Abstract
Allopolyploidy involves the fusion of genomes from different lineages through hybridization and chromosome doubling. However, detecting early allopolyploidy events and understanding the specific subgenomic evolution that contributes to the origin of adaptive innovations for species radiation can be challenging. Here, we present genomes representing all three subfamilies of Salicaceae, a woody model clade, and collect epigenetic and transcriptomic samples. We reveal one shared ancient allopolyploidy event involving Populus, Salix and two sister genera, followed by contrasted karyotypic and subgenomic evolution. The specific evolution sets the stage for the origin of novel photoperiod responses, flowering phenology and small-hairy seeds in the highly speciose Populus and Salix compared with their species-depauperate sister genera, which may lead to their ecological adaptation and radiation. Our findings underscore the role of ancient allopolyploidization and specific subgenomic evolution in driving both innovation and species diversification at deep nodes of the plant tree of life.
Similar content being viewed by others
Introduction
Polyploidy, or whole-genome duplication (WGD), is common in flowering plants and has played a significant role in their evolution1,2,3. Initially, WGDs result in massive genetic redundancy that allows subfunctionalization of duplicated genes4 and gives rise to the development of novel traits and adaptations5,6,7,8,9,10,11,12. One particularly consequential instance may have been the massive independent WGD events that took place near the Cretaceous-Paleocene (K-Pg) boundary around 60 million years ago (Mya). These events likely contributed to the survival of angiosperms during the mass extinction event at this stage13,14,15. Polyploids can arise through allopolyploidy or autopolyploidy, which involve the merging of genomes between distinct species or between plants within the same species6. However, identifying ancient allopolyploidy that underwent rediploidization and distinguishing the subgenomes is challenging due to the often extensive chromosome rearrangements and the possible extinction of parental lineages6,16,17. Previous studies on allopolyploids have primarily focused on recently formed polyploids or those with limited diploidization, such as wheat, cotton, and bamboo18,19,20, where subgenomes remain readily distinguishable. In contrast, rediploidized allopolyploids, such as Brassica, maize, and Papaver16,21,22, require inference of subgenomes based on residual divergences in gene content, expression biases, and repetitive DNA signatures. However, these methods struggle when ancient allopolyploids exhibit highly homogenized subgenomes. While subgenome-aware analyses in Malvaceae23 have successfully identified multiple ancient allopolyploidization events, the systematic characterization of these subgenomes, particularly regarding the establishment of subgenome dominance and subsequent functional specialization, remains to be fully elucidated. Therefore, despite reports suggesting that allopolyploids may have higher ecological adaptability and evolutionary potential relative to their progenitors and can lead to adaptive radiation24,25,26,27,28, the details of the contrasted subgenomic evolution of once ancient allopolyploids and their contributions to radiations with innovative traits, extensive species diversification and obvious niche shifts in higher taxonomic clades remains scarce29.
Ancestral karyotype reconstruction serves as a fundamental approach for elucidating chromosomal evolutionary trajectories, providing critical insights into polyploidization events and structural rearrangements that shape genome diversity. While the previous approaches for inferring ancestral karyotype rely on identifying conserved ancestral genomic segments through collinearity analysis of extant species30, the recently developed WGDI pipeline31 has emerged as a powerful tool that identifies full protochromosomes by detecting shared intact chromosomes or chromosome-like syntenic blocks across divergent lineages. This framework enables tracing genomic evolutionary dynamics through three major types of interchromosomal rearrangements: reciprocal translocations (RTA), end-to-end joining (EEJ), and nested chromosome fusions (NCF), offering a comprehensive approach to decode karyotype evolution from full ancestral karyotype to modern genomes. To date, the WGDI framework has been successfully applied in reconstructing ancestral karyotypes for diverse plant lineages including Malvaceae, Brassicaceae and core eudicots23,32,33, providing unprecedented insights into plant genome evolution.
The Salicaceae is widely recognized as a model system for studies of woody plant molecular mechanisms, speciation, sex chromosomes, phenotypic variation, and ecosystem services34. This family comprises three subfamilies (Samydoideae, Scyphostegioideae, and Salicoideae) with ~56 genera35 (Supplementary Table 1). Speciation rates are highly elevated in two genera of the Salicoideae, Populus and Salix36, which consist of ~100 and ~500 species as the well-known poplars and willows, respectively. Species in these two genera have successfully adapted to diverse environments across the Northern Hemisphere and played crucial roles in temperate and arctic forest ecosystems34,37,38. Traits including catkins, dehiscent capsules, hairy seeds, and early spring blooming possibly facilitated their rapid colonization of higher latitudes and an extensive radiation. In contrast, two other closely related monospecific and dispecific sister genera, Idesia and Itoa, are found in subtropical or tropical regions and have contrasting traits such as capsules or berry fruits, and glabrous seeds39,40,41,42 (Fig. 1a, data from https://www.gbif.org/). Consequently, these genera have long been considered attractive model systems for studying trait innovation and species radiations in woody plants43,44,45. However, although the Populus trichocarpa genome established a base chromosome number of 10 for the ancestral genus45, the broader ancestral karyotype and patterns of chromosomal evolution in the Salicaceae remain unresolved. Previous studies reported that Populus and Salix underwent a ‘salicoid’ WGD event around the K-Pg boundary45,46,47, which has been interpreted as an autopolyploidization event based on the high similarity between subgenomes48,49, though allopolyploid origin has also been proposed50. In addition, the potential involvement of this event in other Salicaceae genera remains uncertain due to limited sampling and lack of genomic evidence beyond chromosome counts41. These knowledge gaps constrain our understanding of the role of this WGD event on trait innovation and radiations within the Salicaceae.
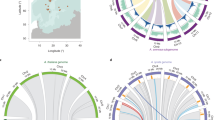
a The geographical distributions of the ten genera in Salicaceae in our study and illustrations depicting the morphological characteristics of the species. The maps were generated using the “maps” and “ggplot2” packages in R. a-j correspond to the respective branches in Fig. 1b. b The phylogenetic relationships and divergence times of species in the Salicaceae, using Passiflora edulis, Passiflora organensis, Arabidopsis thaliana, Rosa chinensis and Vitis vinifera as outgroups. Bootstrap support values at each node are all 100. Arrows indicate calibration nodes. The program collects 20,000 samples and displays the posterior mean divergence time for each node, along with the corresponding 95% confidence interval (CI). The CIs are shown next to the nodes and represented by light blue error bars. The red star represents the ‘salicoid’ WGD event, and the purple star indicates Dianyuea-specific WGD. c The chromosome number, genome size (top), contig N50 (top), repetitive sequences content and types (bottom), and the number of genes (bottom) for each species on the evolutionary tree. The images on the right correspond to branches a-j in Fig. 1b, depicting their flowers and fruits.
Here, we select species representing all three subfamilies and conduct comparative analysis to gain a better understanding of the origin and potential phenotypic effects of this WGD event (Supplementary Table 1). Our study aims to uncover the WGD-derived subgenomic evolution that leads to the emergence of unique photoperiod responses, flowering phenology, and small hairy seeds in Populus and Salix when compared with the other species-depauperate genera, especially their sister genera Idesia and Itoa. The results reveal that these four genera originated from the common ‘salicoid’ WGD event, which is an allopolyploidy event involving the extinction of one parental lineage and subsequent divergent evolution of subgenomes. We further discover that the dynamic gene retention following allopolyploidization, along with lineage-specific expression divergence between subgenomes may facilitate innovative but contrasting phenotypic traits and ecological niches among these genera.
Results
Chromosome-scale genome assemblies of nine Salicaceae species
We combined 94.0-256.7× Nanopore long reads, 54.1-157.0× Illumina short reads, and 118.5-344.1× high-throughput chromosome conformation capture (Hi-C) reads (Supplementary Table 2) to assemble eight chromosomal-level genomes of Salicaceae species from different genera, including seven species from the subfamily Salicoideae (Dovyalis caffra, Scolopia chinensis, Xylosma longifolia, Flacourtia jangomas, Itoa orientalis, Idesia polycarpa, and Salix rehderiana) and one species from the subfamily Samydoideae (Casearia decandra) (Fig. 1b and Supplementary Table 1). Using 45.3× PacBio HiFi reads and 102.3× Hi-C data, we also produced a chromosome-scale assembly for Dianyuea turbinata, a species from the subfamily Scyphostegioideae (Fig. 1b and Supplementary Table 2). These newly assembled genomes varied significantly in size, with D. turbinata (4183 Mb; n = 18 + 1B) and S. chinensis (274 Mb; n = 11) having the largest and smallest genomes, respectively (Fig. 1c and Supplementary Table 2). The contig N50 ranged from 4.2 Mb to 65.4 Mb, and the scaffold N50 ranged from 17.7 Mb to 235.1 Mb. A total of 92.51% to 99.93% of the assembled genome sequences were anchored onto 9 to 21 pseudochromosomes, and the Hi-C heatmaps showed strong interaction signals along the main diagonal, indicating high contiguity and accuracy of the assemblies (Supplementary Fig. 1).
To evaluate assembly completeness, we first performed read mapping analysis by aligning sequencing reads against the assembled genomes. Both Illumina short reads and Nanopore/HiFi long reads showed high alignment rates exceeding 95.96%, with coverage rates ranging from 94.13% to 99.88% and 99.50% to 99.98%, respectively (Supplementary Table 3). Similarly, transcriptomic data from each species also exhibited high mapping rates to their corresponding genomes, ranging from 94.77% to 99.45% (Supplementary Data 1). BUSCO (Benchmark Universal Single Copy Ortholog) assessment revealed high assembly completeness, with all species exceeding 95.5% (Supplementary Table 4). Additionally, the nucleotide-level precision of the assemblies, estimated through the number of homozygous SNPs and InDels, reached 99.65% to 100%; and the Merqury QV scores ranged from 25.6 to 52.2, corresponding to assembly accuracy of 99.73% to 100% (Supplementary Table 3). Overall, these results demonstrated the high quality of our chromosome-scale genome assemblies.
Repeat annotation revealed a strong positive correlation between genome size and repeat content (Supplementary Data 2). For instance, I. polycarpa (genome size ~1.2 Gb) contains ~59% (~716.5 Mb) repetitive sequences, primarily due to the expansion of Gypsy transposable elements (TEs). Interestingly, a similar Gypsy-dominated repeat expansion pattern was also observed previously in Populus euphratica51,52. Notably, D. turbinata exhibits an extraordinary repeat content (~84.81%) and an exceptionally large genome, characterized by simultaneous expansions of Gypsy retrotransposons, unclassified LTR elements and DNA transposons, a pattern distinct from other species (Supplementary Figs. 2 and 3). In total, we identified 20,859 to 43,735 high-confidence protein-coding genes per species (Supplementary Table 5), with 83.67% to 96.37% showing transcriptomic support (Supplementary Data 3). BUSCO assessment further confirmed the annotation quality, demonstrating 94.8% to 98.2% completeness (Supplementary Table 4).
Redefining the phylogenetic position of the ‘salicoid’ WGD event
Using 267 single-copy genes from 19 species identified by BUSCO and OrthoFinder methods, we reconstructed a high-resolution phylogenetic tree (Fig. 1b), which matches a previous topology derived from complete plastomes53. The topology remained the same when STAG was used for all orthogroups containing multi-copy genes identified by OrthoFinder (see Methods). This tree confirmed that the three subfamilies were monophyletic. Within the Salicoideae, we identified two clades: one that consisted of taxa with n = 19-21 chromosomes (Clade I) and another with fewer chromosomes (n = 9-11; Clade II). The genera Itoa and Idesia were successive sisters to Populus and Salix in Clade I, while Dovyalis was sister to the remaining species in Clade II. Our estimates suggest that the Salicaceae diverged from its sister outgroup, Passiflora, around 97 Ma (95% confidence intervals: 88 − 109 Ma), and began to diversify around 79 Ma (69 − 90 Ma). The Scyphostegioideae and Salicoideae subfamilies diverged around 67 Ma (58 − 77 Ma), and Clade I and Clade II of the Salicoideae subfamily diverged around 56 Ma (47 − 65 Ma) (Fig. 1b).
By comparing the chromosome and gene numbers of these genomes, we inferred that the ‘salicoid’ event may be restricted to Clade I species. To test this, we evaluated synonymous substitutions per synonymous site (Ks) for paralogs in each genome (Supplementary Fig. 4a). The results indicated that while all these species experienced an ancient WGD (the core-eudicot-common γ event), D. turbinata and Clade I species exhibited additional recent WGD events, with D. turbinata showing a distinct, significantly lower Ks peak compared to Clade I species (Supplementary Fig. 4a), suggesting independent polyploidization origins. In addition, after correcting for unequal substitution rates among species, we confirmed that the Ks peaks corresponding to the Clade I-specific WGD were greater than those between orthologs within Clade I species, indicating the WGD occurred before their diversification (Supplementary Fig. 5). This was further supported by the well-preserved collinear relationships within Clade I species (2:2) and between Clade I and Clade II species (2:1) (Supplementary Figs. 4b, 4c and 6). Taken together, these results consistently suggest that the ‘salicoid’ WGD event occurred on the ancestral branch of Clade I species.
We next predicted the present distribution of the ten genera in the Salicaceae family using ecological niche modelling, based on available distribution data for all species within each genus (Supplementary Figs. 7 and 8 and Supplementary Data 4 and Supplementary Table 6). The results revealed that the four genera within Clade I, which underwent the ‘salicoid’ WGD, are predominantly distributed in the Northern Hemisphere, while the remaining lineages primarily occur in the Southern Hemisphere. Significant niche differentiation was observed both between these major clades and among the four genera within Clade I. Unlike Itoa and Idesia, which are restricted to subtropical regions, the genera Populus and Salix have expanded to higher latitudes, including temperate and polar regions.
Karyotypic evolution and chromosomal rearrangement
Using shared intact chromosomes and/or chromosome-like syntenic blocks31 among these species, we reconstructed the ancestral Salicaceae karyotype (ASK), which consists of 11 intact protochromosomes (Fig. 2 and Supplementary Figs. 9–11; see Methods), rather than the previously inferred chromosome base number of 1045. Drawing from the ‘salicoid’ WGD and genome rearrangement events, we further traced their evolutionary relationships (Fig. 2), which align closely with the phylogenetic analyses (Fig. 1b). Notably, despite multiple lineage-specific chromosomal rearrangements in D. turbinata, its identical subgenome karyotypes indicate that the Dianyuea-specific WGD was likely an autopolyploidization event unaccompanied by interchromosomal rearrangements (Fig. 2). In comparison, few fusion/fission events occurred in species from Clade II, which did not undergo the ‘salicoid’ WGD. Thus, in Clade II most protochromosomes were preserved intact, with the exception of at least four chromosomal rearrangements (Fig. 2 and Supplementary Figs. 12a and 12b): (i) protochromosome 8 underwent a RTA with protochromosome 11 at the ancestral node of Clade II, (ii) protochromosome 2 underwent an EEJ with protochromosome 3 to form chromosome 2 of D. caffra, (iii) protochromosome 2 independently underwent a RTA with protochromosome 4 to form the contemporary karyotype structure of S. chinensis, and (iv) protochromosome 1 subsequently underwent an EEJ to form chromosome 1 of X. longifolia and F. jangomas. We inferred that the ancestral karyotype of Salicoideae Clade I species had a base number of 21 after the ‘salicoid’ WGD, achieved through an EEJ fusion between duplicated protochromosomes 5 and 9, with at least two subsequent inversions (Fig. 2 and Supplementary Fig. 12c). Additionally, we predicted multiple RTA events between several protochromosomes. Interestingly, these genome-wide reorganizations were fully preserved in I. polycarpa without other large fusion/fission events, indicating that their common ancestor already possessed a relatively stable karyotype during the rediploidization process. In contrast, our results indicated that I. orientalis underwent significant changes through multiple chromosomal rearrangements, resulting in a base chromosome number of 20 (Fig. 2). Ancestors of Populus and Salix also experienced a series of chromosome rearrangements, forming an ancient fused chromosome through successive NCFs of duplicated protochromosomes 3, 7 and 11 (Supplementary Fig. 12d). This newly fused chromosome subsequently underwent two independent RTA events: one with protochromosome 1 in Populus to generate extant chromosomes 1 and 4, and a second with protochromosome 5 in Salix to generate extant chromosomes 4 and 16. This evolutionary trajectory resulted in a 19-chromosomes karyotype in Populus and Salix (Fig. 2).
The evolutionary relationships were constructed based on the shared polyploidy events and chromosome fusions with the shortest steps. The inferred ancestral karyotype contains 11 protochromosomes, marked with different colors. Leaf nodes show the modern karyotypes of species, and chromosomal changes leading to the formation of these modern karyotypes, including RTA, EEJ and NCF, are marked on the ancestral nodes of the phylogenetic tree. Among them, the ancestor of Clade II experienced a RTA, resulting in the karyotype of ancestor A, and subsequently hybridized with ancestor B of another branch (which is now extinct), leading to an allopolyploidization event, which is indicated by a red star. The Dianyuea-specific WGD event (autopolyploidization) is marked by a purple star. Chromosomes from ancestor B are distinguished by white asterisks in the center of the chromosomes. The dashed lines indicate unresolved relationships for karyotypes of existing species. Source data are provided as a Source Data file.
The ‘salicoid’ WGD event is a cryptic allopolyploidization
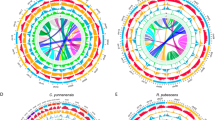
Combining the correspondence between ASK and extant chromosomes of each Salicaceae species, as well as phylogenetic analysis and subgenome division (Supplementary Fig. 13; see Methods), we found strong evidence to support that the ‘salicoid’ WGD is a cryptic allopolyploidization event. Specifically, according to the reconstructed protochromosomes (ASK1-11), we identified 1545 full homologous gene groups (HGGs), among which all Clade I species retained paralogous genes from the ‘salicoid’ WGD, while other species retained the orthologous genes (Supplementary Fig. 14). There were 81-248 full-HGGs across the 11 protochromosomes. Phylogenetic tree using the ASTRAL coalescent method showed that genes from Clade II were more closely related to one copy of ‘salicoid’ paralogs in Clade I species than the other copy (Fig. 3a, topologies ‘q1’ and ‘q2’ in Fig. 3b). This relationship was supported by 86.7% to 94.1% of full-HGGs on all 11 protochromosomes and 82.8% to 86.3% when considering partial-HGGs, where at least one species in Clade I retained ‘salicoid’ paralogs (Fig. 3b and Supplementary Fig. 14). Consistently, we found that a RTA event between protochromosomes 8 and 11 that occurred in the ancestor of Clade II species was also present in only one chromosome in all Clade I species, further supporting their allopolyploid origin (Fig. 2).
a The evolutionary relationships and divergence time estimates of the two subgenomes of Clade I species relative to Clade II, D. turbinata and C. decandra. The subgenome closer to Clade II is designated as A, while the other subgenome is named B. The 95% confidence intervals (CI) for divergence times are shown next to the nodes and represented by light purple error bars. Red and purple stars indicate ‘salicoid’ allopolyploidization and D. turbinata-specific autopolyploidization events, respectively. b The frequency of three topologies (q1-q3) around internal branches (marked with a red star in Fig. 3a) of ASTRAL species trees for each protochromosome (ASK 1-11) in the datasets Full-HGGs and Full-/Partial-HGGs. c The collinearity between the two subgenomes of Clade I species (here I. orientalis, I. polycarpa, P. trichocarpa and S. purpurea), where the inner circle is the A subgenome and the outer circle is the B subgenome. The color of each chromosome indicates its ancestral origin. d The comparison of the corrected molecular evolutionary rates for Clade I species using the two-tailed Mann-Whitney U test. ****p < 0.0001. All pairwise species comparisons showed p < 2.2e-16, except among the three Salix species (Scha-Spu: 0.83, Scha-Sre: 0.13, Spu-Sre: 0.82). The data points for each species from left to right are 11344, 11344, 11099, 10912, 12160, 9466, 10466, 10056. Boxplot elements are as follows: centerline, median; box limits, first and third quartiles; whisker, 1.5x interquartile range. All species names are abbreviations. Ipo I. polycarpa, Ior I. orientalis, Pal P. alba var. pyramidalis, Peu P. euphratica, Ptr P. trichocarpa, Scha S. chaenomeloides, Spu S. purpurea, Sre S. rehderiana. Source data are provided as a Source Data file.
Based on their phylogenetic relationships with Clade II, we divided the genome of Clade I species into two subgenomes: the A subgenome consisted of chromosomes with genes closely related to Clade II, while the B subgenome was composed of the paralogous chromosomes from the ‘salicoid’ WGD (Fig. 3c and Supplementary Fig. 15). As expected, Ks distribution between Clade II species and A subgenome of Clade I species was generally smaller than that between Clade II and B subgenome, but both A and B subgenomes were similar in genetic distance to D. turbinata (or C. decandra in Supplementary Fig. 16), a degree of divergence consistent with phylogenetic relationships under allopolyploidization. Phylogenetic trees reconstructed using the A and B subgenomes by coalescent and concatenated methods revealed the same topologies with high support (Supplementary Fig. 17). These results indicate that the ‘salicoid’ event in Clade I species was an allopolyploidization event with one parent being the common ancestor of Clade II species.
To identify the second possible parent of the ‘salicoid’ allopolyploidization, we assembled the draft genomes of four additional species from the subfamily Salicoideae (including Azara serrata, Prockia crucis, Banara guianensis and Homalium cochinchinense). These draft genomes are fragmented but, based on gene prediction, cover most of the coding regions (Supplementary Table 7). Therefore, we added gene sequences from these four more genera to the HGGs and constructed coalescent and concatenated trees (Supplementary Fig. 18). In both trees, these new genera from Salicoideae clustered within Clade II, while the two subgenomes of D. turbinata clustered together (autopolyploidization) as sister to both subgenomes of Clade I species, indicating that none of these species was direct donors of the B subgenome. Since these species represent all three subfamilies of Salicaceae53,54, we inferred that the parental donor of the B subgenome is likely extinct. Moreover, we inferred that the two parental lineages diverged for ~8 million years prior to the ‘salicoid’ WGD based on our estimates that the A and B subgenomes diverged about 66 Ma and Clade II diverged from the A subgenomes about 58 Ma (Fig. 3a). Finally, based on Ks values corrected for the ‘salicoid’ WGD, rates of molecular evolution were the slowest in I. polycarpa, followed by I. orientalis and Populus, while Salix evolved fastest (Fig. 3d).
Subgenome dominance and lineage-specific gene retention
Subgenome evolution after polyploidization plays a crucial role in reducing gene redundancy and increasing subsequent potential for trait innovation and adaptation4,12,55. Our analysis of 23 Salicaceae genomes/subgenomes showed that the A subgenome exhibited evolutionary dominance over the B subgenome in Clade I species. First, we found that after the ‘salicoid’ event, the rate of gene retention was reduced significantly in Clade I species and was higher for the A subgenome than the B subgenome (Fig. 4a). Second, the B subgenome showed lower gene expression levels than the A subgenome in all Clade I species (Fig. 4b). This may be related to higher repeat content surrounding genes in the B subgenome (Supplementary Fig. 19a), although there was no significant difference in the expansion of TEs between subgenomes A and B overall (Supplementary Fig. 19b). Third, genes in the A subgenome had slightly but significantly higher chromatin accessibility levels (Supplementary Fig. 19c and Supplementary Data 5) and slightly lower methylation levels than those in the B subgenome (Supplementary Fig. 19d, e and Supplementary Data 5), suggesting that the differentiation between the A and B subgenomes is primarily reflected in sequence variation rather than in epigenetic landscapes. Fourth, protein evolution rate (dN/dS) was significantly higher in the B subgenome, indicating relaxed purifying selection (Fig. 4c). Finally, through an analysis of the contribution of subgenome-biased gene retention to lineage evolution, we found that genes specifically retained in the A subgenome of all Clade I species (n = 958) were enriched in functions such as ‘chromosome segregation’, ‘double-strand break repair’, ‘meiotic cell cycle process’, and ‘spindle organization’ (Fig. 4d). In contrast, genes uniquely retained in the B subgenome (n = 558) did not exhibit enrichment in any specific functional categories. These findings suggest that the A subgenome may have played a pivotal role in facilitating and coordinating chromosome recombination during the rediploidization process, while gene retention in the B subgenome appears to be more random and functionally diverse.
a The percentage of gene retention in the D. turbinata, Clade II species and two subgenomes of Clade I species. A total of 183 sliding windows were used to assess gene retention. b Gene expression in the C. decandra, D. turbinata, Clade II, and two subgenomes of Clade I species. c The rates of protein evolution (dN/dS) of the 1809 A and B subgenome branches calculated using codeml. a−c The A and B subgenomes are represented in blue and green, respectively. Significant values of the two-tailed Mann-Whitney U test for the two subgenomes are shown above. Boxplot elements are as follows: centerline, median; box limits, first and third quartiles; whisker, 1.5x interquartile range. All species names are abbreviations. Cde C. decandra, Dtu D. turbinata, Dca D. caffra, Sch S. chinensis, Xlo X. longifolia, Fja F. jangomas, Ipo I. polycarpa, Ior I. orientalis, Pal P. alba var. pyramidalis, Peu P. euphratica, Ptr P. trichocarpa, Scha S. chaenomeloides, Spu S. purpurea, Sre S. rehderiana. d Enriched GO terms (q-value < 0.05) for genes specifically retained in the A subgenome of all Clade I species (A), as well as duplicated gene pairs commonly retained in four genera (Node I: Itoa, Idesia, Populus and Salix), three genera (Node II: Idesia, Populus and Salix), and two genera (Node III: Populus and Salix), respectively. These three categories (Node I-III) correspond to the three ancestral nodes in Fig. 4e. e The number of duplicated gene pairs specifically retained in four genera (Node I), three genera (Node II), and two genera (Node III), and the microsynteny visualization of gene FD identified in the Node II. Blue and green rectangles represent forward and reverse genes, respectively. Only one copy of the duplicated gene was retained in the A subgenome of Itoa, with the lost gene indicated by a cross, while both copies were retained in Idesia, Populus, and Salix. Source data are provided as a Source Data file.
Moreover, beyond subgenome dominance, retention of both the subgenome A and B gene copies may also exhibit lineage-specific dynamics over evolutionary time, and may have laid the groundwork for adaptation to novel environments9. To explore this, we further analyzed gene retention at three key evolutionary nodes in Clade I (Fig. 4e). We identified 1989 WGD-derived gene pairs retained across all Clade I species (Node I). These genes are functionally associated with ‘regulation of photoperiodism, flowering’, ‘inflorescence development’, ‘regulation of seed development’, ‘trichome morphogenesis’, ‘cold acclimation’ and ‘defense response’ (Fig. 4d). The retention of key genes related to photoperiod-flowering (such as FT, CIB1 and HUA2)56,57,58,59, cold acclimation and freezing tolerance (MYB15, ADF5 and GALS2)60,61,62, and trichome development (GIS3, ZFP6 and TOE1)63,64,65 (Supplementary Fig. 20a, b) may have facilitated the divergence of Clade I species from their ancestral lineage by enabling adaptation to northern regions with variable photoperiods and temperatures. Consistent with this, 402 pairs of genes with similar functions were specifically retained in Node II representing the deciduous lineages of Idesia, Populus and Salix that are distinct from the evergreen lineage Itoa (Fig. 4d), and included FD, NF-YB3, SPL4, SPL5 and ELF966,67,68,69, which are related to photoperiodic flowering (Fig. 4e, Supplementary Fig. 20c, d). Additionally, at Node III, representing the Populus and Salix lineages that are widely distributed in the Northern Hemisphere, we identified 129 uniquely retained gene pairs (Fig. 4e). These genes are involved in specific functions like ‘maintenance of floral organ identity’, ‘seed trichome differentiation’, ‘red or far-red light signaling pathway’ and ‘regulation of plant-type cell wall cellulose biosynthetic process’ (Fig. 4d), such as REV70 related to the formation of inflorescence and floral meristem, and CSLD171 related to the development of root hairs and female gametophytes (Supplementary Figs. 20e, f). Overall, the dynamic patterns of gene retention in these lineages suggest that novel photoperiod, flowering, inflorescence, and seed traits have fostered the diversification of Clade I species.
Lineage-specific subgenomic expressions contributed to innovation in multiple traits of Populus and Salix
Expression divergence of paralogs plays a pivotal role in driving species radiation and evolutionary innovation after polyploidization72,73. To investigate the association between subgenome expression patterns and lineage diversification in the Salicaceae, we focused on gene pairs retained across all Clade I species. Transcriptome sequencing and cluster analysis of male and female flowers, as well as fruits at various developmental stages in Itoa, Idesia and Populus (Supplementary Data 5), revealed that the expression biases between subgenomes A and B for gene pairs is largely consistent across tissues and developmental stages within the same species (Fig. 5a). However, significant differences were observed between species. Specifically, among the 1747 gene pairs examined, only 85 (C1) and 51 (C2) gene pairs showed the same expression bias towards subgenomes A and B, respectively, across the three lineages. In contrast, the vast majority (C4-C15, ~80%) of gene pairs showed clear differences in expression divergence among lineages (Fig. 5a).
a Cluster results of expression divergence of duplicated gene pairs between the A and B subgenomes during different developmental stages of female and male flowers and fruits in I. orientalis (Ior), I. polycarpa (Ipo) and P. deltoides (Pde). The subgenome expression divergence was represented by the TPM ratio (A - B)/(A + B), where a ratio > 0 indicates expression dominance in the A subgenome, while a ratio < 0 indicates expression dominance in the B subgenome. The heatmap results are shown on the left, and the mean and standard deviation of the TPM ratio for each sample are displayed on the right. Each sample has three biological replicates. b The expression levels of duplicated gene pairs related to the development of traits in different tissues and developmental stages. These genes are distributed in four clusters, including C1, which is more highly expressed in the A subgenome across all species, and clusters (C4-C6) with lineage-specific expression divergence in Populus. The circle size and color are positively correlated with the expression levels. c Expression of FT1 and FT2 in leaves and buds/shoot apex of I. orientalis (Ior), I. polycarpa (Ipo) and P. deltoides (Pde) from late autumn to early spring. Relative fold change in expression levels of FT1 and FT2 relative to the lowest expression (TPM) within a tissue are shown. The results for P. deltoides (Pde) are adapted from previous studies74,79. d Overall rosette leaf phenotype of A. thaliana WT (wild-type) plant, 35S:ZFP6, and 35S:GIS3 transgenic plants. Arrows indicate the 4th rosette leaf. Bar = 1 mm. e Phenotypes and trichome distribution on the 4th rosette leaves of A. thaliana. Bar = 1 mm. f Statistical analysis of trichome density on the 4th rosette leaves of A. thaliana. Each line included five biological replicates. Data are presented as mean ± SD, and p-values from two-tailed t-tests are shown. g VISTA sequence conservation plot of the Populus-Salix specific CNE around ZFP6 and GIS3, using S. rehderiana as a reference. The A and B subgenomes of Clade I species are represented in blue and green, respectively. All species names are the same as abbreviations in Fig. 4. h The Dual-luciferase assay revealed four CNEs could significantly improve the expression level of luciferases and have potential enhancer activities. CNE3 and CNE4 were tested with 3 and 6 replicates, respectively, while the others were tested with 4 replicates. Data are presented as mean ± SD, and p-values from two-tailed t-tests are shown. Source data are provided as a Source Data file.
Among the clusters with consistent expression biases, gene pairs in C1, with higher expression in the subgenome A (Fig. 5a), were significantly enriched in functions related to reproductive transitions, seed maturation and stress responses (Supplementary Data 6), including genes such as FT, HDA6, AP2, BBX21, GALS2 and transcription factors belonging to WRKY family56,62,74,75,76,77,78 (Fig. 5b). Conversely, gene pairs in C2, with higher expression in the subgenome B, were mainly associated with regulation of abscisic acid signaling and gene expression regulation (Supplementary Data 6). These results suggest the significant role of the subgenome A in regulating reproductive and growth traits in these species. For example, the WGD-derived paralogs of flowering-related gene FT exemplified functional divergence: FT2 in subgenome A was highly expressed in our examined transcriptomes, while FT1 from subgenome B was rarely expressed (Fig. 5b), supporting a role of FT2 in promoting flower and fruit development. Interestingly, previous studies on poplar have shown that FT1 is expressed in cold-exposed buds during winter (mainly January and February), triggering the transition from vegetative meristems to the reproductive phase, while FT2 is primarily expressed during the growing season, regulating vegetative growth. These spatiotemporal shifts in expression are essential for the annual growth cycle of poplar74,79. To further explore the evolutionary insights into the functional divergence of this gene pair, we analyzed their expression patterns in leaves and buds (or shoot apex) from I. orientalis and I. polycarpa across late autumn to early spring, encompassing the critical winter period (Fig. 5c). The results showed that in leaves, both FT1 and FT2 were expressed during winter in I. orientalis, while only FT1 was expressed in I. polycarpa and poplar. In buds or shoot apex, FT1 was exclusively expressed in poplar, whereas FT2 was specifically expressed throughout winter in I. orientalis and only in late winter in I. polycarpa (Fig. 5c). This unique functional divergence of FT genes in poplar is consistent with its phase change from vegetative to reproductive growth during the transition from active growth to winter dormancy, enabling earlier flowering and providing a reproductive advantage in spring. In contrast, the co-expression of FT1 and FT2 in I. orientalis likely correlates with its evergreen and non-dormant traits, while the expression patterns in I. polycarpa appear linked to the initiation of reproductive growth after spring warming. The lineage-specific divergence in FT expression has likely promoted the variation in flowering phenology among these lineages.
Furthermore, we found that the gene pairs with Populus-specific expression divergence (C4-C6 in Fig. 5a) were significantly enriched in functions related to ‘photoperiodism, flowering’, ‘regulation of circadian rhythm’, ‘flower development’, ‘regulation of seed development’ and ‘trichome morphogenesis’ (Supplementary Data 6). Among these, we identified several genes involved in the regulation of flower development, including FIL, PUB13, BRM80,81,82, as well as ENO2 and XYL183,84, which are associated with seed development and seed size determination. Additionally, we also identified ZFP6 and GIS3, a pair of WGD-derived genes previously implicated in the regulation of trichome development in A. thaliana63,64. Both genes exhibited significantly higher expression levels in Populus (especially in fruit and seed development) compared to Idesia and Itoa (Fig. 5b). To further investigate their functional roles, we independently expressed Populus ZFP6 and GIS3 in A. thaliana under the CaMV 35S promoter. We then mainly examined their phenotypic changes in leaves as found before63,64. Transgenic lines with overexpression of either Populus gene exhibited approximately two-fold increases in trichome density compared to wild-type plants (Fig. 5d–f, Supplementary Figs. 21a, b and Supplementary Table 8), demonstrating their conserved role as positive regulators of trichome development. Notably, we identified four Populus-Salix specific conserved noncoding elements (CNEs) in their promoter regions (Fig. 5g). Reporter assays indicated that these CNEs possess potential enhancer activities, likely contributing to their elevated expression during seed trichome development in the Populus-Salix lineage (Fig. 5h and Supplementary Table 9). In summary, these findings suggest that the lineage-specific divergence in subgenomic gene expression significantly contributed to the evolution of distinct flowering phenology and the development of small, hairy seeds in Populus and Salix. These traits likely played critical roles in enabling their rapid colonization and extensive radiation in high-latitude regions.
Discussion
Ancient allopolyploidization events are often obscured by large-scale subgenome reshuffling. Although the ancient WGD event in Salicaceae has been previously investigated, most former studies concentrated exclusively on Populus and Salix45. The limited taxon sampling and high subgenome similarity have resulted in ongoing debate about whether the ‘salicoid’ WGD originated through autopolyploidy or allopolyploidy48,49,50. Furthermore, identification of the phylogenetic placement of this event has been hindered by limited genomic resources, forcing reliance on chromosome number comparisons41. In contrast to earlier studies that used distantly related Vitis (Order Vitales) as an outgroup or inferred WGD origins from transcriptome-based phylogenies48,50, we assembled nine high-quality genomes representing all three Salicaceae subfamilies (order Malpighiales). Combining karyotype reconstruction with subgenome-phased phylogenetics of closely related lineages, we provide clear evidence that the ancient polyploidization event, shared by the genera Populus and Salix, as well as their sister genera Idesia and Itoa in the Salicaceae, was caused by hybridization between two closely related lineages with a divergence time of ~8 million years. The high genomic similarity between these parental lineages obscured detection of this cryptic allopolyploidization in the absence of ancestral lineage information, accounting for the previous misclassification of the ‘salicoid’ WGD as an autopolyploid event48. Our analysis showed that one of the parental genomes is represented by the Clade II lineage of the Salicoideae subfamily, while the other parental lineage is now extinct. Based on this, we successfully separated the genomes of these polyploidized species into two distinct subgenomes, advancing Salicaceae WGD research to subgenome resolution and expanding the taxonomic scope beyond Populus and Salix. Moreover, while a prior study based solely on subgenomes in Populus inferred 10 ancestral chromosomes45, we extended ancestral karyotype reconstruction to deeper phylogenetic nodes, establishing an 11-protochromosome ancestral karyotype for Salicaceae. We also detected unique chromosomal fusions specific to Itoa, Idesia and Populus-Salix. The lineage-specific karyotypic evolution is consistent with the phylogenomic tree and intact chromosomes are widespread and shared by all three lineages (Fig. 2), indicating that despite the 58 million years since allopolyploidization and subsequent species diversification, a considerable number of chromosomes have maintained relatively stable organization. Therefore, the Salicaceae, along with other lineages including Gossypium and related Malvaceae, Brassicaceae, and various subclades of Poaceae23,85,86, provides a potential model to investigate the long-term consequences of allopolyploidization and the subsequent evolution of subgenomes (Fig. 6). This is in contrast to recent polyploids that have not undergone rediploidization or species diversification, or ancient polyploids with unknown parental lineages87,88.
The model shows three subfamilies of the Salicaceae, with an ancestral lineage (Ancestor A) in subfamily Salicoideae hybridized with another extinct ancestral lineage (Ancestor B). The resulting hybrid underwent rediploidization and gradually diverging into four lineages with subgenomic dominance. Duplicated genes underwent dynamic changes in retention and divergence of subgenome expression patterns, setting the stage for trait innovation in these lineages. Among these, the Populus-Salix lineage developed innovative traits related to seeds and flowering, which facilitated its radiation.
Our study shows that subgenome A exhibited evolutionary dominance across Clade I, regardless of the degree of chromosomal rearrangements in descendant lineages. This dominance was manifested as biased gene retention, gene expression, and evolutionary rates. Consistent with previous speculation on the role of paleopolyploidy in the adaptive evolution of angiosperms15, we found that genes associated with environmental responses, particularly those involved in photoperiod regulation, flowering cycles and cold adaptation, were preferentially retained following allopolyploidization. However, although we observed the same dominance of subgenome A over subgenome B, the retained genes and expression divergence exhibited significant variation between three lineages, Itoa, Idesia and Populus-Salix. Importantly, these lineage-specific traits, such as the distinct flowering phenology and highly-effective seed dispersal facilitated by hairs, align with the expansion of the Populus-Salix lineage37,38,42 to new highly seasonal temperate, boreal, and arctic ecological niches in the Northern Hemisphere (Fig. 6). In contrast, the two other lineages, Idesia and Itoa, are adapted to subtropical environments and exhibit different patterns of gene retention and expression. This suggests that the ancestral polyploidization created the foundation for key innovations and diversification in the Salicaceae and provides an important example of its impact in angiosperms10. Notably for our study, the functional characterization of genetic variants in Salicaceae faces inherent challenges due to their prolonged life cycles and extended juvenile phases, necessitating reliance on A. thaliana as a heterologous system. In many cases, this approach may not properly replicate woody plant biology, constraining our understanding of these variants’ precise roles in trait evolution and adaptation. In addition, although our study encompasses representatives from all three Salicaceae subfamilies, broader taxonomic sampling will be essential to fully elucidate how this ancient allopolyploidization event facilitated the remarkable ecological diversification. Future research should expand taxonomic sampling to enhance the phylogenetic framework needed to identify genetic variants underlying trait innovation in Salicaceae, validate their molecular functions, and link them to ecological adaptation.
Overall, our study provides insights into the distinct evolutionary trajectories of subgenomes following allopolyploidy, which underlie the diverse traits observed in the Salicaceae, an economically and ecologically significant model clade for woody plants. These evolutionary patterns likely extend to other major taxonomic lineages influenced by recurrent paleopolyploidization events in both plants (e.g., Brassicaceae, Poaceae, Asteraceae and Fabaceae)89,90,91,92,93 and animals (e.g., hexapods, salmonids)94,95. Despite their prevalence, comprehensive analyses of ancient polyploidization events and the long-term evolutionary dynamics of their subgenomes remain scarce. The integrative framework developed here, coupled with rapidly accumulating genomic resources across these taxa, provides a robust foundation to examine how ancient polyploidy and subgenome evolution shape adaptive radiation and biodiversity patterns throughout the tree of life. Crucially, our findings underscore the need to distinguish between autopolyploidization and allopolyploidization, and to account divergent subgenomic evolution, which can promote innovation and species diversification in deep evolutionary time.
Methods
Sample collection and genome sequencing
Plant material of Casearia decandra, Dovyalis caffra, Scolopia chinensis, Xylosma longifolia and Flacourtia jangomas were collected in XiShuangBanNa Tropical Botanical Garden (Mengla, China), Itoa orientalis and Idesia polycarpa were collected in Chengdu, and Salix rehderiana was collected in Minya Konka of China, respectively. Fresh leaves were collected, and high-quality genomic DNA was extracted using the QIAGEN Genomic DNA extraction kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Three approaches were employed in DNA sequencing. First, genomic DNA was size-selected using the BluePippin system (sage Science), processed following the protocol of Ligation Sequencing Kit (LSK108), and sequenced using the Oxford Nanopore Technology sequencer. Sequencing was performed on the PromethION platform, and base calling was carried out using Guppy v3.2.8. Second, Paired-end libraries were constructed according to the manufacturer’s protocols and sequenced using the Illumina HiSeq 2500 System. Third, Hi-C (high-throughput chromatin conformation capture) libraries were prepared by chromatin extraction and digestion and DNA ligation, purification, fragmentation, and sequenced on an MGISEQ-200096. For Dianyuea turbinata, the fresh leaves were collected in Dehong Dai and Jingpo Autonomous Prefecture, Yunnan, and the HiFi SMRTbell library was constructed using the SMRTbell Express Template Prep Kit v2.0 (Pacific Bioscience) and sequencing was carried out using the PacBio Sequel II platform (Berry Genomics, Beijing, China). Hi-C library was further constructed and sequenced using the same methodology described above. In addition, the dried leaves of Azara serrata, Prockia crucis, Banara guianensis and Homalium cochinchinense were used to extract DNA, construct Illumina paired-end libraries, and perform sequencing.
Genome assembly and annotation
Nanopore long reads were de novo assembled using the Nextdenovo v2.2.0 (https://github.com/Nextomics/NextDenovo). The initial assemblies were further corrected and polished using the program NextPolish v1.0 (https://github.com/Nextomics/NextPolish), by mapping the filtered Nanopore and Illumina reads to the genome using Minimap2 v2.1797 and BWA v0.7.1798. The redundant haplotypes and low-quality contigs were identified and removed from the assemblies using the Purge Haplotigs v1.1.3 pipeline99. PacBio HiFi reads were used to perform de novo genome assembly for D. turbinata with Hifiasm v0.14100. Finally, contigs were clustered, ordered and anchored to the pseudochromosomes by LACHESIS101 or YaHS102 using validly mapped Hi-C reads, and manually corrected in JuiceBox v1.11.08103. In D. turbinata, a very short chromosome was assembled, accounting for only about 1/17 the length of the other chromosomes and exhibiting poor synteny with other species. This was consistent with the karyotype observed in cytological analyses41. Therefore, it was considered a B chromosome and was excluded from subsequent analyses. In addition, Illumina reads of A. serrata, P. crucis, B. guianensis and H. cochinchinense were assembled using Platanus v1.2.4104 by implementing ‘assemble’, ‘scaffold’ and ‘gap_close’ program.
We utilized a combination of homolog-based and de novo approaches to annotate repetitive elements. RepeatMasker v.4.0.7105 was firstly used to perform homolog prediction based on the Repbase database106. Next, RepeatModeler v.1.0.11107 was used to perform de novo prediction of repeat sequence features and the results were then utilized by RepeatMasker v.4.0.7105 to identify and classify repeat elements. Gene models were predicted based on de novo prediction, homologous identification and transcript data. In brief, Augustus v3.2.3108 was used for de novo prediction of protein-coding genes. For homologous identification, we mapped the protein sequences of six published genomes (P. trichocarpa45, P. pruinosa109, P. alba var. pyramidalis110, S. suchowensis46, S. purpurea111, and Arabidopsis thaliana112) onto the genomes using TBLASTN v2.6.0113 and then used GENEWISE v2.4.1114 to predict gene structures. RNA transcripts were used to predict gene models with PASA v2.3.3115. Finally, all of the predictions were integrated using EvidenceModeler v1.1.1116 to generate consensus gene sets.
To assess the quality of the assemblies, Illumina, Nanopore/HiFi, and RNA reads were aligned to the genome assembly using BWA v0.7.1798, Minimap2 v2.1797, and HISAT2 v2.1.0117, respectively. Sequence accuracy was assessed using Merqury v1.3118 and Illumina data. Assembly and annotation completeness was assessed with BUSCO (Benchmarking Universal Single-Copy Orthologs) v3.0119. In addition, the RNA sequencing reads used for annotation were assembled using Trinity v2.8.4 with default parameters. Redundancy in transcript sequences was reduced using CD-HIT v4.6.8120 (similarity >95%) and candidate coding regions were identified with TransDecoder v5.5.0121. The obtained unigenes were also used for subsequent evaluation of transcriptome-based gene prediction by calculating the percentage with >50% sequence coverage by these unigenes.
Phylogenetic analysis
We conducted a phylogenomic analysis for nine sequenced species and three poplar (P. trichocarpa45, P. alba var. pyramidalis110 and P. euphratica51) and two willow species (S. chaenomeloides122 and S. purpurea123), using Passiflora edulis124, Passiflora organensis125, Arabidopsis thaliana112, Rosa chinensis126 and Vitis vinifera127 as outgroups. We constructed two phylogenetic datasets using different strategies: targeted identification of phylogenomic markers (BUSCO) and de novo inference (OrthoFinder). Conserved single-copy genes were identified by BUSCO analyses with the embryophyta_odb10 dataset (1614 BUSCOs)119, resulting in 204 single-copy orthologs retained across 19 species. OrthoFinder v2.3.11128 was used to de novo identify orthologous sequences shared among species with default parameters, resulting in 106 single-copy orthologs. To reconstruct a high-resolution phylogenetic tree, we merged the two datasets and obtained a total of 267 single-copy orthologs. For all orthologs, protein sequences were aligned using MAFFT v7.313129, and converted to codon alignments using PAL2NAL130. A maximum likelihood (ML) phylogenetic tree was then constructed using RAxML v8.2.11131 with the GTR + gamma model and 1000 bootstrap replicates. Additionally, STAG from the OrthoFinder pipeline was also used to infer a species tree based on all orthogroups (including multi-copy genes) identified by OrthoFinder. The topologies of the phylogenetic trees constructed by the two methods were the same.
The divergence times among species were estimated using the MCMCtree program132. Three constraints obtained from the TIMETREE database (http://timetree.org/) were used for time calibrations: (1) the divergence between Vitis and Rosa (109 - 124 Mya), (2) the divergence between Arabidopsis and Populus (102 - 113 Mya), (3) the divergence between Populus and Salix (28 - 60 Mya).
Analysis of WGD events
We identified and localized WGD events in Salicaceae by combining intra- and inter-species synteny analysis and Ks distribution. First, we used the BLASTP v2.7.1133 with a cutoff e-value of 1e-5 to align protein sequences within species and between species (P. trichocarpa vs. I. polycarpa, P. trichocarpa vs. I. orientalis). WGDI31 with the ‘-icl’ parameter was used to identify the intergenomic synteny blocks between P. trichocarpa and others, as well as intragenomic synteny blocks within each species. The Ks between collinear genes was estimated by Nei-Gojobori approach in PAML with the parameter ‘-ks’ of WGDI, and the ‘-bk’ parameter was applied to generate a dot plot of collinear genes and Ks values, visualizing intra- and inter-species synteny. Additionally, the Ks peaks were fitted using the ‘-pf’ parameter, and the density distribution curve of Ks was displayed using the ‘-kf’ parameter. The location of the WGD event was identified based on the comparison of the Ks peaks between paralogs within species and orthologs between species. To address potential inaccuracies in detecting WGD events due to differing substitution rates among candidate species, we further applied KsRates v1.1.3 to bring all the distributions to a common Ks scale by compensating for the differences in synonymous substitution rates relative to the focal species, and the rate-adjusted mixed paralog-ortholog Ks distribution was then used to position adjusted WGD events.
Ecological niche modelling
The data collection for each genus was obtained from the Global Biodiversity Information Facility (GBIF: https://www.gbif.org/), including all species within the genus that have available distribution records, regardless of whether they were sequenced in this study. To remove spatial autocorrelation and sampling bias, the obtained distribution data were subjected to 5 km spatial dilution using SDMtoolbox134, and the final distribution points (Salix: 131,974, Populus: 82,944, Idesia: 1183, Itoa: 113, Flacourtia: 2905, Xylosma: 3802, Scolopia: 3689, Dovyalis: 2039, Dianyuea: 24, Casearia: 14,416) were used for Maxent modeling analysis. Environmental layers for 19 bioclimatic variables at current time (1970-2000) were downloaded from the WorldClim v2.1 dataset (http://www.worldclim.com/) at a spatial resolution of 10 arc minutes (Supplementary Data 4). Pairwise correlations were examined for 19 variables within the distribution of each genus. Taxon distributions were reconstructed using variables with a pairwise Pearson correlation coefficient below 0.8 and the most ecological significance. Ecological niche modeling (ENM) was performed using Maxent 3.4.3135 to simulate potentially suitable habitats under the current climate for each genus. The test output of the models was set at 30%. The accuracy of the model was assessed using the area under the curve (AUC) of the receiver operating characteristic (ROC) plot. AUC values above 0.7 were considered indicative of good model performance136. ArcGIS 10.8 was utilized for mapping the suitable distribution range. To examine niche differences, ENMtools137 was employed to calculate niche overlap statistics Schoener’s D138 and Hellinger’s-based I139, with 100 pseudo-replicates. Values of D and I range from 0 (no ecological niche overlap) to 1 (identical ecological niches).
Ancestral karyotype reconstruction
During karyotype evolution, protochromosomes may have fused or remained as independent chromosomes within existing genomes. We first applied the workflow by Sun et al.23, which identifies protochromosomes and reconstructs ancestral karyotypes by searching for intact chromosomes or chromosome-like syntenic blocks shared across different species. A detailed example of the workflow is available at Github (https://github.com/SunPengChuan/wgdi-example/blob/main/Karyotype_Evolution.md).
Specifically, using the Casearia decandra genome as a reference, we aligned the remaining 13 genomes using WGDI31 with the parameter ‘-d’. Synteny blocks shared between independent chromosomes were first searched in all genomes, and synteny blocks of independent chromosomes identified in at least three genera were assumed to represent Salicaceae protochromosomes. For example, protochromosome 6 (ASK6: homologous to Chr6 of Casearia) of the ancestral Salicaceae karyotype is retained as an independent chromosome in Chr6 of Scolopia, Chr5 of Xylosma and Flacourtia, Chr20 of Itoa, Chr5 of Idesia, Chr13 of Populus and Salix (Supplementary Figs. 9 and 10a). Similarly, ASK1, ASK2, ASK4, ASK5, ASK7, ASK9, ASK10 and ASK11 are retained as independent chromosomes in at least three genera. Therefore, these independent chromosomes were extracted as protochromosomes. Next, all identified synteny blocks were removed from fused chromosomes in existing genomes, and the remaining parts were connected as a chromosomes for a new round of exploration. After removing ASK11 (Chr11 of Casearia), Chr7 and Chr10 in Flacourtia were connected, corresponding to partial fragments of Chr3 and Chr8 in Casearia, which also remain intact in many other species and therefore were identified as ASK8 (Supplementary Fig. 9). After further removing ASK8, Chr2 in Flacourtia corresponds to segments of Chr3 and Chr8 in Casearia, which are independent chromosomes in many other species, and therefore was identified as ASK3 (Supplementary Fig. 9). Ultimately, each extant genome had no remaining genomic blocks, and a total of 11 putative protochromosomes were extracted, hypothesized to form the ancestral Salicaceae karyotype (ASK). Additionally, we also used the MLGO web service (http://www.geneorder.org/)140 to infer ancestral genomes based on information from synteny blocks between species and the phylogenetic tree constructed using single-copy orthologs, which also resulted in 11 ancestral chromosomes.
Moreover, to obtain a more complete ancestral karyotype gene set, we expanded protochromosomes based on six species (Clade II, Dianyuea and C. decandra) that had not undergone the second WGD and retained relatively complete ancestral karyotypes (Supplementary Fig. 10b). Specifically, the genomes were aligned to the initial protochromosomes to identify syntenic blocks. If five or fewer gene clusters surrounded by collinear genes on the chromosomes of an existing species corresponded to two ordered ancestral genes on the protochromosomes, these intermediate genes were added between the two ancestral genes to extend the protochromosomes32. This process resulted in 11 putative protochromosomes containing as many genes as possible. The 14 extant genomes were aligned with the expanded protochromosomes, and WGDI with the parameter ‘-km’ was used to determine the karyotype composition from protochromosomes based on syntenic blocks, allowing the inference of chromosome fusion and evolutionary patterns (Supplementary Fig. 11).
Identification of allopolyploidization and subgenomes
We followed the workflow in the Supplementary Fig. 13 to identify polyploidy types and split subgenomes.
(1) We performed synteny analysis of all 14 Salicaceae species with the reconstructed protochromosomes, and performed Ks calculation and information integration of collinearity fragments using the ‘-ks’ and ‘-bi’ programs of WGDI. Since the Clade I species experienced ‘salicoid’ WGD, we first split the two homologous subgenomes of P. alba var. pyramidalis corresponding to each protochromosome according to the collinear fragment information, while the subgenomes of the remaining 7 Clade I species were split according to the collinear relationship with P. alba var. pyramidalis. The split information was added to the integrated collinear fragment information file, and the homologous gene list between the protochromosomes and each species was obtained through the “-a” program. Finally, the homologous genes of all species were merged to obtain the 1:2 homologous gene groups (HGGs) among CladeII/C. decandra and CladeI/D. turbinata. We then classified the HGGs (C1-C12) based on the number of gene copies in each species, with Full-HGGs and Partial-HGGs used for subsequent analysis (Supplementary Fig. 14).
(2) Gene trees were constructed for each Full and Partial-HGG using RAxML v8.2.11131 with the two Passiflora species as outgroups.
(3) The gene trees were utilized by ASTRAL v.5.6.2141 to infer species trees with quartet scores and posterior probabilities for each protochromosome. The polyploidization type was inferred by counting the proportion of different topologies of the gene tree of protochromosome. In addition, GRAMPA v1.4.0, a topology-based gene-tree reconciliation algorithm, was also used to infer the mode of polyploidization, and the optimal tree with the lowest score was consistent with the topology of the ASTRAL tree. Next, according to the topology of the gene trees, the gene closer to Clade II is classified as the A subgenome, and the other paralogous gene belongs to the B subgenome.
(4) Genes from different subgenomes were mapped onto the chromosomes of their respective species, and the genome was split into A and B subgenomes based on the gene locations.
(5–6) The genes belonging to different subgenomes were concatenated, and the concatenated tree was constructed using RAxML v8.2.11131 to further verify the polyploidization type. Divergence times were estimated using a phylogenetic tree constructed from full-HGGs and MCMCTree132 in the PAML package, based on the divergence time between Populus and Salix.
(7) Finally, yn00 function in PAML132 was used to calculate the Ks values between the A subgenome and Clade II/Dianyuea/Casearia, as well as between the B subgenome and Clade II/Dianyuea/Casearia, thereby obtaining the divergence levels.
Because the Clade I species share ‘salicoid’ WGD event, and genes from different subgenomes were identified in each species, we employed relative rate tests to estimate the evolutionary rate (Ks) after the recent WGD event for each species. Since I. orientalis is at the base of the Clade I, we separately calculated the Ks values of the remaining seven species after they diverged from I. orientalis142.
Gene retention and repeat sequence content of subgenomes
Based on homologous genomic data from 14 species, we compared the genomic and subgenomic characteristics of these species. To quantify the gene retention in the Dianyuea, Clade II species, as well as A and B subgenomes of Clade I species, we first selected HGGs that retained C. decandra and then calculated the percentage of gene retention in these genomes, using non-overlapping windows of 100 genes along the protochromosomes. In addition, based on the results from RepeatMasker and RepeatModeler, we computed the repeat sequence content of genes and their surrounding 2k regions in all genomes. We further searched the genome of Clade I species using the LTRharvest143 and LTRdigest144 programs to de novo detect intact LTRs145. The 5′ and 3′ repeats of each LTR were aligned by MUSCLE v3.8.31146 to estimate the substitution rate, and insertion times were finally estimated by assuming a mutation rate of 2.5 × 10−9 per year147.
Sequencing and analysis of transcriptome data
Leaves, buds, shoot apex, flowers, and fruits of Salicaceae species were collected and immediately frozen in liquid nitrogen. Total RNA was extracted from these samples (Supplementary Data 5) and purified using poly-T oligo-attached magnetic beads. All transcriptome libraries were constructed and sequenced on Illumina HiSeq 2000, DNBSEQ-T7 and BGISEQ-500 platforms. Quality-filtered reads were aligned to their own genomes using HISAT2 v2.1.0117, and then the expression levels (TPM) for each gene were calculated and normalized by StringTie v1.3.3b148. Transcriptome data were used in the following studies: (1)The raw expression levels (TPM) in mature, healthy leaves of 14 species were used to evaluate the expression differences between the A and B subgenomes in each Clade I species. (2) Cross-species comparison of subgenome expression bias during flower and fruit development. This involved selecting retained duplicated gene pairs shared by all four genera, adding homologous genes from P. deltoides, and analyzing tissues from different developmental stages of three species (I. orientalis, I. polycarpa, and P. deltoides). For flower tissues, we collected samples at 4-5 developmental stages spanning from initial inflorescence formation to full flower maturation, based on species-specific inflorescence timelines. Similarly, fruit tissues were sampled at 4-5 stages covering early development to maturity. These sampling intervals were evenly distributed across the developmental timeline to ensure comprehensive coverage of subgenomic regulatory dynamics and minimize bias from single-time-point sampling (Supplementary Data 5). To assess subgenome dominance, we calculated the expression ratio between subgenomes A and B using the formula:
where a positive ratio indicates A-subgenome dominance, while a negative value indicates B-subgenome dominance. To mitigate noise from lowly expressed genes, we applied a TPM threshold of 0.5: gene pairs with TPM below 0.5 for both subgenome A and B in all samples were filtered out; for the remaining gene pairs, if both A- and B-subgenome TPM values were <0.5, the ratio was set to 0 (indicating no detectable differential expression). This filtering step reduced artifacts from negligible expression levels. After that, we performed cluster analysis on the expression ratios across species. (3) To investigate seasonal expression dynamics, we analyzed FT1 and FT2 transcript levels (TPM) in leaves, buds and shoot apex of I. orientalis and I. polycarpa across multiple months. Expression fold changes were calculated relative to the lowest TPM value in each tissue to highlight temporal trends. To prevent excessive fold differences caused by extremely low expression, TPM values below 0.1 were set to 0.1 prior to fold-change calculation.
Sequencing and analysis of epigenetic data
ATAC samples were prepared using leaf tissues149. Vazyme TD501 manual was used to build the ATAC-seq library. For whole genome bisulfite sequencing (WGBS), genomic DNA was extracted from leaves with the DNeasy plant mini kit (Qiagen). After DNA fragmentation, end repair, dA addition to the 3’ end, and adapter addition, the DNA was then subjected to bisulfite treatment, purified, and recovered for PCR amplification150. All libraries were sequenced on Illumina HiSeq 2000, DNBSEQ-T7 and BGISEQ-500 platforms. Quality-filtered reads were aligned to the reference genome using Bowtie2 v2.4.1151 (ATAC-seq) and Bismark v0.22.3152 (WGBS), respectively. We extracted the 2k region upstream of each gene in 10 species and divided it into 20 bins. Subsequently, we counted the number of reads in each bin and normalized it to reads per bin per million mapped reads according to the RPKM method153. This was used to assess the chromatin accessibility of the genomes and subgenomes. Meanwhile, methylation levels of the gene body and the flanking 2k regions were determined by dividing the regions into 30 and 20 bins, respectively.
Gene ontology enrichment analysis
Gene ontology (GO) enrichment analysis was performed on each group of genes using the enricher function in the “clusterProfiler” package154 within R software. After p-value correction using the Benjamini-Hochberg method, terms with q-value < 0.05 were selected as the significant functions.
Identification of conserved noncoding elements (CNEs)
We applied AVID v2.1155 to perform alignments of Salicaceae species and detect Populus-Salix specific CNEs using a 100 bp, 70% identity criterion. Finally, the alignments were visualized using VISTA156.
Vector construction, plant transformation and phenotypic characterization
The coding sequences of ZFP6 and GIS3 were cloned into the binary vector pCXSN, under the control of the Cauliflower Mosaic Virus (CaMV) 35S promoter, to generate the constructs 35S:ZFP6 and 35S:GIS3 (Supplementary Table 8). These constructs were introduced into Agrobacterium tumefaciens strain GV3101, which was subsequently used to transform wild-type (WT) A. thaliana (Col-0) using the floral dip method157. Transgenic lines were selected based on hygromycin resistance, and positive transformants were verified by PCR using gene-specific primers (Supplementary Table 8). Quantitative PCR (qPCR) was performed to assess the expression levels of the transgenes, using Arabidopsis ACTIN as an internal reference gene.
Phenotypic characterization was carried out using a stereomicroscope (Zeiss SteREO Discovery V12), focusing on the 4th rosette leaf at the 14-leaf stage. For each gene construct, two independent transgenic lines were selected, with five biological replicates per line. Leaf area was measured using ImageJ software (http://rsbweb.nih.gov/ij/). The total number of trichomes on each leaf was counted, and trichome density was calculated as the number of trichomes per unit leaf area.
Dual-luciferase assay
The synthetic and cloned fragments of the four CNEs were fused with the 35S minimal promoter to drive the LUC expression as the reporters, and the 35S empty vector was used as a control. These constructs were transiently expressed in N. benthamiana leaves. After incubation in the dark for 2 days and the light for 1 day, the enzyme mixture was prepared according to the manufacturer’s instructions in the Dual-Luciferase Reporter Assay System kit (Promega). Firefly (LUC) and Renilla (internal control, REN) luciferase signals were detected using a multimode reader (Synergy H1; BioTek, Winooski, VT). All primers used are listed in Supplementary Table 9.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All raw sequence data, genome assembly and annotation have been deposited in the National Genome Data Center under BioProject accession PRJCA022976. Source data are provided with this paper.
References
Leebens-Mack, J. H. et al. One thousand plant transcriptomes and the phylogenomics of green plants. Nature 574, 679–685 (2019).
Jiao, Y. et al. Phylogenomic analysis reveals ancient genome duplications in seed plant and angiosperm history. Nature 473, 97–100 (2011).
Albert, V. A. et al. The Amborella genome and the evolution of flowering plants. Science 342, 1241089 (2013).
Force, A. et al. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151, 1531–1545 (1999).
Adams, K. L. & Wendel, J. F. Polyploidy and genome evolution in plants. Curr. Opin. Plant Biol. 8, 135–141 (2005).
Doyle, J. J. et al. Evolutionary genetics of genome merger and doubling in plants. Annu. Rev. Genet. 42, 443–461 (2008).
Hegarty, M. J. & Hiscock, S. J. Genomic clues to the evolutionary success of polyploid plants. Curr. Biol. 18, R435–R444 (2008).
Soltis, P. S., Liu, X., Marchant, D. B., Visger, C. J. & Soltis, D. E. Polyploidy and novelty: Gottlieb’s legacy. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 369, 20130351 (2014).
Soltis, P. S., Marchant, D. B., Van de Peer, Y. & Soltis, D. E. Polyploidy and genome evolution in plants. Curr. Opin. Genet. Dev. 35, 119–125 (2015).
Soltis, P. S. & Soltis, D. E. Ancient WGD events as drivers of key innovations in angiosperms. Curr. Opin. Plant Biol. 30, 159–165 (2016).
Van de Peer, Y., Maere, S. & Meyer, A. The evolutionary significance of ancient genome duplications. Nat. Rev. Genet. 10, 725–732 (2009).
Van de Peer, Y., Mizrachi, E. & Marchal, K. The evolutionary significance of polyploidy. Nat. Rev. Genet. 18, 411–424 (2017).
Fawcett, J. A., Maere, S. & Van De Peer, Y. Plants with double genomes might have had a better chance to survive the cretaceous-tertiary extinction event. Proc. Natl. Acad. Sci. USA 106, 5737–5742 (2009).
Vanneste, K., Maere, S. & Van de Peer, Y. Tangled up in two: a burst of genome duplications at the end of the cretaceous and the consequences for plant evolution. Philos. Trans. R. Soc. Lond. B, Biol. Sci. 369, 20130353 (2014).
Wu, S., Han, B. & Jiao, Y. Genetic contribution of paleopolyploidy to adaptive evolution in angiosperms. Mol. Plant 13, 59–71 (2020).
Schnable, J. C., Springer, N. M. & Freeling, M. Differentiation of the maize subgenomes by genome dominance and both ancient and ongoing gene loss. Proc. Natl. Acad. Sci. USA 108, 4069–4074 (2011).
Wendel, J. F., Jackson, S. A., Meyers, B. C. & Wing, R. A. Evolution of plant genome architecture. Genome Biol. 17, 1–14 (2016).
IWGSC. et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361, eaar7191 (2018).
Chen, Z. J. et al. Genomic diversifications of five Gossypium allopolyploid species and their impact on cotton improvement. Nat. Genet. 52, 525–533 (2020).
Ma, P. F. et al. Genome assemblies of 11 bamboo species highlight diversification induced by dynamic subgenome dominance. Nat. Genet. 56, 710–720 (2024).
Wang, X. et al. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43, 1035–1039 (2011).
Zhang, R. G. et al. Subgenome-aware analyses suggest a reticulate allopolyploidization origin in three Papaver genomes. Nat. Commun. 14, 2204 (2023).
Sun, P. et al. Subgenome-aware analyses reveal the genomic consequences of ancient allopolyploid hybridizations throughout the cotton family. Proc. Natl. Acad. Sci. USA 121, e2313921121 (2024).
Barker, M. S., Arrigo, N., Baniaga, A. E., Li, Z. & Levin, D. A. On the relative abundance of autopolyploids and allopolyploids. N. Phytol. 210, 391–398 (2016).
Fowler, N. L. & Levin, D. A. Ecological constraints on the establishment of a novel polyploid in competition with its diploid progenitor. Am. Nat. 124, 703–711 (1984).
Ramsey, J. Polyploidy and ecological adaptation in wild yarrow. Proc. Natl. Acad. Sci. USA 108, 7096–7101 (2011).
Parisod, C. & Broennimann, O. Towards unified hypotheses of the impact of polyploidy on ecological niches. N. Phytol. 212, 540–542 (2016).
Luo, X. et al. Chasing ghosts: allopolyploid origin of Oxyria sinensis (Polygonaceae) from its only diploid congener and an unknown ancestor. Mol. Ecol. 26, 3037–3049 (2017).
Stull, G. W., Pham, K. K., Soltis, P. S. & Soltis, D. E. Deep reticulation: the long legacy of hybridization in vascular plant evolution. Plant J. 114, 743–766 (2023).
Murat, F., Armero, A., Pont, C., Klopp, C. & Salse, J. Reconstructing the genome of the most recent common ancestor of flowering plants. Nat. Genet. 49, 490–496 (2017).
Sun, P. et al. WGDI: A user-friendly toolkit for evolutionary analyses of whole-genome duplications and ancestral karyotypes. Mol. Plant 15, 1841–1851 (2022).
Jiang, X. et al. Chromosome fusions shaped karyotype evolution and evolutionary relationships in the model family Brassicaceae. Nat. Commun. 16, 1–10 (2025).
Wang, Z. et al. A high-quality Buxus austro-yunnanensis (Buxales) genome provides new insights into karyotype evolution in early eudicots. BMC Biol. 20, 216 (2022).
Stettler, R. F. Biology of Populus and its Implications for Management and Conservation, Vol. 542 (NRC Research Press, 1996).
Chase, M. W. et al. When in doubt, put it in flacourtiaceae: a molecular phylogenetic analysis based on plastid rbcL DNA sequences. Kew Bull. 57, 141–181 (2002).
Sanderson, B. J. et al. Phylogenomics reveals patterns of ancient hybridization and differential diversification that contribute to phylogenetic conflict in willows, poplars, and close relatives. Syst. Biol. 72, 1120–1232 (2023).
Argus, G. W. Infrageneric classification of Salix (Salicaceae) in the new world. Syst. Bot. Monogr. 52, 1–121 (1997).
Tuo-Ya, D. Origin, divergence and geographical distribution of Salicaceae. Plant Divers 17, 1 (1995).
Cronk, Q. C., Needham, I. & Rudall, P. J. Evolution of catkins: inflorescence morphology of selected Salicaceae in an evolutionary and developmental context. Front. Plant Sci. 6, 1030 (2015).
Lemke, D. E. A synopsis of Flacourtiaceae. Aliso 12, 29–43 (1988).
Zhang, Z.-S., Zeng, Q.-Y. & Liu, Y.-J. Frequent ploidy changes in Salicaceae indicates widespread sharing of the salicoid whole genome duplication by the relatives of Populus L. and Salix L. BMC Plant Biol. 21, 1–17 (2021).
Steyn, E., Smith, G. & Van Wyk, A. Functional and taxonomic significance of seed structure in Salix mucronata (Salicaceae). Bothalia 34, 53–59 (2004).
Cronk, Q. Plant eco-devo: the potential of poplar as a model organism. N. Phytol. 166, 39–48 (2005).
Jansson, S. & Douglas, C. J. Populus: a model system for plant biology. Annu. Rev. Plant Biol. 58, 435–458 (2007).
Tuskan, G. A. et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313, 1596–1604 (2006).
Dai, X. et al. The willow genome and divergent evolution from poplar after the common genome duplication. Cell Res. 24, 1274–1277 (2014).
Rodgers-Melnick, E. et al. Contrasting patterns of evolution following whole genome versus tandem duplication events in Populus. Genome Res. 22, 95–105 (2012).
Liu, Y. et al. Two highly similar poplar paleo-subgenomes suggest an autotetraploid ancestor of Salicaceae plants. Front. Plant Sci. 8, 252555 (2017).
Wang, J. et al. Polyploidy index and its implications for the evolution of polyploids. Front. Genet. 10, 807 (2019).
Cai, L. et al. Widespread ancient whole-genome duplications in Malpighiales coincide with Eocene global climatic upheaval. N. Phytol. 221, 565–576 (2019).
Zhang, Z. et al. Improved genome assembly provides new insights into genome evolution in a desert poplar (Populus euphratica). Mol. Ecol. Resour. 20, 781–794 (2020).
Wang, Y. et al. Four near-complete genome assemblies reveal the landscape and evolution of centromeres in Salicaceae. Genome Biol. 26, 111 (2025).
Li, M. et al. Intergeneric relationships within the family Salicaceae sl based on plastid phylogenomics. Int. J. Mol. Sci. 20, 3788 (2019).
Shang, C., Liao, S., Guo, Y. J. & Zhang, Z. X. Dianyuea gen. nov.(Salicaceae: Scyphostegioideae) from southwestern China. Nord. J. Bot. 35, 499–505 (2017).
Cheng, F. et al. Gene retention, fractionation and subgenome differences in polyploid plants. Nat. Plants 4, 258–268 (2018).
Kardailsky, I. et al. Activation tagging of the floral inducer FT. Science 286, 1962–1965 (1999).
Kobayashi, Y., Kaya, H., Goto, K., Iwabuchi, M. & Araki, T. A pair of related genes with antagonistic roles in mediating flowering signals. Science 286, 1960–1962 (1999).
Liu, H. et al. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 322, 1535–1539 (2008).
Doyle, M. R. et al. HUA2 is required for the expression of floral repressors in Arabidopsis thaliana. Plant J. 41, 376–385 (2005).
Kim, S. H. et al. Phosphorylation of the transcriptional repressor MYB15 by mitogen-activated protein kinase 6 is required for freezing tolerance in Arabidopsis. Nucleic Acids Res. 45, 6613–6627 (2017).
Zhang, P. et al. Arabidopsis ADF5 acts as a downstream target gene of CBFs in response to low-temperature stress. Front. Cell Dev. Biol. 9, 635533 (2021).
Takahashi, D. et al. Structural changes in cell wall pectic polymers contribute to freezing tolerance induced by cold acclimation in plants. Curr. Biol. 34, 958–968.e5 (2024).
Sun, L. et al. Glabrous Inflorescence Stems3 (GIS3) regulates trichome initiation and development in Arabidopsis. N. Phytol. 206, 220–230 (2015).
Zhou, Z. et al. Zinc Finger Protein 6 (ZFP6) regulates trichome initiation by integrating gibberellin and cytokinin signaling in Arabidopsis thaliana. N. Phytol. 198, 699–708 (2013).
Liu, Y., Yang, S., Khan, A. R. & Gan, Y. TOE1/TOE2 interacting with GIS to control trichome development in Arabidopsis. Int. J. Mol. Sci. 24, 6698 (2023).
Zhu, Y. et al. TERMINAL FLOWER 1-FD complex target genes and competition with FLOWERING LOCUS T. Nat. Commun. 11, 5118 (2020).
Kumimoto, R. W. et al. The Nuclear Factor Y subunits NF-YB2 and NF-YB3 play additive roles in the promotion of flowering by inductive long-day photoperiods in Arabidopsis. Planta 228, 709–723 (2008).
Jung, J.-H., Lee, H.-J., Ryu, J. Y. & Park, C.-M. SPL3/4/5 integrate developmental aging and photoperiodic signals into the FT-FD module in Arabidopsis flowering. Mol. Plant 9, 1647–1659 (2016).
Song, H.-R. et al. The RNA binding protein ELF9 directly reduces suppressor of overexpression of CO1 transcript levels in Arabidopsis, possibly via nonsense-mediated mRNA decay. Plant Cell 21, 1195–1211 (2009).
Zhang, Z. et al. Convergence of the 26S proteasome and the REVOLUTA pathways in regulating inflorescence and floral meristem functions in Arabidopsis. J. Exp. Bot. 62, 359–369 (2011).
Yoo, C.-M., Quan, L. & Blancaflor, E. B. Divergence and redundancy in CSLD2 and CSLD3 function during Arabidopsis thaliana root hair and female gametophyte development. Front. Plant Sci. 3, 111 (2012).
Throude, M. et al. Structure and expression analysis of rice paleo duplications. Nucleic Acids Res. 37, 1248–1259 (2009).
Huminiecki, L. & Conant, G. C. Polyploidy and the evolution of complex traits. Int. J. Evol. Biol. 2012, 292068 (2012).
Hsu, C.-Y. et al. Flowering Locus T duplication coordinates reproductive and vegetative growth in perennial poplar. Proc. Natl. Acad. Sci. USA 108, 10756–10761 (2011).
Yu, C.-W. et al. Histone Deacetylase6 interacts with Flowering Locus D and regulates flowering in Arabidopsis. Plant Physiol. 156, 173–184 (2011).
Bertran Garcia de Olalla, E. et al. Coordination of shoot apical meristem shape and identity by APETALA2 during floral transition in Arabidopsis. Nat. Commun. 15, 6930 (2024).
Zhao, X., Heng, Y., Wang, X., Deng, X. W. & Xu, D. A positive feedback loop of BBX11-BBX21-HY5 promotes photomorphogenic development in Arabidopsis. Plant Commun. 1, 100045 (2020).
Li, P., Li, X. & Jiang, M. CRISPR/Cas9-mediated mutagenesis of WRKY3 and WRKY4 function decreases salt and Me-JA stress tolerance in Arabidopsis thaliana. Mol. Biol. Rep. 48, 5821–5832 (2021).
André, D. et al. Flowering Locus T paralogs control the annual growth cycle in Populus trees. Curr. Biol. 32, 2988–2996. e4 (2022).
Sawa, S., Ito, T., Shimura, Y. & Okada, K. Filamentous Flower controls the formation and development of Arabidopsis inflorescences and floral meristems. Plant Cell 11, 69–86 (1999).
Li, W. et al. The U-Box/ARM E3 ligase PUB13 regulates cell death, defense, and flowering time in Arabidopsis. Plant Physiol. 159, 239–250 (2012).
Zhang, C. et al. Gibberellin signaling modulates flowering via the DELLA-BRAHMA-NF-YC module in Arabidopsis. Plant Cell 35, 3470–3484 (2023).
Liu, Z. et al. ENO2 affects the seed size and weight by adjusting cytokinin content and forming ENO2-bZIP75 complex in Arabidopsis thaliana. Front. Plant Sci. 11, 574316 (2020).
Di Marzo, M. et al. Cell wall modifications by α-XYLOSIDASE1 are required for control of seed and fruit size in Arabidopsis. J. Exp. Bot. 73, 1499–1515 (2021).
Cai, X. et al. Impacts of allopolyploidization and structural variation on intraspecific diversification in Brassica rapa. Genome Biol. 22, 166 (2021).
Marcussen, T. et al. Ancient hybridizations among the ancestral genomes of bread wheat. Science 345, 1250092 (2014).
Ramírez-González, R. et al. The transcriptional landscape of polyploid wheat. Science 361, eaar6089 (2018).
Wang, J. et al. A common whole-genome paleotetraploidization in cucurbitales. Plant Physiol. 190, 2430–2448 (2022).
Guo, X. et al. Linked by ancestral bonds: multiple whole-genome duplications and reticulate evolution in a Brassicaceae tribe. Mol. Biol. Evol. 38, 1695–1714 (2021).
Mandakova, T., Li, Z., Barker, M. S. & Lysak, M. A. Diverse genome organization following 13 independent mesopolyploid events in Brassicaceae contrasts with convergent patterns of gene retention. Plant J. 91, 3–21 (2017).
Zhang, T. et al. Phylogenomic profiles of whole-genome duplications in Poaceae and landscape of differential duplicate retention and losses among major Poaceae lineages. Nat. Commun. 15, 3305 (2024).
Huang, C.-H. et al. Multiple polyploidization events across Asteraceae with two nested events in the early history revealed by nuclear phylogenomics. Mol. Biol. Evol. 33, 2820–2835 (2016).
Zhao, Y. et al. Nuclear phylotranscriptomics and phylogenomics support numerous polyploidization events and hypotheses for the evolution of rhizobial nitrogen-fixing symbiosis in Fabaceae. Mol. Plant 14, 748–773 (2021).
Li, Z. et al. Multiple large-scale gene and genome duplications during the evolution of hexapods. Proc. Natl. Acad. Sci. USA 115, 4713–4718 (2018).
Lien, S. et al. The Atlantic salmon genome provides insights into rediploidization. Nature 533, 200–205 (2016).
Zhang, L. et al. Bioinformatic analysis of chromatin organization and biased expression of duplicated genes between two poplars with a common whole-genome duplication. Hortic. Res. 8, 62 (2021).
Li, H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Roach, M. J., Schmidt, S. A. & Borneman, A. R. Purge Haplotigs: allelic contig reassignment for third-gen diploid genome assemblies. BMC Bioinforma. 19, 1–10 (2018).
Cheng, H., Concepcion, G. T., Feng, X., Zhang, H. & Li, H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat. Methods 18, 170–175 (2021).
Burton, J. N. et al. Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nat. Biotechnol. 31, 1119–1125 (2013).
Zhou, C., McCarthy, S. A. & Durbin, R. YaHS: yet another Hi-C scaffolding tool. Bioinformatics 39, btac808 (2023).
Durand, N. C. et al. Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom. Cell Syst. 3, 99–101 (2016).
Kajitani, R. et al. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res. 24, 1384–1395 (2014).
Chen, N. Using repeat masker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinform. 5, 4.10. 1–4.10. 14 (2004).
Bao, W., Kojima, K. K. & Kohany, O. Repbase update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 6, 1–6 (2015).
Price, A. L., Jones, N. C. & Pevzner, P. A. De novo identification of repeat families in large genomes. Bioinformatics 21, i351–i358 (2005).
Stanke, M. et al. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 34, W435–W439 (2006).
Yang, W. et al. The draft genome sequence of a desert tree Populus pruinosa. Gigascience 6, gix075 (2017).
Ma, J. et al. Genome sequence and genetic transformation of a widely distributed and cultivated poplar. Plant Biotechnol. J. 17, 451–460 (2019).
Zhou, R. et al. Characterization of a large sex determination region in Salix purpurea L.(Salicaceae). Mol. Genet. Gen.293, 1437–1452 (2018).
Iniative, A. G. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815 (2000).
Camacho, C. et al. BLAST+: architecture and applications. BMC Bioinforma. 10, 1–9 (2009).
Birney, E., Clamp, M. & Durbin, R. GeneWise and genomewise. Genome Res. 14, 988–995 (2004).
Campbell, M. A., Haas, B. J., Hamilton, J. P., Mount, S. M. & Buell, C. R. Comprehensive analysis of alternative splicing in rice and comparative analyses with Arabidopsis. BMC Gen.7, 1–17 (2006).
Haas, B. J. et al. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol. 9, 1–22 (2008).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015).
Rhie, A., Walenz, B. P., Koren, S. & Phillippy, A. M. Merqury: reference-free quality, completeness, and phasing assessment for genome assemblies. Genome Biol. 21, 1–27 (2020).
Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V. & Zdobnov, E. M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212 (2015).
Li, W. & Godzik, A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659 (2006).
Haas, B. J. et al. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 31, 5654–5666 (2003).
Wang, D. et al. Repeated turnovers keep sex chromosomes young in willows. Genome Biol. 23, 1–23 (2022).
Zhou, R. et al. A willow sex chromosome reveals convergent evolution of complex palindromic repeats. Genome Biol. 21, 1–19 (2020).
Ma, D. et al. Chromosome-level reference genome assembly provides insights into aroma biosynthesis in passion fruit (Passiflora edulis). Mol. Ecol. Resour. 21, 955–968 (2021).
Costa, Z. P. et al. A genome sequence resource for the genus Passiflora, the genome of the wild diploid species Passiflora organensis. Plant Genome 14, e20117 (2021).
Raymond, O. et al. The Rosa genome provides new insights into the domestication of modern roses. Nat. Genet. 50, 772–777 (2018).
Shi, X. et al. The complete reference genome for grapevine (Vitis vinifera L.) genetics and breeding. Hortic. Res. 10, uhad061 (2023).
Emms, D. M. & Kelly, S. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16, 1–14 (2015).
Katoh, K., Misawa, K., Kuma, K. I. & Miyata, T. MAFFT: a novel method for rapid multiple sequence alignment based on fast fourier transform. Nucleic Acids Res. 30, 3059–3066 (2002).
Suyama, M., Torrents, D. & Bork, P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34, W609–W612 (2006).
Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014).
Yang, Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Brown, J. L. SDM toolbox: a python-based GIS toolkit for landscape genetic, biogeographic and species distribution model analyses. Methods Ecol. Evol. 5, 694–700 (2014).
Phillips, S. J., Anderson, R. P. & Schapire, R. E. Maximum entropy modeling of species geographic distributions. Ecol. Model. 190, 231–259 (2006).
Fielding, A. H. & Bell, J. F. A review of methods for the assessment of prediction errors in conservation presence/absence models. Environ. Conserv. 24, 38–49 (1997).
Warren, D. L. et al. ENMTools 1.0: An R package for comparative ecological biogeography. Ecography 44, 504–511 (2021).
Schoener, T. W. The Anolis lizards of Bimini: resource partitioning in a complex fauna. Ecology 49, 704–726 (1968).
Warren, D. L., Glor, R. E. & Turelli, M. Environmental niche equivalency versus conservatism: quantitative approaches to niche evolution. Evolution 62, 2868–2883 (2008).
Hu, F., Lin, Y. & Tang, J. MLGO: phylogeny reconstruction and ancestral inference from gene-order data. BMC Bioinform. 15, 1–6 (2014).
Zhang, C., Rabiee, M., Sayyari, E. & Mirarab, S. ASTRAL-III: polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinforma. 19, 15–30 (2018).
Ding, Y. et al. Genome structure-based Juglandaceae phylogenies contradict alignment-based phylogenies and substitution rates vary with DNA repair genes. Nat. Commun. 14, 617 (2023).
Ellinghaus, D., Kurtz, S. & Willhoeft, U. LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinforma. 9, 1–14 (2008).
Steinbiss, S., Willhoeft, U., Gremme, G. & Kurtz, S. Fine-grained annotation and classification of de novo predicted LTR retrotransposons. Nucleic Acids Res. 37, 7002–7013 (2009).
Zhou, S. et al. A comprehensive annotation dataset of intact LTR retrotransposons of 300 plant genomes. Sci. Data 8, 174 (2021).
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004).
Ingvarsson, P. K. Multilocus patterns of nucleotide polymorphism and the demographic history of Populus tremula. Genetics 180, 329–340 (2008).
Pertea, M. et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295 (2015).
Wang, W. et al. Chromatin accessibility dynamics insight into crosstalk between regulatory landscapes in poplar responses to multiple treatments. Tree Physiol. 43, 1023–1041 (2023).
Su, Y. et al. Single-base-resolution methylomes of Populus euphratica reveal the association between DNA methylation and salt stress. Tree Genet. Genom. 14, 1–11 (2018).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Krueger, F. & Andrews, S. R. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27, 1571–1572 (2011).
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L. & Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628 (2008).
Yu, G., Wang, L.-G., Han, Y. & He, Q.-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS: J. Integr. Biol. 16, 284–287 (2012).
Bray, N., Dubchak, I. & Pachter, L. AVID: A global alignment program. Genome Res 13, 97–102 (2003).
Frazer, K. A., Pachter, L., Poliakov, A., Rubin, E. M. & Dubchak, I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 32, W273–W279 (2004).
Clough, S. J. & Bent, A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998).
Acknowledgements
This work was supported by the National Key Research and Development Program of China (2021YFD2201100, 2021YFD2200202 and 2016YFD0600101), National Natural Science Foundation of China (31922061, 32271828 and 32071732), Fundamental Research Funds for the Central Universities (2020SCUNL207, SCU2021D006 and 2020SCUNL103), and the US National Science Foundation (1542599). We thank Susanne S. Renner (Department of Biology, Washington University, Saint Louis, USA.) for insightful comments. We thank Yanping Su, Fuchuan Wu (Center for Gardening and Horticulture, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Science), and Lei Zhang (College of Biological Science & Engineering, North Minzu University, Yinchuan, China) for providing plant samples.
Author information
Authors and Affiliations
Contributions
T.M., J.Q.L. and M.O. led the project. T.M., J.Q.L., M.O., D.Y.W. and Z.X.X. conceived and designed the research. D.Y.W., M.M.L., W.L.Y., J.L.Z., L.X.S., P.C.S., Y.L.L., Y.C., J.X.X., Y.B.W., H.H., and J.L.L. performed data analysis. K.C., L.X., A.F., X.N.H. and T.N.L. performed the experiments. D.Y.W., M.M.L., Z.Q.L. and Z.X.X. collected samples. Y.Z.J. and Q.J.H. provided comments for improving the manuscript. D.Y.W. and T.M. drafted the manuscript. J.Q.L. and M.O. edited the manuscript. All authors approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Yuannian Jiao and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wang, D., Li, M., Yang, W. et al. Ancient allopolyploidy and specific subgenomic evolution drove the radiation of poplars and willows. Nat Commun 16, 6881 (2025). https://doi.org/10.1038/s41467-025-62178-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-62178-y