Abstract
Single-stranded DNA-binding proteins (SSBs) protect transiently exposed ssDNA, yet how DNA polymerase (DNAp) displaces them during replication remains unclear. Using single-molecule force spectroscopy, dual-color imaging, and molecular dynamics simulations on bacteriophage T7 DNAp and SSB, we investigated molecular mechanisms underlying SSB displacement. T7 SSB modulates replication in a force-dependent manner: enhancing it at low tension by preventing secondary structures while impeding it at high tension. Dual-color imaging shows SSBs remain stationary as DNAp advances, supporting a sequential displacement model. Molecular dynamics suggests that DNAp actively lowers the SSB dissociation energy barrier through interactions mediated by the SSB C-terminal tail. FRET confirms close protein proximity during encounters. Optimal replication requires SSB saturation of ssDNA, establishing a delicate balance between protection and efficiency. This spatiotemporal coordination between DNAp and SSB is critical for resolving molecular collisions and may represent a general mechanism for resolving molecular collisions, ensuring both processivity and genomic integrity.
Similar content being viewed by others
Introduction
DNA replication relies on the precise orchestration of multiple protein factors on a dynamic template. Single-stranded DNA (ssDNA) intermediates, essential during DNA replication, recombination, and repair, remain vulnerable to enzymatic degradation and secondary structure formation1,2 that can impede polymerase progression and disrupt DNA synthesis3,4. SSB proteins protect these regions by coating ssDNA, preventing hairpin formation5,6 and recruiting replication machinery like DNAp3 and helicase7,8, thereby enhancing polymerization efficiency3,5,6,9,10. However, this protective mechanism creates a paradox: given SSBs’ high affinity for ssDNA11,12,13, how do replicative enzymes efficiently navigate these potential barriers during DNA synthesis? The mechanism by which polymerases overcome or displace SSB-DNA complexes without hindering replication remains a fundamental yet incompletely resolved question in molecular biology.
Single-molecule techniques are powerful tools for studying dynamic biomolecular processes14,15,16 and have deepened our understanding of replication mechanisms and their regulation. Among the two major types of single-molecule approaches, force-based manipulation studies have revealed that mitochondrial SSBs enhance DNA polymerase γ activity when functional interactions between polymerases and SSBs are firmly established, leading to optimal replication rates4. Concurrently, fluorescence-based visualizations have demonstrated diverse consequences of SSB proteins bound to ssDNA when encountering translocating motor proteins on the same DNA track. For example, human replication protein A (RPA) can be displaced or bypassed by helicases like Xeroderma Pigmentosum Group D17; E. coli SSB can be pushed by a translocase18; and Pif1, a helicase from Saccharomyces cerevisiae, can even chemo-mechanically propel a human RPA heterotrimer along ssDNA19. However, the specific dynamics between replicative polymerases and stationary SSBs, such as T7 SSB13 on the lagging strand, remain poorly characterized, particularly regarding how SSB binding and its interaction with DNAp collectively influence replication efficiency.
Molecular dynamics (MD) simulations can provide atomic-level insights into transient molecular events, challenging to capture with current experimental approaches. The complexity of simulating the polymerase-ssDNA complex stems from its size and the lack of precise force fields for protein-DNA interactions, making modeling this crucial biochemical process rare20. Therefore, MD simulation applications to study the dissociation of SSB and its interactions with partners like DNA polymerases and helicases are limited to a few sample systems, such as human RPA and E. coli AlkB21,22. Our recently developed coarse-grained protein model AWSEM has successfully predicted structures and mechanisms in various complex DNA-binding systems23,24,25. Specifically, the benchmarked ssDNA force field from prior work on the simulation of T7 gp4 helicase moving on a long ssDNA has demonstrated AWSEM’s ability to capture the diverse states encountered during translocation’s large-scale motions23.
In this work, we sought to combine these complementary approaches to visualize how DNAp overcomes SSB and resolves molecular collisions on DNA. We first employed an integrated experimental approach, utilizing both force and fluorescence microscopy, to directly monitor the displacement of stationary T7 SSB by replicative T7 DNA polymerase during lagging-strand DNA synthesis. We selected the T7 bacteriophage replication system as our experimental model due to its well-characterized properties and mechanistic similarities to more complex organisms. T7 SSB offers several advantages as a model: unlike the tetrameric E. coli SSB6,11 or the heterotrimeric eukaryotic RPA26,27, T7 SSB binds ssDNA as a monomer with a single binding mode, displaying high affinity and an effective footprint of ~7–10 nucleotides per monomer13. The protein features a conserved oligonucleotide/oligosaccharide-binding fold (OB domain) that ensures stable ssDNA attachment through its positive charge, complemented by a disordered acidic C-terminal tail that mediates critical interactions with replication partners such as T7 DNA polymerase9,28. Single-molecule optical tweezers studies have revealed T7 SSB’s binding dynamics are finely regulated by multiple factors: the protein exhibits minimal diffusion on ssDNA, while template tension, DNA conformation, and the C-terminal tail collectively modulate DNA accessibility during replication13,28,29.
Our findings revealed that T7 SSB modulates the DNA replication rate dependent on ssDNA template conformation. At 10 pN force, when secondary structures can easily form, DNAp accelerates in the presence of SSB. Contrarily, replication decelerates under similar conditions at 20 pN, where minimal hairpin formation is expected. Next, using dual-color imaging, we visualized the dynamic interplay between T7 SSB and T7 DNA polymerase, uncovering that SSBs are displaced sequentially during this process. Furthermore, we examined the spatial proximity between DNA polymerase and SSBs, revealing potential direct interaction between the proteins, as evidenced by FRET signals indicating a “collision” event. The MD simulation revealed that the binding energy of the gp2.5-ssDNA complex is reduced in the presence of DNA polymerase, suggesting an active manner of the displacement. Notably, the electrostatic interaction analysis demonstrated the pivotal role of the gp2.5 C-terminal tail in facilitating SSB dissociation by anchoring to the DNA polymerase’s front basic patch region, thereby enhancing displacement efficiency. Experiments with a C-terminal truncated variant of T7 SSB (mut T7 SSB) further validated the critical role of this interaction for efficient replication. Under various forces, our real-time DNA primer extension assays and single-molecule force spectroscopy demonstrated the necessity of saturated SSB and C-terminal-mediated interactions for optimal replication, since the mutant SSB variant displays a reduced efficiency. Collectively, these findings provide a comprehensive mechanistic understanding of how DNA polymerase interacts with SSBs during lagging-strand synthesis.
Results
Force-dependent regulation of DNA replication by SSB proteins
The binding of T7 SSB to ssDNA reportedly depends on force-induced ssDNA conformations13, suggesting this interaction may modulate DNA replication in a force-dependent manner. We investigated this interplay using high-resolution optical tweezers to measure DNA length changes catalyzed by DNA polymerase during replication under controlled tensions. In our experimental setup, we tethered a DNA template between two optically trapped beads, enabling precise measurement of T7 DNA polymerase activity under physiologically relevant tension and allowing us to explore how varying force influences the interaction between DNA polymerase and SSB proteins (Method, Fig. 1A, B).
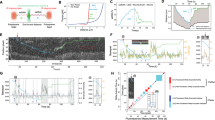
A Experimental setup using dual-optical tweezers. DNA template (~8 kbp) with 25 nt overhang is tethered between optically trapped beads. High tension (45–50 pN) creates ssDNA template via exonuclease activity; reduced tension (10–20 pN) enables polymerization analysis. Inset shows T7 DNAp (PDB: 1T7P) and T7 gp2.5 (PDB: 1JE5) at the ssDNA/dsDNA junction. B Force-dependent changes in end-to-end distance. At 50 pN, exonuclease activity increases the distance. At 10–20 pN, SSB binding and polymerization decrease the distance. Transition from ssDNA (FJC model) to dsDNA (WLC model) enables activity quantification. Change-point detection algorithm determines polymerase/exonuclease activity shifts. C Representative replication trajectories with/without SSB at 10 and 20 pN. Dashed lines: reference rates without SSB. Black arrows: backtracking events. Trajectories include 3–5 s prior to tension transition, aligned to the transition time for comparison. D SSB effects on replication efficiency. At 10 pN: SSB significantly enhances efficiency from 180 ± 10 bp/s (N = 67) to 230 ± 20 bp/s (N = 72, p = 0.0467). At 20 pN: SSB reduces efficiency from 130 ± 10 bp/s (N = 63) to 80 ± 5 bp/s (N = 81, p < 0.0001). E Exonuclease binding probability analysis. Without SSB: higher probability at 20 pN (0.3 ± 0.05, N = 30) than 10 pN (0.2 ± 0.04, N = 35), indicating tension-dependent exo site binding. With SSB: probabilities decrease to 0.06 ± 0.02 (N = 47) at 10 pN and 0.3 ± 0.06 (N = 43) at 20 pN. Results show polymerase site preference. F SSB modulation of replication processivity under varying tensions. At 10 pN: SSB increases processivity from 720 ± 60 bp/event (N = 67) to 1400 ± 100 bp/event (N = 72, p < 0.0001). At 20 pN: decreases from 500 ± 40 bp/event (N = 63) to 380 ± 30 bp/event (N = 81, p = 0.0312). G Average pause duration analysis. Without SSB: decreases from 19 ± 2 s (10 pN, N = 61) to 13 ± 1 s (20 pN, N = 101), possibly due to hairpin impedance at lower force. With SSB: increases to 31 ± 2 s at 10 pN (N = 50, p < 0.0001), remains 15 ± 1 s at 20 pN (N = 99, p = 0.24). H Fluorescently labeled DNAp binding to ssDNA with/without SSB. Only the green channel was displayed. White arrows: DNAp trajectories. I DNAp coating density on ssDNA. Density calculated as the normalized difference between average photon count and background (arbitrary units). With 5 mM Mg²⁺, SSB increases DNAp binding (N = 5–8, p < 0.05). Data presented as mean ± SEM. Statistical comparisons were performed using Welch’s unpaired two-tailed t-tests. Source data are provided as a Source Data file.
To interpret end-to-end distance (EED) changes during replication, we accounted for SSB binding, displacement, polymerization, and exonuclease contributions. At a higher tension of 50 pN, the applied template force leads the DNA polymerase enzyme to primarily act in its exonuclease mode, removing nucleotides from dsDNA to generate ssDNA regions, and thus increasing the EED between the two beads30,31. Lowering tension to 10 or 20 pN encourages SSB binding to ssDNA and initiates replication by DNA polymerase, displacing SSBs and shortening the EED (Fig. 1B, Eq. (1)). We corrected for SSB-induced compaction using empirically determined extension factors obtained from previous work13 and validated in Supplementary Fig. 2. Using this correction, we computed the ssDNA fraction as a function of time based on Eq. (2). To derive replication kinetics with and without SSB under these tensions, we calculated the number of base pairs synthesized as a function of time by multiplying the ssDNA fraction with the total length of the DNA template (Fig. 1C). These replication trajectories (Fig. 1C) provide insights into the dynamic interplay between secondary structure formation, SSB binding, and DNA polymerase replication activities.
We analyzed the EED changes using a change-point detection algorithm to identify transition points in base pair-time traces, which mark shifts into either polymerase or exonuclease activity under 10 or 20 pN (see Methods). Template tensions of 10 and 20 pN were chosen as example force thresholds based on their distinct influences on ssDNA conformation and potential effect on SSB-ssDNA interactions. At 10 pN, conducive to secondary structure formation, the role of SSB proteins is anticipated to be critical. Our data (Fig. 1D), derived from analyses of 30–47 different DNA molecules, indicate that at 10 pN, SSB proteins enhance the replication rate from 180 ± 10 bp/s (mean ± SEM, N = 67) to 230 ± 20 bp/s (N = 72). This increase of ~20% is statistically significant with a p value of 0.0467, confirming previous biochemistry studies under no template tension, which show that SSB proteins bolster DNA replication by precluding the formation of secondary structures5,6,10. Intriguingly, at an increased tension of 20 pN, surpassing the threshold for secondary structure disruption and reducing the likelihood of their formation, the presence of SSB proteins led to a decrease in replication efficiency from 130 ± 10 bp/s (N = 63) to 80 ± 5 bp/s (N = 81) (mean ± SEM, p value <0.0001). To estimate the extent of secondary structure formation under our experimental conditions (5 mM Mg²⁺), we compared force-extension data to the FJC model (Supplementary Fig. 2C). At 10 pN, the ssDNA contour length is reduced by ~20%, indicating substantial secondary structure formation, possibly explaining the statistically significant increase in polymerization rate with the addition of SSB. In contrast, at 20 pN, this reduction diminishes to only ~4%, confirming minimal secondary structure presence. This explains SSB’s dual role—beneficial at lower tensions where secondary structures impede replication, but inhibitory at higher tensions where they become unnecessary obstacles to polymerase progression.
Apart from the SSB’s impact on replication rate, our data also indicate a role of SSB proteins in modulating the binding site preference of DNA polymerases. This preference was quantified by calculating the ratio of exonuclease (exo) events to the sum of exo and polymerase (pol) events for a given replication event (Fig. 1E). Notably, the absence of SSB proteins led to increased backtracking (exo) dynamics in DNA replication (black arrows, Fig. 1C), with a higher probability of DNA polymerase binding at the exonuclease site, particularly at 10 pN (~20%) and 20 pN (~30%). In the presence of SSB proteins, the probability of binding at the exonuclease site was reduced to approximately 10% at 10 pN but was unchanged at 20 pN (~30%). Across all conditions, a preference for binding at the polymerase-active site was observed, with probabilities of binding at the exonuclease site consistently below 50%, supporting the notion that lower tension (<~35 pN) favors replication activity (Fig. 1B)30,31. These findings imply that SSB proteins, by binding to the ssDNA in front of the replication fork, may directly interact with the replicative DNA polymerase, thereby promoting its affinity for the polymerase-active site.
Moreover, the binding site preference of DNAp directly influences replication processivity (Fig. 1F). At a tension of 10 pN, the presence of SSB proteins markedly enhanced processivity, increasing from 720 ± 60 bp/event (N = 67) to 1400 ± 100 bp/ event (N = 72). This enhancement suggests that SSB proteins contribute to replication processivity by both suppressing secondary structures and promoting binding at the polymerase-active site. We note that, the processivity values reported here represent apparent processivity, as rapid exchange of DNAp at the replication fork32 was not considered. Interestingly, when the tension was elevated to 20 pN, the inclusion of SSB proteins led to a decline in replication processivity by around 30%. This can be attributed to the lower replication rate (Fig. 1D) and relatively high probability of exonuclease binding (~20%) and high exonuclease rate (Figs. 1E and S1A) at this tension, which may together interfere with efficient replication.
We further analyzed the average duration of pauses both in the presence and absence of SSB proteins (Methods, Fig. 1G). Without SSB proteins, we observed a decrease in average pause duration from 19 ± 2 s at 10 pN (N = 61) to 13 ± 1 s at 20 pN (N = 101). At the lower tension of 10 pN, the replication process is likely more impeded by hairpin structures. Such structures, which are more prevalent at 10 pN, might stall DNAp, leading to longer pauses. Surprisingly, with SSB proteins, pause durations increased to 31 ± 2 s at 10 pN (N = 50), while at 20 pN the effect of SSB on pause durations was relatively minor (15 ± 1 s, N = 99). The pronounced increase in pause duration at 10 pN suggests that while SSB proteins play a crucial role in facilitating replication by preventing secondary structures, they may also introduce significant roadblocks under certain conditions. DNA polymerase was earlier reported to bind to the ssDNA region32, creating self-imposed roadblocks impeding replication.
To investigate how SSB influences DNAp binding, we monitored DNAp interacting with ssDNA in the presence and absence of SSB (Fig. 1H). SSB addition increased the frequency of DNAp binding events to ssDNA (white arrow, Fig. 1H, I), particularly under our experimental condition of 5 mM Mg²⁺ (N = 5–8 DNA molecules, p < 0.05). This finding, together with our observation of co-localization between DNAp and bound SSB molecules (Supplementary Fig. 1D), suggests that SSB facilitates DNAp recruitment to the DNA template. While our experiments measured binding to general ssDNA regions rather than ss/dsDNA junctions, this recruitment might explain the enhanced replication dynamics we observed under saturating SSB concentrations (Fig. 1E, F). However, this recruitment of DNAp by SSB may not always be productive; SSB’s random positioning on ssDNA can recruit DNAp to non-junction sites, potentially obstructing replication, consistent with increased pausing at 10 pN (Fig. 1G).
DNA polymerase sequentially displaces stationary SSBs
During replication, DNA polymerase must displace bound SSBs to prepare DNA for synthesis. (Fig. 2A). We investigated two possible displacement mechanisms: a sequential “one-by-one” model, where SSBs are gradually removed from ssDNA, and a “whole-train” model, where multiple SSBs are simultaneously pushed forward. Using single-molecule dual-color imaging combined with optical tweezers, we tracked relative movement between T7 DNAp and T7 SSB proteins. Our experimental design used saturated SSB concentrations with a small fraction (~4%) of labeled SSBs as spatial markers. For the “one-by-one” model, we expected a decreasing distance between labeled SSB and DNAp, while the “whole-train” model would show a constant distance over time (Fig. 2A).
A Experimental design combining dual-optical trapping with dual-color imaging to investigate SSB displacement mechanisms during replication. Saturated SSB (150 nM, 4% labeled) tests two displacement models: sequential one-by-one SSB removal versus a collective whole-train pushing mechanism. B Representative mechanical trace illustrating DNAp-SSB interactions with force transition from 45 to 10 pN (dashed line). Junction position derived from independent optical tweezer measurements exploiting elasticity differences between ssDNA and dsDNA, enabling real-time DNAp tracking during replication. C Concurrent confocal kymographs capturing dynamic interactions between fluorescently labeled DNAp (top) and SSB (bottom) for the molecule shown in (B). Yellow line: force transition. Scale: 10 s, 75 nm/pixel. D Composite analysis combining mechanical data from (B) with fluorescence data from (C) to directly observe DNAp-SSB interactions. Only replication processes are displayed for clarity. Fluorescently labeled SSB visualized binding to ssDNA via fluorescence microscopy. Only SSB trajectories >3 pixels were analyzed for reliability. Total analysis: 58 DNAp-SSB interaction traces extracted from 25 unique DNA molecules (from 64 total molecules) exhibiting clear, consistent fluorescence signals. E Three representative distance dynamics between DNA polymerase and SSB during replication. Top panel based on data from (D). Green: processive replication; blue: backtracking; gray: pausing. Time axes rescaled for presentation clarity. F Histogram showing distance changes during displacement events. Analysis limited to first burst segments accounting for potential bidirectional DNAp movement. Negative values indicate backtracking events. Of 58 analyzed events: 15 show DNAp retreat due to backtracking (light blue), 36 indicate DNAp approach through SSB displacement (light green), and seven remain stationary, likely due to pausing. Results strongly support the one-by-one SSB removal model. G T7 SSB diffusion constant calculated using the mean square displacement method reveals predominantly stationary behavior on ssDNA (7 ± 7 × 10⁻⁴ μm²/s). Approximately 10% of measurements showing negative values were excluded as noise artifacts. H Example traces demonstrating displacement of fluorescently labeled SSB by DNAp under 10 pN force. Green line represents the DNAp trajectory. Source data are provided as a Source Data file.
We executed high-resolution mechanical measurements and aligned those data with fluorescence images of DNA polymerase and SSB, to track the relative movement of DNAp and SSB in real-time (Fig. 2B–D). By making use of the differential elasticity between ssDNA and double-stranded DNA (dsDNA), and calibrating the shortening effect of SSB binding to ssDNA (Methods, Supplementary Fig. 2A)13, we can trace the real-time movement of the ssDNA/dsDNA junction, the site where replicative DNAp binds and catalyzes the conversion of ssDNA into dsDNA (Fig. 2B). The solid blue and green lines in Fig. 2B denote examples of backtracking (exo) and replicative (pol) activity, respectively. Note that when we change the template tension from 45 to 10 pN, DNAp replicates ssDNA into dsDNA30,31. Next, we obtained confocal microscopy images of fluorescent DNAp and SSB simultaneously by separating the channels and rectifying signal crosstalk (Fig. 2C). The SSB kymograph was investigated with a trajectory analysis method (Methods) for additional quantification (shown in yellow, Fig. 2D). Subsequently, we superposed the data acquired from DNAp’s position (Fig. 2B) onto the SSB kymograph image to directly track both the DNAp and SSB positions (Figs. 2D and S3). Using this quantitative approach, we can determine the change in distance between replicative DNAp and SSB (Fig. 2E). Notably, due to the potential of DNAp moving toward SSB then reversing (as, for example, the light blue area in Fig. 2E), only the first segment with replication activity was quantified.
In the majority of events (36 out of 58 traces), DNAp moved toward and displaced SSB (Fig. 2F). n some cases (seven out of 58 traces), the DNAp appeared to be stationary, likely due to pausing, while in approximately a quarter of molecules (15 out of 58 traces), DNAp moved away from SSB, attributable to backtracking (Fig. 2F). Importantly, in none of the traces did we see the SSB move directionally in sync with the DNAp. Instead, SSB remains stationary on the ssDNA template while DNAp moves, as evidenced by T7 SSB’s low diffusion constant (7 ± 7 × 10−⁴ μm²/s, mean ± SEM; Fig. 2G) calculated using the mean square displacement method13. This value is comparable to the known static protein EcoRV (1 × 10−⁴ μm²/s)33. Notably, this diffusion coefficient on saturated ssDNA closely matches previous measurements on unsaturated ssDNA (3.9 ± 3.5 × 10⁻⁴ μm²/s)13, indicating T7 gp2.5 is intrinsically immobile rather than constrained by molecular crowding. Unlike E. coli SSB34, T7 gp2.5 exhibits limited mobility, likely due to its monomeric structure preventing the reptation-based movement characteristic of multimeric SSBs. These observations strongly support the sequential one-by-one displacement model, further evidenced by direct visualization of DNAp removing individual SSBs in rare instances (Fig. 2H and Supplementary Fig. 4, N = 2/25 DNA molecules). With our 4% SSB labeling ratio, we observed the expected distribution of fluorescent patterns: isolated single-labeled SSBs (Fig. 2F), multiple labeled proteins (Supplementary Fig. 3D, F), and occasional direct visualization of replicative DNAp removing multiple closely bound SSBs (Fig. 2H).
To determine whether SSB removal results from active DNAp motion or passive SSB dissociation, we compared our previously characterized intrinsic SSB dissociation rate13 with the DNA replication rate. Triple-exponential fitting of dwell times revealed a dominant fast intrinsic off-rate (Koff) of ~2.4 s⁻¹ at 12 pN (Supplementary Data 1). With a replication rate of ~200 nt/s (Fig. 1D), DNAp traverses an SSB footprint (~10 nt) in 0.05 s, potentially displacing ~20 SSB/s. This rate is approximately an order of magnitude faster than spontaneous SSB dissociation. This disparity strongly suggests that DNAp actively displaces SSB during replication rather than SSB passively dissociating. If SSBs were to spontaneously dissociate at such rapid rates, it would compromise their protective role, especially during DNAp’s proofreading activities that generate extensive regions of ssDNA. Therefore, our findings support the hypothesis of active displacement of SSB by DNAp, likely involving direct physical interactions between the two proteins.
Active displacement of SSB by DNA polymerase
The high replication rate of DNAp (Fig. 1D) poses challenges to experimentally capture the structural dynamics of the interaction between DNAp and SSB during replication in real-time. To investigate the structural dynamics in the event of the DNAp-induced SSB displacement from ssDNA, we employed AWSEM MD simulations to explore the intermediate states. T7 SSB has a total of 232 amino acids with a conserved oligosaccharide-oligonucleotide binding fold (OB-fold) that consists of a five-stranded anti-parallel β barrel capped by an α-helix on one end (αA)35. Since the most flexible C-terminal region (residue 206–232) and the linker region (residue 79–88) are missing in the current available template (PDB ID: 1JE5), we modeled the full-length structure using Alphafold236 (Fig. 3A). The C-terminal is positioned in close contact with the positive binding groove in gp2.5 in the Alphafold2 model. The ability to predict residue contacts with state-of-the-art accuracy using Alphafold2 provides us with a promising inference of the apo state of gp2.5 without DNA.
A Structural details of the predicted full-length SSB monomer using AlphaFold2 and Modeller. Gray: template 1JE5 regions; orange: positive binding groove; red: AlphaFold2 C-terminal; blue: Modeller C-terminal. B Free energy profile for SSB binding to ssDNA process using umbrella sampling simulations. Profile reveals three distinct local minima characterizing SSB-ssDNA binding: bound state (1.3–2.0 nm), intermediate state (~2.7 nm), and dissociated state (3.5–4.0 nm). C Representative structure of intermediate state from (B). ssDNA (blue) and C-terminal tail (red) positioned at opposite sides of positive binding groove, indicating competitive binding scenario between C-terminal and ssDNA for groove occupancy, stabilizing intermediate state. D Two-dimensional free energy profile of SSB-ssDNA binding incorporating electrostatic interactions between the positive binding groove and ssDNA as an additional coordinate. Representative structures from each local energy basin are shown. E Structural details of DNAp-thioredoxin-SSB complex bound to DNA using AlphaFold2-predicted SSB positioned based on 6P7E template. Green: full-length DNAp structure; blue: thioredoxin; gray/orange/red: SSB majority/positive binding groove/C-terminal tail, respectively. dsDNA with 5’ ssDNA overhang displayed. F Two-dimensional free energy profile of SSB-ssDNA binding in the presence of DNAp-trx complex. Bound state becomes relatively unstable compared to SSB alone, with the system trapped in a dissociated state. High electrostatic effects from SSB C-terminal promote dissociation. G Top: Schematic explaining FRET proximity measurement setup. Bottom: Representative replication activity from mechanical measurements showing base pair changes over time. Regions with constant base pairs (pausing) and reductions (backtracking) correlate with FRET signal regions (light blue shaded areas from panel H). H Representative kymograph of DNAp-SSB interaction. Top: SSB fluorescence; middle: DNAp fluorescence (green excitation only); bottom: merged image. Yellow line: real-time DNAp position from mechanical measurements. Images processed to reduce background noise, correct signal crosstalk, and filtered for clarity. I Analysis of eight individual FRET events selected from 16 DNA molecules based on fluorescence signal quality. Corresponding base pair-time traces extracted with FRET signal regions highlighted (light blue). FRET signals were detected during pausing events (6/8 cases) and backtracking events (2/8 cases), not observed during active replication, supporting the direct physical interaction model.
We first explored the SSB binding dynamics on ssDNA. Umbrella sampling was used to systematically sample the conformational space, facilitating the subsequent analysis of the free energy landscape. The one-dimensional (1D) free energy profile revealed three distinct states characterizing the SSB-ssDNA binding process (Fig. 3B), leveraging the center of mass (COM) distance between the protein and ssDNA as the primary collective variable. In the initial bound state, this distance is at 1.3–2.0 nm, while the distance in the intermediate state fluctuats around 2.7 nm and finally increases to 3.5–4.0 nm in the dissociated state. The hierarchically-clustered heatmap and dendrogram indicate two main clusters for the bound state structures (Supplementary Fig. 5A, B). Further examination of these structures unveiled two predominant forms of the C-terminal tail, characterized by disorder and an anti-parallel beta-sheet conformation (Supplementary Fig. 5C, D). Notably, as the COM distance between SSB and ssDNA increased, competition arose between the C-terminal region and the ssDNA for occupancy within the positively charged binding groove, leading to the stabilization of an intermediate state. We extracted the representative structures of the three largest clusters from the clustermap of all structures in the local basin of intermediate state (Supplementary Fig. 6). From these structures, we observed the ssDNA was displaced by the C-terminal of the SSB (Fig. 3C). The ssDNA shifted from the center of the binding groove toward the one side of the SSB while the C-terminal of SSB attacks from the other side and occupied the binding groove, indicate a transition of the binding process. A morphing movie among those three representative structures suggests a possible trajectory (Supplementary Movie 1). We also extended the 1D profile to a two-dimensional one with an additional axis showing the strength of electrostatics interactions between positive binding groove and ssDNA (Fig. 3D). Notably, the observed energy change of approximately 5 kcal/mol that the SSB needs to pass to reach the dissociated state from the bound state in the 2D free energy profile aligns with the experimentally derived average binding free energy of −4.7 kcal/mol (Supplementary Data 1). This simulation agrees well with the recent report that SSB remains essentially immobile on ssDNA rather than diffusing with frequent hops on and off the ssDNA13.
Next, we computed the 2D free energy profile for T7 SSB dissociation induced by DNAp during replication. The structure of the full-length T7 SSB predicted from Alphafold2 was assembled with the DNAp-primer/template DNA complex (PDB ID: 6P7E) with a short peptide that mimics the C-terminal of other components37 (Fig. 3E). To characterize how the interaction between gp2.5 C-terminus and DNAp facilitates the gp2.5 dissociation from the bound ssDNA, we performed an umbrella sampling simulation to explore the energy difference among local states during the stripping process (Fig. 3F). By comparing the 2D free energy profile with that obtained from T7 SSB alone (Fig. 3D), we observed that the bound state becomes relatively unstable in the presence of DNAp. The free energy profile indicates that the complex quickly becomes trapped in the dissociated state, possibly due to the interaction between DNAp and SSB. From the representative structures of the dissociated state, we observed that the SSB was stripped from ssDNA while maintaining its connection with DNAp through the C-terminal region (Fig. 3F). This suggests that DNAp likely facilitates SSB removal in an active manner by altering the free energy required for SSB displacement from ssDNA via the C-terminus of SSB.
This active displacement model revealed by MD simulation predicts there is a possible direct physical contact between DNAp and SSB. Indeed, direct contacts between the SSB C-terminal and DNAp, as well as the SSB positive binding groove and ssDNA are observed in some of the representative structures from the movie snapshots (Supplementary Fig. 7). Considering the FRET detection range (<~10 nm)38,39 and the size of T7 SSB and T7 DNAp, estimated to be around 435 and 7 nm40, respectively, this physical contact could result in a possible FRET signal. Therefore, we aimed to experimentally search for the FRET signals using single-molecule FRET and precise force measurements. If the active displacement hypothesis holds, we would anticipate detecting FRET signals indicative of direct interaction as DNAp displaces SSB during replication. The rapid replication rate that should result in up to 20 SSBs being displaced per second, coupled with the relatively low temporal resolution of confocal scanning (~0.7 s per scan along the DNA template), however, posed challenges in precisely discerning FRET signals during the course of processive replication. Recognizing these constraints, we exploited the strengths of this approach by measuring the spatial proximity of these proteins during DNA replication under a tension of 20 pN. This force was chosen because DNA polymerase is more likely to backtrack under such conditions (Fig. 1E), yet T7 SSB still effectively binds to ssDNA13. Higher tension results in reduced T7 SSB binding, while lower tension decreases backtracking likelihood. With our chosen force, the fluorescently labeled DNAp can synthesize dsDNA and occasionally backtrack (Fig. 1C, E), creating free ssDNA on the DNA template for SSB to rebind from solution. These backtracking events with subsequent SSB rebinding and potential for pausing thus provide valuable conditions to have both physical proteins very close on the DNA, allowing us to measure the physical distance between DNAp and SSB.
Our approach used the fluorescently labeled DNAp-SNAP-Surface® 549 (green), approaching the fluorescently labeled SSB-Atto647N (red) bound to ssDNA (Fig. 3G, top). We assumed that by exclusively exciting DNAp with green light, we would detect SSB fluorescence only when the intermolecular distance falls within the FRET detection range (typically less than ~10 nm)38,39. This methodology enabled us to estimate the relative proximity of the two proteins and probe molecular events in detail. A representative dataset displays the fluorescent trajectories of T7 SSB and T7 DNAp with only the green laser exciting DNAp is shown in Fig. 3H. Upon correcting for the background noise and signal crosstalk, we identified a transient red SSB signal (Fig. 3H, top panel). The merged kymograph (Fig. 3H, bottom panel) further highlights instances of DNAp approaching SSB on ssDNA during replication. Simultaneously, we were able to analyze the replication activity corresponding to these events (Fig. 3G) using data on DNA elongation (EED). The mechanical measurements revealed that whenever we observe a FRET signal (light blue shaded area), the base pair count remained relatively constant, indicating pausing, or decreases, which suggests backtracking. We note that not every pause or backtrack event produced a FRET signal, possibly due to the re-association of unlabeled SSB to ssDNA, and the rapid exchange of labeled DNA polymerase with unlabeled DNA polymerase from the solution32.
Figure 3I presents selected traces with FRET signals. Most signals occurred during DNAp pausing (75%; N = 6), with some during backtracking (25%, N = 2), possibly indicating new SSB binding to ssDNA generated during backtracking. No FRET signals were detected during active replication. These FRET signals support our active SSB displacement model by confirming that DNAp and SSB come into close proximity. Our measurements provide relative distance changes rather than absolute distances, as our FRET pair (SNAP-Surface® 549 and Atto647N) may introduce uncertainties due to dipole alignment variability. While temporal resolution limitations prevented capturing displacement during processive replication, the observed proximity during pauses and backtracking provides compelling evidence for direct physical interactions in our proposed mechanism.
The C-terminal tail and saturated SSB states: essential for DNA replication dynamics
The C-terminal tail of SSB for effective DNA replication has been reported earlier41,42, yet the precise mechanism and the intricate dynamics of SSB interaction with T7 DNAp and its subsequent impact on the replication process remain unclear. Free energy profiles have demonstrated that the local minimum is related to the dissociated state in the DNAp and SSB complex (Fig. 3F). Here we constructed the free energy profile of the C-terminal truncated variant of T7 gp2.5 (mut T7 SSB) binding to the ssDNA during DNA replication using MD simulation (Fig. 4A). The blue-colored local energy minima shift from the dissociated state (the position of the right side as indicated in Fig. 3D) to the bound or intermediate state side (the position of the left side as indicated in Fig. 3D). This MD simulation revealed that the presence of the C-terminal facilitated the energy transition from bound to dissociated states, suggesting the DNAp might strip SSB of the DNA using the contact with its C-terminal. Moreover, removing the C-terminal significantly weakened the intermediate state due to the C-terminal’s role as an anchor for binding the front basic patch region of DNAp37, which contains four positively charged residues (K587, K589, R590, and R591), facilitating strong electrostatic interactions with the C-terminal of SSB. A zoom-in view of the representative structure from the dissociated state in the DNAp-SSB energy profile shows the strong contact between the basic patch region and the C-terminal of SSB. The distance between R590 in DNAp and E214 in SSB is 5.6 Å, suggesting that a strong electrostatic interaction forms. The flanks of the basic patch region, K594 and R599 may also help the binding process of SSB, since R599 also contributes to the electrostatics effects with SSB E222 (Fig. 4B). Unbiased simulations comparing full-length gp2.5 and the Δ21C mutant with ssDNA highlighted the detachment dynamics, with the mutant displaying increased ssDNA affinity over time (Fig. 4C). The trajectory movies of the unbiased DNAp-SSB runs (Supplementary Movie 2) and the DNAp-SSBΔ21C runs (Supplementary Movie 3) provides insight into how DNAp actively strips SSB on the DNA replication.
A Two-dimensional free energy profile of T7 SSB-Δ21C (C-terminal truncated mutant) binding to ssDNA in the presence of DNAp-trx complex. Energy barrier between states increased from ~3 kcal/mol (wild-type) to ~10 kcal/mol, hindering state transitions and demonstrating the C-terminal tail’s role in facilitating displacement. B Detailed view of C-terminal SSB binding to DNAp during MD simulation. Orange: coarse-grained ssDNA segments; red: SSB C-terminal tail; green: DNAp front basic patch region. Key positive residues (K587, K589, R590, R591, K594, and R599) colored blue. Critical electrostatic interactions shown: R590-E214 distance (5.6 Å) and R599-E232 distance (8.4 Å), demonstrating strong binding between the basic patch and acidic C-terminal. C Distance variation between DNA and SSB during unbiased simulations comparing DNAp+gp2.5 (blue) versus DNAp+gp2.5-Δ21C (orange). Full-length gp2.5 shows progressive detachment from ssDNA over simulation time, contrasting with persistent binding of the Δ21C mutant, confirming the C-terminal’s role in displacement facilitation. D, E Real-time primer extension assays with escalating wild-type (D) and mutant (E) SSB concentrations (0–3000 nM). Black arrows indicate minimum fluorescence intensity marking transition from polymerase-dominated to exonuclease-dominated phases. N = 3 replicates per concentration. F Quantified polymerase activity as a function of varying SSB concentrations. Activity estimated from initial linear curve section (~1.5 min) representing polymerase-dominated phase. Wild-type SSB increases replication rate (~20% enhancement from 0 to 3000 nM), while mutant SSB decreases the rate (~25% reduction), with effects apparent only above 150 nM. N = 3 replicates per concentration. G, J Single-molecule force spectroscopy analysis of replication rates at 10 pN (G) and 20 pN (J) with varying SSB concentrations (10, 30, 150 nM). Dashed lines: reference rates without SSB. H, K Exonuclease binding probability analysis at 10 pN (H) and 20 pN (K) forces. Probability calculated as the ratio of exonuclease events to total polymerase plus exonuclease events per DNA molecule. Wild-type SSB promotes polymerase mode; mutant favors exonuclease mode. I, L Average pause duration comparison at 10 pN (I) and 20 pN (L) forces with varying SSB concentrations. Wild-type SSB increases pausing at low forces due to recruitment effects. Mutant SSB increases pausing at all concentrations, demonstrating roadblock formation without productive displacement. Data presented as mean ± SEM. Source data are provided as a Source Data file.
Further, we examined the C-terminal’s role in modulating polymerase activity across varying SSB-DNA saturation levels using real-time primer extension assays43. By employing real-time primer extension assays enabled by a 5′-end fluorophore, quenched upon nucleotide incorporation, we monitored the kinetics of DNA synthesis in real-time (Supplementary Fig. 8A, B). Wild-type SSB addition accelerated the polymerization rate, as evidenced by a shortened polymerization phase with increasing SSB concentrations (leftward shift of black arrow, Fig. 4D), suggesting a shorter time to complete the DNA synthesis. In contrast, the mutant SSB showed a slightly reduced polymerization rate (Fig. 4E). We then compared the relative polymerase activities, varied with SSB concentration, by reporting initial rates (RFU/s) as a approximation measured during the polymerase-dominated phase (Methods, Fig. 4F). These results demonstrate that the presence of wt T7 SSB increases the replication rate (increased by ~20% from 0 nM to 3000 nM wt T7 SSB), while the presence of mutant SSB leads to a decreased rate (decrease by ~25% from 0 nM to 3000 nM mut T7 SSB), corroborating our single-molecule studies (Fig. 1C).
Notably, this effect becomes apparent only at concentrations exceeding 150 nM (Fig. 4F). Given the 7–10 nt footprint per wild-type T7 SSB molecule13 and our ssDNA template with a 17 nt overhang (Methods), our results indicate saturation occurs near a 2:1 ratio of SSB to DNA, aligning with the requirement of 150 nM SSB for saturating 70 nM ssDNA. This suggests SSB may “pull” DNAp forward during replication under saturated conditions, while the tailless SSB mutant hinders replication, confirming the C-terminal tail’s crucial role in facilitating DNAp progression along the ssDNA template. Despite stronger DNA binding, mutant SSB still undergoes slow spontaneous dissociation, explaining partial rather than complete inhibition in primer extension assays at high mutant concentrations (Fig. 4D–F), indicating that while the C-terminal tail significantly enhances displacement efficiency, it is not absolutely essential for all SSB removal.
As T7 DNA polymerase possesses an intrinsic exonuclease domain40 for proofreading, our real-time assay trace showed subsequent increases in fluorescence intensity, indicating exonuclease activity (Fig. 4D, E). Control experiments demonstrated relatively stable fluorescence signals when only SSB was added to DNA, though higher SSB concentrations correlated with decreased overall fluorescence (Supplementary Fig. 8C–E), possibly due to SSB binding to ssDNA causing fluorescence quenching. Importantly, this control experiment enables reliable quantification of SSB effects on DNA polymerase activity.
To further elucidate the dynamic mechanism of DNAp interacting with the C-terminal truncated SSB under physiologically relevant tensions, we employed force spectroscopy to measure the interaction between DNAp and mut T7 SSB at varying concentrations and compared the dynamics with that of the T7 SSB. Our previous findings13 have indicated that T7 SSB reaches saturation on DNA binding at concentrations exceeding 50 nM in single-DNA molecule assays. We examined the replication and exonuclease activities of DNAp under three distinct SSB concentration regimes: low (10 nM), semi-saturated (30 nM), and fully saturated (150 nM), utilizing single-molecule, high-resolution optical tweezers to track DNA length alterations.
At a force of 10 pN, the replication rate was enhanced with fully saturated wt T7 SSB compared to replication without SSB (230 ± 20 bp/s v.s. 180 ± 10 bp/s, horizontal dashed line from Fig. 1D added as a reference, Fig. 4G, mean ± SEM, p = 0.046), while this enhancement was not clearly observed at low or semi-saturated SSB levels (120 ± 50 bp/s, and 120 ± 30 bp/s, respectively, mean ± SEM, p > 0.05). In contrast, mutant SSB significantly reduced polymerase rate at 30 nM and 150 nM concentrations (p < 0.001, Supplementary Data 2), consistent with the inhibition observed at saturating concentrations (>150 nM) in primer extension assays (Fig. 4F).
We quantified the likelihood of polymerase engaging in exonuclease activity (Fig. 4H, reference dashed line from Fig. 1E), revealing that wt T7 SSB, particularly at saturated levels, preferentially positions the polymerase in polymerase state (0.229 ± 0.04 at 0 nM v.s. 0.058 ± 0.02 at 150 nM, mean ± SEM, p = 0.001). In contrast, the C-terminal tail-lacking mutant SSB favors exonuclease state binding (0.229 ± 0.04 at 0 nM v.s. 0.548 ± 0.1 at 150 nM, mean ± SEM, p = 0.017). This effect can be attributed to two factors: the C-terminal domain’s interaction with DNAp, possibly through the trx domain from DNAp, facilitating DNAps’ forward movement and reducing proofreading occurrences, and wt T7 SSBs’ reduced DNA affinity, minimizing replication hindrance. The T7 mut SSB, on the other hand, binds to ssDNA longer but fails to interact constructively with DNAp, acting primarily as a roadblock. This suggests DNAp might be only able to advance when mutant SSB disassociates, as further evidenced by increased pausing times in mutant SSB (Fig. 4I) (19 ± 1 s at 0 nM v.s. 60 ± 6 s at 150 nM, mean ± SEM, p < 0.001). The presence of wt T7 SSB also increases pausing times of DNA polymerase (Fig. 4I, reference dashed line from Fig. 1G). This supports the hypothesis that SSB bound to ssDNA recruits DNA polymerase to the DNA template, creating a roadblock that causes the replicative polymerase to pause (Fig. 1H). Interestingly, lower concentrations of SSB lead to longer pauses (40 ± 10 s at 10 nM) compared to higher concentrations, which result in shorter pauses (30 ± 2 s at 150 nM), as shown in Fig. 4I. This observation might be due to the fact that at lower SSB concentrations, the protein is located at random positions on the ssDNA, which can randomly recruit DNA polymerase to bind to the ssDNA and cause longer pauses. In contrast, at higher, saturating SSB concentrations, there is less available space for the polymerase to bind, leading to shorter pausing durations.
Under conditions promoting less secondary structure (20 pN), both the presence of wt T7 SSB and mut T7 SSB decreased replication rates (Fig. 4J), consistent with earlier findings in Fig. 1. Assessing exonuclease site binding probabilities revealed that mutant SSB presence increases the likelihood of exonuclease state binding with rising SSB concentrations (Fig. 4K) (0.308 ± 0.05 at 0 nM vs. 0.6 ± 0.1 at 150 nM, p = 0.016).
These results indicate that at increased tension, any SSB presence acts as a blockade, leading to lower replication rates and higher exonuclease state binding probability. This is likely due to the reduced need to compete with the secondary structure of ssDNA. At lower wt T7 SSB concentrations, the protein is more likely to recruit and stabilize DNA polymerase on the ssDNA, forming a self-imposed roadblock that further reduces the replication rate and increases the exonuclease state probability. This is further supported by the analysis of average pausing durations (Fig. 4L), where lower wt T7 SSB concentrations resulted in longer pauses compared to no SSB (28 ± 3 s at 10 nM v.s. 13 ± 1 s at no SSB, p < 0.001), while saturated wt T7 SSB levels (150 nM) produced pause times comparable to those observed without SSB (15 ± 1 s, p = 0.24). Interestingly, the mutant SSB binds longer to ssDNA but fails to interact constructively with DNA polymerase, primarily acting as a roadblock. Higher concentrations of the mutant SSB thus increased the DNA polymerase blockage times (18 ± 1 s at 10 nM vs. 45 ± 5 s at 150 nM, p < 0.001), demonstrating a concentration-dependent effect.
Discussion
Genomic DNA is a crowded molecular environment where DNA motor proteins frequently collide. A notable example is the encounter between replicative DNAp and high-affinity SSBs coating ssDNA during DNA replication. Our study investigated the molecular mechanisms by which DNAp overcomes these SSBs, revealing a sequential and active displacement model where SSBs are displaced one after another through functional domain interaction. This coordinated process not only enables efficient SSB removal but also promotes each SSB encounter to enhance overall replication efficiency, suggesting an evolved synergy between these seemingly antagonistic proteins.
Our single-molecule dual-color imaging experiments provide compelling evidence for this mechanism. By directly tracking DNAp and fluorescently labeled T7 gp2.5, we observed that DNAp consistently advances toward and removes stationary SSBs, with no indication of forward movement by the SSBs themselves. Measurements of the diffusion constant of T7 SSB on ssDNA further confirm their relative immobility on the template, consistent with prior findings13, and indicate that SSBs are not easily pushed forward. Moreover, high template occupancy restricts lateral diffusion, and the absence of free ssDNA ends in Okazaki fragments precludes displacement by pushing. Together, these observations argue strongly against a model in which SSBs are collectively pushed forward by the polymerase, contrasting with alternative mechanisms proposed in other systems, such as translocases advancing along ssDNA by pushing SSBs forward18. An alternative “dynamic rearrangement” model, where SSBs transiently dissociate and rebind or undergo bidirectional sliding, resulting in net displacement or redistribution without full eviction, might also be possible. Such a mechanism may be present in systems with highly mobile SSBs44. However, our kymograph analyses show no evidence of directed sliding or significant relocalization during DNAp progression, therefor in the T7 gp2.5 system this is unlikely to happen.
Mechanistically, the limited mobility and moderate dissociation rate (~2.4 s⁻¹) of gp2.5 necessitate active displacement by DNA polymerase. Both previous41,42 and current data support a model in which this displacement is driven by direct interaction between DNAp and the acidic C-terminal tail of gp2.5. This interaction likely facilitates removal by altering the local energy landscape of SSB-ssDNA binding. Functionally, the sequential displacement mechanism ensures that SSBs remain stably bound to ssDNA until directly evicted by the advancing DNAp, providing continuous protection against nucleases and suppressing secondary structure formation1,2. This coordination reduces the risk of replication errors and promotes genomic stability. The diversity of SSB properties across species—ranging in diffusion behavior, binding modes, and physiological roles—suggests that their interactions with DNA translocases are system-specific. In contrast to more dynamic SSB systems that allow sliding or exchange17,18,19,44, T7 gp2.5 requires a direct and active removal strategy tailored to its immobile, high-affinity binding behavior.
Using AWSEM coarse-grained MD simulations, we modeled the DNAp-SSB interaction, revealing strong electrostatic interactions between the basic patch on the front of DNAp and the acidic C-terminal tail of SSB. Our experimental findings, using single-molecule FRET and high-resolution force measurements, suggest direct contact and regulatory mechanisms between these proteins. The observed FRET signals imply direct interactions facilitating the displacement of SSB by DNAp during replication. Notably, this displacement does not perturb the overall structure of DNAp’s interaction with thioredoxin (trx), in line with conclusions from prior research45. The highly negative charge of SSB’s C-terminus, akin to the PEST sequence in the C-terminus of IκBα, suggests a universal mechanism among DNA regulatory proteins46,47, involving molecular stripping facilitated by electrostatic interaction. This interaction possibly alters the binding affinity of SSB, leading to its dissociation and facilitating the forward movement of DNAp. Disruptions to this interplay, as observed with tailless SSB, result in diminished DNA replication rates.
Our single-molecule investigations reveal that SSB proteins play a dual role in DNA replication, dependent on the ssDNA conformation regulated by biologically relevant forces. Under low-tension conditions (forces <~15 pN), where ssDNA is prone to forming secondary structures, SSBs prevent these formations, smoothing the DNA template for replication and increasing processivity, aligning with previous studies4,10. Furthermore, the electrostatic interaction between the acidic C-terminal tail of SSB and the basic patch on DNAp increases the replication rate, potentially by facilitating the forward movement of DNAp. The increased probability of DNAp binding in the polymerase-active direction in the presence of SSB indicates that SSBs promote DNAp’s active conformation, likely through direct physical interactions. These single-molecule observations are consistent with prior in vitro ensemble studies conducted under low- or no-tension conditions, which have demonstrated a notable acceleration in DNA replication in the presence of SSBs3,9,10,48,49,50. The observed increase in replication rate with wild-type T7 SSB, particularly at saturating concentrations, suggests that each individual SSB molecule assists in facilitating DNAp progression along ssDNA. This facilitation is likely mediated by the C-terminal tail of SSB, as mutants lacking this domain did not enhance the replication rate41,42. According to the proposed active displacement mechanism, DNAp advancement coordinates with SSB dissociation by lowering the energy barrier, effectively regulating SSB removal during replication.
Conversely, at higher tensions (>~20 pN), where secondary structure formation is less likely, SSBs function as physical barriers, reducing replication efficiency. SSBs possibly act as weak barriers under all conditions, but at low forces, their beneficial role in preventing larger hairpin structures outweighs this obstruction. At high forces, where hairpin formation is suppressed, the obstruction from SSBs becomes more significant. Additionally, excessive recruitment of DNAp due to SSBs positioned along ssDNA can create self-imposed obstacles, leading to extended pause durations during replication. The force-dependent impact of SSBs implies a regulatory mechanism mediated by biological tension—possibly induced by other molecular motors or DNA constraints51—allowing cells to modulate replication dynamics in response to cellular conditions or stress. Our findings align with similar observations in E. coli6 and mitochondrial4 systems, affirming the integral role of SSBs in modulating replication efficiency.
Positioning our findings within the broader biological context highlights the critical role of SSBs in DNA replication dynamics. Yet, various potential discrepancies exist between our experimental conditions and the in vivo situation. Our research focused on an in vitro phage system, which may oversimplify cellular replication complexities. In vivo, replication involves intricate systems, potentially including hybrid machinery from both viral and host cell replisomes. Due to limited temporal resolution in confocal fluorescence microscopy, we did not address how rapid DNAp exchange is affected by SSB displacement. The low throughput of single-molecule assays also limited our survey of other SSBs. Future studies should extend investigations to other replicative systems, such as eukaryotes with distinctive SSB diffusive properties. Such studies would more closely mimic in vivo conditions, broadening the applicability of our findings and potentially uncovering universal principles of DNA replication regulation.
Methods
DNA template construction, protein purification and labeling
The pKYB1 vector (~8.3 kb) DNA construct with biotin labels at both ends was generated using established protocols52. Recombinant T7 DNA polymerase (T7 gp5 fused with trxA) was expressed, purified, and labeled with SNAP-Surface® 549. Pure protein and labeled protein were aliquoted, flash-frozen, and stored at −80 °C. Wild-type T7 gp2.5 were purified and labeled with Atto647N using a protocol based on previous literature53.
Single molecule setup
Single-molecule experiments were performed on a LUMICKS C-Trap instrument, which combines three-color confocal fluorescence microscopy and dual-trap optical tweezers with a five-channel microfluidic flow cell. A 1064-nm fiber laser was used via a water-immersion microscope objective to generate two orthogonally polarized optical traps. The biotinylated pKYB1 DNA construct was tethered between two 1.76-μm streptavidin-coated microspheres (Spherotech Inc) in situ within the flow cell. A single DNA molecule was detected and confirmed through a change in the F-x curve. Orthogonal channels 4 and 5 were used as protein loading and/or experimental imaging chambers as described for each assay. All experiments were performed at room temperature with force data collected at 50 kHz. Fluorescently labeled DNA polymerase was excited at 532 nm, and fluorescently labeled T7 SSB was excited at 638 nm, and to optimize imaging data over a long measurement period, the protein was scanned with a pixel dwell time of 1–2 ms, laser power of 1.2 mW (30%), and scanning interval of 8 ms unless stated otherwise.
Experimental assay
T7 DNA polymerase concentration was set at 30 nM with a labeling efficiency of ~60%. T7 gp2.5 concentration was set at 150 nM with a labeling efficiency of ~4%. The fluorescently labeled SSBs, mixed with unlabeled SSBs, served as markers to determine the relative distance between the replicative DNA polymerase and the SSB proteins.
To prevent nonspecific binding, the flow cell was cleaned with bleach and passivated overnight with 0.1% BSA and 0.5% Pluronic. The experiments were conducted in a standard measurement buffer composed of 20 mM Tris-HCl, pH 7.5, 100 mM NaCl, 5 mM MgCl2, 1 mM DTT, and 0.02% BSA.
Single DNA molecules were captured and moved into a protein channel containing DNA polymerase and SSB. The activity of the DNA polymerase was studied by monitoring the change in DNA extension at a constant tension of 40–50 pN. This tension was used to digest ~5 kbp of dsDNA and create a long section of ssDNA as a template for polymerization activity. Tension was then decreased to 10–20 pN to measure polymerization activity in the presence or absence of SSB.
Real-time DNA primer extension assay
The activity of T7 DNA polymerase was measured in the presence or absence of wild-type T7 single-stranded DNA-binding protein (T7 gp2.5, wt T7 SSB) and a deletion mutant lacking 21C-terminal residues (T7 gp2.5-Δ21C, mut T7 SSB) using a 45-nt long DNA template annealed to a 28-nt primer strand. The carboxyfluorescein (6-FAM) moiety was located at the 5′ end of the template strand. Template strand: 5′/6-FAM/- CCCCCCCCCATGCATGCGCACCTAAAGTTGGGAGTCCTTCGTCCTA-3′. Primer strand: 5′- TAGGACGAAGGACTCCCAACTTTAGGTG-3′. Reactions were performed in a final volume of 10 μl with 25 μm of each dNTP in buffer 20 mM Tris-HCl, pH 7.5, 50 mM NaCl, 3 mM MgCl2, 1 mM DTT, 0.05% BSA, and 0.05% Tween-20. Reactions were initiated by the addition of 70 nM labeled DNA to 30 nM DNA polymerase premixed with the indicated concentration of wt T7 SSB or mut T7 SSB and measured in a 348-well plate using a BMG Labtech Pherastar FS plate reader during 30 min at 25 °C.
The polymerase activity was estimated from the initial linear section of the curve (approximately the first 1.5 min), representing the polymerase-dominated phase where DNA synthesis actively occurs. The slope derived from this fit gives an estimate of the initial polymerase activity in RFU/s. Data analysis was performed using self-written python script and GraphPad Prism software.
Single-molecule experiment data analysis
All data obtained from C-trap is in. tdms format and is analyzed using custom-written Python scripts. Details for access is provided in Code availability section.
Analysis of ssDNA/dsDNA junction trajectory
In our investigation, we manipulated the template tension using an optical tweezer system. The T7 DNA polymerase was tailored to function as an exonuclease at tensions ranging from 40 to 50 pN, producing partially ssDNA, and to execute polymerization at tensions between 10 and 20 pN, facilitating the synthesis of dsDNA. Owing to the catalytic activity of DNA polymerase, the end-to-end distance (EED), measured directly between two optically entrapped beads, comprising both ssDNA and dsDNA, was consistently monitored over time.
We applied a composite of the freely jointed chain (FJC) and worm-like chain (WLC) models to determine the proportions of ssDNA and dsDNA present within the EED, respectively. Given the elasticity differences between ssDNA (described by an FJC model54) and dsDNA (described by an extensible WLC model55), the ssDNA percentage can be derived using the equation:
To adjust for the changing ssDNA length during DNA replication involving saturated SSB binding, we employ a correction factor. This factor accommodates the transient shortening of ssDNA caused by SSB binding during DNA polymerase activity.
The correction formula applied is:
Here, “EED” represents end-to-end distances between the two optically trapped beads, while “FJC” and “WLC” denote the freely jointed chain54 and worm-like chain55 models, respectively. “F” corresponds to the applied force. The correction factor accounts for the ssDNA length alteration when fully coated by SSB proteins, estimated as 0.246 µm under 10 pN and 0.177 µm under 20 pN with gp2.5 protein (Supplementary Fig. 2). This methodology refines our ssDNA length estimates during replication, enhancing the precision of DNA replication dynamics analysis.
The dsDNA percentage can be calculated as
The ssDNA/dsDNA junction position can be determined by assuming that ssDNA appears on the top half of the kymograph:
If the dsDNA appears on the top half of the kymograph, the junction position can be derived as:
This method facilitates high-resolution, real-time tracking of DNAp’s position, even amidst sporadic dissociation of the fluorescently labeled DNAp.
Analysis of single-molecule basepair-time traces
We employed a pwlf Python library56 to fit piecewise linear functions to single-molecule base pair-time traces and detect the most likely change points, which signify shifts in polymerase (pol) or exonuclease (exo) activities. To reduce noise, the traces were initially processed with a Savitzky–Golay (SG) filter. The piecewise linear function, adapted from previous publications56 with minor modifications, was then used to detect these change points, allowing us to mark the steps or change points in the base pair-time traces. Subsequently, we quantified the processivity, velocity, and duration of each identified segment.
One crucial aspect of our study involves the detection and characterization of pausing events, denoted by temporary halts in polymerase activity that yield steady basepair values over a significant period. The “noise threshold” is determined as the standard deviation of basepair-time traces in the absence of protein under the tensions under investigation, estimated by employing the median absolute deviation (MAD) of sequential data points and amplified by a constant factor (1.4826). This method facilitates a robust distinction between inherent fluctuations (noise) and genuine pausing events (signal). The “significant period” is deduced from the known exonuclease rate of T7 DNAp (~100 bp/s) and replication rate (~200 bp/s), indicative of the standard duration necessary for the polymerase to add a base pair under our experimental parameters. We define a pause as an event where the actual process duration is ten times greater than the expected time, which, for simplicity, is defined to occur at 10 bp/s under 50 pN and 20 bp/s under 20 or 10 pN.
Analysis of fluorescent protein movement using kymographs
The acquired kymographs were analysed with a custom-written tracking program in Python using lumicks.pylake package. In brief, the line identification algorithm57 was employed to identify the centers of lines by leveraging local geometric principles, while the greedy algorithm was used for initial feature point detection and line tracing58,59.
Analysis of SSB diffusion using kymographs
The diffusion constant of T7 SSB was calculated using the mean square displacement (MSD) method, exploiting the extracted fluorescence trajectory, as illustrated by the yellow line in Fig. 2D. The computed diffusion constant of 7 ± 7 × 10−4 μm2/s (mean ± SEM) (Fig. 2H), in agreement with our recent report13, substantiates that the SSB is essentially immobile on ssDNA. By comparison, the known static protein EcoRV was previously determined in the same lab33 to exhibit an apparent diffusion constant of 1 × 10−4 µm2/s with a localization accuracy value (~10 nm, employing integration times of 1 s), slightly lower than that determined in our experiments. This supports the hypothesis that T7 SSB binds at a fixed position and does not diffuse.
Correlation between ssDNA/dsDNA junction with fluorescently labeled DNA polymerase
Alongside a distance-time curve, we recorded the fluorescence trajectory from labeled proteins, enabling real-time tracking of DNA polymerase activity at the ssDNA/dsDNA junction. This was achieved by overlaying the calculated ssDNA/dsDNA junction movement, based on earlier calculation, with the DNAp trajectory in the kymograph. We adjusted for variations in starting time and position between the optical tweezer and fluorescence microscopy systems. Fluorescence intensity was extracted along the junction over time, focusing on the green channel to reduce signal crosstalk. A 5-pixel-wide box was used to capture photons within the point-spread function, ensuring optimal signal collection. An optimal overlap between the calculated ssDNA/dsDNA junction and the fluorescent trajectory was determined by seeking the maximum local intensity. We recalibrated the x-offset and y-offset based on these findings.
Determination of the distance between DNAp and SSB
The optimized DNAp trace was overlaid with the fluorescently labeled SSB. To enable further quantification, the SSB trace was isolated and subjected to trajectory analysis, considering only those SSB trajectories which spanned more than three pixels (~1 s). These fluorescently captured trajectories were subsequently aligned with the DNAp trajectories, calculated from the ssDNA/dsDNA junction. Following this alignment, the distance between the DNAp and the SSB, was measured. In addition to these distance measures, we also computed the duration, rate, and diffusion constant of SSB to further illuminate the underlying mechanisms of the DNAp-SSB interaction.
Error discussions on tracking the real-time interaction between DNAp and SSB
Potential sources of error were carefully considered and mitigated in tracking the interaction between DNAp and SSB, which involved tracking the DNAp position, detecting SSB trajectories, and overlapping the optimized DNAp and SSB traces. First, although optimal overlapping of the calculated ssDNA/dsDNA junction and the fluorescent trajectory was attempted, the DNAp track could occasionally deviate from the true trace due to local fluctuations in the fluorescence signal or instrumental noise. Errors may also arise from inaccuracies in estimating the bead size (Supplementary Fig. 2B). Calibration procedures were implemented to minimize these errors. Second, in detecting SSB trajectories, systematic errors arising from the point-spread function and diffraction limit were addressed by employing a line identification algorithm and a greedy algorithm for initial feature point detection and line tracing (Methods). Background subtraction and signal-to-noise ratio optimization techniques were also applied to enhance the accuracy of SSB trajectory detection. Third, in overlapping the optimized DNAp and SSB traces, slight misalignments could occur due to the use of different dyes and excitation lasers (532 and 638 nm, respectively) for labeling DNAp and SSB, leading to variations in optical properties and potential crosstalk between fluorescence channels. Appropriate measures, such as spectral unmixing and channel compensation, were taken to minimize these effects. By acknowledging and addressing these potential sources of error, we aimed to minimize their impact, thereby enhancing the reliability and reproducibility of our findings on the determination of the distance between DNAp and SSB.
Coarse-grained force field for protein-ssDNA system
The latest iteration of the protein folding force field, known as the associative memory, water-mediated, structure and energy model (AWSEM), builds upon a sequence of models refined through energy landscape theory and a quantitative machine learning approach for protein folding. AWSEM represents each amino acid residue using three actual atoms (CA, CB, and O) and three virtual sites (C, N, H, with exceptions for proline and glycine). The AWSEM-Suite, an updated version of AWSEM, has demonstrated its predictive accuracy in the CASP13 protein structure prediction competition, achieving top-three server predictions in several instances60.
In our previous work on the helicase gp4-ssDNA system, we have extended AWSEM to a transferable protein-DNA model with a modified 3SPN.2 C force field that have shown the physical properties of ssDNA23,61. Our coarse-grained DNA model incorporates detailed features of DNA structure, including specific hydrogen bonding and base stacking interactions characteristic of the B-DNA form. Similar to AWSEM, the DNA force field uses three beads to indicate the phosphate, sulfur, and nitrogen base atom group.
For the details of the AWSEM and ss-3SPN2 energy composition, please refer to ref. 25. Briefly, AWSEM is powered by a combination of biophysical and bioinformatically derived energy terms. The backbone term constrains the peptide backbone via a harmonic potential and includes excluded volume interactions among all atomic centers. The contact term captures both direct residue-residue contacts and water/protein-mediated interactions over slightly longer distances. The hydrogen bond term and the burial term are many-body interaction terms that capture the local environment. The modified 3SPN.2C force field is derived from experimental B-DNA form characteristics and allows flexibility to represent dynamic movements. The interaction between protein and DNA includes both electrostatic and excluded volume terms.
Construct the initial structures for MD simulation
The currently only available gp2.5 structure in PDB is 1JE5, which lacks the most flexible C-terminal region (residue 203–232) and the linker region (residue 79–88)35. Deep-learning-based protein structure prediction model Alphafold2 and Modeller version 9.23 were used to construct the full-length gp2.5 structure36,62. AlphaFold2 is a state-of-the-art transformer-based model developed by DeepMind that can predict protein structures from amino acid sequences with atomic-level accuracy. In contrast, Modeller represents the traditional template-based modeling tool for protein structure that achieves good performance. Since the C-terminal is tightly bind to the positive binding groove in the Alphafold2-predicted structure, a short simulation with external force to expel the C-terminal tail from the groove was did to probe an initial state that ssDNA have balanced probability to bind.
For the gp2.5-gp5-ssDNA complex, we choose structure PDB ID 6P7E as a template because this structure contains a peptide DTDF that mimics the C-terminal tail of gp437. By aligning the C-terminal of the gp2.5 structure to the short peptide and extending the 5’ end of the ssDNA with poly-A, we create an initial complex that gp2.5 closes to the longer ssDNA chain. We also use the inferred structures from SAXS data in Supplementary Fig. 7 from ref. 37.
Free energy calculations based on the umbrella sampling technique
Umbrella sampling is an enhanced sampling technique that could force the exploration of regions of state space that would otherwise have insufficient sampling. It uses a series of independent windows along a selected collective variable, which serves as a continuous parameter to describe the system from a higher-dimensional space to model a conformational transition63. In the current system, we use the center of mass between gp2.5 and the first 20 nucleotides of the ssDNA as one metric, while the electrostatics between the charged residues in the positive binding pocket of gp2.5 (residue 72–94, 113–125, 157–165) and ssDNA as the second metric. For the DNAp-SSB complex, we used the distances between the COM of the SSB and the COM of the first 21 base pairs of the 5’ overhang ssDNA as the biasing coordinates to build free energy profiles. No external bias was added to the interaction of DNAp and T7 SSB.
The harmonic biasing potential used for constant temperature umbrella sampling simulations for 8 million steps was scaled to 10 kcal/mol. The biasing center values were chosen to be equally spaced from 12.5 to 27 Å with an increment of 0.5 Å and 50 windows. The initial structures for umbrella sampling were picked as the complex structure described in the above section. The weighted histogram analysis method (WHAM) is used to reconstruct the unbiased free energy landscapes from the umbrella sampling data64. An additional dimension related to the specific conformation changes of interest was added for plotting the 2D free energy profile.
Protein remote homology detection
The protein remote homology detection is conducted by HHpred (https://toolkit.tuebingen.mpg.de/tools/hhpred)65. The default structural/domain databases (PDB_mmCIF70_8_Mar) was used to perform the search.
HHpred simplifies sequence database searching and structure prediction, offering a user-friendly experience akin to BLAST or PSI-BLAST while significantly enhancing sensitivity in identifying distant homologs. HHpred scans alignment databases such as Pfam or SMART, streamlining search results into sequence families rather than individual sequences. The method operates on the comparison of profile hidden Markov models (HMMs), contrasting with traditional sequence search methods that scan databases like UniProt or NR.
Structure clustering and heatmap analysis
The structure frames that fall within the local energy basin area during the umbrella sampling were extracted based on the free energy profile. The Qw value was used to measure the pairwise distance between two structures, with a detailed explanation provided in ref. 60. After calculating the Qw value for all structures in the local basin, the hierarchically-clustered heatmap and dendrogram were computed using the clustermap function in the seaborn package and the dendrogram function in the scipy package, respectively. The Y axis indicates the distance of each branch, as computed by the scipy.cluster.hierarchy.linkage package. The center of the largest cluster was then determined and manually picked out for further analysis.
Quantification and statistical analysis
The number of molecules or events analyzed is indicated in the text or figure legends. Errors reported in this study represent the standard error of the mean (SEM), unless otherwise stated. Statistical significance was assessed using multiple approaches: (1) Welch’s unpaired two-tailed t-tests (not assuming equal variances), (2) bootstrap resampling tests (10,000 iterations), and (3) permutation tests (10,000 permutations). Statistical significance was established at p < 0.05. For pairwise comparisons between SSB conditions, all three statistical methods were applied to ensure robust determination of significance. These analyses were performed using Python 3.x with SciPy, NumPy, and Pandas libraries. Complete statistical results, including mean differences, p values, and sample sizes for all comparisons, are provided in the Supplementary Data 2. Source code is included in the Code availability section. All experiments were independently repeated at least three times with similar results. Representative results are shown in figures.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The datasets generated and/or analyzed during the current study are available from Figshare (https://doi.org/10.6084/m9.figshare.28355369). Source data are provided with this paper.
Code availability
The custom-written Python scripts for analyzing basepair-time traces, for analyzing the displacement of SSB and for analyzing the real-time DNA primer extension data are available at GitHub (https://github.com/longfuxu/Interplay_Between_DNAPol_and_SSB) under the MPL-2.0 license. The script for analyzing the MD simulation result and the weighted histogram analysis method (WHAM) are available at GitHub (https://github.com/CryoSky/MD_simulation_DNAp_SSB). The repository includes an example dataset, an example Jupiter notebook, along with a detailed README file for instructions on installation and usage.
References
Bercy, M. & Bockelmann, U. Hairpins under tension: RNA versus DNA. Nucleic Acids Res. 43, 9928–9936 (2015).
Cocco, S., Monasson, R. & Marko, J. F. Force and kinetic barriers to unzipping of the DNA double helix. Proc. Natl Acad. Sci. USA 98, 8608–8613 (2001).
Ghosh, S., Hamdan, S. M. & Richardson, C. C. Two modes of interaction of the single-stranded DNA-binding protein of bacteriophage T7 with the DNA polymerase-thioredoxin complex. J. Biol. Chem. 285, 18103–18112 (2010).
Cerron, F. et al. Replicative DNA polymerases promote active displacement of SSB proteins during lagging strand synthesis. Nucleic Acids Res. 47, 5723–5734 (2019).
Shereda, R. D., Kozlov, A. G., Lohman, T. M., Cox, M. M. & Keck, J. L. SSB as an organizer/mobilizer of genome maintenance complexes. Crit. Rev. Biochem. Mol. Biol. 43, 289–318 (2008).
Antony, E. & Lohman, T. M. Dynamics of E-coli single stranded DNA binding (SSB) protein-DNA complexes. Semin. Cell Dev. Biol.86, 102–111 (2019).
Lee, J. B. et al. DNA primase acts as a molecular brake in DNA replication. Nature 439, 621–624 (2006).
Johnson, D. S., Bai, L., Smith, B. Y., Patel, S. S. & Wang, M. D. Single-molecule studies reveal dynamics of DNA unwinding by the ring-shaped T7 helicase. Cell 129, 1299–1309 (2007).
Hernandez, A. J. & Richardson, C. C. Gp2.5, the multifunctional bacteriophage T7 single-stranded DNA binding protein. Semin. Cell Dev. Biol. 86, 92–101 (2019).
Ciesielski, G. L. et al. Mitochondrial single-stranded DNA-binding proteins stimulate the activity of DNA polymerase γ by organization of the template DNA. J. Biol. Chem. 290, 28697–28707 (2015).
Roy, R., Kozlov, A. G., Lohman, T. M. & Ha, T. Dynamic structural rearrangements between DNA binding modes of E. coli SSB protein. J. Mol. Biol.369, 1244–1257 (2007).
Pant, K., Karpel, R. L., Rouzina, I. & Williams, M. C. Salt dependent binding of T4 gene 32 protein to single and double-stranded DNA: single molecule force spectroscopy measurements. J. Mol. Biol.349, 317–330 (2005).
Xu, L. et al. Regulation of T7 gp2.5 binding dynamics by its C-terminal tail, template conformation and sequence. Nucleic Acids Res. 51, 6540–6553 (2023).
Joo, C., Balci, H., Ishitsuka, Y., Buranachai, C. & Ha, T. Advances in single-molecule fluorescence methods for molecular biology. Annu. Rev. Biochem. 77, 51–76 (2008).
Bustamante, C. J., Chemla, Y. R., Liu, S. & Wang, M. D. Optical tweezers in single-molecule biophysics. Nat. Rev. Methods Prim. 1, 25 (2021).
Heller, I., Hoekstra, T. P., King, G. A., Peterman, E. J. G. & Wuite, G. J. L. Optical tweezers analysis of DNA–protein complexes. Chem. Rev. 114, 3087–3119 (2014).
Honda, M., Park, J., Pugh, R. A., Ha, T. aekjipand & Spies, M. Single-molecule analysis reveals differential effect of ssDNA-binding proteins on DNA translocation by XPD helicase. Mol. Cell35, 694–703 (2009).
Sokoloski, J. E., Kozlov, A. G., Galletto, R. & Lohman, T. M. Chemo-mechanical pushing of proteins along single-stranded DNA. Proc. Natl Acad. Sci. USA 113, 6194–6199 (2016).
Mersch, K. N., Sokoloski, J. E., Nguyen, B., Galletto, R. & Lohman, T. M. Helicase” activity promoted through dynamic interactions between a ssDNA translocase and a diffusing SSB protein. Proc. Natl Acad. Sci. USA 120, e2216777120 (2023).
Yoo, J., Winogradoff, D. & Aksimentiev, A. Molecular dynamics simulations of DNA–DNA and DNA–protein interactions. Curr. Opin. Struct. Biol. 64, 88–96 (2020).
Mishra, G., Bigman, L. S. & Levy, Y. ssDNA diffuses along replication protein A via a reptation mechanism. Nucleic Acids Res. 48, 1701–1714 (2020).
Tököli, A. et al. Structural adaptation of the single-stranded DNA-binding protein C-terminal to DNA metabolizing partners guides inhibitor design. Pharmaceutics 15, 1032 (2023).
Jin, S. et al. Computationally exploring the mechanism of bacteriophage T7 gp4 helicase translocating along ssDNA. Proc. Natl Acad. Sci. USA 119, e2202239119 (2022).
Li, C. et al. Structural basis of DNA polymerase θ mediated DNA end joining. Nucleic Acids Res. 51, 463–474 (2023).
Chen, X., Jin, S., Chen, M., Bueno, C. & Wolynes, P. G. The marionette mechanism of domain–domain communication in the antagonist, agonist, and coactivator responses of the estrogen receptor. Proc. Natl Acad. Sci. USA 120, e2216906120 (2023).
Fanning, E., Klimovich, V. & Nager, A. R. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 34, 4126–4137 (2006).
Pokhrel, N. et al. Dynamics and selective remodeling of the DNA binding domains of RPA. Nat. Struct. Mol. Biol. 26, 129–136 (2019).
Kim, Y. T. & Richardson, C. C. Acidic carboxyl-terminal domain of gene-2.5 protein of bacteriophage-T7 is essential for protein-protein interactions. J. Biol. Chem. 269, 5270–5278 (1994).
Shokri, L. et al. Kinetics and thermodynamics of salt-dependent T7 gene 2.5 protein binding to single- and double-stranded DNA. Nucleic Acids Res. 36, 5668–5677 (2008).
Wuite, G. J. L., Smith, S. B., Young, M., Keller, D. & Bustamante, C. Single-molecule studies of the effect of template tension on T7 DNA polymerase activity. Nature 404, 103 (2000).
Hoekstra, T. P. et al. Switching between exonucleolysis and replication by T7 DNA polymerase ensures high fidelity. Biophys. J. 112, 575–583 (2017).
Xu, L., Halma, M. T. J. & Wuite, G. J. L. Mapping fast DNA polymerase exchange during replication. Nat. Commun. 15, 5328 (2024).
Candelli, A., Wuite, G. J. L. & Peterman, E. J. G. Combining optical trapping, fluorescence microscopy and micro-fluidics for single molecule studies of DNA–protein interactions. Phys. Chem. Chem. Phys. 13, 7263–7272 (2011).
Zhou, R. et al. SSB functions as a sliding platform that migrates on DNA via reptation. Cell 146, 222–232 (2011).
Hollis, T., Stattel, J. M., Walther, D. S., Richardson, C. C. & Ellenberger, T. Structure of the gene 2.5 protein, a single-stranded DNA binding protein encoded by bacteriophage T7. Proc. Natl Acad. Sci. USA 98, 9557–9562 (2001).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Foster, B. M. et al. Combined solution and crystal methods reveal the electrostatic tethers that provide a flexible platform for replication activities in the bacteriophage T7 replisome. Biochemistry 58, 4466–4479 (2019).
Agam, G. et al. Reliability and accuracy of single-molecule FRET studies for characterization of structural dynamics and distances in proteins. Nat. Methods 20, 523–535 (2023).
Son, H., Mo, W., Park, J., Lee, J.-W. & Lee, S. Single-molecule FRET detection of sub-nanometer distance changes in the range below a 3-nanometer scale. Biosensors10, 168 (2020).
Doublié, S., Tabor, S., Long, A. M., Richardson, C. C. & Ellenberger, T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 Å resolution. Nature 391, 251 (1998).
Marintcheva, B., Qimron, U., Yu, Y., Tabor, S. & Richardson, C. C. Mutations in the gene 5 DNA polymerase of bacteriophage T7 suppress the dominant lethal phenotype of gene 2.5 ssDNA binding protein lacking the C-terminal phenylalanine. Mol. Microbiol. 73, 323–323 (2009).
Ghosh, S., Marintcheva, B., Takahashi, M. & Richardson, C. C. C-terminal phenylalanine of bacteriophage T7 single-stranded DNA-binding protein is essential for strand displacement synthesis by T7 DNA polymerase at a nick in DNA. J. Biol. Chem. 284, 30339–30349 (2009).
Toste Rêgo, A., Holding, A. N., Kent, H. & Lamers, M. H. Architecture of the Pol III–clamp–exonuclease complex reveals key roles of the exonuclease subunit in processive DNA synthesis and repair. EMBO J. 32, 1334–1343 (2013).
Bhattacharyya, B. et al. Structural mechanisms of PriA-mediated DNA replication restart. Proc. Natl Acad. Sci. USA 111, 1373–1378 (2014).
Mohan, M., Pandya, V. & Anindya, R. Escherichia coli AlkB and single-stranded DNA binding protein SSB interaction explored by molecular dynamics simulation. J. Mol. Graph. Model. 84, 29–35 (2018).
Potoyan, D. A., Zheng, W., Ferreiro, D. U., Wolynes, P. G. & Komives, E. A. PEST control of molecular stripping of NFκB from DNA transcription sites. J. Phys. Chem. B 120, 8532–8538 (2016).
Potoyan, D. A., Zheng, W., Komives, E. A. & Wolynes, P. G. Molecular stripping in the NF-κB/IκB/DNA genetic regulatory network. Proc. Natl Acad. Sci. USA 113, 110–115 (2016).
Williams, A. J. & Kaguni, L. S. Stimulation of Drosophila mitochondrial DNA polymerase by single-stranded DNA-binding protein. J. Biol. Chem. 270, 860–865 (1995).
Korhonen, J. A., Pham, X. H., Pellegrini, M. & Falkenberg, M. Reconstitution of a minimal mtDNA replisome in vitro. EMBO J. 23, 2423–2429 (2004).
Kim, Y. T., Tabor, S., Churchich, J. E. & Richardson, C. C. Interactions of gene 2.5 protein and DNA polymerase of bacteriophage T7. J. Biol. Chem. 267, 15032–15040 (1992).
Bannister, A. J. & Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 21, 381–395 (2011).
Candelli, A. et al. A toolbox for generating single-stranded DNA in optical tweezers experiments. Biopolymers 99, 611–620 (2013).
Shokri, L., Marintcheva, B., Richardson, C. C., Rouzina, I. & Williams, M. C. Single molecule force spectroscopy of salt-dependent bacteriophage T7 gene 2.5 protein binding to single-stranded DNA. J. Biol. Chem. 281, 38689–38696 (2006).
Smith, S. B., Cui, Y. & Bustamante, C. Overstretching B-DNA: the elastic response of individual double-stranded and single-stranded DNA molecules. Science 271, 795–799 (1996).
Gross, P. et al. Quantifying how DNA stretches, melts and changes twist under tension. Nat. Phys. 7, 731–736 (2011).
Jekel, C. F. & Venter, G. pwlf:a python library for fitting 1D continuous piecewise linear functions. https://github.com/cjekel/piecewise_linear_fit_py (2019).
Steger, C. An unbiased detector of curvilinear structures. IEEE Trans. Pattern Anal. Mach. Intell. 20, 113–125 (1998).
Sbalzarini, I. F. & Koumoutsakos, P. Feature point tracking and trajectory analysis for video imaging in cell biology. J. Struct. Biol. 151, 182–195 (2005).
Mangeol, P., Prevo, B. & Peterman, E. J. G. KymographClear and KymographDirect: two tools for the automated quantitative analysis of molecular and cellular dynamics using kymographs. Mol. Biol. Cell 27, 1948–1957 (2016).
Jin, S. et al. Protein structure prediction in CASP13 using AWSEM-suite. J. Chem. Theory Comput. 16, 3977–3988 (2020).
Freeman, G. S., Hinckley, D. M., Lequieu, J. P., Whitmer, J. K. & de Pablo, J. J. Coarse-grained modeling of DNA curvature. J. Chem. Phys. 141, 165103 (2014).
Webb, B. & Sali, A. Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinformatics54, 5.6.1–5.6.37 (2016).
Liao, Q. in Computational Approaches for Understanding Dynamical Systems: Protein Folding and Assembly (eds Strodel, B. & Barz, B) (Academic Press, 2020).
Gallicchio, E., Andrec, M., Felts, A. K. & Levy, R. M. Temperature weighted histogram analysis method, replica exchange, and transition paths. J. Phys. Chem. B 109, 6722–6731 (2005).
Zimmermann, L. et al. A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J. Mol. Biol. 430, 2237–2243 (2018).
Acknowledgements
We thank Seyda Aca and Sandrine D’Haene for assistance with protein purification and DNA construction, Noémie Danné for help with implementing the step-fitting algorithm. We thank C.C. Richardson for providing us with the gp2.5-Δ21C samples from his lab. We thank Chen-Yu Lo, Yang Gao, and Iddo Heller for insightful discussion. We also thank Julia Bakx for assistance with the experiments, and Erwin Peterman for critical reading and feedback on this manuscript. This work was financially supported by a PhD fellowship from China Scholarship Council (to L.X., funding No. 201704910912), the European Union H2020 Marie-Sklowdowska Curie International Training Network AntiHelix (to G.J.L.W., funding No. 859853), and the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program MONOCHROME (to G.J.L.W., funding No. 883240). This work is also supported by the Center for Theoretical Biological Physics, sponsored by NSF Grant PHY-2019745 (to S.J. and P.G.W). Additionally, we recognize the D.R. Bullard Welch Chair at Rice University, Grant C-0016 (to P.G.W.). Mia Urem is co-funded by the PPP allowance for the project POLSTOP2 made available by Health Holland, Top Sector Life Sciences & Health, to stimulate public-private partnerships.
Author information
Authors and Affiliations
Contributions
L.X., S.J., and G.J.L.W. conceptualized the research. L.X. prepared protein samples, collected and analyzed the single-molecule data; S.J. implemented the MD simulation and analyzed the data; L.X., M.U., and M.H.L. collected and analyzed the real-time DNA primer extension data. S.-J.L. provided purified gp2.5-Δ21C and tested their biochemical activity; L.X., S.J., and X.C. wrote the original manuscript; L.X., S.J., P.G.W., and G.J.L.W. edited the manuscript; G.J.L.W. and P.G.W. supervised the project; the manuscript was read and confirmed by all the listed authors.
Corresponding authors
Ethics declarations
Competing interests
The combined optical tweezers and fluorescence technologies used in this article are patented and licensed to LUMICKS B.V., in which G.J.L.W. declares a financial interest. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Shixin Liu and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Xu, L., Jin, S., Urem, M. et al. DNA polymerase actively and sequentially displaces single-stranded DNA-binding proteins. Nat Commun 16, 7431 (2025). https://doi.org/10.1038/s41467-025-62531-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-62531-1