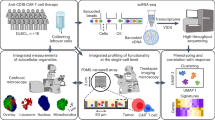
Abstract
Reports of secondary malignancies after chimeric antigen receptor (CAR)-T and possible CAR-T derived malignant transformation necessitate caution. Here we describe a patient with diffuse large B-cell lymphoma who developed new lymphadenopathy 2.5 years after CAR-T in the context of COVID-19 infection with histopathologic features consistent with T-cell lymphoma (TCL). Deep molecular interrogation with genomic sequencing and single-cell spatial transcriptomics reveals a highly proliferative clonal T-cell population co-expressing CD4 and CD8 with biallelic TCR rearrangement and no evidence of the CAR construct. The expanded clonotype displayed T follicular helper (TFH) cell transcriptomic programs and occupies immune-excluded spatial niches within the lymph node, supportive of TFH-like neoplastic T cell behavior. Remarkably, the lymphadenopathy spontaneously resolved on interval imaging. Our data underscore the need for better understanding of post-CAR-T clonal T-cell lymphoproliferative disorders to avoid unnecessary treatment and higher specificity in diagnostic methods for TCL.
Similar content being viewed by others
Introduction
The clinical successes of chimeric antigen receptor (CAR) T cells for treatment of lymphoma, leukemia and myeloma have transformed the therapeutic landscape for B-cell malignancies, rendering long-term remissions in patients with few alternatives1,2,3. Despite these clear successes, therapy-related toxicities pose short- and long-term risks for patients following CAR-T, including immune-mediated toxicities after infusion, infections due in part to ongoing B cell aplasia, and secondary malignancies4.
In November 2023, the FDA issued a warning regarding the risk of T-cell malignancies deriving from the CAR-T manufacturing process5. A low overall risk of secondary malignancy after CAR-T therapy (3–16%) has been reported6,7,8. The majority of these do not appear to result from malignant transformation from CAR-T insertional mutagenesis7,9,10. A recent analysis highlighted the overall rarity of these secondary events (6.5%), with very few cases of TCL. One such case was an Epstein-Barr Virus (EBV)-positive TCL arising 54 days after CAR-T cell infusion (axicabtagene ciloleucel [axi-cel]) in a patient with EBV+ diffuse large B-cell lymphoma (DLBCL)11. A separate T-cell lymphoma was described following anti-BCMA CAR-T therapy (ciltacabtagene autoleucel [Cilta-cel]) for multiple myeloma (MM), wherein the CAR transgene had integrated into the PBX2 gene12. Still another indolent CD4+ CAR-derived TCL in the small intestine was recently reported, wherein whole-genome sequencing (WGS) identified a lentiviral insertion site in the SSU72 gene13. A more recent report described an aggressive CAR+ peripheral T cell lymphoma arising after treatment with tisagenlecleucel for treatment of primary central nervous system lymphoma, which also harbored mutations in TET2 and DNMT3A, suggesting clonal hematopoiesis had contributed to its development14.
Other case reports have noted unexpected effects on cellular function as a result of CAR-T integration in unusual contexts. In one instance, CAR-T insertion into a single leukemic cell promoted early relapse and therapeutic resistance in a patient with B-acute lymphoblastic leukemia (ALL)15. In another, exuberant proliferation and activity of a CAR-T cell in which the CAR transgene had disrupted the TET2 gene conferred enhanced anti-tumor function16. While the benefits offered by CAR-T therapy in inducing enduring remissions in a substantial proportion of patients are largely felt to outweigh these potential long-term risks associated with treatment17, further work is needed to better characterize these cases of post-CAR-T suspected malignancies.
In this work, we analyze our clinical cohort of patients treated with CAR-T who later presented with possible TCL. We identify three potential cases of TCL, one of which we further characterize by WGS and spatial transcriptomic and T cell receptor (TCR) sequencing to identify putative drivers of proliferation.
Results
Clinical cases of possible post-CAR-T lymphoma
We systematically assessed our clinical database of patients with non-Hodgkin lymphoma (NHL), MM, and B-cell ALL receiving one of the six FDA-approved CAR-T products to quantify the incidence of secondary malignancy at our center from 2017 to 2023 with a focus on possible T-cell lymphomas. From 626 total patients, we identified 43 cases of secondary malignancy (6.9%, n = 12 myelodysplastic syndrome [MDS]/acute myeloid leukemia [AML], n = 28 solid tumors, n = 3 possible TCL); six cases were in MM patients and 37 in NHL (Fig. 1). Of the possible TCLs, one was a bona-fide case bearing the CAR transgene after Cilta-cel, as described above13. Of the remaining two cases, one occurred in a patient treated with Cilta-cel for MM who presented with an 8 mm facial lesion approximately 6 months after CAR-T. Biopsy revealed a population of lymphocytes positive for CD2, CD5, CD3, CD8, MUM-1, TCR-BF1, Granzyme B, and Perforin with Ki-67 of 100%, and negative for CD20, CD138, CD30, CD4, EBV, and TCR-Delta. This was diagnosed as Stage I peripheral TCL. Remarkably, the lesion spontaneously resolved after biopsy and the patient received no TCL therapy.
583 patients did not develop secondary malignancy after CAR-T (gray box). Left pie chart—breakdown of primary disease of these 583 patients. Right pie chart—breakdown of CAR-T product received by these 583 patients. Red box—43 patients developed secondary malignancy after CAR-T, with 3 TCL cases, 12 myelodysplastic syndrome (MDS)/acute myeloid leukemia (AML), 28 solid tumor. Colors indicate CAR-T product each patient received.
The second case was an 80-year-old man treated with third-line Axi-cel for transformed follicular lymphoma (FL). The full clinical course leading up to CAR-T is described in Fig. 2a (also see “Methods”). The patient achieved a complete metabolic response on the first restaging PET/CT 30 days after CAR-T, which continued for 2.5 years. Two years after his CAR-T cell infusion, the patient developed COVID-19 infection (despite 6 total vaccinations) with a protracted course, complicated by recurrent superimposed bacterial pneumonias. In the midst of these infections, he presented with weight loss and fatigue, prompting a PET/CT scan, which revealed a highly FDG-avid enlarged right cervical lymph node (LN), concerning for disease relapse or new malignancy (Fig. 2b).
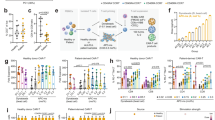
a The patient was originally diagnosed with stage III, grade 1–2 follicular lymphoma (FL) 9 years prior to CAR-T. Upon progression, he was treated with rituximab and achieved a partial response (PR) for 2.5 years before he relapsed with transformed disease. He was then treated with rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone (R-CHOP) for 6 cycles with a complete response (CR) lasting 3 years. He was treated on a clinical trial (NCT03636503) with rituximab/avelumab/utomilumab combination immunotherapy, with a PR followed by progression after 6 months. He then received CAR-T therapy as third-line therapy. Yellow shading indicates time prior to CAR-T, blue shading indicates time post-CAR-T. b PET/CT image of the new lymphadenopathy presenting 2.5 years after CAR-T. c Representative immunohistochemistry images of T- and B-cell markers of the from a single core lymph node biopsy. d PET/CT image of spontaneous regression of the lymphadenopathy 6 weeks after initial scan.
Hematoxylin and eosin (H&E) stain of a core needle biopsy demonstrated architecture effaced by an infiltrate of medium to large cells with irregular/folded nuclei, condensed chromatin, variably prominent nucleoli, and scant eosinophilic cytoplasm with small, scattered lymphocytes in the background (Fig. 2c). By immunohistochemical staining, the abnormal appearing cells expressed mature T-cell markers (CD3, CD2, CD5, CD7), with many positive for both CD4 and CD8, and were positive for TCR-BF1. Scattered cells were also positive for MUM-1 as well as BCL-6, Stathmin, PD-1, and BCL-2 suggestive of a TFH subtype. The abnormal lymphoid cells were negative for CD10, CD30, ALK-1, CD20, PAX5, CD79a, CD21, and C-MYC. Only rare cells stained positive for TCR-Delta. By Ki-67 staining, the cellular proliferation rate was 40–50%, and EBV in situ hybridization was negative. Altogether, the morphologic and immunophenotypic profile was thought to be most in keeping with a TFH TCL. Moreover, TCR gene rearrangement studies demonstrated a clonal process with a Vɣ1-8 primer, supporting a diagnosis of malignancy. Clinical flow cytometry demonstrated sample viability of <5%, limiting interpretation. Given the patient’s history, the highest concern was for TCL arising from a CAR-T cell. Although the biopsy was diagnostic for TCL, the clinical scenario of lymphadenopathy arising in the context of COVID-19 infection and bacterial pneumonia raised the possibility that this represented an infectious or inflammatory phenomenon, and the patient had no other signs or symptoms of progressive malignancy. Thus, the decision was made to observe with short-interval repeat imaging. Indeed, PET/CT 6 weeks later demonstrated spontaneous near-complete resolution of the cervical lymphadenopathy with complete resolution by 3 months (Fig. 2d).
Genomic characterization of possible TCL
To determine the cellular etiology and molecular characteristics of this aberrant T-cell population, remaining cervical LN formalin-fixed paraffin embedded (FFPE) sections and genomic DNA derived from peripheral blood mononuclear cells (PBMCs) obtained 12 months post-CAR-T (“normal”) were evaluated by WGS. To assess for the presence of the CAR construct, we aligned to a custom reference genome including the axi-cel sequence18,19 (“Methods”), and identified zero reads mapping to the axi-cel construct with high mapping confidence (MAPQ ≥ 30). We estimate the sequencing depth provides 95% power to detect reads mapping to the scFv region of the CAR construct (which is distinctly mappable from the human genome) even if the CAR were present in as few as ~3.4% of cells (“Methods”). Thus observing no mapping reads indicates that the clonal T-cell population was not derived from a CAR-T cell. Copy number analyses revealed no large-scale aberrations, but focal deletions at the TCR alpha, beta, and gamma loci were estimated to be present in 67% of cells by ABSOLUTE. MiXCR20 identified two rearranged clonotypes at each of the three loci (Supplementary Data 1).
The spontaneous resolution of lymphadenopathy raised the possibility that this clonal population did not represent TCL, despite the aberrant histologic appearance and phenotype (e.g., CD4+ CD8+ double positive), apparent clonality, and presence of TFH markers, the combination of which are considered diagnostic of TCL21. Of 4584 total mutations (19 nonsilent variants, Supplementary Data 2) that were LN-specific (i.e., not in normal PBMCs), one was identified in TET2, which was a frameshift mutation found in exon 3 resulting in a premature stop codon. While not seen in our normal sample, which was collected at 1-year post-CAR-T, this TET2 mutation was found in the clinical next-generation sequencing panel performed on PBMCs collected three months after the new lymphadenopathy presented, raising the possibility of clonal evolution or clonal hematopoiesis of indeterminate potential. Nevertheless, at such a high allele frequency, it is likely that the mutation is present in the atypical T cells in addition to the myeloid compartment and could be a driver of abnormal T cell proliferation. Of the other 18 non-silent mutations, none have been previously linked to the development of TCL or T cell lymphoproliferative neoplasms. A point mutation was observed in HIP3K, a kinase that can bind to Fas, leading to phosphorylation of Fas-associated death domain (FADD), which plays a key role in T cell apoptosis and proliferation22,23. While HIP3K mutations have not been specifically linked to TCLs, dysregulation of FADD phosphorylation has been implicated in the proliferation capacity of T cell lymphoblastic lymphoma24 and could be a potential mutation of interest for future study.
Spatial transcriptomic interrogation of nodal pathology
To more deeply interrogate the clonality, phenotype, and spatial microenvironment of this hyperproliferative T-cell population, we performed single-nuclei spatial transcriptomics on a fresh-frozen core needle biopsy of the suspicious LN with Slide-tags25. Previously, Slide-tags enabled spatial profiling of nuclei with 3’ capture chemistry of mRNA transcripts. However, 3’ capture chemistry poses challenges for TCR sequencing since the variable region is at the 5’ end of the TCR mRNA transcript. For improved recovery of TCR sequences, we adapted Slide-tags to be compatible with 5’ capture chemistry (“Methods”). Slide-tags 5’ snRNA-seq revealed a LN composition dominated by transcriptionally abnormal CD4+ CD8+ double positive T-cells, making up 51% of profiled nuclei (Fig. 3a). The rest of the LN was composed of 20% macrophages, 10% regulatory T-cells, 5% fibroblasts, 5% CD4+ T-cells, and smaller proportions of CD8+ T-cells, NK cells, endothelial cells, and dendritic cells. No B cells were detected. The CD4+ CD8+ T-cells were distributed across the full area of the profiled LN in a spatially disorganized manner, uncharacteristic of normal LN architecture (Fig. 3b). To examine these T-cell phenotypes, we quantified the usage of T-cell gene expression programs defined from a diverse collection of healthy, COVID-19, cancer, and autoimmunity patient samples (Fig. 3c and Supplementary Data 3)26. The double positive T-cells scored highly for TFH and T peripheral helper cell program usage. Differential gene expression analysis between the double positive T cell population and all other profiled T cells revealed an enrichment for genes involved in proliferation, hypoxia, and stress response, further supporting an abnormal cellular phenotype (Fig. 3d). Concordant with the WGS analysis, we mapped the snRNA-seq results to a custom reference containing the axi-cel construct and did not detect any spatially mapped nuclei containing at least one count of an axi-cel mapped read (“Methods”). Additionally, qPCR to detect the CAR construct sequence in the Slide-tags library from the biopsy was negative for transgene amplification (Supplementary Data 4).
a UMAP embedding of Slide-tags 5’ snRNA-seq transcriptome profiles, colored by cell type assignment. b Spatial mapping of snRNA-seq profiles, split by cell type. c T-cell gene expression program average usage scores. d Volcano plot of differentially expressed genes in the CD4+ CD8+ T cell population compared with all other T cells using a two-sided Wilcoxon rank-sum test with adjustment for multiple testing using the Bonferroni correction. Select genes are highlighted. e Size of beta chain clonotypes by T-cell population. Each color represents a different clonotype. The two beta chain sequences belonging to the most expanded clonotype are written in white text. f Module scoring of viral reactivity transcriptional signature by T-cell population. g Spatial neighborhood enrichment heatmap where Z scores denote the magnitude of enrichment or depletion between a given center cell type and neighbor cell type. Z scores are capped at 9 and −9 for visualization purposes.
The majority of CD4 or CD8 single positive T-cells belonged to single member or small-sized clonotypes (Fig. 3e). In marked contrast, the CD4+ CD8+ T-cells belonged to a highly expanded clonotype, consisting of two alpha chains (CASPGGLTGGGNKLTF, CALSHPFRNSGNTPLVF) and two beta chains (CASSLVVWGRGLNEQFF, CASSQQDSRNTIYF), suggestive of an atypical biallelic rearrangement of both alpha and beta chains (Supplementary Data 5)27,28,29. Examining public CDR3α and CDR3β TCR clonotypes, we identified 6 alpha chain TCRs and 2 beta chain TCRs with reported specificity to viral antigens (Supplementary Data 6 and “Methods”), but the highly expanded clonotype did not match any known viral-reactive TCRs. Double positive T-cells scored lowly on a transcriptional signature for viral antigen reactivity, despite the lymphadenopathy arising during COVID-19 infection (Fig. 3f). The highly clonal nature of the aberrant CD4+CD8+ double positive population supports the notion of TCL, though may also be consistent with a non-malignant lymphoproliferative process.
Finally, we leveraged the spatial data to assess the immune microenvironment surrounding the expanded double positive T-cell population. While double positive T cells spatially clustered with each other, their neighborhoods were depleted of DCs, CD8+ T cells, CD4+ Treg cells, endothelial cells, and fibroblasts (Fig. 3g). We observed a weak enrichment for macrophages and pDCs in the neighborhoods of double positive T cells. Overall, the aberrant T cell population exists in immune-excluded spatial niches.
Discussion
In the current case, pathology review seemingly confirmed a diagnosis of TFH TCL based on conventional immunohistochemical definitions and T-cell clonality. Thus, immediate treatment with TCL-directed therapy would have been reasonable, given the presumptive diagnosis of post-CAR-T TCL. Yet the spontaneous resolution of the lesion, arising in the context of COVID-19 infection and superimposed bacterial pneumonia, argues against this and raises the alternative possibility of a reactive nodal process, where initiation of treatment would have been unnecessary. Alternatively, this case may have been a true TCL with an unusual indolent course that spontaneously remitted in a post-infectious setting. Our understanding of post-CAR-T lymphoproliferative processes is evolving as more cases arise, and conventional pathologic diagnosis of TCL may overestimate the true burden of aggressive post-CAR-T lymphomas. This may have the unintended downstream effect of preventing patients from having access to this potentially curative therapy due to referral biases stemming from concerns about rates of secondary malignancy. Further accounting of similar cases with an in-depth study is needed to better define the nature of these lymphoproliferative neoplasms and assess the risks in the context of the established benefits of CAR-T therapy.
This clinical and pathologic discordance further underscores that current diagnostics have inherent limitations, highlighting the need for more precise definitions of malignancy and raising the notion that the clinical course should be carefully considered in diagnosis. One intriguing possibility is that this case could represent a more indolent T cell neoplasm. Though TET2 mutations have been implicated in T-cell clonal proliferation16, our WGS and snRNA-seq data suggest these mutations were not unique to the clonal T-cell population. However, given the high allele frequency, it is likely that the TET2 mutation was present in the aberrant T cell population and contributed to the clonal proliferation and subsequent clinical presentation (Supplementary Fig. 1). Single-cell spatial profiling confirmed one clonotype dominated the biopsied node, displayed a highly proliferative TFH-like transcriptomic phenotype, and occupied immune-excluded spatial niches at the time of biopsy, supportive of neoplastic T cell behavior. Nevertheless, the lesion resolved post-biopsy. Perhaps robust immunosurveillance in the setting of resolution of an infectious process could result in immune-mediated regression of the lesion. Our findings underscore the need for caution when T-cell lymphoproliferative disorders are identified post-CAR-T and diagnosed as TCL. Current diagnostic criteria may be insufficient when evaluating a patient with a complex immune milieu after CAR-T cell therapy. New diagnostic categories may be needed as we continue to learn more about these post-CAR-T phenomena, particularly in the context of COVID-19 and other viral infections. Careful clinicopathologic correlation is essential when evaluating patients with unusual presentations after CAR-T cell therapy, where clonal lymphoproliferative disorders may instead follow an indolent course without requiring treatment.
Methods
Patient characteristics and clinical outcomes
The research was conducted in accordance with the Declaration of Helsinki. Patients gave written informed consent to research and sample banking protocols approved by the Dana-Farber/Harvard Cancer Center Institutional Review Board. For the index patient described above, this patient was originally diagnosed with stage III, grade 1–2 FL 9 years prior to CAR-T therapy. Upon disease transformation to DLBCL, immunohistochemical studies demonstrated that atypical cells were B cells positive for BSAP, CD10, Bcl-2, and Bcl-6. The atypical cells demonstrated scattered c-Myc positivity (40–50% with variable intensity), scattered/focal MUM-1 positivity, and scattered/variable intensity reactivity for CD21. The atypical lymphoid cells were negative for EBV by in situ hybridization (EBER) and for NKX3.1 by immunohistochemistry. Next-generation sequencing was not available on the original lymphoma. He was treated with rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone (R-CHOP) for six cycles with a complete response (Fig. 2a). His FL relapsed within three years and he was treated on a clinical trial (NCT03636503) with rituximab/avelumab/utomilumab combination immunotherapy, with a PR followed by progression at 6 months. He then received CAR-T therapy as third-line therapy. Five days prior to CAR-T, the patient was treated with lymphodepleting chemotherapy consisting of cyclophosphamide 500 milligrams per meter squared (mg/m2) and fludarabine 30 mg/m2 for 3 days. On day 0, he was infused with 2 × 106 CAR-T cells per kilogram of body weight. His immediate post-CAR-T course was complicated by cytokine release syndrome (CRS, beginning on day +2 after CAR-T infusion, maximum grade 1 by ASTCT and Lee scoring systems30,31, treated with steroids and tocilizumab) and neurotoxicity (i.e., immune effector cell-associated neurotoxicity syndrome [ICANS], beginning day +4, maximum grade 3 by CTCAE and grade 3 by ASTCT32, treated with steroids and anakinra). Prior to his diagnosis of COVID-19 and recurrent pneumonias, the patient did not have significant leukopenia/lymphopenia.
Histopathology evaluation
Histopathologic analysis of the cervical LN biopsy was performed by hematopathologists at the Department of Pathology at the Brigham and Women’s Hospital. H&E-stained and immunohistochemical stain slides of the cervical LN biopsy were prepared at the time of diagnostic workup at the Immunohistochemistry Laboratories in the Department of Pathology at the Brigham and Women’s Hospital. A second core biopsy was obtained at the same time and flash frozen in Tissue-Tek O.C.T. Compound (Sakura) and cryopreserved until the time of spatial transcriptional analysis (see below). For TCL, the following antigens were evaluated: CD2, CD3, CD5, CD5, CD7, CD8, BLC6, Stathmin, PD-1, MUM1, BCL2, CD30, ALK-1, CD10, CD20, PAX5, CD79a, CD21, C-MYC, TCR-BF1, TCR-Delta, and Ki-67. In situ hybridization for EBV-encoded RNA was performed and found to be negative. Representative histopathological microphotographs were collected for publication.
Bulk WGS
“Tumor” DNA was extracted from 12 sections of the remaining FFPE core LN biopsy. Genomic DNA was extracted from PBMC that were collected 12 months after CAR-T infusion. DNA was sequenced at the Broad Institute (Cambridge, MA), as previously described, at a depth of 60× for the “tumor” sample and 30× for the “normal” sample33. Bam files were input into our standard characterization pipeline, which includes MuTect for SNV calling34, Strelka for indel calling35, and HapASeg for copy number segmentation. DeTIN was used to rescue mutations observed at low frequency in the normal36. Additional filtering for artifactual mutations was performed using BLAT, a panel of normals, and filtering for oxidative damage37.
Power to detect at least one read mapping to the axi-cel scFv was calculated under Poisson assumptions as (1):
Where \(\lambda\)autosomal = 0.3 is the number of read pairs per base observed in autosomal regions, L = 743 is the length of the scFv, rCAR = ½ is the copy ratio of the construct compared to autosomal regions assuming a single integration event, and \(\rho\) is the fraction of cells with an integrated CAR construct.
Rapid heme panel
Assessment of hematologic malignancy-associated DNA mutations was performed as part of routine clinical care at the time of the suspicious LN using the rapid heme panel developed at the Dana-Farber Cancer Institute and Brigham and Women’s Hospital, an amplicon-based targeted next-generation sequencing platform designed to cover 95 genes in hotspot regions of oncogenes and coding regions of tumor suppressor genes38. The rapid heme panel was performed on DNA extracted from PBMCs.
Slide-tags 5’ snRNA-seq
Slide-tags were performed as previously described25, with the below modifications for compatibility with 5’ snRNA-seq. Capture sequence on spatial bead barcode oligonucleotides was replaced with the reverse complement of the 10× Genomics template switch oligonucleotide sequence (5’-CCCATATAAGAAA-3’). A 20 µm fresh-frozen tissue section from the LN biopsy was melted onto a 5.5 mm square bead array. The spatial tagging procedure and nuclei preparation were performed using our standard protocol. For the 5’ snRNA-seq profile, 38.7 µl of counted nuclei was loaded into the 10× Genomics Chromium controller using the Chromium Next GEM Single Cell 5’ Kit v2 (10× Genomics, PN-1000263). The Chromium Next GEM Single Cell 5’ v2 Cell Surface Protein User Guide CG000330 Rev F was followed with small modifications. During step 2.2a, 1 μl of 0.3 μM spike-in primer (5′-GTGACTGGAGTTCAGACGT-3′) was added and cDNA primers (PN-2000089) were used in place of Feature cDNA Primers 4 (PN-2000277). Spatial barcode indexing was performed similarly to step 6.1, but Dual Index Plate TT Set A was used in place of Dual Index Plate TN Set A. We performed 13 cycles of PCR according to step 6.1 conditions. A double sided SPRI (0.6× then 1.2×) was conducted for post sample index PCR size selection. We collected Slide-tags 5’ snRNA-seq on two serial sections. The transcriptomic profiles consist of nuclei from both sections. Spatial positions are shown from the section with the most nuclei.
Slide-tags data preprocessing
We used Cell Ranger (v.6.1.2)39 mkfastq (10× Genomics) to generate demultiplexed FASTQ files from the raw sequencing reads. We aligned these reads to human GRCh38 and the Ensembl 100 annotation, supplemented with sequences for both the axi-cel and tisa-cel constructs18 and quantified gene counts as UMIs using Cell Ranger count (10× Genomics). We used CellBender v.0.2.0 for background noise correction and cell calling40, setting --expected-cells to the number of Cell Ranger cell calls and --total-droplets-included to 40,000. Spatial data processing was performed as described in our original Slide-tags manuscript25.
T-cell gene expression program analysis
T-cell function is largely governed by the induction of gene expression programs. To explore the gene expression programs activated by the T-cells in our sample, we used T-CellAnnoTator (TCAT)26. TCAT uses nonnegative least squares regression to quantify the usage of a predefined set of context-diverse T-cell programs on our single-nucleus transcriptome data. We computed the mean usage scores of relevant programs for T-cell populations in the biopsied LN. We report the top 100 genes correlated with each program in Supplementary Data 3.
T-cell receptor sequencing
Alpha and beta chain TCR sequences were enriched from the 5’ snRNA-seq cDNA libraries using the Chromium Single Cell Human TCR Amplification Kit (PN-1000252). TCR sequences were called using the cellranger vdj pipeline.
Public TCR analysis
Human CDR3α and CDR3β sequences from public TCRs reactive to common viruses, including Influenza A, CMV, EBV, and SARS-CoV-2, were obtained from the VDJdb database (vdjdb.cdr3.net, accessed on 2024-09-11). Direct amino acid sequence matching of alpha and beta chains from the 5’ snRNA-seq data to the public TCRs was assessed.
Viral reactivity signature scoring
T-cell transcriptomic profiles were scored on a set of genes associated with virus-specific T-cells using the AddModuleScore function in Seurat41.
Neighborhood enrichment permutation testing
To assess spatial neighborhood enrichment of pairwise cell type combinations, we used a permutation testing framework. For each center cell type, we calculated the average proportion of cells within a 200 µm radius belonging to the queried neighbor cell type. To establish a null distribution, we permuted cell type labels 1000 times and recomputed the average proportions for the queried neighbor cell type. A Z score for the cell type combination was then computed by comparing the observed average proportion to the null distribution. This procedure was repeated for each pairwise cell type combination.
TET2 mutation identification from snRNA-seq reads
TET2 p.776Wfs*4 mutant reads were counted from the snRNA-seq BAM file and the consensus genotype of each UMI was determined. Cell barcodes with at least one variant-containing UMI were considered mutant. Cell barcodes with no variant-containing UMIs but at least one UMI spanning the queried sequence position were considered wild-type. All other cell barcodes were not assigned a genotype.
qPCR for the CAR transcript
The 5’ snRNA-seq cDNA library was obtained for qPCR to detect the CAR transcript. cDNA was synthesized from RNA isolated from an infusion product bag washing from a patient who received CAR-T therapy42 as a positive control and from healthy donor T cells as a negative control. Primer sets were designed to target FMC6343 to detect the CAR transcript and the housekeeping gene GAPDH (ThermoFisher). qPCR was performed as previously described44.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Single cell transcriptome, TCR, and WGS data will be submitted to NCBI’s Database of Genotypes and Phenotype (dbGaP; https://www.ncbi.nlm.nih.gov/gap) under accession code phs002922.v3.p1. Source data are provided with this paper.
Code availability
Code for processing spatial sequencing libraries is available at GitHub (https://github.com/broadchenf/Slide-tags).
References
Neelapu, S. S. et al. Five-year follow-up of ZUMA-1 supports the curative potential of axicabtagene ciloleucel in refractory large B-cell lymphoma. Blood 141, 2307–2315 (2023).
San-Miguel, J. et al. Cilta-cel or standard care in lenalidomide-refractory multiple myeloma. N. Engl. J. Med. 389, 335–347 (2023).
Maude, S. L. et al. Tisagenlecleucel in children and young adults with B-cell lymphoblastic leukemia. N. Engl. J. Med. 378, 439–448 (2018).
Cappell, K. M. et al. Long-term follow-up of anti-CD19 chimeric antigen receptor T-cell therapy. J. Clin. Oncol. 38, 3805–3815 (2020).
Center for Biologics Evaluation & Research. FDA investigating serious risk of T-cell malignancy following BCMA-directed or CD19-directed autologous chimeric antigen receptor (CAR) T cell immunotherapies. U.S. Food and Drug Administration https://www.fda.gov/vaccines-blood-biologics/safety-availability-biologics/fda-investigating-serious-risk-t-cell-malignancy-following-bcma-directed-or-cd19-directed-autologous (2023).
Ghilardi, G. et al. T cell lymphoma and secondary primary malignancy risk after commercial CAR T cell therapy. Nat. Med. 30, 984–989 (2024).
Steffin, D. H. M. et al. Long-term follow-up for the development of subsequent malignancies in patients treated with genetically modified IECs. Blood 140, 16–24 (2022).
Chong, E. A., Ruella, M., Schuster, S. J. & Lymphoma Program Investigators at the University of Pennsylvania Five-year outcomes for refractory B-cell lymphomas with CAR T-cell therapy. N. Engl. J. Med. 384, 673–674 (2021).
Micklethwaite, K. P. et al. Investigation of product-derived lymphoma following infusion of piggyBac-modified CD19 chimeric antigen receptor T cells. Blood 138, 1391–1405 (2021).
Bishop, D. C. et al. Development of CAR T-cell lymphoma in 2 of 10 patients effectively treated with piggyBac-modified CD19 CAR T cells. Blood 138, 1504–1509 (2021).
Hamilton, M. P. et al. Risk of second tumors and T-cell lymphoma after CAR T-cell therapy. N. Engl. J. Med. 390, 2047–2060 (2024).
Harrison, S. J. et al. CAR+ T-cell lymphoma post ciltacabtagene autoleucel therapy for relapsed refractory multiple myeloma. Blood 142, 6939–6939 (2023).
Ozdemirli, M. et al. Indolent CD4+ CAR T-cell lymphoma after cilta-cel CAR T-cell therapy. N. Engl. J. Med. 390, 2074–2082 (2024).
Kobbe, G. et al. Aggressive lymphoma after CD19 CAR T-cell therapy. N. Engl. J. Med. 391, 1217–1226 (2024).
Ruella, M. et al. Induction of resistance to chimeric antigen receptor T cell therapy by transduction of a single leukemic B cell. Nat. Med. 24, 1499–1503 (2018).
Fraietta, J. A. et al. Disruption of TET2 promotes the therapeutic efficacy of CD19-targeted T cells. Nature 558, 307–312 (2018).
Levine, B. L. et al. Unanswered questions following reports of secondary malignancies after CAR-T cell therapy. Nat. Med. 30, 338–341 (2024).
Haradhvala, N. J. et al. Distinct cellular dynamics associated with response to CAR-T therapy for refractory B cell lymphoma. Nat. Med. 28, 1848–1859 (2022).
Kochenderfer, J. N. et al. Construction and preclinical evaluation of an anti-CD19 chimeric antigen receptor. J. Immunother. 32, 689–702 (2009).
Bolotin, D. A. et al. MiXCR: software for comprehensive adaptive immunity profiling. Nat. Methods 12, 380–381 (2015).
Du, J. et al. Precise diagnosis and targeted therapy of nodal T-follicular helper cell lymphoma (T-FHCL). Front. Oncol. 13, 1163190 (2023).
Rochat-Steiner, V. et al. FIST/HIPK3: a Fas/FADD-interacting serine/threonine kinase that induces FADD phosphorylation and inhibits fas-mediated Jun NH(2)-terminal kinase activation. J. Exp. Med 192, 1165–1174 (2000).
Osborn, S. L. et al. Fas-associated death domain (FADD) is a negative regulator of T-cell receptor-mediated necroptosis. Proc. Natl Acad. Sci. USA 107, 13034–13039 (2010).
Marín-Rubio, J. L. et al. Deregulated FADD expression and phosphorylation in T-cell lymphoblastic lymphoma. Oncotarget 7, 61485–61499 (2016).
Russell, A. J. C. et al. Slide-tags enables single-nucleus barcoding for multimodal spatial genomics. Nature 625, 101–109 (2024).
Kotliar, D. et al. Reproducible single cell annotation of programs underlying T-cell subsets, activation states, and functions. Preprint at bioRxiv https://doi.org/10.1101/2024.05.03.592310 (2024).
Schuldt, N. J. & Binstadt, B. A. Dual TCR T Cells: Identity Crisis or Multitaskers?. J. Immunol. 202, 637–644 (2019).
Jang, H. J. et al. Dual receptor T cells mediate effective antitumor immune responses via increased recognition of tumor antigens. J. Immunother. Cancer 11, e006472 (2023).
Muhowski, E. M. & Rogers, L. M. Dual TCR-expressing T cells in cancer: how single-cell technologies enable new investigation. Immunohorizons 7, 299–306 (2023).
Lee, D. W. et al. ASTCT consensus grading for cytokine release syndrome and neurologic toxicity associated with immune effector cells. Biol. Blood Marrow Transplant. 25, 625–638 (2019).
Neelapu, S. S. et al. Chimeric antigen receptor T-cell therapy - assessment and management of toxicities. Nat. Rev. Clin. Oncol. 15, 47–62 (2018).
Lead Organizations: NCI Network Trial Development and Conduct. https://ctep.cancer.gov/protocoldevelopment/electronic_applications/docs/CTCAE_v5_Quick_Reference_8.5x11.pdf.
Parry, E. M. et al. Evolutionary history of transformation from chronic lymphocytic leukemia to Richter syndrome. Nat. Med. 29, 158–169 (2023).
Cibulskis, K. et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 31, 213–219 (2013).
Kim, S. et al. Strelka2: fast and accurate calling of germline and somatic variants. Nat. Methods 15, 591–594 (2018).
Taylor-Weiner, A. et al. DeTiN: overcoming tumor-in-normal contamination. Nat. Methods 15, 531–534 (2018).
Costello, M. et al. Discovery and characterization of artifactual mutations in deep coverage targeted capture sequencing data due to oxidative DNA damage during sample preparation. Nucleic Acids Res. 41, e67 (2013).
Kluk, M. J. et al. Validation and implementation of a custom next-generation sequencing clinical assay for hematologic malignancies. J. Mol. Diagn. 18, 507–515 (2016).
Zheng, G. X. Y. et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049 (2017).
Fleming, S. J. et al. Unsupervised removal of systematic background noise from droplet-based single-cell experiments using CellBender. Nat. Methods 20, 1323–1335 (2023).
Oliveira, G. et al. Phenotype, specificity and avidity of antitumour CD8+ T cells in melanoma. Nature 596, 119–125 (2021).
Maurer, K. et al. Baseline immune state and T-cell clonal kinetics are associated with durable response to CAR-T therapy in large B-cell lymphoma. Blood 144, 2490–2502 (2024).
Schubert, M.-L. et al. Assessment of CAR T cell frequencies in Axicabtagene Ciloleucel and Tisagenlecleucel patients using duplex quantitative PCR. Cancers 12, 2820 (2020).
Penter, L. et al. Integrative genotyping of cancer and immune phenotypes by long-read sequencing. Nat. Commun. 15, 32 (2024).
Acknowledgements
We thank Doreen Hearsey, Allison Anderson and all staff from the Ted and Eileen Pasquarello Tissue Bank in Hematologic Malignancies for excellent technical support with banking of clinical samples. C.J.W. is a member of the Parker Institute for Cancer Immunotherapy at DFCI. Her work is supported, in part, by the Parker Institute for Cancer Immunotherapy. C.J.W. is also the Lavine Family Chair for Preventive Cancer Therapies at DFCI. C.J.W. and F.C. are supported by The Mark Foundation. K.M. is supported by the Lubin Family Foundation Scholar Award. F.C. acknowledges funding from the NCI (R01 CA276865, R33CA291199) and the NYSCF. F.C. is an NYSCF Roberston Investigator. We thank the patients for generous contribution of clinical data and research samples for this study. We are thankful to T. Coorens, V. Shanmugam, C.K. Hahn, D. Fisher, P. Armand, and R.J. Soiffer for helpful discussions and feedback.
Author information
Authors and Affiliations
Contributions
K.M., J.A.W., C.J.W., F.C., and C.J. conceived and designed the study. K.M., H.B., M.M., L.L., and J.D.C. identified and obtained patient samples and relevant clinical information. K.M., H.B., G.P., B.W., P.D.M., J.P.L., and C.J. interpreted patient data. K.M., J.A.W., E.L., and A.N. performed experiments. K.M., J.A.W., N.J.H., L.G., Q.G., M.B., and G.G. designed, performed, and interpreted data analysis. J.A.W., J.S., S.A., and V.K. developed Slide-tags 5’ snRNA-seq. All authors participated in manuscript writing and review and provided final approval of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
C.J. reports consultancy for Kite/Gilead, BMS/Celgene, Novartis, Instil Bio, ImmPACT Bio, Caribou Bio, Miltenyi, Ipsen, ADC Therapeutics, Abbvie, AstraZeneca, Morphosys, Synthekine, and Sana, and research funding from Kite/Gilead and Pfizer. C.J.W. is an equity holder of BioNTech, Inc., receives research funding from Pharmacyclics, and is on the SAB of Repertoire, Adventris, Nature’s Toolbox, Aethon Therapeutics. F.C. is an academic founder of Curio Bioscience and Doppler Biosciences, and a scientific advisor for Amber Bio. F.C.'s interests were reviewed and managed by the Broad Institute in accordance with their conflict-of-interest policies. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Mark Hamilton and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Maurer, K., Weir, J.A., Nagler, A. et al. A clonally expanded nodal T-cell population diagnosed as T-cell lymphoma after CAR-T therapy. Nat Commun 16, 7462 (2025). https://doi.org/10.1038/s41467-025-62709-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-62709-7