH-fed liver (Fig. 4h). KCs are usually located in the sinusoidal space attached to the endothelial cells with clustering of fenestrae in sieve plates5. Therefore, ballooned HEPs are able to form pseudosynaptic clefts with KCs through the endothelial fenestrae. This spatial proximity could effectively activate mGluR5 in KCs by released glutamate. To demonstrate this concept, we co-cultured KCs with EtOH-fed HEPs in the presence of 20 mM EtOH. Direct co-culture of KCs with HEPs showed increased expression of Cybb, and Cxcl1, which induced further release of lactate dehydrogenase (LDH) from damaged HEPs into media than indirectly co-cultured KCs (Fig. 4i). Together, these data indicated that mGluR5 activation in KCs might be facilitated by glutamate release through the pseudosynapse formation between ballooned HEPs and KCs.
Genetic and pharmacologic inhibition of mGluR5 in KCs attenuates ASH
To clarify the role of mGluR5 in KCs, we generated KC-specific mGluR5 depleted murine model (Supplementary Fig. 6a) using Clecf4-cre or Lyz2-cre mice38. After confirming up-regulation of mGluR5 in Grm5fl/fl KCs by 2w-EtOH intake (Supplementary Fig. 6b), Grm5fl/fl crossed with Clecf4-cre (KCΔGrm5) mice were subjected to chronic-plus-binge drinking. Regardless of the glutamate concentration, serum levels of ALT and AST were decreased with the hepatic frequency of neutrophils in KCΔGrm5 mice compared to those of controls (KCfl/fl) (Fig. 5a, b). Accordingly, perivenous hepatic injuries were notably improved in KCΔGrm5 mice, although EtOH-mediated ALDH4A1 and CYP2E1 expressions were not different in both mice (Fig. 5c). In parallel, isolated KCs from KCΔGrm5 mice exhibited much less expression of Cxcl1, Il1b, and Cybb mRNAs and ROS production than those of KCfl/fl mice (Fig. 5d, e). Next, we further explored the effect of mGluR5 in KCs on HEPs. Medium LDH level was increased when co-cultured with KCs of KCfl/fl mice but not with those of KCΔGrm5 mice even though EtOH exposure induced similar glutamate release in media (Fig. 5f). In addition, KCs of KCΔGrm5 mice showed decreased expression of Cxcl1 and Cybb (Fig. 5g).
a Measurement of serum ALT, AST, TG, and glutamate levels in KCfl/fl and KC-specific mGluR5 knockout mice (KCΔGrm5) fed with 2-week EtOH plus binge drinking (n = 6/group). b Representative flow cytometry panels and bar graphs indicate frequencies of the neutrophils, macrophages, and eosinophils of liver MNCs from KCΔGrm5 mice compared to KCfl/fl (n = 6/group). c Representative H & E and immunostaining of ALDH4A1 and CYP2E1 in liver sections from KCfl/fl and KCΔGrm5 mice. Scale bar, 50 μm. d Relative mRNA expression of Cxcl1, Ccl2, Il1b, and Cybb in isolated KCs from KCfl/fl and KCΔGrm5 mice (n = 4/group). e ROS generation in isolated KCs from KCfl/fl and KCΔGrm5 mice (n = 4/group). f Protocol schematic of in vitro co-culture system of HEPs and KCs. Measurement of glutamate (n = 4/group) and LDH in co-culture media (n = 3/group). g Relative mRNA expression of Cxcl1 and Cybb in co-cultured KCs (n = 4/group). h Measurement of serum ALT, AST, TG, and glutamate levels following intraperitoneal injection of VEH or mGluR5 antagonist (MPEP; 10 mg/kg) for three consecutive days prior to sacrifice (n = 5/group). i Representative H&E stains in liver sections. Scale bar, 50 μm. j ROS generation in isolated KCs from VEH or MPEP-treated mice. k Relative mRNA expression related to inflammatory response in isolated KCs (n = 4/group) from MPEP-treated mice. Statistical comparisons were made using one-way ANOVA with Tukey’s multiple comparisons test (f) or two-tailed unpaired t-test (a, b, d, g, h, k). Data are presented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001.
We also confirmed similar results using myeloid cell-specific mGluR5 KO (MyelΔGrm5) mice crossed with Lyz2-cre mice. CHPG-induced mGluR5 protein level and Cybb expression were absent or unchanged in Myelfl/fl and MyelΔGrm5 neutrophils unlike KCs (Supplementary Fig. 6c, d). MyelΔGrm5 mice exposed to 2w-EtOH + Binge showed alleviated liver injury and reduced hepatic infiltration of neutrophils compared to Myelfl/fl mice (Supplementary Fig. 6e–g). Isolated KCs from MyelΔGrm5 mice showed a considerable decrease of Cybb, Cxcl1, Ccl2, and Il1b expression with no appreciable effects on HEP injury (e.g. lower LDH levels) compared to KCs of Myelfl/fl mice (Supplementary Fig. 6h, i).
Next, we investigated whether pharmacologic inhibition of mGluR5 attenuates ASH progression. Treatment of MPEP, a selective mGluR5 antagonist, serologically and histologically improved ASH (Fig. 5h, i). In addition, isolated KC from MPEP-treated mice showed less ROS production and gene expression of Cybb, Cxcl1, and Il1b than those of controls (Fig. 5j, k). Together, these data indicate that mGluR5 inhibition in KCs attenuates the development of ASH through ROS production and inflammatory responses.
mGluR5 induces NOX2 activation in KCs
mGluR5 activation stimulates protein kinase C (PKC)39 and PKC activates NOX2 to produce ROS in monocytes/macrophages40. Thus, we explored NOX2-mediated ROS and its role in ASH using mice with KC-specific depletion of NOX2 (KCΔCybb and MyelΔCybb). Similar to KCΔGrm5 mice, EtOH-mediated liver injury was significantly attenuated in KCΔCybb and MyelΔCybb mice at perivenous area compared to their controls (Fig. 6a, b and Supplementary Fig. 7a, b). In parallel, hepatic frequencies of neutrophils and macrophages were decreased and the expression of pro-inflammatory genes such as Cxcl1, Il1b, and Tnf were down-regulated in KCs of KCΔCybb and MyelΔCybb mice (Fig. 6c, d and Supplementary Fig. 7c). Neutrophils play critical roles in liver injury and inflammation in ASH19. Thus, we tested whether the conditioned media (CM) from co-cultured HEPs and KCs stimulate neutrophils. CM from co-cultured WT KCs up-regulated the expression of Tnf, Il1b, and Elane in neutrophils, whereas CM from mGluR5-deficient KC did not (Fig. 6e). These data suggested that mGluR5 stimulation activates NOX2 in KCs, which further recruited and facilitated neutrophils to induce hepatocyte injury.
a Measurement of serum ALT levels in KC from myeloid cell-specific Cybb knockout mice (KCΔCybb and MyelΔCybb) fed with 2-week EtOH plus binge drinking (n = 6/group). b Representative H&E stains in liver sections. Scale bar, 50 μm. c Representative flow cytometry panels (left) and bar graphs (right) showing frequencies of neutrophils in liver MNCs from Cybb knockout mice (KCΔCybb and MyelΔCybb) compared to floxed mice (n = 6/group). d mRNA expression levels of Cxcl1, Il1b and Tnf in isolated KCs (n = 4/group). e Generation of conditioned media (CM) by co-culturing WT HEPs with KCΔfl/fl or KC ΔCybb with 50 mM EtOH for 24 h, followed by culturing isolated neutrophils with CM for 12 h and analyzing Tnf, Il1b, and Elane mRNA expression (n = 4/group). f Relative mRNA expressions of Prkca, Prkcb, and Prckg in isolated KCs from KCΔCybb and KCΔGrm5 mice (n = 4/group). g Gene ontology analysis and mRNA expression of protein kinase C (PKC)-related genes in KCs from Myelfl/fl and MyelΔCybb mice. Green dots indicate the PKC-related gene sets. h mRNA expression of Prkca, Prkcb, and Prckg in isolated KCs treated with 100 μM MSG (30 min) or 10 μM CHPG (10 min) (n = 4 /group). i Measurement of ALT levels following intraperitoneal injection of VEH or PKC inhibitor (PKCi; chelerythrine chloride; 10 mg/kg) for 3 consecutive days before sacrifice (n = 7/group). j, k Representative of ROS generation and mRNA expression in isolated KCs from VEH or PKCi-treated mice (n = 4/group). l H&E and immunostaining of ALDH4A1 and CYP2E1 in liver sections. Scale bar, 50 μm. m Western blot analysis for the downstream signaling pathways of mGluR5 in isolated KCs from KCfl/fl, KCΔGrm5, and KCΔCybb mice. Statistical comparisons were made using one-way ANOVA with Tukey’s multiple comparisons test (e), multiple comparison by two-stage step-up method of Benjamini, Krieger and Yekutieli. F (d, f, h), or unpaired t-test (a, c, i, k). Data are presented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001. Source data are provided as a Source Data file.
We further investigated whether mGluR5 activates NOX2 through PKC in KCs. In qRT-PCR analysis, we found that not only mGluR5-depleted KCs (KCΔGrm5 and MyelΔGrm5) but also NOX2-depleted KCs (KCΔCybb and MyelΔCybb) showed significantly decreased expression of Prkca, Prkcb, or Prkcg mRNA levels (Fig. 6f and Supplementary Fig. 7d). Similarly, in bulk RNA-seq analysis of KCs from Myelfl/fl and MyelΔCybb mice, most mRNA levels of conventional PKC genes (Prkca, Prkcb, and Prkcg) were considerably down-regulated in KCs of MyelΔCybb mice (Fig. 6g and Supplementary Fig. 7e). In addition, CHPG or MSG-mediated mGluR5 activation in KCs increased PKC-related gene expressions (Fig. 6h), suggesting the regulatory roles of PKC in mGluR5-mediated NOX2 activation. In vivo, pharmacologic inhibition of PKC (PKCi; chelerythrine chloride) showed a significant decrease in serum ALT level and ROS production in KCs (Fig. 6i, j). qRT-PCR revealed that PKCi decreased mRNA levels of Cybb, Cxcl1, and Il1b, whereas Grm5 expression was not affected in KCs (Fig. 6k). Additionally, isolated KCs treated with PKCi and MPEP suppressed PKC expression (Supplementary Fig. 7f). In H&E and immunostainings, liver injury was substantially attenuated and NOX2 expression was prominently decreased in CLEC4F-positive KCs in PKCi-treated mice (Fig. 6l and Supplementary Fig. 7g). Besides PKC, mGluR5 activates other signaling pathways such as ERK or PI3K/AKT41 and ROS generated by alcohol intake stimulates ERK activation and TNF-α production in KCs42. Similarly, decreased phosphorylation of PI3K and unchanged phosphorylation of AKT were observed in both KCs of KCΔGrm5 and KCΔCybb mice compared to their controls, whereas phosphorylation of ERK1/2 was notably decreased only in KCs of KCΔGrm5 with lower levels of NOX2 protein (Fig. 6m), suggesting mGluR5-mediated ERK1/2 activation in KCs. Collectively, these results indicated that mGluR5 activation is closely associated with PKC/NOX2-mediated ROS production and ERK-mediated inflammatory responses in KCs.
Glutamate and VGLUT3 correlate with ASH progression in patients
Prompted by the above observations in mice, we investigated the clinical significance and relationship of glutamate and VGLUT3 in patients with ALD. As shown in Supplementary Table 1, we classified ALD patients with or without liver cirrhosis (LC) and compared the plasma glutamate levels among ALD patients without LC, as there was no difference between healthy controls and ALD patients with LC10. Glutamate level was significantly elevated in ASH patients compared to non-ASH patients (Fig. 7a). Importantly, plasma glutamate level was positively correlated with levels of ALT, AST, and GGT in patients without LC, and among them, patients with higher steatosis grade or ductular proliferation showed higher level of glutamate than their controls (Fig. 7b). These data suggest that elevated glutamate levels in ALD patients may be associated with liver injury and inflammation. To clarify whether elevated plasma glutamate is a cause of ALD or a result of liver damage, we further examined the expression of glutamate-related genes in samples of AFL patients. In parallel with the results in mice, mRNA expressions of SLC17A8, SLC1A2, AHR, and NFE2L2 were increased in patients with AFL compared to the controls (Supplementary Fig. 8a and Supplementary Table 2), which were dramatically correlated with each other in patients with AFL (Supplementary Fig. 8b). Consistently, the expressions of CYP2E1, NRF2, AHR, and EAAT2 were considerably increased in perivenous HEPs of AFL and ASH livers compared to controls, whereas elevated VGLUT3 expression was slightly decreased in HEPs of ASH liver compared to HEPs of AFL (Fig. 7c), suggesting release of glutamate vesicles in ASH liver. Moreover, SLC17A8, AHR, and NFE2L2 mRNAs were significantly elevated in freshly isolated human HEPs (hHEPs) after EtOH treatment (Fig. 7d). Furthermore, relative mRNA levels of GRM5, PPKCA, PPKCB, and CYBB in isolated human KCs were significantly increased by exposure to mGluR5 agonists (CHPG or MSG) (Fig. 7e and Supplementary Fig. 8c). All these data indicate that chronic alcohol consumption increases glutamate vesicles and their release may stimulate mGluR5 in KCs to enhance liver injury in ALD patients.
a Strategy for classifying patients with alcohol-related liver disease (ALD) involved in the study. Plasma glutamate levels were measured in ALD patients (n = 16) with and without alcohol-related steatohepatitis (ASH). b Correlative analysis of plasma glutamate levels with AST, ALT, GGT, steatosis grade, and ductular proliferation in ALD patients without liver cirrhosis (LC) (n = 16). Correlations were assessed by linear regression model. c Representative H&E and immunostaining of CYP2E1, NRF2, AHR, EAAT2, and VGLUT3 in control, alcohol-related fatty liver (AFL), and ASH patients. Scale bar, 50 μm. d Relative mRNA expression of SLC17A8 (n = 3/group), SLC1A2, AHR, and NFE2L2 in isolated human hepatocytes (hHEPs) treated with 20 mM EtOH for 24 h (n = 6/group). e Relative mRNA expression of GRM5, CYBB, PRKCA, and PRKCB in isolated human Kupffer cells (hKCs) treated with 10 μM CHPG for 10 min (n = 3/group). Statistical comparisons were made using two-tailed unpaired t-test. Data are presented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001. Source data are provided as a Source Data file.
Discussion
KCs are the major cell type that promotes ASH by LPS-mediated stimulation of TLR4 after chronic alcohol consumption1. In chronic EtOH feeding plus binge drinking that mimics ASH in humans, massive infiltration of neutrophils plays a decisive role in ASH development through pro-inflammatory cytokines and ROS production7,19. Moreover, HEPs regulate the activation of KCs and neutrophils in ASH by expressing inflammatory mediators including chemokines (e.g. CXCL1), or releasing damage-associated molecular patterns (DAMPs) such as mitochondrial DNA and high mobility group box 143. However, rather than the role of each cell type alone, interactions among KCs, neutrophils, and HEPs may have a stronger impact on ASH development. Here, we unveiled the detailed molecular mechanism of hepatic glutamate vesicle formation and release that drives hepatocyte damage and neutrophil migration through mGluR5/NOX2-mediated ROS production and CXCL1 expression in KCs, respectively. More strikingly, we demonstrated that ballooning of HEPs results in pseudosynapse formation between HEPs and KCs, a prerequisite for mGluR5 activation in KCs.
To relieve the threat of alcohol poisoning, perivenous HEPs quickly detoxify alcohol by alcohol metabolizing enzymes including CYP2E1, but they undergo regeneration due to inevitable HEP damage by alcohol metabolites such as acetaldehyde and ROS1. Interestingly, to compensate for this loss, the liver selects a unique regenerative mechanism rather than repairment. However, it is indispensable that damaged hepatocytes are exactly targeted and quickly eliminated for the homeostasis of hepatic function. An interesting study reported that infused DAMPs released from repeated thawing-induced necrotic cells stimulate TNF-α production in KCs that mediates CXCL1 expression in HEPs for neutrophil mobilization44, suggesting a critical role of resident KCs in the elimination of damaged cells. To our surprise, considering that 17 mM EtOH (0.08 g/dl) is a binge status in human blood45, there was little damage to HEPs even at 100 mM EtOH exposure in vitro. However, we found that co-cultured KCs increased hepatic LDH release and CXCL1 expression after EtOH exposure. This strongly indicated that interaction between them has critical impacts on HEP damage and neutrophil mobilization in ASH development. In line with this, we demonstrated a more precise elimination mechanism, in which injured hepatocytes directly communicate with neighboring resident KCs by releasing glutamate.
Hepatic glutamate is produced and metabolized by diverse pathways depending on the liver zonation9,46. Interestingly, recent studies suggested that blood glutamate concentration positively correlates with inflammatory injury or steatosis in ALD10,47, but it is still unclear where glutamate comes from and how it is released. In our previous report, chronic EtOH intake enhanced ALDH4A1-mediated glutamate production for the compensation of GSH shortage but it suppressed glutamine synthetase specifically in perivenous HEPs10. In addition to up-regulation of ALDH4A1, we found that the expressions of NRF2 and AHR, which cross-regulate various xenobiotic metabolism26,48, were increased to regulate the expression of EAAT2 and VGLUT3 at perivenous HEPs which are co-expressed in murine and human livers of ALD. Moreover, serum level of AHR ligands such as Trp, Kyn, and IAA27 were increased after alcohol intake, and in situ AHR stimulation demonstrated the increased expression of EAAT2 and VGLUT3 in perivenous HEPs, indicating that gut microbiota-mediated AHR ligands might regulate hepatic glutamate metabolism and its neurological signaling. However, if secretion by VGLUT3 and uptake by EAAT2 occur simultaneously in perivenous hepatocytes, mGluR5 activation of KCs at the pseudosynapse may not occur. This is likely because the increase in VGLUT3 expression is much greater than that of EAAT2 (Fig. 2f). As a result, after secretion, sufficient glutamate is released and then reabsorbed by EAAT2. Nonetheless, further study is needed to explore this matter. Regarding to VGLUT3, intriguingly, this study marks the first discovery of the expression and function of VGLUT3 in hepatocytes. Notably, multiple copies (around 10) of either VGLUT1 or VGLUT2 vesicle are present in synaptic vesicles, but a single copy of VGLUT may be sufficient for vesicular glutamate loading49,50. Further study on the VGLUT3 copy number in hepatocytes is also necessary. Furthermore, as intracellular Ca2+ signaling mediates SNARE complex to induce exocytosis of glutamate vesicles in neurons51,52, binge drinking also rapidly changed hepatic intracellular Ca2+ level within 15 min and triggered the exocytosis of glutamate vesicles by regulating the SNARE complex. This in turn caused the disappearance of Ca2+ storage and VGLUT3 vesicles at perivenous HEPs and increased blood level of glutamate immediately after binge (before liver injury). Therefore, if hepatic changes in glutamate vesicles and Ca2+ level can be detected by non-invasive methods, they may be powerful candidates for early diagnostic biomarkers of ALD.
KCs attach to the luminal surface of LSECs and interact with adjacent HEPs through the space of Disse by extending their microvilli into LSEC’s fenestrae5,53. However, the detailed mechanism of cell-to-cell interaction between HEPs and KCs remains unclear in ASH. Ballooning of HEPs is a representative pathological feature instigated by alcohol-mediated disruption of the cytoskeletal structure and membrane Na2+ transporters36,37. Interestingly, in this study, electron microscopy revealed that alcohol-mediated HEP ballooning reduced the distance between HEPs and KCs, facilitating close contact. This results in HEP damage and neutrophil migration by the production of ROS and the expression of TNF-α and IL-1β in KCs. Genetic and pharmacologic inhibitions of VGLUT3 and mGluR5 attenuated KC-mediated HEP damage at the perivenous area. These findings strongly suggest that HEP ballooning is a phenotype shift for cell-to-cell communication and also imply that forming a synapse-like structure is inevitably required to activate mGluR5 in KCs. This topographical proximity may provide further chance for other cell-to-cell interactions through glutamate signaling pathways in different liver diseases. Additionally, hepatic parenchymal or other nonparenchymal cells express various glutamate receptors or transporters10,11. Therefore, further investigation into the unexplored metabolic synapse among hepatic cells is needed, utilizing techniques such as live-cell imaging, fluorescence resonance energy transfer, or proximity ligation assays. Similarly, in the brain, mGluR5 in the processes of astrocytes senses synaptic glutamate release from hippocampal neurons in close proximity54. In adipose tissue, sympathetic nerve-associated macrophages are also closely localized around the neurons to import and degrade norepinephrine by its transporter (SLC6A2) and enzyme (monoamine oxidase A)55. Therefore, non-neuronal cells may also detect and respond to neurotransmitters (e.g. glutamate) through physical contact with neurotransmitter-producing cells. However, the detailed mechanism in the formation of the pseudosynapse needs to be further studied.
In summary, we suggest a unique elimination mechanism of stressed HEPs after alcohol intake, in which perivenous HEPs release glutamate to increase ROS production in neighboring KCs through mGluR5 (Supplementary Fig. 9). Specifically, alcohol-induced ballooning of HEPs creates a ‘pseudosynapse’ that facilitates cell-to-cell interaction with KCs, suggesting the existence of novel metabolic synapse involved in the pathogenesis of ALD. Consistently, our findings were observed in ALD patients, representing a positive correlation between glutamate and inflammatory markers. Therefore, the present study provides insights and molecular targets for the ALD treatment.
Methods
Mice
The Institute Animal Care and Use Committee (IACUC) in Korea Advanced Institute of Science and Technology (KAIST, Republic of Korea) approved all animal experimental protocols. The mice were managed on the regular light-dark cycle (12 h light/12 h dark) in a specific pathogen-free facility (SPF) in KAIST. The temperature and humidity of SPF were maintained at 24 °C and 40–60%. WT mice with a C57BL/6J background were purchased from KAIST SPF facility. The 8-week-old male mice were fed with the Lieber-DeCarli liquid alcohol diet (#710260, Dyets Inc) or control diet for the indicated periods. Alcohol feeding was started 5 days of the acclimatizing period, and the sacrifice was performed 6 h after binge drinking (4 g/kg body weight). Diet intake and changes in body weight were daily monitored.
Clec4f-cre mice (Stock No. 033296) and Grm5fl/fl mice (Stock No. 028626) were purchased from Jackson Laboratory (Bar Harbor, ME). Cybbfl/fl mice (Stock No. EM07096) were purchased from the European Mouse Mutant Archive and LysM-cre mice were kindly provided by Dr. Heung Kyu Lee (KAIST). To generate the KC-specific mGluR5 or CYBB KO mice, Grm5fl/fl mice or Cybbfl/fl mice were crossed and backcrossed with Clec4f-cre or LysM-cre mice, respectively. For pharmacological inhibition of mGluR5 and PKC, the mice were treated with MPEP hydrochloride (#1212, Tocris; 10 mg/kg body weight, i.p., consecutive daily injection for 3 days before sacrifice) and Chelerythrine chloride (#1330, Tocris; 10 mg/kg body weight, i.p., consecutive daily injection for 3 days before sacrifice), respectively.
Human patients
Human samples of plasma and liver tissue in ALD patients according to various disease statuses were provided by Seoul Metropolitan Government Seoul National University Boramae Medical Center (Seoul, South Korea). The present study was approved by the Institutional Review of Board (IRB No. 16-2013-45) and complied with the ethical guidelines of the World Medical Association Declaration of Helsinki. Patients with a drinking history above the excessive amount (>40 g/day in men and 20 g/day in women) were included in the cohort. Next, the patients were divided into 4 subgroups based on the clinical definition (people with excessive alcohol use without liver disease; AWLD, alcohol-related fatty liver; AFL, alcohol-related steatohepatitis; ASH, alcohol-related liver cirrhosis; LC). The exclusion criteria of the patients were as follows; patients with viral liver disease or other diseases related to increased glutamate levels such as HIV infection, cancer, or migraine. The baseline clinical characteristics are described in Supplementary Table 1.
Surgically resected fresh liver tissues from non-tumorous and non-cirrhotic regions of patients without alcohol intake history were provided from a separate cohort of the Department of Surgery, Chungnam National University Hospital (Daejeon, South Korea). Those tissues were used for qRT-PCR or isolation of primary human HEPs and KCs by perfusion with the collagenase solution. Institutional Review of Board in Chungnam National University Hospital authorized the ethical approval to use human samples in research (IRB No. 2016-03-020-030). The standardized informed consents were obtained from all patients involved in this study.
Primary cell cultures
The primary HEPs, KCs, HSCs, and liver MNCs in mice were isolated with differential centrifugation using density gradient reagents such as Opti-prep (Sigma-Aldrich) and Percoll (Sigma-Aldrich) following the previous report10. To briefly summarize the protocols, the portal vein was cannulated with a 22 G angiocatheter (Jelco) and perfused with EGTA solution (0.34 mM Na2HPO4, 140 mM NaCl, 0.44 mM KH2PO4, 5.4 mM KCl, 25 mM Tricine, 0.5 mM EGTA, pH 7.2) to extract blood in the liver. Next, collagenase perfusion buffer (0.075 % of collagenase type I diluted in the HBSS buffer containing DNase I) was infused at 42 °C. After the complete circulation, whole liver was extracted and further digested in the shaking incubator (90 rpm, 20 min) at 37 °C. Digested liver tissues were filtered with cell strainers (70 μm) to remove undigested debris.
Filtered cell suspensions were centrifuged at 50 × g for 5 min to isolate HEPs, and HEPs were purified by further centrifuge with 50% Percoll solution (GE Healthcare). The diameters of HEPs were analyzed by Countess 3 Automated Cell Counters (Thermo Fisher Scientific). To separate hepatic nonparenchymal cells, supernatant solutions were centrifuged at 650 × g for 10 min. Furthermore, 11.5 % and 20% of Opti-prep gradient solution (Sigma-Aldrich) were used to isolate KCs and HSCs by the centrifuge at 1800 × g for 17 min. Isolated cells were purified by flow cytometric sorting with cell-specific characteristics (F4/80highCD11bintCLEC2+TIM4− in KCs and DAPI+ in HSCs)56,57. Additionally, the expression of Clec4f and Timd4 in isolated KCs were confirmed by single-cell RNA sequencing analysis58. On the other hand, the pellet of nonparenchymal cells was suspended in Percoll gradient solution and further centrifuged at 1800 × g for 10 min to isolate liver MNCs. Liver MNCs were subsequently resuspended in saline after lysis of red blood cells.
To isolate primary HEPs and KCs from human liver tissues, non-tumorous and non-cirrhotic regions of surgically resected liver specimens (about 15 g) were provided by the Department of Surgery, Chungnam National University Hospital (Daejeon, South Korea). Visible hepatic vessels in liver specimens were cannulated with an angiocatheter and infused with EGTA and collagenase solutions. The remaining protocols were the same as the isolating methods in mice. For in vitro experiments, isolated cells were seeded in cell culture plates coated with collagen. Cells were experimented in RPMI (#LM-011-01, Welgene) or DMEM (#LM001-11, Welgene) supplemented with 1% penicillin-streptomycin (Thermo Fisher Scientific) and 10% FBS (Welgene), and incubated in a humidified incubator with 5% CO2 at 37 °C. In several in vitro experiments, cultured cells were treated with ethanol (diverse concentrations, Millipore), ITE (0.1 μM or 1 μM, Tocris), N-acetyl-L-cysteine (0.1 mM or 1 mM, Sigma-Aldrich), CHPG (5 μM or 10 μM, Tocris), and monosodium glutamate (20 μM or 50 μM, Sigma-Aldrich) according to the indicating experiments.
Biochemical measurement
Mice serum was obtained from the peripheral blood and analyzed for the measurement of ALT, AST, TG, and TC. The supernatant of co-cultured media was analyzed to evaluate LDH concentration. VetTest Chemistry Analyzer (IDEXX Laboratories) was used for the measurements according to the manufacturer’s instructions.
Histological analyses
To obtain consistent findings from each sample, identical regions of the medial and left lobes of the liver in all mice were used for the histological analyses. Tissue specimens were fixed using 10% neutral buffered formalin (Sigma-Aldrich) overnight. After deparaffinization and rehydration, the tissues were cut with 4 μm thickness and stained with hematoxylin (Sigma-Aldrich) and eosin (Biognost) solution. Light microscopy (Olympus BX51) was used to capture images, and DP2-BSW software was used for imaging analyses. All stained liver tissues were carefully evaluated a pathologist who has a degree of Korean Certificate of Veterinary Pathology.
Immunostaining
Paraffin-embedded sections of liver tissues from mice or human samples were utilized to analyze immunostaining. After deparaffinization and rehydration, the samples were immersed in 10 mM citrate buffer (pH 6.0), and antigen retrieval was performed using the microwave for 5 min. Tissues were incubated in 10% donkey serum for 1 h to block unspecific bindings. Primary antibodies (1:50 ~ 1:200 in PBS containing 0.1 % tween-20) were incubated overnight at 4 °C. For immunohistochemistry stains, samples were incubated with anti-Rabbit IgG (Vector Laboratories) or anti-Mouse IgG (Vector Laboratories) at room temperature for 1 h. Then, DAB substrate kit (Vector Laboratories) was used to develop reactions according to the manufacturer’s instruction, and slides were covered with Balsam (Sigma-Aldrich). For immunofluorescent stains, samples were incubated with Alexa Fluor® 488- or Alexa Fluor® 594-conjugated secondary antibodies at room temperature for 1 h, and covered with DAPI mounding solution (Abcam). Images were captured using Olympus BX51 microscopy with a CCD camera (Olympus, Tokyo, Japan). DP2-BSW software was used for imaging analyses.
Tissue expansion
Mouse liver was sequentially perfused with PBS and 4% paraformaldehyde (PFA) solution in PBS. The liver sample was dissected into smaller lobes. To preserve the tissue, the sample was immersed and gently shaken in 4% PFA solution overnight at 4 °C. The sample was further shaken at room temperature for 12 h. To wash the PFA solution, the sample was immersed in PBS and gently shaken at 4 °C and room temperature for 12 h in each step. Next, the tissue was immersed in eMAP monomer solution (30% acrylamide, 10% sodium acrylate, 0.1% bisacrylamide, and 0.03% VA-044) and incubated overnight at 4 °C for homogenous infiltration of the hydrogel monomers. The tissue with the eMAP monomer solution was moved to a cartridge. Sealing the cartridge with a coverslip and nail polish, the sample underwent gelation at 37 °C for 1 day. The tissue sample was then vibratome-sliced at 150-μm thickness. After immersing the tissue sample in PBS with 0.1% Triton X-100 and 0.02% sodium azide (PBST), it was stained with the primary antibody (1:300 dilution) overnight at 37 °C and then washed with PBST three times for 8 h. Then, the sample was stained with the secondary antibody (1:300 dilution), DAPI, and lectin-DyLight 594 overnight at 37 °C. The sample was washed with PBST three times for 8 h. After washing the sample once with PBS, it was immersed in distilled water (DW) for expansion and prepared for imaging. The sample was imaged for hepatic VGLUT3 using a confocal scanning microscope (Olympus, FV3000) with 10X/0.4-numerical aperture (NA) and 20X/0.8-NA objectives (UPLXAPO10X, UPLXAPO20X). DAPI, VGLUT3, and lectin were detected at 405 nm, 488 nm, and 594 nm, respectively.
Transmission electron microscopy
Liver tissues from Pair- or EtOH-fed mice were fixed with 2% glutaraldehyde-paraformaldehyde in 0.1 M PBS overnight at 4 °C. Samples were then washed with PBS and post-fixed with 1% osmium tetroxide for 2 h. The fixed tissues were dehydrated in the graded ethanol, substituted with propylene oxide, and finally embedded in EMbed-812 resin. Next, the polymerization was performed at 65 °C for 24 h. Tissue sections with 70 nm were made using an ultramicrotome (Leica, EM UC7). Images were captured with a transmission electron microscope (Joel) at 80 kV acceleration voltage.
Quantitative PCR
Total RNA was extracted from liver tissues or isolated cells using RNeasy Mini Kit (Qiagen) or TRIzol reagent (Thermo Fisher Scientific) in accordance with the manufacturer’s instructions. The same amount of RNA was reverse-transcribed to cDNA using ReverTra Ace® qPCR RT Master Mix with gDNA Remover (Toyobo). qRT-PCR was performed using SYBR Green Real-time PCR Master Mix (Toyobo). To standardize the relative expression levels of interested genes, the expression levels of 18S rRNA (in mice) and ACTB (in humans) were used. The information of the primer pair is provided in Supplementary Table 3.
RNA-seq library preparation and sequencing (QuantSeq)
For the RNA-seq analyses, total RNA was extracted as described above from liver tissues or isolated KCs using TRIzol reagent (Thermo Fisher Scientific). For control and test RNAs, the library was constructed using QuantSeq 3’ mRNA-Seq Library Prep Kit (Lexogen, Inc., Austria) in accordance with the manufacturer’s instructions. To briefly summarize, 500 ng of total RNA per sample was prepared for analysis. An oligo-dT primer containing an Illumina-compatible sequence at its 5’ end was hybridized to the corresponding RNA, and reverse transcription was subsequently performed. Then, after the RNA template was degraded, the synthesis of the second strand was initiated by the random primers containing an Illumina-compatible linker sequence at its 5’ end. The purification of the double-stranded library was conducted using magnetic beads to remove all reaction components. Next, the library was amplified to add the complete adapter sequences required for the cluster generation. The finished library is purified from PCR components. High-throughput sequencing was performed as single-end 75 sequencing using NextSeq 550 (Illumina, Inc., USA).
RNA-seq analyses
Bowtie2 was used to align QuantSeq 3’ mRNA-Seq reads. Bowtie2 indices were made from the genome assembly sequence or the representative transcript sequences for the alignment to the genome and transcriptome. The alignment file was applied for assembling transcripts, detecting differential expression of genes, and estimating abundances. Differential expressions of genes were determined according to the counts from unique and multiple alignments with the coverage in Bedtools. The RC (Read Count) data were further processed based on TMM + CPM normalization method using EdgeR within R (R development Core Team, 2020) and Bioconductor. Gene classification was established based on databases in DAVID (http://david.abcc.ncifcrf.gov/) and MEDLINE (http://www.ncbi.nlm.nih.gov/).
Flow cytometry analyses
The isolated liver MNCs or KCs were first incubated with CD16/CD32 (mouse Fc blocker) (BD Bioscience), and LIVE/DEAD™ fixable aqua dead cell stain kit for 405 nm excitation (Thermo Fisher Scientific) was applied to identify dead or live cells. Then, the cells were stained with fluorescence-tagged antibodies with anti-mouse eFluor 450-conjugated CD45 (Thermo Fisher Scientific), APC-Cy7-conjugated CD11b (BD Biosciences), FITC-conjugated F4/80 (eBioscience), PE-Cy7-conjugated Ly-6G (BD Biosciences), and PE-conjugated Siglec-F (BD Biosciences). Cytofix/Cytoperm (BD Biosciences) was used for the intracellular staining of mGluR5. Alex 647-conjugated anti-mGluR5 antibody (Abcam) with 1:100 dilution, along with an isotype control antibody (Abcam), were used to identify the expression of mGluR5 in KCs from wild type and KC-specific mGluR5-depleted mice. Stained cells were read with LSRFortessa X-20 (BD Bioscience). The frequencies of neutrophils (CD11b+Ly6G+), eosinophils (CD11b+SiglecF+), macrophages (F4/80intCD11bhigh), and KCs (F4/80highCD11bintCLEC2+TIM4−) were analyzed with the FlowJo software (version 10.81; FlowJo LLC) through pseudo-color analysis plot. Further specific information on the above antibodies validated by manufacturers is listed in Supplementary Table 4.
Measurement of reactive oxygen species
Isolated KCs were stimulated with 10 μM CHPG or 100 μM MSG for 10 min or 30 min, respectively. The cells were incubated with fluorescent surface antibodies for the identification of KCs. Then, KCs were stained with ROS detection reagent, 5-(and-6)-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate, acetyl ester (CM-H2DCFDA) (Invitrogen), on 37 °C for 10 min, whereas PBS was treated in a negative control. Fluorescence excitation was at 492–495 nm, and the emission was at 517–527 nm. ROS was measured by monitoring FITC with LSRFortessa X-20 (BD Bioscience). Histograms of relative fluorescent intensity were used to compare the ROS production of each group. Measurement of ROS in cultured cells was evaluated by Microplate Reader (VersaMax).
Western blot analyses
Proteins from frozen isolated cells or liver tissues were extracted with the RIPA lysis buffer (10% glycerol, 10% SDS, 1 mM Na3VO4, 1 mM PMSF, 150 mM NaCl, 30 mM Tris, pH 7.5) containing phosphatase inhibitor and protease cocktail (Thermo Fisher Scientific). The proteins were separated with 10% SDS-polyacrylamide gel electrophoresis, and the nitrocellulose membrane (#88018, Thermo Fisher Scientific) was used for the transfer of proteins from the gel. After the transfer of the proteins, the nitrocellulose membrane was blocked by 5% skim milk solution for 1 h at room temperature. Then, the membranes were incubated with primary antibodies (1:500 ~ 1:2000 in PBS containing 0.1% tween-20) overnight at 4 °C. On the next day, the secondary antibodies were incubated for 1 h at room temperature. SuperSignalTM West Pico PLUS Chemiluminescent substrate (Thermo Fisher Scientific) was used to detect immunoreactive bands, and the bands were captured by ImageQuantTM LAS 4000 (GE Healthcare). All immunoblot bands were selected based on molecular weight and protein levels were standardized with the expression of β-actin for each sample. Specifically, the VGLUT3 antibody was validated using liver tissues lacking hepatocyte-specific VGLUT3. All information of primary and secondary antibodies validated by the manufacturers is provided in Supplementary Table 4.
In situ closed liver perfusion
The WT mice were subject to the in situ closed liver perfusion system with chemical reagents as previously described28. In brief, the mouse was anesthetized by a mixture of ketamine and rompun, and the liver was exposed through laparotomy. The portal vein of mice was cannulated with an angiocatheter and intrahepatic inferior vena cava (IVC) was cut. To extract the blood in the liver, pre-warmed PBS buffer was infused for 5 min, and IVC was subsequently clamped. After hepatic blood was totally extracted as identified through discoloration of the liver, the chest was opened to expose the right atrium of the heart. Next, the suprahepatic IVC was cannulated with another angiocatheter by puncturing through the right atrium. A body of 1 ml syringe was connected to the catheter, which was cannulated into suprahepatic IVC, to work as a reservoir. Once the closed hepatic circuit was established, DMEM (#LM001-11, Welgene) with vehicle (PBS) or ITE (1 μM, Tocris) was infused into the closed circuit and maintained for 2 h.
Isolation of neutrophils
Isolation of neutrophils was performed based on the protocol modified from the previous report59. At first, the bone marrow in a mouse femur was flushed with PBS using a 1 ml syringe. RBC lysis buffer (BioLegend) was used to remove RBC, and cells were centrifuged at 1800 × g for 15 min at 4 °C. Whole bone marrow cells were resuspended in PBS, and carefully added on the Percoll density gradients. Cells were centrifuged at 1000 × g for 20 min at 4 °C, and separated into 3 bands according to the differentiation. Matured and segmented neutrophils were located on the lowest layer.
Microvesicle isolation
Isolation of MVs was performed according to the protocol modified from the previous report60. First, HEPs were isolated by liver perfusion from Pair- or EtOH-fed mice, and HEPs were mechanically lysed using homogenizers (Benchmark). Next, stepwise centrifugations were designed to sequentially remove the objects according to each step (50 × g for HEPs, 300 × g for nonparenchymal cells, 3000 × g for dead cells, and 13,000 × g for cell debris). MVs were finally obtained by the ultracentrifugation at 100,000 × g at 4 °C.
Glutamate measurement
Freshly frozen liver tissues were weighed (about 30 mg each) and deproteinized using a homogenizer in 100% methanol. For the analysis of mice serum, 100 μl of each sample was mixed with 400 μl of 100% methanol. A total of 500 μl of supernatant from the co-cultured sample was mixed with 500 μl of 100% methanol. The samples were centrifuged at 10,000 × g for 15 min at 4 °C and supernatant was collected. Solutions in the samples were evaporated by SpeedVac System, and the dried agents were resuspended into 50 μl of 0.1 N HCl solution. Then, diluted samples were subjected to high-performance liquid chromatography (HPLC) to measure glutamate concentrations. Agilent 1200 HPLC machine (Agilent) was used for the measurement, and ZORBAX Eclipse-AAA (4.6 × 75 mm, 3.5 μm) was selected as a chromatographic column. Forty mM NaH2PO4 (5.5 g NaH2PO4 dissolved in 1 L DW, pH 7.8) was in mobile phase A, while 45:45:10 (v/v/v) acetonitrile: methanol: water was in mobile phase B. Mobile phase B gradually increased from 0 to 57% between 0 and 9.8 min, and from 57 to 100% between 9.8 and 10 min. Therefore, phase B remained at 100% between 10 and 11.5 min, and finally, it decreased from 100% to 0% between 11.5 and 13 min. Mobile phase B remained at 0% between 13 and 15 min. The temperature of the column was maintained at 35 °C. The wavelength of the diode array detector was 338 nm, and reference wavelength was 390 nm. One nmol/μl of amino acid standard (Agilent) was used as the standard control before sample analyses. The areas of the sample peaks that indicate glutamates were measured to estimate glutamate concentrations compared to the area of the amino acid standard.
Custom-built laser-scanning intravital confocal microscopy
For the intravital liver imaging, a previously demonstrated custom-built video-rate laser-scanning confocal microscopy system was used61. As excitation sources for multi-color fluorescence imaging, four continuous-wave laser modules with the output wavelengths of 405, 488, 561, and 640 nm (Coherent OBIS, Cobolt MLD, Cobolt Jive, and Cobolt MLD) were utilized. The excitation lights were combined by dichroic beam splitters (FF593-Di03, Di01-R405, FF520-Di02, Semrock) and then delivered to a multi-edge dichroic beam splitter (Di01-R405/488/561/635, Semrock). 2D laser beam scanning in the raster pattern was achieved by a fast rotating 36 facets polygonal mirror scanner (MC-5, Lincoln Laser) providing 17.28 kHz lateral scanning in the X-axis and a galvanometer mirror scanner (6230H, Cambridge Technology) providing 30 Hz vertical scanning in the Y-axis. Then, the scanning laser beams were delivered to the back aperture of a commercial objective lens (16X CFI LWD Plan Fluorite Objective, 0.80 NA, 3.0 mm WD, Nikon) providing the FOV of 640 × 640 μm with 1024 × 1024 pixels in the image. The multi-color fluorescent signals from the imaging samples were epi-detected and separated from the excitation laser beams by the central dichroic beam splitters. Next, the multi-color fluorescence signals were split into the single-color signal by dichroic beam splitters on the PMT side (FF484-Di01, FF560-Di01, FF649-Di01, Semrock) and detected by photomultiplier tubes (PMT; R9110, Hamamatsu) through the bandpass filters (FF01-442/46, FF02-525/50, FF01-600/37, FF01-685/40, Semrock). Electronic outputs from the PMTs were digitized by a four-channel frame grabber (Solios, Matrox) with the sampling rate of 10 MHz. The video-rate images with the frame rate of 15 Hz and size of 1024 × 1024 pixels were displayed and recorded in real-time by custom-written software using Matrox Imaging Library (MIL9, Matrox).
Intravital liver imaging
Mice were anesthetized with an intramuscular injection of the mixture of Zoletil (30 mg/kg) and Xylazine (10 mg/kg). After anesthetization, an intravital liver imaging followed the previously described steps62. An abdominal hair was shaved by using a hair clipper and hair removal cream. A small incision with the size of 10 mm was made on the skin and peritoneum. Then, the left lobe of the liver was carefully exposed. The cover glass attached with a silicone rubber heater and mini-probe thermometer was placed on the exteriorized liver to monitor and maintain the temperature of local liver tissue constant at 36 °C during the intravital imaging. Additionally, a commercial homeothermic heating pad (RightTemp Jr., Kent Scientific, USA) was used to maintain the body temperature at 36 °C during the intravital imaging. A wet gauze soaked with warmed saline was placed between the liver and adjacent tissue to reduce motion artifacts. To avoid dehydration of the surface of the exteriorized liver, warmed saline was continuously supplied to the gauze during the imaging. During the intravital imaging, the level of anesthetization was periodically monitored by pinching the toe. When the movement was observed, half of the initial dose of Zoletil and Xylazine mixture was administered through an intramuscular injection.
For calcium (Ca2+) imaging, Fluo-4 AM dye (#F14201, Invitrogen) was used. This dye was serially dissolved in 4 μl of DMSO and 25 μl of PBS. Calcium signals in HEPs were fluorescently labeled with this solution by the tail vein injection immediately before the imaging. To visualize hepatic vasculature, lectin DyLight 649 (#DL-1178-1, Vector laboratories) was intravenously injected, which fluorescently labeled endothelial cells of the whole body in a systemic manner with Hoechst 33342 solution (#62249, Thermo Scientific) for the cellular nuclear staining. Acquisition of all images was performed in the same imaging condition with the same laser power (405 nm; 0.56 mw, 488 nm; 6.8 mw, 640 nm; 10.2 nm) and objective lens. Acquired images and movies were quantitatively analyzed using commercial image analysis software, IMARIS (Oxford Instruments) and ImageJ (NIH). The HEP with above 15.0 μm of the estimated X-Y diameter in the “Spot” function of the IMARIS program was considered. The filter type was selected as “quality” (lower threshold: 4.0, upper threshold: 15.0 in ROI). The number of the HEPs was automatically detected. Atypical signals and noise were removed by comparing the Hoechst nuclear staining. Fluo-4 AM signal intensity was calculated by using the ImageJ program. HEPs (above >20) were serially and manually selected from the middle of the central vein. The signal intensity values were calculated in the “Histogram” function of the ImageJ.
Statistical analysis and reproducibility
All data were statistically analyzed using Prism version 8.0 (GraphPad Software), and data were presented as the mean ± SEM. In mice experiments, researchers involved in the present study were not completely blinded. However, researchers were blinded in the analysis of human subjects. The inclusion and exclusion criteria of human samples are described in the Experimental Model and Subject Details part. For in vivo experiments, mice with severe problems (i.e., death after binge drinking or body weight loss of more than 30%) were excluded from further analyses. All immunostaining and immunoblotting results were independently repeated at least three times with similar results. The unpaired Student’s t test was used to analyze the statistical significance between the two groups. One-way analysis of variance (ANOVA) with Tukey’s and Dunnett’s test was applied for the multiple comparisons. A p value < 0.05 was defined as having statistical significance.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Raw RNA-seq data generated in this study have been deposited in the NCBI Sequence Read Archive (SRA) under the BioProject accession codes PRJNA917332 and PRJNA917826. The scRNA-seq data from other studies used in this work are available in the GEO database under accession code GSE84498. Source data are provided with this paper.
References
Seitz, H. K. et al. Alcoholic liver disease. Nat. Rev. Dis. Prim. 4, 16 (2018).
Singal, A. K. & Mathurin, P. Diagnosis and treatment of alcohol-associated liver disease: a review. JAMA 326, 165–176 (2021).
Lucey, M. R., Mathurin, P. & Morgan, T. R. Alcoholic hepatitis. N. Engl. J. Med. 360, 2758–2769 (2009).
Krenkel, O. & Tacke, F. Liver macrophages in tissue homeostasis and disease. Nat. Rev. Immunol. 17, 306–321 (2017).
Smedsrod, B. et al. Cell biology of liver endothelial and Kupffer cells. Gut 35, 1509–1516 (1994).
Nagy, L. E. The role of innate immunity in alcoholic liver disease. Alcohol Res. 37, 237–250 (2015).
Chang, B. et al. Short- or long-term high-fat diet feeding plus acute ethanol binge synergistically induce acute liver injury in mice: an important role for CXCL1. Hepatology 62, 1070–1085 (2015).
Roh, Y. S., Zhang, B., Loomba, R. & Seki, E. TLR2 and TLR9 contribute to alcohol-mediated liver injury through induction of CXCL1 and neutrophil infiltration. Am. J. Physiol. Gastrointest. Liver Physiol. 309, G30–G41 (2015).
Brosnan, M. E. & Brosnan, J. T. Hepatic glutamate metabolism: a tale of 2 hepatocytes. Am. J. Clin. Nutr. 90, 857S–861S (2009).
Choi, W. M. et al. Glutamate signaling in hepatic stellate cells drives alcoholic steatosis. Cell Metab. 30, 877–889.e877 (2019).
Choi, W. M. et al. Metabotropic glutamate receptor 5 in natural killer cells attenuates liver fibrosis by exerting cytotoxicity to activated stellate cells. Hepatology 74, 2170–2185 (2021).
Storto, M. et al. Selective blockade of mGlu5 metabotropic glutamate receptors is protective against acetaminophen hepatotoxicity in mice. J. Hepatol. 38, 179–187 (2003).
Ferrigno, A. et al. MPEP attenuates intrahepatic fat accumulation in obese mice. Int. J. Mol. Sci. 24, 6076 (2023).
Gheni, G. et al. Glutamate acts as a key signal linking glucose metabolism to incretin/cAMP action to amplify insulin secretion. Cell Rep. 9, 661–673 (2014).
Morimoto, R. et al. Secretion of L-glutamate from osteoclasts through transcytosis. EMBO J. 25, 4175–4186 (2006).
El Mestikawy, S., Wallen-Mackenzie, A., Fortin, G. M., Descarries, L. & Trudeau, L. E. From glutamate co-release to vesicular synergy: vesicular glutamate transporters. Nat. Rev. Neurosci. 12, 204–216 (2011).
Shigeri, Y., Seal, R. P. & Shimamoto, K. Molecular pharmacology of glutamate transporters, EAATs and VGLUTs. Brain Res. Brain Res. Rev. 45, 250–265 (2004).
Bertola, A., Mathews, S., Ki, S. H., Wang, H. & Gao, B. Mouse model of chronic and binge ethanol feeding (the NIAAA model). Nat. Protoc. 8, 627–637 (2013).
Bertola, A., Park, O. & Gao, B. Chronic plus binge ethanol feeding synergistically induces neutrophil infiltration and liver injury in mice: a critical role for E-selectin. Hepatology 58, 1814–1823 (2013).
Halpern, K. B. et al. Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature 542, 352–356 (2017).
Ku, T. et al. Multiplexed and scalable super-resolution imaging of three-dimensional protein localization in size-adjustable tissues. Nat. Biotechnol. 34, 973–981 (2016).
Park, J. et al. Epitope-preserving magnified analysis of proteome (eMAP). Sci. Adv. 7, eabf6589 (2021).
Sakae, D. Y. et al. Differential expression of VGLUT3 in laboratory mouse strains: Impact on drug-induced hyperlocomotion and anxiety-related behaviors. Genes Brain Behav. 18, e12528 (2019).
Kenison, J. E. et al. Tolerogenic nanoparticles suppress central nervous system inflammation. Proc. Natl. Acad. Sci. USA 117, 32017–32028 (2020).
Henry, E. C., Welle, S. L. & Gasiewicz, T. A. TCDD and a putative endogenous AhR ligand, ITE, elicit the same immediate changes in gene expression in mouse lung fibroblasts. Toxicol. Sci. 114, 90–100 (2010).
Shin, S. et al. NRF2 modulates aryl hydrocarbon receptor signaling: influence on adipogenesis. Mol. Cell Biol. 27, 7188–7197 (2007).
Kim, Y. S. et al. Induction of the hepatic aryl hydrocarbon receptor by alcohol dysregulates autophagy and phospholipid metabolism via PPP2R2D. Nat. Commun. 13, 6080 (2022).
Choi, W. M. et al. Experimental applications of in situ liver perfusion machinery for the study of liver disease. Mol. Cells 42, 45–55 (2019).
Ni, Y. & Parpura, V. Dual regulation of Ca2+-dependent glutamate release from astrocytes: vesicular glutamate transporters and cytosolic glutamate levels. Glia 57, 1296–1305 (2009).
Araque, A., Li, N., Doyle, R. T. & Haydon, P. G. SNARE protein-dependent glutamate release from astrocytes. J. Neurosci. 20, 666–673 (2000).
Bartlett, P. J. et al. Chronic alcohol feeding potentiates hormone-induced calcium signalling in hepatocytes. J. Physiol. 595, 3143–3164 (2017).
Wang, G., Memin, E., Murali, I. & Gaspers, L. D. The effect of chronic alcohol consumption on mitochondrial calcium handling in hepatocytes. Biochem. J. 473, 3903–3921 (2016).
Amaya, M. J. & Nathanson, M. H. Calcium signaling in the liver. Compr. Physiol. 3, 515–539 (2013).
Hof, T. et al. Transient receptor potential channels in cardiac health and disease. Nat. Rev. Cardiol. 16, 344–360 (2019).
Gee, K. R. et al. Chemical and physiological characterization of fluo-4 Ca(2+)-indicator dyes. Cell Calcium 27, 97–106 (2000).
Meijer, A. J. Hepatocyte swelling: techniques and effects on metabolism. In The Hepatocyte Review (Berry, M. N & Edwards, A. M.) 147–167 (Springer, 2000).
Shepard, B. D. & Tuma, P. L. Alcohol-induced alterations of the hepatocyte cytoskeleton. World J. Gastroenterol. 16, 1358–1365 (2010).
Sakai, M. et al. Liver-derived signals sequentially reprogram myeloid enhancers to initiate and maintain Kupffer cell identity. Immunity 51, 655–670.e658 (2019).
Conn, P. J., Battaglia, G., Marino, M. J. & Nicoletti, F. Metabotropic glutamate receptors in the basal ganglia motor circuit. Nat. Rev. Neurosci. 6, 787–798 (2005).
Cathcart, M. K. Regulation of superoxide anion production by NADPH oxidase in monocytes/macrophages: contributions to atherosclerosis. Arterioscler Thromb. Vasc. Biol. 24, 23–28 (2004).
Ledonne, A. & Mercuri, N. B. Insights on the functional interaction between group 1 metabotropic glutamate receptors (mGluRI) and ErbB receptors. Int. J. Mol. Sci. 21, 7913 (2020).
Thakur, V., Pritchard, M. T., McMullen, M. R., Wang, Q. & Nagy, L. E. Chronic ethanol feeding increases activation of NADPH oxidase by lipopolysaccharide in rat Kupffer cells: role of increased reactive oxygen in LPS-stimulated ERK1/2 activation and TNF-alpha production. J. Leukoc. Biol. 79, 1348–1356 (2006).
Gao, B., Ahmad, M. F., Nagy, L. E. & Tsukamoto, H. Inflammatory pathways in alcoholic steatohepatitis. J. Hepatol. 70, 249–259 (2019).
Su, L. et al. Kupffer cell-derived TNF-alpha promotes hepatocytes to produce CXCL1 and mobilize neutrophils in response to necrotic cells. Cell Death Dis. 9, 323 (2018).
Abuse, N.I.o.A. & Alcoholism. NIAAA council approves definition of binge drinking. NIAAA Newsl. 3, 3 (2004).
Kim, H. H., Choi, S. E. & Jeong, W. I. Oxidative stress and glutamate excretion in alcoholic steatosis: metabolic synapse between hepatocyte and stellate cell. Clin. Mol. Hepatol. 26, 697–704 (2020).
Tominaga, T. et al. Clinical significance of measuring plasma concentrations of glutamine and glutamate in alcoholic liver diseases. Alcohol Alcohol Suppl. 1a, 103–109 (1993).
Köhle, C. & Bock, K. W. Activation of coupled Ah receptor and Nrf2 gene batteries by dietary phytochemicals in relation to chemoprevention. Biochem. Pharm. 72, 795–805 (2006).
Takamori, S. et al. Molecular anatomy of a trafficking organelle. Cell 127, 831–846 (2006).
Daniels, R. W. et al. A single vesicular glutamate transporter is sufficient to fill a synaptic vesicle. Neuron 49, 11–16 (2006).
Mielnicka, A. & Michaluk, P. Exocytosis in astrocytes. Biomolecules 11, 1367 (2021).
Parpura, V. & Zorec, R. Gliotransmission: exocytotic release from astrocytes. Brain Res. Rev. 63, 83–92 (2010).
McCuskey, R. S. & McCuskey, P. A. Fine structure and function of Kupffer cells. J. Electron Microsc. Tech. 14, 237–246 (1990).
Panatier, A. et al. Astrocytes are endogenous regulators of basal transmission at central synapses. Cell 146, 785–798 (2011).
Pirzgalska, R. M. et al. Sympathetic neuron-associated macrophages contribute to obesity by importing and metabolizing norepinephrine. Nat. Med. 23, 1309–1318 (2017).
Guo, W. et al. Notch signaling regulates macrophage-mediated inflammation in metabolic dysfunction-associated steatotic liver disease. Immunity 57, 2310–2327.e2316 (2024).
Peiseler, M. et al. Kupffer cell-like syncytia replenish resident macrophage function in the fibrotic liver. Science 381, eabq5202 (2023).
Kim, H. H. et al. Catecholamine induces Kupffer cell apoptosis via growth differentiation factor 15 in alcohol-associated liver disease. Exp. Mol. Med. 55, 158–170 (2023).
Kumar, S. et al. Functional and molecular characterization of NOS isoforms in rat neutrophil precursor cells. Cytom. A 77, 467–477 (2010).
Ishiguro, K., Yan, I. K. & Patel, T. Isolation of tissue extracellular vesicles from the liver. J. Vis. Exp. https://doi.org/10.3791/58649 (2019).
Ahn, J. et al. In vivo longitudinal depth-wise visualization of tumorigenesis by needle-shaped side-view confocal endomicroscopy. Biomed. Opt. Express 10, 2719–2729 (2019).
Moon, J. et al. Intravital longitudinal imaging of hepatic lipid droplet accumulation in a murine model for nonalcoholic fatty liver disease. Biomed. Opt. Express 11, 5132–5146 (2020).
Acknowledgements
We thank Drs. Kunos and Gao for critical reading and comments on improving the manuscript. This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT): RS-2021-NR059919 (W.I.J.), 2021R1A2C2005820 (W.K), RS-2021-NR056442 (W.K.), RS-2022-NR067269 (W.I.J), RS-2023-00223831 (W.I.J.).
Author information
Authors and Affiliations
Contributions
K.Y., W.K., and W.I.J. designed the whole research and wrote the manuscript. K.K. contributed to manuscript editing and review. K.Y., K.K., T.R., H.H.K., Y.R.S., S.E.C., M.J.K., K.P.S.C., E.L. and K.W.L. performed animal experiments and data analyses. K.Y., K.K., J.J., and P.K. performed intravital liver imaging. K.Y., Y.S.K. and T.K. performed liver tissue expansion and imaging. K.Y., H.J. and K.T.N. conducted electron microscopy. K.Y., H.S.E., S.H.K. and W.K. contributed to human hepatic cell isolation and the analyses of human samples. K.Y. and K.K. generated conditional knockout mice. G.L. and D.C. performed metabolite measurements. W.K. and W.I.J. provided important advice and funding acquisition for the experiments.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Salah El Mestikawy and the other, anonymous, reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yang, K., Kim, K., Ryu, T. et al. Binge drinking triggers VGLUT3-mediated glutamate secretion and subsequent hepatic inflammation by activating mGluR5/NOX2 in Kupffer cells. Nat Commun 16, 5546 (2025). https://doi.org/10.1038/s41467-025-60820-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-60820-3