Abstract
Doxycycline pre-exposure prophylaxis (doxyPrEP) has shown potential in preventing bacterial sexually transmitted infections, but the impact on the microbiome is unknown. This study assessed rectal microbiome changes over 48 weeks in 41 participants on HIV PrEP (tenofovir disoproxil fumarate/emtricitabine) enrolled in an open-label, randomized pilot trial comparing immediate (100 mg PO daily started immediately and continued to week 48) versus deferred doxyPrEP (100 mg PO daily starting at week 24, continued to week 48) in HIV-negative gay and bisexual men (Clinical Trial #: NCT02844634). Primary study outcomes included feasibility, adherence, and tolerability of the dual PrEP regimen, while exploratory outcomes included rectal microbiome changes. We performed 16S rRNA sequencing from participants that collected baseline, week 24, and week 48 samples. Microbial composition did not significantly change over time in either study arm as measured by individual taxa levels, or alpha and beta diversity at the genus level. A slight decrease ( < 10%) in alpha diversity was observed at the phylum level in the immediate arm, but not the deferred arm. This study shows doxyPrEP use results in minimal compositional changes in the microbiome over 12 months. Further research is needed to explore the impact of doxycycline for STI prevention on microbiome function and antimicrobial resistance.
Similar content being viewed by others
Introduction
Globally, sexually transmitted infections (STI) continue to increase significantly, notably the bacterial STIs syphilis, chlamydia and gonorrhea1. STIs often disproportionately burden key populations, including gay, bisexual and other men who have sex with men (GBM), along with transgender women2. Three recent clinical trials in GBM and transgender women have demonstrated the efficacy of doxycycline taken as post-exposure prophylaxis (doxyPEP) for STI prevention (when compared to standard of care), demonstrating an overall reduction in incidence ~70%3,4,5. Subsequent analyses of ‘real-world’ doxyPEP use have demonstrated reductions in chlamydia and syphilis of 50–80%, with no to modest reductions in gonorrhea6,7. Daily doxycycline taken as STI pre-exposure prophylaxis (doxyPrEP) has also shown promise for STI prevention, though these were smaller pilot studies with small participant numbers8,9,10. Doxycycline is a tetracycline antibiotic that has been in use since the 1960s, and is safe, inexpensive, generally well-tolerated, and is a recommended treatment for both syphilis and chlamydia11. DoxyPEP and doxyPrEP represent a tremendous advancement in STI prevention, and based on the clinical trial data, doxyPEP is already being formally recommended by various organizations, with formal guidelines published by the US Centers for Disease Control and Prevention12.
Though doxyPEP remains the intervention with more evidence for STI prevention, clinical trials examining doxyPrEP have been ongoing for a decade, demonstrating promising efficacy data as well8,9,10. Both interventions are currently being compared in a large, multi-center Canadian non-inferiority clinical trial (NCT04762134), which will comparatively examine efficacy, tolerability, and preferences. As with HIV PrEP, where the concept of choice between different modalities (e.g., daily vs. on-demand; oral vs. injectable) is key to uptake, adherence and ultimately efficacy13, the same can likely be said about doxycycline for STI prevention. Additionally, one cannot assume that one will necessarily be preferred over the other by patients, as evidenced by previous work demonstrating high acceptability of both doxyPEP and doxyPrEP among GBM14, and the ongoing uncertainty around comparative tolerability and acceptability between doxyPEP and doxyPrEP.
Despite the clear efficacy data, many unresolved concerns exist, key amongst these are the impacts of doxyPEP and doxyPrEP on antimicrobial resistance and the human microbiome15. The human microbiome refers to the collection of microorganisms performing essential functions for immune development, defense against pathogens, and nutrient metabolism and plays a key role in diverse health conditions, including asthma, obesity, inflammatory bowel disease, and allergies16,17,18. Tetracyclines—and specifically doxycycline—appear to be relatively safe for the gut microbiome, based on their low likelihood of causing Clostridioides difficile infection19,20, the main cause of healthcare-associated diarrhea, which results from antibiotic-associated perturbations in the intestinal microbiome and a major cause of morbidity and mortality worldwide21. Few studies have directly examined the impact of doxycycline on the gut microbiome, and these have generally included few samples from a small number of participants. One such study examined gut microbiome changes in stool samples from 10 individuals administered doxycycline and probiotics versus an equal number of controls receiving probiotics alone22. In those receiving doxycycline, a reduction in microbial diversity was seen, notably a decrease in Bifidobacterium, an important gut commensal. The treatment group also had a higher proportion of tetracycline-resistant bacteria than the control group. Some studies using in vitro methodologies have been variable, where some have shown little-to-no changes in microbial diversity23 while others have shown changes to important gut microbiota such as Lactobacillaceae, Bacteroidaceae and Enterobacteriaceae24. A recent sub-analysis of samples from 89 participants of the US-based DoxyPEP trial4 showed that doxycycline use did not measurably impact the relative abundance, alpha or beta diversity in the rectal microbiome, but did increase the proportion of tetracycline antimicrobial resistance genes in rectal microbiota after 6 months of doxy-PEP use25. These microbiome changes were more pronounced in individuals who were using 25 doses of 200 mg or more in a 6-month time period, suggesting that dosage is an important factor. Antibiotic exposures for doxyPrEP in comparison to doxyPEP for STI prevention would likely be higher given the daily dosing regimen, though a very frequent doxyPEP user (e.g., using 4 weekly, 200mg doses) could theoretically surpass the weekly dosage of a daily user (i.e., 7 × 100 mg). For example, in the US DoxyPEP trial4 where microbiome effects were observed equates to an average doxycycline exposure of 4 × 200 mg/month (~800 mg/month), in comparison to a lower daily dosage (100 mg/daily, 3000 mg/month) but higher frequency and duration used in a recently published doxyPrEP study26. This may result in differential impacts on the gut mucosal microbiome between doxyPrEP and doxyPEP strategies.
Though work is currently ongoing in the abovementioned doxyPEP clinical trials to characterize microbiome changes in study participants, to our knowledge, there is no published data on microbiome impacts from doxycycline prophylaxis used for STI prevention. As the broader rollout of doxyPEP occurs, and as alternative doxycycline-based STI prevention options like doxyPrEP are explored, it is critical that these yet-unknown impacts are further characterized in order to better inform the appropriate implementation of these novel prevention tools. Studying microbiome changes with doxyPrEP could be further informative for doxyPEP strategies, as it provides additional, longer-term data on potential impacts of doxycycline that differ in dosing levels and dosing frequency.
In this study, we systematically characterize the compositional microbiome of participants on HIV PrEP enrolled in a previously published26 doxycycline PrEP study over a 48-week study period. Here we show minimal changes in the rectal microbiome composition of study participants using doxyPrEP over 12 months.
Results
Rectal microbiome composition and diversity of DuDHS study participants
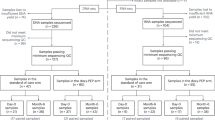
A total of 41 participants, who had been enrolled and followed-up in the dual daily HIV and STI PrEP (DuDHS) trial10, were included in microbiome study as outlined in the Materials and Methods section (Fig. 1). The DuDHS study was an open-label randomized trial of immediate (100 mg PO daily started immediately and continued to week 48) versus deferred doxyPrEP (100 mg PO daily starting at week 24, continued to week 48) in HIV-negative GBM and transgender women receiving daily tenofovir dispoproxil fumarate/emtricitabine (TDF/FTC) for HIV PrEP. Baseline characteristics of study participants in the microbiome study are presented in Table 1. The median age of participants in the immediate and deferred arms was 32 (interquartile range [IQR]: 30–37) and 37 (IQR: 26–44), respectively. Of the 39 (95.1%) participants who identified as cisgender males. Fourteen (50.0%) and 19 (67.9%) participants had previous gonorrhea and chlamydia, respectively. Immediate and deferred arm participants had a median of 6.5 and 9 sexual partners, respectively, in the preceding six months. Concomitant non-doxycycline antibiotic exposures were reported in 11 individuals during the study, 5 in the deferred arm and 6 in the immediate arm. There were no statistically significant differences in self-reported adherence between treatment arms for either doxycycline or TDF/FTC over time (data not shown).
There were three different timepoints when rectal swab samples were collected in the DuDHS study: baseline, week 24 and week 48. At week 24, the immediate arm would have received 24 weeks of doxyPrEP, whereas the deferred arm would have received none. At week 48, the immediate and deferred arms would have received 48 and 24 weeks, respectively, of doxyPrEP. Rectal swab specimens were not available from 11 participants of the original trial for analysis as outlined in the Materials and Methods. We characterized the microbiome composition of study participants by 16S rRNA sequencing that completed all 3 timepoints in the study, which included 22 participants in the immediate arm and 19 in the deferred doxycycline study arms. Each sample was run in duplicate to calculate the average taxa abundance for downstream analysis.
A total of 311 distinct taxa were identified in rectal swab samples (Supplementary Data 1). The most abundant taxa detected across all samples included Finegoldia (mean 11.8%), followed by Prevotella (mean 9.5%), Corynebacterium (mean 7.8%), and Streptococcus (mean 6.5%) (Fig. 2A). The proportion of major taxa within each study participant is shown in Fig. 2A, where most individuals showed relative similarity in bacterial composition during the study period. An examination of beta-diversity, which compares similarity or dissimilarity between microbial communities, showed almost complete overlap of immediate and deferred doxyPrEP study arms with respect to bacterial composition at the baseline visit (R2 = 0.81%, effect size of study arm differences) (Fig. 2B). A univariate analysis did not identify any differences in the proportions of individual microbial taxa between study arms at baseline (Fig. 2C, FDR > 0.05; Supplementary Data 2). The overall alpha diversity between study arms was also similar (Shannon’s H: Deferred: 2.88 vs Immediate: 2.74, p = 0.91) (Fig. 2D). We also evaluated differences in higher order classifications as an exploratory analysis. Alpha-diversity between study arms at baseline was also consistent when comparing at the genus, family, order, class, and phylum levels (Supplementary Fig. 1A–E). Age of study participants did not associate with alpha diversity (R2 correlation coefficient = 0.000008, p = 0.956) nor any individual taxa (FDR > 0.05). The number of sexual partners and sexual frequency did not associate with alpha diversity (R2 < 0.001, p = 0.940 and 0.998, respectively). However, the number of sexual partners and sexual frequency correlated with increased abundance of 9 lower abundance taxa (<0.001% by proportion), including Phascolarctobacterium and Closteridiales_Incertae_Sedis_XIII_unclassified with number of partners (R2 = 0.45 and 0.41, FDR adj p < 0.05, respectively), and Gemmatimonas, Pseudonocardia, Lachnoanerobaculum, Brachyspira, Cerasicoccus, Veillonella, and Stomatobaculum with sexual frequency (R2 > 0.39 to 0.75, FDR adj. p < 0.05). The effect of study arm assignment, age, number of sexual partners, sexual frequency, and other concomitant antibiotics (non-doxycycline) contributed minimally to overall compositional differences in the microbiome (<4%) compared to inter person differences of 68.8% (Fig. 2E). Overall, study participants that were randomized to each study arm were not significantly different with respect to microbial taxa, diversity, or composition of the rectal microbiome, and the effects of sex behaviors were restricted to some lower abundance taxa.
The composition of the rectal microbiome was determined using 16S rRNA sequencing. A Microbial composition of rectal samples of each study participant in each study arm (immediate (n = 22) and deferred (n = 19)) at their baseline visit (BL), week 24, and week 48. Different colors represent the predominant microbial taxa. Average plots of each study arm are shown on the right. B Beta-diversity of baseline microbiome profiles of study participants in each study arm (Bray–Curtis distance metric). Two-sided PERMANOVA p value is shown along with an R2 value to reflect the effect size. C Differential abundance analysis of individual taxa in each study arm at baseline. Blue dots represent those P < 0.05 although none passed FDR. Each dot represents the average abundance value from two technical replicates from a single participant. Differences between groups were determined by Mann–Whitney U test (two-sided) adjusting for multiple comparisons. D Alpha diversity (Shannon’s H) of each study arm at baseline visit. Each data point represents the average of two technical replicates from each study participant. Differences between groups were determined by Mann–Whitney U test (two-sided) adjusting for multiple comparisons. Median and interquartile (IQR) ranges are shown. E Study participant covariates and effect size they have on beta diversity (R2 values shown as determined by PERMANOVA).
Rectal microbiome changes with daily doxycycline
We first evaluated whether there were any changes in microbiome diversity over time in each study arm. There were no intra-person changes in alpha diversity in either the immediate or deferred doxycycline study arms between baseline, week 24, and week 48, when comparing bacterial taxa at the genus and family level (Fig. 3A, B). However, in the immediate arm there was a 7.9% (p = 0.039), 6.2% (p = 0.046), and a 9.9% (p = 0.0019) decrease in alpha-diversity at the order, class, and phylum level (Fig. 3C–E). There were no changes in alpha diversity in the deferred arm.
Rectal microbiome composition was evaluated from rectal swab samples collected at baseline, 24 and 48 weeks after randomization to either the immediate (n = 22) or deferred (n = 19) doxyPrEP study arms. A comparison of alpha diversity measures (Shannon’s H) over time in each study arm is shown at the genus (A), family (B), order (C), class (D), and phylum level (E). Box plots show the median, upper and lower quartiles. Differences between follow-up timepoints were determined using Wilcoxon signed rank tests (two-sided) in paired comparisons, and across all three timepoints using Kruskal–Wallis test.
We next compared changes in beta diversity (microbial composition) in study participants. A heatmap in Fig. 4A shows the top 20 taxa identified by abundance in each study participant sample in the study. These taxa did not cluster by study arm, study timepoint, age, sex behaviors, or concurrent non-doxycycline antibiotic exposures. Clusters were largely driven by the relative abundance of Prevotella, Finegoldia, Ruminococcaceae, Faecalibacterium, and other high-abundance taxa. We did not observe any differences in beta diversity in the deferred or immediate study arms at week 24 or week 48 when compared to baseline. Study participants showed little change in community composition between timepoints, as shown in Fig. 4B–E (p > 0.05). The amount of variance explained by treatment arm on beta diversity at week 24 and 48 was 1.47% to 1.64% in the deferred arm, and 1.03% to 0.679% in the immediate arm, respectively. A comparison of beta-diversity between arms at weeks 24 and 48 also did not show any significant differences in microbiome composition (R2 = 1.11% and 1.30% variance between groups, p = 0.88 and p = 0.82, respectively) (Fig. 4F, G). Overall, these influences are considered very weak (<2%)27, and the composition of microbial communities in study participants remained relatively similar over the course of the study.
Rectal microbiome composition at baseline, 24 and 48 weeks after randomization to either the immediate or deferred doxycycline study arms. A A heatmap showing top 20 most abundant taxa in all study participant samples. Study arm, study timepoint, sex behaviors, age, and concomitant antibiotic usage (non-doxycycline) are overlaid on the top. Clustering is largely driven by predominant taxa. A comparison of beta diversity measures (Bray–Curtis) over time within each study arm (B–E) and between study arms (F–G) are shown. The distance between triangles and circles reflects the relative change in microbiome composition, and degrees of significance represented by two-sided PERMANOVA p values. An R2 value is provided to show the effect size. BL Baseline.
We next investigated whether there were changes in the abundance of individual taxa within each study arm. We did not observe any significant changes in microbial taxa abundances in either the immediate or deferred study arm over the study period of 24 and 48 weeks that passed a false discovery rate by Wilcoxon signed rank tests (Fig. 5A–D). To ensure robustness of these analyses, we further validated these findings using a consensus approach applying three different differential abundance testing methods tests, including Wilcoxon rank sign tests, ANCOM-II, and ALDEx228 in an unadjusted analysis (Supplementary Data 3), and ANCOM-II and a linear mixed effects model for adjusted analysis (Supplementary Data 4). Adjusted models included participant age, sex partners, sex frequency, and concomitant non-doxycycline antibiotics usage during the trial (Supplementary Data 5). There were no taxa that met an FDR-adjusted significance threshold in 2 or more methods as differentially abundant in either study arm. As an exploratory analysis, we relaxed our selection criteria to include any taxa identified by any two tests below a p value of 0.05 in the same comparison for further examination. We identified 5 taxa that met this consensus criteria in the unadjusted analysis, including Fusobacterium, Campylobacter, Prevotellaceae, Intestinibacter, and Solobacterium (Supplementary Data 3). However, in an adjusted analysis (Supplementary Data 4), Fusobacterium, Prevotellaceae, and Solobacterium, Intestinibacter, and Lactonifactor were no longer significant (p < 0.05) when adjusting for sex behaviors in a multivariable model. Campylobacter was the only taxon that met this non-FDR cutoff threshold in two tests (p < 0.05) in the adjusted analysis. Graphical visualization of Campylobacter showed this taxon to decrease at week 24 and at week 48 (97.3% and 81.8% lower, respectively p < 0.05) in the immediate arm, but differences were not observed in the deferred arm at any timepoint (Supplementary Fig. 2) but rather increased in abundance at week 24 and 48 (186% and 153% higher, respectively). These changes seemed to be driven by a few individuals with higher levels at the baseline visit in the immediate arm. Overall, these taxa changes did not appear consistent with effects expected with the same dosage exposure of doxycycline at week 24 of the immediate arm and week 48 of the deferred arm. These observations were consistent when comparing at higher classification levels, including the genus, family, order, class, and phylum level in the deferred (Supplementary Fig. 3) or immediate arm (Supplementary Fig. 4). Overall, these data did not indicate any large changes in individual taxa with doxycycline PrEP.
Differential abundance analysis of microbial taxa for each participant was performed between timepoints in each study arm using Wilcoxon signed-rank tests, adjusted for multiple comparisons. Volcano plots show relative abundance of individual microbial taxa differences (genus) along the x axis, with the p value statistic along the y axis. Shown are analysis for A Deferred arm, baseline versus week 24; B deferred arm, baseline versus week 48; C immediate arm, baseline versus week 24; D Immediate arm, baseline versus week 48. Dots in blue are those that were below a p value significance threshold of 0.05, although none passed a 5% FDR. BL Baseline.
Discussion
In this study, we characterized the compositional rectal microbiome of GBM enrolled in a study of dual HIV PrEP and doxyPrEP for bacterial STI prevention, with a focus on comparing the immediate and deferred doxyPrEP arms. We show that there were no differences or changes in the composition of major microbial taxa in study participants over a 48-week period in either study arm. Further, we show that there is no difference in the microbial composition of study participants between the deferred and immediate study arms at weeks 24 and 48, the timepoint where the immediate arm has been on the drug for 6 months, but the deferred arm has not yet seen the drug. While there was a minor decrease (<10%) of alpha-diversity at the order, class, and phylum level at week 48 in the immediate arm, there was not observed in the deferred arm nor at the individual taxa level. These results are consistent with and build upon the recently published work from the DoxyPEP study, demonstrating no differences in alpha- or beta-diversity over a six-month period in a sample of those using doxyPEP versus those not on doxyPEP25, and add further longer-term data extended to 48 weeks. A key difference between the aforementioned doxyPEP study and the DuDHs study is that the DuDHS participants had higher exposure to doxycycline (100 mg daily, 3000 mg monthly) in comparison to doxyPEP participants (i.e., median of 800 mg monthly)4, which would presumably have a larger effect on microbiome composition. However, changes in composition were not observed in DuDHS participants, and the calculated impact on variance on microbiome beta diversity was <2%, which is considered minor. Our study is among the first to provide key data on microbiome impacts from doxycycline prophylaxis, and to our knowledge, is the first to provide data on doxyPrEP, and the first to provide outcomes longitudinally to 48 weeks.
Gut microbiome studies of GBM populations have observed differences in the predominance of certain microbial taxa. The predominant microbiota in DuDHS study participants includes both Prevotella and Finegoldia, both of which have been observed in other studies of GBM. Prevotella belongs to the Bacteroidota phylum and is common on mucosal surfaces including the gut, oral compartment, and vagina. The higher prevalence of Prevotella in the DuDHS cohort is comparable to other studies of GBM study populations in the U.S., Europe, and Kenya29,30,31 and has been linked to sex preferences and receptive anal intercourse31. Prevotella has effects on gut mucosal immunity, including increased innate immune activation, mucosal inflammation, and T cell activation32,33. Finegoldia is a gram-positive anaerobic cocci and commonly found on skin and mucosal surfaces, including the gastrointestinal tract and oral cavity. Certain species such as F. magna are considered opportunistic pathogens, as it has been isolated in various infections including those of wounds, bacterial vaginosis, pneumonia, and others34,35,36,37,38. F. magna is also pro-inflammatory and can activate neutrophils39. Finegoldia has also been shown to be predominant in other studies of GBM40. We also identified nine low-abundance taxa to be positively associated with increased sexual frequency and the number of partners, consistent with changes in the microbiome with sex behaviors reported in other studies mentioned above. Therefore, the microbiome composition of DuDHS study participants is consistent with other observations in microbiome studies of GBM, with higher abundances of microbiota that have been linked with mucosal inflammation.
It is important to highlight the limitations of this study. The measurements of the microbiome by 16S rRNA sequencing are limited to bacterial composition and proportional value estimates of bacterial abundance. Also, the function of the microbiome, including metabolic by-products such as small metabolites, functional pathways, which are important for gut health and immunity, are not captured in our analyses, and this may be altered with doxyPrEP. Also, 16S rRNA sequencing is limited to genus-level specificity, so we are unable to determine whether there are changes in individual species or strains of bacteria, which can vary considerably in their functional properties. Finally, antimicrobial resistance is a major concern with doxyPrEP and doxyPEP strategies, and a recently published study showed increased tetracycline antimicrobial resistance genes in the rectal microbiome after 6 months of doxy-PEP use25, and antimicrobial resistance gene expression was more pronounced in individuals who took more than 800 mg/month of doxycycline which is lower than the 3000 mg/month in DuDHS study participants which presumably may have a greater effect on gene expression. However, it is unclear if these gene expression differences are leading to antimicrobial resistance, and follow-up studies will be important to evaluate the relevance of these findings in this context. In the recently published DuDHS study26, doxycycline antimicrobial resistance was assessed by culturing commensals from nasal swabs isolates from study participants, and doxycycline-resistant S. aureus was identified in six individuals, with five in the immediate arm. While this suggested a potential increase in antimicrobial resistance over time, this data is not sufficient to understand the implications of doxyPrEP on antimicrobial resistance. Our analysis of rectal swab specimens in this study did not include culturing methods for antimicrobial resistance, nor did we capture sequence data on antimicrobial resistance genes, and this would be an important next step in studying the safety of doxyPrEP. Furthermore, as our study design included microbiome data from 41 participants, we were only powered to detect moderate effects of >50% reduction of major bacterial taxa detected in DuDHS study participants, and smaller differences may be apparent but are not observable with our sample size. Finally, this study focused on the effects on the rectal microbiome and did not include molecular analysis from other mucosal sites, such as the nasal, oral, or other compartments, which are also important for maintaining mucosal health and immune function. It may be that microbiomes across different mucosal sites differ in sensitivity to doxyPrEP with respect to composition, diversity, or antimicrobial resistance. Future studies evaluating the effect of doxycycline for STI prevention that include more diverse sampling strategies in their study design would be better able to address these questions.
In conclusion, we show that doxycycline 100 mg daily as doxyPrEP does not have a major effect on gut microbiome composition over a period of 48 weeks, supporting the safety profile of doxycycline for PrEP. These data are important to advance our understanding of how doxyPrEP interventions may impact the human microbiome. Further research and monitoring of the impact of doxyPrEP on the gut microbiome and antimicrobial resistance are warranted to guide the safe implementation of doxyPrEP for STI prevention.
Methods
Ethics approval and consent to participate
The DuDHS trial received approval from the University of British Columbia Research Ethics Board (REB approval H16-01935), the Case Western Reserve University IRB (STUDY20200059), and all participants provided informed consent. The study was registered with ClinicalTrials.gov (NCT02844634).
Study design, participants, and sample collection
A total of 41 participants, who had been enrolled and followed up in the Dual Daily HIV and STI PrEP (DuDHS) trial, were included in the microbiome study. The Dual Daily HIV and STI PrEP (DuDHS) study is an open-label randomized trial of immediate versus deferred doxyPrEP in HIV-negative GBM and transgender women receiving daily tenofovir disoproxil fumarate/emtricitabine (TDF/FTC) for HIV PrEP. The DuDHS study recruited 52 sexually active adult participants with a syphilis diagnosis in the last three years from a single clinical site in Vancouver, Canada, from February 2018 to May 2019. All participants received 48 weeks of TDF/FTC for HIV PrEP, and were randomized 1:1 to the immediate doxyPrEP arm (i.e., doxycycline 100 mg PO daily started immediately and continued to week 48) or the deferred doxyPrEP arm (i.e., doxycycline 100 mg PO daily starting at week 24, continued to week 48). Primary study outcomes included feasibility, adherence, and tolerability of the dual PrEP regimen, while exploratory outcomes included changes to the rectal microbiome. Of the 52 participants in the trial, 7 individuals did not complete the study (moved (n = 3), lost interest in the study (n = 2), lost to follow up, n = 3); and specimens were not available for four participants. Samples unavailable were higher in the Deferred study arm (15.3% Immediate (n = 4) vs. Deferred 26.9% (n = 7)). Rectal swabs were collected at baseline, week 24, and week 48 post-randomization for rectal microbiome analysis by 16S rRNA sequencing. Swabs were stored until transfer to the laboratory and cryopreserved at −70 °C.
The 16S rRNA experiments and data generation were performed blinded, such that the participant samples were indistinguishable to technicians performing the 16S rRNA experiments, and those generating the final dataset were unaware of the study-group assignments until after study completion. A number generator was used to randomize rectal swab samples for sample preparation steps as well as analysis sequence by 16S rRNA sequencing.
Bacterial DNA isolation
Rectal swabs were first transferred to an Eppendorf tube, immersed in 250 μL of PBS and shaken for 10 min at 4 °C. Swabs were then placed in a Spin x centrifuge unit (Corning Costar 9031, without membrane) and centrifuged at 10,000 × g for 5 min at 4 °C. The supernatant was spun a second time at 23,000 × g for 30 min at 4 °C to further pellet out bacteria. DNA extraction was performed on the entirety of the pellet and 250 μL of eluate according to Qiagen PowerLyzer PowerSoil kit instructions. Briefly, samples underwent five cycles of bead beating (45 sec at 6.5 m/s). Zymo mock community and negative water controls were processed alongside samples.
16S rRNA sequencing
Each sample was run in duplicate. DNA was amplified using the following primer sequences targeting the V4 region of the 16S rRNA gene: 5′-GTG CCA GCM GCC GCG GTA A−3′ for the forward primer and 5’-GGA CTA CHV GGG TWT CTA AT-3’ for the reverse primer. The PCR reaction was performed in a reaction well with 1 μl template DNA, 1 μl of each forward and reverse primer (0.5 μM), 6.25 μL UltraPure™ DNase/RNase-free distilled water (Thermo Fisher, Mississauga, ON), 2.5 μL PCR Gold Buffer (10×), 2.5 μL MgCl2 (25 mM), 0.5 μL dNTPs (10 mM), and 0.25 μL AmpliTaq Gold™ Taq DNA polymerase (5 U/μL) (Thermo Fisher, Mississauga, ON). DNA was denatured at 95 °C for 3 min, then amplified in 25 cycles as follows: 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s. A final extension was performed at 72 °C for 5 min. Amplicons were purified using Ampure XP beads (Beckman Coulter, Mississauga, ON), and purification was confirmed using a QIAXcel Advanced Instrument (QIAGEN). DNA was quantified using a Qubit 2.0 fluorometer (Life Technologies Inc., Burlington, ON), and the library was normalized to 4 nM. The final pooled DNA was diluted to 4 pM with a 10% spike-in of 12.5 pM PhiX. Samples were prepared and run on the Illumina MiSeq following the manufacturer’s protocol, using 500-cycle v2 PE reagents, yielding 2 × 250 bp paired-end reads (Illumina Inc., San Diego, CA, USA).
16S rRNA data processing
Sequence reads were analyzed using the Mothur pipeline (v1.39.5). Briefly, paired-end reads for each sample were assembled into contigs and primer sequences were trimmed; the maximum contig length was set to 420 bp. Contig alignment was performed using the SILVA reference alignment database (v132), following which sequences were classified using a Bayesian classifier and the Ribosomal Database Project (RDP) taxonomy database (v16). Phylotype classification was used. Average abundances for each taxon were calculated from duplicate samples for downstream analysis. Taxa that were detected at higher average levels in the negative water controls relative to samples were identified as contaminants and monitored during differential abundance analyses (Supplementary Data 6). Taxa with <0.02% average proportions across all samples were binned to “other” for compositional figures.
Data and statistical analysis
Beta diversity was calculated using the Bray–Curtis distance metric, and the distance between groups was assessed using PERMANOVA (adonis2) with the vegan package in R, and the effect size of the F-ratio was quantified as the coefficient of determination (R2). Principal Coordinate Analysis (PCoA) was used to visualize Bray–Curtis distance matrix. Alpha diversity was assessed using Shannon’s H diversity index. For differential abundance analysis of individual taxa, we utilized a consensus approach employing three different differential abundance testing methods, including Wilcoxon signed rank tests, ANCOM-II, and ALDEx2 as previously described28. Comparisons between groups were performed using Mann–Whitney U tests. ANCOM-II and linear mixed effects models were used for adjusted analyses. All p values were adjusted for multiple hypothesis testing using the Benjamini–Hochberg correction (FDR = 5%). Visualizations were created in R (ggplot2, 3.5.1).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Code availability
All code used for analyses is publicly available. 16S rRNA data processing included Mothur (v1.39.5) https://mothur.org/41, SILVA (v132) reference alignment database https://www.arb-silva.de/42, and Ribosomal Database Project (RDP) taxonomy database (v16)43. Data analysis and plots were generated using R (4.3.2): R Core Team (2023). ‘R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria https://www.R-project.org/. The R packages utilized in these analyses included: Ggplot2 (3.5.1) H. Wickham. ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag New York, 2016. (https://ggplot2.tidyverse.org/authors.html#citation); Ggpubr (0.6.0) Kassambara A (2023). ggpubr: ‘ggplot2’ Based Publication Ready Plots. https://rpkgs.datanovia.com/ggpubr/; R package version 0.6.0, https://CRAN.R-project.org/package=ggpubr; Vegan (2.6-8) https://vegandevs.github.io/vegan/authors.html#citation. Oksanen J et al. (2024). vegan: Community Ecology Package. R package version 2.6-8, https://CRAN.R-project.org/package=vegan; stats (Base R package 4.3.2) https://CRAN.R-project.org/package=ggpubr.
References
Rowley, J. et al. Chlamydia, gonorrhoea, trichomoniasis and syphilis: global prevalence and incidence estimates, 2016. Bull. World Health Organ 97, 548–562P (2019).
Jansen, K. et al. STI in times of PrEP: high prevalence of chlamydia, gonorrhea, and mycoplasma at different anatomic sites in men who have sex with men in Germany. BMC Infect. Dis. 20, 110 (2020).
Molina, J. M. et al. Post-exposure prophylaxis with doxycycline to prevent sexually transmitted infections in men who have sex with men: an open-label randomised substudy of the ANRS IPERGAY trial. Lancet Infect. Dis. 18, 308–317 (2018).
Luetkemeyer, A. F. et al. Postexposure doxycycline to prevent bacterial sexually transmitted infections. N. Engl. J. Med 388, 1296–1306 (2023).
Molina, J. M. et al. Doxycycline prophylaxis and meningococcal group B vaccine to prevent bacterial sexually transmitted infections in France (ANRS 174 DOXYVAC): a multicentre, open-label, randomised trial with a 2 x 2 factorial design. Lancet Infect. Dis. 24, 1093–1104 (2024).
Traeger, M. W. et al. Doxycycline postexposure prophylaxis and bacterial sexually transmitted infections among individuals using HIV Preexposure prophylaxis. JAMA Intern. Med. 185, 273–281 (2025).
Sankaran, M. et al. Doxycycline postexposure prophylaxis and sexually transmitted infection trends. JAMA Intern. Med. 185, 266–272 (2025).
Grennan T, T. D. et al. Presented at: 25th International AIDS Conference. (Munich, Germany, 2024).
Bolan, R. K. et al. Doxycycline prophylaxis to reduce incident syphilis among HIV-infected men who have sex with men who continue to engage in high-risk sex: a randomized, controlled pilot study. Sex. Transm. Dis. 42, 98–103 (2015).
Grennan, T. et al. A pilot, randomized controlled trial of Dual Daily HIV and sexually transmitted infection pre-exposure prophylaxis using tenofovir disoproxil fumarate/emtricitabine and doxycycline in gay, bisexual and other men who have sex with men and transgender women: the DuDHS Study. Clin. Infect. Dis. 30, ciaf043 (2025).
Peyriere, H., Makinson, A., Marchandin, H. & Reynes, J. Doxycycline in the management of sexually transmitted infections. J. Antimicrob. Chemother. 73, 553–563 (2018).
Bachmann, L. H. et al. CDC Clinical Guidelines on the use of doxycycline postexposure prophylaxis for bacterial sexually transmitted infection prevention, United States, 2024. MMWR Recomm. Rep. 73, 1–8 (2024).
Gravett, R. M. et al. Preferences for monthly oral PrEP over other PrEP modalities among a national sample of gay, bisexual, and other men who have sex with men in the United States. J. Acquir. Immune Defic. Syndr. 99, 128–137 (2025).
Fusca, L. et al. High interest in syphilis pre-exposure and post-exposure prophylaxis among gay, bisexual and other men who have sex with men in Vancouver and Toronto. Sex. Transm. Dis. 47, 224–231 (2020).
Mayer, K. H., Traeger, M. & Marcus, J. L. Doxycycline postexposure prophylaxis and sexually transmitted infections. JAMA 330, 1381–1382 (2023).
Belkaid, Y. & Hand, T. W. Role of the microbiota in immunity and inflammation. Cell 157, 121–141 (2014).
Patrick, D. M. et al. Decreasing antibiotic use, the gut microbiota, and asthma incidence in children: evidence from population-based and prospective cohort studies. Lancet Respir. Med. 8, 1094–1105 (2020).
Schwartz, D. J., Langdon, A. E. & Dantas, G. Understanding the impact of antibiotic perturbation on the human microbiome. Genome Med. 12, 82 (2020).
Tariq, R. et al. Low risk of primary clostridium difficile infection with tetracyclines: a systematic review and metaanalysis. Clin. Infect. Dis. 66, 514–522 (2018).
Xu, D. et al. Why does doxycycline pose a relatively low risk for promotion of clostridioides difficile infection?. Pathog. Immun. 7, 81–94 (2022).
Buddle, J. E. & Fagan, R. P. Pathogenicity and virulence of Clostridioides difficile. Virulence 14, 2150452 (2023).
Saarela, M. et al. Tetracycline susceptibility of the ingested Lactobacillus acidophilus LaCH-5 and Bifidobacterium animalis subsp. lactis Bb-12 strains during antibiotic/probiotic intervention. Int. J. Antimicrob. Agents 29, 271–280 (2007).
Ahn, Y. et al. In vitro test systems to determine tetracycline residue binding to human feces. Regul. Toxicol. Pharm. 99, 105–115 (2018).
Moura, I. B. et al. Profiling the effects of systemic antibiotics for acne, including the narrow-spectrum antibiotic sarecycline, on the human gut microbiota. Front. Microbiol. 13, 901911 (2022).
Chu, V. T. et al. Impact of doxycycline post-exposure prophylaxis for sexually transmitted infections on the gut microbiome and antimicrobial resistome. Nat. Med. 31, 207–217 (2024).
Grennan, T.M.S. et al. A pilot, randomized controlled trial of Dual Daily HIV and sexually transmitted infection pre-exposure prophylaxis using tenofovir disoproxil fumarate/emtricitabine and doxycycline in gay, bisexual and other men who have sex with men and transgender women: the DuDHS study. Clin Infect Dis In press (2025).
Cohen, J. Statistical Power Analysis for the Behavioral Sciences (2nd ed.). (Hillsdale, NJ: Lawrence Erlbaum Associates, Publishers., 1988).
Nearing, J. T. et al. Microbiome differential abundance methods produce different results across 38 datasets. Nat. Commun. 13, 342 (2022).
Gebrebrhan, H. et al. Rectal microbiota diversity in Kenyan MSM is inversely associated with frequency of receptive anal sex, independent of HIV status. AIDS 35, 1091–1101 (2021).
Li, S. X. et al. Gut microbiota from high-risk men who have sex with men drive immune activation in gnotobiotic mice and in vitro HIV infection. PLoS Pathog. 15, e1007611 (2019).
Noguera-Julian, M. et al. Gut microbiota linked to sexual preference and HIV infection. EBioMedicine 5, 135–146 (2016).
Vujkovic-Cvijin, I. et al. HIV-associated gut dysbiosis is independent of sexual practice and correlates with noncommunicable diseases. Nat. Commun. 11, 2448 (2020).
Dillon, S. M. et al. Gut dendritic cell activation links an altered colonic microbiome to mucosal and systemic T-cell activation in untreated HIV-1 infection. Mucosal Immunol. 9, 24–37 (2015).
Anagnostakos, K., Grzega, C., Sahan, I., Geipel, U. & Becker, S. L. Occurrence of rare pathogens at the site of periprosthetic hip and knee joint infections: a retrospective, single-center study. Antibiotics 10, 882 (2021).
Basu, P., Williams, A., O’Brien, M. T., Brouns, M. & Edwards, P. A case of Finegoldia magna (formerly Peptostreptococcus magnus) infection mimicking disseminated malignancy. Int. J. Infect. Dis. 53, 12–14 (2016).
Hosseini Dehkordi, S. H. & Osorio, G. Case of pacemaker pocket infection caused by Finegoldia magna. Anaerobe 47, 135–136 (2017).
Parisio, E. M. et al. Epidemiology and antibiotic susceptibility profiles of obligate anaerobes in a hospital of central Italy during a one-year (2019) survey. Anaerobe 78, 102666 (2022).
Muzny, C. A. et al. Identification of key bacteria involved in the induction of incident bacterial vaginosis: a prospective study. J. Infect. Dis. 218, 966–978 (2018).
Neumann, A., Bjorck, L. & Frick, I. M. Finegoldia magna, an anaerobic gram-positive bacterium of the normal human microbiota, induces inflammation by activating neutrophils. Front. Microbiol. 11, 65 (2020).
Cook, R. R. et al. Alterations to the gastrointestinal microbiome associated with methamphetamine use among young men who have sex with men. Sci. Rep. 9, 14840 (2019).
Schloss, P. D. & Westcott, S. L. Assessing and improving methods used in operational taxonomic unit-based approaches for 16S rRNA gene sequence analysis. Appl. Environ. Microbiol. 77, 3219–3226 (2011).
Quast, C. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596 (2013).
Cole, J. R. et al. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 42, D633–D642 (2014).
Acknowledgements
We would like to thank the participants of the DuDHS study for making this study possible. This study was funded by Gilead Sciences Inc. (grant IN-CA-276-4012 awarded to MWH), with additional funding from and Canadian Institutes of Health Research (CIHR) grant (#365358 awarded to T.G.) and the Pacific Public Health Foundation (formerly, BC Center for Disease Control Foundation for Public Health). This study is also supported, in part, by the Canadian Institutes of Health Research Innovative Biomedical and Clinical HIV/AIDS Research Team Grant (ADB: HB3-164066). Dr. Burgener receives salary support from the National Institutes of Health, including the Rustbelt Center for AIDS Research (2P30 AI036219-26A) and the CWRU Center for Excellence on Substance Use and HIV (5P30DA054557). Dr. Burgener also receives funding support from the European Union’s Horizon 2020 research and innovation program under grant agreement No. 847943 (The Mistral project). We would also like to thank the JCWilt Infectious Disease Research Center for technical support. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
T.G, M.H. and A.D.B. conceived the microbiome sub-study. A.L. and M.D. performed the experimental analysis of swab specimens and generated 16S rRNA data. S.K., M.D., L.N.R., A.L., L.M. and A.D.B. performed analysis and interpreted the data. T.G., M.H., J.W., T.T., S.M., A.G. and J.E. managed the study cohort and clinical trial. S.K., M.D., T.G. and A.D.B. wrote the manuscript. All authors contributed to the review and editing of the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Knodel, S., Main, L., DeLeon, M. et al. Impact of doxycycline pre-exposure prophylaxis (doxyPrEP) for sexually transmitted infections on the microbiome of men who have sex with men on HIV PrEP. Nat Commun 16, 6143 (2025). https://doi.org/10.1038/s41467-025-61426-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-61426-5