Abstract
Drosophila germ granules are enriched with mRNAs critical for development. Within them, mRNAs cluster through intermolecular interactions that may involve base pairing. Here we apply in silico, in vitro and in vivo approaches to examine the type and prevalence of these interactions. We show that RNA clustering can occur without extended sequence complementarity (stretches of six or more continuous complementary bases) and that mRNAs display similar level of foldedness within germ granules as outside. Our simulations predict that clustering is driven by scattered, surface-exposed bases, enabling intermolecular base pairing. Notably, engineered germ granule mRNAs containing exposed GC-rich complementary sequences within stem loops located in the 3′ untranslated region promote intermolecular interactions. However, these mRNAs are also expressed at lower levels, leading to developmental defects. Although germ granule mRNAs contain numerous GC-rich complementary sequences, RNA folding renders them inaccessible for intermolecular base pairing. We propose that RNA folding restricts intermolecular base pairing to maintain proper mRNA function within germ granules.
Similar content being viewed by others
Introduction
RNA granules are a type of biomolecular condensates that are enriched with proteins and RNAs and inextricably linked with post-transcriptional gene regulation1,2. RNA granules have been observed across species3,4,5,6,7,8, and often appear structured, with their proteins9,10,11,12,13,14,15 and RNAs3,16,17,18,19,20 condensing into clusters, which are composed of multiple macromolecules that assemble through intermolecular interactions, in vivo and in vitro.
Protein conformation and protein-protein interactions within protein clusters have been extensively studied21,22,23,24. Depending on the species, some proteins, like Poly(A)-binding protein (Pab1) and globular proteins, partially or completely unfold during condensation21,24. Others, such as Fused in Sarcoma and androgen receptor activation domain, transition to a more compact and folded state22,25. Regardless of their conformations, both in vitro and in vivo, these clusters require multivalent interactions driven by charge-charge, cation-π, π-π, dipole-dipole and hydrophobic interactions, facilitated by diverse amino acid sequence compositions23,26,27.
However, our understanding of RNA-RNA interactions and RNA conformation within RNA clusters remains limited and is mainly derived from in vitro studies. In these studies, in vitro transcribed RNAs self-assemble into visible clusters upon the addition of salts and crowding reagents. Notably, clustering occurs in the absence of other cellular components16,19,20,28,29, demonstrating the potential of RNAs to interact with each other. Like clustering of proteins, RNA clustering requires interaction multivalency30 and may involve sequence complementarity. For example, in the filamentous fungus Ashbya gossypii, the co-clustering of BNI1 and SPA2 mRNAs is facilitated by base pairing of exposed complementary sequences (“zipcodes”) shared between the two transcripts located in five distinct RNA regions28.
To facilitate RNA clustering, intermolecular base pairing may require melting of the RNA structure. Notably, repeat RNAs that cause human neurodegenerative disorders31,32,33 contain an array of exposed and repeated GC-rich sequences that induce RNA condensation in vitro and in vivo16. Computer simulations of repeat RNAs revealed that in the process of condensation, the hairpin structures formed by GC-rich repeats transition into an unfolded state34. This unwinding facilitates intermolecular base pairing, increases the multivalency of interactions and results in the formation of an extended interaction network. Similarly, guanidine riboswitches29 melt their secondary structures to augment intermolecular base pairing in silico or in vitro. Finally, melting of the secondary structures of antisense non-coding RNAs (ncRNAs) is thought to enable intermolecular base pairing with their sense mRNA partners, thereby facilitating enrichment of ncRNAs in stress granules20. These studies illustrate that multivalent intermolecular base pairing could be required for the formation of RNA clusters.
In vivo, mRNAs critical for Drosophila development enrich in germ granules located at the posterior of the developing embryo, where they are post-transcriptionally regulated35. Within these granules, the mRNAs form clusters that contain multiple transcripts derived from the same gene. In contrast, outside of the granules, these mRNAs are mostly single transcripts17,36. Notably, different RNA clusters do not mix with each other, and their interactions are instead homotypic in nature17,18,37. The mRNA concentration within these clusters is remarkably high (5–15 µM)36 suggesting dense packing of mRNAs, and potential involvement of intermolecular base pairing in their formation.
Our previous study using chimeric experiments and stochastic super-resolution microscopy (STORM) on germ granule mRNAs failed to detect dependence of RNA clustering on a particular RNA zipcode36. These findings differ from observations of BNI1 and SPA2 mRNAs, as well as the Drosophila oskar (osk), bicoid (bcd) mRNAs and human immunodeficiency virus (HIV) genomes, whose clustering relies on specific, often GC-rich complementary sequences (CSs) that are present as exposed loops on top of stems36,38,39,40,41,42,43,44,45. This structural arrangement facilitates recognition among mRNAs, leading to stable intermolecular base pairing and RNA clustering36,38,39,40,41,42,43,44,45. These results highlight the distinctive nature of intermolecular base pairing in Drosophila germ granules, setting it apart from the mechanisms observed in BNI1, SPA2, osk, bcd mRNAs and HIV genomes.
In this study, we characterize the type and prevalence of intermolecular interactions that occur within RNA clusters in Drosophila germ granules. While these interactions undoubtedly involve protein-protein, protein-RNA and RNA-RNA interactions, here we focus specifically on intermolecular base pairing. Importantly, the mechanisms resulting in the specificity for homotypic RNA clustering are unclear. However, we speculate that base pairing interactions that help build RNA clusters may also aid in specifying their composition.
Our work revealed that in vitro, within a protein-free environment, intermolecular base paring in RNA clusters does not require sequence complementarity of 6 or more consecutive nucleotides, which we refer to as extended complementarity. Furthermore, the foldedness of mRNAs within and outside of germ granules is similar. Finally, our in silico data predicted that the tendency of RNAs to engage in intermolecular interactions is independent of high sequence complementarity or significant unwinding of secondary structure. This crucial result could explain the lack of dependence of RNA clustering on a particular RNA sequence in germ granules we reported previously36. Finally, engineered germ granule mRNAs, with exposed GC-rich CSs, presented within stem loop structures, induce persistent base pairing in vitro and enhanced intermolecular interactions in vivo but also disrupt normal fly development. Notably, while flies expressing nos with stem loop structures exhibited major developmental defects, those with exposed GC-rich CSs within the stem loops displayed exacerbated phenotypes. Our study, from multiple perspectives, reveals that germ granule mRNAs employ RNA folding as a mechanism to fend off potential detrimental effects of stem loops and exposed GC-rich CSs, while facilitating multivalent intermolecular interactions and RNA clustering. Therefore, RNA folding may not only mediate the nature and prevalence of intermolecular interactions within RNA clusters but also ensure optimal gene functionality within the germ granules.
Results
Stable dimerization of shu-top and shu-bottom RNAs relies on extensive sequence complementarity in vitro
To begin characterizing the type and prevalence of intermolecular base pairing in RNA clusters, we first examined the type of base pairing that is required for stable RNA dimerization in vitro. To this end, we used the split broccoli system coupled with the chemical reporter DFHBI-1T46. This system involves top and bottom RNAs, each carrying half of the broccoli aptamer. When these two RNA segments dimerize, they form a stable broccoli structure detectable by fluorescence emitted by DFHBI-1T intercalated between base-paired top and bottom RNAs (Fig. 1a)46. We reasoned that the observation of a strong fluorescent signal generated by the broccoli system will report on intermolecular base pairing driven by extensive sequence complementarity during dimerization.
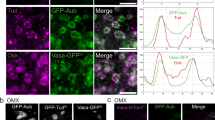
a Schematic and predicted structures of shu-top and shu-bottom RNAs. shu-top and shu-bottom RNAs contain split-broccoli sequences top (blue) and bottom (orange), and their intermolecular base pairing is recognized by DFHBI-1T (green or gray star) (see Supplementary Data S1 for sequences). The predicted structures were drawn by FORNA48. b In vitro dimerization of shu-top and shu-bottom RNAs. Top panel: RNA; bottom panel: DFHBI-1T signal. The ladder was run on a different gel. c Schematics of RNA clustering of shu-top and shu-bottom RNAs that is independent (i) or dependent (ii) of base pairing generated by extensive sequence complementarity or significant structural melting. The two models are distinguished by the absence (gray circle (i)) or presence (green circle (ii)) of DFHBI-1T fluorescence. d, e In vitro RNA clusters (magenta) formed of only shu-top or shu-bottom RNAs (negative controls), co-folded (positive control) or separately folded shu-top or shu-bottom RNAs (e) in the presence of DFHBI-1T (green). The DFHBI-1T images in (d, e) were normalized using the parameters of the co-folded DFHBI-1T image (see also Supplementary Fig. S1d for images normalized to the DFHBI-1T signal of shu-top). Initial RNA concentrations: 3.2 μM (negative controls), 1.6 μM for each RNA in co-folding and separate folding. Scale bars: 7.5 μm. f Normalized intensity of DFHBI-1T by RNA intensities (Supplementary Fig. S1a–c) from outside (gray) or inside (green) in RNA clusters per condition. n = 30 RNA clusters for each condition. Data: Mean ± SEM. n.s.: not significant. p-values (unpaired t test; two-tailed): 0.30 (shu-top), 0.47 (shu-bottom), 0.30 (co-folding) and 0.089 (separate folding). See also Supplementary Fig. S1 and Supplementary Data S1. Source data are provided as a Source Data file.
To place the broccoli aptamer in the sequence context of a germ granule mRNA, we inserted top and bottom RNAs into the 3′UTR of the Drosophila shutdown (shu) RNA, creating shu-top and shu-bottom (Fig. 1a and Supplementary Data S1). While shu accumulates in the germ granules, it does not form clusters within them36. In addition, shu-top and shu-bottom did not form homodimers in vitro (Fig. 1b), allowing us to study the formation of the broccoli aptamer without inference from the shu 3′UTR sequence.
RNAcofold47 predicted the correct broccoli structure when shu-top and shu-bottom were co-folded, while RNAfold47 predicted that top and bottom segments base paired intramolecularly when they were folded separately (Fig. 1a, the predicted structures were drawn using FORNA48). To validate the RNAcofold and RNAfold predictions that shu-top and shu-bottom form a correct broccoli structure detectable by DFHBI-1T, we used non-denaturing RNA gel electrophoresis following established protocols39,41. To this end, we heat-denatured and then co-folded shu-top and shu-bottom RNAs and observed a dimerized broccoli RNA accompanied by a strong DFHBI-1T fluorescence on the gel (Fig. 1b). In contrast, when we mixed shu-top and shu-bottom RNAs that were folded separately, the two RNAs did not dimerize and showed no DFHBI-1T fluorescence signal. Notably, the absence of DFHBI-1T fluorescence was similar to the one recorded for separately folded shu-top and shu-bottom, which were loaded individually on the gel as negative controls (Fig. 1b). Lack of a visible dimer band in samples containing only shu-top and shu-bottom RNAs, or when the two RNAs were folded separately and afterwards mixed, revealed that even though shu-top and shu-bottom RNAs shared extensive sequence complementarity, folded shu-top and shu-bottom RNAs lacked the potential for stable dimerization even at high RNA concentrations (3.2 μM) (Fig. 1b).
RNA clustering of shu-top and shu-bottom can occur without extensive sequence complementarity in vitro
We then examined if clustering of shu-top and shu-bottom relies on base pairing among complementary sequences, accompanied by unfolding of the RNA structure as observed for repeat RNAs16,34. To this end, we fluorescently labeled shu-top and shu-bottom RNAs and adapted existing in vitro RNA clustering protocols20,28. We hypothesized that if RNA clustering of shu-top and shu-bottom does not require intermolecular base pairing generated by extensive sequence complementarity, then the levels of DFHBI-1T fluorescence would be similar inside and outside the shu-top and shu-bottom clusters. This result would indicate that the RNA clusters do not promote the formation of additional broccoli structures (Fig. 1ci). However, if clustering of shu-top and shu-bottom RNAs relied on the intermolecular base pairing driven by extensive sequence complementarity, then we expect to observe a much higher DFHBI-1T fluorescence inside the clusters compared to outside (Fig. 1cii).
We first estimated the DFHBI-1T to shu-top/shu-bottom RNA dimer ratio to be approximately 30:1 in our reactions. This ratio ensured a large excess of the fluorescent reporter compared to the dimerized RNA and saturated detection of broccoli structures49 (“Methods”). Therefore, the intensity of the DFHBI-1T fluorescence should be scaled with the prevalence of broccoli structure.
To establish the baseline for the DFHBI-1T fluorescence inside and outside of RNA clusters, we first measured DFHBI-1T fluorescence in RNA clusters formed only by shu-top or shu-bottom, our negative controls (Fig. 1d, green signal). Importantly, the raw DFHBI-1T fluorescence outside (Supplementary Fig. S1a) and inside (Supplementary Fig. S1b) of RNA clusters for both RNAs was minimal compared to the positive control (see below) (Fig. 1d and Supplementary Fig. S1d). After normalizing the fluorescence to their respective RNA intensity inside and outside the clusters (Fig. 1d, magenta signal, Supplementary Fig. S1c; “Methods”), we did not observe significant difference in DFHBI-1T fluorescence between inside and outside the clusters for either negative control (Fig. 1f). This suggests that the increased DFHBI-1T intensity in RNA clusters formed by the two negative controls was primarily due to the higher RNA concentration within clusters, leading to an increased background signal.
As a positive control, we induced clustering of shu-top and shu-bottom RNAs that were first co-folded and generated stable RNA dimers (Fig. 1b). As anticipated, we observed a 30- to 89-fold increase in DFHBI-1T fluorescence outside the clusters, and a 64- to 80-fold increase inside the clusters, compared to shu-top or shu-bottom, respectively (Supplementary Fig. S1a, b). However, after normalization to the RNA intensity (Fig. 1d and Supplementary Fig. S1c), we observed no significant difference in DFHBI-1T levels between inside and outside of clusters (Fig. 1f). Therefore, the increased RNA concentration within the observable clusters generated by co-folded shu-top and shu-bottom RNAs did not stimulate the formation of additional broccoli dimers.
Finally, we examined DFHBI-1T levels in clusters formed by mixed but separately folded shu-top and shu-bottom RNAs. After normalization to the RNA intensity (Fig. 1d and Supplementary Fig. S1c), we observed no significant difference between the DFHBI-1T levels inside and outside the clusters (Fig. 1f), which also remained similar to the ones recorded for the negative controls. Thus, these results revealed that no additional broccoli structure formed between shu-top and shu-bottom when the two separately folded RNAs clustered. Moreover, we observed no significant differences in the size (Supplementary Fig. S1e) or morphology of RNA clusters (Fig. 1d, e) generated by co-folded and separately folded RNAs. However, based on the increase in RNA intensity, co-folded shu-top and shu-bottom RNAs formed denser RNA clusters (Supplementary Fig. S1c). Collectively, our data suggest that in vitro and in a protein-free environment, the clustering of shu-top and shu-bottom RNAs does not require intermolecular base pairing driven by extensive sequence complementarity nor significant RNA unfolding to expose sequences to accommodate such interactions.
The foldedness of 3′ untranslated regions of germ granule mRNAs is similar inside and outside of germ granules
Given our result with shu-top and shu-bottom RNAs, we reasoned that the absence of stable intermolecular interactions between mRNAs in RNA clusters in germ granules we recorded previously36, may also be due to RNA folding. Such folding may be independent of specific structures formed upon mRNA localization to germ granules. To test this hypothesis, we examined the base accessibility to interactions of germ granule mRNAs using dimethyl sulfate mutational profiling with sequencing (DMS-MaPseq) and probed the folded state of an RNA inside and outside of germ granules. DMS preferentially modifies accessible and unpaired adenosines and cytosines, which creates a mutational profile during reverse transcription50,51.
Exposure to DMS fragments RNA, making it susceptible to loss during subsequent germ granule purification. In addition, the vitelline membrane surrounding the embryos prevents the delivery of DMS. To circumvent these issues, we probed RNA with DMS using isolated germ granules. We purified germ granules marked by a fluorescently tagged core germ granule protein Vasa (Vasa:GFP) from embryo lysates, as done previously52,53. This approach yielded a pellet with germ granule-bound mRNAs as well as a soluble fraction that contained mRNAs outside the germ granules (Supplementary Fig. S1fi, ii). Importantly, germ granules resisted treatment with 10% DMS (Supplementary Fig. S1fii). Moreover, germ granule mRNAs nanos (nos), polar granule component (pgc), and germ-cell-less (gcl) exhibited 114, 117, and 88-fold enrichment in the pellet relative to the soluble fraction, respectively (Supplementary Fig. S1gi–iii), indicating that we were probing the structure of germ granule-bound mRNAs rather than the structure of mRNAs outside the granules.
We specifically focused on the 3′ untranslated regions (UTRs) of nos, pgc and gcl because these regions drive the localization of nos, pgc and gcl to germ plasm54,55. Furthermore, since germ granule mRNAs translate while associated with germ granules54,56,57,58,59,60, we anticipated that the RNA folding and the intermolecular interactions would be minimally perturbed by ribosomes within the 3′UTRs.
Focusing first on the 3′UTR of unlocalized nos, we recapitulated the previously characterized structure of its translational control element (TCE) (Fig. 2a)61,62,63. This result, together with the fact that DMS efficiently recapitulates the structure of the yeast 18S rRNA to 94% accuracy in vivo51, demonstrated the effectiveness of the DMS-MaPseq in probing the base accessibility of germ granule mRNAs in a proteinaceous environment in vivo.
a Predicted secondary structure of nos TCE outside the germ granules with a mapped normalized DMS reactivity generated by DMS-MaPseq. The normalized DMS reactivities were based on an average of two biological replicates (see Supplementary Fig. S1h, i for the correlation coefficients between the two replicates). b An example of the predicted secondary structure of nos 3′UTR outside the germ granules with mapped reactivity by DMS-MaPseq (an average of two replicates is shown). Blue dashed box: nos TCE. c Correlation of the normalized DMS reactivities between inside (y-axis) and outside of germ granules (x-axis). Each purple dot represents the DMS reactivity of the same probed base on nos 3′UTR recorded inside (y-axis) and outside of germ granules (x-axis). d The ratio of the normalized DMS reactivity between inside and outside of the germ granules for the same probed bases on nos 3′UTR. Black dotted line: no change in DMS reactivity. Blue dots and lines: ratio of DMS reactivities between inside and outside of germ granules for the same probed bases. Orange dashed lines represent thresholds with a p-value < 5.68 × 10−5 (adjusted for multiple hypothesis testing; two-tailed; see “Methods”), at which the nucleotides are significantly more exposed inside (above 1) or outside the granules (below 1). Gray blocks: no data due to gaps in PCR amplification (base positions: 595–657) or primer attachment (base positions: 1–22 and 847–880). The bases, which were significantly more exposed inside the granules, were labeled. e The genotypes of the female flies that laid the eggs for the experiments shown in eii (i). Schematic of a nos mRNA fused with a nos localization element + 2 and an antisense nos 3′UTR and a chimeric mRNA containing a CDS of IRFP fused with WT nos 3′UTR. The CDS of both mRNAs were hybridized with spectrally distinct smFISH probes (green for nos ORF and magenta for IRFP ORF) (ii). The two mRNAs display low co-localization in germ granules (iii), with a PCC(Costes) of 0.32 ± 0.01 (iv). n = 14 embryos. Data: Mean ± SEM. f Endogenous nos hybridized with spectrally-distinct smFISH probes (green and magenta), which alternately hybridized with the nos ORF(i), displaying high co-localization (ii), with a PCC(Costes) of 0.93 ± 0.01 (iii). n = 7 embryos. Data: Mean ± SEM. Scale bars: in all images 2 μm. This experiment served as a positive control to establish the PCC(Costes) threshold for high colocalization. It was conducted in a wild-type (WT) background. See also Supplementary Figs. S1–S3 and Supplementary Data S2 and S3. Source data are provided as a Source Data file.
We performed two biological replicates of DMS-MaPseq for RNA fractions from both outside and inside the germ granules. The Pearson correlation coefficients (r) between the two replicates of nos, pgc and gcl 3′UTRs outside of germ granules were 0.93, 0.97 and 0.99, respectively (Supplementary Fig. S1h), while inside of germ granules they were 0.99, 0.98 and 0.89 for nos, pgc and gcl 3′UTRs, respectively (Supplementary Fig. S1i). These results demonstrated a high biological reproducibility among DMS-MaPseq experiments.
We used DMS reactivity data to compare the foldedness of nos, pgc and gcl 3′UTRs inside and outside of germ granules. The average DMS reactivity of each probed base of nos, pgc, and gcl 3′UTRs inside the germ granules showed remarkable similarity to the average reactivity of the same base recorded for these mRNAs outside the germ granules (correlation coefficients of 0.92, 0.98, 0.82, respectively; Fig. 2b, c and Supplementary Fig. S2a–d, Supplementary Data S2). In addition, the predicted normalized ensemble diversity (NED) values for the 3′UTRs inside germ granules (0.16, 0.29 and 0.16, respectively) were almost identical to those outside (0.17, 0.34 and 0.19, respectively) (Supplementary Fig. S3). Thus, upon localization to germ granules, the diversity of possible RNA structures for the nos, pgc, and gcl 3′UTRs is largely unchanged compared to their structural diversity outside granules.
Next, we calculated the ratio of DMS signals of mRNAs inside and outside the germ granules to compare the base accessibility of the same probed bases. We observed no significant change in DMS reactivity in the nos 3′UTR, except for a few nucleotide bases scattered across the 3′UTR and at the AU-rich 3′ end (Fig. 2d). We found a similar result for the pgc 3′UTR (Supplementary Fig. S2e).
Notably, we recorded a more significant change in DMS reactivity in the first 300 nts of the gcl 3′UTR (Supplementary Fig. S2fi). However, these changes primarily occurred at the level of individual bases rather than continuous regions (Supplementary Fig. S2fii). These subtle changes in DMS reactivity may have led to slight adjustments in the base accessibility of the 3′UTRs of nos, pgc, and gcl upon localization, potentially influencing their predicted secondary structures (Supplementary Fig. S3). For example, the AU-rich 3′ end on the nos 3′UTR (base positions: 657–847 with 81% AU content) showed more changes in DMS reactivities between inside germ granules and outside (Fig. 2d), which may lead to different predicted secondary structures (Supplementary Fig. S3ai, aii; green lines). Together, our results suggest that nos, pgc, and gcl 3′UTRs are folded to a similar degree in both environments (Fig. 2c and Supplementary Fig. S2a–d).
To substantiate our findings further, we co-expressed two distinct mRNAs that exhibited 499 nts of perfect complementary in their 3′UTRs. Both mRNAs contained the + 2-localization element derived from nos 3′UTR, ensuring localization of both transcripts64 (Fig. 2ei, eii; “Methods”).
We reasoned that if base pairing between sense and antisense sequences occurred between the two transcripts, then they would assemble into the same cluster. This process would require the structure of the two RNAs to unfold to accommodate intermolecular base pairing. However, upon co-expression, the two mRNAs enriched in the same granule but formed distinct clusters (PCC(Costes) r: 0.32 ± 0.01; Fig. 2eiii, eiv). For comparison, we observed a high co-localization for a doubly-stained WT nos mRNA (PCC (Costes) r: 0.93 ± 0.01; Figure 2fi–iii and Supplementary Data S3). Therefore, our data suggest that in vivo, the 3′UTRs of mRNAs with high sequence complementarity have limited capacity for extensive intermolecular base pairing, possibly due to the folded nature of sense and antisense sequences. Contributing to de-mixing of sense and antisense mRNAs, our previous work36 suggests that the distinct global properties of these mRNAs may further promote their segregation into spatially distinct clusters, in addition to their folded RNA structures.
In silico model predicts that intermolecular base pairing in RNA clusters is driven by regions with low sequence complementarity
Our experiments showed that the foldedness of nos, pgc, and gcl 3′UTRs is similar inside and outside of germ granules (Figs. 1, 217,36);. However, the ability of RNAs to form clusters in the absence of other cellular components in vitro (Fig. 1d;16,20,28,29,30,65,66) nevertheless highlights their capacity for intermolecular interactions, including base pairing, to achieve multivalency. These interactions, which may occur on the nucleotide level, are challenging to examine directly using microcopy, in vitro RNA clustering assays and DMS-MaPseq probing.
To assess whether intermolecular base pairing can occur in RNA clusters and to explore its prevalence and properties, we simulated the tertiary structures of nos, pgc, and gcl 3′UTRs and their homodimers using our previously established minimal coarse-grained single-interaction site (SIS) model34. In this study, the SIS model is not intended to define precise in vivo structures of the nos, pgc, and gcl 3′UTRs, as it does not account for proteins, stacking interactions, or other cellular components that contribute to RNA structures, including other regions of the mRNA. Therefore, the simulated RNA structures may not be directly comparable to those predicted by DMS-MaPseq (see “Methods”).
Nonetheless, the simulation provides a useful framework for investigating modes of intermolecular base pairing that may be difficult to access experimentally beyond extended sequence complementarity for RNA clustering. Notably, its simplified environment makes RNA more accessible, thus enhancing the detection of interactions that may be masked in vivo. Importantly, the model also reproduced the known structure of the in vitro transcribed HIV U4/6 core region, supporting its ability to capture RNA folding under simplified conditions to some extent (Supplementary Fig. S4a–c67).
We first modeled the 3D structures of nos, pgc and gcl 3′UTR monomers. Our simulation showed that nos, pgc and gcl 3′UTRs adopt many distinct secondary and tertiary structures with different energy states (Fig. 3a, b and Supplementary Fig. S4d–g), revealing the structural heterogeneity of these RNAs in silico.
a Examples of simulated structures of monomeric nos 3′UTR at different energy states with an accompanying free energy spectrum, where each blue line represents a cluster of secondary structures generated in the simulation. Shown is a 3D conformation of nos 3′UTR corresponding to the ground state (G = 0 kcal/mol). b Intramolecular base pairing (red lines) within nos 3′UTR in a monomer state. c Simulated base accessibility of each nucleotide of nos 3′UTR in a monomer state (top; blue line) and homodimer state (bottom; magenta and green lines). d Correlation of base accessibility between homodimer and monomer state of nos 3′UTR, with a Pearson correlation coefficient (r) of 0.92. e Intramolecular base pairing (red lines) within nos 3′UTR in a homodimer state. f Simulated nos 3′UTR homodimer (magenta and green). g Intermolecular base pairing (red lines) between two nos 3′UTRs is shown in (i), with a magnified view of example interacting sequences in (ii) (yellow and purple boxes). Note that the sequence orientation in the purple box differs for visualization purposes: in (ii), it is shown from 3′ to 5′, whereas in (i), it is displayed from 5′ to 3′. The sequences within these boxes are connected by intermolecular base pairing interactions (lines). The base pairing probabilities, represented by a red color gradient, are derived from an ensemble of homodimer simulations. Therefore, some bases on one strand may pair with multiple bases on the other strand. See also Supplementary Figs. S4–S6. Source data are provided as a Source Data file.
Next, we simulated intermolecular base pairing for nos, pgc and gcl 3′UTR homodimers. Here, a homodimer refers to two RNA molecules that interact with each other using one or more base pairs without a predefined structure, interaction strength, duration, or the involvement of a particular RNA sequence. We assumed RNA concentrations for nos, gcl and pgc 3′UTRs of 10.7 µM, 29.4 and 50.7 µM, respectively, which were similar to the concentrations reported for these RNAs within the RNA clusters in vivo (“Methods”36). While in vivo, RNA clusters often contain more than 2 mRNAs, we reasoned that a minimal, homodimer system should capture the type of intermolecular interactions occurring within more complex, higher-order RNA clusters. Notably, we detected minimal changes in base accessibility between monomeric and homodimeric states of nos, pgc and gcl 3′UTRs (r = 0.92, 0.73 and 0.81, respectively) (Fig. 3c, d and Supplementary Fig. S4h–k), which implied the absence of extensive disruption of RNA folding upon homodimerization. In support of this observation, 99%, 95% and 97% of the base pairing within homodimers remained intramolecular for nos, pgc and gcl 3′UTR, respectively (Fig. 3e, f and Supplementary Fig. S5a–d).
We observed that intermolecular base pairing within the nos, pgc and gcl 3′UTR homodimers was predominantly driven by discontinuous stretches of bases from regions with low sequence complementarity (fewer than 6 consecutive bases) and low base pairing probability (Fig. 3gi, gii and Supplementary Fig. S5ei–fii). Specifically, in nos 3′UTR homodimers, only 5 out of 22 interacting bases at the 5′ end of nos 3′UTR had a base pairing probability close to 0.4, while the remaining interacting bases had a base pairing probability of ~ 0.1 (Fig. 3gi). Similar results were also observed in pgc and gcl 3′UTR homodimers (Supplementary Fig. S5ei, fi). Because the likelihood of RNA unfolding and maintaining the same base pairing interactions in a homodimer is predicted to be low, we surmise that the formation of higher-order oligomers (e.g., trimers, tetramers) with this type of interaction would be even less likely. This is because this process requires three or more structured RNA molecules to partially unfold, exposing single strands for stable intermolecular base pairing, an energetically unfavorable process. Consequently, the likelihood of oligomerization driven by high sequence complementarity could be even lower than that of dimerization68.
In contrast to osk, bcd and HIV, which use a single GC-rich complementary sequence (CSs) to establish stable dimerization (66% (bcd)39,44 to 100 % (osk, HIV)) recoverable by the RNA gel38,40,41,42,43, our simulation showed that none of the sequences that engaged in intermolecular base pairing of nos, pgc and gcl 3′UTRs resembled the GC-rich CSs found on osk, bcd and HIV (Fig. 3gi, gii and Supplementary Fig. S5ei–fii). To determine whether the CSs on osk, bcd and HIV are specially designed to support a stable RNA dimerization or if any CSs of similar length would achieve the same outcome, we modified the HIV SL1 stem-loop and replaced its dimerization CS with CSs of the same length but varying GC content. We then inserted these modified stem-loops into the shu 3′ UTR, which is a dimerization inert RNA (Supplementary Fig. S6ai, ii). After in vitro transcription, we evaluated the dimerization of RNAs on a non-denaturing agarose gel following established protocols39,41. We observed strong dimerization only for CSs with 100% GC content (Supplementary Fig. S6ai, ii and Supplementary Data S1). Among the three tested CSs with 66% GC content, only one (GUGCAC) dimerized, while others, including those with lower GC contents, did not (Supplementary Fig. S6ai, ii and Supplementary Data S1). In addition, the concatenation of shorter (< 6nts) CSs with 100% GC content or AU-rich CSs, with a length of 14 nts, which is longer than any of the interacting AU-rich regions on nos, pgc and gcl 3′UTRs (Fig. 3gi and Supplementary Fig. S6ei, fi), did not promote stable dimerization (Supplementary Fig. S6b, c and Supplementary Data S1). These data establish that the length and GC content of a CS were the primary determinants of stable intermolecular base pairing in vitro. Consistent with these observations, our simulations showed that none of the nos, pgc and gcl 3′UTRs sequences engaged in intermolecular base pairing were driven by GC-rich CSs found in osk, bcd and HIV or their dimerization-competent variants (Fig. 3g and Supplementary Figs. S5e, f, S6a).
Taken together, our in silico experiments revealed that intermolecular base pairing within nos, pgc and gcl 3′UTR homodimers is driven by regions with low sequence complementarity. Moreover, the probability that the same base pairing interactions persist and recur in different simulated RNA structures is low.
Exposed GC-rich complementary sequences enhance interactions among engineered mRNAs in Drosophila Ras cells
osk, bcd, and HIV RNAs form dimers through exposed GC-rich CSs36,38,39,40,41,42,43,44. However, our in silico model, together with our previous work36 revealed that germ granule mRNAs do not employ specific RNA zipcodes for clustering (Fig. 3, S4, S5). To investigate the potential impact of exposed GC-rich CSs on germ granule mRNAs, we inserted four stem loop structures into shu 3′UTR, a dimerization-inert RNA (Supplementary Fig. S6aii), to enhance mRNA potential for intermolecular interactions (Supplementary Fig. S7ai). These stem loop structures exposed GC-rich CSs derived from the HIV RNA43,69,70 or the nos 3′UTR (Supplementary Fig. S7ai and Supplementary Data S1), hereon referred to as HIV and nos, respectively. As a control, we also generated a construct that contained a non-complementary sequence in the loop (termed non) (Supplementary Fig. S7ai and Supplementary Data S1). Therefore, non, HIV, and nos shared the same four stems but differed in the sequences exposed within the loops. Using in vitro transcribed RNAs and non-denaturing agarose gels, we confirmed that HIV and nos dimerized while non RNA did not (Supplementary Fig. S7aii).
To test whether HIV and nos CSs enhance intermolecular interactions of RNAs in vivo, we inserted these constructs into the non-coding LaczA and LaczB RNA sequences71, which allowed co-localization analysis of the two transcripts in Drosophila Ras cells using single molecule fluorescent in situ hybridization (smFISH) (Supplementary Fig. S7b–d). Importantly, in these experiments, an increased co-localization reveals heightened interaction potential among RNAs regardless of whether it is driven by protein-protein, protein-RNA or RNA-RNA interactions.
Applying Pearson’s Correlation Coefficient (PCC) approach, which reflects the linear relationship between intensities of fluorescently-labeled LaczA and LaczB RNAs, we demonstrated that LaczA and LaczB that contained HIV and nos CSs better co-localized (r(PCC): 0.57 ± 0.03 and 0.50 ± 0.02, respectively) than those that contained non-sequences (r(PCC): 0.33 ± 0.02) (Supplementary Fig. S7f). Importantly, this co-localization could not be explained by differential expression among the constructs (Supplementary Fig. S7g, h). Instead, these data confirmed that exposed GC-rich CSs promoted interactions among LaczA and LaczB RNAs in cells.
To verify that the increased co-localization we observed was not unique to HIV or nos CSs, we also generated constructs whose loops contained GC-rich sense and antisense sequences and inserted them into LaczA and LaczB RNAs (Supplementary Fig. S7e). As recorded for HIV and nos CSs, sense and antisense LaczA and LaczB RNAs better co-localized (r(PCC): 0.49 ± 0.03) than those with non-sequences, irrespective of their gene expression levels (Supplementary Fig. S7f–h). Therefore, GC-rich CSs promoted interactions among RNAs in Drosophila Ras cells.
Exposed GC-rich complementary sequences enhance interactions among engineered nos mRNAs in embryos
To examine the effect of exposed GC-rich CSs in flies, we inserted the HIV, nos and non constructs into 3′UTR of nos gene using CRISPR/Cas9-PhiC31 approach (termed nos-HIV, nos-nos and nos-non, respectively) (Fig. 4a). After eight rounds of crosses with balancer flies to remove possible off-target effects induced by guide RNAs, we crossed nos-non, nos-HIV, and nos-nos flies with nosBN flies, which limited expression of WT nos to early oogenesis72, and thus allowed examination of only the edited nos alleles during embryogenesis (Fig. 4bi, ii). Using DNA gel electrophoresis and the cDNA products extracted from eggs laid by nos-non/nosBN, nos-HIV/nosBN, and nos-nos/nosBN females, we determined that the modified nos transcripts were spliced normally (Fig.4biii and Supplementary Data S4). In addition, qRT-PCR analysis revealed that the 3′UTR processing of the nos-non, nos-HIV, and nos-nos mRNAs was similar (Fig. 4ci–cii and Supplementary Data S4).
a Schematics of the endogenous nos containing four stems with non-palindromic (nos-non), HIV- (nos-HIV) or nos-derived (nos-nos) palindromes in its 3′UTR (demarcated with an orange arrow). b Schematics of unspliced or spliced nos coding sequence (CDS) where the primer targeted regions were shown in magenta arrows (i). Schematic of the crossing scheme with the nos-non experiment shown as an example (ii). nos PCR products from the cDNAs (100 ng total input) of the embryos laid by WT/nosBN (top), nos-non/nosBN, nos-HIV/nosBN and nos-nos/nosBN flies (bottom). Three lanes per each genotype represent three biological replicates (iii). c Schematic of the primer targeting regions (1, 2, 3 and 4) on nos-non, which are also used for WT nos, nos-HIV and nos-nos (iii). The expression levels of each region are determined by qRT-PCR (ii). For biological replicates, n = 6 and 3 embryo samples for regions 1 and 3, 2 and 4, respectively. Data: Mean ± SEM. n.s.: not statistically significant. p-values: 0.0022 (region 1: nos-non/nosBN vs. nos-HIV/nosBN; Mann-Whitney; two-tailed; **), 0.94 (region 1: nos-non/nosBN vs. nos-nos/nosBN; Mann-Whitney; two-tailed), 0.10 (region 2: nos-non/nosBN vs. nos-HIV/nosBN; Mann-Whitney; two-tailed), 0.70 (region 2: nos-non/nosBN vs. nos-nos/nosBN; Mann-Whitney; two-tailed), 0.080 (region 3: nos-non/nosBN, nos-HIV/nosBN and nos-nos/nosBN; Kruskal-Wallis) and 0.30 (region 4: nos-non/nosBN, nos-HIV/nosBN and nos-nos/nosBN; Kruskal-Wallis). d Schematic of the Drosophila embryo and mRNA clusters in its germ granules. Created in BioRender. Trcek, T. (2025) https://BioRender.com/xdghx4d. e smFISH of nos-non, nos-HIV, and nos-nos mRNAs in soma of the embryos laid by nos-non/nosBN, nos-HIV/nosBN and nos-nos/nosBN females (top). Heat maps were generated based on the number of mRNAs per cluster (bottom). Scale bars: 5 μm. f Concentrations of nos-non (n = 7 embryos), nos-HIV (n = 6 embryos) and nos-nos (n = 6 embryos) mRNAs in soma. Data: Mean ± SEM. n.s.: not statistically significant. p-value: 0.24 (nos-non/nosBN, nos-HIV/nosBN and nos-nos/nosBN; Kruskal-Wallis). g Number of nos mRNAs per cluster in soma of the embryos laid by nos-non/nosBN (n = 7 embryos), nos-HIV/nosBN (n = 6 embryos) and nos-nos/nosBN (n = 6 embryos) flies. Data: Mean ± SEM. h smFISH of nos-non, nos-HIV, and nos-nos mRNAs in the germ plasm of the embryos laid by nos-non/nosBN, nos-HIV/nosBN and nos-nos/nosBN females (top). Heat maps were generated based on the number of mRNAs per cluster (bottom). Scale bars: 20 μm. i Concentrations of nos-non (n = 6 embryos), nos-HIV (n = 6 embryos) and nos-nos (n = 6 embryos) mRNAs in the germ plasm. Data: Mean ± SEM. n.s.: not statistically significant. p-values: 0.026 (nos-non/nosBN vs. nos-HIV/nosBN; Mann-Whitney; two-tailed; *) and 0.0022 (nos-non/nosBN vs. nos-nos/nosBN; Mann-Whitney; two-tailed; **). j Number of nos mRNAs per cluster in the germ plasm of the embryos laid by nos-non/nosBN (n = 6 embryos), nos-HIV/nosBN (n = 6 embryos) and nos-nos/nosBN (n = 6 embryos) flies. Data: Mean ± SEM. See also Supplementary Fig. S7 and Supplementary Data S4. Source data are provided as a Source Data file.
However, compared to unedited nos expressed in WT/nosBN eggs, the mRNA levels of nos-non, nos-HIV, and nos-nos were reduced by approximately 22-fold (Fig. 4biii; ci, cii). These data revealed that the changes to the 3′UTR interfered with the expression of nos mRNA (Fig. 4a; “Methods”). Therefore, in our experiments, the phenotypes of nos-non served as the baseline against which we evaluated the effects of HIV and nos GC-rich CSs on the gene expression of the engineered nos mRNAs.
Using smFISH, we first examined the clustering ability of nos-non, nos-HIV, and nos-nos mRNAs in embryos. Previous work showed that WT nos forms clusters only in germ granules (Fig. 4d)17,18,36,37. In contrast, only 46 ± 3.1% nos-HIV and 61 ± 6.0% nos-nos mRNAs appeared as single mRNAs outside of germ granules compared to 81 ± 1.9% detected for nos-non mRNA (Fig. 4e–g). Notably, the somatic expression of the three nos constructs was comparable (Fig. 4f), indicating that the somatic increase in RNA clustering of nos-HIV and nos-nos was not due to differences in mRNA concentration.
In addition, nos-HIV and nos-nos mRNAs exhibited a 1.9 and 2.3-fold higher enrichment in the germ plasm, respectively, compared to nos-non mRNAs (Fig. 4h–j). Moreover, only 38 ± 5.8% and 25 ± 3.2% for nos-HIV and nos-nos mRNAs, respectively, were single transcripts in germ plasm compared to 78 ± 5.4% detected for nos-non mRNAs (Fig. 4j). However, in the germ plasm, 85% of nos-HIV clusters and 92% of nos-nos clusters contained only up to five mRNAs per cluster, compared to unedited, WT nos, where 81% of clusters contained more than five mRNAs per cluster (Supplementary Fig. S7i)17. This difference is primarily due to the significantly lower expression levels of nos-HIV and nos-nos compared to WT (Fig. 4cii).
Together, these data demonstrated that mRNAs produced by all three CRISPR alleles, nos-non, nos-HIV and nos-nos, exhibited comparable efficiencies in splicing and 3′UTR processing. However, notably, nos-HIV and nos-nos mRNAs clustered better inside and outside of germ granules and better localized to them compared to nos-non mRNA.
Embryos expressing nos with stem loop structures in its 3′UTR exhibit reduced Nanos protein levels
Using western blot analysis, we observed that embryos laid by nos-non/nosBN, nos-HIV/nosBN, nos-nos/nosBN flies exhibited significantly reduced Nanos protein levels compared to those laid by nos-non/WT, nos-HIV/ WT, nos-nos/WT flies, which expressed one unedited nos allele (Fig. 5a, b). Notably, while their Nanos protein levels were comparable to each other and to the negative control nosdef/nosBN (Fig. 5a, b), which does not express nos mRNA during late oogenesis and embryogenesis72, the nos mRNA levels in these embryos were much higher than those recorded for nosdef/nosBN (Fig. 5c).
a Western blot using anti-Nanos and anti-β-actin (loading control) from the embryos laid by nos-non/WT, nos-HIV/ WT, nos-nos/WT, nos-non/nosBN, nos-HIV/nosBN, nos-nos/nosBN and nosdef/nosBN flies. The small amount of Nanos protein detected in nosdef/nosBN embryos likely carried over from early oogenesis, during which nosBN is expressed. b Quantification of Nanos protein levels normalized to β-actin. Four biological replicates were used. For biological replicates, n = 4 embryo samples for each condition. Data: Mean ± SEM. n.s.: not significant. p-values: 0.45 (nos-non/WT, nos-HIV/WT and nos-nos/WT; Kruskal-Wallis), 0.92 (nos-non/nosBN, nos-HIV/nosBN, nos-nos/nosBN and nosdef/nosBN; Kruskal-Wallis) and 0.029 (vs. nos-non/WT: nos-non/nosBN, nos-HIV/nosBN, nos-nos/nosBN and nosdef/nosBN; Mann-Whitney; two-tailed; *). c The levels of nos mRNAs from embryos collected in (b). The data from embryos laid by nos-non/nosBN, nos-HIV/nosBN, and nos-nos/nosBN flies were also used in Figure 4cii. For biological replicates, n = 3 biological embryo samples for nos-non/WT, nos-HIV/WT, nos-nos/WT and nosdef/nosBN. n = 6 embryo samples for nos-non/nosBN, nos-HIV/nosBN and nos-nos/nosBN. Data: Mean ± SEM. n.s.: not significant. p-values: 0.54 (nos-non/WT, nos-HIV/WT and nos-nos/WT; Kruskal-Wallis) and 0.024 (vs. nosdef/nosBN: nos-non/nosBN, nos-HIV/nosBN, nos-nos/nosBN; Mann-Whitney; two-tailed; *). d Representative images of cuticle preparations of embryos laid by nosBN/WT (n = 98 embryos), nos-non/WT (n = 116 embryos), nos-non/nosBN (n = 209 embryos), nos-HIV/nosBN (n = 132 embryos), nos-nos/nosBN (n = 188 embryos) and nosdef/nosBN flies (n = 120 embryos). Orange arrows point to abdominal segments. The percentages show the proportion of observed embryos exhibiting the specified phenotypes. Scale bars: 100 μm. See also Supplementary Fig. S8. Source data are provided as a Source Data file.
Embryos expressing nos with stem loop structures in its 3′UTR exhibit impaired embryogenesis
Nanos protein is essential for abdominal patterning and egg hatching of the embryo73,74. Given that the embryos laid by nos-non/nosBN, nos-HIV/nosBN and nos-nos/nosBN expressed low levels of Nanos protein (Fig. 5a, b), it is possible that the western blot was not sufficiently sensitive to detect subtle differences among them. To this end, we examined the phenotypes generated by three CRISPR nos alleles by assessing the abdominal segmentation using cuticle preparation and egg hatching rates, as described previously52,75.
We observed that control eggs laid by nosBN/WT females exhibited eight abdominal segments (Fig. 5d) and an average egg hatching rate of 86.2% ± 1.7% (Supplementary Fig. S8ai), consistent with the phenotype recorded for WT flies52. In contrast, eggs laid by nos-non/nosBN females formed between one and three abdominal segments (Fig. 5d), which also hatched less efficiently (2.7% ± 0.6% (n = 2280 eggs) (Supplementary Fig. S8aii)). Therefore, embryos expressing nos carrying stem loops within its 3′ UTR alone exhibited reduced nos mRNA and protein expression (Fig. 5a, c), which in turn significantly impaired fly embryogenesis.
Embryos expressing nos with exposed GC-rich complementary sequences within stem loop structures in its 3′UTR exhibit exacerbated phenotypes
We observed that 83% and 85% of the nos-HIV/nosBN and nos-nos/nosBN embryos formed only one abdominal segment, respectively, while the remaining embryos had no abdominal segments. This was better than nosdef/nosBN embryos, which did not form segments (Fig. 5d)72. Importantly, these phenotypes cannot be explained by the derepression of unlocalized nos, as this mutation allows the formation of a WT number of abdominal segments but leads to anterior defects61,76. Moreover, nos-HIV/nosBN and nos-nos/nosBN embryos hatched 13- and 24-times less efficient than nos-non/nosBN embryos (Supplementary Fig. S8aii) (0.20 ± 0.13% (n = 2568 eggs) and 0.11 ± 0.06% (n = 1698 eggs), respectively).
In addition, smFISH analysis revealed a similar posterior enrichment of pgc and gcl mRNAs in eggs laid by WT/nosBN, nos-non/nosBN, nos-HIV/nosBN and nos-nos/nosBN females (Supplementary Fig. S8b, c). Therefore, the phenotypes we observed were specific to localized nos-HIV/nosBN, nos-nos/nosBN and nos-non/nosBN mRNA and did not reflect a broader impairment of the germ granule function.
Together, our data revealed that a reduced mRNA and protein expression of the nos gene, triggered by insertion of stem loop structures, impaired embryonic development. However, embryos expressing nos with exposed GC-rich CSs within the stem loop structures in its 3′UTR exhibited more severe phenotypes.
Ovaries expressing nos with stem loop structures and GC-rich complementary sequences in its 3′UTR exhibit impaired germline stem cell maintenance
Nanos protein is also required for the maintenance of germline stem cells (GSCs) during oogenesis77,78. To investigate whether nos-non, nos-HIV and nos-nos had similar effects on female GSCs in ovaries, we crossed nos-deficient flies (nosdef), which lacked the nos gene, with nos-HIV, nos-nos, and nos-non flies. We first compared the ovary morphology of nos-HIV/nosdef, nos-nos/nosdef, and nos-non/nosdef females to their sibling controls (termed nos-HIV/WT, nos-nos/WT, nos-non/WT, respectively) at 3, 9, and 14 days after females were enclosed. While siblings formed round and fecund ovaries (Fig. 6a and Supplementary Fig. S8di–diii), ovaries of nos-non/nosdef females exhibited an intermediate phenotype, with 69%, 72% and 71% of the examined ovaries being smaller than the ovaries of siblings aged 3, 9 and 14 days (Fig. 6bi and Supplementary Fig. S8di). Strikingly, 100% and 81% of ovaries from nos-HIV/nosdef and nos-nos/nosdef 3-day-old females already displayed severe morphological defects, respectively (Fig. 6a, bii, biii) while their siblings in nos-HIV/WT and nos-nos/WT females had normal ovary morphology (Supplementary Fig. S8dii, iii). Therefore, mRNAs derived from the three CRISPR alleles were not acting as dominant negative mutations.
a Images of WT, intermediate and severe phenotypes of ovary morphology. Scale bars: 200 μm. b Percentages of examined ovaries with a WT, intermediate and severe phenotypes in nos-non/nosdef (n = 29, 25 and 24 ovaries) (i), nos-HIV/nosdef (n = 29, 26 and 22 ovaries) (ii) and nos-nos/nosdef (n = 31, 23 and 26 ovaries) (iii) females aged 3, 9 and 14 days (D3, D9 and D14, respectively). c Immunofluorescent images of germaria of nos-non/nosdef (i). Asterisks: GSCs. Blue: DNA/DAPI; green: germ cells immunostained with anti-Vasa antibody; magenta: spectrosomes immunostained with anti-1B1 antibody. Scale bars: 18 μm. Percent of germaria with 0, 1 or 2-3 GSCs in nos-non/nosdef (ii). n = 6, 7 and 7 germaria for D3, D9 and D14, respectively. d Immunofluorescent images of germaria of nos-HIV/nosdef (i). Asterisks: GSCs. Blue: DNA/DAPI; green: germ cells immunostained with anti-Vasa antibody; magenta: spectrosomes immunostained with anti-1B1 antibody. Scale bars: 18 μm. Percent of germaria with 0, 1 or 2-3 GSCs in nos-HIV/nosdef (ii). n = 7, 6 and 6 germaria for D3, D9 and D14, respectively. e Immunofluorescent images of germaria of nos-nos/nosdef (i). Asterisks: GSCs. Blue: DNA/DAPI; green: germ cells immunostained with anti-Vasa antibody; magenta: spectrosomes immunostained with anti-1B1 antibody. Scale bars: 18 μm. Percent of germaria with 0, 1 or 2-3 GSCs in nos-nos/nosdef (ii). n = 6, 6 and 7 germaria for D3, D9 and D14, respectively. f Aberrant germ cell morphology (green) in germaria derived from nos-HIV/nosdef and nos-nos/nosdef females at D9. In each condition, 1 out of 6 examined ovaries exhibited an apoptotic phenotype. Scale bars: 18 μm. See Supplementary Fig. S8. Source data are provided as a Source Data file.
To further examine the GSCs in the germaria of the three nos constructs, we performed immunostaining using anti-Vasa and anti-1B1 to mark germ cells and GSCs, respectively79. While 83%, 71% and 57% of nos-non/nosdef germaria had at least 1 GSC (Fig. 6ci, ii and Supplementary Fig. S8ei, ii; cells marked with asterisks), 86%, 67%, and 83% nos-HIV/nosdef germaria showed a significant depletion of GSCs in 3, 9- and 14-day-old females, respectively (Fig. 6di, ii and Supplementary Fig. S8fi, ii; cells marked with asterisks). Similarly, nos-nos/nosdef germaria displayed an attenuated GSC depletion compared to the nos-non/nosdef germaria and the nos-nos/WT sibling controls (Fig. 6ei, ii and Supplementary Fig. S8gi, ii; cells marked with asterisks). In addition, some nos-HIV/nosdef and nos-nos/nosdef germaria also presented aberrant germ cell morphology (Fig. 6f). Notably, these exacerbated phenotypes observed in nos-HIV/nosdef and nos-nos/nosdef were consistent with those recorded in flies lacking Nanos expression77,78. Together, these data indicated that ovaries expressing nos with stem loop structures in its 3′UTR exhibited GSC depletion, while those with exposed GC-rich CSs within the stem loops displayed more severe phenotypes, consistent with the observations recorded for the embryos.
GC-rich complementary sequences are embedded within the RNA structure in germ granule mRNAs
The additional developmental phenotypes triggered by exposed GC-rich CSs prompted us to examine the prevalence of these sequences in germ granule mRNAs. CSs consist of palindromes, inverted repeats (IRs) and sense-antisense sequences, which can base pair intermolecularly or intramolecularly to form RNA structures (Fig. 7a).
a Types of CSs that enable intermolecular and intramolecular base pairing between mRNAs. b Alignment and abundance of total (magenta boxes) and GC-rich CSs, which had a minimal length of 6 nts in nos, pgc, and gcl 3′UTRs. c Predicted secondary structures of nos 3′UTR with mapped total CSs (magenta, left) or GC-rich CSs with a minimum of 6 nucleotides (magenta, right) outside of the germ granules. The RNA structures were generated based on two DMS-MaPseq replicates. The green sequences (“CGGCCG” and “CUCGAG”) and lines were nos CSs used in nos-nos experiments. A zoom-in image of “CUCGAG” demonstrating that this sequence is partially embedded within the RNA structure. d Predicted secondary structures of pgc 3′UTR with mapped total CSs (magenta, left) or GC-rich CSs with a minimum of 6 nucleotides (magenta, right) outside of the germ granules. The RNA structures were generated based on two DMS-MaPseq replicates. e Predicted secondary structures of gcl 3′UTR with mapped total CSs (magenta, left) or GC-rich CSs with a minimum of 6 nucleotides (magenta, right) outside of the germ granules (i). An example of IR in which half of the sequence was base-paired intramolecularly (ii). The RNA structures were generated based on two DMS-MaPseq replicates. f Proposed model depicting that mRNAs remain folded within mRNA clusters in germ granules (panel 1). The interactions among germ granule mRNAs in clusters are controlled by RNA folding. These interactions are mainly driven by regions with low complementarity and exhibit low probability for sustained interactions (panel 2). Flies expressing germ granule mRNAs with stem loop structures (blue) exhibit major defects in embryogenesis and the maintenance of female germline stem cells (panel 3). Notably, those with exposed GC-rich CSs (red) within the stem loops display exacerbated phenotypes and enhanced intermolecular interactions among the mRNAs (panel 4). See also Supplementary Data S5–S7.
We set out to identify CSs that were similar to those that drive the dimerization of osk, bcd and HIV RNAs38,39,40,41,42,43,44. Since these three RNAs employ a palindrome of 6-nt (osk, HIV)38,40,41,42,43 or an IR of 12 nt (bcd)39,44 and given that only GC-rich CS drive stable dimerization (Supplementary Fig. S6ai, ii), we limited our search to CSs with a minimal length of 6 nts and 50% GC-content.
Our analysis revealed that the 3′UTRs of nos, pgc, and gcl were replete with CSs. Specifically, they contained 259, 27, and 53 CSs with a minimum length of 6 nts, respectively, of which 9, 3 and 11 had a GC content larger than 50%, respectively (Fig. 7b and Supplementary Data S5–S7). However, mapping of these CSs onto the 3′UTR structures of unlocalized nos, pgc, and gcl determined by the DMS-MaPseq (Fig. 2b and Supplementary Fig. S2a, b) revealed that all GC-rich CSs were embedded within the RNA structure (Fig. 7c–ei). The secondary structures predicted by our DMS-MaPseq analysis indicated that often, a portion of a CS was exposed within the RNA loop, while the remainder was base paired intramolecularly (Fig. 7eii). This configuration makes it highly unlikely that these CSs could base pair intermolecularly. Notably, the CGGCCG of nos used in nos-nos was completely embedded within the WT nos 3′UTR structure, while CUCGAG was partially embedded (Fig. 7c; green lines). In addition, the AU-rich 3′ end, which exhibited higher DMS reactivities inside the germ granules than outside (Fig. 2d), contained a pair of GC-rich inverted repeats (CUGGCG and CGCCAG). However, these sequences base paired intramolecularly both inside and outside the germ granules (Supplementary Fig. S3a, magenta).
Together, our analysis revealed that germ granule mRNAs contain many GC-rich CSs, which, when exposed, may be competent to form stable intermolecular base pairing in vitro. However, in vivo, these sequences were embedded within the RNA structure, thus rendering them inaccessible to intermolecular interactions. Thus, using a variety of methods, we have discovered an organizing principle in which the RNA structure shields germ granule mRNAs against potential detrimental effects of exposed GC-rich CSs, thereby preserving normal mRNA function and fly development.
Discussion
In this study, we investigated the type and prevalence of intermolecular base pairing in RNA clusters of Drosophila germ granules. We showed that in vitro and in the absence of proteins, RNAs can form clusters without relying on intermolecular base pairing driven by extensive sequence complementarity. We further demonstrated that the foldedness of mRNAs in germ granules remains similar to outside, where these mRNAs exist as single transcripts in vivo (Figs. 1, 2, 7f17,36). Importantly, our simulations predicted that within RNA clusters, intermolecular base pairing is driven by scattered and discontinuous stretches of bases, resulting from regions of low sequence complementarity. Moreover, the probability that the same base pairing interactions persist and recur is low (Fig. 3g and Supplementary Figs. S5e, f, 7f). Finally, engineered germ granule mRNAs with exposed GC-rich CSs presented within stem loops induce persistent base pairing in vitro (Supplementary Fig. S7a) and enhanced intermolecular interactions in vivo (Fig. 4e–j and Supplementary Fig. S7b–d). However, these mRNAs also exhibit reduced mRNA and protein expression, preventing normal fly development (Figs. 4–6 and Supplementary Fig. S8). Notably, while flies expressing nos with stem loop structures exhibited major developmental defects (Figs. 5, 6 and Supplementary Fig. S8), those with exposed GC-rich CSs within the stem loops displayed exacerbated phenotypes (Figs. 5, 6). Although germ granule mRNAs contain numerous GC-rich CSs (Fig. 7b) that could potentially engage in intermolecular base pairing in vivo, these sequences are sequestered within the RNA structure (Fig. 7c–eii). Thus, our findings underscore the protective role of germ granule mRNA folding in mitigating the potentially adverse effects of stem loop structures and exposed GC-rich CSs, thus preserving gene functionality in densely packed environments like germ granules (Fig. 7f).
RNA base pairing with low sequence complementarity may enable dynamic behavior of mRNAs within germ granules
Previous studies have shown that specific RNA sequences and motifs promote clustering28,38,39,40,41,42,43,44. For example, clustering of BNI1 and SPA2 mRNAs depends on intermolecular base pairing of exposed CSs28. In addition, repeat RNAs and riboswitches often rely on significant structural changes to facilitate clustering29,34. In contrast to these models, our data suggest that Drosophila germ granule mRNAs cluster through a different mechanism. Their clustering does not require specific sequence motifs or extensive structural melting for intermolecular base pairing.
Specifically, our simulations predict that base pairing among germ granule mRNAs is driven by scattered and discontinuous stretches of bases exposed on the surface of folded RNAs. While infrequent, these interactions may nevertheless provide multivalency of interactions and stabilize the RNA cluster. Importantly, our model predicts that the probability of the same intermolecular base pairing recurring within RNA clusters is low, suggesting that these interactions are malleable, which may support the formation of dynamic RNA clusters. This mode of interaction stands in direct contrast to the one observed in clusters composed of RNAs with repeat sequences, where RNAs within clusters readily unfold and expose single-stranded regions68, accommodating extended intermolecular base pairing34.
In addition, our in silico simulations reveal that the 3′UTRs of germ granule mRNAs adopt multiple secondary conformations (Fig. 3a, b and Supplementary Fig. S4d–g), contributing to the diversity of tertiary folding. This finding aligns with decades of RNA structure research80,81,82,83,84,85.
An important implication of these findings is that conformational heterogeneity enables multiple interaction combinations among mRNAs (Fig. 3g and Supplementary Fig. S5e, f). The multitude of these combinations, along with base pairing among regions with low sequence complementarity could serve two key purposes. First, it maintains multivalency for intermolecular interactions without melting of RNA structures within the dense granule environment. This could be crucial for preserving the functionality of germ granule mRNAs, including translation and localization86. Second, it prevents sustained intermolecular base pairing from any single structural conformation, thereby avoiding a network of stable RNA interactions and unfolded RNA structures, which could lead to detrimental cellular outcomes. Together, our study suggests that conformational heterogeneity ensures interaction multivalency within clusters while providing structural flexibility to the RNA, enabling it to dynamically respond to regulation in germ granules. This balance between the strength and frequency of intermolecular base pairing may be crucial for preventing the formation of pathological RNA aggregates observed in RNAs with expanded nucleotide repeats16,34 while enabling mRNAs to congregate and maintain their dynamic functions.
Intermolecular base pairing may help generate compositional specificity of RNA clusters
In Drosophila, individual germ granules contain multiple clusters derived from different genes. However, these clusters are homotypic and do not mix with each other17,18,36,37. In this study, we found that the foldedness of germ granule mRNAs is similar within their respective clusters compared to outside, and that intermolecular base pairing through low sequence complementarity may contribute to the multivalency required for clustering.
The mechanisms driving the formation of mRNA homotypic clusters in germ granules remained poorly understood. De-mixing of polymers into separate phases relies on intermolecular interactions among the macromolecules and their surrounding solution2,87. Therefore, it is plausible that intermolecular base pairing, as predicted by our in silico models, may also help provide the specificity for homotypic RNA clustering. However, in the complex environment of the embryo, specificity is likely governed by a combination of interactions, involving both proteins and RNAs. These predictions align with our previous findings, where we showed that the homotypic specificity of RNA clusters in Drosophila is dictated by the global properties of messenger ribonucleoproteins rather than specific mRNA sequences or features36.
Cellular importance of regulating stem loop structures containing exposed GC-rich complementary sequences
Strikingly, the defects in gene expression induced by the inserted stems containing exposed GC-rich CSs in engineered nos mRNAs are not observed in osk, bcd, and HIV RNAs38,39,40,41,42,43,44. These defects may result from an initial decrease in mRNA levels that subsequently leads to lower protein production or from a direct translational repression.
One possible explanation is that mRNAs containing stem-loop structures and GC-rich CSs may experience transcriptional repression or nuclear retention, similar to what has been observed for the trinucleotide repeat RNAs16,88.
Another possible explanation is that the RNA-binding proteins associated with these sequence motifs in osk, bcd, and HIV RNAs shield them from being recognized as abnormal RNAs, whereas the engineered nos lacks these protein protectors. Such a protective mechanism could apply broadly within the cellular context and is not restricted to germ granules or a specific cell type. The Drosophila Staufen (Stau) is an important regulator for osk mRNA localization and translation89, as well as bcd mRNA localization90. The exposed GC-rich CS within a stem loop in the osk mRNA 3′UTRs is predicted to bind Stau91,92. Similarly, the exposed GC-rich CS within a stem loop in the bcd 3′UTR overlaps with a Stau-binding motif90. These structural motifs not only enhance intermolecular mRNA base pairing but also serve as scaffolds to recruit RNA-binding proteins essential for post-transcriptional regulation.
In the case of HIV, the SL1 stem loop is bound by the nucleocapsid domain of the Gag protein, which facilitates genome dimerization for packaging in vivo93. In addition to stabilizing intermolecular base pairing between HIV genomic RNAs, Gag multimerizes to encapsulate the genome into virions, protecting the HIV genomes from host degradation and enhancing viral infectivity94. Therefore, one possible explanation is that flies may lack RNA-binding proteins that recognize these motifs on the engineered nos mRNAs, leading the embryo to perceive them as abnormal and subsequently downregulate their gene expression.
It is also unclear how flies expressing nos with exposed GC-rich CSs within stem loops generate exacerbated phenotypes (Figs. 5, 6). In embryos, nos translates only upon localization to germ granules56,57,60,95 and the Nanos protein produced is required for abdominal patterning of the embryo. Therefore, as shown previously, the efficiency of abdominal segmentations and egg hatching correlates with Nanos protein levels produced at the posterior pole73,74. Interestingly, nos-HIV and nos-nos mRNAs localize 1.9 to 2.3-fold better than nos-non mRNA (Fig. 4i), respectively, indicating that they should produce 1.9 to 2.3-fold more Nanos protein than nos-non, respectively. However, western blot analysis failed to reveal differences in protein levels among the three CRISPR lines. Since these protein levels were close to the detection limit (Fig. 5a, b), it is possible that the sensitivity of the western blot analysis was insufficient to capture finer differences in protein levels generated by the three CRISPR lines.
Two models may explain the exacerbated phenotypes by GC-rich CSs, which will be the focus of future studies. First, GC-rich CSs promote RNA clustering outside of germ granules (Fig. 4e–g). These sequences could form stable intermolecular interactions through base pairing, protein binding to exposed GC-rich CSs, or a combination of both. Since GC-rich CSs were engineered at the end of the nos 3′UTR, these interactions may interfere with the translation machinery, potentially by impeding ribosome release from the mRNA and its subsequent recycling. Alternatively, the exposed GC-rich CSs may recruit regulatory proteins that obstruct ribosome progression or recruitment on the mRNA.
RNA clustering has been observed in germ granules in C. elegans and zebrafish3,96 as well as P-bodies in C. elegans97. Given that the core functional and structural principles are shared among these RNA granules1,8, our findings suggest that RNAs in these granules might be similarly compacted. This compaction could provide a mechanism by which the folding of mRNA regulates intermolecular base pairing among diverse granule mRNAs. Interestingly, mRNAs become compacted upon localization to stress granules98,99. Therefore, along with RNA helicases19, this structural compaction could further regulate intermolecular base pairing within stress granules and protect mRNA functional integrity for the time when cellular stress is over and when stress granule mRNAs return to their normal cellular functions. Collectively, our findings highlight the role of RNA structure in safeguarding mRNAs across cellular environments as well as the role of proteins in organizing mRNAs in cells.
Methods
Fly stocks
Fly stocks and crosses were maintained at 25 °C on standard cornmeal/agar media. To test the base pairing capacity of mRNAs in vivo, we first crossed flies that expressed two nos-chimeras. The first chimera, termed antisense nos 3′UTR, expressed the endogenous nos gene fused with the 3′UTR composed of the + 2 localization element derived from the nos 3′UTR95 and antisense sequence of nos 3′UTR (region spanning nucleotides 658 to 159). This chimera was generated by initially inserting two ATTP sites into the last intron of the nos gene using CRISPR/Cas9 genome editing, which allowed subsequent site-specific recombination of ATTB-flanked DNA via PhiC31 recombination by Fungene, as described previously100. This approach enabled the reconstitution of the nos CDS and the simultaneous replacement of its 3′UTR with an RNA sequence of choice. The second chimera was transgenically expressed using a reporter mRNA fused with a CDS of a far-red fluorescent protein (IRFP670) and the 3′UTR of WT nos36. Its expression, driven by maternal alpha tubulin (matα) promoter, was induced by the Gal452. nos-antisense nos 3’UTR and IRFP-nos 3’UTR were co-expressed in nosBN flies, which lack nos mRNA expression during late oogenesis and embryogenesis72. This experimental setup allows smFISH probes targeting the nos open reading frame (ORF) to specifically detect only the nos with the antisense 3′UTR.
The nos-non, nos-HIV and nos-nos, which contain the edited nos 3′UTRs (Supplementary Data S1), were generated using CRISPR/Cas9 followed by PhiC31 recombination as described above. This process resulted in the replacement of the WT 3′ with the edited nos 3′UTRs, while the nos promoter, 5′UTR and its coding region remained unchanged compared to the WT endogenous sequences. After eight rounds of background crossing with balancer flies and subsequent removal of balancer chromosomes, these three constructs were crossed with nosdef 78 to study the effects on female GSCs. For embryo studies, the flies expressing these three constructs were crossed with nosBN 72, ensuring that the only source of nos mRNAs and Nos proteins during embryogenesis came from the three edited nos constructs. Sex was not considered in the study design because all ovaries and embryos were derived from female flies. Moreover, the embryos used were at early developmental stages, when determining the sex of individual embryos is not feasible.
Drosophila cell culturing
Drosophila Ras-attP-L1 cell line (DGRC Stock 249) was maintained at 25 °C as described previously101. Schneider’s insect medium (Sigma: S0146) supplemented with 10% FBS (Thermo Fisher: 26140079) and 1X penicillin-streptomycin (Thermo Fisher: 15070063) was used to maintain the cell line.
In vitro RNA transcription
DNA templates containing the T7 promoter for in vitro transcription were ordered from IDT and amplified by Q5 high-fidelity DNA polymerase (NEB: M0492L). The amplified DNA was purified with the Zymo DNA clean & concentrator kit (Zymo: D4003T). For in vitro RNA clustering assays, transcription templates ranging from 200–500 ng were transcribed in vitro using the MEGAscript T7 transcription Kit (Thermo Fisher: AM1333). 20 μL of transcription reaction was carried out for 6 hours (hrs) at 37 °C. Following the transcription reaction, each reaction mixture was treated with 1 μL of TURBO DNase (Thermo Fisher: AM2238) for 15 mins at 37 °C. The reaction was stopped with the ammonium acetate stop solution from the kit following the manufacturer’s instructions. Then the in vitro transcribed RNAs were purified using phenol:chloroform extraction (Thermo Fisher: 15593031) following the manufacturer’s protocol. The purified RNAs were stored in isopropanol (Thermo Fisher: 278475-1 L) at − 20 °C for a maximum of one month to prevent degradation.
In vitro intermolecular RNA base pairing assays and gel electrophoresis
The protocol was adapted from refs. 39,41. In vitro transcribed RNAs suspended in isopropanol were centrifuged at 14,000 × g for 15 mins at 4 °C to precipitate the RNAs. The RNA pellet was washed twice with 80% RNase-free ethanol (Thermo Fisher: BP2818500) by centrifugation at 14,000 × g for 2 mins at 4 °C. The RNA pellet was then air-dried and resuspended in 20 μL of nuclease-free water (Thermo Fisher: AM9937) in a 1.5 mL test tube on ice. The concentration and quality of the RNA were accessed using a NanoDrop spectrometer.
RNA samples resuspended in water were denatured at 90 °C for 2 mins, and afterward the sample tubes were immediately placed on the ice for at least 15 mins. For a reaction using one RNA sample, 32 pmol of denatured RNA was aliquoted into a 200 μL PCR tube and mixed with 2 μL of 5X RNA gel refolding buffer (50 mM sodium cacodylate (pH 7.5) (Electron Microscopy Science: 11654), 300 mM KCl (Sigma: 60128-250G-F) and 5 mM MgCl2 (Sigma: M1028-100ML) and nuclease-free water added to the final volume of 10 μL. The mixture was refolded at RT for 1.5 hrs. For a rection using two RNA species, 16 pmol of each RNA sample was mixed with 1 μL of 5X RNA gel refolding buffer and nuclease-free water to reach a final volume of 5 μL, and was refolded separately at RT for 1 hr. Then the two refolded RNA samples were combined and incubated for an additional 30 mins at RT. A 2% agarose gel supplemented with 1X SYBR safe (Thermo Fisher: S33102) and gel running buffer (0.5x TAE (Thermo Fisher: 15558042), 0.1 mM MgCl2) was prepared and prechilled at 4 °C. To each refolding reaction, 1.9 μL of formamide-free loading dye (Thermo Fisher: R0611) was added. After loading all samples and the RNA ladders (NEB: N0362S) onto the gel, the gel was run for 2 hrs at 70 V and at 4 °C to separate RNA populations. The gel was imaged using a ChemiDoc imaging system. All RNA gel experiments were repeated at least twice using independently prepared RNA samples. The list of RNA sequences used for RNA gel electrophoresis is reported in Supplementary Data S1.
RNA clustering reaction
This protocol was modified based on refs. 20,28. On the day of the experiment, a 10X RNA cluster refolding buffer (200 mM KCl, 100 mM MgCl2, and 100 mM Tris (pH 7.0) (Millipore: 648314-100 ML)) was freshly prepared and stored at RT. A 1 mL aliquot of filtered 100 mM spermine-tetrahydrochloride (Sigma: S1141-1G) in nuclease-free water and 50% PEG8000 (NEB: B1004SVIAL) were thawed on ice. Note that the 50% PEG8000 was used within 2 months of opening to ensure consistent results due to potential degradation.
The preparation of shu-top and shu-bottom RNA samples was similar to the one used for the in vitro intermolecular RNA base pairing assays and gel electrophoresis. However, the in vitro transcription reaction contained Cy5-labeled UTPs (aminoallyl-UTP-Cy5) (Jena Bioscience: NU-821-CY5). The concentration of the labeled UTP was adjusted to ensure an average of 3 labeled uracils per RNA.
For the separate folding condition, 32 pmol of each shu-top and shu-bottom RNAs were separately denatured at 90 °C for 2 mins and the sample tubes were immediately placed on ice for 15 mins. For each RNA sample, 1 μL of the 10X RNA cluster refolding buffer and nuclease-free water were added to the RNAs to reach a final volume of 7 μL for each RNA sample. The folding reaction was incubated at RT for 1 hr. Afterward, the folded shu-top and shu-bottom RNAs were combined. 6 μL of 1:2 (vol:vol) 100 mM spermine and 50% PEG8000 premix were added and thoroughly mixed due to the stickiness of PEG8000, resulting in a final volume of 20 μL with each RNA 1.6 μM per reaction.
For the co-folding condition, 32 pmol of each shu-top and shu-bottom RNAs were denatured at 90 °C for 2 mins together and the sample tube was immediately placed on ice for 15 mins. The clustering reaction, which had a final volume of 20 μL with each RNA 1.6 μM per reaction, was the same as the one for separate folding. The sequences of top and bottom RNAs are listed in Supplementary Data S1.
Fluorescence assays to detect intermolecular base pairings in in vitro RNA clusters
To prepare the staining solution, 780 μL of nuclease-free water was added to 5 mg lyophilized DFHBI-1T (LUCERNA: 410-5MG), resulting in a stock DFHBI-1T staining solution with a concentration of 20 mM. The solution was stored at − 20 °C. Next, 2 μL of 1 mM DFHBI-1T was mixed into the final 20 μL clustering reaction to reach 0.1 mM working concentration, and the reaction was incubated in the dark for 4 hrs at RT. For co-folding and separate folding, the ratio of DFHBI-1T (0.1 mM): shu-top (1.6 μM): shu-bottom (1.6 μM) is 62.5:1:1. Because a base-paired top and bottom can form a broccoli structure with two intercalated DFHBI-1T molecules49, we estimate an approximate 30-fold excess of DFHBI-1T to the potential broccoli structures in our experiments. Afterwards, 8–10 clusters were randomly selected for imaging using VT-iSIM with a 100 × 1.5 NA oil immersion objection and a z-series of 27 slices with a step size of 150 nm. Two experimental replicates were performed for each condition and time point.
In addition, DFHBI-1T and its derivatives exhibit low background fluorescence in the absence of RNA aptamer102. Therefore, the low non-zero intensity observed in shu-top and shu-bottom (Supplementary Fig. S1a, b, d) is likely due to the inherent background fluorescence of the dye rather than specific binding to these RNA sequences. Furthermore, the study from which our top and bottom sequences were derived also reported minimal fluorescence signals when the sequences were expressed individually46. This observation is consistent with our findings, as we also noted a slight increase in DFHBI-1T fluorescence in the negative controls. This indicates that DFHBI-1T can emit low levels of fluorescence independent of the presence of broccoli structure or complementary base pairing.
To detect intermolecular base pairing by RNA gel electrophoresis, 16 pmol of each Cy5 (Jena Bioscience: NU-821-Cy5) labeled shu-top and shu-bottom RNAs were used following the protocol described for in vitro intermolecular RNA base pairing assays and gel electrophoresis. After the electrophoresis was complete, the gel was stained with 5 μM DFHBI-1T at RT for 15 mins and then imaged using Amersham™ Typhoon™ 5 scanner (GE Healthcare) with 488 and Cy5 filters as described before46.
Embryo collection and germ granule isolation
Embryos were collected as described previously103. The granule isolation was adapted from ref. 52. Approximately fifty caged flies were allowed to lay eggs at 25 °C on a fresh apple juice agar plate supplemented with yeast paste for 1.5 hrs. About 20 μL of embryos were collected in 1X PBS in a 1.5 mL test tube. After the embryos settled at the tube bottom, the 1X PBS was replaced with 150 μL of freshly made 1X cold lysis buffer (0.34 M sodium cacodylate (pH 7.5), 6 mM MgCl2, 1X complete mini EDTA-free protease inhibitor (Millipore: 11836170001) and 1 U/μL RNase inhibitor (Thermo Fisher: 10777019) adapted from ref. 104. The embryos were then lysed in lysis buffer in the presence of 20 μL of 0.1 mm glass beads (Millipore: G1145-10G) using a cordless homogenizer for 2 mins at RT. The lysate was clarified by centrifugation at 2000 × g for 2 mins and the supernatant, which was separated from the debris and the beads, was transferred to a new test tube. The supernatant was then centrifuged again at 10,000 × g for 15 mins at 4 °C, and about 120 μL of the soluble fraction without touching the bottom pellet was transferred to a new test tube. This soluble fraction was re-centrifuged at 10,000 ×g for 15 mins at 4 °C. About 100 μL of the final clarified soluble fraction, which represented the fraction outside the germ granules, was transferred to a new test tube. The pellet in the initial sample tube, which represented the germ granule fraction, was washed three more times with 100 μL of 1X cold lysis buffer and centrifuged at 10,000 × g for 5 mins each time at 4 °C. Finally, the pellet was suspended in 100 μL of 1X cold lysis buffer. Both fractions were temporally stored on ice before proceeding to the DMS treatment.
DMS treatment and total RNA isolation
DMS modifies accessible adenines and cytosines at their Watson–Crick base-pairing positions105. When adenines and cytosines are not engaged in hydrogen bonding, DMS methylates the N1 position of adenines and the N3 position of cytosines105. For DMS-MaPseq, these methylated bases introduce mismatches during reverse transcription using thermostable group II reverse transcriptase (TGIRT) enzymes50. As a result, hydrogen bonding, primarily from base pairing, protects RNA from DMS methylation. The mutation signals generated by DMS reflect the accessibility of bases for pairing, which could be used for the prediction of RNA secondary structures.
Given the technical challenges of DMS penetration into embryos, we performed DMS-MaPseq on isolated germ granules instead. However, it is possible that the DMS signals from these isolated granules do not fully reflect the intramolecular or intermolecular RNA base pairing present in intact embryos. The experimental procedures were adapted from refs. 50,104. Before DMS treatment, the sample tubes containing the isolated germ granule fraction and the fraction outside the germ granules were incubated at 26 °C for 10 min. The samples were then treated with 2.5% DMS (Millipore: D186309-5ML) and incubated on a thermomixer at 800 rpm and 26 °C for 5 min, followed by immediate addition of 60 μL of 100% 2-mercaptoethanol (Millipore: 444203) to stop the reaction. Next, 500 μL of TRIzol (Thermo Fisher: 15596026) was added to the samples, and total RNA was purified using the Zymo Direct-zol RNA miniprep kit (Zymo: R2050). The purified RNA was eluted in water and stored at −80°C. For each sample replicate, at least 7 rounds of egg collection and granule isolation were performed. The total RNA samples were pooled and concentrated using the Zymo RNA Clean & Concentrator Kit (Zymo: R1014) to obtain sufficient RNA material. The RNA samples were then treated with TURBO DNase and reverse transcribed using gene-specific reverse primers and TGIRT-III (InGex), as described previously50. The regions of interest for DMS structural probing, with a size of approximately 200 nts, were PCR amplified using Q5 high-fidelity polymerase. The resulting DNA fragments were extracted using SizeSelect 2% precast gel (Thermo Fisher: G661012) on the E-Gel Power Snap Electrophoresis Device and stored at − 20 °C. The primer sequences used in DMS-MaPseq and the reactivity profiles are listed in Supplementary Data S2.
DNA library preparation and sequencing
The concentration of each extracted DNA fragment was determined using Qubit 4 fluorometer with 1X dsDNA HS assay kit (Thermo Fisher: Q32854). For each library preparation, 100 ng of pooled DNA fragments were used as input and prepared using the NEBNext Ultra II DNA library prep kit for Illumina (NEB: E7645S) and its protocol. The prepared DNA libraries were sent for sequencing using the MiSeq system by the Johns Hopkins Genetic Core Facility.
RNA isolation and qRT-PCR
About fifty caged flies were allowed to lay eggs at 25 °C on a fresh apple juice agar plate with a scope of yeast paste for 1.5 hrs. The embryos were collected, homogenized in Trizol, and stored at − 80 °C until the next step. Total RNA was extracted using chloroform (Thermo Fisher: C298-500) and precipitated in isopropanol following the TRIzol Reagent User Guide. Next, about 2 μg of total RNA was treated with RQ1 RNase-Free DNase (Promega: M6101) following the manufacturer’s instructions. cDNA synthesis was performed using SuperScriptTM III reverse transcriptase (Thermo Fisher: 18080093) following the manufacturer’s protocol. For each 10 μL of qRT-PCR reaction, approximately 100 ng of cDNA, 1.5 μL of 1 μM forward and reverse primers, and 5 μL iTaq universal SYBR® Green supermix (Bio-Rad: 1725122) were added. Quantitative PCR analysis was performed using a CFX Opus 96 Real-Time PCR System from Bio-Rad. The primer sequences are listed in Supplementary Data S2 and S4.
smFISH in fly embryos
The embryo collection was carried out as described before103. Until the smFISH experiments, the embryos were stored in 100% methanol at 4 °C. To label individual mRNAs, commercially available Stellaris probes were used. Each set of probes consisted of 30 to 48, 20-nt DNA oligos designed with the default setting on Stellaris Probe Designer. Each oligo was covalently conjugated with either CAL Fluor 590 or Quasar 670. In-house labeling of smFISH probes involved designing the probe set with Stellaris Probe Designer and ordering the oligos from IDT. Subsequently, the oligos were modified using terminal deoxynucleotidyl transferase (Thermo Fisher: EP0161) and amino-11-ddUTP (Lumiprobe: 15040), followed by covalent conjugation with AF488 (Lumiprobe: 21820), AF568 (Lumiprobe: 24820) or AF647 (Lumiprobe: 26820) NHS esters as previously described106. Hybridization of mRNAs with smFISH was performed as previously described103. The sequences of smFISH probes are listed in Supplementary Data S3 or adapted from previous17,36.
Simulation of RNA monomers and intermolecular base pairing in RNA dimers
We performed all simulations using a modified version of the Single Interaction Site (SIS) model with the RNA energy function described previously34. The simulations were performed on Graphics Processing Units (GPUs) using a custom OpenMM code107 to enhance sampling of the conformational space. We used low-friction Langevin dynamics, in which the viscosity of water was reduced by a factor of 100108. Even for the SIS model for RNA, the simulations are computationally extensive, thus requiring the simulated tempering method to ensure that the conformational space is sampled exhaustively109. The trajectories were analyzed using the Multistate Bennett Acceptance Ratio (MBAR) to calculate all properties of interest110. All simulations were performed with 1 M NaCl, where electrostatic interactions are weak, and only base pairing interactions dominate111.
Monomer simulations: A single RNA molecule in the extended conformation was initially placed in a simulation box (the size of the box is much larger than the RNA size and does not play any role in determining RNA structures. Simulations were performed for 5 × 109 time steps, in which the first 5 × 108 steps were discarded, which ensures that only equilibrated structures are used in computing various quantities of interest.
Homodimer simulations: Two representative snapshots from monomer simulations were randomly picked and placed in a sphere of radius R. The two RNAs were constrained inside the sphere by defining the following potential for any particles:
where \({r}_{i}\) is the distance between particle i and the sphere center. R was chosen big enough so that the two RNAs have enough space to adopt extended conformations, but also small enough to prevent the two chains from drifting away from each other. Simulations were then conducted for \(5\times {10}^{9}\) time steps.
We set R equal to 42, 25, and 30 nm for nos, pgc, and gcl 3′UTRs, respectively. For comparison, the radii of gyration of nos, pgc, and gcl 3′UTRs are ~ 8.5, 6.0 and 7.0 nm, respectively. The two RNA molecules, therefore, occupy only a small fraction of the volume inside the sphere and have a small chance of touching the boundary. Even if they reach the boundary, the value of k is small, 1 kcal/mol.A2, to minimally perturb the RNA structures and dynamics. To compare, the strength of an A-U base pair is ~ 4.5 kcal/mol, and the GC base pair is ~ 6.5 kcal/mol. Therefore, the use of the constraint leaves a minimal impact on RNA structures and their interactions.
Base pairing criteria: We used the energy of our base pair potential to determine whether the two bases form a stable pair. For a single snapshot, a base pair was considered stable if its energy, Ubp, was less than − 3kT. In such cases, the probability of forming that base pair was assigned a value of 1, \(\gamma=1\,\). Otherwise, it was assigned a value of 0, \(\gamma=0\,\). The ensemble average probability for each base pair was then calculated using the MBAR formalism. This procedure was repeated for every base pair formed during the simulations.
Base accessibility calculations: Base accessibility of the nucleotide i is defined as \({H}_{i}={S}_{i}{\gamma }_{i}\), where \({S}_{i}\) is the solvent-accessible surface area of nucleotide i (calculated using the Lee-Richards algorithm112 in FreeSASA113. \({\gamma }_{i}\) adopts two values: 0-if the nucleotide i is involved in base pairing and 1-otherwise, as described above. In addition to the RNA secondary structure (reflected in \({\gamma }_{i}\)), the accessibility of a base also depends on the RNA tertiary structure (reflected in \({S}_{i}\)). If a base is deeply buried in the core of the RNA, its accessibility is small. On the other hand, if the base is located near the RNA periphery, its accessibility is higher.
Clustering of secondary structures: RNA conformations are grouped based on the secondary structures as follows. Each conformation i is fully specified by a set of base pairs \({B}_{i}=\left\{\left\{a,b\right\},\ldots \right\}\), where a and b are the two nucleotides that base pair to each other. The similarity between the two conformations i and j are calculated using the Jaccard distance between the two sets \({B}_{i}\) and \({B}_{j}\) as:
The Jaccard distance considers the difference in sizes of two sets and is bounded between 0 and 1. Thus, if the two conformations share no base pair, then \({d}_{J}\left({B}_{i},{B}_{j}\right)=1\), whereas \({d}_{J}\left({B}_{i},{B}_{j}\right)=0\) if \({B}_{i}={B}_{j}\).
We then generated the distance matrix J, where \({J}_{{ij}}={d}_{J}\left({B}_{i},{B}_{j}\right)\) for the pair i and j. J was then used as the input to HDBSCAN114, which is a density-based clustering algorithm, to extract clusters of RNA conformations. The free energies of the clusters were subsequently calculated using MBAR.
Limitations of the SIS model: We used the SIS model simulations to examine the types and prevalence of intermolecular base pairing that could occur within RNA clusters, complementing the observations made from DMS-MaPseq analysis of RNAs isolated from germ granules. However, the structural data generated by these two approaches are not strictly comparable due to the distinct environments experienced by the RNAs in each method. Moreover, it is unclear if the base accessibility obtained in simulations and the structural aspects inferred from experiments are proportional to each other. Therefore, it is difficult to directly compare the structural results from DMS-MaPseq experiments and simulations.
The current SIS model simulations do not account for proteins, RNA modifications and other cellular components, which could significantly influence the RNA folding in vivo. In addition, we focused on modeling 3′UTRs rather than full-length mRNAs, which may adopt different structures when associated with translating ribosomes. Another limitation is that the SIS model, as currently formulated, does not incorporate base stacking interactions, which are likely the dominant contributor to RNA secondary structure stability.
Regarding pseudoknot prediction, our simulations estimated a higher proportion of base pairs forming pseudoknots than the average of ~ 1.5% reported previously115. While this may reflect an overestimation, it is important to note that pseudoknot abundance varies substantially across RNA types and lengths116,117. Thus, a more systematic and quantitative evaluation of pseudoknot prevalence, especially in mRNA-derived sequences, remains an important area for future investigation.
Lastly, while the SIS model can simulate base accessibility in a simplified in silico environment, future work is needed to establish its broader applicability to general RNA species beyond repeat RNAs34,68,118 and the HIV U4/6 core.
The code to perform RNA simulations for this study is available on GitHub (https://github.com/tienhungf91/RNA_llps) and archived on Zenodo with a DOI number (https://doi.org/10.5281/zenodo.15844243).
Calculation of normalized ensemble diversity (NED) values
The ensemble diversity values for nos, pgc, and gcl 3′UTRs were calculated using RNAfold47, which was set to use RNA parameters (Turner model, 2004119) at 26 °C, incorporating SHAPE reactivity data obtained from DMS-MaPseq. To calculate NED, the ensemble diversity value was divided by the length of the 3′UTR, as described in ref. 120.
Transfection for Drosophila Ras cells
Drosophila Ras cells were seeded on the chambered cell culture slides (Grace Bio-Labs: 103510) on the day of transfection. 200 ng of plasmids containing LacZA and LacZB sequences, each tagged with HIV, nos, non, sense or antisense CSs and fused with metallothionein promoters121, were transfected into the cells using effectene transfection reagent (Qiagen: 301425). After 24 hrs, 0.5 mM copper sulfate (Sigma: C8027-500G) was added to the cell culture to induce the expression of LacZA and LacZB RNAs. After 24 hrs of induction, the cells were fixed with 4% paraformaldehyde (Electron Microscopy Sciences: 15713) for 10 mins and subsequently washed with 1X PBS. To detect LacZA and LacZB RNAs, HCR™ commercially designed probes and RNA-FISH protocol (Molecular Instruments) for adherent cells were used following the manufacturer’s instructions.
Cuticle preparation of Drosophila embryos
A previously published protocol (https://krauselab.ccbr.utoronto.ca/file/Cuticle_preps.html) with slight modifications was used. In short, embryos were collected on apple juice-agar plates and aged for 24 hrs at 25 °C. They were subsequently dechorionated using 50% bleach for 1-2 mins and rinsed through a Nitex nylon screen. After rinsing with water twice, the screen was dipped into a scintillation vial containing 5 mL of heptane (Millipore: HX0078-1) and 5 mL of methanol (Sigma: 34860-4L-R) to facilitate the embryos to slide off from the screen. The vial was then capped and shaken vigorously for 10 s to devitellinize the embryos. An additional 5 mL of methanol was added, and the majority of the embryos were settled at the bottom of the methanol phase. A P-1000 tip was used to transfer the embryos into a 1.5 mL tube. Once all embryos were collected, they were placed on a glass slide. After the evaporation of the methanol, approximately 100 μL of Hoyer’s medium, supplemented with 20% lactic acid (Sigma: 252476-100 G), was dropped onto the embryos, which were then covered with a coverslip. The samples were incubated overnight in a 65 °C oven for tissue clearing. The slides were sealed with nail polish on the following day and stored at room temperature. Images of the cleared samples were taken using a darkfield microscope (AmScope: T340-TK-LED) at a magnification of 400X with a digital camera.
Egg hatching assays
Approximately 20 non/nosBN, HIV/nosBN or nos/nosBN virgin females were crossed with 10 WT (W1118) young males. The crosses were maintained at 25 °C for 3 days on standard cornmeal/agar media supplemented with yeast powder. Then the flies were caged and supplied with a fresh apple juice plate containing a dollop of yeast paste for 24 hrs at 25 °C and allowed to lay eggs. Afterward, the plate was collected, and the number of eggs on it was counted and replaced with a new apple juice plate. The plate was then kept at 25 °C for an additional 24 h to count the number of hatched eggs. The hatching rate was determined by dividing the number of hatched eggs by the total number of eggs on the same plate.
Western blot analysis
The embryos were collected and flash-frozen using liquid nitrogen. The samples were stored at − 80 °C until the next step. The samples were homogenized in 100 μL of cold lysis buffer (150 mM sodium chloride, 50 mM pH 8.0 Tris-HCL, 1% Triton-X100, 0.5% sodium deoxycholate, and 0.1% SDS). After incubating on ice for 20 mins, the lysates were centrifuged at 15,000 × g for 20 mins at 4 °C. The resulting supernatants were transferred to new 1.5 mL tubes, and the protein concentrations were quantified using the PierceTM BCA protein assay kit (Thermo Fisher: 23227) and Nanodrop, following the manufacturer’s protocol. About 20–50 μg of total proteins were mixed with 1X Laemmli (Bio-Rad: 1610747) and 50 mM DTT (Thermo Fisher: R0861), resulting in a total final volume of 30 µL. The samples were then boiled at 95 °C for 5 mins. Once cooled, they were loaded into a 7.5% Criterion™ TGX Stain-Free™ Protein Gel (Bio-Rad: 5671024). 10 µL Precision Plus Protein™ WesternC™ blotting standards (Bio-Rad: 1610376) was used as ladders. The gel was run in 1X Tris-glycine SDS running buffer (2.5 mM Tris, 19.2 mM glycine (Sigma: G8898-500G), 0.01% SDS (Thermo Fisher: 28364)) at 150 V for 1.5 hrs at 4 °C. After electrophoresis, the bands were transferred to a Trans-Blot Turbo Midi 0.2μm Nitrocellulose membrane (Bio-Rad: 1704159) using the Trans-Blot Turbo transfer system and Bio-Rad mixed molecular weight protocol. The membrane was then blocked in 5% blotting grade non-fat dry Milk (Bio-Rad: 1706404) in PBST (1x PBS (Thermo Fisher: 70011044), 0.1% Tween-20 (Sigma: 655204)) for 1 h at RT on a rocker. Next, the membrane was incubated overnight at 4 °C with primary, mouse anti-β-actin antibody (Abcam: ab8224), which was diluted in 1% blotting grade non-fat dry milk/PBST to 1:2000. After incubation, the membrane was washed three times with PBST and incubated with secondary, goat anti-mouse IgG (HRP) antibody (Abcam: ab6789), which was diluted in 1% blotting grade non-fat dry milk/PBST to 1:10000 for 2 h. The membrane was washed five times with PBST. The signal was developed using SuperSignalTM western dura extended duration substrate (Thermo Fisher: 34075). The membrane was imaged by ChemiDoc MP with an exposure time of 10–60 ms. To detect Nanos protein expression on the same membrane, the previous primary and secondary antibodies were removed by western blot stripping buffer (Thermo Fisher: 21059) following the manufacturer’s instructions. Rabbit anti-Nanos antibody (a gift from Nakamura Lab) (1:1000)122, and goat anti-rabbit IgG (HRP) antibody (Abcam: ab6721) (1:10000), were then used to detect Nanos following the protocol described for detecting β-actin. Finally, for each protein, two to three biological replicates per sample were run on the same western blot. The analysis was performed following the guidelines from ImageJ User Guide-30.13 Gels.
Immunostaining of fly ovaries
The females were fed with yeast powder the day before the dissection. The ovaries were dissected in Schneider’s insect medium supplied with 200 μg/mL insulin (Sigma: I5500-500MG) at RT. The tissues were fixed in 4% paraformaldehyde in PBTx (1XPBS, 0.1% Triton-X100 (Millipore: TX1568-1) for 20 mins at RT and then washed twice in PBTx, each time for 10 min. Subsequently, the tissues were blocked overnight at 4 °C in BBTx (1XPBTx, 0.5% BSA (Millipore: A3294-50G), 2% NGS (Abcam: ab7481)). Rabbit anti-Vasa antibody (RRID: AB2940894)52 and mouse anti-1B1 antibody (DSHB: AB528070) were diluted in BBTx to 1:500 and 1:50, respectively. They were added to tissues and incubated overnight at 4 °C. Afterward, the tissues were washed twice in PBTx for 10 mins at RT and then treated with goat anti-rabbit IgG Alexa488 antibody (Thermo Fisher: A-11070) and goat anti-mouse IgG Alexa568 antibody (Thermo Fisher: A-11004), which were diluted in BBTx to 1:1000, for 4 h at RT. The tissues were then washed twice in PBTx for 10 mins at RT. To stain the DNA, 1 μg/mL DAPI (Sigma: 10236276001) diluted in PBTx was added to the samples and incubated for 5 mins at RT. Finally, the samples were mounted on slides using ProLong™ glass antifade mountant (Thermo Fisher: P36980) and cured overnight at RT before imaging.
Microscopy and deconvolution
Images were acquired with a vt-instant Structured Illumination Microscope (vt-iSIM; BioVision Technologies) equipped with the 405 nm 100 mW, 488 nm 150 mW, 561 nm 150 mW, 642 nm 100 mW, and 445 nm 75 mW lasers, two ORCA-Fusion sCMOS cameras and the Leica HC PL APO 63x/1.30 GLYC CORR CS2, HC PL APO 63x/1.40 OIL CS2 and HC PL APO 100x/1.47 OIL CORR TIRF objectives as described before52. Images were acquired in three dimensions (3D) and then deconvolved using Huygens (Scientific Volume Imaging).
Identification of CSs
The identification of CSs involves scanning through all possible pairs of starting positions, considering the forward direction of sequence 1 and the reverse direction of sequence 2. Sequences 1 and 2 were identical in order to identify the CSs which were self-complementary. Starting with each possible position in sequence 1, the program checks for complementary nucleotides in the reverse direction of sequence 2. If a match is found between a nucleotide in sequence 1 and its corresponding nucleotide in sequence 2, the program extends the length of the complementary sub-sequence by one, continuing the process for subsequent nucleotides. This extension continues until a mismatch occurs, at which point the program records the CS if the length and/or the GC content passes the threshold. The program then moves on to the next pair of starting positions and repeats the process until all possible positions are evaluated. The identified CSs on 3′UTRs of nos, pgc and gcl are listed in Supplementary Data S5–S7, respectively.
The code for identifying complementary sequences (CSs) from this study is available on GitHub (https://github.com/AnneyYeZiqing/sticky_finder) and archived on Zenodo with a DOI number (https://doi.org/10.5281/zenodo.15802323)123.
Quantifying the level of fold enrichment in in vitro RNA clusters
The images were imported into FIJI/Image J124, and the channels of DFHBI-1T and RNA were split. A 5 × 5 pixel rectangle was used to measure the levels of the integrated density inside and outside the assemblies. For each image, three different regions were randomly selected using a 5 × 5 pixel rectangle. The levels of the integrated density inside and outside the clusters were determined in the RNA channel. The fold enrichment was calculated as Integrated Density (inside)/ Integrated Density (outside).
Quantifying the level of intermolecular base pairing in in vitro RNA clusters
Same as above, three different regions inside and outside the assemblies were randomly chosen and measured. The background signal was determined by the dye-only condition without the addition of RNAs. The normalized integrated density of the selected region was calculated as the Integrated Density of the dye (selected region)-Integrated Density (dye background)/Integrated Density of RNA (selected region).
Quantifying the size of in vitro RNA clusters
The cluster images were imported into FIJI/Image J124. The 3D Objects Counter plugin was used to quantify the size of the RNA clusters, with a minimum size filter of 2. The intensity threshold was set automatically. The number of object voxels was measured to determine the sizes of the RNA clusters. Fifteen images, each with the same Z-step size (300 nm), identical imaging depth and region size, were analyzed for each condition.
Quantifying mRNA fold enrichment in isolated germ granules
Germ granules were isolated as described above and previously52. However, from the last washing step, 100 μL of the soluble fraction in 1X cold lysis buffer was collected, and the pellet was suspended in 100 μL 1X cold lysis buffer. The total RNA from these two fractions was extracted by the Zymo Direct-zol RNA Miniprep kit and used for cDNA synthesis as described previously (see “Methods” Details: RNA isolation and qRT-PCR). For each qRT-PCR reaction of each sample, 100 ± 10 ng of cDNA was used as input. Two biological replicates with three technical replicates were analyzed. To calculate the fold enrichment of a particular gene (here termed X), \({2}^{-({{\rm{Cq}}}\left({{\rm{gene}}}\; {{\rm{X}}}\; {{\rm{in}}}\; {{\rm{soluble}}}\right)-{{\rm{Average}}}\; {{\rm{of}}}\; {{\rm{Cq}}}\left({{\rm{gene}}}\; {{\rm{X}}}\; {{\rm{in}}}\; {{\rm{soluble}}}\right))}\) was used for calculating soluble fractions. \({2}^{-({{\rm{Cq}}}\left({{\rm{gene}}}\; {{\rm{X}}}\; {{\rm{in}}}\; {{\rm{pellets}}}\right)-{{\rm{Average}}}\; {{\rm{of}}}\; {{\rm{Cq}}}\left({{\rm{gene}}}\; {{\rm{X}}}\; {{\rm{in}}}\; {{\rm{soluble}}}\right))}\) was used for pellets. Primers are listed in Supplementary Data S2.
Analysis of DMS-MaPseq data
The raw fasta files were analyzed using the Detection of RNA folding Ensembles using Expectation-Maximization (DREEM) as previously described67. The code for this analysis can be found at https://codeocean.com/capsule/0380995/tree. Two biological replicates were used. For DMS-MaPseq outside the germ granules, Pearson correlation coefficients (r) are 0.93, 0.97 and 0.99 between the replicates of nos, pgc and gcl 3′UTRs, respectively. For DMS-MaPseq within the germ granules, r = 0.99, 0.98 and 0.89 in nos, pgc and gcl 3′UTRs, respectively.
Averages of the two replicates were taken for subsequent analysis. The profiles of average reactivity (K = 1 in DREEM) were used to determine the structuredness of CSs using RNAprobing47, which was set to use Diegan et al.125 and 26 °C. Predicted secondary structures were visualized by FORNA48 and VARNA126. To compare the similarity in DMS profiles between inside and outside the germ granules, the reactivity of each informative nucleotide for each RNA sequence was correlated between the inside and outside of the germ granules. Pearson’s Correlation coefficient was then calculated. In addition, to compare changes in the DMS reactivity of each nucleotide between inside and outside the germ granules, the DMS reactivity of each nucleotide inside the germ granules was divided by the corresponding value outside the granules. The significant thresholds were calculated based on the standard deviation and average of the ratios from two DMS-MaPseq replicates for both inside and outside the granules. To account for multiple hypothesis testing, we set the p-value threshold as 0.05 divided by the length of nos, pgc and gcl 3′UTRs, which are 880, 401 and 524 nts, respectively. This resulted in p-value thresholds of 5.68 × 10−5 for nos, 1.25 × 10−4 for pgc, and 9.54 × 10−5 for gcl. The corresponding Z-scores were calculated based on these p-values (two-tailed). The threshold lines were then determined using the formula ± (Z-score × standard deviation) + average. Ratios of DMS reactivities that fall outside the orange dashed lines are considered significant.
Quantifying the co-localization of germ granule mRNA assemblies by PCC(Costes) and PCC
Analyses were performed using PCC(Costes) and PCC co-localization ImageJ plugin127 as described before36.
qRT- PCR analysis of transcripts in Ras cells and nos-non, nos-HIV and nos-nos flies
For Ras cells, Drosophila Act5C was used as a control to calculate the relative transcript levels of LacZA and LacZB, which was \({2}^{-({{\rm{Cq}}}\left({LacZA\; or\; LacZB}\right)-{{\rm{Average}}}\; {{\rm{of}}}\; {{\rm{Cq}}}\left({Act}5C\right))}\). Then the data was normalized to non.
For fly embryos, Drosophila Gapdh2 was used as a control to calculate the relative transcript levels, which was \({2}^{-({{\rm{Cq}}}\left({{\rm{region}}}\; {{\rm{of}}}\; {{\rm{interest}}}\right)-{{\rm{Average}}}\; {{\rm{of}}}\; {{\rm{Cq}}}\left({Gapdh2}\right))}\). Then the data were further normalized to nos-non. Primers are listed in Supplementary Data S4.
Quantifying mRNA concentration and the number of mRNAs per cluster in embryos
Analyses were performed using the Airlocalize spot detection algorithm128 as described before36.
Western blot analysis
Different samples were run on the same western blot with four biological replicates. The analysis was performed following the guidelines from ImageJ User Guide-30.13 Gels. Nanos protein levels were normalized to β-actin, which served as a loading control.
Counting female GSCs
Female GSCs were counted based on the morphology of the anti-1B1 staining and juxtaposition to the terminal filament as described previously79.
Quantifying fold enrichment of pgc and gcl mRNAs at embryo posterior
Hybridization of mRNAs with smFISH was performed as previously described103, and the probe sequences for pgc and gcl mRNAs were adapted from ref. 17. Embryos containing both posterior localized mRNAs and unlocalized mRNA fractions were imaged using a 63X oil immersion objective. To determine the fold enrichment, a region of interest (50 × 50 pixels) was selected in both the localized and unlocalized areas, and the mean intensities were measured using ImageJ. The fold enrichment was calculated by dividing the mean intensity of the localized mRNA region by that of the unlocalized mRNA region.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
DMS-MaPseq data have been deposited in the Sequence Read Archive (SRA) under accession numbers SRR34411012, SRR34411013, SRR34411014 and SRR34411015 and can be accessed at: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1288049. Microscopy images and associated data are available in the BioImage Archive under accession number S-BIAD2144 at: https://www.ebi.ac.uk/biostudies/bioimages/studies/S-BIAD2144. Other data supporting the findings of this study are available within the paper and its Supplementary Information. Source data are provided in this paper.
Code availability
Codes to perform RNA simulations are available at https://github.com/tienhungf91/RNA_llps and archived on Zenodo (https://doi.org/10.5281/zenodo.15844243). Codes for identifying CSs are available at https://github.com/AnneyYeZiqing/sticky_finder and archived on Zenodo (https://doi.org/10.5281/zenodo.15802323).
References
Tian, S., Curnutte, H. A. & Trcek, T. RNA Granules: a view from the RNA perspective. Molecules 25, 3130 (2020).
Banani, S. F., Lee, H. O., Hyman, A. A. & Rosen, M. K. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298 (2017).
Eno, C., Hansen, C. L. & Pelegri, F. Aggregation, segregation, and dispersal of homotypic germ plasm RNPs in the early zebrafish embryo. Dev. Dyn. 248, 306–318 (2019).
Protter, D. S. W. & Parker, R. Principles and properties of stress granules. Trends Cell Biol. 26, 668–679 (2016).
Riggs, C. L., Kedersha, N., Ivanov, P. & Anderson, P. Mammalian stress granules and P bodies at a glance. J. Cell Sci. 133, jcs242487 (2020).
Sagan, S. M. & Weber, S. C. Let’s phase it: viruses are master architects of biomolecular condensates. Trends Biochem. Sci. 48, 229–243 (2023).
Voronina, E., Seydoux, G., Sassone-Corsi, P. & Nagamori, I. RNA granules in germ cells. Cold Spring Harb. Perspect. Biol. 3, a002774 (2011).
Chiappetta, A., Liao, J., Tian, S. & Trcek, T. Structural and functional organization of germ plasm condensates. Biochem. J. 479, 2477–2495 (2022).
Yang, P. et al. G3BP1 is a tunable switch that triggers phase separation to assemble stress granules. Cell 181, 325–345 e28 (2020).
Smith, J. et al. Spatial patterning of P granules by RNA-induced phase separation of the intrinsically-disordered protein MEG-3. Elife 5, e21337 (2016).
Bose, M., Lampe, M., Mahamid, J. & Ephrussi, A. Liquid-to-solid phase transition of oskar ribonucleoprotein granules is essential for their function in Drosophila embryonic development. Cell 185, 1308–1324 (2022).
Chau, B. A., Chen, V., Cochrane, A. W., Parent, L. J. & Mouland, A. J. Liquid-liquid phase separation of nucleocapsid proteins during SARS-CoV-2 and HIV-1 replication. Cell Rep. 42, 111968 (2023).
Patel, A. et al. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162, 1066–1077 (2015).
Wang, A. et al. A single N-terminal phosphomimic disrupts TDP-43 polymerization, phase separation, and RNA splicing. EMBO J. 37, e97452 (2018).
Feric, M. et al. Coexisting liquid phases underlie nucleolar subcompartments. Cell 165, 1686–1697 (2016).
Jain, A. & Vale, R. D. RNA phase transitions in repeat expansion disorders. Nature 546, 243–247 (2017).
Trcek, T. et al. Drosophila germ granules are structured and contain homotypic mRNA clusters. Nat. Commun. 6, 7962 (2015).
Little, S. C., Sinsimer, K. S., Lee, J. J., Wieschaus, E. F. & Gavis, E. R. Independent and coordinate trafficking of single Drosophila germ plasm mRNAs. Nat. Cell Biol. 17, 558–568 (2015).
Tauber, D. et al. Modulation of RNA condensation by the DEAD-box protein eIF4A. Cell 180, 411–426 (2020).
Van Treeck, B. et al. RNA self-assembly contributes to stress granule formation and defining the stress granule transcriptome. Proc. Natl. Acad. Sci. USA 115, 2734–2739 (2018).
Chen, R. et al. HDX-MS finds that partial unfolding with sequential domain activation controls condensation of a cellular stress marker. Proc. Natl. Acad. Sci. USA 121, e2321606121 (2024).
Sahin, C. et al. Mass Spectrometry of RNA-binding proteins during liquid-liquid phase separation reveals distinct assembly mechanisms and droplet architectures. J Am Chem Soc 145, 10659–10668 (2023).
Brangwynne, C. P., Tompa, P. & Rohit, V. Polymer physics of intracellular phase transitions. Nat. Phys. 11, 899–904 (2015).
Ruff, K. M. et al. Sequence grammar underlying the unfolding and phase separation of globular proteins. Mol. Cell 82, 3193–3208 (2022).
Basu, S. et al. Rational optimization of a transcription factor activation domain inhibitor. Nat. Struct. Mol. Biol. 30, 1958–1969 (2023).
Schmit, J. D., Feric, M. & Dundr, M. How hierarchical interactions make membraneless organelles tick like clockwork. Trends Biochem. Sci. 46, 525–534 (2021).
Shin, Y. & Brangwynne, C. P. Liquid phase condensation in cell physiology and disease. Science 357, eaaf4382 (2017).
Langdon, E. M. et al. mRNA structure determines specificity of a polyQ-driven phase separation. Science 360, 922–927 (2018).
Poudyal, R. R., Sieg, J. P., Portz, B., Keating, C. D. & Bevilacqua, P. C. RNA sequence and structure control assembly and function of RNA condensates. RNA 27, 1589–1601 (2021).
Bevilacqua, P. C., Williams, A. M., Chou, H. L. & Assmann, S. M. RNA multimerization as an organizing force for liquid-liquid phase separation. RNA 28, 16–26 (2022).
Miller, J. W. et al. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 19, 4439–4448 (2000).
Haeusler, A. R. et al. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature 507, 195–200 (2014).
Renton, A. E. et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72, 257–268 (2011).
Nguyen, H. T., Hori, N. & Thirumalai, D. Condensates in RNA repeat sequences are heterogeneously organized and exhibit reptation dynamics. Nat. Chem. 14, 775–785 (2022).
Trcek, T. & Lehmann, R. Germ granules in Drosophila. Traffic 20, 650–660 (2019).
Trcek, T. et al. Sequence-independent self-assembly of germ granule mRNAs into homotypic clusters. Mol. Cell 78, 941–950 (2020).
Niepielko, M. G., Eagle, W. V. I. & Gavis, E. R. Stochastic seeding coupled with mRNA self-recruitment generates heterogeneous drosophila germ granules. Curr. Biol. 28, 1872–1881 (2018).
Jambor, H., Brunel, C. & Ephrussi, A. Dimerization of oskar 3’ UTRs promotes hitchhiking for RNA localization in the Drosophila oocyte. RNA 17, 2049–2057 (2011).
Ferrandon, D., Koch, I., Westhof, E. & Nusslein-Volhard, C. RNA-RNA interaction is required for the formation of specific bicoid mRNA 3’ UTR-STAUFEN ribonucleoprotein particles. EMBO J. 16, 1751–1758 (1997).
Clever, J. L., Wong, M. L. & Parslow, T. G. Requirements for kissing-loop-mediated dimerization of human immunodeficiency virus RNA. J. Virol. 70, 5902–5908 (1996).
Marquet, R. et al. Dimerization of human immunodeficiency virus (type 1) RNA: stimulation by cations and possible mechanism. Nucleic Acids Res. 19, 2349–2357 (1991).
Mujeeb, A., Clever, J. L., Billeci, T. M., James, T. L. & Parslow, T. G. Structure of the dimer initiation complex of HIV-1 genomic RNA. Nat. Struct. Biol. 5, 432–436 (1998).
Paillart, J. C., Skripkin, E., Ehresmann, B., Ehresmann, C. & Marquet, R. A loop-loop “kissing” complex is the essential part of the dimer linkage of genomic HIV-1 RNA. Proc. Natl. Acad. Sci. USA 93, 5572–5577 (1996).
Wagner, C. et al. Dimerization of the 3’UTR of bicoid mRNA involves a two-step mechanism. J. Mol. Biol. 313, 511–524 (2001).
Bose, M., Rankovic, B., Mahamid, J. & Ephrussi, A. An architectural role of specific RNA-RNA interactions in oskar granules. Nat. Cell Biol. 26, 1934–1942 (2024).
Alam, K. K., Tawiah, K. D., Lichte, M. F., Porciani, D. & Burke, D. H. A fluorescent split aptamer for visualizing RNA-RNA assembly in vivo. ACS Synth. Biol. 6, 1710–1721 (2017).
Lorenz, R. et al. ViennaRNA Package 2.0. Algorithms Mol. Biol. 6, 26 (2011).
Kerpedjiev, P., Hammer, S. & Hofacker, I. L. Forna (force-directed RNA): Simple and effective online RNA secondary structure diagrams. Bioinformatics 31, 3377–3379 (2015).
Shanaa, O. A., Rumyantsev, A., Sambuk, E. & Padkina, M. In vivo production of RNA aptamers and nanoparticles: problems and prospects. Molecules 26, 1422 (2021).
Zubradt, M. et al. DMS-MaPseq for genome-wide or targeted RNA structure probing in vivo. Natp Methods 14, 75–82 (2017).
Rouskin, S., Zubradt, M., Washietl, S., Kellis, M. & Weissman, J. S. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature 505, 701–705 (2014).
Curnutte, H. A. et al. Proteins rather than mRNAs regulate nucleation and persistence of Oskar germ granules in Drosophila. Cell Rep. 42, 112723 (2023).
Kistler, K. E. et al. Phase transitioned nuclear Oskar promotes cell division of Drosophila primordial germ cells. Elife 7, e37949 (2018).
Rangan, P. et al. Temporal and spatial control of germ-plasm RNAs. Curr. Biol. 19, 72–77 (2009).
Gavis, E. R. & Lehmann, R. Localization of nanos RNA controls embryonic polarity. Cell 71, 301–313 (1992).
Dahanukar, A. & Wharton, R. P. The Nanos gradient in Drosophila embryos is generated by translational regulation. Genes Dev. 10, 2610–2620 (1996).
Gavis, E. R. & Lehmann, R. Translational regulation of nanos by RNA localization. Nature 369, 315–318 (1994).
Gavis, E. R., Lunsford, L., Bergsten, S. E. & Lehmann, R. A conserved 90 nucleotide element mediates translational repression of nanos RNA. Development 122, 2791–2800 (1996).
Ramat, A., Haidar, A., Garret, C. & Simonelig, M. Spatial organization of translation and translational repression in two phases of germ granules. Nat. Commun. 15, 8020 (2024).
Chen, R., Stainier, W., Dufourt, J., Lagha, M. & Lehmann, R. Direct observation of translational activation by a ribonucleoprotein granule. Nat. Cell Biol. 26, 1322–1335 (2024).
Crucs, S., Chatterjee, S. & Gavis, E. R. Overlapping but distinct RNA elements control repression and activation of nanos translation. Mol. Cell 5, 457–467 (2000).
Dahanukar, A., Walker, J. A. & Wharton, R. P. Smaug, a novel RNA-binding protein that operates a translational switch in Drosophila. Mol. Cell 4, 209–218 (1999).
Smibert, C. A., Wilson, J. E., Kerr, K. & Macdonald, P. M. smaug protein represses translation of unlocalized nanos mRNA in the Drosophila embryo. Genes Dev. 10, 2600–2609 (1996).
Gavis, E. R., Curtis, D. & Lehmann, R. Identification of cis-acting sequences that control nanos RNA localization. Dev. Biol. 176, 36–50 (1996).
Eisenberg, H. & Felsenfeld, G. Studies of the temperature-dependent conformation and phase separation of polyriboadenylic acid solutions at neutral pH. J. Mol. Biol. 30, 17–37 (1967).
Parker, D. M., Tauber, D. & Parker, R. G3BP1 promotes intermolecular RNA-RNA interactions during RNA condensation. Mol. Cell 85, 571–584 (2024).
Tomezsko, P. J. et al. Determination of RNA structural diversity and its role in HIV-1 RNA splicing. Nature 582, 438–442 (2020).
Maity, H., Nguyen, H. T., Hori, N. & Thirumalai, D. Odd-even disparity in the population of slipped hairpins in RNA repeat sequences with implications for phase separation. Proc. Natl. Acad. Sci. USA 120, e2301409120 (2023).
Dirac, A. M., Huthoff, H., Kjems, J. & Berkhout, B. Regulated HIV-2 RNA dimerization by means of alternative RNA conformations. Nucleic Acids Res. 30, 2647–2655 (2002).
St Louis, D. C. et al. Infectious molecular clones with the nonhomologous dimer initiation sequences found in different subtypes of human immunodeficiency virus type 1 can recombine and initiate a spreading infection in vitro. J. Virol. 72, 3991–3998 (1998).
Trcek, T., Sato, H., Singer, R. H. & Maquat, L. E. Temporal and spatial characterization of nonsense-mediated mRNA decay. Genes Dev. 27, 541–551 (2013).
Wang, C., Dickinson, L. K. & Lehmann, R. Genetics of nanos localization in Drosophila. Dev. Dyn. 199, 103–115 (1994).
Wang, C. & Lehmann, R. Nanos is the localized posterior determinant in Drosophila. Cell 66, 637–647 (1991).
Lehmann, R. & Nusslein-Volhard, C. The maternal gene nanos has a central role in posterior pattern formation of the Drosophila embryo. Development 112, 679–691 (1991).
Dold, A. et al. Makorin 1 controls embryonic patterning by alleviating Bruno1-mediated repression of oskar translation. PLoS Genet. 16, e1008581 (2020).
Gavis, E. R., Chatterjee, S., Ford, N. R. & Wolff, L. J. Dispensability of nanos mRNA localization for abdominal patterning but not for germ cell development. Mech. Dev. 125, 81–90 (2008).
Bhat, K. M. The posterior determinant gene nanos is required for the maintenance of the adult germline stem cells during Drosophila oogenesis. Genetics 151, 1479–1492 (1999).
Forbes, A. & Lehmann, R. Nanos and Pumilio have critical roles in the development and function of Drosophila germline stem cells. Development 125, 679–690 (1998).
de Cuevas, M. & Spradling, A. C. Morphogenesis of the Drosophila fusome and its implications for oocyte specification. Development 125, 2781–2789 (1998).
Bonilla, S. L., Vicens, Q. & Kieft, J. S. Cryo-EM reveals an entangled kinetic trap in the folding of a catalytic RNA. Sci. Adv. 8, eabq4144 (2022).
Cruz, J. A. & Westhof, E. The dynamic landscapes of RNA architecture. Cell 136, 604–609 (2009).
Ganser, L. R., Kelly, M. L., Herschlag, D. & Al-Hashimi, H. M. The roles of structural dynamics in the cellular functions of RNAs. Nat. Rev. Mol. Cell Biol. 20, 474–489 (2019).
Pan, J., Thirumalai, D. & Woodson, S. A. Folding of RNA involves parallel pathways. J. Mol. Biol. 273, 7–13 (1997).
Rook, M. S., Treiber, D. K. & Williamson, J. R. Fast folding mutants of the Tetrahymena group I ribozyme reveal a rugged folding energy landscape. J. Mol. Biol. 281, 609–620 (1998).
Spitale, R. C. & Incarnato, D. Probing the dynamic RNA structurome and its functions. Nat. Rev. Genet. 24, 178–196 (2023).
Mortimer, S. A., Kidwell, M. A. & Doudna, J. A. Insights into RNA structure and function from genome-wide studies. Nat. Rev. Genet. 15, 469–479 (2014).
Hyman, A. A., Weber, C. A. & Julicher, F. Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 30, 39–58 (2014).
Khristich, A. N. & Mirkin, S. M. On the wrong DNA track: Molecular mechanisms of repeat-mediated genome instability. J. Biol. Chem. 295, 4134–4170 (2020).
Micklem, D. R., Adams, J., Grunert, S. & St Johnston, D. Distinct roles of two conserved Staufen domains in oskar mRNA localization and translation. EMBO J. 19, 1366–1377 (2000).
Ferrandon, D., Elphick, L., Nusslein-Volhard, C. & St Johnston, D. Staufen protein associates with the 3’UTR of bicoid mRNA to form particles that move in a microtubule-dependent manner. Cell 79, 1221–1232 (1994).
Laver, J. D. et al. Genome-wide analysis of Staufen-associated mRNAs identifies secondary structures that confer target specificity. Nucleic Acids Res. 41, 9438–9460 (2013).
Mohr, S. et al. Opposing roles for Egalitarian and Staufen in transport, anchoring and localization of oskar mRNA in the Drosophila oocyte. PLoS Genet. 17, e1009500 (2021).
Chen, J. et al. Visualizing the translation and packaging of HIV-1 full-length RNA. Proc. Natl. Acad. Sci. USA 117, 6145–6155 (2020).
Murphy, R. E. & Saad, J. S. The interplay between HIV-1 gag binding to the plasma membrane and Env incorporation. Viruses 12, 548 (2020).
Bergsten, S. E., Huang, T., Chatterjee, S. & Gavis, E. R. Recognition and long-range interactions of a minimal nanos RNA localization signal element. Development 128, 427–435 (2001).
Parker, D. M. et al. mRNA localization is linked to translation regulation in the Caenorhabditis elegans germ lineage. Development 147, dev186817 (2020).
Cardona, A. H. et al. Self-demixing of mRNA copies buffers mRNA:mRNA and mRNA:regulator stoichiometries. Cell 186, 4310–4324 (2023).
Adivarahan, S. et al. Spatial organization of single mRNPs at different stages of the gene expression pathway. Mol. Cell 72, 727–738.e5 (2018).
Khong, A. & Parker, R. mRNP architecture in translating and stress conditions reveals an ordered pathway of mRNP compaction. J. Cell Biol. 217, 4124–4140 (2018).
Venken, K. J. et al. MiMIC: a highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nat. Methods 8, 737–743 (2011).
Manivannan, S. N. et al. Targeted integration of single-copy transgenes in Drosophila melanogaster tissue-culture cells using recombination-mediated cassette exchange. Genetics 201, 1319–1328 (2015).
Song, W., Strack, R. L., Svensen, N. & Jaffrey, S. R. Plug-and-play fluorophores extend the spectral properties of Spinach. J. Am. Chem. Soc. 136, 1198–1201 (2014).
Trcek, T., Lionnet, T., Shroff, H. & Lehmann, R. mRNA quantification using single-molecule FISH in Drosophila embryos. Nat. Protoc. 12, 1326–1348 (2017).
Dulk, S. L. & Rouskin, S. Probing RNA structure with dimethyl sulfate mutational profiling with sequencing in vitro and in cells. J. Vis. Exp. https://doi.org/10.3791/64820 (2022).
Wells, S. E., Hughes, J. M., Igel, A. H. & Ares, M. Jr. Use of dimethyl sulfate to probe RNA structure in vivo. Methods Enzymol. 318, 479–493 (2000).
Gaspar, I., Wippich, F. & Ephrussi, A. Enzymatic production of single-molecule FISH and RNA capture probes. RNA 23, 1582–1591 (2017).
Eastman, P. et al. OpenMM 7: Rapid development of high performance algorithms for molecular dynamics. PLoS Comput. Biol. 13, e1005659 (2017).
Honeycutt, J. D. & Thirumalai, D. The nature of folded states of globular proteins. Biopolymers 32, 695–709 (1992).
Marinari, E. & Parisi, G. Simulated tempering: a new monte carlo scheme. Europhys. Lett. 19, 451–458 (1992).
Shirts, M. R. & Chodera, J. D. Statistically optimal analysis of samples from multiple equilibrium states. J. Chem. Phys. 129, 124105 (2008).
Debye, P. & Hückel, E. Zur Theorie der elektrolyte. I. gefrierpunktserniedrigung und verwandte erscheinungen. Phys. Z. 24 (1923).
Lee, B. & Richards, F. M. The interpretation of protein structures: estimation of static accessibility. J. Mol. Biol. 55, 379–400 (1971).
Mitternacht, S. FreeSASA: An open source C library for solvent accessible surface area calculations. F1000Res. 5, 189 (2016).
Campello, R. J. G. B., Moulavi, D. & Sander, J. in Advances in Knowledge Discovery and Data Mining 160–172 (2013).
Mathews, D. H. & Turner, D. H. Prediction of RNA secondary structure by free energy minimization. Curr. Opin. Struct. Biol. 16, 270–278 (2006).
Danaee, P. et al. bpRNA: large-scale automated annotation and analysis of RNA secondary structure. Nucleic Acids Res. 46, 5381–5394 (2018).
Mathews, D. H., Sabina, J., Zuker, M. & Turner, D. H. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288, 911–940 (1999).
Aierken, D. & Joseph, J. A. Accelerated simulations reveal physicochemical factors governing stability and composition of RNA clusters. J. Chem. Theory Comput. 20, 10209–10222 (2024).
Mathews, D. H. et al. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl. Acad. Sci. USA 101, 7287–7292 (2004).
Ma, W., Zhen, G., Xie, W. & Mayr, C. In vivo reconstitution finds multivalent RNA-RNA interactions as drivers of mesh-like condensates. Elife 10, e64252 (2021).
Bunch, T. A., Grinblat, Y. & Goldstein, L. S. Characterization and use of the Drosophila metallothionein promoter in cultured Drosophila melanogaster cells. Nucleic Acids Res. 16, 1043–1061 (1988).
Hanyu-Nakamura, K., Matsuda, K., Cohen, S. M. & Nakamura, A. Pgc suppresses the zygotically acting RNA decay pathway to protect germ plasm RNAs in the Drosophila embryo. Development 146, dev167056 (2019).
Ziqing Ye, S. T. Controlling intermolecular base pairing in Drosophila germ granules by mRNA folding and its implications in fly development Zenodo, https://doi.org/10.5281/zenodo.15802323 (2025).
Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9, 671–675 (2012).
Deigan, K. E., Li, T. W., Mathews, D. H. & Weeks, K. M. Accurate SHAPE-directed RNA structure determination. Proc. Natl. Acad. Sci. USA 106, 97–102 (2009).
Darty, K., Denise, A. & Ponty, Y. VARNA: Interactive drawing and editing of the RNA secondary structure. Bioinformatics 25, 1974–1975 (2009).
Bolte, S. & Cordelieres, F. P. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224, 213–232 (2006).
Lionnet, T. et al. A transgenic mouse for in vivo detection of endogenous labeled mRNA. Nat. Methods 8, 165–170 (2011).
Acknowledgements
We would like to thank Drs. Anthony Leung and Sarah Woodson for the critical reading of our manuscript. We would like to thank Dr. Akira Nakamura for sharing the Nanos protein antibody with us and Dr. Elizabeth Gavis for sharing Hoyer’s medium with us. D.T.’s research was supported by grants CHE 2320256 and F-0019 from the National Science Foundation and the Welch Foundation, respectively. This research was supported by the NIGMS R35GM142737 grant awarded to T.T.
Author information
Authors and Affiliations
Contributions
S.T. wrote the initial draft of the manuscript, with all the authors participating in the revision and editing of the manuscript. S.T. and T.T.: conceptualization. S.T., H.N. and Z.Y.: investigation and analysis. D.T. and H.N.: In silico modeling/analysis of monomers and homodimers. S.R.: Training with DMS-MaPseq technology.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Christine Roden, Gian Gaetano Tartaglia, and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Tian, S., Nguyen, H., Ye, Z. et al. Controlling intermolecular base pairing in Drosophila germ granules by mRNA folding and its implications in fly development. Nat Commun 16, 8135 (2025). https://doi.org/10.1038/s41467-025-62973-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-62973-7