Abstract
Copy number control of DNA and centrosomes is essential for accurate genetic inheritance. DNA replication and centrosome duplication have been recognized as parallel key events for cell division. Here, we discover that the DNA replication machinery directly regulates the licensing and execution processes of centrosome duplication to prevent centrosome amplification. We find that the microcephaly protein DONSON couples DNA replication initiation with Cdc6 translocation to centrosomes. The Cdc6 signal prevents the precocious occurrence of centriole disengagement, the licensing step for centrosome duplication. During DNA replication, DONSON inhibits replisome disassembly by interacting with the CMG helicase, maintaining the intrinsic S/G2 checkpoint signal that blocks centriole-to-centrosome conversion, the execution step for centrosome duplication. Disruption of these dual signals causes precocious centrosome duplication and chromosome mis-segregation, observed in DONSON patient cells. Our results reveal that the DNA replication machinery not only duplicates genetic material but also controls the system for its accurate segregation.
Similar content being viewed by others
Introduction
The timing of DNA replication and centrosome duplication must be tightly connected with the progression of the cell cycle in animal cells in order to maintain the stability of the genome. For the accurate inheritance of genetic material by daughter cells, the initiation and completion of DNA replication must occur prior to mitosis1. During mitosis, the duplicated chromosomes are equally segregated into the two daughter cells by a bipolar spindle that is organized by two centrosomes2. The centrosomes undergo duplication after chromosome segregation so that the number of centrosomes per cell remains two throughout the cell cycle. Precocious duplication of centrosomes prior to chromosome segregation results in the formation of a multipolar spindle with three or more centrosomes3 and thereby chromosome mis-segregation, which is a major cause of developmental disorders and tumorigenesis in humans4. The sequential ordering of cell cycle events is under the control of cell cycle machinery, which depends on cyclin-dependent kinase (CDK) activity5. However, it remains unclear whether there exists a direct coordination between DNA replication and centrosome duplication in the cell cycle progression.
Cells begin the cell cycle with two centrosomes, each of which consists of a single centriole surrounded by a protein matrix called pericentriolar material (PCM)6 (Supplementary Fig. 1a). The PCM harbors protein complexes pivotal for nucleating microtubules, which enables centrosomes to function as a principal microtubule organizing center (MTOC)7. During S phase, when DNA is replicated, a new centriole grows orthogonally from each pre-existing centriole (centriole duplication)4. The newly formed daughter centrioles remain engaged with their mother centrioles (centriole engagement) until late mitosis8. Following chromosome segregation in late mitosis, the engagement between mother and daughter centrioles in each centrosome is dissolved (centriole disengagement)9. Disengaged daughter centrioles subsequently undergo conversion into new centrosomes (centriole-to-centrosome conversion) through PCM acquisition, depending on the activity of the mitotic kinase, Polo-like kinase 1 (Plk1)10. Centriole disengagement is a crucial prerequisite for the ensuing process of centriole-to-centrosome conversion and the replication of daughter centrioles in the next cell cycle. Therefore, centriole disengagement is widely recognized as the crucial step that licenses centrosome duplication. Considering that centriole-to-centrosome conversion results in the duplication of functional centrosomes, it can be regarded as the execution process of centrosome duplication.
DNA replication begins at multiple loci termed replication origins. In late M and G1 phases, these origins acquire the competence to initiate DNA replication by forming a pre-replication complex (pre-RC), including the MCM motor of replicative helicase (CMG helicase)11,12. Recent studies revealed that the formation of the CMG helicase is mediated by Downstream Neighbor of SON (DONSON), which delivers accessory proteins to the MCM motor13,14,15,16. Thereafter, the CMG helicase unwinds double-stranded DNA, and two replisomes traverse in opposite directions, copying DNA as they progress17. After the convergence of two replisomes on an identical DNA stretch, the CMG helicase is removed from the chromatin, leading to the disassembly of the whole replisome18. The termination of DNA replication triggers the initiation of the G2 phase, preparing cells for entry into mitosis. Recent studies revealed that ATR and Chk1 suppress the expression of a mitotic gene network during DNA replication to prevent premature mitotic entry with under-replicated DNA19,20. This mechanism is therefore called “intrinsic S/G2 checkpoint”, although how the checkpoint is maintained during DNA replication remains unclear.
Microcephalic primordial dwarfism (MPD) describes a broad spectrum of clinically related genetic disorders that are characterized by the presence of microcephaly alongside intrauterine and postnatal growth restriction21. It has been postulated that the abnormal neurodevelopment exhibited by the affected patients often stems from the disruption of the cell cycle of neural progenitor cells (NPCs)22. With the identification of genes encoding centrosomal proteins as causative genes of MPD23, it is presumed that abnormalities in the number of centrosomes and related mitotic defects contribute to the disruption of NPCs. Although mutations in genes encoding components of pre-RC, CMG helicase, and DNA damage response have also been found in MPD patients24, it remains poorly understood whether the causes of the MPD phenotype can be explained solely by defects in DNA processing.
In this study, we uncovered dual roles of the replisome component DONSON in the centrosome duplication cycle. First, DONSON mediates the translocation of Cdc6 to centrosomes upon the initiation of DNA replication. Remarkably, the presence of the DNA replication licensing factor Cdc6 at centrosomes prevents the precocious occurrence of centriole disengagement, the licensing step for centrosome duplication. Second, during the process of DNA replication, DONSON also sustains the activity of the intrinsic S/G2 checkpoint, which suppresses precocious activation of Plk1. Therefore, in cells depleted of DONSON, the disengaged centrioles are converted to centrosomes by the precociously activated Plk1 already in the S phase. The resulting duplicated centrosomes provoke multipolar spindle formation in mitosis, leading to chromosome segregation errors. Overall, these findings lead us to propose that the DNA replication machinery transmits the two signals, the Cdc6 signal and the intrinsic S/G2 checkpoint signal, via a crucial mediator, DONSON, to the centrosome, suppressing unscheduled centrosome duplication and thereby ensuring successful segregation of the replicated chromosomes.
Results
A targeted siRNA screen identified DONSON and Cdc6, MPD-related DNA replication factors, as regulators of centrosome number
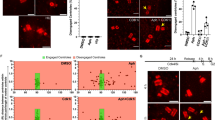
Mutations in genes related to DNA replication have been identified as causative factors for MPD24 (Fig. 1a). Of note, structural and numerical abnormalities in the centrosome are also linked to the cause of MPD21,22,23 (Fig. 1a). Therefore, we decided to examine if these MPD genes had any potential involvement in centrosome biogenesis. To this end, we visualized centrosomes by immunostaining of a centrosome marker, CEP192, in cells transfected with siRNA targeting the mRNA of each MPD gene. Among 12 MPD genes implicated in DNA replication (ATR, Cdc6, Cdc45, Cdt1, DNA2, DONSON, GMNN, MCM5, Orc1, Orc4, Orc6, RBBP8), we found that depletion of Cdc6 or DONSON caused a significant increase in the number of Cep192 foci (>2) per cell (Fig. 1b, c). The same defect in centrosome number was observed in different human cell lines treated with the DONSON siRNA (Supplementary Data Fig. 1b, c). The efficacy or the specificity of Cdc6 and DONSON knockdown was confirmed by immunoblotting (Supplementary Fig. 1d, e) or rescue experiments (Supplementary Fig. 1f, g), respectively. Next, we utilized hTERT-immortalized fibroblast cell lines derived from two patients with different DONSON mutations25 (Patients 2 and 6; Fig. 1d, Supplementary Fig. 1h). Immunofluorescence analysis revealed that excess Cep192 foci were present in the patient-derived cell lines and rescued by the exogenous expression of DONSON wild-type (WT) (Patient 2: ~8% and Patient 6: ~11% in control cells, compared with ~1% in those cells exogenously expressing DONSON, Fig. 1e, f, Supplementary Fig. 1i, j), confirming the specificity for the centrosome amplification phenotype provoked by dysfunction of DONSON. Taken together, these data suggest that the phenotype of centrosome amplification could be a cellular pathology associated with MPD.
a List of genes involved in microcephalic primordial dwarfism (MPD) based on previous studies22. The genes involved in MPD are mostly classified as centrosomal proteins, DNA replication components, or a regulator for DNA damage response. b The number of Cep192 foci upon siRNA treatment against the indicated proteins. c, Histograms represent the frequency of interphase cells with >2 Cep192 foci observed in (b). n = 3 independent experiments, 50 cells for each. d Schematic of the DONSON gene, indicating mutations observed in cells from MPD patient 2 (P2)25. The genomic structure is based on the longest ORF, containing ten coding exons (white rectangles) (NCBI NM_017613.3). e The number of Cep192 foci in hTERT-immortalized fibroblasts derived from the MPD patient 2. Fibroblasts were infected with pMSCV-empty-vector or pMSCV-DONSON and immunostained with antibodies against Cep192 (red). Arrowheads indicate centrosomes. f Histograms represent the frequency of interphase cells with the indicated phenotypes observed in (e). n = 3 independent experiments, 50 cells for each. g The number of centrin and Cep192 foci in G1 or S/G2 phase upon DONSON or Cdc6 depletion. HeLa cells were treated with siRNA treatment against the indicated proteins for 48 h and 10 μM EdU for 30 min. CENP-F and EdU double-negative cells were classified as G1 phase cells, and others as S or G2 phase cells. Arrowheads indicate centrioles. h Histograms represent the frequency of cells with the indicated phenotype observed in (g). n = 3 independent experiments, 30 cells for each. i Time-lapse observation of centriole disengagement in DONSON-depleted cells. HeLa GFP-centrin-1 cells were visualized every 20 min for 48 h after 24 h treatment with siRNA and 3 h treatment with 100 nM SiR-DNA. Arrowheads indicate engaged centriole pairs, and left–right arrows indicate precociously disengaged centrioles. j Cumulative scatterplot indicates the duration from nuclear envelope breakdown (NEBD) to centriole disengagement observed in (i). k Schematic model of the phenotypes of DONSON or Cdc6 depletion. All scale bars, 5 μm. Values are mean percentages ± s.d. Tukey’s multiple comparison test was used in c, and a two-tailed, unpaired Student’s t-test was used in (f) to obtain P-value. Throughout all the figures in this study, the single-channel images were generated by displaying only the signal from one channel extracted directly from the original merged representative image, without any adjustment to intensity or contrast. The insets represent magnified views of regions taken directly from either the merged image or the corresponding single-channel image, also without any further modification. The insets are displayed in the following order from top to bottom: merged, green, red, and cyan. All source data are provided as a Source Data file.
Recent studies reported that DONSON is required for DNA replication initiation in Caenorhabditis elegans and Xenopus laevis13,14,15,16. Consistent with this, our flow cytometric analyses with synchronous human cells revealed that depletion of DONSON efficiently suppressed the entry into S phase. However, at least under this experimental condition, a very gradual entry of the cells into S phase was detectable, albeit with a significant delay (Supplementary Fig. 1k)25. In addition, centrosome amplification was not induced by inhibition of DNA replication initiation with Cdc7 inhibitor or by arresting cells in the S phase with Emi1 siRNA (Supplementary Fig. 1l–p). These data suggest that centrosome amplification in DONSON-depleted cells was not induced by the defects in DNA replication initiation or cell cycle progression.
The centrosome is composed of one or two centrioles (mother and daughter centrioles) surrounded by pericentriolar material (PCM). Each daughter centriole is engaged with the mother centriole orthogonally until late mitosis. After mitotic exit, mother and daughter centrioles are dissociated (centriole disengagement), and disengaged daughter centrioles are subsequently converted into new centrosomes (centriole-to-centrosome conversion) through PCM acquisition. In general, centrosome amplification can arise from either cytokinesis failure, centriole overduplication, or precocious centriole disengagement. While cells that failed cytokinesis in the previous cell cycle have four centrosomes in G1 phase, as their cellular components are doubled, this was rarely observed in cells depleted of DONSON or Cdc6 (Fig. 1g, h). This result indicates that the centrosome amplification is not due to a failure of cytokinesis. We then investigated the number of centrioles, the core structures of the centrosomes, in DONSON or Cdc6-depleted cells. Two mother centrioles within a cell undergo duplication in S phase, resulting in the presence of two pairs of engaged centrioles4. Newly duplicated centrioles remain engaged as a pair with their mother centrioles until late mitosis, a mechanism preventing their precocious conversion into centrosomes9. In DONSON or Cdc6-depleted cells, during G1 phase, the number of centrioles, as indicated by the centriolar protein centrin, did not differ from that in control cells (Fig. 1g, h). Intriguingly, in the S/G2 phase, both centriole amplification and unpaired centrioles were frequently observed in DONSON or Cdc6-depleted cells, unlike in the control cells (Fig. 1g, h, Supplementary Fig. 2a–c). Both phenotypes were considered to potentially result from a single cause, precocious centriole disengagement. Our live imaging analysis of cells stably expressing GFP-centrin-1 revealed that depletion of DONSON or Cdc6 causes paired centrioles precociously to separate from each other during interphase (Fig. 1i, j, Supplementary Fig. 2d and Supplementary Movie 1). These separated centrioles in DONSON-depleted cells were predominantly marked by C-Nap1, a protein loaded to disengaged centrioles26,27 (Supplementary Fig. 2e, f), and the centrosome marker Cep192 (Fig. 1g), indicating that the supernumerary centrosome phenotype was due to precocious disengagement of the newly formed centrioles followed by their conversion to centrosomes. Regarding the positional relationship of centriole pairs during interphase, a previous study has reported that in cells experiencing mild replication stress, the angle between centriole pairs changes due to prolongation of the G2 phase28. We also examined centriole pairs under the same conditions. While changes in the angle between centriole pairs, which is typically around 90 degrees, were observed, no significant increase in the distance between the two centrioles was detected (Supplementary Fig. 2g–i). In contrast, when cells were treated with siDONSON or siCdc6, precocious centriole disengagement was induced, characterized by the separation of paired centrioles with a sufficient distance. This phenotype likely arises via a mechanism distinct from that in the previous research. Taken together, these results indicate that DONSON and Cdc6 maintain centriole engagement to avoid precocious centrosome duplication during interphase (Fig. 1k).
DONSON regulates the translocation of Cdc6 from the nucleus to the centrosomes for the maintenance of centriole engagement in interphase
We next investigated how DONSON and Cdc6 regulate centriole engagement in interphase. First, we determined the localization pattern of DONSON and Cdc6 exogenously expressed in HeLa cells using a doxycycline-inducible expression system. We found that mNG-DONSON localized in the nucleus but not at the centrosomes during interphase (Fig. 2a). Meanwhile, mNG-Cdc6 localized in the nucleus in G1 phase and was then translocated to the cytoplasm and the centrosomes in the S/G2 phase (Fig. 2a) as previously reported29,30. To investigate the exact timing of the Cdc6 translocation, cells were synchronized in G1 phase using Cdc7 inhibitor PHA767491 and released into the S phase. We observed that endogenous Cdc6 is removed from the nucleus and translocated to the cytoplasm and centrosomes immediately after DNA replication initiation (Fig. 2b, Supplementary Fig. 3a–c). Interestingly, we found that the centrosomal translocation of Cdc6 is dependent on the nuclear protein DONSON. Depletion of DONSON significantly reduced the signal intensity of endogenous Cdc6 at centrosomes in S phase (Fig. 2c, d), without decreasing the amount of Cdc6 in mRNA levels, total protein expression levels, or protein expression levels in the cytosol (Supplementary Fig. 3d, e). Remarkably, overexpression of mNG-Cdc6, which enables centrosomal localization of mNG-Cdc6 even upon DONSON depletion, significantly reduced the frequency of precocious centriole disengagement in the S/G2 phase in DONSON-depleted cells (Fig. 2e, f, Supplementary Fig. 3f). In contrast, overexpression of mNG-DONSON did not rescue the phenotype in Cdc6-depleted cells (Supplementary Fig. 3g, h). These data suggest that DONSON acts upstream of Cdc6 in the nucleus to regulate the translocation of Cdc6 to the centrosomes for maintenance of centriole engagement in S/G2 phase.
a Subcellular localization of overexpressed DONSON and Cdc6 in G1 and S/G2 phases. HeLa-Tet3G cells were treated with 1 μg/mL Doxycycline (Dox) for 24 h. b HeLa cells were fixed 24 h after synchronization in G1 phase using 10 µM PHA767491 (Cdc7i) or 4 h after release. c Cdc6 localization at centrioles in S phase in control or DONSON-depleted cells. d Quantification of the normalized signal intensity of Cdc6 at each centriole observed in (c). n = 50 from three independent experiments. e Inducible expression of Cdc6 upon DONOSN depletion. HeLa-Tet3G-mNG-Cdc6 cells were treated with siRNA for 48 h, followed by 1 μg/mL Dox treatment for 24 h. f Histograms represent the frequency of interphase cells with > 2 C-Nap1 foci observed in (e). n = 3 independent experiments, 30 cells for each. g List of proteins previously reported to be involved in centriole engagement or disengagement. h Precocious centriole disengagement was inhibited upon co-depletion of Cdh1 and DONSON/Cdc6. i, j Histograms represent the frequency of interphase cells with >2 C-Nap1 foci observed in (h). n = 3 independent experiments, 50 cells for each. k Cdh1 localization at centrosomes in cells treated with siControl or siCdc6. l Inducible expression of Cdc6 upon DONOSN depletion. HeLa-Tet3G-mRuby3-Cdc6 cells were treated with siRNA for 48 h, followed by 1 μg/mL Dox treatment for 24 h. m Quantification of the normalized signal intensity of Cdh1 at each centriole with the indicated treatment. n = 50 from three independent experiments. n Schematic illustration of the phenotype of DONSON depletion. The depletion of DONSON leads to the mislocalization of Cdc6 at the centrosomes during the S phase, where Cdc6 inhibits the premature recruitment of Cdh1. All scale bars, 5 μm. Values are mean percentages ± s.d. Tukey’s multiple comparison test was used in (i), Two-tailed, unpaired Student’s t-test was used in (f) and (j), Mann–Whitney U test was used in (d), and Kruskal–Wallis test was used in (m) to obtain P-value. Source data are provided as a Source Data file.
We then sought to clarify the relationship between the DONSON-Cdc6 axis and the known regulators of centriole engagement and disengagement (Fig. 2g). While centriole engagement in interphase is maintained by Cep57 and its paralog Cep57L131 (Supplementary Fig. 3i), these two proteins were localized at all of the disengaged centrioles in DONSON-depleted cells (Supplementary Fig. 3j–m). This was also true for PCNT, a PCM component necessary for centriole engagement in mitosis32,33,34 (Supplementary Fig. 3n). Therefore, we next tested the requirement of the known factors positively regulating the disengagement process (Plk1, ESPL1/Separase, Cdc20, Cdh1/Fzr1; Fig. 2g)8,35,36 in the phenotype resulting from DONSON/Cdc6 depletion. Notably, precocious centriole disengagement, in DONSON or Cdc6-depleted cells, was suppressed only by co-depletion of Cdh1 (Fig. 2h–j, Supplementary Fig. 3o, p). The signal of endogenous Cdh1 at the centrosomes appeared at low levels in interphase and greatly increased in mitosis37, which indicates an inverse correlation with the localization pattern of Cdc6 at centrosomes (Fig. 2k). We therefore hypothesized that the DONSON-Cdc6 axis suppresses precocious Cdh1 loading at the centrosomes during interphase. Indeed, depletion of DONSON or Cdc6 significantly increased the signal intensity of Cdh1 at the centrosomes in interphase (Fig. 2k, l). Furthermore, the phenotype of excess Cdh1 loading at the centrosome in DONSON-depleted cells was substantially rescued by overexpression of mRuby3-Cdc6 (Fig. 2l, m). Taken together, these data indicate that the replisome component DONSON mediates the centrosomal translocation of Cdc6 upon the initiation of DNA replication to suppress precocious accumulation of the disengagement factor Cdh1 at the centrosomes, thus maintaining centriole engagement in interphase (Fig. 2n).
DONSON suppresses Plk1 activation to prevent precocious maturation of daughter centrioles during S phase
During late mitosis, disengaged daughter centrioles are converted to centrosomes by recruiting PCM components and centriole duplication factors in a Plk1-dependent manner10,38,39. Consistently, precociously disengaged centrioles in Cep57/Cep57L1 co-depleted cells acquire PCM components only after G2 phase31 when Plk1 starts to be activated40, which we also observed in Cdc6-depleted cells (Supplementary Fig. 4a, b). In contrast, in DONSON-depleted cells, the major PCM component PCNT was localized at most of the disengaged centrioles already in S phase (Fig. 3a, b). Microtubule regrowth assay also revealed that these disengaged daughter centrioles had the ability to nucleate microtubules during S phase (Fig. 3c). Moreover, Cep152, a scaffold protein required for centriole duplication41,42,43, and HsSAS-6, a procentriole marker44, were also recruited to most of the disengaged daughter centrioles in DONSON-depleted cells (Supplementary Fig. 4c–f), indicating that the disengaged centrioles harbored the ability to duplicate again.
a Recruitment of PCNT to disengaged daughter centrioles in S phase after siRNA treatment against the indicated proteins. After 32 h of siRNA transfection, cells were treated with EdU for 30 min to classify cells in S phase. b Histograms represent the frequency of cells with more than two PCNT-positive centrioles in S phase observed in (a). n = 3 independent experiments, 30 cells for each. c Ectopic MTOC activity of precociously disengaged daughter centrioles in S phase in DONSON-depleted cells. HeLa cells were treated with siControl, siCep57/Cep57L1, or siDONSON for 32 h and followed by 10 µM nocodazole treatment for 3 h. After nocodazole treatment, the cells were cold-treated for 1 h, followed by 30-s incubation at 37 °C. d Phosphorylation of Plk1 at T210 residue at centrosomes of WT cells in G1, S, and G2 phases. e Phosphorylation of Plk1 at T210 residue in S phase upon DONSON depletion. f Quantification of the normalized signal intensity of phosphorylated Plk1 (T210) at each centriole observed in (e). n = 50 from three independent experiments. g Recruitment of γ-Tubulin to disengaged daughter centrioles upon Plk1 inhibition in DONSON-depleted cells. HeLa cells were treated with siControl or siDONSON for 48 h, followed by treatment with DMSO or 100 nM BI2536 (Plk1i) for 24 h. After siDONSON treatment, Plk1 activity remains high, preventing linker formation between disengaged centrioles. In contrast, DONSON-depleted cells with Plk1 inhibition retain the linker after disengagement, keeping centrioles closely spaced. Disengagement is defined as >2 C-Nap1 foci. h, i Histograms represent the frequency of the interphase cells with the indicated phenotypes observed in (g). n = 3 independent experiments, 50 cells for each. j The basal Chk1 activity upon DONSON depletion. Representative immunoblot analysis of whole cell extracts for Chk1 and p-Chk1 (S296) from HeLa cells treated with siControl or siDONSON for 48 h. HSP90 was used as a loading control. k Overexpression of a constitutively active mutant of Chk1 (Chk1-L449R construct) upon DONSON depletion. l Histograms represent the frequency of cells with more than two γ-Tubulin-positive centrioles observed in (k). n = 3 independent experiments, 30 cells for each. m Schematic model of the phenotypes of DONSON depletion. All scale bars, 5 μm. Values are mean percentages ± s.d. Tukey’s multiple comparison test was used in (h) and (i), a two-tailed, unpaired Student’s t-test was used in (l), and Mann–Whitney U test was used in (f) to obtain the P-value. Source data are provided as a Source Data file.
Based on the above observations, we assumed that depletion of DONSON may induce precocious Plk1 activation, leading to the centriole-to-centrosome conversion of disengaged daughter centrioles in S phase. Since activation of Plk1 requires phosphorylation on T21045, we monitored the presence of T210-phosphorylated Plk1 (pT210-Plk1) at disengaged centrioles in DONSON-depleted S-phase cells. While the pT210-Plk1 signal was hardly detectable at the centrosomes, prior to G2 entry in control cells (Fig. 3d), we detected a substantial increase of pT210-Plk1 at most of the disengaged centrioles even in S phase upon DONSON depletion (Fig. 3e, f). The increased amount of pT210-Plk1 in DONSON-depleted cells was also confirmed by western blotting (Supplementary Fig. 4g). Moreover, treatment with a Plk1 inhibitor significantly reduced the number of disengaged daughter centrioles co-localizing with the major PCM component γ-tubulin in DONSON-depleted cells, while it had no significant effect on the frequency of precocious centriole disengagement (siDONSON: C-Nap1: ~24%, γ-tubulin: ~24%, siDONSON + Plk1i: C-Nap1: ~22%, γ-tubulin: ~5%, Fig. 3g–i), indicating that Plk1 activity is required for the centrosome conversion of disengaged daughter centrioles upon DONSON loss.
A previous study reported that in unperturbed cells, inhibition of Checkpoint-kinase 1 (Chk1) causes precocious Plk1 activation in S phase46. Indeed, Chk1 inhibition significantly increased pT210-Plk1 at centrioles without affecting the population of cells in S phase (Supplementary Fig. 4h–j). Moreover, it has been reported that DONSON activates the ATR-Chk1 pathway in the presence of DNA replication stress25. We therefore monitored Chk1 auto-phosphorylation at S296, which mirrors the intrinsic activation of Chk120. We found that Chk1 phosphorylation at S296 was detectable even in unperturbed cells, but significantly reduced upon DONSON depletion (Fig. 3j, Supplementary Fig. 4k). Moreover, the expression of a constitutively active mutant of Chk1 (Chk1-L449R)47 efficiently rescued the phenotype of increased centrosome number resulting from DONSON depletion (Fig. 3k, l). In contrast, Chk1 inhibition significantly increased the centriole-to-centrosome conversion of disengaged daughter centrioles in Cep57/Cep57L1-depleted S-phase cells (Supplementary Fig. 4l, m). Furthermore, inhibition of Ataxia-telangiectasia mutated and Rad3-related (ATR), which is known to act upstream of Chk1, also induced precocious Plk1 activation at centrosomes during S phase without affecting the population of cells in S phase (Supplementary Fig. 4j, n, o). Overall, these findings indicate that DONSON suppresses unscheduled Plk1 activation via the ATR-Chk1 pathway to avoid precocious centriole-to-centrosome conversion of disengaged daughter centrioles (Fig. 3m).
DONSON maintains the intrinsic S/G2 checkpoint by ensuring the stability of ETAA1
The ATR-Chk1 pathway negatively regulates the S/G2 transition program and thereby prevents precocious mitotic entry in unperturbed cells, which is called “intrinsic S/G2 checkpoint”19 (Fig. 4a). Plk1 is known to promote mitotic entry. We therefore assumed that the Plk1 activity is one of the processes regulated by the intrinsic S/G2 checkpoint. To investigate whether DONSON is generally involved in the intrinsic S/G2 checkpoint, we monitored the processes regulated by the checkpoint. CyclinB1, which forms a complex with CDK1 and functions as a master regulator of mitosis48, starts accumulating in the cytoplasm in G2 phase (Fig. 4b). As is the case with ATR or Chk1 inhibition19 (Fig. 4c, d), depletion of DONSON led to precocious CyclinB1 accumulation during S phase (Fig. 4e, f). In addition, ultrafine anaphase DNA bridges (UFBs), a hallmark of under-replicated DNA in mitosis49,50 and an indicator of the dysfunction of the intrinsic S/G2 checkpoint19, were observed with higher frequency in DONSON-depleted cells (Supplementary Fig. 5a, b). Overall, these data indicate that DONSON enforces the intrinsic S/G2 checkpoint during S phase.
a A schematic of the intrinsic S/G2 checkpoint based on previous studies19,20,73. b The subcellular localization pattern of CyclinB1 of WT cells in G1, S, and G2 phases. c The expression pattern of CyclinB1 in S phase upon Chk1- or ATR-inhibition. HeLa cells were treated with 250 nM CHIR-124 (Chk1i) or 2 μM VE-821 (ATRi) for 6 h and EdU for 30 min to classify cells in S phase. d Histograms represent the frequency of the S phase cells with the indicated phenotypes observed in (c). n = 3 independent experiments, 30 cells for each. e The expression pattern of CyclinB1 in S phase after siRNA treatment against the indicated proteins. f Histograms represent the frequency of the S phase cells with the indicated phenotypes observed in (e). n = 3 independent experiments, 30 cells for each. g Representative immunoblot analysis of whole cell extracts for the indicated proteins from HeLa cells treated with siControl or siDONSON for 48 h. HSP90 was used as a loading control. h Representative immunoblot analysis of whole cell extracts for ETAA1 upon treatment with a proteasome inhibitor in DONSON-depleted cells. HeLa cells were treated with siControl or siDONSON for 48 h and 20 μM MG132 for 6 h. α-Tubulin was used as a loading control. i Ubiquitination of ETAA1 upon DONSON depletion. HeLa-Tet3G mNG-ETAA1 cells were treated with siControl or siDONSON and doxycycline (Dox) for 48 h, transfected with a HA-ubiquitin expressing plasmid for 24 h, and treated with 20 μM MG132 for 6 h. mNG-ETAA1 was immunoprecipitated with mNG-beads. Immunoprecipitates were blotted with HA antibody to detect ubiquitylated ETAA1. IP, immunoprecipitation; WCE, whole cell extract. j, l Inducible expression of ETAA1 in DONSON-depleted cells. HeLa-Tet3G mNG-ETAA1 cells were treated with siControl or siDONSON following Dox treatment for 24 h. Arrowheads indicate centrosomes. k, m Histograms represent the frequency of S-phase cells with the indicated phenotypes observed in (j) and (l), respectively. n = 3 independent experiments, 30 cells for each. All scale bars, 5 μm. Values are mean percentages ± s.d. Tukey’s multiple comparison test was used in (f), and a two-tailed, unpaired Student’s t-test was used in (k) and (m) to obtain P-value. Source data are provided as a Source Data file.
We then investigated the mechanism of how DONSON enforces the intrinsic S/G2 checkpoint. Intriguingly, we found that depletion of DONSON significantly decreased the total amount of ETAA1, an activator of ATR in the intrinsic S/G2 checkpoint19 (Fig. 4e–g). Conversely, doxycycline-induced overexpression of mNG-DONSON resulted in an increase of the total amount of ETAA1 (Supplementary Fig. 5c). Since there was no significant change in the mRNA levels of ETAA1 between control and DONSON-depleted cells (Supplementary Fig. 5d), we assumed that DONSON protects ETAA1 from protein degradation. The reduction in the amount of ETAA1 was efficiently recovered by the addition of the proteasome inhibitor MG132 in DONSON-depleted cells (Fig. 4h). Consistently, we also observed that depletion of DONSON significantly increased poly-ubiquitination of ETAA1 when overexpressed with HA-tagged ubiquitin and in the presence of MG132 (Fig. 4i). However, the interaction between DONSON and ETAA1 was not detected by co-immunoprecipitation assay (co-IP) (Supplementary Fig. 5e), suggesting that DONSON indirectly suppresses the ubiquitination of the ETAA1 protein, protecting it from proteasome-dependent degradation. We therefore examined whether the phenotype provoked by DONSON depletion could be rescued by exogenous overexpression of ETAA1. As expected, the ETAA1 overexpression significantly reduced precocious CyclinB1 accumulation (Fig. 4j, k) and centrosome amplification (Fig. 4l, m) during S phase in DONSON-depleted cells. Overall, we conclude that DONSON maintains the intrinsic S/G2 checkpoint to prevent precocious Plk1 activation, through the protection of ETAA1 from protein degradation.
DONSON suppresses unscheduled CMG disassembly to enforce the intrinsic S/G2 checkpoint
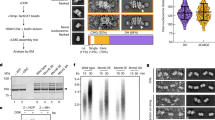
We then sought to investigate the mechanism of how DONSON stabilizes the ETAA1 protein. Given that ETAA1 was exclusively expressed during S phase and degraded in G2 and other cell cycle phases (Fig. 5a), we hypothesized that ETAA1 expression is maintained while DNA synthesis is still underway, and that ETAA1 degradation is triggered by the termination of DNA replication. The CMG helicase, which is composed of Cdc45, MCM2-7, and GINS1-451, could be a key component for the regulation of ETAA1 expression because it is present as a complex during DNA replication but disassembles at its termination. This disassembly of the CMG helicase is driven by the p97 ATPase52,53, which requires ubiquitination of MCM7, a component of the CMG helicase complex54,55 (Fig. 5b). During DNA replication, the ubiquitination of MCM7 is counteracted by USP7, a replisome-enriched deubiquitinase56, which prevents unscheduled disassembly of the CMG helicase. Notably, inhibition of p97 and USP7 increased and decreased ETAA1 expression levels, respectively (Fig. 5c) without affecting the population of S-phase cells (Supplementary Fig. 6a). This indicates that the stability of the CMG helicase is closely linked to the amount of ETAA1. Furthermore, p97 inhibition significantly recovered the amount of ETAA1 in DONSON-depleted cells (Fig. 5d), indicating that DONSON suppresses precocious ETAA1 degradation by maintaining the stability of the CMG helicase until DNA replication termination. To support this hypothesis, we employed a detergent-based extraction of non-chromatin-bound proteins from the nucleus57,58 (Supplementary Fig. 6b) and monitored the presence of the CMG helicase complex on chromatin in S phase. As expected, the amount of chromatin-bound Cdc45 was significantly decreased in DONSON-depleted cells (Fig. 5e, f). Consistently, biochemical cell fractionation revealed that overexpression of DONSON significantly increased the amount of chromatin-bound Cdc45 and ETAA1 (Supplementary Fig. 6c). Recent studies demonstrated that DONSON is required for the initial assembly of the CMG helicase in Caenorhabditis elegans and Xenopus laevis13,14,15,16. Consistently, in our experimental condition, in which DONSON expression was partially suppressed with siRNA in human cells, the initiation of DNA replication was significantly inhibited but DNA replication still proceeded very gradually (Supplementary Fig. 1k). This might be the reason why the precocious disassembly of the CMG helicase was detectable in DONSON-depleted cells released into S phase (Fig. 5g, h). Consistently, depletion of DONSON led to a precocious accumulation of p97, an executor of CMG disassembly, on chromatin during S phase (Fig. 5i, j, Supplementary Fig. 6d, e), and the decrease in the amount of chromatin-bound Cdc45 in DONSON-depleted cells was recovered by p97 inhibition (Fig. 5k, l). To investigate whether precocious CMG disassembly compromises the activity of the intrinsic S/G2 checkpoint in DONSON-depleted cells, we monitored the phosphorylation of Plk1 at T210 at centrioles upon p97 inhibition. We found that precocious Plk1 phosphorylation was suppressed by p97 inhibition in DONSON-depleted S-phase cells (Fig. 5m, n). Taken together, we conclude that DONSON suppresses unscheduled CMG disassembly and thereby enforces the intrinsic S/G2 checkpoint during DNA replication (Fig. 5o).
a Protein expression levels of ETAA1 in G1, S, G2, and M phases of WT cells. HeLa cells were synchronized in G2 phase with 10 µM RO-3306 (Cdk1i), released, and harvested at the indicated time points. b A schematic of the DNA replication termination based on previous studies. c Representative immunoblot analysis of whole cell extracts for ETAA1 from HeLa cells treated with 20 μM P22077 (USP7i) or 10 µM NMS-873 (p97i) for 6 h. α-Tubulin was used as a loading control. d Representative immunoblot analysis of whole cell extracts for ETAA1 from HeLa cells treated with siControl or siDONSON for 48 h, followed by treatment with DMSO or 10 µM NMS-873 (p97i) for 6 h. e The amount of chromatin-bound Cdc45 in S phase upon DONSON depletion. HeLa cells were treated with siControl or siDONSON for 48 h, and soluble proteins were pre-extracted by PBS containing 0.2% Triton X-100 on ice for 10 s before fixation. f Quantification of the normalized signal intensity of chromatin-bound Cdc45 in S phase observed in (e). n = 50 from three independent experiments. g The amount of chromatin-bound Cdc45 in S phase upon DONSON depletion. HeLa cells were treated with siControl or siDONSON for 48 h and synchronized at the G1/S boundary with 6 µM Aphidicolin for 24 h, released and harvested at the indicated time points. Soluble proteins were pre-extracted by PBS containing 0.2% Triton X-100 on ice for 10 s before fixation. h Quantification of the normalized signal intensity of Cdc45 in the chromatin observed in (g). n = 50 from three independent experiments. i The amount of chromatin-bound p97 in S phase upon DONSON depletion. HeLa cells were treated with siControl or siDONSON for 48 h, and soluble proteins were pre-extracted by CSK buffer on ice for 20 s before fixation. j Quantification of the normalized signal intensity of chromatin-bound p97 in S phase observed in (i). n = 50 from three independent experiments. k The amount of chromatin-bound Cdc45 in S phase upon DONSON depletion. HeLa cells were treated with siDONSON for 48 h, and with or without NMS-873 for 6 h, and soluble proteins were pre-extracted by PBS containing 0.2% Triton X-100 on ice for 10 s before fixation. l Quantification of the normalized signal intensity of Cdc45 in the chromatin in S phase observed in (k). n = 50 from three independent experiments. m Phosphorylation of Plk1 at T210 residue in S phase. HeLa cells were treated with siDONSON for 48 h, and with or without NMS-873 for 6 h before fixation. n Quantification of the normalized signal intensity of phosphorylated Plk1 (T210) at each centriole observed in (m). n = 50 from three independent experiments. o Schematic model of the phenotypes of DONSON depletion. During DNA replication, ETAA1 localized to chromatin through its interaction with the single-stranded DNA-binding protein RPA67. Depletion of DONSON causes precocious CMG disassembly and following ETAA1 degradation in S phase. All scale bars, 5 μm. Values are mean percentages ± s.d. Mann–Whitney U test was used in (f), (j), (l), and (n), and the Kruskal–Wallis test was used in (h) to obtain the P-value. Source data are provided as a Source Data file.
The binding of DONSON to MCM3 blocks the loading of the complex destroying the CMG helicase during DNA replication
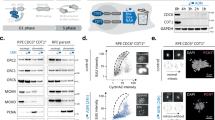
We next set out to gain insight into the mechanism by which DONSON protects the CMG helicase complex from disassembly. To explore whether DONSON directly associates with components of the CMG helicase, we performed an in silico screen for potential interactors of DONSON among CMG components by using the AlphaFold-Multimer (AF-M) algorithm59. The predicted aligned error (PAE) value provided by the AF-M algorithm gives the measure of a distance error for every pair of residues in the predicted protein structure. A low value of PAE (e.g., less than 5 angstroms) means that the prediction of the positional relationship between two amino acid residues is reliable. Thus, in cases where there is a direct binding between two domains, the PAE values between amino acid residues present in both domains tend to be low. Based on comprehensive analyses of the PAE values for the combination of DONSON and all 11 components of the CMG helicase, MCM3 and GINS4 were predicted to directly interact with DONSON (Fig. 6a, b, Supplementary Fig. 7a, b)13,14,15,16, while the N-terminus of GINS4, including the predicted DONSON-binding site (a.a. 19–21), did not adopt a specific structure in Cryo-EM structure of human DNA replisome60 (Supplementary Fig. 7c). We next validated the result of AF-M by a co-immunoprecipitation (co-IP) assay in human HEK293T cells. Indeed, DONSON was found to interact with MCM3, but not MCM6 (Supplementary Fig. 7d). This interaction is highly dependent on the alpha-helix structure (amino acids 354–375) of DONSON, which was predicted by the AF-M algorithm to be in close proximity to MCM3 (Fig. 6b, c). Furthermore, to better mimic the structural state of MCM3 during DNA replication, the interaction of DONSON with MCM3 was examined using the AF-M algorithm under conditions where MCM3 formed a complex with MCM5 and MCM7, which directly interact with MCM3 within the CMG complex. Even in this scenario, DONSON exhibited a similar binding mode to MCM3, and the potential formation of interaction interfaces with MCM5 and MCM7 was also suggested (Supplementary Fig. 8a).
a The Predicted Aligned Error (PAE) plot of the AlphaFold-Multimer (AF-M)-predicted DONSON-MCM3 and DONSON-MCM6 complex structure. b Ribbon representation of the predicted DONSON-MCM3 complex structure corresponding to (a). c HEK293T cells co-expressing MCM3-3×FLAG and HA-DONSON or the indicated deletion mutant were immunoprecipitated with FLAG antibodies. d A schematic of MCM7 poly-ubiquitination mediated by Cul2-LRR1 E3-ubiquitin ligase based on the previous studies. e The Predicted Aligned Error (PAE) plot of the AlphaFold-Multimer (AF-M)-predicted complex structure of MCM3/5/7-LRR1-ELOB-ELOC-Cul2-RBX1 with or without DONSON. f Representative structural models of the predicted protein complexes corresponding to the PAE plots in (e). All proteins are indicated in different colors. In the protein complex on the right, which lacks DONSON, the primary ubiquitination sites of MCM7, K26 and K27 (light blue), can be in close proximity (approximately 50 Å) to the C-terminal region of Cul2 and RBX1. g The interaction between endogenous MCM3 and LRR1 upon DONSON depletion. HeLa cells were treated with siControl or siDONSON for 48 h, and endogenous MCM3 was immunoprecipitated with an antibody against MCM3. h The amount of chromatin-bound LRR1 in S phase upon DONSON depletion. HeLa cells were treated with siControl or siDONSON for 48 h, and soluble proteins were pre-extracted by CSK buffer on ice for 10 s before fixation. i Quantification of the normalized signal intensity of chromatin-bound LRR1 in S phase observed in (h). n = 50 from three independent experiments. j Inducible expression of RNAi-resistant mNG-DONSON or the indicated deletion mutant upon DONSON depletion. k Quantification of the normalized signal intensity of phosphorylated Plk1 (T210) at each centriole with the indicated treatment. n = 50 from three independent experiments. All scale bars, 5 μm. Values are mean percentages ± s.d. Mann–Whitney U test was used in (i) and the Kruskal–Wallis test was used in (k) to obtain the P-value. Source data are provided as a Source Data file.
During DNA replication termination, LRR1, which forms an E3 ligase complex with Cul2, ELOB, ELOC, and RBX1, and mediates ubiquitination of MCM7 for CMG disassembly (Fig. 6d)61,62,63. It has been revealed that LRR1 predominantly binds to MCM3 while also forming interaction interfaces with MCM5 and MCM761,62. We therefore hypothesized that DONSON interacts with the MCM3/5/7 complex to counteract the binding of LRR1 to the MCM3/5/7 complex for stabilizing the CMG helicase until replication termination. To investigate this hypothesis, firstly, the binding of the ubiquitin ligase complex LRR1/ELOB/ELOC/Cul2/RBX1 to the MCM3/5/7 complex was analyzed using the AF-M algorithm. As with previous reports, the PAE values predicted that this ubiquitin ligase complex interacts with the MCM3/5/7 complex through LRR1 (Fig. 6e). Indeed, when the complex structure prediction is validated by the AF-M, LRR1 interacts with the MCM3/5/7 complex in a fixed position (Supplementary Fig. 8b). In contrast, while the N-terminus of Cul2 adopts a relatively fixed position, the arm region of Cul2 exhibits flexible localization, resulting in a wide swing in the predicted position of the C-terminus (Supplementary Fig. 8d). We assume that this flexibility is essential for the conjugation of long polyubiquitin chains to MCM7, which is required for CMG disassembly. Consistent with this, the C-terminus of Cul2 and RBX1 can be predicted to be in close proximity to the primary ubiquitylation sites on MCM7 (Fig. 6f). Subsequently, the effect of DONSON on the binding of this ubiquitin ligase complex to the MCM3/5/7 complex was evaluated. Importantly, in the presence of DONSON, its predominant interactions with the MCM3/5/7 complex hindered LRR1 from binding to its original interface on the MCM3/5/7 complex, potentially resulting in the displacement of LRR1 from the complex (Supplementary Fig. 8c). This prediction was supported by the high reproducibility of the changes in PAE values for the interactions between LRR1 and the MCM3/5/7 complex under conditions with and without DONSON (Fig. 6e). Furthermore, due to its inability to adopt a specific position relative to the MCM3/5/7 complex, Cul2 is unable to come in close proximity to the defined primary ubiquitylation sites on MCM7 (Fig. 6f and Supplementary Fig. 8e). A possible explanation for this outcome is the relatively close proximity of the binding sites of DONSON and LRR1 on MCM3. The bulky size of the ubiquitin ligase complex involving LRR1 could lead to competitive interactions between DONSON and the ubiquitin ligase complex for binding to the MCM3/5/7 complex.
To verify these results, we performed immunoprecipitation assays and confirmed that depletion of DONSON significantly increased endogenous interaction between MCM3 and LRR1 (Fig. 6g). In line with this, depletion of DONSON significantly increased the loading of LRR1 onto chromatin during S phase (Fig. 6h, i). Conversely, overexpression of DONSON significantly decreased the endogenous interaction between MCM3 and LRR1, while overexpression of DONSON Δ354–375 mutant did not (Supplementary Fig. 7e). Given that the CMG disassembly mediated by LRR1 is crucial for ETAA1 degradation, which in turn is responsible for Plk1 inactivation (Figs. 4 and 5), we further investigated the impact of the interaction between DONSON and MCM3 on the activation of Plk1 during S phase. As shown in Fig. 3E, DONSON depletion led to the precocious activation of Plk1 during S phase. This defect was rescued by overexpression of DONSON wild-type but not by the Δ354–375 mutant, which lacks the ability to interact with MCM3 (Fig. 6j, k). Overall, these data suggest that DONSON suppresses precocious loading of the LRR1 E3-ubiquitin ligase complex to the CMG helicase and thereby suppresses precocious Plk1 activation during S phase.
DONSON coordinates DNA and centrosome replication cycles and thereby ensures proper chromosome segregation
We next analyzed the impact of DONSON depletion on mitotic cell division by performing live-cell imaging with HeLa cells stably expressing GFP-centrin-1. DONSON-depleted cells frequently exhibited mitotic defects, such as chromosome misalignment with pseudo-bipolar spindle and multipolar spindle formation64 (Fig. 7a, b and Supplementary Movies 2–4), that are generally observed in cells with centrosome amplification3. Also, these cells were often associated with lagging chromosomes (Supplementary Fig. 9a, b). Moreover, immunostaining of patient-derived cell lines with DONSON mutations revealed similar defects in spindle formation and chromosome segregation (Fig. 7c, Supplementary Fig. 9c). These abnormalities in spindle formation could arise from precociously disengaged daughter centrioles, which had acquired ectopic MTOC activity already in S phase (Fig. 3).
a Time-lapse observation of mitotic cell division in DONSON-depleted cells. HeLa GFP-centrin-1 cells were visualized every 20 min for 48 h after 24 h treatment with siRNA and 3 h treatment with 100 nM SiR-DNA. Left–right arrows indicate precociously disengaged centrioles. b Histograms represent the frequency of cells in mitosis with the indicated phenotypes observed in (a). n = 3 independent experiments, 30 cells for each. c Mitotic cell division of hTERT-immortalized fibroblasts derived from the patients with mutations in DONSON. Fibroblasts were infected with pMSCV-empty-vector or pMSCV-DONSON and immunostained with antibodies against Cep192 (red). d, f Inducible expression of ETAA1 upon DONSON depletion. HeLa-Tet3G mNG-ETAA1 cells were treated with siControl or siDONSON following Dox treatment for 24 h. e, g Histograms represent the frequency of cells in mitosis with the indicated phenotypes observed in (d) and (f), respectively. n = 3 independent experiments, 30 cells for each. h A speculative model showing how DNA replication machinery exerts direct control over the licensing and execution of centrosome duplication. All scale bars, 5 μm. Values are mean percentages ± s.d. Two-tailed, unpaired Student’s t-test was used in (b), (e), and (g) to obtain the P-value. Source data are provided as a Source Data file.
Finally, we explored whether the precocious centrosome conversion of disengaged centrioles during S phase contributed to the mitotic abnormalities observed in DONSON-depleted cells. Overexpression of ETAA1, which can suppress the precocious conversion (Fig. 4l, m), significantly decreased the fraction of mitotic cells with multipolar spindles (Fig. 7d, e) and chromosome segregation errors in DONSON-depleted cells (Fig. 7f, g). This finding showing a harmful effect of the precocious conversion aligns with the evidence that when cells enter mitosis soon after the precocious disengagement, the disengaged daughter centrioles cannot acquire MTOC activity strong enough to solely form a spindle pole36. Taken together, this study reveals that the DNA replication machinery via DONSON ensures mitotic spindle formation with unreplicated centrosomes to maintain genetic stability (Fig. 7h).
Discussion
Central to the process of life is the faithful replication of genetic material and its segregation into offspring cells65. Previously, these two processes were thought to be independently regulated under the control of the cell cycle machinery. However, our study has shown that the DNA replication machinery not only directs the copying of genetic material but also directly controls the system responsible for its accurate inheritance by daughter cells (Fig. 7h). The DNA replication machinery transmits two distinct signals associated with the initiation and progression of DNA replication to centrosomes preventing their precocious duplication until the segregation of replicated chromosomes is complete. Disruptions in this mechanism can lead to abnormal chromosome segregation due to excess centrosomes in the mitotic spindle, which could be a potential cause of MPD.
DNA and centrosomes, both responsible for the inheritance of genetic information, have a strictly regulated number of copies per cell. Alongside DNA, centrioles, the structural basis of the centrosomes, are the only other cellular components that replicate once per cell cycle using themselves as a template4. Ensuring this one-time replication is crucial for maintaining the correct copy number of both DNA and centrioles. For a new copy to be generated, the replicated copy unit must first detach from its template. Therefore, mechanisms that prevent the separation of the template and the newly formed copy serve as safeguards against further replication within the same cell cycle. In the case of the centrioles, this detachment process, known as centriole disengagement, occurs at the end of mitosis9. This is a prerequisite for the replicated centrioles to convert into functional centrosomes, representing the execution process of centrosome duplication66. Consequently, the disengagement process serves as the licensing step for both centriole and centrosome duplication. In DNA replication, an analogous detachment process occurs when the CMG helicase separates the newly synthesized strand from the template strand during S phase67. This separation is similarly prerequisite for the execution process of DNA replication. Therefore, the fundamental principles governing DNA and centrosome replication remain notably similar, suggesting that evolution has developed parallel strategies for controlling the copy number of these essential cellular components.
In this study, we discovered that Cdc6, a key regulator of the licensing process in DNA replication11, plays a crucial role in preventing the precocious disengagement of centrioles at centrosomes (Fig. 2). It is therefore conceivable that Cdc6 acts as a regulator of the licensing steps in both DNA replication and centrosome duplication. Our study further revealed that, by translocating this common licensing regulator from the DNA replication origins to the centrioles through the function of DONSON, the DNA replication machinery couples the initiation of DNA replication with the suppression of centriole disengagement. This coupling is critical for the timing difference between DNA and centrosome replication in the cell cycle, ensuring the formation of bipolar spindles with the correct number of centrosomes (Fig. 7). In future studies, it will be an important challenge to elucidate how DONSON regulates the translocation of Cdc6 to the centrosome at the initiation of DNA replication.
Our study reveals that human DONSON stabilizes the replisome during ongoing DNA replication by suppressing precocious loading of the Cul2-LRR1 E3 ligase complex to the CMG helicase and thus preventing its disassembly (Figs. 5 and 6), which normally should occur at the termination of DNA replication. The stabilization of the replisome by DONSON is crucial for activating the S/G2 checkpoint by maintaining the protein level of ETAA1, an activator of ATR19,68 (Fig. 5). The activated S/G2 checkpoint, through Chk1, inhibits the activation of Plk1 at centrosomes (Figs. 3 and 4). This inhibitory signaling transmitted from the replisome to centrosomes is crucial to suppress the precocious occurrence of the Plk1-mediated centrosome conversion of daughter centrioles, the execution process of centrosome duplication. Our study, therefore, indicates that the DNA replication machinery continuously couples the progression of DNA replication with suppression of the execution process of centrosome duplication.
Our research demonstrates that two signals from the DNA replication machinery, Cdc6 translocation and the S/G2 checkpoint, prevent the precocious occurrence of licensing and execution of centrosome duplication, respectively. Simultaneous disruption of the two signals causes both the disengagement of centrioles and their conversion to centrosomes prematurely during the DNA replication phase. The excess centrosomes that arise from this situation lead to the formation of a multipolar spindle in mitosis, resulting in abnormalities of chromosome segregation (Fig. 7). Importantly, centrosome amplification has been reported to induce microcephaly, characterized by a depletion of neural progenitor cells, as observed in patients suffering from MPD21,22,23. Hence, the uncoupling of DNA and centrosome replications, stemming from mutations in genes encoding components of the DNA replication machinery such as DONSON, could be considered a contributing factor in the onset of MPD disorders. Overall, this study illuminates a direct control mechanism for centrosome duplication by the DNA replication machinery and underscores the vital role of this hierarchical regulatory system not only in the faithful inheritance of genetic materials but also in the developmental processes of individual organisms.
Methods
Cell culture and transfection
HeLa, U2OS, PANC-1, and HEK293T cells were obtained from the ECACC (European Collection of autuhenticated Cell Cultures), and RPE-1 cells were obtained from the ATCC (American Type Culture Collection). HeLa cells and U2OS cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM), supplemented with 10% fetal bovine serum (FBS) and 100 μg/ml penicillin–streptomycin at 37 °C in a 5% CO2 atmosphere. RPE-1 cells were cultured in DMEM / Ham’s F-12, supplemented with 10% fetal bovine serum (FBS) and 100 μg/ml penicillin–streptomycin at 37 °C in a 5% CO2 atmosphere. PANC-1 cells were cultured in RPMI 1640, supplemented with 10% fetal bovine serum (FBS) and 100 μg/ml penicillin–streptomycin at 37 °C in a 5% CO2 atmosphere. hTERT-immortalized fibroblasts derived from patients with DONSON mutations were infected with pMSCV-empty-vector or pMSCV-DONSON and were cultured in Dulbecco’s modified Eagle’s medium (DMEM), supplemented with 10% fetal bovine serum (FBS) and 100 μg/ml penicillin–streptomycin at 37 °C in a 5% CO2 atmosphere. Transfection of siRNA, sgRNA, or DNA constructs into HeLa, U2OS, and RPE-1 cells was conducted using Lipofectamine RNAiMAX (Life Technologies) or Lipofectamine 2000 (Life Technologies), respectively. Unless otherwise noted, the transfected cells were analyzed 48–72 h after transfection with siRNA and 24–36 h after transfection with DNA constructs.
Generation of stable cell lines
The tetON system was used to control protein expression. HeLa-tetR cell line was generated according to the manufacturer’s protocol (Retro-X Tet-On 3G Inducible Expression System User Manual, Clontech Laboratories, Inc.). To generate Dox-inducible stable cell lines, retroviral-mediated integration was used. For the production of viral particles, the gene of interest was cloned into pRetroX-TRE3G (Clontech) and co-transfected with the envelope vector pCMV-VSV-G (Addgene) to a HEK293-based retroviral packaging cell line (GP2-293, Clontech). Media were changed 24 h after transfection, and the virus-containing media was harvested 48 h after transfection and filtered using a 0.45 µM filter (Millipore). The filtered virus medium was supplemented with fresh media, FBS, and 4 µg/ml Polybrene (Nacalai Tesque). HeLa-tetR cells pre-seeded in a 6-well plate were infected by adding the virus-containing solution three times every 4–6 h. Cells were split 24 h after the first transduction and put under puromycin selection after 48 h.
Plasmids
Complementary DNA (cDNA) encoding DONSON isoform (NCBI NP_060083.1), Cdc6 isoform (NCBI NP_001245.1), Chk1 isoform (NCBI NP_001107593.1), ETAA1 isoform (NCBI NP_061875.2), MCM3 isoform (NCBI NP_002379.4), MCM6 isoform (NCBI NP_005906.2) were amplified from cDNA library of HeLa cells and subcloned into p3×FLAG-CMV14 (Addgene), pCMV5-HA (Addgene) or pRetroX-TRE3G (Clontech). The Chk1-L449R mutant constructs and DONSON Δ354–375 mutant constructs were created using PrimeSTAR mutagenesis basal kit (TaKaRa) and In-Fusion HD cloning kit (Clontech) according to the manufacturer’s protocol. pcDNA-HA-Ubiquitin was a gift from Drs. Jun Hamazaki and Shigeo Murata (University of Tokyo, Tokyo, Japan). Sequence information of the primers used for plasmid construction is listed in Supplementary Table 1.
Antibodies
The following primary antibodies were used in this study:
rabbit antibodies against Cep192 (Bethyl Laboratories, A302–324A, IF 1:1000), C-Nap1 (Proteintech, 14498-1-AP, IF 1:1000), CP110 (Proteintech, 12780-1-AP, IF 1:300), Cdt1 (Abcam, ab202067, IF 1:500), Cdc6 (Abcam, ab109315, WB 1:100), RFP-tag (MBP, PM005, IF 1:1000), Cep57 (GeneTex, GTX115931, IF 1:1000), Cep57L1 (Proteintech, 24957-1-AP, IF 1:500), γ-Tubulin (Merck, T5192, IF 1:1000), PCNT (Abcam, ab4448, IF 1:2000), Cep152 (Bethyl Laboratories, A302–480A, IF 1:1000), Cep97 (Novus Biologicals, NPB1-83591 IF 1:500), CENP-F (Bethyl Laboratories, A301–617 A IF 1:1000), Phospho-Chk1 (S296) (Abcam, ab79758, WB 1:100), CyclinB1 (Cell Signaling Technology, 4138, IF 1:200, WB 1:1000), BLM (Abcam, ab2179, IF 1:250), ATR (Bethyl Laboratories, A300-137A, WB 1:500), ETAA1 (Abcam, ab192402, WB 1:100), TopBP1 (Abcam, ab2402, WB 1:1000), mNeonGreen-tag (Cell Signaling Technology, 53061, WB 1:100), p97 (VCP; GeneTex, GTX101089, IF 1:1000), Cdc45 (Cell Signaling Technology, 11881, IF 1:200, WB 1:500), LRR1 (Atlas antibodies, HPA069364, IF 1:200, WB 1:1000) and MCM3 (Cell Signaling Technology, 4012, WB 1:1000), Emi1 (Proteintech, 10872-1-AP, WB 1:1000); mouse antibodies against FLAG-tag (Merck, F1804, IF 1:1000, WB 1:1000), centrin-2 (Merck, 20H5, IF 1:1000), PCNA (Santa Cruz Biotechnology, sc-56, IF 1:1000, WB 1:500), CENP-F (Mitosin; BD Biosciences Pharmingen, 610768, IF 1:1000), Acetylated-Tubulin (Abcam, Ab179484, IF 1:300), Cdc6 (Santa Cruz Biotechnology, sc-13136, IF 1:200), HSP90 (BD Transduction Laboratories, 610418, WB 1:5000), Cdh1 (Fzr1; Abcam, ab89535, IF 1:300), HsSAS-6 (Santa Cruz Biotechnology, sc-81431, IF 1:1000), EB1 (BD Transduction Laboratories, 610534, IF 1:500), Phospho-Plk1 (T210) (Abcam, ab39068, IF 1:300, WB 1:200), Chk1 (Santa Cruz Biotechnology, sc-8408, WB 1:1000), α-Tubulin (Merck, DM1A, IF 1:1000, WB 1:5000), HA-tag (BioLegend, 901501, WB 1:1000), mNeonGreen-tag (Proteintech, 32F6, IF 1:500), Cyclin-E1 (Cell Signaling Technology, 4129, WB 1:1000), Cyclin-A2 (Cell Signaling Technology, 4656, WB 1:1000), Histone H3 (Santa Cruz Biotechnology, sc-517576, WB 1:5000), LaminB1 (Santa Cruz Biotechnology, sc-374015, WB 1:100) and γ-Tubulin-Alexa Fluor 647 (Abcam, ab191114, IF 1:500); rat antibodies against centrin-2 (BioLegend, 698602, IF 1:500) and PCNA (Abcam, ab252848, IF 1:2000).
The following secondary antibodies were used: Alexa Fluor 488 goat anti-mouse IgG (H + L) (Molecular Probes, A-11001, 1:1000), Alexa Fluor 568 goat anti-rabbit IgG (H + L) (Molecular Probes, A-11011, 1:1000), Alexa Fluor 647 goat anti-rat IgG (H + L) (Invitrogen, A21247, 1:1000), llama/alpaca anti-mouse IgG-X2 Abberior Star 635P (Progen, 1A23, IF, 1:500) for IF; Goat polyclonal antibodies-horseradish peroxidase against mouse IgG (Promega, W402B, 1:5000) and rabbit IgG (Promega, W401B, 1:5000) for WB.
RNA interference
The following siRNAs were used: Silencer Select siRNA (Life Technologies) against DONSON (#1: s26833, #2: s26834), Cdc6 (#1: s2744, #2: s2745), Orc1 (s9893, s9894), Orc4 (s9898, s9899), Orc6 (s532063, s532064), Cdt1 (s532008, s532009), GMNN (s27306, s27307), Cdc45 (s15830, s15831), MCM5 (s8595, s8596), ATR (s536, s57272), RBBP8 (s11849, s11850), DNA2 (s4173, s4174), Separase (ESPL1; s18686), Cdc20 (s2747), Cdh1 (Fzr1; s27992), Cep57 (s18692), Cep57L1 (s226224), TopBP1 (s21823), ETAA1 (s29018), and negative control (4390843). Unless otherwise noted, DONSON #2 and Cdc6 #1 were used in this study.
Single-guide RNA (sgRNA)
HeLa cells stably expressing Cas9 (HeLa-Cas9) that were generated in the previous study34 were used in this study. sgRNA oligos targeting DONSON (Fw, 5′-GTAGCCAAGACAAAGCAGGG-3′; Rv, 5′-CCCTGCTTTGTCTTGGCTAC-3′) and Cdc6 (Fw, 5′-GTTTACCCAGACGTTTCCTG-3′; Rv, 5′-CAGGAAACGTCTGGGTAAAC-3′ccccccccccccccccccccccccccc) were transcribed in vitro with the HiScribe T7 transcription kit (New England Biolabs) and purified using the RNA Clean and Concentrator kit (Zymo Research).
Drug treatment
The following chemicals were used in this study: Aphidicolin (Sigma, A0781), SiR-DNA (Spirochrome, CY-SC007), Doxycycline (Merck, D9891), PHA767491 (Selleck, S2742), BI2536 (AdooQ, A10134), Nocodazole (Wako, 31430-18-9), CHIR-124 (Cayman Chemical Company, 16553), VE-821 (Selleck, S8007), MG132 (Wako, 135-18453), RO-3306 (Sigma, SML0569), P22077 (Calbiochem, 662142) and NMS-873 (ab270807). For determination of S phase cells, cells were treated with 10 μM EdU for 30 min before fixation unless otherwise stated. The fixed cells were stained using the Click-iT EdU Alexa Fluor 555/647 imaging kit (Life Technologies) according to the manufacturer’s recommendations.
Immunostaining and microscopy
For immunofluorescence analysis, the cells cultured on coverslips (Matsunami, 15 mm, thickness No. 1_0.13-0.17 mm) were fixed using −20 °C methanol for 7 min or 4% paraformaldehyde (PBS solution) for 10 min at room temperature and washed with PBS unless otherwise noted. The cells were permeabilized after fixation with PBS/0.05% Triton X-100 (PBS-X) for 5 min three times and incubated for blocking with 1% BSA in PBS-X for 20 min at room temperature (RT). The cells were incubated with primary antibodies for 24 h at 4 °C, washed with PBS-X three times, and incubated with secondary antibodies for 1 h at RT. The cells were thereafter washed with PBS-X twice, stained with 0.2 μg/ml Hoechst 33258 (DOJINDO) in PBS for 5 min at RT, washed again with PBS-X, and mounted onto glass slides. Counting the number of immunofluorescence signals was performed using an Axioplan2 fluorescence microscope (Carl Zeiss) with a ×63 or ×100/1.4 NA plan-APOCHROMAT objective and DeltaVision Personal DV-SoftWoRx system (Applied Precision) equipped with a CoolSNAP CH350 CCD camera. Confocal microscopy images were taken by the Leica TCS SP8 HSR system equipped with a Leica HCX PL APO ×63/1.4 oil CS2 objectives and excitation wavelengths 405, 488, and 561 nm. To obtain high-resolution images, the pinhole was adjusted at 0.5 airy units. Scan speed was set to 200 Hz in combination with a five-fold line average in 856 × 856 format (pixel size 43 nm). The images were collected at 130 nm z steps. For deconvolution, Huygens Essential software (SVI; Scientific Volume Imaging) was used. STED microscopy images were acquired using the Leica TCS SP8 STED 3× system with a Leica HC PLAPO 100×/1.40 oil STED WHITE objective and a 660 nm gated STED laser.
RNA sequencing
Total RNAs were extracted from HeLa cells. Sequencing libraries were constructed using a SMART-Seq HT PLUS kit (TaKaRa) according to the manufacturer’s instructions, and then were run on the Illumina NovaSeq 6000 system with 2 × 100 bp read length.
Pre-extraction of soluble proteins
To pre-extract soluble proteins, cells were treated with CSK buffer (25 mM HEPES (pH 7.4), 50 mM NaCl, 1 mM EDTA, 3 mM MgCl2, 300 mM sucrose, 1/1000 Protease Inhibitor Cocktail (Nacalai Tesque) and 0.5% Triton X-100) or PBS containing 0.2% Triton X-100 and 1/1000 Protease Inhibitor Cocktail on ice for 1 min, washed with PBS for three times before fixation.
Reverse transcription and real-time qPCR
Total RNA (1 μg) isolated from cells with the use of TRIZOL reagent (Thermo Fisher Scientific) was subject to reverse transcription using Quantiscript Reverse Transcription Kit (QIAGEN), and the resulting cDNA was subjected to real-time qPCR analysis with TB Green PCR Master Mix (TaKaRa) and specific primers in a StepOnePlus Real-Time PCR system (Applied Biosystems). The amount of each mRNA was normalized by that of GAPDH mRNA.
Live imaging
Cell Voyager CQ1 (Yokogawa Electric Corp.), equipped with a ×40 oil immersion objective lens and a stage incubator for a 35-mm dish, was used for live-cell imaging. HeLa cells stably expressing GFP-centrin-1 or HeLa Flp-In/T-Rex cells expressing GFP-DONSON were cultured on 35 mm glass-bottom dishes (Greiner-bio-one, #627870) at 37 °C in a 5% CO2 atmosphere. Before imaging, cells were treated with siRNAs or 1 μg/mL Doxycycline for 24 h and with 100 nM SiR-DNA for 3 h. Cells were visualized every 10 min over 24 h or 20 min over 48 h by a Back-illuminated sCMOS camera. The images were collected at 1.2 μm z steps. Maximum projections were generated using ImageJ (National Institutes of Health).
Flow cytometric analysis
Cells were trypsinized and fixed in 70% cold ethanol at −20 °C for >3 h. The fixed cells were then washed with PBS and incubated with Muse Cell Cycle Reagent (Cytek Biosciences, MCH100106) at room temperature in the dark for 30 min. The DNA contents of the cells were then measured using Muse Cell Analyzer (Merck Millipore).
Western Blotting
Protein samples were loaded onto 5–12% acrylamide gels with SDS–PAGE and transferred onto Immobilon-P membranes (Merck). The membranes were blocked with PBS-Tween20 (0.02%) (PBS-T) containing 5% milk powder for 10 min at room temperature. The membranes were then probed with the primary antibodies for 1 h at room temperature, followed by incubation with their respective horseradish peroxidase-conjugated secondary antibodies (Promega) for 30 min at room temperature. Washes were performed in PBS-T. For detection of phosphorylated proteins, membranes were blocked with Blocking One-P (Nacalai Tesque) and washed in TBS containing 0.02% Tween (TBS-T). The signal was detected with Chemi Doc XRS+ (Bio-Rad). Unless otherwise specified, the experiments of western blotting were repeated at least two times.
Immunoprecipitation
IP of mNeonGreen-tagged proteins was performed using the mNeonGreen-Trap Agarose Kit (Proteintech, NTAK-20) according to the manufacturer’s protocol. Note that, in this experiment, immunoprecipitation was performed under denaturing conditions.
For IP of FLAG-tagged proteins, cells were lysed in lysis buffer (20 mM Tris/HCl pH 7.5, 50 mM NaCl, 1% Triton X-100, 5 mM EGTA, 1 mM DTT, 2 mM MgCl2, 25 U/mL Benzonase, and 1/1000 protease inhibitor cocktail) and centrifuged at 15,000 rpm for 10 min. 10% of the volume was kept for input controls. The remaining supernatant was incubated with equilibrated FLAG antibody-conjugated M2 agarose gel (Merck) for 2 h at 4 °C.
To assess the interaction of endogenous MCM3 and LRR1, cells were lysed in RIPA buffer (10 mM Tris/Cl pH 7.6, 150 mM NaCl, 0.5 mM EDTA, 0.1% SDS, 1% Triton X-100 and 1% deoxycholate) supplemented with 2 mM MgCl2, 25 U/mL Benzonase and 1/1000 protease inhibitor cocktail and centrifuged at 21,130 g for 10 min. 10% of the volume was kept for input controls. The remaining supernatant was mixed with 25 µl of equilibrated beads (nProtein A Sepharose 4 Fast Flow; GE, 17-5280-01) coupled to 5 µg anti-MCM3 antibody. The beads were washed three times, resuspended with 2×SDS sample buffer, and then denatured at 95 °C for 5 min.
Biochemical cell fractionation
Whole cell extracts were obtained by sonication in SDS sample buffer (Nacalai Tesque). To fractionate cells into cytosol (S2), soluble nuclear (S3), and chromatin (P3) fractions, cells were suspended in buffer A (10 mM HEPES, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 0.1% Triton X-100, 1 mM DTT, 1:1000 protease inhibitor cocktail, and 1:1000 phosphatase inhibitor cocktail (Nacalai Tesque)) and incubated on ice for 5 min. Nuclei were collected by low-speed centrifugation for 4 min at 1200 × g. The supernatant, containing soluble cytosolic proteins (S2), was collected and centrifuged for 10 min at 15,000 × g to remove cell debris and insoluble material. The nuclei were washed once with buffer A and lysed in buffer B (3 mM EDTA, 0.2 mM EGTA, 1 mM DTT, 1:1000 protease inhibitor cocktail, and 1:1000 phosphatase inhibitor cocktail (Nacalai Tesque)). Insoluble chromatin was collected by centrifugation for 4 min at 1800 × g, and the supernatant containing soluble nuclear protein was collected (S3). The chromatin pellet was washed twice in buffer B and pelleted again. The final pellet (P3) was resuspended and sonicated in 1×SDS sample buffer (Nacalai Tesque).
Protein structure prediction
The predicted structures of the human DONSON-CMG component complex and the human DONSON-MCM3/5/7-LRR1-ELOB-ELOC-Cul2-RBX1 complex were obtained using AlphaFold-based methods. For the DONSON-CMG complex, the amino acid sequences were submitted to ColabFold: AlphaFold2 with MMseqs2Colab (https://colab.research.google.com/github/sokrypton/ColabFold/main/AlphaFold2.ipynb)69 using the default settings. For the DONSON-MCM3/5/7-LRR1-ELOB-ELOC-Cul2-RBX1 complex, the sequences were submitted to AlphaFold3 using the AlphaFold server (https://alphafoldserver.com/) with the default settings70. The molecular graphics of the predicted protein complexes were visualized using UCSF Chimera and UCSF ChimeraX software71,72.
Statistical analysis and reproducibility
Two-tailed, unpaired Student’s t-tests were performed using Microsoft Excel software. Tukey’s multiple comparison tests, Kruskal–Wallis tests, and Mann–Whitney U tests were performed using GraphPad Prism 9.0 software. All data shown are mean ± S.D. Statistical tests and sample sizes are described in each Figure legend. p-values are indicated in each figure. In this study, each experiment showing only representative images was repeated independently with similar results at least twice.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data used to generate graphs and all pictures showing uncropped gels or blots accompany this manuscript in the source data file. All other data supporting the findings of this study are available from the corresponding author on reasonable request. The RNA-seq data analyzed in this study have been deposited in the DDBJ database under the accession number PRJDB20465. Source data are provided with this paper.
References
Bell, S. P. & Dutta, A. DNA replication in eukaryotic cells. Annu. Rev. Biochem 71, 333–374 (2002).
Prosser, S. L. & Pelletier, L. Mitotic spindle assembly in animal cells: a fine balancing act. Nat. Rev. Mol. Cell Biol. 18, 187–201 (2017).
Srsen, V. & Merdes, A. The centrosome and cell proliferation. Cell Div. 1, 1–5 (2006).
Nigg, E. A. & Holland, A. J. Once and only once: mechanisms of centriole duplication and their deregulation in disease. Nat. Rev. Mol. Cell Biol. 19, 297–312 (2018).
Ding, L. et al. The roles of cyclin-dependent kinases in cell-cycle progression and therapeutic strategies in human breast cancer. Int. J. Mol. Sci. 21, 1–28 (2020).
Conduit, P. T., Wainman, A. & Raff, J. W. Centrosome function and assembly in animal cells. Nat. Rev. Mol. Cell Biol. 16, 611–624 (2015).
Bornens, M. Centrosome composition and microtubule anchoring mechanisms. Curr. Opin. Cell Biol. 14, 25–34 (2002).
Tsou, M. F. B. & Stearns, T. Mechanism limiting centrosome duplication to once per cell cycle. Nature 442, 947–951 (2006).
Tsou, M. F. B. et al. Polo kinase and separase regulate the mitotic licensing of centriole duplication in human cells. Dev. Cell 17, 344–354 (2009).
Wang, W. J., Soni, R. K., Uryu, K. & Tsou, M. F. B. The conversion of centrioles to centrosomes: essential coupling of duplication with segregation. J. Cell Biol. 193, 727–739 (2011).
Bleichert, F., Botchan, M. R. & Berger, J. M. Mechanisms for initiating cellular DNA replication. Science 355, eaah6317 (2017).
Chang, F. et al. Cdc6 ATPase activity disengages Cdc6 from the pre-replicative complex to promote DNA replication. Elife 4, 1–14 (2015).
Lim, Y. et al. In silico protein interaction screening uncovers DONSON’s role in replication initiation. Science 381, eadi3448 (2023).
Xia, Y. et al. DNSN-1 recruits GINS for CMG helicase assembly during DNA replication initiation in Caenorhabditis elegans. Science 87, 149–200 (2023).
Hashimoto, Y., Sadano, K., Miyata, N., Ito, H. & Tanaka, H. Novel role of DONSON in CMG helicase assembly during vertebrate DNA replication initiation. EMBO J. 42, 1–17 (2023).
Cvetkovic, M. A. et al. The structural mechanism of dimeric DONSON in replicative helicase activation. Mol. Cell 83, 4017–4031 (2023).
Burgers, P. M. J. & Kunkel, T. A. Eukaryotic DNA replication fork. Annu. Rev. Biochem. 86, 417–438 (2017).
Dewar, J. M. & Walter, J. C. Mechanisms of DNA replication termination. Nat. Rev. Mol. Cell Biol. 18, 507–516 (2017).
Saldivar, J. C. et al. An intrinsic S/G 2 checkpoint enforced by ATR. Science 810, 806–810 (2018).
Michelena, J., Gatti, M., Teloni, F., Imhof, R. & Altmeyer, M. Basal CHK1 activity safeguards its stability to maintain intrinsic S-phase checkpoint functions. J. Cell Biol. 218, 2865–2875 (2019).
Klingseisen, A. & Jackson, A. P. Mechanisms and pathways of growth failure in primordial dwarfism. Genes Dev. 25, 2011–2024 (2011).
Jayaraman, D., Bae, B.-I. & Walsh, C. A. The genetics of primary microcephaly. Annu. Rev. Genom. Hum. Genet. 19, 177–200 (2018).
Farcy, S., Hachour, H., Bahi-Buisson, N. & Passemard, S. Genetic primary microcephalies: when centrosome dysfunction dictates brain and body size. Cells 12, 1–34 (2023).
Tingler, M., Philipp, M. & Burkhalter, M. D. DNA Replication proteins in primary microcephaly syndromes. Biol. Cell 114, 143–159 (2022).
Reynolds, J. J. et al. Mutations in DONSON disrupt replication fork stability and cause microcephalic dwarfism. Nat. Genet. 49, 537–549 (2017).
Mayor, T., Stierhof, Y. D., Tanaka, K., Fry, A. M. & Nigg, E. A. The centrosomal protein C-Nap1 is required for cell cycle-regulated centrosome cohesion. J. Cell Biol. 151, 837–846 (2000).
Yang, J., Adamian, M. & Li, T. Rootletin interacts with C-Nap1 and may function as a physical linker between the pair of centrioles/basal bodies in cells. Mol. Biol. Cell 17, 1033–1040 (2006).
Dwivedi, D., Harry, D. & Meraldi, P. Mild replication stress causes premature centriole disengagement via a sub-critical Plk1 activity under the control of ATR-Chk1. Nat. Commun. 14, 6088 (2023).
Xu, X. et al. DNA replication licensing factor Cdc6 and Plk4 kinase antagonistically regulate centrosome duplication via Sas-6. Nat. Commun. 8, 15164 (2017).
Kim, G. S., Kang, J., Bang, S. W. & Hwang, D. S. Cdc6 localizes to S- and G2-phase centrosomes in a cell cycle-dependent manner. Biochem. Biophys. Res. Commun. 456, 763–767 (2015).
Ito, K. et al. Cep57 and Cep57L1 cooperatively maintain centriole engagement during interphase to ensure proper centriole duplication cycle. J. Cell. Biol. 220, e202005153 (2021).
Matsuo, K. et al. Kendrin is a novel substrate for separase involved in the licensing of centriole duplication. Curr. Biol. 22, 915–921 (2012).
Lee, K. & Rhee, K. Separase-dependent cleavage of pericentrin B is necessary and sufficient for centriole disengagement during mitosis. Cell Cycle 11, 2476–2485 (2012).
Watanabe, K., Takao, D., Ito, K. K., Takahashi, M. & Kitagawa, D. The Cep57-pericentrin module organizes PCM expansion and centriole engagement. Nat. Commun. 10, 931 (2019).
Hatano, T. & Sluder, G. The interrelationship between APC/C and Plk1 activities in centriole disengagement. Biol. Open 1, 1153–1160 (2012).
Prosser, S. L., Samant, M. D., Baxter, J. E., Morrison, C. G. & Fry, A. M. Oscillation of APC/C activity during cell cycle arrest promotes centrosome amplification. J. Cell Sci. 125, 5353–5368 (2012).
Gupta, A., Tsuchiya, Y., Ohta, M., Shiratsuchi, G. & Kitagawa, D. NEK7 is required for G1 progression and procentriole formation. Mol. Biol. Cell 28, 2123–2134 (2017).
Fu, J. et al. Conserved molecular interactions in centriole-to-centrosome conversion. Nat. Cell Biol. 18, 87–99 (2016).
Izquierdo, D., Wang, W. J., Uryu, K. & Tsou, M. F. B. Stabilization of cartwheel-less centrioles for duplication requires CEP295-mediated centriole-to-centrosome conversion. Cell Rep. 8, 957–965 (2014).
Lindqvist, A. et al. Polo-like kinase-1 is activated by Aurora A to promote checkpoint recovery. Nature 455, 119–123 (2008).
Hatch, E. M., Kulukian, A., Holland, A. J., Cleveland, D. W. & Stearns, T. Cep152 interacts with Plk4 and is required for centriole duplication. J. Cell Biol. 191, 721–729 (2010).
Cizmecioglu, O. et al. Cep152 acts as a scaffold for recruitment of Plk4 and CPAP to the centrosome. J. Cell Biol. 191, 731–739 (2010).
Sonnen, K. F., Gabryjonczyk, A. M., Anselm, E., Nigg, E. A. & Stierhof, Y. D. Human cep192 and cep152 cooperate in plk4 recruitment and centriole duplication. J. Cell Sci. 126, 3223–3233 (2013).
Leidel, S., Delattre, M., Cerutti, L., Baumer, K. & Gönczy, P. SAS-6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nat. Cell Biol. 7, 115–125 (2005).
Jang, Y. J., Ma, S., Terada, Y. & Erikson, R. L. Phosphorylation of threonine 210 and the role of serine 137 in the regulation of mammalian polo-like kinase. J. Biol. Chem. 277, 44115–44120 (2002).
Lemmens, B. et al. DNA replication determines timing of mitosis by restricting CDK1 and PLK1 activation. Mol. Cell 71, 117–128.e3 (2018).
Han, X. et al. Conformational change of human checkpoint kinase 1 (Chk1) induced by DNA damage. J. Biol. Chem. 291, 12951–12959 (2016).
Akopyan, K. et al. Assessing kinetics from fixed cells reveals activation of the mitotic entry network at the S/G2 transition. Mol. Cell 53, 843–853 (2014).
Mankouri, H. W., Huttner, D. & Hickson, I. D. How unfinished business from S-phase affects mitosis and beyond. EMBO J. 32, 2661–2671 (2013).
Chan, Y. W., Fugger, K. & West, S. C. Unresolved recombination intermediates lead to ultra-fine anaphase bridges, chromosome breaks and aberrations. Nat. Cell Biol. 20, 92–103 (2018).
Dewar, J. M., Budzowska, M. & Walter, J. C. The mechanism of DNA replication termination in vertebrates. Nature 525, 345–350 (2015).
Maric, M., Mukherjee, P., Tatham, M. H., Hay, R. & Labib, K. Ufd1-Npl4 recruit Cdc48 for disassembly of ubiquitylated CMG helicase at the end of chromosome replication. Cell Rep. 18, 3033–3042 (2017).
Sonneville, R. et al. CUL-2LRR-1 and UBXN-3 drive replisome disassembly during DNA replication termination and mitosis. Nat. Cell Biol. 19, 468–479 (2017).
Maric, M., Maculins, T., De Piccoli, G. & Labib, K. Cdc48 and a ubiquitin ligase drive disassembly of the CMG helicase at the end of DNA replication. Science 346, 1253596 (2014).
Moreno, S. P., Bailey, R., Campion, N., Herron, S. & Gambus, A. Polyubiquitylation drives replisome disassembly at the termination of DNA replication. Science 346, 477–481 (2014).
Galarreta, A. et al. USP7 couples DNA replication termination to mitotic entry. Preprint at bioRxiv https://doi.org/10.1101/305318 (2018).
Toledo, L. I. et al. ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell 155, 1088 (2013).
Forment, J. V. & Jackson, S. P. A flow cytometry-based method to simplify the analysis and quantification of protein association to chromatin in mammalian cells. Nat. Protoc. 10, 1297–1307 (2015).
Evans, R. et al. Protein complex prediction with AlphaFold-multimer. Preprint at bioRxiv https://doi.org/10.1101/2021.10.04.463034 (2022).
Jones, M. L., Baris, Y., Taylor, M. R. G. & Yeeles, J. T. P. Structure of a human replisome shows the organisation and interactions of a DNA replication machine. EMBO J. 40, 1–23 (2021).
Jenkyn-Bedford, M. et al. A conserved mechanism for regulating replisome disassembly in eukaryotes. Nature 600, 743–747 (2021).
Fab, Y. et al. Lrr1-mediated replisome disassembly promotes DNA replication by recycling replisome components. J. Cell. Biol. 220, e202009147 (2021).
Dewar, J. M., Low, E., Mann, M., Räschle, M. & Walter, J. C. CRL2Lrr1 promotes unloading of the vertebrate replisome from chromatin during replication termination. Genes Dev. 31, 275–290 (2017).
Fuchs, F. et al. Clustering phenotype populations by genome-wide RNAi and multiparametric imaging. Mol. Syst. Biol. 6, 1–13 (2010).
Blow, J. J. & Tanaka, T. U. et al. The chromosome cycle: coordinating replication and segregation. EMBO Rep. 6, 1028–1034 (2005).
Ito, K. K. et al. Multimodal mechanisms of human centriole engagement and disengagement. EMBO J. 44, 1294–1321 (2025).
Costa, A. & Diffley, J. F. X. The initiation of Eukaryotic DNA replication. Annu. Rev. Biochem. 91, 107–131 (2022).
Haahr, P. et al. Activation of the ATR kinase by the RPA-binding protein ETAA1. Nat. Cell Biol. 18, 1196–1207 (2016).
Mirdita, M. et al. ColabFold: making protein folding accessible to all. Nat. Methods 19, 679–682 (2022).
Abramson, J. et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 630, 493–500 (2024).
Pettersen, E. F. et al. UCSF Chimera - a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Meng, E. C. et al. UCSF ChimeraX: Tools for structure building and analysis. Protein Sci. 32, e4792 (2023).
Achuthankutty, D. et al. Regulation of ETAA1-mediated ATR activation couples DNA replication fidelity and genome stability. J. Cell Biol. 218, 3943–3953 (2019).
Acknowledgements
We thank Mariya Genova for the proofreading of the manuscript, Drs. Jun Hamazaki and Shigeo Murata at the Graduate School of Pharmaceutical Sciences at the University of Tokyo for providing pcDNA-HA-Ubiquitin, and the Kitagawa lab members for technical support and helpful discussions. This work was supported by JSPS KAKENHI grants (Grant numbers: 18K06246, 19H05651, 20K15987, 20K22701, 21H02623, 22H02629, 22K20624, 23K14176, 23H02627, 16H06279 (PAGS)) from the Ministry of Education, Science, Sports and Culture of Japan, the PRESTO program (JPMJPR21EC) and the CREST program (JPMJCR22E1) of the Japan Science and Technology Agency, Takeda Science Foundation, The Uehara Memorial Foundation, The Research Foundation for Pharmaceutical Sciences, Koyanagi Zaidan, The Kanae Foundation for the Promotion of Medical Science, Kato Memorial Bioscience Foundation, Naito Foundation, Heiwa Nakajima Foundation, Sumitomo Foundation, Inamori foundation, and Tokyo Foundation for Pharmaceutical Sciences. G.S.S. is supported by a Cancer Research UK (CR-UK) programme grant (C17183/A23303).
Author information
Authors and Affiliations
Contributions
K.M., K.K.I., S.H., and D.K. designed the study. K.M., K.K.I., K.N., A.S., and K.T. performed experiments. K.M., K.K.I., S.H., and D.K. analyzed data. A.T. ran RNA-sequencing. G.S.S. generated the DONSON patient cell lines. K.W., M.F., S.Y., and T.C. provided suggestions. K.M., S.H., and D.K. wrote the manuscript. All authors contributed to discussions and manuscript preparation.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Patrick Meraldi and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Matsuhashi, K., Ito, K.K., Nagai, K. et al. The DNA replication machinery transmits dual signals to prevent unscheduled licensing and execution of centrosome duplication. Nat Commun 16, 7799 (2025). https://doi.org/10.1038/s41467-025-63002-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-63002-3