Abstract
The subcellular positioning of organelles is critical to their function and is dynamically adapted to changes in cell morphology. Yet, how cells sense shifts in their dimensions and redistribute organelles accordingly remains unclear. Here we reveal that cell-size-scaling of mitochondria distribution and function is directed by polarised trafficking of mRNAs. We identify a 29 bp 3’UTR motif in mRNA encoding TRAK2, a key determinant of mitochondria retrograde transport, that promotes cell-size-dependent targeting of TRAK2 mRNA to distal sites of cell protrusions. Cell-size-scaled mRNA polarisation in turn scales mitochondria distribution by defining the precise site of TRAK2-MIRO1 retrograde transport complex assembly. Consequently, 3’UTR motif excision perturbs size-regulated transport and eradicates scaling of mitochondria positioning, triggering distal accumulation of mitochondria and progressive hypermotility as cells increase size. Together, our results reveal an RNA-driven mechanistic basis for the cell-size-scaling of organelle distribution and function that is critical to homeostatic control of motile cell behaviour.
Similar content being viewed by others
Introduction
The precise regulation of organelle size, number and positioning is fundamental to the control of cell function1,2,3,4. In the case of mitochondria, tight control of mitochondrial biogenesis, turnover and distribution directs core cellular processes as diverse as cell metabolism, survival and fate determination5,6,7. As cells grow and/or change shape, the demand on organelles shifts, as well as the subcellular positions that they must assume. Consequently, in the face of cell size alterations, cells reduce/expand and/or reposition their organellar pool as appropriate to maintain normal physiological function1,2,3,4. To achieve this, cells employ a suite of cell-size-scaling rules that robustly scale organelle dynamics with cell size change1,2. Yet, despite this phenomenon being a focus of research for over a century8,9,10, a mechanistic basis for cell-size-scaling remains largely elusive, especially regarding the cell-size-dependent re-distribution of organelles.
The appropriate subcellular positioning of organelles is largely directed by motor-driven cytoskeletal transport3,4. For mitochondria, long-range trafficking in mammalian cells is predominantly mediated by microtubules and driven by either the plus-end directed kinesin-1 or the minus-end directed cytoplasmic dynein-1 (dynein) family of motor proteins6,11. These motors are recruited to mitochondria via the trafficking kinesin protein (TRAK)/Milton family of motor-adaptor proteins that connect kinesin-1/dynein to the outer mitochondrial membrane protein, Miro12,13,14,15,16,17,18. Whilst Milton was first identified in Drosophila, its mammalian orthologues TRAK1 and TRAK2 share highly conserved functions in mitochondrial transport. In particular, TRAK1 promotes bidirectional transport via interaction with kinesin-1 and dynein, whereas TRAK2 is thought to predominantly mediate minus-end directed trafficking due to preferential interaction with dynein and its partner complex, dynactin18,19,20, although this remains a topic of debate21,22. As mitochondria are the powerhouse of cells23, this cytoskeletal trafficking is fundamental to the spatial control of energy production. Consequently, tight regulation of mitochondria transport, among other functions, drives polarised energy release during cell migration24,25,26,27,28,29,30,31,32, fuels local remodelling of neurons33,34, and determines the fitness of daughter cells following mitosis35,36,37. Moreover, this transport is cell-size-responsive, enabling redistribution of mitochondria as cells shift size38. Without such cell-size-scaling mechanisms, even small changes in cell geometry could profoundly impact positioning-dependent mitochondrial functions. Yet, how cells sense their dimensions and adjust mitochondrial trafficking and distribution accordingly remains unclear.
The localisation of mRNAs to distinct subcellular sites is critical to the spatial control of gene expression and function39. Of note, we and others recently revealed that mRNA encoding the key mitochondria motor-adaptor, TRAK2, is robustly targeted to distal sites of cell protrusions in diverse cell types40,41,42. Indeed, upon reanalysis of our previous characterisation of protrusion-enriched RNAs40, we find that TRAK2 is unique amongst mRNAs encoding mitochondria trafficking machinery (for example, TRAK1, MIRO1 and MIRO2), which are not localised to protrusions and display unbiased cellular distributions (Supplementary Fig. 1). To achieve this, TRAK2 mRNA contains a conserved 3’UTR GA-rich motif, an RNA element that acts to drive RNA-binding protein (RBP)-mediated transport of a suite of mRNAs to the plus-end of microtubules40,41,43,44,45. In contrast, searching for the presence of this RNA motif in TRAK1 using the Find Individual Motif Analysis (FIMO) programme46 confirmed that TRAK1 does not contain any GA-rich motifs. These observations suggest that mRNA localisation modulates a specific but unknown attribute of TRAK2 function. Considering that these GA-rich targeting motifs operate in multiple cell types and are highly conserved between species47, it is surprising that their function remains largely unexplored. However, recent work is starting to build a consensus view that these GA-rich elements spatially modulate protein-protein interactions by defining the precise site of nascent protein production and co-translational protein complex assembly45,48,49. Despite these observations, the function of TRAK2 mRNA localisation and any role in the control of mitochondrial dynamics remains unknown.
Motile cells exhibit a broad range of cell shape dynamics and adopt distinct morphologies depending on their substrate and mode of migration50. For example, primary endothelial cells exhibited a rounded morphology with multiple lamellipodia when migrating on a glass substrate but adopted highly elongated uniaxial shapes on cell-derived matrix (CDM) (Supplementary Fig. 2a–c), similar to endothelial cells in vivo51. Consequently, adaptation to substrate composition generates cells that vary widely in both morphometric and migratory characteristics (Supplementary Fig. 2c–e). This broad morphological variation exists even within a cell population on a single substrate, with cells on CDM exhibiting a ninefold variance in aspect ratio, 6-fold differences in circularity and threefold variation in protrusion length (Supplementary Fig. 2c). To deal with this broad diversity in shape, cells must reduce/expand and/or reposition organelles as appropriate to match their geometry1,2,3,4. Yet, how cells sense their dimensions and act accordingly remains unclear.
Here, using single-molecule mRNA imaging and endogenous gene editing, we show that TRAK2 mRNA targeting functions to modulate the cell-size-scaling of mitochondria distribution and function. We show that the targeting of TRAK2 mRNA to distal sites of cell protrusions is itself cell-size-dependent and acts to define the precise site of TRAK2-MIRO1 retrograde transport complex assembly. As such, following broad shifts in cell size, scaled targeting of TRAK2 mRNA governs cell-size-dependent re-distribution of mitochondria to elegantly maintain appropriate mitochondrial positioning. Consequently, excision of a 29 bp 3’UTR motif in TRAK2 is alone sufficient to eradicate cell-size-scaling of both mitochondria positioning and cell motility. Taken together, these data define an RNA-mediated mechanistic basis for cell-size-scaling of organelle distribution that directs homoeostatic control of cell migration.
Results
TRAK2 mRNA localisation scales with shifts in cell size
We previously identified a cluster of five mRNAs (TRAK2, KIF1C, RAB13, RASSF3 and NET1) that are highly polarised at the leading edge of endothelial cell (EC) protrusions40. Using single-molecule RNA imaging, we observed that shifts in cell geometry were tightly associated with changes in the polarised subcellular distribution of these mRNAs (Fig. 1; Supplementary Fig. 3), hinting at a role for mRNA trafficking in cell-size-sensing. In particular, we found that all five mRNAs displayed significant substrate-dependent subcellular polarisation in ECs (Fig. 1a–i; Supplementary Fig. 3a–h). TRAK2, RASSF3, RAB13, KIF1C and NET1 were all consistently diffusely enriched at the leading edge of lamellipodium of cells on glass, however, on CDM, displacement of transcripts at the tip of protrusions appeared even more acute (Fig. 1a, d, g; Supplementary Fig. 3a, d, g, h). Quantification of the position of the mRNA centre of mass (CoM) relative to the nucleus confirmed that all five mRNA populations were displaced further away from the nucleus in cells on CDM versus glass (Fig. 1b, e, h; Supplementary Fig. 3b, e). Indeed, a tight correlation between the positioning of the mRNA CoM and the length of cell protrusions indicated that targeting of these mRNAs scaled with shifts in protrusion size (Fig. 1c, f, i; Supplementary Fig. 3c, f). In contrast, positioning of a diffuse non-targeted control transcript, GAPDH, was significantly less impacted by changes in cell morphology (Fig. 1c, f, i; Supplementary Fig. 3c, f). Moreover, ACTB, an mRNA that is targeted to protrusions via distinct mechanisms52, exhibited mRNA CoM values that were indistinguishable from GAPDH control (Fig. 1j–l). Thus, active targeting mechanisms underpin cell-size-scaled positioning of TRAK2, RASSF3, RAB13, KIF1C and NET1 mRNAs as cells change shape. Importantly, we also observed that each of these mRNAs exhibited distinct scaling properties (Fig. 1m; Supplementary Fig. 3i). When the position of the RNA CoM was normalised to protrusion length, it was noted that mRNAs encoding RASSF3 and RAB13 became increasingly more polarised as protrusions elongate (Fig. 1m). In contrast, localisation of the CoM for TRAK2 mRNA was remarkably robust to morphological change, being consistently maintained at a position ~60% along the length of cell protrusions, independent of shifts in protrusion size (Fig. 1m, n). As such, whereas the RNA centre of mass for RASSF3 and RAB13 is sensitive to shifts in cell size, the precise subcellular distribution of TRAK2 mRNA is highly robust to changes in size scale. This ability of mRNA trafficking to be cell-size-scaled and demarcate a fixed position in cells of differential length hinted that TRAK2 mRNA may play a key role in the cellular response to shape change.
a, d, g, j smFISH detection of TRAK2, RASSF3, RAB13, and ACTB mRNAs in exemplar ECs cultured either on glass or CDM (red circles indicate distinct mRNA spots; arrowheads indicate mRNA accumulation at distal sites in protrusions; dashed line indicates cell outline). b, e, h, k Quantification of the distance that the RNA centre of mass (CoM) sits from the nucleus for the indicated mRNAs when ECs were cultured either on glass or CDM (n = 19 cells glass, 11 cells CDM for b, n = 24 cells glass, 9 cells CDM for e, n = 20 cells glass, 11 cells CDM for h, n = 21 cells glass, 12 cells CDM for k; two-tailed Mann–Whitney test, ***P = 0.0002 for e <0.0001 for h, k, **P = 0.0053). c, f, i, l Plots comparing the distance that the RNA CoM sits from the EC nucleus versus protrusion length for the indicated mRNAs (n = 30 cells for c, n = 33 cells for f, n = 31 cells for i, n = 33 cells for l; two-tailed Pearson’s correlation coefficient, magenta asterisks, ***P ≤ 0.0001 for all; two-sided analysis of covariance, black asterisks, ***P = 0.0018 versus GAPDH for c <0.0001 for f, i; ns P = 0.8874 versus GAPDH). m Plots comparing the distance that the RNA CoM sits from the EC nucleus normalised to protrusion length versus protrusion length for the indicated mRNAs (n = as for c, f, i, l; two-tailed Pearson’s correlation coefficient, magenta asterisks, *P = 0.0225 for RASSF3, 0.0161 for RAB13, ns P = 0.3506 for ACTB, 0.8563 for TRAK2; two-sided analysis of covariance, black asterisks, ***P = 0.0012 versus GAPDH for RAB13, <0.0001 versus GAPDH for TRAK2 and RASSF3, ns P = 0.1304 versus GAPDH). n Positioning of TRAK2 mRNA as cells shift in size. For analyses in c, f, i, l, m data for cells cultured on glass and CDM were pooled. Data are mean ± s.e.m. (b, e, h, k, m). a, d, g, j scale bars, 10 µm. Source data is provided as a Source Data file.
TRAK2 modulates the cell-size-scaling of mitochondria positioning
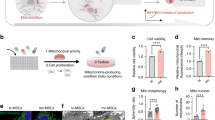
TRAK2 protein is a well-established cytoskeletal adaptor that modulates motor-dependent mitochondria positioning18. Considering that the subcellular positioning of TRAK2 mRNA was size-dependent (Fig. 1a–c, m, n), we speculated that TRAK2 may be a previously unknown determinant of cell-size-scaled mitochondria positioning. To address this hypothesis, TRAK2 expression was perturbed using siRNAs (Fig. 2a) and the impact on mitochondria positioning assessed following culture of endothelial cells on glass or CDM (Fig. 2b–f). In particular, as TRAK2 is known to promote retrograde transport of mitochondria18,21,22, we determined if loss of TRAK2 promoted mitochondria accumulation at distal sites, indicative of reduced retrograde movement. Whereas, for cells on glass, we observed no significant differences in the distal localisation of mitochondria (Fig. 2b, c), on CDM, siTRAK2 cells consistently accumulated mitochondria specifically at distal-most sites in protrusions when compared to controls (siCTRL; 0–10 µm region; Fig. 2d–f). This 2-fold enrichment of mitochondria in siTRAK2 cells was not indirect due to broad changes in the morphology or length of protrusions (Fig. 2g, h), or perturbation of the microtubule cytoskeleton (Supplementary Fig. 4a, b), indicating that TRAK2 is indeed required for retrograde mitochondria trafficking. However, the presence of this phenotype only in siTRAK2 cells cultured on CDM, which are longer than cells cultured on glass (Supplementary Fig. 2c), suggested that TRAK2-mediated mitochondria transport was highly sensitive to the uniaxial elongation of cells. Indeed, quantification of the enrichment of mitochondria at the distal-most leading edge of siTRAK2 cells revealed a clear size-dependent progressive accumulation of mitochondria as protrusions increase in length (Fig. 2i). Together, these data confirm that TRAK2 influences cell-size-scaling of mitochondria spatial positioning, acting to maintain an appropriate subcellular distribution as cells change morphology.
a Representative western blot for TRAK2 and β-actin in ECs treated with either control (siCTRL) or TRAK2-targeting (siTRAK2) siRNAs and densitometric analysis of the ratio of TRAK2:β-Actin levels (n = 3 independent experiments, two-tailed unpaired t test, **P = 0.0051). b Representative images of MitoTracker-labelled mitochondria in siCTRL- or siTRAK2-treated ECs cultured on glass (brackets indicate the distal-most 10 µm and 20 µm of protrusions; dashed line indicates cell outline as defined by F-actin labelling). c Quantification of mitochondria levels at the indicated positions of EC protrusions following culture on glass and treatment with siCTRL or siTRAK2 (data are normalised to siCTRL, n = 17 cells siCTRL, n = 11 cells siTRAK2, two-tailed Mann–Whitney test, ns P = > 0.2441). d, e Representative images of MitoTracker-labelled mitochondria in whole protrusions (d) or the distal-most 10 µm of protrusions (e) of siCTRL- or siTRAK2-treated ECs cultured on CDM (brackets indicate the distal-most 10 µm, 20 µm, and 30 µm of protrusions; dashed line indicates cell outline as defined by F-actin labelling). f Quantification of mitochondria levels at the indicated positions of EC protrusions following culture on CDM and treatment with siCTRL or siTRAK2 (data are normalised to siCTRL, n = 19 cells siCTRL, n = 18 cells siTRAK2, unpaired Kruskal–Wallis test and Dunn multiple comparison test, ***P = 0.0003, ns P ≥ 0.6664). g Quantification of protrusion morphometrics (area, aspect ratio, and circularity) of ECs cultured on CDM and treated with siCTRL or siTRAK2 (n = 19 cells siCTRL, n = 18 cells siTRAK2, two-tailed Mann–Whitney test, ns P ≥ 0.2844). h Quantification of EC protrusion length following culture on glass or CDM and treatment with siCTRL or siTRAK2 (n = 17 cells siCTRL glass, n = 11 cells siTRAK2 glass, n = 19 cells siCTRL CDM, n = 18 cells siTRAK2 CDM, two-tailed Mann–Whitney test, **P = 0.0018, ***P ≤ 0.0001, ns P = 0.2100 for CDM siCTRL vs CDM siTRAK2, 0.4581 for glass siCTRL vs glass siTRAK2). i Plot comparing mitochondria levels at the indicated positions of EC protrusions following treatment with siCTRL or siTRAK2 (data normalised to siCTRL, n = 36 cells siCTRL, n = 29 cells siTRAK2, two-tailed unpaired t test, *P = 0.0125 for 40–60 µm, 0.0329 for 60–80 µm, **P ≤ 0.0023 for 50–70 µm, 0.001 for >70 µm, ns P = 0.4211 for 20–40 µm, 0.4112 for 30–50 µm; two-sided analysis of covariance, **P = 0.002 versus siCTRL). For analyses in i, data for cells cultured on glass and CDM were pooled. Data are mean ± s.e.m. (a, c, f–i). For b, d scale bars, 10 µm. e Scale bar, 5 µm. Source data are provided as a Source Data file.
A 29 bp motif directs cell-size-scaling of TRAK2 mRNA distribution
To determine if the cell-size-dependent trafficking of TRAK2 mRNA mechanistically underpins TRAK2-mediated size-scaling of mitochondria distribution, we aimed to specifically manipulate mRNA localisation. We and others previously identified a conserved GA-rich 3’UTR motif element that is necessary and sufficient to drive localisation of mRNAs to distal sites of cell protrusions40,45. Such localisation elements are often repeated multiple times in 3’UTRs, with TRAK2 containing three such motif repeats40. Hence, we first identified which of these GA-rich motifs was critical to the targeting of TRAK2 mRNA to protrusions. To achieve this, we used the MS2-MCP reporter construct system to expand our previous analysis of the TRAK2 3’UTR40. With this tool, a non-targeted HBB coding sequence was tagged with 24x bacteriophage-derived MS2 hairpin repeats, which act as a high-affinity binding site for the MS2-capping protein (MCP). Co-expression of MCP-nlsGFP can then be used to visualise the subcellular localisation of MS2-tagged RNAs. Fusion of the HBB-MS2 construct with wild-type (Wt) or motif-excised TRAK2 3’UTRs thus enabled the function of each motif to be determined (Supplementary Fig. 5a). In controls lacking a 3’UTR fusion, the nuclear localisation tag of MCP-nlsGFP restricted localisation to the nucleus (Supplementary Fig. 5b, c). In contrast, fusion with the Wt TRAK2 3’UTR, a truncated 3’UTR containing all three G-motifs (1-1280 bp), or a truncated 3’UTR lacking the central 374-403bp G-motif all promoted robust GFP accumulation at distal sites of cell protrusions (Supplementary Fig. 5b, c). However, removal of just the 114–143 bp proximal-most G-motif was sufficient to eradicate protrusion-targeting properties of the 3’UTR (Supplementary Fig. 5c). Hence, this single 29bp G-motif acted as a critical localisation element driving polarised mRNA trafficking.
To assess the endogenous function of this 29 bp motif, CRISPR-Cas9 was used to excise the 114–143 bp region from the endogenous TRAK2 3’UTR. Guide RNAs were designed to flank the 29bp G-motif (Fig. 3a) and CRISPR-Cas9 ribonucleoprotein complexes nucleofected into endothelial cells alongside a GFP expression vector. Single GFP-positive cells were isolated, expanded, sequenced, and subjected to PCR to identify clones containing homozygous G-motif deletion (ΔTRAK2; Fig. 3a, b; Supplementary Fig. 6a, b). Control Wt clones were also created upon nucleofection of ribonucleoprotein complexes without guide RNA (Supplementary Fig. 6a). Single-molecule imaging of endogenous mRNAs in cells on glass revealed that loss of the G-motif efficiently eradicated polarised targeting of TRAK2 to protrusions (Supplementary Fig. 6c–e). TRAK2 mRNA was visibly depolarised and more proximally distributed in ΔTRAK2 versus Wt cells (Supplementary Fig. 6c). Moreover, the polarisation index53 of TRAK2 mRNA was significantly lower in ΔTRAK2 cells, and became indistinguishable from the diffuse control transcript, GAPDH (Supplementary Fig. 6d). Likewise, TRAK2 mRNA dispersion index53 was also significantly lower in ΔTRAK2 cells (Supplementary Fig. 6e). In contrast, GAPDH mRNA polarisation and dispersion were unchanged (Supplementary Fig. 6c–e). Importantly, excision of the G-motif also had no impact on TRAK2 mRNA spot or TRAK2 protein levels, indicating that loss of polarised mRNA transport did not perturb mRNA/protein turnover dynamics (Supplementary Fig. 6f, g). Thus, the 29bp G-motif is a critical modulator of TRAK2 spatial distribution that promotes polarised mRNA trafficking to distal sites of cell protrusions.
a Sequences of TRAK2 3’UTRs from Wt and ΔTRAK2 ECs indicating the location of gRNA target (red), NGG PAM (blue), and G-motif sequences (magenta). b PCR of the TRAK2 3’UTR demonstrating a band shift in ΔTRAK2 ECs (n = 3 experimental repeats). c, d smFISH co-detection of TRAK2 and GAPDH mRNAs in Wt (c) or ΔTRAK2 (d) ECs (red circles indicate mRNA spots; arrowheads indicate distal mRNA accumulation; brackets indicate distal-most 10 µm, 20 µm, and 30 µm of protrusions; dashed line indicates cell outline). e Number of TRAK2 mRNA spots in Wt and ΔTRAK2 ECs (n = 31 cells Wt, n = 28 cells ΔTRAK2, two-tailed Mann–Whitney test, ns P = 0.1130). f, g Relative distribution of TRAK2 mRNAs (f) or GAPDH mRNAs (g) in Wt and ΔTRAK2 EC protrusions (n = 31 cells Wt TRAK2, n = 28 cells ΔTRAK2 TRAK2, n = 26 cells Wt GAPDH, n = 25 cells ΔTRAK2 GAPDH, two-tailed Mann–Whitney test, ***P ≤ 0.0001, **P ≤ 0.0096 for 10–20 µm in f, 0.0044 for 0–10 µm in f, ns P = 0.2848 in f, 0.591 for proximal in g, 0.556 for 20–30 µm in g, 0.3945 for 10–20 µm in g, 0.8702 for 0–10 µm in g). h Distance the RNA CoM sits from the nucleus for GAPDH and TRAK2 mRNAs in Wt and ΔTRAK2 ECs (n = 29 cells Wt GAPDH, 26 cells ΔTRAK2 GAPDH, n = 32 cells Wt TRAK2, 37 cells ΔTRAK2 TRAK2, two-tailed Mann–Whitney test, ***P ≤ 0.0001, ns P = 0.2793). i Protrusion length in Wt and ΔTRAK2 ECs (n = 31 cells Wt, n = 28 cells ΔTRAK2, two-tailed Mann–Whitney test, ns P = 0.3282). j Distance the RNA CoM sits from the nucleus normalised to protrusion length for GAPDH and TRAK2 mRNAs in Wt and ΔTRAK2 ECs (n = 33 cells Wt GAPDH, 30 cells ΔTRAK2 GAPDH, n = 32 cells Wt TRAK2, 37 cells ΔTRAK2 TRAK2, two-tailed unpaired t test, ***P ≤ 0.0001, ns P = 0.8949). k Plots comparing the distance that the RNA CoM sits from the EC nucleus versus protrusion length for GAPDH and TRAK2 mRNAs in either Wt or ΔTRAK2 ECs (n = at least 26 cells, two-sided analysis of covariance, ***P ≤ 0.0001 versus GAPDH, ns P = 0.3410 versus GAPDH). l Plot comparing the distance that the TRAK2 RNA CoM sits from the nucleus in Wt and ΔTRAK2 ECs with protrusion length (n = 32 cells Wt, n = 37 cells ΔTRAK2, two-tailed unpaired t test, ***P = 0.0002 for 50–70 µm, <0.0001 for 60–80 µm, **P = 0.0067, ns P = 0.1523 for 20–40 µm, 0.0753 for 30–40 µm, 0.0530 for 40–60 µm Wt versus ΔTRAK2; two-sided analysis of covariance, magenta asterisk, ***P = 0.0005 Wt versus ΔTRAK2). All cells cultured on CDM except k, l where data for cells cultured on glass and CDM were pooled. Data are mean ± s.e.m. (e–j, l). c, d Scale bar, 10 µm. Source data are provided as a Source Data file.
Similar to cells cultured on glass, we noted that TRAK2 mRNA was also significantly depolarised in ΔTRAK2 cells cultured on CDM (Fig. 3c, d). Again, excision of the G-motif had no impact on total mRNA spot levels (Fig. 3e), however, polarised enrichment of TRAK2 spots at the distal-most 0–10 µm leading edge of cell protrusions was entirely lost upon G-motif excision (Fig. 3c, d, f). Consequently, TRAK2 mRNA was significantly more proximally located in ΔTRAK2 cells (Fig. 3d, f), and instead resembled the non-polarised distribution of the diffuse control mRNA, GAPDH (Fig. 3g). Likewise, quantification of the position of the mRNA CoM relative to the nucleus in Wt and ΔTRAK2 cells also confirmed that mutant TRAK2 mRNA was significantly less polarised and was positioned closer to the nucleus, being indistinguishable from GAPDH controls (Fig. 3h). Loss of the G-motif had no impact on the length of cell protrusions, indicating that TRAK2 mRNA distribution does not impact cell size control and that loss of mRNA polarisation was not an indirect consequence of any reduction in protrusion length (Fig. 3i). Thus, when the position of the TRAK2 mRNA CoM was normalised to protrusion length, the CoM in ΔTRAK2 cells was no longer maintained at ~60% along the length of cell protrusions, was much more proximally localised and again was indistinguishable from GAPDH controls (Fig. 3j). In contrast to TRAK2, positioning of the CoM of another G-motif-containing mRNA, RAB13, was unperturbed in ΔTRAK2 cells, confirming specificity of this approach (Supplementary Fig. 6h). Most importantly, G-motif excision indeed significantly perturbed the cell-size-scaling properties of TRAK2 mRNA distribution. In particular, the robust correlation between positioning of the mRNA CoM and the length of cell protrusions was significantly attenuated in ΔTRAK2 cells, to now resemble GAPDH controls (Fig. 3k). Moreover, this resulted in a significant difference in TRAK2 mRNA distribution when protrusions extended beyond ~60 µm in length in ΔTRAK2 cells (Fig. 3l). Overall, these data reveal that the 29 bp 3’UTR G-motif underpins cell-size-dependent positioning of TRAK2 mRNA, and that the impact of motif excision was progressively more severe as protrusions elongated past a threshold length of ~60 µm.
TRAK2 mRNA directs the cell-size-scaling of mitochondria distribution
Next, we determined if the loss of cell-size-dependent TRAK2 mRNA distribution had any impact on mitochondria positioning. First, we observed that excision of the G-motif was itself sufficient to phenocopy TRAK2 knockdown (Fig. 2d–f), resulting in robust accumulation of mitochondria at the distal-most sites of ΔTRAK2 cell protrusions when compared to Wt control cells on CDM (0–10 µm region; Fig. 4a–c). Again, this near twofold enrichment of mitochondria was not indirect due to broad changes in the morphology or length of cell protrusions on CDM, which remained unchanged (Fig. 4d). Likewise, the microtubule cytoskeleton was not perturbed in ΔTRAK2 cells (Supplementary Fig. 7a, b). Moreover, further analysis revealed a clear interrelationship between TRAK2 mRNA and mitochondria positioning (Fig. 4e). In Wt cells, an increase in TRAK2 mRNA spot numbers at the distal sites of cell protrusions was coupled with a decrease in mitochondria enrichment at these sites. In contrast, absence of TRAK2 mRNAs at distal sites in ΔTRAK2 cell protrusions was linked to elevated mitochondria enrichment relative to Wt controls (Fig. 4e). Similar to the knockdown of TRAK2 (Fig. 2b–f), this mis-localisation of mitochondria was specific to ΔTRAK2 cells cultured on CDM, as no differences in mitochondria enrichment were observed in cells on glass (Fig. 4f), again suggesting a cell-size-dependent phenotype. Indeed, quantification of the enrichment of mitochondria at the distal-most leading edge of ΔTRAK2 cells confirmed size-dependent progressive accumulation of mitochondria as protrusions increase in length (Fig. 4g). Strikingly, a significant ectopic accumulation of mitochondria was only observed in cells with protrusions longer than ~60 µm in length (Fig. 4g), mirroring the size threshold at which TRAK2 mRNA distribution is significantly perturbed in ΔTRAK2 cells (Fig. 3l). Furthermore, if cells cultured on CDM were subdivided into populations with either short (<60 µm; Fig. 4h) or long (>60 µm; Fig. 4i) protrusions, significant ectopic accumulation of mitochondria was only observed in ΔTRAK2 mutant cells with the longest protrusions (Fig. 4i). Consequently, size-scaled TRAK2 mRNA trafficking critically directs the cell-size-dependent positioning of mitochondria during cell migration, with excision of the TRAK2 G-motif triggering a near threefold accumulation of mitochondria at the leading edge of elongated cells (Fig. 4i).
a, b Images of MitoTracker-labelled mitochondria in whole protrusions (a) or the distal-most 10 µm of protrusions (b) of Wt or ΔTRAK2 ECs cultured on CDM (brackets indicate distal-most 10 µm, 20 µm and 30 µm of protrusions; dashed line indicates cell outline). c Mitochondria levels at the indicated positions of Wt or ΔTRAK2 EC protrusions following culture on CDM (data is normalised to Wt, n = 29 cells Wt, n = 32 cells ΔTRAK2, unpaired Kruskal–Wallis test and Dunn multiple comparison test, **P = 0.0042, ns P ≥ 0.9999). d Quantification of protrusion morphometrics (area, aspect ratio, circularity, and protrusion length) of Wt or ΔTRAK2 ECs cultured on CDM (n = 29 cells Wt, n = 32 cells ΔTRAK2, two-tailed Mann–Whitney test, ns P ≥ 0.1109). e Plots comparing the number of TRAK2 mRNA spots (grey data points and dashed line) and mitochondria levels (magenta data points and dashed line) at the indicated distal-most positions of Wt or ΔTRAK2 EC protrusions (n = 31 cells Wt TRAK2, 28 cells ΔTRAK2 TRAK2, 29 cells Wt mitochondria, 32 cells ΔTRAK2 mitochondria, two-tailed Mann–Whitney test, **P = 0.0030 for % of total mitochondria in Wt, ns P = 0.4115 for % of total mitochondria in ΔTRAK2). f Mitochondria levels at the indicated positions of Wt or ΔTRAK2 EC protrusions following culture on glass (data is normalised to Wt, n = 17 cells Wt, n = 19 cells ΔTRAK2, two-tailed Mann–Whitney test, ns P ≥ 0.2711). g Plots comparing mitochondria levels in the distal-most 10 µm of protrusions in Wt or ΔTRAK2 ECs with protrusions of the indicated length (data is normalised to Wt, n = 48 cells Wt, n = 46 cells ΔTRAK2, two-tailed Mann–Whitney test, *P = 0.0494 for 50–70 µm, 0.0293 for >70 µm, **P = 0.0067, ns P = 0.3426 for 20–40 µm, 0.0919 for 30–50 µm, 0.0786 for 40–60 µm; two-sided analysis of covariance, magenta asterisk, **P = 0.0083 Wt versus ΔTRAK2). For analyses in (g) data for cells cultured on glass and CDM were pooled. h, i Mitochondria levels in protrusions of Wt or ΔTRAK2 ECs with protrusions either <60 µm in length (h) or >60 µm in length (i) and following culture on CDM (data is normalised to Wt, n = 15 cells Wt <60 µm, n = 13 cells ΔTRAK2 < 60 µm, n = 14 cells Wt >60 µm, n = 19 cells ΔTRAK2 > 60 µm, unpaired Kruskal–Wallis test and Dunn multiple comparison test, ***P = 0.0009, ns P = > 0.4018). Data are mean ± s.e.m. (c–i). a scale bars, 10 µm. b scale bar, 7.5 µm. Source data are provided as a Source Data file.
TRAK2 mRNA trafficking modulates cell-size-scaling of cell motility
Considering that a single G-motif in the 3’UTR of TRAK2 modulated the spatial distribution of mitochondria, we aimed to define the functional significance of this observation. Tight control of mitochondria spatial positioning at the leading edge of migrating cells is critical to the control of motile cell behaviour24,25,26,27,28,29,30,31,32. As such, we hypothesised that ectopic accumulation of mitochondria at distal sites of ΔTRAK2 cells may increase local mitochondrial activity. To test this, we quantified mitochondrial membrane potential upon staining of Wt and ΔTRAK2 cells with the cell-permeant dye, tetramethylrhodamine methyl ester (TMRM)30. Consistent with increased mitochondrial function at the leading edge, we observed that the ratio of TMRM to mitochondria staining was significantly enhanced at distal sites of cell protrusions in ΔTRAK2 versus Wt cells (Fig. 5a–c). Thus, not only did mitochondria accumulate at distal regions of ΔTRAK2 cells, but these mitochondria also exhibited enhanced activity versus those in Wt cells. Considering the key role of local mitochondria activity in driving cell migration24,25,26,27,28,29,30,31,32, we predicted that this ectopic accumulation of active mitochondria at distal sites may deregulate the migration of cells as they change shape. To explore this, we quantified key motile characteristics upon live imaging of Wt and ΔTRAK2 cells cultured on CDM. Consistent with a key role for distally localised mitochondria in the production of ATP that fuels migration, ΔTRAK2 cells that accumulated mitochondria at their leading edge exhibited a marked increase in velocity, as well as the accumulated distance and Euclidean distance migrated versus Wt controls (Fig. 5d–g). In contrast, the directionality of ΔTRAK2 cells remained equivalent to Wt cells (Fig. 5h), indicating an increase in inherent motility, but not directional sensing, of migrating cells upon excision of the TRAK2 G-motif. Importantly, this enhanced motility of ΔTRAK2 cells was again cell-size-dependent (Fig. 5i). When Wt control populations were subdivided into cells with either short (<60 µm average length) or long (>60 µm average length) protrusions, cells exhibited equivalent motility independent of any differences in size. However, when ΔTRAK2 populations were likewise subdivided, cells with short protrusions (<60 µm average length) displayed motility identical to Wt controls, whereas it was exclusively the cells with long protrusions (>60 µm average length) that were significantly more motile (Fig. 5i). Further analysis revealed that the velocity of Wt cells was remarkably robust to broad changes in cell length (Fig. 5j). In stark contrast, the motility of ΔTRAK2 cells was no longer robust to shifts in cell size, instead being significantly correlated with changes in protrusion length (Fig. 5k). Comparison of cell motility rates further confirmed that Wt cells exhibited remarkable homoeostatic control of cell velocity, maintaining a constant rate of motility independent of shifting cell size (Fig. 5l). However, upon excision of the TRAK2 G-motif, this homoeostatic control was lost, with ΔTRAK2 cells displaying progressive hypermotility as they increased in size (Fig. 5l). Importantly, a significant increase in the motility of ΔTRAK2 versus Wt cells was only observed in cells with protrusions longer than ~60 µm in length (Fig. 5l), again mirroring the size threshold at which cell-size-dependent TRAK2 mRNA and mitochondria distribution are significantly perturbed upon G-motif excision (Figs. 3l and 4g). Together, these data revealed that cell-size-scaled TRAK2 mRNA distribution acts as a previously unknown homoeostatic control mechanism that maintains a consistent rate of cell motility in the face of dynamic changes in cell size.
a, b Representative images of MitoTracker- and TMRM-labelled mitochondria in protrusions of Wt (a) or ΔTRAK2 (b) ECs (brackets indicate the distal-most 10 µm, 20 µm and 30 µm of protrusions; dashed line indicates cell outline as defined by wide-field imaging). c Quantification of the ratio of TMRM to MitoTracker staining levels at the indicated positions of Wt or ΔTRAK2 EC protrusions (n = 14 cells Wt, n = 15 cells ΔTRAK2, unpaired Kruskal–Wallis test and Dunn multiple comparison test, *P = 0.0411, ***P = 0.0003, ns P ≥ 0.9999). d–f Quantification of the velocity (d, n = 45 cells Wt, 209 cells ΔTRAK2), accumulated distance (e, n = 91 cells Wt, 209 cells ΔTRAK2), Euclidian distance (f, n = 91 cells Wt, 209 cells ΔTRAK2) of Wt and ΔTRAK2 ECs cultured on CDM (two-tailed Mann–Whitney test, ***P ≤ 0.0001 for all). g, Rose plots of Wt and ΔTRAK2 motile EC trajectories when cultured on CDM (n = 91 cells Wt, n = 208 cells ΔTRAK2). h Quantification of the directionality of Wt and ΔTRAK2 ECs cultured on CDM (n = 91 cells Wt, n = 208 cells ΔTRAK2, two-tailed Mann–Whitney test, ns P = 0.0704). i Quantification of the velocity of non-elongated (<60 µm protrusions) and elongated (>60 µm protrusions) Wt and ΔTRAK2 ECs cultured on CDM (n = 80 cells Wt <60 µm, n = 171 cells Wt >60 µm, n = 130 cells ΔTRAK2 < 60 µm, n = 180 cells ΔTRAK2 > 60 µm, two-tailed Mann–Whitney test, ***P ≤ 0.0001 for all, ns P = 0.2786 for Wt <60 µm vs Wt >60 µm, 0.2971 for Wt <60 µm vs ΔTRAK2 < 60 µm). j, k Plots comparing the velocity of cells versus protrusion length in Wt (j) and ΔTRAK2 (k) ECs (n = 251 cells Wt, n = 310 cells ΔTRAK2, two-tailed Pearson’s correlation coefficient, ns P = 0.8546,***P ≤ 0.0001; r = 0.3676). l Plots comparing the velocity of Wt or ΔTRAK2 ECs with protrusions of the indicated length (n = 251 cells Wt, n = 310 cells ΔTRAK2, two-tailed Mann–Whitney test,***P = 0.0007 for 50–70 µm, <0.0001 for 60–80 µm and >70 µm, ns P = 0.5453 for 20–40 µm, 0.765 for 30–50 µm, 0.175 for 40–60 µm Wt versus ΔTRAK2; two-sided analysis of covariance, magenta asterisk, ***P ≤ 0.0001 Wt versus ΔTRAK2). Data are mean ± s.e.m. (c–f, h, i, l). Source data are provided as a Source Data file.
TRAK2 RNA localisation spatially modulates TRAK2-MIRO1 interaction
Whilst data presented above confirmed that TRAK2 mRNA distribution fundamentally modulates cell-size-scaling of both mitochondria distribution and cell motility, a mechanistic basis for these observations remained unclear. However, recent work suggests that G-motif-mediated subcellular targeting of mRNAs defines the precise site of nascent protein production to modulate co-translational protein complex assembly and/or association of newly translated protein with specific interaction partners45,48,49. Considering that binding of TRAK2 to the mitochondrial membrane protein, MIRO1, is critical to dynein-mediated retrograde mitochondria transport12,13,14,15,16,17,18,19,20,21,22 (Fig. 6a), we tested whether TRAK2 mRNA polarisation modulated the association of TRAK2 with MIRO1. Contrary to a role for TRAK2 mRNA targeting in defining the frequency of TRAK2-MIRO1 interactions, co-immunoprecipitation studies revealed that excision of the TRAK2 G-motif had no impact on association of TRAK2 with MIRO1 (Fig. 6b). However, use of proximity ligation assays (PLA) to define the precise site of close TRAK2-MIRO1 interaction (i.e., both proteins being <40 nm apart) revealed that TRAK2 mRNA localisation acts to precisely spatially modulate the association of TRAK2 with MIRO1 (Fig. 6c–g). As expected, PLA signal was not detected in control cells lacking either primary antibody against TRAK2 or MIRO1 (Fig. 6c). In comparison, PLA using antibodies against both TRAK2 and MIRO1 uncovered that interaction of these proteins was present all along the length of Wt cells (Fig. 6c). Quantification confirmed that TRAK2-MIRO1 association was not biased to any position and was equally distributed across the length Wt cells (Fig. 6d). In contrast, PLA revealed that excision of the TRAK2 G-motif significantly displaced the positioning of TRAK2-MIRO1 interactions, predominantly restricting them to the proximal-most aspect of migrating ΔTRAK2 cells (Fig. 6c, e). Changes in PLA foci positioning were not due any indirect effect on cell protrusion length (Fig. 6f) and were driven by a significant re-distribution of TRAK2-MIRO1 associations towards the cell body (Fig. 6g). Moreover, consistent with no impact on the frequency of interaction between TRAK2 and MIRO1, there was no difference in the total number of PLA foci detected in Wt versus ΔTRAK2 cells (Fig. 6h). Hence, we propose that TRAK2 mRNA targeting to the leading edge of migrating cells functions to specifically promote distal interactions of TRAK2 and MIRO1, ensuring robust cell-size-scaled retrograde transport of mitochondria and homoeostatic control of motile behaviour. In comparison, excision of the TRAK2 G-motif almost appears to exclusively restrict TRAK2-MIRO1 association to the proximal aspect of migrating cells, leading to distal accumulation of mitochondria and progressive ectopic hypermotility as cells increase size (Fig. 6i). Taken together, these results define an RNA-based mechanistic principle for cell-size-dependent regulation of both mitochondria distribution and cell migration, which has fundamental implications for our understanding the control of mitochondria function and cell behaviour in diverse tissue contexts.
a Schematic of the interactions between Dynein, TRAK2 and MIRO1 underpinning retrograde transport of mitochondria along microtubules. Figure adapted from Fenton et al.22 (https://doi.org/10.1038/s41467-021-24862-7) b Western blot of TRAK2 and MIRO1 following immunoprecipitation using anti-TRAK2 or control IgG antibodies in Wt or ΔTRAK2 ECs and densitometric analysis of the ratio of MIRO1:TRAK2 levels (n = 3 independent experiments, two-tailed Mann–Whitney test, ns P = 0.7000). c Proximity ligation assay (PLA) to detect sites of interaction between TRAK2 and MIRO1 in protrusions of Wt or ΔTRAK2 ECs cultured on CDM. Control PLA was performed in Wt ECs in the absence of primary antibody (arrowheads indicate sites of PLA foci indicative of TRAK2-MIRO1 interaction, brackets and arrows indicate position of PLA foci along protrusions). d, e Proportion of PLA foci detected at the indicated positions of protrusions in either Wt (d) or ΔTRAK2 (e) ECs (n = 4 experiments for Wt, 67 cells total, n = 4 experiments for ΔTRAK2, 67 cells total, ordinary one-way ANOVA with Tukey’s multiple comparisons test, **P = 0.0062 for 10–30%, 0.0021 for 30–50%, 0.0042 for 50–70%, 0.0031 for 70–90%, 0.0023 for 90–100% versus 0–10%, ns P = 0.9989 for 10–30%, 0.9416 for 30–50%, 0.9771 for 50–70%, >0.9999 for 70–90%, 0.9993 for 90–100% versus 0–10%). f Length of protrusions in Wt or ΔTRAK2 ECs that were used for PLA (n = 90 cells Wt, n = 77 cells ΔTRAK2, two-tailed Mann–Whitney test, ns P = 0.3919). g Position of individual PLA foci along the length of protrusions in Wt or ΔTRAK2 ECs (n = 124 cells Wt, n = 50 cells ΔTRAK2, two-tailed Mann–Whitney test, **P = 0.0060). h Total number of PLA foci in Wt or ΔTRAK2 ECs. Control PLA was performed in Wt ECs in the absence of primary antibody (n = 144 cells Wt, n = 116 cells ΔTRAK2, n = 51 cells control, unpaired Kruskal–Wallis test and Dunn multiple comparison test, ***P ≤ 0.0001 for Wt or ΔTRAK2 versus control, ns P ≥ 0.9999). i Schematic of the hypothesised role of TRAK2 mRNA localisation in the distal interaction of TRAK2 and MIRO1 and modulation of the cellular distribution of mitochondria. It is well-established that kinesin-TRAK1-MIRO1 and dynein-TRAK2-MIRO1 interactions promote anterograde and retrograde transport of mitochondria, respectively11,12,13,14,15,16,17,18,19,20,21,22. We find that TRAK2 mRNA spatially modulates TRAK2-MIRO1 interactions to support cell-size-scaling of mitochondria distribution. Thus, in elongated cells, mitochondria accumulate at distal sites. Data are mean ± s.e.m. (b, d–h). Cells were cultured on CDM. Scale bars, 10 µm. Source data are provided as a Source Data file.
Discussion
Cells exhibit a wide diversity of shapes and undergo broad shifts in morphology during dynamic processes such as cell migration50. To match these changes in geometry, cells precisely adjust the size, number and positioning of organelles to maintain normal physiological function1,2,3,4. Yet, the mechanisms by which cells sense shifts in their dimensions and tune organelle properties accordingly remain largely unclear. Here, we identify an unexpected role for mRNA transport in the cell-size scaling of organelle behaviour. In particular, we show that trafficking of TRAK2 mRNA, driven by a single 29 bp 3’UTR motif, fundamentally underpins cell-size-dependent control of mitochondria positioning and function. Considering the large number of mRNAs known to be spatially targeted in cells40,41,42,43,44,45 and the broad conservation of this phenomenon between diverse cell types47, it seems unlikely that this phenomenon is restricted to just TRAK2. Indeed, the protein product of another targeted mRNA explored in this study, RASSF3 (Fig. 1d–f, m), was very recently shown to both directly interact with MIRO1 and to impact the subcellular positioning of mitochondria and peroxisomes54. Hence, cell-size-dependent spatial targeting of RASSF3 likely also directs organelle dynamics. Furthermore, both RAB13 and KIF1C mRNAs, which are co-trafficked with TRAK2 and RASSF3 (Fig. 1a–l; Supplementary Fig. 3a–f), encode translated protein products that modulate the trafficking of other key organelles, such as endosomes55,56. Thus, our work hints that the use of cell-size-dependent mRNA trafficking may be a widespread phenomenon underpinning the size-scaling of diverse organelle dynamics.
Numerous G-motif-containing polarised mRNAs have now been identified40,41,42,43,44,45, yet the function of their subcellular trafficking remains largely unclear. Recent work has elegantly shown that subcellular targeting of some of these mRNAs facilitates interaction of nascent protein with specific binding partners45,48,49. Yet here we show that TRAK2 mRNA trafficking has no impact on the magnitude of TRAK2 association with its key binding partner MIRO1. In contrast, we find that mRNA transport is essential to the spatial control of TRAK-MIRO1 protein complex assembly. Following excision of the 29bp G-motif in ΔTRAK2 cells, interactions between TRAK2 and MIRO1 are no longer uniformly positioned along the length of the cell but instead predominantly restricted to the proximal zone of protrusions. Thus, TRAK2 mRNA transport acts to define the spatial domain of protein-protein interactions and the distal-most extent to which mitochondria can be retrieved by TRAK2-MIRO1-mediated retrograde transport. Consequently, excision of a single 29 bp 3’UTR motif in TRAK2 disrupts homoeostatic control of organelle positioning to trigger progressive distal accumulation of mitochondria as cells increase in size. However, these observations do not entirely rule out an additional role for mRNA trafficking as a facilitator of interaction with specific TRAK2-binding partners. For example, recruitment of the co-factor Lissencephaly-1 (LIS1) to the TRAK2-dynein-dynactin complex was recently proposed to bias retrograde TRAK2 transport in single-molecule assays using cellular extracts22. Considering that LIS1 localises to the leading edge of migrating cells57, it is indeed feasible that TRAK2 mRNA transport to distal sites may aid recruitment of LIS1. However, given that TRAK2-MIRO1 still robustly accumulates at proximal sites in ΔTRAK2 cells, consistent with minus-end-directed transport, any impact of a failure to interact with LIS1 on retrograde transport is likely minimal. Despite these observations, it is still possible that ΔTRAK2 mutation further impacts mitochondrial transport by spatially modulating interaction with other unidentified TRAK2-binding partners, in addition to MIRO1. As such, future work should fully define (1) the wider spatial interactome of TRAK2, (2) how this is broadly directed by TRAK2 mRNA localisation, and (3) the impact of any other identified mRNA-mediated protein interactions on mitochondrial motility. Indeed, if other mRNA-regulated TRAK2-binding partner interactions were identified, this may suggest much broader roles for mRNA transport in the control of mitochondrial dynamics. Intriguingly, we find that mRNA encoding another microtubule transport protein, KIF1C, is also trafficked to distal sites in a cell-size-dependent manner (Supplementary Fig. 3a–c, g). KIF1C is a member of the kinesin superfamily of proteins that classically promote microtubule plus-end-directed anterograde transport. However, unusual amongst kinesins, KIF1C is also known to direct retrograde microtubule transport58,59. Whilst recent work reveals that disruption of KIF1C mRNA trafficking perturbs cell migration49, it remains unclear if this impacts the spatial domain of KIF1C-binding partner interactions, KIF1C-dependent retrograde transport and/or organelle dynamics. As such, it is exciting to speculate a broadly conserved role for trafficking of KIF1C and other mRNAs in the spatial control of retrograde trafficking complex assembly and function.
Importantly, here we define a previously unidentified RNA-based mechanism underpinning homoeostatic control of mitochondria distribution and cell motility. Redistribution of mitochondria to the leading edge of migrating cells generates subcellular energy gradients that bias ATP levels at lamellipodia24,25,26,27,28,29,30,31,32. Thus, tight control of mitochondria positioning is a critical determinant of motile cell behaviour24,25,26,27,28,29,30,31,32. Here we find that migrating cells sense shifts in their dimensions and can adjust mitochondria positioning appropriately to maintain a remarkably consistent velocity despite broad changes in cell shape. Moreover, we reveal that TRAK2 mRNA trafficking fundamentally directs this cell-size-dependent homoeostatic control. Consequently, excision of a single 29 bp TRAK2 G-motif eradicates cell-size scaling, triggering progressive mitochondria misplacement and hypermotility as cells increase in size. Others have observed that shifts in cell shape have no impact on cell motility60, and now we provide a potential mechanistic basis for this phenomenon. Moreover, our work identifies that TRAK2 mRNA dynamics act as a previously unappreciated key control point modulating mitochondria distribution and function. Indeed, previous work has shown that post-translational modification of TRAK2 via glycosylation can dynamically tune mitochondria positioning in response to metabolic change61. Regulated shifts in TRAK2 mRNA positioning and/or abundance would likewise enable rapid modulation of mitochondria spatial distribution. Indeed, our single-molecule analyses reveal surprisingly low TRAK2 mRNA spot number (~5 per cell), indicating that mitochondrial dynamics would be highly sensitive to even subtle changes in mRNA levels or stability. Yet, whether such shifts in TRAK2 mRNA dynamics are linked to switches in invasive cell state and/or could be therapeutically manipulated to predictably modulate invasive cell behaviour in disease remains an open question. Likewise, it remains unclear how modulation of TRAK2 mRNA levels and/or localisation influences other critical aspects of mitochondria dynamics, such as fission, fusion and mitophagy. However, what is clear is that the RNA-binding protein (RBP) machinery that directs TRAK2 microtubule plus-end-directed transport must also be a key determinant of cell-size-scaled mRNA transport. Several RBPs are implicated in trafficking of G-motif-containing mRNAs, including APC and UNC41,44, suggesting that the use of distinct RBPs and/or post-translational modification of RBP function could be employed to fine-tune size-scaled responses in distinct tissue contexts. Taken together, this work sheds light on a previously unappreciated role for mRNA transport in the cell-size-dependent modulation of organelle dynamics that has wide implications for understanding the control of mitochondrial function and motile cell behaviour in health and disease.
Methods
Cell culture, transfections, and cell-derived matrix production
All cells were maintained at 37 °C, 5% CO2, with regular passages. HUVEC (PromoCell, Cat. No. C-12200) and hCMEC/d3 (Poller et al., 2008; Merc, Cat.No. SCC066) were cultured on 0.1% gelatin (Millipore) coated T75 flasks (Corning) in endothelial cell basal medium 2 (EBM-2, Promocell) with an additional supplement pack (5% FCS, EGF (5 ng ml−1), VEGF (0.5 ng ml−1), FGF2 (10 ng ml−1), long R3 insulin growth factor-1 (20 ng ml−1), hydrocortisone (0.2 μg ml−1), and ascorbic acid (1 μg ml−1)). 50 mg ml−1 gentamycin (Sigma) and 250 μg ml−1 amphotericin (Sigma) were also added to the culture media. All HUVEC experiments were conducted with cells between passages 3–6. Mouse Balb/c brain endothelioma cells (b.End5, ATCC; Merc, Cat. No. 96091930) and Telomerase-Immortalised Fibroblasts (TIFs; Gift from Prof. P.T. Caswell, University of Manchester) were cultured in Dulbecco's Modified Eagle Medium (DMEM, Sigma) supplemented with 10% FCS and 10 U ml−1 Penicillin-Streptomycin. All cells were detached using Trypsin-EDTA (Sigma) and were split before reaching 90% confluency, with media being changed every 2 days.
Gene knockdown was achieved using ON-TARGETplus non-targeting or gene-specific SMARTpool siRNAs (Horizon). 0.3 μM siRNA was diluted in 200 μl Opti-MEM containing 1.5% v/v GeneFECTOR (Venn Nova). This mix was then added to a well of a sixwell plate containing 1 ml Opti-MEM and the sample was incubated at 37 °C for 3 h. Opti-MEM was then replaced with endothelial growth media and samples were allowed to recover for 48–72 h.
Cell-derived matrix (CDM) was produced according to previously described protocols (Cukierman et al., 2001; Franco-Barraza et al., 2016). Briefly, 0.2% gelatin-coated (Sigma) cell culture dishes or coverslips were fixed with 1% Glutaraldehyde (Sigma), before quenching with 1 M Glycine (Sigma) and equilibration with DMEM. TIFs were seeded at full confluency and grown for 8 days with 25 μg ml−1 Ascorbic Acid (Sigma), changing the media every 2–3 days. TIFs were then denuded with 20 mM NH4OH, 0.5% Triton-X-100 in PBS. Finally, samples were treated with 10 μg ml−1 DNAse I (Roche) before seeding of endothelial cells.
For MS2 transfections, 1×105 b.End5 cells were cultured in 35 mm round glass-bottom dishes (Greiner CellView) and transfected with the following: 0.5 μg pcDNA3.1-Lyn-Cherry, 1 μg pCS2-MCP-GFPnls, 1 μg different versions of pcDNA3.1-HBB-24xMS2SL-3’UTR. Lipofectamine 3000 (Thermo) was used following manufacturer’s instructions. Cells were incubated for 48 hours before microscopic analysis.
Generation of CRISPR-Cas9 mutant cell lines
crRNAs were designed to target sequences immediately surrounding the minimal localisation elements in the TRAK2 3’UTRs using the online IDT Alt-R CRISPR-Cas9 design tool to minimise off-target effects (Supplementary table 1). IDT Alt-R CRISPR-Cas9 protocol was followed mostly according to manufacturer’s instructions. Briefly, 200 μM each crRNA was hybridised to 200 μM tracrRNA by heating to 95 °C in a thermal cycler and allowing to cool to room temperature. RNP complexes were formed by mixing 120pmol crRNA:tracrRNA duplex with 104 pmol Alt-R Cas9 in PBS and incubating at room temperature for 20 min. RNP complexes for generating wt cell lines contain no crRNA:tracrRNA duplexes.
RNP complexes were added to 5 × 105 hCMEC/d3 cells along with 2 μg pMAX-GFP plasmid (Lonza) and transfection was carried out using a Nucleofector 2b (Lonza) according to the manufacturer’s instructions. Cells were then cultured for 72 hours before individual GFP-expressing clones were sorted into wells of a 96-well plate using a FACSAria Fusion cell sorter (BD Biosciences). Clonal populations were screened for genomic lesions using gene-specific primers to amplify regions in the 3’UTRs by PCR (Supplementary Table 1). Two Wt and four ΔTRAK2 lines were created and used in combination for each experiment.
smFISH and fluorescence labelling
Endothelial cells on glass coverslips or CDM were fixed in methanol-free 4% formaldehyde and permeabilised in 70% ethanol. Stellaris single-molecule fluorescence in situ hybridisation (smFISH) gene-specific probe sets (Supplementary Table 2), conjugated to Quasar 570/670 fluorophores, were designed using an online tool (Biosearchtech). Predesigned GAPDH reference probe sets are purchased directly. smFISH probe hybridisation was carried out as described previously40. Briefly, samples were washed in wash buffer (2× SSC, 10% formamide) before incubation overnight at 37 °C with smFISH probe sets diluted in hybridisation buffer (2× SSC, 10% formamide, 10% w/v Dextran Sulphate). Samples were then washed twice in wash buffer at 37 °C for 30 min. To co-label nuclei and F-actin, samples were further incubated with 5ug ml−1 DAPI (Sigma) in PBS, and with 165 nM AlexaFluor 488/647 Phalloidin in PBS (Thermo Fisher Scientific), for 5 mins at room temperature. To label mitochondria, MitoTracker was added to the cell media at 250 nM for 30 min. Fresh media were then added, and cells were incubated for 2 hrs before fixation. To label microtubules, cells were fixed in 4% PFA at room temperature for 10 min, then permeabilised in ice-cold methanol at 4 °C for 20 min. Cells were then stained with anti-alpha tubulin antibody (Abcam ab7291) 1:100, followed by a goat anti-mouse AlexaFluor 568-conjugated secondary antibody (1:500, Thermo Fisher Scientific). All fixed samples were mounted using Prolong Gold (Thermo Fisher Scientific). For TMRM staining, cells were seeded onto glass-bottom dishes in EBM-2 1–2 days before imaging.
Growth media was replaced with fresh media containing 200 nM Mitotracker Green FM 9074 (Cell Signalling Technology) and 1× Image-iT TMRM Reagent (Invitrogen). Cells were incubated for 30 min and then washed 1× with PBS before fresh media was added prior to imaging.
Plasmid construction
All plasmids for in vitro MS2 experiments were generated as described previously40. Briefly, 3’UTR sequences were amplified by PCR using sequence-specific primers (Supplementary table 1) from human genomic DNA using MyTaq Red Mix (Bioline) according to manufacturer’s instructions. Site-directed mutagenesis was used to create precise deletions in 3’UTRs. Primers for SDM were designed using Agilent QuickChange II design software. Phusion (NEB) was then used with the following reaction conditions: denaturation 95 oC for 30 s, annealing 55 °C for 1 m 30 s, extension 72 °C for 6 m for 18 cycles. Reactions were then treated with DpnI (NEB) for a minimum of 1 hr at 37 °C. 3’UTR sequences were then cloned into a pcDNA3.1-HBB-24xMS2SL-MCS expression vector using the NheI and XhoI/ApaI restriction sites in the multiple cloning site40.
Microscopy
Unless otherwise stated, fluorescent microscopy data were obtained using an Olympus IX83 inverted microscope using Lumencor LED excitation, a ×100/1.40 UplanSApo objective and a Sedat QUAD filter set (Chroma 89000). The images were collected using an R6 (Q-imaging) CCD camera with a Z optical spacing of 0.2 μm. Raw images were then deconvolved using the Huygens Pro software (SVI), and maximum intensity projections of these deconvolved images are shown in the results. To image microtubules, cell were imaged using a Zeiss LSM 980 Airyscan microscope using a Zeiss Plan-APOCHROMAT ×63/1.4 Oil Ph3 objective and a Z-stack spacing of 0.5 µm. For TMRM staining, images were acquired on an Eclipse Ti inverted microscope (Nikon) using a ×20/0.45 SPlan Fluor or ×40/0.6 SPlan Fluor objective, Nikon filter sets for brightfield, GFP and mCherry and a pE-300 LED (CoolLED) fluorescent light source. Throughout imaging, cells were maintained at 37 °C and 5% CO2. Images were then collected using a Retiga R6 (Q-Imaging) camera with a Z optical spacing of 0.5 μm. Cell motility tracking data was obtained using an Eclipse Ti inverted microscope (Nikon) using a ×10/0.45 Plan Fluor (Ph1DLL) objective, and a pE-300 LED (CoolLED) fluorescent light source. Imaging software NIS Elements AR.46.00.0. Point visiting was used in combination with laser-base autofocus to allow multiple positions to be imaged within the same time-course, and cells were maintained at 37 °C and 5% CO2. The images were collected using a Retiga R6 (Q-Imaging) camera.
Immunoprecipitation
ECs were trypsinized, pelleted, washed twice in PBS, pelleted and then lysates prepared upon resuspension in 500 µl ice-cold NP-40 lysis buffer (20 mM Tris-HCl pH 8, 137 mM NaCl, 10% glycerol, 1% Nonidet P-40, 2 mM EDTA, 1:100 protease inhibitor cocktail). Lysates were rotated for 1 hr at 4 °C, centrifuged at 13,500 × g for 30 min and supernatant collected. 10 µg of anti-TRAK2 antibody or 10 µg control IgG antibody was then added to EC lysates and incubated at 4 °C overnight with mixing. 0.25 mg Protein A/G Magnetic Beads (Pierce) were washed in NP-40 lysis buffer, then the antibody/lysate mixture was added and incubated at room temperature for 1 hr with mixing. Beads were then collected, washed twice in NP-40 lysis buffer and then once in purified water. Beads were finally collected, incubated with 40 μl of 1× Laemmli buffer, mixed on a rotor for 15 min at room temperature and then supernatant collected in preparation for western blotting.
Western blotting
Western blotting was performed using either whole cell protein samples or immunoprecipitation supernatant. Whole cell protein was extracted using RIPA buffer (25 mM Tris–HCl pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate and 0.1% SDS) containing 1:100 Protease Inhibitor Cocktail. Protein concentration was quantified using Pierce BCA Protein Assay Kit (Thermo Fisher Scientific) before denaturation with Laemmli Buffer (250 mM Tris–HCl pH 6.8, 2% SDS, 10% glycerol, 0.0025% bromophenol blue, 2.5% β-mercaptoethanol) at 95 °C for 5 min. Proteins were separated using 10% Mini-PROTEAN TGX precast protein gels (Bio-Rad). Trans-Blot Turbo transfer system (Bio-Rad) was used to transfer proteins to PVDF membranes using manufacturer's guidelines. Membranes were then blocked for 1 hr at room temperature in 2.5% BSA (Sigma) in TBS containing 0.1% Tween-20. Primary antibodies were diluted in blocking buffer and incubated overnight at 4 oC. Membranes were then washed with TBS containing 0.1% Tween-20 before incubation with secondary antibodies diluted in blocking buffer for 1 hr at room temperature, followed by more washes. Pierce SuperSignal West Atto Western blotting substrate (Thermo Fisher Scientific) was used to develop chemiluminescent signal, which was then detected digitally using a ChemiDoc MP Imager (Bio-Rad).
Antibodies
Primary and secondary antibodies were used at the following concentrations: rabbit anti-TRAK2 (1:1000 Western blot, Proteintech, 13770-1), mouse anti-TRAK2 (1:500 Western blot, Thermo Fisher Scientific, MA5-27606), mouse anti-MIRO1 (1:200 Western blot, proximity ligation assay, Abcam, CL1083), rabbit anti-β-actin (1:1000 Western blot, Cell Signalling technology, 4967), rabbit polyclonal anti-TRAK2 (1:50 Immunoprecipitation, 1:200 proximity ligation assay, Thermo Fisher Scientific, PA5-34889), mouse anti-alpha tubulin (1:100 Abcam ab7291), rabbit IgG (1:50 Immunoprecipitation, Proteintech, 30000-0-AP), goat anti-mouse AlexaFluor 568 (1:500 Immunofluorescence, Thermo Fisher Scientific, A-21043), goat anti-rabbit AlexaFluor 488 (1:500 Immunofluorescence, Thermo Fisher Scientific, A-11008).
Image analysis
RNA spot counts were obtained using Fuji (2.14.0/1.54j) plugin FindFoci62. This tool uses an automated Otsu thresholding algorithm for detecting peaks with intensity above the background. RNA spots with an intensity peak above the threshold were then counted and highlighted with red circles in images. RNA polarisation measurement was obtained for background-subtracted smFISH images using the previously described Polarisation and Dispersion Index53. The Polarisation Index assesses the extent to which an mRNA is either centrally localised or peripherally polarised within a defined cell shape. Here, if an mRNA is polarised, the centroid of the mRNA should be distinct from the centroid of the cell. The Polarisation Index thus defines the displacement vector pointing from the cell centroid to the mRNA centroid. This polarisation vector is then divided by the radius of gyration of the cell, which is calculated by the root-mean-square distance of all pixels within an image of a cell from the cell centroid. Hence, the Polarisation Index defines the polarisation of the mRNA normalised to the size and the elongation of a cell. The Dispersion Index determines the extent to which an mRNA is either clustered together or uniformly distributed within a defined cell shape. Here, mRNA dispersion is quantified by determining the second moment μ2 of RNA positions, which is dictated by the shape and size of the cell and mRNA distribution. Within a cell of defined shape, the Dispersion Index calculates the second moment μ2 for both the test mRNA and a hypothetical mRNA of uniform distribution. Consequently, dividing the test mRNA second moment μ2 by the second moment μ2 of the hypothetical uniform mRNA thus normalises the test mRNA dispersion to cell shape. Alternatively, RNA polarisation was also measured by calculating the XY coordinates of the centre of mass (CoM) of the RNA fluorescence (using the Analyse menu in Fuji (2.14.0/1.54j)) and determining the Euclidean distance of the RNA CoM from the nearest edge of the nucleus. For some analyses, the distance that the RNA centre of mass (CoM) sits from the EC nucleus was also normalised to protrusion length. For quantification of the fluorescence intensity of labelled mitochondria, sum intensity projections of raw image files were generated in Fuji (2.14.0/1.54j), cell outlines and other regions of interest (ROI) were defined using the polygon tool and integrated density of fluorescence of ROIs and background measured, along with the ROI area. Cell motility tracking in CDM was performed manually using the MTrackJ plugin for Fuji (2.14.0/1.54j). Randomly selected cells were tracked through each frame of the time-lapse movies. Quantification of cells on glass was achieved using the Chemotaxis and Migration Tool (Ibidi). For cells both in CDM and on glass, directionality was calculated as the displacement of a cell from point A to B, divided by the total length of the path taken to get there. EC morphometrics (aspect ratio, circularity and protrusion length) were quantified using Fuji (2.14.0/1.54j). For analysis of motile cell protrusions, firstly, these protrusions were identified as the cell protrusion exhibiting either the highest enrichment of localised mRNAs, polarised perinuclear enrichment of mitochondria or enrichment of TRAK2-MIRO1 PLA foci. As cells were predominantly uniaxially elongated, motile protrusions were usually easily distinguishable as the longest protrusion extended by a cell. To quantify protrusion length, the distance from the nuclear envelope to the leading edge of the cell was then determined.
Proximity ligation assays
ECs cultured on CDM in glass-bottomed dishes were first fixed in 4% PFA for 10 min, washed twice for 5 min in PBS and then permeabilised in 0.1% PBS Triton-X for 30 min at room temperature. Proximity ligation assays were then performed using the Duolink PLA fluorescence kit (Merck), as per manufacturer's guidelines. Of note, the volume of incubations was set at 120 µl per glass-bottomed dish, and primary antibodies were incubated overnight at 4 °C. Moreover, prior to mounting, samples were incubated with Phalloidin-Alexafluor 488 (1:40 dilution) for 20 min.
Statistics & reproducibility
All statistical analyses were performed using GraphPad Prism 10 (v.10.23(347)) software. No statistical methods were used to predetermine sample sizes. Normality testing (Shapiro–Wilk) was performed to determine if datasets were suitable for parametric or non-parametric statistical tests. All data are presented as mean ± s.e.m., unless stated otherwise. Either a two-tailed unpaired Student’s t-test or a two-tailed Mann–Whitney test was used for the comparison of two normally distributed or non-normally distributed datasets, respectively. An ordinary one-way ANOVA with Tukey’s multiple comparisons test was used to compare more than two normally distributed datasets. To compare more than two non-normally distributed datasets, an unpaired Kruskal–Wallis test and Dunn multiple comparison test were used. Data correlation was tested via the Pearson’s correlation coefficient. Differences between linear regressions were assessed via a two-sided analysis of covariance. Error bars, P values, n numbers and statistical tests used are reported in the figures and/or figure legends. All statistical tests were performed on datasets acquired from at least two independent experiments. For each analysis, measurements were taken from distinct samples.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data generated or analysed during this study are included in this published article (and its supplementary information files). Source data are provided with this paper.
References
Rafelski, S. M. & Marshall, W. F. Building the cell: design principles of cellular architecture. Nat. Rev. Mol. Cell Biol. 9, 593–602 (2008).
Marshall, W. F. Scaling of subcellular structures. Annu Rev. Cell Dev. Biol. 36, 219–236 (2020).
van Bergeijk, P., Hoogenraad, C. C. & Kapitein, L. C. Right time, right place: probing the functions of organelle positioning. Trends Cell Biol. 26, 121–134 (2016).
Kroll, J. & Renkawitz, J. Principles of organelle positioning in motile and non-motile cells. EMBO Rep. 25, 2172–2187 (2024).
Detmer, S. A. & Chan, D. C. Functions and dysfunctions of mitochondrial dynamics. Nat. Rev. Mol. Cell Biol. 8, 870–879 (2007).
Shah, M., Chacko, L. A., Joseph, J. P. & Ananthanarayanan, V. Mitochondrial dynamics, positioning and function mediated by cytoskeletal interactions. Cell Mol. Life Sci. 78, 3969–3986 (2021).
Zhang, H., Menzies, K. J. & Auwerx, J. The role of mitochondria in stem cell fate and aging. Development 145, dev143420 (2018).
Noel, J. S., Dewey, W. C., Abel, J. H. Jr & Thompson, R. P. Ultrastructure of the nucleolus during the Chinese hamster cell cycle. J. Cell Biol. 49, 830–847 (1971).
Posakony, J. W., England, J. M. & Attardi, G. Mitochondrial growth and division during the cell cycle in HeLa cells. J. Cell Biol. 74, 468–491 (1977).
Conklin, E. G. Cell size and nuclear size. J. Exp. Zool. 12, 1–98 (1912).
Boldogh, I. R. & Pon, L. A. Mitochondria on the move. Trends Cell Biol. 17, 502–510 (2007).
Babic, M. et al. Miro’s N-terminal GTPase domain is required for transport of mitochondria into axons and dendrites. J. Neurosci. 35, 5754–5771 (2015).
Brickley, K. & Stephenson, F. A. Trafficking kinesin protein (TRAK)-mediated transport of mitochondria in axons of hippocampal neurons. J. Biol. Chem. 286, 18079–18092 (2011).
Glater, E. E., Megeath, L. J., Stowers, R. S. & Schwarz, T. L. Axonal transport of mitochondria requires milton to recruit kinesin heavy chain and is light chain independent. J. Cell Biol. 173, 545–557 (2006).
Guo, X. et al. The GTPase dMiro is required for axonal transport of mitochondria to Drosophila synapses. Neuron 47, 379–393 (2005).
Russo, G. J. et al. Drosophila Miro is required for both anterograde and retrograde axonal mitochondrial transport. J. Neurosci. 29, 5443–5455 (2009).
Stowers, R. S., Megeath, L. J., Gorska-Andrzejak, J., Meinertzhagen, I. A. & Schwarz, T. L. Axonal transport of mitochondria to synapses depends on milton, a novel Drosophila protein. Neuron 36, 1063–1077 (2002).
van Spronsen, M. et al. TRAK/Milton motor-adaptor proteins steer mitochondrial trafficking to axons and dendrites. Neuron 77, 485–502 (2013).
Henrichs, V. et al. Mitochondria-adaptor TRAK1 promotes kinesin-1 driven transport in crowded environments. Nat. Commun. 11, 3123 (2020).
Lopez-Domenech, G. et al. Miro proteins coordinate microtubule- and actin-dependent mitochondrial transport and distribution. EMBO J. 37, 321–336 (2018).
Canty, J. T., Hensley, A., Aslan, M., Jack, A. & Yildiz, A. TRAK adaptors regulate the recruitment and activation of dynein and kinesin in mitochondrial transport. Nat. Commun. 14, 1376 (2023).
Fenton, A. R., Jongens, T. A. & Holzbaur, E. L. F. Mitochondrial adaptor TRAK2 activates and functionally links opposing kinesin and dynein motors. Nat. Commun. 12, 4578 (2021).
Nunnari, J. & Suomalainen, A. Mitochondria: in sickness and in health. Cell 148, 1145–1159 (2012).
Mosier, J. A., Fabiano, E. D., Ludolph, C. M., White, A. E. & Reinhart-King, C. A. Confinement primes cells for faster migration by polarizing active mitochondria. Nanoscale Adv. 6, 209–220 (2023).
Garde, A. et al. Localized glucose import, glycolytic processing, and mitochondria generate a focused ATP burst to power basement-membrane invasion. Dev. Cell 57, 732–749 e737 (2022).
Kelley, L. C. et al. Adaptive F-actin polymerization and localized ATP production drive basement membrane invasion in the absence of MMPs. Dev. Cell 48, 313–328 e318 (2019).
Cunniff, B., McKenzie, A. J., Heintz, N. H. & Howe, A. K. AMPK activity regulates trafficking of mitochondria to the leading edge during cell migration and matrix invasion. Mol. Biol. Cell 27, 2662–2674 (2016).
Caino, M. C. et al. PI3K therapy reprograms mitochondrial trafficking to fuel tumor cell invasion. Proc. Natl. Acad. Sci. USA 112, 8638–8643 (2015).
Desai, S. P., Bhatia, S. N., Toner, M. & Irimia, D. Mitochondrial localization and the persistent migration of epithelial cancer cells. Biophys. J. 104, 2077–2088 (2013).
Zhao, J. et al. Mitochondrial dynamics regulates migration and invasion of breast cancer cells. Oncogene 32, 4814–4824 (2013).
Campello, S. et al. Orchestration of lymphocyte chemotaxis by mitochondrial dynamics. J. Exp. Med. 203, 2879–2886 (2006).
Schuler, M. H. et al. Miro1-mediated mitochondrial positioning shapes intracellular energy gradients required for cell migration. Mol. Biol. Cell 28, 2159–2169 (2017).
Spillane, M., Ketschek, A., Merianda, T. T., Twiss, J. L. & Gallo, G. Mitochondria coordinate sites of axon branching through localized intra-axonal protein synthesis. Cell Rep. 5, 1564–1575 (2013).
Li, Z., Okamoto, K., Hayashi, Y. & Sheng, M. The importance of dendritic mitochondria in the morphogenesis and plasticity of spines and synapses. Cell 119, 873–887 (2004).
Moore, A. S. et al. Actin cables and comet tails organize mitochondrial networks in mitosis. Nature 591, 659–664 (2021).
Hinge, A. et al. Asymmetrically segregated mitochondria provide cellular memory of hematopoietic stem cell replicative history and drive HSC attrition. Cell Stem Cell 26, 420–430.e426 (2020).
Katajisto, P. et al. Stem cells. Asymmetric apportioning of aged mitochondria between daughter cells is required for stemness. Science 348, 340–343 (2015).
O’Toole, M., Latham, R., Baqri, R. M. & Miller, K. E. Modeling mitochondrial dynamics during in vivo axonal elongation. J. Theor. Biol. 255, 369–377 (2008).
Medioni, C., Mowry, K. & Besse, F. Principles and roles of mRNA localization in animal development. Development 139, 3263–3276 (2012).
Costa, G., Bradbury, J. J., Tarannum, N. & Herbert, S. P. RAB13 mRNA compartmentalisation spatially orients tissue morphogenesis. EMBO J. 39, e106003 (2020).
Arora, A. et al. High-throughput identification of RNA localization elements in neuronal cells. Nucleic Acids Res. 50, 10626–10642 (2022).
Pichon, X. et al. The kinesin KIF1C transports APC-dependent mRNAs to cell protrusions. RNA 27, 1528–1544 (2021).
Chrisafis, G. et al. Collective cancer cell invasion requires RNA accumulation at the invasive front. Proc. Natl. Acad. Sci. USA 117, 27423–27434 (2020).
Mili, S., Moissoglu, K. & Macara, I. G. Genome-wide screen reveals APC-associated RNAs enriched in cell protrusions. Nature 453, 115–119 (2008).
Moissoglu, K. et al. RNA localization and co-translational interactions control RAB13 GTPase function and cell migration. EMBO J. 39, e104958 (2020).
Grant, C. E., Bailey, T. L. & Noble, W. S. FIMO: scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018 (2011).
Goering, R., Arora, A., Pockalny, M. C. & Taliaferro, J. M. RNA localization mechanisms transcend cell morphology. Elife 12, e80040 (2023)
Gasparski, A. N. et al. mRNA location and translation rate determine protein targeting to dual destinations. Mol. Cell 83, 2726–2738.e2729 (2023).
Norris, M. L. & Mendell, J. T. Localization of Kif1c mRNA to cell protrusions dictates binding partner specificity of the encoded protein. Genes Dev. 37, 191–203 (2023).
Bodor, D. L., Ponisch, W., Endres, R. G. & Paluch, E. K. Of cell shapes and motion: the physical basis of animal cell migration. Dev. Cell 52, 550–562 (2020).
Costa, G. et al. Asymmetric division coordinates collective cell migration in angiogenesis. Nat. Cell Biol. 18, 1292–1301 (2016).
Zhang, H. L. et al. Neurotrophin-induced transport of a beta-actin mRNP complex increases beta-actin levels and stimulates growth cone motility. Neuron 31, 261–275 (2001).
Park, H. Y., Trcek, T., Wells, A. L., Chao, J. A. & Singer, R. H. An unbiased analysis method to quantify mRNA localization reveals its correlation with cell motility. Cell Rep. 1, 179–184 (2012).
Singh, S., Bernal Astrain, G., Hincapie, A. M., Goudreault, M. & Smith, M. J. Complex interplay between RAS GTPases and RASSF effectors regulates subcellular localization of YAP. EMBO Rep. 25, 3574–3600 (2024).
Sahgal, P. et al. GGA2 and RAB13 promote activity-dependent beta1-integrin recycling. J. Cell Sci. 132, jcs233387 (2019).
Theisen, U., Straube, E. & Straube, A. Directional persistence of migrating cells requires Kif1C-mediated stabilization of trailing adhesions. Dev. Cell 23, 1153–1166 (2012).
Grabham, P. W., Seale, G. E., Bennecib, M., Goldberg, D. J. & Vallee, R. B. Cytoplasmic dynein and LIS1 are required for microtubule advance during growth cone remodeling and fast axonal outgrowth. J. Neurosci. 27, 5823–5834 (2007).
Siddiqui, N. et al. PTPN21 and Hook3 relieve KIF1C autoinhibition and activate intracellular transport. Nat. Commun. 10, 2693 (2019).
Kendrick, A. A. et al. Hook3 is a scaffold for the opposite-polarity microtubule-based motors cytoplasmic dynein-1 and KIF1C. J. Cell Biol. 218, 2982–3001 (2019).
Fraley, S. I. et al. A distinctive role for focal adhesion proteins in three-dimensional cell motility. Nat. Cell Biol. 12, 598–604 (2010).
Pekkurnaz, G., Trinidad, J. C., Wang, X., Kong, D. & Schwarz, T. L. Glucose regulates mitochondrial motility via Milton modification by O-GlcNAc transferase. Cell 158, 54–68 (2014).
Herbert, A. D., Carr, A. M. & Hoffmann, E. FindFoci: a focus detection algorithm with automated parameter training that closely matches human assignments, reduces human inconsistencies and increases speed of analysis. PLoS ONE 9, e114749 (2014).
Acknowledgements
We thank the University of Manchester Bioimaging Core, Biological Services and Flow Cytometry Facilities for technical support. We also thank members of the S.P. Herbert and R. Das laboratories for discussion. This work was supported by a fellowship grant from the Wellcome Trust (219500/Z/19/Z) to S.P.H. and a BBSRC DTP PhD studentship to J.J.B.
Author information
Authors and Affiliations
Contributions
S.P.H. conceived and designed the project. J.J.B. performed experimental studies with assistance from G.E.H., R.V., G.C. and H.E.L. S.P.H. supervised the study. The manuscript was written by S.P.H. and J.J.B.
Corresponding author
Ethics declarations
Competing interests
Authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bradbury, J.J., Hulmes, G.E., Viswanathan, R. et al. mRNA trafficking directs cell-size-scaling of mitochondria distribution and function. Nat Commun 16, 7029 (2025). https://doi.org/10.1038/s41467-025-61940-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-61940-6