Abstract
Radiation-induced liver disease (RILD) severely impairs the outcome of patients receiving irradiation (IR); however, its underlying mechanism remains unknown. GSDMD drives the progression of pyroptosis, and can be induced by IR in the gut and bone marrow, but its role in RILD remains unknown. Here we show that GSDMD is significantly upregulated and positively correlated with RILD severity in a mouse model. Hepatocytes are identified as critical pyroptotic cells in RILD thorough scRNA-seq, immunofluorescence and fluorescence-activated cell sorting analysis. Functional and mechanistic analysis using Gsdmd knockout (Gsdmd∆Hep) mice and cell models. Mechanistically, GSDMD is indispensable for triggering hepatocyte pyroptosis and initiating the activation of transcription factor STAT5A, which subsequently promoted CXCL1 expression to recruit neutrophil into liver to accelerate the severity of RILD. We also discovered that pharmacological targeting GSDMD and its downstream CXCL1 effectively alleviated RILD. Together, our study demonstrates that GSDMD as therapeutic targets to improve RILD.
Similar content being viewed by others
Introduction
The liver, a pivotal organ with multifarious functions, manifests as a radiosensitive entity warranting particular attention1. Exposure to ionizing irradiation, such as nuclear and radiological incidents2, total body irradiation (TBI)3 before allogeneic stem cell transplantation, and radiotherapy for tumors4,5,6,7 in the upper abdomen (liver, biliary, and pancreas) and thoracic (right lower lung, distal esophagus, and breast), can result in radiation-induced liver disease (RILD). RILD manifests as acute liver injury in the short-term after RT followed by chronic progression to liver fibrosis8,9, while no clinic effective drugs to alleviate it. Patients will irreversibly develop into cirrhosis, liver failure and even death. Pathophysiologically, radiation results in the increase of inflammatory cytokines and recruitment of immune cells, ultimately leading to tissue fibrosis and liver dysfunction10. However, the underlying mechanism of immune regulation in RILD remains unclear.
Pyroptosis, a form of inflammatory cell death, is crucial in the immune environment. During pyroptosis, inflammasome activation by canonical (NOD-like receptor [NLR], absent in melanoma 2 [AIM2], and caspase-1) or noncanonical (caspase-4/5 in humans or caspase-11 in mice) pathways leads to the cleavage of GSDMD. GSDMD-N then forms pores in the membrane and results in cell swelling, a burst of inflammatory cytokines and cytosolic content release and ultimately cell lysis. Therefore, GDSMD, as a key executioner of pyroptosis and cytokine release, is thought to be a crucial mediator in shaping the inflammatory environment involved in the progression of multiple liver diseases11,12,13,14. Recently, Xiao et al.15 reported that TBI could induce GSDMD-FL/N increase in the gut and bone marrow of mice, which were positively correlated with the severity of bone loss, but the role of GSDMD in this process remains unclear. However, Wu et al.16 revealed that there were no significant increase of GSDMD-FL/N in lung tissues after chest irradiation. These discoveries suggest that the existence and effects of GSDMD are complicated and vary in different radiation-induced tissue injuries. However, the occurrence of GSDMD-executed pyroptosis in RILD and its impact on the expression and secretion of inflammatory factors still remain elusive. Therefore, further investigation is warranted to elucidate the role of GSDMD-executed pyroptosis in RILD. Additionally, it is crucial to explore other uncharted functions of GSDMD in the context of RILD, considering that cleavage-independent functions of gasdermin proteins have been reported to influence the growth of pancreatic adenocarcinoma and the progression of asthma17,18.
In the present study, we unveil the intriguing upregulation of GSDMD-FL/N and the emergence of a pyroptotic phenotype in irradiated hepatocytes, shedding light on the molecular mechanisms underlying RILD. Through its targeted interaction with STAT5A, GSDMD facilitated the phosphorylation of STAT5A, leading to its translocation from the cytoplasm to the nucleus. This dynamic process played a critical role in mediating the expression of CXCL1, initiating a cascade of events within the liver microenvironment. GSDMD-N played a pivotal role in the secretion of CXCL1, attracting neutrophils to the liver and exacerbating the severity of RILD. We mitigated neutrophil recruitment and alleviated RILD in mice by blocking STAT5A/CXCL1/neutrophil pathway. Furthermore, our promising findings indicate that pharmacological inhibition of GSDMD offers a potent approach to suppress the progression of RILD. Collectively, our discoveries highlight the exciting potential of targeting GSDMD as a therapeutic strategy to prevent RILD.
Results
GSDMD-dependent hepatocyte pyroptosis occurs in RILD
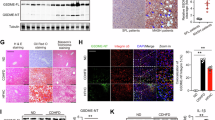
To determine whether pyroptosis triggered by GSDMD occurs in RILD, we next confirmed the expression pattern of GSDMD in RILD mice. As shown in Supplementary Fig. 1A, we subjected mice to a universal short-term (ST) and long-term (LT) RILD modeling protocol19,20. First, we observed that GSDMD-FL and GSDMD-N levels were robustly elevated in the irradiated murine liver tissue in both the ST-RILD and LT-RILD groups (Fig. 1A). Moreover, hepatic GSDMD-N levels were closely correlated with the degree of liver injury (serum ALT and AST), liver function (serum ALB), and liver fibrosis (fibronectin, Supplementary Fig. 1B-C). These results suggested that GSDMD and its pyroptosis-triggerred N-terminal fragment are upregulated in liver tissues during RILD.
A Immunoblots of GSDMD-FL/N in livers from GsdmdFL/FL mice after short-term (ST-RILD; 30 Gy, 7 days) or long-term (LT-RILD; 6 Gy × 3, 10 weeks) irradiation (n = 3, Ctrl; n = 5, ST/LT). B Gsdmd, Il1b and other pyroptosis-related genes expression in each immune cluster of Ctrl and IR groups. Y axis indicates log-normalized expression. C Representative low-magnification (Scale bars, 100×, 200 μm) and blow-up magnification (Scale bars, 400×, 50 μm) images of GSDMD-FL staining in Ctrl, ST-RILD and LT-RILD mice (five fields were observed per liver, n = 5/group). ****p = 5E-10, Ctrl vs ST; ****p = 3.499E-05, Ctrl vs LT. D Immunofluorescence visualization of GSDMD-FL (green), DAPI (blue), HNF4a/CD11b/CD3/CK19/ɑ-SMA (red) in liver sections from Ctrl and IR-treated mice (ST- and LT-RILD, n = 5/group). Up, representative images (Scale bars, 200×, 100 μm); Down, statistical analysis of the change of the cells co-stained with GSDMD-FL (five fields were observed per liver, n= 5roup). HNF4a: ****p = 2.063E-09, Ctrl vs ST; ****p = 7.967E-07, Ctrl vs LT; CD3: ****p = 3.230E-07, Ctrl vs ST; CK19: **p = 0.008, Ctrl vs ST; ****p = 2.740E-07, Ctrl vs LT; E GSDMD-N protein expressions from hepatocyte, monocyte-granulocyte, HSC, T cell and sinusoidal endothelial cell (SEC) were detected by immunoblot analysis. F Immunoblot analysis of GSDMD-FL and GSDMD-N expression from GsdmdFL/FL primary mouse hepatocytes exposed to 8 Gy irradiation at different times. G–I GsdmdFL/FL and Gsdmd∆Hep hepatocytes 24 h after 8 Gy irradiation, cell morphology was observed under microscopy (black indicates bubble-like morphology) (G), and cell viability was assessed by LDH assay (H) and propidium iodide (PI) staining (I) (Each sample was from 3 biologically independent replicates for E-I). **p = 0.007, GsdmdFL/FL-Ctrl vs IR; *p = 0.02, GsdmdFL/FL-IR vs Gsdmd∆Hep-IR; ****p = 1.177E-06, GsdmdFL/FL-Ctrl vs IR; ****p = 1.456E-06, GsdmdFL/FL-IR vs Gsdmd∆Hep-IR; P values were calculated using one-way ANOVA with Tukey’s multiple comparison test (C, D, H) and two-way ANOVA following Fisher’s LSD test for multiple comparisions (I). Data are mean ± SEM. No data were excluded from the analyses. Source data are provided as a Source Data file.
Previous studies have shown that GSDMD-dependent pyroptosis widely occurs in immune cells21,22. To identify which immune cell type is involved in pyroptosis after irradiation in an unbiased and precise manner, we performed comprehensive single-cell RNA sequencing (scRNA-seq) of whole liver tissue from control (ctrl) and ST-RILD mice. Within the immune compartments, we compared the expression of Gsdmd and other pyroptosis-associated gene markers, including Il1b, Il18, Casp1, Caspase4/11, Nlrp3, Nod1, and Nod2 (Fig. 1B). Unexpectedly, Gsdmd was rarely elevated in hepatic immune cells after IR. Under physiological conditions, Gsdmd was slightly expressed in monocytes and Kupfer cells and barely expressed in neutrophils, dendritic cells (DCs), B cells, and T/NK cells. Moreover, no noticeable increase in the expression of Gsdmd or other pyroptosis-associated genes were observed in these cells after IR. Since pyroptosis can also occur in hepatocytes rather than immune cells in steatohepatitis and CCL4-induced liver injury23, we assumed that hepatocytes might be the major pyroptotic cells in RILD, with strong activation of GSDMD. Indeed, a robust increase in GSDMD-FL expression was preferentially found in the parenchymal cells of RILD mouse livers (Fig. 1C). Further immunofluorescence analysis was performed to clearly determine the GSDMD expression changes in hepatocytes and other cells. As we speculated, remarkable coexpression of GSDMD with hepatocellular nuclear factor 4-alpha (HNF4ɑ, a marker of hepatocytes) was observed in both ST-RILD and LT-RILD livers (Fig. 1D), while CD11b+ monocytes, CD3+ T cells, CK19+ cholangiocytes and α-SMA+hepatic stellate cells (HSCs) displayed low expression of GSDMD. To accurately confirm the occurrence of pyroptosis in hepatocyte, we isolated primary hepatocytes and other non-parenchymal cells (NPC) from the liver of ctrl and ST-RILD mice and compared their GSDMD-N levels by western blot (WB) analysis (Fig. 1E). Hepatocytes also had the highest GSDMD-N level after irradiation compared with other cells, further indicating that hepatocytes are critical pyroptotic cells in RILD.
Subsequently, to determine whether irradiation induced hepatocyte pyroptosis, primary hepatocytes were isolated from mice and irradiated with 8 Gy irradiation to establish ex vivo radiation models (Supplementary Fig. 1D). Indeed, irradiation could aggravate hepatocyte injury, manifested as decreased ALB and increased ALT and AST levels 24 h after IR (Supplementary Fig. 1E). Besides, the levels of the key pyroptotic proteins GSDMD-FL/N increased from 12 h to 24 h after irradiation (Fig. 1F). We also investigated another gasdermin family member, GSDME, which can be cleaved by caspase-3 to induce pyroptosis following chemotherapy24. In hepatocytes post-irradiation, we examined the levels of GSDME and caspase-3. Although pro-caspase-3 increased after irradiation, there was no significant difference in cleaved caspase-3 and GSDME-N (Supplementary Fig. 1F). Compared to nonirradiated hepatocytes, irradiated hepatocytes also exhibited other late-stage features of pyroptosis: cell swelling with a balloon-shaped protrusion emerging from the plasma membrane by microscopy (Fig. 1G), increased LDH levels in the supernatant (Fig. 1H) and decreased cell viability (Fig. 1I). These results confirmed that hepatocyte pyroptosis is activated after IR. To further investigate the requirement of GSDMD for hepatocyte pyroptosis, primary hepatocytes from GsdmdFL/FL and Gsdmd∆Hep mice were subjected to irradiation. Compared to GsdmdFL/FL hepatocytes, Gsdmd∆Hep hepatocytes were resistant to irradiation-mediated pyroptosis (Fig. 1F–I). Altogether, these results show that GSDMD is required for triggering pyroptosis in irradiated hepatocytes.
Collectively, these data demonstrated that GSDMD is required for driving hepatocyte pyroptosis in RILD.
GSDMD deficiency ameliorates acute liver injury and chronic fibrosis in RILD
To further investigate the role of GSDMD-dependent hepatocyte pyroptosis in RILD, we generated RILD models with GsdmdFlox/Flox (GsdmdFL/FL) and Gsdmd∆Hep mice (Supplementary Fig. 2A). No significant difference in weight loss was found between these two groups of mice with ST-RILD (Supplementary Fig. 2B), while Gsdmd∆Hep mice with LT-RILD displayed less weight loss and regained body weight more quickly after irradiation, in contrast to the GsdmdFL/FL-IR mice (Supplementary Fig. 2C). Moreover, in ST-RILD mice, liver tissues of GsdmdFL/FL-IR mice manifested increased steatosis, while GSDMD deficiency markedly mitigated this pathological damage (Fig. 2A). In LT-RILD mice, the livers of GsdmdFL/FL-IR mice exhibited apparent scattered nodules (Fig. 2B), notable hepatic stellate cell (HSC) activation (α-SMA+), obvious extracellular matrix (ECM) deposition (collagen+ and fibronectin+) (Fig. 2C–E), and enhanced expression of the profibrotic markers Col1a1, Tgfb1, and Timp1 (Fig. 2F), indicating that chronic liver fibrosis was induced by irradiation. However, all of these fibrotic changes were notably suppressed in the context of GSDMD deficiency. In addition, in both ST- and LT-RILD mice, the absence of GSDMD also led to a striking decrease in serum ALT and AST and an improvement in ALB, reflecting enhanced liver function (Fig. 2G). Collectively, these results suggest that GSDMD deficiency exerts a vital inhibitory effect on both early IR-induced hepatic injury and advanced liver fibrosis.
A Representative high magnification images of Hematoxylin Eosin staining (HE) staining in GsdmdFL/FL and Gsdmd∆Hep mice (n = 5/group) treated with or without one single 30 Gy hepatic radiation. (Scale bars, 200×, 200 μm, 400×, 50 μm). B-D Representative liver macroscopic image (B), ɑ-SMA staining (C) and sirius-red staining (D) in GsdmdFL/FL or Gsdmd∆Hep mice in LT-RILD. Scale bars, 400×, 50 μm; 200×, 100 μm. Statistical analysis of ɑ-SMA and collagen area were present in the right, 5 fields were observed per mice (n = 5/group). GsdmdFL/FL-IR vs Gsdmd∆Hep-IR: ɑ-SMA, ***p = 0.0001; collagen area, ***p = 0.0004. E Immunoblot analysis of liver protein expression of COL1A1, Fibronectin and ɑ-SMA from GsdmdFL/FL and Gsdmd∆Hep mice (n = 1, no IR; n = 3, GsdmdFL/FL-IR; n = 5, Gsdmd∆Hep-IR). F Key hepatic profibrogenic genes of Col1a1, Tgfb1 and Timp1 in GsdmdFL/FL and Gsdmd∆Hep mice treated with IR (n = 3/group, Ctrl; n = 5/group, LT). GsdmdFL/FL-IR vs Gsdmd∆Hep-IR: Col1a1, p = 0.0008; Tgfb1, p = 0.0003; Timp1, p = 0.0003. G Serum levels of liver function markers ALT, AST and ALB were determined in Ctrl-, ST- and LT-RILD groups (n = 3/group, Ctrl; n = 5/group, ST/LT). ST-GsdmdFL/FL vs Gsdmd∆Hep: ALT, p = 2.087E-06; AST, p = 6.926E-05; ALB, p = 0.002. LT-GsdmdFL/FL vs Gsdmd∆Hep: ALT, p = 1.857E-05; AST, p = 4.838E-05; ALB, p = 1.273E-05. H Supernatant levels of liver function markers ALT, AST and ALB were determined in hepatocytes with and without IR treatment (Each sample was from 3 biologically independent replicates). ALT, p = 1.156E-06; AST, p = 2.89E-07; ALB, p = 0.002. Data are expressed as mean ± SEM. p values were calculated using two-way ANOVA with Fisher’s LSD multiple comparisions test (C, D, F–H). No data were excluded from the analyses. Source data are provided as a Source Data file.
To further determine the pathogenic role of GSDMD in RILD, AAV9-GSDMD-FL was administered to Gsdmd∆Hep mice to restore GSDMD expression prior to IR (Supplementary Fig. 2D–E). GSDMD-FL re-expression did not affect weight loss in Gsdmd∆Hep ST-RILD mice (Supplementary Fig. 2F) but resulted in a significant increase in hepatic histology damage and liver injury (Supplementary Fig. 2G–H). These results confirmed that upregulation of GSDMD accelerated the progression of RILD.
Similar protection from GSDMD deficiency was observed in an ex vivo hepatocyte irradiation model. Compared to GsdmdFL/FL hepatocytes, Gsdmd∆Hep hepatocytes released higher levels of ALB and lower levels of ALT and AST after IR (Fig. 2H). Overall, these data suggested that GSDMD contributes to the progression of RILD probably depending on GSDMD driving hepatocyte pyroptosis.
GSDMD deficiency inhibits the recruitment of neutrophils to the irradiated liver
GSDMD influenced the progression of diseases by modulating inflammatory responses11,25,26,27. Therefore, to explore whether GSDMD aggravates RILD by affecting the hepatic immune environment, we set out to obtain single-cell suspensions of liver tissue from GsdmdFL/FL-Ctrl, GsdmdFL/FL-IR, and Gsdmd∆Hep-IR mice, followed by scRNA-seq to characterize immune cell profiles (Fig. 3A). As shown in Fig. 3B, hepatic immune cells were visualized by uniform manifold approximation and projection (UMAP). Compared to the GsdmdFL/FL-Ctrl, the hepatic immune cell composition of GsdmdFL/FL-IR mice underwent significant changes, shown as a decrease in dendritic cells, T/B cells, and natural killer cells, a slight increase in Kupffer cells, and a striking elevation in neutrophils, suggesting an important role of neutrophils in RILD. Intriguingly, this IR-induced neutrophil infiltration of the liver was markedly reduced after in the absence of GSDMD (Fig. 3C), indicating that GSDMD might contribute to neutrophil recruitment under IR.
A, B Workflow and Proportion plot of immune cells. A Livers from GsdmdFL/FL -Ctrl, GsdmdFL/FL-ST and Gsdmd∆Hep-ST group were used. 7 days after the last 30 Gy irradiation, the livers were digested and 10 × Genomics scRNA-seq was used to profile the cells; B UMAP plot of liver single CD45+ cells from three group mice, colored by the group. C Colors indicate the cell type annotated using Single R and manual annotation and validated by cell-type-specific gene expression. D Flow Cytometric analysis of liver-infiltrated neutrophils (FVD-CD45+CD11b+Ly6G+) of GsdmdFL/FL and Gsdmd∆Hep mice. GsdmdFL/FL vs Gsdmd∆Hep: ST, p = 3.1E-09; LT, p = 0.002. E Neutrophil infiltrationwas analyzed by immunohistology staining (left, scale bars, 200×, 50 μm) with antibodies against MPO, and following morphometric quantification (right). D, E n = 3/group, Ctrl; n = 5/group, ST/LT. GsdmdFL/FL vs Gsdmd∆Hep: ST, p = 1.218E-07; LT, p = 2.86E-07. F, G Detection of the neutrophil infiltration in liver of Gsdmd∆Hep mice treated with or without AAV9-GSDMD-FL adeno-associated virus. F Representative plots (left) and statical analysis (right) of the percentage of CD11b+Ly6G+ cells with the flow cytometry. **p = 0.005. G MPO staining images (left) and quantification (right). ****P < 0.0001. F, G n = 4/group. H Quantification of neutrophil migration in response to GsdmdFL/FL and Gsdmd∆Hep-hepatocytes pretreated with irradiation or not. Each sample was from 3 biologically independent replicates. ***P = 0.0003. I The depletion effect of neutrophils in liver were tested by flow cytometry in ST-RILD mice. ****P < 0.0001. J Representative liver HE images from ST-RILD mice (Scale bars, 200×, 50 μm). K Serum levels of ALT, AST, and ALB.****P < 0.0001. n = 5 mice/group for I–K. p values were using two-way ANOVA with Fisher’s LSD multiple comparisions test (D, E, H) and unpaired two-sided Student’s t test (F, G and I). No data were excluded from the analyses. Data are expressed as mean ± SEM. Source data are provided as a Source Data file.
Decreased liver infiltration of neutrophils in Gsdmd∆Hep mice were then confirmed by flow cytometry analysis (Fig. 3D). Moreover, myeloperoxidase (MPO)-positive active neutrophils were also significantly accumulated in both ST-RILD and LT-RILD GsdmdFL/FL mouse livers, as detected by IHC, and were markedly reduced by knocking out Gsdmd (Fig. 3E). Likewise, restoration of GSDMD expression by administration of AAV9-GSDMD-FL to Gsdmd∆Hep-ST-RILD mice significantly increased hepatic neutrophil infiltration, with levels similar to those in GsdmdFL/FL-ST-RILD mice (Fig. 3F–G).
Next, we wondered if GSDMD-mediated hepatocyte pyroptosis facilitated the recruitment of neutrophils. Purified neutrophils were then cocultured with non-irradiated or irradiated hepatocytes from GsdmdFL/FL or Gsdmd∆Hep mice, followed by counting of migratory neutrophils (Supplementary Fig. 3A-B). In the case of equivalent cells, the recruitment of neutrophils was significantly increased by coculture with GsdmdFL/FL-IR hepatocytes but notably decreased when cocultured with Gsdmd∆Hep-IR hepatocytes (Fig. 3H).
To further explore the role of neutrophils in promoting RILD in vivo, we depleted neutrophils using a combination of anti-Ly6G Ab and anti-rat kappa light chain Ab (referred to as “Combo”) (Supplementary Fig. 3C and Fig. 3I). Neutrophil depletion resulted in significant improvements in hepatic histology damage and liver injury, but did not affect body weight (Fig. 3I–K, and Supplementary Fig. 3D). These findings confirm that neutrophils recruited by GSDMD accelerate the progression of RILD.
The activation of HSCs is critical for the progression of fibrosis in RILD while little is known about pyroptotic hepatocyte impact on HSCs. We designed both contact and non-contact co-culture experiments of hepatocytes with HSCs (Supplementary Fig. 3E). The results show that there was no difference between the two approaches in terms of having no effect on HSCs activation (Supplementary Fig. 3F–G).
Collectively, our data illustrated that GSDMD-mediated hepatocyte pyroptosis facilitates neutrophil infiltration into the liver.
Selectively CXCL1 neutralization effectively attenuated the progression of GSDMD driving RILD through suppressing neutrophil recruitment
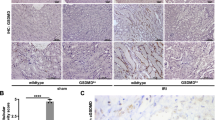
Given that neutrophils are not liver-resident immune cells and require chemokines for their trafficking25,27,28,29,30, we hypothesized that GSDMD might regulate neutrophil recruitment by affecting chemokine expression. Therefore, RNA-seq of GsdmdFL/FL-IR and Gsdmd∆Hep-IR mouse livers was performed to screen neutrophil trafficking-associated chemokines regulated by GSDMD. Notably, we found that CXCL1 was the most significantly downregulated chemokine in the livers of Gsdmd∆Hep-IR mice compared with GsdmdFL/FL-IR mice (Fig. 4A). To validate whether GSDMD impacts CXCL1 expression, hepatic CXCL1 in GsdmdFL/FL and Gsdmd∆Hep mice before and after IR was examined at the mRNA and protein levels. We discovered that CXCL1 was maintained at a low expression level in both GsdmdFL/FL and Gsdmd∆Hep mice under steady-state conditions and was markedly increased in GsdmdFL/FL mice after radiotherapy, while Gsdmd∆Hep mice exhibited notable resistance to this upregulation (Fig. 4B–C). Moreover, further re-expressing GSDMD-FL in Gsdmd∆Hep mice effectively restored IR-induced hepatic CXCL1 upregulation (Fig. 4D) implying CXCL1 should act as a downstream factor of GSDMD during the progression of RILD.
A Volcano plot depiction of neutrophil chemokine genes between GsdmdFL/FL and Gsdmd∆Hep mice (n = 5/group). B Detection of the hepatic mRNA level of Cxcl1 in GsdmdFL/FL and Gsdmd∆Hep mice (n = 5/group) accepted or were free of 30 Gy or 6 Gy×5 IR. ***p = 0.001, ST: GsdmdFL/FL-Ctrl vs Gsdmd∆Hep; *p = 0.01, LT: GsdmdFL/FL-Ctrl vs Gsdmd∆Hep. C Representative immunohistochemistry images of CXCL1 in liver tissue of Ctrl and IR group from GsdmdFL/FL and Gsdmd∆Hep mice (Scale bars, 40×, 200 μm; 400×, 50 μm, left). Statical analysis of CXCL1 expression were calculated from five fields per liver (n = 5/group, right). ****P < 0.0001. D Representative liver CXCL1 staining from ST-RILD mice treated with AAV9-control and AAV9-GSDMD-FL adeno virus (Scale bars, 40×, 200 μm, left). Statical analysis of CXCL1 expression were calculated from five fields per liver (n = 4/group, right). ****P < 0.0001. E Detection of the neutrophil infiltration in liver of mice treated with isotype IgG antibody and anti-CXCL1 antibody. The percentage of cells with CD11b+Ly6G+ with flow cytometry (left) and its statical analysis were presented in both ST- and LT-RILD cohort (right; n = 3 mice/group, ST; n = 5 mice/group, LT). **p = 0.008, ST: isotype IgG vs anti-CXCL1 Ab; *p = 0.03, LT: isotype IgG vs anti-CXCL1 Ab. F Representative liver HE images from ST-RILD mice (Scale bars, 200×, 50 μm). G–I Representative liver macroscopic images (G), ɑ-SMA staining (H, p = 0.0002) and sirius-red staining (I, p = 0.0039) in cohort of LT-RILD mice. (Scale bars, 400×, 50 μm). Statistical analysis of ɑ-SMA and collagen area were present in the right, 5 fields were observed per mice. (F-I, n = 3/group). J Hepatic mRNA levels of Col1a1 (p = 0.0169), Tgfb1 (p = 0.044) and Timp1 (p = 0.0478) from LT-RILD mice. K Imunoblot analysis of COL1A1, Fibronectin and ɑ-SMA. n = 5/group for J-K. L Serum levels of ALT, AST, and ALB (n = 3/group, isotype-ST; n = 5/group, anti-CXCL1 Ab and isotype-LT). ST-isotype IgG vs anti-CXCL1 Ab: ALT, p = 8.27E-07; AST, p = 0.000219; ALB, p = 0.0068. LT-isotype IgG vs anti-CXCL1 Ab: ALT, p = 9.72E-07; AST, p = 1.52E-05; ALB, p = 0.0016. p values were using two-way ANOVA with Fisher’s LSD multiple comparisions test (C, L) and unpaired two-sided Student’s t test (B, D, E and H–J). Data are expressed as mean ± SEM. No data were excluded from the analyses. Source data are provided as a Source Data file.
To further investigate the role of CXCL1 in RILD through influencing neutrophil infiltration in vivo, GsdmdFL/FL-IR mice were administered either a control immunoglobulin G (IgG) antibody or an anti-CXCL1 antibody (anti-CXCL1 Ab), which effectively neutralized CXCL1 in RILD models (Supplementary Fig. 4A–B). Flow cytometry analysis confirmed that CXCL1 inhibition resulted in twofold and fourfold decreases in liver neutrophil infiltration in the ST-RILD and LT-RILD mouse models, respectively (Fig. 4E). Likewise, MPO+ active neutrophils also decreased significantly after blocking CXCL1, as indicated by IHC (Supplementary Fig. 4C).
Similar to the observation in Gsdmd∆Hep mice, neutralizing CXCL1 had no effect on weight loss in ST-RILD mice but alleviated weight loss in the LT-RILD group (Supplementary Fig. 4D–E). Furthermore, compared to the IgG-treated mice, neutralization of CXCL1 resulted in reduced liver steatosis and congestion in ST-RILD mice and attenuated liver fibrosis in LT-RILD mice (Fig. 4F–K). In addition, notable suppression of liver injury and a significant improvement in liver function in both the ST-RILD and LT-RILD mouse models were observed in mice treated with anti-CXCL1 Ab (Fig. 4L).
Taken together, these results suggest that selectively inhibiting CXCL1 can effectively suppress the progression of GSDMD driving RILD through the blockade of neutrophil recruitment.
GSDMD regulates hepatocytes release CXCL1 to recruit neutrophil
To further explore the mechanism of GSDMD regulating CXCL1 to recruit neutrophils during RILD, we first determine the cellular resource of CXCL1. Previous studies have shown that hepatic CXCL1 is mostly released from hepatocytes and HSCs in steatohepatitis and CCL4-induced liver fibrosis29,31. However, our immunofluorescence results demonstrated that CXCL1 was expressed rarely in HSCs (α-SMA+) but mostly in hepatocytes (HNF4α+) in RILD mice (Fig. 5A). This observation was then validated in in vitro experiments. Double immunostaining of CXCL1 and GSDMD-FL revealed that radiation-induced upregulation of GSDMD-FL in hepatocytes was accompanied by a significant elevation of CXCL1 (Fig. 5B). Because GSDMD cleavage mediated the release of cell contents and simultaneously initiated cell death, we investigated whether GSDMD-N mediated CXCL1 release before cell death after irradiation exposure. After 12 h after irradiation, primary hepatocytes showed no significant increased PI uptake, indicating the absence of notable cell death at the same time point (Fig. 5C). While, the CXCL1 levels in the supernatants increased after irradiation in GsdmdFL/FL hepatocytes but decreased in Gsdmd∆Hep hepatocytes (Fig. 5D), suggesting a role for GSDMD-N in CXCL1 release. Next, we investigated whether GSDMD-N facilitated the secretion of cytosolic CXCL1. As the last step of GSDMD cleave requires the “scissors”-caspase and caspase-1 activation is reported to be governed by AIM2, which sense double-strand DNA damage after irradiation32, we also detected irradiation can activate caspase-1 in hepatocytes (Fig. 5E). To further define GSDMD-N function in controlling CXCL1 release, we used caspase-1 inhibitor Ac-YVAD-cmk to prevent the formation of GSDMD-N. To differentiate between intracellular and extracellular CXCL1 levels, we measured intracellular CXCL1 to quantify its retention within the cells, while supernatant CXCL1 levels were measured via ELISA to represent extracellular release. The results showed that following irradiation, CXCL1 release into the supernatant was elevated (Fig. 5F). However, pre-treatment with Ac-YYAD-cmk effectively inhibited CXCL1 release into the supernatant, while intracellular CXCL1 levels increased, indicating that CXCL1 was retained within the cells (Fig. 5E–F). Together, these results indicated irradiation-induced GSDMD-N promoted the secretion of CXCL1.
A Immunofluorescence visualization of CXCL1 (green), HNF4α (red), ɑ-SMA (red) in liver sections from Ctrl and IR-treated mice (n = 5/group). (Scale bars, 200×, 100 μm). B Double-immunostaining for CXCL1 and GSDMD-FL in primary hepatocytes 24 h after irradiation. Confocal analysis demonstrates coexpression of CXCL1 and GSDMD-FL in hepatocytes receiving radiation. Scale bars, 200×, 50 μm (n = 5/group). C Cell viability was assessed by Annexin-V+/PI+. D ELISA assay of CXCL1 release in culture supernatants from GsdmdFL/FL and Gsdmd∆Hep hepatocyte accepted irradiation or not. IR: GsdmdFL/FL vs Gsdmd∆Hep, p = 0.0004. C, D each sample was from 3 biologically independent replicates. E Immunoblot analysis of caspase-1, cleaved-caspase-1, GSDMD-FL/N and CXCL1 expression from GsdmdFL/FL primary mouse hepatocytes treated or untreated with 20 μM Ac-YVAD-cmk, analyzed at 12 h following 8 Gy irradiation (3 biologically independent replicates). F ELISA assay of CXCL1 release in culture supernatants from hepatocytes pretreated without or with 20 μM Ac-YVAD-cmk. IR: Vehicle vs Ac-YVAD-cmk, p = 1.42E-07. G, H Quantification of neutrophil migration. Quantification of neutrophil migration in response to irradiated GsdmdFL/FL hepatocytes pretreated with anti-CXCL1 antibody or not (G), p = 0.003; Quantification of neutrophil migration in response to irradiated Gsdmd∆Hep hepatocytes pretreated with CXCL1 RP or not (H), p = 0.002. F–H, n = 3 biologically independent mice per group. P values were using two-way ANOVA with Fisher’s LSD multiple comparisions test (D, F) and unpaired two-sided Student’s t test (C and G, H). No data were excluded from the analyses. Data are expressed as mean ± SEM. Source data are provided as a Source Data file.
To clarify that GSDMD regulate hepatocyte after irradiation can recruit neutrophils via CXCL1, GsdmdFL/FL hepatocytes administered an anti-CXCL1 Ab showed a significantly reduced ability to recruit neutrophils (Fig. 5G). In contrast, the addition of the recombinant CXCL1 protein (CXCL1 Rp) to Gsdmd∆Hep hepatocytes restored the chemotactic ability of neutrophils (Fig. 5H). In vivo, restoration of GSDMD in Gsdmd∆Hep mice by injecting AAV9-GSDMD-FL, administration of CXCL1-Ab also can inhibit the neutrophil infiltration into liver (Supplementary Fig. 5A–C). Afterwards, early radiation-induced liver injury was reversed (Supplementary Fig. 5D–F). Collectively, these data provide compelling evidence for the regulatory influence of GSDMD on hepatocytes, thereby driving the recruitment of neutrophil through the enhanced expression of CXCL1 upon irradiation.
STAT5A blockade effectively curbs the progression of RILD by suppressing CXCL1 and neutrophil recruitment
Next, we explored the mechanism GSDMD regulate CXCL1. Through comprehensive analysis utilizing the Gene-Hancer database within the GeneCards Suite (http://www.genecards.org/), we scrutinized the regulatory elements encompassing the promoter and enhancer regions of CXCL1. Leveraging integrative analysis with transcriptomic sequencing (GsdmdFL/F vs Gsdmd∆Hep), we unearthed seven putative transcription factors (Fig. 6A), under the influence of GSDMD, that exhibit the potential to bind to the promoter or enhancer regions of CXCL1. Among the examined transcriptional regulators, STAT5A exhibited the most prominent differential expression between GsdmdFL/FL and Gsdmd∆Hep mice (Fig. 6B). To further elucidate the effects of STAT5A activation on RILD, GsdmdFL/FL mice were treated with vehicle or IST5-002, an inhibitor that strongly suppresses STAT5A nuclear translocation and phosphorylation (P-STAT5A) (Supplementary Fig. 6A). Indeed, the administration of IST5-002 resulted in a significant reduction in P-STAT5A in irradiated livers, as assessed by WB (Supplementary Fig. 6B and C). Compared to vehicle control, IST5-002 significantly curbed acute liver injury in ST-RILD mice and reduced weight loss and liver fibrosis in LT-RILD mice (Supplementary and Fig. 6D–I), accompanied by improved liver function, as indicated by serum ALT, AST, and ALB levels (Supplementary Fig. 6J). Moreover, STAT5A inhibition treatment reduced CXCL1 expression and subsequent neutrophil infiltration in vivo (Supplementary Fig. 6K–L).
A Identification of promoters and enhancers of CXCL1 with integrative analysis of transcriptomic sequencing and database within the GeneCards Suite (http://wwww.genecards.org/). B Heatmap of expression of the indicated genes from (A) (n = 5/group). C, D Primary hepatocytes were pretreated with or without 25 μM STAT5A inhibitor IST5-002 for 6 h prior to 8 Gy irradiation. After 24 h, the translocation inhibited ability were determined by the WB assay (C); the culture medium level of CXCL1 was assayed by ELISA (D). Vehicle vs IST5: Ctrl, p = 0.008; IR, p = 4.98E-06. E Luciferase activity assay was performed to measure the binding of STAT5A at the GsdmdFL/FL or the promoter region with mutations (Mut) of Cxcl1 gene with or without IR in HEK 293 T cell. WT-Ctrl vs WT-IR: p = 6.66E-13. F Quantification of neutrophil migration in response to irradiated hepatocytes pretreated with IST5-002 or not. Vehicle vs IST5: Ctrl, p = 0.003; IR, p = 1.71E-05. G Immunoblot analysis of STAT5A and P-STAT5A from GsdmdFL/FL and Gsdmd∆Hep primary hepatocyte exposed to 8 Gy irradiation. H Protein level of STAT5A and P-STAT5A in cytoplasmic extracts and nuclear extracts from 8 Gy irradiated GsdmdFL/FL and Gsdmd∆Hep primary hepatocytes were determined by Immunoblot. I Co-IP analysis for physical interaction of GSDMD and STAT5A in hepatocytes. J ChIP assays were performed to detect the promoter sites of STAT5A binding to Cxcl1. IgG as a negative control for ChIP assay. GsdmdFL/FL: IgG vs stat5a, p = 1.12E-08; stat5a: GsdmdFL/FL vs Gsdmd∆Hep, p = 1.75E-08 C-J, n = 3 biologically independent mice per group. K, L Immunoblot analysis of STAT5A and P-STAT5A from GsdmdFL/FL and Gsdmd∆Hep mice (n = 1, no IR; n = 5, IR). M Immunoblot analysis of STAT5A and P-STAT5A of IR-Gsdmd∆Hep mice administered with AAV9-Control and AAV9-GSDMD-FL (n = 4/group). p values were using two-way ANOVA with Fisher’s LSD multiple comparisions test (D-F and J). No data were excluded from the analyses. Data are expressed as mean ± SEM. Source data are provided as a Source Data file.
Next, we sought to elucidate the role of STAT5A as a regulator of CXCL1. First, IST5-002 significantly reduced the production of CXCL1 in irradiated hepatocytes (Fig. 6C, D). To investigate the regulatory role of STAT5A in Cxcl1 transcription, we constructed luciferase reporter plasmids containing cloned Stat5a binding sites within the Cxcl1 promoter region. We observed an interferon gamma-activated site (GAS)-like core sequence (TTCT/CNA/GGAA) in promoter of mouse Cxcl1 gene (Supplementary Fig. 6M). Strikingly, upon irradiation, we observed a substantial increase in luciferase activity specifically at the STAT5A binding sites. Notably, this effect was markedly diminished when the STAT5A binding site was mutated, highlighting the critical involvement of STAT5A in driving Cxcl1 transcription in response to irradiation (Fig. 6E). In addition, the application of IST5-002 to irradiated hepatocytes effectively inhibited the migration of neutrophils (Fig. 6F). These results indicated that STAT5A regulated the recruitment of neutrophils in a CXCL1-dependent manner.
GSDMD deficiency inhibits the activation of the STAT5A/CXCL1/neutrophil pathway
We next examined the role of GSDMD in regulating STAT5A activation in RILD. In vitro, irradiation-treated hepatocytes exhibited enhanced STAT5A activation, evidenced by the elevated P-STAT5A levels (Fig. 6G). Notably, the absence of Gsdmd also attenuated the nuclear translocation of P-STAT5A induced by irradiation, as observed through nuclear protein analysis (Fig. 6H). Moreover, co-immunoprecipitation (coIP) assay confirmed that GSDMD and STAT5A could directly interact with each other (Fig. 6I). Subsequently, we conducted chromatin immunoprecipitation (ChIP) assays, which revealed that depletion of GSDMD significantly impeded the binding of STAT5A to the promoter region of the Cxcl1 gene (Fig. 6J). This finding underscores the critical involvement of STAT5A in GSDMD-mediated CXCL1 expression. Intriguingly, we also observed that GSDMD deficiency markedly suppressed STAT5A activation in livers affected by RILD (Fig. 6K, L), whereas re-expression of GSDMD-FL reinstated the levels of P-STAT5A in Gsdmd∆Hep mice (Fig. 6M). Collectively, these findings highlight the role of GSDMD in regulating CXCL1 expression through enhancing STAT5A phosphorylation and nuclear translocation.
In response to extracellular stress signals, such as irradiation, several downstream signaling pathways, including extracellular signal-regulated kinases (ERKs), p38 MAP kinase, and c-Jun N-terminal kinase (JNK), are recruited and activated33,34,35. To elucidate the key pathways involved in the upregulation of GSDMD expression, we conducted further investigations. Comparative analysis revealed that, in irradiated hepatocytes, only the phosphorylation of ERK was significantly elevated when compared to the vehicle group. Moreover, the inhibition of ERK using GDC-0994 resulted in a substantial decrease in GSDMD expression (Supplementary Fig. 6N–P). These findings highlight the predominant role of the ERK pathway in mediating the irradiation-induced increase in GSDMD expression.
The therapeutic potential of the GSDMD inhibitor disulfiram in preventing RILD
To determine whether GSDMD-mediated pyroptosis acts as a key therapeutic target, disulfiram (DSF), a drug approved by the FDA for inhibiting pyroptosis, was administered to RILD mice. In RILD, GsdmdFL/FL mice were treated with vehicle ctrl or DSF according to the protocol shown in Supplementary Fig. 7A. Indeed, the administration of DSF resulted in a significant reduction in GSDMD-FL/N in irradiated livers, as assessed by WB and IHC (Fig. 7A and Supplementary Fig. 7B). Moreover, DSF treatment reduced the activation of STAT5A, the expression of CXCL1, and subsequent neutrophil infiltration (Fig. 7A–D), further confirming the molecular mechanisms of GSDMD in RILD, as we discussed above. Of note, in contrast to treatment with the vehicle ctrl, DSF administration significantly curbed acute liver injury in ST-RILD mice and reduced weight loss and liver fibrosis in LT-RILD mice, which was accompanied by an improvement in liver function, as determined by serum ALT, AST, and ALB levels (Fig. 7E–J, and Supplementary Fig. 7C–D).
A Hepatic protein levels of GSDMD-FL, GSDMD-N, STAT5A and P-STAT5A between DSF and vehicle groups were evaluated by Immunoblot. B Representative staining of CXCL1 in liver tissue from vehicle and DSF-treated mice (Scale bars, 40×, 200 μm). n = 4/group, Vehicle-ST; n = 3/group, Vehicle-LT; n = 5/group, DSF. ST-Vehicle vs DSF: p = 1.64E-07; LT-Vehicle vs DSF: p = 4.38E-07. C, D The neutrophil infiltration was evaluated by flow cytometry (C) and MPO immunostaining (D) in vehicle and DSF-treated mice (Scale bars, 200×, 50 μm). (n = 4 mice/group, vehicle; n = 3 mice/group, ST-vehicle; n = 5 mice/group, DSF-LT). CD11b + : ST-Vehicle vs DSF: p = 2.77E-07; LT-Vehicle vs DSF: p = 0.002. E Representative images of HE staining in vehicle or DSF-treated ST-RILD mice. Scale bars, 100×, 50 μm. F–H Representative liver macroscopic images (F), ɑ-SMA staining (G) and sirius-red staining (H) in cohort of LT-RILD mice (Scale bars, 200×, 50 μm). Two-sided and Student’s t test. G-H, n = 3 mice/group, vehicle; n = 5 mice/group, DSF. I Immunoblot analysis of COL1A1, Fibronectin and ɑ-SMA. J Serum levels of ALT, AST, and ALB. n = 5 mice/group, vehicle; n = 6 mice/group, DSF. ST-Vehicle vs DSF: ALT, p = 2.49E-08; AST, p = 2.49E-08; ALB, p = 0.0004. LT-GsdmdFL/FL vs Gsdmd∆Hep: ALT, p = 7.76E-05; AST, p = 7.76E-05; ALB, p = 1.44E-05. K Immunoblot analysis of GSDMD-FL/N, STAT5A and P-STAT5A. L-M Supernatants from vehicle or disulfiram pre-treated primary hepatocyte were collected for LDH (L **** p = 3.6E-05) and CXCL1assay (M, **** p = 6.35E-05). n = 3 biologically independent mice per group for (K–M). p values were using two-way ANOVA with Fisher’s LSD multiple comparisions test (B, C, J, L, M) and unpaired two-sided Student’s t test (D and G-H). No data were excluded from the analyses. Data are expressed as mean ± SEM. Source data are provided as a Source Data file.
The pyroptosis-inhibitory and radioprotective roles of DSF were also noted in irradiated hepatocytes ex vivo. DSF treatment markedly reduced GSDMD-FL/N expression and LDH levels (Fig. 7K, L and Supplementary Fig. 7E). In addition, STAT5A activation and CXCL1 release were both inhibited in DSF-treated hepatocytes (Fig. 7K, M). Ultimately, DSF-treated hepatocytes were protected from radiation toxicities (Supplementary Fig. 7F). These findings suggest that DSF, a pyroptosis inhibitor, is a potential therapeutic strategy for the management of RILD in cancer patients.
Discussion
RILD not only harms the public’s health but greatly hinders radiotherapy’s application8,9. Unfortunately, underlying mechanisms and therapeutic agents were both unrevealed in RILD. In this study, we elucidated that irradiation triggers elevated expression of GSDMD in hepatocytes, thereby exacerbating RILD progression. With the use of genetically and pharmacologically modified models, our data found that IR mediated upregulation of GSDMD activated STAT5A to drive CXCL1 expression while GSDMD-N, a mature form of GSDMD, formed a pore to release the accumulated CXCL1 in the hepatocytes exposed to IR. As a neutrophil chemokine, CXCL1 from hepatocytes recruits neutrophil into liver and promotes RILD progression. Therefore, GSDMD emerges as a pivotal determinant in the regulation of RILD, underscoring its potential as a promising therapeutic target to impede the progression of this condition.
GSDMD is composed of ≈480 amino acids and widely expresses in many organs11,26,36. Previous studies have revealed that GSDMD plays an important role in maintaining homeostasis37, eliminating pathogen infection38,39,40, and regulating the development of diseases13,30,41,42. GSDMD is inactive in steady state, and its function mainly depend on GSDMD-N, which performs a pore-forming function through oligomerization in the plasma membrane. Membrane perforation leads to the release of many cytokines, which activate downstream pathways. Wang et al.37 reported that GSDMD-N pores promote mucin granule secretion in the gut epithelium by mediating Ca2+ entry and help maintain gut homeostasis. In bacterial infection, GSDMD-N drives pyroptosis of infected cells and leads to the release of intracellular bacteria followed by neutrophil-mediated killing38,43. Moreover, the release of IL-33, IL-1β and IL-18 through the pores formed by GSDMD-N also aggravates the progression of allergic airway inflammation42, septic shock44, and colitis45. Hence, GSDMD-FL/N are regarded as critical proteins regulating homeostasis and the outcome of disease. A previous study reported that TBI increased gut and bone marrow expressed GSDMD-FL/N, which correlated positively with the severity of bone loss15. However, the effect of GSDMD-N executed pyroptosis and mediated inflammatory factors release in RILD remains unclear. Here, we revealed that IR mobilize GSDMD-N expression and the pyroptosis in hepatocytes. Of note, the level of GSDMD-N positively correlated with the severity of RILD, and inhibition of GSDMD could effectively alleviate the progression of RILD.
Although the primary finding of this study is the impact of hepatocyte pyroptosis on RILD, it is important to note that radiation can induce various forms of cell death beyond apoptosis. These additional forms of cell death also contribute to radiation injury to some extent. Specifically, DNA damage from irradiation may initiate pyroptosis46 and apoptosis, while reactive oxygen species47 from irradiation are likely initial triggers of ferroptosis in radiation-induced intestine injury (RIII). Furthermore, mitochondrial dysfunction is known to contribute to the development of necroptosis48. These cell death, while distinct, can occur across multiple organs and are not tissue-specific. Notably Zhang et al.46 reported that both pyroptosis and ferroptosis were induced simultaneously in intestinal injury due to irradiation. Recent findings indicate that the effective irradiation dose for inducing pyroptosis and ferroptosis ranges from 10 Gy to 15 Gy over 3-6 days in RIII48,49. In contrast, Xu et al. used a higher dose of 25 Gy50, administered at a later time point, to induce necroptosis. The induction of pyroptosis, ferroptosis, and necroptosis by ionizing radiation varies with both the dosage and the timing of exposure. While these cell death pathways have long been considered to function in parallel with little or no overlap, it is currently clear that apoptosis, necroptosis, and pyroptosis are tightly connected and can cross-regulate each other51,52. Our current study focuses on the role of pyroptosis in exacerbating the severity of RILD, without delineating the impacts of other cell death mechanisms on radiation-induced liver damage. This limitation points to the need for further research to clarify the roles and interrelations of different cell death mechanisms in RILD.
GSDMD-N mediated pyroptosis has been well documented in different cell types and promotes the release of multiple cytokines. Recent studies have demonstrated that GSDMD-N promotes macrophages pyroptosis and release regulator factors, which induces coagulation, leading to disseminated intravascular coagulation in sepsis53,54. GSDMD-driven pyroptosis in myeloid cells also plays a pivotal role in the pathogenesis of hepatic ischemia–reperfusion injury13. Although the role of pyroptosis in immune cells has been extensively studied, its implications in hepatocytes and the resulting impact on the immune microenvironment remain largely unexplored. Wang et al. reported that a natural compound could alleviate CCL4-induced liver injury by inhibiting the pyroptosis of hepatocytes, but the underlying mechanism is uncertain55. In this study, we identified hepatocyte as the critical pyroptotic cell type in RILD. In addition, we also demonstrated that the impact of GSDMD-N in remodeling immune environment via its pore-forming activity. In brief, upon IR, CXCL1 were released from hepatocytes via GSDMD-N cleaved by caspase-1 and recruited neutrophils to change the immune environment of liver.
Our study also has found cleavage-independent function of GSDMD, where it activates the transcription factor STAT5A to drive CXCL1 expression. In support of this cleavage-independent function, other gasdermin family proteins were reported to have cleavage-independent function to regulate the downstream genes. Huang et al. reported that GSDME mediates the transcription factor to enter the nucleus where it promotes the expression of downstream genes17. GSDMB is also found to be localized in the nucleus and regulates the expression of genes by acting as a transcriptional coactivation or enhancer18. These findings highlight the need for further investigation to fully understand the diverse roles of GSDMD.
We also aim to evaluate the efficacy of a protective drug for RILD. Disulfiram is known to be approved by FDA to inhibit GSDMD-N mediated pore formation without affecting the processing of caspase-1/11-GSDMD in a macrophage pyroptosis model induced by LPS/nigericin after 1 h of stimulation. Our findings are not isolated in demonstrating that DSF impacts GSDMD expression. Several studies56,57,58,59 have reported that DSF can inhibit both GSDMD-FL and GSDMD-N expression. We hypothesize that the discrepancy between our results and those of Xia et al. may be due to the longer observation period in our study. While Xia et al. observed effects over a short duration, we extended our observations to 24 h, which might explain the differences in findings. Our experiments confirmed that DSF reduces GSDMD-FL and GSDMD-N levels over an extended period, suggesting a time-dependent effect on GSDMD processing. In addition, other side effects induced by antitumor therapy, including chemotherapy60,61, were reported to be related to GSDMD-mediated pyroptosis, which indicates that pyroptosis is a potential therapeutic target in alleviating the side effects of other anti-tumor therapies in cancer patients. Moreover, DSF has been reported to be effective against diverse cancer types in preclinical studies through p97/Nuclear protein localization protein 4 homolog (NPL4) pathway62,63,64. These results suggest that DSF may not only alleviate RILD, but also further enhance the tumor control rate.
STAT5A is a member of STAT family, a group of latent transcription factors that are activated in response to various cytokine signaling pathways. The predominant mechanism by which STAT5 influences neutrophils, primarily through the action of granulocyte colony-stimulating factor (G-CSF), is by modulating the transcription of genes associated with cell survival and proliferation65. In contrast, STAT3, another member of the STAT family, has been reported to be essential for the chemotaxis of mature neutrophils towards CXCR2 ligands, including CXCL266,67. Our luciferase activity assays and CXCL1 ELISA measurement provide evidence that STAT5A is indispensable for the transcription of cxcl1 following irradiation. Additionally, we discovered that STAT5A plays a crucial role in neutrophil chemotaxis. This collective evidence demonstrates that, as a transcription factor, STAT5A also mediates the recruitment of neutrophils through CXCL1.
In conclusion, our study demonstrated that irradiation induced GSDMD activation in hepatocytes, which plays a crucial role in RILD progression. GSDMD induces pyroptotic phenotype and increased synthesis and release of CXCL1 in hepatocytes. Mechanistically, GSDMD promotes STAT5A into the nucleus which subsequently increased CXCL1 synthesis and GSDMD-N supports CXCL1 release from forming pores. Afterwards, as a chemokine, CXCL1 acts to recruit neutrophils into liver to ultimately aggravate the severity of RILD. Both gene knockout and pharmacological inhibition of GSDMD effectively ameliorate RILD. These findings identify a potential target for the development of effective strategies for treating patients with RILD.
Mehthods
Animal studies
Hepatocyte-specific Gsdmd∆Hep were generated by breeding GsdmdFL/FL with Alb-cre mice on the C57BL/6 J background (GemPharmatech, Nanjing, China). Mice were genotyped with tail DNA using a Mouse Direct PCR kit (Vazyme, PD101-01). Detailed primer sequences were listed in Supplementary Table 1. All animals were acclimatized for 1 week before experiments and housed in a specific pathogen-free environment with a 12-h light-dark cycle and permitted ad libitum consumption of water and a standard chow diet unless otherwise stated. This research was approved by the Institutional Care and Animal Use Committee of Sun Yat-sen University Cancer Center (20110 M).
Mouse model of RILD
Male GsdmdFL/FL and Gsdmd∆Hep mice, aged between seven to ten weeks, were carefully anesthetized with an intraperitoneal injection of pentobarbital (50 mg/kg). A single fraction of 30 Gy hepatic radiation was administered to establish the short-term (ST) radiation-induced liver disease (RILD) model. To explore the long-term (LT) side effects of RILD, mice received five fractions of 6 Gy hepatic radiation every two days and were allowed to spontaneously recover for a period of 10 weeks.
Neutrophils were depleted by daily intravenous injection of anti-Ly6G antibody (25 μg/mouse, i.v.) (clone 1A8, #BP0075-1), combined with anti-rat kappa immunoglobulin (50 μg/mouse, i.v.) (clone MAR 18.5, #BE0122) injected every other day68. CXCL1 was blocked by administration of a mouse CXCL1 neutralizing antibody (100 μg/mouse, i.v.) (R&D systems, Clone 48415) 15 min before irradiation as described previously69. For LT-RILD, mice were injected additionally with CXCL1 neutralizing antibody (25 μg/mouse, i.v.) weekly after the last irradiation. Control mice were injected with the same dosage rat isotype IgG control antibody.
For ST-RILD, GsdmdFL/FL mice were given intraperitoneal injection of IST5-002 (30 mg/kg, MCE), disulfiram (100 mg/kg, MCE) or vehicle 15 min before the first radiotherapy session, continuing for 7 days. For LT-RILD, in addition to the injection of IST5-002 (30 mg/kg) were injected 15 min before radiotherapy and weekly after the end of radiotherapy. Disulfiram (100 mg/kg) was administered 15 min before radiotherapy, followed by weekly injections of 50 mg/kg after the completion of radiotherapy. IST5-002 and disulfiram were dissolved in 10% DMSO and 100% corn oil, while control mice were given vehicle (containing only 10% DMSO and 100% corn oil).
Adeno-associated virus 9 (AAV9) vector that expressed GSDMD-FL (AAV9-GSDMD-FL) and control (AAV9-Control) were purchased from Genechem Company (Shanghai, China). The virus solution was diluted by PBS and then injected into Gsdmd∆Hep mice through the tail vein at a dose of 1×1011 v.g./mouse. The mice were subjected to 30 Gy hepatic irradiation and sacrificed at day 7.
Biochemical measurement
Serum and supernatant levels of alanine aminotransferase (ALT), aspartate aminotransferase (AST), Albumin (ALB) and Lactic dehydrogenase (LDH) were measured with the kits according to the manufacturer’s instructions (Nanjing Jiancheng Bioengineering Institute, Nanjing, China). We acknowledge Biorender.com for providing icons of illustrations.
Immunoblot
Cytoplasmic and nuclear proteins’ extraction were performed according to the manufacturer’s instructions (Minute™ Cytoplasmic and Nuclear Fractionation kit, sc-003). The total protein was extracted from primary hepatocytes and liver tissues as previously described70. Total protein concentration was determined using the Pierce®BCA Protein Assay kit (Thermo Scientific). Equal amounts of protein per sample were separated on 10% SDS-PAGE and transferred to polyvinylidene difluoride membranes. The membranes were blocked with 5% nonfat milk for 1 h at room temperature, followed by probing with corresponding primary antibody over night at 4 °C. Then, the membranes were incubated with HRP-conjugated secondary antibodies for 1 h at room temperature. The specific bands were detected using ECL western blotting substrate (Merck millipore).
qRT-PCR Analysis
Total RNA was extracted from liver tissues using TRIzol reagent (Thermo) according to the manufacturer’s protocol (Invitrogen, USA) and subjected to complementary DNA synthesis. Reverse transcription products of different samples were amplified by LightCycle480 II (Roche) using QPCR Master Mix (LightCycler 480 SYBR Green I Master, Roche) according to the manufacturer’s instructions. Sequences of primers used in this study are summarized in Supplementary Table 1.
Histology, immunohistochemistry (IHC) and immunofluorescence
Connective tissue stain (Sirius Red), Hematoxylin-eosin (HE), and IHC were performed in formalin-fixed paraffin-embedded liver sections or snap-frozen liver pieces, as described previously70. Detailed information about antibodies used is summarized in Supplementary Table 2.
Single cell RNA-sequencing and data analysis
Liver preparation into single cell suspension
To prepare single cell suspensions, mice liver was minced and digested using Collagenase II (Worthington Biochemical Corporation) and DNase (Sigma) and processed through the gentle MACS Dissociator (Miltenyl) utilizing a mouse liver protocol for processing. Following tissue dissociation, cells were passed through a 70 mm filter (Falcon, Corning), washed, and counted with a Cellometer Auto 2000 (Nexcelom).
Single Cell RNA-Sequencing
Single-cell suspensions were loaded onto a 10× Chromium Controller (10× Genomics) and library preparation was performed according to the manufacturer’s instructions for the Single Cell 3’ Library Gel Based-In-Emulsions (10× Genomics). Sequencing was performed on the Illumina HiSeq XTEN platform (Illumina, 150-bp paired-end protocol), according to the manufacturer’s protocol.
Data processing and clustering
Read processing was performed using the 10× Genomics workflow. Briefly, the Cell Ranger Single-Cell Software Suite v3.0.1 was used for demultiplexing, barcode assignment, and unique molecular identifier (UMI) quantification (https://software.10xgenomics.com/single-cell/overview/welcome). The following single-cell data analysis and visualization were mostly performed in Python (version 3) with some analysis done in R (version 4.3.2) with ggplot2 (version 3.3.2).
Scrublet was used to identify potential doublets in each sample, and, after that, cells with scrublet score > 0.4 were filtered out as doublets. Next, additional filtering was performed to discard potential empty droplets and doublet cells using the custom thresholds for number of genes and unique molecular identifiers (UMI) counts: for cells (min 500 and max 5000 genes, min 500 counts and max 50,000 counts). Cells with more than 10% mitochondrial genes expressed were removed as potential low-quality cells.
Scanpy Python package (version 1.9.2) was used to load the cell-by-gene count matrix and perform processing according to the standard pipeline with modifications. Marker genes were identified using different approaches. t-test was applied to identify differentially expressed genes in the given cluster as compared to the rest using sc.tl.rank_gene_groups (method = ‘t-test_overestim_var’, corr_method = ‘benjamini-hochberg’); obtained P values were corrected using the Benjamini–Hochberg method.
ELISA assay of GSDMD-N and CXCL1
The concentration of GSDMD-N in the serum of human were determined by the use of ELISA kits (Jianglai Biological). CXCL1 in the culture medium of hepatocytes was measured by ELISA assay (ThermoFisher) according to the manufacturer’s instruction.
Hepatocyte and neutrophil isolation
Primary hepatocytes were freshly isolated from adult GsdmdFL/FL or Gsdmd∆Hep mouse (20–25 g) according to a modified two-step collagenase perfusion method71. Briefly, the liver was perfused in situ under anesthesia with perfusion buffer (0.5 mM EDTA, 25 mM HEPES in no Ca2+, no Mg2+ and no phenol red HBSS) for 3–5 min at 37 °C followed by perfusion with 25 μg/mL liberase in digestion Solution (25 mM HEPES in HBSS with Ca2+, Mg2+ and phenol red) for 3–5 min Hepatocytes were released in Williams’E solution and filtered through a sterile nylon mesh (70 µm). The cell suspension was enriched and purified by low-speed centrifugation (50 g) and Percoll solution. The hepatocytes were cultured in Williams’ E medium with 10% fetal bovine serum (FBS). After 3 h the medium was replaced with fresh, prewarmed culture medium. Hepatocyte viability was determined by trypan blue exclusion and only preparations with a viability > 90% were used.
Neutrophils were isolated from bone marrow of 7-10 week C57BL/6 N mouse (20–25 g) according to previously published protocol72. Briefly, femur and tibia were placed in a perforated 600ul tube, and after high-speed centrifugation (15000 g) for 15 s at 4 °C, the bone marrow was collected through the hole into a 1.5 mL centrifuge tube below. And then the erythrocytes were removed by resuspension with 0.2% sodium chlorie (NaCl) and 1.6% NaCl. Overlay the bone marrow cell suspension on the top of the density gradient (overlay 3 ml of Histopaque 1077 [Sigma-Aldric] on the 3 mL of Histopaque 1119 [Sigma-Aldric]) and centrifuge for 30 min at 2000 rpm at 25 °C without brake. The neutrophils were collected at the interface of Histopaque 1119 and Histopaque 1077 layers. Washed and cultured with 1640 medium supplemented with 10% FBS and antibiotics.
FACS sorting analysis
The non-parenchymal cells (NPC) were collected from flow-cytometric sorting according to the protocol provided by Manco et al.73. In brief, mice livers were perfused as mentioned above. The cell suspension was enriched and purified by 50 g centrifugation, and the NPCs were collected from supernatant and blocked by bovine serum and then incubated by indicated antibodies. We used Biosciences influx machine to isolate liver sinusoidal endothelial cells (SEC, CD32+CD11b-UV-PI-), monocyte-granulocyte (CD11b+CD32-UV-PI-), hepatic stellate cells (UV+CD11b-CD32-PI-) and T cell (CD3+CD45+PI-).
Hepatocytes treatment in vitro
Freshly isolated murine hepatocytes cells were treated with GDC-0994 (1 μM, MCE), or JNK-IN-8 (1 μM, MCE), or SB 203580 (1 μM, MCE), or Ac-YVAD-cmk (10 μM, MCE), or IST5-002 (20 μM, MCE), or CXCL1 Ab (5 μg/mL, R&D systems), or CXCL1 recombinant protein (200 ng/mlL NovoProtein), or disulfiram (20 μM, MCE) for 6 h or 24 h, and then exposed into 8 Gy irradiation.
Co-culture system of Neutrophils or LX-2 with hepatocytes
Neutrophil migration ability was determined by 24-well transwell migration assay. After 24 h, 1 × 105 hepatocytes received radiation in the bottom chamber, 3 − 4 × 105 neutrophils were added in the top chamber (3 μm pore size). Cells were incubated for additional 6 h at 37 °C. Cells and medium from the lower chamber were harvested for the Ly6G-FITC incubation, and then followed by flowcytometry analysis. The migrated neutrophils were calculated by flowcytometry.
With the same cell numbers, non-irradiated or pre-irradiated hepatocytes (1 × 105) were co-cultured with HSCs (1 × 105, 1:1) in both contact and non-contact manners for 3 days. All cells in the lower chamber in contact manner and cells in above chamber in non-contact manners were all collected and for WB analysis.
Luciferase reporter assays
Luciferase assays were performed as described previously74. Cxcl1 promoter DNA was cloned into luciferase reporter and transfected into HEK293T cell using Lipofectamine 3000 according to the manufacturer’s instructions (Invitrogen, L3000015). The luciferase activity was analyzed by Dual-Luciferase Reporter Assay System (Promega, E1910).
Co-immunoprecipitation (Co-IP)
For co-immunoprecipitation, cells were lysed with lysis buffer and the supernatant was incubated with antibody (anti-GSDMD, anti- STAT5A and then participitated with protein A/G-agarose beads. The beads were washed with and boiled to isolate the protein. Further analysis was carried out by SDS-PAGE gels electrophoresis separation.
Chromatin immunoprecipitation (ChIP) assay
ChIP assay was conducted according to the manufacturer’s protocol (Millipore). In brief, cells (1.5 × 106) were plated into 6 cm culture dishes with or without IR, and samples were harvested for cross-linking and sheared by sonication 24 h later. The resultant chromatin fraction was immunoprecipitated using or negative control anti-IgG (Sigma) antibody. PCR amplification of purified DNA fragments was performed with the indicated specific primers. Forward primer: 5’-GCGTTAATCTGCCACACTCA; Reverse primer: 5’-GAACTGGTTAGAGGCTCTGA.
Plasmid transfection
GSDMD-FL plasmid were purchased from Genechem Company (Shanghai, China). The constructed plasmid was amplified and confirmed by sequencing. Adult mouse hepatocytes were then transfected for 6 h using Lipo 3000 reagent (ThermoFisher, L3000015) according to the manufacturer’s instructions.
Flowcytometry
Single cells of liver were obtained according to previously published protocol with some modifications75. Briefly, tissues passed through a 70 μm cell strainer in 1640 contain 2% FBS and were digested with 0.625 mg/mL collagenase D at 37 °C agitation (200 rpm) for 30 min. The cell suspension was centrifuged at 50 g for 1 min to pellet the hepatocytes. The supernatant enriched liver leukocytes were centrifuged at 700 g for 10 min. The resulting pellet containing leukocytes was resuspended in 3 mL of ACK lysis buffer. Cells were then washed and ready for staining. Cells were then stained by mouse FVD-ef780, CD45-ef450, CD4-BV510, CD8-PE, CD11b-SB600, F4/80-APC, Ly6G-FITC antibody. Flow cytometry data were acquired with Thermofisher (Invitrogen attune nxt) and analyzed by FlowJo software (BD, San Jose, CA).
RNA-Sequencing for Liver Tissue
RNA was extracted from the liver of LT-RILD GsdmdFL/FL and Gsdmd∆Hep mice (n = 5). Total RNA was isolated and purified using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s procedure. After purification and validation, the cDNA libraries were constructed using a SuperScript™ II Reverse Transcriptase Kit (Invitrogen, 18064014) and sequenced using illumina Novaseq™ 6000 following the vendor’s recommended protocol. RNA-seq FASTQ files were processed and evaluated through the Fastp software (https://github.com/OpenGene/fastp). After that, HISAT2 was run (https://ccb.jhu.edu/software/hisat2) to map reads to the reference genome of Homo sapiens. Then, all transcriptomes from all samples were merged to reconstruct a comprehensive transcriptome. After the final transcriptome was generated, StringTie and was used to estimate the expression levels of all transcripts. The differentially expressed mRNAs were selected with fold change > 2 or fold change < 0.5 and with parametric F-test comparing nested linear models (P value < 0.05) by R package edgeR.
Quantification and statistical analysis
All experiments were performed with at least three biological repeats. Analyses were conducted using Graphpad Prism 9.0 software. Results are expressed as the mean ± SEM as indicated, and analysed using Student’s t-test. Comparison of more than two groups was performed by one-way ANOVA with Dunnett’s multiple comparisons test. Groups with two variables were compared with two-way ANOVA with Tukey’s multiple comparisons test. Mann-Whitney test were used to evaluate whether there were any statically difference in terms of continuous variables. The correlations were detected using Spearman correlation. P < 0.05 was considered statistically significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The scRNA-seq and bulk RNA-seq data generated in this study have been deposited in the GEO database under accession codes GSE271691 and GSE277923, respectively. These datasets are publicly available without restriction. Source data are provided with this paper.
References
Li, T., Cao, Y., Li, B. & Dai, R. The biological effects of radiation-induced liver damage and its natural protective medicine. Prog. Biophys. Mol. Biol. 167, 87–95 (2021).
Kumar, P., Wang, P., Farese, A. M., MacVittie, T. J. & Kane, M. A. Metabolomics of multiorgan radiation injury in non-human primate model reveals system-wide metabolic perturbations. Health Phys. 121, 395–405 (2021).
Wong, J. Y. C., Filippi, A. R., Dabaja, B. S., Yahalom, J. & Specht, L. Total body irradiation: guidelines from the international lymphoma radiation oncology group (ILROG). Int J. Radiat. Oncol. Biol. Phys. 101, 521–529 (2018).
Le Pechoux, C. et al. Postoperative radiotherapy versus no postoperative radiotherapy in patients with completely resected non-small-cell lung cancer and proven mediastinal N2 involvement (Lung ART): an open-label, randomised, phase 3 trial. Lancet Oncol. 23, 104–114 (2022).
Ren H., et al. Intraoperative radiotherapy vs concurrent chemoradiotherapy in the treatment of patients with locally advanced pancreatic cancer. Pancreatology. https://doi.org/10.1016/j.pan.2021.04.007 (2021).
Chino, J. et al. The ASTRO clinical practice guidelines in cervical cancer: Optimizing radiation therapy for improved outcomes. Gynecol. Oncol. 159, 607–610 (2020).
Kim, N. et al. Stereotactic body radiation therapy vs. radiofrequency ablation in Asian patients with hepatocellular carcinoma. J. Hepatol. 73, 121–129 (2020).
Kim, J. & Jung, Y. Radiation-induced liver disease: current understanding and future perspectives. Exp. Mol. Med 49, e359 (2017).
Guha, C. & Kavanagh, B. D. Hepatic radiation toxicity: avoidance and amelioration. Semin Radiat. Oncol. 21, 256–263 (2011).
Fischietti, M. et al. Low Radiation environment switches the overgrowth-induced cell apoptosis toward autophagy. Front Public Health 8, 594789 (2020).
Xu, B. et al. Gasdermin D plays a key role as a pyroptosis executor of non-alcoholic steatohepatitis in humans and mice. J. Hepatol. 68, 773–782 (2018).
Lv X., et al. Gasdermin D-mediated pyroptosis suppresses liver regeneration after 70% partial hepatectomy. Hepatol Commun. 9, 2340–2353 (2022).
Li, J. et al. Blocking GSDMD processing in innate immune cells but not in hepatocytes protects hepatic ischemia-reperfusion injury. Cell Death Dis. 11, 244 (2020).
Chen, G. et al. ALKBH5-Modified HMGB1-STING Activation contributes to radiation induced liver disease via innate immune response. Int J. Radiat. Oncol. Biol. Phys. 111, 491–501 (2021).
Xiao, J. et al. Radiation causes tissue damage by dysregulating inflammasome-gasdermin D signaling in both host and transplanted cells. PLoS Biol. 18, e3000807 (2020).
Wu, D. M. et al. Increased susceptibility of irradiated mice to Aspergillus fumigatus infection via NLRP3/GSDMD pathway in pulmonary bronchial epithelia. Cell Commun. Signal 20, 98 (2022).
Lv, J. et al. Gasdermin E mediates resistance of pancreatic adenocarcinoma to enzymatic digestion through a YBX1-mucin pathway. Nat. Cell Biol. 24, 364–372 (2022).
Das, S. et al. GSDMB induces an asthma phenotype characterized by increased airway responsiveness and remodeling without lung inflammation. Proc. Natl Acad. Sci. USA 113, 13132–13137 (2016).
Du, S. et al. DNA sensing and associated type 1 interferon signaling contributes to progression of radiation-induced liver injury. Cell Mol. Immunol. 18, 1718–1728 (2021).
Kim, J., Wang, S., Hyun, J., Guy, C. D. & Jung, Y. Hedgehog Signaling is Associated with Liver Response to Fractionated Irradiation in Mice. Cell Physiol. Biochem 40, 263–276 (2016).
Shi, Y. et al. Host Gasdermin D restrains systemic endotoxemia by capturing Proteobacteria in the colon of high-fat diet-feeding mice. Gut Microbes Jan.13, 1946369 (2021).
Ma, C. et al. Gasdermin D in macrophages restrains colitis by controlling cGAS-mediated inflammation. Sci. Adv. 6, eaaz6717 (2020).
Ding, C. et al. A Cell-type-resolved Liver Proteome. Mol. Cell Proteom. 15, 3190–3202 (2016).
Wang, Y. et al. Chemotherapy drugs induce pyroptosis through caspase-3 cleavage of a gasdermin. Nature 547, 99–103 (2017).
Karmakar, M. et al. N-GSDMD trafficking to neutrophil organelles facilitates IL-1beta release independently of plasma membrane pores and pyroptosis. Nat. Commun. 11, 2212 (2020).
Shi, H. et al. GSDMD-Mediated Cardiomyocyte Pyroptosis Promotes Myocardial I/R Injury. Circ. Res 129, 383–396 (2021).
Sollberger G., et al. Gasdermin D plays a vital role in the generation of neutrophil extracellular traps. Sci Immunol. https://doi.org/10.1126/sciimmunol.aar6689 (2018).
Mills, E. L. et al. UCP1 governs liver extracellular succinate and inflammatory pathogenesis. Nat. Metab. 3, 604–617 (2021).
Chang, B. et al. Short- or long-term high-fat diet feeding plus acute ethanol binge synergistically induce acute liver injury in mice: an important role for CXCL1. Hepatology 62, 1070–1085 (2015).
Jiang K., et al. Gasdermin D inhibition confers antineutrophil-mediated cardioprotection in acute myocardial infarction. J Clin Invest. https://doi.org/10.1172/JCI151268 (2022).
Shi W. P., et al. CD147 Promotes CXCL1 Expression and Modulates Liver Fibrogenesis. Int. J. Mol. Sci. 10, 19 (2018).
Hu, B. et al. The DNA-sensing AIM2 inflammasome controls radiation-induced cell death and tissue injury. Science 354, 765–768 (2016).
Su, A. R. et al. BX-795 inhibits HSV-1 and HSV-2 replication by blocking the JNK/p38 pathways without interfering with PDK1 activity in host cells. Acta Pharm. Sin. 38, 402–414 (2017). Mar.
Ala, M. et al. Sildenafil improves radiation-induced oral mucositis by attenuating oxidative stress, NF-kappaB, ERK and JNK signalling pathways. J. Cell Mol. Med 26, 4556–4565 (2022).
Liu, J. et al. 3,4,5-O-tricaffeoylquinic acid alleviates ionizing radiation-induced injury in vitro and in vivo through regulating ROS/JNK/p38 signaling. Environ. Toxicol. 37, 349–361 (2022).
Burdette, B. E., Esparza, A. N., Zhu, H. & Wang, S. Gasdermin D in pyroptosis. Acta Pharm. Sin. B 11, 2768–2782 (2021).
Zhang, J. et al. Epithelial Gasdermin D shapes the host-microbial interface by driving mucus layer formation. Sci. Immunol. 7, eabk2092 (2022).
Shi, J. et al. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 526, 660–665 (2015).
Aglietti, R. A. & Dueber, E. C. Recent insights into the molecular mechanisms underlying pyroptosis and gasdermin family functions. Trends Immunol. 38, 261–271 (2017).
Luan, J. & Ju, D. Inflammasome: a double-edged sword in liver diseases. Front Immunol. 9, 2201 (2018).
Silva, C. M. S. et al. Gasdermin D inhibition prevents multiple organ dysfunction during sepsis by blocking NET formation. Blood 138, 2702–2713 (2021).
Chen, W. et al. Allergen protease-activated stress granule assembly and gasdermin D fragmentation control interleukin-33 secretion. Nat. Immunol. 23, 1021–1030 (2022).
Miao, E. A. et al. Caspase-1-induced pyroptosis is an innate immune effector mechanism against intracellular bacteria. Nat. Immunol. 11, 1136–1142 (2010).
Kang, R. et al. Lipid Peroxidation Drives Gasdermin D-Mediated Pyroptosis in Lethal Polymicrobial Sepsis. Cell Host Microbe 24, 97–108 e4 (2018).
Ma, C. Y. D. et al. Gasdermin D in macrophages restrains colitis by controlling cGAS-mediated inflammation. Sci. Adv. 6, eaaz6717 (2020).
Zhang, Y. et al. STING-Dependent Sensing of Self-DNA Driving Pyroptosis Contributes to Radiation-Induced Lung Injury. Int J. Radiat. Oncol. Biol. Phys. 117, 928–941 (2023).
Zhang, F. et al. Activation of pyroptosis and ferroptosis is involved in radiation-induced intestinal injury in mice. Biochem Biophys. Res Commun. 631, 102–109 (2022).
Yi, J. et al. PTPMT1 protects cardiomyocytes from necroptosis induced by gamma-ray irradiation through alleviating mitochondria injury. Am. J. Physiol. Cell Physiol. 324, C1320–C1331 (2023).
Wu, D. et al. Protective Effects of Flagellin A N/C Against Radiation-Induced NLR Pyrin Domain Containing 3 Inflammasome-Dependent Pyroptosis in Intestinal Cells. Int J. Radiat. Oncol. Biol. Phys. 101, 107–117 (2018).
Xu, Y. et al. Nrf2 alleviates radiation-induced rectal injury by inhibiting of necroptosis. Biochem Biophys. Res Commun. 554, 49–55 (2021).
Bertheloot, D., Latz, E. & Franklin, B. S. Necroptosis, pyroptosis and apoptosis: an intricate game of cell death. Cell Mol. Immunol. 18, 1106–1121 (2021).
Ruera, C. N. et al. Coexistence of apoptosis, pyroptosis, and necroptosis pathways in celiac disease. Clin. Exp. Immunol. 214, 328–340 (2023).
Yang, X. et al. Bacterial endotoxin activates the coagulation cascade through gasdermin D-dependent phosphatidylserine exposure. Immunity 51, 983–996 e6 (2019).
Wu, C. et al. Inflammasome activation triggers blood clotting and host death through pyroptosis. Immunity 50, 1401–1411 e4 (2019).
Wang, J. et al. Direct inhibition of GSDMD by PEITC reduces hepatocyte pyroptosis and alleviates acute liver injury in mice. Front Immunol. 13, 825428 (2022).
Deng, W. et al. Disulfiram suppresses NLRP3 inflammasome activation to treat peritoneal and gouty inflammation. Free Radic. Biol. Med 152, 8–17 (2020).
Hu, S., Wang, L., Xu, Y., Li, F. & Wang, T. Disulfiram attenuates hypoxia-induced pulmonary hypertension by inhibiting GSDMD cleavage and pyroptosis in HPASMCs. Respir. Res. 23, 353 (2022).
Wu, J. et al. Treatment of Severe Acute Pancreatitis and Related Lung Injury by Targeting Gasdermin D-Mediated Pyroptosis. Front Cell Dev. Biol. 9, 780142 (2021).
Liu, M. et al. Caffeic acid, but not ferulic acid, inhibits macrophage pyroptosis by directly blocking gasdermin D activation. MedComm (2020) 4, e255 (2023).
Li, Y. et al. Activation of GSDMD contributes to acute kidney injury induced by cisplatin. Am. J. Physiol. Ren. Physiol. 318, F96–F106 (2020).
Tavakoli Dargani, Z. & Singla, D. K. Embryonic stem cell-derived exosomes inhibit doxorubicin-induced TLR4-NLRP3-mediated cell death-pyroptosis. Am. J. Physiol. Heart Circ. Physiol. 317, H460–H471 (2019).
Skrott, Z. et al. Alcohol-abuse drug disulfiram targets cancer via p97 segregase adaptor NPL4. Nature 552, 194–199 (2017).
Iljin, K. et al. High-throughput cell-based screening of 4910 known drugs and drug-like small molecules identifies disulfiram as an inhibitor of prostate cancer cell growth. Clin. Cancer Res. 15, 6070–6078 (2009).
Chen, D., Cui, Q. C., Yang, H. & Dou, Q. P. Disulfiram, a clinically used anti-alcoholism drug and copper-binding agent, induces apoptotic cell death in breast cancer cultures and xenografts via inhibition of the proteasome activity. Cancer Res 66, 10425–10433 (2006).
Kimura, A. et al. The transcription factors STAT5A/B regulate GM-CSF-mediated granulopoiesis. Blood 114, 4721–4728 (2009).
Li, H. S. & Watowich, S. S. Innate immune regulation by STAT-mediated transcriptional mechanisms. Immunol. Rev. 261, 84–101 (2014).
Nguyen-Jackson, H., Panopoulos, A. D., Zhang, H., Li, H. S. & Watowich, S. S. STAT3 controls the neutrophil migratory response to CXCR2 ligands by direct activation of G-CSF-induced CXCR2 expression and via modulation of CXCR2 signal transduction. Blood 115, 3354–3363 (2010).
Boivin, G. et al. Durable and controlled depletion of neutrophils in mice. Nat. Commun. 11, 2762 (2020).
Su, L. et al. Kupffer cell-derived TNF-alpha promotes hepatocytes to produce CXCL1 and mobilize neutrophils in response to necrotic cells. Cell Death Dis. 9, 323 (2018).
Wei, G. et al. Synthetic human ABCB4 mRNA therapy rescues severe liver disease phenotype in a BALB/c.Abcb4(-/-) mouse model of PFIC3. J. Hepatol. 74, 1416–1428 (2021).
Charni-Natan, M. & Goldstein, I. Protocol for primary mouse hepatocyte Isolation. STAR Protoc. 1, 1000s86 (2020).
Swamydas, M. & Lionakis, M. S. Isolation, purification and labeling of mouse bone marrow neutrophils for functional studies and adoptive transfer experiments. J. Vis. Exp. 10, e50586 (2013).
Manco, R. et al. Reactive cholangiocytes differentiate into proliferative hepatocytes with efficient DNA repair in mice with chronic liver injury. J. Hepatol. 70, 1180–1191 (2019).
Wang, X. et al. UDP-glucose accelerates SNAI1 mRNA decay and impairs lung cancer metastasis. Nature 571, 127–131 (2019).
Lee, J., Byun, J., Shim, G. & Oh, Y. K. Fibroblast activation protein activated antifibrotic peptide delivery attenuates fibrosis in mouse models of liver fibrosis. Nat. Commun. 13, 1516 (2022).
Acknowledgements
This work is supported by National Natural Science Foundation of China (NSFC) (No. 82072029, 82103771), the top-notch young talents of the national “Ten Thousand Program” (No.2019-04), Shenzhen Science and technology (No. JCYJ20240813150317023), and Research Start-up Fund of Post-doctoral of SAHSYSU (00302593108). We acknowledge BioRender.com for providing icons of illustrations.
Author information
Authors and Affiliations
Contributions
Z.P., Yi.Z., and X.W. contributed to the design and data interpretation. Z.P., Yi.Z., X.W. contributed to revise article critically for important intellectual content and final approve the version to be published. A.D., G.W., Y.D. and Z.L. contributed to data generation and manuscript draft. Yuhao Tang, Yunyan.Ling, Shuping Li and Yong Chen contributed to data generation.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Liying Li, Kyle C. Cuneo, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Dong, A., Wei, G., Liang, Z. et al. Gasdermin D aggravates a mouse model of radiation-induced liver disease by promoting chemokine secretion and neutrophil recruitment. Nat Commun 16, 6064 (2025). https://doi.org/10.1038/s41467-025-61397-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-61397-7