Abstract
Mono-methylation of histone H4 lysine 20 (H4K20me1) regulates DNA replication, cell cycle progression and DNA damage repair. How exactly H4K20me1 regulates these biological processes remains unclear. Here, we report that an evolutionarily conserved tandem Tudor domain (TTD) in BAHCC1 (BAHCC1TTD) selectively reads H4K20me1 for facilitating replication origin activation and DNA replication. Our biochemical, structural, genomic and cellular analyses demonstrate that BAHCC1TTD preferentially recognizes H4K20me1 to promote the recruitment of BAHCC1 and its interacting partners, notably Mini-chromosome Maintenance (MCM) complex, to replication origin sites. Combined actions of the H4K20me1-reading BAHCC1 and the H4K20me2-reading Origin Recognition Complex (ORC) ensure genomic loading of MCM for replication. Depletion of BAHCC1, or disruption of the BAHCC1TTD:H4K20me1 interaction, reduces H4K20me1 levels and MCM loading, leading to defects in replication origin activation and cell cycle progression. In summary, this study identifies BAHCC1TTD as an effector transducing H4K20me1 signals into MCM recruitment to promote DNA replication.
Similar content being viewed by others
Introduction
All of the four possible methylation states of histone H4 lysine 20, namely, non-, mono-, di- and tri-methylation (H4K20me0, H4K20me1, H4K20me2 and H4K20me3), have been suggested to be critical for regulating a wide variety of biological processes, including DNA replication, chromosome segregation, cell division, transcription, DNA damage repair, genome integrity, amongst others1,2,3,4,5,6,7. Notably, different H4K20 methylation states display quite different genomic distribution throughout the cell cycle, indicating their distinct functionalities1,5,6,7,8. For example, the unmodified H4K20me0 shows the highest level during the S phase, followed by gradual methylation during DNA replication5. Meanwhile, H4K20me1 accumulates during the replication and reaches the highest level at the G2 and M phases8,9,10. Such a cell cycle-dependent change of H4K20me1 is induced at least partly by dynamic and complex interplays between the H4K20me1-specific writers, such as SET domain containing 8 (SET8; also known as KMT5A, SETD8 and PR-Set7), and the H4K20me1 erasers, such as PHD Finger Protein 8 (PHF8; also known as KDM7B)11,12,13,14,15. Furthermore, another set of enzymes, Suppressor Of Variegation 4-20 Homolog 1 (SUV4-20H1; also known as KMT5B) and Suppressor Of Variegation 4-20 Homolog 2 (SUV4-20H2; also known as KMT5C), convert H4K20me1 to the highly methylated H4K20me2 and H4K20me3, which are relatively stable across the cell cycles but increase when cells enter quiescence4,16,17.
Exquisite mechanisms exist to ensure the accurate and orderly execution of DNA replication from genomic sites termed replication origins, which involves the complicated interplays among the replication initiators and loaders, replisome machinery, and various other regulatory factors18,19,20,21,22. Origin recognition and licensing, which occur strictly during the late mitosis and G1 phase, is one of the most critical steps during DNA replication, which relies at least partly on Origin Recognition Complex (ORC) for the chromatin recruitment and loading of Mini-chromosome Maintenance (MCM) complex, a ring-shaped helicase19,20,21. Density of the chromatin-associated ORC and MCM complexes is positively correlated with the replication initiation and timing, while the appropriate chromatin states, including chromatin accessibility and the cell cycle phase-associated H4K20 methylation, play critical roles in regulating the replication complex loading and origin licensing and activation1,2,3,4,6,7,22,23,24. For instance, H4K20me3, a histone modification enriched in heterochromatin, facilitates the genomic binding and recruitment of Origin Recognition Complex-Associated (ORCA; also known as LRWD1), which acts as a replication origin licensing facilitator to ensure proper replication from the late-firing origins associated with heterochromatin25. Meanwhile, H4K20me2, which is read by the Bromo-adjacent Homology (BAH) domain of ORC1 (ORC1BAH), provides a critical mechanism for recruiting ORC. Mis-regulation of this pathway due to germline mutation of ORC1 leads to the development of Meier-Gorlin syndrome2. H4K20me2 constitutes an overwhelming majority (~80%) of all H4K20 sites at any given time26; however, only a very limited number of potential replication origins are eventually selected and used27, indicating the existence of additional molecular determinants for origin recognition and/or activation. Indeed, a recent study showed that H2A.Z in the nucleosome provides a signal for recruiting SUV4-20H enzymes to deposit H4K20me2, thereby licensing and activating early-replication origins28. Evidence also points to a critical role of H4K20me1 in the regulation of MCM loading13,29, and to date, only the indirect effects of H4K20me1, such as chromatin compaction and its influence on H4K20me2 or H4K20me34,17,30,31, were proposed to contribute to replication regulation. A reader specific to H4K20me1 remains to be identified and a regulatory mechanism directly linking H4K20me1 to genome duplication remains to be elucidated. In this study, we identify BAHCC1TTD as a highly selective reader for H4K20me1, which transduces H4K20me1 signals into the recruitment of MCM to promote efficient DNA replication.
Results
The S phase-enriched BAHCC1 regulates DNA replication in a BAH-independent manner
Previously, we have shown that the BAH Domain And Coiled-Coil Containing 1 (BAHCC1) protein harbors a C-terminal BAH module (BAHCC1BAH, Fig. 1a) to read H3K27me3 for mediating gene repression32. Knockdown (KD) of BAHCC1 not only altered target gene expression32 but also significantly delayed the cell cycle progression, as assessed by S phase-associated incorporation of 5-ethynyl-2′-deoxyuridine (EdU) into the newly synthesized DNA (Supplementary Fig. 1a-c, KD versus WT). Notably, such a BAHCC1 loss-associated cell cycle phenotype cannot be recapitulated by an engineered H3K27me3-binding-defective mutation of BAHCC1BAH, Y2533A32 (hereinafter referred to as a BAH-dead mutant; Supplementary Fig. 1a-c, Y2533A versus WT), suggesting a BAH-independent role of BAHCC1 in replication regulation. Next, we assessed the BAHCC1 level across different cell cycle stages using the synchronized HeLa and U2OS cells. Following a time course post-release of synchronized cells from the thymidine block-mediated arrest in early S phase, we used both flow cytometry-based DNA content analysis (Supplementary Fig. 1d) and the expression level assessment of mitotic cyclins, namely, cyclin E1 (CCNE1) and cyclin B1 (CCNB1) that mediate the G1/S and G2/M transition respectively (Supplementary Fig. 1e-f), to define the cell phases. We observed BAHCC1 to be relatively higher in G1-S phase, albert a notable expression across almost all cell stages (Supplementary Fig. 1e-f). In consistence, more chromatin binding of BAHCC1 was detected in S phase in both HeLa (Fig. 1b) and U2OS cells (Supplementary Fig. 1g). Together, we showed that BAHCC1, a chromatin-bound factor enriched in G1/S phase cells, significantly contributes to DNA replication in a BAH-independent fashion.
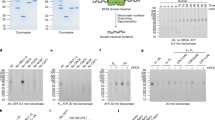
a Domain architecture of human BAHCC1 and mouse Bahcc1. TTD, tandem-Tudor domain; BAH, Bromo adjacent homology. b Time-course measurement for chromatin association of the indicated protein or histone modification in HeLa cells, released from a double thymidine block-induced early S arrest. Cell cycle phase and time post-release are indicated on the top. Experiment was repeated twice with similar results. c Alignment of BAHCC1TTD amino acid sequences from different species. Homo, Homo sapiens; Mus, Mus musculus; Bov, Bovine; Zeb, Zebrafish; Dme, Drosophila melanogaster. d Histone peptide arrays probed with the purified recombinant protein of GST-Bahcc1TTD, followed by α-GST immunoblot. Red boxes show the position of peptides with H4K20 methylation, with red color-highlighted peptides corresponding the strongest binding in each box. ac, acetylated; me1, mono-methylated; me2a, asymmetrically di-methylated; me2s, symmetrically di-methylated; me3, tri-methylated. e Pull-down for GST-53BP1TTD (top) or GST-Bahcc1TTD (middle) using biotinylated peptide with the indicated H4K20 methylation (lanes 1–4), followed by α-GST immunoblot. Bottom panel shows the peptide staining. f Isothermal Titration Calorimetry (ITC) assay measuring the affinity of Bahcc1TTD binding to histone peptide with the indicated H4K20 methylation. Kd, dissociation constant; n.m., not measurable. Source data are provided as a Source Data file.
A cryptic tandem Tudor domain (TTD) in BAHCC1 (BAHCC1TTD) is a highly selective reader of H4K20me1
While the molecular basis underlying the BAHCC1-mediated promotion of replication remains unclear, we noted that BAHCC1 harbors an evolutionarily conserved TTD module (BAHCC1TTD; Fig. 1a,c). Tudor belongs to the so-called Royal superfamily domains, some of which were reported to bind specific histone modifications, such as the TTD of P53-binding protein 1 (53BP1TTD), a reader of H4K20me233,34. To assess potential histone-binding activity of BAHCC1TTD, we probed a histone peptide array with the recombinant protein of murine Bahcc1TTD, and strikingly, we found that it specifically bound the H4K20me1-containing peptides (Fig. 1d and Supplementary Fig. 2a). We verified the Bahcc1TTD:H4K20me1 interaction by individual peptide pull-down and used 53BP1TTD as a control, which revealed their respective preferential binding to H4K20me1 and H4K20me2 (Fig. 1e). The H4K20me1-specific binding by Bahcc1TTD was further evaluated by Isothermal Titration Calorimetry (ITC)— Bahcc1TTD binding to the H4K20me1-containing peptide exhibits a dissociation constant (Kd) of 40.4 µM while it displayed much weaker binding to H4K20me2 (with a Kd of >800 µM) and showed no appreciable binding to H4K20me0 or H4K20me3 (Fig. 1f; Supplementary Fig. 2b; Table 1). Meanwhile, the binding between Bahcc1TTD and other histone lysine mono-methylation, including H3K4me1, H3K9me1, H3K27me1, H3K36me1 and H3K79me1, was all much weaker (Supplementary Fig. 2c; Table 1). It is worth noting that the TTD domain of winged eye35 (WgeTTD), the Drosophila orthologue of BAHCC1, also exhibits a preferential binding of H4K20me1 (Supplementary Fig. 2a, d, and e; Table 1). Altogether, an evolutionarily conserved TTD module of BAHCC1 demonstrates a direct and highly selective interaction with H4K20me1 over the rest H4K20 methylation states.
Structural basis of the Bahcc1TTD:H4K20me1 interaction
To understand the molecular underpinning of Bahcc1TTD-mediated H4K20me1 recognition, we solved the co-crystal structure of murine Bahcc1TTD in complex with the H4(14-25)K20me1 peptide at a resolution of 1.90 Å (Fig. 2a, b; Supplementary Table 1). Each asymmetric unit contains two Bahcc1TTD molecules, with the H4(14-25)K20me1 peptide bound to one but not the other due to crystal packing. At each Bahcc1TTD domain, the two Tudor subdomains (designated as Tudor-N and Tudor-C, respectively) pack against each other through extensive van der Waals contacts (Fig. 2a, b). Tudor-N is composed of four twisted anti-parallel β-strands flanked by a short α-helix on one side, while Tudor-C is formed by a five-stranded β-sheet barrel (Fig. 2a). The side chain of H4K20me1 inserts into a hydrophobic ‘pocket’ lined by the side chains of three aromatic residues (W1990, Y1998 and F2020) and two aspartates (D2022 and D2024) of Tudor-C, with its two mono-methylammonium protons hydrogen bonded to the side chain carboxylates of Bahcc1 D2022 and D2024 (Fig. 2c). Hydrogen bonding of the latter two acidic residues in the ‘cage’ (D2022 and D2024) with H4K20me1 provides a mechanism for the H4K20me1 selectivity, a binding principle as illustrated previously2,36,37. Meanwhile, the side chain of H4 R23, positioned in parallel to that of H4K20me1, forms hydrogen bonds with the backbone of Bahcc1 Y1198 and H2027 as well as a cation-π interaction with the indole ring of Bahcc1 W1990 (Fig. 2c, d). Furthermore, the backbone carbonyl oxygen of H4 V21 forms a hydrogen bond with the side chain Nε1 atom of Bahcc1 W1990, while its side chain engaging in van der Waals contacts with the side chain of Bahcc1TTD S1991 (Fig. 2c). Together, a series of interactions underpin the sequence-specific recognition of H4K20me1. All the Bahcc1TTD residues forming the H4K20me1-binding ‘cage’ are highly conserved through evolution (Figs. 1c, 2d). To assess the significance of these ‘cage’ residues, we performed ITC to measure the H4K20me1 binding by the ‘cage’ mutants of Bahcc1TTD, in which W1990, Y1998, D2022 and D2024 were each replaced by alanine. We found that the Bahcc1TTD:H4(14-25)K20me1 binding was severely reduced or largely abolished by these ‘cage’ residue mutations (Fig. 2e; Supplementary Fig. 3a; Table 1). These observations established the molecular basis underlying specific recognition of H4K20me1 by Bahcc1TTD.
a Ribbon diagram of murine Bahcc1TTD bound to the H4(14-25)K20me1 peptide (yellow stick). The two subdomains of TTD, designated as Turdo-N and Tudor-C, are colored in green and blue, respectively. b–d Electrostatic surface of murine Bahcc1TTD (b) and close-up view (c) of Bahcc1TTD bound to the H4(14-25)K20me1 peptide, with the H4K20me1-binding pocket highlighted in d (with the corresponding ‘cage’ residue of human BAHCC1TTD shown in parentheses). The hydrogen bonds in (c) and (d) are depicted in dashed lines. e Summary of ITC measurements for the binding between the indicated Bahcc1TTD mutant and H4K20me1 peptide. f Co-immunoprecipitation (Co-IP) for binding of endogenous BAHCC1 with the indicated histone species in HeLa cells. Experiment was repeated twice with similar results. g Correlation analysis of BAHCC1 CUT&RUN dataset with histone modification, including the indicated ChIP-seq data of H4K20 methylation28 or ENCODE histone modification profiles. h Heat-maps showing CUT&Tag signals of BAHCC1, H4K20me1, H4K20me2 and H3K4me1 (after normalization to spike-in control signals), ± 5 kb from the centers of BAHCC1 peaks in HeLa cells. Color scale was quantified using the median of Fragments Per Kilobase per Million (FPKM) at the called peaks, with blue and yellow indicating high and low signals, respectively. i Boxplot comparing the BAHCC1 CUT&Tag signals at chromatin regions with H4K20me1 relative to H4K20me2 or H3K4me1. RPKM, reads per kilobase per million mapped reads. P-value is generated by unpaired two-tailed Mann-Whitney U test. Center lines represent the medians, edges of the box indicate the 25th and 75th percentiles, and whiskers indicate the 5% minima and 95% maxima.
To verify the BAHCC1:H4K20me1 interaction in cells, we first performed co-immunoprecipitation (coIP) in Hela cells and detected strong association of BAHCC1 with histones harboring H4K20me1, less so for H4K20me2, and no detectable interaction with other tested histone lysine mono-methylation such as H3K4me1, H3K36me1 and H3K27me1 (Fig. 2f). As previously reported32, BAHCC1BAH binds to the histones carrying H3K27me3 but not H4K20me3 (Fig. 2f). Although the BAHCC1TTD:H4K20me2 interaction (with a measured Kd of >800 uM; Table 1) was much weaker than the BAHCC1TTD:H4K20me1 interaction, H4K20me2 is significantly more abundant in cells than H4K20me1 and detected in ~80% of histone H4 at any given cell cycle stage. Additionally, a recent profiling of H4K20 methylation showed that about 54% of genomic sites with H4K20me1 overlap those with H4K20me228, while another mass spectrometry-based study of immunoaffinity-purified mono-nucleosomes reported the pairing of H4K20me1 with H4K20me2 in asymmetrically modified mono-nucleosomes in different cell models38. High H4K20me2 abundance in cells, as well as its coexistence with H4K20me1 genome-wide, may explain the observed H4K20me2 association in the BAHCC1 coIP experiments. In agreement with this notion, correlational studies using histone modification profiles showed high colocalization of BAHCC1 with H4K20me1, and relatively less so with H4K20me2, using the publicly available chromatin immunoprecipitation sequencing (ChIP-seq) data28 (Fig. 2g) or our own Cleavage Under Targets and Tagmentation (CUT&Tag) data (Fig. 2h-i). Conceivably, H4K20me1 can be generated from H4K20me2 due to the action of demethylases39 and conversely, H4K20me1 can be converted to H4K20me2 due to the action of SUV4-20H methylases4,17. Towards this end, we employed A-196, a potent and selective inhibitor of SUV420H1 and SUV420H240, to remove the global H4K20me2 and H4K20me3, leading to a concomitant increase of H4K20me1 in HeLa cells (Supplementary Fig. 3b, left). Following A-196 treatment, we detected a concurrent increase of BAHCC1 in the chromatin-bound cell fraction (Supplementary Fig. 3b, right); furthermore, CUT&Tag of H4K20me1 and BAHCC1 in HeLa cells after the A196 treatment detected global increase of H4K20me1 and concurrently elevated BAHCC1 binding at same sites (Supplementary Fig. 3c, d), which again points to the BAHCC1:H4K20me1 interaction in cells.
Altogether, BAHCC1 preferentially binds H4K20me1 in vitro and in cells, in agreement with the molecular basis revealed by structural studies of the Bahcc1:H4K20me1 co-crystal.
BAHCC1TTD maintains the cellular level of lowly methylated H4K20
To further dissect the biological function of the H4K20me1-binding BAHCC1TTD, we utilized CRISPR-cas9-based editing to generate HeLa cells with homozygous H4K20me1-interaction-defective mutation at BAHCC1TTD (BAHCC1Y1993A, equivalent to murine Bahcc1Y1998A in Fig. 2d; hereinafter referred to as a TTD-dead mutant) (Supplementary Fig. 4a, b, TTD mutant versus WT). Out of the BAHCC1TTD-edited cells, we additionally identified lines with the small deletion-caused BAHCC1 frameshift, leading to BAHCC1 truncation, which we designated as BAHCC1 knockout (KO; Supplementary Fig. 4a, b, KO versus WT). Co-IP in HeLa cells showed that the association between the TTD-dead mutant (Y1993A) of BAHCC1 and H4K20me1 was disrupted, compared to WT control, whereas association between such a TTD-dead mutant and H3K27me3 remained intact (Fig. 3a), indicating a division of labor in reading distinct histone modifications by different BAHCC1 modules. Interestingly, we consistently observed global levels of H4K20me1 to be decreased in cells bearing BAHCC1 KO or BAHCC1TTD-dead mutation, relative to WT, while the global levels of other histone methylations such as H4K20me3 and H3K27me3 were unaffected (Fig. 3b). Previously, we generated mouse embryonic fibroblast cells (MEFs) carrying homozygous H3K27me3-binding-defective mutation of Bahcc1BAH (Y2537A as the BAH-dead mutant32; Supplementary Fig. 4c), in which the H4K20me1 level was unaffected compared to WT (Fig. 3c, right). Here, we also generated MEFs with homozygous H4K20me1-binding-defective mutation at Bahcc1TTD (Y1998A as the TTD-dead mutant; Supplementary Fig. 4d) and found that, relative to WT, these Bahcc1-TTDY1998A-bearing MEFs displayed the decreased H4K20me1 and, to a lesser extent, decreased H4K20me2, without affecting global levels of H4K20me3 or H3K27me3 (Fig. 3c, left). Independent results of HeLa and MEF cells thus consistently pointed to specific interplay between BAHCC1TTD and the lowly methylated H4K20, notably H4K20me1.
a Co-IP for the interaction between the indicated histone species and endogenous BAHCC1, either wild-type (WT; left) or with an engineered H4K20me1-binding-defective mutation (Y1993A or TTD-dead; right), in HeLa cells. Experiment was repeated independently twice with similar results. b, c Immunoblotting for the total level of the indicated histone methylation in HeLa (b) and MEF (c) cells bearing the BAHCC1 KO or mutation (TTD-dead: Y1993A of human BAHCC1TTD, equivalent to Y1998A of murine Bahcc1TTD; or BAH-dead: Y2537A of murine Bahcc1BAH), compared to respective WT cells. Experiment was repeated twice with similar results. d, e Analytical flow cytometry-based assessment of H4K20me1 intensity (y-axis in d) and its quantification (e) across different cell cycle phases, determined by EdU labeling and DAPI staining of HeLa cells with BAHCC1 KO or the BAHCC1TTD-dead mutation (Y1993A), compared to WT. Error bars in (e) indicate mean ± s.d.; Two-tailed unpaired t-test: ****, P < 0.0001; in the panels from left to right: n = 6,185 (G1), 6,908 (S), 1,518 (G2/M), 6,253 (G1), 6,191 (S), 1,387 (G2/M), 5,580 (G1), 6,865 (S), and 1,778 (G2/M). f, g Heatmaps (top panels) and box plots (bottom panels) showing the H4K20me1 CUT&RUN (f) or CUT&Tag (g) signals (after normalization to spike-in control signals) at the genomic regions ± 5 kb from the centers of H4K20me1 peaks, in HeLa cells bearing BAHCC1 KO or the BAHCC1TTD-dead mutation (Y1993A), compared to WT. Color scale was quantified using the median of FPKM at the called peaks, with blue and yellow indicating high and low signals, respectively. P-values are analyzed by two-tailed Wilcoxon test. Center lines represent the medians, edges of the box indicate the 25th and 75th percentiles, and whiskers indicate the 5% minima and 95% maxima. h Flow cytometry analysis for cell cycle progression of HeLa cells with BAHCC1 KO or the TTD-dead (Y1993A) mutation, compared to WT, post-release from the early S arrest. Source data are provided as a Source Data file.
Next, we further assessed the H4K20 methylation levels at single-cell resolution by using flow cytometry. Compared to WT HeLa cells, those bearing BAHCC1 KO or BAHCC1-TTDY1993A mutant showed the significantly decreased H4K20me1 (Fig. 3d, e) and H4K20me2 (Supplementary Fig. 5a, b) across all cell cycle phases including G1, S and G2/M. Consistently, relative to WT controls, MEFs bearing the TTD-dead mutant of Bahcc1 also displayed the exactly same decreased patterns of H4K20me1 (Supplementary Fig. 5c, d) and H4K20me2 (Supplementary Fig. 5e, f). In contrast, no decreases (or even a very mild increase) of H4K20me1 (Supplementary Fig. 5g, h) and H4K20me2 (Supplementary Fig. 5i, j) were observed in MEFs with the Bahcc1 BAH-dead mutation (Y2537A) compared to WT.
Lastly, we conducted genome-wide mapping of H4K20me1 and corroborated its significant decrease in HeLa cells bearing BAHCC1 KO or the TTDY1993A mutation, compared to WT, with the more dramatic decrease seen in BAHCC1-KO cells, as assessed by independent approaches, either Cleavage Under Targets and Release Using Nuclease (CUT&RUN, Fig. 3f) or CUT&Tag (Fig. 3g). Notably, genomic mapping using HeLa cells with the BAHCC1-TTDY1993A mutation versus WT also detected a decrease of H4K20me2 (Supplementary Fig. 6a), presumably due to a decrease of H4K20me1, the precursor of H4K20me2. In agreement with the BAHCC1TTD:H4K20me1 engagement, the chromatin association of BAHCC1TTD-dead mutant was significantly decreased, compared to WT, as assessed by CUT&RUN and CUT&Tag (Supplementary Fig. 6b, c).
Collectively, these results unveiled a role for BAHCC1TTD in regulating the homeostasis of lowly methylated H4K20.
The BAHCC1TTD:H4K20me1 interaction facilitates the timely cell cycle progression
H4K20 methylation regulates cell cycle progression and DNA replication3,13. The H4K20 methylation decrease caused by the BAHCC1TTD-dead mutation prompted us to analyze the cell cycle progression in mutant cells. Relative to WT HeLa cells, those bearing the BAHCC1 KO or TTDY1993A homozygous mutation both showed the significantly delayed cell cycle progression following the release from early S arrest (Fig. 3h; Supplementary Table 2). Likewise, MEFs bearing the Bahcc1 TTDY1998A mutation versus WT controls also displayed the delayed cell cycle progression following release from serum starvation (Supplementary Fig. 6d). These observations support that the BAHCC1-mediated H4K20me1 readout is important for proper cell cycle progression.
BAHCC1 interacts with the MCM complex
To further dissect the mechanism underlying the BAHCC1-mediated regulation of cell cycle progression, we conducted Biotin Identification (BioID), a proximity-dependent interactome assay41,42, and identified the BAHCC1-associated proteins (Supplementary Fig. 7a; Supplementary Data 1). Interestingly, we identified several MCM complex subunits from the BAHCC1 BioID experiments using two different cell lines, HeLa and U2OS (Fig. 4a), in addition to the HDAC:Sin3 complex (such as Sin3A, Sin3B, HDAC1 and HDAC2) that was previously reported to the BAHCC1-interacting partner32,43,44. The association with MCM complex immediately suggested a mechanism for the cell cycle progression defect observed with the BAHCC1TTD-mutant cells and therefore, we explored it further. Here, we first verified the association between endogenous BAHCC1 and the MCM complex subunits, MCM2 and MCM3, by coIP (Supplementary Fig. 7b). Co-IP using the cell sample treated with a non-specific nuclease, benzonase, also detected association between exogenous BAHCC1 and MCM3 or ORC1, an ORC subunit required for the MCM loading, after their co-expression in HeLa cells, suggesting a DNA-independent interaction (Fig. 4b; Supplementary Fig. 7c). To further assess such an interaction, we reconstituted the system by tethering BAHCC1, which was fused to the DNA-binding domain of GAL4 (GAL4DBD) in a doxycycline-inducible expression vector, onto the genomic location integrated with the 5×GAL4-binding sites and a downstream luciferase reporter in MEFs (Supplementary Fig. 7d). Following doxycycline-induced expression of GAL4DBD-BAHCC1 in cells (Supplementary Fig. 7e, top), we detected the expected GAL4DBD-BAHCC1 recruitment to the 5×GAL4-binding sites, compared to mock without doxycycline treatment (Fig. 4c, FLAG panel: + versus -). Concurrently, we observed the significant enrichment of endogenous MCM2 at same 5×GAL4-binding sites after induction of GAL4DBD-BAHCC1 (Fig. 4c, MCM panel: + versus -). Next, we repeated this assay by co-expressing HA-tagged MCM3 together with either GAL4DBD only or the GAL4DBD-BAHCC1 fusion (Supplementary Fig. 7e). While both GAL4DBD and GAL4DBD-BAHCC1 showed the expected targeting to 5×GAL4-binding sites (Supplementary Fig. 7f, P1-P3 versus a negative control [NC] locus), only in the presence of GAL4DBD-BAHCC1, and not GAL4DBD, did we observe significant enrichment of endogenous MCM2 and exogenously introduced HA-MCM3 at same sites (Supplementary Fig. 7g, h, red versus blue). Furthermore, we detected the decreases in chromatin binding of MCM2 and MCM3 in HeLa cells bearing the BAHCC1TTD-dead mutant (Y1993A) or BAHCC1 KO, compared with WT (Fig. 4d, e; Supplementary Fig. 7i), corresponding to the impaired BAHCC1 genome binding (Supplementary Fig. 6b, c). It appeared that the decrease of MCM2 chromatin occupancy was more severe in cells with the BAHCC1TTD-dead mutation than those with BAHCC1 KO (Fig. 4d), indicating that BAHCC1Y1993A mutant may act in a dominant-negative fashion to sequestrate critical partners such as MCM. Together, these observations support a model that BAHCC1 interacts with MCM complex to facilitate its optimal chromatin recruitment.
a BioID-based identification of BAHCC1-associated proteins in HeLa and U2OS cells. BioID/Control shows ratio of results from biotin-treated versus vehicle-treated cells, with cut-off set to the log2-transformed value of fold change (FC) above 1. MCM and HDAC complex subunits are highlighted in red and blue, respectively. b Co-IP for Flag-BAHCC1 interaction with HA-MCM3 or HA-ORC1 in HeLa cells, without (left) or with benzonase treatment (right). c ChIP-qPCR for GAL4DBD-BAHCC1-Flag (α-Flag) and MCM2 binding at the 5×GAL4-binding sites in MEFs, after the GAL4DBD-BAHCC1-Flag expression induction by doxycycline (dox) treatment (+) versus mock (-). Mock-treated cells and a negative control (NC) locus serve as cell and locus controls, respectively. ChIP signals, presented as means ± SD, were normalized to those of 1% of input (n = 3 replicated experiments). Two-tailed unpaired t-test was conducted. Exact P-values are labeled. d, e Heatmaps (top) and box plots (bottom) showing CUT&Tag signals of MCM2 (d) and CUT&RUN signals of HA-MCM3 (e), ± 5 kb from the peak centers in HeLa cells with BAHCC1 KO or BAHCC1TTD-dead mutation, compared to WT (after normalization to spike-in control signals). Color scale was quantified using the median of FPKM at peaks, with blue and yellow indicating high and low signals, respectively. f Venn diagram showing overlap between the BAHCC1 and HA-tagged ORC1 peaks in HeLa cells. g–j Box plots showing the binding signals of BAHCC1 (g), H4K20me1 (h), HA-MCM3 (i) and MCM2 (j) at genomic regions bound by BAHCC1 only or HA-ORC1 only or co-bound by both in HeLa cells with WT BAHCC1 (left) or the TTD-dead/Y1993A mutation (right). k Integrated Genomics Viewer (IGV) views of BAHCC1, HA-ORC1, MCM2, HA-MCM3 and H4K20me1/2/3 at MYC in HeLa cells with WT BAHCC1 or BAHCC1TTD-dead mutation. For (d, e) and (g−j) P-values are analyzed by two-tailed Wilcoxon test for paired comparison or two-tailed Mann-Whitney U test for unpaired comparison. Center lines represent the medians, edges of the box indicate the 25th and 75th percentiles, and whiskers indicate the 5% minima and 95% maxima. Source data are provided as a Source Data file.
The BAHCC1TTD:H4K20me1 interaction facilitates MCM loading
To further analyze the regulatory role of BAHCC1 during DNA replication, we defined genome-wide binding sites of BAHCC1, as well as the ORC1 subunit of ORC as a marker of potential sites of MCM recruitment and loading45,46,47, in HeLa cells (Fig. 4f). First, a set of genomic sites showed overlapping binding of BAHCC1 and ORC1, which we termed BAHCC1:ORC1-cobound regions (Fig. 4f). In agreement with a role of the BAHCC1TTD:H4K20me1 interaction, both BAHCC1 and H4K20me1 levels were significantly higher at regions bound by BAHCC1 only or cobound by both BAHCC1 and ORC1, when compared with those bound by ORC1 alone and not BAHCC1 (Fig. 4g, h, left panels of WT). Meanwhile, the ORC1 binding level was higher at regions bound by ORC1 alone, compared to the other two above sets of genomic sites (Supplementary Fig 7j). Importantly, we observed the MCM2 and MCM3 binding at those BAHCC1:ORC1-cobound regions to be significantly more enriched, than those seen at sites with BAHCC1 or ORC1 alone (Fig. 4i, j, red in left panels of WT). In the cells carrying the BAHCC1TTD-dead mutation, genome-wide binding of BAHCC1, H4K20me1, MCM2 and MCM3 were all significantly decreased, including significant loss of MCM2 and MCM3 from the BAHCC1:ORC1-cobound regions (Fig. 4g–j, see right panels of Y1993A). We made similar observations at individual replication origins previously mapped nearby or within the genes of MYC, TOP1 and MCM413,48,49 (Fig. 4k; Supplementary Fig. 8). These results collectively support a crucial role of the BAHCC1TTD:H4K20me1 interaction for optimal MCM loading at BAHCC1/ORC1-cobound chromatin regions, indicating cooperation between BAHCC1 and ORC1 for facilitating replication origin licensing and activation.
We next examined MCM’s chromatin association in cells subject to detergent and salt extraction. Compared to WT, HeLa cells bearing either the BAHCC1 KO or BAHCC1TTD-dead mutation (Y1993A) showed defects in MCM chromatin association and loading, as assessed by immunoblotting after cell fractionation (Fig. 5a). Here, MCM2 and MCM3 are markers of the full MCM2-7 complex and we infer MCM loading as resistance to both detergent and high salt extraction. Similarly, we observed the impaired MCM chromatin association and loading in MEFs bearing the Bahcc1TTD-dead mutation (Fig. 5b). In contrast, no changes in MCM chromatin association were observed in MEFs with the Bahcc1BAH-dead mutation (Y2735A), relative to WT (Supplementary Fig. 9a). These results indicated a requirement of BAHCC1TTD, and not the C-terminal BAHCC1BAH, for efficient MCM loading, in line with the preferential H4K20me1 readout by BAHCC1TTD. The significantly decreased MCM loading was further quantified at single-cell levels and confirmed by flow cytometry-based assessment of MCM2 in both HeLa (Fig. 5c, d) and MEF cells (Fig. 5e; Supplementary Fig. 9b). Furthermore, we examined the MCM loading factor and observed the impaired chromatin association of ORC4, a marker of ORC, in HeLa cells bearing BAHCC1TTD-dead mutation or BAHCC1 KO relative to WT, as assessed by flow cytometry (Fig. 5f, g). By immunoblotting, we found the chromatin association of CDC6 or CDT1 unaltered (Supplementary Fig. 9c). Collectively, we conclude that the H4K20me1:BAHCC1TTD interaction functions to promote DNA replication, at least partly, through chromatin association of MCM.
a, b Immunoblotting to assess the chromatin association and loading of MCM2 and MCM3 in HeLa (a) or MEF cells (b) bearing the BAHCC1TTD-dead mutation (Y1993A for HeLa or Y1998A for MEF) or BAHCC1 KO, compared to WT. MCM loading is inferred from resistance to high salt extraction. GAPDH serves as a chromatin fractionation control and histone H3 as a loading control. c Flow cytometry-based assessment of MCM2 chromatin binding (y-axis) across different cell cycle stages, as determined by EdU incorporation and DNA content (DAPI intensity, x-axis) in HeLa cells with BAHCC1 KO (right) or its TTD-dead mutation (Y1993A; middle), relative to WT (left). Blue, MCM2 positive/EdU-negative (G1 phase); Orange, MCM2 positive/EdU positive (S phase); Gray, MCM2 negative nuclei (mainly G2/M). d Quantification of the intensities of MCM2 chromatin binding in the origin licensing phase (G1) of HeLa cells with BAHCC1 KO (right) or its TTD-dead mutation (Y1993A; middle), relative to WT (left). Error bars indicate mean ± SD; Two-tailed unpaired t-test: ****P < 0.0001; from left to right: rep1, n = 2,936 (WT), 2,766 (Y1993A) and 2,737 (KO); rep2, n = 2,974 (WT), 2,955 (Y1993A) and 2,750 (KO); rep3: n = 4,261 (WT), 2,495 (Y1993A) and 3,235 (KO). e Chromatin binding of MCM2 (y-axis) shown as a function of DAPI intensity (x-axis) and cell cycle stages, discriminated by EdU labeling in MEFs bearing either WT or TTD-dead (Y1998A) Bahcc1. f Chromatin-associated ORC4 levels (y-axis) shown as a function of DAPI intensity (x-axis) and cell cycle stages, discriminated by EdU labeling in HeLa cells bearing either WT (left) or TTD-dead (Y1993A; middle) BAHCC1 or its KO (right). Green and purple colors refer to cells with loaded ORC4 in G1 and S phases, respectively, while gray color designates cells without loaded ORC4. g Quantification of the intensities of chromatin-bound ORC4 in G1 phase cells shown in (f). Error bars indicate mean ± s.d.; Two-tailed unpaired t-test: ****P < 0.0001; from left to right: rep1, n = 3,920 (WT), 3,382 (Y1993A) and 4,096 (KO); rep2, n = 3,617 (WT), 2,803 (Y1993A) and 2,590 (KO). Source data are provided as a Source Data file.
The BAHCC1TTD:H4K20me1 interaction is required for proper S-phase progression
Given the observed defects in MCM complex loading, we further analyzed the rate of DNA synthesis by EdU pulse-labeling and found it to be impaired in HeLa cells bearing the BAHCC1TTD-dead mutation (Y1993A) or BAHCC1 KO, relative to WT (Fig. 6a, b). Likewise, we quantified EdU incorporation defects in MEFs bearing BAHCC1TTD-dead mutation (Y1998A) compared to WT cells (Fig. 6c, d). The BAHCC1TTD-dead mutant, but not BAHCC1BAH-dead mutant, recapitulated the EdU incorporation defects seen with BAHCC1 KO, in both Jurkat and MEFs (Supplementary Fig. 1b, c and Supplementary Fig. 9d, e versus Fig. 6a–d), highlighting a specific role for BAHCC1TTD in facilitating DNA replication. Previously, it has been demonstrated that the appearance of PCNA foci marks the onset of the S phase50 and once the PCNA foci disappear, the cell progresses into G2 until nuclear envelope breakdown in M phases50. We next took this strategy to conduct the single cell-based live imaging assays by introducing a mTurquoise2-tagged PCNA51 into MEFs, either WT or with the Bahcc1TTD-defective mutation. Here, we observed that the Bahcc1TTD-defective MEFs displayed the significantly prolonged S phase, but not G1 or G2/M phase, and a corresponding lengthening of the entire cell cycle in these mutant cells (Fig. 6e–g). Additionally, it has been reported that PCNA is loaded as an integral component of active replication forks52, and the DNA-bound PCNA is therefore a quite reliable marker of the relative number of active replication forks, irrespective of fork speed. Next, we used flow cytometry to quantify the chromatin-bound PCNA level in the S phase. Here, our results revealed the globally decreased PCNA loading in HeLa cells with BAHCC1 KO or BAHCC1TTD-defective mutation relative to WT controls (Fig. 6h, i), indicating the broadly impaired replication origin activation in S phase cells with the BAHCC1 loss-of-function. This result is consistent with the MCM loading defect in these cells (Fig. 5a) and is a logical consequence of fewer licensed origins. Together, optimal replication origin activation and S-phase progression require the BAHCC1TTD-mediated preferential reading of lowly methylated H4K20, notably, H4K20me1.
a, b Flow cytometric analysis of DNA synthesis by EdU pulse labeling and DNA content (a) and quantification of EdU incorporation in S phase (b) using HeLa cells with BAHCC1 KO (right) or its TTD-dead/Y1993A mutation (middle), relative to WT (left). Two-tailed unpaired t-test in (b): ****P < 0.0001; n = 4,800 (WT), 4,053 (Y1993A) and 3,805 (KO) S phase cells determined by flow cytometry. Values represent means ± SD. c, d Flow cytometric analysis of EdU labeling (c) and quantification of EdU labeling in S phase (d) using MEFs bearing WT Bahcc1 (left) or its TTD-dead mutant (right; Y1998A). Two-tailed unpaired t-test in (d): ****P < 0.0001; n = 2,549 (WT) and 3,166 (Y1998A) S phase cells determined by flow cytometry. Values represent means ± SD. e Representative live-cell imaging views of MEFs expressing mTurquoise-tagged PCNA to mark the boundaries of S phase by PCNA localization. One representative cell from each population of MEFs bearing either WT (top) or TTD-dead/Y1998A-mutated Bahcc1 (bottom) is shown. Bar, 10 μm. f, g Quantification of the total cell cycle length (f) and the length of the indicated cell cycle phases (g) based on live cell imaging. WT (blue), n = 40; TTD-dead/Y1998A-mutated (red), n = 35. Values represent means ± SD. Two-tailed unpaired t-test was conducted (P = 0.008). h, i Representative flow cytometry analysis (h) and quantification (i) of the chromatin-loaded PCNA levels in the S phase of HeLa cells with WT BAHCC1, BAHCC1 KO or the BAHCC1TTD-defective mutation (Y1993A). The x-axis and y-axis in (h) measured the DNA content and chromatin-bound PCNA levels, respectively, with S phase cells showing in purple. S phase cells from three independent experiments were quantified in (i). One-tailed paired t-test was conducted using WT as controls (n = 3 biological replicates per group). *P ≤ 0.05; ***P ≤ 0.001. Values represent means ± SD. j A working model showing that H4K20me1:BAHCC1TTD-mediated and H4K20me2:ORC1BAH-mediated signaling axes promote the optimal MCM loading for efficient replication origin activation. Source data are provided as a Source Data file.
Discussion
Different H4K20 methylation states and their specific readers play crucial roles in a variety of biological processes, including DNA replication and stress response, transcription, DNA damage repair, chromosome segregation and genome integrity. Previously, it has been reported that H4K20me2 and H4K20me3 serve as docking sites, respectively, for the BAH domain of ORC12 and the WD40 domain of ORCA/LRWD125, which recruit and/or stabilize the chromatin binding of replication initiators and loader machineries for ensuring appropriate replication firing and timing. Additionally, Long et al. showed that H2A.Z directly recruits the SUV4-20H enzymes to deposit H4K20me2, thus licensing and activating the early-replication origins28. During genomic DNA replication, the unmodified H4K20me0 of new histones is recognized by its specific readers such as the Ankyrin Repeat domain of TONSL5 and BARD153, which act to recruit DNA damage repair complexes (namely, TONSL–MMS22L and BARD1–BRCA1) for monitoring the replicative stress and ensuring genome stability5,53. In the literature, studies investigating the roles of H4K20me1 versus H4K20me2 during the DNA replication are quite limited, with the former often considered as an intermediate state for generating H4K20me2. Partly, this limitation is due to a lack of knowledge regarding specific readers that discriminate H4K20me1 over the highly methylated H4K20. Consequently, the direct roles for H4K20me1 in regulating DNA replication and cell cycle progression remain elusive to date.
In this study, we fill in the above gap by demonstrating BAHCC1 to be a high-affinity, high-selectivity reader for H4K20me1 and a crucial regulator of cell replication. BAHCC1, a chromatin-associated factor expressed across almost all cell cycle stages, is particularly enriched in G1/S phase cells and required for efficient execution of DNA replication. Such a regulation of the replication process relies on BAHCC1TTD, which preferentially reads H4K20me1, and not BAHCC1BAH, another BAHCC1 module that functions as a H3K27me3-specific reader32. Our structural and biophysical analyses also revealed the molecular underpinnings for the BAHCC1TTD-directed sensing of H4K20me1. The sequence specificity for BAHCC1TTD:H4K20me1 interaction is underpinned by the surface concaves flanking the H4K20me1 pocket, which anchor the side chains of the H18, V21 and R23 residues in H4 (Fig. 2b, c). Additionally, a set of residues form the binding ‘cage’ to directly engage H4K20me1 (Fig. 2d); here, hydrogen bonding between the H4K20me1 sidechain and the two conserved acidic ‘cage’ residues (i.e., D2022 and D2024 in murine Bahcc1TTD) can sterically exclude the highly methylated H4K20, resembling what was previously reported for other monomethyl-lysine readers36,37, which provides an explanation for the observed H4K20me1 preference. Strikingly, BAHCC1 KO and the BAHCC1TTD-defective point mutation resulted in a global decrease of H4K20me1, a phenomenon not observed with the H3K27me3-binding-defective mutation of BAHCC1BAH. Furthermore, the unbiased interactome study of BAHCC1 revealed the interaction with MCM complex and consistently, our genomic profiling showed that the level of MCM’s chromatin recruitment is positively correlated with the presence of BAHCC1 (a preferential reader of H4K20me1) and ORC1 (a preferential reader of H4K20me2), thereby connecting both H4K20me1 and H4K20me2 directly with the origin licensing and replication initiation. Importantly, cells harboring the BAHCC1TTD-defective mutation exhibit defects in the chromatin recruitment and loading of BAHCC1 and MCM complexes, leading to the broadly impaired replication origin activation in S phase cells, as assessed by genomic loading of PCNA, a quite reliable indicator for the number of active replication forks52. Based on the observations of this study and previously published works2,13,15,25,28,29, we favor a model (Fig. 6j) that (1) during replication, BAHCC1 is essential for maintaining global H4K20me1, which is deposited in the previous M phase by the writer SET8, and a direct BAHCC1TTD:H4K20me1 interaction functions to facilitate the chromatin association and/or loading of MCM complex; (2) acting in parallel with such a BAHCC1TTD-directed signaling is that the maintained H4K20me1 marks are further catalyzed by SUV4-20H enzymes (activity of which is facilitated by the presence of the histone variant H2A.Z in the nucleosomal context28,54) to generate H4K20me2 and H4K20me3, which directly recruit ORC12 and ORCA/LRWD125, respectively, as well as their associated ORC and MCM complexes; (3) due to both signaling axes of H4K20me1:BAHCC1TTD:MCM and H4K20me2-H2A.Z:ORC1BAH/ORC:MCM, the optimal density of origin licensing complexes is achieved at those BAHCC1/ORC1-cobound chromatin regions, thus permitting the fruitful replication initiation and firing.
The observation that the H3K27me3-binding-defective mutation of BAHCC1BAH does not cause the DNA replication defects points to a division of labor in reading distinct histone modifications by different BAHCC1 modules. To test this notion, we purified a recombinant BAHCC1 protein fragment covering dual TTD and BAH domains (termed BAHCC1BAH-TTD), which exhibits the expected strong binding to H4K20me1 peptide and H3K27me3 peptide (Table 1 and Supplementary Fig. 10a). Here, we further measured binding of BAHCC1BAH-TTD to a set of in vitro reconstituted mono-nucleosomes, either un-modified or modified with either H3Kc27me3 alone (with c indicating methylation mimetic55), H4Kc20me1 alone or both modifications (Supplementary Fig. 10b). Biolayer interferometry (BLI)-based measurement showed that binding of BAHCC1BAH-TTD to the H3Kc27me3/H4Kc20me1 dually modified mono-nucleosomes is stronger than binding to either one of the two singly modified mono-nucleosomes and is ~2.5-folds stronger than binding to unmodified ones (Supplementary Fig. 10c). The binding differences seen in this nucleosomal system are modest when compared to what was detected using histone H4 peptides, which is most likely due to the existence of DNA and the use of mono-nucleosomes: besides BAHCC1TTD- and BAHCC1BAH-mediated histone interactions, BAHCC1 may interact with DNA, which is widely observed for various histone readers (such as Bromo, Tudor and Chromo domains)56,57,58,59. Overall, these results support a notion that BAHCC1TTD and BAHCC1BAH may direct the readout of differential histone modifications, either independently or in an additive fashion. We focus this current study on the BAHCC1TTD:H4K20me1 engagement during replication in G1/S phases, in which PRC2, the H3K27me3 writer, was reported to exhibit the weakest genomic binding and low activity60. It is also worth mentioning that H4K20me1 represents a multifunctional histone modification—in several model systems, H4K20me1 was shown to colocalize with H3K27me361,62 and/or facultative heterochromatin63, leading to chromatin compaction39,64, while H4K20me1 was also reported to mark the actively transcribed regions31, the sites generally lacking H3K27me3. In agreement, our genomic profiling of HeLa cells detected the genomic regions showing H4K20me1 positivity but lacking H3K27me3, as well as those positive for both H4K20me1 and H3K27me3 (Supplementary Fig. 10d, H4K20me1 versus H3K27me3 in top left panel). Thus, we speculate that dual BAHCC1TTD and BAHCC1BAH modules may elicit context-dependent regulatory effects, which await investigation in future.
Furthermore, genomic binding of BAHCC1 exhibits a positive correlation with both H4K20me1 and PHF8, the H4K20me1 demethylase exhibiting high activity in G1/S15, at the genome-wide scale (Supplementary Fig. 10d; with Pearson correlation scores measured to be 0.57-0.61 and 0.62-0.63, respectively). In agreement with such co-occupancy patterns, we detected overall decrease of H4K20me1 at the BAHCC1-bound genomic regions upon KO of BAHCC1 or introduction of the H4K20me1-interaction-defective mutation at BAHCC1TTD, indicating a role for BAHCC1 in sustaining the H4K20me1 homeostasis. Conceivably, the BAHCC1TTD-mediated engagement of H4K20me1 may protect it from demethylation by PHF8 (which is highly active in G1/S15) and/or additional methylation by SUV4-20H. The decrease of H4K20me1, the substrate of SUV4-20H, can additionally lead to decrease of H4K20me2 as the cell cycle progresses. Requirement of the BAHCC1TTD:H4K20me1 binding for maintenance of H4K20me1 is reminiscent of what was reported for Mixed Lineage Leukemia 1 (MLL1, also known as KMT2A), which employs a CXXC domain to protect unmethylated CpG DNA from methylation65. Further study is warranted to examine the involvement of numerous potential demethylases and methylases that could influence H4K20me1/2 homeostasis under the content studied herein. The collective effects of H4K20me1 and H4K20me2 perturbations may better explain the observed defects in MCM and ORC loading upon BAHCC1 loss-of-function. Please note that BAHCC1 is preferentially expressed in G1/S phase cells whereas the activity of SET8 (the H4K20me1 writer) exhibits a quite different dynamics, with the induced H4K20me1 peaking during G2/M1,3,4. SET8 was also reported to regulate chromatin compaction during mitosis30,31,64, and H4K20me1 in mitotic cells was reported to be recognized by the non-structural maintenance of chromosomes (SMC) condensin II subunits15. These observations point to a BAHCC1-independent role for SET8 and H4K20me1 during mitosis. Together, the interplays among different H4K20 methylation states and their readers, erasers and writers are rather complex and context-dependent (e.g. the origin licensing in G1/S phases, as well as the chromatin compaction and chromosome segregation during mitosis).
While our in vitro biochemistry and co-crystal structure data strongly support the high H4K20me1 selectivity, we also found BAHCC1 to be associated with H4K20me2 based on coIP and histone modification profiling results in cells, which can be explained by the below reasons. On one hand, such association may come from a co-occurrence pattern of H4K20me1 and H4K20me2 genome-wide; for instance, the mass spectrometry-based profiling of immunoaffinity-purified mono-nucleosomes and genomic mapping of H4K20 methylation reported the coexistence of H4K20me1 with H4K20me228,38. On the other hand, we detected a low-affinity in vitro binding of BAHCC1TTD to H4K20me2, with a Kd measured to be over 800 μM. Given that H4K20me2 is detected in about 80% of H4 molecules in any given cell stage and more abundant than H4K20me1, it remains a possibility that a weak BAHCC1TTD binding to H4K20me2 can further promote the tethering of BAHCC1 and partners to replication origins (see a dashed line in Fig. 6j), albeit to an arguably lesser extent than contribution of the BAHCC1TTD:H4K20me1-based force (which, in an in vitro system, is about 20-folds stronger than BAHCC1TTD binding to H4K20me2; Table 1).
A key finding of this work is to mechanistically connect BAHCC1TTD, a preferential reader of H4K20me1, with the replication origin activation and cell cycle progression. It unveils a previously unknown layer of replication regulation and adds to the reported modulatory mechanisms directed by ORC1 (a preferential reader of H4K20me2)2, H2A.Z (a facilitator of H4K20me2 deposition)28,54, ORCA/LRWD1 (a binder of H4K20me3-containing nucleosome and ORC complex)25,66, amongst others. However, the limitations of this work exist. As mentioned above, a low-affinity interaction between BAHCC1TTD and H4K20me2 may contribute to the observed defects upon mutation of BAHCC1TTD, which awaits the detailed study. Additionally, this work assessed the steady state levels of H4K20 methylation after BAHCC1 loss-of-function and future effort shall be directed to tease out the regulatory roles for BAHCC1 in distinct chromatin environment, such as euchromatin (early origins) versus heterochromatin regions (late origins), as well as the cell stage-specific modulation of H4K20 methylation dynamics.
Methods
Plasmids
The plasmids for bacterial expression of the BAHCC1TTD protein domain, which includes the amino acids 1904-2044 of murine Bahcc1 and its Drosophila homolog domain, the amino acids 1094-1264 of winged eye (wge)35, were generated by subcloning the corresponding cDNA fragment into a modified pRSFDuet vector containing a His6-SUMO tag or a GST-fusion vector (GE Life Science; GEX-4T1 and GEX-6P2) and used as before32,67,68. A bacterial expression vector containing 53BP1TTD (amino acids 1235–1709) was a gift of S. Frye and used as described before69. To generate the BAHCC1 protein segment harboring both the TTD and BAH domains (BAHCC1TTD-BAH), we constructed a cDNA fragment encoding BAHCC1 residues 1899-2638 but with internal deletion of residues 2070-2493 to minimize proteolytic degradation during protein purification, followed by insertion into an in-house bacterial expression vector containing an N-terminal His6-MBP tag.
A full-length cDNA of human BAHCC1 (NM_001291324.2) was synthesized by BGI Geneland/Qinglan Inc. Synonymous mutations were introduced into this cDNA for creating restriction enzyme sites to facilitate cDNA subcloning and mutagenesis. The synthesized full-length BAHCC1 cDNA, together with an in-frame fused EGFP tag at the N-terminus and a 3 × FLAG tag at the C-terminus, was cloned into a PiggyBac (PB) vector of PB-EF1α-MCS-IRES-Neo (System Biosciences #PB533A-2), creating the PB-EF1α-GFP_BAHCC1_3×FLAG-IRES-Neo plasmid. To generate a doxycycline (dox)-inducible expression construct of BAHCC1, we replaced the EF1α promoter in PB-EF1α-GFP_BAHCC1_3×FLAG-IRES-Neo with a tight TRE promoter from pCW-Cas9 (Addgene #50661) and then, a reverse Tet repressor (rTetR) element from pCW-Cas9 was inserted between the UbC promoter and the IRES2 element, which created the dox-inducible PB-TRE-EGFP_BAHCC1_3×FLAG-rTetR-IRES-Neo plasmid. An in-frame fusion of BAHCC1 with either BirA* (for biotin identification [BioID] experiments) or the GAL4 DNA-binding domain (GAL4_DBD; for targeting BAHCC1 to the genomic sites with upstream activating sequence [UAS]) was generated by replacing the N-terminal EGFP in the above dox-inducible PiggyBac-BAHCC1 plasmid with PCR-amplified BirA* or GAL4_DBD, which created the dox-inducible plasmids of PB-TRE-BirA*_BAHCC1_3 × FLAG-rTetR-IRES-Neo and PB-TRE-GAL4_DBD_BAHCC1_3 × FLAG-rTetR-IRES-Neo. The plasmids of dox-inducible pCW-3 × HA-ORC1-blasticidin and pCW-3 × HA-MCM3-blasticidin were made by replacing Cas9 in pCW-Cas9 with the cDNA fragment of ORC1 (NM_004153.4) and MCM3 (NM_002388.6) respectively, with a 3 × HA tag fused to the N-terminus and with the puromycin cassette replaced by blasticidin. A lentiviral plasmid containing the mTurquoise-tagged PCNA was used as described before51. A fragment covering the 5×UAS sites and a downstream luciferase gene was amplified by PCR from a pGL2-SV40-LUC plasmid (Addgene, cat# 26280) and then inserted into the PB-CMV-MCS-EF1α-Puro plasmid (System Biosciences, cat# PB510B-1) to replace the CMV promoter, which produces a PiggyBac-based delivery system for genomic insertion of the GAL4_DBD/5×UAS-luciferase reporter (termed PB-5×GAL4-LUC-puro).
Cell culture and stable cell line generation
Cells used in this study included HeLa (American Tissue Culture Collection [ATCC], CCL-2), U2OS (ATCC, HTB-96) and 293FT (Thermo Fisher Scientific, R70007), which were cultured in the DMEM base medium supplemented with 10% of fetal bovine serum (FBS) and 1% of penicillin and streptomycin. Mouse embryonic fibroblast cells (MEFs) were cultured in DMEM base medium supplemented with 20% of FBS and 1% of penicillin and streptomycin as recommended70. Lentivirus or retrovirus containing 3×HA-MCM3, 3 × HA-ORC1 or mTurquoise-PCNA was prepared in 293FT cells and used for cell infection to generate the stable cell lines as described before32,71,72. PiggyBac-based genomic insertion was conducted by co-transfection of a transposase plasmid (System Biosciences, cat# PB200PA-1) and a PiggyBac (PB) elements-containing vector (such as the above PB plasmid with tagged BAHCC1, or a GAL4_DBD/5×UAS-luciferase reporter) into cells (such as HeLa or MEFs), followed by drug selection and/or generation of clonal cell lines.
Cell synchronization and release
For synchronization of HeLa cells in early S phase, we employed a double thymidine block protocol15,73. In brief, HeLa cells were first incubated with culture medium containing 2 mM thymidine (Sigma, cat# T9250) for 12 h, then washed with sterile 1×PBS for three times, and incubated with regular medium for 11 h, which was then followed by a second block step with 2 mM thymidine for 12 h. At this point, cells were synchronized and released from the block into S phase upon cultivation in normal medium. The released cells were collected at different timepoints (0–24 h) for downstream analyses, which included propidium iodide (PI) staining and flow cytometry, RT-qPCR of cell cycle markers such as Cyclin E1 (CCNE1) and Cyclin B1 (CCNB1), and western blotting (WB). Synchronization of U2OS cells was conducted using a single thymidine block protocol, while synchronization of MEFs used a serum starvation protocol74. For the latter, MEFs were cultured in serum-free medium for 3 days and at this point, most cells were in G0 phase. Then, these synchronized cells were released by cultivation in normal FBS-containing medium and collected at different timepoints for downstream analyses.
Antibodies
A home-made α-BAHCC1 antibody was previously generated and used as before32. Detailed information for the antibodies used for WB, co-immunoprecipitation (co-IP), chromatin immunoprecipitation (ChIP), flow cytometry, Cleavage Under Targets & Release Using Nuclease (CUT&RUN), and Cleavage Under Targets and Tagmentation (CUT&Tag) is provided in Supplementary Data 2.
Chemical compounds
The SUV420H1 and SUV420H2 selective inhibitor, A-196 (Sigma, SML1565), was dissolved in dimethylsulfoxide (DMSO) as 5 mM stock solution and used as before67. A three-day treatment with 5 µM A-196, relative to DMSO, was applied for HeLa cells in this study.
DNA content analysis
Cells were fixed and stained with propidium iodide (Sigma) to quantify the DNA content for cell cycle analysis. Briefly, cells were trypsinized and counted using a BioRad automated cell counter. 500,000 live cells were transferred to a 4 mL flow cytometry tube and pelleted. Next, they were washed with PBS and fixed in 80% methanol at -20 °C for at least 3 h or overnight. Following fixation, the cells were washed and resuspended in 1 mL 1 × PBS plus 0.5% of BSA. RNase (100 ug/mL; Sigma Aldrich) and propidium iodide (25 ug/mL) was added and the cells were incubated in the dark for 1 h at room temperature. DNA content was detected using a Thermo Fisher Attune NxT flow cytometer (available in the UNC flow cytometry core facility), followed by analysis with Flowjo software.
5-ethynyl-2’-deoxyuridine (EdU) labeling analysis
Cells were incubated with 10 μM EdU (provided in the commercial Click-iT Imaging Kits, Invitrogen, cat# C10337; 30 min for Hela cells and 2 h for MEFs) before collection. Then, cells were washed twice with 1 × PBS and fixed in 4% formaldehyde for 15 min at room temperature, followed by incubation with 0.5% Triton X-100 and 1% BSA in 1 × PBS for 20 min at room temperature. EdU labeling was conducted using the commercial Click-iT Imaging Kit (Invitrogen) according to the manufacturer’s instructions. Finally, EdU-labeled cells were resuspended in 1 × PBS with 1 μg/mL DAPI (Life Technologies) and incubated for 1 h at room temperature. Cells were analyzed with the Thermo Fisher Attune NxT flow cytometer, followed by analysis with Flowjo software.
Preparation of chromatin-associated cell fractions
Preparation of the chromatin-associated fraction of cells was conducted as before71. Briefly, cells were collected and washed with cold 1×PBS and then resuspended in the CSK-100 buffer (10 mM PIPES pH 7.0, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2 and 0.5% Triton X-100; freshly supplemented with the inhibitor cocktail of protease and phosphatase [Roche]), followed by incubation on ice for 20 min and centrifugation at 3,000 rpm for 15 min. After the supernatant was removed, the remaining cell pellet was washed once with the ice-cold CSK-100 buffer and collected as the chromatin-bound cell fraction for immunoblotting. For assaying the chromatin-loaded fraction of MCMs, we followed the same preparation protocol as above, except that the higher salt CSK-300 buffer (10 mM PIPES pH 7.0, 300 mM NaCl, 300 mM sucrose, 3 mM MgCl2 and 0.5% Triton X-100; supplemented with inhibitors of protease and phosphatase) was used instead.
Co-immunoprecipitation (Co-IP)
Collected cells were resuspended in cell lysis buffer (50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 2 mM EDTA, 0.1% SDS and 1% Triton X-100; freshly supplemented with inhibitors of protease and phosphatase [Roche]). The total cell lysate was subjected to brief sonication and high-speed centrifugation to remove cell debris. For benzonase nuclease treatment, 1 μl benzonase ( ≥ 250 units/μL; Sigma-Aldrich, cat# E1014) was added into the cell lysate after brief sonication, followed by incubation on ice for 1 h. The cell supernatant was collected after centrifugation and incubated with primary antibodies on a rotator overnight at 4 °C. Then, 50 μL of Dynabeads (Invitrogen) was added and incubated at 4 °C for additional 2 h. Beads were washed five times with lysis buffer, and resuspended in 2× protein sample loading buffer. Samples were boiled at 90 °C for 10 min before separation by SDS-PAGE gels and WB.
Protein expression and purification
As mentioned above, a gene fragment encoding murine Bahcc1TTD (amino acids 1904-2044) or Drosophila WgeTTD (amino acids 1094-1264) was subcloned into a modified pRSFDuet vector containing a His6-SUMO tag, followed by transformation using the Escherichia coli BL21(DE3) RIL cells. Likewise, a bacterial expression plasmid containing an N-terminal His6-MBP tag and a construct harboring BAHCC1’s dual TTD and BAH domains (His6-MBP-BAHCC1TTD-BAH) was used for bacterial transformation. The transformed BL21(DE3) RIL cells were first grown at 37 °C. After the cell density reached an optical density at 600 nm of 0.8, the temperature was lowered to 16 °C. For protein expression in bacteria, cells were induced with 0.4 mM isopropyl β-d-thiogalactopyranoside (IPTG) at 20 °C for 16 h. The cells were then collected, lysed and the insoluble pellet was removed by centrifugation. Subsequently, the proteins were purified by Ni2+-NTA affinity chromatography. As of His6-SUMO tagged TTD proteins, the affinity-purified sample was cleaved by ULP1 and dialyzed against a buffer containing 25 mM Tris, pH 7.5, 300 mM NaCl; next, the proteins were then subjected to a second Ni2+-NTA column to remove the His6-SUMO tag and ULP1 protease, and then were concentrated and subjected to size-exclusion chromatography (Superdex 200 16/60, GE) equilibrated with 25 mM Tris, pH 7.5, 100 mM NaCl and 2 mM DTT. As of the His6-MBP-tagged BAHCC1TTD-BAH protein, the affinity-purified sample was first subject to ion-exchange chromatography on a Heparin column (Cytiva Inc) and size-exclusion chromatography on a Hiload 16/60 Superdex 200 pg column (Cytiva Inc); then, the purified protein samples were concentrated in 25 mM Tris-HCl (pH 7.5), 100 mM NaCl, 5% glycerol and 5 mM dithiothreitol (DTT), and stored at −80 °C before use. Lastly, the His6-MBP tag was removed from the purified His6MBP-BAHCC1TTD-BAH fusion protein via proteolytic cleavage with Tabacco Etch Virs (TEV) protease and nickel affinity chromatography, followed by size-exclusion chromatography on a Hiload 16/60 Superdex 200 pg column.
All the mutants of Bahcc1TTD were generated by direct mutagenesis and purified as described above. For structural determination, a L1962M/L2018M double mutation was introduced into mBahcc1 to allow selenomethionine-labeling of the protein. Recombinant protein of 53BP1TTD was generated and used as described before69.
Assays for reader protein binding by histone peptide arrays and peptide pull-down
Assaying the binding specificity of the reader proteins was performed using the Modified™ Histone Peptide Array (Active Motif, cat# 13005) according to the vendor’s user manuals. Pull-down using the biotinylated histone peptide was performed as previously described32,75. Two versions of biotinylated H4 peptides, either amino acids 11 to 30 or 11 to 27 of H4, which carry unmodified H4K20, H4K20me1, H4K20me2 or H4K20me3, were used in independent experiments and produced same results. In brief, each one of the individual biotinylated histone peptides was incubated with Pierce™ High Capacity NeutrAvidin™ Agarose (Thermal Fisher, cat# 29204) for 4 h at room temperature, followed by extensive washing to eliminate the unbound peptide. Then, 20 µL of the prepared peptide:Avidin resin complexes were incubated with 20 µg of the purified GST fusion protein in the pull-down buffer (20 mM HEPES [pH 7.9], 200 mM KCl, 0.05% of IGEPAL CA-630 and 5% of glycerol), supplemented with 1 mM DTT, 0.1 mM PMSF and protease inhibitor cocktail [Roche]. After extensive washing, the GST protein bound to resin was released and collected after boiling in protein loading buffer for 5 min and, after brief centrifugation, the supernatant was collected and separated by SDS–PAGE for either western blotting with α-GST antibody or for silver staining.
Isothermal titration calorimetry (ITC) measurements
Both murine Bahcc1TTD (amino acids 1904-2044) and histone peptides were dialyzed at 4 °C against a buffer containing 20 mM Tris-HCl (pH 7.5), 50 mM NaCl and 0.5 mM DTT. For all the peptides, a C-terminal tyrosine was introduced for the purpose of spectrophotometric measurement. Protein and peptides concentrations were determined based on their respective ultraviolet absorption at 280 nm wavelength. ITC measurements were carried out at 7 °C on a MicroCal iTC200 system (GE Healthcare), including a total of 15−17 injections with spacing of 180 s and a reference power of 5 μcal/s. The subsequent ITC data were processed with software ORIGEN (MicroCal) using a one-site fitting model.
CUT&RUN and CUT&Tag
CUT&RUN76,77 was performed as we have previously described42,72 using the commercially available kit (EpiCypher CUTANA™ pAG-MNase for ChIC/CUT&RUN, cat# 15-1116). Briefly, 0.5 million cells were collected and used for CUT&RUN, and 0.3 ng of sonicated chromatin from Drosophila S2 cells was added into each sample for spike-in normalization. CUT&RUN was conducted as before42,72 and DNA was purified by using the Monarch DNA Cleanup Kit (NEB, cat# T1030S) according to the manufacturer’s instructions. Finally, multiplexed CUT&RUN libraries were prepared using the NEB Ultra II DNA Library Prep kit (E7645).
CUT&Tag was performed using the previously published protocol78,79 and the commercially available CUTANA™ pAG-Tn5 kit (EpiCypher, cat# 15-1017). In brief, 0.1 million cells were collected and incubated in NE buffer (20 mM HEPES-KOH pH 7.9, 10 mM KCl, 0.1% Triton X-100, 20% Glycerol, 0.5 mM Spermidine, and 1x Roche cOmplete™ Protease Inhibitor Cocktail) for 10 min on ice to isolate the nuclei. For spike-in control normalization, 2% of MEFs cells (murine) were added to HeLa cells (human) for each sample. ConA beads (Bangs Laboratories, BP531) were activated in bead activation buffer (20 mM HEPES pH 7.9, 10 mM KCl, 1 mM CaCl2, and 1 mM MnCl2) and then incubated with the nuclei for 10 min at room temperature. The ConA beads-nuclei slurry was resuspended in 50 μL cold Antibody150 Buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM Spermidine, 1x Roche cOmplete™ Protease Inhibitor Cocktail, 0.03% Digitonin, and 2 mM EDTA) and incubated overnight at 4 °C with 0.5 μg of the primary antibodies against the protein of interest. The following day, the beads-nuclei slurry was resuspended in cold Digtonin150 buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM Spermidine, 1x Roche cOmplete™ Protease Inhibitor Cocktail, and 0.03% Digitonin) and incubated with 0.5 μg of the secondary antibodies (anti-rabbit secondary antibody, ABIN101961) for 30 min at room temperature. After two washes with cold Digtonin150 buffer, the beads-nuclei slurry was resuspended in 50 μL of the cold Digtonin300 buffer (20 mM HEPES pH 7.5, 300 mM NaCl, 0.5 mM Spermidine, 1x Roche Protease Inhibitor Cocktail, and 0.01% Digitonin) and incubated with 2.5 μL CUTANA™ pAG-Tn5 for 1 h at room temperature. The beads-nuclei complexes were then washed twice with Digtonin300 buffer, followed by incubation with 50 μL Tagmentation buffer (Digitonin300 buffer plus 10 mM MgCl2) for 1 h at 37 °C. 10 μL SDS Release buffer (10 mM TAPS pH 8.5 and 0.1% SDS) was added into the beads after washing with 50 μL TAPS buffer (10 mM TAPS pH 8.5 and 0.2 mM EDTA), followed by incubation for 1 h at 58 °C in a thermocycler. Finally, 30 μL SDS Quench buffer (0.67% Triton-X 100 in molecular-grade water) was added and the resulting CUT&Tag DNA was purified by using NEB Monarch DNA Cleanup Kit followed by multiplexed library preparation.
All the multiplexed libraries of CUT&RUN or CUT&Tag were subjected to pair-end deep sequencing on a NextSeq500 platform (available from shared facilities of the university).
Analysis of CUT&RUN and CUT&Tag datasets
CUT&RUN76,77 and CUT&Tag78,79 data were analyzed as previously described42,72,80. Fastq files were mapped to the reference genome (either hg19, mm10 or dm3) using bowtie2.3.581. Normalization against the spike-in control signals was conducted as we recently described82,83. The non-primary alignments, PCR duplicates, or blacklist regions were removed from the aligned data by using Samtools (v1.9), Picard ‘MarkDuplicates’ function (v2.20.4), and bedtools (v2.28.0), respectively. Peak calling was conducted by MACS2 (macs2 callpeak -f BAMPE -g hs/mm –keep-dup 1)84. Bigwig files, heatmaps and averaged plotting of CUT&RUN and CUT&Tag signals were generated by deeptools (v3.3.0)85. The Integrative Genomics Viewer (IGV) was used for viewing the generated bigwig files.
Reverse transcription followed by quantitative PCR (RT-qPCR) and chromatin immunoprecipitation (ChIP) followed by quantitative PCR (ChIP-qPCR)
RT-qPCR and ChIP-qPCR were conducted as we described before42,86. The RT-qPCR and ChIP-qPCR primer information is provided in Supplementary Data 2.
Crystallization and structure determination
The Bahcc11904-2044:H4(14-25)K20me1 complex was crystallized using the sitting-drop vapor diffusion method at 20 °C by mixing 2 μL of Bahcc11904-2044:H4(14-25)K20me1 (1.3 mM: 3 mM) with 4 μL of reservoir solution containing 100 mM MES (pH 6.5), 10 mM Li2SO4, and 8% PEG20000. Crystals grew into full size in 2 days. The reservoir solution supplemented with 15% glycerol was used as cryo-protectant when the crystals were flash-frozen in liquid nitrogen. The diffraction data were collected on the beamline 5.0.1. at the Advanced Light Source (ALS), Lawrence Berkeley National Laboratory. The data were indexed, integrated and scaled by HKL2000 package87. The structure was solved using the Se-SAD method with the data collected for the Bahcc1TTD L1962M/L2018M mutant. The initial model was built using the Autosol module in the PHENIX software package88. Subsequently, structural refinement was carried out using the data collected for the native crystals of wild-type Bahcc1TTD. Iterative cycles of model rebuilding and refinement were performed using COOT89 and PHENIX, respectively. The same R-free test set was used throughout the refinement. The statistics for data collection and structural refinement is summarized in Supplementary Table 1.
CRISPR–cas9-based genomic editing for site-specific mutation of BAHCC1TTD and for BAHCC1 knockout (KO)
The Alt-R CRISPR-Cas9 System (Integrated DNA technologies [IDT] Inc.) in combination with a single-stranded oligodeoxynucleotide donor (ssODN) was used to introduce a point mutation into the genomic region coding for BAHCC1TTD, by using a previously described protocol32. For the Alt-R CRISPR-Cas9 System, the trans-activating CRISPR RNA (tracrRNA) labeled with ATTO 550, the S.p. HiFi Nuclease V3 and electroporation enhancer were all purchased from IDT, and the crRNA targeting the site to be mutated (Y1993 in BAHCC1TTD for HeLa cells) and the ssODN were both custom-designed and synthesized by IDT. Three phosphorothioate bonds were added at the 5′- and 3′-ends of ssODN to stabilize the donor oligonucleotides and to ensure more efficient homology-directed DNA damage repair. Synonymous mutations were introduced at the same time to prevent recutting the target sequence after the desired point mutation was introduced. Sequence information for the crRNA and ssODN are listed in Supplementary Data 2. Preparation and delivery of the CRISPR-cas9-ribonucleoprotein (RNP) complex and ssODN into cells were conducted according to the manufacturer’s protocol. Briefly, tracrRNA and crRNA were mixed at a 1:1 ratio and heated at 95 °C for 5 min. Then, the heated mixture was cooled in the dark at room temperature to form the crRNA:tracrRNA duplex. Next, the S.p. HiFi Cas9 Nuclease was added to the diluted duplex followed by 10−20 min’ incubation to form the RNP complex (Alt-R CRISPR-Cas9 crRNA:tracrRNA and S.p. HiFi Cas9 Nuclease). The RNP complexes were further mixed with ssODN and electroporation enhancer and delivered into Hela cells with an Amaxa nucleofector machine and the Cell Line Nucleofector Kit R (Lonza, VCA-1001). After 36 h, ATTO 550-positive cells were sorted by FACS (conducted by the UNC flow cytometry core facility) and split into 96-well plates with one cell per well. Finally, clonal lines derived from single-cell clones with homozygous BAHCC1Y1993A mutation (defined as TTD-mutated), or with the damaging mutations such as frameshift caused by deletion (defined as BAHCC1 KO), were identified by PCR-base genotyping and then, by direct Sanger sequencing of PCR products. MEFs carrying murine Bahcc1 TTD mutation (Y1998A) were derived with the below reagents. The guide RNA targeting the murine Bahcc1Y1998 codon in exon 23 was cloned into a T7 promoter vector followed by in vitro transcription (protospacer sequence 5’- gTCCCGGTGTCTATATCC-3’; the lower-case g represents a heterologous guanine added for T7 in vitro transcription) and silica spin column purification. SNP complex used for editing of murine Bahcc1 gene contained Cas9 protein, the above guide RNA and donor oligonucleotide designed to facilitate homologous recombination to introduce the Y1998A mutation (i.e. TAT above to GCC [corresponding to GGC below]; with donor sequence as 5’- GCCTCAGCCCTAGTCCCGGCCTCGAGCATGCTTACCTCGAACCACATTGCCTGGGGCTAGACACCGGGACTTCTGACTCCAGTAGGCACAGACCCGGGTACCAGGTGGAA-3’). MEFs carrying wildtype (WT) or BAH-mutated (Y2537A) Bahcc1 were generated and maintained as described before32. Before use, all the WT and mutant cell lines were verified by genotyping and Sanger sequencing to ensure cell identities.
FACS-based analyses of H4K20 methylation and chromatin-bound PCNA, ORC and MCM complex
Cells were incubated with 10 μM EdU in the culture medium (30 min for Hela cells, and 2 h for MEFs) and then collected. Then, cells were washed twice with 1 × PBS and fixed in 4% formaldehyde for 15 min at room temperature. EdU-labeling was performed as described above. The α-H4K20me1 (Abcam cat# ab9051) or α-H4K20me2 (Abcam cat# ab9052) antibodies, which were diluted 1:200 in 1 × PBS containing 0.1% of IGEPAL CA-630 and 1% of BSA, were then added onto EdU-labeled cells and mixed, followed by incubation in the dark at 37 °C for 1 h (or overnight at 4 °C). Next, cells were washed once with 1×PBS with 0.1% IGEPAL CA-630 and 1% BSA and incubated with Alexa Fluor 594-labeled anti-rabbit secondary antibody (Invitrogen, A-21207; 1:500 dilution) for 1 h at 37 °C in the dark. Finally, cells were resuspended in 400 μL 1 × PBS containing 1 μg/mL DAPI, 1% BSA, 0.1% IGEPAL CA-630 and 100 μg/mL RNase. After incubation in the dark for 1 h at 37 °C, cells were analyzed with the Thermo Fisher Attune NxT flow cytometer.
Analysis of chromatin-bound MCM, ORC and PCNA was performed as previously described90. Briefly, cells were treated with 10 μM EdU (Santa Cruz Biotechnology, cat# 69075-47-4) before collection (30 min for Hela cells, 2 h for MEFs). Cells were then trypsinized and collected, then lysed on ice for 8 min in the CSK buffer (10 mM Pipes pH 7.0, 300 mM sucrose, 100 mM NaCl, and 3 mM MgCl2) with 0.5% triton X-100 with a cocktail of protease/phosphatase inhibitors (Roche). After washing once with 1 × PBS containing 1% BSA, cells were then fixed in 1×PBS containing 4% paraformaldehyde for 15 min at room temperature. For EdU detection, samples were incubated in 1×PBS containing 1 mM CuSO4, 1 μM Alexa 647-azide (Life Technologies) and 100 mM ascorbic acid for 30 min at room temperature in the dark. Then, cells were labeled in primary antibody and incubated in the dark at 37 °C for 1 h. Next, cells were labeled with secondary antibody and incubated in the dark at 37 °C for 1 h Finally, cells were resuspended in 1×PBS containing 1% BSA, 0.1% NP-40, 1 μg/mL DAPI and 100 μg/mL RNAse A and incubated in the dark overnight at 4 °C, followed by analysis using an Attune NxT flow cytometer (Thermo Fisher Scientific).
Collected data were analyzed by using the FCS Express 7 software (De Novo Software). Control samples were prepared by omitting primary antibody or EdU labeling to define the thresholds of detection. Primary antibodies used were the following: α-MCM2 (used at 1:100; BD Biosciences, cat# 610700), α-PNCA (used at 1:100; Santa Cruz Biotechnology, cat# sc-25280) and α-ORC4 (used at 1:50; Santa Cruz Biotechnology, cat# 136331), and secondary antibody was Donkey anti-mouse Alexa 488-labeled secondary antibody (used at 1:1,000, Jackson Immuno Research, cat# 715-545-150). The gating strategy is provided in Supplementary Fig. 11.
Live cell imaging
The pLenti-mTurquoise-PCNA51 plasmid was stably introduced via viral transfection into MEFs containing WT Bahcc1 or TTD-mutated Bahcc1Y1998A. Cells expressing PCNA-mTurquoise were plated on fibronectin (1 ug/cm2; Sigma)-coated F1141 glass-bottom plates (Cellvis) with FluoroBrite™ DMEM (Invitrogen) supplemented with 10% of FBS, 4 mM l-glutamine and antibiotics (penicillin and streptomycin). Fluorescence images were acquired using a Nikon Ti Eclipse inverted microscope with Plan Apochromat dry objective lenses 20x (NA 0.75). Images were captured using an Andor Zyla 4.2 sCMOS detector with 12 bit resolution. Autofocus was provided by the Nikon Perfect Focus System (PFS), and a custom enclosure (Okolabs) was used to maintain constant temperature (37 °C) and atmosphere (5% CO2) in a humidified chamber. The CFP filter set was from Chroma, 436/ 20 nm; 455 nm; 480/40 nm (excitation; beam splitter; emission filter). Images were collected every 10 min using NIS-Elements AR software. No photobleaching or phototoxicity was observed in cells imaged by this protocol.
Individual cells were segmented and tracked in time-lapse movies by a user-assisted approach as previously described51. In brief, all movies were pre-processed using rolling ball background subtraction. Individual cells in the movie were manually tracked using in-house developed ImageJ scripts. Using user-defined tracks, nuclear regions of interest (ROI) were segmented automatically based on intensity of PCNA followed by separation of touching nuclei by a watershed algorithm. In case of failed segmentation, the user could manually define polygons as replacement ROIs.
Cell cycle phases were separated according to the distribution of PCNA fluorescent reporter. When no PCNA foci are detected after division, the cell is in G1 phase. The appearance of PCNA foci marks the onset of the S phase. Once the foci disappear, the cell progresses into G2 until nuclear envelope breakdown in M phases.
BioID
BioID, a proximity-dependent biotinylation labeling for the identification of associated proteins, was conducted as previously described41,86,91. First, doxycycline-inducible PiggyBac vector PB-TRE-BirA*_BAHCC1_3×FLAG-rTetR-IRES-Neo was introduced into HeLa and U2OS cells, followed by establishment of stable cell lines by neomycin selection. The stable cell lines were subjected to treatment of 0.5 μg/mL doxycycline into the culture medium, either simultaneously added with or without 50 μM biotin (with the latter serving as BioID controls), for inducing the expression of BirA*_BAHCC1_3×FLAG for 24 h. Then, cells were collected and washed twice with cold 1×PBS. The cell pellets were resuspended in 1 mL RIPA lysis buffer (10% glycerol, 25 mM Tris-HCl pH 8.0, 150 mM NaCl, 2 mM EDTA, 0.1% SDS, 1% NP-40, and 0.2% sodium deoxycholate; freshly supplemented with inhibitors of protease and phosphatase). After a mild sonification, the lysates were incubated with 1 μl of benzonase (Sigma-Aldrich, E1014) on ice for 1 h. Then, centrifugation was performed at the maximum speed for 30 min at 4 °C, and the supernatant was collected and incubated with Neutravidin beads (Thermo Fisher, 29204) overnight at 4 °C. The Neutravidin beads were then sequentially washed twice with RIPA buffer and TAP lysis buffer (10% glycerol, 350 mM NaCl, 2 mM EDTA, 0.1% NP-40, 50 mM HEPES, pH 8.0). Finally, the beads were washed three times with ABC buffer (50 mM ammonium bicarbonate, pH 8.0) and subjected to mass spectrometry-based analysis.
Mass spectrometry-based protein identification from BioID
Mass spectrometry-based analysis were performed as previously described42,86. BioID samples were eluted from beads by adding 50 μL 2× Laemmli buffer (Boston Bioproducts) and heated at 95 °C for 5 min. 50 μL total of each sample was separated by SDS-PAGE using a 4-20% Tris-glycine wedge well gel (Invitrogen) and stained with Coomassie blue. Each SDS-PAGE gel lane was sectioned into 12 segments of equal volume. In-gel trypsin digestion was applied to each segment as following: Gel slices were resuspended in 50% methanol (Fisher) and 50 mM ammonium bicarbonate (Sigma-Aldrich) for destaining. Then reduction was performed in 10 mM Tris [2-carboxyethyl] phosphine (Pierce) followed by alkylation in 50 mM iodoacetamide (Sigma-Aldrich). Gel slices were then dehydrated in acetonitrile (Fisher), followed by incubation with 100 ng porcine sequencing grade modified trypsin (Promega) in 50 mM ammonium bicarbonate (Sigma-Aldrich) for 12–16 h at 37 °C. Peptide products were then acidified in 0.1% formic acid (Pierce). Tryptic peptides were separated by reverse phase XSelect CSH C18 2.5 um resin (Waters) on an in-line 150 × 0.075 mm column using a nanoAcquity UPLC system (Waters). Elution of peptides were conducted in a 30 min gradient with buffer A:B ratio from 97:3 to 67:33 (buffer A: 0.1% formic acid, 0.5% acetonitrile; buffer B: 0.1% formic acid, 99.9% acetonitrile). Eluted peptides were ionized by electrospray (2.15 kV) followed by MS/MS analysis using higher-energy collisional dissociation (HCD) on an Orbitrap Exploris 480 mass spectrometer (Thermo) in top-speed data-dependent mode. MS data were acquired using the FTMS analyser in profile mode at a resolution of 240,000 over a range of 375 to 1,500 m/z. Following HCD activation, MS/MS data were acquired using the ion trap analyser in centroid mode and normal mass range with precursor mass-dependent normalized collision energy between 28.0 and 31.0. Proteins were identified by searching the UniProtKB database restricted to homo sapiens using Mascot (Matrix Science) with a parent ion tolerance of 3 ppm and a fragment ion tolerance of 0.5 Da, fixed modifications for carbamidomethyl of cysteine, and variable modifications for oxidation on methionine and acetyl on N termini. Scaffold (Proteome Software) was used to verify MS/MS-based peptide and protein identifications. Peptide identifications were accepted if they could be established with <1.0% false discovery by the Scaffold Local false discovery rate algorithm. Protein identifications were accepted if they could be established with <1.0% false discovery and contained at least two identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm41. Proteins were filtered out if they had a spectral count <8 in all sample groups and the counts were normalized to log2-normalized spectral abundance factor (NSAF) values. Significant interacting proteins were defined with a cut-off of a log2-transformed fold change above 1 in the experimental versus control samples.
Bio-layer interferometry (BLI) binding assay
Nucleosome core particles (NCPs) were prepared following previously reported protocols92,93. In essence, Xenopus laevis histone proteins, which include H3 (WT or C110S/K27C mutant), H4 (WT or K20C mutant), H2A and H2B, were produced from E. coli BL21 (DE3) RIL cells, followed by reconstitution of histone octamers and NCPs. 5’-biotin labeled Widom 601 DNA94 was prepared by PCR amplification using forward primer: 5’-biotin- CTCTCTCCGTAAACATGCTGGAGAA-3’ and reverse primer 5’-GCGGACGGATAACCGATCGGATGTATATATCTGACA-3’. All nucleosomes were confirmed by 5% TBE (Tris-borate-EDTA) native gel with SYBR Gold staining. Chemical modifications of H3 and H4 proteins to produce methyl-lysine analogs of H3K27me3 (H3Kc27me3) and/or H4K20me3 (H4Kc20me3) were performed as previously described55 and confirmed by mass spectrometry.
The binding affinity of MBP-tagged BAHCC1TTD-BAH protein for nucleosomes, unmodified or modified with H4Kc20me0, H4Kc20me1, H4Kc20me2, H3Kc27me3, or H4Kc20me1/H3Kc27me3, were measured at 30 °C by BLI on a gator prime instrument (Gatorbio, USA) at a shaking speed of 1000 rpm, using MBP alone (control) or MBP-BAHCC1TTD-BAH and NCPs. The binding buffer contains 20 mM Tris-HCl (pH 7.5), 50 mM NaCl, 1 mM DTT, 0.01% Tween-20 and 0.01% NP-40. Prior to the experiments, all biosensors were equilibrated in the binding buffer for 10 min. Assembled NCPs were diluted to 10 nM and immobilized on streptavidin biosensors (Gatorbio, USA) until the shift signal reached a level of ~0.8. After a 120 s baseline step in binding buffer, the ligand-loaded biosensors were incubated with various concentrations (250 to 2000 nM) of MBP or MBP-BAHCC1TTD-BAH protein solution for 120 s to measure the association kinetics, and then incubated with the binding buffer for 600 s to measure the dissociation kinetics. Data were aligned and baseline corrected and analyzed using the Gatorbio data analysis software. The data were fitted using 1:1 mode and the results were plotted in GraphPad Prism 9.1.0. Experiments were independently performed twice to ensure consistency.
Statistics and reproducibility
Experimental data are presented as mean ± s.d. of three independent experiments unless otherwise noted. Statistical analysis was performed using unpaired two-sided Student’s t-test when comparing two sets of data with assumed normal distribution. We used a two-tailed Wilcoxon test to generate the p-values for comparisons of signals from deep-sequencing data. All statistical analysis and visualizations were performed using GraphPad or Excel. Statistical significance levels are denoted as following: *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. No statistical methods were used to predetermine sample size. All data from representative experiments (such as western blot images) were independently repeated at least twice with similar results.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Next-generation sequencing data sets have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE213114 and GSE293272. Published available datasets used in this study include ChIP-seq of H4K20 methylation in HeLa cells, GSM3983187 (H4K20me1), GSM3983189 (H4K20me2), and GSM3983191 (H4K20me3), and in HeLa-S3 cells ENCFF669QWC (H3K4me1), ENCSR000AOE (H3K4me2), ENCSR340WQU (H3K4me3), ENCFF626BOH (H3K9me2), ENCFF761QZP (H3K9me3), ENCFF000BCD (H3K36me3), ENCFF265XYQ (H3K79me1), and ENCFF432DSJ (H3K79me2). Coordinates and structural factors for the Bahcc1 TTD-H4K20me1 complex have been deposited in the PDB under accession code 8YE8. The mass spectrometry proteomics data from the BioID experiments have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD036436 [http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=036436]. Source data are provided with this paper.
References
Beck, D. B., Oda, H., Shen, S. S. & Reinberg, D. PR-Set7 and H4K20me1: at the crossroads of genome integrity, cell cycle, chromosome condensation, and transcription. Genes Dev. 26, 325–337 (2012).
Kuo, A. J. et al. The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier-Gorlin syndrome. Nature 484, 115–119 (2012).
Jorgensen, S., Schotta, G. & Sorensen, C. S. Histone H4 lysine 20 methylation: key player in epigenetic regulation of genomic integrity. Nucleic acids Res. 41, 2797–2806 (2013).
Beck, D. B. et al. The role of PR-Set7 in replication licensing depends on Suv4-20h. Genes Dev. 26, 2580–2589 (2012).
Saredi, G. et al. H4K20me0 marks post-replicative chromatin and recruits the TONSL-MMS22L DNA repair complex. Nature 534, 714–718 (2016).
van Nuland, R. & Gozani, O. Histone H4 lysine 20 (H4K20) methylation, expanding the signaling potential of the proteome one methyl moiety at a time. Mol. Cell. Proteom.: MCP 15, 755–764 (2016).
Corvalan, A. Z. & Coller, H. A. Methylation of histone 4’s lysine 20: a critical analysis of the state of the field. Physiol. Genomics 53, 22–32 (2021).
Wu, S. & Rice, J. C. A new regulator of the cell cycle: the PR-Set7 histone methyltransferase. Cell cycle 10, 68–72 (2011).
Houston, S. I. et al. Catalytic function of the PR-Set7 histone H4 lysine 20 monomethyltransferase is essential for mitotic entry and genomic stability. J. Biol. Chem. 283, 19478–19488 (2008).
Rice, J. C. et al. Mitotic-specific methylation of histone H4 Lys 20 follows increased PR-Set7 expression and its localization to mitotic chromosomes. Genes Dev. 16, 2225–2230 (2002).
Fang, J. et al. Purification and functional characterization of SET8, a nucleosomal histone H4-lysine 20-specific methyltransferase. Curr. Biol. CB 12, 1086–1099 (2002).
Nishioka, K. et al. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol. cell 9, 1201–1213 (2002).
Tardat, M. et al. The histone H4 Lys 20 methyltransferase PR-Set7 regulates replication origins in mammalian cells. Nat. Cell Biol. 12, 1086–1093 (2010).
Oda, H. et al. Regulation of the histone H4 monomethylase PR-Set7 by CRL4(Cdt2)-mediated PCNA-dependent degradation during DNA damage. Mol. cell 40, 364–376 (2010).
Liu, W. et al. PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature 466, 508–512 (2010).
Schotta, G. et al. A chromatin-wide transition to H4K20 monomethylation impairs genome integrity and programmed DNA rearrangements in the mouse. Genes Dev. 22, 2048–2061 (2008).
Evertts, A. G. et al. H4K20 methylation regulates quiescence and chromatin compaction. Mol. Biol. cell 24, 3025–3037 (2013).
Bleichert, F., Botchan, M. R. & Berger, J. M. Mechanisms for initiating cellular DNA replication. Science 355, eaah6317 (2017).
On, K. F., Jaremko, M., Stillman, B. & Joshua-Tor, L. A structural view of the initiators for chromosome replication. Curr. Opin. Struct. Biol. 53, 131–139 (2018).
Bleichert, F. Mechanisms of replication origin licensing: a structural perspective. Curr. Opin. Struct. Biol. 59, 195–204 (2019).
Costa, A. & Diffley, J. F. X. The Initiation of Eukaryotic DNA replication. Annu. Rev. Biochem. 91, 107–131 (2022).
Creager, R. L., Li, Y. & MacAlpine, D. M. SnapShot: Origins of DNA replication. Cell 161, 418–418 e411 (2015).
Kirstein, N. et al. Human ORC/MCM density is low in active genes and correlates with replication time but does not delimit initiation zones. Elife 10, e62161 (2021).
Miotto, B., Ji, Z. & Struhl, K. Selectivity of ORC binding sites and the relation to replication timing, fragile sites, and deletions in cancers. Proc. Natl Acad. Sci. USA 113, E4810–E4819 (2016).
Brustel, J. et al. Histone H4K20 tri-methylation at late-firing origins ensures timely heterochromatin replication. EMBO J. 36, 2726–2741 (2017).
Paquin, K. L. & Howlett, N. G. Understanding the histone DNA repair code: H4K20me2 makes its mark. Mol. Cancer Res. 16, 1335–1345 (2018).
Wang, W. et al. Genome-wide mapping of human DNA replication by optical replication mapping supports a stochastic model of eukaryotic replication. Mol. cell 81, 2975–2988 e2976 (2021).
Long, H. et al. H2A.Z facilitates licensing and activation of early replication origins. Nature 577, 576–581 (2020).
Hayashi-Takanaka, Y. et al. Chromatin loading of MCM hexamers is associated with di-/tri-methylation of histone H4K20 toward S phase entry. Nucleic Acids Res. 49, 12152–12166 (2021).
Centore, R. C. et al. CRL4(Cdt2)-mediated destruction of the histone methyltransferase Set8 prevents premature chromatin compaction in S phase. Mol. cell 40, 22–33 (2010).
Shoaib, M. et al. Histone H4K20 methylation mediated chromatin compaction threshold ensures genome integrity by limiting DNA replication licensing. Nat. Commun. 9, 3704 (2018).
Fan, H. et al. BAHCC1 binds H3K27me3 via a conserved BAH module to mediate gene silencing and oncogenesis. Nat. Genet. 52, 1384–1396 (2020).
Botuyan, M. V. et al. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell 127, 1361–1373 (2006).
Bunting, S. F. et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141, 243–254 (2010).
Katsuyama, T. et al. Involvement of winged eye encoding a chromatin-associated bromo-adjacent homology domain protein in disc specification. Proc. Natl Acad. Sci. USA 102, 15918–15923 (2005).
Li, H. et al. Structural basis for lower lysine methylation state-specific readout by MBT repeats of L3MBTL1 and an engineered PHD finger. Mol. Cell 28, 677–691 (2007).
Taverna, S. D., Li, H., Ruthenburg, A. J., Allis, C. D. & Patel, D. J. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat. Struct. Mol. Biol. 14, 1025–1040 (2007).
Voigt, P. et al. Asymmetrically modified nucleosomes. Cell 151, 181–193 (2012).
Brejc, K. et al. Dynamic control of X chromosome information and repression by a histone H4K20 demethylase. Cell 171, 85–102 e123 (2017).
Bromberg, K. D. et al. The SUV4-20 inhibitor A-196 verifies a role for epigenetics in genomic integrity. Nat. Chem. Biol. 13, 317–324 (2017).
Roux, K. J., Kim, D. I., Burke, B. & May, D. G. BioID: A screen for protein-protein interactions. Curr. Protoc. Protein Sci. 91, 19 23 11–19 23 15 (2018).
Li, J. et al. ZMYND11-MBTD1 induces leukemogenesis through hijacking NuA4/TIP60 acetyltransferase complex and a PWWP-mediated chromatin association mechanism. Nat. Commun. 12, 1045 (2021).
Sardiu, M. E. et al. Suberoylanilide hydroxamic acid (SAHA)-induced dynamics of a human histone deacetylase protein interaction network. Mol. Cell. Proteom. MCP 13, 3114–3125 (2014).
Banks, C. A. S. et al. A structured workflow for mapping human sin3 histone deacetylase complex interactions using halo-mudpit affinity-purification mass spectrometry. Mol. Cell. Proteom.: MCP 17, 1432–1447 (2018).
Bowers, J. L., Randell, J. C., Chen, S. & Bell, S. P. ATP hydrolysis by ORC catalyzes reiterative Mcm2-7 assembly at a defined origin of replication. Mol. cell 16, 967–978 (2004).
Fernandez-Cid, A. et al. An ORC/Cdc6/MCM2-7 complex is formed in a multistep reaction to serve as a platform for MCM double-hexamer assembly. Mol. cell 50, 577–588 (2013).
Yardimci, H. & Walter, J. C. Prereplication-complex formation: a molecular double take?. Nat. Struct. Mol. Biol. 21, 20–25 (2014).
Miotto, B. & Struhl, K. HBO1 histone acetylase is a coactivator of the replication licensing factor Cdt1. Genes Dev. 22, 2633–2638 (2008).
Miotto, B. & Struhl, K. HBO1 histone acetylase activity is essential for DNA replication licensing and inhibited by Geminin. Mol. cell 37, 57–66 (2010).
Zerjatke, T. et al. Quantitative cell cycle analysis based on an endogenous all-in-one reporter for cell tracking and classification. Cell Rep. 19, 1953–1966 (2017).
Grant, G. D., Kedziora, K. M., Limas, J. C., Cook, J. G. & Purvis, J. E. Accurate delineation of cell cycle phase transitions in living cells with PIP-FUCCI. Cell cycle 17, 2496–2516 (2018).
Moldovan, G. L., Pfander, B. & Jentsch, S. PCNA, the maestro of the replication fork. Cell 129, 665–679 (2007).
Nakamura, K. et al. H4K20me0 recognition by BRCA1-BARD1 directs homologous recombination to sister chromatids. Nat. Cell Biol. 21, 311–318 (2019).
Huang, L. et al. Structural insight into H4K20 methylation on H2A.Z-nucleosome by SUV420H1. Mol. cell 83, 2884–2895 e2887 (2023).
Simon, M. D. et al. The site-specific installation of methyl-lysine analogs into recombinant histones. Cell 128, 1003–1012 (2007).
Musselman, C. A. et al. Binding of PHF1 Tudor to H3K36me3 enhances nucleosome accessibility. Nat. Commun. 4, 2969 (2013).
Kalra, P., Zahid, H., Ayoub, A., Dou, Y. & Pomerantz, W. C. K. Alternative mechanisms for DNA engagement by BETbromodomain-containing proteins. Biochemistry 61, 1260–1272 (2022).
Connelly, K. E. et al. Engagement of DNA and H3K27me3 by the CBX8 chromodomain drives chromatin association. Nucleic acids Res. 47, 2289–2305 (2019).
Zhen, C. Y. et al. Live-cell single-molecule tracking reveals co-recognition of H3K27me3 and DNA targets polycomb Cbx7-PRC1 to chromatin. Elife 5, e17667 (2016).
Asenjo, H. G. et al. Polycomb regulation is coupled to cell cycle transition in pluripotent stem cells. Sci. Adv. 6, eaay4768 (2020).
Keniry, A. & Blewitt, M. E. Chromatin-mediated silencing on the inactive X chromosome. Development 150, dev201742 (2023).
Tjalsma, S. J. D. et al. H4K20me1 and H3K27me3 are concurrently loaded onto the inactive X chromosome but dispensable for inducing gene silencing. EMBO Rep. 22, e51989 (2021).
Sato, Y. et al. Visualizing histone H4K20me1 in knock-in mice expressing the mCherry-tagged modification-specific intracellular antibody. Histochem Cell Biol. 162, 41–52 (2024).
Vielle, A. et al. H4K20me1 contributes to downregulation of X-linked genes for C. elegans dosage compensation. PLoS Genet 8, e1002933 (2012).
Erfurth, F. E. et al. MLL protects CpG clusters from methylation within the Hoxa9 gene, maintaining transcript expression. Proc. Natl Acad. Sci. USA 105, 7517–7522 (2008).
Sahu, S., Ekundayo, B. E., Kumar, A. & Bleichert, F. A dual role for the chromatin reader ORCA/LRWD1 in targeting the origin recognition complex to chromatin. EMBO J. 42, e114654 (2023).
Ren, W. et al. DNMT1 reads heterochromatic H4K20me3 to reinforce LINE-1 DNA methylation. Nat. Commun. 12, 2490 (2021).
Ren, W. et al. Direct readout of heterochromatic H3K9me3 regulates DNMT1-mediated maintenance DNA methylation. Proc. Natl Acad. Sci. USA 117, 18439–18447 (2020).
Perfetti, M. T. et al. Identification of a fragment-like small molecule ligand for the methyl-lysine binding protein, 53BP1. ACS Chem. Biol. 10, 1072–1081 (2015).
Xu, J. Preparation, culture, and immortalization of mouse embryonic fibroblasts. Curr. Protoc. Mol. Biol. 28, 28 21 (2005).
Yu, X. et al. A selective WDR5 degrader inhibits acute myeloid leukemia in patient-derived mouse models. Sci. Transl. Med. 13, eabj1578 (2021).
Wang, J. et al. EZH2 noncanonically binds cMyc and p300 through a cryptic transactivation domain to mediate gene activation and promote oncogenesis. Nat. cell Biol. 24, 384–399 (2022).
Chen, G. & Deng, X. Cell synchronization by double thymidine block. Bio. Protocol 8, e2994 (2018).
Thwaites, M. J., Coschi, C. H., Isaac, C. E. & Dick, F. A. Cell synchronization of mouse embryonic fibroblasts. Methods Mol. Biol. 1342, 91–99 (2016).
Cai, L. et al. An H3K36 methylation-engaging Tudor motif of polycomb-like proteins mediates PRC2 complex targeting. Mol. cell 49, 571–582 (2013).
Skene, P. J., Henikoff, J. G. & Henikoff, S. Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nat. Protoc. 13, 1006–1019 (2018).
Skene, P. J. & Henikoff, S. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. eLife 6, e21856 (2017).
Kaya-Okur, H. S., Janssens, D. H., Henikoff, J. G., Ahmad, K. & Henikoff, S. Efficient low-cost chromatin profiling with CUT&Tag. Nat. Protoc. 15, 3264–3283 (2020).
Kaya-Okur, H. S. et al. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun. 10, 1930 (2019).
Fan, H. et al. A conserved BAH module within mammalian BAHD1 connects H3K27me3 to Polycomb gene silencing. Nucleic acids Res. 49, 4441–4455 (2021).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Ahn, J. H. et al. The phenylalanine-and-glycine repeats of NUP98 oncofusions form condensates that selectively partition transcriptional coactivators. Mol. cell 85, 708–725 e709 (2025).
Lu, J. et al. Structure-guided functional suppression of AML-associated DNMT3A hotspot mutations. Nat. Commun. 15, 3111 (2024).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Ramirez, F. et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016).
Ahn, J. H. et al. Phase separation drives aberrant chromatin looping and cancer development. Nature 595, 591–595 (2021).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 276, 307–326 (1997).
Adams, P. D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D. 58, 1948–1954 (2002).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. 60, 2126–2132 (2004).
Matson, J. P. et al. Rapid DNA replication origin licensing protects stem cell pluripotency. eLife 6, e30473 (2017).
Roux, K. J., Kim, D. I. & Burke, B. BioID: a screen for protein-protein interactions. Curr. Protoc. protein Sci. 74, 19 23 11–19 23 14 (2013).
Dyer, P. N. et al. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 375, 23–44 (2004).
Zhang, W., Sankaran, S., Gozani, O. & Song, J. A Meier-Gorlin syndrome mutation impairs the ORC1-nucleosome association. ACS Chem. Biol. 10, 1176–1180 (2015).
Lowary, P. T. & Widom, J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 276, 19–42 (1998).
Acknowledgements
We thank E. Julien, S. Frye, B. Strahl and M. Emanuele for graciously providing reagents used in this study. We thank the Wang, Song, Cook, Cai and Tackett laboratory members for helpful discussion and technical support. We also thank the core facilities at University of North Carolina (UNC) at Chapel Hill, Duke University and University of Arkansas for Medical Sciences (UAMS), including Flow Cytometry Core, Tissue Culture Facility and Genomics Core for their professional assistance of this work and thank staff members at the Advanced Light Source (DE-AC02-05CH11231), Lawrence Berkeley National Laboratory for access to X-ray beamlines. The Berkeley Center for Structural Biology is supported in part by the National Institutes of Health, the National Institute of General Medical Sciences and the Howard Hughes Medical Institute. The cores affiliated to UNC Cancer Center are supported in part by the UNC Lineberger Comprehensive Cancer Center Core Support Grant P30-CA016086 and cores affiliated to Duke Cancer Institute funded by grant 1S10RR027867-01 for the confocal systems and by the NCI Cancer Center Support Grant (CCSG) award P30CA014236. This work was supported by National Institutes of Health grants R01-CA215284, R01CA271603 and R01CA268519 to G.G.W.; R01-CA262903 to L.C; R35GM119721 to J.S,; R24GM137786 and P20GM121293 and R01CA236209 to A.J.T.; R35GM141833 to J.G.C. G.G.W. was a Leukemia and Lymphoma Society (LLS) Scholar and an American Cancer Society Scholar.
Author information
Authors and Affiliations
Contributions
D.L., Z-M.Z., L.M., Y.Y., Y.G., S.G.M., J.C., D.F.A., A.K., A.J.S., R.D.E., S.D.B. and L.C. performed experiments. D.L., Y.Y., Y.G., D.F.A, and A.K. designed the research, performed biological experiments and interpreted data under the supervision of L.C. and G.G.W. Z-M.Z. and J.C. performed structural and biophysical analyses under the supervision of J.S. L.M. conducted the chromatin-fractionation flow cytometry and live cell imaging under the supervision of J.G.C. S.G.M., A.J.S., R.D.E. and S.D.B. performed proteomic analysis under the supervision of A.J.T. D.L., Z-M.Z., L.M., J.G.C., J.S., and G.G.W. wrote the manuscript with the inputs from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, D., Zhang, ZM., Mei, L. et al. BAHCC1 binds H4K20me1 to facilitate the MCM complex loading and DNA replication. Nat Commun 16, 5502 (2025). https://doi.org/10.1038/s41467-025-61284-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-61284-1