Abstract
JAK (Janus Kinase) inhibitors, such as ruxolitinib, were introduced a decade ago for treatment of myeloproliferative neoplasms (MPN). To evaluate ruxolitinib’s impact on MPN clonal evolution, we interrogate a myelofibrosis patient cohort with longitudinal molecular evaluation and discover that ruxolitinib is associated with clonal outgrowth of RAS pathway mutations. Single-cell DNA sequencing combined with ex vivo treatment of RAS mutated CD34+ primary patient cells, demonstrates that ruxolitinib induces RAS clonal selection both in a JAK/STAT wild-type and hyper-activated context. RAS mutations are associated with decreased transformation-free and overall survival only in patients treated with ruxolitinib. In vitro and in vivo competition assays demonstrate increased cellular fitness of RAS-mutated cells under ruxolitinib or JAK2 knock-down, consistent with an on-target effect. MAPK pathway activation is associated with JAK2 downregulation resulting in enhanced oncogenic potential of RAS mutations. Our results prompt screening for pre-existing RAS mutations in JAK inhibitor treated patients with MPN.
Similar content being viewed by others
Introduction
Targeted therapies have emerged over the last two decades as a fundamental component of the anti-cancer therapeutic arsenal. Their use is often associated with resistance mechanisms mainly driven by cancer cell adaptability1. In the case of chronic myeloid leukemia, for example, despite the efficacy of BCR-ABL1 tyrosine kinase inhibitors, resistance mechanisms driven by the selection of on-target ABL1 mutated clones appear in a subset of patients2. Given the intra-patient heterogeneity of cancer, clonal selection of pre-existing resistant subclones without second-site mutations in the targeted protein also emerged as a potential resistance mechanism. Indeed, FLT3 secondary mutations are infrequently found in patients developing secondary resistance to the FLT3 inhibitors gilteritinib and crenolanib. Acquired resistance is mediated by subclonal selection of RAS/MAPK activating mutations3,4. Similarly, MEK1 and NRAS mutated clonal selection underlies resistance to the RAF inhibitor vemurafenib in melanoma5,6. Modulation of clonal architecture thus appears as a major resistance mechanism, creating new oncogenic dependencies and therapeutic opportunities across many cancer types. A better understanding of treatment-induced clonal evolution is needed to improve cancer patients’ outcomes.
JAK-targeted therapies, such as the first-in-class JAK1/2 ATP competitive inhibitor ruxolitinib7,8,9, were introduced a decade ago for the treatment of myeloproliferative neoplasms (MPN) driven by JAK/STAT signaling activating mutations in JAK210, CALR11 or MPL12. Additional mutations in genes involved in the regulation of epigenetic (ASXL1, EZH2, TET2, DNMT3A, IDH1/2), mRNA splicing (SRSF2, SF3B1, U2AF1, ZRSR2), gene transcription (TP53, NFE2, IKZF1) or non-JAK/STAT signaling pathways (NRAS, KRAS, CBL)13,14 complement the MPN molecular landscape. Several studies highlighted the role of such mutations in modifying patient prognosis15,16, leading to their implementation in advanced prognostic scoring systems17. MPNs are considered pre-leukemic, and clonal evolution with sequential acquisition of mutations is likely to participate in the transition towards a more aggressive disease13,18,19. In addition to high molecular risk (HMR) mutations (ASXL1, EZH2, IDH1/2, SRSF2, U2AF1), reported to be associated with adverse outcomes, mutations in TP53 are also frequently associated with the occurrence of secondary AML13,20. No clear mechanism explaining the selection of mutated clones has been identified to date.
Despite significant quality of life improvement with decreased symptoms and reduced splenomegaly, the effects of ruxolitinib on disease modification and long-term patient outcome remain controversial21. Although JAK2 mutations within the kinase domain have been described as driving ruxolitinib resistance in vitro, we and others failed to identify second-site on-target mutations involving JAK2 in patients22,23,24. MPNs inter-patient molecular landscape is highly heterogeneous, with some patients harboring a unique clone, while others carrying a number of genetically diverse sub-clones15,18. Longitudinally, the emergence of a new clone or an increase in the allele frequency of pre-existing clones, sometimes at the expense of other sub-clones, is observed13. We thus hypothesized that modulation of clonal architecture upon ruxolitinib selective pressure could not only hamper the clinical efficacy of JAK inhibitors, but also adversely modulate the natural history of MPNs.
In this study, to evaluate the effect of JAK inhibitors on MPN clonal evolution and its potential impact on clinical outcome, we interrogate a cohort of patients with myelofibrosis and validate our findings using in vitro and in vivo MPN models. We reveal a selection of RAS mutations upon ruxolitinib exposure, negatively impacting clinical outcome. Mechanistically, this effect results from the enhanced oncogenic potential of RAS-mutated clones after inactivation of the JAK/STAT pathway. Using MPN as a dynamically evolving model, our results reflect the major challenge of selectively targeting heterogeneous and adaptive diseases such as cancer.
Results
Ruxolitinib treatment is associated with the accumulation of RAS pathway mutations in patients with myelofibrosis
To evaluate the impact of ruxolitinib treatment on clonal evolution, we collected clinical and molecular data across a cohort of 143 patients with the myelofibrosis MPN subtype. Within this monocentric cohort, 72 patients were treated with ruxolitinib. Patients’ characteristics are presented in Table S1. Ruxolitinib-treated patients had more constitutional symptoms, higher IPSS and DIPSS scores, and a higher proportion of HMR mutations (Table S1). Non-ruxolitinib-treated patients were diagnosed between 1990 and 2019, while ruxolitinib-treated patients were diagnosed between 1994 and 2018. 73% (n = 52/71) and 47% (n = 34/72) patients were diagnosed after ruxolitinib EMA approval for myelofibrosis treatment (in 2012 or later), in the non-ruxolitinib and ruxolitinib-treated groups, respectively. Treatment of patients not exposed to ruxolitinib is detailed in Table S2. Molecular data obtained using a Next Generation Sequencing (NGS) panel targeting 36 genes involved in myeloid diseases, detailed in the methods section, is presented in Supplementary Data 1.
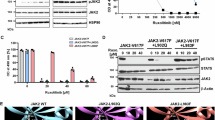
First, we focused on the 73 myelofibrosis patients for whom longitudinal molecular evaluation was available. We analyzed molecular evolution on all patients exposed (n = 45) or not exposed (n = 28) to ruxolitinib between baseline and follow-up molecular evaluation (Fig. 1A). After a median molecular follow-up of 21 months IQR[13; 44] in the ruxolitinib exposed population, a total of 53 mutations were newly detected among 26 patients. 28% (n = 15 /53) of these mutations occurred within genes involved in the RAS pathway (NRAS, KRAS, CBL) (Fig. 1B). Additionally, if we only focus on the 22 patients for whom the baseline NGS was performed prior to ruxolitinib initiation (n = 22/45), 37% of the newly detected mutations involved RAS pathway genes after a median ruxolitinib exposure time of 16 months IQR[12; 28] (Fig. S1A). In the non-ruxolitinib exposed population, 12 patients accumulated a total of 14 mutations over a molecular follow-up of 25 months IQR[15; 55]. Only one of these mutations (7%) occurred in RAS pathway genes (Fig. 1C). Using a COX regression analysis, ruxolitinib exposure was associated with RAS mutations accumulation (HR 9.8 CI95%[1.23; 78.89], p = 0.031) (Fig. 1Dand Table S3). Conversely, age and disease aggressiveness markers including an intermediate-2 or high DIPSS prognosis score at time of molecular evaluation and presence of HMR (ASXL1, EZH2, SRSF2, U2AF1 or IDH1/2) mutations, were not associated with RAS pathway mutations accumulation (Table S3).
A Flow-chart depicting the patients included in the study according to their exposure to ruxolitinib treatment and molecular data availability. B, C Pie Charts depicting the additional mutations longitudinally newly identified in patients with myelofibrosis exposed (n = 45) (B) or not exposed (n = 28) (C) to ruxolitinib between baseline and follow-up molecular evaluation. D Cumulative Hazard curve of the acquisition of RAS pathway mutations for ruxolitinib-treated patients (n = 45) compared to non-ruxolitinib-treated patients (n = 28) with molecular follow-up available. Two-sided COX proportional hazards regression was used for comparing the groups. P-value reported in the figure. E, FRAS mutations variant allele frequency longitudinal evolution in patients with myelofibrosis exposed (n = 45) (E) or not exposed (n = 28) (F) to ruxolitinib between baseline and follow-up molecular evaluation. Variant allele frequency was considered zero when mutations were absent at baseline or follow-up molecular evaluation. Statistical significance determined using two-sided Mann-Whitney test in comparison to baseline molecular evaluation. P-values reported in the figure. G, H Oncoprints (G) and lollipop plots (H) showing RAS pathway mutations in patients treated with ruxolitinib (n = 45) compared to patients not treated with ruxolitinib (n = 28). N-term = N-terminal domain 1, EF = EF hand-like domain, SH2 = SH2-like domain, ZF=Zinc finger, C3HC4 type (RING finger), U = UBA/TS-N domain. Source data are provided as a Source Data file.
We then compared RAS mutations variant allele frequency between baseline and follow-up molecular evaluations according to patients’ exposures to ruxolitinib. RAS mutations variant allele frequency significantly increased at follow-up molecular evaluation only in patients exposed to ruxolitinib (Fig. 1E, F). Accordingly, at last molecular evaluation, RAS pathway mutations were more frequently present in patients treated with ruxolitinib, including patients harboring multiple RAS-mutated clones (Fig. 1G, H). Importantly, while 10 ASXL1 mutations (n = 10 /53, 19%) were newly detected among ruxolitinib exposed patients in comparison with only 1 acquired ASXL1 mutation (n = 1/ 14, 7%) in the non-ruxolitinib exposed population (Fig. 1B, C); ASXL1 mutations variant allele frequency did not increase at follow-up molecular evaluation (Fig. S1B, C). This observation suggests a specific effect of ruxolitinib on RAS pathway mutations clonal selection.
RAS pathway mutations adversely impact MPN patient prognosis in the context of ruxolitinib treatment
Because clonal selection of additional mutated clones is usually associated with worse prognosis in myelofibrosis, and RAS pathway mutations have been associated with poor prognosis and poor ruxolitinib treatment response in MPN25,26, we next aimed to evaluate the clinical impact of ruxolitinib-induced RAS mutant clonal selection. We interrogated our cohort of 143 patients with myelofibrosis according to their exposure to ruxolitinib (Fig. 2A). Within a median follow-up of 7.0 years, 13 and 7 patients experienced a transformation of their MPN to MDS/AML, and 26 and 9 patients died, among ruxolitinib treated (n = 72) and not treated (n = 71) patients respectively.
A Flow-chart depicting the patients included in the study according to their exposure to ruxolitinib treatment prior to their last NGS molecular evaluation. B, C Kaplan-Meier curves of overall survival (left) or transformation-free survival (right), for ruxolitinib treated (B) or non-ruxolitinib-treated (C) patients according to the presence of RAS pathway mutations. Two-sided COX proportional hazards regression analysis was used to compare the groups. P-value reported in the figure. D Pie Chart depicting the proportion of AML/MDS transformations among RAS-mutated patients, treated (n = 8/17) or not with ruxolitinib (n = 1/6). E, F Evolution of RAS (E) and ASXL1 (F) mutations variant allele frequency (VAF) between two time points: prior to ruxolitinib initiation and at time of AML/MDS transformation, for RAS-mutated MPN patients treated or not with ruxolitinib. Data was available for n = 5/8 ruxolitinib-treated patients and n = 1/1 non-ruxolitinib-treated patients. Source data are provided as a Source Data file.
To identify variables associated with Overall survival (OS) and Transformation Free Survival (TFS), we performed a Cox model univariate analysis including age, DIPSS at molecular evaluation, presence of RAS pathway mutations, but also potential confounding molecular variables such as the presence of HMR (ASXL1, EZH2, SRSF2, U2AF1 or IDH1/2) mutations. In the overall cohort, presence of RAS mutations was associated with decreased OS and TFS in univariate analysis (HR 3.2 CI95%[1.6; 6.4], p = 0.001 and 5.2 CI95%[2.1; 12.5], p < 0.001). RAS mutational status remained significantly associated with TFS but not with OS in the multivariate analysis (3.0 CI95%[1.2; 7.8], p = 0.023 and p = 0.0884) (Fig. S1D and Tables S4-5). Whereas the presence of RAS mutations was independently associated with decreased transformation free (HR 6.6 CI95%[1.9; 23.1], p = 0.003) and overall (HR 3.1 CI95%[1.3; 7.1], p = 0.008) survival in ruxolitinib treated patients (Fig. 2B and Tables S6, 7), such mutations did not influence clinical outcome of MPN patients not treated with ruxolitinib (Fig. 2C). In the ruxolitinib treated cohort, median OS and TFS were respectively 18.51 years and not reached in the absence of RAS pathway mutations, and 7.63 years and 7.54 years when a RAS pathway mutation was present. Mean ruxolitinib treatment duration until transformation was 44.7 months Range[4.2; 81.5] in the global cohort, 33.5 months Range[4.2; 81.5] for patients harboring a RAS mutation, and 62.5 months Range[36.7; 77.3] for patients without RAS mutation. Given the intrinsic biases associated with comparing ruxolitinib-treated and non-treated patients, due to the clinical use of ruxolitinib in patients with more aggressive disease, we also evaluated the impact of RAS mutational status on OS and TFS after stratification according to the DIPSS score. Importantly, the presence of RAS mutations remained associated with a poorer OS and TFS among patients harboring an intermediate-2/high risk score when exposed to ruxolitinib (HR 3.8 CI95%[1.3; 11.0], p = 0.012 and HR 6.2 CI95%[1.6; 24.2], p = 0.009) (Fig. S1E). Conversely, these associations were not significant among intermediate-2/high risk score patients not exposed to ruxolitinib (HR 1.6 CI95%[0.2; 14.3], p = 0.682 for OS and HR 2.0 CI95%[0.2; 19.5], p = 0.559 for TFS). A low number of RAS mutations and death/transformation events precluded a similar analysis among DIPSS low/intermediate-1 patients.
Among ruxolitinib treated patients harboring RAS pathway mutations, 47.06% (n = 8/17) transformed to AML/MDS, while only 16.67% (n = 1/6) of RAS mutated patients not treated with ruxolitinib developed an acute transformation (Fig. 2D). Variant allele frequency of the RAS mutated clones increased at time of transformation for all ruxolitinib treated patients with available molecular evaluation (n = 5 patients), while it did not increase in the RAS mutated patient who transformed in the absence of ruxolitinib treatment (Fig. 2E). The molecular landscape at time of transformation was available for 8 patients and is presented in Fig. S1F according to ruxolitinib treatment status. While 7 of these 8 patients harbored an ASXL1 mutation at the time of transformation, including the patient who transformed in the absence of ruxolitinib exposure, the variant allele frequency of ASXL1 mutations only increased in 1 patient (Fig. 2F). This finding argues again for the specific effect of ruxolitinib on RAS pathway mutations selection.
Causes of death in non-ruxolitinib-treated patients included hemorrhage, infection, and global condition deterioration, with 4 patients who died in an AML/MDS disease stage, while 5 died in chronic MPN (Table S8).
Taken together, our results suggest that ruxolitinib-induced selection of RAS pathway mutations represents an unfavorable prognostic factor associated with poor clinical outcome, including both a decreased OS and increased MDS/AML transformation risk. We also unmask a differential prognosis associated with RAS mutations according to ruxolitinib treatment status, suggesting that ruxolitinib exposure might increase RAS-mutated clones oncogenic fitness.
Ruxolitinib treatment positively selects RAS mutant clones both in the context of JAK/STAT hyper-activation and with wild-type (WT) JAK/STAT
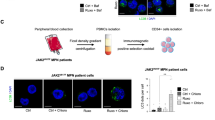
To evaluate the mechanistic link between ruxolitinib exposure and RAS pathway mutations selection, we next investigated whether ruxolitinib positively selects RAS mutant clones through ex vivo treatment of primary human CD34+ hematopoietic cells sorted from peripheral blood mononuclear cells of patients with RAS-mutated MPN (n = 6). RAS mutation allele frequency increased across all six patients upon in vitro ruxolitinib treatment (Fig. 3A). In contrast, driver mutations’ (JAK2, CALR, or MPL) allele frequencies decreased upon ruxolitinib exposure in five out of six patients, suggesting that RAS-mutated clones might not always be present within the MPN driver clone (Fig. 3A). Evolution of the variant allele frequencies of the remaining mutations are presented in Fig. S2. Of note, although no mutation was found in the RAS pathway regulator PTPN11 across our cohort, ruxolitinib ex vivo treatment of our primary patient samples revealed a PTPN11Q510E low VAF mutation that was not detected prior to ruxolitinib (Patient#2, Fig. S2). To study the clonal architecture of these samples, we performed single-cell DNA sequencing using a custom-made targeted panel. RAS mutations were either present within the MPN driver clone, or in a clone without a driver mutation, suggesting that ruxolitinib-induced RAS clonal selection can occur both in a JAK/STAT hyper-activated or WT context (Fig. 3B). Furthermore, longitudinal molecular evaluation of two of these patients treated with ruxolitinib for their MPN showed an increase in the allele frequency of RAS mutations over time, confirming in vivo in patients the impact of ruxolitinib treatment on RAS mutations selection. Notably, in one of these patients chronic MPN transformed into a secondary myelodysplastic syndrome harboring a dominant KRAS mutated clone (Fig. 3C).
A Graphical representation of the allele burden of RAS mutations and driver mutations detected 10 days after DMSO or ruxolitinib (20 nM) in vitro treatment of CD34+ hematopoietic cells derived from six MPN patients harboring RAS mutations. B Clonal architecture of CD34+ hematopoietic cells derived from targeted single-cell DNA sequencing of samples from four patients with MPN harboring RAS mutations. Each circle represents a single clone with the corresponding mutated genes below (Het = heterozygous, Hom = homozygous). Mutations acquired at each step are written in red. Red circles correspond to RAS-mutated clones. C Fish Plots depicting clonal evolution between MPN diagnosis and last molecular evaluation for two patients with MPN treated with ruxolitinib. RAS-mutated clones are highlighted in red. Source data are provided as a Source Data file.
MAPK pathway activation confers a fitness advantage and increased clonogenic potential to JAK2 V617F, CALR del52 and WT hematopoietic cells upon ruxolitinib exposure
To further validate the effects of ruxolitinib on Jak2WT hematopoietic cells, we next performed competitive co-culture experiments using murine lineage negative (Lin−) bone marrow cells isolated from CD45.1/2 Jak2WT NrasG12D transgenic mice or from syngeneic CD45.1 Jak2WT NrasWT C57BL/6 mice. Starting from a 50/50 or a 20/80 Nras mutant to WT cellular ratio, we observed a time-dependent enrichment in mutant cells at the expense of WT cells, suggesting that NrasG12D cells are less sensitive than normal cells to the JAK inhibitor's anti-proliferative effect (Fig. 4A, S3A). Indeed, when untreated, NrasWT cells had a higher proliferation rate than NrasG12D cells. However, while NrasWT cells were sensitive to ruxolitinib treatment, NrasG12D cells displayed an increased proliferation rate after ruxolitinib exposure in comparison with non-treated NrasG12D cells (Fig. 4B). Consistently, an in vivo bone marrow transplantation competition experiment further validated the increased fitness advantage of CD45.1/2 Jak2WT NrasG12D cells in comparison with CD45.2 Jak2V617F NrasWT cells upon ruxolitinib selective pressure. A mixture of 10% Jak2WT NrasG12D lineage negative cells and 90% Jak2V617F NrasWT cells were co-transplanted within lethally irradiated secondary C57BL/6 recipient mice, to mimic the clonal selection situation represented by Patient#1 in Fig. 3C, in which the RAS mutation was present in a non-driver subclone in competition with the main RASWTJAK2V617F driver clone. Ruxolitinib treatment was initiated 4 weeks after bone marrow transplantation (Fig. 4C). CD45.1/2 and CD45.2 peripheral blood flow cytometry monitoring showed an increased proportion of NrasG12D mutant cells at the expense of NrasWT cells in the ruxolitinib-treated group in comparison with vehicle-treated mice (Fig. 4D).
A Percentage of CD45.1/2 NrasG12D cells among CD45.1/2 NrasG12D and CD45.1 NrasWT Lin− murine cells (50/50 ratio) treated with DMSO or ruxolitinib (0.25 µM). B Proliferation curves of NrasG12D or NrasWT Lin− cells after DMSO or ruxolitinib (0.25 µM). A, B Mean of n = 2 biological replicates. C In vivo NrasG12D competition model. Created in BioRender59. D Percentage of CD45.1/2 NrasG12D cells in peripheral blood of mice transplanted with 10% CD45.1/2 Jak2WT NrasG12D and 90% CD45.2 Jak2V617F NrasWT Lin− cells (n = 4 per group). Ruxolitinib (90 mg/kg twice daily) was started 4 weeks post-transplantation. E In vivo NrasQ61K competition model. Created in BioRender59. F Percentage of GFP+ NrasQ61K Jak2V617F cells in the bone marrow of mice transplanted with Jak2V617F cells expressing a GFP+ NrasQ61K vector (n = 5 per group). Ruxolitinib (90 mg/kg twice daily) was started 3 weeks post-transplantation. G, H Colony formation assay for cKit+ bone marrow cells from Jak2WT (G) or Jak2V617F (H) mice expressing Empty or NrasQ61K vectors. Colony number after at least 6 days of DMSO or ruxolitinib (1 µM) (n = 4 biological replicates). I Western blot for the indicated proteins in HEL cells expressing GFP, Empty or NRASQ61K vectors, treated 24 h with ruxolitinib (3 µM). J In vitro competition model. Created in BioRender59. K Percentage of GFP+ and GFP−_Empty or GFP−_NRASQ61K HEL or UKE-1 cells. 24 days after ruxolitinib (3 µM and 15 µM for HEL and UKE-1, respectively), the indicated cells were treated 3 days with ruxolitinib or ruxolitinib (Ruxo.) and trametinib (Trame.) (10 µM and 0.1 µM for HEL and UKE-1, respectively) (n = 3 biological replicates). L Western blot for the indicated proteins in Ba/F3 MPL-CALRWT and MPL-CALRdel52 cells expressing Empty or NRASQ61K vectors, treated 24 h with ruxolitinib (75 nM). M Percentage of Crimson+ and Crimson−_Empty or Crimson−_NRASQ61K Ba/F3 MPL-CALRWT and MPL-CALRdel52 cells after ruxolitinib (n = 3 biological replicates). Statistical significance using two-tailed Mann–Whitney (D, F) or Welch’s t-test (H, K, M). Experiments(I, L) were performed twice with similar results. Error bars represent mean ± SEM. P-values in the figure. Source data provided as Source Data file. Schemas created using BioRender.
To extend our findings to the Jak2V617F hematopoietic cellular context, thus mimicking patients for whom RAS pathway mutations are harbored within the JAK/STAT activated MPN driver clone such as Patient#4 in Fig. 3C, we performed a second in vivo competition experiment. cKit+ murine myeloid cells isolated from NrasWT Jak2V617F transgenic mice were transduced with an exogenous GFP-NrasQ61K-expressing vector and transplanted into lethally-irradiated secondary C57BL/6 recipient mice. Ruxolitinib treatment was started 3 weeks after bone marrow transplantation (Fig. 4E). In vivo GFP monitoring through bone marrow biopsy, revealed an increased NrasQ61K cellular proportion in mice treated with ruxolitinib, in comparison with vehicle-treated control mice (Fig. 4F).
To further evaluate the impact of ruxolitinib on the oncogenic potential of RAS clones, we next sought to study its effects on the clonogenic potential of NrasQ61K cells. We thus performed a colony formation assay on cKit+ murine cells, isolated from Jak2WT or Jak2V617F C57BL/6 mice, transduced with an empty or a NrasQ61K expressing vector. While non-RAS mutant cells were sensitive to ruxolitinib with a serially decreased clonogenic potential, ruxolitinib did not decrease Jak2WT NrasQ61K cells clonogenic potential, and even increased NrasQ61K cells clonogenic potential in a Jak2V617F cellular context (Fig. 4G, H).
To further evaluate the effects of ruxolitinib treatment on human RAS-mutated cells, we transduced HEL and UKE-1 JAK2V617F human myeloid cell lines with an empty or an NRASQ61K encoding vector. Activation of the MAPK pathway upon exogenous NRASQ61K expression and inhibition of the JAK/STAT pathway after ruxolitinib treatment were validated by western blot. Ruxolitinib treatment for 24 h also resulted in decreased ERK phosphorylation in a RAS WT context (Fig. 4I), in line with the role of JAK kinases in activating not only the STAT pathway but also parallel cell survival signals including the MAPK pathway14. First, we evaluated the effects of ruxolitinib treatment on each cell type through a growth inhibition experiment. While RAS WT cell growth was impaired after 6 days of ruxolitinib treatment, NRASQ61K cells were resistant to JAK/STAT inhibition, in both HEL and UKE-1 cell lines (Fig. S3B). Using our set of transduced cells, we then designed an in vitro competition assay. Empty vector or NRASQ61K expressing cells were put in competition with GFP expressing cells at a 20/80 ratio, prior to treatment with either DMSO or ruxolitinib (Fig. 4J). The proportion of GFP+ cells was evaluated by flow cytometry every 12 days for a total of 24 days post-ruxolitinib treatment initiation. Ruxolitinib treatment did not strongly impact the empty vector / GFP control cells ratio. Compared to empty vector cells, we observed a decrease of untreated NRASQ61K cellular fitness, consistent with the previously reported growth arrest of RAS-mutated cells through oncogene-induced senescence27. Interestingly, this phenotype was not only reverted by ruxolitinib treatment, but we observed a time-dependent increase of the NRASQ61K / GFP cellular ratio under ruxolitinib treatment, suggesting that RAS-mutant cells are more resistant to JAK/STAT pathway inhibition than RAS wild-type cells. To confirm that this resistant phenotype is dependent on mutant RAS induced MAPK pathway activation, we exposed our NRASQ61K / GFP cellular mix previously treated with ruxolitinib, to either DMSO, ruxolitinib or a combination of ruxolitinib and the MEK1/2 inhibitor trametinib. Trametinib treatment partially reverted the ruxolitinib-induced NRASQ61K cellular fitness increase, suggesting that the MAPK pathway inhibition re-sensitizes NRASQ61K cells to ruxolitinib (Fig. 4K). These results are in line with the previously described synergistic effects of JAK/STAT and MEK/ERK inhibition in MPN murine models28.
Importantly, NrasG12D, NRASG12V, and KRASG13D HEL expressing cells were also resistant to JAK/STAT inhibition (Fig. S3C, D), and ruxolitinib exposure resulted in the positive selection of NRASG12V and KRASG13D HEL expressing cells (Fig. S3E), highlighting a similar impact on different RAS isoforms and mutations.
Additionally, to evaluate the effect of ruxolitinib on the clonal selection of RAS mutations within a CALR mutated context, we transduced Ba/F3 cells expressing exogenous human MPL and CALRWT or CALRdel5229 with an empty or a NRASQ61K encoding vector (Fig. 4L). While RAS WT cell growth was impaired after 3 days of ruxolitinib treatment, Ba/F3 MPL-CALRWT-NRASQ61K and Ba/F3 MPL-CALRdel52-NRASQ61K cells were resistant to JAK/STAT inhibition (Fig. S3F). Ruxolitinib treatment also resulted in the positive selection of Ba/F3 MPL-CALRWT-NRASQ61K and Ba/F3 MPL-CALRdel52-NRASQ61K cells in our in vitro competition assay (Fig. 4M).
Finally, to further evaluate the specificity of ruxolitinib-mediated RAS clonal selection in vitro, we assessed the effect of ruxolitinib exposure on ASXL1 mutations selection. Given the controversy around the gain- or loss-of-function impact of ASXL1 mutations in myeloid malignancies30, we developed two complementary strategies to evaluate the impact of ruxolitinib on clonal selection of HEL cells: 1) overexpression of an ASXL1G646Wfs *12 mutation (Fig. S4A–C), or 2) ASXL1 knock-down using 2 shRNAs targeting ASXL1 (Fig. S4D–F). In both cases, ruxolitinib exposure only mildly remodeled the fitness of the ASXL1 clone (Fig. S4A–F), suggesting a specific effect of ruxolitinib on RAS pathway-mutated clones.
Taken together, our results argue that MAPK pathway activation in the context of RAS pathway mutations drives ruxolitinib resistance, enabling cellular escape from JAK/STAT pathway growth inhibition and a subsequent fitness advantage over RAS WT clones.
JAK2 Inhibition mediates ruxolitinib’s enhancement of RAS mutant cell fitness and increased clonogenic potential
As an orthogonal approach to evaluate the direct effect of ruxolitinib-induced JAK2 inhibition on RAS clonal selection, we next used shRNA-mediated Jak2 suppression. First, we transduced 32D murine cells with a CRIMSON, an empty, a GFP-NrasQ61K or a GFP-NrasQ61K-shJak2 encoding vector. A total of 10 shRNAs targeting Jak2 were used in a pooled fashion. NRAS overexpression and Jak2 knockdown were validated by qPCR (Fig. 5A). While NrasQ61K cellular viability was decreased in comparison with non-mutant cells, Jak2 knockdown fully reverted this effect, enabling RAS mutant cells to increase their proliferative capacity (Fig. 5B).
A qRT-PCR for Nras and Jak2 expression level in murine 32D cells infected with an Empty, a NrasQ61K or a NrasQ61K-shJak2 encoding vector. Statistical significance was determined using a two-tailed Welch’s t-test. Error bars represent the mean of n = 4 biological replicates ± SEM. B Growth inhibition of 32D murine cells expressing an Empty, a NrasQ61K or a NrasQ61K-shJak2 encoding vector 6 days after GFP sorting. Statistical significance was determined using a two-tailed Welch’s t-test. Error bars represent the mean of n = 6 biological replicates ± SEM. C Model of the in vitro NrasQ61K-shJak2 murine myeloid cells competition assay. Created in BioRender59. D Percentage of GFP+_CRIMSON− 32D cells expressing an Empty, a NrasQ61K or a NrasQ61K_shJak2 encoding vector 3 days after sorting. Statistical significance was determined using two-tailed Welch’s t-test. Error bars represent mean of n = 3 biological replicates ± SEM. E–F Colony formation assay of Jak2WT (E) or Jak2V617F (F) C57BL/6 primary bone marrow c-Kit+ murine cells expressing an Empty, a NrasQ61K or a NrasQ61K-shJak2 encoding vector at least 6 days after GFP sorting. Statistical significance was determined using a two-tailed Welch’s t-test. Error bars represent the mean of n = 4 biological replicates ± SEM. G Genetic variants association with resistance to the JAK2 inhibitors ruxolitinib (n = 497), momelotinib (n = 476) and fedratinib (n = 93) according to the Beat AML v2 human primary patient AML dataset. Data is presented as volcano plots for gene variants' effect size (Glass) on the x-axis versus –log10(P-value) on the y-axis. Significance was set at – log10(P-value)>1.3, based on the two-tailed Student t-test with Welch’s correction, with variants having a negative effect size associated with drug sensitivity (red), while those having a positive effect size associated with drug resistance (blue). P-values are reported in the figure. Source data provided as a Source Data file.
Using our set of transduced cells, we then performed an in vitro competition experiment between CRIMSON expressing cells and either empty, GFP-NrasQ61K or GFP-NrasQ61K-shJak2 expressing cells, at an 80/20 cellular ratio, respectively (Fig. 5C). CRIMSON and GFP markers allowed cellular proportion monitoring by flow cytometry. NrasQ61K expression reduced cellular fitness in comparison with non-Ras mutant cells, while GFP-NrasQ61K-shJak2 cells had a partially restored cellular fitness, suggesting that Jak2 knockdown increased Ras mutant cells fitness (Fig. 5D).
To further validate the effect of JAK2 impairment, we performed a colony formation assay on cKit+ murine cells obtained from Jak2WT or Jak2V617F transgenic mice, transduced with an empty, a NrasQ61K or a NrasQ61K-shJak2 expressing vector. While the colony-formation ability of NrasQ61K cells decreased over serial replating in comparison with empty vector transduced cells, JAK/STAT signaling pathway impairment through Jak2 knock-down restored NrasQ61K cells colony-formation ability (Fig. 5E–F). Conversely, Jak2 knock-down reduced NrasWT cells’ colony-formation ability (Figs. S5A, B).
These results are consistent with the effects of ruxolitinib treatment in increasing RAS-mutated cell fitness in vivo and increasing their clonogenic and stemness potential in vitro.
Finally, to gain more confidence in this JAK2-mediated on-target effect, we evaluated the effect of other JAK2 inhibitors harboring diverse specificities against the other JAK family members in primary AML samples. To identify specific mutational backgrounds associated with JAK2 inhibitor response in myeloid malignancies, we interrogated the Beat AML patient cohort31. Notably, KRAS and NRAS genetic alterations were among the top scoring variants associated with resistance to ruxolitinib, but also to momelotinib and fedratinib, two more recently FDA approved JAK2 inhibitors, thus arguing for an on-target therapeutic class effect (Fig. 5G).
Altogether, our results confirm that ruxolitinib-mediated RAS clonal selection is not due to an off-target effect, but rather to a specific JAK2 mediated JAK/STAT pathway inhibition.
MAPK pathway activation is associated with JAK2 impairment resulting in RAS-mutated cells release from oncogene-induced senescence and increased oncogenic potential
Finally, we sought to further understand the oncological ontogeny of this seemingly paradoxical cooperation between activation of MAPK signaling and inhibition of the JAK/STAT oncogenic pathway. Indeed, exploring this oncogenic cooperation could help further decipher the enigmatic positive impact of JAK/STAT signaling pathway inhibition on RAS mutant cell proliferation.
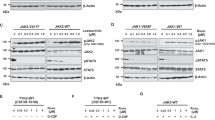
We thus explored the effects of MAPK activation on JAK2 expression. NRASQ61K overexpression in both HEL and UKE-1 JAK2V617F mutated cell lines resulted in the decrease of JAK2 mRNA and protein levels (Fig. 6A, B). This effect was also observed in a JAK2WT cellular context (Fig. 5A). To further evaluate the relevance of this divergent transcriptional regulation, we evaluated whether JAK/STAT down-regulation correlated with MAPK pathway activation in the Beat AML patient cohort31. Patients were classified as JAK2 “low” or JAK2 “high” according to their JAK2 mRNA expression level (Fig. S6A) and their enrichment for three previously reported gene signatures: JAK2 direct targets established from JAK2 shRNA knock-down in the HEL cell line32 and JAK2V617F homozygous or heterozygous mutants established in MPN primary patient samples32 as evaluated by single sample Gene Set Enrichment Analysis (ssGSEA) (Fig. S6B). Next, we performed GSEA to identify differentially expressed gene sets between the JAK2 “low” (n = 60) and JAK2 “high” (n = 64) groups. We found that RAS/MAPK gene sets are enriched in the JAK2 “low” patient samples (Fig. 6C–E and S6C), in line with our in vitro findings in HEL and UKE-1 cell lines under ruxolitinib treatment. Interestingly, along with the RAS/MAPK enrichment in the JAK2 “low” group, we also observed a significant enrichment of cell-cycle related gene sets (Fig. 6C–E and S6D). Accordingly, ruxolitinib treatment enabled the cycling of NRASQ61K cells as shown by propidium iodide-based cell cycle analysis. Indeed, in comparison with the DMSO control condition, a higher proportion of NRASQ61K ruxolitinib-treated cells were in the G2/M phase, arguing for a released cycling checkpoint responsible for increased RAS mutant cells proliferation upon JAK/STAT pathway inhibition (Fig. 6F). Additionally, an EdU incorporation assay showed increased cell cycling upon ruxolitinib exposure in an NRASQ61K context in contrast to the effect of ruxolitinib on RASWT cells (Fig. 6G). Given the potential for RAS oncogene-induced senescence to hamper RAS oncogenesis27, we next investigated whether the impact of JAK/STAT pathway inhibition on NRASQ61K expressing HEL and UKE-1 cell lines cycling and fitness, could potentially be due to a release from the senescent cellular state caused by the NRASQ61K mutation. As expected, NRASQ61K mutant cells displayed a highly senescent phenotype based on a β-galactosidase staining assay. Intriguingly, the number of NRASQ61K senescent cells was decreased in the context of 6 days of ruxolitinib treatment in both HEL and UKE-1 cell lines (Fig. 6H, I). Finally, we evaluated whether extrinsically overexpressing the constitutively active form of JAK2 (JAK2V617F) could impair NRASQ61K mutant cell growth. Indeed, when the JAK/STAT pathway was further activated in the HEL cell line, NrasQ61K leukemic cells growth was impaired in comparison with NrasQ61K mutant cells in the absence of JAK2V617F exogenous overexpression (Fig. 6J and S7). Altogether, our results suggest that JAK/STAT pathway inhibition might be necessary to fully enable RAS-induced oncogenic potential, thus explaining the divergent transcriptional regulation of JAK2 and RAS.
ANRAS and JAK2 expression level qRT-PCR in Empty or NRASQ61K HEL and UKE-1 cells. Error bars represent mean of n = 3 (HEL) and n = 4 (UKE-1) biological replicates ± SD. B Western blot for indicated proteins, in Empty or NRASQ61K HEL and UKE-1. The experiment was performed twice with similar results. C, D Gene Set Enrichment Analysis (GSEA) for the AML dataset Beat_AML_v2, across the collection of MSigDB_v7.4 Hallmark (50) and additional RAS/MAPK (24) and cell cycle (11) gene sets. Data represented as volcano plots (C) of –log10(p-value + 0.0001) versus the Normalized Enrichment Score (NES) for each gene set. NES using Kolmogorov-Smirnov enrichment test. Gene sets related to RAS/MAPK and Cell Cycle highlighted in red and blue respectively. Gray dots indicate all other. Representative GSEA plots (D) for gene sets related to RAS/MAPK and Cell Cycle. E Heatmaps depicting relative gene expression changes for leading edge genes of the top RAS/MAPK and Cell Cycle enriched gene sets. Depleted and enriched genes are respectively in blue and red. Row-normalized data. F Propidium Iodide cell cycle analysis of Empty or NRASQ61K HEL and UKE-1 cells after 11 or 9 days of ruxolitinib (3 and 15 µM, respectively). Cells in sub G1 not represented. Mean of n = 2 biological replicates. G EdU incorporation of Empty or NRASQ61K HEL and UKE-1 cells after 12 days of ruxolitinib (3 and 15 µM, respectively). Error bars represent the mean of n = 3 biological replicates ± SEM. H, I Beta-galactosidase staining for Empty or NRASQ61K HEL and UKE-1 cells after 6 days of ruxolitinib (3 and 15 µM, respectively). Quantification (H) and representative images (I). Error bars represent mean of n = 2 (HEL) and n = 3 (UKE-1) biological replicates ± SD. J Proliferation curves of HEL cells expressing Empty, NrasQ61K, JAK2V617F or NrasQ61K and JAK2V617F vectors. Error bars represent mean ± SD of n = 10 (day 0), n = 8 (days 3, 6), n = 4 (day 9) biological replicates. Statistical significance using two-tailed Welch’s t-test (A, F–H, J). P-values are reported in the figure. Source data provided as a Source Data file.
Discussion
Our findings strongly argue for a role for JAK2 inhibition in the selection of RAS pathway mutated clones in MPNs. This observation was suggested by Mylonas et al., who evaluated clonal evolution across 15 JAK inhibitor-treated patients with myelofibrosis33. However, the absence of a non-treated control group did not allow the exclusion of RAS mutations acquisition as part of the natural history of myelofibrosis, thus precluding any association with ruxolitinib exposure. As in patients with AML treated with FLT3 inhibitors, the emergence of RAS pathway mutated clones in ruxolitinib-treated patients could be due to a specific resistance induced by the upregulation of RAS signaling 3. In fact, JAK2 downstream effects are not only driven by STAT3/5 phosphorylation, but also by the activation of complementary signaling pathways such as PI3K/AKT and MAPK/ERK14. In line with this, both intrinsic and extrinsic MAPK activating mechanisms have been reported as limiting ruxolitinib treatment efficacy in in vitro and in vivo MPN models28,34. Indeed, PDGFRA remains activated under ruxolitinib, leading to ERK activation in JAK2V617F and MPLW515L MPN models28. Additionally, upon phosphorylation of the splicing factor YBX1 by mutated JAK2, YBX1 intrinsically mediates MNK1 mRNA splicing, resulting in ERK phosphorylation and survival of JAK2-mutated cells34. Combinatorial therapeutic options with MEK and ERK inhibitors have thus emerged to increase ruxolitinib’s efficacy28,34,35 (NCT04097821). In line with this, the ectopic expression of NRASG12V reduced the sensitivity of JAK2V617F-mutated cell lines to the BAD-dependent pro-apoptotic effects of ruxolitinib36. Complementing these findings, our results further demonstrated the ability of RAS signaling activation to confer a constitutive resistance mechanism to JAK inhibitors within the MPN driver clone but also within neighboring non-driver clones. Importantly, while our primary patient and in vitro data points towards a similar RAS clonal selection across patients harboring the three MPN driver mutations, the majority of our validation experiments were conducted within a JAK2V617F mutational context and further experimental data is required to more precisely evaluate the interaction between RAS and CALR/MPL mutations.
In addition to the ruxolitinib resistance driven by the intrinsic MAPK activation, our findings suggest that JAK2 downregulation could be required in RAS-mutated cells to finely tune the needed MAPK oncogene dosage, highlighting the fact that hematopoietic cells might only tolerate a certain amount of MAPK. This is reminiscent of the “Goldilocks principle” of oncogene pathways, where an ideal amount of MAPK is required for oncogenesis37. Accordingly, our findings show that further activation of the MAPK pathway through constitutive activation of JAK2 results in an anti-oncogenic effect, while further JAK2 impairment through exposure to ruxolitinib results in an increase in the oncogenic potential of RAS-mutated clones. In line with this concept, our data shows that RAS mutant clonal selection is faster in a non-JAK/STAT hyper-activated cellular context. Although an oncogene overdose principle has been documented for several oncogenic pathways, including MAPK, PI3K/AKT and WNT38, its mechanistic underpinnings remain poorly understood. A few studies suggested increased oncogene-induced stress as the main driver of this phenomenon39,40, in line with our findings of JAK/STAT pathway inhibition releasing NRAS-mutated cells from oncogene-induced senescence. Another mechanistic explanation has been recently reported in B cell leukemia, where the analysis of 1148 leukemia genomes revealed that STAT5 and ERK activating oncogenic lesions are almost mutually exclusive and segregate to competing clones41. STAT5 and ERK induce opposing transcriptional programs implicated in the pro-B to pre-B cellular transition. Therefore, experimental reactivation of the impaired pathway comes at the expense of the oncogenic potential of the driver oncogene. Our results complement these findings and highlight in patients the counter-intuitive oncogenic risk of impairing oncogenic pathways in specific mutational contexts.
The requirement of JAK inhibition for RAS-induced oncogenesis, as shown here in myeloid malignancies, might also be investigated in tumors with frequent RAS pathway gene mutations, such as pancreatic, skin, and lung cancers42. Interestingly, tumor progression has hampered the clinical development of ruxolitinib in several solid tumors (NCT01562873, NCT00638378, NCT02117479, NCT02119676, NCT02119650)43. Additionally, ruxolitinib treatment has been recently associated with an increased incidence of aggressive non-melanoma skin cancers44. In light of our results, the high dependency of cutaneous squamous cell carcinomas on MAPK pathway activation suggests a similar oncogenic mechanism potentially resulting in increased fitness of RAS-mutated skin cells. Targeting specific oncogenic pathways could therefore represent an extrinsic path to increased oncogenesis, reflecting the major challenge of targeting heterogeneous and adaptive diseases such as cancer. Our results warrant the search for such oncogenic interactions in order to improve our approach to the application of targeted therapies to cancer, as well as to benign disorders where patients may carry phenotypically silent somatic mutations45.
In terms of clinical implications for the management of patients with MPN, our results suggest that ruxolitinib-induced selection of RAS-mutated clones might negatively impact prognosis in patients with myelofibrosis, particularly in the DIPSS intermediate-2/high risk group. NRAS and KRAS genes were identified three decades ago as important oncogenes in hematopoietic malignancies, harboring recurrent activating mutations46,47,48,49,50. Importantly, although our study highlights the impact of ruxolitinib on clonal selection of diverse RAS mutant cells, isoform- and mutation-specific differences in protein structure and signaling are described, and further studies are required to more precisely identify potential differences upon ruxolitinib exposure51. NRAS and KRAS mutations have only been recently associated with a poorer outcome and treatment resistance in MPN25,26,52. In our study, we confirmed the poor prognostic value of RAS pathway mutations in myelofibrosis. However, we showed that poorer OS and TFS related to these mutations are mainly observed in patients treated with ruxolitinib, while OS and TFS did not seem to be statistically impacted in patients that did not receive JAK2 inhibitor therapy. This observation suggests that the selective pressure induced by ruxolitinib could adversely impact patient outcome. Importantly, long-term findings from ruxolitinib phase 3 clinical trials did not report a higher incidence of secondary AML in the ruxolitinib-treated group7,8,53,54. However, the re-analysis of long-term clinical trials harbors a number of biases driven by the fact that these studies were not designed for long-term randomized groups’ comparison. Cross-over to the ruxolitinib arm, absence of longitudinal NGS molecular follow-up and the low rate of RAS mutations and leukemic transformation events in MPN, could have masked such observations. Given the low number of patients with RAS pathway mutations in the non-ruxolitinib treated patient group and in the DIPSS low/intermediate-1 risk categories in our study, our clinical findings need to be confirmed in larger external cohorts and across all DIPSS categories. Additionally, our results argue for an on-target JAK2 inhibition therapeutic class effect, suggesting similar findings could be observed in the clinical setting of more recently approved JAK2 inhibitors such as momelotinib, fedratinib and pacritinib. Although our results strongly suggest that JAK2 inhibitors may favor the emergence of RAS pathway mutant clones, only a subset of patients harbor such clones (24% versus 8% of ruxolitinib-treated and non-treated patients, respectively). In the context of such diseases with very limited therapeutic options, JAK2 inhibitors remain a key option in the MPN therapeutic arsenal with clear benefit for symptom management and potential survival benefits for a subset of patients.
Detailed information on ruxolitinib dose was not available for our retrospective cohort, precluding analysis of its potential impact on clinical outcomes. This should be further evaluated in future studies to inform the management of patients harboring RAS pathway mutations. Among the 8 patients with newly acquired RAS pathway mutations within the ruxolitinib-treated cohort, 6 initially responded to ruxolitinib treatment (5 clinical improvement (CI), 1 partial response (PR)), while 2 did not respond (1 stable disease (SD), 1 progressive disease (PD)). Among these patients, 5 continued on ruxolitinib treatment after identification of the RAS mutation while ruxolitinib was interrupted for the 3 others. Given the retrospective nature of our study, patients’ treatment was not specifically adjusted after identification of RAS pathway mutations as the impact of ruxolitinib on RAS-mutated patient prognosis was not known at the time. Our findings argue for the implementation of sensitive molecular testing for early detection of RAS-mutated clones prior to JAK2 inhibitor initiation. Additionally, regular molecular follow-up followed by treatment adjustment, when possible, might also be considered. Treatment adaptation could include rationally designed combinatorial therapeutic strategies currently being evaluated in clinical trials.
In conclusion, our translational study revealed a selection of RAS mutations upon ruxolitinib exposure, negatively impacting clinical outcomes in patients with MPN. The enhanced oncogenic potential of RAS mutations after inactivation of the oncogenic JAK/STAT pathway illuminates an intriguing paradoxical oncogenic mechanism, highlights the complexity of the cancer combinatorial mutational landscape and challenges our current approach to targeted therapy use in cancer. Clinically, our results underscore the importance of monitoring adaptive responses to JAK inhibitors and considering multidrug combination approaches for patients with RAS mutated MPN. More broadly, in the context of a growing number of emerging JAK inhibitors with many novel therapeutic indications for both malignant and benign diseases, our findings highlight the importance of careful monitoring for the emergence of RAS-driven malignancies among patients treated with JAK inhibitors.
Methods
Study approvals
Patients’ study
The study was performed in accordance with the ethical guidelines of the Declaration of Helsinki, and was approved by our institutional review board (APHP Paris Nord IRB00006477, CER-2020-55). Patients provided informed consent for molecular and clinical data analysis. A unique anonymized database was established and housed in a secured system, meeting the security standards required by the protection of personal data law promulgated on 20/06/2018 in FRANCE. A diagnosis of Compliance and Security Research was carried out and approved by the data protection reference department of Saint-Louis Hospital.
Animal study
The French National Ethics Committee on Animal Care reviewed and approved all mouse experiments described in this study (APAFIS #34469-2021122222426491 v3). Housing conditions within Saint-Louis research institutes’ animal facility satisfy the French National Ethics Committee on Animal Care requirements.
Cell culture
Cell lines
32D cell line was a kind gift of Dr Iannis Aifantis to Camille Lobry (Saint-Louis Research Institute, Paris, FRANCE). HEL cell line was purchased from the American Type Culture Collection. UKE-1 cell line was a kind gift of Dr Walter Fiedler (University Hospital Eppendorf, Hamburg, GERMANY). Ba/F3 cells expressing exogenous MPL and CALRWT or CALRdel52 (Ba/F3 MPL-CALRWT and Ba/F3 MPL-CALRdel52 cell lines) were previously published29. All post-MPN AML cell lines were confirmed by STR genotyping. HEL cells were maintained in RPMI 1640 (Gibco) supplemented with 1% penicillin-streptomycin (Gibco) and 10% fetal bovine serum (Sigma-Aldrich) at 37 °C with 5% CO2. UKE-1 cells were maintained in IMDM (Gibco) supplemented with 1% penicillin-streptomycin (Gibco), Hydrocortisone (1µmol/L), and 20% fetal bovine serum (Sigma-Aldrich) at 37 °C with 5% CO2. 32D cells were maintained in RPMI 1640 (Gibco) supplemented with 1% penicillin-streptomycin (Gibco), 10% fetal bovine serum (Sigma-Aldrich), and 1 ng/ml of mIL-3 (Peprotech) at 37 °C with 5% CO2. Ba/F3 cells were maintained in RPMI 1640 (Sigma-Aldrich) supplemented with 10% fetal bovine serum (Sigma-Aldrich), 1% penicillin-streptomycin (Gibco,) and 3 ng/mL of mIL-3 (Peprotech). HEK-293T cells were maintained in DMEM (Gibco) supplemented with 1% penicillin-streptomycin (Gibco) and 10% fetal bovine serum (Sigma-Aldrich).
Primary cells
Primary CD34+ cells were magnetically sorted (EasySep Human CD34 Pos Selection Kit, Cat #17856, Stemcell Technologies) from primary MPN patients’ peripheral blood mononuclear cells (PBMC) and cultured in CTS™ StemPro™ HSC Expansion Medium (Thermo Fisher) with the appropriate cocktail of cytokines: IL-3 (50 ng/L), SCF (50 ng/L), TPO (100 ng/L), IL-6 (50 ng/L), FLT3-Ligand (100 ng/L) (Peprotech). This experimental workflow was specifically developed by our team to pre-clinically study drug effects on primary CD34+ cells extracted from MPN patients.
To obtain NrasG12D primary hematopoietic murine cells, total bone marrow cells were isolated from femurs and tibias of 6 to 12-week-old female hMRP8-NrasG12D mice55 and B6.SJL-PtprcaPepcb/BoyCrl Ly5.1 congenic mice (Charles River Laboratories), by flushing from the bone cavity with ice-cold PBS using a 21 G x 1.5 needle, then passed through a 70 µM cell strainer to obtain a single cell suspension, followed by red blood cell lysis (Qiagen). Lineage-negative (Lin-) cells were then magnetically sorted (EasySep Mouse Hematopoietic Progenitor Cell Isolation, Cat #19856 A, Stemcell technologies), and maintained in StemSpan SFEM (StemCell Technologies) supplemented with 1% penicillin-streptomycin (Gibco) and 20 ng/ml mIL-3, 100 ng/ml mFLT3-ligand, and 100 ng/ml mSCF (Peprotech).
To obtain Jak2V617F and Jak2WT primary hematopoietic murine cells, total bone marrow cells were isolated from femurs, spine and tibias of 6 to 12 week-old Wild-Type C57BL/6JOlaHsd (Envigo) or Vav-cre X Floxed-Jak2V617F C57BL/6 mice56, by bone crushing then passed through a 70 µM cell strainer to obtain a single cell suspension, followed by red blood cell lysis (Sigma-Aldrich). c-Kit+ cells were then magnetically sorted (CD117 MicroBeads, mouse, Cat #130-091-224, Miltenyi Biotec) and maintained in StemSpan SFEM (StemCell Technologies) supplemented with 1% penicillin-streptomycin (Gibco) and 10 ng/ml mIL-3, 10 ng/ml IL-6, 25 ng/ml mFLT3-ligand, and 25 ng/ml mSCF (Peprotech).
Methylcellulose colony formation assay
Eight thousand c-Kit+ primary murine cells were plated in four replicates into semisolid methylcellulose medium (MethoCult M3534, StemCell Technologies) supplemented with 25 ng/ml mFLT3-ligand. After at least 6 days, the colony number was counted. The remaining cells of the same condition were then resuspended, pooled, and washed once in PBS, prior to their replating at 8000 cells or a lower number if <8000 cells per replicate were recovered.
RNA extraction and qRT-PCR analysis
RNA was extracted using RNeasy Mini Kit (Qiagen) and retrotranscription was performed using SuperScript IV Reverse Transcriptase (Invitrogen) per manufacturer’s protocols. Real-time qPCR was performed using TaqMan (Applied Biosystems) or KAPA SYBR® FAST (KAPABIOSYSTEMS) protocols provided by the supplier. Data was analyzed using the QuantStudio real time PCR instrument and software (Thermofischer).
The list of probes and primers used for real time qPCR is provided in Table S9.
Growth measurement
To assess growth, cells were plated in 384-well cell-culture coated white plates. ATP content was then measured using CellTiter-Glo (Promega) per the manufacturer’s instructions. Briefly, CellTiter-Glo reagent was added to each well and incubated at room temperature for 25 min prior to luminescence reading at the indicated time points.
Western Blotting
Cells were lysed in Cell Signaling Lysis Buffer (Cell Signaling Technology), Halt™ Protease and Phosphatase Inhibitor Cocktail (Thermofischer), resolved by gel electrophoresis using Bolt 4%–12% Bis-Tris gels (Invitrogen), transferred to PVDF membranes (Merck), and blocked for 1 h in 3% BSA (Sigma-Aldrich). Blots were incubated with primary antibodies, followed by secondary antibodies. Bound antibodies were detected using Pierce™ ECL or SuperSignal West Pico PLUS Chemiluminescent Western Blotting Substrates (Thermofischer). Blots were developed through radiographic exposition on CL-XPosure films (Thermofisher), and scanned with a CanonScan LiDE 300 scanner. Uncropped and unprocessed scans are supplied the Source Data file.
The list of antibodies used for western blotting is provided in Table S10.
Beta galactosidase assay
Beta galactosidase level was assessed using a Senescence β-Galactosidase Staining Kit (9860, Cell Signaling) following the manufacturer’s recommendations57.
Flow cytometry and cell sorting
For flow cytometry analysis, cells were washed with PBS-0.5% BSA-2 mM EDTA before analysis of at least 5000 cells for each condition on BD FACSCanto II or BD LSRFortessa instruments (BD Biosciences). Murine hematopoietic chimerism analysis was performed on the indicated days using FITC anti-mouse CD45.1 (Cat 110706, Biolegend) and PE anti-mouse CD45.2 antibodies (Cat #109808, Biolegend) staining. Murine peripheral blood cells were collected and washed with PBS-0.1% BSA-2 mM EDTA before staining for 30 min at 4 °C with the corresponding cell surface antibodies. Then, cells were lysed for 10 min with red blood cell lysis buffer (Qiagen) and washed twice with PBS-0.1%BSA-2 mM EDTA prior to FACS analysis. Data was analyzed using the Diva (Becton Dickinson) or FlowJo softwares.
For cellular sorting, cells were washed with PBS-0.5% BSA-2 mM EDTA and resuspended in PBS-0.5% BSA-2 mM EDTA, prior to flow cytometry sorting of the cellular populations expressing each fluorescent marker of interest using a BD FACSAria II instrument (BD Biosciences).
Cell cycle analysis
For propidium iodide staining, cells were harvested at the indicated time points, washed in PBS and fixed in ice cold 70% ethanol for a minimum of 12 h. Cells were then washed in PBS, resuspended in PBS and incubated for 5 min at 4 °C with RNAse A (30 mg/ml, Sigma), prior to their incubation for 30 min in propidium iodide (1 mg/ml, Invitrogen). FACS analysis was performed on a BD FACSCanto II instrument (BD Biosciences) directly after incubation.
For the EdU assay, cells were treated with 10 µM EdU during 2 h, before being washed with PBS, fixed and permeabilized (Cytofix/Cytoperm kit #554714, BD Biosciences), at the indicated time points. Cells were then processed per manufacturers’ instructions (EdU Click-iT™ Alexa Fluor™ 647, Cat C10340, ThermoFisher). FACS data acquisition and analysis were performed on a BD FACSCanto II instrument (BD Biosciences) and using FlowJo software.
Chemicals
Ruxolitinib and Trametinib were purchased from MedChemExpress.
Plasmids, shRNA Constructs and cell infection
pLEX-FHH-IRES-Puro (Empty Vector) and pLEX-FHH-NRASQ61K-IRES-Puro (NRASQ61K Vector) were a gift from Paul Khavari (Addgene # 120568 and # 120570). pLenti-PGK-GFP-Puro (w509-5) (GFP Vector) was a gift from Eric Campeau & Paul Kaufman (Addgene # 19070). pLenti-PGK-Hygro-DEST-w530-1 (Empty Vector) and pLenti-PGK-NRASG12V were a gift from Eric Campeau, Paul Kaufman & Daniel Haber (Addgene # 19066 and # 35632). MSCV-IRES-GFP (Empty Vector), MSCV-IRES-mCherry FP (mCherry Vector) and MSCV-IRES-GFP ASXL1 (ASXL1G646Wfs*12 Vector) were a gift from Tannishtha Reya, Dario Vignali & Anjana Rao (Addgene # 20672, # 52114 and # 81021). pcw107 (Empty Vector) and JAK2 (V617F)-pcw107-V5 (JAK2V617F Vector) were a gift from John Doench, Kris Wood & David Sabatini (Addgene # 62511 and # 64610).
Crimson and NrasQ61K vectors were designed by cloning crimson and NrasQ61K cassettes in place of the native eGFP sequence of the SGEN vector (Addgene # 111171), using the AscI and XhoI restriction sites, and by replacing the NeoR-encoding sequence by an eGFP cassette using the BspEI and SalI restriction sites. NrasG12D vector was established by cloning an NrasG12D cassette using XhoI and EcoRI restriction sites into a pMIG empty plasmid in which the native eGFP cassette was replaced by a Crimson encoding sequence through NcoI and PacI restriction sites. KRASG13D vector was established by substituting the native eGFP cassette of the SGEN vector (Addgene # 111171) by a KRASG13D encoding cassette, using AscI and XhoI restriction sites.
shRNA constructs targeting Jak2 and ASXL1 – sequences listed below – were cloned respectively into the SGEN vector (Addgene # 111171) or the LENC vector (Addgene # 111163), as XhoI–EcoRI fragments, which were generated by amplifying 97-mer oligonucleotides (Invitrogen) using 5′miRE-XhoI (TGAACTCGAGAAGGTATATTGCTGTTGACAGTGAGCG) and 3′miRE-EcoRI (TCTCGAATTCTAGCCCCTTGAAGTCCGAGGCAGTAGGC) primers and the Vent polymerase kit (Invitrogen) with the following conditions: 50 μl reaction containing 0.05 ng oligonucleotide template, 1× Vent buffer, 0.3 mM of each dNTP, 0.8 μM of each primer, and 1.25 U Vent polymerase; cycling: 94 °C for 3 min; 35 cycles of 94 °C for 30 s, 54 °C for 30 s, and 75 °C for 20 s; 75 °C for 5 min. For our shJak2 experiments, a total of 10 shRNAs targeting Jak2 were used in a pooled fashion.
The list of shRNA sequences is provided in Table S11.
For virus production 18 × 106 HEK-293T cells were plated in 15-cm plates and transfected with 13.5 μg DNA for each lentiviral vector and with 11µG of PAX2 and 5.5 µg of VSVG packaging vectors. Viral supernatants were harvested 72 h later and filtered using 0.45-μm filters.
Human and murine cell lines were spin-infected for 2 h at 37 °C with 3 ml of lentiviral supernatant, 25 mM HEPES (Sigma-Aldrich) and 8 μg/ml polybrene (Sigma-Aldrich), and selected 48 to 72 h later with 1 μg/ml or 2 µg/ml of puromycin (Invivogen) for UKE-1 and HEL cells respectively, or sorted according to the expressed fluorescent marker.
Primary murine cells were spin-infected for 2 h at 37 °C with 4 ml lentiviral supernatant, 25 mM HEPES (Sigma-Aldrich) and 8 μg/ml polybrene (Sigma-Aldrich), on RetroNectin coated plates following manufacturer’s recommendations (Takara Bio).
In vivo transplantation
Lethally irradiated (9 Gy) 8-week-old C57BL/6 female mice were tail-vein injected with a total of 2×106 Lin− Jak2WT NrasG12D and Jak2V617F NrasWT hematopoietic cells obtained as described above, at a 10/90 cellular ratio, respectively. Four weeks post-transplantation, mice were randomized to be treated with Ruxolitinib (90 mg/kg, oral gavage, twice daily) or vehicle (0.5% methylcellulose (w/v) and 0.1% Tween 80) for 28 days. Peripheral blood mononuclear cells were collected by submandibular bleeding at the indicated time points.
Lethally irradiated (2 ×4.5 Gy) 6-week-old C57BL/6 male mice were tail-vein injected with 1×106 Jak2V617F c-Kit+ hematopoietic cells obtained as described above and lentivirally transduced with the SGEN-eGFP-NrasQ61K overexpressing vector. 0.3×106 total murine wild-type support bone marrow was concomitantly injected in each mouse. Three weeks post-transplantation, mice were randomized to be treated with Ruxolitinib (90 mg/kg, oral gavage, twice daily) or vehicle (5% dimethyl acetamide, 0.5% methylcellulose), using a 4 weeks ON / 2 weeks OFF treatment schedule for up to 12 weeks total. Bone marrow cells were collected by bone marrow biopsy performed on mice femurs, at the indicated time points.
Mice were euthanized if they exhibited signs of moribund condition, which included hunched posture, reduced activity, labored breathing and weight loss ( > 15%) as humane endpoint.
Patients’ population
A total of 143 consecutive patients from whom a next generation sequencing (NGS) molecular analysis was performed at diagnosis and/or during follow-up were diagnosed with primary or secondary myelofibrosis (MF) according to WHO criteria and followed in Saint-Louis hospital between January 2011 and November 2019. Among these patients, 73 patients had a longitudinal molecular follow-up defined as at least 2 NGS evaluations, and 72 patients received Ruxolitinib treatment prior to their last NGS molecular evaluation.
Clinical and molecular characteristics at time of diagnosis and during follow-up were collected from medical charts and electronic medical records. Prognostic scores were evaluated both at diagnosis (IPSS) and at time of molecular evaluation (DIPSS).
Patient samples NGS molecular data acquisition
We used a capture-based custom next-generation sequencing (NGS) panel (Sophia Genetics) targeting 36 myeloid genes (ABL1; ASXL; BRAF; CALR; CBL; CCND2; CEBPA; CSF3R; CUX1; DNMT3A; ETNK1; ETV6; EZH2; FLT3; HRAS; IDH1; IDH2; IKZF1; JAK2; KIT; KRAS; MPL; NFE2; NPM1; NRAS; PTPN11; RUNX1; SETBP1; SF3B1; SH2B3; SRSF2; TET2; TP53; U2AF1; WT1; ZRSR2) (Table S12). Libraries were prepared using 200 ng of DNA extracted from whole blood or CD34+ patient-derived cells (Qiagen). Sequencing was then performed on a MiSeq instrument (Illumina). Bioinformatics were carried out at Sophia Genetics (Switzerland) using the SOPHIA DDM software and significant variants were retained with a sensitivity of 1%. Cbioportal oncoprinter was used to generate oncoprint plots (https://www.cbioportal.org/oncoprinter). Fishplots depicting phylogenetic trajectories were inferred from single-cell DNA sequencing combined with longitudinal NGS analysis, and generated using “fishplot” R package (version 4.1.1 for Mac). Mutations detected by NGS and not detected in our single cell DNA analysis (either absent at time of single-cell sequencing or not included in our single-cell DNA sequencing panel) were not included in the Fishplots representation to avoid incorrect clonal inference.
Patient samples single-cell DNA sequencing
Primary CD34+ cells from primary MPN patients’ peripheral blood were resuspended in Tapestri cell buffer (Mission Bio) and quantified using an automatic cell counter (Biorad). Single cells (3000-4000 cells per μL, >80% viable) were encapsulated using a Tapestri microfluidics cartridge (Mission Bio), lysed, and barcoded. Barcoded samples were then subjected to targeted polymerase chain reaction (PCR) amplification of a custom 96 amplicons covering 17 genes known to be mutated in MPN (ASXL1; CALR; CBL; DNMT3A; EZH2; IDH1; IDH2, JAK2; KRAS; MPL; NFE2; NRAS; SF3B1; SRSF2; TET2; TP53; U2AF1). PCR products were removed from individual droplets, purified with Ampure XP beads (Beckman Coulter), and used as a template for PCR to incorporate Illumina i5/i7 indices. PCR products were purified a second time, quantified via an Agilent Bioanalyzer, and pooled to be sequenced. Library pools were sequenced on using NextSeq 550 instrument (Illumina). Fastq files were processed using the Tapestri Pipeline for cell calling and using Genome Analysis Toolkit 4/haplotype caller for genotyping. The Tapestri Insights software was used to further filter variants, and samples were included if they harbored 3 or more protein-encoding, nonsynonymous/insertion/deletion variants and >1000 cells with definitive genotype for all protein-coding variants within the sample. We next sought to define genetic clones, which we identified as cells that possessed identical genotype calls for the protein-encoding variants of interest. Importantly, almost 95% of the panel amplicons in each sample had sufficient coverage to annotate variants. The estimated median allele dropout rate was 7.54% (IQR, 5.6% to 9.3%). From the variants annotated for each sample, we first removed those with low quality ( < 30% in the Tapestri Insights software) and low frequency ( < 0.5% cells). As a result, we detected a total of 17 different variants passing the pre-filtering step across the 4 patients, with a median of 4.5 variants per patient (range from 3.0–5.0). All variants detected in bulk whole-blood NGS analysis below the 0.5% threshold were not considered in the single-cell analysis.
Gene variants and drug effects associations
The Beat AML v2 cohort31 was interrogated to evaluate single-nucleotide variants (silent variants excluded) association with resistance to the JAK2 inhibitors ruxolitinib (497 samples with genotypes), momelotinib (476 samples with genotypes) and fedratinib (93 samples with genotypes). Drug AUC data is available from https://biodev.github.io/BeatAML2/ and drug effect visualization from the Vizome platform (http://vizome.org/). Data is presented as volcano plots for gene variants effect size (Glass) on the x-axis vs. –log10(P-value) on the y-axis. Significance was set at – log10(P-value)>1.3, with variants having a negative effect size associated with drug sensitivity, while those having a positive effect size associated with drug resistance.
Gene Set enrichment Analysis
To accurately define patients harboring low vs. high JAK/STAT pathway activation among the 707 Beat AML v2 cohort patients for which bulk RNA-seq expression data is publicly available31, we classified patients based both on their level of JAK2 expression, and on their single sample GSEA scores for JAK2 direct targets and JAK2V617F mutant gene signatures32. The functional associations of the molecular phenotypes were explored with the single sample GSEA (ssGSEA) method58 based on the Bioconductor GSVA v1.40.1 implementation. ssGSEA is an extension of Gene Set Enrichment Analysis that calculates separate enrichment scores for each pairing of a sample and gene set. Each ssGSEA enrichment score represents the degree to which the genes in a particular gene set have coordinately increased or decreased expression within a sample. JAK2 “low” and “high” groups were defined based on the consensus conditions between i) JAK2 expression z-scores across the full cohort (<-0.5 or >0.5 respectively), and ii) at least two of the three ssGSEA scores for shJAK2 vs. GFP32, JAK2V617F homozygous vs. Normal and/or JAK2V617F heterozygous vs. Normal gene signatures32 ( < -0.5 or >0.5 respectively). Beat AML v2 JAK2 “low” (n = 60) and “high” (n = 64) groups definition was validated for JAK2 expression (log2(TPM + 1)) and association with JAK2 gene target and mutated gene set signatures through Genome-wide Gene Set Enrichment Analysis for enrichment in expression changes. The Gene Set enrichment analysis (GSEA) v4.2.0 software was used to identify functional associations of molecular phenotypes with gene sets included in the MSigDB v7.4 database and the National Cancer Institute’s RAS Initiative (https://www.cancer.gov/research/key-initiatives/ras) (Supplementary Data 2). The goal of GSEA was to identify the gene sets that are distributed at the top or at the bottom of the ranked list of genes based on the Kolmogorov-Smirnov enrichment test. Gene sets with absolute Normalized Enrichment Score (NES) ≥ 1.3, a nominal p-value ≤ 0.05 and an FDR ≤ 0.25 for the Kolmogorov-Smirnov test were considered significant hits. The results were visualized on volcano plots for the normalized enrichment score (NES) vs. -log10(p-value) and on GSEA plots. Heatmaps depicting relative gene expression changes for leading edge genes for the top enriched gene sets were also represented.
Statistics
In vitro and in vivo statistical analysis
Statistical analysis was performed using PRISM 8.0.1 (GraphPad). Data were analyzed using a nonparametric Mann-Whitney test (with the assumption of no Gaussian distribution of the group) or parametric Welch’s correction (with the assumption that both groups of data are sampled from Gaussian populations), unless otherwise specified, and the threshold of significance (α) was set at 0.05.
Patients’ cohort statistical analysis
Continuous variables are reported as medians and interquartile ranges (IQR), while categorical variables are reported as numbers and proportions. Comparison of continuous and categorical variables between subgroups was performed by Mann-Whitney test and Fisher’s exact test, respectively.
RAS pathway mutations cumulative hazard was estimated by the Nelson-Aalen method and compared by cause-specific hazard Cox model. Time to mutation was calculated from the date of first available NGS evaluation to the last available NGS molecular assessment. Ruxolitinib treatment was considered a time-dependent covariate. OS and transformation-free survival (TFS) were estimated using the Kaplan-Meier method. OS was measured from the date of MF diagnosis until death, censoring patients still alive at the date of last follow-up and patients undergoing Hematopoietic Stem Cell Transplantation (HSCT) at the time of transplantation. TFS was measured from the date of MF diagnosis until the date of the transformation (AML or MDS), censoring patients still alive without transformation at the date of last follow-up and patients undergoing HSCT at the time of transplantation. A total of eight patients (5/71 in the non-ruxolitinib-treated group and 3/72 in the ruxolitinib-treated group) had HSCT.
Univariate and multivariate analyses assessing the impact of categorical and continuous variables on “RAS pathway” mutations acquisition, OS or TFS were performed using Cox regression models. The proportional hazard assumption was validated. All significant variables (p < 0.05) were included in the multivariate analysis. Hazard ratios (HR) are given with 95% confidence interval (CI). Statistical analyses were performed using the STATA software (STATA 15.1 Corporation, College Station, TX).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Human single cell DNA sequencing data generated in this study are available at the Sequence Read Archive (SRA) repository of the National Center for Biotechnology Information (NCBI). The accession number for these SRA data is PRJNA1222460 and data can be accessed through the following link: https://www.ncbi.nlm.nih.gov/sra/PRJNA1222460. Supplementary information, including Supplementary Figs. and legends, Supplementary Tables, Supplementary Data and Source data are provided with this paper. Sources for reagents and cells are indicated in the Materials and Methods section. No custom code was generated in the course of this study. Source data are provided with this paper.
References
Labrie, M., Brugge, J. S., Mills, G. B. & Zervantonakis, I. K. Therapy resistance: opportunities created by adaptive responses to targeted therapies in cancer. Nat. Rev. Cancer 22, 323–339 (2022).
Shah, N. P. et al. Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell 2, 117–125 (2002).
McMahon, C. M. et al. Clonal selection with RAS pathway activation mediates secondary clinical resistance to selective FLT3 inhibition in acute myeloid leukemia. Cancer Discov. 9, 1050–1063 (2019).
Zhang, H. et al. Clinical resistance to crenolanib in acute myeloid leukemia due to diverse molecular mechanisms. Nat. Commun. 10, 244 (2019).
Wagle, N. et al. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J. Clin. Oncol. 29, 3085–3096 (2011).
Nazarian, R. et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature 468, 973–977 (2010).
Verstovsek, S. et al. A double-blind, placebo-controlled trial of ruxolitinib for myelofibrosis. N. Engl. J. Med 366, 799–807 (2012).
Harrison, C. et al. JAK inhibition with ruxolitinib versus best available therapy for myelofibrosis. N. Engl. J. Med 366, 787–798 (2012).
Quintás-Cardama, A. et al. Preclinical characterization of the selective JAK1/2 inhibitor INCB018424: therapeutic implications for the treatment of myeloproliferative neoplasms. Blood 115, 3109–3117 (2010).
James, C. et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature 434, 1144–1148 (2005).
Nangalia, J. et al. Somatic CALR mutations in myeloproliferative neoplasms with nonmutated JAK2. N. Engl. J. Med 369, 2391–2405 (2013).
Pikman, Y. et al. MPLW515L is a novel somatic activating mutation in myelofibrosis with myeloid metaplasia. PLoS Med 3, e270 (2006).
Lundberg, P. et al. Clonal evolution and clinical correlates of somatic mutations in myeloproliferative neoplasms. Blood 123, 2220–2228 (2014).
Vainchenker, W. & Kralovics, R. Genetic basis and molecular pathophysiology of classical myeloproliferative neoplasms. Blood 129, 667–679 (2017).
Grinfeld, J. et al. Classification and personalized prognosis in myeloproliferative neoplasms. N. Engl. J. Med 379, 1416–1430 (2018).
Guglielmelli, P. et al. The number of prognostically detrimental mutations and prognosis in primary myelofibrosis: an international study of 797 patients. Leukemia 28, 1804–1810 (2014).
Guglielmelli, P. et al. MIPSS70: Mutation-enhanced international prognostic score system for transplantation-age patients with primary myelofibrosis. J. Clin. Oncol. 36, 310–318 (2018).
Beer, P. A. et al. Two routes to leukemic transformation after a JAK2 mutation-positive myeloproliferative neoplasm. Blood 115, 2891–2900 (2010).
Ortmann, C. A. et al. Effect of mutation order on myeloproliferative neoplasms. N. Engl. J. Med 372, 601–612 (2015).
Harutyunyan, A., Klampfl, T., Cazzola, M. & Kralovics, R. p53 lesions in leukemic transformation. N. Engl. J. Med 364, 488–490 (2011).
Barosi, G. & Gale, R. P. Does ruxolitinib really prolong survival in individuals with myelofibrosis? The never-ending story. Blood Adv. 6, 2331–2333 (2022).
Deshpande, A. et al. Kinase domain mutations confer resistance to novel inhibitors targeting JAK2V617F in myeloproliferative neoplasms. Leukemia 26, 708–715 (2012).
Weigert, O. et al. Genetic resistance to JAK2 enzymatic inhibitors is overcome by HSP90 inhibition. J. Exp. Med 209, 259–273 (2012).
Andreoli, A. et al. Clinical resistance to ruxolitinib is more frequent in patients without MPN-associated mutations and is rarely due to mutations in the JAK2 kinase drug-binding domain. Blood 122, 1591–1591 (2013).
Santos, F. P. S. et al. Prognostic impact of RAS-pathway mutations in patients with myelofibrosis. Leukemia 34, 799–810 (2020).
Coltro, G. et al. RAS/CBL mutations predict resistance to JAK inhibitors in myelofibrosis and are associated with poor prognostic features. Blood Adv. 4, 3677–3687 (2020).
Serrano, M., Lin, A. W., McCurrach, M. E., Beach, D. & Lowe, S. W. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88, 593–602 (1997).
Stivala, S. et al. Targeting compensatory MEK/ERK activation increases JAK inhibitor efficacy in myeloproliferative neoplasms. J. Clin. Invest 129, 1596–1611 (2019).
Marty, C. et al. Calreticulin mutants in mice induce an MPL-dependent thrombocytosis with frequent progression to myelofibrosis. Blood 127, 1317–1324 (2016).
Kitamura, T. ASXL1 mutations gain a function. Blood 131, 274–275 (2018).
Burd, A. et al. Precision medicine treatment in acute myeloid leukemia using prospective genomic profiling: feasibility and preliminary efficacy of the Beat AML Master Trial. Nat. Med 26, 1852–1858 (2020).
Rampal, R. et al. Integrated genomic analysis illustrates the central role of JAK-STAT pathway activation in myeloproliferative neoplasm pathogenesis. Blood 123, e123–133 (2014).
Mylonas, E. et al. Single-cell analysis based dissection of clonality in myelofibrosis. Nat. Commun. 11, 73 (2020).
Jayavelu, A. K. et al. Splicing factor YBX1 mediates persistence of JAK2-mutated neoplasms. Nature 588, 157–163 (2020).
Brkic, S. et al. Dual targeting of JAK2 and ERK interferes with the myeloproliferative neoplasm clone and enhances therapeutic efficacy. Leukemia 35, 2875–2884 (2021).
Winter, P. S. et al. RAS signaling promotes resistance to JAK inhibitors by suppressing BAD-mediated apoptosis. Sci. Signal 7, ra122 (2014).
Das Thakur, M. et al. Modelling vemurafenib resistance in melanoma reveals a strategy to forestall drug resistance. Nature 494, 251–255 (2013).
Chang, L. et al. Systematic profiling of conditional pathway activation identifies context-dependent synthetic lethalities. Nat. Genet 55, 1709–1720 (2023).
Hong, A. et al. Exploiting drug addiction mechanisms to select against MAPKi-resistant melanoma. Cancer Discov. 8, 74–93 (2018).
Kong, X. et al. Cancer drug addiction is relayed by an ERK2-dependent phenotype switch. Nature 550, 270–274 (2017).
Chan, L. N. et al. Signalling input from divergent pathways subverts B cell transformation. Nature 583, 845–851 (2020).
Bailey, M. H. et al. Comprehensive characterization of cancer driver genes and mutations. Cell 173, 371–385.e18 (2018).
Hurwitz, H. et al. Ruxolitinib + capecitabine in advanced/metastatic pancreatic cancer after disease progression/intolerance to first-line therapy: JANUS 1 and 2 randomized phase III studies. Invest N. Drugs 36, 683–695 (2018).
Lee, G. H. et al. Cutaneous squamous-cell carcinoma after treatment with ruxolitinib or belumosudil. N. Engl. J. Med. 389, 188–190 (2023).
Cook, E. K., Luo, M. & Rauh, M. J. Clonal hematopoiesis and inflammation: Partners in leukemogenesis and comorbidity. Exp. Hematol. 83, 85–94 (2020).
Janssen, J. W. et al. RAS gene mutations in acute and chronic myelocytic leukemias, chronic myeloproliferative disorders, and myelodysplastic syndromes. Proc. Natl Acad. Sci. USA 84, 9228–9232 (1987).
Padua, R. A. et al. RAS mutations in myelodysplasia detected by amplification, oligonucleotide hybridization, and transformation. Leukemia 2, 503–510 (1988).
Nakagawa, T. et al. Multiple point mutation of N-ras and K-ras oncogenes in myelodysplastic syndrome and acute myelogenous leukemia. Oncology 49, 114–122 (1992).
Braun, B. S. et al. Somatic activation of oncogenic Kras in hematopoietic cells initiates a rapidly fatal myeloproliferative disorder. Proc. Natl Acad. Sci. USA 101, 597–602 (2004).
Zhang, J. et al. Oncogenic Kras-induced leukemogeneis: hematopoietic stem cells as the initial target and lineage-specific progenitors as the potential targets for final leukemic transformation. Blood 113, 1304–1314 (2009).
Prior, I. A., Lewis, P. D. & Mattos, C. A comprehensive survey of Ras mutations in cancer. Cancer Res 72, 2457–2467 (2012).
O’Sullivan, J. M. et al. RAS-pathway mutations are common in patients with ruxolitinib refractory/intolerant myelofibrosis: molecular analysis of the PAC203 cohort. Leukemia https://doi.org/10.1038/s41375-023-02027-3 (2023).
Harrison, C. N. et al. Long-term findings from COMFORT-II, a phase 3 study of ruxolitinib vs best available therapy for myelofibrosis. Leukemia 30, 1701–1707 (2016).
Verstovsek, S. et al. Long-term treatment with ruxolitinib for patients with myelofibrosis: 5-year update from the randomized, double-blind, placebo-controlled, phase 3 COMFORT-I trial. J. Hematol. Oncol. 10, 55 (2017).
Kogan, S. C. et al. The PEBP2betaMYH11 fusion created by Inv(16)(p13;q22) in myeloid leukemia impairs neutrophil maturation and contributes to granulocytic dysplasia. Proc. Natl Acad. Sci. USA 95, 11863–11868 (1998).
Mullally, A. et al. Physiological Jak2V617F expression causes a lethal myeloproliferative neoplasm with differential effects on hematopoietic stem and progenitor cells. Cancer Cell 17, 584–596 (2010).
Dagher, T. et al. JAK2V617F myeloproliferative neoplasm eradication by a novel interferon/arsenic therapy involves PML. J. Exp. Med 218, e20201268 (2021).
Barbie, D. A. et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 462, 108–112 (2009).
Cassinat, B. https://BioRender.com/sbbc7tu (2025).
Acknowledgements
The authors thank the clinical care team of the Comprehensive Myeloproliferative neoplasms Center for samples and data collection, and the staff of the cellular biology laboratory for excellent technical assistance. We are indebted to Veronique Montcuquet, Niclas Setterblad, Christelle Doliger, and Claire Maillard from the Saint-Louis Research Institute Core Facility for their technical support. The authors also thank the French Intergroup for Myeloproliferative neoplasms (FIM) for insightful discussions. This work was supported by an “Association Laurette Fugain” (to J.J.K. and L.B.), a “Fédération Leucémie Espoir” (to J.J.K. and L.B.), a “Fondation ARC pour la recherche sur le cancer” (to J.J.K.), an “INCa Prev-Bio” (to J.J.K. and L.B.), a “CCA-INSERM-Bettencourt” (to L.B.,) and an “ATIP-Avenir / Ligue National Contre le Cancer” (to L.B.) funding. L.B. is supported by the ERC Starting program (101117339) and an Emergence Ville de Paris grant. N.K. is supported by the “Fondation pour la Recherche Médicale - FRM” / “Fondation Capucine”. K.S. was supported by the National Cancer Institute R35 CA210030. This work was supported by the Groupe Francophone des Myelodysplasies (to R.A.P.). P.G. was supported by the Chinese Scholarship Council (CSC N°201706180057). A.P. is supported by the ERC Starting and Consolidator programs (758848 and 101088563). L.B., C.L., and A.P. are supported by the SIRIC InsiTu program (INCa-DGOS-INSERM-ITMO Cancer_18008) and the IHU France 2030 “Leukemia Institute Paris Saint-Louis” (ANR-23-IAHU-0005).
Author information
Authors and Affiliations
Contributions
N.K., N.M., and B.R.: formal analysis, validation, investigation, visualization, methodology, writing original draft. G.A.: conceptualization, methodology, investigation, formal analysis, visualization. H.P., E.V., R.D.O., R.M., C.C., and N.G.: investigation, methodology, resources. F.G.: conceptualization, methodology. L.P.Z.: visualization. S.Ga., P.G., B.M., and F.L.: methodology, investigation. J.S.D., N.P., W.V., E.R., R.A.P., C.M., I.P., and S.Gi.: resources, methodology. C.L. and K.S.: conceptualization, investigation, methodology. A.P.: conceptualization, validation, investigation, methodology, visualization. J.J.K. and B.C.: conceptualization, formal analysis, methodology, resources, supervision, funding acquisition, visualization, writing original draft. L.B.: conceptualization, investigation, formal analysis, methodology, resources, supervision, funding acquisition, visualization, writing original draft, project administration. All co-authors reviewed, edited and critically discussed the manuscript.
Corresponding author
Ethics declarations
Competing interests
L.B. received research funding from Gilead and Pfizer for unrelated projects, and personal fees from BMS, Novartis and GSK outside of the submitted work. N.G. received personal fees from Novartis, Abbvie and Astra Zeneca outside of the submitted work. K.S. is on the SAB and has stock options in Auron Therapeutics and received grant funding from Novartis and KronosBio on topics unrelated to this manuscript. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Frederik Damm, Leonidas Platanias and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Maslah, N., Kaci, N., Roux, B. et al. JAK2 inhibition mediates clonal selection of RAS pathway mutations in myeloproliferative neoplasms. Nat Commun 16, 6270 (2025). https://doi.org/10.1038/s41467-025-60884-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-60884-1