Abstract
Conserving crop wild relatives (CWR) in their natural environments, together with the complex communities of microorganisms that live with them, could lay the foundation to unlock novel mechanisms for crop resilience and new strategies for achieving food security.
Similar content being viewed by others
Introduction
An immense diversity of soil microorganisms plays a pivotal role in maintaining the vitality of terrestrial plant communities, including both cultivated crops and their wild relatives (crop wild relatives, CWR). These microorganisms are essential because they drive soil organic matter turnover, reinforce soil structure, enhance nutrient cycling, increase nutrient availability to plants, and suppress pathogen proliferation. By promoting soil stability and fertility, they create a foundation for resilient and productive ecosystems, making them indispensable for sustainable food production.
With this in mind, plant-associated microbes are increasingly recognized as a “second plant genome” due to their fundamental roles for the plant with respect to nutrition, water uptake, defense, and resilience against environmental stressors1. Crop growth and quality are strongly linked to the microbiome’s functions2. The establishment of these plant-microbe associations is governed by a complex interplay of geographic, topographic, soil, and host plant characteristics3,4. Additionally, plants actively shape their interactions with soil microorganisms through the production of secondary metabolites, the selective secretion of root exudates, and root architecture, effectively recruiting and cultivating specific microbial communities in and around their root systems5,6. As increasing numbers of plant species face global extinction7,8, a frequently overlooked consequence is the “invisible extinction” of their associated microbial biodiversity. In undisturbed ecosystems—from grasslands and tundras to savannas and rainforests—plant and microorganisms engage in complex, co-evolved relationships that enable ecosystems to withstand environmental fluctuations. However, anthropogenic disturbances, including climate change and large-scale intensive agriculture, threaten these critical plant-microbe interactions. The loss of (unexplored) microbial diversity could eliminate key plant-microbe synergies before their potential applications in agriculture are fully realized. For instance, fungal networks and actinorhizal plants, vital for soil health and ecosystem balance, are under threat in Central Asia and North-eastern India9,10, jeopardizing microbes essential for revegetation and soil detoxification. From this, it follows that the decline of CWR-associated microbes may compromise agricultural sustainability by diminishing traits that contribute to crop resilience and productivity, highlighting the critical role of natural microbial diversity in agroecosystems.
Domestication of wild plant species, combined with modern agricultural practices such as irrigation, fertilization, pesticide application as well as the cultivation of plants beyond their native ranges, often in pure stands in a narrow crop rotation or even in monoculture, has altered plant-microbe equilibrium that evolved over millennia. In addition, it is important to remember that crop plants are now being grown in regions well removed from the habitat of their progenitors. Post-domestication selection has been largely based on yield, and this has likely resulted in the selection of desirable plant genotypes adapted to the local microbial populations. This will include resistance or tolerance to harmful soil microbes and nematodes, and interactions that optimize utilization of the local soils. Emerging evidence shows that domesticated plants, regardless of their specific domestication pathways or geographic origins, exert a strong influence on plant-associated microbiome assembly and function. As a result, their associated microbial communities differ from those of their wild progenitors4,11,12 through the loss, gain, or replacement of microbial taxa, depending on the ecological processes governing microbiome assembly13. These shifts in microbiome assembly can have profound consequences for plant performance, as changes in microbial composition and function affect nutrient uptake and disease resistance14,15. For instance, in Phaseolus vulgaris (common bean), domestication has positively affected the microbiota: breeding for Fusarium resistance has promoted the recruitment of microbial taxa capable of producing antifungal compounds16. A recent large exploration of tetraploid wheat revealed that domesticated plants selectively recruit soil keystone bacteria that carry nitrogen fixation genes and organic nitrogen ammonification genes to sustain plant N uptake and plant growth14.
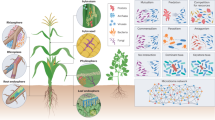
However, numerous studies have reported a consistent decline in the diversity of potentially beneficial microbes associated with domesticated crops compared to their wild progenitors, including key species such as wheat, maize, soybean, and rice17,18,19. In this context, it is plausible that soil microorganisms that co-evolved with CWR may benefit crops but have not persisted in modern agroecosystems due to the new environments where these major crops are now grown and the impact of tillage, and high-input agricultural practices. This includes the disruption of mycorrhizal associations which, in many situations, can support soil health and plant nutrition20 (Fig. 1). For instance, an extensive experimental study on 27 herbaceous crops and their CWR revealed a significant reduction in mycorrhizal symbiosis following domestication and genetic improvement, with wild relatives exhibiting greater affinity for arbuscular mycorrhizal associations than modern varieties21. This phenomenon has been partially attributed to fertilizer inputs reducing the necessity or even the benefit of mycorrhizal interactions for plant growth. Furthermore, management practices in intensive agricultural systems—including conventional tillage, pesticide, and fertilized use—can reduce the biomass and diversity of mycorrhizal fungi, thereby impairing their capacity to promote crop growth and facilitating phosphorus uptake22. Moreover, in domesticated wheat, a functional shift in the microbiome led to (i) depleted bacterial biocontrol capacities against pathogenic fungi and (ii) altered bacterial gene expression related to nitrogen and phosphorus cycling compared to its wild ancestors20. Interestingly, multidisciplinary studies on the soil microbiome associated with five wild and seven domesticated rice accessions reported that domesticated rice alters the rhizosphere microbiome, reducing nitrogen fixation and increasing nitrous oxide emissions19,23. Overall, while domestication has sometimes enabled crops to recruit beneficial microbes, its broader legacy includes the erosion of ancient plant-microbe partnerships. Therefore, there is an opportunity for crop-specific strategies to harness microbial diversity for sustainable agriculture.
a Managed agricultural ecosystems (e.g., pure stand of wheat) exhibit lower biodiversity, fewer ecological interactions, and a weak metabolic circular economy (MCE), where nutrient and energy flow cycles from root to shoot, into the environment, and back again are less efficient. b In contrast, in situ cultivation of CWR in nature-near ecosystems supports a rich diversity of microbes and variety of life-forms, including microfauna with stronger MCE and ecological interactions. Created with BioRender.com.
This Perspective positions CWR as “guardians” of adaptive microbial diversity, highlighting their potential to enhance the resilience and sustainability of cropping systems (Figs. 1 and 2). Given their close genetic relationship to cultivated crops, CWR may host microorganisms that promote agronomically beneficial traits, particularly stress resilience and resource-use efficiencies3,24. Such microorganisms, that have co-evolved with their host plants, represent a largely untapped frontier for nature-based solutions in agriculture. Preserving the ecosystems where these plant-microbe interactions persist is essential for enabling future research into their potential applications. By safeguarding these natural habitats, we not only protect vital microbial diversity but also open opportunities to enhance plant resilience and productivity through the integration of CWR-associated microorganisms. Indeed, the significance of these plant-microbe interactions extends beyond individual plant health, influencing entire ecosystems and agricultural sustainability. This concept aligns with and expands upon the established One Health framework, recognizing the interconnectedness of soil, plant, animal, and human health, highlighting the critical role of microbial populations in maintaining ecosystem integrity25 (Fig. 2).
The need for conservation, research, and translation
Awareness is growing of the role of CWR as reservoirs of diverse, agriculturally relevant microbiomes4,24,26,27. However, significant challenges remain in effectively disseminating this knowledge, particularly in integrating it into policy frameworks. More broadly, recent publications have highlighted the need to explicitly integrate soil microbiomes into the One Health framework25 and emphasize that “soil biodiversity requires policy without borders”28. In pursuit of these objectives, several large-scale initiatives are dedicated to collecting, conserving, and monitoring soil microbial communities. These efforts include the African Microbiome Initiative, the Australian Microbiome Initiative, the China Soil Microbiome Initiative, SoilBON, the European LUCAS soil survey, Dig Up Dirt | Find Your Friendly Fungi, the Global Fungi Initiative, the Earth Microbiome Project, the Society for the Protection of Underground Networks or the UK Crop Microbiome CryoBank.
Despite their recognized importance, global initiatives have primarily focused on collecting CWR for ex-situ conservation29 or protecting them in their natural habitats through in situ conservation30. However, no coordinated efforts have yet been established to conserve both CWR and their associated microbiomes at a global scale, leaving a critical gap in biodiversity conservation strategies. Of note, in situ conservation is essential for preserving the co-adaptive relationships between CWR and their microbiomes while enabling the ongoing evolution of these associations in response to climate change27. The importance of in situ conservation is underscored by the recognition that specific edaphic (soil-related) and climatic conditions inherent to CWR habitats significantly influence the composition of their associated soil microbiota. Recent studies have demonstrated that in situ CWR spanning diverse ecoregions, from forests to deserts, exhibit remarkable adaptability to environmental stressors, owing to their diverse genetic makeup and associated microbes27. Thus, CWR may function as reservoirs of adaptive microbial diversity. Therefore, this Perspective advocates for the establishment of in situ “CWR Biodiversity Sanctuaries”—conservation hubs intended to safeguard both CWR and their associated microbiomes within dynamically evolving ecosystems. These sanctuaries should encompass adequately sized protected areas and, for their long-term sustainability, must be co-designed, developed, and maintained with the full participation of indigenous and local communities. A good example of the integrated conservation and management of CWRs in their local agro-ecological context is the Parque de La Papa in Peru, which is managed by local communities in association with the Association for Nature and Sustainable Development, ANDES.
Beyond conservation, further research is required to advance understanding of the identity, function, and ecological interactions of CWR-associated microbial communities, particularly their roles in biogeochemical cycling, plant disease mitigation, and the enhancement of soil structure and nutrient availability within resilient and ecologically integrated cultivation systems that rely on less synthetic fertilizers and agrochemicals. These efforts can be supported through the ex-situ conservation of soil microbial populations (e.g., in archived freeze-dried soil samples) through the sampling of topsoil (0–30 cm depth), which is rich in nutrients, soil organic substance, roots, and microorganisms. However, the recovery of viable microbes from these samples may be limited due to the low cultivability of many soil microorganisms.
Bridging in situ and ex-situ conservation efforts should promote the development of a research pipeline aimed at translating fundamental research findings into improved agronomic practices. These advancements would strengthen microbiome-crop symbioses and integrate the benefits of CWR-derived microbial communities into sustainable agricultural systems. A compelling example of leveraging intrinsic CWR attributes to enhance plant-microbial interactions is the transfer of a genetic region from Leymus racemosus, a CWR of wheat, to elite wheat varieties31. The introgression of this chromosomal region significantly reduced the abundance of ammonia-oxidizing bacteria through the exudation of compounds that inhibit nitrifying bacteria, thereby mitigating nitrogen loss and greenhouse gas emissions.
Expanding on this idea, a key strategy is to identify specific genomic regions in CWR that control interactions between plants and microbes, as well as essential agronomic traits. In particular, traits that influence root system development and function are of central importance. Characteristics like lateral root growth, root hair density, and the composition of root exudates directly impact the microbial communities in the rhizosphere. These traits determine how effectively plants acquire nutrients, attract beneficial microbes, and activate defense mechanisms against pathogens. Additionally, targeting genomic regions associated with plant-microbe signaling pathways, nutrient uptake efficiency, resilience to abiotic stress, and disease resistance can further enhance crop improvement efforts32. For instance, several quantitative trait loci (QTLs) have been identified in barley, millet, rice, soybean, wheat, and rapeseed that are functionally significant for root traits and plant-microbe interactions, providing specific targets for breeding programs33,34,35. Evaluating CWR of diverse crop species for favorable alleles at key genetic loci, including microbiome-regulating host genes, termed M genes—will accelerate trait discovery and enable the breeding of crops predisposed to fostering beneficial microbial associations in the soil36, thereby integrating conservation efforts with gene-guided microbiome engineering for sustainable crop resilience.
This approach could be further refined by analyzing CWR root exudate profiles, which shape soil microbial communities by selectively promoting beneficial microorganisms. Identifying key compounds in CWR root exudates could reveal strategies to enhance microbial selection in agricultural soils. The synthesis of such compounds—either selectively or broadly—could offer a means to manipulate soil microbiomes37, fostering beneficial microbial communities that support soil fertility, plant resilience, and disease resistance38.
Additionally, the assembly of synthetic microbial communities (SynComs) to improve plant health and soil quality is another promising approach39. In this method, beneficial microbes from natural communities are combined, permitting the retention of useful microbes that are also good colonizers, while eliminating potentially harmful components of the microbiome. Identifying key microbial taxa associated with CWR—particularly those contributing to abiotic stress tolerance, nutrient acquisition, disease suppression, and overall plant resilience—could provide a foundation for targeted microbial inoculation strategies. By isolating and characterizing these beneficial microbes from CWR species, tailored SynComs could be developed and tested in conjunction with modern crops.
Essentials for worldwide conservation initiatives
One of our biggest challenges is to establish pathways and pipelines to use CWR-associated microbes. We know the potential impact of their deployment is large from the examples provided above. However, these successes have been largely serendipitous rather than strategically planned. Consider what could be achieved if we were armed with well-characterized conservation sites coupled with a detailed knowledge of the resident microbial populations, and their changes over time. If we further link this to an understanding of genomic regions in the CWR that are associated with beneficial changes in the associated microbiomes, then we can start to select crop varieties that are best able to develop and maintain desirable microbial associations. Although it will be some time before the systematic use of CWR-microbe genetics will be routine, it is a worthy and achievable objective. Therefore, urgent conservation efforts are required as CWRs and their microbiomes face escalating threats in their natural habitats7,8. To address this, we propose the immediate launch of an internationally integrated, multi-location research initiative. The following section outlines the key considerations for designing such an initiative.
Consultation and partnerships
A major international initiative focused on the conservation, basic, and translational research on CWR and their associated microbes necessitates a thoughtful, inclusive, and multilateral discourse. This discourse must ensure the acknowledgment of the rights, cultures, and knowledge of local communities, promote their active participation, and respect applicable legislation regarding genetic resources and associated traditional knowledge. A critical initial step in conserving the diversity and genetic resources associated with CWR entails the establishment and routine updating of a comprehensive “red list” that systematically documents CWR species worldwide, prioritizing those requiring urgent conservation measures. Field expeditions will play a vital role in assessing in situ populations of CWR, facilitating precise identification and characterization of their geographical distributions. This “red list” would serve as a foundational tool to guide collaborative efforts, enabling international organizations to align their expertise and resources toward the most pressing conservation priorities. Several international organizations are well-positioned at the interface of agriculture and the environment to coordinate an extensive knowledge network among stakeholders, societies, and nations. These include the UN Global Soil Partnership (GSP), the Intergovernmental Technical Panel on Soils (ITPS), the Intergovernmental Panel on Climate Change (IPCC), the Consultative Group for International Agricultural Research (CGIAR), the Global Crop Diversity Trust (Crop Trust), and the International Platform for Biodiversity and Ecosystem Services (IPBES). Involvement from these organizations may be required in some capacity. Operating at the science-policy-society interface, these organizations can offer valuable recommendations to initiate and guide future policy actions.
Financing
The urgency of conserving CWR-associated soil microbiomes calls for strong in situ and ex-situ strategies and broad collaboration. However, securing adequate financial resources remains a critical challenge for implementing such initiatives on a global scale. While exact funding requirements depend on scope and geographic focus, past large-scale efforts provide a useful benchmark. For example, the CWR Project at the Crop Trust allocated $50 million over 10 years, and CGIAR genebanks require ~$25 million annually. Given the added complexity of microbiome conservation, a truly global initiative would likely require funding at least on the scale of these efforts, if not significantly more. To ensure long-term sustainability, significant funding should be mobilized from a diverse array of sources, including public and private sectors, global research initiatives, biodiversity conservation programs, climate action funds, and development agendas. Particularly relevant are funding mechanisms aligned with the Kunming-Montreal Global Biodiversity Framework, the United Nations Sustainable Development Goals (SDGs), and the European Union´s Green Deal objectives.
Sharing and access to data and biological materials
Conserving, classifying, and providing access to microbial biodiversity—especially strains, germplasm (CWR), and biological data like sequenced genomes—remains a significant challenge. Facilitating access to these resources and sharing associated genetic information is of utmost importance. For instance, the exchange of germplasm played a key role in protecting global sorghum production from a severe aphid infestation40. Some of the aforementioned organizations could oversee these initiatives, storing and disseminating plant, soil, and microbial samples along with genetic sequences through a multisectoral, interdisciplinary approach. An automated system to flag pathogens in samples for deposition in public repositories, such as the International Nucleotide Sequence Database Collaboration, could provide policymakers with timely insights into potential microbial threats. Identifying harmful microbes in samples will be essential for ensuring the safety of conserved and distributed materials. Collaborative international CWR initiatives, particularly those exploring plant-associated soil microbial diversity, should enable access while adhering to benefit-sharing standards, including those for digital sequence information41.
Equally important is the standardization of protocols and metadata collection, coupled with streamlined processes for data storage, accessibility, and sharing41. This ensures compliance with the principles of Findability, Accessibility, Interoperability, and Reusability (FAIR)42, thereby enhancing data discoverability, transparency, and reproducibility in scientific research. Standardized data formats are also vital for facilitating the application of Artificial Intelligence (AI) in predictive analytics. AI can analyze large datasets to identify patterns, assess risks, and inform conservation strategies for CWR and their microbiomes. AI tools can assist in the identification, mapping, and monitoring of CWR populations, prioritizing conservation efforts, and tracking changes over time. Machine learning algorithms can further analyze the complex interactions between CWR and their microbiomes, yielding insights to guide the development of more resilient, productive crops. Consequently, data standardization is essential for optimizing AI functionality, enabling seamless analysis of extensive datasets.
Data availability
All the information supporting this article is available within the article.
References
Roy, S. & Müller, L. M. A rulebook for peptide control of legume–microbe endosymbioses. Trends Plant Sci. 27, 870–889 (2022).
Song, Y. et al. Seed tuber microbiome can predict the growth potential of potato varieties. Nat. Microbiol. 10, 28–40 (2024).
Yue, H. et al. Host genotype-specific rhizosphere fungus enhances drought resistance in wheat. Microbiome 12, 44 (2024).
Bulgarelli, D. et al. Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host Microbe 17, 392–403 (2015).
Francioli, D., Strack, T., Dries, L., Voss-Fels, K. P. & Geilfus, C. Roots of resilience: optimizing microbe-rootstock interactions to enhance vineyard productivity. Plants People Planet 7, 524–535 (2024).
Gifford, M. L., Xu, G., Dupuy, L. X., Vissenberg, K. & Rebetzke, G. Root architecture and rhizosphere–microbe interactions. J. Exp. Bot. 75, 503–507 (2024).
Khoury, C. K. et al. Crop wild relatives of the United States require urgent conservation action. Proc. Natl. Acad. Sci. USA 117, 33351–33357 (2020).
Goettsch, B. et al. Extinction risk of Mesoamerican crop wild relatives. Plants People Planet 3, 775–795 (2021).
Jonathan Moens. How a few bags of dirt could help make the planet more resilient to climate change. (The Washington Post, 2023).
Bhattacharyya, P. N. et al. Frankia-actinorhizal symbiosis: a non-chemical biological assemblage for enhanced plant growth, nodulation and reclamation of degraded soils. Symbiosis 92, 1–26 (2024).
Kim, H., Lee, K. K., Jeon, J., Harris, W. A. & Lee, Y.-H. Domestication of Oryza species eco-evolutionarily shapes bacterial and fungal communities in rice seed. Microbiome 8, 20 (2020).
Abdullaeva, Y. et al. Domestication impacts the wheat-associated microbiota and the rhizosphere colonization by seed-and soil-originated microbiomes, across different fields. Front. Plant Sci. 12, 806915 (2022).
Soldan, R., Fusi, M., Cardinale, M., Daffonchio, D. & Preston, G. M. The effect of plant domestication on host control of the microbiota. Commun. Biol. 4, 936 (2021).
Yue, H. et al. Plant domestication shapes rhizosphere microbiome assembly and metabolic functions. Microbiome 11, 70 (2023).
Carrillo, J., Ingwell, L. L., Li, X. & Kaplan, I. Domesticated tomatoes are more vulnerable to negative plant–soil feedbacks than their wild relatives. J. Ecol. 107, 1753–1766 (2019).
Mendes, L. W., Raaijmakers, J. M., de Hollander, M., Mendes, R. & Tsai, S. M. Influence of resistance breeding in common bean on rhizosphere microbiome composition and function. ISME J. 12, 212–224 (2018).
Hetrick, B., Wilson, G. & Cox, T. Mycorrhizal dependence of modern wheat varieties, landraces, and ancestors. Can. J. Bot. 70, 2032–2040 (1992).
Porter, S. S. & Sachs, J. L. Agriculture and the disruption of plant–microbial symbiosis. Trends Ecol. Evol. 35, 426–439 (2020).
Chang, J. et al. Domesticated rice alters the rhizosphere microbiome, reducing nitrogen fixation and increasing nitrous oxide emissions. Nat. Commun. 16, 2038 (2025).
Abdullaeva, Y. et al. Domestication caused taxonomical and functional shifts in the wheat rhizosphere microbiota, and weakened the natural bacterial biocontrol against fungal pathogens. Microbiol. Res. 281, 127601 (2024).
Martín-Robles, N. et al. Impacts of domestication on the arbuscular mycorrhizal symbiosis of 27 crop species. N. Phytol. 218, 322–334 (2018).
Edlinger, A. et al. Agricultural management and pesticide use reduce the functioning of beneficial plant symbionts. Nat. Ecol. Evol. 6, 1145–1154 (2022).
Chang, J. et al. Nitrogen, manganese, iron, and carbon resource acquisition are potential functions of the wild rice Oryza rufipogon core rhizomicrobiome. Microbiome 10, 196 (2022).
Pérez-Jaramillo, J. E., Carrión, V. J., de Hollander, M. & Raaijmakers, J. M. The wild side of plant microbiomes. Microbiome 6, 1–6 (2018).
Singh, B. K., Yan, Z.-Z., Whittaker, M., Vargas, R. & Abdelfattah, A. Soil microbiomes must be explicitly included in One Health policy. Nat. Microbiol. 8, 1367–1372 (2023).
Raaijmakers, J. M. & Kiers, E. T. Rewilding plant microbiomes. Science 378, 599–600 (2022).
Fernández-Alonso, M. J. et al. Soil biogeography at the habitats of origin of major crops. (Research Square, 2023).
van der Putten, W. H. et al. Soil biodiversity needs policy without borders. Science 379, 32–34 (2023).
Dempewolf, H. et al. Adapting agriculture to climate change: a global initiative to collect, conserve, and use crop wild relatives. Agroecol. Sustain. Food Syst. 38, 369–377 (2014).
Vincent, H. et al. Modeling of crop wild relative species identifies areas globally for in situ conservation. Commun. Biol. 2, 136 (2019).
Bozal-Leorri, A. et al. Biological nitrification inhibitor-trait enhances nitrogen uptake by suppressing nitrifier activity and improves ammonium assimilation in two elite wheat varieties. Front. Plant Sci. 13, 1034219 (2022).
Freschet, G. T. et al. Root traits as drivers of plant and ecosystem functioning: current understanding, pitfalls and future research needs. N. Phytol. 232, 1123–1158 (2021).
Liu, T. et al. Rhizosheath formation and involvement in foxtail millet (Setaria italica) root growth under drought stress. J. Integr. Plant Biol. 61, 449–462 (2019).
Mwafulirwa, L. et al. Identification of barley genetic regions influencing plant–microbe interactions and carbon cycling in soil. Plant Soil 468, 165–182 (2021).
Ahmad, N. et al. Quantitative trait loci mapping reveals important genomic regions controlling root architecture and shoot biomass under nitrogen, phosphorus, and potassium stress in rapeseed (Brassica napus L.). Front. Plant Sci. 13, 994666 (2022).
Cernava, T. Coming of age for microbiome gene breeding in plants. Nat. Commun. 15, 6623 (2024).
König, A. et al. Reverse microdialysis: a window into root exudation hotspots. Soil Biol. Biochem. 174, 108829 (2022).
Nannipieri, P. et al. Legacy effects of rhizodeposits on soil microbiomes: a perspective. Soil Biol. Biochem. 184, 109107 (2023).
Martins, S. J. et al. The use of synthetic microbial communities to improve plant health. Phytopathology® 113, 1369–1379 (2023).
Muleta, K. T. et al. The recent evolutionary rescue of a staple crop depended on over half a century of global germplasm exchange. Sci. Adv. 8, eabj4633 (2022).
Halewood, M., Bagley, M. A., Wyss, M. & Scholz, A. H. New benefit-sharing principles for digital sequence information. Science 382, 520–522 (2023).
Wilkinson, M. D. et al. The FAIR guiding principles for scientific data management and stewardship. Sci. Data 3, 1–9 (2016).
Acknowledgements
We apologize to those whose work was not cited due to space and reference count limitations. M.W. is thankful to the Alexander von Humboldt Foundation for the postdoctoral fellowship. S.G.T. is supported by the US Department of Energy, Office of Science, Biological and Environmental Research award DE-SC0021369. The views and opinions expressed in this paper are solely those of the individual authors and do not necessarily reflect the views or positions ofthe individual author’s employer, institution, or other associated entity.
Author information
Authors and Affiliations
Contributions
M.W., S.R.M., D.F., S.G.T., D.M., C.K.K., L.H.R., H.D., P.L., and C.-M.G. contributed equally to the conceptual development, writing, and critical revision of the manuscript. All authors approved the final version.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Christina Birnbaum and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Waqas, M., McCouch, S.R., Francioli, D. et al. Blueprints for sustainable plant production through the utilization of crop wild relatives and their microbiomes. Nat Commun 16, 6364 (2025). https://doi.org/10.1038/s41467-025-61779-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-61779-x