Abstract
Induced proximity by molecular glues refers to strategies that leverage the recruitment of proteins to facilitate their modification, regulation or degradation. As prospective design of molecular glues remains challenging, unbiased discovery methods are necessary to discover new chemical targets. Here we establish a high throughput affinity proteomics workflow leveraging E3 ligase activity-impaired CRBN-DDB1ΔB in cell lysates for the unbiased identification of molecular glue targets. By mapping the interaction landscape of CRBN-binding molecular glues, we unveil 298 protein targets and demonstrate the utility of enrichment methods for identifying targets overlooked by established methods. We use a computational workflow to estimate target confidence and perform biochemical and structural validation of uncharacterized neo-substrates. We further identify a lead compound for the previously untargeted non-zinc finger PPIL4 through a biochemical screen. Our study provides a comprehensive inventory of targets chemically recruited to CRBN and delivers a robust and scalable workflow for identifying drug-induced protein interactions in cell lysates.
Similar content being viewed by others
Introduction
Targeted Protein Degradation (TPD) represents a promising therapeutic approach to remove disease-associated proteins from cells1,2. The process of TPD involves hijacking the ubiquitylation machinery for the covalent attachment of ubiquitin molecules to a desired protein of interest, which in turn leads to degradation by the proteasome3,4. Ubiquitin-mediated TPD utilizes two types of small molecules, molecular glues5 and heterobifunctional degraders (also known as PROteolysis Targeting Chimeras, or PROTACs)3, both of which chemically induce ternary complex formation between a protein target and a ubiquitin E3 ligase, followed by proximity-driven ubiquitylation and subsequent degradation.
Despite the rapid growth of TPD as a therapeutic strategy, the discovery and development of effective degraders remains challenging. Heterobifunctional degraders rely on a linker to connect two binding warheads: one for the ligase and one for the protein of interest3,6,7. Although this modular design offers the flexibility to target any protein with a known binder, the resulting molecules often possess poor drug-like properties due to their large size. Molecular glues present an alternative due to their small size and improved drug-like properties. However, they lack binding to their target protein alone and instead enhance protein-protein interactions (PPI) between a ligase and substrate, therefore, rational design of molecular glues is far more challenging8. Over the last decade, the discovery of molecular glue degraders heavily relied on serendipity through phenotypic screening of large libraries of molecules and retrospective identification of their degradation targets. Although FDA-approved molecular glue degraders exist, including the immunomodulatory drugs (IMiDs) thalidomide, lenalidomide, and pomalidomide, they have all been characterized as molecular glues in retrospect after their serendipitous discovery. IMiD molecules bind to CRBN, a substrate receptor of the CUL4-RBX1-DDB1-CRBN (CRL4CRBN) E3 ligase5,9,10,11,12,13 creating a favorable surface for noncanonical proteins (neo-substrates) to bind for induced degradation. Since this discovery, significant efforts into the design and screening of unique IMiD analogs have revealed up to 50 neo-substrates in the public domain, all carrying a glycine-containing β-hairpin structural degron5,11,14,15,16,17,18,19,20,21,22. Remarkably, previously reported computational modeling of the AlphaFold2 (AF2) structures available in the Protein Data Bank (PDB) suggests that we are just scraping the surface of what is targetable by these molecules14.
Given the mechanism of action (MoA) of degraders, global proteome profiling has proven to be an effective tool for the identification of protein degradation targets19,23,24,25. Using this approach, the target space of degraders for multiple therapeutic target classes have been extensively mapped for tractable targets, including kinases23,26,27, bromo-domains28,29,30, HDACs31, and zinc finger (ZF) transcription factors19. Although this method has greatly expanded the repertoire of known targets, limited sensitivity has restricted the ability to identify proteins with low expression levels without screening libraries of cell lines or using target enrichment methods. This approach also remains blind to a key aspect of these molecules: the identification of proteins that are recruited to the ligase but do not ultimately get degraded. Such “non-degrading glue” targets may be subject to poor lysine accessibility, lack of degradative ubiquitin chain formation32, high deubiquitinase activity, poor proteasome access, or other resistance mechanisms. Nevertheless, these substrates still represent important therapeutic targets if these factors can be overcome to convert silent molecular glues into molecular glue degraders or functional modulators of the target.
Methods to identify chemically induced protein-protein interactions include immunoprecipitation mass spectrometry (IP-MS)33 and proximity labeling approaches coupled to mass spectrometry34,35. IP-MS approaches have been employed for the identification of direct protein interactions, whereas proximity labeling approaches are commonly employed for the mapping of proximity interactomes in cells and in vivo34,36,37,38, enabling the identification of protein interactions within a 10-30 nm radius of the epitope-tagged protein of interest34,36,37,39. Although these in-cell methods have demonstrated successful identification of chemically induced interactions, they often require extensive fine-tuning of various factors, including noise, sensitivity, variability, and scalability.
In this study, we establish a simple, robust, and sensitive workflow involving spike in of recombinant, activity-impaired CRBN-DDB1ΔB and degrader molecules in cell lysates to facilitate compound-induced complex formation for high-throughput discovery of degrader-induced protein-protein interactions and downstream development into selective tools and therapeutic candidates. This method allowed us to build a comprehensive inventory of 298 distinct protein targets recruited to CRBN, including many uncharacterized zinc finger (ZF) transcription factors and proteins from various target classes, including RNA-recognition motif (RRM) domain proteins. We evaluate the binding potential of these targets through structural alignment with IMiD-bound CRBN and performed biochemical and structural validation studies on a series of non-ZF targets. Furthermore, screening a library of ~ 6000 IMiD analogs against a non-ZF target, PPIL4, identified a selective lead degrader molecule, thereby presenting a blueprint for the effective target-centric discovery of molecular glue degraders. Finally, we have updated our open-access proteomics portal (https://v2.dfci-fischerlab.com/) to present this quantitative interactome data along with compound structures as a public resource.
Results
Unbiased identification of degrader-induced interactors in-lysate
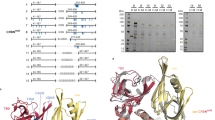
To establish a workflow for the identification of chemically induced protein-protein interactions, we set out to simplify traditional IP-MS methods. We hypothesized that we could create a controlled environment with reduced biological variability and enhanced scalability by establishing a workflow in cell lysate using spiked in recombinant protein as the bait. Our workflow harnesses small-molecule degrader-induced ternary complex formation in cell lysate using recombinant FLAG-tagged CRBN in complex with DDB1, excluding the BPB domain (∆B), which prevents CUL4 interaction to inhibit ubiquitylation of the recruited target. After incubation, we enrich with a highly selective antibody for the FLAG epitope tag followed by label free quantitative proteomic assessment to identify interactors (Fig. 1a). To benchmark and explore the viability of this approach for identification of protein-protein interactions, we selected two representative degrader molecules that have been thoroughly profiled in published reports – pomalidomide, an IMiD molecular glue19,40 and SB1-G-187, a kinase-targeted heterobifunctional degrader26 (Fig. 1b). To determine the optimal quantification approach for this proteomics workflow, we compared TMT and label-free quantification using whole cell lysates treated with pomalidomide or SB1-G-187. Label free quantification effectively identified published degrader targets, justifying its use in the subsequent enrichment studies (Supplementary Fig. 1a). We then proceeded to perform enrichment proteomics to profile these two molecules across two different cell lines, MOLT4 and Kelly, selected for their orthogonal expression profiles and broad coverage of known CRBN neo-substrates including IKZF1/3 (MOLT4) and SALL4 (Kelly)19. The pomalidomide screen revealed 11 different enriched proteins across these two cell lines (9 in MOLT4 and 4 in Kelly cells), which revealed three unfamiliar targets, ASS1, ZBED3, and ZNF219 (Fig. 1c, Supplementary Fig. 1b, c and Supplementary Data 1). We then validated recruitment of these targets to CRBN using dose-response immunoblot or TR-FRET analysis (Supplementary Fig. 1d, e).
a Schematic representation of the first-generation enrichment-based quantitative proteomics workflow established for target enrichment and identification. Created in BioRender. Donovan, K. (2025) https://BioRender.com/zf0fy5z. b Chemical structures of degraders – Pomalidomide (molecular glue) and SB1-G-187 (heterobifunctional). c Scatterplots depicting relative protein abundance following Flag-CRBN-DDB1ΔB enrichment from in-lysate treatment with 1 µM Pomalidomide and recombinant Flag-CRBN-DDB1ΔB spike in. Left: MOLT4 cells and Right: Kelly cells. Scatterplots display fold change in abundance to DMSO. Significant changes were assessed by a two-sided moderated t-test as implemented in the limma package86 with log2 FC shown on the y-axis and log10 P-value on the x-axis. Values determined by n = 3-4 independent replicates of each treatment group. d Venn diagram showing unique and overlapping hits for pomalidomide found in our enrichment study and in publicly available whole cell degradation data. e As in (c), but with 1 µM SB1-G-187 treatment. f As in (d), but with SB1-G-187 treatment.
To assess the overlap of these enriched targets with published degradation data for pomalidomide, we performed a hit comparison with publicly available global proteomics data (https://proteomics.fischerlab.org/), which includes ten independent pomalidomide treatments spanning HEK293T, Kelly and MOLT4 cell lines (Fig. 1d). Like our enrichment data, the global degradation data also maps 11 targets as degradable, however only 4 of these targets (IKZF1, IKZF3, ZFP91 and SALL4) overlap with those that we see enriched in this dataset. The SB1-G-187 kinase degrader screen identified 18 enriched targets across the two cell lines (16 in MOLT4 and 7 in Kelly cells), including multiple non-protein kinase targets, which raised the question of how these proteins are being recruited to CRBN by a kinase degrader (Fig. 1e, Supplementary Fig. 1f, g and Supplementary Data 1). Assessment of the non-kinase targets revealed that several are known to form complexes with different kinases, such as TAB1 and TAB2, which form a functional kinase complex with MAP3K741,42, and UNC119, which binds to myristoylated SRC to regulate cellular localization43. This data suggests that these non-kinases are being indirectly recruited to CRBN through piggybacking on their biological complex binding partners. Of the other recruited non-kinase targets, ZBED3 is also identified in the pomalidomide treatment, suggesting recruitment through the IMiD handle of the degraded, and SDR39U1 was reported as a non-kinase target in a compound-based affinity profiling study of kinase inhibitor probes44. Next, we assessed the differences and overlap in hits between publicly available degradation data in MOLT4 and Kelly cells for the kinase-targeted heterobifunctional degrader, SB1-G-187, and enrichment data (Fig. 1e). We found 6 overlapping hits - all protein kinases - including CDK1, IRAK1, LCK, LYN, MAP3K7, and SRC. Like the pomalidomide data, we observe similar numbers of proteins identified in either degradation data (15) or enrichment data (12), demonstrating that these two methods complement each other to expand the target scope of these molecules. Together, the data collected for these two degrader molecules demonstrate the value of our workflow for identifying chemically induced protein-protein interactions invisible to degradation assays, while also highlighting opportunities for improving sensitivity.
Mapping the interactomes of IMiD molecular glue degraders
Next, using the functional enrichment method as a basis, we set out to optimize and address the critical need for sensitivity and high throughput. IP-MS experiments typically require labor-intensive sample preparation steps, which create a significant source of variability and lead to high background and false positive rates while also placing limits on the number of samples that can be prepared in parallel. To address these limitations, we automated the enrichment and sample preparation procedures to enable effective mapping of interaction targets across libraries of molecules at scale. We incorporated a cost effective Opentrons OT-2 liquid handling platform to automate the sample preparation process from addition of all immunoprecipitation components to tryptic digestion for 96 samples in parallel (Fig. 2a). To address throughput and depth of the proteomics workflow, we took advantage of recent updates in instrumentation (timsTOF Pro2, Bruker) and acquisition methods (diaPASEF)45 that enable robust and high sensitivity sampling of peptides in complex samples (Fig. 2a). In contrast to the data-dependent acquisition (DDA) data collected in our proof-of-concept analysis (Fig. 1), data-independent acquisition (DIA) measures peptides by systematically sampling all precursor ions within a specified m/z range regardless of their abundance which enhances the reproducibility and depth of peptide coverage to allow for accurate and reliable quantification.
a Schematic representation of the second-generation enrichment-based quantitative proteomics workflow established for target enrichment and identification. Created in BioRender. Donovan, K. (2025) https://BioRender.com/s22eej5. b Chemical structures of the 20 CRBN-based degraders profiled in this study. c Scatterplot depicting relative protein abundance following Flag-CRBN-DDB1ΔB enrichment from in-lysate treatment with degrader and recombinant Flag-CRBN-DDB1ΔB spike in. Scatterplot displays fold change in abundance to DMSO. Significant changes were assessed by a two-sided moderated t test as implemented in the limma package86 with log2 FC shown on the y-axis and log10 P-value on the x-axis. Values determined by n = 4 independent replicates of each treatment group. Scatterplots for all 21 treatments across MOLT4 and Kelly cells can be found in separate PDF’s “Supplementary Figs. 2, 3”, a representative example for a single treatment (Pomalidomide, 1 µM) is displayed here. d The number of independent IPs for which enrichment was observed for each target. The plot is displaying targets enriched > 3 times. The inset displays the top 20 frequently enriched target proteins. e Venn diagram showing unique and overlapping hits found in our enrichment study and in published literature. f Donut chart representing the proportions of enriched proteins contained within the Top 10 different superfamily categories. g Donut chart representing the proportions of enriched proteins contained within the Top 10 different domain categories.
Work over the last several years has led to the identification of a growing list of ~ 50 neo-substrates that are recruited to CRBN by IMiD analogs for chemically induced degradation14. Validated targets include a large number of C2H2 zinc finger ZF transcription factors such as IKZF1/35, ZFP9118, or SALL415,19, but only a few non-ZF proteins such as G1 to S phase transition protein 1 (GSPT1)11 and casein kinase 1 alpha (CSNK1A1)12,17. These targets do not possess any similarity, but instead all share a common CRBN binding structural motif consisting of an 8-residue loop that connects the two strands of a β-hairpin and has a glycine at the sixth position (G-loop)11,12,18. A recently reported analysis of available AlphaFold2 (AF2) predicted structures for proteins in the human proteome uncovered over 2500 proteins that harbor a G-loop potentially compatible with IMiD-recruitment to CRBN, with C2H2 ZF proteins revealing themselves as the most prevalent domain class, aligning with the dominance that this class has amongst the experimentally confirmed targets14. Due to the extensive range of proteins that are predicted to be chemically recruitable to CRBN, we asked how many of these proteins are already targeted by existing chemistry, but not yet identified due to the lack of sensitivity of existing methods. To explore the range of proteins chemically recruited to CRBN, we screened a curated library of 20 different IMiD analogs through our automated lysate-based IP workflow (Fig. 2b). We assembled this library of compounds to incorporate a broad range of IMiD-based scaffolds including the parental FDA-approved IMiDs (thalidomide, lenalidomide, pomalidomide)46,47, where there is a high value to identifying unique targets for drug repurposing efforts. We included a series of IMiD analogs that are undergoing clinical trials (CC-220, CC-92480, CC-90009)48,49,50 and also molecules that have demonstrated promiscuity (FPFT-2216, CC-122)51,52. Finally, we included a series of in-house synthesized scaffolds developed in the context of targeting Helios (IKZF2)53 or part of an effort to diversify IMiDs with the addition of fragments on an extended linker. We screened this library at 1 µM concentration across MOLT4 and Kelly cell lines (including a second 5 µM concentration for pomalidomide) and identified proteins that were enriched in the degrader compared to DMSO control IP treatment (Fig. 2c, Supplementary Fig. 2, 3 and Supplementary Data 2, 3). Using significance cutoffs of fold change (FC) > 1.5, P-value < 0.001, and combining the data from both cell lines, we identified a total of 298 enriched proteins (Supplementary Data 4). We rationalized that the likelihood of observing the same proteins enriched as false positives across multiple treatments with similar IMiD analog molecules is low, and therefore used ‘frequency of enrichment’ as a measure of confidence. We observed 102 proteins enriched in at least three independent IPs, and each of the top 5 proteins (PATZ1, ZBED3, WIZ, IKZF2, and ASS1) enriched in more than 20 independent IPs across the database (Fig. 2d and Supplementary Data 4). Surprisingly, although published reports have confirmed degradation of PATZ1, WIZ, and IKZF2, none of these top 5 enriched proteins regularly feature amongst those proteins that we commonly see reported in existing unbiased screens of IMiD-based molecules, indicating the orthogonal data generated by this profiling method can identify targets that might otherwise be overlooked. ZBED3 and ASS1 showed frequent enrichment across our database without any prior reporting of degradation, even at concentrations up to 5 µM of pomalidomide (Supplementary Fig. 1h and Supplementary Data 4), suggesting examples of targets that are chemically glued to CRBN but lack productive degradation54, thus emphasizing the benefit of alternative binding-focused approaches for target identification. Also, important to note is that the additional targets identified in this study are not only targets of new IMiD analogs but are also identified as targets of IMiDs in clinical trials and with FDA approval.
To assess the fraction of newly identified IMiD targets, we compared the 298 enriched proteins to a list of literature reported targets and discovered an overlap of only 28 targets. We identified 270 novel targets and found only 22 targets that were reported in the literature but not identified as hits in our study (Fig. 2e and Supplementary Data 4)14. Considering the prevalence of C2H2 ZF transcription factors amongst reported IMiD targets, we asked whether this dominance holds true across our extended list of targets. To assess this, we extracted superfamily, family, and domain information from curated databases including InterPro55,56, Uniprot57, and Superfamily58 to categorize the targets based on studied features (Fig. 2f, g, Supplementary Data 4). Indeed, of the 298 targets identified, after C-terminal domain classification, the C2H2 ZF superfamily represents the largest segment, accounting for > 14% of the targets in the top 10 enriched superfamilies. This is followed by RNA-recognition motif domain proteins (RRM, > 13%) and nucleotide-binding alpha-beta plait domain superfamilies (α-β plait domain, > 12%). Notably, protein kinase-like domain proteins also features on this top 10 list of superfamilies (kinase-like domain, > 6.4%) which aligns with our knowledge that kinases can be targets of IMiD molecular glues (eg, CSNK1A1 or WEE1)12,17,59 and suggests that molecular glues may be a viable alternative to PROTACs, which are widely explored for kinase targeting. Exploration of the top 10 domain classifications across the dataset shows a similar trend with C2H2 ZF, RRM, ZF, protein kinases, and BTB/POZ domains showing the highest representation across the targets identified (Fig. 2g).
This dataset builds upon previous identifications of protein kinases as targets of IMiD molecules17,22,60, and further extends the kinase list, adding CDK7, IRAK1, and TBK1 as putative molecular glue targets. It also broadens the scope of tractable targets by introducing multiple protein families as targetable by CRBN-based molecular glues, illustrating the extensive potential of these molecules. Through the application of our unbiased target enrichment workflow, we have significantly increased the number of experimentally detected IMiD targets, expanding beyond the C2H2 ZF protein family to a wide range of protein families, including protein kinases and proteins involved in RNA metabolism.
G-loop alignments as a computational filter for neo-substrate compatibility
To validate the 270 previously unreported targets, we sought to establish a computational screening pipeline to score the compatibility of targets for recruitment to CRBN. Structural studies on IMiD-mediated CRBN neo-substrates, both natural and designed, have established the common G-loop motif that is recognized by the CRBN-IMiD complex12,61. We used MASTER62 to mine the AF2 database63 for proteins containing G-loops with similar backbone architecture to the G-loop in known neo-substrate CSNK1A1 (PDB: 5FQD, aa35-42), resulting in a set of 46,040 loops across 10,926 proteins with an R.M.S.D cutoff of less than 2 Å (Fig. 3a).
a Schematic representation of the computational workflow established for AF2 G-loop binding compatibility with CRBN-IMiD. b Heatmap displaying the log2 fold change (log2 FC) of significant (P-value < 0.001) molecular glue dependent ZF targets in MOLT4 cells. White space in the heatmap corresponds to log2FC = 0 or no quantification. Previously reported targets are marked with a blue dot, newly reported targets are marked with a green dot, and targets with a structural G-loop are marked with a gray dot. Significant changes were assessed by a two-sided moderated t test as implemented in the limma package86. Values determined by n = 4 replicates of each treatment group. c As in (b), but with Kelly cells. d Venn diagram showing unique and overlapping ZF hits comparing MOLT4 and Kelly cell targets. e Stacked bar plot showing the proportion of targets complexed and degraded by each of the indicated IMiD molecules. “not seen” indicates enriched targets were not quantified in global proteomics studies. “seen, degraded” indicates enriched targets quantified and reported as degraded in global proteomics. “seen, not degraded” indicates enriched targets were quantified but not degraded in global proteomics19. f Pie chart displays the IMiD-grouped data from (e).
Due to structural constraints, not all these proteins are compatible with CRBN. To identify CRBN-compatible proteins, we first extracted domains containing the G-loops based on domain definitions from DPAM, a tool that parses domains from AF models based upon predicted aligned errors (PAE) and evolutionary classification64. Next, we aligned the domains to our reference CSNK1A1-IMiD-CRBN-DDB1ΔB structure based on the G-loop and calculated a clash score. We used the van der Waals force term for interchain contacts in Rosetta’s low-resolution mode65 to obtain a side-chain independent clash estimate. Out of 16 known neo-substrates with validated G-loops (Supplementary Data 5 and Supplementary Figs. 4 and 5), 15 had clash scores below 2, while ZNF65440 had a score of 172, indicating a minor clash. The clash was caused by a low confidence region in the AF2 structure and could be resolved by relaxing the complex with Rosetta (Supplementary Fig. 6a)66. On the other hand, a protein with no evidence supporting it being a neo-substrate, PAAF1, had a major clash with a score of 1551 which could not be resolved by relaxation (Supplementary Fig. 6b). Based on these examples, and analysis of the clash scores of all hit proteins containing a clear structural G-loop in AF2 (Supplementary Fig. 6c), we filtered out domains with scores greater than 200 resulting in a list of 16,901 loops across 7111 proteins with nonexistent, or marginal clashes with CRBN (Fig. 3a and Supplementary Data 5).
Zinc finger transcription factors enriched among targets
Of the 298 total enriched candidates,199 were found to have a clear structural G-loop, with 162 having a clash score below 200. Given the high proportion of ZF proteins identified as targets across this enrichment database (Fig. 2f), we mapped the fold change in enrichment for proteins with an annotated ZF domain across all 20 degraders for both MOLT4 (Fig. 3b) and Kelly cells (Fig. 3c). Across these two cell lines, we identified 20 previously reported and 27 new neo-substrates as chemically recruited to CRBN. We then used our G-loop database to inform on which of these targets have a tractable G-loop and found that only five of the 47 identified targets do not contain a structural G-loop (Fig. 3c and Supplementary Data 5). Given what we know about the recruitment and binding of CRBN neo-substrates, targets usually bind through a dominant structural hairpin. Since we do not have validated degron information for all these ZF targets, we assumed that the G-loop with the lowest clash score has the highest likelihood to bind and therefore proceeded with the evaluation of a single G-loop for each target. To gauge how the clash scores for these ZF targets compare to all hits in the G-loop database, we compared the clash scores for our ZF targets to those of all hits (Supplementary Fig. 6c), demonstrating a pronounced trend towards lower scores for ZF targets, suggesting fewer unfavorable interactions (Supplementary Fig. 4, 5). Notably, when we explored the ZF hits with higher clash scores (> 10) and > 3 hit frequency, we realized that almost all of these have a reported association with at least one of the validated hits – ZMYND8 (cs 455, binds to ZNF687), and RNF166 (cs 17, binds to ZNF653/ZBTB39/ZNF827) – which also offers the possibility that these proteins could be collateral targets, recruited via piggybacking on their binding partners, the direct binders (Supplementary Data 4). Finally, we compared ZF targets across the two cell lines as an additional means for validation, and found 10 overlapping proteins, 5 of which are novel recruited targets (ZBED3, MNAT1, MTA2, ZBTB44, TRIM28) (Fig. 3d).
There are many factors to take into consideration when looking to predict target degradability, such as ternary complex formation26,31,67 and target ubiquitylation12,32,68,69,70, and multiple studies have placed an emphasis on exploring their role in driving productive degradation71,72. For degrader-induced degradation to occur, a ternary complex consisting of ligase-degrader-target needs to form for proximity-mediated ubiquitin transfer to the target protein. Because ternary complex formation is necessary for successful protein degradation, we set out to explore the relationship between ternary complex formation and degradation for ZF targets identified in this study. We focused our evaluation on the parental IMiD molecules, which have been subjected to degradation target profiling using unbiased global proteomics analysis across a panel of four cell lines (SK-N-DZ, Kelly, MM.1S, hES)19. Comparison of the enriched ZF targets to the published degradation data shows a consistent trend across the three IMiDs, where only ~ 30% of the enriched targets that were quantified in global proteomics studies were degraded, with ~ 60% of the targets quantified but not reported as degraded (Fig. 3e and Supplementary Data 5). The data was then grouped to allow a global comparison of the enriched versus degraded IMiD targets. The comparison revealed that of the 31 ZF targets enriched across these three molecular glues, only 11 of the 29 proteins quantified in global proteomics experiments were found to be degraded (Fig. 3f and Supplementary Data 5). 18 proteins were quantified in global proteomics but were not identified as degraded. This prompted us to question whether these targets were resistant to degradation by IMiDs and their analogs, or if they were not identified as degraded due to experimental limitations such as inadequate sensitivity to detect minor changes in protein abundance, rapid protein turnover, or suboptimal experimental conditions. We found that although several of the targets (WIZ, PATZ1, ZNF687, ZMYM2, and HIC2) were not reported as degraded in Donovan et al.19, they have since been reported as degraded in other published studies40,73,74, confirming that IPs provide a complementary approach able to overcome limitations in sensitivity. The absence of degradation data for the remaining targets could imply that these targets are resistant to degradation, or similar to the above proteins, the appropriate degradation experiment has yet to be performed. These data demonstrate that our IP workflow provides a significant advantage over global proteomics analysis by enabling selective isolation and enrichment of targets that may be below the change in abundance threshold for consistent identification with global proteomics approaches.
IMiD derived molecular glues recruit hundreds of non-zinc finger proteins
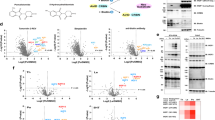
The largest target class of CRBN neo-substrates today are ZF- containing proteins, however, of the ~ 20,000 proteins in the human proteome, ZF containing proteins only make up a relatively small proportion with about ~ 1700 ZF proteins reported75. So far, only a handful of targets are reported to lack a ZF motif, which includes GSPT111, CSNK1A112,17, PDE6D76, and RAB2876. With this in mind, we examined our list of targeted proteins with a focus on those that do not contain a reported ZF domain and found 251 non-ZF proteins enriched across the IP dataset (Fig. 4a and Supplementary Data 4). These non-ZF proteins include a wide range of families such as protein kinases (IRAK1, TBK1, CDK7), RNA recognition motif proteins (ELAVL1, PPIL4, CSTF2, RBM45), metabolic enzymes (ASS1, PAICS, ACLY, CS, ACADVL), translational proteins (MARS1, ETF1, EEF1E1, EIF4B) and more spanning different biological pathways. To assist in establishing confidence in some of these targets, we performed a comparison of the non-ZF targets enriched in the two tested cell lines and found 39 targets were identified in both MOLT4 and Kelly cells, including the four above mentioned targets (Fig. 4a, b). We then assessed the AF2 structures of each of these 39 proteins and found that almost all of them (33/39) contain a structural G-loop (Fig. 4b, Supplementary Fig. 5 and Supplementary Data 5).
a Venn diagram showing unique and overlapping non-ZF hits comparing MOLT4 and Kelly cell targets. b Heatmap displaying the log2 fold change (log2 FC) for the 39 overlapping hits from (a). White space in the heatmap corresponds to log2FC = 0 or no quantification. Previously reported targets are marked with a blue dot, newly reported targets are marked with a green dot, and targets with a structural G-loop are marked with a gray dot. Significant changes were assessed by a two-sided moderated t test as implemented in the limma package86. Values determined by n = 4 replicates of each treatment group. c TR-FRET with titration of FPFT-2216 or pomalidomide to N-terminally biotinylated FL PDE6D, DTWD1, PPIL4 or RAB28 at 20 nM, incubated with terbium-streptavidin at 2 nM to monitor binding to GFP-CRBN-DDB1∆B at 200 nM. Values were determined by technical replicates of n = 2. d Immunoblots of ubiquitylation assay establishing PDE6D, DTWD1, and PPIL4 as FPFT-2216 and pomalidomide-induced neo-substrates of CRBN. e Structural G-loop alignment of AF2 PPIL4, PDE6D, and DTWD1 with CRBN-DDB1ΔB (PDB ID 5FQD, 6UML). The corresponding clash score is displayed. f Cryo-EM 3D reconstruction of PPIL4-RRM bound in ternary complex with FPFT-2216-CRBN-DDB1 FL, and PDE6D bound with FPFT-2216-CRBN-DDB1∆B. Maps are postprocessed with DeepEMhancer93. Inset of each shows the potential binding mode of action of FPFT-2216 engaging PPIL4 or PDE6D via neo-substrate G-loop and its interacting residues.
Given the large number of non-ZF targets identified in this study and the lack of emphasis in the public domain with regard to non-ZF CRBN neo-substrates, we selected a series of non-ZF proteins for further experimental validation. Three of these four selected proteins were identified as enriched in both cell lines (Fig. 4b), whereas DTWD1 was only identified in Kelly cells. Firstly, to demonstrate that these neo-substrates are directly recruited to CRBN, we examined ternary complex formation using recombinant purified proteins. Using two of the more promiscuous molecular glues, pomalidomide and FPFT-2216, we tested previous reported degradation targets PDE6D, RAB28, and DTWD1, along with a newly discovered target PPIL4. Indeed, PDE6D, DTWD1, and PPIL4 formed compound dependent ternary complex with CRBN at varying effective concentrations (Fig. 4c). However, RAB28, which was previously reported to be degraded by IMiDs19 and FPFT-221676, did not show any evidence for direct binding to CRBN using purified proteins. Since RAB28 has previously been reported as a CRBN neo-substrate and consistently scored across our enrichment study, we explored whether there was any evidence suggesting that RAB28 could be a collateral target. Exploration of protein-protein interaction databases, including BioPlex33 and STRING-DB77 revealed that RAB28 is known to bind to two validated IMiD-CRBN neo-substrates, PDE6D and ZNF653 (Supplementary Data 4), suggesting that RAB28 is likely an indirectly recruited target. These data demonstrate that in addition to identifying direct binders, we can also identify indirect binding partners that may be simultaneously recruited together with direct binding neo-substrates.
As targeted protein degradation requires not only recruitment to CRBN but also CRBN-mediated ubiquitin transfer for degradation, we monitored whether the recruited proteins can be ubiquitylated by CRL4CRBN. In vitro ubiquitylation assays showed robust ubiquitin modification on all 3 recruited non-ZF proteins in the presence of pomalidomide or FPFT-2216 (Fig. 4d). In addition, all three of these targets were degraded in response to IMiD treatment as observed by global proteomics analysis (Supplementary Fig. 6d and Supplementary Data 5). Using structural G-loop alignments, we then assessed the potential for each of these three proteins to bind to IMiD-CRBN and found that all three proteins had a G-loop with a clash score of < 200 (Fig. 4e). However, the aligned clash score for DTWD1 was relatively high and approaching the upper 200 thresholds (cs 198). We performed relaxation with Rosetta and found that this reduced the clash score to 1.58 by allowing minor shifts in the overall conformation while retaining the structural G-loop (Supplementary Fig. 6e). This process demonstrates that in some cases, clash scores can be relieved through minor structural rearrangements using Rosetta relax.
To expand our understanding of the recruitment of non-ZF targets, we determined cryo-EM structures of CRBN-DDB1ΔB-FPFT-2216 bound to PPIL4 and PDE6D, respectively (Fig. 4f, Supplementary Fig. 7 and Table 1). The complex structures were both refined to a global resolution of around 3.4 Å, and the quality of the resulting maps were sufficient to dock the complex components, but the flexibly tethered PPIL4 resulted in a lower local resolution. We were able to observe PPIL4 engagement with FPFT-2216-CRBN via its Gly278 harboring G-loop, as expected from the G-loop alignment, as well as for PDE6D via its Gly28 G-loop. Furthermore, overall density allowed fitting of FPFT-2216 in bulk, showing that the glutarimide ring engages CRBN’s binding pocket in a similar manner to other IMiD molecular glues. The triazole interacts with the backbone of the G-loop, and the methoxythiophene moiety potentially contacts both the PPIL4 backbone of the G-loop and Arg273. This suggests that the triazole and the methoxythiophene moieties could provide specificity elements to FPFT-2216-mediated neo-substrate recruitment. The methoxythiophene moiety also engaged Arg23 of PDE6D, indicating that FPFT-2216 potentially derives specificity in engaging an arginine residue from its neo-substrates. Analysis of the non-ZF targets of FPFT-2216 revealed several other proteins harboring an arginine or a lysine residue at this sequence location (PDE6D, SCYL1, RBM45, PPIL4). Finally, we compared the experimental structure to the AF2 predicted G-loop aligned structure of PPIL4 (Supplementary Fig. 6f). The G-loop aligned structure of PPIL4 presented a clash score of 3.38, which showed the C-terminal region of CRBN around residue Arg373 to have a minor clash with PPIL4’s loop harboring residue Val250. Although the low resolution permitted only backbone-level fitting of PPIL4, we observed that the cryo-EM structure revealed a small shift in the RRM domain of PPIL4 to accommodate this minor clash suggested in the G-loop aligned structure while retaining overall conformational similarity of the G-loop (Supplementary Fig. 6f). Meanwhile, PDE6D retained overall similar conformation with minor shifts that did not alter the interaction with CRBN (Supplementary Fig. 6g, h).
Together, these data demonstrate that RRM domain-containing proteins represent an additional class of proteins targetable through CRBN-dependent molecular glues. Using structural modeling, we increase confidence in these targets while also providing a reminder that structural analysis and AF2 predicted structures are static models, and although they provide excellent structural guidance, we need to keep in mind that proteins in solution are flexible and dynamic.
Discovery of a selective molecular glue for PPIL4
While the proteomics-based screening workflow identifies novel putative CRBN targets and provides initial chemical matter, it does not necessarily provide the best starting point for developing a chemical probe or therapeutic due to the limited number of molecules screened. We hypothesized that this limitation could be overcome by following up proteomics screening with a target-centric screen of a larger CRBN binder library to identify the optimal chemical starting point. To test this, we set out to identify PPIL4 targeting molecular glues with improved selectivity and lacking the triazole moiety. We employed an IMiD molecular glue library consisting of ~ 6000 compounds of various IMiD analogs that were either synthesized in-house or purchased externally. We screened this library against PPIL4 using TR-FRET to measure compound-induced PPIL4 recruitment to CRBN (Fig. 5a). TR-FRET ratios were obtained by incubating the library with GFP-fused CRBN-DDB1∆B and Tb-labeled streptavidin coupled to biotinylated PPIL4. The library was compared relative to the positive control, whereby the 520/490 ratio of FPFT-2216 at 10 µM was normalized as 1, and compounds were tested at 1.66 µM or 3.33 µM to find hits with equal or improved efficacy in directly recruiting PPIL4 to CRBN-DDB1∆B. We were able to narrow down the library to two molecules that performed similar or better than FPFT-2216 (Fig. 5b). These lead compounds were subject to a full titration to assess recruitment efficacy by TR-FRET. Ultimately, after recognizing one of the two hits was due to autofluorescence, we were able to identify a molecule, Z6466608628, that produced a higher 520/490 ratio, and a better EC50 of 0.34 µM compared to FPFT-2216, measured at 1.05 µM in this experiment (Fig. 5c, d).
a Schematic of the high-throughput TR-FRET screening workflow used to screen > 6000 IMiD analogs. Created in BioRender. Donovan, K. (2025) https://BioRender.com/uwuaveg. b TR-FRET: Normalized 520/490 ratio for each of the > 6000 compounds derived from IMiD molecules combined from the Gray/Fischer laboratories and those purchased from Enamine with GFP-CRBN-DDB1∆B at 50 nM, biotinylated PPIL4 at 20 nM, and terbium-streptavidin at 2 nM. c TR-FRET: titration of FPFT-2216 and lead compounds to GFP-CRBN-DDB1∆B at (200 nM), biotinylated PPIL4 at 20 nM, and terbium-streptavidin at 2 nM. Values were determined by technical replicates of n = 2. d Chemical structures of FPFT-2216 alongside new lead compound from (b) and (c). e Scatterplot depicting relative protein abundance following Flag-CRBN-DDB1ΔB enrichment from Kelly cell in-lysate treatment with FPFT-2216 (left) and Z6466608628 (right) and recombinant Flag-CRBN-DDB1ΔB spike in. Scatterplot displays fold change in abundance to DMSO. Significant changes were assessed by a two-sided moderated t test as implemented in the limma package86 with log2 FC shown on the y-axis and log10 P-value on the x-axis. f. Scatterplot depicting relative protein abundance following Z6466608628 treatment in MOLT4 cells. Significant changes were assessed by a two-sided moderated t test as implemented in the limma package86 with log2 FC shown on the y-axis and log10 P-value on the x-axis. g Docking model of CRBN-Z6466608628-PPIL4 showing the terminal nitrogen’s interaction with CRBN Glu377. h TR-FRET comparing Z6466608628 and its structural analogs’ ability to recruit PPIL4 to CRBN. GFP-CRBN-DDB1∆B at (200 nM), biotinylated PPIL4 at 50 nM, and terbium-streptavidin at 2 nM were used. Values were determined by technical replicates of n = 2. i Compound structures tested, aligned with their strength for inducing ternary complex formation.
To test the efficacy and selectivity of our lead compound, we first performed IP-MS in comparison with FPFT-2216 in Kelly cell lysate. While FPFT-2216 recruited numerous proteins, Z6466608628 selectively recruited PPIL4, along with its known binding partner DHX40 (Fig. 5e and Supplementary Data 5). Global quantitative proteomics in MOLT4 cells then confirmed that Z6466608628 can induce selective, but modest downregulation of PPIL4 (Fig. 5f and Supplementary Data 5). We then performed western blot analysis to confirm dose-dependent depletion of PPIL4 (Supplementary Fig. 8a) and in vitro ubiquitylation over time (Supplementary Fig. 8b). Despite effective ubiquitylation, the data show incomplete degradation of cellular PPIL4 even at the high 10 µM treatment concentration. PPIL4 is known as a regulator of the spliceosome, and the structure of PPIL4 bound to the BACR spliceosome is reported78. Aligning PPIL4 bound to the BACR spliceosome to our CRBN-FPFT-2216-PPIL4 ternary complex shows that the conformation of PPIL4 bound to the spliceosome would occlude PPIL4 recruitment to CRBN through molecular glues (Supplementary Fig. 8c), which would also hinder its ubiquitylation. This provides one of many potential explanations as to why PPIL4 is not completely degraded even with higher doses of compound.
The identification of a selective PPIL4 recruiter/degrader from a single hit in a screen of over 6000 compounds is remarkable. This led us to explore whether analyzing similar scaffolds could provide insights into the structure-activity relationship. Z6466608628 contains an extended phenylpiperazine group with a backbone of lenalidomide. While it is difficult to compare Z6466608628 with FPFT-2216 due to its largely different pharmacophore, molecular docking of Z6466608628 potentially points to why this molecule, which has an extended phenylpiperazine group with a lenalidomide backbone, outperforms lenalidomide (Fig. 5g). The phenylpiperazine group may interact with CRBN’s Glu377, potentially stabilizing its interaction with CRBN by promoting a rigid conformation of the phthalimide, which could then engage with the G-loop backbone or residue Trp275 of PPIL4. We created a pool of related compounds that alter the end terminal basic nitrogen that interacts with CRBN’s Glu377 and assessed their ability to recruit PPIL4 to CRBN. The data reveals that modification at this site on the molecule leads to a significant loss of binding by TR-FRET (Fig. 5h, i). These data collectively demonstrate the complete workflow, starting from the identification of a novel non-ZF target PPIL4 in an affinity proteomics screen, to the discovery of a PPIL4-selective molecular glue that would serve as an excellent lead molecule for structural optimization.
Discussion
Targeted protein degradation and induced proximity are part of a rapidly expanding field focused on the development of small molecules that leverage induced neo-protein-protein interactions to drive pharmacology. In this study, we develop and showcase a workflow for high-sensitivity, unbiased target identification of degraders and non-degrading molecular glues, identifying more than 290 targets recruited to CRBN by IMiD-like molecules. We demonstrate that this approach to target identification can reveal critical insights and targets that are missed by traditional screening methodologies and provide a blueprint for a discovery workflow from screening to optimization and structure-guided design of molecular glue degraders.
Thalidomide and its derivatives, lenalidomide and pomalidomide (IMiDs), have had a checkered past. These molecules have been in use for a variety of indications, on and off, since the 1950’s and have experienced perhaps the greatest turnaround in drug history. From devastating birth defects to effective hematological cancer therapy, and more recently, significant investment in utilizing these molecules for TPD-based therapeutics. While a decade of research has slowly uncovered around 50 reported neo-substrates of IMiD’s, thousands of proteins harbor structural G-loops that have the potential for recruitment to CRBN by IMiD molecules. Our simple, cost-effective, and highly scalable unbiased screening workflow combines whole cell lysate with recombinant Flag-CRBN and degraders to enrich target binders from the complex proteome. Through an IMiD-analog screen across two cell lines, we mapped a significant expansion of the neo-substrate repertoire by identifying 298 proteins recruited to CRBN, with 270 of these being novel targets. Unlike many current high-throughput screening workflows that focus on the endpoint – degradation, this workflow allows us to explore the fundamental first step of proximity induced degradation – recruitment, where we are now able to identify targets that are directly or indirectly recruited to CRBN. This sensitive workflow sheds light onto a previously unchartered element of the molecular glue mechanism of action and establishes insights into how and why certain molecular glues may exhibit higher efficacy than others. We discovered targets recruited to CRBN that are resistant to degradation, providing numerous examples of targets being glued to CRBN without productive degradation54. Exploration of two of these targets, ASS1 and ZBED3, does not offer any clues as to why they are not degraded, since both have reported ubiquitylated sites79. Numerous possibilities exist, from these targets being tightly preoccupied by other binding partners, geometric constraints leading to inaccessibility of lysines, removal of ubiquitylation by deubiquitinases, or preclusion of the catalytic sites due to size and shape preventing active ubiquitylation. It is important to note that the non-degrading functions of these molecular glues may have interesting degradation-independent pharmacology that have not yet been investigated, thus providing an opportunity for future experimental research.
The comprehensive G-loop database provided us with prefiltered insights as to whether these targets have the potential to be recruited to CRBN through the currently established mechanism of G-loop binding. However, although most targets identified in this study do have a structural G-loop, we do have numerous instances of proteins that do not harbor a G-loop. Some of these targets do have a structurally similar hairpin motif but are lacking the ‘essential’ glycine in position six. Whereas other targets did not have this structural motif at all. These findings indicate the potential for alternative recruitment mechanisms, such as proteins piggybacking on a direct G-loop carrying target. This concept of collateral (or bystander) targeting was also demonstrated in a study exploring HDAC degradability, where it was found that both HDACs and their known complex binding partners can be degraded31. Alternatively, and perhaps more intriguingly, the potential for recruitment of proteins through a distinct structural motif suggesting there may be uncharacterized binding mechanisms that are pending discovery. The potential capacity for IMiDs to yield interfaces favorable for the recruitment of various structural motifs would considerably expand the diversity of CRBN neo-substrates and broaden therapeutic applications80.
Amongst the targets identified in this study, we not only discovered many C2H2 ZF transcription factor targets but also extended targets beyond C2H2 ZF proteins, into additional classes of proteins such as those containing RNA recognition motif (RRM) domains and kinase domains, confirming that CRBN is an incredibly versatile ligase and very well suited to hijacking for TPD applications. We reveal 251 non-ZF targets, a dramatic increase in the breadth and number of proteins targeted by CRBN from the currently reported targets of less than a dozen. Binding data using TR-FRET on a selection of these targets validates their direct interaction mechanism, and structural characterization further corroborates this binding while validating the generated G-loop alignment database as a tool to assist prioritization of targets using clash score assessment. Using the accumulative data, we selected a non-ZF neo-substrate, PPIL4, for additional screening to illustrate the utility of this workflow for prioritization efforts. After a biochemical ternary complex recruitment screen of around 6000 IMiD analogs, we selected a single hit compound and used affinity proteomics to confirm selective recruitment of PPIL4 to CRBN. A small SAR library around this lenalidomide-based scaffold suggests the importance of the phenylpiperazine group for stabilizing the interaction with CRBN and enabling selective recruitment of PPIL4. Genomic studies have reported that PPIL4 is essential for brain-specific angiogenesis and has implications in intracranial aneurysms81, and is known to regulate the catalytic activation of the spliceosome78. Thus, this molecular glue could be of great interest to target the splicing pathway, in relation to intracranial aneurysms, or in other contexts.
We believe our optimized workflow and comprehensive data package, along with outlining specific applications of these, provides a valuable resource for the chemical biology, drug discovery, and induced proximity communities. Importantly, the workflow is neither limited to CRBN nor to TPD but rather can be applied to any induced proximity application. We expect the enrichment workflow will provide a blueprint for expansion into target identification for induced proximity platforms, as well as further expansion of targets for protein degraders beyond molecular glues. Through initial scouting efforts on heterobifunctional degraders and additional ligases, we are confident there are many exciting discoveries to be made with already existing chemistry, and we envision this as an evolving resource where we will continue to release data as it becomes available.
Methods
Materials & correspondence
Small molecules described in this study will be made available on request, upon completion of a Materials Transfer Agreement. Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Eric S. Fischer (Eric_Fischer@DFCI.HARVARD.EDU).
Mammalian cell culture
MOLT4 (ATCC: CRL-1582) and Kelly (Sigma: 92110411) cells were cultured in RPMI-1640 media supplemented with 10% fetal bovine serum and 2 mM L-glutamine and grown in a 37 °C incubator with 5% CO2.
Insect cell culture
High Five insect cells were cultured at 27 °C in SF-4 Baculo Express ICM medium (BioConcept) in suspension. Sf9 insect cells were cultured at 27 °C in ESF 921 medium (Expression Systems).
Immunoprecipitation
A total of 1 × 107 cells per IP were collected and lysed in lysis buffer (50 mM Tris pH 8, 200 mM NaCl, 2 mM TCEP, 0.1% NP-40, 10 units turbonuclease/200 µL buffer, 1x cOmplete protease inhibitor tablet, Sigma-Aldrich/5 mL buffer) and sonicated on ice for 5 rounds of 2 s followed by 10 s pauses at 25% amplitude. After centrifugation clarification, the lysate was transferred to new lobind tubes. 20 µg of Flag-CRBN-DDB1ΔB, 1 µM of MLN4924 and CSN5i-3 (neddylation trap)82, and 1 µM of selected degraders (purchased from MedChemExpress or obtained from the Bradner or Gray labs) or DMSO vehicle control were added to each lysate and incubated with end-over-end rotation for 1 h in the cold room. 20 µL of pre-washed and resuspended M2-Flag magnetic bead slurry was added to each IP and incubated with end-over-end rotation for 1 h in the cold room. Beads were washed with wash buffer (50 mM Tris pH 8, 2 mM TCEP, 0.1% NP-40, 1x cOmplete protease inhibitor tablet/5 mL buffer) containing the respective compounds, followed by a specific elution method for the next application (immunoblot or mass spectrometry).
Samples prepared on the OT2 followed the same procedures as above, but using scaled-down equivalents of reagents. After the three detergent washes described above, the samples were washed an additional three times with non-detergent buffer (50 mM Tris pH 8, 2 mM TCEP, 1x cOmplete protease inhibitor tablet/5 mL buffer) containing the required compounds prior to appropriate elution described below.
Immunoblot
IP samples were eluted by resuspension in SDS sample buffer and heated at 95 °C for 5 min. Samples were run on 4-20% Mini-PROTEAN TGX Precast Protein gels (Bio-Rad). Protein was transferred to PVDF membranes using the iBlot 2.0 dry blotting system (Thermo Fisher Scientific). Membranes were blocked with Intercept blocking solution (LiCor), washed three times with PBST and incubated with primary antibodies (Anti-ASS1 1:1000, Cell Signaling Technology, Cat# 70720, Anti-PPIL4 1:500, Sigma, Cat# HPA031600, Anti-Strep II 1:4000, AbCam, ab76950) diluted using Intercept® T20 (PBS) Antibody Diluent, overnight at 4 °C, followed by three washes with PBST and incubation with secondary antibodies (Anti-Rabbit IgG 1:4000, Licor, Cat# 92632211) for 1 h in the dark. The membrane was washed three final times in PBST and imaged on the Odyssey LCx imaging system (LiCor).
Cloning and protein expression
All proteins are derived from human origin sequences. ZBED3 (residues 39–108), ZNF219 (residues 53–110) were cloned into pGEX4T1-TEV vectors with C-terminal Avi-Strep fusion. These were expressed in LOBSTR BL21(DE3) E. coli, purified by GST-affinity, followed by liberation of the GST-tag by overnight TEV protease incubation, ion exchange chromatography, and size exclusion chromatography into a final buffer of 25 mM HEPES, 200 mM NaCl, and 1 mM TCEP, pH 7.5. UBE2D3, UBE2G1, and UBE2M were cloned into pGEX4T1-TEV vectors, and NEDD8 was cloned into pGEX4T1-3C vector. Purification of these proteins were performed as described above. APPBP1-UBA3 was cloned into a pET-based vector and expressed in LOBSTR BL21(DE3) E. coli, purified by nickel affinity, ion exchange chromatography, and size exclusion chromatography into a final buffer of 25 mM HEPES, 200 mM NaCl, and 1 mM TCEP, pH 7.5. Ubiquitin was cloned into a pET3a vector and was expressed in Rosetta BL21(DE3) E. coli. Ubiquitin was purified by SP Sepharose resin at pH 4 and was subject to size exclusion chromatography into a final buffer of 25 mM HEPES, 150 mM NaCl, 1 mM TCEP, pH 7.5. PDE6D, DTWD1, PPIL4, PPIL4 RRM (residues 240–318), and RAB28 were cloned into pAC8-Strep-Avi vectors. Baculovirus was generated in Spodoptera frugiperda cells, and proteins were expressed in Trichoplusia ni cells. These were purified by Strep affinity, followed by ion exchange chromatography and size exclusion chromatography into 25 mM HEPES, 200 mM NaCl, and 1 mM TCEP, pH 7.5. GST-TEV-CRBN, GST-TEV-eGFP-CRBN, DDB1, GST-TEV-UBA1, CUL4A (residues 38-C), RBX1 (residues 5-C) were cloned into pLIB vectors. His-TEV-DDB1∆B (∆residues 396–705 with a GNGNSG linker) and Flag-Spy-CRBN were cloned into a pAC8 vector. CRBN and DDB1 were coexpressed in the following pairs, CRBN-DDB1, CRBN-DDB1∆B, Flag-CRBN-DDB1∆B, and eGFP-CRBN-DDB1∆B, purified by GST-affinity, followed by TEV cleavage overnight, ion exchange chromatography, and size exclusion chromatography into 25 mM HEPES, 150 mM NaCl, 1 mM TCEP, pH 7.5. CUL4A and RBX1 were coexpressed, CUL4-RBX1 and UBA1 were purified as described above.
Time-resolved fluorescence resonance energy transfer (TR-FRET)
Ternary complex formation of CRBN-DDB1, neo-substrate, and compound was measured by TR-FRET. 20 nM or 50 nM biotinylated neo-substrate, 2 nM Tb (CoraFluor-1)-labeled Streptavidin (R&D Systems, Cat#7920/20U), and 200 nM eGFP-CRBN-DDB1∆B were added in assay buffer (25 mM HEPES, 100 mM NaCl, 0.5% Tween-20 (Sigma-Aldrich, Cat#9005-64-5), and 0.5% BSA (Cell Signaling, Cat#9998S)). This reaction mix was added to a 384 well microplate, and the compound was titrated to indicated concentrations using a D300e Digital Dispenser (HP). Reaction was incubated at room temperature for 1 h and was measured using a PHERAstar FS microplate reader (BMG Labtech). The 520 nm/490 nm signal ratio was measured to calculate ternary complex formation, and datapoints were plotted to calculate EC50 values by using agonist versus response variable slope (four parameter model) in Graphpad Prism (v10.1.1).
In-vitro ubiquitylation assay
Ubiquitylation of neo-substrates was performed by premixing 500 nM neddylated CRL4CRBN, 2 µM UBE2D3, 2 µM UBE2G1, 200 nM UBA1, 500 nM Strep tagged-neo-substrate, and 10 µM compound in assay buffer (25 mM HEPES, 100 mM NaCl, 10 mM MgCl2, 5 mM ATP, pH 7.5). This mixture was incubated for 15 min on ice, and the reaction was started by adding 60 µM ubiquitin at room temperature. The sample was quenched by the addition of SDS sample buffer, and reaction products were separated on 4-20% SDS-PAGE gels. The assay was analyzed by immunoblotting as described above. Neddylation of CUL4-RBX1 was performed through incubation of 12 µM CUL4-RBX1, 1 µM UBE2M, 0.2 µM APPBP1-UBA3, 25 µM NEDD8 at room temperature for 10 min in assay buffer (25 mM HEPES, 100 mM NaCl, 10 mM MgCl2, 5 mM ATP, pH 7.5)83. Reaction was quenched by the addition of 10 mM DTT and was purified by size exclusion chromatography into buffer containing 25 mM HEPES, 150 mM NaCl, 1 mM TCEP, pH 7.5.
Cryo-EM sample preparation and Data processing
10 µM CRBN-DDB1∆B, 20 µM PPIL4 RRM or PDE6D, and 100 µM FPFT-2216 were incubated on ice for 30 min in buffer containing 25 mM HEPES, 150 mM NaCl, 1 mM TCEP, pH 7.5. The protein complex was diluted to 1.5 µM final concentration before application to grids. Quantifoil UltraAuFoil grids (R0.6/1) were glow discharged at 20 mA for 2 min. Prior to complex application, 4 µL of 10 µM CRBN-agnostic IKZF1 (residues 140–196, harboring mutants Q146A, G151N) was applied to saturate the air-water interface84,85. After 1 min, the grid was blotted from the back, and complex was applied, and immediately plunged into liquid ethane.
The dataset was collected on a Talos Arctica at 200 kV equipped with a Gatan K3 direct detector in counting mode. Movies were collected with a total dose of 54 e−/Å2 over 40 frames, at 1.1 Å/pixel with a nominal magnification of 36,000x, with a defocus range of − 0.8 µm to − 2.0 µm.
Sample preparation for immunoprecipitation mass spectrometry (IP-MS)
Immunoprecipitation (IP) was performed as described above with three or four replicates for each treatment. Controls included a DMSO vehicle control and pomalidomide as a positive control since it has an established target profile. After the final wash step, samples were eluted using 0.1 M Glycine-HCl, pH 2.7. Tris (1 M, pH 8.5) was added to the elution to reach a pH of 8. Samples were then reduced with 10 mM TCEP for 30 min at room temperature, followed by alkylation with 15 mM iodoacetamide for 45 min at room temperature in the dark. Alkylation was quenched by the addition of 10 mM DTT. Proteins were isolated by methanol-chloroform precipitation (only for manual IPs. Automated IPs undergo three non-detergent washes instead). The protein pellets were dried and then resuspended in 50 μL 200 mM EPPS pH 8.0. The resuspended protein samples were digested with 2 μg LysC and 1 μg Trypsin overnight at 37 °C. Sample digests were acidified with formic acid to a pH of 2-3 prior to desalting using C18 solid phase extraction plates (SOLA, Thermo Fisher Scientific). Desalted peptides were dried in a vacuum-centrifuged and reconstituted in 0.1% formic acid for LC-MS analysis.
Whole cell treatment and sample preparation for global proteomics
Experiment wp-esf_118 (TMT quantification) included treatment of MOLT4 cells with DMSO vehicle control (n = 3) or 1 µM FPFT-2216 (n = 3) for 5 hrs. Experiment wp-esf_588 (label free quantification) included treatment of MOLT4 cells with DMSO vehicle control (n = 4) or 1 µM of Z6466608628 (n = 2) for 5 hrs. All cells were harvested by centrifugation, washed twice with PBS, and snap frozen in liquid nitrogen.
Cells were lysed by the addition of lysis buffer (8 M urea, 50 mM NaCl, 50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (EPPS) pH 8.5, Protease and Phosphatase inhibitors) followed by manual homogenization by 20 passes through a 21-gauge (1.25 in. long) needle or homogenization by bead beating (BioSpec) for three repeats of 30 s at 2400 strokes/min. Lysate was clarified by centrifugation and protein quantified using Bradford (Bio-Rad, Cat#500-0205) assay. 50–100 µg of protein for each sample was reduced, alkylated, and precipitated using methanol/chloroform. In brief, four volumes of methanol were added to the cell lysate, followed by one volume of chloroform, and finally three volumes of water. The mixture was vortexed and centrifuged at 14,000 x g for 5 min to separate the chloroform phase from the aqueous phase. The precipitated protein was washed with three volumes of methanol, centrifuged at 14,000 x g for 5 min, and the resulting washed precipitated protein was allowed to air dry. Precipitated protein was resuspended in 4 M urea, 50 mM HEPES pH 7.4, buffer for solubilization, followed by dilution to 1 M urea with the addition of 200 mM EPPS, pH 8. Proteins were digested for 12 h at room temperature with LysC (1:50 ratio), followed by dilution to 0.5 M urea and a second digestion step was performed by addition of trypsin (1:50 ratio) for 6 h at 37 °C or urea diluted to 1 M urea and digested with the addition of LysC (1:50; enzyme:protein) and trypsin (1:50; enzyme:protein) for 12 h at 37 °C.
Label free quantitative mass spectrometry with DDA and data analysis
Sample digests were acidified with formic acid to a pH of 2-3 before desalting using C18 solid phase extraction plates (SOLA, Thermo Fisher Scientific). Desalted peptides were dried in a vacuum-centrifuged and reconstituted in 0.1% formic acid for liquid chromatography-mass spectrometry analysis.
Data were collected using an Orbitrap Exploris 480 mass spectrometer (Thermo Fisher Scientific) equipped with a FAIMS Pro Interface and coupled with a UltiMate 3000 RSLCnano System. Peptides were separated on an Aurora 25 cm × 75 μm inner diameter microcapillary column (IonOpticks), and using a 60 min gradient of 5–25% acetonitrile in 1.0% formic acid with a flow rate of 250 nL/min. Each analysis used a TopN data-dependent method. The data were acquired using a mass range of m/z 350–1200, resolution 60,000, 300% normalized AGC target, auto maximum injection time, dynamic exclusion of 30 sec, and charge states of 2–6. TopN 40 data-dependent MS2 spectra were acquired with a scan range starting at m/z 110, resolution 15,000, isolation window of 1.4 m/z, normalized collision energy (NCE) set at 30%, standard AGC target, and the automatic maximum injection time.
Proteome Discoverer 2.5 (Thermo Fisher Scientific) was used for.RAW file processing and controlling peptide and protein level false discovery rates, assembling proteins from peptides, and protein quantification from peptides. MS/MS spectra were searched against a SwissProt human database (January 2021) with both the forward and reverse sequences, as well as known contaminants such as human keratins. Database search criteria were as follows: tryptic with two missed cleavages, a precursor mass tolerance of 10 ppm, fragment ion mass tolerance of 0.03 Da, static alkylation of cysteine (57.0215 Da) and variable oxidation of methionine (15.9949 Da), N-terminal acetylation (42.0106 Da), and phosphorylation of serine, threonine, and tyrosine (75.966 Da). Peptides were quantified using the MS1 Intensity, and peptide abundance values were summed to yield the protein abundance values. Resulting data was filtered to only include proteins that had a minimum of 3 counts in at least 3 replicates of each independent comparison of the treatment sample to the DMSO control. Protein abundances were globally normalized using in-house scripts in the R framework (R Development Core Team, 2014). Proteins with missing values were imputed by random selection from a Gaussian distribution either with a mean of the non-missing values for that treatment group or with a mean equal to the median of the background (in cases when all values for a treatment group are missing). Significant changes comparing the relative protein abundance of the treatment to DMSO control comparisons were assessed by a two-sided moderated t-test as implemented in the limma package within the R framework86.
Label free quantitative mass spectrometry with diaPASEF and data analysis
Sample digests were acidified with formic acid to a pH of 2-3 before desalting using C18 solid phase extraction plates (SOLA, Thermo Fisher Scientific). Desalted peptides were dried in a vacuum-centrifuged and reconstituted in 0.1% formic acid for liquid chromatography-mass spectrometry analysis.
Data were collected using a TimsTOF Pro2 (Bruker Daltonics, Bremen, Germany) coupled to a nanoElute LC pump (Bruker Daltonics, Bremen, Germany) via a CaptiveSpray nano-electrospray source. Peptides were separated on a reversed-phase C18 column (25 cm × 75 μM ID, 1.6 μM, IonOpticks, Australia) containing an integrated captive spray emitter. Peptides were separated using a 50 min gradient of 2–30% buffer B (99.9% acetonitrile, 0.1% formic acid) with a flow rate of 250 nL/min and column temperature maintained at 50 °C.
To perform diaPASEF, the precursor distribution in the DDA m/z-ion mobility plane was used to design an acquisition scheme for DIA data collection, which included two windows in each 50 ms diaPASEF scan. Data was acquired using sixteen of these 25 Da precursor double window scans (creating 32 windows) which covered the diagonal scan line for doubly and triply charged precursors, with singly charged precursors able to be excluded by their position in the m/z-ion mobility plane. These precursor isolation windows were defined between 400 - 1200 m/z and 1/k0 of 0.7–1.3 V.s/cm2.
The diaPASEF raw file processing and controlling peptide and protein level false discovery rates, assembling proteins from peptides, and protein quantification from peptides was performed using library-free analysis in DIA-NN 1.887. Library free mode performs an in-silico digestion of a given protein sequence database alongside deep learning-based predictions to extract the DIA precursor data into a collection of MS2 spectra. The search results are then used to generate a spectral library, which is then employed for the targeted analysis of the DIA data searched against a SwissProt human database (January 2021). Database search criteria largely followed the default settings for directDIA including: tryptic with two missed cleavages, carbamidomethylation of cysteine, and oxidation of methionine, and precursor Q-value (FDR) cut-off of 0.01. The precursor quantification strategy was set to Robust LC (high accuracy) with RT-dependent cross-run normalization. Resulting data was filtered to only include proteins that had a minimum of 3 precursor counts in at least 4 replicates of each independent comparison of the treatment sample to the DMSO control. Protein abundances were globally normalized using in-house scripts in the R framework (R Development Core Team, 2014). Proteins with missing values were imputed by random selection from a Gaussian distribution either with a mean of the non-missing values for that treatment group or with a mean equal to the median of the background (in cases when all values for a treatment group are missing). Significant changes comparing the relative protein abundance of the treatment to DMSO control comparisons were assessed by a two-sided moderated t-test as implemented in the limma package within the R framework86.
TMT quantitative global proteomics
Anhydrous ACN was added to each digested peptide sample from above to a final concentration of 30%, followed by the addition of Tandem mass tag (TMT) reagents at a labeling ratio of 1:4 peptide:TMT label. TMT labeling occurred over a 1.5 h incubation at room temperature, followed by quenching with the addition of hydroxylamine to a final concentration of 0.3%. Each of the samples were combined using adjusted volumes and dried down in a speed vacuum, followed by desalting with C18 SPE (Sep-Pak, Waters). The sample was offline fractionated into 96 fractions by high pH reverse-phase HPLC (Agilent LC1260) through an aeris peptide xb-c18 column (phenomenex) with mobile phase A containing 5% acetonitrile and 10 mM NH4HCO3 in LC-MS grade H2O, and mobile phase B containing 90% acetonitrile and 5 mM NH4HCO3 in LC-MS grade H2O (both pH 8.0). The resulting 96 fractions were recombined in a non-contiguous manner into 24 fractions and desalted using C18 solid phase extraction plates (SOLA, Thermo Fisher Scientific), followed by subsequent mass spectrometry analysis.
Data were collected using an Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific, San Jose, CA, USA) coupled with a Proxeon EASY-nLC 1200 LC system (Thermo Fisher Scientific, San Jose, CA, USA). Peptides were separated on a 50 cm x 75 μm inner diameter EasySpray ES903 microcapillary column (Thermo Fisher Scientific). Peptides were separated over a 190 min gradient of 6–27% acetonitrile in 1.0% formic acid with a flow rate of 300 nL/min. Quantification was performed using an MS3-based TMT method as described previously88. The data were acquired using a mass range of m/z 340 – 1350, resolution 120,000, AGC target 5 × 105, maximum injection time 100 ms, dynamic exclusion of 120 s for the peptide measurements in the Orbitrap. Data-dependent MS2 spectra were acquired in the ion trap with a normalized collision energy (NCE) set at 35%, AGC target set to 1.8 × 104 and a maximum injection time of 120 ms. MS3 scans were acquired in the Orbitrap with HCD collision energy set to 55%, AGC target set to 2 × 105, maximum injection time of 150 ms, resolution at 50,000 and with a maximum synchronous precursor selection (SPS) precursors set to 10.
TMT quantitative global proteomics LC-MS data analysis
Proteome Discoverer 2.2 (Thermo Fisher Scientific) was used for.RAW file processing and controlling peptide and protein level false discovery rates, assembling proteins from peptides, and protein quantification from peptides. The MS/MS spectra were searched against a SwissProt human database (January 2021) containing both the forward and reverse sequences. Searches were performed using a 20 ppm precursor mass tolerance, 0.6 Da fragment ion mass tolerance, tryptic peptides containing a maximum of two missed cleavages, static alkylation of cysteine (57.0215 Da), static TMT labeling of lysine residues and N-termini of peptides (229.1629 Da), and variable oxidation of methionine (15.9949 Da). TMT reporter ion intensities were measured using a 0.003 Da window around the theoretical m/z for each reporter ion in the MS3 scan. The peptide spectral matches with poor quality MS3 spectra were excluded from quantitation (summed signal-to-noise across channels < 110 and precursor isolation specificity < 0.5), and the resulting data was filtered to only include proteins with a minimum of 2 unique peptides quantified. Reporter ion intensities were normalized and scaled using in-house scripts in the R framework89. Statistical analysis was carried out using the limma package within the R framework86.
Searching for CRBN-compatible G-loops in the AlphaFold2 database
Using MASTER v1.662, the AlphaFold2 human database (v4)63 comprising 23,391 protein structures was queried for 8-residue loops with backbone root-mean-squared-deviation less than 2 Å to CSNK1A1 residues 35–42 (extracted from PDB 5FQD)12. Loops not having a glycine at the sixth residue were filtered out, resulting in a set of 46,040 G-loops from 10,926 proteins. Next, domains containing the G-loops were extracted using domain definitions from DPAM64 and aligned to the CSNK1A1 reference loop and CRBN from 5FQD. To estimate backbone clashes, each structure was coarse-grained to represent the side chain as a pseudoatom and scored using Rosetta v13.365. Coarse-graining the structure makes the score rotamer-independent. The interchain_vdw term of the score function was used as a clash score and represents a modified Lennard-Jones 6–12 potential that penalizes atoms overlapping at the interface90. Domains with a clash score of greater than 200 were filtered out, resulting in a list 16,901 loops from 7,111 proteins.
Relieving minor clashes with Rosetta refinement
For select domains with low clash scores, clashes were relieved by relaxing the neo-substrate with Rosetta FastRelax65,66 while holding the G-loop in place. No energy minimization or rotamer optimization was performed on CRBN. Rigid body translation between CRBN and the neo-substrate was disabled. Of the 10 independent trajectories run, the one with the lowest total score was selected, and then the clash score was calculated as above.
Compound docking
The missing loops of the cryo-EM structure of the ternary complex (CRBN-FPFT-2216-PPIL4) were built using PRIME from schrodinger suite (2024v3)91, and the completed structure was prepared using the default process of the proteinprep protocol. The hit compound Z6466608628 was docked into the complex using the InducedFit92 program with hydrogen bonding constraints on the glutarimide with CRBN. The resulting top 5 poses were visually inspected, and the putative binding mode was chosen based on experimental SAR and binding score.
Statistical analysis and hit frequency calculation
Statistical methods are described in the according figure legends and methods. Cryo-EM statistics are based on the gold-standard FSC = 0.143 to determine resolution. For quantitative proteomics experiments, statistical analysis was carried out by two-sided moderated t test using the limma package within the R framework86. Limma employs an empirical Bayes approach to shrink the protein-wise variance toward a common prior distribution. The prior distribution used is empirically estimated from the data.
The frequency of enrichment was determined by counting the number of times each protein was determined to be a “hit” (> 1.5 fold upregulated and P-Value < 0.001) across the defined dataset, which included the diaPASEF data collected for all 20 molecules across both Kelly and MOLT4 cell lines.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The raw proteomics datasets generated during this study are available at PRIDE accessions: PXD054862, PXD054860, PXD054732, PXD054761; PXD054764; PXD055258. Proteomics data generated during this study are available in our custom online database webtool: https://v2.dfci-fischerlab.com/. Cryo-EM data are available from the Electron Microscopy Data Bank and the RCSB Protein Data Bank under the accession code numbers: EMD-47268, EMD-47269, EMD-47270 [https://www.ebi.ac.uk/pdbe/entry/emdb/EMD-47260], PDB-9DWV [https://doi.org/10.2210/pdb9DWV/pdb], PDB-9DWW [https://doi.org/10.2210/pdb9DWW/pdb]. The source data underlying Figs. 4, 5, Supplementary Figs. 1,8 are provided in the Source Data file. Source data are provided in this paper.
Code availability
The code necessary to reproduce the statistical analysis for quantitative proteomics can be found at: https://doi.org/10.5281/zenodo.15741629.
References
Bondeson, D. P. & Crews, C. M. Targeted protein degradation by small molecules. Annu. Rev. Pharmacol. Toxicol. 57, 107–123 (2017).
Neklesa, T. K., Winkler, J. D. & Crews, C. M. Targeted protein degradation by PROTACs. Pharmacol. Ther. 174, 138–144 (2017).
Sakamoto, K. M. et al. Protacs: chimeric molecules that target proteins to the Skp1–Cullin–F box complex for ubiquitination and degradation. Proc. Natl. Acad. Sci. 98, 8554–8559 (2001).
Komander, D. & Rape, M. The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 (2012).
Kronke, J. et al. Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science 343, 301–305 (2014).
Winter, G. E. et al. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science 348, 1376–1381 (2015).
Bondeson, D. P. et al. Catalytic in vivo protein knockdown by small-molecule PROTACs. Nat. Chem. Biol. 11, 611 (2015).
Wang, B., Cao, S. & Zheng, N. Emerging strategies for prospective discovery of molecular glue degraders. Curr. Opin. Struct. Biol. 86, 102811 (2024).
Ito, T. et al. Identification of a primary target of thalidomide teratogenicity. Science 327, 1345–1350 (2010).
Fischer, E. S. et al. Structure of the DDB1-CRBN E3 ubiquitin ligase in complex with thalidomide. Nature 512, 49–53 (2014).
Matyskiela, M. E. et al. A novel cereblon modulator recruits GSPT1 to the CRL4(CRBN) ubiquitin ligase. Nature 535, 252–257 (2016).
Petzold, G., Fischer, E. S. & Thoma, N. H. Structural basis of lenalidomide-induced CK1alpha degradation by the CRL4(CRBN) ubiquitin ligase. Nature 532, 127–130 (2016).
Lu, G. et al. The myeloma drug lenalidomide promotes the cereblon-dependent destruction of Ikaros proteins. Science 343, 305–309 (2014).
Oleinikovas, V., Gainza, P., Ryckmans, T., Fasching, B. & Thomä, N. H. From thalidomide to rational molecular glue design for targeted protein degradation. Annu. Rev. Pharmacol. Toxicol. 64, 291–312 (2024).
Matyskiela, M. E. et al. SALL4 mediates teratogenicity as a thalidomide-dependent cereblon substrate. Nat. Chem. Biol. 14, 981–987 (2018).
Sievers, Q. L., Gasser, J. A., Cowley, G. S., Fischer, E. S. & Ebert, B. L. Genome-wide screen identifies cullin-RING ligase machinery required for lenalidomide-dependent CRL4(CRBN) activity. Blood 132, 1293–1303 (2018).
Kronke, J. et al. Lenalidomide induces ubiquitination and degradation of CK1alpha in del(5q) MDS. Nature 523, 183–188 (2015).
An, J. et al. pSILAC mass spectrometry reveals ZFP91 as IMiD-dependent substrate of the CRL4(CRBN) ubiquitin ligase. Nat. Commun. 8, 15398 (2017).
Donovan, K. A. et al. Thalidomide promotes degradation of SALL4, a transcription factor implicated in Duane Radial Ray syndrome. Elife 7, https://doi.org/10.7554/elife.38430 (2018).
Yamamoto, J. et al. ARID2 is a pomalidomide-dependent CRL4CRBN substrate in multiple myeloma cells. Nat. Chem. Biol. 16, 1208–1217 (2020).
Renneville, A. et al. Avadomide induces degradation of ZMYM2 fusion oncoproteins in hematologic malignancies. Blood Cancer Discov. 2, 250–265 (2021).
Li, L. et al. A cereblon modulator CC-885 induces CRBN-and p97-dependent PLK1 degradation and synergizes with volasertib to suppress lung cancer. Mol. Ther. Oncolytics 18, 215–225 (2020).
Huang, H. T. et al. A chemoproteomic approach to query the degradable kinome using a multi-kinase degrader. Cell Chem. Biol. 25, 88–99 (2018).
Sathe, G. & Sapkota, G. P. Proteomic approaches advancing targeted protein degradation. Trends Pharmacol. Sci. 44, 786–801 (2023).
King, E. A. et al. Chemoproteomics-enabled discovery of a covalent molecular glue degrader targeting NF-κB. Cell Chem. Biol. 30, 394–402 (2023).
Donovan, K. A. et al. Mapping the degradable kinome provides a resource for expedited degrader development. Cell 183, 1714–1731 (2020).
Bondeson, D. P. et al. Lessons in PROTAC design from selective degradation with a promiscuous warhead. Cell Chem. Biol. 25, 78–87 (2018).
Nowak, R. P. et al. Plasticity in binding confers selectivity in ligand-induced protein degradation. Nat. Chem. Biol. 14, 706–714 (2018).
Li, Y.-D. et al. Template-assisted covalent modification underlies activity of covalent molecular glues. Nat. Chem. Biol. 20, 1640–1649 (2024).
Hsia, O. et al. Targeted protein degradation via intramolecular bivalent glues. Nature 627, 204–211 (2024).
Xiong, Y. et al. Chemo-proteomics exploration of HDAC degradability by small molecule degraders. Cell Chem. Biol. 28, 1514–1527 (2021).
Huang, H.-T. et al. Ubiquitin-specific proximity labeling for the identification of E3 ligase substrates. Nat. Chem. Biol. 20, 1227–1236 (2024).
Huttlin, E. L. et al. The BioPlex network: a systematic exploration of the human interactome. Cell 162, 425–440 (2015).
Cho, K. F. et al. Proximity labeling in mammalian cells with TurboID and split-TurboID. Nat. Protoc. 15, 3971–3999 (2020).
Roux, K. J., Kim, D. I., Burke, B. & May, D. G. BioID: a screen for protein-protein interactions. Curr. Protoc. protein Sci. 91, 19.23. 11–19.23. 15 (2018).
Kim, D. I. et al. Probing nuclear pore complex architecture with proximity-dependent biotinylation. Proc. Natl. Acad. Sci. USA 111, E2453–E2461 (2014).
Kim, D. I. et al. An improved smaller biotin ligase for BioID proximity labeling. Mol. Biol. cell 27, 1188–1196 (2016).
Kido, K. et al. AirID, a novel proximity biotinylation enzyme, for analysis of protein–protein interactions. Elife 9, e54983 (2020).
Martell, J. D. et al. Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy. Nat. Biotechnol. 30, 1143–1148 (2012).
Sievers, Q. L. et al. Defining the human C2H2 zinc finger degrome targeted by thalidomide analogs through CRBN. Science 362, https://doi.org/10.1126/science.aat0572 (2018).
Roh, Y. S., Song, J. & Seki, E. TAK1 regulates hepatic cell survival and carcinogenesis. J. Gastroenterol. 49, 185–194 (2014).
Shibuya, H. et al. TAB1: an activator of the TAK1 MAPKKK in TGF-β signal transduction. Science 272, 1179–1182 (1996).
Garivet, G. et al. Small-molecule inhibition of the UNC-Src interaction impairs dynamic Src localization in cells. Cell Chem. Biol. 26, 842–851 (2019).
Golkowski, M. et al. Rapid profiling of protein kinase inhibitors by quantitative proteomics. Medchemcomm 5, 363–369 (2014).
Meier, F. et al. diaPASEF: parallel accumulation–serial fragmentation combined with data-independent acquisition. Nat. Methods 17, 1229–1236 (2020).
Knight, R. In Seminars in oncology. (Elsevier, 2005).
D’Amato, R. J., Loughnan, M. S., Flynn, E. & Folkman, J. Thalidomide is an inhibitor of angiogenesis. Proc. Natl. Acad. Sci. USA 91, 4082–4085 (1994).
Lonial, S. et al. Iberdomide plus dexamethasone in heavily pretreated late-line relapsed or refractory multiple myeloma (CC-220-MM-001): a multicentre, multicohort, open-label, phase 1/2 trial. Lancet Haematol. 9, e822–e832 (2022).
Richardson, P. G. et al. Mezigdomide plus dexamethasone in relapsed and refractory multiple myeloma. N. Engl. J. Med. 389, 1009–1022 (2023).
Surka, C. et al. CC-90009, a novel cereblon E3 ligase modulator, targets acute myeloid leukemia blasts and leukemia stem cells. Blood, J. Am. Soc. Hematol. 137, 661–677 (2021).
Gemechu, Y. et al. Humanized cereblon mice revealed two distinct therapeutic pathways of immunomodulatory drugs. Proc. Natl. Acad. Sci. USA 115, 11802–11807 (2018).
Hagner, P. R. et al. CC-122, a pleiotropic pathway modifier, mimics an interferon response and has antitumor activity in DLBCL. Blood, J. Am. Soc. Hematol. 126, 779–789 (2015).
Wang, E. S. et al. Acute pharmacological degradation of Helios destabilizes regulatory T cells. Nat. Chem. Biol. 17, 711–717 (2021).
Costacurta, M. et al. Mapping the IMiD-dependent cereblon interactome using Bio ID-proximity labelling. FEBS J. 291, 4892–4912 (2024).
Jones, P. et al. InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240 (2014).
Paysan-Lafosse, T. et al. InterPro in 2022. Nucleic Acids Res. 51, D418–D427 (2023).
UniProt: the universal protein knowledgebase in 2023. Nucleic Acids Res. 51, D523–D531 (2023).
Gough, J., Karplus, K., Hughey, R. & Chothia, C. Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J. Mol. Biol. 313, 903–919 (2001).
Razumkov, H. et al. Discovery of CRBN-dependent WEE1 molecular glue degraders from a multicomponent combinatorial library. J. Am. Chem. Soc. 146, 31433–31443 (2024).
Zhao, M. et al. Cereblon modulator CC-885 induces CRBN-dependent ubiquitination and degradation of CDK4 in multiple myeloma. Biochem. Biophys. Res. Commun. 549, 150–156 (2021).
Furihata, H. et al. Structural bases of IMiD selectivity that emerges by 5-hydroxythalidomide. Nat. Commun. 11, 4578 (2020).
Zhou, J. & Grigoryan, G. Rapid search for tertiary fragments reveals protein sequence–structure relationships. Protein Sci. 24, 508–524 (2015).
Varadi, M. et al. AlphaFold Protein Structure Database in 2024: providing structure coverage for over 214 million protein sequences. Nucleic Acids Res. 52, D368–D375 (2024).
Zhang, J., Schaeffer, R. D., Durham, J., Cong, Q. & Grishin, N. V. DPAM: A domain parser for AlphaFold models. Protein Sci. 32, e4548 (2023).
Leman, J. K. et al. Macromolecular modeling and design in Rosetta: recent methods and frameworks. Nat. Methods 17, 665–680 (2020).
Maguire, J. B. et al. Perturbing the energy landscape for improved packing during computational protein design. Proteins Struct. Funct. Bioinform. 89, 436–449 (2021).
Rodriguez-Rivera, F. P. & Levi, S. M. Unifying catalysis framework to dissect proteasomal degradation paradigms. ACS Cent. Sci. 7, 1117–1125 (2021).
Barroso-Gomila, O. et al. BioE3 identifies specific substrates of ubiquitin E3 ligases. Nat. Commun. 14, 7656 (2023).
Mukhopadhyay, U. et al. A ubiquitin-specific, proximity-based labeling approach for the identification of ubiquitin ligase substrates. Sci. Adv. 10, eadp3000 (2024).
Liwocha, J. et al. Mechanism of millisecond Lys48-linked poly-ubiquitin chain formation by cullin-RING ligases. Nat. Struct. Mol. Biol. 31, 378–389 (2024).
Bai, N. et al. Modeling the CRL4A ligase complex to predict target protein ubiquitination induced by cereblon-recruiting PROTACs. J. Biol. Chem. 298, https://doi.org/10.1016/j.jbc.2022.101653 (2022).
Békés, M., Langley, D. R. & Crews, C. M. PROTAC targeted protein degraders: the past is prologue. Nat. Rev. Drug Discov. 21, 181–200 (2022).
Yamanaka, S. et al. A proximity biotinylation-based approach to identify protein-E3 ligase interactions induced by PROTACs and molecular glues. Nat. Commun. 13, 183 (2022).
Yu, H. H. et al. Single subunit degradation of WIZ, a lenalidomide-and pomalidomide-dependent substrate of E3 ubiquitin ligase CRL4CRBN. Preprint at https://doi.org/10.1101/595389 (2019).
Cassandri, M. et al. Zinc-finger proteins in health and disease. Cell Death Discov. 3, 1–12 (2017).
Teng, M. et al. Development of PDE6D and CK1α degraders through chemical derivatization of FPFT-2216. J. Med. Chem. 65, 747–756 (2021).
Szklarczyk, D. et al. The STRING database in 2023: protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 51, D638–D646 (2023).
Schmitzová, J., Cretu, C., Dienemann, C., Urlaub, H. & Pena, V. Structural basis of catalytic activation in human splicing. Nature 617, 842–850 (2023).
Hornbeck, P. V. et al. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 43, D512–D520 (2015).
Petzold, G. et al. Mining the CRBN target space redefines rules for molecular glue–induced neosubstrate recognition. Science 389, eadt6736 (2025).
Barak, T. et al. PPIL4 is essential for brain angiogenesis and implicated in intracranial aneurysms in humans. Nat. Med. 27, 2165–2175 (2021).
Reichermeier, K. M. et al. PIKES analysis reveals response to degraders and key regulatory mechanisms of the CRL4 network. Mol. Cell 77, 1092–1106 (2020).
Scott, D. C. et al. Structure of a RING E3 trapped in action reveals ligation mechanism for the ubiquitin-like protein NEDD8. Cell 157, 1671–1684 (2014).
Mercer, J. A. et al. Continuous evolution of compact protein degradation tags regulated by selective molecular glues. Science 383, eadk4422 (2024).
Watson, E. R. et al. Molecular glue CELMoD compounds are regulators of cereblon conformation. Science 378, 549–553 (2022).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Demichev, V., Messner, C. B., Vernardis, S. I., Lilley, K. S. & Ralser, M. DIA-NN: neural networks and interference correction enable deep proteome coverage in high throughput. Nat. Methods 17, 41–44 (2020).
McAlister, G. C. et al. MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal. Chem. 86, 7150–7158 (2014).
R: A language and environment for statistical computing (R Foundation for Statistical Computing, 2014).
Marze, N. A., Roy Burman, S. S., Sheffler, W. & Gray, J. J. Efficient flexible backbone protein–protein docking for challenging targets. Bioinformatics 34, 3461–3469 (2018).
Jacobson, M. P. et al. A hierarchical approach to all-atom protein loop prediction. Proteins: Struct. Funct. Bioinform. 55, 351–367 (2004).
Miller, E. B. et al. Reliable and accurate solution to the induced fit docking problem for protein–ligand binding. J. Chem. Theory Comput. 17, 2630–2639 (2021).
Sanchez-Garcia, R. et al. DeepEMhancer: a deep learning solution for cryo-EM volume post-processing. Commun. Biol. 4, 874 (2021).
Acknowledgements
We thank Sarah Dixon-Clarke, Moritz Hunkeler, Talya Levitz, Tony Zhang, and members of the Fischer and Gray labs for helpful discussions, reagents, and support. We thank the Harvard Cryo-EM Center for Structural Biology for support on data collection. Financial support for this work was provided by the National Institutes of Health (R01CA214608 and R01CA262188, both to E.S.F.). K.B. is a Meghan E. Raveis Fellow of the Damon Runyon Cancer Research Foundation (DRG-2514-24). Hojong.Y is supported by the National Institutes of Health grant (K99CA287069). Figures 1a, 2a and 5a were created in Biorender.
Author information
Authors and Affiliations
Contributions
K.B. designed experiments, performed structural work and biochemical assays, performed immunoblots, analyzed data, interpreted results, wrote the manuscript. R.J.M. initiated the study, designed proteomics experiments, performed proteomics experiments, developed IP-MS automation, supervised proteomics, and analyzed data. S.S.R.B. performed computational alignment analysis, interpreted results. J.W.B. initiated the study and performed biochemical experiments. Hojong Y designed proteomics experiments. R.J.L. wrote the proteomics analysis code. J.K.R. performed proteomics experiments. D.M.A. performed proteomics experiments. M.L. performed immunoblots. Hong Y. performed TR-FRET. S.O. performed TR-FRET. Y.X. analyzed the compound SAR. J.C. performed compound docking and analyzed compound SAR. A.L.V. synthesized molecules. A.M.S. developed IP-MS automation, analyzed data, and supervised proteomics. N.S.G. supervised experiments. K.A.D. conceived the study, designed experiments, developed IP-MS automation, supervised proteomics, analyzed data, interpreted results, wrote the manuscript, and supervised the study. E.S.F. conceived the study, developed IP-MS automation, interpreted results, supervised, and funded the study. All authors read, edited, and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
E.S.F. is a founder, scientific advisory board (SAB) member, and equity holder of Civetta Therapeutics, Proximity Therapeutics, Stelexis Biosciences, Neomorph, Inc. (also board of directors), and Anvia Therapeutics (also board of directors). He is an equity holder and SAB member for Avilar Therapeutics, Photys Therapeutics, and Ajax Therapeutics, and an equity holder in Lighthorse Therapeutics, and CPD4 (also board of directors). E.S.F. is a consultant to Novartis, EcoR1 Capital, Odyssey, and Deerfield. The Fischer lab receives or has received research funding from Deerfield, Novartis, Ajax, Interline, Bayer, and Astellas. K.A.D. receives or has received consulting fees from Kronos Bio and Neomorph Inc. N.S.G. is a founder, science advisory board member (SAB), and equity holder in Syros, C4, Allorion, Lighthorse, Inception, Matchpoint, Shenandoah (board member), Larkspur (board member), and Soltego (board member). The Gray lab receives or has received research funding from Novartis, Takeda, Astellas, Taiho, Jansen, Kinogen, Arbella, Deerfield, Springworks, Interline, and Sanofi. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Jun-Seok Lee, Markus Schirle, Hyun Kyu Song, and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Baek, K., Metivier, R.J., Roy Burman, S.S. et al. Unveiling the hidden interactome of CRBN molecular glues. Nat Commun 16, 6831 (2025). https://doi.org/10.1038/s41467-025-62099-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-62099-w
This article is cited by
-
Unbiased mapping of cereblon neosubstrate landscape by high-throughput proteomics
Nature Communications (2025)