Abstract
Embryo assessment in in vitro fertilization (IVF) involves multiple tasks—including ploidy prediction, quality scoring, component segmentation, embryo identification, and timing of developmental milestones. Existing methods address these tasks individually, leading to inefficiencies due to high costs and lack of standardization. Here, we introduce FEMI (Foundational IVF Model for Imaging), a foundation model trained on approximately 18 million time-lapse embryo images. We evaluate FEMI on ploidy prediction, blastocyst quality scoring, embryo component segmentation, embryo witnessing, blastulation time prediction, and stage prediction. FEMI attains area under the receiver operating characteristic (AUROC) > 0.75 for ploidy prediction using only image data—significantly outpacing benchmark models. It has higher accuracy than both traditional and deep-learning approaches for overall blastocyst quality and its subcomponents. Moreover, FEMI has strong performance in embryo witnessing, blastulation-time, and stage prediction. Our results demonstrate that FEMI can leverage large-scale, unlabelled data to improve predictive accuracy in several embryology-related tasks in IVF.
Similar content being viewed by others
Introduction
The success of in vitro fertilization (IVF) hinges on the accurate assessment and selection of viable embryos1,2. However, current diagnostic tools and practices encounter several challenges, including high costs, lack of standardization, and varying regulations concerning preimplantation genetic testing for aneuploidy (PGT-A) across different countries. Standardization in embryo assessment involves establishing consistent and uniform protocols for evaluating embryo quality and viability across diverse clinical settings. Presently, variations in diagnostic tools, scoring systems, and embryologist interpretations lead to inconsistencies in embryo selection, which can adversely affect IVF success rates and patient outcomes. These limitations highlight the urgent need for a more efficient, non-invasive, and affordable approach to embryo assessment. Such advancements could significantly enhance IVF success rates and accessibility, reduce the emotional and financial strain on patients, and minimize risks associated with IVF, such as multiple pregnancies and their complications3,4. Therefore, developing solutions that address these challenges is essential for advancing reproductive health outcomes.
Recent advancements in artificial intelligence (AI) have aided in multiple IVF tasks, including predicting the morphology and ploidy status of embryos, critical factors for a successful procedure. Models like STORK and ERICA use deep learning to analyze embryo morphology from images to do specific downstream tasks5,6,7. While these models show promise, they are often limited by their focus on specific developmental stages and reliance on both image-based data and embryologist input. These limitations introduce bias as well as a less seamless process for integrating AI into clinics. Addressing these gaps, BELA (Blastocyst Evaluation Learning Algorithm) was developed, which predicts ploidy status through using a multitask learning approach on sequences of time-lapse images, without any embryologist input. This approach not only predicts quality scores but also uses these scores to determine ploidy status, enabling a more comprehensive and objective analysis of embryo development. BELA has demonstrated superior performance with an AUC of 0.76, surpassing models that rely on manual embryologist scoring. However, BELA faces challenges, notably in ensuring accuracy dependent on the quality and diversity of training data. Moreover, BELA is still limited to only predicting embryo quality scores and ploidy status8.
Foundation models in computer vision are large-scale models pre-trained on extensive datasets, enabling them to generalize across various tasks. These models, typically deep neural networks, learn a broad range of features during pre-training, which can be fine-tuned for specific applications9. The self-supervised learning paradigm is central to their effectiveness, allowing the models to leverage vast amounts of unlabeled data by creating pretext tasks, such as image inpainting or contrastive learning. Vision Transformers (ViTs), a prominent foundation model architecture, utilize a transformer-based approach instead of traditional convolutional neural networks (CNNs)10. ViTs split images into patches, linearly embed these patches, and process the sequence using transformer layers. This method allows ViTs to capture long-range dependencies and complex patterns within images. The key advantage of ViTs lies in their ability to handle large-scale data and perform well on diverse vision tasks after fine-tuning. The promise of foundation models in computer vision includes improved performance on various tasks, reduced need for labeled data, and enhanced adaptability to new domains. These models have demonstrated state-of-the-art results in image classification, object detection, and segmentation, making them invaluable tools in fields requiring robust and scalable image analysis solutions. In the field of IVF, Wang et al. developed IVFormer as a backbone for various IVF-related tasks; however, its utility is constrained by limited training dataset diversity, and the absence of a publicly available model11.
In this study, we utilized the Vision Transformer masked autoencoder (ViT MAE), which uses self-supervised learning (SSL) to reconstruct the original image from a masked input12. The ViT MAE uses an encoder-decoder structure to learn important features about the dataset, which allows the model to perform better reconstruction. By learning domain-specific information through self-supervision, the ViT MAE can then be used for downstream tasks in a supervised manner. ViT MAEs have been applied to a range of tasks in previous literature, demonstrating its versatility and effectiveness. Zhou et al. developed RETFound, built on a ViT MAE architecture, which was employed for various retinal disease-related tasks13. Liu et al. explored the use of ViT MAE for general domain medical tasks by training it on 2.5 million unlabeled images from various modalities (CT, MR, PET, X-rays, and ultrasound)14. Their findings highlighted the model’s capacity to achieve high performance compared to benchmarks.
In this paper, we present FEMI (Foundational IVF Model for Imaging), a foundation model trained on ~18 million time-lapse embryo images.
Results
FEMI Pre-training
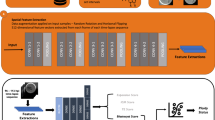
FEMI is trained using a ViT MAE backbone architecture as shown in Fig. 1. To train and test FEMI, we compiled a diverse dataset of 17,968,959 time-lapse images sourced from multiple clinics. These included 1998 time-lapse sequences from Weill Cornell Medicine (WCM) captured using the Embryoscope (ES), 841 sequences from WCM (2020) using the Embryoscope+ (ES + ), 543 sequences from IVI RMA Valencia captured with the Embryoscope + , 869 sequences from IVF Florida using the Embryoscope + , and 4860 sequences from WCM post-2021 using the Embryoscope + . More information about these datasets is shown in Supplemental Table 1. Additionally, two public datasets were incorporated: 704 time-lapse sequences from the University Hospital of Nantes and 2344 blastocyst images from clinics across Europe15,16. For FEMI’s training dataset, we selected time-lapse images taken after 85 h post-insemination (hpi) at z-axis depths ranging from −30 to +30. These images predominantly featured the embryo slide background. To enhance feature learning, images were tightly cropped around the embryos. This was facilitated by an embryo segmentation model we developed based on the InceptionV3 architecture. The segmentation model was trained on a dataset that consisted of embryo image-mask pairs where the masks contained whole embryo segmentation. Details on this model are in the Methods section. For each image, the segmentation model generated a mask, within which a circular embryo shape was identified via contour detection. A bounding box was then drawn around this detected shape, and the image was cropped and resized to 224 × 224 pixels for SSL input. A ViT MAE model pre-trained on the ImageNet-1k dataset was further pre-trained on our collection of 17,968,959 time-lapse images for 800 epochs with early stopping, to learn in-domain imaging features. The time-lapse image dataset was divided into an 80% training and 20% validation split, treating each image as an independent sample.
This training resulted in the IVF foundation model, FEMI, (Fig. 1) from which the encoder is subsequently utilized for various downstream tasks. Detailed methodologies of this pre-training phase are provided in the Methods section. We evaluated FEMI on several clinically relevant tasks, including ploidy prediction, blastocyst quality scoring, embryo component segmentation, embryo witnessing, blastulation time prediction, and stage prediction. Task-specific layers were appended to the encoder as required. For most tasks, the input consisted solely of single-embryo time-lapse images. However, for blastocyst quality scoring and ploidy prediction, the model also processed sequences of time-lapse images (video input). Additionally, for ploidy prediction, maternal age was incorporated as a feature due to its demonstrated predictive value. A variety of models were trained for comparison across these downstream tasks, detailed in the Methods section and in Supplemental Table 2. Benchmark models included both traditional supervised architectures (VGG16, ResNet-RS, EfficientNet V2, ConvNeXt, CoAtNet, MoViNet) and models pre-trained via self-supervision (ImageNet ViT MAE, Swin Transformer, I-JEPA, MEDSAM). Each task’s dataset was partitioned into training and held-out test sets, with the training data further split into training and validation segments through 4-fold cross-validation. Model performances were averaged across the held-out test set and any task-specific external validation datasets, maintaining consistent data splits across all model architectures.
Downstream tasks
Ploidy prediction
Ploidy status, indicating whether an embryo is chromosomally normal (euploid) or abnormal (aneuploid), is a critical factor in selecting embryos for implantation. Aneuploid (ANU) embryos are further classified into single aneuploid (one chromosomal aberration) and complex aneuploid (CxA) (multiple chromosomal aberrations). Ploidy status is a critical factor in embryo selection as it directly influences the potential for successful implantation and ongoing pregnancy. Euploid (EUP) embryos, which have the correct number of chromosomes, exhibit higher implantation rates and lower risks of miscarriage compared to aneuploid embryos, which contain chromosomal abnormalities. Accurate prediction of ploidy status enables the selection of embryos with the highest viability1. Traditionally, ploidy assessment is performed post-blastocyst development through a biopsy for PGT-A1,2,3. Despite its diagnostic value, PGT-A is costly and considered unethical in some regions due to its invasive nature. We explore the capability of FEMI in predicting embryo ploidy using non-invasive methods. First, FEMI’s performance was assessed on image-based ploidy predictions using single images captured at 110 hpi. We then expanded the approach to include sequences of images (video input) from 96 to 112 hpi, a period identified by previous research as important for ploidy determination8. Incorporating sequences of time-lapse images into ploidy prediction allows FEMI to capture dynamic developmental processes and morphological changes over time, which are critical indicators of embryo viability and chromosomal normality8. Furthermore, we incorporated maternal age into models, considering its established predictive relevance for ploidy status. Advanced maternal age is associated with an increased likelihood of chromosomal abnormalities1,8. FEMI was evaluated on two classification tasks: distinguishing between euploid and aneuploid embryos, and between euploid and complex aneuploid embryos. The latter distinction is particularly important as complex aneuploid embryos are less likely to result in successful implantations compared to single aneuploid embryos, which may still lead to viable pregnancies despite chromosomal aberrations.
To fine-tune and evaluate our models, we utilized datasets with ploidy-labeled embryos, including Weill ES, 2020 Weill ES + , Spain ES + , Florida ES + , and 2021+ Weill ES + . The collective dataset for euploid vs. aneuploid comprised 6285 embryos. For complex aneuploidy, we removed the Spain ES+ dataset as it did not contain the information to discriminate between different types of aneuploids. This resulted in 4436 embryos for euploid vs. complex aneuploid. For each task, 25% of each dataset was reserved as a held-out test set, where each embryo was treated as an independent sample. The remaining data underwent four-fold cross-validation for model training, and performance metrics were aggregated across all folds. Model efficacy was primarily assessed using the area-under-the-receiver-operator-curve (AUROC).
We compared the performance of FEMI on ploidy prediction tasks against various benchmark models. In the supervised learning category, models such as VGG16, ResNet101-RS, EfficientNet V2, ConvNext, and CoAtNet were trained on each task. Additionally, for video-based ploidy prediction, a MoViNet model, pre-trained on ImageNet-1k, was also employed. FEMI was further benchmarked against three architectures that underwent self-supervised learning: a ViT MAE model solely pre-trained on ImageNet-1k, a Swin Transformer, and an Image-based Joint-Embedding Predictive architecture. The latter two, referred to as IVF SWIN and I-JEPA, respectively, were pre-trained on ImageNet-1k and further refined through self-supervised learning using the same 18 million images from the IVF domain that trained FEMI. For image and image+age inputs, we also compared FEMI’s performance to a previously published model, STORK-A. STORK-A utilizes a ResNet18 architecture as its backbone, and we evaluate four versions of it, image and image+age inputs for both EUP vs. ANU and EUP vs. CxA classification.
Figure 2 shows the performance of all models on all ploidy prediction tasks. For image-only euploid vs. aneuploid classification, FEMI significantly outperforms all comparison models in three datasets, whereas in the other two datasets, there is a non-significant increase in performance from FEMI. FEMI can achieve performances of greater than 0.70 AUROC at certain datasets (2020 Weill ES + − 0.71 ± 0.01, Spain ES + − 0.75 ±.01), with only image data. With the inclusion of maternal age, FEMI is significantly better in the 2020 Weill ES + , Spain ES + , and 2021+ Weill ES+ datasets and comparable at Weill ES and Florida ES + . For video-only models, FEMI outperforms all comparison models significantly in all datasets. However, with the inclusion of maternal age, FEMI only has non-significant performance increases compared to other models across all datasets.
Dot plots represent model performance based on AUROC for ploidy prediction on ES (Embryoscope) and ES+ (Embryoscope + ) microscopes. Performances are aggregated across 4 replicates (four-fold cross validation) (n = 4). Euploid vs. aneuploid classification: a image input; Weill ES, p = 0.84; 2020 Weill ES + , p = 2.1e-24; Spain ES + , p = 2.3e-24; Florida ES + , p = 0.69; 2021+ Weill ES + , p = 3.2e-21. b image + age input; Weill ES, p = 0.07; 2020 Weill ES + , p = 1.7e-16; Spain ES + , p = 2.9e-19; Florida ES + , p = 0.14; 2021+ Weill ES + , p = 4.5e-14. c video input; Weill ES, p = 2.6e-23; 2020 Weill ES + , p = 6.3e-25; Spain ES + , p = 4.1e-27; Florida ES + , p = 1.7e-19; 2021+ Weill ES + , p = 1.0e-22 d video + age input; Weill ES, p = 0.79; 2020 Weill ES + , p = 0.99; Spain ES + , p = 0.99; Florida ES + , p = 0.69; 2021+ Weill ES + , p = 0.99. For euploid vs. complex aneuploid classification: e image input; Weill ES, p = 2.7e-20; 2020 Weill ES + , p = 9.2e-25; Florida ES + , p = 4.3e-21; 2021+ Weill ES + , p = 4.8e-26. f image + age input; Weill ES, p = 8.6e-10; 2020 Weill ES + , p = 0.99; Florida ES + , p = 2.0e-11; 2021+ Weill ES + , p = 6.6e-12. g video input; Weill ES, p = 1.7e-17; 2020 Weill ES + , p = 3.4e-22; Florida ES + , p = 0.99; 2021+ Weill ES + , p = 1.2e-22. h video + age input; Weill ES, p = 0.07; 2020 Weill ES + , p = 0.73; Florida ES + , p = 0.58; 2021+ Weill ES + , p = 0.99. Asterisks represent statistical significance with p < 0.05, where statistical significance is determined by performing a one-way ANOVA test followed by a Tukey HSD test. For all subplots, error bars show mean values +/- SEM. Source data are provided as a Source Data file.
We also explore euploid vs. complex aneuploid tasks, shown in Fig. 2. In general, model performances across architectures are better in these tasks compared to euploid vs. aneuploid due to complex aneuploid embryos being a more extreme case of aneuploidy and having more distinct morphology than single aneuploid embryos. For the image-only setting, FEMI does significantly better than comparison models on all datasets. When maternal age is added, FEMI performs significantly better in the Weill ES, Florida ES + , and 2021+ Weill ES+ datasets. For the video-only setting, we see FEMI significantly outperforms comparison models in three datasets, Weill ES, 2020 Weill ES + , and 2021+ Weill ES + . Interestingly, we can achieve a 0.80 ± 0.01 AUROC in the 2021+ Weill ES+ dataset, with just video input and no maternal age. With the inclusion of maternal age, FEMI achieves an AUROC of greater than 0.85 in both the Weill ES and 2020 Weill ES+ datasets. We investigated the performance of FEMI across various age groups defined by the Society for Assisted Reproductive Technology (SART). Performances for Image, Image+Age, Video, Video+Age models across all test sets are shown in Supplemental Tables 3 and 4. In general, FEMI shows consistent performance across age groups both with and without the inclusion of maternal age. FEMI also outperforms logistic regression models trained only on maternal age, shown in Supplemental Table 5. We also show FEMI’s area under the precision-recall curve (AUPRC) for EUP vs. CxA tasks in Supplemental Table 6 as the task has a slight class imbalance.
We investigated FEMI’s performance in ploidy prediction, particularly in scenarios involving low-quality embryos. Using Weill ES, Weill ES + , and 2021+ Weill ES+ datasets, we analyzed ploidy prediction across specific blastocyst quality score ranges: high (3-5), medium (6-9), and low (10-14). For 320 low-quality embryos in EUP vs. ANU image-only prediction, FEMI achieved an AUC of 0.677 ± 0.0346, significantly outperforming the next best model, ResNet-RS, which had an AUC of 0.602 ± 0.0172 (p < 0.01). These results demonstrate FEMI’s superior accuracy in predicting ploidy under conditions of low embryo quality.
Model interpretation for ploidy prediction
We investigated what image features FEMI used for performing downstream tasks, specifically in the case of ploidy prediction. We use Score-CAM on the final block of FEMI’s encoder to visualize the salient portions of the image that the model used for classification (Fig. 3)17. We notice that the model primarily uses boundaries of the cells within the embryos and portions of the inner cell mass and trophectoderm. These features correspond with general characteristics that embryologists look at when determining the quality of an embryo. For ploidy prediction, the quality of an embryo is a strong indicator of any abnormalities8. For example, if an embryo has not expanded to the blastocyst state by a certain time point, it is likely delayed, potentially due to chromosomal abnormalities.
Embryo quality score prediction
During the process of embryo development, embryologists assess and grade embryo quality, typically on Day 5 or Day 6 post-fertilization. These grades serve as indicators of embryo quality, assisting in the ranking and selection of embryos for transfer. However, grading systems are not standardized and can vary widely between different clinics and even among embryologists within the same clinic. Consequently, there is significant interest in developing an AI model that can be personalized to individual scoring systems. In this study, we evaluated the performance of FEMI using a scoring system outlined by Zhan et al., which is based on the widely recognized Gardner grading system18,19. The scoring system by Zhan et al. has been shown to correlate with key clinical outcomes such as ploidy, implantation, and fetal heart rates, thereby serving as a robust indicator of embryo quality. This system, employed at Weill Cornell Medicine (WCM) and subsequently referred to as the WCM scoring system, comprises several components: an Expansion Score, an Inner Cell Mass (ICM) Score, and a Trophectoderm (TE) Score, each ranging from 1 to 4. The overall blastocyst score (BS), which serves as a comprehensive quality metric, is calculated by summing these scores and adding a day-specific value (an addend of 0 for Day 5 biopsies and 2 for Day 6 biopsies), resulting in a total score range from 3 to 14. We trained FEMI to predict each of these four scores as part of a regression task to assess how effectively the model can learn and replicate the WCM scoring system. To train our models, we utilized a subset of the Weill ES dataset, which includes embryo quality scores for 1798 embryos. We designated 25% of this dataset as an internal test set, and the remainder was used to train models using 4-fold cross-validation. Each embryo was treated as an independent sample. Additionally, two external datasets employing the same scoring system, 2020 Weill ES+ (841 embryos with scores) and 2021+ Weill ES+ (2,668 embryos with scores), were used to evaluate model performance.
We further assessed FEMI’s performance using the scoring system employed by IVF Florida, a variant of the Zhan et al. system. The Florida scoring system omits the use of +/- gradings, leading to a less granular scoring range compared to the Zhan et al. system (Supplemental Table 7). For IVF Florida, our models were trained solely to predict the overall BS, ranging from 3 to 14. For the IVF Florida dataset, which comprises scores for 869 embryos, we similarly allocated 25% as a held-out test set and used the remainder for training through 4-fold cross-validation. Model performance was evaluated based on this test set.
In addition to image inputs, we explored the integration of video inputs for quality prediction tasks, specifically using sequences captured between 96 - 112 hpi. FEMI’s performance was compared against several pre-trained architectures, including VGG16, ResNet101-RS, EfficientNet V2, ConvNext, CoAtNet, and MoViNet for video classification. Models were also benchmarked against ImageNet-1k ViT MAE, IVF SWIN, and I-JEPA. Performance across all tasks was measured using mean absolute error.
Figure 4 shows the performance of all models on WCM quality scores. For both image and video inputs, FEMI significantly outperforms other models in multiple datasets on overall BS and inner cell mass score prediction. Supplemental Fig. 1 shows the correlation between blastocyst scores predicted by FEMI and the ground truth. For the expansion score, FEMI once again significantly outperforms all other models in both image and video inputs, except within the 2020 Weill ES+ and 2021+ Weill ES+ datasets for the image setting. For the trophectoderm score, FEMI significantly outperforms all models in the 2021+ Weill ES+ dataset and are comparable in other datasets in the image setting. In the video setting, FEMI significantly outperforms other models in the 2020 Weill ES+ and 2021+ Weill ES+ datasets. Supplemental Figure 2 shows the performance of image and video models for the Florida scoring system. For both input types, FEMI significantly outperforms comparison models.
Mean absolute error is used to measure performance on ES (Embryoscope) and ES+ (Embryoscope + ) microscopes. Performances are aggregated across 4 replicates (four-fold cross validation) (n = 4). Asterisks represent statistical significance with p < 0.05, where statistical significance is determined by performing a one-way ANOVA test followed by a Tukey HSD test. For blastocyst score: a image input; Weill ES, p = 0.41; 2020 Weill ES + , p = 1.6e-21; 2021+ Weill ES + , p = 1.1e-24. b video input; Weill ES, p = 0.26; 2020 Weill ES + , p = 0.18; 2021+ Weill ES + , 1.7e-9. For expansion score: c image input; Weill ES, p = 2.8e-18; 2020 Weill ES + , p = 0.99; 2021+ Weill ES + , p = 2.5e-20. d video input; Weill ES, p = 7.9e-26; 2020 Weill ES + , p = 0.23; 2021+ Weill ES + , p = 4.5e-17. For inner cell mass score: e image input; Weill ES, p = 0.99; 2020 Weill ES + , p = 1.4e-22; 2021+ Weill ES + , p = 1.7e-28. f video input; Weill ES, p = 2.7e-15; 2020 Weill ES + , p = 3.6e-21; 2021+ Weill ES + , p = 1.2e-27. For trophectoderm score: g image input; Weill ES, p = 0.34; 2020 Weill ES + , p = 0.36; 2021+ Weill ES + , p = 1.5e-33. h video input; Weill ES, p = 0.86; 2020 Weill ES + , p = 2.5e-37; 2021+ Weill ES + , p = 3.7e-39. For all subplots, error bars show mean values +/- SEM. Source data are provided as a Source Data file.
We evaluated FEMI’s performance in quality scoring across low, medium, and high-quality embryos. While FEMI’s performance on medium-quality embryos is comparable to that of other models, it significantly outperforms competitors on both low and high-quality embryos (p < 0.05). Specifically, FEMI achieves mean absolute errors of 2.04 ± 0.017 for low-quality embryos and 0.941 ± 0.028 for high-quality embryos, compared to 2.28 ± 0.033 and 1.17 ± 0.032, respectively, for the next best model, EfficientNet-V2. These results indicate that FEMI performs better in data regimes with fewer representative samples.
Embryo component segmentation
Segmentation of various blastocyst components such as the trophectoderm, zona pellucida (ZP), and inner cell mass is a critical task that facilitates both visualization and downstream analytical processes. By segmenting these regions of interest, it allows the further analysis of key morphological components that could affect embryo viability. In this study, we explored the capability of FEMI to perform segmentation on images of blastocysts. For this purpose, we utilized a publicly available dataset from Simon Fraser University, which includes 274 embryos20. Segmentations and masks for a sample image are shown in Supplemental Fig. 3. We reserved 25% of this dataset as a test set and employed the remaining data for model training through 4-fold cross-validation. Given the dataset’s limited size, data augmentation techniques were implemented to enhance the robustness of the training data. FEMI was adapted for segmentation tasks using the UNETR decoder architecture, as described by Hatamizadeh et al.21. This adaptation involved integrating three decoders, each with skip connections, into FEMI’s encoder. Each decoder was specifically trained to segment one of the three blastocyst components: TE, ZP, or ICM. We benchmarked FEMI against several models, including a U-Net with an ImageNet-1k VGG16 encoder, a UNETR utilizing an ImageNet-1k ViT encoder, and Segment Anything in Medical Images (MedSAM). Detailed descriptions of the implementations for these benchmark models are provided in the Methods section of the manuscript. The effectiveness of each model was assessed using the Dice score, which can evaluate the similarity between a predicted segmentation mask and the ground truth segmentation mask.
Figure 5a shows the performances of all models on the segmentation of the three embryo components. While FEMI does not significantly outperform comparison models in any of the three components, we note that FEMI does have a non-significant increase in Dice score.
Performances on ES (Embryoscope) and ES+ (Embryoscope + ) microscopes for each task. Performances are aggregated across 4 replicates (four-fold cross validation) (n = 4). Asterisks represent statistical significance with p < 0.05, where statistical significance is determined by performing a one-way ANOVA test followed by a Tukey HSD test. a Segmentation performance for each embryo component via Dice Score. Inner Cell Mass, p = 0.53; Zona Pellucida, p = 0.26; Trophectoderm, p = 0.06. b Embryo witnessing performance via F1 score across all datasets. Weill ES, p = 0.99; 2020 Weill ES + , p = 8.9e-22; Spain ES + , p = 1.7e-10; Florida ES + , p = 1.9e-25; 2021+ Weill ES + , p = 1.7e-25. c Blastulation time prediction performance using mean absolute error for Weill ES, Spain ES + , and 2020 Weill ES+ datasets. Weill ES, p = 0.81; 2020 Weill ES + , p = 1.3e-39; Spain ES + , p = 1.8e-30. For all subplots, error bars show mean values +/- SEM. Source data are provided as a Source Data file.
Embryo witnessing
The IVF process is susceptible to human errors, particularly during steps such as embryo biopsy, which can lead to mismatches when embryos are reintroduced into time-lapse imaging systems. To mitigate these risks, some clinics implement embryo witnessing procedures, where an embryologist manually verifies the identity of an embryo before and after critical steps22. While effective, this approach is labor-intensive. Previous research has utilized the ViT MAE architecture for embryo witnessing23. However, these studies were limited by a smaller dataset (20,000 images) and confined the identification process to a narrow timeframe (105 hpi - 110 hpi). In this study, we expand the scope of embryo witnessing by investigating whether FEMI can distinguish an embryo’s image at 112 hpi from its image at 96 hpi. This scope allows us to investigate the boundaries of FEMI’s capacity for witnessing. To train the witnessing model, we employed the FaceNet framework, using the encoder from FEMI as a feature extractor. The model was trained on triplets generated through semi-hard mining, where a typical triplet comprises an anchor image (e.g., a 96 hpi image of embryo A), a positive image (e.g., a 112 hpi image of embryo A), and a negative image (e.g., a 112 hpi image of embryo B). For model evaluation, features were extracted from both a 96 hpi and a 112 hpi image using the trained witnessing model. The embryos are considered to be the same if the Euclidean distance between their feature embeddings falls below a pre-defined threshold, established from the validation datasets. An equal number of positive and negative pairs were created for each test set to ensure balanced performance metrics.
To train our models for embryo witnessing, we utilized a subset of the Weill ES dataset consisting of 1998 embryos, reserving 25% as an internal test set. The remaining data was used for model training using 4-fold cross-validation. Additionally, four external datasets, which adhere to the same scoring system, were employed to evaluate model performance: Spain ES+ (890 embryo pairs), Florida ES+ (1680 embryo pairs), 2020 Weill ES+ (1960 embryo pairs), and 2021+ Weill ES+ (6000 embryo pairs). Model performance was assessed using the F1 Score, a metric that considers both the precision and the recall of the test to compute the score. A correct classification occurs when the model accurately identifies a pair of images as depicting the same embryo or correctly discerns pairs depicting different embryos. For the task of embryo witnessing, FEMI’s efficacy was benchmarked against several models pre-trained on ImageNet-1k, including VGG16, ResNet101-RS, EfficientNet V2, ConvNext, and CoAtNet. We also compared FEMI’s performance against two additional architectures, ImageNet-1k ViT MAE and IVF SWIN, to establish its relative proficiency in accurate embryo identification across various model frameworks.
Figure 5b shows the performance of all models for embryo witnessing. FEMI outperforms all comparison models in all datasets except the Weill ES dataset. Previous papers have achieved ~99% accuracy in witnessing, but these models had very narrow time identification spans (e.g. 105 - 110 hpi). We show > 90% F1 Score even after expanding the time scope to 96 - 112 hpi, where the embryo goes through multiple distinct changes as it develops into a mature blastocyst. We also explore if the FEMI embryo witnessing model can be used to cluster time-lapse images by source embryo. Supplemental Fig. 4 plots the first two principal components of the embeddings generated from ten randomly selected embryos from 2021+ Weill ES + . Embeddings were generated from a fully trained FEMI embryo witnessing model. The results indicate that images can indeed be grouped by source embryo, which may have downstream benefits beyond witnessing.
Blastulation time prediction
Predicting the blastulation time (tB), or the time at which an embryo develops into a blastocyst, is useful for embryologists both for assessing embryo quality and for planning subsequent visualization processes. In this study, we explored whether FEMI could accurately predict the hours post-insemination at which an embryo begins to form a blastocyst. This task was formulated as a regression problem, where the input was an image of the embryo at 72 hpi (end of Day 3, prior to blastocyst stage) and the label was the difference in hours between tB and 72 hpi. To train our models, we utilized a portion of the Weill ES dataset, which includes tB annotations for 1935 embryos. We reserved 25% of this dataset as an internal test set, and the remaining data was employed for training using 4-fold cross-validation. Performance was also evaluated using two external datasets that adhere to the same scoring system: Spain ES+ (531 embryos with tB) and 2020 Weill ES+ (983 embryos with tB). Model performance was assessed using the mean absolute error. For the blastulation time prediction task, FEMI’s effectiveness was compared against several models pre-trained on ImageNet-1k, including VGG16, ResNet101-RS, EfficientNet V2, ConvNext, and CoAtNet. Additionally, FEMI was benchmarked against the ImageNet-1k ViT MAE, IVF SWIN, and I-JEPA, to evaluate its predictive capabilities relative to other leading model architectures in accurately determining blastulation time.
Figure 5c shows the performance of all models for blastulation time prediction. FEMI significantly outperforms other models in the Spain ES+ and 2020 Weill ES+ datasets and performs comparably in Weill ES. Specifically, FEMI achieves an mean absolute error of 7.14 ± 0.13 hpi on Weill ES, 5.93 ± 0.05 on Spain ES + , and 6.28 ± 0.13 on 2020 Weill ES + .
Stage prediction
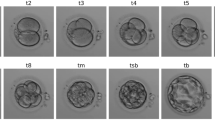
Accurate staging of embryos is crucial for monitoring developmental progression and optimizing outcomes in IVF procedures. Traditional manual staging by embryologists is time-consuming and subject to inter-observer variability. Automated models can provide consistent and efficient stage classification, enhancing decision-making in clinical settings. We evaluated FEMI’s performance in classifying embryos into twelve developmental stages (Fig. 6a), comparing it with several benchmark models: Embryovision, VGG16, ResNet101-RS, EfficientNet V2, ConvNext, CoAtNet, and ViT MAE ImageNet. Embryovision is a pipeline wherein a step is specifically designed for stage classification, which we use an additional baseline24. Embryonic development is a continuous process, and images often capture embryos in transitional stages. Discrete classification may not adequately represent the subtle morphological changes occurring between stages. Therefore, for FEMI and the other benchmark models (excluding Embryovision), we treated stage classification as a regression task. This approach allows the models to predict fractional stages, providing a more nuanced understanding of embryo development. After training, we binned the regression outputs into integer classes corresponding to the 12 defined stages for evaluation purposes. For more details about the methodology, see Methods. We assessed the models using top-1 accuracy, top-2 accuracy, Quadratic Weighted Kappa (QWK), and Spearman rank correlation to capture both exact matches and the quality of ordinal predictions (Fig. 6b).
Asterisks represent statistical significance with p < 0.05, where statistical significance is determined by performing a one-way ANOVA test followed by a Tukey HSD test. a Representative image sample for each of the 12 stages of development was collected from 12 different embryos. b Performances of FEMI and competitor models across multiclass classification metrics. Performances are aggregated across 4 replicates (four-fold cross validation) (n = 4). Top 1 Accuracy, p = 0.54; Top 2 Accuracy, p = 1.5e-54; Quadratic Weighted Kappa (QWK), p = 1.8e-58; Spearman Rank Correlation, p = 3.7e-56. Error bars show mean values +/- SEM. c Confusion matrix showing performance of FEMI across development stages. Source data are provided as a Source Data file.
FEMI outperformed the benchmark models across most metrics. Specifically, FEMI achieved a top-1 accuracy of 60.31%, comparable to Embryovision’s 60.58%, and surpassed the performances of the other models. Notably, FEMI was able to achieve comparable performance to Embryovision without using multiple focal planes. The top-2 accuracy of FEMI was 90.79%, significantly better than Embryovision’s 88.36%, indicating that even when the exact stage was not predicted, the model’s second choice was often correct, reflecting its ability to closely approximate the true developmental stage. The higher QWK score of FEMI (96.10% compared to Embryovision’s 94.75%) demonstrates better agreement with the true stages, accounting for the ordinal nature of the classification task. Additionally, FEMI’s Spearman rank correlation coefficient was 96.07%, suggesting a strong monotonic relationship between the predicted and actual stages. FEMI performed less optimally at predicting stages from the 3-cell to morula stages due to increased label noise; the ground truth labels during transitional periods are more difficult to assign accurately (Fig. 6c). These results indicate that treating stage classification as a regression problem captures the continuous progression of embryo development more effectively than discrete classification. FEMI’s superior performance underscores the advantage of leveraging self-supervised learning on large-scale, unlabeled data to capture complex developmental features.
Discussion
In this study, we introduced FEMI, a foundation model based on the ViT MAE architecture, trained on approximately 18 million time-lapse images of embryos. FEMI was evaluated across multiple clinically relevant tasks, demonstrating significant improvements in embryo assessment accuracy compared to existing models. Notably, FEMI achieved a marked enhancement in ploidy prediction, with AUROC values exceeding 0.70 in several datasets, specifically achieving an AUROC of 0.75 in the Spain ES+ dataset using only image data. On discriminating complex aneuploidy from euploid embryos, FEMI achieves over 0.85 AUROC with the inclusion of maternal age in multiple datasets. This performance is critical as accurate ploidy status prediction is a cornerstone in selecting embryos with the highest potential for successful implantation. The application of FEMI in clinical settings could revolutionize the process of embryo selection in IVF treatments by providing a standardized, non-invasive, and efficient methodology for assessing embryo viability. This shift could not only lower the costs associated with embryo selection by allowing embryologists to de-prioritize embryos predicted to be aneuploidy. Deprioritizing embryos predicted to be aneuploid is crucial as aneuploid embryos are associated with lower implantation rates and a higher risk of miscarriage compared to euploid embryos. Similarly, in embryo quality scoring, FEMI consistently outperformed traditional and other AI-based models, particularly in predicting the ICM score and overall blastocyst score from both image and video inputs. The model’s ability to accurately replicate the scoring systems used in different clinical settings, such as those at Weill Cornell Medicine and IVF Florida, underscores its potential to standardize and enhance the embryo selection process in IVF clinics globally. Furthermore, by automating the grading process, FEMI could help eliminate subjective biases and variability among embryologists, leading to more objective and reproducible assessments. The application of FEMI to the task of embryo component segmentation illustrated its capacity to perform detailed morphological analyses, important for automating and refining the assessment of embryo development stages. Although FEMI did not significantly outperform all comparison models in segmentation tasks, its competitive performance suggests it as a valuable tool for clinical and research applications in embryology. Continuing from the segmentation, FEMI’s application in embryo witnessing and blastulation time prediction further exemplifies its utility in embryological assessments. In embryo witnessing, a task vital for ensuring the accuracy of embryo handling and identification, FEMI demonstrated superior performance, achieving F1 scores over 90% in distinguishing embryos across distant time points (96 hpi to 112 hpi). This capability is particularly important in high-throughput clinical environments where manual witnessing is prone to errors and resource-intensive. In the task of predicting blastulation time, FEMI’s predictions were impressively precise, with mean absolute errors as low as 5.93 hpi in the Spain ES+ dataset. The accuracy in predicting the transition to the blastocyst stage provides embryologists with critical information to optimize the timing of interventions and improve the selection process for embryo transfer. There is also substantial potential in applying FEMI to other aspects of reproductive medicine, such as predicting implantation potential, live birth, and other clinically relevant tasks. These tasks require task-specific labels. Clinics that have access to these labels and want to train a generalizable prediction model could use FEMI as the backbone. Moreover, the performances shown here for downstream tasks are likely not the ceiling for FEMI and could be further increased with a larger labeled dataset.
Compared to other deep-learning-based models, FEMI’s self-supervised learning framework allows it to excel in tasks where traditional supervised models falter due to the variability and limited size of labeled training datasets. For example, the Vision Transformer architecture of FEMI has proven more adaptable than models like VGG16 or EfficientNet V2, particularly in complex tasks like video-based ploidy prediction and blastulation time prediction, where both spatial and temporal features play an important role. These comparisons underscore FEMI’s capacity to not only match but exceed the performance of existing technologies in embryo assessment, promising to set new standards in the accuracy and objectivity of IVF treatments. The generalizability of FEMI across various datasets highlights its robustness and adaptability to different clinical environments. FEMI demonstrated strong performance across multiple clinics with varied imaging protocols, as evidenced by its superior results in tasks such as ploidy prediction and embryo quality scoring across datasets from Weill Cornell Medicine, IVF Florida, and clinics across Europe. This broad applicability suggests that FEMI can be effectively integrated into diverse clinical workflows without the need for extensive customization.
Models for embryo selection are generally classified into one-step and two-step approaches25. One-step models directly predict clinical outcomes, such as embryo viability or implantation potential, from raw embryo images using deep learning methods. While efficient, these models often operate as black boxes, limiting their interpretability. In contrast, two-step models first annotate specific embryonic features—either manually by embryologists or automatically using AI—and subsequently use these annotated features to rank or predict clinical outcomes. This separation enhances transparency and interpretability, as the decision-making process is based on identifiable and clinically relevant features. FEMI bridges these paradigms by enabling both annotation and outcome prediction within a unified framework which can be used to extract both comprehensive morphological features and utilize them to predict various clinical outcomes. This integrated approach combines the efficiency of one-step models with the interpretability benefits of two-step models, ensuring that predictions are both accurate and transparent.
Despite its wide applicability, FEMI’s training and validation have inherent limitations. The model’s performance is contingent upon the quality and diversity of the dataset used. The current dataset, while extensive, is predominantly sourced from high-resource settings, which may not capture the variability found in lower-resource environments. This limitation could potentially affect the model’s performance and generalizability in globally diverse clinical settings. With a larger dataset, more computational resources will be needed to train the model, further limiting the size of the training dataset. The time-lapse images themselves are limited due to their inherent spatial and temporal resolutions. Furthermore, each downstream task presents distinct limitations. For ploidy prediction, we excluded mosaic embryos and faced challenges due to insufficient labeled data for training models to differentiate specific types of aneuploidies. Our ploidy prediction downstream tasks also only use input up to 112 hpi. Recent research has shown that slower-developing blastocysts, such as Day 6/7 blastocysts with blastulation occurring after 130 hpi may lead to euploid embryos26. However, training on these late stage embryos may present challenges due to limited time-lapse image series that extend to Day 7. That said, FEMI, as a foundational model, is designed to be adaptable and extensible. While our current training primarily focuses on data up to 112 hpi, the model architecture allows future researchers to fine-tune FEMI with additional datasets that include later time points. It is important to note that our ploidy prediction models are not intended to replace PGT-A. In the realm of quality score estimation, the ground truth labels may reflect biases influenced by embryologist input. Nevertheless, this study demonstrates that FEMI can effectively learn and replicate established scoring systems, allowing users to adapt FEMI for their specific datasets. The embryo component segmentation task also suffers from a small dataset. Future advancements in embryo component segmentation could benefit from integrating true 3D modeling approaches. Utilizing 3D time-lapse imaging would allow for more accurate and consistent segmentation of the ICM and TE by capturing the complete spatial context of the blastocyst. Moreover, for segmentation and stage prediction tasks, the same datasets were used for both training and testing due to the limited availability of labeled data. This may impact the ability to test the generalizability of FEMI in these specific tasks. Lastly, the labels used for predicting blastulation time may suffer from intra-observer variability in recording morphokinetics. Despite these issues, our results affirm FEMI’s effectiveness in these tasks as a proof of concept.
The development of a foundation model for IVF time-lapse imaging is a significant advancement in reproductive medicine. This model will serve as a crucial decision support tool for clinicians, enhancing their ability to select the most viable embryos. Furthermore, by making FEMI available to the broader scientific community, it can be adapted and utilized for various research purposes and clinic-specific needs, fostering greater collaboration and innovation in the field. To ensure that FEMI provides tangible benefits in clinical settings, we plan to implement randomized controlled trials in the future. These trials will compare FEMI-assisted embryo selection with standard manual selection techniques, aiming to assess improvements in IVF success rates and overall embryo assessment accuracy. This approach will enable us to robustly evaluate FEMI’s performance and ensure its reliability and effectiveness as a decision-support tool in reproductive medicine.
Methods
The study was performed in accordance with relevant guidelines and regulations. The study was approved by the Institutional Review Board at Weill Cornell Medicine (numbers 1401014735 and 19–06020306) and by the IVI Valencia Institutional Review Board (number 1709-VLC-094-MM). IRB determined that this research meets the exemption requirements at HHS 45 CFR 46.104(d) and is secondary research for which consent is not required. A waiver of informed consent was granted from the IRB as the images were de-identified for this retrospective review of clinical data. The embryo imaging was performed as part of the standard care procedure during the preimplantation and IVF cycle. No discarded embryos were used. In this study, information, which may include information about biospecimens, is recorded by the investigator in such a manner that the identity of the human subjects cannot readily be ascertained directly or through identifiers linked to the subjects. Moreover, the investigators do not contact the subjects, and the investigators will not re-identify the subjects. As such, informed consent was not obtained, and participants did not receive compensation for the study.
Datasets and preprocessing
We curated images from multiple public and private datasets for training FEMI. Attributes of each of the dataset can be found in Supplemental Table 1. While each embryo has associated time-lapse images, not all embryos have associated clinical information like PGT-A, morphokinetics, blastocyst scores, etc. These annotations are specific to each dataset, and within each dataset, some samples may be missing the annotations. For training FEMI via SSL, a combined 17,968,959 images were processed, spanning −30 to 30 focal depths, and cropped using the mask from the embryo segmentation model. Focal planes used for model training were defined as z = 0 μm, representing the equatorial plane of the embryo, and z = ±30 μm, providing additional depth perspectives. The z = 0 μm plane was automatically determined by the Embryoscope+ incubator’s automated focusing system, with no manual adjustments made during culture or re-insertion of the dish. Including images from z = ±30 μm allows the model to capture different morphological features across various depths, thereby enhancing its ability to learn comprehensive embryo characteristics. For the training of FEMI, images after 85 hpi were used. The selection of images taken after 85 hpi aligns with the morula stage of embryo development, a phase characterized by significant morphological changes. This timepoint is chosen because most clinically relevant tasks, such as ploidy prediction and embryo quality scoring, are conducted after the morula stage. Additionally, embryos exhibit more complex features post-morula, providing a richer dataset for the foundation model to learn from. Contributing to this dataset is 4,521,481 images from Weill ES (25.16 %), 2,080,483 images from 2020 Weill ES+ (11.57 %), 1,962,797 images from Florida ES+ (10.92 %), and 9,022,769 images from 2021+ Weill ES+ (50.21 %). We also had 36,722 images from Spain ES + , 342,363 images from the Hospital of Nantes, and 2344 images from European clinics which was a total of ~1% of the overall dataset.
An embryo segmentation model was trained to crop embryos from the petri dish to maximize information within the image space for FEMI to learn about embryos better. An InceptionV3 U-Net was trained using 30 image-mask pairs of whole-embryo segmentations. 80% of the dataset was used for training the model, with the rest being used for validation. To make the model more robust, we added data augmentations to the training data, including random cropping, flipping, rotation, and translation. This segmentation model was then used to create masks for the images curated for FEMI SSL training. Before generating segmentation masks, each image undergoes an automatic brightness and contrast adjustment to standardize lighting conditions and enhance the visibility of embryo structures. This process is implemented using an adaptive algorithm that first converts colored images to grayscale. The algorithm then computes the grayscale histogram and identifies the intensity levels corresponding to the central 80% of the pixel distribution by excluding the top and bottom 10% of pixel intensities. Based on these thresholds, the algorithm calculates scaling factors for brightness (β) and contrast (α) to adjust the image intensity values uniformly across all images. After generating a segmentation mask for each input image, circular contours were detected in the image using OpenCV. A bounding box is drawn around the most circular contour, which is likely the embryo. The image is then cropped around the bounding box and resized to 224 ×224 for input into FEMI. Input images are then normalized using ImageNet mean and standard deviation parameters, consistent with the training recipe for ViT MAE architectures.
FEMI - ViT Masked Autoencoder
Weights from ImageNet-1k pre-trained masked ViT MAE (encoder and decoder) were downloaded from (https://github.com/facebookresearch/mae) and converted to TensorFlow. Weights that were trained with normalized pixel loss were used as previous studies have suggested that using normalized pixel loss leads to better performance on downstream tasks. The encoder is a large vision transformer with 24 transformer blocks and an embedding vector size of 1,024. The decoder is a smaller vision transformer with 8 transformer blocks and an embedding vector size of 512. Unmasked patches of size 16 × 16 are inputted into the encoder, which then projects them into feature vectors of size 1024. These vectors undergo processing through the encoder’s 24 transformer blocks, each incorporating multiheaded self-attention and a multilayer perceptron, to generate high-level features. The decoder, in turn, inputs these features along with masked dummy patches and reconstructs the image patch using a linear projection. The primary objective during model training is the reconstruction of time-lapse images from highly masked versions. As suggested by He et al, only cropping augmentations were used when training FEMI. 80% of 17,968,959 images were used for training, and the remaining was used for validation. Ablation experiments with different training dataset sizes for FEMI were studied with predicting ploidy status as the downstream task (Supplemental Fig. 5). Their experiments suggested the ViT-Large architecture approached a ceiling in performance with ~18 million training data points. A mask ratio of 0.75 was used. FEMI was trained for 800 epochs with early stopping (patience: 20 epochs), halting training and restoring weights to the lowest validation loss if the validation loss failed to decrease within 20 epochs. To train FEMI, we use AdamW optimizer with a custom learning rate schedule, which has 20 epochs of warmup followed by cosine decay. A batch size of 256 per device was used with a learning rate of 1.5 ×10−4.
Benchmark models
Supervised learning models
To compare FEMI against traditional supervised learning on downstream tasks, we explored the use of multiple models, specifically the VGG16, ResNet101-RS, EfficientNet V2, ConvNext, CoAtNet, and MoViNet architectures. The VGG16 architecture is one of the earliest large-scale convolutional architectures used for image classification, but can readily be adapted for various other tasks. Despite its age, the VGG16 model still performs well in multiple image-based medical tasks27. More recently, ResNet101-RS,EfficientNet V2, ConvNext, and CoAtNet models have become state-of-the-art models for supervised learning on image classification tasks28,29,30,31. These models enhance their predecessors through various means, such as the addition of residual connections, attention layers, etc., and have garnered competitive performances on the ImageNet-1k benchmark. For this study, we use the encoders from each of these architectures, pre-trained on ImageNet-1k, and add downstream layers, similar to what we do to the FEMI encoder. The MoViNet architecture is a video classification architecture that only takes in videos as inputs32. It has superior performance to many commonly used video classification models, such as 3D Convolutional models, I3D, and Video Vision Transformers, and is significantly less data hungry. Because some of the downstream tasks we explored could be adapted to a video input, we also looked to compare FEMI with MoViNet on these tasks. Similar to the other architectures, we use a Kinetics-600 pre-trained MoViNet encoder for downstream tasks, which use the MoViNet, where downstream layers can be adapted for both classification and regression. For ploidy prediction, we also explore a previously published model, STORK-A. STORK-A utilizes a ResNet18 architecture as its backbone and was trained on static time-lapse images at 110 hpi. For EUP vs. ANU, STORK-A was trained on a total of 10,378 time-lapse images (aneuploids (n = 5953) and euploids (n = 4425)). For EUP vs. CxA, STORK-A was trained on 7,434 time-lapse images (complex aneuploids (n = 3009) and euploids (n = 4425)).
SSL models
We also benchmarked FEMI against models pre-trained via self-supervision. Our first comparison was the ImageNet-1k ViT MAE, where the ViT MAE was only trained using the ImageNet-1k dataset and not the time-lapse image dataset curated for FEMI. For multiple downstream tasks, we also compared the performance of FEMI to a Swin Transformer V1, trained on both ImageNet-22k and time-lapse images33. The weights for the Swin Transformer were downloaded from (https://github.com/microsoft/Swin-Transformer) and trained on the 17,968,959 time-lapse images FEMI was trained on. The images went through the same processing as they did for FEMI. The Swin Transformer was trained for 800 epochs with early stopping. We use AdamW optimizer with a custom learning rate schedule, which has 20 epochs of warmup followed by cosine decay. A batch size of 256 per device was used with a learning rate of 1.5e-4. We also compare FEMI to another state-of-the-art SSL architecture, I-JEPA, trained on the same 18 million embryo image dataset. I-JEPA is a self-supervised learning architecture recently released by Meta that has outperformed many comparison SSL architectures in natural images. Similar to the Swin transformer, weights for I-JEPA (VIH-H trained on ImageNet-1k) were downloaded from (https://github.com/facebookresearch/ijepa) and further pretrained on 17,968,959 time-lapse images34. I-JEPA was pretrained for 100 epochs using the AdamW optimizer with a custom learning rate schedule, which has 10 epochs of warmup followed by cosine decay. A batch size of 128 per device was used with a learning rate of 2e-3. For the embryo component segmentation downstream tasks, we also compare FEMI to MedSAM35. To develop MedSAM, the Segment Anything Model (SAM) was fine-tuned on various anatomical structures. Broadly, the model uses a ViT encoder to extract image features, a prompt encoder for integrating user interactions (bounding boxes), and a mask decoder. In this study, the bounding box is the entire image.
Adaptation for downstream tasks
Hyperparameter tuning and fine-tuning strategy
To ensure a fair comparison across FEMI and all benchmark models, we performed a systematic hyperparameter search for each architecture using a common range of learning rates (e.g., 1e-4 to 1e-2), batch sizes (from 8 to 64 depending on memory constraints), and optimizers (AdamW/Adam). For each model and each task, a small-scale grid search was performed to identify the optimal hyperparameters, using our cross-validation procedure on the training folds and selecting the setting with the best mean performance on the validation folds. We applied the same procedure to determine which layers to freeze vs. train in each model. Early layers were typically frozen, while deeper layers (responsible for more task-specific features) were fine-tuned. For FEMI (ViT MAE backbone), we found that freezing all but the final block improved performance consistently across downstream tasks (e.g., ploidy prediction, quality scoring). Benchmark models such as VGG16, ResNet101-RS, EfficientNet V2, ConvNext, CoAtNet, and MoViNet underwent the same tuning steps, in which we froze the backbone’s initial layers and only trained a subset of top blocks after verifying that this resulted in stable, optimal performance. This identical strategy for FEMI and all comparator architectures ensures our performance comparisons are not biased by divergent tuning practices.
Ploidy prediction
In this study, we study ploidy prediction in multiple ways. The input to the model could be image-only, image+age, video-only, or video+age. In addition, the task could be to discriminate between euploidy and any aneuploid embryos, or between euploid and complex aneuploid embryos. The image input is the time-lapse image from 110 hpi. The video input is a time-lapse sequence from 96 - 112 hpi. Rajendran et al. identified the period from 96 to 112 hpi as critical for ploidy determination due to significant morphological changes that occur during this timeframe, which are indicative of chromosomal abnormalities8. This finding informed the decision to incorporate sequences of time-lapse images within this period for ploidy prediction tasks. For the single image input, 110 hpi was used as there is a larger volume of data at this specific time point, which facilitates more effective training and enhances the model’s predictive performance. For image-only tasks, we add a fully-connected layer with sigmoid activation to the encoder (from FEMI or benchmark models). For video-only tasks, apart from the MoViNet architecture, each frame of the video is passed through the encoder. These frame features are then passed to a bi-directional long short-term memory (LSTM) layer with hierarchical attention. The output of the attention layer is passed to a fully-connected layer with sigmoid activation. For the MoViNet benchmark, the entire video sequence is processed by the encoder and outputs a feature embedding that is processed by a classifier layer. For models where maternal age is included, maternal age is first passed through a dense layer and then concatenated with either the encoder output in image models or the output of the attention layers in video models.
All embryos classified as mosaic have been removed from this analysis, based on previous studies. For euploid vs. complex aneuploid, single aneuploid embryos were removed. The task is framed as binary classification. To reduce overfitting, we incorporate label smoothing. We use binary cross-entropy loss and AdamW as the optimizer. The batch size for image models is 32 and for video models, 8. We train the model for 100 epochs with early-stopping. We use a ReduceLROnPlateau scheduler, with a base learning rate of 1e-3. We only fine-tuned specific layers of the encoder, dependent on the architecture, to maximize performance. Specifically for FEMI, we found that only fine-tuning the last hidden layer and freezing all earlier layers performed the best for ploidy prediction.
Blastocyst quality scoring
For quality scores, we explored both image and video inputs, similar to ploidy prediction. The architectures for quality scoring are the same as those used for ploidy prediction, except that scoring is framed as a regression task, which requires a linear activation on the final fully-connected layer. Scores were scaled to a range of between 0 - 1, which was achieved by dividing the overall blastocyst scores by 14, and dividing the sub-scores (Expansion Score, ICM Score, and TE Score) by 4. We use logcosh loss and AdamW as the optimizer. The batch size for image models is 32 and for video models, 8. We train the model for 100 epochs with early-stopping. We use a ReduceLROnPlateau scheduler, with a base learning rate of 1e-3. Like ploidy prediction, we only fine-tuned specific layers of the encoder. For FEMI, we only fine-tuned the last hidden layer.
Embryo component segmentation
In this study, we explored the segmentation of three embryo components. For FEMI, VGG16, and ImageNet-1k ViT MAE, we used an architecture consisting of one encoder and three decoders, one for each component. For FEMI and ImageNet-1k ViT MAE, we use a UNETR architecture to perform segmentation21. Briefly, a UNETR architecture uses a ViT as the encoder with skip connections to the decoder from specific hidden layers of the encoder. For the VGG16 model, we use a U-Net architecture for segmentation. Our final benchmark model is a pre-trained MedSAM. Because MedSAM requires a bounding box in addition to the image as input, we opted to build three different MedSAM models, one for each component. Each MedSAM model was trained to segment one component, and the bounding box provided enclosed the entire image. We added data augmentations to the training data, including flipping, rotation, zooming, and translation. The model was trained for 200 epochs with early-stopping. We use a ReduceLROnPlateau scheduler, with a base learning rate of 1e-3. All layers of all models were unfrozen during training.
Blastulation time prediction
For blastulation time prediction, we explore an image input, where the image is taken from 72 hpi. This time point was chosen as it is well before embryos develop into a blastocyst. We use the same architecture as that used in quality score estimation, as we frame blastulation time prediction as a regression task. The labels were the difference in time (in hours) between 72 hpi and the time to the start of blastulation. The values were scaled by dividing by 150. We use logcosh loss, AdamW as the optimizer, and a batch size of 32. We train the model for 100 epochs with early-stopping. We use a ReduceLROnPlateau scheduler, with a base learning rate of 1e-3. We only fine-tuned specific layers of the encoder. For FEMI, we only fine-tuned the last hidden layer.
Embryo witnessing
For embryo witnessing, we train the model using a FaceNet framework36. Specifically, the model architecture utilizes an encoder, followed by a L2 normalization layer. In this study, the encoder is from FEMI, ImageNet-1k ViT MAE, ResNet101-RS, EfficientNet V2, ConvNext, CoAtNet, and IVF SWIN. To train the model, we use a triplet loss algorithm. Triplet loss involves three data points— an anchor (A), a positive (P), and a negative (N). The anchor and the positive belong to the same class, while the negative belongs to a different class. The goal of the loss function is to ensure that the distance between the anchor and the positive is less than the distance between the anchor and the negative by some margin. In particular, we use triplet semi-hard loss, which refers to the method of negative selection during the training process. A negative is considered semi-hard if it is harder than the easiest negatives (those that are already further away from the anchor than the positive) but not as challenging as the hardest negatives (those that are closer to the anchor than the positive). The pairs consisted of embryos from 96 and 112 hpi, where the model learned to identify if a pair contains images from the same embryo. We use AdamW as the optimizer, and a batch size of 144. We train the model for 100 epochs with early-stopping. We use a ReduceLROnPlateau scheduler, with a base learning rate of 1e-2. We only fine-tuned specific layers of the encoder. For FEMI, we only fine-tuned the last hidden layer.
Stage prediction
We utilized a dataset comprising 78,000 time-lapse images captured at various developmental stages, with each of the 12 stages represented by an equal number of samples. For each embryo, images were acquired across multiple focal planes to capture comprehensive morphological details. We compared eight models for stage classification: VGG16, ResNet101-RS, EfficientNet V2, ConvNext, CoAtNet, ViT MAE pre-trained on ImageNet, Embryovision, and FEMI. Embryovision is a previously published model tailored for embryo stage classification, which we adapted to classify 12 stages24. It utilizes a ResNeXt101 backbone and processes three focal planes (z = 0, +15, –15) of a single image as input. Embryovision was trained as a multiclass classification model using categorical cross-entropy loss. For the other seven models (VGG16, ResNet101-RS, EfficientNet V2, ConvNext, CoAtNet, ViT MAE ImageNet, and FEMI), we used only the z = 0 focal plane image and modeled the task as a regression problem. Embryonic development is a continuous process, and images may capture embryos transitioning between stages. Treating stage classification as a regression task allows models to learn and predict fractional stages, reflecting the continuous nature of development. This approach captures subtle morphological changes between stages, potentially leading to more accurate and informative predictions. After training, we binned the continuous outputs into integer classes corresponding to the 12 stages for evaluation. For the regression-based models, we used the log-cosh loss function, which is robust to outliers and provides smooth gradients. The models were trained using the Adam optimizer with an initial learning rate of 1e-3 for 50 epochs. For FEMI, we fine-tuned only the last hidden layer to leverage the pre-trained features while reducing overfitting. Early stopping was employed based on the validation loss to prevent overfitting. For Embryovision, we maintained its original multiclass classification framework without modifications to its training procedure. We evaluated model performance using top-1 accuracy, top-2 accuracy, Quadratic Weighted Kappa (QWK), and Spearman rank correlation. These metrics provide a comprehensive assessment of both exact stage predictions and the models’ ability to capture the ordinal relationships between developmental stages.
Computational resources
We used 10 NVIDIA RTX A6000 GPUs to train FEMI. FEMI took about four months to train. For downstream models, we used a combination of A6000 GPUs and A100 GPUs. Training times (on one A100 GPU) for downstream tasks are shown in Supplemental Table 2.
Statistics and reproducibility
Relevant metrics were calculated for each task. We calculated the mean and standard deviation over all four folds of cross-validation for each task and each architecture. The standard error is calculated by dividing the standard deviation by the square root of the sample size, or 4. For each task, we performed a one-way ANOVA test followed by a Tukey’s Honestly Significant Difference (HSD) test to determine if FEMI was significantly better than all other models. A p-value < 0.05 was considered significant. Sample sizes for datasets were determined based on the maximum usable subset available, after all exclusion criteria were applied to embryos. These exclusion criteria included embryos with a mosaic PGT-A status, and embryos with missing information (specific to the downstream task) such as blastocyst score, ploidy status, and maternal age. Randomization was introduced into experimentation through four-fold cross-validation in all relevant comparisons. The investigators were not blinded to allocation during experiments and outcome assessment.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The WCM, IVF Florida, and IVI RMA embryo time-lapse imaging datasets were not collected as part of this study, and were analyzed retrospectively. The embryo-imaging datasets are available under restricted access owing to reasonable privacy and security concerns. Researchers can request access to the named institutions which will be evaluated on a case-by-case basis. Any requests should be sent to N.Z. (nizanin@med.cornell.edu). Requests will receive a response within a week. The public imaging datasets we used are available at https://doi.org/10.5281/zenodo.6390798 and https://github.com/software-competence-center-hagenberg/Blastocyst-Dataset. The current FEMI trained model is available for academic and non-profit use. They can be accessed at https://huggingface.co/ihlab/FEMI. Source data are provided with this paper.
Code availability
The code used to develop the model, perform the analyses and generate results in this study is publicly available and has been deposited in https://github.com/ih-lab/FEMI (10.5281/zenodo.15490833), under the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International Public License37.
References
Niakan, K. K., Han, J., Pedersen, R. A., Simon, C. & Pera, R. A. Human pre-implantation embryo development. Dev. (Camb., Engl.) 139, 829–841 (2012).
Niederberger, C. et al. Forty years of IVF. Fertil. Steril. 110, 185–324.e5 (2018).
Greco, E. et al. Preimplantation genetic testing: Where we are today. Int. J. Mol. Sci. 21, 4381 (2020).
Zhang, Y. X. et al. The pregnancy outcome of mosaic embryo transfer: A prospective multicenter study and meta-analysis. Genes 11, 973 (2020).
Chavez-Badiola, A. et al. Embryo ranking intelligent classification algorithm (ERICA): Artificial intelligence clinical assistant predicting embryo ploidy and implantation. Reprod. biomedicine online 41, 585–593 (2020).
Barnes, J. et al. A non-invasive artificial intelligence approach for the prediction of human blastocyst ploidy: a retrospective model development and validation study. Lancet Digital health 5, e28–e40 (2023).
Khosravi, P. et al. Deep learning enables robust assessment and selection of human blastocysts after in vitro fertilization. npj Digit. Med. 2, 21 (2019).
Rajendran, S. et al. Automatic ploidy prediction and quality assessment of human blastocysts using time-lapse imaging. Nat. Commun. 15, 7756 (2024).
Moor, M. et al. Foundation models for generalist medical artificial intelligence. Nature 616, 259–265 (2023).
Dosovitskiy, A. et al. An Image is Worth 16x16 Words: Transformers for Image Recognition at Scale. International Conference on Learning Representations (2021).
Wang, G. et al. A generalized AI system for human embryo selection covering the entire IVF cycle via multi-modal contrastive learning. Patterns (N. Y., N. Y.) 5, 100985 (2024)
He, K. et al. Masked autoencoders are scalable vision learners. CVPR (p./pp. 15979-15988),: IEEE. ISBN: 978-1-6654-6946-3 (2022).
Zhou, Y. et al. A foundation model for generalizable disease detection from retinal images. Nature 622, 156–163 (2023).
Liu, Z., et al. VISION-MAE: A foundation model for medical image segmentation and classification. CoRR, abs/2402.01034 (2024).
Gomez, T. et al. A time-lapse embryo dataset for morphokinetic parameter prediction. Data brief. 42, 108258 (2022).
Kromp, F. et al. An annotated human blastocyst dataset to benchmark deep learning architectures for in vitro fertilization. Sci. data 10, 271 (2023).
Wang, H. et al. Score-cam: Score-weighted visual explanations for Convolutional Neural Networks. 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW). https://doi.org/10.1109/cvprw50498.2020.00020 (2020).
Zhan, Q. et al. Blastocyst score, a blastocyst quality ranking tool, is a predictor of blastocyst ploidy and implantation potential. FS Rep. 1, 133–141 (2020).
Gardner D. K., Schoolcraft W. B. In vitro culture of human blastocyst. In: Jansen R., Mortimer D., eds. Towards Reproductive Certainty: Infertility and Genetics Beyond. Parthenon Press; Carnforth: 1999. pp. 377–388.
P. Saeedi, D. Yee, J. Au and J. Havelock, Automatic identification of human blastocyst components via texture, in IEEE transactions on biomedical engineering, vol. 64, pp. 2968-2978, Dec. (2017).
Hatamizadeh, A. et al. (2022). UNETR: Transformers for 3D medical image segmentation. WACV (p./pp. 1748-1758),: IEEE. ISBN: 978-1-6654-0915-5
Forte, M. et al. Electronic witness system in IVF-patients perspective. J. Assist. Reprod. Genet. 33, 1215–1222 (2016).
Liu, M. et al. WISE: whole-scenario embryo identification using self-supervised learning encoder in IVF. J. Assist. Reprod. Genet. 41, 967–978 (2024).
Leahy, B. D. et al. Automated measurements of key morphological features of human embryos for IVF. Medical image computing and computer-assisted intervention: MICCAI. Int. Conf. Med. Image Comput. Computer-Assist. Intervention 12265, 25–35 (2020).
Lee, T., Natalwala, J., Chapple, V. & Liu, Y. A brief history of artificial intelligence embryo selection: from black-box to glass-box. Hum. Reprod. (Oxf., Engl.) 39, 285–292 (2024).
Hernandez-Nieto, C. et al. What is the reproductive potential of day 7 euploid embryos? Hum. Reprod. (Oxf., Engl.) 34, 1697–1706 (2019).
Wang, L. et al. Trends in the application of deep learning networks in medical image analysis: Evolution between 2012 and 2020. Eur. J. Radiol. 146, 110069 (2022).
Bello, I. et al. Revisiting ResNets: Improved training and scaling strategies. In Ranzato M., Beygelzimer A., Dauphin Y. N., Liang P. & Vaughan J. W. (eds.), NeurIPS (p./pp. 22614-22627) (2021).
Tan, M. & Le, Q. V. EfficientNetV2: Smaller models and faster training. In Meila M. & Zhang T. (eds.), ICML (p./pp. 10096-10106),: PMLR. (2021).
Liu, Z. et al. A ConvNet for the 2020s. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) (11976–11986). IEEE. https://doi.org/10.1109/CVPR52688.2022.01167 (2022).
Dai, Z., Liu, H., Le, Q. V. & Tan, M. CoAtNet: Marrying convolution and attention for all data sizes. Adv. Neural Inf. Process. Syst. 35, 3965–3977 (2021).
Kondratyuk, D. et al. MoViNets: Mobile video networks for efficient video recognition. CVPR (p./pp. 16020-16030),: Computer Vision Foundation / IEEE (2021).
Liu, Z. et al. Swin Transformer: Hierarchical vision transformer using shifted windows (cite arxiv:2103.14030) (2021).
Assran, M. et al. Self-supervised learning from images with a joint-embedding predictive architecture, in Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, 15619–15629 (2023).
Ma, J. et al. Segment anything in medical images. Nat. Commun. 15, 654 (2024).
Schroff, F., Kalenichenko, D. & Philbin, J. Facenet: A unified embedding for face recognition and clustering. Proceedings of the IEEE conference on computer vision and pattern recognition (p./pp. 815-823) (2015).
Rajendran, S. et al. ih-lab/FEMI, https://doi.org/10.5281/zenodo.15490833 (2025).
Acknowledgements
This study is supported by an NIGMS Maximizing Investigators’ Research Award (MIRA) R35GM138152 to I.H. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. I.H. is also supported by an Irma Hirschl Career Scientist Award for this project. SR would like to acknowledge the support from the Tri-Institutional Training Program in Computational Biology and Medicine (CBM) funded by the NIH grant 1T32GM083937. S.R. would also like to acknowledge funding from the National Science Foundation Graduate Research Fellowship.
Author information
Authors and Affiliations
Contributions
S.R. and I.H. conceived the study. S.R., E.R., W.P. and I.H. conceived the method and designed the algorithmic techniques. S.R. wrote the codes and performed the computational analysis with input from E.R., W.P., and I.H. Q.Z., J.E.M., Z.R. and N.Z. provided the Weill Cornell datasets and labeled images. M.M. provided the Spain dataset. K.A.M. provided the Florida dataset. S.R. drafted the manuscript with input from O.E., Q.Z. and I.H. All the authors read the paper and suggested edits. I.H. supervised the project.
Corresponding author
Ethics declarations
Competing interests
O.E. is a scientific adviser for, and an equity holder in, Freenome, Owkin, Volastra Therapeutics, OneThree Biotech, Genetic Intelligence, Acuamark DX, Harmonic Discovery, and Champions Oncology, and has received funding from Eli Lilly, Johnson & Johnson–Janssen, Sanofi, AstraZeneca, and Volastra. N.Z. is a paid consultant for AIVF and Fairtility, and is on the advisory board of, and has equity in, Alife Health. I.H. is a consultant and is on the advisory board of Noor Sciences. M.M. received speaker fees from Merck, Vitrolife, Ferring, Theramex, and Gideon Richter. K.A.M. serves as a paid consultant and advisory board member for Fairtility and Alife Health (holding equity), and as a scientific board member for Genomic Prediction and Igenomix. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Akira Funahashi, who co-reviewed with Yusuke Hiki and Takashi Morikura, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Rajendran, S., Rehani, E., Phu, W. et al. A foundational model for in vitro fertilization trained on 18 million time-lapse images. Nat Commun 16, 6235 (2025). https://doi.org/10.1038/s41467-025-61116-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-61116-2