Abstract
Intestinal tuft cells are epithelial sentinels that trigger host defense upon detection of parasite-derived compounds. While they represent potent targets for immunomodulatory therapies in inflammation-driven intestinal diseases, their functioning and differentiation are poorly understood. Here, we reveal common intermediary transcriptomes among the previously described tuft-1 and tuft-2 subtypes in mouse and human. Tuft cell subtype-specific reporter knock-ins in organoids show that the two subtypes reflect successive post-mitotic maturation stages within the tuft cell lineage. In vitro stimulation with interleukin-4 and 13 is sufficient to fuel the generation of new Nrep+ tuft-1 cells, arising from tuft precursors (tuft-p). Subsequently, changes in crypt-villus signaling gradients, such as BMP, and cholinergic signaling, are required to advance maturation towards Chat+ tuft-2 phenotypes. Functionally, we find chemosensory capacity to increase during maturation. Our tuft subtype-specific reporters and optimized differentiation strategy in organoids provide a platform to study immune-related tuft cell subtypes and their unique chemosensory properties.
Similar content being viewed by others
Introduction
Tuft cells are solitary chemosensory epithelial cells that respond to specific environmental stimuli by secretion of effector molecules that initiate appropriate changes in the immune state and physiology of their surroundings. Although they are rare, they attract significant interest now that increasing amounts of evidence underscore their crucial role at the interface between the intestinal epithelial barrier and the immune system, making them interesting targets for therapeutic immunomodulation1,2,3. In the intestine, tuft cells detect parasitic helminths and Tritrichomonas protists and secrete the cytokine interleukin-25 (IL-25) to initiate the type 2 immune response required for parasitic clearance, characterized by increased mucus production (weep) and muscle contractility (sweep)4,5,6. More specifically, IL-25 activates innate lymphoid cells (ILC2s) in the lamina propria and triggers the release of type 2 cytokines, including interleukin-13 (IL-13). IL13, together with IL4, acts on epithelial progenitors and biases their differentiation towards goblet and tuft cells, resulting in a tuft-ILC2 feedforward circuit that leads to a strong increase in tuft cell numbers, up to as much as 8% in the case of a helminth infection4.
In addition to remodeling of the intestinal epithelium through the tuft-ILC2 circuit, tuft cells also signal directly to nearby epithelial cells. For instance, tuft cells are the only epithelial cells that express choline acetyltransferase (ChAT), the enzyme catalyzing acetylcholine biosynthesis. In turn, tuft cell-derived acetylcholine induces fluid secretion from enterocytes by stimulating apical Cl- ion release, further contributing to the ‘weep’ into the intestinal lumen7. Similarly, prostaglandin D2 secreted by activated tuft cells can increase mucus secretion from goblet cells8.
Like most epithelial cell types of the small intestine, tuft cells arise from Lgr5+ stem cells at the crypt base and migrate up towards the tip of the villus where they are eventually shed into the intestinal lumen9,10. Following the discovery of tuft cells as important sentinels and signal relays, multiple studies have shed light on the mechanisms of tuft lineage specification. For instance, the transcription factor POU class 2 homeobox 3 (POU2F3) was found to be a master regulator of tuft cell fate and other transcription factors, such as GFI1B and SOX4, were found to be important for tuft lineage commitment5,11,12. Interestingly, while loss of NOTCH signaling followed by expression of atonal bHLH transcription factor 1 (Atoh1) is essential for differentiation towards most secretory cell types (goblet, enteroendocrine and Paneth), the role of NOTCH signaling and Atoh1 in tuft cell differentiation seems to be more complex. Multiple studies describe Atoh1-independent tuft cell populations in the small intestine, while there is evidence for Atoh1-dependent populations in the colon9,10,11,12,13.
Despite these insights into early tuft cell fate specification, much less is known about the signals that further advance tuft cell differentiation and maturation, nor how functionality relates to transcriptional heterogeneity that was observed within the intestinal tuft cell pool14. Indeed, two different types of tuft cell have been identified by single-cell RNA sequencing: tuft-1 cells with a neuron-related transcriptional program and tuft-2 cells with an immune-related transcriptional program. Computational analysis of spatial transcriptional programs along the crypt-villus axis, suggested zonation of these two tuft cell types with tuft-2 markers expressed higher up towards the tip of the villus and tuft-1 markers towards the bottom of the villus15. While a tuft-1 specific function in the small intestine has yet to be found, sensing of the bacterial metabolite N-undecanoylglycine through vomeronasal receptor VMN2R26, has been mainly attributed to tuft-2 cells8. Although multiple metabolite GPCRs, such as Ffar3 and Tas1rs, as well as orphan GPCRs, are specifically expressed by intestinal tuft cells14, tuft cell activation by ligand stimulation has only been established for the succinate receptor SUCNR1 and the aforementioned vomeronasal receptor VMN2R268,16.
Here, we analyze single-cell intestinal tuft cell transcriptomes from mouse and human to reveal conserved proliferative tuft cell precursors (tuft-p), as well as abundant intermediaries of the post-mitotic tuft-1 and tuft-2 cell states. Moreover, new organoid models with tuft cell subtype-specific reporter knock-ins show that treatment with IL-4 and IL-13 leads to increased tuft cell numbers, but solely generates tuft-1 states. Adaptation of the crypt-like organoid medium to better mimic the villus environment, where tuft-2 cells reside in vivo, facilitates transitioning of post-mitotic immature tuft-1 states to mature tuft-2 states. Practically, our optimized organoid differentiation strategy provides tuft cells that closely resemble their functional counterparts in vivo and are amenable for in-depth cell biological studies, such as real-time imaging of tuft cell morphological and chemosensory properties.
Results
Gradual Chat expression marks a continuum of zonated tuft cell states on the crypt-villus axis
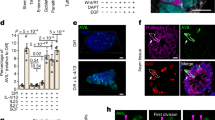
To characterize intestinal tuft cell heterogeneity with high-resolution, we performed single-cell RNA sequencing on a FACS-enriched tuft cell population isolated from ChatBAC-eGFP mice where tuft cells are labeled with enhanced green fluorescent protein (eGFP), based on tuft cell-specific epithelial expression of choline acetyltransferase (Chat)17 (Fig. 1a, and Supplementary Fig. 1a, b). Unsupervised clustering resulted in three clusters (Fig. 1a), with the two largest identified clusters showing high similarity to the previously described tuft-1 and tuft-2 populations14 (Fig. 1b). The third and smallest cluster robustly expressed general tuft cell markers such as Dclk1, Trpm5 and Avil, but did not show strong expression of genes related to tuft cell chemosensory or immunological function such as Sucnr1, Chat, and Il25 (Fig. 1c). Differential expression analysis indicated that this cluster separated from the other two clusters based on gene expression that is also detected in other cellular lineages, such as Aldob and Dmbt1 (enterocytes), Muc3 (Goblet cells) or Reg3b and Reg3g (Paneth cells), leading to a relatively low tuft-specific signature score (Supplementary Fig. 1c, d, and Supplementary Data 1). Moreover, components required for MHCII-dependent antigen presentation, such as Cd74, H2-Ab1 and H2-Aa, earlier found to be expressed by intestinal stem cells18, were expressed in this cluster (Supplementary Fig. 1d). Last, we observed high expression of proliferation markers Mcm2, Mki67, Top2a and Pcna (Fig. 1c) and a large proportion of this cluster was predicted to be in S/G2/M phase of the cell cycle (Supplementary Fig. 1e). Collectively, these data suggest that the third cluster represents immature early-state tuft precursors (tuft-p). We validated the existence of the three transcriptomic tuft cell states by integration with other scRNA-seq datasets of the mouse small intestinal epithelium14,19,20 (Fig. 1d). Affirmatively, we found tuft-p, tuft-1 and tuft-2 clusters among the more than 2000 tuft cell profiles in the integrated dataset, supported by differential expression of their respective transcriptional signatures (Fig. 1e, and Supplementary Fig. 1f).
a Left: zoom-in of intestinal villus where ChatBAC-eGFP marks solitary tuft cells (white), with epithelial membrane (b-catenin, red) and nuclear staining (blue). Middle: FACS strategy for scRNA-seq of GFP+ cells from ChatBAC-eGFP mice (n = 3). Right: UMAP with unsupervised clustering of 443 ChatBAC-eGFP+ tuft cells. Tuft-p: tuft-precursors. Illustration from NIAID NIH BIOART Source bioart.niaid.nih.gov/bioart/279. b Violin plots depicting distribution of tuft-1 and tuft-2 signature scores14 per cluster. c Single-cell dot plot showing expression of tuft cell and proliferation markers per cluster. d Left: UMAP of integrated single-cell transcriptomic datasets of mouse small intestinal epithelium. Cells are colored by study origin. Right: zoom-in of tuft cell cluster (~2000 cells). EE: enteroendocrine cells. Sec. prog.: secretory progenitors. e Relative expression of tuft-p, tuft-1 and tuft-2 signatures (top 100 differentially expressed genes) overlayed on UMAP of integrated tuft cell cluster and as violin plots (insets). Signatures were extracted from ChatBAC-eGFP+ tuft clusters in (a). f Relative expression of Dclk1 and Chat overlayed on UMAP of integrated tuft cell cluster and as violin plots (insets). g Violin plot depicting distribution of relative Chat expression for all cells per cluster (ANOVA p = 1.85 × 10−15, Tukey HSD test). h Fluorescent image of ChatBAC-eGFP mouse small intestine stained for DCLK1 (red), GFP (green) and nuclei (blue). White arrows: DCLK1+ tuft cells. i Quantification of frequency of Chat+ cells among DCLK1+ cells for indicated regions on the crypt-villus axis (n = 3 mice, 267 DCLK1+ tuft cells, ANOVA P = 0.0002, Tukey HSD test). j Representative image showing lower expression of ChatBAC-eGFP in tuft cells in crypt regions. White dashed line: crypt-villus junction. k Zonation profile of tuft-p, tuft-1 and tuft-2 clusters (panel a). Boxplots show center of mass on the crypt-villus axis for the top 100 cluster-specific genes featured in Manco et al. dataset15. Boxes in Tukey box-and-whisker plot represent interquartile range (IQR, Q1 (25th percentile) to Q3 (75th percentile)), central lines mark median values and whiskers indicate outlier boundaries (1.5x IQR) (ANOVA P < 2 × 10−16, Tukey HSD test). l Barplot depicting frequency of EdU+ cells, split by Chat phenotype (Chat+ or Chat-), among DCLK1+ cells in the crypt (n = 3 mice, 112 DCLK1+ tuft cells, t-test). m Heatmap showing co-expression of tuft-1 and tuft-2 marker genes (genes with adjusted P value < 0.01 are shown, Wilcoxon rank-sum test with Bonferroni correction). Cells from the post-mitotic tuft-1 and tuft-2 clusters (panel a) are ordered by the ratio of tuft-1 and 2 scores14, indicated at the top of the heatmap. Relative Dclk1 and Chat expression per cell are shown above the heatmap.
Next, we evaluated Chat expression within the tuft cell pool and noticed substantial heterogeneity, while general tuft cell marker Dclk1 was uniformly expressed (Fig. 1f). Specifically, Chat was most prominently expressed in tuft-2 cells, which is corroborated by differential expression levels in our scRNA-seq dataset of sorted ChatBAC-eGFP+ cells (Fig. 1g). We validated heterogeneous Chat expression within the tuft cell pool by immunofluorescence against DCLK1 and GFP on intestinal tissue sections of the ChatBAC-eGFP mice. This confirmed Chat expression to be confined to a subset of DCLK1+ tuft cells (Fig. 1h), mostly residing on the villus (Fig. 1i), which was also found by a recent report7. This could be a consequence of a gradual increase in Chat expression as tuft cells migrate along the crypt-villus axis, which is supported by higher GFP signal in cells on the villus compared to the crypt (Fig. 1j).
To test this, we determined the localization of the three tuft cell subtypes from our ChatBAC-eGFP scRNA-seq dataset. We selected the top marker genes of each cluster and plotted the center of mass of their expression on the crypt-villus axis, as determined by Manco et al.15 (Fig. 1k). Additionally, we calculated single-cell signature scores along the zones of the crypt-villus axis, to further support regional abundance predictions for the three tuft types15 (Supplementary Fig. 1g). This resulted in a zonated pattern, with the tuft-p cells predicted to be closest to the crypt, an intermediate pattern for tuft-1 cells, and the tuft-2 cells located highest up towards the villi tips (Fig. 1k, Supplementary Fig. 1g). Considering the differential Chat expression in the three tuft cell subtypes (Fig. 1g), such zonated localization is in line with the increase in ChatBAC-eGFP+ cells towards the villus, observed in our immunofluorescence experiments (Fig. 1h–j). We further validated confinement of the proliferative tuft-p state to the crypt region by assessing EdU incorporation in DCLK1+ cells after a 4-hour pulse (Fig. 1l, and Supplementary Fig. 1h). These experiments also indicated that proliferation primarily precedes emergence of Chat expression (Fig. 1l), in line with low Chat expression in tuft-p cells (Fig. 1g).
Taking into account the zonated localization of the post-mitotic tuft-1 and tuft-2 cells on the crypt-villus axis, we ranked cells from these clusters based on a tuft-2/tuft-1 signature score and visualized expression patterns of tuft-1 and tuft-2 marker genes in a heatmap (Fig. 1m). Notably, this revealed a gradual transition of tuft-1 to tuft-2 transcriptomic profiles via intermediary phenotypes encompassing mixed expression of both tuft-1 and tuft-2 markers. Crucially, this continuum of cell states and absence of binary confinement, suggests that the two previously reported tuft-types represent sequential differentiation states along the crypt-villus axes. As expected, Chat expression levels gradually increased from tuft-1 to tuft-2 states (Fig. 1m), in line with the predominant localization of ChatBAC-eGFP+ cells towards the higher part of the villus (Fig. 1i), while Dclk1 expression levels were high among all post-mitotic tuft cells (Fig. 1m). In sum, these descriptive in vivo data reveal early tuft precursors in the crypt and a continuum of tuft-1 and tuft-2 transcriptomic states along the crypt-villus axis, which are marked by gradually increasing Chat expression.
Small intestinal tuft cell phenotypes are conserved between mouse and human
To investigate if intestinal tuft cell phenotypes are conserved between mouse and human, we reanalyzed a recently published single-cell transcriptomic dataset of the human small intestinal epithelium, containing ~800 tuft cells20 (Fig. 2a). Unsupervised re-clustering of the tuft cells in the dataset resulted in three clusters, which we labeled tuft-p, tuft-1 and tuft-2 (Fig. 2b) based on expression of the mouse tuft cell signatures (Supplementary Fig. 2a) and expression of proliferation markers (Fig. 2c, and Supplementary Fig. 2b). Recently, similar tuft subtypes were found in human organoids and intestinal tissue21, including a proliferative state (tuft-3), akin to the tuft-p cells described here.
a Schematic and UMAP of published single-cell transcriptomics dataset of human small intestine20. Cell types are annotated by color. EE: enteroendocrine cells. Abs. prog.: absorptive progenitors. Sec. prog.: secretory progenitors. Illustration from NIAID NIH BIOART Source bioart.niaid.nih.gov/bioart/232. b Unsupervised clustering of 844 human tuft cells20 identifies three separate tuft cell populations. c Single cell dot plot showing expression of tuft cell and proliferation markers per cluster. Average expression is represented by dot color while the percentage of expressing cells is denoted by the dot size. d Lollipop plot showing GO-term enrichment analysis (clusterProfiler; Biological Process) of human tuft-1 and tuft-2 expression profiles. Dot size represents the number of genes within the gene set, stalk length corresponds to significance (one-sided Fisher’s exact test). e Heatmap showing co-expression of tuft-1 and tuft-2 marker genes in human post-mitotic tuft cells (genes with adjusted P value < 0.01 are shown, Wilcoxon rank-sum test with Bonferroni correction). Transcriptomic profiles are ordered by their ratio of human tuft-1 and 2 signature scores, indicated at the top of the heatmap. AVIL and CHAT expression per cell is shown above the heatmap. f Zonation profile of human tuft-1 and tuft-2 populations from (b). Center of mass on crypt-villus axis22 for the top 50 cluster-specific genes is shown. Boxes in Tukey box-and-whisker plot represent interquartile range (IQR, Q1 (25th percentile) to Q3 (75th percentile)), central lines mark median values and whiskers indicate outlier boundaries (1.5*IQR) (ANOVA P = 0.002, Tukey HSD test). g Differential expression analysis between tuft-1 and tuft-2 subtypes in mouse and human (Wilcoxon rank-sum test). Tones of gray represent significance level. Tuft-specific genes, shared between human and mice, are highlighted in red. Contour plot indicates density distribution of these shared tuft-specific genes.
Next, we determined human tuft-1 and tuft-2 signatures and performed GO-term enrichment analysis (Fig. 2d). Akin to mouse, this showed immune-related programs to be enriched in human tuft-2 cells, while human tuft-1 cells showed enrichment of neuron-related programs. Also in the human setting, CHAT was highest expressed in tuft-2 cells whilst pan-tuft cell markers like AVIL were uniformly expressed, and intermediary tuft phenotypes with cells expressing both tuft-1 and tuft-2 markers could be observed (Fig. 2e). Moreover, previous spatial transcriptomics data22 indicate that human tuft cells show a similar zonation profile along the crypt-villus axis as observed in mice, with tuft-2-specific markers found predominantly higher on the crypt-villus axis than tuft-1-specific markers (Fig. 2f).
To perform a more comprehensive cross-species comparison of human and murine tuft cells, we established a core tuft cell expression profile shared between both species and determined differentially expressed genes between the tuft-1 and tuft-2 clusters within each species (Fig. 2g). Most tuft cell-specific genes were expressed in tuft-2 cells in both mouse and human, suggesting that the tuft-2 state is stronger conserved between mouse and human. Vice versa, most genes that show higher expression levels in tuft-1 cells are not unique to the tuft lineage. Taken together, these analyses show similar transcriptomic profiles for tuft cell subtypes in mice and human and suggest a conserved tuft cell differentiation trajectory in which tuft cell precursors transition through an early tuft-1 phenotype into a mature tuft-2 phenotype.
Tuft subtype-specific reporter knock-ins enable identification of tuft-1 and tuft-2 transcriptional states in vitro
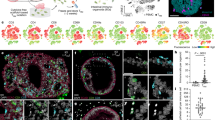
To experimentally investigate the differentiation trajectories that give rise to the continuum of tuft-1 and tuft-2 states and evaluate their chemosensory capacity, we designed tuft subtype-specific reporter organoids. This in vitro system enables functional investigation of rare tuft cells in a controlled environment and allows for on demand enrichment of tuft cell numbers and real-time imaging of their differentiation trajectories. We first established small intestinal organoids from ChatBAC-eGFP mice, in which spontaneous differentiation towards GFP+ tuft cells was occasionally observed in the standard culture medium (Fig. 3a). Notably, the fraction of GFP+ tuft cells barely increased in organoids after a commonly applied treatment with IL-4 and IL-134,5,23,24 (Fig. 3b, c). Like in vivo, GFP+ cells in organoids represented a subset of DCLK1+ tuft cells, recapitulated preferential localization to the villus compartment (Fig. 3d, e) and co-expressed mature tuft-2 markers, such as CD45 and FOLR1 (Supplementary Fig. 3). In sum, these data suggest that ChatBAC-eGFP in organoids, like in vivo, predominantly marks a subset of tuft cells that is enriched for mature tuft-2 markers.
a Representative phase contrast image with fluorescent GFP (green) overlay of small intestinal organoid derived from ChatBAC-eGFP mouse (n = 50 organoids examined). Illustration from NIAID NIH BIOART Source bioart.niaid.nih.gov/bioart/279. b Treatment regimen for tuft cell induction with IL-4 and IL13. E: EGF, N: Noggin, R; R-spondin, IF: immunofluorescence. c Flow cytometry analysis of GFP(Chat)+ cells in organoids cultured in ENR with or without IL-4 and IL-13 according to the regimen shown in (b) (n = 3 independent experiments, t-test). E: EGF, N: Noggin, R; R-spondin. d Quantification of GFP(Chat)+ cells among DCLK1+ cells in crypt and villus region of organoids assessed by immunofluorescence following cytokine treatment as in (b). e Representative image of ChatBAC-eGFP (top, green) organoid following cytokine treatment as in (b) and stained for DCLK1 (bottom, blue). White arrows: DCLK1+ tuft cells. White dashed line: crypt-villus junction (n = 148 DCLK1+ cells examined). f Vulcano plot showing differentially expressed genes (Wilcoxon rank-sum test) between tuft-1 and tuft-2 clusters from the integrated mouse in vivo single-cell transcriptomic dataset (Fig. 1d). Genes selected for reporter knock-ins are highlighted in red. Point size represents tuft specificity (see “Methods”). g Relative expression levels of general tuft cell marker Dclk1 and selected reporter genes in all tuft cells from the integrated mouse in vivo single-cell transcriptomics dataset (Fig. 1d). Cells are ranked based on their normalized tuft-2/1 signature score ratio. h Representative fluorescence image of RNAscope FISH on mouse small intestinal tissue (n = 4) targeting Nrep (green), Gng13 (magenta), Chat (red) and Folr1 (yellow). Bottom: overview image showing entire crypt-villus units with nuclei (blue) and Gng13 (magenta). Top: zoom-ins showing expression of the four markers at four positions along the crypt-villus axis. i Location of cells expressing indicated tuft cell markers on crypt-villus axis. Numbers of analyzed cells are indicated (n = 2 mice). Histograms of counts and normalized density plots are shown. j Co-expression of indicated tuft cell markers along the crypt-villus axis. Boxes in Tukey box-and-whisker plot represent interquartile range (IQR, Q1 (25th percentile) to Q3 (75th percentile)), central lines mark median values and whiskers indicate outlier boundaries (1.5x IQR) (n = 101 cells from 2 mice, ANOVA P = 5.8 × 10−8, Tukey HSD test). k Schematic of knock-in template integrated at C-terminus of reporter genes. NLS: nuclear localization signal. mScarlet-I: monomeric red fluorescent protein. DTR: diphtheria toxin receptor. pA: polyadenylation signal. l Top: Confocal brightfield images of tuft subtype-specific reporter knock-in organoids, overlayed with fluorescent ChatBAC-eGFP signal (green). Bottom: fluorescent overlays of ChatBAC-eGFP+ tuft cells (white) with indicated tuft-subtype reporters (red) and nuclear stain (blue) (n = 20 organoids examined per knock-in). m Experimental setup of reporter lines, culture conditions and flow cytometry to enrich for fluorescent tuft cells prior to plate-based single-cell transcriptomic analysis. n UMAP with unsupervised clustering of organoid single-cell transcriptomic dataset, containing cells from all three knock-in tuft subtype reporter lines and both medium conditions. o Heatmap showing co-expression of tuft-1 and tuft-2 marker genes in organoid tuft cells (genes with adjusted P value < 0.01 are shown, Wilcoxon rank-sum test with Bonferroni correction). Organoid tuft cell transcriptomic profiles are ordered by the ratio of mouse tuft subtype signature scores (tuft-2 score / tuft-1 score). Original tuft subtype (yellow: tuft-1; brown: tuft-2) and normalized fluorescent mScarlet-I and eGFP signals of cells during sorting are indicated for each cell at the top of the heatmap. p Contour plot showing density of tuft-1 and tuft-2 signature scores14 ratios for tuft cells from the integrated in vivo dataset (Fig. 1d, red), as well as the organoid dataset (Fig. 3n, black), indicating underrepresentation of mature tuft-2 cells in vitro. q Violin plots of normalized fluorescent signal for each reporter per indicated cell cluster (ANOVA P < 0.001, ns not significant, *** adjusted P value < 0.0001, * adjusted P value < 0.05, exact P values are included in the Source Data, Tukey HSD test). r Normalized mScarlet-I and eGFP fluorescent signal intensities measured during cell sorting, superimposed on UMAP of organoid single-cell transcriptomic dataset.
Next, we used our integrated in vivo single-cell transcriptomic datasets of mouse small intestinal epithelium to identify more markers that can discriminate between tuft-1 and tuft-2 cells and are not expressed in other epithelial lineages (Fig. 3f). We selected Nrep as a marker gene that demarcates tuft-1 transcriptomic states (Fig. 3f, g, Supplementary Fig. 4a, b). Vice versa, we picked Folr1, which shows highest expression in mature tuft-2 states (Fig. 3f, g, and Supplementary Fig. 4a, b). In addition, we chose Gng13 as an exclusive tuft cell marker, which shows strong expression in tuft-1 and intermediary phenotypes (Fig. 3f, g, and Supplementary Fig. 4a, b). Using RNAscope, we confirmed confined expression of the tuft-1 marker Nrep in the crypt, tuft-2 markers Chat and Folr1 on the villus and Gng13 along the entire crypt-villus axis (Fig. 3h, i). Differentially zonated co-expression of combinations of these markers supported transitioning from tuft-1 to tuft-2 phenotypes on the villus (Fig. 3j, and Supplementary Fig. 4c). Independent genetic knock-ins were integrated at the STOP-codon of these genes and contained a fluorescent mScarlet reporter (monomeric RFP variant) and diphtheria toxin receptor (DTR) that both become separated from the tuft cell markers during translation of the P2A peptides25 (Fig. 3k, and Supplementary Fig. 4d–g). We generated all three reporter lines in the ChatBAC-eGFP background, such that the eGFP signal can be leveraged as a reference to cross-compare the new tuft subtype-specific markers (Fig. 3l). Last, we confirmed confinement of our markers to DCLK1+ tuft cells and zonated localization of tuft-1 and tuft-2 markers to the organoid crypt and villus compartments, respectively (Supplementary Fig. 4h, i).
A prerequisite to study mature tuft cell functioning in organoids, is to confirm that tuft cell phenotypes are accurately recapitulated. To examine the degree of similarity between in vivo and in vitro-derived tuft cells, we sorted the various fluorescent cell populations from our organoids for scRNA-seq (Fig. 3m, and Supplementary Fig. 5a). The ensuing dataset contained >500 tuft cells (Fig. 3n) with tuft-p, tuft-1 and tuft-2 phenotypes that closely resemble their in vivo counterparts as illustrated by strong enrichment of their respective signatures and marker genes (Supplementary Fig. 5b–d). Resemblance between in vivo and in vitro-derived tuft cells is further demonstrated by the many tuft cells that showed an intermediary phenotype with mixed expression of both tuft-1 and tuft-2 markers (Fig. 3o). Cytokine treatment did not induce alternative tuft phenotypes that were not present in the standard culture medium, as illustrated by mixing of tuft cells derived from either medium on the UMAP and a lack of medium-specific tuft clusters (Supplementary Fig. 5e). For a direct comparison, we plotted tuft-1 and tuft-2 signature scores from tuft cells originating from either the integrated in vivo dataset (Fig. 1d) or our in vitro organoid dataset (Fig. 3n). Despite a strong overlap, most in vitro tuft cells were representative of the tuft-1 phenotype (Fig. 3p), presumably a consequence of the crypt-inspired signaling factors in the organoid medium.
Next, we evaluated if our tuft cell subtype-specific reporters reliably indicate the intended tuft cell transcriptomic states. To this end, we superimposed the signal intensities of the fluorescent reporters, measured during cell sorting, on the single-cell transcriptomics data (Fig. 3o, q, r). GFP signal from the ChatBAC-eGFP was most dominant in cells with a high tuft-2 signature score (Fig. 3o, q, r). In contrast, for the tuft-1 marker Nrep we found high mScarlet signals in tuft-1 cells, which extended to cells with intermediary tuft phenotypes (Fig. 3o, q, r). As expected, Gng13 shows a gradual increase of mScarlet signal starting in tuft-1 cells and becoming more abundant towards intermediary and tuft-2 expression profiles (Fig. 3o, q, r). Folr1 marks cells with the highest tuft-2 signature scores (Fig. 3o, q, r). For all markers, we found strong concordance between fluorescent signal and gene expression (Fig. 3o, q, r, Supplementary Fig. 5f). Notably, despite being predicted as tuft-specific by the in vivo single-cell atlases (Supplementary Fig. 4b), Folr1 turned out to be less exclusive for the tuft lineage than the other markers (Fig. 3q, r, and Supplementary Fig. 5g). Nonetheless, Folr1-driven mScarlet expression is a reliable indicator of tuft-2 phenotypes within the tuft cell compartment and can therefore be used in combination with Chat to identify the most mature tuft cells (Fig. 3o, q, r, and Supplementary Fig. 5g). Together, these data show that tuft cell phenotypes, including intermediary states, are accurately recapitulated in organoids and can be readily identified by our tuft type-specific reporters.
Cytokines drive the generation of new tuft-1 cells but are insufficient to advance their maturation
To study type 2 cytokine-induced tuft cell differentiation we characterized the frequency of tuft phenotypes in each of the three reporter lines under standard culture conditions (ENR) and upon tuft cell induction with the cytokines IL-4 and IL-13 (IL4 + IL13) (Fig. 4a). In agreement with our earlier observation that treatment with cytokines leads to a general increase in DCLK1+ tuft cell numbers but not Chat+ cells (Fig. 3b–d), our knock-ins unequivocally demonstrate that the cytokines induce a strong increase in tuft-1 cells (Fig. 4b, c; mScarlet+ cells in the Nrep and Gng13 reporter lines), but not tuft-2 cells (Fig. 4b, c; mScarlet+ cells in the Folr1 reporter line and ChatBAC-eGFP+ cells). Exposure to NOTCH inhibition, WNT stimulation, or longer treatment with cytokines, yielded similar skewed tuft subtype induction, with high frequency of tuft-1 cells and virtually no tuft-2 cells (Supplementary Fig. 6a, b). These observations are in stark contrast to type 2 cytokine-induced tuft cell hyperplasia in vivo, where both post-mitotic tuft cell subtypes increase in numbers, but most prominently the tuft-2 cells14,24 (Supplementary Fig. 6c–e). Importantly, this reveals that the commonly applied in vitro differentiation strategy of type 2 cytokine addition to standard crypt-like organoid medium, induces generation of new tuft cells, but predominantly of the immature tuft-1 subtype.
a Schematic of experimental setup showing organoid knock-in reporters, treatment regimen and readout strategies (timelapse microscopy and flow cytometry). ENR: EGF, Noggin and R-spondin medium. b Representative flow cytometry analysis of the three indicated tuft subtype-specific mScarlet-reporter lines and the parental ChatBAC-eGFP line (parental). All lines were treated with IL-4 and IL-13 for 4 days, 3 days after seeding in ENR medium as illustrated in (a). Percentages of the fluorescent populations are indicated in each plot. Individual panels contain combined data from 3 independent experiments, 5000 cells are shown per plot. c Flow cytometry quantifications of tuft-subtype reporter frequencies (%) upon induction with indicated cytokines as in (a) (n = 5 (Nrep), 3 (Gng13 and Folr1) and 8 (Chat) independent experiments, t-test). d Microscopic stills from live imaging experiments demonstrating the temporal dynamics of tuft subtype reporters. Left: confocal brightfield image of organoids at the start of imaging, overlayed with fluorescent signals of indicated reporters. Right: stills of fluorescence channels (mSarlet-I: red, eGFP: green) from timepoints preceding and following co-expression of fluorescent markers to visualize differential dynamics with respect to ChatBAC-eGFP. Cell that shows co-expression is indicated with a white arrowhead throughout imaging timepoints. e Quantification of mScarlet-I emergence relative to ChatBAC-eGFP expression for the three indicated tuft sutype-specific reporter knock-ins. f Top: Representative confocal brightfield image with overlay of indicated fluorescent reporter. Bottom: Location distribution of tuft-reporter+ cells along the crypt-villus axes within organoids scored at emergence of reporter+ cells versus the end of imaging procedure.
Since both the in vivo and in vitro single-cell transcriptome datasets indicate existence of intermediary tuft cell phenotypes, we treated organoids with cytokines to induce tuft cell specification and tracked the dynamics of tuft cell identity by means of fluorescent reporters, using confocal live-cell imaging (Fig. 4a). First, we noticed clear differences in temporal expression patterns for the tuft-1 and tuft-2 markers within organoids (Fig. 4d, e). Using ChatBAC-eGFP as a reference signal, we observed that tuft-1 marker Nrep and tuft1/2 marker Gng13 become active ahead of Chat expression, while the late tuft-2 marker Folr1 becomes active in cells that were already positive for ChatBAC-eGFP (Fig. 4d, e). In agreement with the lack of binary confinement between tuft cell subtypes, these temporal expression patterns are indicative of a linear differentiation trajectory where tuft-1 cells transition into tuft-2 cells through an intermediary state. This is further supported by the fact that tuft-1 marker expression predominantly emerged in the crypt compartment of organoids, while tuft-2 markers appeared in the villus compartment (Fig. 4f).
Importantly, while type 2 cytokines elicit a massive increase in tuft-1 cells, this was not a consequence of tuft-1 proliferation, as we observed virtually no division of cells positive for our reporters in the time-lapse experiments. To confirm that our early tuft-1 marker Nrep marks post-mitotic tuft cells, we performed live imaging with fluorescent anti-CD24, a known tuft cell marker7,26 that is already expressed in tuft-p cells (Supplementary Fig. 7a). Indeed, anti-CD24 fluorescence was generally detectable before Nrep expression emerged (Supplementary Fig. 7b) and marked divisions more frequently (Supplementary Fig. 7c–e). Notably, most CD24+ (tuft-p) cells exhibited one round of division, similar to what was observed for precursor cells of other mouse small intestinal secretory cell types27. In the rare case of an Nrep+ cell division, Nrep expression was typically initiated just prior to division (Supplementary Fig. 7d, e), while Chat+, Gng13+ or Folr1+ divisions were never observed. In conclusion, these experiments show that our tuft cell subtype-specific reporter knock-ins mark successive stages of maturation of post-mitotic cells committed to the tuft cell lineage. Moreover, in vitro treatment with IL-4 and IL-13 induces a strong increase in the generation of tuft-1 states, but is insufficient to efficiently advance their differentiation to the more mature tuft-2 cell states found on the villus.
Crypt-villus signaling gradients advance maturation from tuft-1 to tuft-2 states
Since tuft cell differentiation in cytokine-exposed organoids (IL-4 and IL-13) predominantly resulted in generation of tuft-1 phenotypes (Nrep and Gng13 Fig. 4c) but not tuft-2 phenotypes (Chat and Folr1 Fig. 4c), we set out to identify signals that may stimulate the transitioning of tuft-1 cells into mature tuft-2 cells. In vivo, tuft-2 markers are found predominantly on the villus (Figs. 1h–k and 3i, j), which is reflected in our reporter organoids by a higher incidence of Chat+ and Folr1+ tuft-2 cells in the villus compartment of organoids (Fig. 4f, >50 cells tracked). Therefore, we adjusted the cytokine differentiation regimen (Fig. 5a, #1) to better represent a villus environment, starting with the depletion of stem cell-niche inspired growth factors EGF, Noggin, and R-spondin (Fig. 5a, #2). Next, we screened for additional factors that are more abundant in the villus environment, such as WNT5A, NRG1 and BMP4, and factors that are produced by tuft cells and could potentially act in an autocrine manner, such as BMP2, acetylcholine and IL25 (Fig. 5a, #3). Subtraction of the stem cell niche factors (ENR) from the medium resulted in higher frequencies of ChatBAC-eGFP+ cells (Fig. 5b, #2). Moreover, addition of villus and tuft cell produced factors further increased the ChatBAC-eGFP+ cell population (Fig. 5b, #3), with BMPs and acetylcholine eliciting the largest effects (Supplementary Fig. 8a). Indeed, according to the single-cell transcriptome datasets, BMP receptors are expressed by all tuft cell subtypes and the BMP response genes Id1 and Id3 are higher expressed in tuft-2 cells (Supplementary Fig. 8b), suggestive of a role for BMP signaling in differentiation to mature tuft-2 states. Concordantly, in Bmpr1a depleted intestinal epithelia28 only the tuft-2, but not the tuft-1, transcriptomic signature is significantly reduced (Supplementary Fig. 8c).
a Experimental setup to promote tuft cell maturation with villus-inspired medium. ChatBAC-eGFP reporter organoids were pretreated with IL-4 and IL-13 on the 3rd day after seeding in ENR (EGF, Noggin and R-spondin) medium. On the 4th day, factors were added to, or depleted from, the medium. Frequency of GFP(Chat)+ tuft-2 induction was tested at day 7 with flow cytometry. b Percentage of ChatBAC-eGFP+ tuft-2 cells in organoids treated with culture medium (ctrl, n = 11) or IL-4 and IL-13 in presence (#1, n = 4) or absence (#2, n = 7) of crypt or villus/autocrine-inspired signaling factors (#3, n = 6) (ANOVA P = 6.8 × 10−7, Tukey HSD test). c Representative flow cytometry analysis of fluorescent population frequencies in indicated reporter organoids treated with crypt (#1) or villus-inspired medium (#3) after IL-4 and IL-13 pretreatment. 5000 cells are shown per plot. d Percentage of mScarlet+ (left) and mScarlet+GFP+ (right) cells in indicated organoid reporter lines treated with crypt or villus-inspired medium (condition #1 vs #3, Fig. 5a, b) measured by flow cytometry (n = 7 (Nrep), 5 (Gng13) and 6 (Folr1) independent experiments, t-test). e Representative fluorescence image of ChatBAC-eGFP;NrepP2A-mScarlet-I organoids treated with crypt or villus-inspired medium regimen (condition #1 vs #3, Fig. 5a, b). White arrows: ChatBAC-eGFP+ cells. f Microscopic stills from live imaging experiments of ChatBAC-eGFP;NrepP2A-mScarlet-I organoids in villus-inspired medium (condition #3 of Fig. 5a, b). Left: confocal brightfield image of organoid at the start of imaging. Right: stills of fluorescence channels (mSarlet-I: red, eGFP: green) from timepoints preceding and following co-expression of fluorescent markers. Cell that shows co-expression is indicated with a white arrowhead throughout imaging timepoints (n = 2 independent experiments). g Average mScarlet-I signal over time in ChatBAC-eGFP;NrepP2A-mScarlet-I organoids treated with crypt or villus-inspired medium regimen (condition #1 vs #3, Fig. 5a, b). Shading in plot represents SEM. Number of cells comprising each graph are indicated. h Barplot showing fraction of mScarlet-I+ cells in ChatBAC-eGFP;NrepP2A-mScarlet-I organoids with stable/rising or declining fluorescence signal in crypt or villus medium with cytokines (IL4 + IL13). Related to g. Number of cells comprising each graph are indicated. i Experimental setup for tuft cell subtype-specific depletion with diphtheria toxin (DT). Organoids were treated as in condition #3 of Fig. 5a, b and DT was added with every medium change. j Percentage of mScarlet+ cells in indicated reporter lines and parental no knock-in control (ChatBAC-eGFP; no mScarlet-I or DTR) after differentiation during depletion with DT (n = 3 independent experiments, t-test). k Percentage of GFP+ cells in indicated reporter lines and parental no knock-in control (ChatBAC-eGFP; no mScarlet-I or DTR) after differentiation during depletion with DT (n = 3 independent experiments, ANOVA P = 0.04, Tukey HSD test). l RNA velocity-based trajectory inference with single cell transcriptomes from organoids treated with ENR, NR + IL4 + IL13 or villus-inspired medium superimposed on UMAP. Direction of arrows predict unidirectional differentiation from tuft-1 to tuft-2 transcriptomic states. m Schematic depicting linear model of intestinal tuft cell differentiation. Tuft-p: tuft precursors; IL4: interleukin-4; IL13: interleukin-13; ACh: acetylcholine; BMPs: bone morphogenetic proteins.
To test the effect of our villus-inspired maturation medium on the transitioning of tuft-1 to tuft-2 phenotypes, we evaluated their frequencies in our reporter organoids. Besides an overall increase in Nrep, Gng13 and Folr1 positive cells, this showed a clear shift in the maturation status of the tuft cell populations indicated by the emergence of double positive Nrep+Chat+ and Gng13+Chat+ tuft-2 cells as well as the most mature Chat+Folr1+ tuft-2 cells (Fig. 5c–e). Using flow cytometry time course experiments we observed these double positive mature tuft-2 phenotypes to arise late during the differentiation regimen (day 7), while an increase in Nrep+ tuft-1 cells was already observable two days prior (day 5) (Supplementary Fig. 9a). Concordantly, time-lapse imaging showed that, also in the villus-inspired maturation medium, Chat+ tuft-2 cells originate from Nrep+ tuft-1 cells (Fig. 5f). Notably, switching to villus-inspired medium substantially changed single-cell expression dynamics, eliciting more transient Nrep expression (Fig. 5g, h), akin to what is observed in vivo (Fig. 3i, j). This decrease in Nrep expression coincides with initiation of Folr1 expression (Supplementary Fig. 9b), which is in line with these two markers labeling successive early and late stages of tuft cell differentiation, respectively.
To evaluate if the transient tuft-1 state is mandatory or facultative during tuft cell differentiation we used diphtheria toxin (DT) mediated subtype ablation via the DT receptor that was simultaneously knocked-in with the mScarlet reporters (Fig. 3k, Supplementary Fig. 4d). Depletion of (Nrep+) tuft-1 cells with DT during our optimized differentiation regimen (Fig. 5i, j), blocked the generation of all tuft-2 cells (ChatBAC-eGFP+) (Fig. 5k, and Supplementary Fig. 9c, d). In contrast, under continuous depletion of late tuft-2 phenotypes (ChatBAC-eGFP+ and Folr1+), early tuft-2 cells (ChatBAC-eGFP+, but Folr1-) did arise (Fig. 5j, k, and Supplementary Fig. 9c, d). These ablation experiments demonstrate that Chat+ tuft-2 cells cannot arise from an alternative cellular source other than Nrep+ tuft-1 cells, in the situation of cytokine-induced tuft cell hyperplasia.
Lastly, we performed scRNA-seq on reporter organoids treated with our villus-inspired maturation medium to confirm a shift in maturation states and exclude induction of alternative non-physiological transcriptomic states. Unsupervised clustering with our previous organoid dataset showed mixing of tuft cells derived from the different medium conditions on the UMAP (Supplementary Fig. 10a). Furthermore, tuft-1 and tuft-2 signature scores were similar in crypt and villus-inspired medium conditions (Supplementary Fig. 10b). This indicates that, while the maturation medium had strong effects on the frequency of tuft-2 cells (Fig. 5c, d), it did not induce tuft cell transcriptomic states that were previously not observed. Moreover, trajectory inference through RNA velocity analysis corroborated the unidirectional differentiation trajectory from tuft-p to tuft-1 to tuft-2, which we repeatedly observed in our live imaging experiments (Fig. 5l). Collectively, these experiments show that crypt-villus signaling gradients control the transitioning of tuft-1 to tuft-2 phenotypes (Fig. 5m).
An organoid-based platform for functional characterization of tuft cell properties
Our tuft-type reporter organoids in combination with the optimized tuft maturation medium provide a powerful in vitro platform to study the basic cell biology that governs mature tuft cell functioning. To demonstrate its utility, we first set out to investigate the dynamics of the typical bottle-shaped tuft cell morphology29 (Fig. 6a), which features distinctive membrane characteristics. In addition to the well-known robust protrusions at the apical surface that form the tuft of the tuft cell, EM studies have also identified the presence of lateral microvilli as well as large basolateral extensions30, which are speculated to be involved in cell-cell communication with neighboring cells29,31,32,33,34,35. Indeed, within our live-cell imaging recordings, we observed long dynamic protrusions. Subsequent imaging of Chat+ cells in combination with a genetically encoded nuclear marker (H2B-mScarlet) confirmed that these dynamic protrusions were basolateral extensions (Fig. 6b, and Supplementary Fig. 11, Supplementary Movie 1), which presented with dynamic growth and shrink rates of more than 10 µm per h (Fig. 6c). Moreover, long protrusions (~20 µm, up to 6 h lifetime) showed more stability than short ones (5 µm, one h lifetime) (Fig. 6d). Basal protrusions were also observed in Nrep+ tuft-1 cells (Fig. 6e). These novel experimental opportunities highlight how future studies may improve our limited understanding of tuft cell morphological changes and its relation to functioning.
a Microscopic images of ChatBAC-eGFP+ cell in organoid showing typical tuft cell morphology (observed in 3 independent experiments). Left: overview confocal brightfield image overlayed with GFP fluorescence channel. Right: zoom-in of indicated region. White dotted line outlines ChatBAC-eGFP+ tuft cell. b Dynamic protrusions in Chat+ tuft-2 cells that can span several neighboring cells. Merge of fluorescence channels is shown: green, eGFP; red, nuclei visualized with H2B-mScarlet-I. Three stills of indicated timepoints are shown. White triangles with letters indicate protrusions. c. Protrusion lengths over time. Triangles with letters indicate single protrusion tracks corresponding to protrusions shown in microscopy images of (b). d Relationship between maximum protrusion length and protrusion lifetime. e Microscopic stills from live imaging of NrepP2A-mScarlet-I-nls organoids stained with anti-CD24, visualizing membrane protrusions of Nrep+ tuft-1 cells. Merge of fluorescence channels is shown: cyan, anti-CD24; red, NrepP2A-mScarlet-I-nls. White triangles with letters indicate protrusions. Three different examples of Nrep+ cells with dynamic protrusions are shown. f Relative expression level of tuft-specific GPCRs within indicated cell types, as extracted from the mouse in vivo integrated single-cell dataset (Fig. 1d) and organoids (Fig. 3n). g Experimental setup for live imaging experiments to monitor tuft cell activation. Tuft-subtype reporter organoids expressing the Tq-Ca-FLITS calcium biosensor (left, representative image of 10 examined organoids) are re-plated to form 2D monolayers that can be apically stimulated and are compatible with high temporal resolution imaging (right). h Fluorescent image of 2D monolayer of reporter organoid with CellMask deep red incubation to label cell boundaries (membranes, white) and mScarlet-I reporter signal to label tuft identity. Image is prior to stimulation with cis-epoxysuccinic acid (cESA) (n = 3 independent experiments). i Fluorescent intensity fluctuations of Tq-Ca-FLITS biosensor within the field-of-view cell monolayer as shown in (d), following stimulation with cESA at t = 0. Time (seconds) post cESA exposure is indicated at the top of each panel. White outline indicates Nrep+ tuft cell. j Single-cell traces of Tq-Ca-FLITS intensity fluctuations within monolayer of (e). Traces are colored by mScarlet-I signal measured in the corresponding cell. k Responsiveness of tuft-reporter positive cells following stimulation with 1.5 mM cESA. Per bar, each point represents a separate experiment and is colored for the medium used (number of cells comprising each bar and p-values are indicated, t-test). l As in panel d, but now monolayer of ChatBAC-eGFP reporter organoid prior exposure to propionate (n = 2 independent experiments). m As in panel e, but Tq-Ca-FLITS intensity fluctuation post propionate exposure. Time since propionate addition is indicated at the top of each panel. Arrow indicates ChatBAC-eGFP+ tuft cell. n Single-cell traces of Tq-Ca-FLITS intensity fluctuations within monolayer of panel i. Traces are colored by GFP signal measured in the corresponding cell. o Responsiveness of non-tuft cells (mScarlet-I− GFP−) in the proximity of the ChatBAC-eGFP+ cell shown in (h). Number of cells comprising each bar are indicated at the top of each bar. Results of one representative experiment are shown.
Next, we investigated tuft cell chemosensing that, similar to cells in the taste buds, involves GPCR-mediated signaling to control intracellular calcium pulses and subsequent membrane depolarization. Therefore, we complemented our tuft-subtype reporter organoids with a genetically encoded calcium biosensor (Tq-Ca-FLITS36), to visualize and measure the GPCR-mediated activation potential of putative stimuli (Supplementary Fig. 12a). We extracted tuft-specific GPCRs from our high-resolution integrated in vivo scRNA-seq dataset (Fig. 6f). Reassuringly, the resulting list contained GPCRs that were previously found to be tuft-specific, such as Sucnr1 and Vmn2r26, as well as some novel tuft-specific GPCRs, like Gabbr1 and Gpr18. Most of the tuft-specific GPCRs recapitulated their in vivo expression pattern in organoids, with expression generally being highest in tuft-2 cells (Fig. 6f). To develop a screening setup that enables apical stimulation and single-cell measurements of calcium influx in real-time, we converted calcium biosensor-expressing reporter organoids into 2D monolayers37,38 [see “Methods”] (Fig. 6g). We first exposed these monolayers to the succinate mimetic cis-epoxysuccinic acid (cESA), a known stimulus (Fig. 6h). As expected, we observed calcium spikes within a minute after administration of the ligand (Fig. 6i). While tuft cells were the first to respond to the stimulus (Fig. 6j), calcium spikes did not remain exclusive to tuft cells and were similar in strength between tuft and non-tuft cells (Supplementary Fig. 12b). This propagation of calcium pulses to nearby non-tuft cells is analogous to what has recently been observed in the trachea39. Concordant with the highest Sucnr1 expression in tuft-2 cells (Fig. 6f), we observed the highest response probability in cells positive for tuft-2 markers (Fig. 6k). As a proof-of-principle of our functional platform to study chemosensing and screen for novel stimuli, we next tested responsiveness to the FFAR3 ligand propionate, a microbiota-derived short chain fatty acid which is speculated but never formally shown to act directly on tuft cells. Stimulation with propionate readily triggered calcium spikes in mature Chat+ tuft cells (Fig. 6l–n), which propagated to nearby non-tuft cells (Fig. 6n, o). Unlike stimulation with the succinate-mimetic cESA, Nrep+ tuft-1 cells showed no response to propionate (Supplementary Fig. 12c), underscoring the importance of studying chemosensing in fully mature functional tuft-2 cells.
Discussion
Tuft cells in the intestine function as epithelial orchestrators of host defense against luminal parasites5,6,7,17. Beyond their role during infections, alterations in intestinal tuft cell numbers and function are associated with immune-related diseases such as inflammatory bowel disease, coeliac disease and neoplasias40. Despite scientific and clinical interest in tuft cells, their rare occurrence in vivo and the absence of accurate in vitro models, have proven to be practical limitations that hamper investigation of the cell biological processes underlying tuft cell functioning. Here, we unlock organoids as an experimental model system to study tuft cell functioning in vitro. We show that the various transcriptomic states of tuft cells in vivo and their interdependencies are truthfully reflected in organoids, optimize culture conditions to enrich for the most mature tuft cell states, and showcase experimental opportunities to study the dynamics of distinctive tuft cell morphology, behavior and chemosensing properties at high resolution.
In our in-depth characterization of in vivo tuft cell phenotypes, upfront enrichment of Chat+ cells prior to single-cell transcriptomic profiling ensured a substantial number of high-quality tuft cell profiles. Through analysis of multiple scRNA-seq datasets from mouse, organoids and human, we consistently find three tuft cell subtypes marked by pan-tuft cell markers (e.g., Dclk1 in mice and POU2F3 and AVIL in mice and human). In addition to the earlier reported tuft-1 and tuft-2 transcriptomic states within the tuft cell pool14, we also document precursor tuft cells (tuft-p) with proliferative capacity that resemble the recently documented proliferative tuft cells in human organoids (there called tuft-3)21. Moreover, our data revealed frequent presence of intermediate tuft cell profiles with co-expression of both tuft-1 and tuft-2 markers. This continuum of transcriptomic states, with no clear demarcation between the two previously described tuft-1 and tuft-2 types, argues against an early binary bifurcation to either of two mature tuft cell types during differentiation. Using tuft subtype-specific reporter knock-ins in organoids, we investigated directionality over the spectrum of tuft profiles by real-time tracking of cell identity and showed that cytokine-induced tuft cell differentiation follows a linear trajectory from a tuft-p to a tuft-1 to a tuft-2 state. This indicates that the transcriptional programs in the tuft cell subtypes are expressed sequentially within the same cell and not by two distinct mature tuft cell subtypes and explains the earlier reported spatial preference of the tuft-1 and tuft-2 cells to the crypt and villus compartments, respectively15. Previous timelapse imaging of organoids from a dual Trpm5GFP:Adgrg2mCherry reporter mouse, in combination with histological analysis of intestinal tissues, also showed emerging expression of tuft-2 marker Adgrg2 in cells that already expressed general tuft cell marker Trpm5, providing independent support for our observations41.
Using our organoid reporters, we show that tuft cell maturation is stimulated by BMP signaling, which also supports functional zonation of enterocytes, goblet cells and enteroendocrine cells along the crypt-villus axis28,42. Previously, BMP2 signaling originating from tuft cells was shown to prevent de novo tuft cell specification from stem cells located at crypt bottoms24. This does not contradict our finding that BMP ligands, increasingly expressed by the intestinal stroma towards villus tips, promote maturation of already existing tuft cells. Similarly, microbiota-derived butyrate has been shown to limit tuft cell specification from stem cells23, but its effect on tuft cells that populate villi is unknown.
While ChatBAC-eGFP was thought to label all cells committed to the tuft cell lineage within the small intestine17, our transcriptomic datasets and organoid timelapse measurements indicate that Chat expression levels emerge in late tuft-1 cells and keep rising during tuft cell maturation. Even though we included cells with minor Chat expression levels in our analyses, our study mainly provides insights into the consecutive steps of tuft cell differentiation and maturation. In contrast, many studies have focused on the initial steps of stem cell specification towards the tuft lineage9,10,11,12,13, and recently the capacity of tuft cells to act as reserve stem cells21. While our data is less suited to investigate early fate decisions, we did observe a significant fraction of tuft cells with a progenitor phenotype. These tuft-p cells expressed pan-tuft cell markers, as well as markers of proliferation and other epithelial lineages, hinting at recent multilineage potential. Consequently, single marker genes specific to the tuft-p cells were not identified. This prohibited their genetic labeling with knock-in reporters in organoids and prevented investigations into how tuft-p cells are generated.
We found few phenotypic traits in tuft-1 cells that are unique to the tuft cell lineage, yet absent in tuft-2 cells. Rather they seem to express most tuft-specific genes at a lower level when compared to tuft-2 cells, akin to a more immature state. Of the few tuft-specific genes that showed higher expression in tuft-1 than in tuft-2, Nrep was the most prominent marker being exclusive to tuft-1 cells in both mice and human. Alternatively, most tuft-specific genes, and in particular GPCRs that play a central role in chemosensing, as well as immune effector molecules, showed increasing expression levels from tuft-1 to tuft-2 in both mouse and human. Furthermore, our organoid models allowed us to show that both tuft-1 and tuft-2 cells generate highly dynamic basolateral protrusions of several tens of microns long. For future studies, it will be intriguing to investigate how diverse input signals are processed by tuft cells, how they are communicated to other cells, and the relationship between dynamic morphology and function.
Understanding the unique cell biology that underlies tuft cell functioning is of high interest to identify therapeutic targets to intervene in immune-related disorders and infections in the intestinal tract. The findings of our study show high similarity between the mature tuft cell phenotypes in mice, organoids and human. Moreover, mouse organoids are fully complementary with in vivo models and will be instrumental to generate mechanistic insights that can then be tested in physiological settings. Our comprehensive characterization and comparison of tuft cell phenotypes in vivo and in organoids, paired with reporter models and optimized differentiation strategies, established a functional platform to address the poorly understood mechanisms of perception, processing and transmission of chemosensory signals by tuft cells.
Methods
Animals
ChatBAC-eGFP mice were obtained from the Jackson Laboratory and housed at the Hubrecht institute and Netherlands Cancer Institute mouse facilities under specific pathogen-free (SPF) conditions. For all experiments, adult mice at least 6 weeks of age, of either sex, were used. Animal experiments were approved by the IvD HI-KNAW and Netherlands Cancer Institute animal welfare committees and conducted according to relevant guidelines and regulations.
Visualization of Chat BAC-eGFP in mouse small intestinal tissue
Slide preparation for single-cell-resolution imaging of fluorescent proteins in their three-dimensional near-native environment (Fig. 1a, j) was performed as in Snippert et al.43 In short, intestines were fixed in 4% paraformaldehyde (PFA) at room temperature for 20 min and washed in cold PBS. 1 cm2 of intestinal wall was transferred to a mold. Four percent low melting point agarose (40 °C) was added and allowed to cool on ice. Once solid, a vibrating microtome was used to make semi-thick sections (150 μm). Sections were stained with anti-mouse β-catenin (Sigma C2206, 1:200), overnight at 4 °C with mild rocking. After washing, sections were incubated with AF568 goat anti-rabbit secondary antibody (Invitrogen A11036, 1:500) and counterstained with Hoechst 33342 (ThermoFischer Scientific 62249, 1:2000). Sections were embedded in Vectashield (Vector Laboratories) after a final washing step. Image acquisition was performed on a Leica SP8 WLL scanning confocal microscope (40x-water-N.A.1.1) with Leica Application Suite X. Post-acquisition image processing and analysis were performed with FIJI ImageJ 2.16.0/1.54 g.
EdU labeling and immunofluorescence of mouse small intestinal tissue
To label cells in S phase, 1 mg of EdU (200 μl in PBS) was injected intraperitoneally 4 h prior to euthanasia. Mouse intestines were excised, rinsed with PBS, opened longitudinally, and fixed overnight in 1% PFA. Following fixation, intestines were washed with PBS, rolled into Swiss rolls, and embedded in parrafin for formalin-fixed parrafin embedded (FFPE) tissue blocks or OCT compound (frozen at −80 °C) for cryosectioning. FFPE samples (Fig. 1h) were sectioned and slides were stained with anti-DCLK1 (Abcam ab37994, 1:1000) and anti-GFP (Aves GFP-1020, 1:4000). OCT-embedded tissues (Fig. 1i, l) were cryosectioned and stained with anti-DCLK1 (Abcam EPR6085, 1:1000) and anti-GFP (Abcam EPR14104, 1:1000). EdU-positive cells were visualized using the Click-iT reaction, according to the manufacturer’s instructions (Click-iT EdU Imaging Kit, Thermo Fisher Scientific/Invitrogen C10340). Secondary antibodies included: Donkey anti-goat IgG Alexa Fluor 488 (A-11055, 1:1000), Donkey anti-rabbit IgG (H + L) Alexa Fluor 568 (A-10042, 1:1000) and Alexa Fluor 647 from the Click-iT reaction (C10340, 1:200). DAPI (1:000) was used for nuclear counterstaining. Cryosections were mounted using antifade mounting medium (Vectashield, Vector Laboratories). Imaging was performed with an Olympus VS200 slide scanner (FFPE) or a Leica TCS SP8 confocal microscope (cryosections; 25x water immersion objective; HC FLUOTAR L, N.A. 0.95 W, VISIR, 0.17 mm cover glass, FWD 2.4 mm). Post-acquisition analysis and quantification were performed with QuPath 0.5.1.
RNAscope fluorescent in situ hybridization assay
Prior to FISH, small intestinal tissues were fixed and embedded as described above to make FFPE tissue blocks. FFPE blocks from ChatBAC-eGFP mice or wildtype C57BL/6 mice (acknowledgements) were cut (5 μm slice thickness) and stained with probes targeting Nrep (Biotechne 562221), Gng13 (Biotechne 462531-C4), Chat (Biotechne 408731-C3) and Folr1 (Biotechne 575401-C2) using the RNAscope Multiplex Fluorescent Detection Kit v2 (Biotechne 323110) according to the manufacturer’s instructions. In brief, paraffin sections were deparaffinized, treated with hydrogen peroxide for 10 min and boiled in target retrieval buffer for 15 min before a 30-min protease treatment. Probes were amplified and detected using fluorescent probes based on opal dyes. Slides were counterstained with DAPI for 30 s, mounted using ProLong Gold Antifade Mountant (Thermo Fisher scientific) and imaged on an Olympus VS200 slide scanner. Post-acquisition analysis and quantification were performed with QuPath 0.5.1.
Organoid derivation and culture
Small intestinal crypts were isolated as described previously44. In brief, mouse small intestines were excised, rinsed with PBS and incubated with DPBS containing 5 mM EDTA for 40 min at 4 °C on a carousel to extract crypts. Crypts were pelleted by centrifugation of the supernatant, washed with advanced DMEM (Gibco) and plated in drops of Matrigel (Corning) to form organoids. Organoids were derived and cultured with ENR medium: advanced DMEM/F12 (Gibco) supplemented with 1% Penicillin/Streptomycin (P/S, Lonza), 1% HEPES buffer (Gibco) and 1% GlutaMAX (Gibco), 5% R-spondin conditioned medium (in-house production), 10% Noggin conditioned medium (in-house production), 1x B27 (Invitrogen), 1.25 mM N-acetylcysteine (Sigma-Aldrich) and 50 ng/ml EGF (Invitrogen). Organoids were maintained at 37 °C with 5% CO2 and passaged weekly through fragmentation by shear-stress (pipetting). Medium was refreshed every 2 days.
Organoid tuft cell differentiation
3D organoid cultures were subjected to differentiation regimens as indicated. In general, tuft cell specification was induced on the 3rd day after passaging with ENR medium supplemented with recombinant murine IL-4 and IL-13 (10 ng/ml each, Immunotools). Thereafter, tuft cell maturation was facilitated by removal of EGF, Noggin and R-spondin from the medium and addition of one of the following or a combination thereof on the 4th day after passaging: recombinant murine IL-25 (20 ng/ml, Immunotools), recombinant human BMP2 (20 ng/ml, Immunotools), recombinant human BMP4 (20 ng/ml, Immunotools), recombinant human NRG1 (20 ng/ml, R&D systems), recombinant murine WNT5a (20 ng/ml, Biotechne) and acetylcholine chloride (100 μM, Sigma Aldrich). For investigation of alternative tuft cell inducing factors (Supplementary Fig. 6a, b), DAPT (5μM Sigma D5942) and Wnt-surrogate FC fusion protein (0.5 nM, U-Protein Express) were used. For DT depletion experiments, medium was supplemented with 50 ng/ml Diphtheria toxin (Sigma D0564). Organoids were harvested for downstream analysis on the 7th day after passaging unless otherwise stated.
Tissue and organoid dissociation for flow cytometry and sorting
For isolation of tuft cells from mouse tissue, small intestines were excised, rinsed with PBS and incubated for 10 min in PBS containing 100 μM DTT. Next, the small intestines were cut into smaller fragments, transferred into DPBS containing 5 mM EDTA and spun on a carousel for 40 min at 4 °C. Organoids were extracted from Matrigel with cold advanced DMEM. To make single-cell suspensions, tissue fragments or organoids were trypsinized for 5 min at 37 °C with TypLE containing 10 μM Y-27632 Rho-kinase inhibitor (Gentaur) to inhibit anoikis. Trypsinized single-cell preparations were washed with advanced DMEM, filtered (40 μm cell strainer) and resuspended in advanced DMEM containing 10 μM Y-27632. In case of immunofluorescence antibody stainings, single-cell suspensions were stained for 30 min on ice with APC anti-mouse EpCAM (Biolegend 118213, 1:100) and AF700 anti-mouse CD45 (Biolegend 103127, 1:100). Cells were stained briefly with DAPI prior to flow cytometry.
Flow cytometry
Single-cell suspensions were prepared as described above. Flow cytometry measurements were performed on a BD FACSCelesta CellAnalyzer. Single live cells (DAPI negative) were gated and GFP and mScarlet-I fluorescence was measured in the FITC-A and PE-A channels, respectively. Gates were set based on negative control samples. Flow cytometry data was analyzed and visualized using BD FACSdiva software 9.2 and the flowCore (2.14.0) and CytoExploreR (1.1.0) R packages.
Plate-based scRNA-seq
Single-cell suspensions were prepared as described above. Viable single cells (DAPI negative) were sorted (BD FACSAria III) into 384-well cell-capture plates from Single Cell Discoveries, which contain a 50 nl droplet of well-specific barcoded primers and 10 µl of mineral oil (Sigma M8410). After sorting, plates were briefly centrifuged at 500 × g and then kept on dry ice till further storage at –80 °C. Single-cell RNA sequencing was performed by Single Cell Discoveries according to an adapted version of the SORT-seq protocol45 with primers described in van den Brink et al.46. Cells were heat-lysed at 65 °C followed by cDNA synthesis. After second-strand cDNA synthesis, all the barcoded material from one plate was pooled into one library and amplified using in vitro transcription (IVT). Following amplification, library preparation was performed following the CEL-Seq2 protocol47 to prepare a cDNA library for sequencing using TruSeq small RNA primers (Illumina). The DNA library was sequenced by paired-end sequencing on an Illumina Nextseq™ 500, high output, with a 1 × 75 bp Illumina kit (read 1: 26 cycles, index read: 6 cycles, read 2: 60 cycles).
scRNA-seq analysis
For alignment of reads, an adopted version of the nf-core scrnaseq pipeline (2.4.0)48 was used (https://github.com/gowanaka/nf-core-scrnaseq). In brief, STARsolo (2.7.10b) was used to align reads to a custom mm10 transcriptome including eGFP and P2A-mScarlet-I-P2A-DTR transgenes and ERCC spike-ins. Following mapping, count matrices were generated with STARsolo (2.7.10b). Gene expression was analyzed using Seurat (5.0.1)49. Cells with 30% mitochondrial content, <25% exogenous ERCC spike-in content and >1000 detected genes were selected for downstream analysis. Mitochondrial transcript counts were removed prior to count normalization and scaling by the Seurat NormalizeData and ScaleData functions, respectively. Seurat CCA-integration was performed to eliminate technical plate-based batch effects and subsequently unsupervised clustering was used to cluster cells according to the standard Seurat workflow. Gene expression signature scores were calculated with the Seurat AddModuleScore function. Differential expression analysis was performed with the FindAllMarkers function. Tuft specificity scores for Fig. 3f were calculated as log10((1/average expression in non-tuft epithelial cells)+1). Integration with published single-cell transcriptomic datasets (Haber et al. GSE92332, Zwick et al. GSE201859 and Böttcher et al. GSE152325) was performed with the Seurat package (harmony integration) according to the standard workflow. Clusters were annotated according to the cell type annotations included with the published datasets. GO-term enrichment analysis was performed with clusterProfiler (4.8.3)50. RNA velocity analysis was performed with scVelo (0.3.2)51.
Analysis of published RNA sequencing data
Transcript counts from mice infected with Nippostrongylus brasiliensis and Bmpr1a knockout mice were obtained from E-MTAB-918324 and GSE19400428, respectively. The R package DESeq2 (1.40.2)52 was used to perform differential expression analysis, followed by gene set enrichment analysis (GSEA) with clusterProfiler (4.8.3)50 and visualization with enrichplot (1.20.3).
Generation of organoid knock-ins
Organoid knock-ins were generated by in-trans paired nicking (ITPN) as described in Bollen and Hageman et al25. (Supplementary Fig. 4d–g). Cas9 D10A nickase (addgene #48141) locus-specific expression vectors were generated according to published protocols53. For transfection, organoids were trypsinized to cell clumps containing ~5 cells (1 × 106 cells total) and coelectroporated with 15 μg DNA and 5 ug of Cas9 D10A nickase and targeting vector using the NEPA21 Super Electroporator (Nepagene) following described conditions54. Electroporated cell clumps were plated in Matrigel overlayed with ENR culture medium, which was supplemented with 10 μM Y-27632 Rho-kinase inhibitor (Gentaur) and 0.25 nM Wnt surrogate-FC fusion protein (U-Protein Express) for the first 3 days. Targeted cells were selected using 2 μg/ml puromycin (Sigma) and maintained as polyclonal populations. Proper integration of the knock-in constructs was validated by targeted PCR of genomic organoid DNA (isolated with QIAamp DNA Micro Kit from Qiagen, according to the manufacturer’s instructions) using the primers listed in Supplementary Table 1, followed by Sanger sequencing of the PCR product.
Lentiviral transduction of organoids
Organoids were transduced with lentivirus encoding hEF1a-Tq-Ca-FLITS or H2B-mScarlet-I followed by an IRES and a blasticidin resistance cassette. In brief, organoids were incubated in trypsin at 37 °C to make a suspension of cell clumps containing ~5–10 cells. Cell clumps were transduced by spinoculation for 1 h at room temperature and then plated in Matrigel overlayed with ENR culture medium, which was supplemented with 10 μM Y-27632 Rho-kinase inhibitor (Gentaur) and 0.25 nM Wnt surrogate-FC fusion protein (U-Protein Express) for the first 3 days. Transduced organoids were selected using 3 μg/ml blasticidin (InvivoGen) and maintained as polyclonal populations.
Calcium imaging of 2D organoid monolayers
For calcium imaging of 2D monolayers, organoids were fragmented and seeded on polyacrylamide gels (18 kPa stiffness) that had been coated overnight with 100 µg/ml Laminin (Sigma-Aldrich L2020) and 250 µg/ml Collagen I (First Link (UK)) as in Pérez-González et al.37. After seeding, organoid fragments were overlayed with ENR medium (described above), which was supplemented with 1x N2 Supplement (Thermo Scientific), 10 ng/ml human-FGF2 (Peprotech), 3 µM Chiron (Bio-Connect), 10 µM Y-27632 and 100 ug/ml primocin (InvivoGen), and contained a higher amount of R-spondin conditioned medium (20%). Chiron and Y-27632 were removed from the medium one day later. For differentiation of 2D monolayers, IL-4 and IL-13 (10 ng/ml each) were added 2 days before imaging, followed by addition of BMP2 and BMP4 (20 ng/ml each) in combination with removal of Noggin from the medium on the day prior to imaging. Timelapse imaging was performed 3-4 days after seeding of organoid crypts, when monolayers formed mature crypt-villus compartments, on a Leica SP8 WLL scanning confocal microscope (40x-water-N.A.1.1, 37 °C, 6% CO2) with Leica Application Suite X at 1 frame per second using resonant scanning (8000 Hz). The genetically encoded Tq-Ca-FLITS36 calcium biosensor was excited with an ultraviolet laser at 405 nm, to minimize bleed through of EGFP in the Tq-Ca-FLITS channel, and emission was measured at 445–505 nm. Organoid monolayers were stimulated during imaging with cis-epoxysuccinic acid (cESA, 1.5 mM, Fischer Scientific) or propionate (2 mM, Sigma-Aldrich P1880), as indicated. CellMask Deep Red (1:20000, Invitrogen, 640 nm excitation and 655–710 nm emission) was added 30 min prior to imaging to stain plasma membranes and enable post-acquisition cell segmentation. Before acquisition of each timelapse, EGFP (488 nm excitation and 495–545 nm emission) and mScarlet-I (570 nm excitation and 590-630 nm emission) signals were measured to determine tuft reporter status per cell in the field of view.
Live organoid timelapse imaging
For live imaging, organoids were mechanically fragmented and seeded in cold BME (Trevigen) in a four-well chambered cover glass (#1.5 high-performance cover glass, Cellvis). After seeding, wells were placed on a cold block for 10 min allowing the organoids to sink to a position close to the glass, as described earlier27. Organoids were cultured for 2 to 3 days in ENR medium until clear crypt-villus structures were apparent. Next, IL4 and IL13 (10 ng/ml each) were added to the medium and organoids were imaged for 3 days, starting on the 1st or 5th day after interleukin treatment to capture transitioning fluorescence phenotypes of all tuft reporters. For live staining of CD24, APC anti-CD24 (ThermoFischer 17-0242-80) was used. Imaging was performed at 37 °C and 5% CO2 with a scanning confocal microscope, using either the Leica TCS SP8 with a 40× water immersion objective (numerical aperture, 1.10) or the Nikon A1R MP with a 40x oil immersion objective (NA = 1.30). The voxel size used was 0.4 × 0.4 × 2 micron for both microscopes. The maximum time resolution used was 24 min per frame.
Timelapse analysis
All tracking and timelapse analyses were performed using OrganoidTracker 2.055. To measure marker intensities, the average fluorescent intensity around the nucleus center was measured using a Gaussian kernel with a width of 1.5 pixels. To avoid measuring bleed through from the green (ChatBAC-eGFP) channel we subtracted the green channel from the red channel (all other markers) such that there was no signal in the cytoplasm, where Nrep/Gng13/Folr1P2A-mScarlet-I-nls marker signal should be absent. Tuft cell locations were determined by evaluation of transmission signal. Cells in the spherical end of organoid buds were categorized as located in the crypt, while cells that fitted into a sphere drawn around the central villus region were deemed to be in the villus. Cells in the narrow region between crypt and villus compartment were deemed to be in the neck region. The crypt and the neck region were combined when analyzing the colocalization of ChAT and DCLK1 (Fig. 3d). For the analysis of the relative timing of marker expression and the locations of emergence, data from at least three replicates was pooled. For the NrepP2A-mScarlet-I signal in villus-inspired and standard crypt medium, a representative organoid was analyzed.
Automated tuft cell protrusion analysis
To track protrusions, we first extracted a 3D cell mask by thresholding on the ChatBAC-eGFP signal. We then performed binary erosion and dilation operations to extract the mask of the cell body. The difference between the cell body and the full cell mask gave us the protrusions. Protrusion tips were defined by the pixel in the protrusion that was furthest away from the cell body. Protrusions were linked by connecting protrusions to the protrusion with which they shared maximum overlap in the next frame. If there was little or zero overlap (for instance if protrusions were very thin or small), protrusions were connected with the tips closest to each other. Using the nuclear marker, tuft cells were automatically rotated such that the epithelial plane was in XY. For this, we locally fitted an ellipsoid to the nuclear signal, so that the two major axes defined a plane tangential to the epithelial layer. These could then be used to find the proper rotation needed.
Immunostaining of organoids
Immunofluorescence of organoids was performed as in Dekkers et al.56. Briefly, organoids were harvested from Matrigel droplets with cold advanced DMEM/F12 (Gibco) supplemented with 1% Penicillin/Streptomycin (P/S, Lonza), 1% HEPES buffer (Gibco) and 1% GlutaMAX (Gibco) and pelleted by centrifugation. Following fixation with 4% paraformaldehyde in PBS at 4 °C for 45 min, organoids were permeabilized with 0.1% (vol/vol) PBS-Tween and blocked with PBS containing 0.2% bovine serum albumin and 0.1% Triton X-100 (4 °C for 15 min). Primary antibodies (anti-mouse CD45, 1:200, Biolegend 103102; anti-mouse Folr1, 1:100, R&D Systems AF6936-SP) were incubated overnight in blocking buffer at 4 °C with mild rocking. After washing in blocking buffer (4x), secondary antibodies (AF568 donkey anti-Sheep, Invitrogen A21099, 1:500; AF568 goat anti-rat, Invitrogen A11077, 1:500) were incubated, together with Hoechst 33342 (ThermoFischer Scientific 62249, 1:2000), in the same manner. Finally, organoids were washed in blocking buffer (4x) and mounted on glass slides in a fructose-glycerol clearing solution (60% (vol/vol) glycerol and 2.5 M fructose). Image acquisition was performed on a Leica SP8 WLL scanning confocal microscope (40x-water-N.A.1.1) with Leica Application Suite X. Post-acquisition image processing and analysis were performed with FIJI ImageJ 2.16.0/1.54 g.
Quantification and statistics
Statistical analysis was performed as noted in figure legends using R (R base, ggplot2 (3.5.1), ggpubr (0.6.0) and Seurat packages (5.0.1)). All experiments were performed in multiple distinct replicates. Data distribution was assumed to be normal, but this was not formally tested. Statistical tests were two-tailed Student’s t-tests unless otherwise stated. For comparisons between more than two sample groups, one-way ANOVA was performed, using Tukey HSD for post-hoc analysis. Data are presented as mean +/- standard deviation, unless otherwise stated in the figure legend. For single-cell dot plots, average expression is represented by dot color and percentage of expressing cells is denoted by the dot size.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Organoid and primary tissue single-cell RNA sequencing data generated in this study (including clustered Seurat objects as featured in the figures) can be accessed at GSE276950. Source data are provided with this paper.
References
Kotas, M. E., O’Leary, C. E. & Locksley, R. M. Tuft cells: context- and tissue-specific programming for a conserved cell lineage. Annu. Rev. Pathol. 18, 311–335 (2023).
Banerjee, A. et al. Succinate produced by intestinal microbes promotes specification of tuft cells to suppress ileal inflammation. Gastroenterology 159, 2101–2115 (2020).
O’Keefe, R. N. et al. A tuft cell - ILC2 signaling circuit provides therapeutic targets to inhibit gastric metaplasia and tumor development. Nat. Commun. 14, 6872 (2023).
Howitt, M. R. et al. Tuft cells, taste-chemosensory cells, orchestrate parasite type 2 immunity in the gut. Science (1979) 351, 1329–1333 (2016).
Gerbe, F. et al. Intestinal epithelial tuft cells initiate type 2 mucosal immunity to helminth parasites. Nature 529, 226–230 (2016).
von Moltke, J., Ji, M., Liang, H.-E. & Locksley, R. M. Tuft-cell-derived IL-25 regulates an intestinal ILC2-epithelial response circuit. Nature 529, 221–225 (2016).
Billipp, T. E. et al. Tuft cell-derived acetylcholine promotes epithelial chloride secretion and intestinal helminth clearance. Immunity 57, 1243–1259.e8 (2024).
Xiong, Z. et al. Intestinal Tuft-2 cells exert antimicrobial immunity via sensing bacterial metabolite N-undecanoylglycine. Immunity 55, 686–700.e7 (2022).
Gerbe, F. et al. Distinct ATOH1 and Neurog3 requirements define tuft cells as a new secretory cell type in the intestinal epithelium. J. Cell Biol. 192, 767–780 (2011).
Westphalen, C. B. et al. Long-lived intestinal tuft cells serve as colon cancer-initiating cells. J. Clin. Invest 124, 1283–1295 (2014).
Bjerknes, M. et al. Origin of the brush cell lineage in the mouse intestinal epithelium. Dev. Biol. 362, 194–218 (2012).
Gracz, A. D. et al. Sox4 promotes Atoh1-independent intestinal secretory differentiation toward tuft and enteroendocrine fates. Gastroenterology 155, 1508–1523.e10 (2018).
Herring, C. A. et al. Unsupervised trajectory analysis of single-Cell RNA-Seq and imaging data reveals alternative tuft cell origins in the gut. Cell Syst. 6, 37–51.e9 (2018).
Haber, A. L. et al. A single-cell survey of the small intestinal epithelium. Nature 551, 333–339 (2017).
Manco, R. et al. Clump sequencing exposes the spatial expression programs of intestinal secretory cells. Nat. Commun. 12, 3074 (2021).
Nadjsombati, M. S. et al. Detection of succinate by intestinal tuft cells triggers a type 2 innate immune circuit. Immunity 49, 33–41.e7 (2018).
Ndjim, M. et al. Tuft cell acetylcholine is released into the gut lumen to promote anti-helminth immunity. Immunity 57, 1260–1273.e7 (2024).
Biton, M. et al. T helper cell cytokines modulate intestinal stem cell renewal and differentiation. Cell 175, 1307–1320.e22 (2018).
Böttcher, A. et al. Non-canonical Wnt/PCP signalling regulates intestinal stem cell lineage priming towards enteroendocrine and Paneth cell fates. Nat. Cell Biol. 23, 23–31 (2021).
Zwick, R. K. et al. Epithelial zonation along the mouse and human small intestine defines five discrete metabolic domains. Nat. Cell Biol. 26, 250–262 (2024).
Huang, L. et al. Tuft cells act as regenerative stem cells in the human intestine. Nature 634, 929–935 (2024).
Harnik, Y. et al. A spatial expression atlas of the adult human proximal small intestine. Nature 632, 1101–1109 (2024).
Eshleman, E. M. et al. Microbiota-derived butyrate restricts tuft cell differentiation via histone deacetylase 3 to modulate intestinal type 2 immunity. Immunity 57, 319–332.e6 (2024).
Lindholm, H. T. et al. BMP Signaling in the intestinal epithelium drives a critical feedback loop to restrain IL-13-driven tuft cell hyperplasia. Sci. Immunol. 7, eabl6543 (2022).
Bollen, Y. et al. Efficient and error-free fluorescent gene tagging in human organoids without double-strand DNA cleavage. PLoS Biol. 20, e3001527 (2022).
Feng, X. et al. Tuft cell IL-17RB restrains IL-25 bioavailability and reveals context-dependent ILC2 hypoproliferation. Nat. Immunol. 26, 567–581 (2025).
Zheng, X. et al. Organoid cell fate dynamics in space and time. Sci. Adv. 9, eadd6480 (2023).
Beumer, J. et al. BMP gradient along the intestinal villus axis controls zonated enterocyte and goblet cell states. Cell Rep. 38, 110438 (2022).
Silverman, J. B., Vega, P. N., Tyska, M. J. & Lau, K. S. Intestinal tuft cells: morphology, function, and implications for human health. Annu Rev. Physiol. 86, 479–504 (2024).
Hoover, B. et al. The intestinal tuft cell nanostructure in 3D. Sci. Rep. 7, 1652 (2017).
Saqui-Salces, M. et al. Gastric tuft cells express DCLK1 and are expanded in hyperplasia. Histochem Cell Biol. 136, 191–204 (2011).
Cheng, X., Voss, U. & Ekblad, E. Tuft cells: Distribution and connections with nerves and endocrine cells in mouse intestine. Exp. Cell Res 369, 105–111 (2018).
Bezençon, C. et al. Murine intestinal cells expressing Trpm5 are mostly brush cells and express markers of neuronal and inflammatory cells. J. Comp. Neurol. 509, 514–525 (2008).
Krasteva, G. et al. Cholinergic chemosensory cells in the trachea regulate breathing. Proc. Natl Acad. Sci. USA 108, 9478–9483 (2011).
Ting, H.-A. & von Moltke, J. The immune function of tuft cells at gut mucosal surfaces and beyond. J. Immunol. 202, 1321–1329 (2019).
van der Linden, F. H. et al. A turquoise fluorescence lifetime-based biosensor for quantitative imaging of intracellular calcium. Nat. Commun. 12, 7159 (2021).
Pérez-González, C. et al. Mechanical compartmentalization of the intestinal organoid enables crypt folding and collective cell migration. Nat. Cell Biol. 23, 745–757 (2021).
Thorne, C. A. et al. Enteroid monolayers reveal an autonomous WNT and BMP circuit controlling intestinal epithelial growth and organization. Dev. Cell 44, 624–633.e4 (2018).
Perniss, A. et al. A succinate/SUCNR1-brush cell defense program in the tracheal epithelium. Sci. Adv. 9, eadg8842 (2023).
Hendel, S. K. et al. Tuft cells and their role in intestinal diseases. Front Immunol. 13, 822867 (2022).
Grunddal, K. V. et al. Adhesion receptor ADGRG2/GPR64 is in the GI-tract selectively expressed in mature intestinal tuft cells. Mol Metab 51, (2021).
Beumer, J. et al. Enteroendocrine cells switch hormone expression along the crypt-to-villus BMP signalling gradient. Nat. Cell Biol. 20, 909–916 (2018).
Snippert, H. J., Schepers, A. G., Delconte, G., Siersema, P. D. & Clevers, H. Slide preparation for single-cell-resolution imaging of fluorescent proteins in their three-dimensional near-native environment. Nat. Protoc. 6, 1221–1228 (2011).
Sato, T. et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 (2009).
Muraro, M. J. et al. A single-cell transcriptome atlas of the human pancreas. Cell Syst. 3, 385–394.e3 (2016).
van den Brink, S. C. et al. Single-cell sequencing reveals dissociation-induced gene expression in tissue subpopulations. Nat. Methods 14, 935–936 (2017).
Hashimshony, T. et al. CEL-Seq2: sensitive highly-multiplexed single-cell RNA-Seq. Genome Biol. 17, 77 (2016).
Peltzer, A. et al. nf-core/scrnaseq: 2.5.1. https://doi.org/10.5281/zenodo.10554425 (2024).
Hao, Y. et al. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat. Biotechnol. 42, 293–304 (2024).
Wu, T. et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innov. (Camb. (Mass.)) 2, 100141 (2021).
Bergen, V., Lange, M., Peidli, S., Wolf, F. A. & Theis, F. J. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat. Biotechnol. 38, 1408–1414 (2020).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Ran, F. A. et al. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013).
Fujii, M., Matano, M., Nanki, K. & Sato, T. Efficient genetic engineering of human intestinal organoids using electroporation. Nat. Protoc. 10, 1474–1485 (2015).
Kok, R. N. U. et al. OrganoidTracker: efficient cell tracking using machine learning and manual error correction. PLoS One 15, e0240802 (2020).
Dekkers, J. F. et al. High-resolution 3D imaging of fixed and cleared organoids. Nat. Protoc. 14, 1756–1771 (2019).
Acknowledgements
We gratefully acknowledge all lab members for reagents, suggestions, and discussions and the UMCU Flow Core Cytometry Facility and L. Kleij for support with flow cytometry and microscopy experiments. We thank Prof. Dr. Gadella (van Leeuwenhoek Centre for Advanced Microscopy, Swammerdam Institute for Life Sciences, University of Amsterdam) for providing the Tq-Ca-FLITS plasmid and Dr. van Es (Hubrecht Institute, KNAW, Utrecht) for providing wildtype C57BL/6 small intestinal FFPE tissues. This work is funded by the ‘Organoids in time’ (OCENW.GROOT.2019.085) and Gravitation programme (IMAGINE!; 024.005.009) from the Netherlands Organization for Scientific Research (NWO). This work is part of the Oncode Institute, which is partly financed by the Dutch Cancer Society.
Author information
Authors and Affiliations
Contributions
Conceptualization, J.R.B.d.A. and H.J.G.S.; Methodology, J.R.B.d.A., J.H.H. and R.M.H.; Investigation, J.R.B.d.A., J.H.B., M.A.B., V.E.G., H.B., T.V., I.V-K., M.C.H., I.J. and D.L.; Formal analysis, J.R.B.d.A. and M.A.B.; Data curation, S.R.B.; Dynamic protrusion analysis, M.A.B.; Visualization, J.R.B.d.A. and M.A.B.; Writing – original draft and revised version, J.R.B.d.A. and H.J.G.S.; Resources, M.G.; Supervision, H.J.G.S., J.S.v.Z. and S.J.T.; Funding acquisition, H.J.G.S., J.S.v.Z., S.J.T., J.v.R. and H.C.
Corresponding author
Ethics declarations
Competing interests
H.C. is the head of Pharma Research and Early Development at Roche, Basel, and holds several patents related to organoid technology. The full disclosure is given at https://www.uu.nl/staff/JCClevers/. The other authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Alain Chariot, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Buissant des Amorie, J.R., Betjes, M.A., Bernink, J.H. et al. Intestinal tuft cell subtypes represent successive stages of maturation driven by crypt-villus signaling gradients. Nat Commun 16, 6765 (2025). https://doi.org/10.1038/s41467-025-61878-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-61878-9