Abstract
ADAR1 is an RNA editing enzyme which prevents autoimmunity by blocking interferon responses triggered by cytosolic RNA sensors, and is a potential target in immuno-oncology. However, predictive biomarkers for ADAR1 inhibition are lacking. Using multiple in vitro and in vivo systems, we show that BRCA1/2 and ADAR1 are synthetically lethal, and that ADAR1 activity is upregulated in BRCA1/2-mutant cancers. ADAR1 depletion in BRCA1-mutant cells causes an increase in R-loops and consequently, an upregulation of cytosolic nucleic acid sensing pattern recognition receptors (PRR), events which are associated with a tumor cell-autonomous type I interferon and integrated stress response. This ultimately causes autocrine interferon poisoning. Consistent with a key role of R-loops in this process, exogenous RNase H1 expression reverses the synthetic lethality. Pharmacological suppression of cell-autonomous interferon responses or transcriptional silencing of cytosolic nucleic acid sensing PRR are also sufficient to abrogate ADAR1 dependency in BRCA1-mutant cells, in line with autocrine interferon poisoning playing a central part in this synthetic lethality. Our findings provide a preclinical rationale for assessing ADAR1-targeting agents in BRCA1/2-mutant cancers, and introduces a conceptually novel approach to synthetic lethal treatments, which exploits tumor cell-intrinsic cytosolic immunity as a targetable vulnerability of cancer cells.
Similar content being viewed by others
Introduction
Adenosine Deaminase Acting on RNA 1 (ADAR1) catalyzes the conversion of adenosines to inosines in double-stranded RNA (dsRNA) substrates – a post-transcriptional process referred to as “A-to-I RNA editing”1. ADAR1 is part of a family of three paralogs that share a common structural backbone, consisting of two or three N-terminal dsRNA-binding domains and one C-terminal catalytic deaminase domain. ADAR1 uniquely harbors two Z-DNA binding domains, conferring the ability to recognize the left-handed helical variant of DNA (Z-DNA) in a sequence-independent manner. ADAR1 exists as two isoforms: (i) a full-length, interferon-stimulated ADAR1p150 isoform predominantly localized in the cytoplasm (which potentially shuttles to and from the nucleus); and (ii) an N-terminally truncated ADAR1p110 isoform exclusively localized in the nucleus1.
The canonical function of ADAR1 is editing of dsRNA species derived primarily from non-coding genomic regions (notably introns and 3′-UTRs harboring Alu elements) and, less frequently, from protein-coding regions of mRNAs2. By carrying out this activity, cytoplasmic ADAR1p150 prevents autoimmune responses to endogenous transcripts by limiting the extent of dsRNA sensing mediated by pattern recognition receptors (PRR) such as RIG1, MDA5, LGP2, PKR, or ZBP13. ADAR1, particularly ADAR1p110 isoform, has also been found to maintain genome stability by promoting the resolution of R-loop at telomeres, stalled replication forks and DNA double-strand breaks (DSBs)4,5,6. These ostensibly distinct functions of ADAR1 link the DNA damage response (DDR) and anti-tumor immunity7,8.
ADAR1 is an attractive therapeutic target in immuno-oncology, on the basis that inhibiting ADAR1 could increase tumor immunogenicity by suppressing RNA editing9,10. Previous studies have proposed that triple-negative breast cancer (TNBC) cells with a heightened interferon-stimulated gene signature are sensitive to ADAR1 gene silencing11,12,13 as are tumor cells with elevated levels of dsRNA species14,15. However, as yet, no clinically-actionable predictive biomarkers for ADAR1 inhibition sensitivity have been discovered.
In this work, we uncover a robust and penetrant synthetic lethality (SL)16 between BRCA1/2 and ADAR1. In contrast to other BRCA1/2 SL effects – such as that mediated by PARP inhibitors which relies on DNA damage, cGAS-STING pathway activation and T-cell-mediated adaptive immune response17,18, we show that the BRCA1/2–ADAR1 SL is a tumor cell-autonomous effect not only caused by R-loops, but importantly, by a resultant PRR activation leading to autocrine interferon poisoning.
Results
A BRCA1/2–ADAR1 synthetic lethality exists both in vitro and in vivo
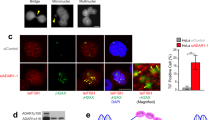
Our prior work, and that of others, has shown that BRCA1/2-mutant (BRCAm) tumor cells display elevated cGAS-STING signaling that can be exacerbated by exposure to PARP inhibitors (PARPi)17,18,19,20,21. Although this relationship has been established using both in vitro and in vivo models of cancer, there is no evidence of BRCAm tumor cells being reliant upon cGAS-STING signaling for survival, and inactivation of cGAS-STING signaling only alters the response to PARPi in vivo in presence of a functional immune system and not in vitro. This indicates that the contribution of cGAS-STING signaling to the BRCA1/2–PARPi SL is not tumor cell-autonomous (i.e., it relies on non-tumor cell lineages) and is best explained by cGAS-STING signaling activating a T-cell-mediated immune response17,18. We hypothesized that, unlike cGAS or STING, other nucleic acid sensing pattern recognition receptors (PRR) might be essential for BRCAm tumor cells. To assess this, we carried out a focused small-interfering RNA (siRNA) screen to identify synthetic lethal (SL) interactions between nucleic acid sensing PRR and BRCA1. In this screen, we used a BRCA1-isogenic system consisting of the BRCA1-mutant (c.2288delT, p.N723fsX13), SUM149 triple-negative breast cancer (TNBC) cell line (SUM149 BRCA1-Mut) and a previously described daughter clone, SUM149 BRCA1-Rev, with a secondary reversion mutation in BRCA1 (c.[2288delT;2293del80]) that restores BRCA1 function22 and PARPi resistance (Fig. 1A, and Supplementary Fig. 1A). Cells were reverse transfected with a siRNA library targeting 18 PRR-encoding genes and cell viability was estimated six days later (Fig. 1B). siRNA targeting ADAR1, but none of the other PRR-targeting siRNAs, elicited BRCA1 SL (Fig. 1C; median surviving fraction of 20.4% in SUM149 BRCA1-Mut vs. 96.7% in SUM149 BRCA1-Rev cells; P < 0.0001, two-way ANOVA).
A Schematic showing experimental design for the generation of SUM149 BRCA1-isogenic cell lines. B, C A focused siRNA screen (B) identifies BRCA1–ADAR1 synthetic lethality in the SUM149 BRCA1-isogenic model (C). Box-and-whiskers indicate median, lower and upper quartiles, and the min to max range; n = 4 biological replicates, two-way ANOVA post hoc Šidák’s test. P value, ****< 0.0001. D, E Clonogenic survival of SUM149 BRCA1-Mut and BRCA1-Rev cells transfected with ADAR1 siRNA (P, Pool; #1; #2). siCTRL, non-targeting, negative control siRNA; siPLK1, PLK1-targeting, positive control siRNA. Violin plots indicate median, lower and upper quartiles; N = 6 values from individual wells, representative of n = 3 biologically-independent experiments, two-way ANOVA post hoc Dunnett’s test. P values, *=0.0196, ****< 0.0001. F, G Clonogenic survival of SUM149 BRCA1-Mut and BRCA1-Rev cells transfected with ADAR1 sgRNA (#1; #2; #3; #4). sgCTRL, non-targeting, negative control sgRNA. Violin plots indicate median, lower and upper quartiles; N = 3 values from individual wells, representative of n = 3 biologically-independent experiments, two-way ANOVA post hoc Dunnett’s test. P values, **=0.0015, ****< 0.0001. Source data are provided as a Source Data file. Elements of panels A and B were provided by Servier Medical Art (https://smart.servier.com/) and BioRender (https://www.biorender.com/), licensed under CC BY 4.0 (https://creativecommons.org/licenses/by/4.0/).
In validation experiments, we found that the transfection of ADAR1-targeting siRNA SMARTPool (siADAR1-P; a pool of four different siRNAs targeting ADAR1, which was used in the screen) as well as two individual ADAR1 siRNAs (siADAR1-#1 and -#2, deconvoluted from the pool) silenced both ADAR1p110 and ADAR1p150 isoforms (Supplementary Fig. 1B) and selectively inhibited clonogenic survival of SUM149 BRCA1-Mut cells when compared to SUM149 BRCA1-Rev cells (Fig. 1D, E; and Supplementary Fig. 1C, D). To eliminate the possibility of an off-target effect of the siRNAs, we carried out two orthogonal experimental approaches to target ADAR1 in SUM149 BRCA1-isogenics: (i) CRISPR-Cas9 mutagenesis of ADAR1 using four different single-guide RNAs (sgRNAs); and (ii) transcriptional silencing of ADAR1 using a doxycycline-inducible short-hairpin RNA (shRNA). Both approaches depleted ADAR1 levels and led to a reduction in clonogenic survival, cell viability and cell proliferation of SUM149 BRCA1-Mut cells, while SUM149 BRCA1-Rev cells were less affected (Fig. 1F, G; and Supplementary Fig. 1E–L). The extent of these effects was proportionate to the level of ADAR1 silencing (Supplementary Fig. 1I–L), suggesting a correlation between cell survival and residual ADAR1 expression in BRCA1-mutant cells.
Many SL effects have incomplete penetrance, i.e., are limited to a small number of models, fail to operate in molecularly-diverse backgrounds and/or are often restricted to specific cancer histotypes16. To explore whether the BRCA1–ADAR1 SL effect was private to SUM149 cells or more penetrant, we assessed the effects of siRNA- or CRISPR-Cas9-mediated targeting of ADAR1 in ten additional independent models (Fig. 2A): (i) four isogenic systems of BRCA1 deficiency, including mouse (Mus musculus, Mm) embryonic fibroblasts (MEFs)23, human retinal pigment epithelial cells (RPE1)24, mouse mammary carcinoma cells (4T1)25 or mouse ovarian carcinoma cells (ID8)26; and (ii) a molecularly diverse, non-isogenic panel of six human TNBC cell lines with/without endogenous BRCA1 mutations (BRCA1-wildtype: MDA-MB-231, CAL51, CAL120, Hs578T; BRCA1-mutant: MDA-MB-436, HCC1937). The homologous recombination (HR) status of these cell lines was confirmed by assessing PARPi sensitivity (Supplementary Figs. 1–2). In all models, BRCA1-mutant cells were significantly more sensitive to ADAR1 silencing than BRCA1-wildtype cells (Fig. 2B–D; and Supplementary Fig. 1N; Supplementary Fig. 2). In assessing whether ADAR1 SL effects extended to BRCA2-mutant cells, we found that ADAR1 silencing elicited SL in two different BRCA2-isogenic systems: human BRCA2-knockout colorectal carcinoma cells (DLD1)27 and mouse Brca2-knockout 4T1 cells25 (Fig. 2D; and Supplementary Fig. 2G–J, 3A-F). We also confirmed the BRCA1/2–ADAR1 SL effect in isogenic ADAR1-wildtype or -knockout human embryonic kidney cells (HEK293T) subjected to BRCA1 or BRCA2 silencing28 (Fig. 2D–F).
A Schematic describing the isogenic and non-isogenic cell line models used throughout the study. B, C Clonogenic survival of MEF Brca1-wildtype (WT) and Brca1-mutant (Δ11) cells transfected with a concentration range (nM) of Adar1 siRNA. Violin plots indicate median, lower and upper quartiles; N = 6 values from individual wells, representative of n = 3 biologically-independent experiments, two-way ANOVA post hoc Dunnett’s test. P values, *=0.0402, **=0.0017, ****< 0.0001. D Heatmap showing surviving fractions elicited by ADAR1 suppression in models evaluated in Figs. 1, 2 and Supplementary Fig. 1–3 (blue, SF > 0.8; red, SF < 0.8). E, F Cell survival of HEK293T ADAR1-wildtype (WT) and ADAR1-knockout (KO) cells transfected with a concentration range (nM) of BRCA1 (E) or BRCA2 (F) siRNA. Box-and-whiskers indicate median, lower and upper quartiles, and the min to max range; N = 4 values from individual wells, representative of n = 3 biologically-independent experiments, two-way ANOVA post hoc Dunnett’s test. P values, **[ADAR1-WT, siBRCA2 2 nM]=0.001 (F), ***[ADAR1-KO, siBRCA1 2 nM]=0.0009 (E), ***[ADAR1-KO, siBRCA1, 4 nM]=0.0005 (E), ****<0.0001. G–J Representative images and quantifications of morphological phenotypes (G, I) and acridine orange staining (H, J) in zebrafish embryos subjected to morpholino (MO)-mediated knockdown of brca2 and/or adar1. Dead embryos are indicated with an asterisk (G). White arrows indicate acridine orange-positive cells on images taken within a defined region along the anterior-posterior axis (H); scale bar, 500 µm. Percentages of morphological phenotypes (I) were calculated based on N = 149 [no MO], N = 95 [control MO], N = 109 [brca2 MO], N = 97 [adar1 MO] and N = 174 [brca2/adar1 MOs] values from individual embryos, representative of n = 3 biologically-independent clutches. Violin plots indicate median, lower and upper quartiles; N = 31 [no MO], N = 33 [control MO], N = 25 [brca2 MO], N = 23 [adar1 MO] and N = 24 [brca2/adar1 MOs] values from individual embryos, representative of n = 3 biologically-independent experiments, Kruskal-Wallis test post hoc Dunn’s test. P value, ****<0.0001. siCTRL, non-targeting, negative control siRNA; siPLK1, PLK1-targeting, positive control siRNA. Source data are provided as a Source Data file. Elements of panel A were provided by Servier Medical Art (https://smart.servier.com/), licensed under CC BY 4.0 (https://creativecommons.org/licenses/by/4.0/).
Considering the predominant role of ADAR1p150 isoform in mediating RNA editing29 and averting dsRNA sensing-driven autoimmunity30, we hypothesized that the loss of ADAR1p150 function might be sufficient to elicit SL in BRCAm tumor cells. Short-term survival assays in SUM149 BRCA1-isogenics subjected to transfection with an ADAR1p150-specific siRNA revealed that ADAR1p150 silencing phenocopied the SL elicited by pan-isoform ADAR1 siRNAs in BRCA1-mutant cells (Supplementary Fig. 3G, H). Similar SL effects were also observed in an ADAR1p150-null (p110-intact) HEK293T clone28 subjected to BRCA1 or BRCA2 silencing to the same extent as in isogenic ADAR1-knockout cells (Supplementary Fig. 3I–L), corroborating the existence of a BRCA1/2–ADAR1p150 SL. To further explore the relative contribution of ADAR1 isoforms to the SL, we conducted short-term survival assays in SUM149 BRCA1-isogenic cells subjected to (i) ADAR1 knockdown with an siRNA targeting ADAR1 3’-UTR region and (ii) concomitant exogenous overexpression of ADAR1p110 or ADAR1p150. We found that overexpression of ADAR1p150, but not ADAR1p110, could reverse the BRCA1–ADAR1 SL effects (Supplementary Fig. 3M, N), supporting a key role for ADAR1p150 in this SL. To next investigate whether ADAR1p150 deaminase activity was involved in the SL, we transfected plasmids encoding wildtype ADAR1p150, the deaminase-defective mutant G1007R (p150-ΔCD), or the Z-DNA binding-defective mutant P193A (p150-ΔZα)31 in SUM149 models. We observed that ADAR1p150-ΔZα, but not ADAR1p150-ΔCD, significantly reversed the SL effects (Supplementary Fig. 3O), indicating the essentiality of ADAR1p150 deaminase activity in BRCA1-mutant cells survival.
In mice, constitutive Brca2 deletion is embryonically lethal32, whereas in zebrafish (Danio rerio, Dr), its orthologue brca233 can be inactivated where it causes a HR defect and PARPi sensitivity34,35. Moreover, the zebrafish adar1 gene conserves the A-to-I RNA editing36,37 and protection against aberrant interferon signaling38 functions, similar to human ADAR1. This allowed us to assess the BRCA2–ADAR1 SL in a whole organism system, using a translation-blocking morpholino (MO) approach39 (Fig. 2G–J). In fish injected with either brca2 or adar1 MO alone, a large majority of offspring survived and developed normally (alive / normal morphology: brca2 MO, 93% / 87%; adar1 MO, 86% / 65%; control MO, 96% / 95%; Fig. 2G, I). In contrast, in fish injected concomitantly with adar1 and brca2 MOs, offspring showed a significantly higher percentage of mortality (48%), and all remaining living larvae showed different degrees of abnormal morphology (mild, 11%; severe, 41%; Fig. 2G, I) associated with apoptosis, as assessed by acridine orange staining (Fig. 2H, J).
Altogether, this data showed that the BRCA1/2–ADAR1 SL operates in vitro and in vivo across a variety of model systems (Fig. 2D), and that the concomitant loss of adar1 and brca2 in zebrafish is embryonically lethal.
ADAR1-mediated RNA editing is upregulated in BRCA1/2-mutant cancers
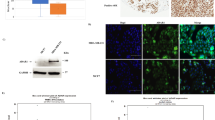
Given the increased reliance of BRCAm cells upon ADAR1 function, we next sought evidence for a dysregulation of ADAR1 activity in BRCAm cancers. To this aim, we conducted a series of analyses assessing ADAR1 expression and activity in patient-derived samples of TNBC and prostate cancer – two of the cancer types in which BRCA1/2 mutations are the most prevalent. First, we built a cohort of 63 patients with TNBC (including 32 BRCA1-wildtype and 31 BRCA1-mutant tumor samples) and optimized an immunohistochemistry assay to evaluate nuclear vs. cytoplasmic ADAR1p150 expression in tumor cells. This revealed that BRCA1-mutant tumors displayed a higher cytoplasmic ADAR1p150 expression (Fig. 3A, B; P = 0.0359, Mann-Whitney U test), while no significant difference was observed regarding ADAR1p150 nuclear expression (Supplementary Fig. 4A; P = 0.3976, not significant). Interestingly, higher cytoplasmic ADAR1p150 expression was also associated with increased tumor-infiltrating lymphocytes, regardless of the BRCA1 gene status (Fig. 3C; Supplementary Fig. 4B–E). As the upregulation of BRCA1/2 synthetic lethal partners is a general feature of HR-defective cancers40, one reasonable assumption is that this higher ADAR1 expression seen in BRCA1-mutant cancers reflects the dependency these cancers have upon ADAR1 function.
A, B Pathological evaluation (A) and representative images (B) of ADAR1p150 cytoplasmic expression according to BRCA1 gene status (BRCA1-wildtype vs. BRCA1-mutant) in a cohort of 63 treatment-naïve triple-negative breast cancer (TNBC) patients. H-score of ADAR1p150 expression (range, 0–300). Violin plots indicate median, lower and upper quartiles; N = 32 [BRCA1-wildtype], N = 31 [BRCA1-mutant] values from individual tumor samples, two-tailed Mann-Whitney U test. P value, *=0.0359. Hematoxylin and eosin (H&E) and ADAR1p150 staining (magnification, ×20) are shown. Scale bars, 50 μm. C Percentage of TILs in TNBC tumors from the cohort described in A, according to cytoplasmic ADAR1p150 expression (based on A; ADAR1p150-low, lower quartile of H-score; ADAR1p150-high, upper quartile of H-score). Violin plots indicate median, lower and upper quartiles; N = 17 [BRCA1-wildtype], N = 15 [BRCA1-mutant] values from individual tumor samples, two-tailed Mann-Whitney U test. P value, **=0.0043. D, E Schematics illustrating the conceptual approach (D) and bioinformatic pipelines (E) used to evaluate A-to-I RNA editing levels from RNA-Seq data. F, G. A-to-I RNA editing levels displayed as RNA editing index (F) or number of RNA editing sites (G) in SUM149 BRCA1-Mut and BRCA1-Rev cells transfected with ADAR1 siRNA (P, Pool; #1). siCTRL, non-targeting, negative control siRNA. Bar plots indicate mean ± SD; n = 3 biological replicates, two-way ANOVA post hoc Tukey’s test. P values, ****< 0.0001. H Schematic of clinical history of the BRCA2-mutant and -revertant patient-derived xenografts (PDXs) MR-0009 and MR-0191; arrows indicate times of tumor biopsies for PDX establishment. Duration of each treatment delivered after diagnosis is indicated in months. Details of the corresponding BRCA2 mutations (germline vs. reversion) are presented to the left. I, J A-to-I RNA editing levels displayed as RNA editing index (I) or number of RNA editing sites (J) in BRCA2-mutant and -revertant PDXs MR-0009 and MR-0191. Bar plots indicate mean, where applicable; N = 1 [MR-0009 BRCA2-Mut], N = 2 [MR-0009 BRCA2-Rev], N = 1 [MR-0191 BRCA2-Mut], N = 1 [MR-0191 BRCA2-Rev] values from individual tumor samples. Source data are provided as a Source Data file. Elements of panel H were provided by Servier Medical Art (https://smart.servier.com/), licensed under CC BY 4.0 (https://creativecommons.org/licenses/by/4.0/).
Previous studies have shown that ADAR1-mediated RNA editing was elevated in breast cancer as compared with matched, normal breast tissue41,42 and compared with other types of cancers41, but the impact of BRCA1/2 mutations on the magnitude of A-to-I RNA editing in cancer remains unknown. To further investigate a functional link between the BRCAm genotype and ADAR1 activity, we sought to measure RNA editing levels (i.e., the frequency of A-to-I editing events in RNA transcripts, used as a surrogate marker of ADAR1 activity) in BRCA1/2-isogenic cell lines or patient-derived samples. To do this, we first generated RNA-Seq data of SUM149 BRCA1-isogenic cells exposed to ADAR1 silencing by transfection of siADAR1-P or siADAR1-#1 (Supplementary Fig. 4F) and used two complementary approaches to identify A-to-I RNA editing events from raw RNA-Seq reads (Fig. 3D, E)43,44. As expected, silencing of ADAR1 caused a dramatic reduction of RNA editing in both BRCA1-mutant and -revertant cells as measured by a decreasing A-to-I RNA Editing Index (REI; Fig. 3F) and fewer A-to-I RNA Editing Sites (RES; Fig. 3G). More importantly, these analyses revealed a significantly higher A-to-I RNA editing activity of BRCA1-mutant cells compared with BRCA1-revertant cells (mean number of 143,169 vs. 62,630 RES, respectively; P < 0.0001, two-way ANOVA; Fig. 3F, G), despite similar ADAR1 protein levels (Supplementary Figs. 1B, D, 4F). This difference was further specific to A-to-I RNA editing, as levels of editing observed for all other types of editing remained extremely low and unchanged (mean number of RES < 1,200 for C-to-T transitions and close to null for all transversions; Supplementary Fig. 4G-P). To assess whether such correlation would also operate in patients, we took advantage of two previously described isogenic patient-derived xenograft (PDX) models of prostate cancer – termed MR-0009 and MR-019145, characterized by the presence of BRCA2 germline and secondary reversion mutations conferring sensitivity or resistance to PARPi, respectively (Fig. 3H). As in SUM149 BRCA1-isogenics, BRCA2-mutant PDXs exhibited a greater REI / more RES than their isogenic BRCA2-revertant counterparts (Fig. 3I, J), despite similar ADAR1 transcript levels (MR-0009, 98.7 vs. 84.5 TPM; MR-0191, 48.4 vs. 50.8 TPM for BRCA2-Mut vs. BRCA2-Rev, respectively).
ADAR1 suppresses DNA damage in BRCA1/2-mutant cells by preventing R-loop accumulation
Since the vast majority of BRCA1/2 SL effects involve DNA repair proteins (e.g., PARP1, POLQ, APEX2, FEN1, CIP2A)46 and result from an impaired DDR47, we hypothesized that the causes of the BRCA1/2–ADAR1 SL may also reside in an inability to process certain types of DNA damage. Indeed, we found that ADAR1 silencing in BRCAm cells of three different isogenic models (SUM149, MEF and 4T1) elicited nuclear γ-H2AX foci (Fig. 4A–D; and Supplementary Fig. 5A–H) and in SUM149 BRCA1-Rev cells elicited RAD51 foci (Supplementary Fig. 5C–E), suggesting that the loss of ADAR1 caused DNA damage mainly in BRCAm cells, and invoked RAD51-mediated HR repair in BRCA1-revertant cells. ADAR1 silencing also caused micronucleation in BRCAm but not in BRCA1-revertant or BRCA1/2-wildtype cells (Fig. 4E-H; Supplementary Fig. 5I), suggesting a selective induction of chromosomal instability in BRCAm cells. We further noted that ADAR1 silencing in BRCA1-mutant cells caused a significant accumulation of RPA foci in S-phase population (Fig. 4I, J; and Supplementary Fig. 5J), increased CHK1 phosphorylation and PARP1 cleavage as assessed by western blot (Fig. 4K, L; and Supplementary Fig. 5K, L), while BRCA1-revertant cells showed little or no change in these marks, suggesting increased replication stress and apoptosis in BRCA1-mutant cells subjected to ADAR1 knockdown.
A–D Representative images and quantifications of γ-H2AX foci (number of γ-H2AX foci per nucleus) in SUM149 BRCA1-Mut and BRCA1-Rev cells (A, B) or MEF Brca1-wildtype (WT) and Brca1-mutant (Δ11) cells (C, D) transfected with ADAR1 siRNA (P, Pool; #1; #2). Violin plots indicate median, lower and upper quartiles; N = 150 values from individual nuclei, representative of n = 3 biologically-independent experiments, two-way ANOVA post hoc Dunnett’s test (B) or Šidák’s test (D). P values, *=0.0101, **=0.0014, ****< 0.0001. E–H Representative images and quantifications of micronuclei (percentage of cells harboring > 1 micronucleus in the assessed population) in SUM149 BRCA1-Mut and BRCA1-Rev cells (E, F) or MEF Brca1-wildtype (WT) and Brca1-mutant (Δ11) cells (G, H) transfected with ADAR1 siRNA (P, Pool; #1; #2). Scatter dot plots indicate mean ± SD; N = 3 values from individual microscopic fields, representative of n = 3 biologically-independent experiments, two-way ANOVA post hoc Dunnett’s test (F) or Šidák’s test (H). P values, **=0.0099, ****< 0.0001. I, J Representative images and quantifications of RPA foci (number of RPA foci per nucleus) in CCNA2-positive SUM149 BRCA1-Mut and BRCA1-Rev cells transfected with ADAR1 siRNA (P, Pool; #1). Violin plots indicate median, lower and upper quartiles; N = 150 values from individual nuclei, representative of n = 3 biologically-independent experiments, two-way ANOVA post hoc Dunnett’s test. P values, ****<0.0001. K, L Western blot of SUM149 BRCA1-Mut and BRCA1-Rev cells (K) or MEF Brca1-wildtype (WT) and Brca1-mutant (Δ11) cells (L) transfected with a concentration range (nM) of ADAR1 siRNA. Data representative of n = 2 biologically-independent experiments. siCTRL, non-targeting, negative control siRNA; siPLK1, PLK1-targeting, positive control siRNA. Source data are provided as a Source Data file.
ADAR1 was recently found to facilitate R-loop resolution via A-to-I editing of RNA:DNA hybrids4,5,6,48. To explore whether an increase in R-loops could explain the BRCA1/2–ADAR1 SL, we overexpressed RNase H1 (RH1), one of the main endonucleases responsible for R-loop degradation. RH1 overexpression in BRCA1-mutant cells reversed their sensitivity to ADAR1 silencing in colony-formation assay (Fig. 5A, B) and to a lesser extent in short-term survival assays (Supplementary Fig. 6A–E). Consistent with recent reports4,6,49, immunofluorescence assays revealed that upon ADAR1 silencing, BRCA1-mutant cells demonstrated a selective increase in the formation of RNase H-sensitive S9.6 foci, and of aberrantly-shaped or ectopic nucleoli (Fig. 5C, D; Supplementary Fig. 6F-J), suggesting an accumulation of R-loops and nucleoli disruption, as previously described in other contexts49. Interestingly, RH1 overexpression also partially reversed the γ-H2AX, RPA foci and micronuclei phenotypes observed in BRCA1-mutant cells transfected with ADAR1 siRNA, and likewise, the RAD51 foci phenotype in BRCA1-revertant cells (Fig. 5E; and Supplementary Fig. 7), implying a contribution of R-loops to the DDR elicited by ADAR1 knockdown.
A, B Clonogenic survival of SUM149 BRCA1-Mut and BRCA1-Rev cells transfected with ADAR1 siRNA (P, Pool; #1; #2) in the context of exogenous overexpression of RNase H1 (RH1). Violin plots indicate median, lower and upper quartiles; N = 6 values from individual wells, representative of n = 3 biologically-independent experiments, two-way ANOVA post hoc Dunnett’s test. P values, *[BRCA1-Mut +RH1, siADAR1-#1] = 0.0109, *[BRCA1-Rev, siADAR1-P] = 0.019, **[BRCA1-Rev, siADAR1-#1] = 0.0084, **[BRCA1-Rev, siADAR1-#2] = 0.0071, ****<0.0001. C, D Representative images and quantifications of R-loops (number of fibrillarin-negative S9.6 foci per nucleus) in SUM149 BRCA1-Mut and BRCA1-Rev cells transfected with ADAR1 siRNA (P, Pool; #1). Violin plots indicate median, lower and upper quartiles; N = 150 values from individual nuclei, representative of n = 3 biologically-independent experiments, two-way ANOVA post hoc Tukey’s test. P values, **=0.0032, ****< 0.0001. E Quantification of γ-H2AX foci in SUM149 BRCA1-Mut and BRCA1-Rev cells transfected with ADAR1 siRNA (P, Pool; #1) in the context of exogenous overexpression of RNase H1 (RH1). Violin plots indicate median, lower and upper quartiles; N = 150 values from individual nuclei, representative of n = 3 biologically-independent experiments, two-way ANOVA post hoc Dunnett’s test. P value, ****< 0.0001. siCTRL, non-targeting, negative control siRNA; siPLK1, PLK1-targeting, positive control siRNA. Source data are provided as a Source Data file.
Altogether, these results indicated that ADAR1 protects cells from R-loop-mediated DNA damage and genomic instability, and that such function is critical for the survival of BRCAm cells.
ADAR1 hedges against pattern recognition receptors-dependent, cell-autonomous interferon poisoning in BRCA1/2-mutant cells
Although R-loops appeared to play a key role in the BRCA1/2–ADAR1 SL, we asked whether other consequences of ADAR1 inhibition may also be involved. As ADAR1 suppresses dsRNA sensing innate immune responses through RNA editing3, and tumor cell lines with an elevated interferon-stimulated gene (ISG) signature are sensitive to ADAR1 silencing11,12,13, we sought to test whether cytosolic nucleic acid sensing might also contribute to the BRCA1/2–ADAR1 SL effects. First, we measured in three different BRCA1/2-isogenic models (SUM149, MEF and 4T1) the expression and/or phosphorylation levels of proteins involved in ADAR1-dependent immunity, including (i) key dsRNA-sensing PRR (RIG1, MDA5, LGP2, PKR); (ii) markers of the integrated stress response (ISR; p-PKR, p-eIF2α); and (iii) markers of the type I interferon response (p-IRF3, p-STAT1). Western blots revealed an increased expression of dsRNA-sensing PRR and phosphorylation of PKR, eIF2α, IRF3 and STAT1 in BRCAm cells but not in their isogenic BRCA1-revertant or BRCA1/2-wildtype counterparts, accompanied by selectively increased transcriptional levels of IFNB1, CCL5, and other ISGs in BRCAm cells of all three models, as assessed by RT-qPCR (Fig. 6A–F; and Supplementary Fig. 8A–E). The latter phenotype was also seen in BRCA1-mutant MDA-MB-436 cells but not BRCA1-wildtype MDA-MB-231 cells (Supplementary Fig. 8F, G). We noted increased IRF3 phosphorylation levels of SUM149 BRCA1-Mut in response to ADAR1 shRNA expression, even in absence of doxycycline (Fig. 6A), suggesting potential promoter leakage of the shRNA in that cell line. Secondly, to further explore the selective activation of an interferon response in BRCA1-mutant cells, we compared the ISG signature of SUM149 BRCA1-Mut and BRCA1-Rev cells following ADAR1 silencing using RNA-Seq. This revealed (i) a constitutively elevated ISG signature in SUM149 BRCA1-Mut cells in absence of ADAR1 silencing (Supplementary Fig. 8H; LFC = 1.66 for BRCA1-Mut vs. BRCA1-Rev, P < 0.0001), and (ii) a selective upregulation of the ISG signature upon ADAR1 silencing in SUM149 BRCA1-Mut cells (Supplementary Fig. 8I; LFC = 0.34 for siADAR1 vs. siCTRL in BRCA1-Mut, P = 0.0029; LFC = 0.16 for siADAR1 vs. siCTRL in BRCA1-Rev, P = 0.0617, not significant), thereby confirming our previous observations (Fig. 6A–F).
A Western blot of SUM149 BRCA1-Mut and BRCA1-Rev cells transduced with a doxycycline-inducible ADAR1-targeting shRNA and exposed to a titration of doxycycline (ng/mL). Data representative of n = 2 biologically-independent experiments. B, C RT-qPCR of IFNB1 (B) and CCL5 (C) mRNAs in SUM149 BRCA1-Mut and BRCA1-Rev cells transfected with a concentration range (nM) of ADAR1 siRNA. IFNB1 and CCL5 mRNAs were analyzed separately relative to GAPDH. Box-and-whiskers show arbitrary units of gene expression normalized to the BRCA1-mutant siCTRL condition; N = 4 values from individual measurements, representative of n = 3 biologically-independent experiments, two-way ANOVA post hoc Dunnett’s test. P values, ***[IFNB1, BRCA1-Rev, siADAR1 1.25 nM]=0.0002, ****< 0.0001. D Western blot of MEF Brca1-wildtype (WT) and Brca1-mutant (Δ11) cells transfected with a concentration range (nM) of Adar1 siRNA. Data representative of n = 2 biologically-independent experiments. E, F RT-qPCR of Ifnb1 (E) and Ccl5 (F) mRNAs in MEF Brca1-wildtype (WT) and Brca1-mutant (Δ11) cells transfected with a concentration range (nM) of Adar1 siRNA. Data presented as in (B, C). P values, ****< 0.0001. G, H Clonogenic survival of SUM149 BRCA1-Mut and BRCA1-Rev cells subjected to co-transfection with ADAR1 siRNA and one of a series of siRNAs targeting pattern recognition receptors. Violin plots indicate median, lower and upper quartiles; N = 6 values from individual wells, representative of n = 3 biologically-independent experiments, two-way ANOVA post hoc Dunnett’s test. P values, ****< 0.0001. I Western blot of SUM149 BRCA1-Mut and BRCA1-Rev cells subjected to co-transfection with ADAR1 siRNA and one of a series of siRNAs targeting pattern recognition receptors. Data representative of n = 3 biologically-independent experiments. J, K Cell survival of SUM149 BRCA1-Mut and BRCA1-Rev cells transfected with a concentration range (nM) of ADAR1 siRNA in the context of exposure to the JAK/STAT pathway inhibitors (JSPi) ruxolitinib (J; 10 µM) or upadacitinib (K; 32 µM). Box-and-whiskers indicate median, lower and upper quartiles, and the min to max range; N = 4 values from individual wells, representative of n = 3 biologically-independent experiments, two-way ANOVA post hoc Dunnett’s test. P values, ****< 0.0001. siCTRL, non-targeting, negative control siRNA; siPLK1, PLK1-targeting, positive control siRNA. Source data are provided as a Source Data file.
We next directly assessed whether the BRCA1/2–ADAR1 SL might depend upon cytosolic nucleic acid sensing, and if so, sought to identify which PRR might be involved. To this aim, we conducted co-silencing experiments in which SUM149 BRCA1-isogenics were transfected with either ADAR1 siRNA and a non-targeting control siRNA or with ADAR1 siRNA and one of a series of siRNAs targeting cytosolic dsRNA sensors (RIG1, MDA5, LGP2, PKR) or the non-canonical RNA:DNA hybrid sensor cGAS. In these assays, we found that the concomitant silencing of ADAR1 with either RIG1, MDA5, LGP2, PKR or cGAS resulted in a significant reversal of ADAR1 SL effects in SUM149 BRCA1-Mut cells (Fig. 6G, H), while the knockdown of each sensor individually had little effect on cell viability in absence of ADAR1 silencing (Supplementary Fig. 8J-M). If the extent of this rescue was modest with MDA5 and RIG1 knockdown, LGP2, PKR or cGAS knockdown elicited a substantial reversal of the SL, akin to the effect of type I interferon receptor (IFNAR1) silencing, used as a positive control (Fig. 6G, H). Consistent with this, we observed that co-transfection of LGP2 siRNA was sufficient to abrogate ADAR1 siRNA-induced phosphorylation of PKR and IRF3, while co-transfection of RIG1 or MDA5 siRNA only partially hindered these effects (Fig. 6I). Co-transfection of PKR siRNA did not affect ADAR1 siRNA-induced phosphorylation of IRF3, in line with the notion that PKR is activated by and acts downstream of type I interferon signaling50. Of note, silencing of cGAS also reduced the expression levels of RIG1, MDA5, and LGP2 while conversely, silencing of either RIG1, MDA5, or LGP2 reduced the expression levels of all three dsRNA sensors (as well as phosphorylation levels of PKR) but not those of cGAS (Fig. 6I). This indicated an interdependence between cytosolic dsRNA, RNA:DNA and DNA sensing pathways as previously described51, and suggested some level of redundancy among these proteins in mediating the BRCA1/2–ADAR1 SL effects.
To corroborate this data, we tested whether increased interferon signaling might also directly contribute to the BRCA1/2–ADAR1 SL. To do so, we evaluated the cytotoxic effects of silencing ADAR1 in presence of non-toxic concentrations of pharmacological inhibitors of the JAK-STAT pathway (JSPi) – namely ruxolitinib (JAK1/2 inhibitor), upadacitinib (JAK1 inhibitor) or deucravacitinib (TYK2 inhibitor), which are commonly used to block downstream interferon signaling52,53. JSPi completely reversed ADAR1 SL effects elicited in BRCA1-mutant SUM149 and MEF cells in short-term survival assays, and these effects further extended to the abrogation of ADAR1-dependent dsRNA-sensing PRR upregulation, PKR and IRF3 phosphorylation (Fig. 6J, K; and Supplementary Fig. 8N, 9A–C). To further test the influence of interferon signaling stimulation on the BRCA1/2–ADAR1 SL, we complemented cell culture media with interferons, and found that non-toxic concentrations of either type I (IFN-α, IFN-β) or type II (IFN-γ) interferons (i) enhanced the cytotoxic effects of ADAR1 silencing in SUM149 BRCA1-Mut cells; and (ii) sensitized SUM149 BRCA1-Rev cells to ADAR1 silencing (Supplementary Fig. 9D–F). We noted that, compared with IFN-γ, type I interferons caused a more profound sensitization of BRCA1-revertant cells, also replicated in BRCA1-wildtype MDA-MB-231 cells (Supplementary Fig. 9G–I).
To investigate the role and biological relevance of PKR activation in the BRCA1/2–ADAR1 SL, we assessed the formation of cytosolic G3BP1 bodies—a marker of stress granules, the prototypical hallmark of ISR—by immunofluorescence in SUM149 BRCA1-isogenics. ADAR1 silencing caused a substantial accumulation of cytosolic G3BP1 bodies selectively in BRCA1-mutant cells, associated with increased colocalizing PKR foci (Supplementary Fig. 9J–L). Based on this, we reasoned that ISR signaling might play a role in BRCA1/2–ADAR1 SL, and next assessed the effect of ISRIB—a pharmacological inhibitor of the ISR54, on the sensitivity of BRCA1-mutant cells to ADAR1 silencing. Short-term survival assays revealed a partial rescue of ADAR1 siRNA-mediated SL effects following exposure to ISRIB in SUM149 BRCA1-Mut cells (Supplementary Fig. 9M), in contrast with the complete rescue observed with JSPi. This was associated with an impaired formation of cytosolic G3BP1 bodies and PKR foci (Supplementary Fig. 9N–P), confirming ISRIB inhibitory activity and suggesting a contribution of PKR-driven ISR to the BRCA1/2–ADAR1 SL.
Since both RH1 overexpression and PRR knockdown reversed the BRCA1/2–ADAR1 SL, we assessed the contribution of R-loops to PRR-driven innate immune responses. To do this, we replicated western blots of dsRNA sensors, type I interferon and ISR markers in SUM149 BRCA1-isogenic cells subjected to (i) ADAR1 silencing and/or (ii) RH1 overexpression and/or (iii) JSPi exposure. Whilst BRCA1-mutant cells were unable to activate a type I interferon response upon ADAR1 silencing when exposed to JSPi (Supplementary Fig. 10A) or co-silencing of RIG1 or LGP2 (Supplementary Fig. 10B), RH1 overexpression also counteracted (albeit partially) the upregulation of PRR, type I interferon and integrated stress response markers elicited upon ADAR1 silencing in BRCA1-mutant cells (Supplementary Fig. 10A, B). Consistent with this, short-term survival assays conducted in the same conditions revealed that RH1 overexpression and exposure to JSPi both contributed to reverse the cytotoxic effects of ADAR1 siRNA in SUM149 BRCA1-Mut cells (Supplementary Fig. 10C).
Altogether, this data supported the idea that increased R-loop burden elicited by ADAR1 suppression associates with PRR-driven cell-autonomous interferon poisoning in BRCAm cells, thereby ultimately causing the BRCA1/2–ADAR1 SL.
Discussion
Here, we uncover the defect in the RNA editing enzyme ADAR1 as a novel genetic vulnerability of BRCA1/2-mutant (BRCAm) cancer cells. Using functional genomics in a panel of diverse BRCA1/2-isogenic model systems, we identify a robust and penetrant synthetic lethal (SL) interaction between BRCA1/2 mutations and the loss of ADAR1, which operates across a variety of molecular backgrounds, cancer histotypes, and species, and also exists reciprocally, in ADAR1-mutated systems (Figs. 1, 2). The evidence that ADAR1 is dysregulated in BRCAm human tumors and PDXs (Fig. 3) supports the translational significance of this SL. Mechanistically, we show that the BRCA1/2–ADAR1 SL relies upon R-loop-associated DNA damage (Figs. 4, 5) and the selective activation of an R-loop- and PRR-driven cell-autonomous innate immune response in BRCAm cells (Fig. 6), providing the first evidence for autocrine interferon poisoning as a mechanism of BRCA1/2 SL46.
We show that when ADAR1 is silenced in BRCA1-mutant cells, overexpression of RNase H1—which reduces R-loop burden— antagonizes ADAR1 SL effects as well as the upregulation of cytosolic nucleic acid sensing PRR, type I interferon and integrated stress response markers, suggesting that R-loops accumulation triggers, directly or indirectly, downstream pattern recognition receptors (PRR) activation and interferon-dependent lethality. Importantly, these findings delineate a distinctive SL mechanism, which differs from those of other BRCA1/2-associated SL (including the BRCA1/2–PARPi SL) in that it depends on cytosolic nucleic acid sensing PRR activation. In line with the recently described immunogenic potential of R-loops55,56,57,58,59,60,61,62, our results indeed support a model (Fig. 7) in which unresolved R-loops elicited by the loss of ADAR1 function4,6,48 activate cytosolic nucleic acid sensing PRR, resulting in a toxic cell-autonomous innate immune response which drives the BRCA1/2–ADAR1 SL. ADAR1p150 plays an essential role in this SL and could be responsible for the suppression of nuclear R-loops in BRCAm cells, consistent with its nucleocytoplasmic distribution63,64. Still, some elements of the BRCA1/2–ADAR1 SL mechanism remain to be explored.
A model for the proposed mechanism driving sensitivity of BRCA1/2-mutant cancers to ADAR1 inhibition. Elements of this figure were provided by Servier Medical Art (https://smart.servier.com/) and BioRender (https://www.biorender.com/), licensed under CC BY 4.0 (https://creativecommons.org/licenses/by/4.0/).
First, the PRR specificity of the BRCA1/2–ADAR1 SL remains unclear. Our results indicate that LGP2, PKR and cGAS silencing most reproducibly rescue ADAR1 SL effects, suggesting a contribution of multiple cytosolic nucleic sensing pathways. Whilst cGAS and PKR are known to detect RNA:DNA hybrids similar to those formed at R-loops55,65, LGP2 has not yet been described to do so, suggesting either that its substrate specificity might be less discrete than expected, or that the activation of some PRR, through crosstalk, causes activation of others. Indeed, considering the interdependence and possible redundancy of these pathways, other PRR might also contribute to these SL effects (e.g., TLR3 or ZBP155,66). Intriguingly, silencing of DHX9 (a PRR which both suppresses cytosolic dsRNA sensing59,67 and bears a nuclear RNA helicase activity against R-loops68) did not elicit BRCA1-dependent SL effects in our original screen, suggesting a possible redundancy of DHX9 function (i) at the nuclear level, with that of other helicases for the resolution of R-loops and/or (ii) at the cytosolic level, with that of other PRR for the suppression of dsRNA sensing-mediated responses.
In this context, the exact nature of cytosolic nucleic acid species triggering cell-autonomous interferon poisoning remains to be defined. Although our data point to a central role of R-loops in the mechanism of this SL, multiple cytosolic nucleic sensing PRR—not restricted to those described as putative RNA:DNA hybrid sensors (i.e., cGAS55,69, TLR355, TLR970 and NLRP371), are activated by ADAR1 silencing in BRCA1/2-mutant cells. Prior work indicates that R-loop-derived cytosolic nucleic acids – including DNA, dsRNA, and RNA:DNA hybrids, can trigger interferon responses via DNA- and dsRNA-sensing PRR55,56,57,58,59,60,61,62, and conversely, that canonical cytosolic DNA and dsRNA sensors can detect RNA:DNA hybrids55,65, suggesting a possible contribution of several cytosolic nucleic acid species to the SL. Besides, others have shown that cytosolic dsRNA arises from R-loop-prone genomic regions, including telomeres (via telomeric repeat-containing RNA, TERRA)4,66,72,73, and micronuclei (via aberrant transcription of chromosomes trapped in micronuclei)48,74. These latter findings could be consistent with our observations that an increased R-loop burden also, in the context of the BRCA1/2–ADAR1 SL, activates dsRNA-sensing PRR, and are further compatible with a possible co-existence of various R-loop-derived immunogenic cytosolic nucleic acids species in response to ADAR1 silencing. Interestingly, our observation that RNase H1 overexpression does not fully abrogate the type I interferon response elicited by ADAR1 silencing in BRCA1-mutant cells is in line with the notion that cytosolic RNA:DNA hybrids originate from a subset of long-lived nuclear R-loops that are partially RNase H-resistant55.
Secondly, the characterization of R-loops as the trigger of ADAR1-dependent SL effects would require further investigation to determine the nature (promoter-paused vs. elongation-associated R-loops75), genomic location (promoter proximal, exonic or non-coding regions) and functional context (co-transcriptional vs. DNA repair-associated R-loops at DSBs76,77,78 or replication forks79) of R-loops involved in the BRCA1/2–ADAR1 SL. The use of orthogonal methods to visualize (e.g., catalytically-inactive mutant RNase H1 protein) and capture both native (e.g., R-loop CUT&Tag) and ex cellulo-isolated R-loops (e.g., DRIP-Seq) would allow a more comprehensive profiling of the R-loop landscape in response to impaired ADAR1 function. Similarly, resolutive methods of detection and analysis of cytosolic RNA:DNA hybrids (e.g., CytoDRIP-Seq55) would be needed to better understand how R-loops control the activation status of DNA- and dsRNA-sensing PRR.
Thirdly, complementary mechanisms might also contribute to this SL. These include the antagonism of ADAR1-mediated recoding activity at specific editing sites80, mitotic failure due to defective ADAR1p150 function in chromosome segregation81, or ZBP1-dependent necroptosis following defective editing of Alu duplex RNA82,83,84. Yet, the complete abrogation of SL effects upon JAK/STAT pathway inhibition suggests a central role of interferon-dependent, “viral mimicry” responses in mediating the BRCA1/2–ADAR1 SL phenotype. Whether these alternative mechanisms may occur synergistically or in a context-specific fashion would deserve further exploration.
Lastly, and most importantly, this SL highlights novel therapeutic opportunities. ADAR1 has previously been proposed as a therapeutic target in TNBC11,12,13 and in tumors harboring elevated levels of dsRNA species14,15 due to enhanced responsiveness to interferon signaling. However, no clinically-actionable biomarker has thus far been identified that would allow cancer patients to be stratified to receive an ADAR1-targeting agent. Here, we identify BRCA1/2 mutations—a routinely-used biomarker in oncology—as a robust determinant of sensitivity to ADAR1 inhibition. The multifaceted nature of mechanisms underlying the BRCA1/2–ADAR1 SL, which are distinct from those underlying sensitivity or resistance to agents targeting BRCA1/2 defects, such as PARPi, chemotherapy or other DDR inhibitors (DDRi), suggests that targeting ADAR1 in platinum- or PARPi-resistant settings might deserve evaluation. Similarly, combining an ADAR1-targeting agent with DDRi in BRCAm cancers, may be complementary to the currently-evaluated DDRi-DDRi combinations, whose tolerability is challenging. Also, whether this SL could be extended to other HR defects, or tumors that harbor a “BRCAness” phenotype, remains to be explored. Still, despite recent attempts to develop ADAR1-targeting agents85,86, very few promising preclinical candidates exist87 and poorly-selective adenosine analogs virtually remain the only commercially-available preclinical compounds88, urging the development of potent, selective ADAR1 small-molecule inhibitors or degraders.
In conclusion, we describe a genetic interaction between BRCA1/2 and ADAR1 that opens novel perspectives towards the development of ADAR1-targeting approaches in immuno-oncology. Once drug-like ADAR1 inhibitors are discovered, these could readily be assessed in clinical trials including BRCAm patients.
Methods
This study complies with all relevant ethical regulations, as detailed below. A complete list of all antibodies, RNAi, CRISPR and RT-qPCR reagents is available in Supplementary Information.
Clinical specimens
Use of breast cancer samples was approved by the institutional ethics review board at Gustave Roussy (CSET, Commission Scientifique des Essais Thérapeutiques), in accordance with the Declaration of Helsinki. Informed consent was obtained from all subjects for the use of their tumor tissue as part of this study. A total of 63 TNBC surgical specimens were collected between January 2005 and September 2023. Samples were routinely fixed immediately after surgery in 10% formalin for 24 h at room temperature. After fixation, samples were dehydrated, incubated in xylene, infiltrated with paraffin, and finally embedded in paraffin.
Animal studies
All animal experiments were conducted in compliance with institutional animal protocols at Inserm, approved by DDPP Val-de-Marne, France, under license number F 94-043-013. AB strain of zebrafish was used in this study. Zebrafish embryos were staged and cared for according to standard protocols89.
Cell lines
BRCA1-isogenic SUM149 cells (described in ref. 22 and90; original source: Asterand) were cultured under normal growth conditions (37 °C, 5% CO2) in Ham’s F-12 medium (Thermo Fisher, #21765037) supplemented with 10% (v/v) heat-inactivated fetal bovine serum (HI-FBS; Sigma Aldrich, #F7524), 5 μg/mL human insulin (Santa Cruz, #sc-360248), 1 μg/mL hydrocortisone (Sigma Aldrich, #H4001) and 1% (v/v) penicillin/streptomycin (P/S; Thermo Fisher, #15140122). MDA-MB-436, MDA-MB-231, CAL51, CAL120, HCC1937 and Hs578T (source: ATCC) were maintained under normal growth conditions in Roswell Park Memorial Institute-1640 (RPMI-1640; Thermo Fisher, #61870044) or high-glucose Dulbecco's Modified Eagle Medium (DMEM; Thermo Fisher, #31966047) supplemented with 10% (v/v) HI-FBS and 1% (v/v) P/S. BRCA1-isogenic TP53‒/‒ RPE1 cells (described in ref. 24; gift from D. Durocher) and BRCA2-isogenic DLD1 cells (source: Horizon Discovery) were maintained under normal growth conditions in DMEM supplemented with 10% (v/v) HI-FBS and 1% (v/v) P/S. Brca1-isogenic mouse embryonic fibroblasts (MEF; generated as previously described23) and Brca1/2-isogenic 4T1 cells (described in ref. 25; gift from R. Samstein) were maintained under normal growth conditions in RPMI-1640 supplemented with 10% (v/v) HI-FBS and 1% (v/v) P/S. Brca1-isogenic ID8 cells (described in ref. 26; gift from I. McNeish) were maintained under normal growth conditions in DMEM supplemented with 10% (v/v) HI-FBS, 1X insulin-transferrin-selenium (ITS; Thermo Fisher, #41400045) and 1% (v/v) P/S. ADAR1-isogenic HEK293T (described in ref. 28; gift from C. Rice) were maintained under normal growth conditions in DMEM supplemented with 10% (v/v) HI-FBS and 1% (v/v) P/S. Mycoplasma testing was performed monthly using the MycoAlert Mycoplasma Detection Kit (Lonza, #LT07-318), and all cell lines were short-tandem-repeat (STR) typed using the GenePrint® 10 System (Promega, #B9510) to confirm identity prior to the study.
Antibodies
Primary antibodies were used for western blotting, immunofluorescence and immunohistochemistry (see Supplementary Table 1 for details). The following secondary antibodies (ThermoFisher) were used for immunofluorescence: Anti-Mouse Alexa Fluor 647 (#A-21422), Anti-Rabbit Alexa Fluor 488 (#A-11034), Anti-Rabbit Alexa Fluor 647 (#A-21244), Anti-Mouse Alexa Fluor 488 (#A-11001).
Drugs and chemicals
The JAK/STAT pathway inhibitors ruxolitinib (INCB-18424; JAK1/2i), upadacitinib (ABT-494; JAK1i) and deucravacitinib (BMS-986165, TYK2i), and the integrated stress response inhibitor ISRIB were purchased from MedChemExpress. The PARPi talazoparib (BMN-673) and olaparib (AZD2281) were purchased from SelleckChem. Inhibitor stock solutions were prepared in DMSO and stored in aliquots at -80 °C.
RNAi and siRNA transfection
To perform siRNA-mediated gene silencing, cells were either (i) forward-transfected i.e., seeded at 60% density in 6-well plate format and transfected 24 h later (for immunofluorescence, western blotting and RT-qPCR); or (ii) reverse-transfected i.e., seeded and transfected in suspension at seeding (in 96-well plate format for short-term survival assays; in 6-well plate format for colony formation assays), with the indicated siRNA using Lipofectamine RNAiMAX (Thermo Fisher, #13778150) and Opti-MEM™ (Thermo Fisher, #31985062) according to manufacturers’ instructions. siRNAs targeting the following genes were used: ADAR1, BRCA1, BRCA2, CGAS, IFNAR1, LGP2, MDA5, PKR, and RIG1 (see Supplementary Table 2 for details). In siRNA co-transfection experiments, equimolar concentrations of each siRNA were used in all conditions. The transfection efficiency was monitored by assessment of cell growth inhibition ( > 95%) in pools of cells concomitantly transfected with a positive control, PLK1-targeting siRNA. Transcriptional silencing efficiency was systematically assessed via evaluation of protein expression by western blotting on pools of concomitantly transfected cells, 72 h after transfection.
siRNA synthetic lethal screen
SUM149 BRCA1-isogenic cells were reverse-transfected with a library of 22 siRNAs, including three non-targeting control siRNAs (siCTRL#1, #2, #3), one lethal control siRNA (siPLK1), and 18 siRNAs targeting genes of interest. The custom library (purchased from Horizon Discovery) was arrayed in 96-well plates and transfections were conducted in quadruplicates, using Lipofectamine RNAiMAX (Thermo Fisher, #13778150) and Opti-MEM™ (Thermo Fisher, #31985062) according to manufacturers’ instructions. The transfection media was removed one day after transfection, and after six subsequent days of continuous culture, cell viability was determined by use of CellTiter-Glo® luminescent reagent (Promega, #G7573). Surviving fractions elicited by each siRNA were calculated compared to the median of non-targeting control siRNA conditions.
CRISPR-Cas9 gene editing and crRNA transfection
To perform CRISPR-Cas9-mediated inactivation of ADAR1, cells were reverse-transfected in 6-well plate format with the indicated Edit-R crRNA (Horizon Discovery; see Supplementary Table 3 for details), Edit-R tracrRNA (Horizon Discovery, #U-002005-50) and recombinant Cas9 protein (15 pmol/well), using Lipofectamine CRISPRmax (Thermo Fisher, #CMAX00015) and Opti-MEM™ (Thermo Fisher, #31985062) according to manufacturers’ instructions. The transfection efficiency was monitored by assessment of cell growth inhibition ( > 95%) in pools of cells concomitantly transfected with a positive control, PLK1-targeting crRNA. Genetic inactivation efficiency was systematically assessed via evaluation of protein expression by western blotting on pools of concomitantly transfected cells, 48 h after transfection.
Virus production
Lentiviral particles were generated by transfecting HEK293T cells, using Lipofectamine 2000 (Thermo Fisher, #11668019), with the expression plasmid of interest and the psPAX2 (Addgene, #12260) and pMD2.G (Addgene, #12259) packaging vectors at a ratio of 1:1:1, respectively. Viral supernatants were collected 3 and 4 days after transfection and filtered through a 0.45 μm strainer. Viral particles were precipitated by incubation of the supernatants O/N at 4 °C with 1X PEG-it Virus Precipitation Solution (System Bioscience, #LV810A-1). The supernatants were then centrifuged at 1500 g for 30 min, and precipitates were resuspended in 2 mL culture media. The resuspended particles were then immediately used for transduction (250 µL per 60 cm2 petri dish) or kept at -80 °C for long-term storage.
Plasmid constructions and stable lentiviral transduction
To generate stable RH1-expressing cells in the SUM149 BRCA1-isogenic, MDA-MB-436 and MDA-MB-231 models, cells were transfected with ppyCAG-RNaseH1-V5 plasmid (Addgene, #111906) with Lipofectamine 2000 (Thermo Fisher, #11668019) according to manufacturer’s instructions. Stable pools of transfectants were generated by selection with hygromycin B (SUM149, 100 μg/mL; MDA-MB-436, 200 μg/mL; MDA-MB-231, 400 μg/mL) and the resultant selected populations were submitted to clonal isolation using the limiting dilution method. Clones were recovered and profiled for RNase H1 or V5 expression by western blot.
To generate stable doxycycline-inducible shADAR1-expressing cells in the SUM149 BRCA1-isogenic model, cells were transduced with a pLKO-Tet-On lentiviral vector (Addgene, #21915) engineered to carry the ADAR1-targeting shRNA sequence 3’-TTACCAAGGCCTGAGATATAACTCGAGTTATATCTCAGGCCTTGGTAA-5’. Stable pools of transductants were generated by selection with puromycin (1 μg/mL), and the resultant selected populations were submitted to clonal isolation using the limiting dilution method. Clones were recovered and profiled for ADAR1 expression by western blot following exposure to doxycycline (100 ng/mL).
Plasmid constructions and transient transfection of ADAR1 mutants
Mutated variants of ADAR1 were generated by site-directed mutagenesis using the Phusion™ Site-Directed Mutagenesis kit (Thermo Fisher, #F541) from plasmids containing wildtype ADAR1p150 (Addgene, #117927) and ADAR1p110 (Addgene, #117928) cDNAs. Catalytically-inactive, RNA editing-defective G1007R31 (p150-ΔCD) and Z-DNA binding-defective P193A31 (p150-ΔZα) mutants of ADAR1p150 were generated.
Short-term survival assays were performed in 96-well plates. On day 0, exponentially-growing SUM149 BRCA1-isogenic cells were seeded at a density of 15,000 cells per well and reverse-transfected in quadruplicates using Lipofectamine 3000 (Thermo Fisher, #L3000015) and Opti-MEM™ (Thermo Fisher, #31985062), with: (i) 100 pg/mL of the indicated wildtype or mutant ADAR1p110 or ADAR1p150 plasmid constructs, or the corresponding empty vector (Addgene, # 117926); and (ii) 5 nM of control, non-targeting siRNA (siCTRL), or an individual siRNA targeting ADAR1 3’UTR. On day 1, the media was removed and replaced with fresh media. On day 6, cell viability was determined by use of CellTiter-Glo® luminescent reagent (Promega, #G7573). Surviving fractions were calculated compared to the non-targeting control siRNA conditions.
Cell-based assays
Cell survival assays
Short-term survival assays were performed in 96-well plates. On day 0, exponentially-growing cells were seeded at a density of 15-25,000 cells per well and reverse-transfected in quadruplicates with the indicated siRNAs using Lipofectamine RNAiMAX (Thermo Fisher, #13778150) and Opti-MEM™ (Thermo Fisher, #31985062) according to manufacturers’ instructions. On day 1, the media was removed and replaced with fresh media (or fresh media containing the indicated small-molecule inhibitor, as appropriate, in the case of exposure to JSPi and ISRIB). On day 6, cell viability was determined by use of CellTiter-Glo® luminescent reagent (Promega, #G7573). Surviving fractions were calculated compared to the non-targeting control siRNA condition.
Colony-formation assays
Clonogenic assays were performed in 6-well plates. On day 0, exponentially-growing cells were seeded at a density of 400-600,000 cells per well and reverse-transfected with the indicated siRNAs using Lipofectamine RNAiMAX (Thermo Fisher, #13778150) and Opti-MEM™ (Thermo Fisher, #31985062) according to manufacturers’ instructions. On day 1, cells were trypsinized, counted, and re-seeded in triplicates at a density of 1,500 cells per well in 6-well plates. On day 10-12, cells were fixed with 0.5% crystal violet in methanol for 20 min Colonies of > 50 cells were counted manually and surviving fractions were calculated compared to the non-targeting control siRNA condition.
Cell proliferation assays
Short-term proliferation assays were performed in 96-well plates, in the same conditions as short-term survival assays. Cell proliferation was measured by use of an Incucyte® SX5 (Sartorius) over a period of 6–10 days after transfection. Growth curves were plotted for each siRNA condition.
Immunoblotting
Cells were lysed in RIPA lysis and extraction buffer (Thermo Fisher, #89900) supplemented with 0.5% phenylmethylsulfonyl fluoride (PMSF; Sigma Aldrich, #93482) and 1% Halt™ protease and phosphatase inhibitor cocktail (Thermo Fisher, #78444). Lysates were generated on ice, and centrifuged 10 min at 16,900 g prior to supernatant collection. Quantification of total protein in supernatants was performed using the Pierce™ BCA protein assay (Thermo Fisher, #23225), and equal amounts of each sample were loaded into NuPAGE™ 4-12% Bis-Tris (Invitrogen, #NP0335BOX) or 3-8% Tris-Acetate precast gels (Invitrogen, #EA0378BOX) and subjected to electrophoresis using NuPAGE™ MOPS SDS (Invitrogen, #NP000102) or Tris-Acetate SDS running buffers (Invitrogen, #LA00401), respectively. After migration, proteins were transferred to a nitrocellulose membrane (iBlot NC Regular Stacks, Invitrogen, #IB23001). 5% bovine serum albumin (BSA; Sigma Aldrich, #A7990C) in Tris-buffered saline (TBS; Euromedex, #ET220-B) supplemented with 0.1% Tween-20 (TBS-T 0.1%; Sigma Aldrich, #P7949) was used to block the membranes at room temperature (RT) for 1 h. Primary antibodies were diluted in 5% BSA in TBS-T 0.1%, and incubated at 4 °C overnight (O/N). The next day, the membranes were washed three times with TBS-T 0.1% for 10 min, followed by incubation with horseradish-peroxidase-conjugated secondary antibodies at RT for 1 h, in 5% milk in TBS-T 0.1%. The membranes were washed again three times with TBS-T 0.1%, and incubated with Clarity ECL substrate (Biorad, #1705060) or Clarity Max ECL substrate (Biorad, #1705062). The membranes were then imaged with a BioRad ChemiDoc XRS+ chemiluminescent detection system.
Immunofluorescence
Immunofluorescence assays were performed in 6-well plates. On day 0, exponentially-growing cells were seeded in triplicates at a density 200-350,000 cells per well on 13 mm coverslips. On day 1, cells were forward-transfected with the indicated siRNAs using Lipofectamine RNAiMAX (Thermo Fisher, #13778150) and Opti-MEM™ (Thermo Fisher, #31985062) according to manufacturers’ instructions. 6-8 h after transfection, the media was removed and replaced with fresh media (or fresh media containing the indicated small-molecule inhibitor, as appropriate). On day 4, cells were either (i) for γ-H2AX, RAD51, RPA, CCNA2, G3BP1 and PKR immunofluorescence, fixed in 4% (v/v) paraformaldehyde (PFA; Euromedex, EM-15710) in PBS for 10 min at RT, washed twice with PBS, and permeabilized with 0.5% (v/v) Triton X-100 (Sigma Aldrich, #X100) in PBS for 10 min; or (ii) for R-loop (S9.6) and fibrillarin immunofluorescence, fixed in ice-cold 100% methanol for 7 min at -20 °C, washed twice with PBS, quenched with 0.1 M glycine (made fresh) for 10 min at RT, washed again twice with PBS, and permeabilized with 0.5% (v/v) Triton X-100 in PBS for 7 min For R-loop (S9.6) and fibrillarin immunofluorescence, RNase H digestion was performed by incubating coverslips with RNase H (New England Biolabs, #M0297) diluted 1:50 in 1X RNase H buffer (New England Biolabs, #B0297) for 5 h at 37 °C. After two additional washes with PBS, cells were blocked with 2% (w/v) BSA, 1% (v/v) FBS in PBS (IFF) for 1 h at RT, followed by incubation with the indicated primary antibodies in IFF at 4 °C overnight. The next day, cells were washed three times with PBS for 10 min, and incubated with the indicated secondary antibodies, DAPI (Thermo Fisher, #62248) and PicoGreen® (Thermo Fisher, #P7581; only when assessing micronuclei) in IFF for 1 h at RT. After three additional washes with PBS for 10 min, coverslips were dried and mounted on microscope slides using Fluoromount™ mounting medium (Sigma Aldrich, #F4680). Slides were imaged at 40× on a digital slide scanner (Hamamatsu).
Analysis of immunofluorescence images
Quantification of the number of micronuclei, γ-H2AX, RAD51, S9.6, RPA, G3BP1, and PKR foci was performed manually under identical microscopy settings between samples, using the OlyVIA image viewer software (Olympus) for visualization. Three independent fields comprising a minimum of 150 cells were used. For quantification of R-loops (S9.6 foci), only nuclear foci outside nucleoli (fibrillarin-positive bodies) were counted. For quantification of RPA foci, only CCNA2-positive cells were used in the analysis.
Immunohistochemistry
For each patient sample, a single representative formalin-fixed paraffin embedded (FFPE) block was selected for the study. FFPE blocks were sectioned (4 μm thick) on a RM2245 microtome (Leica Biosystems) and placed onto histological TOMO® adhesion microscope slides (VWR, #10748-166). ADAR1 and ADARp150 automated immunohistochemistry staining was performed using a BOND RX automated research stainer (Leica Biosystems). After deparaffinization with BOND™ Dewax solution (Leica, #AR9222), epitope retrieval was performed through incubation in BOND™ Epitope Retrieval solution 2 (Leica, #AR9640) at 100 °C, pH 9 for 20 min, and endogenous peroxidase activity was inhibited by treatment with the Peroxide Block reagent of the BOND™ Polymer Refine Detection kit (Leica, #DS9800) for 5 min The slides were then incubated with primary antibodies for 30 min at RT (ADAR1, 1:700; ADARp150, 1:1,000). Detection was performed with a secondary antibody and subsequently revealed with the 3,3′-diaminobenzidine tetrahydrochloride (DAB) chromogenic substrate of the BOND™ Polymer Refine Detection kit (Leica, #DS9800). Counterstaining was performed by incubation with the hematoxylin reagent of the BOND™ Polymer Refine Detection kit (Leica, #DS9800) for 5 min Hematoxylin and eosin (H&E) automated staining was performed using a GEMINI AS (MM France). Tissue sections were deparaffinized in xylene and rehydrated by incubation in serial 100% ethanol baths. The slides were incubated with haemalun for 4.5 min and then with eosin for 2.5 min Dehydration was carried out by 3 successive baths of 100% ethanol followed by xylene. Coverslips were mounted on microscope slides using PERTEX® mounting medium (VWR, #LEIC811).
Pathological assessment of TNBC tumors
Archival samples from treatment-naïve TNBC surgical specimen or biopsies were used to build a cohort of 63 cases (BRCA1-wildtype, N = 32; BRCA1-mutant, N = 31). Blinded histopathological assessment of ADAR1p150 and tumor-infiltrating lymphocytes (TILs) was performed by a senior breast pathologist. For ADAR1p150, expression was evaluated in the nucleus and cytoplasm of tumor cells using an H-score (percentage of stained tumor cells multiplied by each intensity from 0 to 3 + , value from 0 to 300). TILs density was quantified using morphological evaluation of lymphocytes based on a standardized methodology for breast cancer91.
RT-qPCR
RNA was extracted from cells using the RNeasy Mini kit (Qiagen, #74104). DNA contamination was removed using the RNase-Free DNase Set (Qiagen, #79256). RNA was then quantified using NanoDrop™ 2000 spectrophotometer (Thermo Fisher) and diluted to equal concentrations across all samples. Reverse-transcription was performed using the SuperScript VILO cDNA Synthesis kit (Thermo Fisher, #11754250), following the manufacturer’s protocol. The qPCR reactions were performed in 384-well plates with TaqMan™ Fast Advanced Master Mix (Thermo Fisher, # 4444963), and samples were analyzed using a ViiA7 real-time PCR system (Applied Biosystems). Probes against the following transcripts were used (see Supplementary Table 4 for details): CCL5, IFNB1, IFI44, IFIT1, MX1. Results were normalized to GAPDH.
RNA-Seq
RNA was extracted from cells using the RNeasy Mini kit (Qiagen, #74104). DNA contamination was removed using the RNase-Free DNase Set (Qiagen, #79256). Initial quality control and quantification of the RNA material was performed using Qubit RNA HS Assay kit (Thermo Fisher, #Q32852) on a Qubit 2.0 Fluorometer (Thermo Fisher). RNA degradation was determined through evaluation of the RIN, using RNA 6000 Pico kit (Agilent, #5067-1513) on an Agilent 2100 Bioanalyzer System (Agilent). rRNA depletion was conducted using the Illumina® TruSeq Stranded Total RNA Library Prep (Human/Mouse/Rat), and the rRNA-free residue was cleaned up by ethanol precipitation. Library preparation for sequencing was carried out using the NGS Stranded RNA Library Prep Set (Novogene) on the rRNA-depleted RNA, as per manufacturer’s instructions. Briefly, after fragmentation, first-strand cDNA synthesis was performed using random hexamer primers, followed by second-strand cDNA synthesis in which dUTPs were replaced with dTTPs in the reaction buffer. Subsequently, the cDNA fragments were subjected to end repair, A-tailing, adapter ligation, size selection, USER enzyme digestion, amplification, and purification. The completed directional libraries were then quantified with Qubit and qPCR, and checked for size distribution detection using RNA 6000 Pico kit (Agilent, #5067-1513) on an Agilent 2100 Bioanalyzer System (Agilent). Quantified libraries were finally pooled and sequenced on a NovaSeq 6000 S4 (Illumina) using a 150 bp paired-end (PE150) sequencing strategy.
Fastq files were both trimmed and controlled with Fastp92, cutting both front and tail on a window of size 6 and a minimum quality of 10. Reads shorter than 15 bases were taken out, reads with more than 50% of unqualified bases were discarded, and overrepresentation analysis was turned on.
RNA editing analyses
RNA editing analysis was performed by SPRINT44 and RNAEditingIndex43. SPRINT used cleaned fastq files from Fastp, and each edition event was counted from bed output files with R. Bam files from SPRINT were sorted by Samtools93 then used by RNAEditingIndex with default parameters to compute REI scores.
Interferon-stimulated genes (ISG) signature
ISG signature genes were retrieved from Extended Data Fig. 1a of Liu et al.11. The ISG signature was evaluated using RNA-Seq data from SUM149 BRCA1-isogenic cells and GSVA (version 2.0.1)94, with the option "kcdf" set to "Poisson". Statistical tests between conditions were performed by limma (version 3.62.1)95 and heatmap graphs were computed using the ComplexHeatmap R package (version 2.22.0)96,97.
Zebrafish studies
Morpholino (MO) injections
Knockdown of brca2 and adar1 was performed using established translation-blocking antisense brca2 and adar1 morpholinos (MO) with the sequences 3’-TTTCAAACATGCTGCCATGACTGTG-5’98 and 3’-TCCCTCCTCTACCTCTGCTCATAGC-5’36, respectively. A standard, widely-used negative control MO was used as control, with the sequence 3’-CCTCTTACCTCAGTTACAATTTATA-5’ targeting a human beta-globin intron. All MOs were purchased from Gene Tools and injected at the single cell stage into AB wildtype (WT) embryos as follows: 4 ng of brca2 MO, 1 ng of adar1 MO, 6 ng of control standard MO and 4 ng/1 ng of brca2/adar1 MOs per embryo. Embryos were checked at epiboly stage and all unfertilized eggs were discarded, only fertilized eggs were raised and analyzed at 48 h post fertilization (hpf). Injections were performed at least three times with more than 30 embryos per group per experiment.
Phenotype scoring
Embryos were raised and analyzed at 48 hpf for morphological phenotypes, that were scored under a dissecting scope. Embryos were grouped into four categories: normal, mild, severe and dead as shown in the corresponding figure.
Acridine orange staining and analysis
48 hpf embryos were anesthetized with 0.03% tricaine and incubated in fish water containing 5 μM acridine orange (AO) for 20 min in the dark. After wash, they were embedded in 1.4% low melting point agarose, and imaged with a Leica SP8 confocal microscope. Fluorescent corpses were counted blindly within a length of ≈ 600 µm spanning from the first somite and through the yolk sac extension along the anterior-posterior (AP) axis of the embryo. Image analyses and AO-positive cells counting were performed using Image J software.
Statistical analyses
No statistical methods were used to predetermine sample size and experiments were not randomized. Biological replicates are indicated as n and technical replicates as N. All graphs show either mean or median values with error bars (standard deviation, SD) calculated using GraphPad Prism 10 software; 95% confidence intervals were used and significance was considered when *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; ns, not significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Materials availability: All new unique reagents generated in this study are available from the corresponding authors with a completed materials transfer agreement.
Datasets availability: The RNA-Seq data generated as part of this study have been deposited in the European Genome-Phenome Archive (EGA), under accession number EGAS50000000518. The datasets on EGA will be made available to interested researchers under limited access on a project-specific basis, subject to approval by the Gustave Roussy Data Access Committee in compliance with the data access agreement terms. Requests should be directed to the corresponding authors. Upon establishment of the data transfer agreement, EGA data release can be expected within 3 business days. Once access has been granted, the period for which the data can be downloaded is flexible and will be defined according to the downloader’s needs.
The remaining data are available within the article or Supplementary Information. A Source Data file containing the raw data underlying all figure panels, including full uncropped and unprocessed scans of all blots, is provided along with this article. Any additional information required to reanalyze the data reported in this study is available from the corresponding authors upon request. This study does not report original code. Source data are provided with this paper.
References
Eisenberg, E. & Levanon, E. Y. A-to-I RNA editing - immune protector and transcriptome diversifier. Nat. Rev. Genet. 19, 473–490 (2018).
Nishikura, K. A-to-I editing of coding and non-coding RNAs by ADARs. Nat. Rev. Mol. Cell Biol. 17, 83–96 (2016).
de Reuver, R. & Maelfait, J. Novel insights into double-stranded RNA-mediated immunopathology. Nat. Rev. Immunol. 24, 235–249 (2024).
Shiromoto, Y., Sakurai, M., Minakuchi, M., Ariyoshi, K. & Nishikura, K. ADAR1 RNA editing enzyme regulates R-loop formation and genome stability at telomeres in cancer cells. Nat. Commun. 12, 1654 (2021).
Jimeno, S. et al. ADAR-mediated RNA editing of DNA:RNA hybrids is required for DNA double strand break repair. Nat. Commun. 12, 5512 (2021).
Zhang, B. et al. ADAR1 links R-loop homeostasis to ATR activation in replication stress response. Nucleic Acids Res. 51, 11668–11687 (2023).
Chabanon, R. M. et al. Targeting the DNA damage response in immuno-oncology: developments and opportunities. Nat. Rev. Cancer 21, 701–717 (2021).
Kornepati, A. V. R., Rogers, C. M., Sung, P. & Curiel, T. J. The complementarity of DDR, nucleic acids and anti-tumour immunity. Nature 619, 475–486 (2023).
Manguso, R. T. et al. In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature 547, 413–418 (2017).
Ishizuka, J. J. et al. Loss of ADAR1 in tumours overcomes resistance to immune checkpoint blockade. Nature 565, 43–48 (2019).
Liu, H. et al. Tumor-derived IFN triggers chronic pathway agonism and sensitivity to ADAR loss. Nat. Med. 25, 95–102 (2019).
Gannon, H. S. et al. Identification of ADAR1 adenosine deaminase dependency in a subset of cancer cells. Nat. Commun. 9, 5450 (2018).
Kung, C.-P. et al. Evaluating the therapeutic potential of ADAR1 inhibition for triple-negative breast cancer. Oncogene 40, 189–202 (2021).
Mehdipour, P. et al. Epigenetic therapy induces transcription of inverted SINEs and ADAR1 dependency. Nature 588, 169–173 (2020).
Wu, Z. et al. Reprogramming of the esophageal squamous carcinoma epigenome by SOX2 promotes ADAR1 dependence. Nat. Genet. 53, 881–894 (2021).
Ryan, C. J., Bajrami, I. & Lord, C. J. Synthetic Lethality and Cancer - Penetrance as the Major Barrier. Trends Cancer 4, 671–683 (2018).
Pantelidou, C. et al. PARP inhibitor efficacy depends on CD8+ T-cell recruitment via intratumoral STING pathway activation in BRCA-deficient models of triple-negative breast cancer. Cancer Discov. 9, 722–737 (2019).
Ding, L. et al. PARP inhibition elicits STING-dependent antitumor immunity in Brca1-deficient ovarian cancer. Cell Rep. 25, 2972–2980 (2018).
Parkes, E. E. et al. Activation of STING-dependent innate immune signaling by S-phase-specific DNA damage in breast cancer. J. Natl. Cancer Inst. 109, djw199 (2017).
Chabanon, R. M. et al. PARP inhibition enhances tumor cell–intrinsic immunity in ERCC1-deficient non–small cell lung cancer. J. Clin. Invest. 129, 1211–1228 (2019).
Reisländer, T. et al. BRCA2 abrogation triggers innate immune responses potentiated by treatment with PARP inhibitors. Nat. Commun. 10, 3143 (2019).
Dréan, A. et al. Modeling therapy resistance in BRCA1/2-mutant cancers. Mol. Cancer Ther. 16, 2022–2034 (2017).
Callen, E. et al. 53BP1 enforces distinct pre- and post-resection blocks on homologous recombination. Mol. Cell 77, 26–38.e7 (2020).
Noordermeer, S. M. et al. The shieldin complex mediates 53BP1-dependent DNA repair. Nature 560, 117–121 (2018).
Samstein, R. M. et al. Mutations in BRCA1 and BRCA2 differentially affect the tumor microenvironment and response to checkpoint blockade immunotherapy. Nat. Cancer 1, 1188–1203 (2021).
Walton, J. B. et al. CRISPR/Cas9-derived models of ovarian high grade serous carcinoma targeting Brca1, Pten and Nf1, and correlation with platinum sensitivity. Sci. Rep. 7, 16827 (2017).
Zatreanu, D. et al. Polθ inhibitors elicit BRCA-gene synthetic lethality and target PARP inhibitor resistance. Nat. Commun. 12, 3636 (2021).
Chung, H. et al. Human ADAR1 prevents endogenous RNA from triggering translational shutdown. Cell 172, 811–824.e14 (2018).
Sun, T. et al. Decoupling expression and editing preferences of ADAR1 p150 and p110 isoforms. Proc. Natl. Acad. Sci. USA 118, e2021757118 (2021).
Hu, S.B. et al. ADAR1p150 prevents MDA5 and PKR activation via distinct mechanisms to avert fatal autoinflammation. Mol. Cell 83, 3869–3884.e7 (2023).
Rice, G. I. et al. Mutations in ADAR1 cause aicardi-goutières syndrome associated with a type I interferon signature. Nat. Genet. 44, 1243–1248 (2012).
Sharan, S. K. et al. Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature 386, 804–810 (1997).
Howe, K. et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature 496, 498–503 (2013).
Vierstraete, J. et al. Accurate quantification of homologous recombination in zebrafish: brca2 deficiency as a paradigm. Sci. Rep. 7, 16518 (2017).
Vierstraete, J., Fieuws, C., Willaert, A., Vral, A. & Claes, K. B. M. Zebrafish as an in vivo screening tool to establish PARP inhibitor efficacy. DNA Repair 97, 103023 (2021).
Niescierowicz, K. et al. Adar-mediated A-to-I editing is required for embryonic patterning and innate immune response regulation in zebrafish. Nat. Commun. 13, 5520 (2022).
Buchumenski, I. et al. Systematic identification of A-to-I RNA editing in zebrafish development and adult organs. Nucleic Acids Res. 49, 4325–4337 (2021).
Kasher, P. R. et al. Characterization of samhd1 morphant zebrafish recapitulates features of the human type I interferonopathy Aicardi-Goutières syndrome. J. Immunol. Baltim. Md 1950 194, 2819–2825 (2015).
Bill, B. R., Petzold, A. M., Clark, K. J., Schimmenti, L. A. & Ekker, S. C. A primer for morpholino use in zebrafish. Zebrafish 6, 69–77 (2009).
Haider, S. et al. The transcriptomic architecture of common cancers reflects synthetic lethal interactions. Nat. Genet. 57, 522–529 (2025).
Paz-Yaacov, N. et al. Elevated RNA editing activity is a major contributor to transcriptomic diversity in tumors. Cell Rep. 13, 267–276 (2015).
Fumagalli, D. et al. Principles governing A-to-I RNA editing in the breast cancer transcriptome. Cell Rep. 13, 277–289 (2015).
Roth, S. H., Levanon, E. Y. & Eisenberg, E. Genome-wide quantification of ADAR adenosine-to-inosine RNA editing activity. Nat. Methods 16, 1131–1138 (2019).
Zhang, F., Lu, Y., Yan, S., Xing, Q. & Tian, W. SPRINT: an SNP-free toolkit for identifying RNA editing sites. Bioinforma. Oxf. Engl. 33, 3538–3548 (2017).
Bigot, L. et al. Development of Novel Models of Aggressive Variants of Castration-resistant Prostate Cancer. Eur. Urol. Oncol. S2588-9311, 00226–2 (2023).
Schäffer, A. A., Chung, Y., Kammula, A. V., Ruppin, E. & Lee, J. S. A systematic analysis of the landscape of synthetic lethality-driven precision oncology. Med N. Y. N. 5, 73–89.e9 (2024).
Groelly, F. J., Fawkes, M., Dagg, R. A., Blackford, A. N. & Tarsounas, M. Targeting DNA damage response pathways in cancer. Nat. Rev. Cancer 23, 78–94 (2023).
Tang, S., Stokasimov, E., Cui, Y. & Pellman, D. Breakage of cytoplasmic chromosomes by pathological DNA base excision repair. Nature 606, 930–936 (2022).
Bayona-Feliu, A., Barroso, S., Muñoz, S. & Aguilera, A. The SWI/SNF chromatin remodeling complex helps resolve R-loop-mediated transcription-replication conflicts. Nat. Genet. 53, 1050–1063 (2021).
Sadler, A. J. & Williams, B. R. G. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 8, 559–568 (2008).
Cao, X. Self-regulation and cross-regulation of pattern-recognition receptor signalling in health and disease. Nat. Rev. Immunol. 16, 35–50 (2016).
Ding, S. et al. STAG2 deficiency induces interferon responses via cGAS-STING pathway and restricts virus infection. Nat. Commun. 9, 1485 (2018).
Roulois, D. et al. DNA-Demethylating Agents Target Colorectal Cancer Cells by Inducing Viral Mimicry by Endogenous Transcripts. Cell 162, 961–973 (2015).
Maurano, M. et al. Protein kinase R and the integrated stress response drive immunopathology caused by mutations in the RNA deaminase ADAR1. Immunity 54, 1948–1960.e5 (2021).
Crossley, M. P. et al. R-loop-derived cytoplasmic RNA-DNA hybrids activate an immune response. Nature 613, 187–194 (2023).
Mackenzie, K. J. et al. Ribonuclease H2 mutations induce a cGAS/STING-dependent innate immune response. EMBO J. 35, 831–844 (2016).
Chatzidoukaki, O. et al. R-loops trigger the release of cytoplasmic ssDNAs leading to chronic inflammation upon DNA damage. Sci. Adv. 7, eabj5769 (2021).
Zannini, L., Cardano, M., Liberi, G. & Buscemi, G. R-loops and impaired autophagy trigger cGAS-dependent inflammation via micronuclei formation in Senataxin-deficient cells. Cell. Mol. Life Sci. CMLS 81, 339 (2024).
Murayama, T. et al. Targeting DHX9 triggers tumor-intrinsic interferon response and replication stress in small cell lung cancer. Cancer Discov. 14, 468–491 (2024).
Maxwell, M. B. et al. ARID1A suppresses R-loop-mediated STING-type I interferon pathway activation of anti-tumor immunity. Cell S0092-8674, 00451–00453 (2024).
Schumann, T. et al. Deficiency for SAMHD1 activates MDA5 in a cGAS/STING-dependent manner. J. Exp. Med. 220, e20220829 (2023).
Giordano, A. M. S. et al. DNA damage contributes to neurotoxic inflammation in Aicardi-Goutières syndrome astrocytes. J. Exp. Med. 219, e20211121 (2022).
Eckmann, C. R., Neunteufl, A., Pfaffstetter, L. & Jantsch, M. F. The human but not the Xenopus RNA-editing enzyme ADAR1 has an atypical nuclear localization signal and displays the characteristics of a shuttling protein. Mol. Biol. Cell 12, 1911–1924 (2001).
Strehblow, A., Hallegger, M. & Jantsch, M. F. Nucleocytoplasmic distribution of human RNA-editing enzyme ADAR1 is modulated by double-stranded RNA-binding domains, a leucine-rich export signal, and a putative dimerization domain. Mol. Biol. Cell 13, 3822–3835 (2002).
Wang, I. X. et al. Human proteins that interact with RNA/DNA hybrids. Genome Res. 28, 1405–1414 (2018).
Nassour, J. et al. Telomere-to-mitochondria signalling by ZBP1 mediates replicative crisis. Nature 614, 767–773 (2023).
Cottrell, K. A. et al. Induction of viral mimicry upon loss of DHX9 and ADAR1 in breast cancer cells. Cancer Res. Commun. 4, 986–1003 (2024).
Cristini, A., Groh, M., Kristiansen, M. S. & Gromak, N. RNA/DNA Hybrid Interactome Identifies DXH9 as a Molecular Player in Transcriptional Termination and R-Loop-Associated DNA Damage. Cell Rep. 23, 1891–1905 (2018).
Mankan, A. K. et al. Cytosolic RNA:DNA hybrids activate the cGAS –STING axis. EMBO J. 33, 2937–2946 (2014).
Rigby, R. E. et al. RNA:DNA hybrids are a novel molecular pattern sensed by TLR9. EMBO J. 33, 542–558 (2014).
Vanaja, S. K. et al. Bacterial RNA:DNA hybrids are activators of the NLRP3 inflammasome. Proc. Natl. Acad. Sci. USA 111, 7765–7770 (2014).
Balk, B. et al. Telomeric RNA-DNA hybrids affect telomere-length dynamics and senescence. Nat. Struct. Mol. Biol. 20, 1199–1205 (2013).
Arora, R. et al. RNaseH1 regulates TERRA-telomeric DNA hybrids and telomere maintenance in ALT tumour cells. Nat. Commun. 5, 5220 (2014).
Sasaki, N. et al. RNA sensing induced by chromosome missegregation augments anti-tumor immunity. Mol. Cell 85, 770–786.e7 (2025).
Castillo-Guzman, D. & Chédin, F. Defining R-loop classes and their contributions to genome instability. DNA Repair 106, 103182 (2021).
Ouyang, J. et al. RNA transcripts stimulate homologous recombination by forming DR-loops. Nature 594, 283–288 (2021).
Cohen, S. et al. Senataxin resolves RNA: DNA hybrids forming at DNA double-strand breaks to prevent translocations. Nat. Commun. 9, 533–533 (2018).
Hatchi, E. et al. BRCA1 recruitment to transcriptional pause sites is required for R-loop-driven DNA damage repair. Mol. Cell 57, 636–647 (2015).
Promonet, A. et al. Topoisomerase 1 prevents replication stress at R-loop-enriched transcription termination sites. Nat. Commun. 11, 3940 (2020).
Chen, L. et al. Recoding RNA editing of AZIN1 predisposes to hepatocellular carcinoma. Nat. Med. 19, 209–216 (2013).
van Gemert, F. et al. ADARp150 counteracts whole genome duplication. Nucleic Acids Res. 52, 10370–10384 (2024).
Hubbard, N. W. et al. ADAR1 mutation causes ZBP1-dependent immunopathology. Nature 607, 769–775 (2022).
de Reuver, R. et al. ADAR1 prevents autoinflammation by suppressing spontaneous ZBP1 activation. Nature 607, 784–789 (2022).
Jiao, H. et al. ADAR1 averts fatal type I interferon induction by ZBP1. Nature 607, 776–783 (2022).
Mendoza, H. G., Matos, V. J., Park, S., Pham, K. M. & Beal, P. A. Selective inhibition of ADAR1 using 8-azanebularine-modified RNA duplexes. Biochemistry 62, 1376–1387 (2023).
Wang, Z. et al. Structurally specific Z-DNA proteolysis targeting chimera enables targeted degradation of adenosine deaminase acting on RNA 1. J. Am. Chem. Soc. 146, 7584–7593 (2024).
Wang, X. et al. Targeting ADAR1 with a small molecule for the treatment of prostate cancer. Nat. Cancer 6, 474–492 (2025).
Cottrell, K. A., Torres, L. S., Dizon, M. G. & Weber, J. D. 8-azaadenosine and 8-chloroadenosine are not selective inhibitors of ADAR. Cancer Res. Commun. 1, 56–64 (2021).
Kimmel, C. B., Ballard, W. W., Kimmel, S. R., Ullmann, B., & Schilling, T. F. Stages of embryonic development of the zebrafish. Dev. Dyn. Publ. Am. Assoc. Anat. 203, 253–310 (1995).
Pettitt, S. J. et al. Genome-wide and high-density CRISPR-Cas9 screens identify point mutations in PARP1 causing PARP inhibitor resistance. Nat. Commun. 9, 1849 (2018).
Salgado, R. et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an International TILs Working Group 2014. Ann. Oncol. J. Eur. Soc. Med. Oncol. 26, 259–271 (2015).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinforma. Oxf. Engl. 34, i884–i890 (2018).
Danecek, P. et al. Twelve years of SAMtools and BCFtools. GigaScience 10, giab008 (2021).
Hänzelmann, S., Castelo, R. & Guinney, J. GSVA: gene set variation analysis for microarray and RNA-Seq data. BMC Bioinforma. 14, 7 (2013).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Gu, Z., Eils, R. & Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinforma. Oxf. Engl. 32, 2847–2849 (2016).
Gu, Z. Complex heatmap visualization. iMeta 1, e43 (2022).
Kroeger, P. T. et al. The zebrafish kidney mutant zeppelin reveals that brca2/fancd1 is essential for pronephros development. Dev. Biol. 428, 148–163 (2017).
Acknowledgements
We dedicate this work to patients and their families. We particularly thank donators to the Association Ruban Rose and Fondation pour la Recherche Médicale for their financial support, which has been instrumental in the pursuit of this project. We also thank Dr. Charles M Rice and Dr. Robert Samstein for their material contribution. This work was funded by project grants to S.P.V., R.M.C. and P.P. from Institut National du Cancer (INCa, 2023-165-PLBIO23-139), Fondation pour la Recherche sur le Cancer (Fondation ARC, ARCPGA2023010005866_6378), Association Ruban Rose (Prix Ruban Rose Avenir 2023 to R.M.C.) and Fondation pour la Recherche Médicale (FRM, Prix Raymond Rosen 2023 to S.P.V.). This work was also funded by programme grants to S.P.V. from European Research Council (ERC Starting Grant, TargetSWitch, grant no. 101077864), Fondation ARC (PGA1-RF20190208576), INSERM ATIP-Avenir / La Ligue Contre le Cancer 2018, PRT-K20-067 (ARIANES-CTC, funded by INCa and DGOS), and as part as of programme funding to Gustave Roussy from Institut National du Cancer (INCa-DGOS-Inserm_12551 SIRIC2 and INCa-DGOS-Inserm-ITMO Cancer_18002 SIRIC EpiCURE). Some of the work carried out by R.M.C. at The Institute of Cancer Research was funded by a programme grant to C.J.L. from Breast Cancer Now as part of programme funding to the Breast Cancer Now Toby Robins Research Centre.
Author information
Authors and Affiliations
Contributions
Conceptualization and design of the study: R.M.C., C.J.L., S.P-V. Development of methodology: R.M.C., M.A., C.J.L., S.P-V. Acquisition of data: R.M.C., L.S., M.L-T., J.Z., V.K., A.G., M.G., E.E. Analysis and interpretation of data: R.M.C., L.S., M.L-T., M.A., V.K., M.T., C.D., Y-L.L., P.P., C.J.L., S.P.V. Manuscript drafting: R.M.C., C.J.L., S.P-V. Editing and approval of the final submitted manuscript: R.M.C., L.S., M.L-T., M.A., J.Z., V.K., A.G., M.G., E.E., L.B., D.B.K., R.B., S.J.P., T.T-B., R.S., C.M., M.D., A.N.J.T., F.B., Y.L., S.D., M.T., C.D., Y-L.L., B.P., P.P., C.J.L., S.P-V. Study supervision and funding acquisition: R.M.C., P.P., C.J.L., S.P-V.
Corresponding authors
Ethics declarations
Competing interests
R.M.C. makes the following disclosures: is named inventor on a patent describing the use of ADAR1 inhibitors as anti-cancer agents (WO/2024/189433). J.Z. makes the following disclosures: has received research funding and travel subsidies from Menarini. B.P. makes the following disclosures: has received research funding as part of an institutional program from: Astra Zeneca, Daiichi Sankyo, Gilead, Seagen, MSD; receives and/or has received consulting fees from: Astra Zeneca (institutional), Seagen (institutional), Gilead (institutional), Novartis (institutional), Lilly (institutional), MSD (institutional), Pierre Fabre (personal), Daiichi Sankyo (institutional/personal), Olema (institutional); has received travel subsidies from: Astra Zeneca; Pierre Fabre; MSD; Daiichi Sankyo, Pfizer. A.N.J.T. makes the following disclosures: Receipt of grants / research support: AstraZeneca research costs associated with OlympiA, Myriad Genetics research support for TNT Trial, Breast Cancer Now research grant to Institution, CRUK research grant to Institution. Receipt of honoraria or consultation fees: AstraZeneca, Inbiomotion, Gilead, Innovation in Breast Cancer Symposium, SABCS, GBCC, Cancer Panel, Research to Practise, AACR, Aicme, Penn Medicine, PAGE Therapeutics, Ellipses, VHIO, Dana Faber David Livingston Memorial Symposium, Tango Therapuetics, Guardant, CRUK, Neogenomics, Merck. Educational seminar: Guardant. Stock option: InBioMotion. Royalties: Benefits from ICR’s Inventors Scheme associated with patents for use of several PARP inhibitors in DNA repair-deficient cancers held by AstraZeneca with royalty payments to A.N.J.T.’s personal account and research accounts at the Institute of Cancer Research, and to the Institute of Cancer Research. Institutional financial interests: Royalties as above from AstraZeneca. Research costs from AstraZeneca, Myriad Genetics. Non-financial interests: Leadership/Guideline Advisor roles: Director of Breast Cancer Now Research Centre—ICR/KCL, St Gallen Early Breast Cancer Guidelines Panel, ESMO Early Breast Cancer Guidelines Committee, ESMO Scientific Committee, Chair—CRUK Clinical Research Committee, BCRF SAB member, Strategy Committee UK NCRI, Trustee The Cridlan Ross Smith Charitable Trust. C.J.L. makes the following disclosures: receives and/or has received research funding from: AstraZeneca, Merck KGaA, Artios, Neophore; has received consultancy, SAB membership or honoraria payments from: FoRx, Syncona, Sun Pharma, Gerson Lehrman Group, Merck KGaA, Vertex, AstraZeneca, Tango Therapeutics, 3rd Rock, Ono Pharma, Artios, Abingworth, Tesselate, Dark Blue Therapeutics, Pontifax, Astex, Neophore, Glaxo Smith Kline, Dawn Bioventures, Blacksmith Medicines, ForEx; has stock in: Tango, Ovibio, Hysplex, Tesselate, Ariceum. C.J.L. is also named inventor on patents describing the use of DNA repair inhibitors and stands to gain from their development and use as part of the ICR “Rewards to Inventors” scheme and also reports benefits from this scheme associated with patents for PARP inhibitors paid into C.J.L.’s personal accounts and research accounts at the Institute of Cancer Research. C.J.L. is also named inventor on a patent describing the use of ADAR1 inhibitors as anti-cancer agents (WO 2024/189433 A1). S.P.V. makes the following disclosures: has received research funding from Hoffman La Roche and AstraZeneca for unrelated research projects. As part of the Drug Development Department (DITEP), S.P.V. is principal investigator or sub-investigator of clinical trials from Abbvie, Agios Pharmaceuticals, Amgen, Argen-X Bvba, Arno Therapeutics, Astex Pharmaceuticals, Astra Zeneca, Aveo, Bayer Healthcare Ag, Bbb Technologies Bv, Blueprint Medicines, Boehringer Ingelheim, Bristol Myers Squibb, Celgene Corporation, Chugai Pharmaceutical Co., Clovis Oncology, Daiichi Sankyo, Debiopharm S.A., Eisai, Eli Lilly, Exelixis, Forma, Gamamabs, Genentech, Inc., GlaxoSmithKline, H3 Biomedicine, Inc, Hoffmann La Roche Ag, Innate Pharma, Iris Servier, Janssen Cilag, Kyowa Kirin Pharm. Dev., Inc., Loxo Oncology, Lytix Biopharma As, Medimmune, Menarini Ricerche, Merck Sharp & Dohme Chibret, Merrimack Pharmaceuticals, Merus, Millennium Pharmaceuticals, Nanobiotix, Nektar Therapeutics, Novartis Pharma, Octimet Oncology Nv, Oncoethix, Onyx Therapeutics, Orion Pharma, Oryzon Genomics, Pfizer, Pharma Mar, Pierre Fabre, Roche, Sanofi Aventis, Taiho Pharma, Tesaro Inc, and Xencor. S.P.V. has participated to advisory boards for Merck KGaA. S.P.V. is also named inventor on a patent describing the use of ADAR1 inhibitors as anti-cancer agents (WO 2024/189433 A1). All other authors have no conflicts of interest or financial interests to disclose.
Peer review
Peer review information
Nature Communications thanks Priyanka Verma and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chabanon, R.M., Shcherbakova, L., Lacroix-Triki, M. et al. Autocrine interferon poisoning mediates ADAR1-dependent synthetic lethality in BRCA1/2-mutant cancers. Nat Commun 16, 6972 (2025). https://doi.org/10.1038/s41467-025-62309-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-62309-5
This article is cited by
-
ADAR1 is an editor of DNA replication forks
Nature Structural & Molecular Biology (2026)