Abstract
The ability of Staphylococcus aureus to adapt and thrive in diverse host niches adds to the challenge in combating this ubiquitous pathogen. While extensive research has been pursued on the adaptive mechanisms of methicillin-resistant S. aureus (MRSA) in various infection models, a comprehensive analysis of its fitness across different host niches is lacking. In this study, we employ transposon sequencing to analyze the adaptive strategies of MRSA in various infection niches. Our analysis encompasses a cell model that mimics an intracellular niche, human blood, which represents a major extracellular environment as well as a major intermediary route encountered by bacteria during systemic infection, and a male murine sepsis model that recapitulates intra-organ environments. Our findings reveal substantial differences in the genetic determinants essential for bacterial survival in intracellular and blood environments. Moreover, we show that each organ imposes unique growth constraints, thus fostering heterogeneity within the mutant population that can enter and survive in each organ of the mouse. By comparing genes important for survival across all examined host environments, we identify 27 core genes that represent potential therapeutic targets for treating S. aureus infections. Additionally, our findings aid in understanding how bacteria adapt to diverse host environments.
Similar content being viewed by others
Introduction
Bacteremia caused by methicillin-resistant Staphylococcus aureus (MRSA) is a leading cause of human death resulting from bacterial infections, posing a serious threat to public health1,2,3. However, treatment of MRSA infection has been hampered not only by the continual emergence of evolved MRSA strains but also by the acquisition of mechanisms to invade and survive in the intracellular niches of host cells4,5,6. Therefore, identification of the underlying networks and genes required for the intracellular entry and survival of S. aureus can provide essential information for the development of antibacterial therapies.
S. aureus is commonly found as a commensal in different body parts of the human host, but exposure to different niches and conditions can transform it into a devastating pathogen that causes permanent organ damage or death7,8. Previously classified as a strictly extracellular pathogen, S. aureus is now also considered to be able to establish an intracellular infection4,5,6,9. It begins its infection by colonizing various host surfaces and matrices and establishing extracellular infections. Then, it can proliferate and reach the bloodstream of the host and further penetrate major organs such as the liver and lungs; while traveling around the bloodstream, S. aureus can also reach intracellular niches to establish infection. To switch to its pathogenic mode in all these different host niches, S. aureus relies on a fine-tuned network of transcription factors and two-component systems (TCSs). These regulatory systems not only control its ability to adapt to different nutrient environments and cope with different levels of oxidative stress, but also facilitate activation of protective mechanisms, such as biofilm formation, which contribute to its virulence and survival in the host10,11. The ability of S. aureus to switch rapidly from living extracellularly to living intracellularly is also attributed to its metabolic and physiological plasticity, which is controlled by these numerous regulators. Among these regulators, Agr, SaeRS, and the recently discovered SrrAB are thought to be hubs of the regulatory circuitry for S. aureus survival in the host10,12,13. For example, SaeRS governs key reprogramming during infection by signaling for the induction of stress response molecules, cation transporters, and membrane remodeling proteins14,15. These mechanisms of regulation for survival represent potential druggable targets to antagonize MRSA persistence and improve clinical outcomes.
The plasticity of the metabolic and regulatory circuits of S. aureus allows it to efficiently establish infections in almost any niche, whether extracellular, intracellular, or body organ, and renders it capable of quickly switching between niches. Over the past decades, mechanisms governing in vivo infection or intracellular survival of S. aureus have been extensively studied using transcriptomic and genetic profiling16,17,18,19. However, results obtained from one infection model may not necessarily apply to another. To address the need for a comprehensive analysis of adaptive mechanisms required for S. aureus survival in various niches, we investigate in this study how the fitness of S. aureus transposon mutants changes in different niches. We performed transposon sequencing (Tn-seq) on a pooled S. aureus mutant library using a cell infection model that recapitulates an intracellular niche, human blood that represents an extracellular niche as well as a major intermediary route for S. aureus to reach the organs, and a mouse sepsis model to show adaptation mechanisms in major organs involved in infection. We find that most genes and regulons required for intracellular infection are distinct from those required for survival in an extracellular environment. The results from the in vivo sepsis model bear strong similarities to the results for human blood, suggesting that S. aureus is primarily exposed to the extracellular rather than the intracellular environment during sepsis. However, from the sepsis model, we also found that the environment of each organ imposes different constraints on the growth of S. aureus, uniquely shaping the S. aureus population within each organ. Our results suggest that targeting genes essential during infection may combat S. aureus infection and the progression of sepsis, and we describe a core set of genes important for infection in all environments that may be particularly effective therapeutic targets.
Results
Transposon sequencing reveals genetic remodeling important for S. aureus survival in intracellular infection
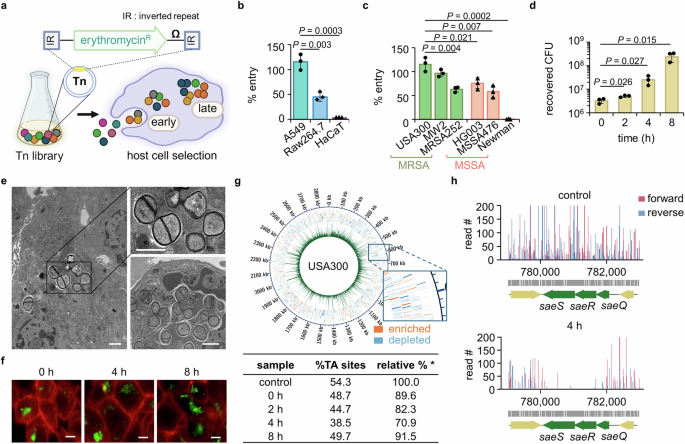
First, we established a cell infection model using a S. aureus transposon (Tn) mutant library and analyzed the genes required for the early and late phases of intracellular infection (Fig. 1a). We wanted a minimal bottleneck effect for our genetic analysis, so we first assessed an infection efficiency of the methicillin-resistant S. aureus (MRSA) strain USA300 in three cell lines relevant to S. aureus infection. A549 cells derived from the lungs allowed entry of almost all bacterial cells, while 50% and less than 10% of the bacteria entered murine macrophage (Raw264.7) cells and human keratinocyte (HaCaT) cells, respectively (Fig. 1b). Based on the finding that A549 cells are permissive to bacterial infection and survival compared to the other cell lines, we selected this cell line for our model. We next examined whether A549 cells are permissive to the entry of several other MRSA strains or methicillin-sensitive S. aureus (MSSA) strains. The MRSA strains USA300 and MW2 demonstrated the highest infection rates, whereas MSSA strain Newman was non-invasive (Fig. 1c). These results show that there can be very large differences in cell infection efficiency among different pathogenic S. aureus strains20. We selected USA300 for genetic profiling because it represents the predominant community-acquired MRSA clone in North America and is therefore of high current interest21,22. We found that USA300 is not only able to efficiently invade A549 cells but also proliferates intracellularly up to 8 h post-infection (Fig. 1d), consistent with previous observations5,23. Interestingly, we did not observe proliferation for the MSSA strain HG003, which is in the same clonal complex as USA300 (Supplementary Fig. 1), showing that USA300 is better adapted for intracellular growth. To confirm that USA300 cells indeed grow and divide intracellularly, we examined infected A549 cells using transmission electron microscopy (Fig. 1e). At 4 h post-infection, we observed multiple S. aureus cells with septa, indicating that these bacteria were in the process of division inside the host cells when the samples were processed. Growth and division in the intracellular environment were also confirmed by fluorescence microscopy using a USA300 strain expressing FtsZ-GFP (Fig. 1f).
a To determine genes essential for the early entry into and late survival in host cells, A549 cells were infected with the USA300 Tn library for 1.5 h, after which extracellular bacteria were removed by gentamicin. Samples collected right after gentamicin were defined as 0 h throughout the manuscript. Image created in BioRender (Lee, W. (2025) https://BioRender.com/bytpvde). Extent of infection (b) of USA300 into host cells (A549, Raw264.7, and HaCaT) and (c) of MRSA and MSSA strains into A549 cells were determined by cell infection assay (\(\%\;{\rm{entry}}=\,\frac{{\rm{CFU}}\; {\rm{recovered}}\; {\rm{at}}\;0\;{\rm{h}}}{{\rm{input}}\; {\rm{CFU}}}\times 100\)). d The CFU were counted at 0–8 h post-infection as a measure of USA300 survival inside the A549 cells. Data in b–d are presented as the mean ± SD (n = 3 biological replicates); P values are calculated by a two-sided, unpaired Student’s t-test. Source data for b–d are available in the Source Data file. e The intracellular bacteria were examined by transmission electron microscopy at 4 h post-infection (scale bar = 1 μm). f Entry and survival of FtsZ-GFP-expressing USA300 in A549 cells were inspected by fluorescence microscopy. The cell membrane and bacteria are shown in red and green, respectively (scale bar = 5 μm). The images in e–f are representative of three independent experiments. g The entire sequenced genome at 0 h was plotted using Circos with the green peaks representing read counts for each TA site and blue and orange outer squares representing genes with counts lower (depleted) and higher (enriched) than those of the control library, respectively. The % TA sites (% TA sites detected out of all TA sites in the genome) show the coverage of the genome by Tn-seq and are presented in a table (\( * \;{\rm{relative}}\%=\,\frac{\%{\rm{TA}}\; {\rm{sites}}\; {\rm{of}}\; {\rm{an}}\; {\rm{experimental}}\; {\rm{sample}}}{\%{\rm{TA}}\; {\rm{sites}}\; {\rm{of}}\; {\rm{a}}\; {\rm{control}}\; {\rm{Tn}}\; {\rm{library}}\;(54.3\%)} \times 100\)). h The read counts generated by sequencing in both forward (red) and reverse (blue) directions for each TA site within the saeQRS locus of the control and 4 h samples were plotted using R.
We next infected the A549 cells with a USA300 Tn mutant library at a multiplicity of infection (MOI) of 10, which resulted in full recovery of input bacteria. The mutant library used in this study is a high-density library containing more than 760,000 Tn mutants made by phage-based transposition24. The number of mutants exceeds the number of TA sites in the S. aureus genome because the library is composed of six bar-coded sublibraries, each produced using a slightly different Tn construct. Some constructs have outward-facing promoters that can minimize the polar effects of insertion on downstream gene expression. Tn mutants recovered right after infection (at 0 h post-infection) were used to identify genes required for the early stage of infection, while Tn mutants recovered at 2, 4, and 8 h post-infection were analyzed to identify genes required for intracellular proliferation (Fig. 1a). A Tn library not exposed to the host cells was used as an input control. After sequencing, the read counts due to the Tn insertions for each gene were mapped to the genome of USA300, generating a complete profile for all time points. The percentage of TA sites containing the Tn insertions was comparable to the input at each time point, suggesting a minimal bottleneck effect (Fig. 1g, table). Moreover, Circos plots of the read counts for each TA site confirm high coverage of Tn mutants across the genome (Fig. 1g and Supplementary Fig. 2, green bars). In these plots, genes for which reads were depleted and enriched compared with the input control are represented in blue and orange (Fig. 1g, magnification), respectively. Among the genes required for intracellular survival (depleted reads compared with the input control), we found SaeRS, a TCS previously shown to be required during host invasion (Fig. 1h), which validates our genetic analysis25.
S. aureus faces nutrient limitation during intracellular infection
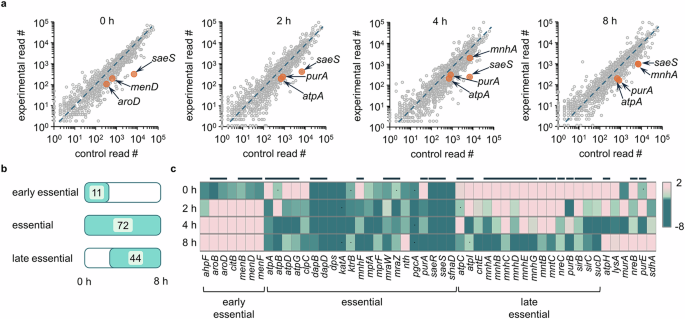
Since we aimed to identify genes required for intracellular infection, we focused on only the depleted genes in the Tn-seq analysis. For this, we plotted the read counts of the genes from each sample against those of the input and set up a significance cut-off as 1.3-fold decrease of log2 fold change with a P value ≤ 0.05 (Fig. 2a). We have excluded the genes with a small read count (<50) in the control input library from our further analysis as done in previous studies26,27. Based on our Tn-seq datasets, we categorized genes as early or late essential based on changes of read counts during an 8-h infection period. A total of 226 genes were identified as essential for intracellular infection from the early stages to the proliferation (late) stages; 11 genes were categorized as early essential, 44 genes as essential for proliferation (late essential), and 72 genes essential for both the early and proliferation stages (Fig. 2b). Genes that were substantially changed and clustered were represented as a heat map (Fig. 2c) with a complete list provided in Supplementary Fig. 3. Importantly, as marked by the black lines above the genes in Fig. 2c, multiple genes belonging to the same functional group are consistently depleted, thereby providing cross-validation for their functional importance.
a Read counts from samples collected at 0 h and during intracellular survival (2–8 h post-infection) were plotted against the read counts of the respective genes of the control (not exposed to host cells) sample. The saeS gene is represented on the graphs as a gene depleted at all time points, while menD and aroD are representative genes depleted at an earlier stage of infection. Genes, such as purA, atpA, and mnhA, represent genes that appear later during the intracellular infection. Source data is available in the Source Data file. b Genes that demonstrated a substantial decrease in their read counts (log2 fold change ≤ −1.3, P value ≤ 0.05) were classified into genes that are entry-specific (early essential; substantially depleted at 0–2 h post-infection), common for entry and post-entry events (essential; substantially depleted at 3 or more time points throughout the experiment (0–8 h)), or post-entry survival-specific (late essential; depleted only at 8 h, 2 + 8 h, or 4 + 8 h post-infection). The numbers of genes in each category are presented in a table. c Representative early essential and late essential genes, and genes essential throughout the infection period are expressed as a heat map. The heat map also includes representative genes that are depleted at one or two time points but do not fall into any of the classification categories. Genes were considered essential if log2 fold change ≤ −1.3 with P value ≤ 0.05; dots on the heat map present genes that meet the log2 fold change criterion but not the P value criterion, and the black lines above the heat map express genes appearing in a gene group.
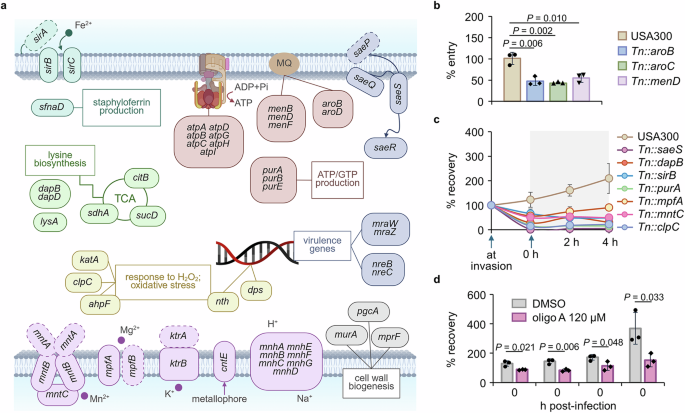
In Fig. 3a, we summarized the sets of genes required for the intracellular infection, which include genes involved in cell wall biogenesis (murA, mprF, and pgcA), pathogenesis (sae, mra and nre loci), nutrient/ion uptake (i.e., mnh, mpf and sir loci), stress response (i.e., clpC and ahpF), energy production (i.e., atp and pur loci), and carbon source utilization (i.e., citB and dap locus). We also included a full list of genes required at each stage of infection by listing all genes that appeared to be required at least at one time point during the infection period (Supplementary Fig. 3). Among these genes, the greatest depletion across all time points was observed in saeS and saeR. The SaeRS TCS responds to host environmental cues (i.e., calprotectin, oxidative stress, and neutrophil proteins) and relays signals to produce virulence factors for adhesion, proteolysis, and hemolysis (i.e., hla)28. Therefore, SaeRS plays a central role in adaptation to the host milieu. Additionally, we identified other transcriptional regulators, nre and mra loci, as essential for intracellular survival. However, we did not observe srrA, which is involved in responding to nitric oxide stress, perhaps because A549 is an epithelial cell line with weak phagocytic activity29. Interestingly, we found groups of genes for which the extent of depletion was not consistent throughout the sampling time points (included in the last category in Supplementary Fig. 3). Among these, murA was depleted at 0 and 4 h but not at 2 and 8 h (Fig. 2c). In addition, menaquinone (MQ) biosynthesis genes such as men and aro loci were substantially depleted in read counts during the early stage rather than the late stages therefore classified as early essential. MQ is electron-charged between ATP synthase and cytochrome oxidase in the ATP generation system of S. aureus30,31. When we analyzed the whole locus of the men operon, most genes were depleted, suggesting a requirement for MQ during the early stage of infection. To validate genes involved in this early stage, we tested individual mutants with the Tn insertion in the identified genes. Mutants of the genes found to be involved in the early phase of infection (i.e., aroB or menD) invaded less into the host cells than the wild-type strain, as shown in Fig. 3b. Although mutants with the Tn insertion at these genes grow slowly (Supplementary Fig. 4a), we do not think slow growth explains these early phase results because other mutants with growth defects did not appear in our Tn-seq results.
a The genes involved in intracellular survival are grouped by their functions, and the selected pathways are presented in a diagram. Genes that were not substantially depleted but are needed as part of the functional group are marked with dotted lines. MQ, menaquinone. Image created in BioRender (Lee, W. (2025) https://BioRender.com/sfrjhgv). The mutants of selected genes were obtained from the Nebraska Tn library and were confirmed for their role in (b) intracellular entry or (c) both entry and post-entry survival by the infection assay (\(\%\;{\rm{entry}}=\,\frac{{\rm{CFU}}\; {\rm{recovered}}\; {\rm{at\;}}0\;{\rm{h}}}{{\rm{input}}\; {\rm{CFU}}}\times 100\); \(\%\;{\rm{recovery}}=\,\frac{{\rm{CFU}}\; {\rm{recovered}}\; {\rm{from}}\; {\rm{cells}}}{{\rm{input}}\; {\rm{CFU}}}\times 100\)). d USA300 pre-exposed to 120 μM of oligomycin A (oligo A), an ATPase inhibitor, was examined for the extent of infection into and proliferation inside the host A549 cells in the presence of 120 μM oligomycin A (\(\%\;{\rm{recovery}}=\,\frac{{\rm{CFU}}\; {\rm{recovered}}\; {\rm{from}}\; {\rm{cells}}}{{\rm{input}}\; {\rm{CFU}}}\times 100\)). Data in b–d are presented as the mean ± SD (n = 3 biological replicates), and the P values are calculated by a two-sided, unpaired Student’s t-test. The source data for b–d are available in the Source data file.
Along with the early essential group of genes, we found another group of genes that were depleted to a greater degree during the late stage of infection or were depleted throughout the 8-h period. These include genes involved in ATP generation (atp and pur loci) and multiple genes related to nutrient uptake: transmembrane transporters (i.e., cntE and mnh locus) that transport metals and ions across the membrane and iron acquisition genes (i.e., sir locus and sfnaD) involved in siderophores or iron acquisition. Finally, we also identified genes related to the oxidative stress response (i.e., nth and katA) and general stress response (i.e., clpC) to be essential for survival in the intracellular milieu (Fig. 3a). These results suggest that intracellular S. aureus faces a severe limitation of nutrients, including essential metal ions. In addition, the essentiality of genes involved in the TCA cycle (i.e., citB and sucD) suggests that S. aureus may become heavily dependent on the TCA cycle and electron transport chain (ETC) to produce ATP, since substrate-level ATP production via glycolysis might become limited due to the limited availability of glucose or other hexose sugar sources while non-carbohydrate carbon sources such as lactate or fatty acid may be more available in the intracellular environment of the host cell. To validate these genes, we chose key genes in each pathway, and mutants of these genes showed no detectable growth defects in TSB medium (Supplementary Fig. 4b) but low fitness when tested individually in the A549 infection assay (Fig. 3c and Supplementary Fig. 4c). Also, the operon analysis showed that many genes required for intracellular invasion and survival were included within the same operon, further validating our Tn-seq analysis (Supplementary Fig. 5). Since we found multiple genes for the ETC including MQ biosynthesis genes, atpA and sucD, we wanted to validate the essentiality of ATP generation via the ETC by testing bacterial survival in A549 cells in the presence of oligomycin A, a macrolide that inhibits the F1F0-ATPase complex by binding to the F0 subunits 6 and 932. As shown in Fig. 3d, at a concentration that affects neither bacteria nor the host A549 cells (Supplementary Fig. 6a-b), bacteria exposed to an ATPase inhibitor demonstrated reduced infection and survival in the intracellular environment.
S. aureus extracellular proliferation in human blood requires different genes from those required for intracellular survival
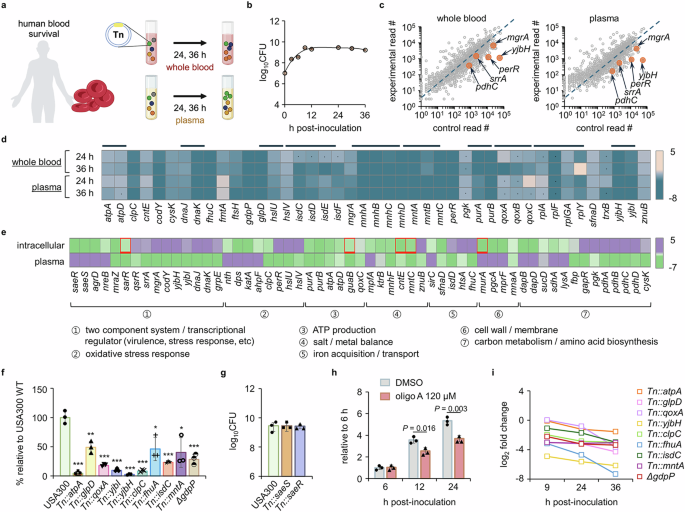
Invasive S. aureus infections result when S. aureus forms colonies on vulnerable bodily surfaces such as abraded skin and somehow reaches the circulatory system, causing bacteremia33. Since blood is considered a major extracellular environment for bacteria during systemic infection, we aimed to understand the physiological and metabolic changes that occur in S. aureus during bacteremia by identifying genes essential for survival in whole blood and plasma using the USA300 Tn library (Fig. 4a). The human blood used in this study was commercially sourced and treated with heparin, so the immune cells are likely mostly deactivated. Utilizing this system, we aimed to study the fitness of mutants in the blood as a host environment with a distinct set of nutrients and carbon sources without rapid clearance by immune cells. We first monitored the growth of USA300 in both human whole blood and plasma by measuring colony forming units (CFU) over 36 h. We found that in both whole blood and plasma, the CFU of USA300 reached a plateau at 9 h (Fig. 4b and Supplementary Fig. 7a), which is comparable to the growth profile in the TSB medium (Supplementary Fig. 4a-b). We used electron microscopy to examine whether the majority of USA300 remained outside of red blood cells (RBC) and found only minor infiltration of USA300 into RBC (Supplementary Fig. 7b).
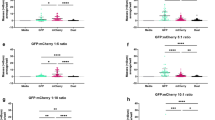
a The USA300 Tn library was cultured in human whole blood and plasma for 24 and 36 h, after which the bacteria were collected and prepared for Tn-seq. Image created in BioRender (Lee, W. (2025) https://BioRender.com/gwn8ee3). b The growth of USA300 was examined for 36 h. c Gene read counts from a 24 h post-inoculation sample were compared to those of the Tn library grown in TSB. Representative genes from the list of substantially depleted genes are presented on the plot. d Representative genes depleted in whole blood and plasma are expressed as a heat map (dots: genes that meet the log2 fold change criterion but not the P value criterion; black lines: gene groups). e Representative genes required for intracellular (2 h) and plasma (36 h) survival are compared (red squares: genes for which the 4 or 8 h post-infection read counts were used since they were either defined as late essential or were not depleted at 2 h but rather at other time points). The mutants of selected genes were validated for their (f) essentiality and (g) non-essentiality for survival in whole blood. The mutants were cultured in whole blood for 24 h, and CFU were counted to measure the survival. * P < 0.05; ** P < 0.01; *** P < 0.001 compared to the USA300 strain. The exact P values are available in the Source Data file. h USA300 pre-exposed to 120 μM of oligomycin A (oligo A) was placed into whole blood in the presence of oligomycin A to investigate the essentiality of ATP production for blood survival. Data in b and f–h are presented as the mean ± SD (n = 3 biological replicates), and the P values are calculated by a two-sided, unpaired Student’s t-test. i The log2 fold change of the mutants of genes essential for blood survival was expressed in a time course up to 36 h post-blood inoculation. Source data for b, c and f–i are available in the Source Data file.
For Tn-seq analysis, we collected S. aureus cells at 24 or 36 h based on the growth profile (Fig. 4b and Supplementary Fig. 7a). This allowed S. aureus to grow for over 10 generations, enabling sufficient time for identification of mutants with lower fitness. We first plotted the read counts from the Tn library cultured in whole blood or plasma against those from TSB media (Fig. 4c and Supplementary Fig. 8a). Primarily, we focused on the depleted genes. A total of 115 genes were found to be essential for survival in the blood (log2 fold change ≤ −1.2; P value ≤ 0.05), and we found no notable differences between genes essential for survival in plasma and those essential for survival in whole blood (Supplementary Fig. 8b). Representative depleted genes are displayed in Fig. 4d. Among these were genes related to metal ion uptake (i.e., isd, mnh, and mnt loci), stress response (i.e., yjb locus, clpC, and trxB), and ATP metabolism (i.e., atp and qox loci). We further compared the depleted genes with those depleted during intracellular infection. We identified distinct genes essential for each growth condition and classified them into seven major functional groups (Fig. 4e). We found that genes encoding major TCSs of S. aureus, including saeRS, agrD, sarR, srrA, and mgrA, are differentially required in the two environments: saeRS is required for intracellular survival, while srrA is necessary for survival in blood, suggesting that different groups of TCSs are required for survival in these two environments. Genes related to oxidative stress responses, such as perR and hslU, were identified only in the blood conditions, suggesting that S. aureus may face more oxidative stress in the bloodstream compared to the intracellular environment of non-phagocytic cells. However, genes responsible for ATP production (i.e., pur and atp loci) and ion/metal balance (i.e., mnh and mnt loci) were depleted in both conditions, suggesting that in both conditions, S. aureus may be exposed to limited nutrients and an environment where ATP is primarily produced through the electron transport chain. Similar to the results from the intracellular invasion, the operon analysis further confirmed that many genes required for survival in blood were within the same operon (Supplementary Fig. 9).
To validate our Tn-seq results, we selected mutants from the seven functional groups and tested them for their growth in blood. As shown in Fig. 4f, these mutants showed lowered fitness in blood. Importantly, we were able to confirm the Tn-seq result for saeS and saeR; as shown in Fig. 4g, we found no growth phenotype of these mutants in blood, suggesting that S. aureus regulates genes independently of SaeRS in blood. Regarding genes related to ATP production, we were also able to validate their requirement using oligomycin A. As shown in Fig. 4h, in the presence of oligomycin A, the growth of USA300 was delayed. Finally, we note that we observed no growth defects of the tested mutants in TSB medium, suggesting that the reduced fitness in blood is not due to the inactivated genes (Supplementary Fig. 4b and 10). Additionally, since the mnh operon appeared to be essential for survival in human blood, we generated ΔmnhF for validation of its essentiality for blood survival. Although the mnhF mutant demonstrated growth defects compared to wild type (Supplementary Fig. 11a), the extent of reduced blood survival within 48 h surpasses the growth retardation (Supplementary Fig. 11b), implying that the reduced fitness is not simply due to slow growth caused by gene inactivation. In addition, the time-dependent decrease in the read counts of the genes provides validation for the essentiality of these genes for blood survival (Fig. 4i).
S. aureus in human blood remodels its metabolism to allow simultaneous utilization of lactate and glucose
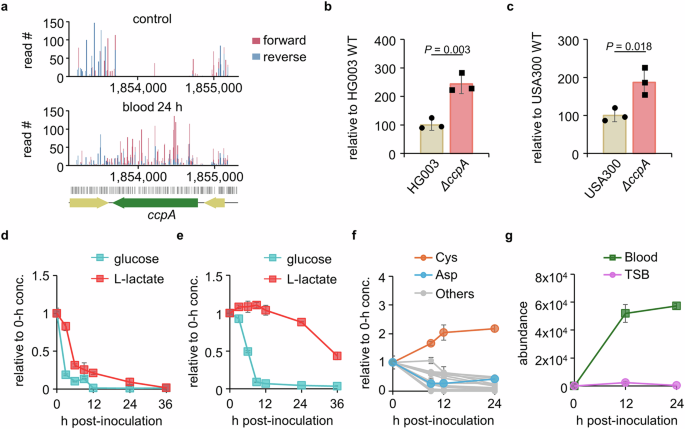
Next, we sought to identify key metabolic pathways involved in bacterial survival and growth in human blood. Since human blood contains distinct nutrients, particularly the available carbon sources, and since carbon source utilization is pivotal for bacterial survival, we have re-evaluated our Tn-seq data from this perspective. Interestingly, in the blood samples we found a substantial enrichment of reads due to the Tn insertions in ccpA, which is involved in catabolic repression in S. aureus (Fig. 5a). We confirmed this enrichment by using a ccpA deletion mutant, which exhibited enhanced growth in human blood (Fig. 5b, c) with the growth profile in TSB shown in Supplementary Fig. 11c, d. CcpA regulates a selective utilization of carbon sources when multiple carbon sources are available34,35. This result led us to investigate the nutrient composition in human blood. We focused on carbon sources that directly enter glycolysis and the TCA cycle, which include amino acids, glucose, and lactate. As previously reported, we found that human blood contains high concentrations of glucose (0.8 mM), lactate (1.3 mM), and amino acids (Supplementary Fig. 12)36. First, we examined the consumption of carbon sources by S. aureus in blood. As shown in Fig. 5d, S. aureus simultaneously consumed lactate and glucose. Typically, bacteria prefer glucose over lactate or acetate when both carbon sources are available, and this is regulated by catabolic repression37,38,39. When we monitored carbon source consumption in TSB media containing both 10 mM glucose and 10 mM lactate, we observed a typical pattern in which S. aureus initially consumed glucose until it was depleted and then began to utilize lactate (Fig. 5e). The simultaneous utilization of carbon sources has also been observed in Mycobacterium tuberculosis40,41. However, unlike M. tuberculosis, S. aureus exhibited the ability to adapt its mode of catabolic repression and shifted to a simultaneous mode only in the host environment, which conferred a fitness advantage.
a The read counts for all TA sites within the ccpA gene detected by sequencing in both forward (red) and reverse (blue) directions were plotted using R. For identification of the role of the ccpA gene for bacterial growth in blood, ΔccpA strains generated in (b) HG003 and (c) USA300 strains were cultured in blood for 24 h and were counted for CFU. WT; wild-type. For the measurement of lactate and glucose consumption of USA300 in blood and TSB, bacteria were cultured in (d) blood or (e) TSB for up to 36 h. At designated time points, blood and TSB samples were centrifuged for the collection of plasma and supernatant, respectively, which were prepared for mass spectrometric analysis. f USA300 was cultured in human whole blood for up to 36 h for the measurement of amino acid consumption. At designated time points, blood was centrifuged for plasma collection and prepared for mass spectrometric analysis (others include Ala, Arg, Asn, Glu, Gln, His, Leu, Lys, Met, Phe, Pro, Thr, Trp, Tyr, and Val). g For the measurement of bacterial bacillithiol, USA300 was inoculated into human whole blood or TSB for 9, 12, or 24 h. At designated time points, samples were centrifuged for the collection of bacterial cells, after which the bacterial cells were quenched, lysed by bead beating, and prepared for mass spectrometric analysis. Bacillithiol levels at each time point were normalized to log10 bacterial CFU. Glucose, lactate, amino acid concentrations, and bacillithiol levels in d–g were measured using LC/MS-TOF, and the abundance of extracted metabolite ion intensities was acquired using Profinder 660. All data in d–g are presented as the mean ± SD (n = 3 biological replicates). Source data is available in the Source Data file.
Additionally, we monitored the concentrations of amino acids during the 24-h blood culture and observed that S. aureus actively consumed all amino acids except cystine (Fig. 5f). Interestingly, we found that S. aureus produced cystine over two-fold compared to the control. In line with this data, we identified substantial depletion of cysK, the final enzyme in cysteine biosynthesis, suggesting the essentiality of this gene for survival in the blood environment (Fig. 4d)42. The cysK is the only gene we found related to amino acid biosynthesis, further validating the results of cystine increase in blood. Interestingly, in blood, we found that genes such as perR, trxB, and hslUV were substantially depleted in the blood environment, indicating that S. aureus experiences oxidative stress in blood. Bacterial thiols act as protective agents against oxidative stress, and in S. aureus these include bacillithiol (N-cysteinyl-α-D-glucosaminyl-L-malate), a hexosamine conjugated to L-cysteine43,44. Indeed, aligned with the cysteine accumulation results, we observed that bacillithiol levels increased sustainably over time in the USA300 strain cultured in blood. However, we found only a minimal increase in the intracellular bacillithiol levels in USA300 during the stationary phase when cultured in TSB (Fig. 5g). Therefore, the depletion of genes related to oxidative stress is consistent with the requirement of cysK and the accumulation of cysteine and bacillithiol in the blood environment. To survive in blood, S. aureus depends on genes related to response to oxidative stress, including those associated with bacterial thiol synthesis. Furthermore, our results indicate that S. aureus adjusts its metabolism to better adapt to the extracellular host environment, allowing for more efficient utilization of available carbon sources and response to oxidative stress.
The genes required for invasion into the liver or lungs show substantial overlap with the genes important for extracellular survival of S. aureus
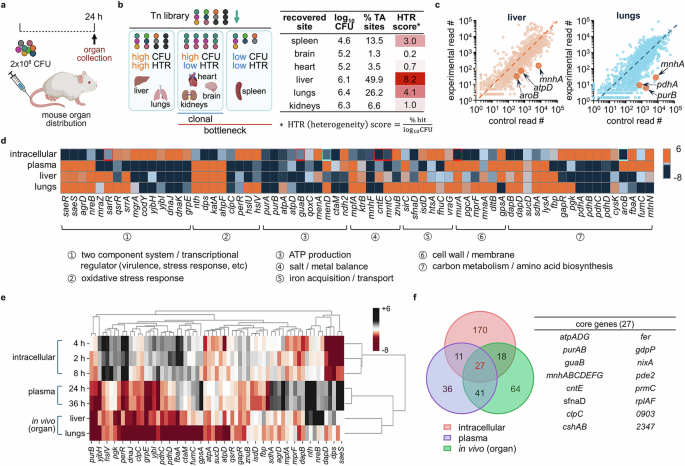
Next, we attempted to identify how S. aureus adapts to different host organs. For this, we decided to use a mouse USA300 infection sepsis model. First, we tested different CFU administered through the tail vein and chose the dose resulting in an 83% survival rate at 48 h post-infection. For Tn-seq, based on the dose, we infected a total of 2 × 108 CFU of the USA300 Tn library, and after 24 h, we separately recovered the surviving Tn mutants from six organs for sequencing (Fig. 6a). Among these six organs, the CFU from the kidneys were the highest, while those of the heart, brain, and spleen were lower, as expected. However, interestingly, in our Tn-seq analysis, we found a very low diversity of Tn mutants in the kidneys. This suggests that the kidneys are relatively hard to infect, but mutants able to do so may encounter a less hostile growth environment than in other organs. In contrast, we recovered a low count of CFU in the spleen, but the mutants were relatively more heterogeneous than in the kidneys, suggesting that there is a lower bottleneck to entry but an unfavorable environment inside the spleen for S. aureus replication. For a numerical representation of these data, we have generated a heterogeneity (HTR) score by normalizing % TA sites (TA sites detected in experimental samples out of all the TA sites in the genome) by the total recovered CFU from each organ (Fig. 6b). Higher HTR means higher diversity of recovered mutants. Using the counts of CFU and HTR scores, we categorized the six organs into three groups: (I) high CFU and high HTR (liver and lungs), (II) high CFU and low HTR (heart, kidneys, brain), and (III) low CFU and low HTR (spleen). Group I includes organs that show high recovery of CFU (log10CFU > 5) and diversity of mutants (HTR score > 4), implying little physical barrier for bacterial invasion and survival. Group II includes organs with high CFU but low HTR (score <4), suggesting that a bottleneck limited mutant entry. Permissive growth conditions after the bottleneck resulted in a clonal mutant population. Group III includes the spleen, which shows low CFU (log10CFU < 5) with low HTR, implying that this organ provides an unfavorable environment for both entry and growth of bacteria. Since bottlenecks limit genetic diversity in the mutant pools for organs in groups II and III, we focused on organs in group I, the liver and lungs, which showed a high CFU with HTR score > 4, for further gene analysis. In addition, because S. aureus is common in lung infections, data generated from the lungs may be of clinical relevance45. Tn data for each organ identified 119 depleted genes for liver and 81 depleted genes for lungs (liver − log2 fold change ≤ −2 with P value ≤ 0.01; lungs − log2 fold change ≤ −4 with P value ≤ 0.01) (Fig. 6c and Supplementary Fig. 13).
a Mice (BALB/c, 8-week-old, male) were infected with 2×108 CFU of the USA300 Tn library via tail vein injection. Mice were euthanized at 24 h post-infection upon which organs were collected and prepared for Tn-seq. Image created in BioRender (Lee, W. (2025) https://BioRender.com/c7082qe). b Log10 of bacterial CFU, % TA sites (TA sites detected in experimental samples out of all the TA sites in the genome), and HTR scores (\(\frac{\%\;{\rm{TA\; sites}}}{{\log }_{10}{\rm{CFU}}}\)) are presented in a table. The organs were classified into three groups (high CFU (log10CFU > 5) – high HTR (HTR > 4); high CFU – low HTR (HTR < 4); low CFU (log10CFU < 5) – low HTR). Image created in BioRender (Lee, W. (2025) https://BioRender.com/ngl11mx). c Gene read counts from lungs and liver were plotted against the read counts of the respective genes of a control Tn library. Representative genes are marked on the plot. Source data is available in the Source Data file. d Genes essential for intracellular survival (2 h), plasma survival (36 h), and survival in organs (both liver and lungs) are compared in a heat map (red squares: genes for which the 4 or 8 h post-infection read counts were used because they were either defined as late essential or were not depleted at 2 h but rather at other time points; blue squares: genes for which the 0 h read counts were used because they were defined as early essential). e Gene clustering analysis was performed using the seaborn, clustermap Python package (v.0.12.2) using all genes from d, but aroB, menA, and menD were excluded since they were early essential genes that do not appear at 2, 4, or 8 h post-infection. f The numbers of genes that are specifically and commonly essential for intracellular, blood, and organ survival are presented in a Venn diagram, and the 27 core genes that are essential for infection in all three host environments (intracellular, blood, and intra-organ) are listed in a table.
As shown in Fig. 6d, in both organs we identified genes encoding TCSs and associated regulators, including mgrA and yjb locus. We also found genes for ATP production (atp locus, qoxC, and ndh2) and genes related to the oxidative stress response (Fig. 6d, groups 2 and 3). Notably, the oxidative stress response genes found to be important in the liver and lungs were different from those involved in intracellular survival. Similarly, TCS SaeRS and cell wall biosynthesis and metabolism-associated genes (mpfA, murA, lysA, and dapB) were depleted only in an intracellular environment but not in blood and intra-organ environments. Most genes depleted in the liver and lungs were shared, but several, such as copZ and gpsA, were depleted only in the lungs (Supplementary Fig. 13), and the essentiality of the functional groups was further validated by the operon analysis (Supplementary Fig. 14). The essentiality of genes was confirmed with the atpD mutant being less infective in both liver and lungs (Supplementary Fig. 15a), while copZ and pyrE genes were only essential for lung or liver infection, respectively (Supplementary Fig. 15b, c). In addition, the mice infected with the atpD mutant showed improved survival compared to the mice infected with wild-type USA300 (Supplementary Fig. 15d), implying that an atp locus could serve as an effective therapeutic target for the treatment of bacterial infection. All these results were obtained with no apparent growth defects in the mutants (Supplementary Fig. 15e). When we compared the genes found to be important in the lungs or liver with those involved in the intracellular and extracellular milieus, we found that the set of genes identified as essential for survival in these organs resembled those required for extracellular survival (Fig. 6d) and as analyzed by the clustering analysis (Fig. 6e). Genomic profiling using the in vivo sepsis model is a conglomeration of results from the blood, tissue, and intracellular bacteria. Therefore, the results from bacteria recovered from organs might be skewed by the bacteria remaining in the extracellular and tissue environments.
All three systems, intracellular, extracellular, and intra-organ, required sets of genes involved in ATP production and maintenance of nutrient and ion balance, resulting in 27 core genes (Fig. 6f). These results indicate that ATP production and nutrient/ion balance are essential for growth in any given environment, despite other differences in each host niche.
Discussion
Our study demonstrates that S. aureus can adapt differentially within various host niches to exploit nutrients and respond to hostile conditions. It does so by employing both metabolic changes and regulators that serve as hubs for signal relay for many virulence-related genes, and this shaping of the metabolic and physiological makeup of S. aureus is crucial for survival and response to host environmental cues. These differential environmental cues vary depending on whether S. aureus is in an intracellular, extracellular, or intra-organ environment.
Here, we used three infection models—a cell culture model, blood, and a mouse model focusing on specific organs—to identify genes necessary for survival within the host. Despite residing on the skin, which constitutes an extracellular environment, S. aureus is fully equipped to thrive intracellularly and within organs9,16,17,18,46. In our Tn-seq analysis using the mouse sepsis model, our approach involved assigning a heterogeneity score to each organ, revealing distinct characteristics for each. For example, S. aureus showed the largest bacterial count but a highly clonal population in the kidneys, suggesting that only certain mutants can infiltrate into the kidneys or, alternatively, that most mutants are filtered out of the kidneys, but a small number of mutants attach to the organ and multiply. On the contrary, in the lungs and liver, where bacteria also grew to a high count, S. aureus showed high heterogeneity, suggesting that these organs are accessible to most mutants. Notably, S. aureus primarily enters the lungs via the respiratory route rather than the bloodstream, but regardless of the mode of entry, the lungs are highly accessible to S. aureus47. Therefore, we conclude that these organs are extremely vulnerable to bacterial infection and provide a favorable niche for bacterial survival. In the spleen, S. aureus shows both low bacterial count and low heterogeneity, which might be expected from an organ rich in macrophages. Unexpectedly, a high bacterial count was found in the brain, an organ that is protected by the blood-brain barrier. This finding suggests that there is a route for S. aureus entry into the brain parenchyma, but as can be seen from the low heterogeneity, only certain mutants can do so. These findings demonstrate the potentially different levels of barriers and protection presented by each organ during systemic S. aureus infections, highlighting distinct environmental characteristics that allow for S. aureus replication. Our results may suggest which organ is most vulnerable during S. aureus sepsis.
By studying the bacterial interaction with human blood and comparing the gene sets essential for survival in whole blood and plasma, we demonstrate that bacteria do not invade the RBC. Since the human blood used in this study was commercially sourced and treated with heparin, with immune cells mostly deactivated, the Tn-seq results for blood can apply to the general extracellular body fluid encountered by the bacteria once inside the host body. We have particularly shown that the essentiality of genes responsible for response to oxidative stress along with the increased production of bacillithiol in S. aureus upon incubation in blood implies the importance of the stress response pathways for survival in the blood environment. In addition, our organ data resembled the extracellular results in blood, suggesting that once inside the organs, a large population of S. aureus resides in an extracellular body fluid rather than inside the cells.
Regulators such as SaeRS, AgrA, SarA, SrrA and MgrA are known to be required during S. aureus infection10,11,12,13. However, our findings suggest that they are differentially required in specific host niches and not consistently across all environments, which should be considered in mapping their signal relay. On the other hand, we found that in all three infection systems, 27 core genes, such as ATP synthase and the mnh antiporters, required for nutrient processing, are crucial for survival within the host. Our study implies that energy metabolism plays an essential role in bacterial survival, regardless of the host niche. Valentino et al. also utilized Tn-seq for the identification of genes essential for survival in skin abscess48. These authors showed that several genes involved in the pyrimidine and purine biosynthesis pathways are critical for survival in skin abscesses; and some of these genes, such as purB and guaB, were also identified as essential in bacterial survival in all biological systems (intracellular, extracellular, and intra-organ) used in the current study. This similarity indicates that energy metabolism is a possible therapeutic target for combating S. aureus infection. It has also been shown that the cydABCD locus and polysaccharide pathway of E. faecalis are crucial for adaptation and survival in the bloodstream49. Therefore, a broader understanding of metabolic reprogramming during infection can be crucial for suppressing the growth of bacterial pathogens in the host environment. A comprehensive and comparative understanding of the underlying mechanisms responsible for the establishment of bacterial infection in different host niches will greatly contribute to the development of efficient treatment strategies.
Methods
Ethics statement and animal information
Mouse experiments were approved by and performed in strict accordance with the institutional guidelines of the Institutional Animal Care and Use Committee of Sungkyunkwan University School of Medicine (approval number: SKKUIACUC2021-11-35-1). All animal experiments were performed in a facility approved for BSL-2 experiments involving a bacterial Tn library. Male, 7-week-old BALB/c mice were purchased from Orient Bio (Seongnam, South Korea) and acclimated to an animal facility isolated for infection studies at Sungkyunkwan University School of Medicine. All mice were maintained under pathogen-free conditions on a 12:12 h light:dark cycle, with free access to facility chow and water. The temperature and humidity of the facility were maintained at 22 °C and 50%, respectively. The experiments performed with human blood were all approved by the Sungkyunkwan University Institutional Review Board (IRB: SKKU 2023-01-023).
Cell lines and culture conditions
The lung epithelial cell line A549 (Korean Cell Line Bank, Seoul, South Korea; #10185) was grown in Roswell Park Memorial Institute 1640 (RPMI 1640) medium (Welgene, Gyeongsan, South Korea) supplemented with 10% fetal bovine serum (FBS; Welgene). The murine macrophage cell line Raw264.7 (Korean Cell Line Bank; #40071) and human keratinocyte cell line HaCaT (Cellular Engineering Technologies, Coralville, IA; #CR1017-500) were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM; Welgene) supplemented with 10% FBS. All cell lines were grown in a humidified 37 °C, 5% CO2 incubator and sub-cultured by trypsinization.
Bacterial strains and culture conditions
Methicillin-resistant S. aureus (MRSA) strains USA300, MW2, and MRSA252, and methicillin-sensitive S. aureus (MSSA) strains HG003, MSSA476, and Newman strains were grown at 30 °C in liquid tryptic soy broth (TSB; Beckton Dickinson, Franklin Lakes, NJ) with shaking or on agarized TSB plates (TSA). Mutant strains obtained from the Nebraska transposon mutant library (NTML) were grown in liquid TSB or TSA with the addition of 10 µg/mL erythromycin (Erm; Sigma-Aldrich, Burlington, MA). The mutant strains used in the study are summarized in Supplementary Table 1, and all mutants were confirmed for the Tn insertion at the target gene using primers (Macrogen, Seoul, South Korea) as detailed in Supplementary Table 2. On the day of an experiment, an overnight bacterial culture was re-inoculated at OD600 = 0.01 and grown until OD600 = 0.4–0.6. The transposon mutant library (Tn library) was created in USA300 by phage-based transposition as previously described24. Library aliquots were stored at −80 °C and on the day of the experiment were grown from OD600 = 0.2 to OD600 = 0.4 for no more than 3 h.
Generation of mutants of S. aureus
For a generation of ΔmnhF and ΔccpA, DNA fragments corresponding to 1 kb upstream and downstream of mnhF and ccpA were amplified with the primer pairs mnhF1/mnhF3 and mnhF2/mnhF4 for mnhF and ccpA1/ccpA3 and ccpA2/ccpA4 for ccpA (Supplementary Table 2). The kanamycin (Km) resistant gene was amplified with the primer pair KmF/KmR. Following purification, the Km-resistant fragment was combined with the mnhF and ccpA fragments using KOD Xtreme Hot Start DNA polymerase (Novagen, Madison, WI) with the primer pairs mnhF5/KmR and mnhF5/mnhF6 for mnhF and ccpA5/KmR and ccpA5/ccpA6 for ccpA (Supplementary Table 2). The combined PCR product was digested with BamHI/HindIII and BamHI/SaI for mnhF and ccpA, respectively, and the resulting products were cloned into pKFC (pKFC-delmnhF and pKFC-delccpA) and transformed into electrocompetent S. aureus RN4220, resulting in the generation of RN4220-ΔmnhF and RN4220-ΔccpA. Homologous recombination was performed to integrate the plasmid into the bacterial chromosome, and the deletion of the target gene was confirmed by sequencing. Confirmed deletions were transduced using φ11 into HG003 and USA300 to generate HG003-ΔmnhF, HG003-ΔccpA, USA300-ΔccpA50. All bacterial strains, primer sequences, and plasmids are summarized in Supplementary Tables 1 and 2.
Determination and validation of genes essential for intracellular infection
Intracellular infection assay
To determine the extent of intracellular infection and survival of S. aureus, host cells were seeded in the T25 cell culture flasks at 3.5 × 106 cells/flask or the 24-well culture plates at 2.5 × 105 cells/well and placed in a humidified 37 °C, 5% CO2 incubator for 48 h. At 1 h before bacterial infection, the culture medium was replaced with fresh medium. The bacterial strains were grown to a log phase and added to the host cell culture medium at a multiplicity of infection (MOI) of 10 as per the result for screening for MOI that resulted in full recovery of input bacteria. After 1.5 h of infection, bacteria-containing culture medium was removed, and host cells were washed five times with sterilized PBS. Afterwards, medium containing 200 µg/mL gentamicin was added for 1 h to remove extracellular bacteria, after which the host cells were again washed 5 times with sterilized PBS. The host cells were immediately (time point designated as 0 h post-infection throughout the manuscript) lysed with 0.1% Triton X-100, and the collected bacteria were spread on TSA plates and grown at 30 °C for counting of bacterial colony-forming units (CFU). For examination of post-entry events, fresh culture medium was added to the flask after the removal of gentamicin-containing medium and washing with PBS, and host cells were lysed at 2, 4, and 8 h post-addition of fresh medium (time points designated as 2, 4, and 8 h post-infection throughout the manuscript). Bacteria were spread on TSA plates and grown at 30 °C for counting of bacterial CFU. The protocol was adapted and modified from previous publications51,52,53.
Tn library infection experiment
For the determination of genes essential for intracellular entry and post-entry survival, an infection assay of the USA300 Tn library into host A549 cells was performed. Host A549 cells were seeded in T25 cell culture flasks at 3.5 × 106 cells/flask, and the Tn library was grown to OD600 = 0.4 as described above. The infection procedure was performed as described above. The host cells were lysed at 0, 2, 4, and 8 h post-entry and spread on TSA+Erm (10 µg/mL) plates and grown at 30 °C. The colonies were then scraped and pooled for preparation for the transposon sequencing (Tn-seq).
Validation assay with mutants
To validate the results obtained from the Tn library infection assay, individual mutants were tested for the extent of their intracellular entry and post-entry survival54. All mutants are listed in Supplementary Table 1 and were confirmed for the Tn insertion at the target gene using primers as detailed in Supplementary Table 2. For validation, host A549 cells were seeded in 24-well culture plates at 2.5 × 105 cells/well, and the rest of the procedure was the same as outlined above for the intracellular infection assay. Upon lysis of host cells at 0, 2, 4, and 8 h post-entry, mutants were spread on TSA plates containing appropriate antibiotics and counted for the number of colonies at each time point.
Confirmation of the effects of an ATPase inhibitor on bacterial intracellular entry and post-entry survival
For the determination of the effects of ATPase inhibition, USA300 was cultured in media containing 120 µM of oligomycin A (Sigma-Aldrich) for 4 h32. Then, bacteria pre-treated with oligomycin A were utilized for the intracellular infection assay. A549 cells were exposed to bacteria pre-treated with oligomycin A, also in the presence of 120 µM oligomycin A during infection and throughout the experiment until A549 cell lysis at various time points. Upon lysis of host cells at 0, 2, 4, and 8 h post-entry, the bacteria were spread on TSA plates and counted for the number of colonies at each time point.
Determination and validation of genes essential for survival in human blood/plasma
Human blood/plasma survival assay
To determine the bacterial growth and survival in human blood, human blood anticoagulated with heparin was purchased from Innovative Research (Novi, MI) through an FDA-approved collection center in the USA. To separate plasma, whole blood was centrifuged at 845 × g at 4 °C for 15 min, after which the supernatant was obtained. Whole blood and plasma were placed into 14-mL round-bottom tubes in 2-mL aliquots. The bacterial strains were grown to a log phase and added to the human whole blood or plasma at a final OD600 = 0.02. Whole blood and plasma were incubated at 37 °C, with shaking at 150 rpm for 24 or 36 h. The samples were then placed in red blood cell (RBC) lysis buffer (10×; 1.5 M NH4Cl, 100 mM NaHCO3, 1 mM EDTA, pH 7.4) and spread on TSA plates for CFU counting.
Tn library infection into human blood and plasma
For the determination of genes essential for the survival of USA300 in human blood, whole blood and plasma were prepared as outlined above and placed into 14-mL round-bottom tubes in 4-mL aliquots. The Tn library was grown to OD600 = 0.4 and added to the human whole blood and plasma at a final OD600 = 0.02. Whole blood and plasma were incubated at 37 °C, with shaking at 150 rpm for 24 and 36 h. After 24 and 36 h of growth, samples were placed on ice. RBC lysis buffer was added at a volume of 4 mL and incubated for 10 min to lyse RBC in whole blood samples. Both whole blood (now with lysed RBC) and plasma samples were centrifuged at 9391 × g at 4 °C for 5 min and washed 3 times with sterilized PBS. The resulting pellet was used for preparation for Tn-seq.
Validation assay with mutants
To validate the genes found essential in the blood and plasma survival assay, individual mutants were tested for the extent of their blood survival. All mutant strains are listed in Supplementary Table 1 and were confirmed for the Tn insertion at the target gene using primers as detailed in Supplementary Table 2. For the validation assay, whole blood was placed into 14-mL round-bottom tubes in 2-mL aliquots into which the mutants were inoculated at an initial OD600 = 0.02. The tubes were placed at 37 °C, with shaking at 150 rpm for 24 h after which the bacteria were spread on TSA plates containing appropriate antibiotics and counted for the number of colonies.
Validation of effects of ATPase inhibition on bacterial blood survival
For the determination of the effects of ATPase inhibition, USA300 was cultured in media containing 120 µM of oligomycin A (Sigma-Aldrich) for 4 h. Then, the bacteria pre-treated with oligomycin A were added to human blood at an OD600 = 0.02, also in the presence of 120 µM oligomycin A. After 6, 12, and 24 h of growth, the bacteria were spread on TSA plates and counted for the number of colonies appearing at each time point.
Determination and validation of genes essential for an intra-organ survival
Tn library infection into the mouse sepsis model
For the identification of genes essential for bacterial survival in the organs, a mouse sepsis model was used55. Animals were injected with a USA300 Tn library at 2 × 108 CFU per mouse by intravenous injection with an injection volume of 100 μL. For this, the Tn library (grown to OD600 = 0.4 as described above) was resuspended in sterilized phosphate-buffered saline (PBS) used as an injection medium. At 24 h post-bacterial injection, animals were euthanized by isoflurane overdose, followed by exsanguination and organ collection for analysis. At collection, the spleen, brain, heart, liver, lungs, and kidneys were homogenized in PBS containing 0.1% Triton X-100 (Sigma-Aldrich), after which the homogenate was spread on TSA+Erm (10 µg/mL) plates and grown at 30 °C for bacterial colony collection for Tn-seq.
Validation assay with NMTL mutants
To validate the results obtained from the Tn-seq of the organs, individual mutants obtained from the NTML were tested for the extent of their survival in the lungs and liver. All mutants are listed in Supplementary Table 1 and were confirmed for the Tn insertion at the target gene using primers as detailed in Supplementary Table 2. Briefly, mice were injected with individual NTML mutants or wild-type USA300 at 2 × 108 CFU/mouse by intravenous injection with an injection volume of 100 μL. At 24 h post-injection, mice were euthanized, and organs (lungs and liver) were collected and homogenized. The samples were spread on TSA+Erm (10 µg/mL) plates and grown at 30 °C after which the numbers of CFU on the plates were counted.
Tn-seq and data analysis
A Tn library grown on TSA+Erm (10 µg/mL) plates obtained from the intracellular infection study, blood/plasma survival study, and the mouse sepsis study was scraped, and genomic DNA was harvested and prepared for deep sequencing as previously outlined24. Illumina high-throughput sequencing was performed on a Hi-Seq2000 or Hi-Seq2500 for 100 cycles with 40% ΦX174 spiked into the sequencing reaction (Macrogen). The sequencing data were trimmed, filtered, and processed using the Galaxy software suite. The chromosome nucleotide FASTA file for USA300-TCH1516 (NC_010079.1) was downloaded from the NCBI Genome database, and the obtained sequencing data were mapped to the genome using Bowtie software. Python scripts were used to compare the number of reads in the control and experimental conditions using the Mann-Whitney U test, correcting for multiple hypothesis testing with the Benjamini-Hochberg procedure. An EL-ARTIST essential gene analysis was done in MATLAB. The essentiality threshold was set to P value ≤ 0.05 after correcting for the false discovery rate. The operon analysis of the Tn-seq data was performed using Rockhopper (v.2.0.3)56.
Transmission electron microscopy
For transmission electron microscopy (TEM) imaging, A549 cells were seeded in Φ15 mm glass-bottom cell culture dishes (Nest Scientific, Rahway, NJ) at a density of 7 × 105 cells/plate and incubated in a humidified 37 °C, 5% CO2 incubator for 48 h. At 1 h before infection, the cell culture medium was changed to fresh medium, after which USA300 (grown to OD600 = 0.4–0.6) was added to the culture medium at an MOI = 10 for infection lasting 1.5 h, followed by washing (5× with sterilized PBS) and 1 h treatment with gentamicin 200 μg/mL. Cells were then again washed 5× with PBS, and fresh RPMI 1640 medium was added, after which cells were placed in a humidified 37 °C, 5% CO2 incubator for 4 h. At 4 h post-incubation, the sample was fixed in 2.5% glutaraldehyde solution for 1 h at room temperature. The blood samples were generated by culturing USA300 in human whole blood from an initial OD600 = 0.02 for 4 and 12 h at 37 °C, with shaking at 150 rpm. At designated time points, samples were centrifuged at 9391 × g at 4 °C for 5 min and washed with sterilized PBS three times. Then, the samples were fixed in 2.5% glutaraldehyde solution for 1 h at room temperature. Both cell and blood samples were then dehydrated with an ethanol series and infiltrated with Spurr’s resin series, after which the samples were polymerized for 8 h at 60 °C. The sample-embedded blocks were cut with a diamond knife on an ultramicrotome (Leica Microsystems, Wetzlar, Germany), and the resulting sections were directly mounted on 150 mesh copper grids. The sections were stained for 20 min with 2% uranyl acetate in 50% methanol, followed by staining with Reynold’s lead citrate for 10 min. The grids were imaged in a bio-imaging core facility of Korea Institute of Science and Technology (KIST) using Tecnai F20 G2 (FEI, Hillsboro, OR) equipped with a RIO 16 camera under the magnification of 17,000× to 65,000×.
Fluorescence microscopy
Preparation of FtsZ-GFP expressing USA300 strain
The pLow-ftsZ-GFP plasmid was inserted into the RN4220 strain by electroporation using a GenePulser cuvette (BioRad, Hercules, CA) at a voltage of 2900 V, 25 μF capacitance, 100 Ω resistance, and 2 mm length57. After electroporation, FtsZ-GFP-expressing RN4220 (RN4220-FtsZ-GFP) colonies were selected on TSA+Erm (10 μg/mL) at 30 °C. The pLow-ftsZ-GFP was then transduced into USA300 using staphylococcal phage ϕ11, generating the FtsZ-GFP-expressing USA300 strain (USA300-FtsZ-GFP). Details of all strains and plasmids can be found in Supplementary Tables 1 and 2.
Fluorescence microscopy
The obtained USA300-FtsZ-GFP was used to visually inspect and analyze bacterial infection into and survival in host cells. For imaging, A549 cells were seeded in Φ15 mm glass-bottom cell culture dishes (Nest Scientific) at 7 × 105 cells/plate and incubated in a humidified 37 °C, 5% CO2 incubator for 48 h. At 1 h before infection, culture medium was changed to fresh medium; and at the time of infection, USA300 (grown to OD600 = 0.4–0.6) was added at an MOI = 10 to the culture medium. After 1.5 h, cells were washed 5× with PBS, followed by exposure to medium containing gentamicin 200 μg/mL. Cells were then washed 5× with sterilized PBS. After being washed with PBS, cells were stained with wheat germ agglutinin Alexa Fluor™ 555 conjugate (Invitrogen, Waltham, MA) for labeling of the cell membrane58. For examination of intracellular survival, fresh RPMI 1640 medium was added, and cells were placed in a humidified 37 °C, 5% CO2 incubator for 4 and 8 h followed by the same staining procedure. All fluorescent images were taken with a Leica DMI8 fluorescence microscope (Leica Microsystems) equipped with a Leica DFC9000 GT VSC-12294 camera under HC PL APO 100× oil immersion lens. Gamma, brightness, and contrast were adjusted identically for the compared image sets using LAS X software (version 3.7.2.22383).
Liquid chromatography mass spectrometry
Extraction of blood metabolites
For determination of metabolites in blank plasma, plasma was separated by centrifugation at 845 × g at 4 °C for 15 min, after which 200 μL of plasma was mixed with 400 μL of cold methanol (v/v ratio 1:2). For determination of plasma metabolites upon consumption by bacteria, bacterial culture was added to 2-mL aliquots of whole blood at an initial OD600 = 0.02. The samples were incubated at 37 °C, with shaking at 150 rpm for various time points from 3 to 36 h. Whole blood to which no bacteria were added was used as the control 0 h sample. At the designated time points, whole blood samples were centrifuged for Z.
Extraction of bacterial bacillithiol
For determination of bacterial bacillithiol levels, USA300 was inoculated into whole blood or TSB at an OD600 = 0.02 with shaking at 150 rpm at 37 °C. At 9, 12, and 24 h post-inoculation, samples were centrifuged at 4 °C, 6010 × g for 6 min for the collection of bacterial cells. Bacterial cells collected at a log phase (OD600 = 0.5) were defined as a 0 h sample. The resulting pellet was quenched by the addition of quenching buffer (40:40:20; Acetonitrile:Methanol:Water). Bacterial cells were lysed by mechanical disruption using 0.1 mm glass beads and a precellys homogenizer (Bertin Technologies, Montigny-le-Bretonneux, France) with 12 cycles of 10,000 rpm, 30 sec cycle. Samples were placed on ice in-between each cycle for the prevention of protein degradation. Lysates were centrifuged at 4 °C, 15,871 × g for 5 min, after which supernatant was added to Spin-X centrifuge tubes (Corning Inc.) and centrifuged at 4 °C, 15,871 × g one more time.
Liquid chromatography mass spectrometry analysis
Blood metabolites and bacillithiol were analyzed with time-of-flight (TOF) mass spectrometry (Agilent 6230, Santa Clara, CA) coupled with a liquid chromatography system (Agilent 1290) operated in both positive and negative modes. The mobile phase consisted of solvent A (ddH2O with 0.2% formic acid) and solvent B (acetonitrile with 0.2% formic acid). The gradient condition was as following: 0–2 min, 85% B; 3–5 min, 80% B; 6–7 min, 75% B; 8–9 min, 70% B; 10–11.1 min, 50% B; 11.1–14 min 20% B; 14.1–24 min 5% B followed by a 10 min re-equilibration period at 85% B at a flow rate of 0.4 mL/min. The abundance of extracted metabolite ion intensities was acquired using Profinder 6.059,60. Bacillithiol levels were all normalized by the bacterial CFU measured for each sample.
Growth curve measurement
For the measurement of growth of all mutant strains, strains (grown to OD600 = 0.4–0.6) were added to TSB in 96-well plates at an initial OD600 of 0.002. Growth was observed using a plate reader (BioTek, Winooski, VT) at 30 °C for 20 to 30 h.
Measurement of bacterial minimum inhibitory concentration
To determine the concentration of oligomycin A that does not exhibit any effect on bacteria, a minimum inhibitory concentration (MIC) measurement was performed. Oligomycin A was serially diluted from 120 μM in a 96-well plate in TSB, in which USA300 was grown from OD600 = 0.002 for 24 h. The final OD600 was measured by the plate reader (BioTek) for the determination of oligomycin A concentration affecting bacterial growth.
Cell cytotoxicity
To determine the concentration of oligomycin A which does not exhibit toxicity on host A549 cells, cell cytotoxicity measurement was performed using the CytoTox-Glo™ assay (Promega, Madison, WI), according to the manufacturer’s instructions61. Briefly, A549 cells were seeded in white-bottom 96-well plates at a density of 1 × 104 cells/well. After 24 h of incubation in a 37 °C, 5% CO2 incubator, cells were treated with 0–120 μM oligomycin A for 8 h, after which a substrate that luminesces upon reaction with intracellular organelles but cannot cross the intact membrane of live cells is added to measure cytotoxicity. The fluorescence intensity was measured using the Synergy HTX multi-mode plate reader (BioTek).
Statistical analyses
Statistical analyses between experimental groups were conducted by a two-sided, unpaired Student’s t-test using Microsoft Excel (v.16.98). The results were expressed as the mean ± SD of 3 biological replicates representative of duplicate or triplicate independent experiments. Differences between groups were inferred as significant for P < 0.05. The exact P values are denoted in the Source Data file and the figures.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All datasets generated during this study are available in the Source Data file. All the raw data generated by the Tn-seq are available in a public repository, the NCBI BioProject database, PRJNA1236917. The raw data generated by mass spectrometry analysis are deposited in the NIH Common Fund’s National Metabolomics Data Repository (NMDR) website, the Metabolomics Workbench, https://www.metabolomicsworkbench.org/ under the Project ID PR002520. The data can be accessed directly via its Project https://doi.org/10.21228/M8WZ6W. Source data are provided with this paper.
Code availability
Codes needed for processing of the sequencing data is available from GitHub (https://github.com/SuzanneWalkerLab/5SATnSeq)26.
References
Kern, W. V. & Rieg, S. Burden of bacterial bloodstream infection-a brief update on epidemiology and significance of multidrug-resistant pathogens. Clin. Microbiol. Infect. 26, 151–157 (2020).
Tong, S. Y., Davis, J. S., Eichenberger, E., Holland, T. L. & Fowler, V. G. Jr Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol Rev. 28, 603–661 (2015).
Collaborators, G. B. D. A. R. Global mortality associated with 33 bacterial pathogens in 2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet 400, 2221–2248 (2022).
Garzoni, C. & Kelley, W. L. Staphylococcus aureus: new evidence for intracellular persistence. Trends Microbiol. 17, 59–65 (2009).
Peyrusson, F. et al. Intracellular Staphylococcus aureus persisters upon antibiotic exposure. Nat. Commun. 11, 2200 (2020).
Garzoni, C. & Kelley, W. L. Return of the Trojan horse: intracellular phenotype switching and immune evasion by Staphylococcus aureus. EMBO Mol. Med. 3, 115–117 (2011).
Turner, N. A. et al. Methicillin-resistant Staphylococcus aureus: an overview of basic and clinical research. Nat. Rev. Microbiol. 17, 203–218 (2019).
Potter, A. D. et al. Host nutrient milieu drives an essential role for aspartate biosynthesis during invasive Staphylococcus aureus infection. Proc. Natl Acad. Sci. USA 117, 12394–12401 (2020).
Lowy, F. D. Is Staphylococcus aureus an intracellular pathogen? Trends Microbiol. 8, 341–343 (2000).
Tiwari, N. et al. The SrrAB two-component system regulates Staphylococcus aureus pathogenicity through redox sensitive cysteines. Proc. Natl Acad. Sci. USA 117, 10989–10999 (2020).
Crosby, H. A. et al. The Staphylococcus aureus ArlRS two-component system regulates virulence factor expression through MgrA. Mol. Microbiol. 113, 103–122 (2020).
Novick, R. P. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48, 1429–1449 (2003).
Munzenmayer, L. et al. Influence of Sae-regulated and Agr-regulated factors on the escape of Staphylococcus aureus from human macrophages. Cell Microbiol. 18, 1172–1183 (2016).
Sun, F. et al. In the Staphylococcus aureus two-component system sae, the response regulator SaeR binds to a direct repeat sequence and DNA binding requires phosphorylation by the sensor kinase SaeS. J. Bacteriol. 192, 2111–2127 (2010).
Mashruwala, A. A., Gries, C. M., Scherr, T. D., Kielian, T. & Boyd, J. M. SaeRS Is responsive to cellular respiratory status and regulates fermentative biofilm formation in Staphylococcus aureus. Infect Immun 85 (2017).
Pastar, I. et al. Intracellular Staphylococcus aureus triggers pyroptosis and contributes to inhibition of healing due to perforin-2 suppression. J. Clin. Invest. 131 (2021).
Flannagan, R. S., Kuiack, R. C., McGavin, M. J. & Heinrichs, D. E. Staphylococcus aureus Uses the GraXRS regulatory system to sense and adapt to the acidified Phagolysosome in Macrophages. mBio 9, e01143–18 (2018).
Nakaminami, H. et al. Efflux transporter of Siderophore Staphyloferrin A in Staphylococcus aureus contributes to bacterial fitness in abscesses and epithelial cells. Infect. Immun. 85, e00358–17 (2017).
Edwards, A. M., Potts, J. R., Josefsson, E. & Massey, R. C. Staphylococcus aureus host cell invasion and virulence in sepsis is facilitated by the multiple repeats within FnBPA. PLoS Pathog. 6, e1000964 (2010).
Strobel, M. et al. Post-invasion events after infection with Staphylococcus aureus are strongly dependent on both the host cell type and the infecting S. aureus strain. Clin. Microbiol. Infect. 22, 799–809 (2016).
Alam, M. T. et al. Transmission and microevolution of USA300 MRSA in U.S. households: evidence from whole-genome sequencing. mBio 6, e00054 (2015).
Tenover, F. C. & Goering, R. V. Methicillin-resistant Staphylococcus aureus strain USA300: origin and epidemiology. J. Antimicrob. Chemother. 64, 441–446 (2009).
Goncheva, M. I., Gibson, R. M., Shouldice, A. C., Dikeakos, J. D. & Heinrichs, D. E. The Staphylococcus aureus protein IsdA increases SARS CoV-2 replication by modulating JAK-STAT signaling. iScience 26, 105975 (2023).
Santiago, M. et al. A new platform for ultra-high density Staphylococcus aureus transposon libraries. BMC Genomics 16, 252 (2015).
Liang, X. et al. Inactivation of a two-component signal transduction system, SaeRS, eliminates adherence and attenuates virulence of Staphylococcus aureus. Infect. Immun. 74, 4655–4665 (2006).
Coe, K. A. et al. Multi-strain Tn-Seq reveals common daptomycin resistance determinants in Staphylococcus aureus. PLoS Pathog. 15, e1007862 (2019).
Santiago, M. et al. Genome-wide mutant profiling predicts the mechanism of a Lipid II binding antibiotic. Nat. Chem. Biol. 14, 601–608 (2018).
Liu, Q., Yeo, W. S. & Bae, T. The SaeRS Two-Component System of Staphylococcus aureus. Genes 7, 81 (2016).
Hawdon, N. A. et al. Cellular responses of A549 alveolar epithelial cells to serially collected Pseudomonas aeruginosa from cystic fibrosis patients at different stages of pulmonary infection. FEMS Immunol. Med. Microbiol. 59, 207–220 (2010).
Wassmann, C. S. et al. The menaquinone pathway is important for susceptibility of Staphylococcus aureus to the antibiotic adjuvant, cannabidiol. Microbiol Res 257, 126974 (2022).
Lee, B. S., Singh, S. & Pethe, K. Inhibiting respiration as a novel antibiotic strategy. Curr. Opin. Microbiol. 74, 102327 (2023).
Devenish, R. J., Prescott, M., Boyle, G. M. & Nagley, P. The oligomycin axis of mitochondrial ATP synthase: OSCP and the proton channel. J. Bioenerg. Biomembr. 32, 507–515 (2000).
Kwiecinski, J. M. & Horswill, A. R. Staphylococcus aureus bloodstream infections: pathogenesis and regulatory mechanisms. Curr. Opin. Microbiol. 53, 51–60 (2020).
Seidl, K. et al. Effect of a glucose impulse on the CcpA regulon in Staphylococcus aureus. BMC Microbiol. 9, 95 (2009).
Xiao, F. et al. A new CcpA binding site plays a bidirectional role in carbon catabolism in Bacillus licheniformis. iScience 24, 102400 (2021).
Psychogios, N. et al. The human serum metabolome. PLoS One 6, e16957 (2011).
Choe, M., Park, Y. H., Lee, C. R., Kim, Y. R. & Seok, Y. J. The general PTS component HPr determines the preference for glucose over mannitol. Sci. Rep. 7, 43431 (2017).
Beisel, C. L. & Afroz, T. Rethinking the hierarchy of sugar utilization in bacteria. J. Bacteriol. 198, 374–376 (2016).
Gorke, B. & Stulke, J. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat. Rev. Microbiol. 6, 613–624 (2008).
de Carvalho, L. P. et al. Metabolomics of Mycobacterium tuberculosis reveals compartmentalized co-catabolism of carbon substrates. Chem. Biol. 17, 1122–1131 (2010).
Borah, K. et al. Metabolic fluxes for nutritional flexibility of Mycobacterium tuberculosis. Mol. Syst. Biol. 17, e10280 (2021).
Benoni, R. et al. Activation of an anti-bacterial toxin by the biosynthetic enzyme CysK: mechanism of binding, interaction specificity and competition with cysteine synthase. Sci. Rep. 7, 8817 (2017).
Linzner, N. et al. Staphylococcus aureus Uses the Bacilliredoxin (BrxAB)/Bacillithiol Disulfide Reductase (YpdA) Redox pathway to defend against oxidative stress under infections. Front Microbiol 10, 1355 (2019).
Perera, V. R., Newton, G. L. & Pogliano, K. Bacillithiol: a key protective thiol in Staphylococcus aureus. Expert Rev. Anti Infect. Ther. 13, 1089–1107 (2015).
Ragle, B. E., Karginov, V. A. & Bubeck Wardenburg, J. Prevention and treatment of Staphylococcus aureus pneumonia with a beta-cyclodextrin derivative. Antimicrob. Agents Chemother. 54, 298–304 (2010).
Huo, S. et al. Overcoming Planktonic and Intracellular Staphylococcus aureus-associated infection with a cell-penetrating peptide-conjugated antimicrobial peptide. ACS Infect. Dis. 6, 3147–3162 (2020).
Parker, D. & Prince, A. Immunopathogenesis of Staphylococcus aureus pulmonary infection. Semin. Immunopathol. 34, 281–297 (2012).
Valentino, M. D. et al. Genes contributing to Staphylococcus aureus fitness in abscess- and infection-related ecologies. mBio 5, e01729–01714 (2014).
Van Tyne, D. et al. Impact of antibiotic treatment and host innate immune pressure on enterococcal adaptation in the human bloodstream. Sci. Transl. Med. 11, eaat8418 (2019).
Lee, W. et al. Antibiotic combinations that enable one-step, targeted mutagenesis of chromosomal genes. ACS Infect. Dis. 4, 1007–1018 (2018).
Truong-Bolduc, Q. C. et al. Role of the Tet38 Efflux Pump in Staphylococcus aureus Internalization and Survival in Epithelial Cells. Infect. Immun. 83, 4362–4372 (2015).
Yang, J., Liang, X. & Ji, Y. The mutated staphylococcal H35A alpha-toxin inhibits adhesion and invasion of Staphylococcus aureus and group A streptococci. Virulence 4, 77–81 (2013).
Yang, J., Liang, X. & Ji, Y. The novel transcriptional regulator SA1804 Is involved in mediating the invasion and cytotoxicity of Staphylococcus aureus. Front. Microbiol. 6, 174 (2015).
Fey, P. D. et al. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. mBio 4, e00537–00512 (2013).
Buras, J. A., Holzmann, B. & Sitkovsky, M. Animal models of sepsis: setting the stage. Nat. Rev. Drug Discov. 4, 854–865 (2005).
Tjaden, B. A computational system for identifying operons based on RNA-seq data. Methods 176, 62–70 (2020).
Liew, A. T. F. et al. A simple plasmid-based system that allows rapid generation of tightly controlled gene expression in Staphylococcus aureus. Microbiology 157, 666–676 (2011).
Legant, W. R. et al. High-density three-dimensional localization microscopy across large volumes. Nat. Methods 13, 359–365 (2016).
Yu, J. S. et al. Potential antimicrobial activity of galloyl-flavonoid glycosides From Woodfordia uniflora against Methicillin-resistant Staphylococcus aureus. Front Microbiol 12, 784504 (2021).
Kim, J. H. et al. Reducing Peptidoglycan crosslinking by chemical modulator reverts beta-lactam resistance in Methicillin-Resistant Staphylococcus aureus. Adv. Sci. 11, e2400858 (2024).
Kesanakurti, D., Chetty, C., Rajasekhar Maddirela, D., Gujrati, M. & Rao, J. S. Functional cooperativity by direct interaction between PAK4 and MMP-2 in the regulation of anoikis resistance, migration and invasion in glioma. Cell Death Dis. 3, e445 (2012).
Acknowledgements
We thank all members of the Wonsik Lee laboratory for their help in the experiments. We also thank the TEM analysis center of KIST for help in TEM imaging. This work was supported by the National Research Foundation (NRF) of Korea (NRF-2020M3A9H5104234, RS-2023-00228746, RS-2022-NR070873, and RS-2025-00555658 to W.L. and NRF-2020R1I1A1A01052803 and RS-2023-00243009 to J.C.). This work was also supported by the National Institutes of Health of the United States (P01AI083214 to S.W). The schematics in Fig. 1a, Fig. 3a, Fig. 4a, Fig. 6a, and Fig. 6b were created using BioRender.
Author information
Authors and Affiliations
Contributions
W.L. and S.W. conceived the project. W.L., S.W., and J.C. designed and coordinated the overall study. The experiments and data analysis were performed by J.C., C.L., I.K., J.K., J.H.K., and T.Y. The manuscript was drafted by W.L., J.C., C.L., D.H., and S.W., and edited by input from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Chang, J., Lee, C., Kim, I. et al. Environmental cues in different host niches shape the survival fitness of Staphylococcus aureus. Nat Commun 16, 6928 (2025). https://doi.org/10.1038/s41467-025-62292-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-62292-x