Abstract
Receptor-interacting protein kinase 3 (RIPK3), a key regulator of necroptosis, is modulated by ubiquitination through various E3 ligases and deubiquitinases. However, the effects of different polyubiquitination processes on RIPK3 and necroptosis remain unclear. Using a proteomic approach, we identify SMAD Ubiquitination Regulatory Factor 1 (SMURF1) and Ubiquitin-specific peptidase 5 (USP5) as crucial regulators of RIPK3 within the necrosome during necroptosis. SMURF1 facilitates K63 polyubiquitination of RIPK3 at lysine 55 and 363, inhibiting necrosome formation and necroptosis. SMURF1 depletion accelerates necroptosis, while the reintroduction of functional SMURF1 reverses this. Conversely, USP5 acts as a deubiquitinase, removing K63 ubiquitin chains and promoting necroptosis. Reducing SMURF1, using a RIPK3 mutant defective in SMURF1-mediated ubiquitination, or overexpressing USP5 enhances necroptosis in leukaemia cells, leading to reduced tumour growth in xenograft models treated with birinapant and emricasan. These findings highlight the opposing regulation of K63-linked polyubiquitination of RIPK3 by SMURF1 and USP5 in necroptosis.
Similar content being viewed by others
Introduction
Necroptosis is a form of programmed cell death distinct from apoptosis1,2,3. It is characterized by features typical of necrosis, such as cell swelling, membrane rupture, and the release of intracellular contents, leading to inflammation1,4,5. Unlike necrosis, which is generally considered an uncontrolled and accidental process, necroptosis is a regulated mechanism that serves as a backup when apoptosis is inhibited, especially during certain infections and pathological conditions6. A key player in the necroptosis pathway is receptor-interacting serine/threonine-protein kinase 3 (RIPK3)3,5. The process is initiated when RIPK3 is activated through upstream signals, particularly those involving death receptors such as tumor necrosis factor receptor 1 (TNFR1)7. RIPK3 interacts with RIPK1 and mixed lineage kinase domain-like protein (MLKL) to form a complex known as the “necrosome”7,8,9,10. This assembly is critical for continuing the necroptotic signaling pathway.
Within the necrosome, RIPK3 phosphorylates MLKL, a pivotal step in the execution of necroptosis9,11. Phosphorylated MLKL oligomerises and translocates to the plasma membrane, where it disrupts membrane integrity, causing ion imbalances and cell swelling. Eventually, the cell membrane ruptures, leading to the release of cellular contents, including damage-associated molecular patterns (DAMPs), which trigger an inflammatory response1,12,13. Necroptosis, facilitated by RIPK3, thus acts as a warning signal to recruit and alert immune cells9,14. While this mechanism can be beneficial in clearing infections and preventing pathogen spread, excessive necroptosis may result in chronic inflammation and tissue damage, contributing to various diseases12,15,16. RIPK3, therefore, plays a dual role as both protector and potential culprit in pathological conditions, depending on the context12,17. Its activity is tightly regulated by multiple checkpoints, including post-translational modifications such as ubiquitination, which control its stability and protein-protein interactions18,19,20. Understanding RIPK3’s role in necroptosis is crucial for developing therapeutic strategies aimed at modulating necroptosis in diseases characterized by aberrant cell death or survival.
Ubiquitination is a post-translational modification process wherein ubiquitin, a small regulatory protein, is covalently attached to the target proteins20,21. This process can tag proteins for degradation via the proteasome, a protein complex that breaks down proteins into peptides. The ubiquitin-proteasome pathway plays a critical role in maintaining cellular homeostasis by removing damaged, misfolded, or unnecessary proteins22,23,24. In addition to protein degradation, ubiquitination can regulate protein activity, localization, and interactions, thereby influencing cellular processes such as DNA repair, cell cycle progression, and immune responses25,26. Ubiquitination occurs in various forms, including monoubiquitination, where a single ubiquitin molecule is attached, and polyubiquitination, where a chain of ubiquitin molecules is linked. The type and linkage of ubiquitin chains, such as K48-linked or K63-linked, determine the fate and function of the substrate protein. K48-linked chains typically signal proteasomal degradation, while K63-linked chains are often involved in non-degradative functions such as signaling and trafficking22,23,24. Ubiquitination is reversible, and deubiquitinating enzymes (DUBs) can remove ubiquitin from substrates, allowing dynamic regulation of protein functions27,28,29,30,31,32. Dysregulation of ubiquitination is implicated in many diseases, including cancer, neurodegenerative disorders, and immune dysfunctions, making components of the ubiquitin system potential therapeutic targets33,34,35.
SMURF1 (SMAD Ubiquitination Regulatory Factor 1) is an E3 ubiquitin ligase involved in the ubiquitination and proteasomal degradation of target proteins36,37. SMURF1, a member of the HECT (Homologous to the E6-AP Carboxyl Terminus) family of E3 ubiquitin ligases, regulates several cellular pathways38. It plays a crucial role in the TGF-β (Transforming Growth Factor-beta) signaling pathway by targeting SMAD proteins, modulating cellular processes such as proliferation and differentiation37,39. SMURF1 also regulates the bone morphogenetic protein (BMP) pathway, thereby influencing bone development and homeostasis40,41. Beyond its role in growth factor signaling, SMURF1 affects cell morphogenesis and migration through the ubiquitination of RhoA, a small GTPase involved in cytoskeletal dynamics36,42. SMURF1 is also implicated in cancer progression, wherein its regulation of TGF-β and other pathways can influence tumor growth and metastasis43. Although extensive research has been conducted on the various roles of SMURF1 in cell death, its specific function in necroptosis remains unclear.
Deubiquitinases (DUBs) are enzymes capable of removing polyubiquitin chains from substrates34. Emerging evidence shows that DUBs play crucial roles in multiple signaling pathways and that their dysregulation is linked to numerous diseases25,44. The human genome contains five subclasses of DUBs, with the ubiquitin-specific protease (USP) subfamily being the largest23,27. Within this subfamily, USP5 (also known as ubiquitin isopeptidase T or ISOT) is implicated in several physiological and pathological processes45,46. USP5 is a deubiquitinating enzyme (DUB) that plays a vital role in the ubiquitin-proteasome system by regulating the ubiquitin cycle. It is primarily localized in the cytosol and nucleoplasm and is widely expressed across tissues, including the testis, cerebral cortex, cerebellum, skin, lung, and gallbladder, indicating its involvement in various physiological processes such as development, DNA repair, stress responses, and immune functions47,48,49,50. However, similar to that of SMURF1, the specific biological function of USP5 in necroptosis remains unclear.
Recent studies have shown that E3 ligases can regulate necroptosis by influencing RIPK3, not in terms of protein stability but protein interactions51,52,53. For example, Parkin, phosphorylated by AMPK, mediates K33-linked polyubiquitination of RIPK3, regulating its interaction with RIPK1 and thereby suppressing necroptosis51. USP22, on the other hand, is a deubiquitinase known to regulate RIPK3 activity, although its specific role in promoting or inhibiting necroptosis remains unclear53. Notably, no study has explored an E3 ligase and a deubiquitinase that modulate RIPK3 ubiquitination at the same sites, which could regulate necroptosis. Although previous research has examined E3 ligases and deubiquitinating enzymes associated with the necrosome, the precise mechanisms by which they regulate the same ubiquitination site on RIPK3, involving the same type of ubiquitin linkage, remain unclear.
In this study, we identify that SMURF1 and USP5 are associated with the necrosome complex, which assembles during necroptosis. SMURF1 deficiency accelerates necroptosis, which can be reversed by reconstituting SMURF1, but not by its E3 ligase-defective mutant (SMURF1 C699A), suggesting that its E3 ligase activity is essential for this function54. Conversely, USP5 deficiency suppresses necroptosis, which can be reversed by reconstituting USP5, but not by its deubiquitination-defective mutant (USP5 C335A), indicating that its deubiquitinase activity is crucial55. Mechanistically, SMURF1 promotes K63-linked polyubiquitination of RIPK3 at lysine 55 and 363, preventing necrosome formation during necroptosis. USP5, in turn, deubiquitinates these SMURF1-mediated K63-linked polyubiquitin chains on RIPK3. Furthermore, the downregulation of SMURF1 or the introduction of a RIPK3 K55/363R mutant, which is defective in SMURF1-mediated polyubiquitination, or the overexpression of USP5, enhances necroptosis in leukemia cells (Molm13 or NB4). This leads to suppressed tumor growth in a xenograft model upon treatment with birinapant and emricasan56,57,58. These findings demonstrate that SMURF1 and USP5 regulate necroptosis through antagonistic modulation of RIPK3 K63-linked polyubiquitination and necrosome complex formation.
Results
SMURF1 and USP5 associate with the necrosome via RIPK3 during TNFα-induced necroptosis
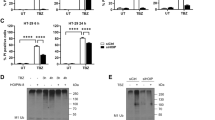
To identify RIPK3-interacting proteins, we transfected HEK 293T cells with either FLAG-MOCK or FLAG-RIPK3 DNA constructs (Fig. 1A), followed by immunoprecipitation using a RIPK3 antibody, and visualized the results with Coomassie Brilliant Blue staining (Fig. S1A). Comparing immunoprecipitates from cell lysates expressing FLAG-MOCK and FLAG-RIPK3 revealed distinct protein bands, prompting us to conduct immunoprecipitation-mass spectrometry to identify these proteins. The mass spectrometry results confirmed known RIPK3-interacting proteins, such as RIPK1, Hsp70, and UCHL37, validating our approach7,44,59,60. Additionally, we identified interacting proteins, with SMURF1, an E3 ligase, and USP5, a deubiquitinase, being of particular interest (Fig. 1B). As these proteins had not been previously associated with RIPK3, we investigated their potential roles in the RIPK3 pathway.
A Method that SDS-PAGE and Coomassie brilliant blue (CBB) staining of RIPK3-immunoprecipitated proteins from HEK 293T cells stably overexpressing FLAG-RIPK3. Icons of this figure created with BioRender.com. B Proteins that we identified by mass spectrometry (MS) analysis. Icons of this figure created with BioRender.com. C HT-29 cells were treated with 20 ng/mL TNFα, 2 μM Birinapant, and 20 μM Z-VAD-FMK (T/Bi/Z) for 2 h and 3 h. After treatment, the cell lysates were immunoprecipitated with RIPK3 antibody and analyzed using immunoblotting. D HT-29 cells were treated with T/Bi/Z for 2 h and 3 h. After treatment, the cell lysates were immunoprecipitated with SMURF1 antibody and analyzed using immunoblotting. E HT-29 cells were treated with T/Bi/Z for 2 h and 3 h. After treatment, the cell lysates were immunoprecipitated with SMURF1 antibody and analyzed using immunoblotting. F HT-29 cells were treated with T/Bi/Z for 3 h. The cell lysates were fractionated according to molecular size by gel-filtration chromatography and examined using immunoblotting. G HT-29 cells were treated with T/Bi/Z for 3 h. After treatment, the cells were fixed and stained using anti-SMURF1 and p-RPIPK3 antibodies, and DAPI. H HT-29 cells were treated with T/Bi/Z for 3 h. After treatment, the cells were fixed and stained using anti-USP5 and p-RIPK3 antibodies, and DAPI. Scale bars = 10 μm.
First, we confirmed that RIPK3 interacts with both SMURF1 and USP5 by performing immunoprecipitation assays in HEK 293 T cells expressing FLAG-RIPK3 and either 3xMYC-SMURF1 or 3xMYC-USP5 (Fig. S1B, C). To further validate the endogenous interactions between RIPK3 and these two proteins, we induced necroptosis in HT-29 colon cancer cells using TNF-α, birinapant, and zVAD-fmk (T/Bi/Z). Immunoprecipitation with anti-RIPK3 antibodies revealed that endogenous SMURF1 and USP5 did not associate with RIPK3 under normal conditions. Interestingly, SMURF1 was recruited to the RIPK3 complex after 2 h of necroptotic stimulation, while USP5 incorporation occurred after 3 h of treatment (Fig. 1C). These results suggest that SMURF1 is recruited to the necrosome early during necroptosis, followed by USP5.
To verify the associations between RIPK3, SMURF1, and USP5 during necroptosis, we performed additional immunoprecipitation assays using anti-SMURF1 and anti-USP5 antibodies. These experiments confirmed that, under necroptotic conditions, both SMURF1 and USP5 co-immunoprecipitated with key necroptosis proteins, including RIPK1, RIPK3, and MLKL, along with their phosphorylated forms: p-RIPK1, p-RIPK3, and p-MLKL (Fig. 1D, E). Interestingly, SMURF1 interacts with necrosome-associated proteins 2 h after necroptosis induction, whereas USP5 associates with necrosome components 3 h post-induction. This sequential interaction pattern indicates that SMURF1 binds to RIPK3 first, followed by USP5 at a later stage. These findings suggest that SMURF1 and USP5 regulate RIPK3 ubiquitination in a stepwise manner, playing distinct roles in the modulation of necroptosis progression. Notably, SMURF1 was immunoprecipitated even in the absence of USP5, whereas USP5 immunoprecipitation required SMURF1, indicating that SMURF1 is required for USP5 to interact with necrosome complexes. It is also likely that USP5 and SMURF1 exert their effects on RIPK3 through a sequential mechanism rather than through direct interaction. Notably, the necrosome complex immunoprecipitated using RIPK3 antibodies showed the presence of SMURF1 and USP5. These data indicate that there might be heterogeneous populations of necrosome complexes integrating either SMURF1 or USP5 (Fig. 1C–E).
To explore the incorporation of SMURF1 and USP5 into the necrosome complex further, we performed gel filtration analysis using HT-29 cells. Consistent with previous studies56,57, necroptosis induction triggered the translocation of RIPK1, RIPK3, and MLKL into a large multiprotein complex. Notably, SMURF1 and USP5 were also detected in this complex, confirming their presence within the necrosome assemblies (Fig. 1F). Additionally, the immunoprecipitation of each fraction with a RIPK3 antibody indicated that SMURF1 and USP5 also interact within the necrosome complexes containing RIPK1, RIPK3 and MLKL (Fig. S1D). This finding demonstrates that SMURF1 and USP5 are incorporated into the necrosome complexes.
Finally, confocal microscopy analysis demonstrated the co-localization of endogenous phosphorylated RIPK3 with SMURF1 or USP5 at the necrosome complex. (Fig. 1G, H). Collectively, these findings indicate that, while SMURF1 and USP5 do not associate with RIPK1, RIPK3, or MLKL under normal conditions, both proteins are incorporated into the necrosome during necroptosis. Additionally, the interaction of SMURF1 and USP5 with RIPK3 occurs sequentially, with SMURF1 binding first, followed by USP5. This suggests a potential regulatory role of SMURF1 and USP5 in modulating the necroptotic pathway.
SMURF1 E3 ligase activity is required to suppress necrosome formation and necroptosis
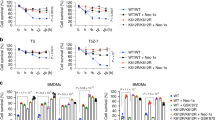
To investigate SMURF1’s role in necroptosis, we established HT-29 cell lines with stable SMURF1 knockdown using shRNAs (Fig. 2A). Under these conditions, no changes were observed in the protein expression levels of RIPK1, RIPK3, or MLKL (Fig. 2A). However, SMURF1 knockdown led to approximately a 40% increase in necroptosis in HT-29 cells treated with T/Bi/Z compared to control cells (Fig. 2B). Similarly, SMURF1 depletion using siRNAs or shRNA in mouse TC-1 or HeLa-RIPK3 cells, respectively, did not affect the expression levels of RIPK1, RIPK3, or MLKL, but increased their sensitivity to necroptotic stimuli by about 30% and 35%, respectively (Fig. S2A, E; Fig. S2B, F). Notably, SMURF1 knockdown promoted the phosphorylation of RIPK1 (pRIPK1), RIPK3 (pRIPK3), and MLKL (pMLKL) without altering their protein levels, indicating an acceleration of necroptosis under SMURF1 depletion (Fig. 2C). Corroborating these findings, SMURF1 depletion enhanced the assembly of necrosome complexes compared to control cells (Fig. 2D). Similarly, in TC-1 and HeLa-RIPK3 cell lines, SMURF1 depletion increased necroptosis, as detected by p-MLKL levels and accelerated necrosome formation (Fig. S2C, D, G, H).
A HT-29 cells were transduced with lentivirus to generate stable SMURF1-knockdown (KD) cell lines. The expression levels of RIPK1, RIPK3, MLKL, and SMURF1 in each cell line were analyzed by western blotting. B HT-29 cells depleted of SMURF1 using the indicated shRNAs were treated with T/Bi/Z in the presence or absence of 0.1 μM GSK'963 for 2.5 h. Flow cytometry was performed by counting 10,000 cells based on FSC (Forwarder Scatter) and SSC (Side Scatter), and calculating the percentage of cells in the Annexin V-FITC positive region. This experiment was independently repeated 3 times. Data are presented as means ± standard deviations (SDs), n = 3. Significance between groups was determined using two-sided Students’ test. C SMURF1 KD and control HT-29 cells were treated with T/Bi/Z to induce necroptosis. GSK'963 was used to confirm that this cell death is necroptosis. Cells were harvested at the indicated time points and analyzed by western blotting. D SMURF1 KD and control HT-29 cells treated with T/Bi/Z were lysed after 2 h. The necrosome was pulled down using an anti-RIPK3 antibody and protein-G agarose beads. The protein-bead conjugate was washed and analyzed by western blotting to assess the assembly of the RIPK1-RIPK3-MLKL complex. The asterisk indicates p-MLKL. E shSMURF1-infected cells, reconstituted with either SMURF1 WT or the E3 ligase-inactive form (C699A), were treated with T/Bi/Z in the presence or absence of 0.1 μM GSK'963 for 2 h. Flow cytometry was performed by counting 10,000 cells based on FSC (Forwarder Scatter) and SSC (Side Scatter), and calculating the percentage of cells in the Annexin V-FITC positive region. This experiment was independently repeated three times. Data are presented as means ± standard deviations (SDs), n = 3. Significance between groups was determined using two-sided Students’ test. F shSMURF1-infected cells, reconstituted with either SMURF1 WT or the E3 ligase-inactive form (C699A), were treated with T/Bi/Z to induce necroptosis. GSK'963 was used to confirm that this cell death is necroptosis. Cells were harvested at the indicated time points and analyzed by western blotting. The asterisk indicates p-RIPK3. G shSMURF1-infected cells, reconstituted with either SMURF1 WT or the E3 ligase-inactive form (C699A), were treated with T/Bi/Z and lysed after 2 h. The necrosome was pulled down using an anti-RIPK3 antibody and protein-G agarose beads. The protein-bead conjugate was washed and analyzed by western blotting to examine the assembly of the RIPK1-RIPK3-MLKL complex.
Given the increase in necroptosis following SMURF1 depletion, we next investigated whether the E3 ligase activity of SMURF1 is necessary for necroptosis suppression. Reconstitution of wild-type (WT) SMURF1 in SMURF1-depleted HT-29 cells restored necroptosis levels to those of control cells. In contrast, reconstitution with the E3 ligase-defective mutant SMURF1 C699A failed to prevent cell death, indicating that SMURF1’s E3 ligase activity is essential for the inhibition of necroptosis (Fig. 2E). Consistent with these data, reconstitution with WT SMURF1 prevented the phosphorylation of RIPK3 and MLKL in response to necroptotic stimuli, and inhibited necrosome complex formation, as detected by immunoprecipitation using anti-RIPK3 antibodies. Conversely, SMURF1 C699A was unable to inhibit RIPK3 and MLKL phosphorylation or necrosome complex formation (Fig. 2F, G).
These results suggest that SMURF1 suppresses necrosome complex formation and necroptosis via its E3 ligase activity without affecting the expression levels of necrosome-associated proteins (Fig. 2F, G). Overall, these observations indicate that SMURF1’s E3 ligase activity is essential for the suppression of necroptosis. Since SMURF1 depletion did not affect the protein levels of RIPK1, RIPK3, or MLKL, it is likely that regulatory ubiquitination processes are involved in suppressing necroptosis.
SMURF1 directs K63-linked polyubiquitination of RIPK3 during TNFα-induced necroptosis, suppressing necroptosis
The identification of SMURF1 as a RIPK3-interacting protein, along with the requirement for its E3 ligase activity to inhibit necroptosis, prompted us to investigate whether SMURF1 mediates non-proteolytic ubiquitination of RIPK3 via direct binding (Fig. 1A–D, A–G).
First, to determine whether SMURF1 interacts with necroptosis-related proteins such as RIPK1, RIPK3, and MLKL, we performed immunoprecipitation after overexpressing these proteins in HEK 293 T cells. The results confirmed that SMURF1 interacts with RIPK1, RIPK3, and MLKL (Fig. 3A). As we previously observed that the E3 ligase activity of SMURF1 influenced necroptosis sensitivity, we conducted ubiquitination assays on RIPK1, RIPK3, and MLKL. SMURF1 was found to specifically ubiquitinate RIPK3, whereas no ubiquitination was detected for RIPK1 and MLKL (Figs. 3B, S3A, B).
A FLAG-RIPK1, RIPK3, MLKL and 3xMYC-SMURF1 were overexpressed in HEK 293T cells via PEI transfection. Cells were lysed and incubated with anti-FLAG antibodies and protein-G agarose beads to pull down 3xMYC-SMURF1.The protein-bead conjugate was washed and analyzed by SDS-PAGE. The immunoprecipitation of the SMURF1 and RIPK1, RIPK3, MLKL were assessed using western blotting. B FLAG-RIPK3 was transfected into HEK 293T cells along with HA-Ub, with or without 3xMYC-SMURF1 expression. After 20 h, cells were lysed in a 1% SDS buffer, and lysates diluted to 0.1% SDS were immunoprecipitated using an anti-FLAG antibody. RIPK3 ubiquitylation was detected by western blotting with an HRP-conjugated anti-HA antibody. C FLAG-RIPK3 (1–296, kinase domain; 294–518, C-term domain; 1–440 and 452–518 ΔRHIM domian) and 3xMYC-SMURF1 were overexpressed in HEK 293T cells via PEI transfection. Cells were lysed and incubated with anti-FLAG antibodies and protein-G agarose beads to pull down 3xMYC-SMURF1. The protein-bead conjugate was washed and analyzed by SDS-PAGE. The immunoprecipitation of the SMURF1 and RIPK3 domains was assessed using western blotting. The schematic diagram of RIPK3 shows the binding domain for SMURF1. Icons of this figure created with BioRender.com. D HA-SMURF1 (1–159, 119–731, 1–345, 346–731, 119–365) and FLAG-RIPK3 were overexpressed in HEK 293 T cells via PEI transfection. Cells were lysed and incubated with anti-HA antibodies and protein-G agarose beads to pull down the specific domain of HA-SMURF1. The protein-bead conjugate was washed and analyzed by SDS-PAGE. The immunoprecipitation of the RIPK3 and SMURF1 domains was assessed using western blotting. The schematic diagram of RIPK3 shows the binding domain for RIPK3. Icons of this figure created with BioRender.com. E HEK 293 T cells were transfected with FLAG-RIPK3, 3xMYC–SMURF1, and HA-Ub along with various ubiquitin mutants. After 20 h, cells were lysed in a 1% SDS buffer, and lysates diluted to 0.1% SDS were immunoprecipitated using an anti-FLAG antibody. RIPK3 ubiquitylation was detected by western blotting with an HRP-conjugated anti-HA antibody. F HEK 293T cells were transfected with FLAG-RIPK3, 3xMYC–SMURF1, and HA-Ub along with the K63R ubiquitin mutant. After 20 h, cells were lysed in a 1% SDS buffer, and lysates diluted to 0.1% SDS were immunoprecipitated using an anti-FLAG antibody. RIPK3 ubiquitylation was detected by western blotting with an HRP-conjugated anti-HA antibody. G To confirm the endogenous ubiquitination status of RIPK3, HT-29 cells expressing shGFP or shSMURF1 (#4 and #8) were treated with T/Bi/Z to induce necroptosis. After 3 h, SMURF1 KD and control HT-29 cells were lysed. Immunoprecipitation was performed using an anti-RIPK3 antibody and protein-G agarose beads. The protein-bead conjugate was washed, and ubiquitination was detected using an anti-Ub-HRP conjugated antibody and an anti-K63-Ub-HRP conjugated antibody.
When RIPK3 is overexpressed in HEK 293T cells, it undergoes auto-phosphorylation, leading to its phosphorylation (Fig. S4A). This process simulates necroptosis progression, enabling phosphorylated RIPK3 to interact with SMURF1, similar to the interaction observed among endogenous proteins in HT-29 cells during necroptosis. (Fig. 1C, D). These findings suggest that RIPK3 phosphorylation plays a crucial role in its interaction with SMURF1. This finding was further supported by the reduced interaction between SMURF1 and the RIPK3 S227A mutant, which cannot undergo phosphorylation, as observed in immunoprecipitation experiments using overexpressed proteins in HEK 293T cells (Fig. S4B)
Next, we dissected the binding domains between SMURF1 and RIPK3. We found that the RIPK3 RIMH (RIP homotypic interaction motif) domain interacted with SMURF1, while the WW domain of SMURF1 was crucial for binding with RIPK3 (Fig. 3C, D).
Further evidence that SMURF1 does not regulate RIPK1 or NF-κB pathways downstream of RIPK1 was provided by experiments in HT-29 cells treated with TNFα (Fig. S5A, B). TNFα treatment did not affect RIPK1-mediated complex I formation or its downstream NF-κB pathway (Fig. S5A, B). This suggests that SMURF1, when incorporated into the necrosome under necroptotic conditions, may specifically mediate the ubiquitination of RIPK3, potentially limiting necrosome complex formation and thereby suppressing necroptosis.
Next, we explored the type of polyubiquitin chain involved in SMURF1-mediated polyubiquitination of RIPK3. Using ubiquitin mutants that contain only one functional lysine residue, we discovered that SMURF1 specifically facilitates K63-linked polyubiquitination of RIPK3 (Fig. 3E). Supporting this observation, SMURF1 was unable to ubiquitinate RIPK3 when the lysine 63 residue of ubiquitin was mutated to arginine (Fig. 3F).
Additionally, we examined the ubiquitination status of endogenous RIPK3 in necroptosis-induced HT-29 cells. Under necroptotic conditions, there was an overall increase in RIPK3 ubiquitination. However, the knockdown of SMURF1 specifically suppressed K63-linked polyubiquitination despite the overall increase in RIPK3 ubiquitination (Fig. 3G).
These findings indicate that SMURF1 promotes K63-linked polyubiquitination of RIPK3, which may suppress necrosome formation and thereby limit necroptosis.
SMURF1 mediates K63-ubiquitination of RIPK3 at lysine 55 and 363 during TNFα-induced necroptosis
To identify the specific lysine residues on RIPK3 targeted for ubiquitination by SMURF1, we mutated individual or combinations of lysine residues previously identified through mass spectrometry as polyubiquitination sites (Fig. 4A)51,52,53,61,62. Each of these lysine residues was mutated to arginine, and the mutants were assessed for their susceptibility to SMURF1-mediated K63-linked polyubiquitination. Among the tested mutants, the K55R and K363R mutants showed resistance to SMURF1-induced ubiquitination (Fig. 4B). Notably, the double mutant K55/363R completely abolished SMURF1-mediated ubiquitination (Fig. 4B).
A This is a schematic diagram illustrating the various ubiquitination enzyme sites on RIPK3 identified to date. Icons of this figure created with BioRender.com. B FLAG-RIPK3 WT, K55R, K197R, K302R, K363R, K364R, K518R, and K55/363R were transfected into HEK 293T cells with HA-Ub, with or without 3xMYC-SMURF1 expression. After 20 h, cells were lysed in a 1% SDS buffer, and lysates diluted to 0.1% SDS were immunoprecipitated using an anti-FLAG antibody. RIPK3 ubiquitination was detected by western blotting with an HRP-conjugated anti-HA antibody. C Adenovirus-mediated expression of RIPK3 WT, K55R, K363R, and K55/363R was induced in HeLa cells, which were then treated with T/Bi/Z to induce necroptosis. After 5 h, the cells were harvested and stained with propidium iodide (PI). Flow cytometry was performed by counting 10,000 cells based on FSC (Forwarder Scatter) and SSC(Side Scatter), and calculating the percentage of cells in the PI(Propidium Iodide) positive region. This experiment was independently repeated 3 times. Data are presented as means ± standard deviations (SDs), n = 3. Significance between groups was determined using two-sided Students’ test. D HeLa cells expressing RIPK3 WT, K55R, K363R, and K55/363R were treated with T/Bi/Z to induce necroptosis at the indicated time points. Cells were harvested at these time points and analyzed by western blotting. The asterisk indicates p-RIPK3. E HeLa cells expressing shGFP, shSMURF1, RIPK3 WT, and RIPK3 K55/363R were treated with T/Bi/Z to induce necroptosis. After 5 h, the cells were harvested and stained with propidium iodide (PI). Flow cytometry was performed by counting 10,000 cells based on FSC (Forwarder Scatter) and SSC (Side Scatter), and calculating the percentage of cells in the PI(Propidium Iodide) positive region. This experiment was independently repeated 3 times. Data are presented as means ± standard deviations (SDs), n = 3. Significance between groups was determined using two-sided Students’ test. F HeLa cells expressing shGFP, shSMURF1, RIPK3 WT, and RIPK3 K55/363R were treated with T/Bi/Z to induce necroptosis. After 4 h, the cells were harvested and analyzed by western blotting.
To further evaluate the functional role of these RIPK3 mutants (K55R, K363R, and K55/363R) in necroptosis, we overexpressed RIPK3 WT, K55R, K363R, or K55/363R in HeLa cells. The results demonstrated that the RIPK3 K55/363R mutant significantly enhanced necroptosis compared to the individual K55R and K363R mutants or RIPK3 WT (Fig. 4C). Consistent with these findings, the K55/363R mutant accelerated the phosphorylation of RIPK1 (p-RIPK1), RIPK3 (p-RIPK3), and MLKL (p-MLKL), key markers of necroptotic signaling (Fig. 4D). Interestingly, the necroptotic sensitivity of cells expressing the RIPK3 K55/363R mutant was similar to that of SMURF1-depleted cells, suggesting that SMURF1-mediated ubiquitination at lysine 55 and 363 plays a suppressive role in necroptosis under TNFα-induced conditions (Fig. 4E, F).
To further investigate how the RIPK3 K55/363R mutant accelerates RIPK3 phosphorylation, we analyzed how K63-linked polyubiquitination affects the mechanism of RIPK3 phosphorylation. Previous studies have shown that RIPK3 can be phosphorylated either through RIPK1-RIPK3 interaction or via the self-assembly of RIPK3 upon necroptosis induction1,3. To explore whether SMURF1 influences these protein interactions, we performed immunoprecipitation experiments. Interestingly, we found that SMURF1 suppressed the interaction between RIPK1 and RIPK3 (Fig. S6A). However, the SMURF1 C699A mutant, which lacks E3 ligase activity, did not interfere with the RIPK1-RIPK3 interaction (Fig. S6A). Similar to the RIPK1-RIPK3 interaction, self-assembly of RIPK3 was also disrupted by the E3 ligase function of SMURF1 (Fig. S6B). Compared to RIPK3 WT, the RIPK3 K55/363R mutant, which cannot undergo SMURF1-mediated K63-linked polyubiquitination, showed an increased extent of interaction with RIPK1 and enhanced self-assembly. Notably, the presence of SMURF1 did not significantly affect this interaction (Fig. S6A, B). These findings indicate that SMURF1 facilitates the K63-linked polyubiquitination of RIPK3 at lysine residues 55 and 363, ultimately suppressing necroptosis by restricting activation of the necroptotic signaling pathway.
USP5 promotes TNFα-induced necroptosis
Since USP5 was identified as a binding partner of RIPK3, we investigated its potential antagonistic role against SMURF1 in necroptosis. Knockdown of USP5 using shRNAs in HT-29 cells did not cause any detectable changes in the protein expression levels of RIPK1, RIPK3, or MLKL (Fig. 5A). However, when necroptosis was induced by T/Bi/Z treatment under USP5 knockdown, cell death decreased by approximately 30% (Fig. 5B). Similarly, USP5 knockdown using siRNA or shRNA in TC-1 or HeLa-RIPK3 cells resulted in a 25% and 20% reduction in necroptosis, respectively, without affecting the expression levels of RIPK1, RIPK3, or MLKL (Fig. S7A, B, E, F). In HT-29 cells, USP5 depletion also led to a reduction in the phosphorylation of RIPK1, RIPK3, and MLKL, indicating a suppression of necroptosis (Fig. 5C).
A HT-29 cells were transduced with lentivirus to generate stable USP5-knockdown (KD) cell lines. The expression levels of RIPK1, RIPK3, MLKL, and USP5 in each cell line were analyzed by western blotting. B HT-29 cells depleted of USP5 using the indicated shRNAs were treated with T/Bi/Z in the presence or absence of 0.1 μM GSK'963 for 3.5 h. Flow cytometry was performed by counting 10,000 cells based on FSC (Forwarder Scatter) and SSC (Side Scatter), and calculating the percentage of cells in the Annexin V-FITC positive region. This experiment was independently repeated 3 times. Data are presented as means ± standard deviations (SDs), n = 3. Significance between groups was determined using two-sided Students’ test. C USP5 KD and control HT-29 cells were treated with T/Bi/Z to induce necroptosis. GSK'963 was used to confirm that this cell death is necroptosis. Cells were harvested at the indicated time points and analyzed by western blotting. D USP5 KD and control HT-29 cells treated with T/Bi/Z were lysed after 3.5 h. The necrosome was pulled down using an anti-RIPK3 antibody and protein-G agarose beads. The protein-bead conjugate was washed and analyzed by western blotting to assess the assembly of the RIPK1-RIPK3-MLKL complex. E shUSP5-infected cells, reconstituted with either USP5 WT or the deubiquitinase-defective mutant USP5 C335A, were treated with T/Bi/Z in the presence or absence of 0.1 μM GSK'963 for 3.5 h. Flow cytometry was performed by counting 10,000 cells based on FSC (Forwarder Scatter) and SSC (Side Scatter), and calculating the percentage of cells in the Annexin V-FITC positive region. This experiment was independently repeated 3 times. Data are presented as means ± standard deviations (SDs), n = 3. Significance between groups was determined using two-sided Students’ test. F shUSP5-infected cells, reconstituted with either USP5 WT or the deubiquitinase-defective mutant USP5 C335A, were treated with T/Bi/Z to induce necroptosis. GSK'963 was used to confirm that this cell death is necroptosis. Cells were harvested at the indicated time points and analyzed by western blotting. The asterisk indicates p-RIPK3 and p-MLKL. G shUSP5-infected cells, reconstituted with either USP5 WT or the deubiquitinase-defective mutant USP5 C335A, were treated with T/Bi/Z and lysed after 3.5 h. The necrosome was pulled down using an anti-RIPK3 antibody and protein-G agarose beads. The protein-bead conjugate was washed and analyzed by western blotting to examine the assembly of the RIPK1-RIPK3-MLKL complex.
Furthermore, immunoprecipitation analyses using anti-RIPK3 antibodies showed a significant reduction in necrosome complex formation under USP5 knockdown (Fig. 5D). Similar effects were observed in TC-1 and HeLa-RIPK3 cells, where a decrease in p-MLKL levels and necrosome formation was also noted under USP5 knockdown (Fig. S7C, D, G, H). Given the observed decrease in necroptosis under USP5 depletion, we further examined whether USP5’s deubiquitinase activity is necessary for modulating necroptosis. When wild-type (WT) USP5 was reintroduced into USP5-depleted HT-29 cells, necroptosis was restored to control levels. In contrast, reconstitution with the deubiquitinase-defective mutant USP5 C335A failed to enhance cell death, indicating that deubiquitinase activity is essential for USP5-mediated necroptosis acceleration (Fig. 5E). Supporting this, reconstitution with WT USP5 restored RIPK3 and MLKL phosphorylation and enhanced necrosome complex formation, as detected by anti-RIPK3 antibodies. On the other hand, reconstitution with USP5 C335A was unable to promote phosphorylation or necrosome complex formation (Fig. 5F, G).
Lastly, USP5 knockdown did not affect RIPK1-mediated complex I formation or its downstream NF-κB signaling pathway, suggesting that USP5 may specifically regulate necroptosis (Fig. S8A, B). Additionally, we assessed the sensitivity of RIPK1-dependent apoptosis in USP5 knockdown HT-29 cells following T/Bi stimulation and found no significant differences compared to the control cells (Fig. S8C, D). These results suggest that USP5 is not involved in RIPK1-dependent complex I signaling or RIPK1-dependent apoptosis. Overall, these findings highlight the critical role of the deubiquitinase activity of USP5 in promoting necroptosis. Notably, the levels of RIPK1, RIPK3, and MLKL remained unaltered regardless of the presence of USP5, which indicated that USP5-mediated necroptosis may be regulated through specific ubiquitination processes.
Antagonistic regulation of RIPK3 K63 polyubiquitination by SMURF1 and USP5
The data presented above demonstrate that SMURF1-mediated ubiquitination of RIPK3 suppresses necroptosis, while USP5-dependent deubiquitination promotes cell death. The next question to address is how these two regulators interact to influence RIPK3 ubiquitination. To investigate the mechanism by which SMURF1 and USP5 associate with RIPK3, we first compared the effects of knocking down USP5, SMURF1, or both in HT-29 cells subjected to necroptotic stimuli. Consistent with previous observations, there were no changes in the protein expression levels of RIPK1, RIPK3, MLKL, SMURF1, or USP5 (Fig. 6A). Necroptotic cell death was similarly promoted in both SMURF1 knockdown and SMURF1/USP5 double knockdown cells, while it was suppressed in USP5 knockdown cells (Fig. 6B). These findings correlate with changes in RIPK3 and MLKL phosphorylation (Fig. 6C).
A HT-29 cells depleted of SMURF1, USP5 and Double Knocked down by the indicated shRNAs. B HT-29 cells depleted of SMURF1, USP5 and Double Knocked down by the indicated shRNAs were treated with T/Bi/Z in the presence or absence of 0.1 μM GSK’963 for 3 h. The cells were harvested and stained with annexin V-FITC. Flow cytometry was performed by counting 10,000 cells based on FSC (Forwarder Scatter) and SSC (Side Scatter), and calculating the percentage of cells in the Annexin V-FITC positive region. This experiment was independently repeated 3 times. Data are presented as means ± standard deviations (SDs), n = 3. Significance between groups was determined using two-sided Students’ test. C Each HT-29 cells depleted of SMURF1, USP5 and Double Knocked down cells were treated with T/Bi/Z to induce necroptosis. GSK’963 (0.1 μM) was used to inhibit necroptosis. Cells were harvested and analyzed using western blotting. D Each HT-29 cells depleted of SMURF1, USP5 and Double Knocked down cells treated with T/Bi/Z were lysed after 3 h. The necrosome was pulled down using an anti-RIPK3 antibody and protein-G agarose beads. The protein-bead conjugate was washed and analyzed by western blotting to assess the assembly of the RIPK1-RIPK3-MLKL complex. E To confirm the endogenous ubiquitination status of RIPK3, HT-29 cells depleted of SMURF1, USP5 and Double Knocked down were treated with T/Bi/Z to induce necroptosis. After 3 h, SMURF1 KD and control HT-29 cells were lysed. Immunoprecipitation was performed using an anti-RIPK3 antibody and protein-G agarose beads. The protein-bead conjugate was washed, and ubiquitination was detected using an anti-Ub-HRP conjugated antibody and an anti-K63-Ub-HRP conjugated antibody.
Next, we examined necrosome complex formation by immunoprecipitating RIPK3 in cells with USP5, SMURF1, or both knocked down. The results showed that necrosome formation was enhanced in SMURF1-deficient cells compared to shGFP HT-29 control cells. In contrast, USP5 depletion led to suppression of necrosome formation (Fig. 6D). Notably, in cells lacking both SMURF1 and USP5, necrosome formation increased compared to USP5-only deficient cells (Fig. 6D). Interestingly, under SMURF1 depletion, USP5 did not associate with complexes immunoprecipitated by RIPK3 antibodies (Fig. 6D). However, under USP5 depletion, SMURF1 remained associated with necrosome complexes (Fig. 6D). This suggests a possible sequential interaction, where SMURF1 associates with the necrosome before USP5 during necroptosis.
Since deubiquitinating enzymes typically remove ubiquitin from substrates ubiquitinated by E3 ligases, we hypothesized that USP5 might antagonistically regulate the K63 polyubiquitination mediated by SMURF1. To determine whether USP5 interacts with necroptosis-related proteins such as RIPK1, RIPK3, and MLKL, we performed immunoprecipitation after overexpressing these proteins in HEK 293 T cells. The results confirmed that USP5 interacted with RIPK1, RIPK3, and MLKL (Fig. S9A). Specifically, USP5 interacted with RIPK3 through the kinase domain of RIPK3 (Fig. S9B). Reciprocally, the USP5 ZnF domain alone was unable to interact with RIPK3 (Fig. S9C). As expected, USP5 overexpression completely inhibited SMURF1-induced K63 polyubiquitination of RIPK3, as shown in the ubiquitination assay (Fig. S10A). USP5 primarily targets unanchored ubiquitin chains by recognizing the free C-terminal diglycine (GG) motif and sequentially cleaving ubiquitin monomers45,46. However, accumulating evidence suggests that USP5 can also act on covalently attached ubiquitin chains, either via direct deubiquitination or by modifying ubiquitin linkage types47,48,49. To experimentally verify whether USP5 directly functions as a deubiquitinase for SMURF1-mediated K63-linked ubiquitination of RIPK3, we transfected HEK 293 T cells with RIPK3, SMURF1, and USP5, followed by immunoprecipitation to isolate the associated proteins. Using these isolated proteins, we performed an in vitro ubiquitination assay with recombinant E1, E2, ATP, and ubiquitin proteins (Fig. S10B). Our findings demonstrated that USP5 directly eliminated SMURF1-mediated ubiquitination from RIPK3, thereby refuting the notion that USP5 exclusively targets untethered polyubiquitin chains. Endogenously, RIPK3 ubiquitination increased with necroptosis progression (Fig. 6E). In SMURF1 or SMURF1/USP5 double knockdown cells, there was an increase in total RIPK3 ubiquitination but a simultaneous decrease in K63-linked polyubiquitination (Fig. 6E). In contrast, USP5 knockdown caused a drastic reduction in total RIPK3 polyubiquitination, while K63-linked ubiquitination persisted significantly (Fig. 6E).
These results suggest that K63-linked polyubiquitination of RIPK3 at lysines 55 and 363 suppresses necroptosis. To further explore this, we introduced RIPK3 WT or RIPK3 K55/363R mutants (defective in SMURF1-mediated polyubiquitination) into HeLa cells to generate HeLa RIPK3 WT or HeLa RIPK3 K55/363R cell lines. Upon treatment with T/Bi/Z, HeLa RIPK3 K55/363R cells exhibited increased cell death compared to HeLa RIPK3 WT cells (Fig. S10C). Notably, while USP5 knockdown protected HeLa RIPK3 WT cells from necroptotic stimuli, this protection was lost in HeLa RIPK3 K55/363R cells (Fig. S10C). Similarly, USP5 knockdown suppressed MLKL phosphorylation in HeLa RIPK3 WT cells, but this effect was absent in HeLa RIPK3 K55/363R cells (Fig. S10D).
Finally, we immunoprecipitated lysates from HeLa RIPK3 WT and K55/363R cells treated with T/Bi/Z using RIPK3 antibodies. In HeLa RIPK3 WT cells, both USP5 and SMURF1 were immunoprecipitated, whereas only SMURF1 was detected in HeLa RIPK3 K55/363R cells (Fig. S10E). These findings are consistent with the results from Fig. 6D, where USP5 was no longer part of necrosome complexes upon SMURF1 depletion. Collectively, these results suggest that ubiquitination at K55/363 suppresses necroptosis, which can be reversed by USP5-mediated deubiquitination. To further analyze the prerequisites for USP5 and RIPK3 binding during necroptosis progression, we conducted an endogenous ubiquitination assay and immunoprecipitation of RIPK3 under necroptosis-inducing conditions, with or without the RIPK3 kinase inhibitor GSK'872 (Fig. S11A, B). Results showed that the K63-linked polyubiquitination of RIPK3 peaked 2 h after necroptosis induction and then decreased at 3 h. Interestingly, when cells were treated with GSK'872, the level of K63-linked polyubiquitination of RIPK3 was abolished at the same time points after necroptosis induction. This indicates that the K63-linked polyubiquitination of RIPK3 requires prior phosphorylation of RIPK3 (Fig. S11A). Similarly, SMURF1 interacted with RIPK3 at 2 h post-necroptosis induction, corresponding to the highest level of K63-linked polyubiquitination of RIPK3. In contrast, at 3 h post-necroptosis induction, USP5 interacted with phosphorylated RIPK3, coinciding with a decrease in K63-linked polyubiquitination. This suggests that USP5 interaction with RIPK3 is linked to a reduction in the degree of K63-linked polyubiquitination (Fig. S11A, B). Furthermore, neither SMURF1 nor USP5 was incorporated into the necrosome complexes when GSK'872 was added along with necroptosis induction (Fig. S11B). These findings indicate that RIPK3 phosphorylation is a prerequisite for its interaction with SMURF1 and USP5. Additionally, USP5 seemed to interact with the necrosome complexes only after SMURF1-mediated polyubiquitination of RIPK3 (Figs. 1E, 6D, S10E). By removing these polyubiquitin chains, USP5 restored necroptosis to its full capacity.
USP5 overexpression inhibited SMURF1-induced K63 polyubiquitination of RIPK3, as confirmed by ubiquitination assays (Fig. S12A, B). Additionally, endogenous RIPK3 ubiquitination increased with the progression of necroptosis, with K63 polyubiquitination specifically increasing upon USP5 knockdown but being suppressed in SMURF1/USP5 double knockdown cells (Fig. 6E). In HeLa RIPK3 K55/363R mutant cells, necroptosis was enhanced and not reversed by USP5 knockdown, further supporting the antagonistic roles of USP5 and SMURF1. To confirm that USP5 specifically targets SMURF1-mediated ubiquitination, we examined whether USP5 can also remove the ubiquitin added by other E3 ligases, such as CHIP, Parkin and TRIM25, which are known to modulate RIPK3 sensitivity to necroptosis. The results revealed that USP5 specifically deubiquitinates RIPK3 ubiquitinated by SMURF1, but not CHIP, Parkin, or TRIM25 (Fig. S12A, B). A previous study demonstrated that USP22 functions as a deubiquitinase for RIPK353. To further investigate this, we examined whether SMURF1 and USP22 have an antagonistic relationship in regulating RIPK3 ubiquitination. However, our results showed that USP22 does not deubiquitinate RIPK3 when ubiquitinated by SMURF1 (Fig. S12C). These findings strongly suggest that the regulatory axis of SMURF1 and USP5 operates independently of the previously identified E3 ligases (CHIP, Parkin, and TRIM25) and deubiquitinases (USP22) in controlling RIPK3 ubiquitination51,53,61. To further confirm that USP5 deubiquitinates RIPK3 at K55 and K363, we examined its interaction with the RIPK3 K55/363R double mutant, which lacks SMURF1-mediated ubiquitination at these sites. The results showed that USP5 did not interact with the K55/363R mutant, indicating that ubiquitination at K55 and K363 is essential for USP5 binding and subsequent deubiquitination. However, when we tested the individual K55R and K363R mutants, their interaction with USP5 was comparable to that of RIPK3 WT, suggesting that USP5 can still recognize and bind to RIPK3 when only one of these lysine residues is mutated under overexpression conditions in HEK 293 T cells (Fig. S12D). Although these single mutants showed reduced levels of K63-linked ubiquitination compared to WT, USP5 was still able to eliminate SMURF1-mediated K63-linked polyubiquitination. This indicates that USP5 functions as a K63-specific deubiquitinase at the K55 and K363 sites of RIPK3 (Fig. S12E). These findings demonstrate that USP5 acts as a positive regulator of necroptosis by removing the inhibitory K63-linked polyubiquitin chains from RIPK3 at lysine 55 and 363, a modification mediated by SMURF1. This underscores the opposing functions of E3 ligases and deubiquitinating enzymes in the regulation of necroptosis.
SMURF1 depletion or overexpression of the RIPK3 K55/363R mutant enhances Birinapant/Emricasan-induced necroptosis in leukemia cells
To explore the impact of SMURF1 on necroptosis in a more physiologically relevant context, we first utilized Molm-13 leukemia cells, which can induce autocrine TNFα and promote necroptosis upon treatment with birinapant (a SMAC mimetic) and emricasan (a pan-caspase inhibitor)56,57,58. SMURF1 knockdown in Molm-13 cells was achieved using shRNA to assess its role in regulating necroptotic sensitivity. Following SMURF1 knockdown, necroptosis was significantly enhanced compared to the control, as evidenced by accelerated cell death and increased phosphorylation of key necroptosis effectors, RIPK3 and MLKL (Fig. 7A–C). These findings suggest that SMURF1 depletion removes its inhibitory effect on necroptosis, resulting in heightened activation of the necroptotic pathway.
A Molm-13 cells were transduced with lentivirus to generate stable SMURF1-KD cell lines. The expression levels of RIPK1, RIPK3, MLKL, and SMURF1 in each cell line were analyzed by western blotting. B Molm-13 depleted of SMURF1 using the indicated shRNAs were treated with Bi/E in the presence or absence of 0.1 μM GSK'963 for 12 h. The cells were harvested and stained with PI. Flow cytometry was performed by counting 10,000 cells based on FSC (Forwarder Scatter) and SSC (Side Scatter), and calculating the percentage of cells in the PI (Propidium Iodide) positive region. This experiment was independently repeated 3 times. Data are presented as means ± standard deviations (SDs), n = 3. Significance between groups was determined using two-sided Students’ test. C SMURF1 KD and control Molm-13 cells were treated with Bi/E to induce necroptosis. GSK'963 was used to confirm that this cell death is necroptosis. Cells were harvested at the indicated time points and analyzed by western blotting. D Tumor xenografts were established by injecting control or SMURF1-knockdown (KD) Molm-13 cell lines into the flanks of mice (n = 6). This experimental setup aimed to examine the effect of SMURF1 depletion on tumor progression and response to treatment. E Following xenograft establishment, the mice received intraperitoneal injections of birinapant (2 mg/kg) and emricasan (1 mg/kg) every 4 days for a total of six treatments. Six independent mice were used for xenograft experiments in each experimental group, and treatments were administered independently for each group. Representative images of the resulting xenografts were taken to visually document tumor development. Tumor growth was systematically measured at 4-day intervals throughout the treatment period to monitor any differences between control and SMURF1 KD groups. Data are presented as means ± standard deviations (SDs). Significance was determined using the two-sided Students’ test. F At the end of the experiment, the tumors were excised, and their masses were recorded to evaluate the impact of SMURF1 depletion on tumor size. The tumor weight was measured from each of the six independent mice in each group. Data are presented as means ± standard deviations (SDs). Significance was determined using the two-sided Students’ test. G The tumors were lysed, and SMURF1 expression was analyzed using western blotting. H Additional tumor xenografts were generated using NB4 cell lines expressing either RIPK3 WT or the RIPK3 K55/363R mutant (n = 6). This setup aimed to assess the role of specific RIPK3 mutations in tumor growth and response to therapeutic agents. I Mice bearing these xenografts were treated with birinapant (2 mg/kg) and emricasan (1 mg/kg) via intraperitoneal injection every 5 days, for a total of eight treatments. Six independent mice were used for xenograft experiments in each experimental group, and treatments were administered independently for each group. Tumor growth was measured at 5-day intervals throughout the course of the experiment to track progression over time in both RIPK3 WT and K55/363R groups. Data are presented as means ± standard deviations (SDs). Significance was determined using the two-sided Students’ test. J Upon completion of the experiment, tumors were dissected, and their weights were measured to quantify differences between groups. The tumor weight was measured from each of the six independent mice in each group. Tumor weights were compared to determine any statistically significant variations attributable to SMURF1 KD or RIPK3 mutations.
Next, we sought to determine whether SMURF1 depletion could enhance necroptosis in an in vivo model and further suppress tumor growth. A xenograft study was conducted using SMURF1-depleted Molm-13 cells injected subcutaneously into nude mice. Mice bearing these subcutaneous tumors were treated with birinapant and emricasan every 4 days to induce necroptosis. While the combination of birinapant and emricasan alone slightly reduced tumor size in control tumors, significant suppression of tumor growth was observed in SMURF1-depleted tumors (Fig. 7D–F). Importantly, untreated SMURF1-depleted tumors did not exhibit a reduction in size, indicating that SMURF1 knockdown alone was insufficient to drive necroptosis. The expression levels of SMURF1 in each xenografted tumor were measured via western blotting (Fig. 7G). These results demonstrate that SMURF1 depletion potentiates the effect of necroptosis-inducing treatments, leading to a pronounced decrease in tumor burden.
In addition to Molm-13 cells, we extended our analysis to NB4 cells, a leukemia cell line that lacks RIPK3 but expresses autocrine TNFα, RIPK1, and MLKL57. (Fig. S13A)
To further confirm the role of SMURF1- and USP5-mediated ubiquitination in regulating necroptosis, we engineered NB4 cells to overexpress either RIPK3 WT or the RIPK3 K55/363R mutant, which is resistant to SMURF1-mediated inhibitory K63- poly ubiquitination. These models allowed us to directly assess the impact of disrupting SMURF1-mediated K63-polyubiquitination on necroptosis. Following treatment with birinapant and emricasan to induce necroptosis, RIPK3 K55/363R-expressing NB4 cells displayed significantly increased necroptosis compared to cells expressing wild-type RIPK3. Enhanced phosphorylation of necroptosis-related proteins, such as MLKL, further confirmed that disrupting SMURF1-mediated ubiquitination sensitizes the cells to necroptotic cell death (Fig. S13B, C).
We further evaluated the impact of SMURF1-mediated regulation of RIPK3 in an in vivo xenograft model using NB4 cells overexpressing either RIPK3 WT or the RIPK3 K55/363R mutant. Without treatment, tumor growth in both RIPK3 WT and RIPK3 K55/363R-expressing cells was similar. Treatment with birinapant and emricasan had little effect on the growth of NB4 cells expressing RIPK3 WT. However, in cells expressing the RIPK3 K55/363R mutant, treatment with these agents induced a marked reduction in tumor size, as measured by both volume and weight (Fig. 7H–J). The expression levels of RIPK3 WT or RIPK3 K55/363R were measured in each xenografted tumor (Fig. S13D).
Furthermore, we investigated whether USP5 overexpression can serve as a therapeutic strategy for tumor elimination via necroptosis. Our results confirmed that USP5 overexpression accelerates necroptosis in HT-29 cell lines (Fig. S14A, B). To further explore this effect, we established NB4 cell lines expressing RIPK3 WT + Mock, RIPK3 WT + USP5, and RIPK3 K55/363R + USP5. The results demonstrated that USP5 and RIPK3 WT overexpression reduced tumor growth and weight by enhancing necroptosis when compared with RIPK3 WT alone (Fig. S115A–D). As expected, there was no difference in tumor sizes when USP5 was overexpressed with RIPK3 WT or the K55/363R mutant (Fig. S15A–D). These findings suggest that USP5 is a positive regulator of RIPK3-mediated necroptosis promotion.
Taken together, these findings provide strong evidence that disrupting SMURF1 and USP5’s inhibitory control over RIPK3 K63-linked polyubiquitination enhances necroptosis, leading to effective tumor suppression.
Discussion
Necroptosis, a form of regulated cell death, plays a critical role not only in maintaining tissue homeostasis but also in triggering inflammatory responses that activate the immune system. This process differs from apoptosis primarily due to its immunogenic nature, driven by the release of pro-inflammatory cytokines12,18. Necroptosis is executed through the phosphorylation of receptor-interacting protein kinase 3 (RIPK3), which activates mixed lineage kinase domain-like pseudokinase (MLKL). MLKL then permeabilises the plasma membrane, leading to the release of intracellular contents, including cytokines and damage-associated molecular patterns (DAMPs). Thus, RIPK3 serves as a key regulator of necroptosis, driving programmed cell death and subsequent immune responses.
In our study, we focused on the roles of SMURF1 and USP5 in regulating necroptosis through their impact on RIPK3. We demonstrated the opposing functions of SMURF1 and USP5, shedding light on the post-translational modifications of RIPK3 and their influence on necroptosis. SMURF1, identified as a crucial E3 ligase, facilitates K63-linked polyubiquitination of RIPK3. This modification appears to suppress necrosome formation, the complex essential for propagating necroptotic signals. Notably, SMURF1 deficiency leads to a marked acceleration of necroptosis, suggesting that K63-linked ubiquitination by SMURF1 serves as a regulatory checkpoint that prevents inappropriate necroptotic signaling under normal conditions. This restraint is crucial for avoiding pathological inflammation and tissue damage. Furthermore, the reversal of necroptosis acceleration by reintroducing functional SMURF1 underscores the importance of its E3 ligase activity. This not only confirms that SMURF1’s ubiquitin ligase function is vital for modulating RIPK3 but also highlights its potential as a therapeutic target in conditions where excessive necroptosis contributes to disease, such as chronic inflammatory and autoimmune disorders.
Conversely, USP5 acts as a deubiquitinase, removing K63-linked polyubiquitin chains from RIPK3 and thereby dampening the necroptotic process. The suppression of necroptosis observed in USP5 deficiency emphasizes its critical role in maintaining balance within the necroptotic pathway. By removing the ubiquitin chains added by SMURF1, USP5 counteracts the stabilization of RIPK3, promoting the disassembly of the necrosome when necessary. The ability to reverse necroptosis suppression by reintroducing functional USP5 further highlights the necessity of its deubiquitinase activity in regulating RIPK3 and necroptosis.
The dynamic interplay between SMURF1 and USP5 offers insight an into a finely tuned regulatory mechanism that controls RIPK3 ubiquitination and necroptosis. Our findings reveal an antagonistic relationship, where SMURF1-mediated ubiquitination and USP5-mediated deubiquitination together regulate necrosome formation and function. This balance ensures that necroptosis is triggered only in response to appropriate signals while preventing excessive cell death and inflammation. To investigate the interactions among RIPK3, SMURF1, and USP5, we performed immunoprecipitation assays in HEK 293 T and HT-29 cells, confirming their incorporation into the necrosome during necroptosis (Figs. 1C–E, 3A and S1B, S1C, S4B, S9A, S12D). Treatment with the RIPK3 kinase inhibitor GSK'872 disrupted their interaction, demonstrating that RIPK3 phosphorylation is essential for SMURF1 and USP5 binding (Fig. S11B). Overall, these findings suggest that the association of SMURF1 and USP5 with RIPK3 is a multi-step process, wherein RIPK3 phosphorylation enhances its interaction with SMURF1. This process is necessary, as phosphorylation-defective RIPK3 is unable to bind to SMURF1 (Fig. S4B). The SMURF1-mediated K63-linked polyubiquitination seems to suppress necroptosis by preventing RIPK3 self-association or its interaction with RIPK1 (Fig. S6A–C). Meanwhile, this polyubiquitination seems to facilitate USP5 recruitment into the necrosome complexes, thereby inducing deubiquitination of RIPK3 and promoting necroptosis (Figs. 1C, E, 5D, 6D and S10E, S11B). More detailed mechanistic studies are required in future to further identify these paradoxical phenomena of pro- or anti- cell death progress revolving around the polyubiquitination modification of RIPK3.
While examining the temporal regulation of RIPK3 ubiquitination, we found that K63-linked ubiquitination peaked at 2 h after necroptosis induction but declined after 3 h, while K27-linked ubiquitination progressively increased. This suggests the occurrence of dynamic and heterogeneous forms of RIPK3 polyubiquitination as necroptosis progresses. While our study highlights SMURF1 and USP5 as key regulators of K63-linked ubiquitination, which has a negative effect on necroptosis, it remains unknown whether other types of polyubiquitination could have different effects on necroptosis. Alternative modifications such as K27, K48, and mixed forms should be carefully examined with matching enzymes to further identify the highly dynamic nature of RIPK3 ubiquitination fine-tuning multiple regulatory pathways (Fig. S16A).
Importantly, our study also explores the therapeutic implications of modulating this pathway. We found that SMURF1 downregulation enhances necroptosis in leukemia cells, correlating with reduced tumor growth in xenograft models treated with birinapant and emricasan. This suggests that targeting components of the ubiquitination machinery can modulate necroptosis for therapeutic benefit, particularly in cancer treatments where apoptosis pathways are compromised. Our results highlight the potential of SMURF1 and USP5 as targets for therapeutic strategies in diseases where necroptosis plays a detrimental role. Modulating these pathways could offer benefits in both chronic inflammatory conditions and cancer, making this an exciting area for further investigation.
Future research is needed to further investigate the upstream mechanisms governing the balance of RIPK3 K63-linked ubiquitination by SMURF1 and USP5 and how this balance promotes or inhibits necroptosis. The interplay between SMURF1 and USP5 reveals a finely tuned regulatory system that controls necroptosis. In physiological conditions, the balance between SMURF1-mediated suppression and USP5-driven promotion of necroptosis may help fine-tune immune responses and maintain tissue homeostasis. Under circumstances where apoptosis is inhibited or insufficient—such as in cells treated with caspase inhibitors—this balance ensures necroptosis can act as an alternative cell death mechanism, allowing for the continued removal of damaged or infected cells and the activation of immune responses. The regulation of cytokine production during necroptosis adds another layer of complexity to this system. Recent studies, including ours, indicate that cytokine production can continue even after cells have lost membrane integrity and committed to necroptosis. This sustained cytokine production, dependent on the maintenance of endoplasmic reticulum integrity, plays a critical role in shaping the immune response to necroptotic cell death. The ability of necroptotic cells to continue producing cytokines after functional death suggests that necroptosis is not merely a passive release of cellular contents but rather a highly regulated process designed to optimize immune activation63.
SMURF1 and USP5 likely play a role in coordinating this extended cytokine production. By suppressing RIPK3 activation, SMURF1 may limit cytokine production, preventing excessive inflammation that could result in tissue damage. In contrast, USP5’s promotion of RIPK3 activation and necroptosis may prolong cytokine production, enhancing immune cell recruitment and the clearance of necroptotic cells. This coordination between cell death and cytokine production ensures that necroptosis not only removes damaged or infected cells efficiently but also signals the immune system to initiate an appropriate inflammatory response.
In cancer, the role of necroptosis is more complex. While necroptosis can promote immune responses leading to tumor clearance, some cancers may exploit SMURF1 to suppress necroptosis and evade immune detection. Inhibiting SMURF1 could restore necroptosis in tumor cells, facilitating their destruction and activating anti-tumor immunity. Conversely, USP5 could be targeted in cancers where excessive necroptosis contributes to a pro-inflammatory tumor microenvironment that supports tumor growth. Understanding the specific roles of SMURF1 and USP5 in different cancer types will be crucial for developing targeted therapies that manipulate necroptosis for therapeutic benefit.
Overall, the findings suggest that targeting SMURF1 or modulating RIPK3 ubiquitination through SMURF1 or USP5 could be a therapeutic approach in necroptosis-based cancer treatment (Figs. 7, S15). Enhancing necroptosis, particularly in cancer cells that evade apoptosis, through SMURF1 inhibition could help overcome resistance to conventional therapies and improve tumor eradication. Future studies are needed to explore the broader applicability of this approach across different cancer types and to develop therapeutic strategies that exploit the necroptotic pathway to combat cancer.
Methods
Cell culture, plasmids, and transfection
HT-29 (human colorectal carcinoma; HTB-38; ATCC, Manassas, VA, USA), TC-1 (mouse lung cancer cell line; CRL-2785; ATCC), and Molm-13 (acute myeloid leukemia; ATCC), NB-4 (acute myeloid leukemia; ATCC) cells were maintained in Roswell Park Memorial Institute (RPMI; HyClone, Chicago, IL, USA) in 5% CO2 at 37 °C. HEK 293T (human embryonic kidney; CRL-3216; ATCC), HeLa (cervical cancer cell line, ATCC) were maintained in Dulbecco’s modified Eagle’s medium (DMEM; HyClone) in 5% CO2 at 37 °C. All media were supplemented with 10% fetal bovine serum (HyClone) and 1% penicillin/streptomycin (Invitrogen, Carlsbad, CA, USA). All cells were tested mycoplasma contamination using e-Myco™ plus Mycoplasma PCR Detection Kit (#25237; Intron, Seongnam, Gyeonggi, South Korea).
pcDNA3-FLAG-RIPK1, RIPK3, MLKL, RIPK3 1-296, 294-518, pCS4-3xMYC-CHIP constructs were generated by subcloning the respective coding sequences into the indicated vectors31,61. pSV-Parkin-myc was provided by H. W. Lee (Yonsei University, Korea). The SMURF1 and USP5 plasmid was purchased from Sinobiological (Beijing, China, #HG13144-NY, MG51371-CF). pcs4-3xMYC-SMURF1 mutants (C699A), pcs4-3xMYC-USP5 mutants (C335A) were generated using site-directed mutagenesis and PCR, respectively. Each domain of SMURF1 plasmid construct were generated using PCR (SMURF1 1-159, 119-731, 1-345, 346-731, 119-365) subcloned into the pcDNA3-HA. Each domain of USP5 plasmid construct and pcDNA3-GFP-USP5 were generated using PCR (USP5 1-289, 1-625, 289-835, 625-835) subcloned into the pcDNA3-HA. The pRK5–HA/Ub construct was provided by Jinho Seo (KRIBB, Korea). The point mutant constructs (K55R, K197R, K302R, K363R, K518R, K55/363R of RIPK3) were generated by site-directed mutagenesis. pcDNA3-FLAG-RIPK3-ΔRHIM were generated using PCR and subcloned into the pcDNA3-FLAG plasmid. GFP-USP22 was provided by Chung, Kwang Chul (Yonsei University, Korea). TRIM25 plasmid was purchased from Sinobiological (Beijing, China, #HG17528-CF) subcloned into pCS4-3xMYC Plasmid. pDEST-LTR-N-FLAG-HA-IRES-puro-USP5 was used for Stable USP5 overexpression HT-29 Cell Lines.
Plasmids were transfected using polyethylenimine (#408727; Sigma-Aldrich, St. Louis, MO, USA). For making Re-constitution of SMURF1 WT, SMURF1 C699A, USP5 WT, USP5 C335A and over expression of RIPK3 WT, K55R, K363R, K55/363R using the adenoviruses were purchased from VectorBuilder (Chicago, IL, USA). The siRNAs used in this study were siSmurf1 (Dharmacon, CO, USA; LQ-040947-00-0005,0007), siUsp5 (#LQ-046492-01-0009,0011; Dharmacon). Lipofectamine RNAiMAX reagent (#13778150; Thermo Fisher, Waltham, MA, USA) was used to transfect the siRNAs according to the manufacturer’s instructions.
Stable cell line generation
pLKO.1 puro-shSMURF1 #4 and pLKO.1 puro-shSMURF1 #8 were generated using oligo annealing and cloning into an empty vector using following primers: shSMURF1 #4F 5′-CCGGCCGAAGGCTACGAACAAAGAACTCGAGTTCTTTGTTCGT AGCCTTCGGTTTTTG-3′, R 5′-AATTCAAAAACCGAAGGCTACGAACAAAGAACTC GAGTTCTTTGTTCGTAGCCTTCGG-3′, shSMURF1 #8 F 5′- CCGGGCATCGAAGTGTC CAGAGAAGCTCGAGCTTCTCTGGACACTTCGATGCTTTTTG-3′, R 5′-AATTCAAA AAGCATCGAAGTGTCCAGAGAAGCTCGAGCTTCTCTGGACACTTCGATGC-3′. pLKO.1 puro-shUSP5 #1 and pLKO.1 puro-shUSP5 #2 were generated using oligo annealing and cloning into an empty vector using following primers: shUSP5 #1 F 5′- CCGGGACC ACACGATTTGCCTCATTCTCGAGAATGAGGCAAAT CGTGTGGTCTTTTTG-3′, R 5′- AATTCAAAAAGACCACACGATTTGCCTCATTCTCG AGAATGAGGCAAATCGTGTGGTC-3′, shUSP5 #2 F 5′- CCGGCGGGCCACGAACAA TAGTTTACTCGAGTAAAC TATTGTTCGTGGCCCGTTTTTG-3′, R 5′- AATTCAAA AACGGGCCACGAACAATA GTTTACTCGAGTAAACTATTGTTCGTGGCCCG-3′.
For virus production, HEK 293T cells were transfected with lentiviral vector and packaging vectors using Lipofectamine 3000 for 48 h, and the supernatants were harvested, filtered, and transferred into HeLa, HT-29, Molm-13, NB-4 cells. Especially, Molm-13 and NB-4 cells are Spin in balanced centrifuge for 2 h at 2400 rpm at 32 °C with virus and discard supernatants. Select cells infected with the virus by treating them with 1–2 μg/mL puromycin for 7 days. After puromycin selection, selection was performed in order to identify SMURF1 and USP5 knockdown cells, verified by immunoblotting.
Immunoprecipitation and immunoblotting
Cells were lysed in a buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.5% Triton X-100, 1 mM EDTA, and a protease inhibitor cocktail. The cell lysates were subjected to immunoprecipitation by incubating with antibodies for 2 h, followed by 2-h incubation with protein G Sepharose (GE Healthcare, Little Chalfont, UK). The immunoprecipitates were then boiled in 2× sample buffer for 5 min. For immunoprecipitation, the following antibodies were used: SMURF1 (#sc-100616; Santacruz), RIPK3 (#13526; Cell Signaling Technology), USP5 (sc-390943; Santacruz), TNFR1 (sc-8436; Santa Cruz), mouse RIPK3 (NBP1-77299; Novus Biologicals), HA (11867431001; Sigma), FLAG (F3165; Sigma), and Anti-FLAG M2 Affinity Gel (A220; Sigma). Normal rabbit IgG (sc-2027) and goat IgG (sc-2028) from Santa Cruz Biotechnology (Santa Cruz, CA, USA), normal mouse IgG (#sc-2025: Santa Cruz) served as controls.
Complexes were purified by immunoprecipitation and followed by immunoblot analysis: RIPK1 (#610459; BD Biosciences), RIPK3 (#13526; Cell Signaling Technology), MLKL (#GTX107538; GeneTex), phospho-RIPK1 Ser166 (#65746; Cell Signaling), phospho-RIPK3 Ser227 (#ab209384; Abcam), phospho-MLKL Ser358 (#91689S; Cell Signaling), mouse RIPK3 (NBP1-77299; Novus Biologicals), mouse MLKL (AP14272b; Abgent), phospho-mouse MLKL Ser 345 (#37333; Cell Signaling), SMURF1 (#sc-100616; Santacruz), USP5 (sc-390943; Santacruz), IKKβ (#2370, Cell Signaling), IκBα (#4814, Cell Signaling), phospho-IκBα (Ser539) (#2859, Cell Signaling), TNFR1 (#AF225;R&D Systems), TRADD (#610572; BD transduction), Ubiqutin (P4D1) mouse mAB(HRP conjugate) (#14049, Cell Signaling), K63-linkage Specific Polyubiquitin (D7A11) Rabbit mAb (HRP Conjugate) (#12930, Cell Signaling), K27-linked Ub (ab181537, Abcam),Actin (#A5316; Sigma-Aldrich), HA-Peroxidase (#12013819001; Sigma-Aldrich), FLAG-Peroxidase (#SAB4200119; Sigma-Aldrich), MYC (#sc-40: Santacruz) were utilized. All antibodies were used at a 1:2000 dilution in 5% BSA in TBST solution.
Chemicals and cell death stimulation
DMSO (#D8418; Sigma-Aldrich), Human TNFα (210-TA-020; R&D Systems), mouse TNFα (14-8321-62; eBioscience), birinapant (S7015; Selleck Chemicals), zVAD-fmk (#FKM001; R&D Systems), RIPK1 inhibitor GSK'963 (AOB9775; Aobious), GSK'872 (530389; Calbiochem), Emricasan (#BML2227; Sigma-Aldrich) for using necroptosis stimulation. To induce necroptosis, HT-29 cells were treated with 20 ng/ml TNF-α, 2 μM birinapant, and 20 μM zVAD-fmk. The HeLa-RIPK3 WT, K55R, K363R, K55/363R cells were treated with 20 ng/ml TNF-α, 2 μM birinapant, and 20 μM zVAD-fmk. The TC-1 cells were treated with 20 ng/ml mouse TNF-α, 2 μM birinapant, 20 μM zVAD-fmk. The Molm-13 cells were treated with 3 μM birinapant and 30 μM emricasan. The NB-4 cells were treated with 2 μM birinapant and 20 μM emricasan. To inhibit RIPK1, 0.1 μM GSK’963 were used.
Flow cytometry
To determine cell death, HT-29 and TC-1 cells using Annexin V and 7-AAD double staining. The treated cells were collected and washed with PBS, then incubated with Annexin V-FITC (556547; BD Biosciences) and 7-AAD (00-6993-50; eBioscience) in Annexin V binding buffer (51-66121E; BD Biosciences) for 15 min. The HeLa-RIPK3 WT, K55R, K363R, K55/363R, Molm-13 and NB-4 cells were using propidium iodide (PI) single staining. The treated cells were collected and washed with PBS, then incubated with PI (P4170; Sigma-Aldrich) for 15 min. Dead cells were identified as the PI-positive population. The stained cells were analyzed by flow cytometry (BD Accuri C6, BD Biosciences). Data analysis was performed using BD Accuri C6 Plus software (BD Biosciences). All cell viability assays were conducted in triplicate, with results presented as the mean ± standard deviation.
Immunofluorescence microscopy
For immunofluorescence staining, HT-29 cells were cultured on coverslips in 12-well plates. After treatment with the indicated reagents, the cells were fixed with 4% paraformaldehyde for 30 min at room temperature. Anti-SMURF1 (#sc-100616; Santa Cruz), USP5 (sc-390943; Santa Cruz), and p-RIPK3 (S227) antibody (ab209384; Abcam) were added to each sample in immunofluorescence blocking buffer (PBS with 3% BSA, 1% saponin, and 1% Triton X-100) and incubated at 4 °C overnight. Each sample was then washed three times with PBS. Alexa Fluor 594-conjugated anti-rabbit (Invitrogen, A-21207) or Alexa Fluor 488-conjugated anti-mouse (Invitrogen, A-11029) were added to each sample in immunofluorescence blocking buffer and incubated at room temperature for 1 h. The nuclei were stained with DAPI for 5 min and analyzed using an LSM980 (Carl Zeiss, Oberkochen, Germany) with the ZEN (blue edition) program.
Gel filtration chromatography analysis
HT-29 cells were treated with hTNFα (20 ng/mL), Birinapant (2 µM), and Z-VAD-FMK (20 µM). Cells were collected from ten 100 mm culture dishes and lysed with 5 mL of lysis buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.5% Triton X-100, and 1 mM EDTA. The lysates were processed using an AKTA Prime Plus and separated on a HiLoad 16/600 Superdex 200 column at a flow rate of 0.4 mL/min (GE Healthcare). Fractions of 2 mL were collected from 42 to 102 mL, and samples were analyzed by western blotting with the indicated antibodies. Molecular weights were determined using gel filtration calibration kits (GE Healthcare).
In vitro ubiquitylation assay
FLAG–RIPK3, HA-SMURF1, and HA-USP5 purified from transfectioned HEK293T and were incubated in 100 ng of an E1 (UBE1; E-305; Boston Biochem), 250 ng of an E2 (UbcH5c; E2-627; Boston Biochem), 5 μg of ubiquitin (Sigma), and 2 mM ATP (Fermentas) in the absence or presence of HA-SMURF1 and HA–USP5 in 40 mM Tris-HCl (pH 7.6), 50 mM NaCl, and 1 mM dithiothreitol for 3 h. After this reaction, the samples were boiled in ×4 sample buffer and analyzed by western blotting using anti-RIPK3, SMURF1, and USP5 antibodies.
In-gel digestion with trypsin and extraction of peptides
The pcDNA3-FLAG or pcDNA3-FLAG-RIPK3 plasmids were transfected into HEK293T cells seeded in twelve 100-mm dishes. After 24 h of transfection, cells were collected from ten 100 mm culture dishes and lysed with 5 mL of lysis buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.5% Triton X-100, and 1 mM EDTA. The cell lysates were subjected to immunoprecipitation by incubating with RIPK3 antibodies for 2 h, followed by a 2-h incubation with protein G Sepharose (GE Healthcare, Little Chalfont, UK). The immunoprecipitates were then boiled in 2× sample buffer for 5 min. For protein visualization, SDS-PAGE and Coomassie Blue staining were performed using Bio-Rad labware (Hercules, CA, USA). Protein bands from SDS-PAGE gels were excised and digested in-gel with trypsin. Briefly, protein bands were excised from stained gels, cut into pieces, and washed for 1 h at room temperature in 25 mM ammonium bicarbonate buffer (pH 7.8) containing 50% (v/v) acetonitrile (ACN). After dehydration in a centrifugal vacuum concentrator (SpeedVac™ SPD1030, Thermo Scientific) for 10 min, the gel pieces were rehydrated in 20 ng of sequencing-grade trypsin solution (Promega, Madison, WI, USA). Following overnight incubation at 37 °C in 25 mM ammonium bicarbonate buffer (pH 7.8), the tryptic peptides were extracted with 100 µL of 1% formic acid (FA) containing 50% (v/v) ACN for 20 min with mild sonication. The extracted solution was then concentrated using a centrifugal vacuum concentrator. Prior to mass spectrometric analysis, the peptide solution was desalted using a reversed-phase column. Briefly, after equilibration with 10 µL of 5% (v/v) formic acid, the peptide solution was loaded onto the column, washed with 10 µL of 5% (v/v) formic acid, and eluted with 8 µL of 70% ACN containing 5% (v/v) formic acid.
Identification of proteins by LC-MS/MS
LC–MS/MS analysis was performed through nano ACQUITY UPLC and LTQ-orbitrap-mass spectrometer (Thermo Electron, San Jose, CA). The column used BEH C18 1.7 μm, 100 μm ×100 mm column (Waters, Milford, MA, USA). The mobile phase A for the LC separation was 0.1% formic acid in deionized water and the mobile phase B was 0.1% formic acid in acetonitrile. The chromatography gradient was set up to give a linear increase from 10% B to 40% B for 16 min, from 40% B to 95% B for 8 min, and from 90% B to 10% B for 11 min. The flow rate was 0.5 µL/min. For tandem mass spectrometry, mass spectra were acquired using data-dependent acquisition with full mass scan (300-2000 m/z) followed by MS/MS scans. Each MS/MS scan acquired was an average of one microscans on the LTQ. The temperature of the ion transfer tube was controlled at 275 °C and the spray was 2.3 kV. The normalized collision energy was set at 35% for MS/MS. The individual spectra from MS/MS were processed using the SEQUEST software (Thermo Quest, San Jose, CA, USA) and the generated peak lists were used to query in house database using the MASCOT program (Matrix Science Ltd., London, UK). We set the modifications of Carbamidomethyl (C), Deamidated (NQ), Oxidation (M) for MS analysis and tolerance of peptide mass was 10 ppm. MS/MS ion mass tolerance was 0.8 Da, allowance of missed cleavage was 2, and charge states (+2, +3) were taken into account for data analysis. We took only significant hits as defined by MASCOT probability analysis.
In vivo ubiquitination assay
Cells were lysed in 6M guanidinium-HCl buffer (pH 8) containing 5 mM N-ethylmaleimide (Sigma-Aldrich) to prevent deubiquitination. To assess endogenous ubiquitination of RIPK3, cells were harvested in PBS containing 10 nM NEM and lysed in 1% SDS by boiling for 10 min. Cell lysates were diluted to 0.1% SDS by adding lysis buffer containing protease inhibitors and NEM, and immunoprecipitated with anti-RIPK3 antibodies, followed by WB. Transferred proteins were denatured by soaking PVDF membranes with 6 M guanidine-HCl containing 20 mM Tris–HCl (pH 7.5), 5 mM mercaptoethanol and 1 mM PMSF for 30 min at 4 °C. Ubiquitinated RIPK3 was detected using HRP-conjugated anti-Ubiqutin (P4D1) mouse mAB(HRP conjugate) (#14049, Cell Signaling), K63-linkage Specific Polyubiquitin (D7A11) Rabbit mAb (HRP Conjugate) (#12930, Cell Signaling). An ubiquitination assay using HA-Ub was also performed by immunoprecipitation under denaturing conditions (1% SDS).
Xenograft study
The subcutaneous tumor xenograft model was established by resuspending 1 × 106 Molm-13 cells expressing shGFP and shSMURF1 #4 and 5 × 105 NB-4 cells expressing RIPK3 WT,RIPK3 K55/363R, RIPK3 WT + MOCK, RIPK3 + USP5, RIPK3 K55/363R + USP5 in 50 µL of PBS and 50 µL of Matrigel matrix (354234, BD Sciences). This mixture was then subcutaneously inoculated into the flanks of 6-week-old female BALB/c nude mice (Narabiotech). They were maintained under specific pathogen-free conditions with 22–26 °C, 40–60% humidity and 12 h light/12 h dark cycle and fed regular diet. Seven days or eight post-inoculation, mice bearing xenograft tumors were intraperitoneally injected with birinapant (2 mg/kg) and emricasan (1 mg/kg), as shown in Fig. 7E, I. On days 27, 45, and 22 after inoculation, the mice were euthanized, and tumor volumes and masses were measured using previously described methods30. Tumor growth was carefully monitored, and mice were sacrificed when the tumor volume reached 1500–2000 mm³. At no point did the tumor volume exceed this limit during the course of the experiments. All animal experiments involving the subcutaneous tumor xenograft model were approved by the Institutional Animal Care and Use Committee of the Laboratory Animal Research at Yonsei University (IACUC-A-202407-1881-01)
Statistics and reproducibility
The student’s t test was used to compare the differences between two independent groups, one-way analysis of variance (ANOVA) and two-way ANOVA were used for comparisons among three or more groups, and the results are presented as the mean values ± SD. All statistical tests were two-tailed. IB images and CBB staining gel were performed at least three times as independent experiments with similar results, and representative images are shown. Statistical analyses were performed using GraphPad Prism version 5.0 (La Jolla, CA, USA). All schematic diagrams in the article were generated by PowerPoint or BioRender.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Source data are provided with this paper. Protein mass spectrometry raw data are publicly available at PeptideAtlas under accession code PASS05893. Source data are provided with this paper.
References
Cho, Y. et al. Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell 137, 1112–1123 (2009).
Vandenabeele, P., Galluzzi, L., Vanden Berghe, T. & Kroemer, G. Molecular mechanisms of necroptosis: an ordered cellular explosion. Nat. Rev. Mol. cell Biol. 11, 700–714 (2010).
Newton, K. et al. Activity of protein kinase RIPK3 determines whether cells die by necroptosis or apoptosis. Science 343, 1357–1360 (2014).
Kaczmarek, A., Vandenabeele, P. & Krysko, D. V. Necroptosis: the release of damage-associated molecular patterns and its physiological relevance. Immunity 38, 209–223 (2013).
Moriwaki, K. et al. The necroptosis adaptor RIPK3 promotes injury-induced cytokine expression and tissue repair. Immunity 41, 567–578 (2014).
Galluzzi, L. et al. Molecular mechanisms of cell death: recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 25, 486–541 (2018).
Newton, K. RIPK1 and RIPK3: critical regulators of inflammation and cell death. Trends Cell Biol. 25, 347–353 (2015).
Pasparakis, M. & Vandenabeele, P. Necroptosis and its role in inflammation. Nature 517, 311–320 (2015).
Rodriguez, D. et al. Characterization of RIPK3-mediated phosphorylation of the activation loop of MLKL during necroptosis. Cell Death Differ. 23, 76–88 (2016).
Martens, S. et al. Sorafenib tosylate inhibits directly necrosome complex formation and protects in mouse models of inflammation and tissue injury. Cell Death Dis. 8, e2904–e2904 (2017).
Murphy, J. M. et al. The pseudokinase MLKL mediates necroptosis via a molecular switch mechanism. Immunity 39, 443–453 (2013).
Zhou, Y. et al. RIPK3 signaling and its role in regulated cell death and diseases. Cell Death Discov. 10, 200 (2024).
Shan, B., Pan, H., Najafov, A. & Yuan, J. Necroptosis in development and diseases. Genes Dev. 32, 327–340 (2018).
Kearney, C. J. & Martin, S. J. An inflammatory perspective on necroptosis. Mol. cell 65, 965–973 (2017).
Ye, K., Chen, Z. & Xu, Y. The double-edged functions of necroptosis. Cell Death Dis. 14, 163 (2023).
Wu X et al. MLKL-mediated endothelial necroptosis drives vascular damage and mortality in systemic inflammatory response syndrome. Cell. Mol. Immunol. 21, 1309–1321 (2024).
Snyder, A. G. et al. Intratumoral activation of the necroptotic pathway components RIPK1 and RIPK3 potentiates antitumor immunity. Sci. Immunol. 4, eaaw2004 (2019).
Seo, J., Nam, Y. W., Kim, S., Oh, D.-B. & Song, J. Necroptosis molecular mechanisms: recent findings regarding novel necroptosis regulators. Exp. Mol. Med. 53, 1007–1017 (2021).
Seo, J. et al. The roles of ubiquitination in extrinsic cell death pathways and its implications for therapeutics. Biochem. Pharmacol. 162, 21–40 (2019).
Mansour, M. A. Ubiquitination: friend and foe in cancer. Int. J. Biochem. Cell Biol. 101, 80–93 (2018).
Hershko, A. & Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 (1998).
Kulathu, Y. & Komander, D. Atypical ubiquitylation—the unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat. Rev. Mol. Cell Biol. 13, 508–523 (2012).
Callis, J. The ubiquitination machinery of the ubiquitin system. Arabidopsis Book 12, e0174 (2014).
Popovic, D., Vucic, D. & Dikic, I. Ubiquitination in disease pathogenesis and treatment. Nat. Med. 20, 1242–1253 (2014).
Liu, J. et al. Targeting the ubiquitin pathway for cancer treatment. Biochim. Biophys. Acta1855, 50–60 (2015).
Vittal, V., Stewart, M. D., Brzovic, P. S. & Klevit, R. E. Regulating the regulators: recent revelations in the control of E3 ubiquitin ligases. J. Biol. Chem. 290, 21244–21251 (2015).
Clague, M. J., Urbé, S. & Komander, D. Breaking the chains: deubiquitylating enzyme specificity begets function. Nat. Rev. Mol. Cell Biol. 20, 338–352 (2019).
Liu, L. et al. UbiHub: a data hub for the explorers of ubiquitination pathways. Bioinformatics 35, 2882–2884 (2019).
Lee, E.-W., Seo, J.-H., Jeong, M.-H., Lee, S.-S. & Song, J.-W. The roles of FADD in extrinsic apoptosis and necroptosis. BMB Rep. 45, 496–508 (2012).
Lee, E.-W. et al. Ubiquitination and degradation of the FADD adaptor protein regulate death receptor-mediated apoptosis and necroptosis. Nat. Commun. 3, 978 (2012).
Seo, J. et al. K6 linked polyubiquitylation of FADD by CHIP prevents death inducing signaling complex formation suppressing cell death. Oncogene 37, 4994–5006 (2018).
Lee, E. et al. USP11-dependent selective cIAP2 deubiquitylation and stabilization determine sensitivity to Smac mimetics. Cell Death Differ. 22, 1463–1476 (2015).
Deng, L., Meng, T., Chen, L., Wei, W. & Wang, P. The role of ubiquitination in tumorigenesis and targeted drug discovery. Signal Transduct. Target. Ther. 5, 11 (2020).
Basar, M. A., Beck, D. B. & Werner, A. Deubiquitylases in developmental ubiquitin signaling and congenital diseases. Cell Death Differ. 28, 538–556 (2021).
Song, L. & Luo, Z.-Q. Post-translational regulation of ubiquitin signaling. J. Cell Biol. 218, 1776–1786 (2019).
Cao, Y. & Zhang, L. A Smurf1 tale: function and regulation of an ubiquitin ligase in multiple cellular networks. Cell. Mol. life Sci. 70, 2305–2317 (2013).
Wrana, J. L. Signaling by the TGFβ superfamily. Cold Spring Harb. Perspect. Biol. 5, a011197 (2013).
Sluimer, J. & Distel, B. Regulating the human HECT E3 ligases. Cell. Mol. Life Sci. 75, 3121–3141 (2018).
Ebisawa, T. et al. Smurf1 interacts with transforming growth factor-β type I receptor through Smad7 and induces receptor degradation. J. Biol. Chem. 276, 12477–12480 (2001).
Murakami, K. & Etlinger, J. D. Role of SMURF1 ubiquitin ligase in BMP receptor trafficking and signaling. Cell. Signal. 54, 139–149 (2019).
Yamashita, M. et al. Ubiquitin ligase Smurf1 controls osteoblast activity and bone homeostasis by targeting MEKK2 for degradation. Cell 121, 101–113 (2005).
Fei, C. et al. Smurf1-mediated Lys29-linked nonproteolytic polyubiquitination of axin negatively regulates Wnt/β-catenin signaling. Mol. Cell. Biol. 33, 4095–4105 (2013).
Xia, Q., Li, Y., Han, D. & Dong, L. SMURF1, a promoter of tumor cell progression?. Cancer Gene Ther. 28, 551–565 (2021).
Lee, C.-S., Kim, S., Hwang, G. & Song, J. Deubiquitinases: modulators of different types of regulated cell death. Int. J. Mol. Sci. 22, 4352 (2021).
Avvakumov, G. V. et al. Two ZnF-UBP domains in isopeptidase T (USP5). Biochemistry 51, 1188–1198 (2012).
Gao, S.-T., Xin, X., Wang, Z. -y, Hu, Y. -y & Feng, Q. USP5: Comprehensive insights into structure, function, biological and disease-related implications, and emerging therapeutic opportunities. Mol. Cell. Probes 73, 101944 (2024).
Meng, J. et al. USP5 promotes epithelial-mesenchymal transition by stabilizing SLUG in hepatocellular carcinoma. Theranostics 9, 573 (2019).
Cai, B. et al. USP5 attenuates NLRP3 inflammasome activation by promoting autophagic degradation of NLRP3. Autophagy 18, 990–1004 (2022).
Lim, K.-H. & Baek, K.-H. Deubiquitinating enzymes as therapeutic targets in cancer. Curr. Pharm. Des. 19, 4039–4052 (2013).
Mann, M. K. et al. Structure–Activity Relationship of USP5 Inhibitors. J. Med. Chem. 64, 15017–15036 (2021).
Lee, S. B. et al. The AMPK–Parkin axis negatively regulates necroptosis and tumorigenesis by inhibiting the necrosome. Nat. Cell Biol. 21, 940–951 (2019).
Onizawa, M. et al. The ubiquitin-modifying enzyme A20 restricts ubiquitination of the kinase RIPK3 and protects cells from necroptosis. Nat. Immunol. 16, 618–627 (2015).
Roedig, J. et al. USP22 controls necroptosis by regulating receptor-interacting protein kinase 3 ubiquitination. EMBO Rep. 22, e50163 (2021).
Wang, M. et al. ATR/Chk1/Smurf1 pathway determines cell fate after DNA damage by controlling RhoB abundance. Nat. Commun. 5, 4901 (2014).
Pan, J. et al. USP5 facilitates non-small cell lung cancer progression through stabilization of PD-L1. Cell Death Dis. 12, 1051 (2021).
Seo, J. et al. Beclin 1 functions as a negative modulator of MLKL oligomerisation by integrating into the necrosome complex. Cell Death Differ. 27, 3065–3081 (2020).
Seong, D. et al. Identification of MYC as an antinecroptotic protein that stifles RIPK1–RIPK3 complex formation. Proc. Natl. Acad. Sci. USA 117, 19982–19993 (2020).
Lee, C.-S., Hwang, G., Nam, Y. W., Hwang, C. H. & Song, J. IKK-mediated TRAF6 and RIPK1 interaction stifles cell death complex assembly leading to the suppression of TNF-α-induced cell death. Cell Death Differ. 30, 1575–1584 (2023).
Chen, T. et al. HSP70 attenuates neuronal necroptosis through the HSP90α-RIPK3 pathway following neuronal trauma. Mol. Biol. Rep. 50, 7237–7244 (2023).
Johnston, A. N. et al. Necroptosis-blocking compound NBC1 targets heat shock protein 70 to inhibit MLKL polymerization and necroptosis. Proc. Natl. Acad. Sci. USA 117, 6521–6530 (2020).
Seo, J. et al. CHIP controls necroptosis through ubiquitylation-and lysosome-dependent degradation of RIPK3. Nat. Cell Biol. 18, 291–302 (2016).
Choi, S.-W. et al. PELI1 selectively targets kinase-active RIP3 for ubiquitylation-dependent proteasomal degradation. Mol. Cell 70, 920–935. e927 (2018).
Choi, M. et al. Immunogenic cell death in cancer immunotherapy. BMB Rep. 56, 275 (2023).
Acknowledgements
This study was supported by a grant from the National Research Foundation of Korea (NRF), funded by the Ministry of Science, ICT, and Future planning (MSIP) (No. NRF-2021R1A6A3A1304015, C.H.Hwang) in part by NRF grant funded by the Korean government (MSIT) (No. NRF-2020R1A5A1019023, J.S.), in part by a NRF grant (No. NRF-2023K2A9A2A1005899911, J.S.), in part by a NRF grant (No. RS-2024-00346972, J.S.), and in part by the Brain Korea 21 (BK21) FOUR program (C.H. Hwang).
Author information
Authors and Affiliations
Contributions
C.H. Hwang conceptualized the study, designed and conducted all experiments, analyzed the data, and wrote the manuscript. M. Lee and J.W. Kim prepared materials and conducted biochemical experiments. Y.W. Nam, G.H. Hwang, and H.S. Ryu performed the xenograft model experiments. J. Seo, E.-W. L. and H.W.K. designed and interpreted the experimental results. J.S. supervised the experiments and revised the manuscript. All authors approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Zhigao Wang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Hwang, C.H., Lee, M., Kim, J.W. et al. Opposing regulation of the K63-linked polyubiquitination of RIPK3 by SMURF1 and USP5 in necroptosis. Nat Commun 16, 7360 (2025). https://doi.org/10.1038/s41467-025-62723-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-62723-9