Abstract
Proximity-inducing modalities that co-opt cellular pathways offer new opportunities to regulate oncogenic drivers. Inspired by the success of proximity-based chimeras in both intracellular and extracellular target space, here we describe the development of LYsosome Membrane TArgeting Chimeras (LYMTACs) as a small molecule-based platform that functions intracellularly to modulate the membrane proteome. Conceptually, LYMTACs are heterobifunctional small molecules that co-opt short-lived lysosomal membrane proteins (LMPs) as effectors to deliver targets for lysosomal degradation. We demonstrate that a promiscuous kinase inhibitor-based LYMTAC selectively targets membrane proteins for lysosomal degradation via RNF152, a short-lived LMP. We extend this concept by showing that oncogenic KRASG12D signaling can be potently inhibited by LYMTACs. Mechanistically, LYMTACs display multi-pharmacology and exert their activity through both target relocalization into the lysosome and degradation. We further generalize LYMTACs across various LMPs and thus offer a platform to access challenging membrane proteins through targeted protein relocalization and degradation.
Similar content being viewed by others
Introduction
Plasma membrane-bound proteins play a central role in cellular signaling, and their deregulation has been implicated in a wide variety of diseases1,2. While the majority of FDA-approved drugs engage the membrane proteome, a considerable proportion of membrane-bound targets remain undruggable by current targeted approaches, which are largely comprised of antibodies and small molecule inhibitors (SMI)3,4,5. Despite the potential clinical benefit in modulating membrane proteins, small molecule and antibody-based therapies often suffer from limitations such as stoichiometric target inactivation, challenges in target engagement due to lack of druggable sites, dose-dependent toxicities, and resistance mechanisms6,7. Hence, alternative therapeutic strategies are needed to expand the addressable pool of membrane targets more effectively. Of particular interest, targeted protein degradation (TPD) by proteolysis targeting chimeras (PROTACs) has demonstrated several advantages over inhibitor-based therapies8,9,10,11,12,13,14. For instance, PROTACs can utilize a broader set of target binding ligands, including silent binders or functional binders. Second, degradation of a target removes the entire protein, including its scaffolding functions, essentially mimicking a genetic knockout. Even though PROTACs have been successful at degrading a wide array of cellular proteins, their utility for degrading membrane proteins has not been fully explored15,16. This is, in part, because membrane proteins are primarily degraded through lysosomal pathways such as receptor-mediated endocytosis and autophagy and may not always be optimally accessible for PROTACs17,18. In recent years, lysosome targeting chimeras (LYTACs), antibody-based PROTACs (AbTACs), and proteolysis targeting antibodies (PROTABs) have emerged as promising proximity-inducing modalities to target secreted and membrane proteins for lysosomal degradation; however, to date, such approaches have mainly relied on the use of antibodies19,20,21,22,23,24. Therefore, we anticipate small molecule-based strategies that function intracellularly for the degradation of membrane proteins through lysosomal machineries could expand the scope of TPD modalities.
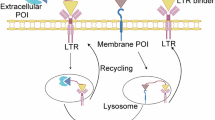
Lysosomal membrane proteins (LMPs) regulate essential cellular processes such as membrane repair, autophagy, phagocytosis, and viral infection25,26. A recent study has demonstrated that a subset of LMPs, such as RNF152 and LAPTM4a, are short-lived and are internalized into the lysosome for degradation in a ubiquitin- and endosomal sorting complexes required for transport (ESCRT)-dependent manner, referred to as lysosomal microautophagy27,28,29. We hypothesized that the rapid turnover and lysosomal proximity of LMPs could be co-opted to regulate the fate of proteins of interest (POIs) using proximity-inducing small molecules. We turned this concept into practice and have developed Lysosome Membrane Targeting Chimeras (LYMTACs), heterobifunctional molecules composed of a POI ligand, a linker, and an LMP ligand (Fig. 1A). Mechanistically, a LYMTAC induces the proximity between a POI and a short-lived LMP, thereby inducing relocalization of the target protein to the lysosomal membrane, promoting its internalization and subsequent degradation. By applying this concept to various membrane kinases and oncogenic KRASG12D, we demonstrate the utility of LYMTACs in targeting membrane proteins for lysosomal clearance.
A Schematic of the LYMTAC concept. A LYMTAC is a heterobifunctional molecule composed of a POI ligand, a linker and an LMP ligand. LYMTACs co-opt short-lived lysosomal membrane proteins as effectors to deliver target proteins for lysosomal degradation. B Cycloheximide (CHX) assay in FLAG-MTH1-RNF152 stably expressing HCT116 cells. Cells were pre-treated with DMSO, 200 nM Bafilomycin A1 (BafA1), or 1 µM ubiquitin E1 inhibitor (TAK-243) for 30 min, followed by co-treatment with 100 µg/mL CHX for 4 h. Cells were harvested at 0 h to include as a control. Cells were lysed and subjected to immunoblotting with MTH1 and vinculin antibodies. Quantified data are representative of two independent experiments. C Structure of LYMTAC-1 (promiscuous kinase inhibitor-PEG2-MTH1 ligand) and PROTAC TL-12-186. D Quantitative proteomics analysis of LYMTAC-1. HEK293 cells stably expressing FLAG-MTH1-RNF152 were treated with either DMSO or 500 nM LYMTAC-1 for 19 h and subjected to global proteomics analysis. Data are representative of three treatment replicates. Differential expression was assessed with the limma moderated t-test (empirical-Bayes framework, two-sided). Reported P values are unadjusted; no correction for multiple comparisons was applied. E Quantitative proteomics analysis of PROTAC (TL-12-186). HEK293 cells stably expressing FLAG-MTH1-RNF152 were treated with either DMSO or 500 nM PROTAC for 19 h and subjected to global proteomics analysis. Data are representative of three treatment replicates. Differential expression was assessed with the limma moderated t-test (empirical-Bayes framework, two-sided). Reported P values are unadjusted; no correction for multiple comparisons was applied. F LYMTAC-1 induced dose-dependent degradation of PTK2. HEK293 cells stably expressing FLAG-MTH1-RNF152 were treated with increasing concentrations of LYMTAC-1 for 19 h and subjected to immunoblotting. Quantified data are representative of three independent experiments. G LYMTAC-1 induced dose-dependent degradation of EPHA2. Quantified data are representative of three independent experiments. H, I HEK293 cells stably expressing MTH1-RNF152 were pre-treated with BafA1 and MG-132 for 30 min, co-treated with DMSO or 500 nM LYMTAC-1 for 19 h, and subjected to immunoblotting with the indicated antibodies. Quantified data are representative of two independent experiments.
In this work, we demonstrate that LYMTACs are capable of exerting pharmacological activity through substrate relocalization and sequestration at the lysosome even in the absence of extensive degradation. This dual mode of action translated into a deeper suppression of KRASG12D signaling and inhibition of cancer cell viability compared to reversible KRAS inhibitors. The scope of this platform is further highlighted by establishing that multiple LMPs, such as RNF152, LAMPT4a, and LAPTM5, can serve as effectors for the LYMTAC technology. We anticipate that the chemical modularity and tunability of small molecule LYMTACs will offer new opportunities to optimize such agents for the efficient degradation of diverse membrane-associated proteins.
Results and discussion
Development of LYMTAC as a modular tool for lysosomal degradation of membrane proteins
To establish the LYMTAC technology, we used RNF152 as our primary LMP effector for proof-of-concept studies. Due to the lack of selective and well-characterized ligands for RNF152, we generated a model system using MTH1-tagged RNF152 (MTH1-RNF152) to allow for induced proximity to the POI using a selective MTH1 binding ligand30. Since MTH1 ligands have been successfully incorporated into heterobifunctional molecules and tested in chemical-induced dimerization studies, we used the MTH1 tag and its ligands to establish the LYMTAC concept31. Importantly, MTH1 tagging did not alter the turnover rate of RNF152 as shown by almost complete degradation of MTH1-RNF152 after only four hours of protein synthesis inhibition by cycloheximide in HEK293 and HCT116 cells (Fig. 1B and Supplementary Fig. 1). Furthermore, this rapid turnover of MTH1-RNF152 was both ubiquitin- and lysosome-dependent as evidenced by inhibition by the ubiquitin E1 inhibitor (TAK-243) and lysosomal V-ATPase inhibitor (Bafilomycin A1) (Fig. 1B).
After establishing the MTH1-RNF152 LMP model cellular system, we probed the impact of LYMTAC on target protein degradation. As such, we devised an untargeted approach using a promiscuous kinase inhibitor as the POI ligand32. We generated a multi-kinase targeting LYMTAC (LYMTAC-1) by linking the promiscuous kinase inhibitor, TL-13-87, to an MTH1-targeting ligand (Fig. 1C). This non-selective kinase inhibitor binds to a vast array of membrane, cytoplasmic, and nuclear protein kinases, thus allowing us to identify the target space that is most readily addressed by LYMTACs. HEK293 cells stably expressing MTH1-RNF152 were treated with LYMTAC-1 and subjected to global proteomics to identify degradable protein targets in an unbiased manner. A total of six protein kinases were significantly downregulated by LYMTAC-1, validating the ability of LYMTACs to drive target protein degradation (Fig. 1D). Strikingly, five out of six kinases that were downregulated by LYMTAC-1 were exclusively integral membrane proteins (EPHA2, INSR, EPHB4, ACVR1, TGFBR2)33,34.
To directly compare the degradable protein pool of pan-kinase LYMTAC to a PROTAC functionalized with the same pan-kinase inhibitor, HEK293 cells were treated with previously reported pan-kinase PROTAC (TL-12-186) and subjected to global proteomics. Consistent with previous reports, the pan-kinase PROTAC mostly induced degradation of cytosolic and nuclear kinases (Fig. 1E)32,35,36. Even though approximately 300 kinases were detected in both experiments, the PROTAC targeted a small fraction of membrane proteins (6 out of 67 downregulated kinases) for degradation. This is in stark contrast to LYMTAC-1, which mainly induced downregulation of membrane protein kinases (5 out of 6 downregulated kinases were membrane kinases) (Supplementary Fig. 2). This includes ACVR1, INSR, and TGFBR2, which are uniquely degraded by LYMTAC-1 and not by TL-12-186. Thus, LYMTACs provide access to a distinct pool of cellular targets. We speculate that the observed substrate selectivity may be due to the distinct pathways co-opted by the two effectors, RNF152 and CRBN, which engage the lysosomal and proteasomal degradation pathways, respectively. By engaging the endo-lysosomal degradation pathway, LYMTACs may be more suitable for degrading membrane proteins, given that membrane proteins are predominantly degraded via the endo-lysosomal pathway.
To corroborate the proteomics results, HEK293 cells stably expressing MTH1-RNF152 were treated with increasing concentrations of LYMTAC-1 and subjected to immunoblotting. Consistent with the proteomics data, LYMTAC-1 induced PTK2 and EPHA2 degradation in a dose-dependent manner, with robust degradation observed at 10 nM (Fig. 1F, G). LYMTAC-1 activity was dependent on RNF152, since the addition of excess MTH1 ligand abrogated PTK2 degradation (Supplementary Fig. 3). Next, we investigated the route of degradation. Cells were pre-treated with either lysosome (Bafilomycin A1) or proteasome (MG-132) inhibitors, and LYMTAC-1-induced degradation of PTK2 and EPHA2 was assessed. LYMTAC-1-induced PTK2 and EPHA2 degradation was rescued by Bafilomycin A1 but not MG-132, consistent with lysosomal-mediated clearance of these membrane proteins, further confirming the anticipated degradation mechanism of LYMTACs (Fig. 1H, I).
LYMTACs induce ubiquitylation and lysosomal degradation of KRASG12D
Having established the ability of LYMTACs to target membrane proteins for degradation, we sought to further investigate the therapeutic potential of these agents in degrading a historically challenging-to-drug protein, KRASG12D. KRASG12D is a membrane-anchored protein that has been associated with the tumorigenic capacity of many cancer types37. We first utilized a chemical genetic system by co-expressing MTH1-RNF152 effector and HiBiT-FKBP12F36V-KRASG12D POI (Fig. 2A)38,39. We chose to first establish this system using dimerization tags in cells not dependent on KRAS signaling for survival. Endogenously tagged HCT116 cells were generated by knocking in HiBiT-FKBP12F36V at the KRAS locus, where gene editing was used to introduce G12D by correcting the G13D mutation that was present naturally in HCT116 cells. For proof of concept, we chose to leverage FKBP12F36V-tagged proteins of interest (POIs) as a suitable target protein given the availability of potent and selective ligands for FKBP12F36V. Additionally, the use of FKBP12F36V-tagging allowed us to quantitatively assess KRASG12D levels and ligand engagement without the complications associated with the use of pharmacologically active inhibitors in cells sensitive to KRASG12D depletion. Next, we designed LYMTAC-2, comprising an FKBP12F36V ligand, a connecting linker, and an MTH1 ligand to dimerize the target and effector proteins (Fig. 2B). In line with our pan-kinase data, RNF152-recruiting LYMTAC-2 induced potent but partial KRASG12D degradation in a dose-dependent manner after 24 h (Fig. 2C). However, LYMTAC-2-induced KRAS degradation was not observed in the absence of MTH1-RNF152, demonstrating that LYMTAC-induced KRAS degradation is dependent on MTH1-RNF152. To further understand the effect of untagged RNF152 (which mimics the endogenous RNF152) on LYMTAC’s function, we overexpressed varying amounts of untagged RNF152 in HCT116 cells and assessed its impact on LYMTAC-induced KRAS degradation in cells stably expressing MTH1-RNF152. Overexpression of RNF152 did not alter the efficiency of LYMTAC-induced KRAS degradation, even at the highest overexpression level (Supplementary Fig. 4).
A Schematic of the chemical genetic system where the target protein is HiBiT-FKBP12F36V-KRASG12D, and the effector is MTH1-RNF152. B Structure of LYMTAC-2 (FKBP12F36V ligand-PEG2-MTH1 ligand). C HiBiT cellular degradation assay in knock-in HCT116 (HiBiT- FKBP12F36V-KRASG12D: (HF-KRAS)) cells in the absence or presence of stably expressing MTH1-RNF152. Cells were treated with increasing concentrations of LYMTAC-2 for 24 h, and HiBiT levels were measured using Nano-Glo® HiBiT lytic reagent. Data are representative of three independent experiments, reported as the mean ± S.E. D HCT116 (HF-KRAS) cells stably expressing FLAG-MTH1-RNF152 were pre-treated with BafA1, MG-132, or TAK-243 for 30 min followed by co-treatment with DMSO or 500 nM LYMTAC-2 for 6 h and subjected to immunoblotting with KRAS and tubulin antibodies. Quantified data are representative of two independent experiments. E KRASG12D ubiquitylation assay. HA-ubiquitin was transfected into HCT116, pre-treated with 200 nM BafA1 for 30 min, then co-treated with DMSO or 1 µM LYMTAC-2 for 4 h. Next, cells were lysed and subjected to immunoprecipitation with anti-HiBiT antibody. Whole cell lysate was used as the input. The respective blots are probed with HA, MTH1, HiBiT, and vinculin antibodies. Data are representative of three independent experiments.
To obtain additional insights on LYMTAC-2-induced KRASG12D degradation, HCT116 cells were pre-treated with inhibitors such as Bafilomycin A1, MG-132, or TAK-243, and then co-treated with LYMTAC-2 and subjected to immunoblotting. LYMTAC-2-mediated KRASG12D degradation was rescued by both ubiquitin E1 (TAK-243) and lysosomal (Bafilomycin A1) inhibitors (Fig. 2D). However, LYMTAC-induced KRASG12D degradation was not rescued by proteasome inhibition. These data indicate that LYMTAC-2-mediated KRASG12D degradation follows the native ubiquitin- and lysosome-dependent turnover of RNF152. RNF152 is an E3 ubiquitin ligase LMP whose rapid internalization and lysosomal degradation are dependent on its own auto-ubiquitylation27. Thus, we speculated that LYMTAC-2 may drive degradation of KRASG12D through RNF152-dependent ubiquitylation of KRASG12D. To this end, we performed a cellular ubiquitylation assay by transiently expressing HA-ubiquitin in HCT116 cells, followed by immunoprecipitation and immunoblotting. Notably, cells treated with LYMTAC-2 revealed rapid KRASG12D ubiquitylation (Fig. 2E). Collectively, our data suggest that LYMTACs can lead to the formation of a productive ternary complex with KRASG12D and RNF152, promote KRASG12D ubiquitylation, and induce lysosome-dependent clearance of KRASG12D.
KRASG12D LYMTACs induce pathway suppression irrespective of KRASG12D degradation
Emerging resistance to KRAS inhibitors (KRASi) is a critical challenge in maximizing the clinical potential of these drugs40,41,42,43. Degradation strategies that remove rather than inhibit oncogenic proteins offer the potential of addressing a number of such resistance mechanisms, which has encouraged our development of proximity-inducing approaches to degrade membrane-associated oncogenic KRAS. Having established and characterized the biochemical activity of KRASG12D LYMTACs, we next investigated the downstream consequences of lysosomal-mediated degradation of KRASG12D. To target endogenous KRASG12D directly, we generated KRASG12D LYMTACs by replacing the FKBP12F36V ligand with a pan-KRAS inhibitor (pan-KRASi) (Fig. 3A, B). Extensive research in the TPD field has defined several common structural features that influence the efficacy of PROTAC design such as exit vector orientation, linker length, linker composition, and E3 ligase ligand44,45,46. X-ray crystallographic characterization of the binding mode of the pan-KRAS inhibitor has suggested attachment of the effector ligand via the fused bicyclo-pyrollidine ring could yield active PROTACs and LYMTACs.
A Schematic of complex formation between KRASG12D and MTH1-RNF152 in the presence of KRAS-targeting LYMTAC. B Structures of the pan-KRAS inhibitor and PROTAC C Structures of LYMTAC-3 and LYMTAC-4. D HiBiT cellular degradation assay in FLAG-MTH1-RNF152-stably expressing, knock-in HCT116 (HiBiT-FKBP12F36V-KRASG12D) cells. Cells were treated with indicated doses of compounds for 24 h and subjected to Nano-Glo® HiBiT lytic assay. Data are representative of three independent experiments, reported as the mean ± S.E. E Ternary complex formation assay, AsPC-1 cells stably expressing FLAG-MTH1-RNF152 were pre-treated with BafA1 for 30 min, followed by co-treatment with DMSO or 1 µM LYMTAC-3 or LYMTAC-4 for 4 h and subjected to immunoprecipitation with anti-FLAG antibody. Whole cell lysate was used as the input. The respective blots are probed with KRAS, MTH1, and vinculin antibodies. Data are representative of two independent experiments. F AsPC-1 cells stably expressing FLAG-MTH1-RNF152 were treated with DMSO or 100 nM of the indicated compounds for 24 h and subjected to immunoblotting with KRAS, pERK, ERK, and vinculin antibodies. Quantified data are representative of two independent experiments. G AsPC-1 cells stably expressing FLAG-MTH1-RNF152 were treated with 100 nM KRAS inhibitor, 100 nM PROTAC, or 100 nM LYMTAC-4 for 6 h. For control experiments with LYMTAC-4, cells were pretreated with MTH1-ligand or BafA1 for 30 min, followed by co-treatment in the absence or presence of 100 nM LYMTAC-4 for 6 h, and subjected to immunoblotting with KRAS, p-ERK, ERK, and vinculin antibodies. Quantified data are representative of two independent experiments. H HEK293 cells stably expressing HiBiT-FKBP12-KRASG12D and FLAG-MTH1-RNF152 were pre-treated with 1 µM TAK-243 for 30 min followed by DMSO or 1 µM LYMTAC-2 treatment for 6 h. The respective blots are probed with KRAS, p-ERK, ERK, and tubulin antibodies. Quantified data are representative of two independent experiments.
First, we screened KRAS LYMTACs with two different linker compositions for cellular HiBiT- FKBP12F36V-KRASG12D degradation using HCT116 cells and compared their efficacy to that achieved through PROTAC-mediated degradation (Fig. 3B, C). While our KRAS PROTAC and two LYMTACs displayed varying levels of KRASG12D degradation, rigid linker-based LYMTAC-4 (which also tethered the MTH1 ligand to the pan-KRAS inhibitor via a shorter linker) displayed improved Dmax (61%) and DC50 (2 nM) values compared to PEG linker-based LYMTAC-3 [Dmax = 48%, DC50 = 25 nM), Fig. 3D). Leveraging the more potent LYMTAC-4 in subsequent experiments, we functionally characterized the effect of LYMTAC-induced KRASG12D degradation in AsPC-1 cells, a relevant KRASG12D mutant pancreatic cancer cell line that relies on this oncogene for cell proliferation and survival47. MTH1-RNF152 was stably introduced into AsPC-1 cells to evaluate ternary complex formation (TCF) by various LYMTACs. The data suggest that LYMTAC-4 induced a more productive ternary complex between KRASG12D and RNF152, compared to LYMTAC-3 (Fig. 3E). Experience with this system further supported the selection of LYMTAC-4 as an appropriate tool molecule for subsequent investigation. To evaluate the cellular effect of LYMTAC-4, AsPC-1 cells were treated with pan-KRASi, PROTAC, or LYMTAC-4 for 24 h and probed for pathway activity. LYMTAC-4 induced KRAS degradation and robust pERK downregulation at 24 h, comparable to pan-KRASi and PROTAC (Fig. 3F). Additionally, to test the ability of KRAS LYMTACs to degrade other KRAS variants, we tested LYMTAC-4 in HCT116 harboring the KRASG13D mutation. Consistent with the ability of the KRAS inhibitor to bind G13D, we observed LYMTAC-4 induced KRASG13D degradation in HCT116 cells (Supplementary Fig. 5).
We next sought to further elucidate LYMTAC mode of action and its dependence on ternary complex formation (TCF). This was investigated by treating AsPC-1 cells with LYMTAC-4 for 6 h in the absence or presence of excessive competing MTH1-ligand or Bafilomycin A1. Excess MTH1 ligand abrogated LYMTAC-induced KRASG12D degradation and p-ERK signaling, demonstrating that TCF is required to induce KRASG12D degradation and inhibit downstream signaling (Fig. 3G). As expected, Bafilomycin A1 treatment rescued LYMTAC-4-induced KRASG12D degradation. Unexpectedly, in Bafilomycin A1-treated cells, LYMTAC-4 still suppressed KRASG12D downstream signaling even in the absence of KRASG12D degradation. To rule out that p-ERK downregulation was due to the pan-KRASi moiety of LYMTAC-4, we stably co-expressed FKBP12F36V-KRASG12D and MTH1-RNF152 in HEK293 cells and engaged FKBP12-KRAS with a non-inhibitory FKBP12 ligand containing LYMTAC-2 (FKBP12 ligand-PEG2-MTH1 ligand). Strikingly, FKBP12-targeting LYMTAC-2 also suppressed p-ERK signaling irrespective of KRAS ubiquitylation and degradation (Fig. 3H). Collectively, these data provide compelling evidence that pathway inhibition is not due to the inhibitory effect of pan-KRASi-based LYMTACs but could be attributed to the ability of LYMTACs to affect target biology beyond target degradation. Overall, the data indicate that LYMTAC-induced KRAS ubiquitylation and degradation are not uniformly required for downstream signaling inhibition, while TCF is critical to suppress p-ERK signaling.
KRAS relocalization and lysosomal degradation using LYMTAC lead to deeper pathway suppression and potent cell killing compared to SMI
The observation that signaling downstream of KRASG12D can be modulated by LYMTACs even in the absence of KRASG12D degradation prompted us to further explore whether quantitative relocalization of KRAS from the plasma membrane to the lysosome could be driving this effect (Fig. 4A). To address this hypothesis, we generated mNeonGreen-KRASWT stably expressing HEK293 cells to allow for subcellular KRAS tracking by confocal microscopy. After generating the cell line, we also verified in a CHX assay that addition of the mNeonGreen tag does not significantly alter the half-life of KRAS (Supplementary Fig. 6). Pre-treatment of these cells with Bafilomycin A1 followed by LYMTAC-4 revealed rapid and marked KRAS relocalization from the plasma membrane followed by co-localization with RNF152 and the lysosomal marker LAMP1 (Fig. 4B). This was specific for LYMTACs, as this phenotype was absent in cells treated with pan-KRASi or PROTAC (Supplementary Fig. 7). We performed time lapse imaging in HEK293 cells to illustrate the kinetics of LYMTAC-4 induced KRAS relocalization. Internalization was evident as early as 30 min and gradually increased in a time-dependent manner (Fig. 4C). We believe that the kinetics of relocalization may depend on several factors, such as the expression levels of MTH1-RNF152 and KRAS in each cell, and the efficiency of ternary complex formation.
A Schematic of LYMTAC-induced KRAS relocalization from plasma membrane to lysosome. B KRAS localization analyzed by confocal microscopy. FLAG-MTH1-RNF152-stably expressing HEK293 mNeonGreen-KRAS WT cells were pre-treated with 200 nM BafA1 for 30 min and then co-treated with DMSO or 1 µM LYMTAC-4 for 4 h. Cells were fixed, permeabilized, and immunostained with FLAG and LAMP1 antibodies, followed by appropriate secondary fluorophore-conjugated antibodies. Nuclei were stained with Hoechst dye. Data are representative of two independent experiments. C Kinetics of LYMTAC-4 induced KRAS relocalization. FLAG-MTH1-RNF152-stably expressing HEK293 mNeonGreen-KRAS WT cells were pre-treated with 200 nM BafA1 for 30 min and then co-treated with 1 µM LYMTAC-4. Live cell images were acquired every 30 min up to 4 h. Puncta formation in each cell is highlighted with orange, green and pink arrows at each time point. Data are representative of two independent experiments. D Washout experiment in AsPC-1 cells stably expressing FLAG-MTH1-RNF152. Cells were treated with 100 nM pan-KRASi and 100 nM LYMTAC-4 for 4 h, washed three times with PBS and replaced with fresh media in the absence or presence of 5 µM MTH1-ligand for 48 h. Quantified data are representative of two independent experiments. E pan-KRASi, PROTAC, and LYMTAC-4 activity in a 5-day cell proliferation assay in AsPC-1 cells stably expressing MTH1-RNF152. Data are representative of three independent experiments, reported as the mean ± S.E.
KRAS is a plasma membrane-anchored protein that activates oncogenic signaling at the membrane via multiple effector proteins48. Various attempts to block KRAS trafficking to the plasma membrane have been made, yet these approaches have been largely unsuccessful49,50. In addition, membrane-bound KRAS is susceptible to pathway rebound driven by upstream receptor tyrosine kinases such as EGFR, ultimately contributing to therapeutic resistance42,51,52. Given that LYMTACs display dual functionality in the inactivation of membrane proteins (i.e., via intracellular relocalization and via subsequent lysosomal degradation), their distinct mechanism of action may offer multiple advantages in overcoming some of the limitations commonly seen with current therapies. We explored this possibility by measuring pathway rebound upon drug withdrawal. AsPC-1 cells were treated with pan-KRASi or LYMTAC-4 for four hours, excess compound was washed away, and fresh media was added for 48 h before analysis. Consistent with the reported KRASi-induced signaling rebound, we observed a p-ERK signal rebound after 48 h in the pan-KRASi-treated samples (Fig. 4D). Strikingly, LYMTAC-4 induced sustained KRASG12D degradation and delayed rebound of p-ERK signaling compared to pan-KRASi (Fig. 4D). Excess MTH1 ligand rescued KRASG12D degradation and p-ERK inhibition, indicating that this effect was on target and dependent on RNF152 (Fig. 4D). Notably LYMTAC-4 suppressed downstream p-ERK signaling similarly to a highly optimized clinically used KRASi, MRTX1133, that does not display p-ERK signaling rebound after washout (Supplementary Fig. 8). Given that LYMTAC-4 maintained prolonged signaling inhibition, we sought to further monitor the effect of LYMTACs on AsPC-1 cell proliferation. Importantly, the depth of pathway suppression also translated phenotypically as LYMTAC-4 demonstrated improved cell killing compared to both KRASi and PROTAC (Fig. 4E). This potent cell killing could be attributed to the multi-pharmacology displayed by LYMTACs via KRAS relocalization, sustained p-ERK inhibition, and lysosomal degradation.
Expansion of the LYMTAC platform to other LMPs
To demonstrate the generality of our platform, we targeted additional LMP effectors beyond RNF152. We focused on LAMPT4a and LAPTM5, two LMPs that associate with the NEDD4-1 E3 ubiquitin ligase at the lysosome membrane through PY motifs (Fig. 5A)27,29. After generating cells stably expressing MTH1-tagged LAPTM4a and LAPTM5, we first performed a cycloheximide assay in HEK293 cells to confirm their short-lived nature. The data indicate that both MTH1-LAPTM4a and MTH1-LAPTM5 are rapidly degraded within four hours in HEK293 cells (Supplementary Fig. 9). Similarly to HEK293 cells, HCT116 cells stably expressing MTH1-tagged LAPTM4a or LAPTM5 showed rapid LMP turnover in a ubiquitin and lysosome-dependent manner (Fig. 5B, C).
A Schematic of Lysosome Membrane Proteins (LMPs). B CHX assay in FLAG-MTH1-LAPTM4a-stably expressing HCT116 cells. Cells were pre-treated with DMSO, 200 nM Bafilomycin A1 (BafA1), or 1 µM E1 inhibitor (TAK-243) for 30 min, followed by co-treatment with CHX for 4 h. Cells were harvested at 0 h to include as a control. Cells were lysed and subjected to immunoblotting with MTH1 and vinculin antibodies. Quantified data are representative of two independent experiments. C CHX assay in FLAG-MTH1-LAPTM5-stably expressing HCT116 cells. Quantified data are representative of two independent experiments. D HiBiT cellular degradation assay in FLAG-MTH1-LAPTM4a- and FLAG-MTH1-LAPTM5-stably expressing HCT116 (knock-in-HiBiT- FKBP12F36V-KRASG12D: (HF-KRAS)) cells. Cells were treated with increasing concentrations of LYMTAC-2 for 24 h, and HiBiT levels were measured using Nano-Glo® HiBiT lytic reagent. Data are representative of three independent experiments and reported as the mean ± S.E. E HCT116 (HF-KRAS) cells stably expressing FLAG-MTH1-LAPTM5 were pre-treated with BafA1, 2 BTZ, or TAK-243 for 30 min, followed by co-treatment with DMSO or 500 nM LYMTAC-2 for 6 h and subjected to immunoblotting with KRAS and tubulin antibodies. Quantified data are representative of two independent experiments. F KRAS ubiquitylation assay. HA-ubiquitin was transfected into HCT116 expressing FLAG-MTH1-LAPTM5, which were pre-treated with 200 nM BafA1 for 30 min, followed by co-treatment with DMSO or 1 µM LYMTAC-2 treatment for 4 h. Next, cells were lysed and subjected to immunoprecipitation with anti-HiBiT antibody. Whole cell lysate was used as the input. The respective blots were probed with HA, MTH1, HiBiT, and vinculin antibodies. Data are representative of two independent experiments. G AsPC-1 cells stably expressing FLAG-MTH1-LAPTM4a were pre-treated with either pan-KRAS inhibitor or MTH1 ligand, followed by DMSO or 100 nM LYMTAC-4 treatment for 6 h. Cells were lysed and subjected to immunoblotting with KRAS, pERK, and tubulin antibodies. Quantified data are representative of two independent experiments.
Having confirmed the fast turnover of MTH1-LAPTM4a and MTH1-LAPTM5, we next asked whether these two LMPs can induce target protein degradation in a LYMTAC-dependent manner. To this end, we utilized HiBit-FKBP12F36V-KRASG12D knock-in HCT116 cells and stably expressed MTH1-LAPTM4a or MTH1-LAPTM5. Consistent with results seen for MTH1-RNF152, both MTH1-LAPTM4a and MTH1-LAPTM5 induced potent KRASG12D degradation in a LYMTAC-2-dependent manner (Fig. 5D), validating these two LMPs as effectors for the LYMTAC technology. To verify the mechanism of action for LAPTM5-based LYMTACs, HCT116 cells were treated with lysosomal (Bafilomycin A1), proteasomal (bortezomib), and ubiquitin E1 (TAK-243) inhibitors prior to LYMTAC-2 treatment and subjected to immunoblotting. Consistent with RNF152-recruiting LYMTACs, LAPTM5-induced KRASG12D degradation was rescued by both lysosome and ubiquitin E1 inhibitors, but not proteosome inhibitor (Fig. 5E). To further understand the ubiquitin-dependent mechanism of action, cellular ubiquitylation assays were performed. As expected, LAPTM5 formed a ternary complex with KRASG12D and induced KRASG12D ubiquitylation in the presence of LYMTAC-2 (Fig. 5F).
Finally, to expand LMPs that can be harnessed for LYMTAC development, we generated MTH1-LAPTM4a expressing AsPC-1 cells and confirmed the rapid degradation of this construct as had been observed with prior LMP (Supplementary Fig. 10). We further confirmed that KRASG12D levels were unaffected by overexpressing either MTH1-RNF152 or MTH1-LAPTM4a in the absence of LYMTAC molecules (Supplementary Fig. 10). Similarly to RNF152, LAPTM4a formed a productive ternary complex with KRASG12D in the presence of LYMTAC-4 (Supplementary Fig. 11). Moreover, LAPTM4a-recruiting LYMTAC-4 induced KRASG12D degradation and p-ERK inhibition in AsPC-1 cells without affecting the levels of MTH1-LAPTM4a (Fig. 5G and Supplementary Fig. 12). To further verify the mechanism of action, cells were treated with either pan-KRASi or MTH1-ligand prior to LYMTAC-4 treatment. The ligand competition data indicate that LYMTAC-induced KRASG12D degradation and p-ERK inhibition are LAPTM4a-dependent and require a ternary complex between KRASG12D and LAPTM4a.
In summary, we describe here a SM-based LYMTAC platform that functions by repurposing short-lived lysosomal membrane protein effectors to induce membrane protein relocalization and subsequent degradation. Using a promiscuous kinase inhibitor-based LYMTAC, we demonstrate that LYMTACs preferentially deliver membrane kinases for lysosomal degradation. The observed substrate selectivity was a differentiating feature of LYMTAC-driven degradation when compared to cytosolic and nuclear protein degradation by a promiscuous kinase inhibitor-based PROTAC. The selectivity of LYMTACs towards membrane-associated proteins is likely explained by their natural turnover in the lysosome and continuous flux in the membrane trafficking pathways that offer opportunities for these membrane proteins to be captured by LMPs. Furthermore, using oncogenic KRASG12D as a therapeutically relevant, membrane-associated target, we demonstrated that LYMTACs induce sustained pathway inhibition and potent cell killing as compared to a KRAS inhibitor. Notably, LYMTAC-induced KRAS relocalization from the plasma membrane to the lysosome is sufficient to suppress downstream signaling. While we observe a lack of complete KRAS degradation (Dmax ~50–60%) with LYMTACs, we observe quantitative relocalization of KRAS from plasma membrane to lysosomes, potent inhibition of p-ERK signaling, and cytotoxicity.
Our proof-of-concept results demonstrate that the LYMTAC technology could be an effective strategy to sequester KRAS away from the plasma membrane. This is reminiscent of proximity-inducing modalities that have recently been shown to modulate target protein via non-degradative mechanisms such as relocalization and sequestration53,54,55,56,57. In addition to the in-depth characterization of RNF152, we validated the LYMTAC technology with other LMPs such as LAPTM4a and LAPTM5, clearly demonstrating the potential compatibility of many more LMPs with LYMTAC technology. Overall, LYMTACs are a small molecule proximity-based platform that function intracellularly to inhibit oncogenic signaling through multi-pharmacology, including relocalization (occupancy-driven pharmacology) and lysosomal degradation (event-driven pharmacology) of membrane proteins (Fig. 6). Other modalities such as LYTACs, PROTABs, and AbTACs have also emerged as novel platforms to regulate the membrane proteome. A potential advantage of LYMTAC compared to existing antibody-based technologies would be that 1) LYMTAC is a small molecule-based platform with dual mechanism of action and engages membrane targets from the cytosolic side, and 2) has the potential to target both integral membrane and membrane-anchored proteins for relocalization and lysosomal degradation. However, compared to LYTACs, LYMTACs are unable to target extracellular proteins for degradation.
While we introduce LYMTAC as a chemical biology-driven proof-of-concept study, these MTH1-derived LYMTAC tool molecules can be utilized in fundamental research to functionally characterize the effect of target protein relocalization and lysosomal degradation. Identification of potent and selective ligands for short-lived LMPs remains a key challenge to advance the LYMTAC technology beyond the chemical biology concept presented here. We speculate that techniques such as DNA-encoded library screening, chemoproteomics, and fragment-based library screening will facilitate the discovery of ligands for multiple lysosomal membrane proteins in the future. Similarly to the challenges encountered with PROTAC delivery in animal models, extensive optimization will be required to deliver LYMTACs into animal models in the future. Future work should focus on understanding the target scope, LMP space (including cell-type specificity), and ligand discovery for LMPs to enable the LYMTAC technology for therapeutic development.
Methods
Cell lines
Human embryonic kidney 293 (HEK293), AsPC-1, and HCT116 cells were obtained from ATCC. HEK293 cells were cultured in DMEM (Gibco) supplemented with 10% (v/v) heat-inactivated fetal bovine serum (FBS) and 1% penicillin/streptomycin (Pen/strep) (v/v). AsPC-1 and HCT116 cells were grown in RPMI media supplemented with 10% FBS and 1% Pen/strep. Jump-In™GripTite™ HEK293 cells (A14150) were obtained from Invitrogen and grown in DMEM Glutamax supplemented with 10% FBS, 0.1 mM NEAA, 25 mM HEPES, 1x Pen/strep, 600 µg/mL Geneticin. Cells were cultured at 37 °C and 5% CO2 in a humidified incubator and regularly screened for mycoplasma contamination.
Antibodies and reagents
Antibodies against MTH1 (43918), FLAG (14793), Vinculin (13901), Tubulin (2144), pERK (9101), ERK (4695), PTK2 (3285), EPHA2 (6997), HA (3724), LAMP1(15665), and GAPDH (2118) were purchased from Cell Signaling Technology. HiBiT antibody (N7200) was obtained from Promega, and KRAS antibody (LS-C175665) was purchased from LSBio. RNF152 antibody (PA5-95817) was purchased from Invitrogen. Alexa Fluor 568 (A11011) and Alexa Fluor 647 (A32728) conjugated secondary antibodies and Hoechst 33342 stain (H3570) were purchased from Invitrogen. Amersham ECL Rabbit IgG, HRP-linked whole secondary Ab (from donkey) (NA934) and Amersham ECL Mouse IgG, HRP-linked whole secondary Ab (from sheep) (NA931) were purchased from Cytiva. RIPA Lysis and Extraction Buffer (89900) and IP-lysis buffer (87787) were purchased from ThermoScientific. Protease inhibitor cocktail (11873580001) was obtained from Roche, and phosphatase inhibitor cocktail (A32957) were obtained from ThermoScientific. 1,10-phenanthroline monohydrate (S5543), N-ethylmaleimide (S3692), and M3814 (S8586) were purchased from Selleckchem. PR-619 (4482) was purchased from Tocris. Nano-Glo® HiBiT lytic detection reagent (N3040) was purchased from Promega, and CyQUANT™ direct cell proliferation assay (C35011) was obtained from Invitrogen. Bafilomycin A1 (54645), MG-132 (2194) and Cycloheximide (2112) were purchased from Cell Signaling Technology. TAK-243 (HY-100487) and Bortezomib (HY-10227) were obtained from MedChemExpress. Puromycin (ant-pr-1), blasticidin (ant-bl-05), and Geneticin (ant-gn-1) were purchased from InvivoGen. Pierce™ Anti-DYKDDDDK (FLAG) Magnetic Agarose beads (A36797) and 16% Formaldehyde (28906) were purchased from ThermoScientific. Protein G Magnetic Sepharose beads (28951379) were obtained from Cytiva. 8-well Ibidi dishes (80826) were purchased from Ibidi, and 4X NuPAGE™ LDS sample buffer (NP0007) was purchased from Invitrogen.
Plasmids
HA-ubiquitin cDNA in pcDNA3.1+ vector was obtained from GenScript. FLAG-MTH1-RNF152, FLAG-MTH1-LAPTM4a, and FLAG-MTH1-LAPTM5 cDNA were cloned into the pGenLenti-puro vector (GenScript). Lentivirus particles were used to transduce HEK293, HCT116, and AsPC-1 cells and generated stable cells expressing different MTH1 fusions.
Generation of endogenously tagged KRASG12D(HiBiT-FKBP12F36V-KRASG12D) in HCT116 cells
HCT116 cells have a heterozygous G13D mutation on the endogenous KRAS locus. To generate HCT116 cells with N-terminal HiBiT-FKBP12F36V-KRASG12D homozygous clones, a cutting site was designed near the endogenous KRAS G12 locus (guide RNA: 5’-aaacttgtggtagttggagc-3’). The HDR template was designed to contain the BSD-P2A-HiBiT-FKBP12F36V insertion and the G12D mutation simultaneously and de novo synthesized at GenScript. To generate the homozygous clones, HCT116 were seeded overnight in 12-well plates at 6×105 cells per well overnight. The next day, cells were transfected with a mixture of 4x plasmids containing guide RNA, CRISPR enzyme, HDR template, and i53 HDR enhancer58. Six hours after transfection, cells were passaged 1:8 into 6 well plates with 2.5 mL culture medium with 1 μM M3814 as another HDR enhancer. One day later, the media was replaced with media containing 2 μg/mL puromycin (the guide RNA plasmid contains a puromycin-resistant gene for selection) and 1 μM M3814 for another 2 days. The media was then replaced with media containing 20 μg/mL blasticidin to select cells with endogenous KRAS tagging for 2 days. The cell pool was further recovered in regular medium for 2 days and were used for single cloning. For single cloning, approximately 500 cells were plated on a 10 cm dish in tissue culture media and allowed to grow for approximately one week. Single colonies were isolated and transferred to a 96-well plate using a stereomicroscope and 20 μL pipette tips. Single clones were identified with genomic PCR and subjected to Sanger sequencing to confirm homozygous HiBiT- FKBP12F36V tagging and G12D mutation at the N-terminus of endogenous KRAS.
Generation of HEK293, HCT116 (KI HiBiT-FKBP12-KRAS), and AsPC-1 cells stably expressing MTH1 fusions
To generate RNF152-expressing cells for global proteomics analysis, HEK293 cells were transduced with lentivirus particles containing FLAG-MTH1-RNF152 in the presence of polybrene (10 µg/mL). The cells were selected using 2 µg/ml puromycin for 2 weeks to generate stable cells, and MTH1-RNF152 expression was verified using immunoblotting. FLAG-MTH1-LAPTM4a and FLAG-MTH1-LAPTM5 stably expressing HEK293 cells were generated, similar to RNF152. To generate effector-expressing HCT116 (KI HiBiT-FKBP12F36V-KRASG12D) cells, cells were separately transduced with lentivirus particles containing FLAG-MTH1-RNF152, FLAG-MTH1-LAPTM4a, and FLAG-MTH1-LAPTM5, selected using 2 µg/mL puromycin for 2 weeks and verified stable expression via immunoblotting. To generate effector-expressing AsPC-1 cells, cells were transduced with lentivirus particles containing FLAG-MTH1-RNF152 or FLAG-MTH1-LAPTM4a, selected using 1.5 µg/mL puromycin for 2 weeks and effector expression was verified via immunoblotting.
Generation of HEK293 cells stably co-expressing HiBiT-FKBP12F36V-KRASG12D and MTH1-RNF152 fusion
To generate HiBiT-FKBP12F36V-KRASG12D stably expressing Jump-In™GripTite™ cells, HiBiT-FKBP12F36V-KRASG12D cDNA was cloned into pJTI™ R4 Dest CMV pA vector. This plasmid was co-transfected with pJTI™ R4 Int vector into Jump-In™GripTite™ HEK293 cells. After transfection, cells were selected using 10 µg/mL blasticidin to obtain single cell clones. HiBiT-FKBP12F36V-KRASG12D expression was verified by immunoblotting and HiBit assay. To generate effector expressing cells, HiBiT-FKBP12F36V-KRASG12D stable cells were transduced with lentivirus particles containing FLAG-MTH1-RNF152 in the presence of 10 µg/mL polybrene. The cells were selected using 2 µg/mL puromycin for 2 weeks to generate stable cells and MTH1-RNF152 expression was verified using immunoblotting.
Generation of HEK293 cells stably co-expressing mNeonGreen-KRASWT and MTH1-RNF152
To generate mNeonGreen-KRASWT stable cells, mNeonGreen-KRASWT cDNA was cloned into pJTI™ R4 Dest CMV pA vector. This plasmid was co-transfected with pJTI™ R4 Int vector into Jump-In™GripTite™ HEK293 cells. After transfection, cells were selected for two weeks using 5 µg/mL blasticidin, and clones were obtained by single-cell sorting. mNeonGreen-KRASWT expression was verified by immunoblotting and microcopy analysis. To generate effector-expressing cells, mNeonGreen-KRAS cells were transduced with lentivirus particles containing FLAG-MTH1-RNF152 in the presence of 10 µg/mL polybrene. The cells were selected using 2 µg/mL puromycin for 2 weeks to generate stable cells, and MTH1-RNF152 expression was verified using immunoblotting.
Cell lysis and immunoblotting
After the desired treatments, cells were washed 2X with PBS, trypsinized, and harvested as cell pellets. Cell pellets were lysed in RIPA buffer supplemented with 1X protease and 1X phosphatase inhibitor cocktail, incubated on ice for 20 min, and the cell lysate was clarified by centrifugation. Lysate was then mixed with 4X NuPAGE™ LDS Sample Buffer supplemented with 10% β-mercaptoethanol, boiled at 95 °C, and proteins were separated on 4–20% Criterion TGX precast gradient gels (Bio-Rad) and transferred to PVDF membrane. Next, the blots were incubated with blocking buffer (5% (w/v) nonfat dry milk in TBST (0.1% Tween-20, 20 mM Tris–HCl pH 7.6, 150 mM NaCl) for 1 h at room temperature, followed by overnight incubation with primary antibodies at 4 °C. After primary antibody incubation, membranes were washed 3X with TBST and incubated with HRP-conjugated secondary antibodies for 1 h at room temperature. Membrane was washed 3X with TBST, developed using ECL or Super Signal West Femto substrate (ThermoFisher), and images were acquired using Bio-Rad ChemiDoc system.
Lysosome membrane protein (LMP) turnover analysis using cycloheximide assays
HCT116 cells stably expressing MTH1-LMP were seeded at a density of 6 × 105 cells per well in a 6-well dish. After 24 h, cells were pre-treated with DMSO, 200 nM Bafilomycin A1, and 1 µM E1 inhibitor (TAK-243) for 30 min, followed by co-treatment with 100 µg/mL cycloheximide for 4 h. Cells were harvested at 0 h to include as a control. After 4 h, cells were harvested and lysed with RIPA buffer supplemented with protease inhibitor cocktail and subjected to immunoblotting with the indicated antibodies.
Quantitative proteomics analysis
HEK293 cells stably expressing MTH1-RNF152 were seeded at a density of 5 × 105 cells per well in a 6-well dish. After 24 h, cells were treated with DMSO, 500 nM LYMTAC-1, and 500 nM PROTAC (TL-12-186) for 19 h. Each treatment was performed in triplicate. Next, cells were washed 1X with PBS, trypsinized, and harvested. Cell pellet was washed 2X with PBS and subjected to global proteomics analysis.
LC-MS sample preparation
Cell pellets were lysed in 100 µL of lysis buffer (2% SDS, 100 mM Tris-HCl) and sonicated using an Abcam PIXUL sonicator (50 Hz, 20 min). The protein concentration of the lysates was determined by the BCA method (Thermo Fischer Scientific 23225). For each sample, 30 µg of protein was reduced and alkylated by adding Bond-Breaker TCEP (Thermo 77720) to a final concentration of 100 mM and chloroacetamide to a final concentration of 500 mM and incubating at 56 °C for 45 min. The samples were then subjected to the SP3 protocol for clean-up. Briefly, 30 µg of protein was bound to >300 µg of beads in 80% (v/v) ethanol for 15 min, and then washed three times with 80% ethanol and once with acetonitrile. The protein-bound beads were digested with 1.2 µg of Trypsin/Lys-C mix in 50 mM TEAB buffer for 18 h at 37 °C. The beads were removed, and the peptides were dried to eliminate TEAB. Before LC-MS analysis, the samples were resuspended in 5% formic acid.
LC-MS analysis method
LYMTAC and PROTAC treatment experiments were performed using Thermo Scientific Vanquish Neo LC with Thermo Scientific Orbitrap Eclipse or Evosep One LC with Bruker TIMS-TOF-HT.
Vanquish neo-orbitrap eclipse LC-MS method
Liquid Chromatography (LC) Conditions: The chromatographic separation was performed using a Thermo Scientific Vanquish Neo LC system with a Bruker PepSep C18 analytical column (15 cm × 150 µm, 1.5 µm particle size). The mobile phases were Solvent A (0.1% formic acid in water) and Solvent B (80% acetonitrile and 0.1% formic acid in water). The flow rate was set to 1.2 µL/min, and the column was equilibrated with 0% Solvent B before sample injection. The gradient elution was programmed as follows: 0% Solvent B from 0 to 1 min, followed by an increase to 2% Solvent B at 1 min. Subsequently, Solvent B was linearly ramped up to 24% over the course of 84 min(1–85 min). After reaching 24%, the percentage of Solvent B was further increased to 40% over the next 14 min (85-99 min). Finally, Solvent B was rapidly increased to 95% from 99 to 100 min to wash the column and held at 95% for 4 min (100–104 min) before the flow was returned to the initial conditions. Column re-equilibration was conducted at 95% Solvent B for an additional 4 min. The total run time for the method was 104 min, including the gradient, column wash, and re-equilibration steps.
Mass Spectrometry (MS) Conditions: Mass spec analysis was performed using a Thermo Scientific Orbirap Eclipse mass spectrometer with a variable window DIA method. The MS1 scans were acquired by Orbitrap at a resolution of 60,000 and a mass range set to m/z 400–1400. The RF lens was set to 60%, and the AGC (Automatic Gain Control) target was normalized to a value of 200%. The absolute AGC value was set at 1.0e6, with a custom maximum injection time of 100 ms.
For the data independent MS/MS (tMS²) analysis, the MS2 scans were acquired in Orbitrap at the resolution of 15,000, the fragmentation of precursors was performed by using high-energy collisional dissociation (HCD) mode at the energy of 33%. The quadrupole was used for ion isolation, with isolation windows defined by a pre-determined table (m/z), the variable window definition is in the supplementary material (Table S1). The mass range for the MS/MS scans was set from m/z 200 to m/* 2000. The RF lens was maintained at 50%, and the AGC target was normalized to 1000%. The absolute AGC value was set at 5.00e5.
Evosep One- TIMS-TOF-HT LC-MS method
Peptide separations were performed using the Evosep One 30SPD method (i.e., a throughput of 30 samples per day) on a Bruker PepSep C18 analytical column (15 cm × 150 µm, 1.5 µm particle size). The TIMS-TOF-HT operated in diaPASEF mode, with ion mobility separation spanning an inverse reduced mobility (1/K0) range of 0.7–1.3 V·s/cm², mass range from 100-1700 m/z. Both the ramp time and accumulation time were set to 75 ms, and 36 scan windows were acquired per cycle, yielding a total cycle time of 0.97 s.
LC-MS data analysis method
Both variable width window DIA and diaPASEF data were searched against an in-house built human spectral library using DIA-NN (v1.8.2) with swiss-prot canonical human reference proteome (20221016), two pass Neural Network was used and with match between run enabled, protein quant results were analyzed through a custom R script and applied statistical modeling and hypothesis testing using the limma package.
HiBiT cellular degradation assay
MTH1-fusions (MTH1-RNF152, MTH1-LAPTM4a, MTH1-LAPTM5) stably expressing HCT116 (knock-in-HiBiT- FKBP12F36V-KRASG12D (HF-KRAS)) cells were seeded at a density of 10,000 cells per well in 96-well plates (Corning 3904). Next day, cells were treated with DMSO or increasing concentrations (2.2 nM to 1000 nM) of KRAS PROTAC, LYMTAC-3, or LYMTAC-4. For HiBiT assays with LYMTAC-2, cells were treated with DMSO or increasing concentrations (0.001 nM to 100 nM) of LYMTAC-2. After 24 h, HiBiT assay was performed using Nano Glo-HiBit lytic reagent according to the manufacturer’s protocol (N3040, Promega). The luminescence readings were recorded using EnVision plate reader under the settings Ultra-Sensitive Luminescence (US LUM) and plotted as a four-parameter non-linear regression curve fit using GraphPad Prism 8.4.3.
Mechanism of action studies using control compounds
HCT116 (HF-KRAS) cells stably expressing MTH1-fusion were seeded at a density of 5 × 105 cells per well in 6-well dishes. Next day, cells were pre-treated with 200 nM BafA1, 2 µM BTZ or 10 µM MG-132, and 1 µM TAK-243 for 30 min and co-treated with DMSO or 500 nM LYMTAC-2 for 6 h. For PTK2 and EPHA2 rescue experiments, HEK293 cells were pre-treated with 200 nM BafA1 and 10 µM MG-132 for 30 min and co-treated with DMSO or 500 nM LYMTAC-1 for 19 h. AsPC-1 cells were pre-treated with 200 nM BafA1 for 30 min, followed by 100 nM LYMTAC-4 co-treatment for 6 h. After treatment, cells were lysed in RIPA buffer and subjected to immunoblotting with the indicated antibodies.
Ligand competition experiments
Cells stably expressing MTH1-fusion were seeded at a density of 5 × 105 cells per well in 6-well dishes. HEK293 cells stably expressing MTH1-RNF152 were pre-treated with 25 µM MTH1-ligand for 30 min, followed by co-treatment with 500 nM LYMTAC-1 for 19 h. AsPC-1 cells were pretreated with 10 µM MTH1-ligand or 10 µM pan-KRAS inhibitor for 30 min, followed by 100 nM LYMTAC-4 co-treatment for 6 h. After treatment, cells were lysed and subjected to immunoblotting with the indicated antibodies.
Ternary complex formation assay
For ternary complex assays, AsPC-1 cells stably expressing FLAG-MTH1-RNF152 or FLAG-MTH1-LAPTM4a were seeded in 10-cm dishes at a density of 5 × 106 cells. Next day, cells were pre-treated with BafA1 for 30 min, followed by co-treatment with DMSO, 1 µM LYMTAC-3, or 1 µM LYMTAC-4 for 4 h. Cells were harvested and lysed in IP-lysis buffer supplemented with protease inhibitor. Next, the cell lysate was subjected to immunoprecipitation with anti-FLAG magnetic beads overnight at 4 °C. Then, beads were washed three times with wash buffer (100 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% Tween-20) and the samples were subjected to SDS-PAGE and Western blotting. The respective blots are probed with the indicated antibodies.
Ubiquitylation assays
To probe LYMTAC-induced KRAS ubiquitylation, HCT116 (HF-KRAS + MTH1 fusion) were seeded in a 10-cm dish at a cell density of 5 × 106 cells. Next day, 4 µg HA-ubiquitin was transfected into HCT116 (HF-KRAS + MTH1 fusion) using Lipofectamine 2000. After 24 h, cells were pre-treated with 200 nM BafA1 for 30 min followed by co-treatment with DMSO or 1 µM LYMTAC-2 for 4 h. Cells were then placed on ice, washed with ice-cold 1X PBS, and lysed in IP lysis buffer supplemented with 1X protease inhibitor cocktail, 5 mM 1,10-phenanthroline monohydrate, 10 mM N-ethylmaleimide, and 20 µM PR-619. Then, the lysate was incubated with 2 µg of anti-HiBiT antibody for 20 min at room temperature. HiBiT antibody incubated lysate was next added to 20 µL of protein G Sepharose magnetic beads and incubated overnight. After incubation, beads were washed 3X with wash buffer, and bound proteins were eluted using 2X NuPAGE™ LDS Sample Buffer supplemented with 10% β-ME. Proteins were separated in SDS-PAGE and immunoblotted with respective antibodies.
Confocal microscopy
For microscopy experiments, HEK293 mNeonGreen-KRASWT cells stably expressing FLAG-MTH1-RNF152 were seeded at a density of 15,000 cells per well in 8-well chambered dishes (Ibidi). Next day, cells were pre-treated with 200 nM BafA1 for 30 min and co-treated with 1 µM LYMTAC-4 for 4 h. Then, cells were washed 2X with PBS and fixed with 4% paraformaldehyde in PBS for 15 min at room temperature. After fixing, cells were permeabilized with 0.2% Triton-X-100 in PBS for 10 min at room temperature. Then, cells were incubated with blocking buffer (3% BSA, 0.2% Triton-X-100 in PBS) for 1 h at room temperature, followed by overnight incubation at 4 °C with FLAG (1:300 dilution) and LAMP1 (1:50 dilution) antibodies in antibody dilution buffer (1% BSA, 0.2% Triton-X-100 in PBS). Next day, cells were washed 3X with PBS at room temperature and incubated with secondary antibodies (1:500 dilution, Alexa Fluor 568 and Alexa Fluro 647) at room temperature for 1 h. Next, cells were washed 3X with PBS and stained with Hoechst stain (3 μg/mL) in PBS for 10 min at room temperature, followed by two PBS washes. The cells were imaged using a 63x/1.30 NA glycerol immersion objective lens on an inverted Leica TCS SP8 confocal microscope and images were analyzed using Leica LAS X software.
Time lapse imaging
HEK293 stably transfected with mNeonGreen-KRASWT and MTH1-RNF152 were seeded in a 35 mm glass-bottom dish with 2 mL Dulbecco’s Modified Eagle’s medium (DMEM; Gibco). After 24 h, cells were treated with 200 nM BafA1 and NucBlue™ Live ReadyProbes™ Reagent for 30 min at 37 °C in 5% CO2 medium. Then the media was replaced with new media (2 mL) containing 1 μM LYMTAC-4 and 200 nM BafA1, and images were acquired every 30 min up to 4 h. The cells were imaged using a 63x/1.30 NA glycerol immersion objective lens on an inverted Leica TCS SP8 confocal microscope, and images were analyzed using Leica LAS X software.
Washout experiment with LYMTAC-4 in AsPC-1
AsPC-1 cells stably expressing RNF152 were seeded at a density of 4 × 105 cells per well in 6-well dishes. After 24 h, cells were treated with DMSO, 100 nM pan-KRAS inhibitor, or 100 nM LYMTAC-4 for 4 h, removed media, washed three times with PBS, and replaced with fresh media for 48 h. For LYMTAC-4 treated wells, fresh media was added in the absence or presence of 5 μM MTH1-ligand for 48 h. After 48 h, cells were lysed and subjected to SDS-PAGE and immunoblotting with respective antibodies.
Cell proliferation assays
AsPC-1 cells stably expressing RNF152 were seeded at a density of 2000 cells per well in 96-well plates. Next day, cells were treated with DMSO and dose titrations (0.5 nM to 1000 nM) of pan-KRASi, KRAS PROTAC, or LYMTAC-4. After five days of treatment, cell viability was measured using CyQUANT™ direct cell proliferation assay reagents following the manufacturer’s protocol (C35011). The fluorescence readings were recorded using EnVision plate reader (settings: FITC_bottom, Ex: FITC FP 480, Em: FITC 535) and plotted as a four-parameter non-linear regression curve fit using GraphPad Prism 8.4.3.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Source data are provided with this paper as a Source Data file. All proteomic data have been uploaded to MaSSIVE repository under the accession: MSV000096874. Source data are provided with this paper.
References
Lin, C.-Y. et al. Membrane protein-regulated networks across human cancers. Nat. Commun. 10, 3131 (2019).
Jelokhani-Niaraki, M. Membrane proteins: structure, function and motion. Int. J. Mol. Sci. 24, 468 (2022).
To’a Salazar, G., Huang, Z., Zhang, N., Zhang, X.-G. & An, Z. Antibody therapies targeting complex membrane proteins. Engineering 7, 1541–1551 (2021).
Hutchings, C. J. Mini-review: antibody therapeutics targeting G protein-coupled receptors and ion channels. Antib. Ther. 3, 257–264 (2020).
Santos, R. et al. A comprehensive map of molecular drug targets. Nat. Rev. Drug Discov. 16, 19–34 (2017).
Rodríguez-Nava, C. et al. Mechanisms of action and limitations of monoclonal antibodies and single chain fragment variable (scFv) in the treatment of cancer. Biomedicines 11, 1610 (2023).
Cruz, E. & Kayser, V. Monoclonal antibody therapy of solid tumors: clinical limitations and novel strategies to enhance treatment efficacy. Biologics: targets Ther. 13, 33–51 (2019).
Sakamoto, K. M. et al. Protacs: chimeric molecules that target proteins to the Skp1-Cullin-F box complex for ubiquitination and degradation. Proc. Natl Acad. Sci. USA 98, 8554–8559 (2001).
Bondeson, D. P. & Crews, C. M. Targeted protein degradation by small molecules. Annu. Rev. Pharmacol. Toxicol. 57, 107–123 (2017).
Nalawansha, D. A. & Crews, C. M. PROTACs: an emerging therapeutic modality in precision medicine. Cell Chem. Biol. 27, 998–1014 (2020).
Guenette, R. G., Yang, S. W., Min, J., Pei, B. & Potts, P. R. Target and tissue selectivity of PROTAC degraders. Chem. Soc. Rev. 51, 5740–5756 (2022).
Samarasinghe, K. T. G. & Crews, C. M. Targeted protein degradation: a promise for undruggable proteins. Cell Chem. Biol. 28, 934–951 (2021).
Winter, G. E. et al. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science 348, 1376–1381 (2015).
Verma, R., Mohl, D. & Deshaies, R. J. Harnessing the power of proteolysis for targeted protein inactivation. Mol. cell 77, 446–460 (2020).
Simpson, L. M. et al. Target protein localization and its impact on PROTAC-mediated degradation. Cell Chem. Biol. 29, 1482–1504.e7 (2022).
Burslem, G. M. et al. The advantages of targeted protein degradation over inhibition: an RTK case study. Cell Chem. Biol. 25, 67–77.e3 (2018).
Sardana, R. & Emr, S. D. Membrane protein quality control mechanisms in the endo-lysosome system. Trends Cell Biol. 31, 269–283 (2021).
Piper, R. C. & Luzio, J. P. Ubiquitin-dependent sorting of integral membrane proteins for degradation in lysosomes. Curr. Opin. Cell Biol. 19, 459–465 (2007).
Nalawansha, D. A., Mangano, K., den Besten, W. & Potts, P. R. TAC-tics for leveraging proximity biology in drug discovery. Chembiochem 25, e202300712 (2024).
Banik, S. M. et al. Lysosome-targeting chimaeras for degradation of extracellular proteins. Nature 584, 291–297 (2020).
Marei, H. et al. Antibody targeting of E3 ubiquitin ligases for receptor degradation. Nature 610, 182–189 (2022).
Cotton, A. D., Nguyen, D. P., Gramespacher, J. A., Seiple, I. B. & Wells, J. A. Development of antibody-based PROTACs for the degradation of the cell-surface immune checkpoint protein PD-L1. J. Am. Chem. Soc. 143, 593–598 (2021).
Caianiello, D. F. et al. Bifunctional small molecules that mediate the degradation of extracellular proteins. Nat. Chem. Biol. 17, 947–953 (2021).
Pance, K. et al. Modular cytokine receptor-targeting chimaeras for targeted degradation of cell surface and extracellular proteins. Nat. Biotechnol. 41, 273–281 (2023).
Schwake, M., Schröder, B. & Saftig, P. Lysosomal membrane proteins and their central role in physiology. Traffic 14, 739–748 (2013).
Xu, J. The role of lysosomal membrane proteins in autophagy and related diseases. FEBS J. 291, 3762-3785 (2024).
Zhang, W. et al. A conserved ubiquitin- and ESCRT-dependent pathway internalizes human lysosomal membrane proteins for degradation. PLoS Biol. 19, e3001361 (2021).
Zhu, L., Jorgensen, J. R., Li, M., Chuang, Y. S. & Emr, S. D. ESCRTs function directly on the lysosome membrane to downregulate ubiquitinated lysosomal membrane proteins. eLife 6, e26403 (2017).
Wang, Y., et al. LAPTM5 mediates immature B cell apoptosis and B cell tolerance by regulating the WWP2-PTEN-AKT pathway. Proc. Natl Acad. Sci. USA 119, e2205629119 (2022).
Veits, G. K. et al. Development of an AchillesTAG degradation system and its application to control CAR-T activity. Curr. Res. Chem. Biol. 1, 100010 (2021).
Mangano, K. et al. VIPER-TACs leverage viral E3 ligases for disease-specific targeted protein degradation. Cell Chem. Biol. 32, 423–433.e9 (2025).
Huang, H. T. et al. A chemoproteomic approach to query the degradable kinome using a multi-kinase degrader. Cell Chem. Biol. 25, 88–99.e6 (2018).
Tapial Mart¡nez, P., L¢pez Navajas, P. & Lietha, D. FAK structure and regulation by membrane interactions and force in focal adhesions. Biomolecules 10, 179 (2020).
Wilson, K., Shiuan, E. & Brantley-Sieders, D. M. Oncogenic functions and therapeutic targeting of EphA2 in cancer. Oncogene 40, 2483–2495 (2021).
Bondeson, D. P. et al. Lessons in PROTAC design from selective degradation with a promiscuous warhead. Cell Chem. Biol. 25, 78–87.e5 (2018).
Donovan, K. A. et al. Mapping the degradable kinome provides a resource for expedited degrader development. Cell 183, 1714–1731.e10 (2020).
Moore, A. R., Rosenberg, S. C., McCormick, F., Malek, S. RAS-targeted therapies. Nat. Rev. Drug Discov. (2021).
Schwinn, M. K. et al. CRISPR-mediated tagging of endogenous proteins with a luminescent peptide. ACS Chem. Biol. 13, 467–474 (2018).
Clackson, T. et al. Redesigning an FKBP-ligand interface to generate chemical dimerizers with novel specificity. Proc. Natl. Acad. Sci. USA 95, 10437–10442 (1998).
Ash, L. J. et al. KRAS: biology, inhibition, and mechanisms of inhibitor resistance. Curr. Oncol. (Tor., Ont.) 31, 2024–2046 (2024).
Huang, L., Guo, Z., Wang, F. & Fu, L. KRAS mutation: from undruggable to druggable in cancer. Signal Transduct. Target. Ther. 6, 386 (2021).
Liu, J., Kang, R. & Tang, D. The KRAS-G12C inhibitor: activity and resistance. Cancer Gene Ther. 29, 875–878 (2022).
Ostrem, J. M. L., Peters, U. & Shokat, K. M. Direct RAS inhibitors turn 10. Nat. Chem. Biol. 20, 1238–1241 (2024).
Zografou-Barredo, N. A., Hallatt, A. J., Goujon-Ricci, J. & Cano, C. A beginner’s guide to current synthetic linker strategies towards VHL-recruiting PROTACs. Bioorg. Med. Chem. 88-89, 117334 (2023).
Békés, M., Langley, D. R. & Crews, C. M. PROTAC targeted protein degraders: the past is prologue. Nat. Rev. Drug Discov. 21, 181–200 (2022).
Bond, M. J., Chu, L., Nalawansha, D. A., Li, K. & Crews, C. M. Targeted degradation of oncogenic KRASG12C by VHL-recruiting PROTACs. ACS Cent. Sci. 6, 1367–1375 (2020).
Gurreri, E. et al. KRAS-dependency in pancreatic ductal adenocarcinoma. mechanisms of escaping in resistance to KRAS inhibitors and perspectives of therapy. Int. J. Mol. Sci. 24, 9313 (2023).
Tran, T. H. et al. KRAS interaction with RAF1 RAS-binding domain and cysteine-rich domain provides insights into RAS-mediated RAF activation. Nat. Commun. 12, 1176 (2021).
Henkels, K. M., Rehl, K. M. & Cho, K. -J. Blocking K-Ras interaction with the plasma membrane is a tractable therapeutic approach to inhibit oncogenic K-ras activity. Front. Mol. Biosci. 8, 673096 (2021).
Haidar, M. & Jacquemin, P. Past and future strategies to inhibit membrane localization of the KRAS oncogene. Int. J. Mol. Sci. 22, 13193 (2021).
Yaeger, R. et al. Molecular Characterization of acquired resistance to KRASG12C-EGFR inhibition in colorectal cancer. Cancer Discov. 13, 41–55 (2023).
Holderfield, M. et al. Concurrent inhibition of oncogenic and wild-type RAS-GTP for cancer therapy. Nature 629, 919–926 (2024).
Raina, K. et al. Regulated induced proximity targeting chimeras—RIPTACs—A heterobifunctional small molecule strategy for cancer selective therapies. Cell Chem. Biol. 31, 1490–1502.e42 (2024).
Ng, C. S. C., Liu, A., Cui, B. & Banik, S. M. Targeted Protein Relocalization via Protein Transport Coupling. Nature 633, 941–951 (2024).
Gibson, W. J. et al. Bifunctional small molecules that induce nuclear localization and targeted transcriptional regulation. J. Am. Chem. Soc. 145, 26028–26037 (2023).
Sadagopan, A. et al. p53 protein abundance is a therapeutic window across TP53 mutant cancers and is targetable with proximity inducing small molecules. bioRxiv, 2024.07.27.605429 (2024).
Schulze, C. J. et al. Chemical remodeling of a cellular chaperone to target the active state of mutant KRAS. Science 381, 794–799 (2023).
Canny, M. D. et al. Inhibition of 53BP1 favors homology-dependent DNA repair and increases CRISPR–Cas9 genome-editing efficiency. Nat. Biotechnol. 36, 95–102 (2018).
Acknowledgments
We thank Ryan Case, Jan Andersson, Olivier Bedel, Rati Verma, Kusal Samarasinghe, Spencer Hill, and Kyle Mangano for helpful discussions. We thank Alex Golkar for supporting final compound characterization. We thank Craig Kiefer for generating graphics for the manuscript. We thank all members of the Induced Proximity Platform for their support.
Author information
Authors and Affiliations
Contributions
D.A.N. and P.R.P. conceived the project and designed experiments. D. A.N., W.D., J.X., S.L., R.G.G., and C.E.S. performed experiments and analyzed data. K.S.A., G.H., G.M., A.T.T., R.P.W., B.A.L., S.A., M.K.A., A.S., and R.K. designed and synthesized compounds. C.W. performed the final compound characterization. D.A.N. wrote the original draft of the manuscript, revised by F.D.S.M., S.K., and P.R.P. and was further revised with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
As indicated in the affiliations, some authors are employees of Amgen. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Markella Konstantinidou and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Nalawansha, D.A., Mazis, G., Husemoen, G. et al. LYMTACs:chimeric small molecules repurpose lysosomal membrane proteins for target protein relocalization and degradation. Nat Commun 16, 7812 (2025). https://doi.org/10.1038/s41467-025-63128-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-63128-4