Abstract
MicroRNA (miRNA) maturation is initiated by the Microprocessor complex, comprising DROSHA and DGCR8, that processes primary miRNAs (pri-miRNAs). Recent studies have identified ERH and SAFB2 as auxiliary factors that enhance the functionality of the Microprocessor. These factors are required for cluster assistance, where optimal pri-miRNAs facilitate the processing of adjacent suboptimal pri-miRNAs. However, the specific action mechanisms of ERH and SAFB2 have not yet been defined. In this study, we found that ERH broadly enhances the processing of pri-miRNAs regardless of their genomic contexts, affecting both stand-alone and clustered ones. Suboptimal hairpins are affected more prominently by ERH knockdown than efficiently processed hairpins. In contrast, SAFB2 specifically supports the processing of suboptimal pri-miRNA hairpins within clusters. This study reveals the distinct roles of ERH and SAFB2 in cluster assistance and presents a new model, in which SAFB2 facilitates the Microprocessor’s transfer between hairpins, while ERH enables the efficient processing of suboptimal pri-miRNAs.
Similar content being viewed by others
Introduction
MicroRNAs (miRNAs) are small non-coding RNAs that play critical roles in post-transcriptional gene regulation1,2. The canonical miRNA pathway involves several key steps3. RNA polymerase II synthesizes long primary transcripts (pri-miRNAs)4,5, which are cleaved to yield precursor miRNAs (pre-miRNAs) by a ribonuclease III enzyme DROSHA6,7. DROSHA interacts with its essential cofactor DiGeorge syndrome critical region gene 8 (DGCR8), together constituting the Microprocessor complex6,8,9,10,11,12. Pre-miRNAs are then exported to the cytoplasm, where they undergo further processing by another ribonuclease III enzyme, DICER, to generate mature miRNA duplexes13,14,15,16,17. The duplex is loaded onto an Argonaute protein. After one strand is removed, the remaining mature miRNA guides the Argonaute protein to target transcripts, resulting in RNA silencing.
The Microprocessor complex serves as a gatekeeper by selectively recognizing and processing pri-miRNAs. A canonical pri-miRNA possesses distinctive structural characteristics, which optimally include a double-stranded stem of ~35 base pairs (bp), an apical loop with more than 10 nucleotides (nt), and a single-stranded 3′ basal segment of 9 nt or longer in the flanking region18,19,20,21. Additionally, specific sequence motifs, such as basal UG, apical UGUG, mismatched GHG (mGHG), bulged GWG (bGWG), DROSHA dsRNA recognition site (DRES), midBMW, and flanking CNNC motifs, contribute to the efficiency and specificity of the cleavage process18,20,21,22,23,24,25. Despite these general features, it is noteworthy that individual pri-miRNAs exhibit large variances in structure and sequences– less than 1% of human pri-miRNAs embody all the optimal structural features25. Thus, ideal pri-miRNA structures are rare, and a substantial fraction of miRNAs are processed either inefficiently and/or inaccurately. Some of these suboptimal pri-miRNAs are rescued by auxiliary factors of the Microprocessor.
The core Microprocessor complex is a heterotrimer consisting of one DROSHA molecule and two DGCR8 molecules6,12. This heterotrimer is sufficient to cleave many pri-miRNAs tested in vitro. The elongated shape formed by DROSHA and DGCR8 allows them to span the entire hairpin structure and accurately measure its stem length and cleave specific sites26,27,28,29,30. DROSHA anchors at the basal side through its alpha-helical hairpin (“belt”) from the PAZ-like domain and the “wedge” loop from the central domain28,30. The two RNase III domains of DROSHA form an intramolecular dimer to constitute a catalytic center to cleave both strands of the hairpin at ~11 bp away from the basal junction (relative to the cleavage site at the 3′ strand)19,26,27,28,29,30. DGCR8 contacts the upper stem region using its two dsRNA-binding domains (dsRBDs), offering a strong affinity to pri-miRNA28. DGCR8 binds to the outer surface of RNase III domains of DROSHA through its C-terminal tail (CTT), which forms an alpha-helix, thereby stabilizing the DROSHA protein. DGCR8 also contains a highly disordered region at the N-terminus31.
Although in vitro studies have demonstrated that DROSHA and DGCR8 alone can cleave pri-miRNAs, several additional proteins, including SRSF3, have been found to associate with the Microprocessor complex and contribute to the processing of a subset of pri-miRNAs18,22,32,33,34,35,36. Furthermore, pri-miRNA processing is influenced by other RNA-regulatory processes, including splicing and transcription-coupled mechanisms37,38,39,40. These findings reveal the complexity of pri-miRNA processing and the interplay between different cellular processes.
An intriguing aspect of pri-miRNA processing is the phenomenon known as “cluster assistance.” Approximately 50% of conserved miRNA genes in the human genome are arranged in tandem within distances of less than 10 kb36. These clustered pri-miRNAs are often transcribed into “polycistronic” transcripts containing multiple hairpins and undergo coordinated processing. While clustered miRNAs can typically produce miRNAs when expressed individually in experimental setups, some hairpins rely on their neighboring pri-miRNAs for processing34,41,42,43,44,45. That is, suboptimal “recipient” hairpins are poorly processed on their own but rescued when located near optimal “helper” hairpins46,47,48,49. This assistance occurs independently of whether the transcription is mediated by RNA polymerase II promoter46. The effect diminishes when the distance between recipient and helper hairpins increases to about 2000 nt46,47. Furthermore, in the absence of neighboring helper hairpins, simply tethering the Microprocessor near the suboptimal hairpin is sufficient to enhance its processing46,47.
Recent studies have identified Enhancer of Rudimentary Homolog (ERH) and Scaffold Attachment Factor B2 (SAFB2) as auxiliary factors required for cluster assistance31,46,48. Depletion of ERH or SAFB2 selectively reduces recipient hairpin processing, while that of the helper hairpins remains unaltered. ERH is a small (~12 kDa), abundant nuclear protein that forms a homodimer and binds to the N-terminus region of DGCR8 (Pro96-Gly139) with a 2:2 stoichiometry31. ERH also interacts with many other proteins, including SNRPD3, CHTOP, and CIZ1, and is thought to facilitate the complex formation of its binding partners50,51,52,53,54. SAFB2 is a large (~107 kDa) nuclear protein implicated in multiple cellular processes, such as chromatin organization, transcriptional regulation, RNA splicing, and stress response55,56. Through its domain spanning amino acids 561 to 726, SAFB2 interacts with the N-terminal region of DROSHA48. This domain also enables the formation of homo- or heteromeric protein complexes and is fully capable of mediating cluster assistance48. ERH and SAFB2 are also known to interact with each other and jointly modulate SR protein phosphorylation57. Given that knockdown of ERH results in changes in miRNA population similar to those in SAFB2-depleted cells, it was proposed that these two proteins work together in cluster assistance31,46,48,57. However, as studies on ERH and SAFB2 have been conducted independently by different research groups, a systematic comparison of their functions and molecular mechanisms remains to be established.
Here, we aim to interrogate the action mechanisms of ERH in regulating pri-miRNA processing. We find that ERH facilitates the processing of pri-miRNA hairpins, regardless of cluster arrangement, indicating its function beyond cluster assistance. Suboptimal hairpins are affected by ERH depletion more prominently than optimal hairpins. In contrast, SAFB2 specifically assists clustered suboptimal pri-miRNAs, suggesting its role in the transfer of the Microprocessor between clustered pri-miRNA hairpins. This study clarifies the distinct roles of two auxiliary cofactors of the Microprocessor and advances our understanding of miRNA maturation.
Results
ERH facilitates pri-miRNA processing irrespective of cluster arrangement
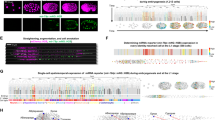
ERH is known to function in cluster assistance46, and accordingly, clustered miRNAs are generally more reduced than stand-alone (monocistronic) miRNAs in ERH-depleted HEK293E cells (Fig. 1A, x-axis)31. Intriguingly, however, some stand-alone miRNAs, such as miR-589 and miR-365a, also show reduced levels in ERH-depleted cells, despite lacking neighboring hairpins. A similar reduction is observed in human mesenchymal stem cells (hMSC) with a deletion in DGCR8 exon 2, which disrupts ERH binding (Fig. 1A, y-axis)58. These findings suggest that ERH may act more broadly than previously anticipated and assist the biogenesis of these stand-alone miRNAs as well as clustered miRNAs. To test this, we performed RT-qPCR following ERH knockdown in HEK293E cells (Fig. 1B). The levels of mature miR-589 and miR-365a decreased, while miR-197 showed a modest increase upon ERH depletion. Moreover, the pri-miRNA levels of miR-589 and miR-365a increased in ERH-depleted cells, indicating that ERH enhances their pri-miRNA processing.
A Scatter plots of log2 fold changes of siERH31 versus DGCR8 ∆ex258. A miRNA is defined as ‘clustered’ if another miRNA gene is found within ±1500 bp in the genome. R is Pearson’s correlation coefficient. B RT-qPCR for the expression levels of pri-miRNAs detected by SYBR Green-based PT-qPCR after knocking down ERH for 4 days, and those of miRNAs were deduced from small RNA-seq. RT-qPCR data shown represent the average of three biological replicate experiments. Error bars indicate the standard error of the mean. Statistical analysis was performed using a one-sided Student’s t-test against the null hypothesis of no change. *P < 0.05; N.S. denotes not significant. RT-qPCR representing relative miRNA abundance level upon ERH knockdown with ectopically expressed pri-miRNA (C) pri-miR-589 or 365a or 197 (n = 4), D pri-miR-589 of varying length and genomic locus (n = 3), E mutant pri-miR-589 or 365a in GNAI3 exon9 (n = 3). After 2 days of ERH knockdown, pri-miRNA expression plasmids were transfected with additional siRNA. RT-qPCR data shown represent the average of biological replicate experiments. Error bars indicate the standard error of the mean. Statistical analysis was performed using a one-tailed Student’s t-test. RT-qPCR representing relative miRNA abundance level upon ERH knockdown with artificial pri-miR-144 ~ 589 plasmids with varying distances between two clustered hairpins. The relative abundance against the “siNC_892nt spacer” sample is illustrated in (F), while the relative abundance of each plasmid is illustrated in (G). After 2 days of ERH knockdown, pri-miRNA expression plasmids were transfected with additional siRNA. The data shown represent the average of three biological replicate experiments. Error bars indicate the standard error of the mean. Statistical analysis was performed using a one-tailed Student’s t-test. All source data are provided as a Source data file.
In light of these unexpected findings, we further sought to investigate these monocistronic miRNAs. To this end, we generated plasmids that encode a miRNA hairpin and its surrounding sequences under the HCMV promoter (Fig.1C and Supplementary Fig. 1B for construct design). These plasmids were transfected into HEK293E cells after siRNA transfection. ERH knockdown resulted in a significant reduction in miR-589 and miR-365a levels from these plasmids, while miR-197 was not influenced by ERH knockdown. Furthermore, ectopic expression of ERH in ERH-depleted cells restored miR-589 and miR-365a levels (Supplementary Fig. 1C). We also made miRNA constructs with the U6 promoter dependent on RNA polymerase III, which gave similar results (Supplementary Fig. 1D), confirming that ERH acts irrespective of the transcription machinery. Together, these results demonstrate that ERH facilitates the processing of these stand-alone pri-miRNA hairpins.
We initially hypothesized that certain cis-acting elements (e.g., pri-miRNA-like hairpin) might be present near the miR-589 and miR-365a hairpins to aid in pri-miRNA processing, in a manner similar to cluster assistance. To test this hypothesis, we made a series of pri-miR-589 plasmids with varying neighboring sequences, 400–1200 nt (Fig. 1D). However, all of these plasmids showed ERH dependence. Furthermore, when we replaced the miR-197 hairpin originally present in GNAI3 exon 2 with the miR-589 or miR-365a hairpin, these constructs also exhibited ERH dependence (Fig. 1E). Thus, the ERH dependence of these stand-alone miRNAs cannot be attributed to adjacent cis-acting elements, suggesting that ERH may act directly on the hairpin itself, irrespectively of a clustered arrangement.
Given ERH’s reported role in cluster assistance, we further explored its impact on miR-589 within a cluster context. For this, we placed the miR-589 hairpin downstream of the miR-144 hairpin. The miR-144 hairpin is an optimal Microprocessor substrate that can assist its suboptimal neighbor, miR-451a, in the pri-miR-144–451a cluster31,46,47. Previous studies indicated that the cluster assistance effect diminishes as the distance between hairpins increases31,46,47. We cloned plasmids with varying spacer lengths (Fig. 1F). miR-589 production was higher when its hairpin was placed closer (92 nt apart) to the miR-144 hairpin than when located farther away (492 nt or 892 nt apart), showing that the miR-589 hairpin can indeed benefit from the cluster assistance mechanism (Fig. 1F, siNC). However, regardless of the hairpin spacing, miR-589 production was reduced by a similar magnitude (3.3–3.7 fold) in ERH-depleted cells (Fig. 1F, siERH). Figure 1G shows the same data with different normalization against the control siRNA. Moreover, a separate series of constructs with miR-144 and miR-451 also showed that ERH facilitates the production of suboptimal miR-451, even when the hairpins are far apart and the cluster assistance effect is weak (Supplementary Fig. 1E, F). Taken together, our observations indicate that ERH does not require a closely clustered arrangement to promote pri-miRNA processing.
ERH promotes suboptimal pri-miRNAs
To investigate what determines ERH dependence, we analyzed the processing efficiency of hundreds of human pri-miRNA hairpins using high-throughput in vitro assays34. Processing efficiencies modestly correlated with changes in cellular miRNA levels upon ERH depletion (Fig. 2A, r = 0.25), suggesting that suboptimal hairpins are more affected by ERH loss. The weak correlation possibly reflects additional influences on mature miRNA levels, such as indirect transcriptional regulation. To directly assess ERH’s effect on processing, we constructed plasmids expressing miRNAs under a common HCMV promoter (Fig. 2B). We tested two poorly processed hairpins (miR-378c and miR-500b) and one efficient hairpin (let-7a), based on in vitro processing data34. Upon ERH knockdown, miR-378c and miR-500b production from these plasmids was markedly reduced, whereas let-7a was largely unaffected (Fig. 2B). Combined with earlier results (Fig. 1C), these findings suggest that suboptimal pri-miRNAs (miR-589, miR-365a, miR-378c, and miR-500b) are critically dependent on ERH, unlike efficiently processed ones (miR-197 and let-7), implying that the suboptimality of the miRNA hairpin may be a main determinant of ERH dependence.
A Scatter plots of log2 fold changes of siERH31 versus in vitro processing efficiency of pri-miRNA hairpins34. r is Pearson’s correlation coefficient. B RT-qPCR showing ERH dependence of ectopically expressed pri-miRNAs. After 2 days of ERH knockdown, pri-miRNA expression plasmids were transfected with additional siRNA. The data shown represent the average of three biological replicate experiments. Error bars indicate the standard error of the mean. Statistical analysis was performed using a one-tailed Student’s t-test. C Schematic diagram of plasmids that modify the hairpin of 451a in the miR-144-451a cluster and RT-qPCR showing ERH dependence of miR-451a mutants47. After 2 days of ERH knockdown, pri-miRNA expression plasmids were transfected with additional siRNA. The data shown represent the average of 4 biological replicate experiments. Error bars indicate the standard error of the mean. Statistical analysis was performed using a one-tailed Student’s t-test. D Schematic diagram of artificial pri-miRNA that differ in the presence or absence of the pri-miRNA motifs and RT-qPCR showing ERH dependence of artificial miRNA20. After 2 days of ERH knockdown, pri-miRNA expression plasmids were transfected with additional siRNA. The data shown represent the average of three biological replicate experiments. Error bars indicate the standard error of the mean. Statistical analysis was performed using a one-tailed Student’s t-test. All source data are provided as a Source data file.
To test this possibility further, we prepared constructs with the miR-451a hairpin and its variants with increased loop size or stem length, which were previously shown to improve processing47 (Fig. 2C, left). While wild-type pri-miR-451a was strongly dependent on ERH, its variants with a larger loop and/or longer stem, which are more efficiently processed, showed reduced sensitivity to ERH depletion (Fig. 2C, right). Of note, because miR-451a is nearly undetectable when expressed from a stand-alone plasmid, this experiment was conducted with clustered constructs that contain the miR-144 hairpin.
Additionally, we created monocistronic constructs using an artificial pri-miRNA, A2, described previously20 (Fig. 2D, left). A2 features a stable stem with perfect Watson-Crick pairs as well as the mGHG, basal UG, apical UGUG, and CNNC motifs. We employed the variants lacking a part or all of these motifs: variant A2.3 lacks the mGHG motif, while A2.2 lacks the UG, UGUG, and CNNC motifs, and A2.1 does not possess any of these sequence motifs. The efficiency of pri-miRNA processing was estimated by normalizing the level of the small RNA produced from the 5’ arm against miR-1 from a separate promoter within the same plasmid. Consistent with the previous report20, A2 and A2.3 showed similar small RNA levels, while A2.1 and A2.2 exhibited lower yields (Fig. 2D). The good substrates, A2 and A2.3, were not significantly affected by ERH depletion, whereas the poor variants, A2.1 and A2.2, exhibited ERH-dependence. These results demonstrate that ERH is specifically required for suboptimal pri-miRNAs, irrespective of its surrounding sequences or helper hairpins.
ERH supports pri-miRNA processing in vitro
To further examine the role of ERH, we conducted in vitro processing assays using cell lysate overexpressing DROSHA and DGCR8 (Fig. 3A). In addition to wild-type (WT) DGCR8, we used two deletion mutants with impaired ERH binding: one with a deletion in the 96–139 aa region (ΔBS) and another lacking the N-terminus (ΔN, 1–275 aa)31. Three pri-miRNAs were utilized as substrates for these assays: miR-144, miR-197, and miR-589 (Fig. 3B). Although the miR-144 and miR-197 hairpins were both unaffected by ERH depletion, they differ significantly in sequence, structure, and processing efficiency. According to SHAPE-Map-determined secondary structures25, the miR-144 hairpin exhibits more favorable structural features, such as a longer and less mismatched stem, compared to the miR-197 hairpin (Fig. 3B). In vitro assays also showed differences in their processing rates, ranking pri-miR-144 higher (59th) compared to pri-miR-197 (267th) and pri-miR-589 (739th)34.
A Schematic diagram of in vitro pri-miRNA processing and DGCR8 variants used in the assay. Repoter RNAs were labeled with radioisotope (RI; ATP-[γ−32P]) at the 5’ end. B Schematic diagram of the structure and motifs of pri-miR-144, pri-miR-197, and pri-miR-589 based on a previous publication25. C Representative gel images from time course in vitro processing of RI-labeled pri-miRNA and quantification of the intensities of the cleaved 5’ fragment of pri-miRNA bands (arrowheads) from two independent replicates. Corresponding rate constants for wild-type DGCR8 (kWT in red), DGCR8 ΔBS (k∆BS in black), and DGCR8 ΔN (k∆N in gray) are indicated. The levels of Microprocessor components were quantified by performing western blot analysis (Supplementary Fig. 2A). D Representative gel images from time course in vitro processing of RI-labeled pri-miRNA and quantification of the intensities of the cleaved 5’ fragment of pri-miRNA bands (arrowheads) from three independent replicates. “wERH” indicates ERH wild-type and ‘mERH’ indicates ERH homodimerization mutant (I5R/L7R). Corresponding rate constants for DGCR8 with wERH (kwERH in red) and DGCR8 with mERH (kmERH in black) are indicated. The levels of Microprocessor components was quantified by performing western blot analysis (Supplementary Fig. 2C). Statistical analysis was performed using a one-tailed Student’s t-test. E Schematic diagram of in vitro pri-miRNA processing with nuclear lysate. F Representative gel images from time course in vitro processing of RI-labeled pri-miRNA with nuclear lysates and quantification of the intensities of the cleaved 5’ fragment of pri-miRNA bands (arrowheads) from three independent replicates. Corresponding rate constants for DGCR8 ΔN (k₁ in gray), wild-type DGCR8 (k₂ in black), and wild-type DGCR8 with exogenous ERH (k₃ in red) are indicated. The levels of Microprocessor components were quantified by performing western blot analysis (Supplementary Fig. 2E). Statistical analysis was performed using a one-tailed Student’s t-test. All source data involving replicate data are provided as a Source data file.
When pri-miR-144 was incubated in cell lysates containing WT DGCR8 or the ΔBS mutant, pre-miR-144 production was comparable between WT and mutants (Fig. 3C, left). In contrast, with less efficiently processed pri-miR-197, deletion mutations impaired pre-miRNA production, with the ΔN DGCR8 mutant showing a stronger defect than the ΔBS mutant (Fig. 3C, right).
Since ERH has other binding partners, such as SNRPD3 and CIZ154,59, which may potentially compete and titrate away ERH, we made an additional DGCR8 variant with ERH fused to the N-terminus (wERH-DGCR8; Fig. 3A). As a control, we introduced a point mutation in ERH, which disrupts its dimerization and activity60 (mERH-DGCR8; Fig. 3A and Supplementary Fig. 2B). Compared to the inactive mutant, wERH-DGCR8 showed enhanced processing efficiency, with a greater effect on pri-miR-197 than pri-miR-144 (Fig. 3D).
Because suboptimal pri-miR-589 and pri-miR-451a were not processed to detectable levels under these conditions (Supplementary Fig. 2D), we improved processing by using nucleus lysates and over-expressing ERH (Fig. 3E). Under these conditions, pri-miR-144 processing was improved in the presence of wild-type DGCR8 and exogenous ERH compared to the ΔN mutant or wild-type DGCR8 alone (Fig. 3F). For pri-miR-197, a more substantial increase in processing was observed, highlighting its stronger dependence on both full-length DGCR8 and ERH. Although pri-miR-589 processing remained inefficient, cleavage was specifically observed in nucleus lysates with wild-type DGCR8 and ERH, indicating that ERH promotes pri-miR-589 processing (Supplementary Fig. 2F). These findings support a model in which ERH, through binding to DGCR8, generally enhances Microprocessor activity, which impacts suboptimal hairpins more strongly than optimal ones.
We also tested a purified recombinant Microprocessor (rMP) with or without recombinant ERH protein (Supplementary Fig. 2G). No differences in processing efficiency of pri-miR-144 and pri-miR-589 were observed depending on ERH under these conditions (Supplementary Fig. 2H–I). It is unclear why purified proteins do not recapitulate ERH’s effects observed in cells and cell lysates. One possibility is that additional cofactor(s) may be required for ERH’s action, which warrants further investigation in future studies.
SAFB2 acts specifically on clustered miRNAs
We next examined SAFB2, which is also known to mediate cluster assistance48. To verify the role of SAFB2 in cluster assistance, we performed RT-qPCR following SAFB2 knockdown in HEK293E cells (Supplementary Fig. 3A). As expected, SAFB2 is required for cluster assistance in the miR-144 ~ 451a cluster located in either intronic or exonic regions (Supplementary Fig. 1D). Consistent results were obtained using a miRNA construct driven by the pol III-driven U6 promoter as well (Supplementary Fig. 1D). These results validate the earlier finding of SAFB2’s role in cluster assistance.
We next investigated whether SAFB2 regulates monocistronic pri-miRNA processing, similarly to ERH. To address this, we re-examined a small RNA sequencing data from previous report48. Intriguingly, in both Ramos and HEK293T cells co-depleted of SAFB and SAFB2, we did not observe strongly affected monocistronic miRNAs (Supplementary Fig. 3B; cutoff: log2 fold change < −1). This finding contrasts with the effect of ERH depletion on some monocistronic miRNAs (Fig. 1A).
To explore the role of SAFB2 in cluster assistance, we utilized plasmids encoding the miR-589 hairpin, which strongly relies on ERH (Fig. 4A). We made a monocistronic plasmid as well as a clustered construct encoding the miR-589 hairpin conjugated to the miR-144 hairpin. The miR-1 hairpin is transcribed from an independent promoter, and the miR-1 level was used for normalization. In stark contrast to ERH knockdown, which affected both monocistronic and clustered forms, SAFB2 depletion reduced miR-589 production exclusively from the clustered construct (Fig. 4A, right and Supplementary Fig. 3C). This result indicates that SAFB2 is specifically required for clustered miRNA processing but not for stand-alone miRNAs.
A RT-qPCR results for ERH or SAFB2 dependence of monocistronic or artificially clustered pri-miR-589. After 2 days of ERH or SAFB2 knockdown, pri-miRNA expression plasmids were transfected with additional siRNA. The data shown represent the average of three replicate experiments. Error bars indicate the standard error of the mean. Statistical analysis was performed using a one-tailed Student’s t-test. B Schematic diagram of dual luciferase for relative pri-miRNA processing assay and results. After 2 days of ERH or SAFB2 knockdown, luciferase plasmids were transfected with additional siRNA. The data shown represent the average of three replicate experiments. Error bars indicate the standard error of the mean. Statistical analysis was performed using a one-tailed Student’s t-test. C Schematic diagram of tethering assay with λN-tagged DGCR8 and RT-qPCR showing the λN-tagged DGCR8 tethering effect and ERH/SAFB2 dependence of pri-miR-144 and pri-miR-451a with 5XBoxB sequence. After 2 days of ERH or SAFB2 knockdown, pri-miRNA expression plasmids and DGCR8 expression plasmids were co-transfected with additional siRNA. The data shown represent the average of three replicate experiments. Error bars indicate the standard error of the mean. Statistical analysis was performed using a one-tailed Student’s t-test. All source data are provided as a Source data file.
We extended our analysis to additional miRNAs known to depend on cluster assistance, including miR-15a ~ 16-1 and miR-425 ~ 19131,46,48. Because their basal levels are high, making it difficult to measure their changes after ectopic expression, we took an orthogonal approach using a dual luciferase reporter system that contains a miRNA hairpin inserted into the 3′ UTR region (Fig. 4B). Drosha-mediated cleavage of the hairpin leads to decreased luciferase expression. Suboptimal miRNA hairpins (miR-15a or miR-425) were inserted into the 3′ UTR of the firefly luciferase-coding sequence, while optimal miRNA hairpins (miR-16-1 or miR-191) were inserted into the 3′ UTR of the renilla luciferase-coding sequence (Fig. 4B, left). Depletion of ERH increased the firefly-to-renilla luciferase ratio, reflecting the reduced processing of the miR-15a hairpin and indicating the role of ERH in the processing of this suboptimal pri-miRNA (Fig. 4B, right). In contrast, SAFB2 knockdown had no effect on luciferase levels, indicating that SAFB2 is not required for miR-15a processing when the hairpin is in a separate transcript from its original partner, miR-16-1. Similar results were obtained with miR-425 (suboptimal) and miR-191 (optimal) (Fig. 4B, right). Collectively, these findings suggest that SAFB2 is not necessary for the processing of suboptimal pri-miRNAs unless the hairpins are in clustered arrangements.
The prevailing model for cluster assistance posits that the optimal “helper” hairpin initially recruits the Microprocessor, increasing the chance of interactions with the “recipient” hairpin. To emulate this process, we used the λN-BoxB system to recruit the Microprocessor to the miR-451a hairpin (Fig. 4C, left). miR-451a production was enhanced when DGCR8 was tethered to the monocistronic miR-451 hairpin via the λN-BoxB interaction, suggesting that high local Microprocessor concentration increases the processing rate of the suboptimal pri-miRNA. Under these conditions, ERH knockdown reduced miR-451 production, showing that ERH is still required to promote processing even at increased local Microprocessor concentrations (Fig. 4C, right). Notably, SAFB2 depletion did not affect this tethering construct, suggesting that SAFB2 may be dispensable once the Microprocessor is stably associated with the transcript. This implies that SAFB2 may contribute to the recruitment step or translocation step between hairpins, whereas ERH is involved in the final step of recipient hairpin processing. Control experiments using the optimal hairpin miR-144 showed no effect of DGCR8 tethering or knockdown of ERH or SAFB2.
Protein interactions among the Microprocessor components and their auxiliary factors
As no direct comparison of ERH and SAFB2 has yet been made under the same experimental conditions, we further investigated how ERH and SAFB2 interact with the Microprocessor complex. Firstly, to examine the SAFB2-DROSHA interaction, we co-expressed Flag-tagged DROSHA and V5-tagged SAFB2 (Fig. 5A, lane 3). DGCR8 C-terminal tail (CTT) was also co-expressed, as it is required to stabilize the DROSHA protein. Anti-Flag antibody co-precipitated V5-SAFB2, validating the SAFB2-DROSHA interaction (Fig. 5A, lane 7). In contrast, when Flag-DGCR8 was co-expressed instead of Flag-DROSHA, SAFB2 was precipitated only weakly (Fig. 5A, lanes 4 and 8), indicating that SAFB2 interacts with DROSHA rather than with DGCR8, corroborating previous reports31,48. Moreover, when we performed co-immunoprecipitation experiments after ERH knockdown, anti-SAFB2 antibody precipitated DROSHA even when ERH level was reduced (Fig. 5B). Therefore, the SAFB2-DROSHA interaction does not strictly rely on ERH. Reciprocally, when SAFB2 was depleted, anti-ERH antibodies co-precipitated DGCR8 protein, demonstrating that the ERH-DGCR8 interaction does not require SAFB2 (Fig. 5C).
A Western blots of co-immunoprecipitation assay with overexpressed DROSHA or DGCR8. DROSHA-Flag was co-transfected with the CTT of DGCR8 for efficient expression. 1% input was loaded, based on the total amount used for immunoprecipitation. B Co-Immunoprecipitation western blots of endogenous SAFB2 following ERH knockdown. Non-targeting siRNA was transfected in the absence of knockdown. Four days after knockdown, cells were harvested for further co-IP experiments. 2.5% input was loaded, based on the total amount used for immunoprecipitation. C Co-Immunoprecipitation western blots of endogenous ERH following SAFB2 knockdown. Non-targeting siRNA was transfected in the absence of knockdown. Four days after knockdown, cells were harvested for further co-IP experiments. 2.5% input was loaded, based on the total amount used for immunoprecipitation. D–E Co-Immunoprecipitation western blots of ectopically expressed SAFB2 core domain. D HEK293E cell E HCT116 parental or DROSHA knock-out cell. 1% input was loaded, based on the total amount used for immunoprecipitation. F Model of interactions among the Microprocessor and auxiliary factors. All data in A–E are from one experiment and are representative of at least two independent replicates with similar results. All source data are provided as a Source data file.
As SAFB2 is known to interact with DROSHA through the 561-726 aa region, we validated this interaction by expressing the SAFB2 fragment fused to the Flag tag (Fig. 5D, Flag-SAFB2561-726). A Myc-tagged SAFB2 fragment was used as a control for the specificity of immunoprecipitation. Flag-SAFB2561-726 specifically co-precipitated ERH, DROSHA, and DGCR8 (Fig. 5D, lane 4), confirming the previous findings that this region is sufficient to mediate SAFB2’s interaction with the Microprocessor48. This is also consistent with prior reports of SAFB2-ERH interaction57, though it remains unclear whether the interaction is direct. This SAFB2-ERH-DGCR8 association persisted in DROSHA knockout cells (Fig. 5E), suggesting that SAFB2 may interact with ERH, which in turn links to DGCR8, independently of DROSHA. Since SAFB2561-726 mediates dimerization or multimerization48, it is possible that SAFB2 fragments form multimers that help recruit DROSHA, ERH, and DGCR8 simultaneously (Fig. 5F).
Discussion
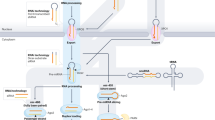
In this study, we uncovered the distinct roles of ERH and SAFB2. ERH enhances the processing of both clustered and stand-alone pri-miRNAs, functioning regardless of the clustered arrangement and surrounding sequences/structures. Optimal substrates can be processed without ERH, likely because their affinity to the Microprocessor is high and cellular Microprocessor activity is normally sufficient to support their processing. In contrast, suboptimal pri-miRNAs–containing short stems or loops or lacking sequence motifs like UG or UGUG–have lower affinity to the Microprocessor and therefore require ERH for efficient processing (Fig. 6A). Unlike ERH, SAFB2 is dispensable for stand-alone miRNAs but is specifically required for clustered miRNAs.
A Working model of ERH-mediated monocistronic pri-miRNA processing. B Working model of ERH/SAFB2-mediated cluster assistance. C Schematic diagram of DGCR8 mRNA. D RT-qPCR for the expression levels of pri-miR-1306, which is DGCR8 mRNA detected by SYBR Green-based qPT-PCR after knocking down ERH for 4 days, and those of miRNAs were deduced from small RNA-seq. RT-qPCR data shown represent the average of three biological replicate experiments. Error bars indicate the standard error of the mean. Statistical analysis was performed using a one-sided Student’s t-test against the null hypothesis of no change. *P < 0.05; **P < 0.01; N.S. denotes not significant. RT-qPCR representing relative miR-1306 abundance level upon (E) ERH knockdown or (F) ERH rescue with ectopically expressed pri-miR-1306. After 2 days of ERH knockdown, pri-miRNA expression plasmids were transfected with additional siRNA. RT-qPCR data shown represent the average of three biological replicate experiments. Error bars indicate the standard error of the mean. Statistical analysis was performed using a one-tailed Student’s t-test. G RT-qPCR for the comparative expression of proteins by ERH or DROSHA knockdown in HEK293E cells. Cells were harvested after knocking down for 4 days. RT-qPCR data shown represent the average of three biological replicate experiments. Error bars indicate the standard error of the mean. Statistical analysis was performed using a one-tailed Student’s t-test. H Western blot for comparative expression of proteins by ERH or DROSHA, DGCR8 knockdown in HEK293E cells (left) and quantification of the intensities of the protein bands from the experiment in panel (right). Cells were harvested after knocking down for 4 days. Quantification data shown represent the average of three biological replicate experiments. Error bars indicate the standard error of the mean. Statistical analysis was performed using a one-tailed Student’s t-test. I Schematic diagrams of homeostatic mechanisms between ERH and DGCR8. All source data are provided as a Source data file.
Our findings indicate that cluster assistance involves at least two distinct steps. Both SAFB2 and ERH are needed for the effective maturation of clustered suboptimal pri-miRNAs, but they have different roles in cluster assistance. Together with the earlier findings31,46,47,48, our current observations suggest the following model (Fig. 6B). First, the helper hairpin recruits the Microprocessor and undergoes rapid processing. Next, SAFB2 mediates interactions between the Microprocessor and the recipient hairpin. Finally, ERH comes into action, enhancing the processing of suboptimal recipient hairpins. While the detailed mechanism by which ERH facilitates processing needs further investigation, previous findings suggest that the ERH dimer binds to the DGCR8 dimer at the N-terminal region of DGCR831, potentially stabilizing the Microprocessor complex in a more active conformation. Structural studies on the full Microprocessor-ERH complex will be critical to advance our understanding of this process.
Regarding SAFB2, we initially hypothesized that SAFB2 multimerizes to bring multiple copies of the Microprocessor together, given that SAFB2561-726 is known to multimerize48,61 and interact with DROSHA and ERH (Fig. 5). However, co-immunoprecipitation assays showed that the DROSHA-DROSHA interaction was not influenced by SAFB2 depletion (Supplementary Fig. 4A, right), refuting this model. Thus, we instead propose that SAFB2 facilitates the transfer of the Microprocessor from the helper hairpin to the recipient hairpin by associating with both the Microprocessor and RNA and thereby retaining the Microprocessor in the vicinity of the recipient hairpin. It remains to be investigated how SAFB2 performs this function and if there are any other cofactors involved in this process.
Our observations of ERH’s role in monocistronic hairpins lead us to revise and expand the concept of ERH-dependent pri-miRNAs. Although clustered miRNAs often rely heavily on ERH (e.g., miR-15-1, miR-425, miR-451a), we also found many monocistronic miRNAs that require ERH (e.g., miR-589, miR-365a, miR-378c, miR-500b). This demonstrates that ERH functions beyond cluster assistance, acting as a general cofactor that enhances pri-miRNA processing. Optimal substrates, which bind the Microprocessor with higher affinity, do not critically depend on ERH, whereas poor substrates are heavily reliant on ERH.
Notably, we found that ERH negatively controls DGCR8 by enhancing the cleavage of the DGCR8 mRNA (Fig. 6C). The DGCR8 mRNA contains two hairpins, miR-3618 and miR-1306, which are cleaved and destabilized by the Microprocessor62,63,64. This mechanism is known to be crucial for the homeostatic control of the Microprocessor level. In ERH-depleted cells, mature miR-1306 levels decrease while uncleaved hairpins accumulate, reflecting reduced processing (Fig. 6D–F). Consequently, ERH depletion leads to increased RNA and protein levels of DGCR8, which in turn stabilizes DROSHA protein through protein-protein interaction and elevates Microprocessor complex levels62,63,64 (Fig. 6G–I). Thus, ERH depletion not only causes an immediate reduction of pri-miRNA processing but also indirectly upregulates Microprocessor activity through this compensation mechanism, partially offsetting the downregulating effects of ERH loss. These findings also suggest that the impact of ERH is likely underestimated in knockdown experiments and that a majority of miRNAs may rely, at least modestly, on ERH for maturation. In addition, this compensatory effect may also explain why some hairpins, such as miR-197, appear to be independent of ERH in cellular knockdown experiments (Fig. 1) but not in in vitro processing assays (Fig. 3), where such compensation is absent.
Our findings carry evolutionary implications. ERH may have broadly facilitated the emergence of miRNA genes, independently of their genomic locations. By enabling fortuitous hairpins to produce enough small RNAs, ERH may have allowed these genes to undergo natural selection and integrate into cellular regulatory networks. SAFB2 may have played a more specific role in supporting the evolution of emergent hairpins near pre-existing miRNA loci, promoting the formation of miRNA operons that are frequently observed in modern animal genomes.
Methods
Cell lines
HEK293E cells (human embryonic kidney 293 EBNA1; authenticated by ATCC STR profiling) were grown in DMEM (Welgene) supplemented with 9.1% fetal bovine serum (Welgene). HCT116 DGCR8 CTT domain knock-out cells65 and HCT116 DROSHA knock-out cells66 (whose parental cells were authenticated by ATCC STR profiling) were cultured in McCoy’s 5 A medium (Welgene) supplemented with 9.1% fetal bovine serum (Welgene).
Plasmids and siRNAs
Sequences of siRNAs and primers used for cloning are provided in Supplementary Table 1.
Small RNA seq data analysis
Small RNA-seq results from previous papers were utilized. Data was uploaded through GEO (GSE116303 for D8ex2, GSE142818 for ERH, GSE141098 for SAFB2). To filter out lowly expressed miRNAs for reliable analysis, the same cutoff from the previous paper was used31. In vitro pri-miRNA processing data are from supplementary data of Kim et al.34. Detailed information on source data is organized in data availability section.
Gene knockdown and miRNA RT-qPCR
siRNA was transfected into 50% confluent HEK293E cells using Lipofectamine 3000 (Thermo Fisher Scientific) following the manufacturer’s instructions. After 48 h, the cells were trypsinized and counted for reverse transfection. For reverse transfection, 40 pmol of siRNA and 2 μg of pri-miRNA expression plasmid were co-transfected into 6E5 cells in a six-well culture plate using 4.5 μl of Lipofectamine 3000. In the case of DGCR8 tethering assay, each 1 μg of DGCR8 plasmid and pri-miRNA expression plasmid was used. After an additional 48 h, RNA was harvested with TRIzol. The level of mature miRNAs was measured with the TaqMan MicroRNA Assays (Thermo Fisher Scientific, #001973 for U6 snRNA, #002676 for miR-144, #001105 for miR-451a, #000497 for miR-197, #002409 for miR-589, #001020 for-miR-365a, #000385 for miR-1, #00377 for-let-7a, #243977 for miR-378c, #243179 for miR-500b) as manufacturer’s instructions. In the case of artificial miRNA from A2, Custom TaqMan Small RNA Assay was customized by the manufacturer to capture mature 5’arm A2 miRNA. Information regarding miRNA qPCR kits can be found in the Supplementary Table 3.
In the case of pri-miRNA plasmids, which do not contain internal control miRNA, the relative accumulation of miRNA was calculated by two separate qPCR results. The level of ectopic pri-miRNA transcripts was measured with RT-qPCR using SYBR green MIX (Thermo Fisher Scientific) (Supplementary Fig. 1B). The cDNAs for the qPCR were synthesized from 500 ng of total RNA using random hexamer primers (Thermo Fisher Scientific) and RevertAid Reverse Transcriptase (Thermo Fisher Scientific). U6 snRNA was used as the internal control for both qPCR. The pri-miRNA expression vector contains an SV40 artificial intron, in which the pri-miRNA sequence is inserted, and the sfGFP coding sequence. Transfection efficiencies between samples were normalized using sfGFP.
ERH rescue assay
ERH rescue assay for pri-miRNA transfection and measurement of mature miRNA is carried out in accordance with previous work31. One microgram of shERH plasmids and 1 μg of wild-type or homodimerization mutants ERH plasmids were co-transfected into 50% confluent six-well culture plates using 6 μl of Fugene HD. After 48 h incubation, cells were trypsinized and counted for reverse transfection.
RI-labeled pri-miRNA preparation
For in vitro transcribed RNAs, DNA templates were prepared with PCR using a forward primer (T7 promoter+gene_specific_F) and reverse primer (gene_specific_R) following Agarose gel purification. Four hundred and four nanograms of DNA template were in vitro transcribed using MEGAscript™ T7 Transcription Kit (AMB13345) as manufacturer’s instruction with slight modification. In the reaction mixture, ATP:GTP:CTP:UTP:α−32PUTP was used 1.5:1.5:1.5:1.5:2 ratio for internal UTP RI-labeling. An equal ratio of NTP was used for pri-miRNA with 5′-end labeling. DNA template was removed using TURBO DNase I (RNase-free). In the case of pri-miRNA with 5′-end RI labeling, in vitro transcribed RNA was treated with CIP (Takara) and further labeled with T4 PNK (Takara) following the manufacturer’s instruction. RI-labeled RNA was analyzed and purified from 6% urea polyacrylamide gel. The gel slices containing RI-labeled RNA were ground using Gel Breaker Tubes (Istbiotech) and centrifugation (20,000 × g, 4 °C for 1 min. The ground gel was incubated with 600 μL of 0.3 M NaCl solution overnight in a rotator. The RNA eluate was clarified with Corning Costar Spin-X Centrifuge Tube Filters (MilliporeSigma) and centrifugation (20,000 × g, 4 °C) for 5 min. The labeled RNA from flowthrough was precipitated with 1 mL of 100% EtOH and 1 μL of GlycoBlue Coprecipitant (Thermo Fisher Scientific) at −80 °C for 2 h. Precipitated RNA was harvested with centrifugation (20,000 × g, 4 °C) for 1 h. RNA pellet was washed twice with 75% EtOH and added TDW to dissolve RNA. RNA quantification was done with Qubit RNA HS (High Sensitivity) Assay Kit (Thermo Fisher Scientific).
Whole-cell lysate preparation
For in vitro processing assay with whole cell lysate, the experiments were carried out in accordance with previous work, incorporating minor adjustments46. Ectopic expression, DROSHA-HA and Flag-DGCR8 plasmids, which are cloned into the pCK expression vector, respectively, were used for transfection. Plasmids were diluted in 500 μL of OPTI-MEM with a 1:3 ratio of plasmid (μg): Fugene HD (μL; Promega) and mixed well. Mixtures were added into 50% confluent HEK293E cells in a 100 mm dish. After 48 h later, the cultured cells were collected by centrifugation (300 × g, 4 °C), and the remaining pellet was washed once with 500 µL of PBS and lysed with 250 µL of reaction buffer, considering the difference of experiment scale. The crude lysate was sonicated and clarified by centrifugation (20,000 × g, 4 °C) for 10 min. Clear nuclear lysate was aliquoted and stored at −80 °C for further use.
Nuclear lysate preparation
For ectopic expression, 4 μg DROSHA-HA and 1 μg Flag-DGCR8 plasmids, which are cloned into the pCK expression vector, respectively, were used for transfection. If necessary, 1 μg ERH or sfGFP in pCK expression plasmids was co-transfected. Plasmids were diluted in 500 μL of OPTI-MEM with a 1:3 ratio of plasmid (ug): Fugene HD (μL; Promega) and mixed well. Mixtures were added into 50% confluent HEK293E cells in a 100 mm dish. After 48 h later, the fractionation procedure for cytoplasm was performed the previously described method40. Briefly, the cultured cells were harvested in ice-cold PBS, and digitonin buffer (150 mM NaCl, 50 mM HEPES at pH 7.5, 150 µg/mL digitonin) was used for the removal of cytoplasmic fraction. The remaining pellet was washed once with 500 µL of PBS, collected by centrifugation (500 × g, 4 °C), and membrane fractions were removed with the subcellular protein fractionation kit for cultured cells (Thermo Fisher Scientific). The remaining pellet was washed once with 500 µL of PBS and lysed with 500 µL of Lysis buffer (500 mM NaCl, 50 mM Tris-HCl pH 7.5, protease inhibitor cocktail (Calbiochem)). The crude lysate was sonicated and clarified by centrifugation (20,000 × g, 4 °C) for 10 min. Clear nuclear lysate was aliquoted and stored at −80 °C for further use.
Recombinant Microprocessor preparation
To purify the recombinant full-length Microprocessor complex, 10 μg DROSHA-HA and 2.5 μg Flag-DGCR8 plasmids, which are cloned into the pCK expression vector, respectively, were used for transfection. Plasmids were diluted in 1 mL of OPTI-MEM with 37.5 μL Fugene HD (Promega) and mixed well. Mixtures were added into 50% confluent HEK293E cells in a 150 mm dish. After 48 h incubation at 37 °C, cells were harvested and lysed in 1 mL of Lysis buffer (500 mM NaCl, 50 mM Tris-HCl pH 7.5, protease inhibitor cocktail (Calbiochem). The crude lysate was sonicated and clarified by centrifugation (20,000 × g, 4 °C) for 10 min. During centrifugation, anti-Flag M2 affinity gel (Sigma-Aldrich) was prepared by washing with Lysis buffer three times. The 1 mL lysate supernatant was incubated with 20 μL pre-washed anti-Flag M2 affinity gel at 4 °C for 1 h 30 min in a rotator. Incubated affinity gel was washed two times with 1 mL of wash buffer (500 mM NaCl, 50 mM Tris-HCl pH 7.5) supplemented with the final 0.1% NP40 and three times with 1 mL of wash buffer. Wash buffer was entirely drained by syringe with a 30 G needle, and 100 μL of Elution buffer (500 mM NaCl, 50 mM Tris-HCl pH 7.5, 1 mg/ml 3XFlag peptide (MilliporeSigma)) was added. The mixture was incubated in the ThermoMixer for 30 min at 4 °C, 1300 rpm shaking. The eluate was collected with a syringe with a 30 G needle and stored at −80 °C for further use.
In vitro processing
For in vitro pri-miRNA processing with recombinant Microprocessor, labeled pri-miRNA was mixed with a nuclear lysate or recombinant Microprocessor in 10 μL buffer containing 100 mM NaCl, 50 mM Tris-HCl pH7.5, 2 mM MgCl2, 1 mM DTT, and 4 U/μL SUPERase inhibitor (Ambion). The mixture was incubated at 37 °C for 1 hr. After the reaction, 9 μL of 2X RNA loading buffer (NEB) and 1 μL of 20 mg/ml Proteinase K (MilliporeSigma) were added into the mixture and incubated at 37 °C for 30 min, then 50 °C for 30 min. In the case of in vitro processing with nuclear lysate, incubated mixtures were subjected to phenol-chloroform extraction followed by ethanol precipitation. Precipitated RNA was dissolved in 10 μL TDW and 10 μL of 2X RNA loading buffer (NEB). The sample was boiled at 95 °C for 5 min before running on the gel. Processed pri-miRNA was analyzed in 6% urea polyacrylamide gel with RNA decade marker (Ambion). Quantification of pri-miRNA processing was done utilizing Multi Gauge V3.0 software (Fujifilm). For time-course experiments, the observed rate constant was obtained by fitting all data from two independent time-course experiments to the equation \(y=1-{{{{\rm{e}}}}}^{-k\times t}\). Among the five time points, the samples from the final time point exhibited fluctuations, thus, data from four time points were employed for the purpose of the fitting.
Dual Luciferase assay
HEK293E cells were transfected with siRNAs incubated for knockdown. After 48 h incubation, cells were trypsinized and counted for reverse transfection. 1.5E5 cells on a 24-well plate were further transfected with 100 nM siRNAs with 100 ng of pmirGLO plasmids by Lipofectamine 3000. After 24 hrs later, cells were lysed and analyzed with the dual-luciferase reporter assay system (Promega) according to the manufacturer’s instructions.
Co-immunoprecipitation with ectopically expressed protein
For ectopic expression of the protein, a total of 5 μg of protein expression plasmid and 15ul of Fugene HD (Promega) were diluted in 500 μl OPTI-MEM. The mixture was incubated for 5 min before adding to 50% confluent HEK293E cells in a 100 mm dish. After 2 days, cells were harvested and lysed in the Lysis buffer (20 mM Tris pH 7.5, 100 mM KCl, 0.2 mM EDTA, 0.1% NP-40, 10% glycerol with 0.1 ng/μl RNase A, and protease inhibitor cocktails (Calbiochem)) by sonication. The cell lysates were collected by centrifugation for 10 min at 20,000 × g, 4 °C. For Flag or HA-tagged protein immunoprecipitation, anti-Flag M2 affinity gel (Sigma-Aldrich) or anti-HA agarose affinity gel (Sigma-Aldrich) was pre-washed three times with 1 ml of the Wash buffer (20 mM Tris pH 7.5, 100 mM KCl, 0.2 mM EDTA, 0.1% NP-40, 10% glycerol). Individual lysates were quantified using the Bradford assay to ensure equal amounts, and equal volumes of lysates were used accordingly. Lysates and beads were incubated at 4 °C for 1–2 h. Incubated beads were washed five times with the Wash buffer. The remaining supernatants were removed using a 30-gauge needle. Beads were resuspended in 1× SDS protein sample buffer (50 mM Tris pH 6.8, 10% Glycerol, 2% SDS, 100 mM DTT, 0.01% bromophenol blue) for western blotting.
Co-immunoprecipitation of endogenous protein
Information of antibodies used in this study is presented in Supplementary Table 2. For endogenous protein immunoprecipitation, HEK293E cells grown on 100 mm dishes were lysed in the Lysis buffer (20 mM Tris pH 7.5, 100 mM KCl, 0.2 mM EDTA, 0.2% NP-40, 10% glycerol, 0.4 ng/μl RNase A and protease inhibitor cocktails (Calbiochem)) and sonicated. Clear lysates were collected by centrifugation for 10 min at 20,000 × g, 4 °C. The antibody was pre-incubated with 20 μl of prewashed protein G Sepharose (GE Healthcare) at 4 °C for 1 h. Antibody-conjugated beads were incubated with lysates at 4 °C for 1–2 h and washed with the Wash buffer (20 mM Tris pH 7.5, 100 mM KCl, 0.2 mM EDTA, 0.2% NP-40, 10% glycerol) 5 times. The remaining supernatants were removed using a 30-gauge needle. Beads were resuspended in 1× SDS protein sample buffer (50 mM Tris pH 6.8, 10% Glycerol, 2% SDS, 100 mM DTT, 0.01% bromophenol blue) for western blotting. In cases involving protein knockdown, 100 μl HEK293E cells were subjected to siRNA knockdown for 48 h. Prior to lysate-bead binding, individual lysates were quantified using the Bradford assay, and equal amounts of lysates were utilized.
Western blotting
Protein samples were boiled at 95 °C for 5 min before separating on Novex WedgeWell 8–16% Tris-Glycine Mini Gels (Thermo Fisher Scientific). Protein samples were transferred to a polyvinylidene fluoride membrane (GE Healthcare). Primary antibodies are typically diluted in a blocking solution (1% nonfat dry milk in TBS-T) with a dilution of 1:1000. The signals were detected using SuperSignal West Pico PLUS Chemiluminescent Substrate (Thermo Fisher Scientific) or SuperSignal West Femto PLUS Chemiluminescent Substrate (Thermo Fisher Scientific). Quantification of protein was done utilizing Multi Gauge V3.0 software (Fujifilm).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Source data are provided with this paper. The small RNA sequencing data with ERH depletion that supports the findings of this study are available in NCBI Gene Expression Omnibus under the accession number: GSE149421 (https://doi.org/10.1093/nar/gkaa827) (ref. 31), GSE142818 (https://doi.org/10.1016/j.molcel.2020.01.026) (Ref. 46). The in vitro pri-miRNA processing data used in this study are available in Supplementary Tables 3 and 6 of “https://doi.org/10.1016/j.molcel.2021.07.002” (ref. 34). The pri-miRNA structure information used in this study are available in Document S2 at https://doi.org/10.1016/j.molcel.2024.02.005 (ref. 25). The small RNA sequencing data with SAFB knockout reanalyzed in this study are available in NCBI Gene Expression Ominbus under the accession number: GSE141098 (https://doi.org/10.1016/j.molcel.2020.05.011) (ref. 48). Source data are provided with this paper.
References
Bartel, D. P. Metazoan microRNAs. Cell 173, 20–51 (2018).
Chen, L.-L. & Kim, V. N. Small and long non-coding RNAs: past, present, and future. Cell 187, 6451–6485 (2024).
Kim, H., Lee, Y.-Y. & Kim, V. N. The biogenesis and regulation of animal microRNAs. Nat. Rev. Mol. Cell Biol. 26, 276–296 (2025).
Lee, Y., Jeon, K., Lee, J.-T., Kim, S. & Kim, V. N. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 21, 4663–4670 (2002).
Cai, X., Hagedorn, C. H. & Cullen, B. R. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 10, 1957–1966 (2004).
Nguyen, T. A. et al. Functional anatomy of the human microprocessor. Cell 161, 1374–1387 (2015).
Lee, Y. et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 425, 415–419 (2003).
Denli, A. M., Tops, B. B. J., Plasterk, R. H. A., Ketting, R. F. & Hannon, G. J. Processing of primary microRNAs by the microprocessor complex. Nature 432, 231–235 (2004).
Gregory, R. I. et al. The Microprocessor complex mediates the genesis of microRNAs. Nature 432, 235–240 (2004).
Han, J. et al. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 18, 3016–3027 (2004).
Landthaler, M., Yalcin, A. & Tuschl, T. The human DiGeorge syndrome critical region gene 8 and Its D. melanogaster homolog are required for miRNA biogenesis. Curr. Biol. 14, 2162–2167 (2004).
Herbert, K. M. et al. A heterotrimer model of the complete Microprocessor complex revealed by single-molecule subunit counting. RNA 22, 175–183 (2016).
Hutvágner, G. et al. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293, 834–838 (2001).
Ketting, R. F. et al. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 15, 2654–2659 (2001).
Bernstein, E., Caudy, A. A., Hammond, S. M. & Hannon, G. J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409, 363–366 (2001).
Grishok, A. et al. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106, 23–34 (2001).
Knight, S. W. & Bass, B. L. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science 293, 2269–2271 (2001).
Auyeung, V. C., Ulitsky, I., McGeary, S. E. & Bartel, D. P. Beyond secondary structure: primary-sequence determinants license pri-miRNA hairpins for processing. Cell 152, 844–858 (2013).
Han, J. et al. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell 125, 887–901 (2006).
Fang, W. & Bartel, D. P. The menu of features that define primary microRNAs and enable de novo design of microRNA genes. Mol. Cell 60, 131–145 (2015).
Kwon, S. C. et al. Molecular basis for the single-nucleotide precision of primary microRNA processing. Mol. Cell 73, 505–518.e5 (2019).
Kim, K., Nguyen, T. D., Li, S. & Nguyen, T. A. SRSF3 recruits DROSHA to the basal junction of primary microRNAs. RNA 24, 892–898 (2018).
Li, S., Nguyen, T. D., Nguyen, T. L. & Nguyen, T. A. Mismatched and wobble base pairs govern primary microRNA processing by human Microprocessor. Nat. Commun. 11, 1–17 (2020).
Nguyen, T. L., Nguyen, T. D., Ngo, M. K., Le, T. N.-Y. & Nguyen, T. A. Noncanonical processing by animal Microprocessor. Mol. Cell 83, 1810–1826.e8 (2023).
Baek, S. C. et al. Structural atlas of human primary microRNAs generated by SHAPE-MaP. Mol. Cell 84, 1158–1172.e6 (2024).
Senturia, R. et al. Structure of the dimerization domain of DiGeorge critical region 8. Protein Sci. 19, 1354–1365 (2010).
Kwon, S. C. et al. Structure of Human DROSHA. Cell 164, 81–90 (2016).
Partin, A. C. et al. Cryo-EM structures of human Drosha and DGCR8 in complex with primary microRNA. Mol. Cell 78, 411–422.e4 (2020).
Jin, W., Wang, J., Liu, C.-P., Wang, H.-W. & Xu, R.-M. Structural Basis for pri-miRNA Recognition by Drosha. Mol. Cell 78, 423–433.e5 (2020).
Garg, A., Shang, R., Cvetanovic, T., Lai, E. C. & Joshua-Tor, L. The structural landscape of Microprocessor-mediated processing of pri-let-7 miRNAs. Mol. Cell 84, 4175–4190.e6 (2024).
Kwon, S. C. et al. ERH facilitates microRNA maturation through the interaction with the N-terminus of DGCR8. Nucleic Acids Res. 48, 11097–11112 (2020).
Guil, S. & Cáceres, J. F. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat. Struct. Mol. Biol. 14, 591–596 (2007).
Michlewski, G. & Cáceres, J. F. Antagonistic role of hnRNP A1 and KSRP in the regulation of let-7a biogenesis. Nat. Struct. Mol. Biol. 17, 1011–1018 (2010).
Kim, K. et al. A quantitative map of human primary microRNA processing sites. Mol. Cell 81, 3422–3439.e11 (2021).
Morlando, M. et al. Primary microRNA transcripts are processed co-transcriptionally. Nat. Struct. Mol. Biol. 15, 902–909 (2008).
Wang, Y., Luo, J., Zhang, H. & Lu, J. MicroRNAs in the same clusters evolve to coordinately regulate functionally related genes. Mol. Biol. Evol. 33, 2232–2247 (2016).
Kataoka, N., Fujita, M. & Ohno, M. Functional association of the Microprocessor complex with the spliceosome. Mol. Cell. Biol. 29, 3243–3254 (2009).
Agranat-Tamir, L., Shomron, N., Sperling, J. & Sperling, R. Interplay between pre-mRNA splicing and microRNA biogenesis within the supraspliceosome. Nucleic Acids Res. 42, 4640–4651 (2014).
Nojima, T. et al. Mammalian NET-seq reveals genome-wide nascent transcription coupled to RNA processing. Cell 161, 526–540 (2015).
Son, S., Kim, B., Yang, J. & Kim, V. N. Role of the proline-rich disordered domain of DROSHA in intronic microRNA processing. Genes Dev. 37, 383–397 (2023).
Donayo, A. O. et al. Oncogenic biogenesis of pri-miR-17∼92 reveals hierarchy and competition among polycistronic microRNAs. Mol. Cell 75, 340–356.e10 (2019).
Truscott, M., Islam, A. B. M. M. K. & Frolov, M. V. Novel regulation and functional interaction of polycistronic miRNAs. RNA 22, 129–138 (2016).
Lataniotis, L. et al. CRISPR/Cas9 editing reveals novel mechanisms of clustered microRNA regulation and function. Sci. Rep. 7, 8585 (2017).
Haar, J. et al. The expression of a viral microRNA is regulated by clustering to allow optimal B cell transformation. Nucleic Acids Res. 44, 1326–1341 (2016).
Vilimova, M. et al. Cis regulation within a cluster of viral microRNAs. Nucleic Acids Res. 49, 10018–10033 (2020).
Fang, W. & Bartel, D. P. MicroRNA clustering assists processing of suboptimal microRNA hairpins through the action of the ERH protein. Mol. Cell 78, 289–302.e6 (2020).
Shang, R. et al. Genomic clustering facilitates nuclear processing of suboptimal Pri-miRNA Loci. Mol. Cell 78, 303–316.e4 (2020).
Hutter, K. et al. SAFB2 enables the processing of suboptimal stem-loop structures in clustered primary miRNA transcripts. Mol. Cell 78, 876–889.e6 (2020).
Shang, R. & Lai, E. C. Parameters of clustered suboptimal miRNA biogenesis. Proc. Natl. Acad. Sci. USA 120, e2306727120 (2023).
Weng, M.-T. et al. Evolutionarily conserved protein ERH controls CENP-E mRNA splicing and is required for the survival of KRAS mutant cancer cells. Proc. Natl. Acad. Sci. USA 109, E3659–E3667 (2012).
Takai, H. et al. 5-hydroxymethylcytosine plays a critical role in glioblastomagenesis by recruiting the CHTOP-methylosome complex. Cell Rep. 9, 48–60 (2014).
Wang, X., Xie, H., Zhu, Z., Zhang, J. & Xu, C. Molecular basis for the recognition of CIZ1 by ERH. FEBS J. 290, 712–723 (2023).
Kozlowski, P. Thirty years with ERH: an mRNA splicing and mitosis factor only or rather a novel genome integrity protector? Cells 12, 2449 (2023).
Street, L. A. et al. Large-scale map of RNA-binding protein interactomes across the mRNA life cycle. Mol. Cell 84, 3790–3809.e8 (2024).
Townson, S. M. et al. SAFB2, a new scaffold attachment factor homolog and estrogen receptor corepressor*. J. Biol. Chem. 278, 20059–20068 (2003).
Oesterreich, S. Scaffold attachment factors SAFB1 and SAFB2: Innocent bystanders or critical players in breast tumorigenesis?. J. Cell. Biochem. 90, 653–661 (2003).
Drakouli, S., Lyberopoulou, A., Papathanassiou, M., Mylonis, I. & Georgatsou, E. Enhancer of rudimentary homologue interacts with scaffold attachment factor B at the nuclear matrix to regulate SR protein phosphorylation. FEBS J. 284, 2482–2500 (2017).
Deng, L. et al. Stabilizing heterochromatin by DGCR8 alleviates senescence and osteoarthritis. Nat. Commun. 10, 3329 (2019).
Pang, K. et al. ERH gene and its role in cancer cells. Front. Oncol. 12, 900496 (2022).
Arai, R. et al. Crystal structure of an enhancer of rudimentary homolog (ERH) at 2.1 Angstroms resolution. Protein Sci. 14, 1888–1893 (2005).
Norman, M., Rivers, C., Lee, Y.-B., Idris, J. & Uney, J. The increasing diversity of functions attributed to the SAFB family of RNA-/DNA-binding proteins. Biochem. J. 473, 4271–4288 (2016).
Han, J. et al. Posttranscriptional crossregulation between Drosha and DGCR8. Cell 136, 75–84 (2009).
Triboulet, R., Chang, H.-M., Lapierre, R. J. & Gregory, R. I. Post-transcriptional control of DGCR8 expression by the Microprocessor. RNA 15, 1005–1011 (2009).
Shenoy, A. & Blelloch, R. Genomic analysis suggests that mRNA destabilization by the microprocessor is specialized for the auto-regulation of Dgcr8. PLoS ONE 4, e6971 (2009).
Nguyen, T. A., Park, J., Dang, T. L., Choi, Y.-G. & Kim, V. N. Microprocessor depends on hemin to recognize the apical loop of primary microRNA. Nucleic Acids Res. 46, 5726–5736 (2018).
Kim, Y.-K., Kim, B. & Kim, V. N. Re-evaluation of the roles of DROSHA, Export in 5, and DICER in microRNA biogenesis. Proc. Natl. Acad. Sci. USA 113, E1881–E1889 (2016).
Acknowledgements
We thank our laboratory members for their support, especially Kijun Kim, Soomin Son, and Young-Yoon Lee for providing the pri-miRNA expression plasmid used in this paper. This work was supported by Institute for Basic Science from the Ministry of Science and ICT of Korea [IBS-R008-D1 to H.J., J.P., and V.N.K.] and by BK21 Research Fellowships from the Ministry of Education of Korea [to H.J. and J.P.].
Author information
Authors and Affiliations
Contributions
H.J. and V.N.K. designed experiments. H.J. and J.P. performed cell-related experiments. H.J. performed the in vitro assay. H.J. and V.N.K. analyzed the data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Jang, H., Park, J. & Kim, V.N. ERH promotes primary microRNA processing beyond cluster assistance. Nat Commun 16, 7913 (2025). https://doi.org/10.1038/s41467-025-63015-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-63015-y