Abstract
Infections by intracellular pathogens often cause insult to host cell DNA, which stimulates responses that ultimately eliminate the damaged cell and hence the microbial niche. p53 is an innate immunity mediator that responds to DNA damage and intracellular infection by transcriptionally activating pathways that arrest the cell cycle, repair DNA, and elicit apoptosis. How pathogens counter p53 are incompletely understood. Here, we demonstrate that the endotheliotropic obligate intracellular bacterium Orientia tsutsugamushi blocks transcription of TP53 to nearly deplete p53 levels. Contrary to the unrestricted proliferation expected based on the transcriptome of p53-deficient infected cells, Orientia arrests the cell cycle at S phase to promote bacterial replication. It protects host endothelial cells from DNA damage even if induced by etoposide and delays genotoxic-dependent apoptosis until late in infection once a high bacterial load has been achieved. TP53 downregulation, protection against genotoxicity, and inhibition of DNA damage-dependent apoptosis are executed by the Orientia nucleomodulatory effector, Ank13. Therefore, O. tsutsugamushi inhibits TP53 expression and genotoxicity to reconfigure the intracellular environment of its host cell into one that favors bacterial replication.
Similar content being viewed by others
Introduction
Intracellular pathogens reshape the host cellular environment into one that supports optimal replication. Intracellular infections are often genotoxic, which elicits DNA damage responses that ultimately trigger apoptosis to eliminate the marred cell along with its microbial inhabitants1,2. As a countermeasure, intracellular microbes must prevent host genome instability and inhibit programmed cell death responses to sustain their niches2. The pathogenic mechanisms that facilitate this evolutionarily conserved survival strategy are inadequately defined.
Known as “the guardian of the genome”, p53 is a tumor-suppressing transcription factor that is conserved among higher eukaryotes and controls the response to DNA damage2,3. Its importance to maintaining cellular homeostasis is underscored by the fact that TP53, the gene that encodes p53, is mutated in more than half of human cancers. Under normal conditions, p53 is marked for degradation by the HDM2 (human double minute 2) E3 ubiquitin ligase. When DNA damage is incurred, p53 is stabilized and activates a transcriptional program that temporarily arrests the cell cycle to allow for DNA repair. If the damage is irreversible, p53 induces senescence, a state of permanent cell cycle arrest where the cell remains viable, or undergoes apoptosis1,2. It is also a central mediator of innate immunity to intracellular microbes4. p53 is therefore an antimicrobial hurdle that these organisms must overcome. Several intracellular pathogens orchestrate p53 degradation5,6,7,8,9,10,11,12,13,14,15,16,17,18,19. Multiple viruses inhibit p53 translation while the cattle parasite, Theileria annulata, blocks p53 translocation into the nucleus20,21,22,23,24,25,26. TP53 is post-transcriptionally repressed by a TLR4-NF-κB-mediated cellular pathway activated in response to lipopolysaccharides from Klebsiella pneumoniae and other Enterobacteria27. Microbial p53 suppression has been repeatedly linked to tumorigenesis1,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30. No pathogen has been shown to repress TP53 transcription.
Orientia tsutsugamushi is a mite-transmitted obligate intracellular bacterium that causes scrub typhus, an emerging potentially fatal human rickettsiosis31,32. Scrub typhus is endemic throughout the Asia-Pacific and occurs in numerous African, Middle Eastern, and South American countries32,33,34,35,36,37. Orientia recently was detected in mites in North Carolina, potentially extending the disease’s geographic footprint to North America38,39. Diagnosis can be difficult, treatment options are limited, and there is no vaccine40,41. Scrub typhus initiates as an acute non-specific febrile illness accompanied by localized vasculitis and has not been linked to cancer. If left untreated or if antibiotic therapy is delayed, severe symptoms, including respiratory distress, renal failure, meningoencephalitis, and myocarditis, ensue. Moreover, widespread vasculitis can lead to hypotensive shock, multiorgan failure, and death41,42. Scrub typhus vasculitis is linked to heavy O. tsutsugamushi burdens of endothelial cells43,44,45,46,47,48. Following endothelial cell invasion, the pathogen replicates to high numbers in the cytosol49,50,51,52,53. By around 72 h post-infection (p.i.) logarithmic replication has ceased and the cell is loaded with bacteria primed for release50,51,53,54. At this time point, DNA damage and apoptosis are first observable, both in infected endothelial cell lines and the endothelium of infected mice43,43,43. High O. tsutsugamushi loads in endothelial cells are also associated with an overzealous inflammatory response that further contributes to the vasculitis and other serious manifestations43,45,46,47,48. These observations suggest that O. tsutsugamushi inhibits DNA damage-induced apoptosis to replicate sufficiently.
Here, we report that O. tsutsugamushi inhibits TP53 transcription to severely reduce p53 levels. Infected endothelial cells exhibit transcriptional profiles consistent with a lack of p53. However, contrary to the uncontrolled proliferation anticipated for such cells, O. tsutsugamushi pauses the cell cycle at S phase to maximize its intracellular growth. It protects endothelial cells from genotoxicity and inhibits DNA damage-induced apoptosis until 72 h p.i. The bacterium’s abilities to downregulate TP53, inhibit DNA damage, and delay genotoxic-dependent apoptosis involve its nucleomodulin, Ank13. These modulatory functions depend on the effector’s nucleotropism and ability to co-opt the host SCF E3 ubiquitin ligase complex. Our findings outline a new mechanism by which a pathogen modulates p53 and its downstream responses to increase host cell longevity and optimize intracellular infection.
Results
O. tsutsugamushi downregulates TP53
The O. tsutsugamushi Ikeda strain, which was originally recovered from a scrub typhus patient in Japan, causes severe disease in humans and is fatal in laboratory mice56,57,58,59,60. Ikeda was used for all experiments and will hereafter be referred to as O. tsutsugamushi or Orientia unless discussed in the context of other strains. The endothelial cell pathways that O. tsutsugamushi infection alters at the transcriptional level are poorly characterized. RNA-seq was performed on paired infected and uninfected EA.hy926 human vascular endothelial cells at 8, 48, and 72 h p.i., timepoints that correspond to lag, mid-exponential, and late-exponential Orientia replication, respectively (Supplementary Fig. 1a)50,53,61. Three biological replicates with similar bacterial loads were evaluated (Supplementary Fig. 1b). Pearson’s correlation coefficient and principal component analyses confirmed that differentially expressed genes (DEGs) could be defined at each timepoint (Supplementary Fig. 1c, d). DEGs were defined as having a -log10(padj) value of ≥ 1.3 and a log2(fold change) of ≥ 1 or ≤ −1 in infected compared to uninfected cells (Supplementary Data 1). At 8, 48, and 72 h p.i., DEGs made up approximately 1%, 13%, and 30% of transcripts detected, respectively (Fig. 1a–c). DEGs were predominantly upregulated at 8 and 48 h p.i., but comparably up- and downregulated at 72 h p.i. (Fig. 1a–c). Only four DEGs were altered at all timepoints (Fig. 1a–d). TP53, which encodes p533, and CLIC1, which contributes to intracellular physiological stability62, were downregulated. BST2 and MX2, which promote antiviral responses63 and innate immunity64, respectively, were upregulated.
a–c Volcano plots of the gene expression profiles for O. tsutsugamushi infected EA.hy926 cells compared to uninfected cells. Each dot corresponds to an individual gene. Differential expression significant analysis followed by the Benjamini and Hochberg’s approach was employed to generated adjusted P values between infected and uninfected samples per timepoint. Teal and red dots indicate differentially expressed genes (DEGs) that are up- or downregulated, respectively. Black dots represent genes that are not differentially expressed. Grey lines delineate DEGs as genes with a significant -log10(padj)value (horizontal lines ≥ 1.3) and log2(fold change) (vertical lines ≥ 1 or ≤ -1). Fractions indicate the numbers of DEGs up- (upper right) or downregulated (upper left) out of the total number of genes identified. d Venn diagram showing the number of unique and shared DEGs at 8 h p.i. (10 total), 48 h p.i. (138 total), and 72 h p.i. (1117 total). Overlapping regions display the number of common DEGs among intersecting groups with TP53, CLIC1, BST2, and MX2 being present at all timepoints and colored to indicate up- (teal) or downregulated (red) expression. e STRING analysis of all DEGs at 48 h p.i. Disconnected nodes were removed, and Markov Clustering was employed to define prominent clusters. Solid lines denote a direct connection between genes. Line thickness indicates the strength of the connection based on existing literature. Dotted lines represent connections between genes that are on the cusp of multiple clusters. Four clusters are defined: Cluster 1 (red) – interferon signaling and ISG15-protein conjugation; Cluster 2 (yellow) – p53-related pathways (TP53 indicated by a purple circle); Cluster 3 (green) – signaling receptors; Cluster 4 (blue) – apolipoproteins. f Bubble plot showing the top 10 GO terms subdivided by biological processes that are associated with Cluster 2. Bubble size correlates with the number of DEGs from the input present in that GO term, bubble color indicates the false discovery rate (FDR), and bubble position on the x-axis shows the strength of the enrichment. g Schematic of up- (teal) or downregulated (red) p53-associated DEGs that negatively (inhibitor lines) or positively (arrows) modulate proliferation and cell cycle, DNA damage response, and apoptosis. Filled in ovals indicate proteins that directly bind p53. Source data are provided as a Source Data file.
DEGs (Supplementary Data 1) were subjected to STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) analysis with Markov clustering to identify host pathways altered at the transcriptional level during infection (Fig. 1e; Supplementary Fig. 2). At 48 h p.i., four distinct clusters were observed. Cluster 1 contained genes associated with interferon signaling and ISG15-(interferon-stimulated gene 15) protein conjugation and produced gene ontology (GO) terms centered around antimicrobial responses (Fig. 1e; Supplementary Data 2). Cluster 2 expanded from TP53 with the top 10 GO terms related to p53-regulated pathways (Fig. 1e, f; Supplementary Data 2). Clusters 3 and 4 contained only two DEGs that encoded signaling receptors and apolipoproteins, respectively. TP53 also shared a connection with DEGs in Clusters 1 and 3 (Fig. 1e; Supplementary Data 2). At 72 h p.i., DEGs were enriched in pathways involved in p53-related DNA damage responses; interferon, IL-10, and NF-κB signaling; eukaryotic translation elongation; the mitotic spindle checkpoint; and the inflammasome (Supplementary Fig. 2; Supplementary Data 2). Cross-referencing DEGs from all timepoints to a comprehensive list of known p53 target genes65 identified a total of 76 that exhibited expression patterns indicative of an absence of p53 (Supplementary Data 3; Supplementary Fig. 3)2,3,66,67,68,69. Of these, 31 aligned with unrestricted proliferation and cell cycle progression, limited DNA damage repair responses, and inhibition of apoptosis (Fig. 1g; Supplementary Fig. 3). Thus, O. tsutsugamushi downregulates TP53 expression, which alters multiple p53-regulated pathways at the transcriptional level.
O. tsutsugamushi rapidly reduces p53 levels
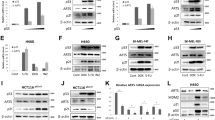
RT-qPCR analysis validated that TP53 expression is downregulated throughout Orientia infection (Fig. 2a). Immunoblot studies detected a sustained near absence of p53 as early as 4 h p.i. (Fig. 2b–e). The O. tsutsugamushi immunodominant outer membrane protein, TSA56 (56-kDa type-specific antigen), served as an infection control, while human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was a loading control for all western blots. Chloramphenicol, an antibiotic that crosses eukaryotic membranes to inhibit bacterial protein synthesis and hence replication, prevented O. tsutsugamushi from lowering p53 levels (Fig. 2f, g). Expression of p21, a p53 target that regulates cell cycle arrest, senescence, and apoptosis70, was reduced throughout infection as early as 24 h p.i. and in a chloramphenicol-sensitive manner (Supplementary Fig. 4). Additionally, scrub typhus clinical isolates UT76, Karp, and Gilliam grow in EA.hy926 cells comparably to Ikeda and impede TP53 and p53 expression throughout infection (Supplementary Fig. 5). Therefore, the conserved ability among O. tsutsugamushi strains to rapidly downregulate TP53 expression correlates with a sustained reduction in p53 that has downstream consequences.
a RNA and b–g whole cell lysates (WCLs) were collected from uninfected and O. tsutsugamushi infected EA.hy926 cells at a–c 8, 24, 48, and 72 h p.i. or d, e every hour for 8 h p.i. and subjected to a RT-qPCR or b,d western blot and c, e densitometry analysis. f, g Uninfected and infected EA.hy926 cells were treated with 34 µg/mL of chloramphenicol (CAM) or vehicle (VEH) control at 2 h p.i. At 72 h p.i., WCLs were collected and f western blot and g densitometry analysis was performed. Blots were probed with antibodies against p53, TSA56, or GAPDH. Red asterisks (*) denote TSA56 in infected samples. Mean normalized ratios ± SEM of a relative TP53 gene expression normalized to GAPDH (n = 3 independent experiments) or c, e, g p53:GAPDH were calculated [n = c 5, e 3, g 4 independent experiments]. Statistical significance was evaluated by One-way ANOVA followed by Tukey’s post hoc test. Statistically significant values (P ≤ 0.05) are displayed. Source data are provided as a Source Data file.
O. tsutsugamushi specifically targets TP53 gene expression
p53 is constitutively expressed to ensure a rapid response to stress. This is counterbalanced by the ubiquitin ligase, HDM2, which polyubiquitinates p53 to target it for proteasomal degradation16,27. Many intracellular pathogens exploit HDM2 to degrade p535,6,7,8,9,10,11,12,13,14,15,16,17,18,19. We investigated if Orientia employs a similar mechanism to target p53. HDM2 transcript and protein were pronouncedly lowered throughout infection (Fig. 3a-c; Supplementary Data 1), suggesting that HDM2 is inessential for O. tsutsugamushi to modulate p53 levels. To confirm that HDM2 activity is unnecessary for Orientia fitness, Nutlin 3a, a steric inhibitor that prevents HDM2 ubiquitination of p5371, was employed to stabilize p53 (Supplementary Fig. 6a). Nutlin 3a had no effect on the O. tsutsugamushi yield (Fig. 3d). Next, p53 levels were assessed in infected and uninfected cells treated with the proteasome inhibitor, bortezomib. Proteasome inhibition was confirmed by increased Nrf1 expression (Fig. 3e)72. p53 levels were lower in infected cells regardless of bortezomib treatment (Fig. 3e, f). Thus, O. tsutsugamushi inhibits p53 expression independent of HDM2 and the proteasome.
a–c RNA and WCLs were collected from uninfected and O. tsutsugamushi infected EA.hy926 cells at 8, 24, 48, and 72 h p.i. and subjected to a RT-qPCR, or b western blot and c densitometry analysis. Blots were probed with antibodies against HDM2, TSA56, or GAPDH. a Relative HDM2 gene expression normalized to GAPDH or c HDM2:GAPDH were calculated. d The O. tsutsugamushi yield was determined for vehicle (VEH) or Nutlin 3a (Nut3a) treated infected EA.hy926 cells. a, c, d Data are means ± SEM [n = a 3, c 4, d 3 independent experiments]. e,f Uninfected or infected EA.hy926 cells were treated with VEH or bortezomib (Bort; 50 nM) for 24 h prior to collection and e western blotting. Blots were probed for p53, Nrf1, TSA56, or GAPDH. Mean normalized ratios ± SEM of f p53:GAPDH were determined by densitometry. Data are means ± SEM (n = 3 independent experiments). g, h WCLs were collected from uninfected and O. tsutsugamushi infected EA.hy926 cells and subjected to g western blot. Blots were probed with antibodies against phosphorylated (p)-PKR (T446), PKR, TSA56, or GAPDH. Mean normalized ratios ± SEM of h p-PKR:GAPDH were determined by densitometry (n = 3 independent experiments). i–l O. tsutsugamushi infected EA.hy926 cells were treated with VEH or C16 (100 nM). i Lysates were collected at 72 h p.i. for qPCR to determine O. tsutsugamushi yield. Data are means ± SEM (n = 3 independent experiments). j–l WCLs were collected at 24 and 72 h p.i. for j western blot and k, l densitometry analysis. Blots were probed with antibodies against p53, p-PKR (T446), PKR, TSA56, or GAPDH. Vertical lines between bands indicate where the blot was cropped. Mean normalized ratios ± SEM of k p-PKR:GAPDH and l p53:GAPDH were calculated [n = k, l 4 independent experiments]. m Schematic of TP53 and its promoters. n,o RNA was collected from uninfected and O. tsutsugamushi infected EA.hy926 cells and subjected to RT-qPCR. Mean normalized ratios ± SEM of relative TP53 n P1 and o P2 gene expression normalized to GAPDH [n = n, o 3 independent experiments]. Red asterisks (*) denote TSA56 in infected samples. Statistical significance was evaluated by One-way ANOVA followed by the post hoc test Tukey’s (black) or Dunnett’s (blue) compared to uninfected VEH control. Statistically significant values (P ≤ 0.05) are displayed. Source data are provided as a Source Data file.
Double-stranded RNA viruses trigger the protein kinase R (PKR) stress response that inhibits host cell translation and thereby p53 expression73,74. PKR is activated by phosphorylation of T44675. During O. tsutsugamushi infection, PKR activation was significantly increased at 24 and 48 h p.i. (Fig. 3g, h). Inhibiting PKR phosphorylation using the small molecule inhibitor, C1676, failed to restore p53 levels in infected cells and even led to a higher bacterial load (Fig. 3i–l). Therefore, despite activating the PKR stress response, Orientia blocks p53 expression in a PKR-independent manner.
TP53 encodes two families of p53 isoforms via alternative promoters, P1 and P2 (Fig. 3m). Canonical TP53 transcription initiated from P1 generates an mRNA that is translated into full-length p53. Non-canonical transcription from P2 produces a transcript encoding either of two truncated isoforms77. P1 and P2 encoded TP53 mRNAs share a 3’-UTR that is critical for their stability. Lipopolysaccharides of enterobacteria with tumorigenic potential destabilize both P1 and P2 TP53 mRNA at the 3’-UTR, which decreases p53 levels and p53-regulated responses27. RT-qPCR conducted with isoform-specific primers found that O. tsutsugamushi prevents TP53 expression from P1 but not P2 (Fig. 3n, o), suggesting that it employs the novel mechanism of targeting the P1 TP53 isoform to block full-length p53 production.
Orientia modulates the cell cycle
p53 deficiency typically leads to uncontrolled proliferation66,67,68, a phenotype that is suggested by the transcriptome of O. tsutsugamushi infected endothelial cells (Supplementary Data 3; Fig. 1g; Supplementary Fig. 3). Accordingly, we assessed proliferation rates of uninfected and infected EA.hy926 cells that had been synchronized at G0/G1 by serum starvation and released when Orientia was added. Immunofluorescence microscopy was used to confirm that ≥ 75% of the cells in infected cultures harbored bacteria (Fig. 4a, b). Starting at 24 h, proliferation rates of infected cultures trended lower and were significantly reduced by two-fold at 48 h (Fig. 4c, d). Hence, despite p53 inhibition, infected cell proliferation is hindered and is most pronouncedly so during O. tsutsugamushi mid-log growth. We then measured cyclin protein turnover to monitor cell cycle progression (Fig. 4e)68. Cyclin D1 and cyclin D3 levels were robustly reduced during infection in a chloramphenicol-sensitive manner (Fig. 4f–h; Supplementary Fig. 7a–c), which indicates cell cycle progression through G1 and correlates with the bacterial protein synthesis-dependent decrease in p53 (Fig. 2f, g)68. In contrast, cyclin E levels were significantly higher at 72 h p.i. (Fig. 4f, i). Cyclin A was slightly elevated at 24 and 48 h p.i. (Fig. 4f, j), while cyclin B1 levels trended lower in infected cells at all time points (Fig. 4f, k). These data indicate that O. tsutsugamushi promotes a pause in the host cell cycle at S phase during its exponential replication.
a–d CellTrace host cell proliferation assay. a Coverslips with O. tsutsugamushi infected EA.hy926 cells were methanol fixed and immunolabeled for TSA56 (green), stained with DAPI (blue), and imaged using immunofluorescent microscopy. b The percentage of infected cells was determined by examining 100 cells at each timepoint (n = 3 independent experiments). Scale bar represents 20 µm. c Host cell proliferation was assessed using flow cytometry. d The fold change in mean fluorescent intensity (MFI; n = 3 independent experiments) was determined by normalizing each sample to the 8 h timepoint. e Schematic of cyclin protein dynamics at different phases of the host cell cycle. The phase of the cell cycle that matches the cyclin profile during O. tsutsugamushi infection is denoted by a red dashed box. f–k EA.hy926 cells were serum starved for 24 h before O. tsutsugamushi infection. At time of infection, serum-free medium was replaced with complete medium. WCLs were collected at 8, 24, 48, and 72 h p.i. and subjected to f western blot followed by densitometric analysis of g Cyclin D1, h Cyclin D3, i Cyclin E (red bars), j Cyclin A, or k Cyclin B1. Red asterisks (*) denote TSA56 in infected samples. The ratio of the densitometric signal for each cyclin relative to GAPDH was compared between infected and uninfected samples to determine the fold change at each timepoint [n = g 5, h 4, i 5, j 4, k 4 independent experiments]. l–n Pharmacologic or siRNA-mediated inhibition of the cell cycle. l Experimental design employed for treatment of uninfected and infected host cells with cell cycle inhibitors. Created in BioRender. Carlyon, J. (2025) https://BioRender.com/m6yrg1o. m EA.hy926 cells were treated with vehicle (VEH), medium without FBS (serum starved; SS; purple), hydroxyurea (HU; 80 µM; red bar), or RO-3306 (9 µM; green) 24 h before infection. Treatment was maintained (closed circle) or removed at 48 h p.i. (open circle). Lysates were collected at 96 h p.i. n EA.hy926 cells were transfected with siNT, siCCNE1, siGMNN, or siCDC25a before being infected with O. tsutsugamushi. Lysates were collected at 72 h p.i. (m-n) O. tsutsugamushi yield was determined by qPCR. Data are means ± SEM [n = m, n 3 independent experiments]. Statistical significance was evaluated by One-way ANOVA followed by Dunnett’s post hoc test to VEH or siNT controls. Statistically significant values (P ≤ 0.05) are displayed. Source data are provided as a Source Data file.
To determine if S phase benefits O. tsutsugamushi fitness, we employed serum starvation to restrict the cell cycle in G0/G178, hydroxyurea to arrest in S phase79, or RO-3306 to halt at the G2/M junction (Fig. 4l)80. The Orientia DNA load was measured at 96 h p.i. Each treatment’s effect on host cell metabolic activity was verified using the MTT (3-[4,5-dimethylthiazol-2-yl]−2,5-diphenyl-2H-tetrazolium bromide) assay81. Serum starvation and hydroxyurea had no effect, whereas RO-3306 reduced metabolic activity by half (Supplementary Fig. 7d), the latter of which is expected during G2/M phase restriction82. Exposure of host cell-free O. tsutsugamushi to hydroxyurea and RO-3306 prior to infection did not impact its subsequent intracellular growth (Supplementary Fig. 7e). Whereas serum starvation and RO-3306 significantly lowered the bacterial yield, Orientia grew just as well in hydroxyurea treated cells as in controls (Fig. 4m). Treatment removal at 48 h p.i. to release cell cycle phase restriction did not bolster bacterial growth. As a complementary approach, cells were treated with siRNA targeting cyclin E to induce G1 arrest, geminin to induce S phase arrest, or cell division cycle 25 A to induce G2 arrest83. Knockdown was confirmed at the time of infection (Supplementary Fig. 7f–k). Cells arrested in G1 and G2/M phase yielded significantly less O. tsutsugamushi, while the loads in cells arrested in S phase and control cells were comparable (Fig. 4n). Together, these results show that O. tsutsugamushi pauses the cell cycle at S phase to maximize its growth. Although the lack of p53 would facilitate transition to S phase, the delay in host cell proliferation observed for infected cells suggests that Orientia employs a p53-independent mechanism to arrest the cell cycle.
O. tsutsugamushi inhibits host cell DNA damage
p53 is induced by DNA damage and is a key regulator of the DNA damage repair response84. Phosphorylation of histone H2.AX is a hallmark of DNA damage85,86. Phosphorylated H2.AX (ƔH2.AX) levels are unaltered in O.tsutsugamushi-infected EA.hy926 cells until 72 h p.i. when they are significantly elevated (Fig. 5a, b). To determine if O. tsutsugamushi inhibits DNA damage, cells were treated with etoposide, which induces irreversible double stranded DNA breaks87,88,89. Treatment was initiated 24 h before each collection timepoint. Etoposide did not affect O. tsutsugamushi growth (Fig. 5c). As expected, etoposide strongly raised p53 and ƔH2.AX levels in uninfected cells (Fig. 5d–f). In infected cells, p53 levels were decreased even in the presence etoposide at all time points. ƔH2.AX levels were also reduced until 72 h p.i. at which time they had risen comparably to those in uninfected cells. Thus, O. tsutsugamushi inhibits DNA damage until late infection.
a,b WCLs were collected from uninfected and O. tsutsugamushi infected EA.hy926 cells at 8, 24, 48, and 72 h p.i. followed by a western blot. Blots were probed with ƔH2.AX, TSA56, and GAPDH. The mean normalized ratio ± SEM of b ƔH2.AX:GAPDH was determined by densitometry analysis (n = 3 independent experiments). c–k EA.hy926 cells were infected and treated with etoposide (Eto; 1 µM; teal) or vehicle (VEH) 24 h prior to collection. c O. tsutsugamushi yield was determined using qPCR. Data are means ± SEM (n = 3 independent experiments). d–f WCLs collected at 24, 48, and 72 h p.i. were subjected to d western blot and e, f densitometry analysis. Blots were probed with p53, ƔH2.AX, TSA56, and GAPDH. g–j RT-qPCR was performed on total RNA collected at 24, 48, and 72 h p.i. Mean normalized ratios ± SEM of e p53:GAPDH or f ƔH2.AX:GAPDH, and relative gene expression of g PMAIP1, h CDKN1A, iTP53I3, j SESN1, and k ZMAT3 normalized to GAPDH were calculated [n = e 3, f 3, g–k 4 independent experiments]. Red asterisks (*) denote TSA56 in infected samples. Statistical significance was evaluated by One-way ANOVA followed by the post hoc test Tukey’s (black) or Dunnett’s (blue) compared to uninfected VEH control. Statistically significant values (P ≤ 0.05) are displayed. Source data are provided as a Source Data file.
DNA damage stabilizes p53 which then activates a subset of DNA repair genes that promote senescence or cell death90,91,92. To further elucidate the effects that O. tsutsugamushi-driven p53 downregulation has on the DNA damage repair response, we treated cells with etoposide or vehicle and used RT-qPCR to measure expression of genes activated by p53 including PMAIP1 (phorbol-12-myristate-13-acetate-induced protein 1), CDKN1A (cyclin dependent kinase inhibitor 1A), TP53I3 (tumor protein p53 inducible protein 3), SESN1 (sestrin 1), and ZMAT3 (zinc finger matrin-type 3). In vehicle-treated infected cells, CDKN1A, TP53I3, SESN1, and ZMAT3 were downregulated at all timepoints while PMAIP1 was upregulated at 72 h p.i. (Fig. 5g-k). In response to etoposide, PMAIP1 expression was more robustly induced in infected cells while CDKN1A, TP53I3, SESN1, and ZMAT3 transcription increased similarly in uninfected and infected cells. Upregulation of CDKN1A, which encodes p21, during treatment with etoposide translated to an increase in p21 levels (Supplementary Fig. 8). Therefore, although O. tsutsugamushi does not induce expression of any of the assayed DNA damage response genes until late infection, it does not repress these genes in response to etoposide-induced genotoxicity.
Orientia inhibits drug-induced apoptosis
Because a major p53 function is to induce apoptosis68, we examined if O. tsutsugamushi infected endothelial cells are recalcitrant to this cell death pathway. Cleaved caspase 3, caspase 7, and PARP1, which are apoptosis hallmarks93, were detected in uninfected cells exposed to etoposide but not in vehicle- or etoposide-treated infected cells (Fig. 6a–d). Staurosporine is a pan-kinase inhibitor that induces apoptosis by p53-dependent and -independent mechanisms that do not directly induce DNA damage94,95,96. O. tsutsugamushi suppressed p53 levels and caspase 3, caspase 7, and PARP1 cleavage in staurosporine treated cells (Supplementary Fig. 9). To account for caspase-independent mechanisms of apoptosis/cell death, we used flow cytometry to assay for annexin V binding to phosphatidylserine on the plasma membrane and uptake of membrane-impermeable propidium iodine (PI). In the absence of infection, etoposide and staurosporine robustly increased numbers of apoptotic/dead cells (Fig. 6e, f). This effect was not observed for infected cells until 72 h p.i. Thus, O. tsutsugamushi inhibits both DNA damage-dependent and -independent apoptosis of endothelial cells until late infection.
a–d Uninfected and infected EA.hy926 were treated with etoposide (Eto) 24 h prior to collection. (a) Western blots of WCLs collected at 24, 48, and 72 h p.i. were probed for procaspase 3 and cleaved caspase 3, procaspase 7 and cleaved caspase 7, full length and cleaved (cPARP1) PARP1, TSA56, and GAPDH. Red asterisks (*) denote TSA56 in infected samples. Mean normalized ratios ± SEM of b Caspase3:GAPDH, c Caspase7:GAPDH, and d cPARP1:GAPDH densitometric signals were calculated [n = b–d 3 independent experiments]. e, f Vehicle (VEH), staurosporine (Stauro)-treated, or Eto-treated uninfected and infected samples were collected at 24, 48, and 72 h p.i. and stained with FITC-Annexin V and propidium iodine (PI). e Samples were subjected to flow cytometry. Quadrants displayed on dot plot of uninfected + VEH-treated sample represent the quadrants across all samples. Quadrant 1 (Q1; FITC-/PI + ) and Q2 (FITC + /PI + ) contain late apoptotic and dead cells, respectively. Q3 (FITC + /PI-) consists of early apoptotic cells and Q4 (FITC-/PI-) contains live cells. f The percentage of apoptotic cells/dead (Q1-3) was determined. The mean percentage of cells ± SEM of each quadrant was calculated (n = 4 independent experiments). Statistical significance was evaluated by One-way ANOVA followed by the post hoc test Tukey’s (black) or Dunnett’s (blue) compared to uninfected VEH control. Statistically significant values (P ≤ 0.05) are displayed. Source data are provided as a Source Data file.
Ank13 deters DNA damage-induced apoptosis
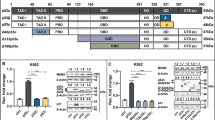
We rationalized that the chloramphenicol-sensitive ability of O. tsutsugamushi to downregulate TP53 involves a bacterial protein that modulates host cell gene expression. Orientia encodes one of the largest cadres of ankyrin repeat (AR)-containing effectors (Anks), most of which exhibit a bipartite architecture consisting of an N-terminal AR domain and a C-terminal F-box56,97,98,99,100. ARs mediate protein-protein interactions, and pathogen acquisition of genes encoding AR proteins is linked to increased virulence99,101,102. The F-box is a eukaryotic-like motif that coopts the host SCF E3 ubiquitin ligase complex and is essential for multiple Orientia Anks to modulate host cell processes49,50,51,98,103. Ank13 is an O. tsutsugamushi nucleomodulin first documented to be expressed during infection of HeLa cells49. It has eight ARs and an F-box (Supplementary Fig. 10a). Its nucleotropism depends on I127 located in AR4. When ectopically expressed in HeLa cells, Ank13 dysregulates the expression of more than 2000 genes in nucleotropism- and F-box-dependent manners49. We confirmed that O. tsutsugamushi expresses ank13 throughout infection of EA.hy926 cells (Supplementary Fig. 10b, c). In the absence of tools for genetically manipulating O. tsutsugamushi104, we determined if Ank13 can downregulate TP53 and p53 expression by transfecting EA.hy926 cells to express GFP-Ank13, nucleotropism-defective GFP-Ank13I127R49, or GFP. Cells were also transfected to express GFP-Ank13-F-boxAAAAA, which cannot bind the SCF complex due to its five F-box residues essential for doing so being replaced with alanine49,98,105. GFP-positive cells were enriched for by fluorescent-activated cell sorting followed by RT-qPCR or immunoblot analysis (Fig. 7a-f). Cells expressing GFP-tagged Ank13 and Ank13-F-boxAAAAA but not Ank13I127R had significantly lower TP53 transcript along with reduced p53 and ƔH2.AX protein levels compared to cells expressing GFP (Fig. 7a-d). Moreover, GFP-Ank13 and GFP-Ank13-F-boxAAAAA but not GFP-Ank13I127R inhibited expression of the TP53 P1 isoform (Fig. 7e), while none altered TP53 P2 isoform expression (Fig. 7f). Thus, ectopically expressed Ank13 phenocopies O. tsutsugamushi infection by specifically inhibiting P1 TP53 isoform expression in a nucleotropism-dependent but F-box-independent manner to reduce p53 levels. The nucleotropism of Ank13 is also key for it to protect endothelial cells from DNA damage. Finally, GFP-Ank13 prevented endothelial cell apoptosis induced by etoposide but not staurosporine and required both its nucleotropism and F-box to do so (Fig. 7g, h). Overall, Ank13 is linked to the ability of O. tsutsugamushi to impede p53 expression, protect against genotoxicity, and inhibit DNA damage-dependent but not -independent apoptosis.
a–f RNA and WCLs were collected from fluorescence-activated cell-sorted EA.hy926 cells expressing GFP (green bar), GFP-Ank13, GFP-Ank13∆F-boxAAAAA, or GFP-Ank13I127R and subjected to a, e, f RT-qPCR and b western blot, respectively. Blots were probed for p53, ƔH2.AX, GFP, and GAPDH. Red asterisks (*) denote p53. Mean normalized ratios ± SEM of aTP53, eTP53 P1, and f TP53 P2 to GAPDH and c p53:GAPDH and d ƔH2.AX:GAPDH densitometric signal ratios were calculated [n = a 3, c 3, d 4, e,f 3 independent experiments]. g, h EA.hy926 cells transfected to express GFP or the indicated GFP-tagged Ank13 proteins were treated with staurosporine (Stauro), etoposide (Eto), or vehicle (VEH) and stained with PE-Annexin V and 7-AAD. g Samples were subjected to flow cytometry. Quadrants displayed on the dot plot of GFP + VEH represent the quadrants for all samples. Quadrant 1 (Q1; FITC-/PI + ) and Q2 (FITC + /PI + ) consists of late apoptotic and dead cells, respectively. Q3 (FITC + /PI-) contains early apoptotic cells. Q4 (FITC-/PI-) contains live cells. h The percentage of apoptotic cells/dead cells (Q1-3) was determined. The fold change in the mean percentage of cells ± SEM were calculated (n = 3 independent experiments). Statistical significance was evaluated by One-way ANOVA followed by Dunnett’s post hoc test for comparison of each sample to GFP control (black) or each sample to VEH-treated counterpart (blue), respectively. Statistically significant values (P ≤ 0.05) are displayed. Source data are provided as a Source Data file.
Discussion
Apoptosis is a key innate defense against intracellular microbes that eliminates the replicative niche and is elicited in response to DNA damage incurred during infection. O. tsutsugamushi is the first pathogen identified to delay DNA damage-dependent apoptosis by blocking TP53 expression. Orientia couples this strategy with protecting its endothelial host cell DNA from insult in a p53-independent manner and modulating the cell cycle to yield an intracellular environment that favors its logarithmic growth. Many pathogens that target p53 do so by directing its proteasomal degradation5,6,7,8,9,10,11,12,13,28. Yet, as demonstrated for Chlamydia trachomatis and Plasmodium yoelii, this strategy is an Achille’s heel that can be nullified using Nutlin 3a as a host-directed therapeutic to restore p53 levels and reduce the pathogen load8,12. Because the Orientia mechanism is HDM2- and proteasome independent, Nutlin 3a would be ineffective against scrub typhus.
The abilities of O. tsutsugamushi to inhibit TP53 expression, protect against host cell genotoxicity, and delay apoptosis are linked, at least in part, to Ank13, as it singularly executes all three promicrobial strategies when ectopically expressed. O. tsutsugamushi-mediated p53 reduction is first observed during the initial hours of infection, which agrees with its high turnover rate and implies the action of a preloaded effector like Ank13. Indeed, because Orientia expresses Ank13 throughout its infection cycle, it would be ready for deployment upon invasion. Ank13 blocks expression of the P1 but not P2 TP53 isoform to recapitulate how Orientia targets TP53 transcription to deplete host cells of p53. The effector does so in a nucleotropism-dependent manner, indicating that it acts in the nucleus to block TP53 at the transcriptional level by binding DNA at the TP53 promoter region, mRNA at the 5’-untranslated region, or a transcription factor. For the latter possibility, because Ank13 TP53 inhibition is F-box-independent, it would impair the target transcription factor without ubiquitinating or directing it for proteasomal degradation. The mechanism(s) by which nucleotropic Ank13 impedes TP53 expression are likely also employed to alter expression of more than 2000 genes49.
O. tsutsugamushi inhibits apoptosis using Ank13-dependent and -independent strategies. Ank13 protects against DNA damage responses induced by cationic lipid transfection or etoposide in a nucleotropism-dependent manner, which phenocopies Orientia prevention of double-stranded DNA breaks during early infection52,55. The ability of Ank13 to impair etoposide-induced apoptosis requires both its nucleotropism and F-box, the latter of which echoes how O. tsutsugamushi Ank5 uses its F-box to orchestrate ubiquitination and proteasomal degradation of NLRC-5 (NOD-like receptor family CARD domain containing 5) to prevent major histocompatibility class I expression50. Ank13 may promote the degradation of one or more host factors that drive apoptosis due to irreparable DNA damage. Ank13 is not involved in the resistance to staurosporine-induced apoptosis exhibited by Orientia-infected cells51, reinforcing that it counters apoptosis exclusively in a genoprotective manner. Rather, resistance to apoptosis elicited by staurosporine is conferred by a subset of five F-box-containing Anks (Ank1, Ank5, Ank6, Ank9, Ank17) that retain Cul1 in the cytoplasm to lower the Cul1:c-MYC intranuclear balance and thereby dysregulate expression of c-MYC target genes involved in apoptosis51. Thus, at least six Orientia Anks transcriptionally inhibit apoptosis. O. tsutsugamushi also exerts an anti-apoptotic effect by impeding intracellular calcium release via an unknown mechanism106.
O. tsutsugamushi reduction of p53 levels promotes an endothelial cell transcriptional program that would normally culminate in uncontrolled cell cycle progression and proliferation. Yet, the pathogen modulates this response by arresting the cell cycle at S phase to promote its logarithmic growth using an undefined mechanism. How might S-phase arrest benefit Orientia? Following invasion, it traffics to the centrosome where it replicates. The resulting bacterial population remains there until exiting to initiate the next infection wave42. Because the centrosome duplicates during G2 and the duplicated centrosomes separate during the G2/M transition107, perhaps Orientia orchestrates the pause at S phase to prevent disruption of its preferred intracellular location for growth. Also, cells in S phase undergo metabolic shifts to support DNA replication and cell division, becoming enriched in amino acids and other intermediates that can feed into the tricarboxylic acid cycle (TCA)108. Conspicuously, O. tsutsugamushi growth corresponds with reductions in both amino acids and TCA intermediates53. Moreover, p53 exerts antimicrobial activity by inhibiting glycolysis and the pentose phosphate pathway, both of which feed into the TCA2,108. By simultaneously downregulating TP53 and arresting the cell cycle at S phase, Orientia may synergistically enrich its intracellular environment with the host metabolites that fuel its growth.
The majority of endothelial cell genes upregulated during O. tsutsugamushi replication include interferon, NF-κB, and IL-10 signaling as well as inflammasome activation. This indicates that antimicrobial responses, many of which are classically defined as antiviral, including programmed cell death, are mounted in response to late-stage Orientia infection and are consistent with observations for scrub typhus patient and animal model studies109,110,111,112,113,114. However, p53 contributes to interferon-mediated innate immunity, and the p53 downstream target, p21, which is also depleted during Orientia infection, is an important interferon-induced antiviral restriction factor4,112. By blocking TP53 expression, O. tsutsugamushi at least partially counters these responses. Furthermore, even though NF-κB responses are transcriptionally upregulated in Orientia-infected cells and loss of p53 can lead to increased NF-κB activity115, the bacterium inhibits NF-κB-dependent gene expression by impairing the transcription factor’s accumulation in the nucleus103. This strategy minimally involves the actions of Orientia Ank1 and Ank6 and bacterial stabilization of the host NF-κB inhibitor, p105103,116. Collectively, this suggests that although O. tsutsugamushi has developed numerous strategies to perturb the host response to infection, it is unable to outcompete proinflammatory responses that lead to apoptosis when bacterial burden is high. Interestingly, the only group of host transcripts downregulated when Orientia load is high is involved in translation elongation. Many intracellular pathogens counteract host translation to prevent proinflammatory protein production117,118,119,120. Previous work showed that O. tsutsugamushi Boryong strain effectors TPR43 and TRP46 inhibit eukaryotic translation elongation121. Also, our lab demonstrated that Ikeda Ank4 and Ank9 disrupt the unfolded protein response122,123, which is directly linked to an inhibition of host translation117,118. Therefore, inhibiting host cell translation might be yet another means by which O. tsutsugamushi facilitates its intracellular survival.
Downregulating TP53 to deplete p53 is a strategy that O. tsutsugamushi Ikeda shares with UT76, Karp, and Gilliam. The complement of Ank genes varies among all sequenced isolates, and of the four examined herein, only Ikeda and UT76 encode Ank1397. Thus, O. tsutsugamushi strains utilize Ank13 and unidentified virulence factors to block TP53 expression. Regardless, the overall conservation of this tactic underscores its importance to Orientia fitness and suggests that p53 suppression during scrub typhus is common. Although largely considered an acute illness31,41,42, limited studies have suggested that scrub typhus can become latent124,125. While cells heavily infected with O. tsutsugamushi that have accumulated DNA damage would presumably be eliminated by apoptosis during acute disease, the intracellular bacterial burden and whether unrepaired genotoxicity is incurred during latent infection are unknown. Given that infectious diseases cause up to 20% of human cancers and O. tsutsugamushi infection can elevate the risk for other serious health conditions later in life1,2,126, it might be worthwhile for longitudinal studies to assess the epidemiological relationship between scrub typhus and subsequent development of cancer.
In summary, O. tsutsugamushi is the first example of a pathogen that lowers p53 host cell levels by downregulating TP53 expression. In the absence of p53, which would be expected to lead to unchecked genotoxicity and cell cycle progression, Orientia modulates both. During its logarithmic replication, it protects against DNA damage, inhibits DNA damage-induced apoptosis, and mediates cell cycle arrest at S phase, the latter of which likely affords the bacterium needed metabolites. The abilities of Orientia to block TP53 expression, mediate genoprotection, and impair DNA damage-dependent apoptosis involve its nucleotropic effector, Ank13. This study furthers understanding of how O. tsutsugamushi and potentially other intracellular pathogens modulate p53 and p53-related pathways to maintain their niches for as long as possible.
Methods
Cultivation of cell lines and O. tsutsugamushi infections
Uninfected HeLa human cervical epithelial cells (CCL-2; American Type Culture Collection [ATCC], Manassas, VA) and EA.hy926 human umbilical vein cells (CRL-2922; ATCC) were maintained as done previously50. HeLa cells infected with O. tsutsugamushi strains Ikeda, UT76, Karp, or Gilliam were maintained as described50,53. To isolate O. tsutsugamushi from infected host cells, media was removed and the cells were washed with 1X phosphate-buffered saline (PBS; 1.05 mM KH2PO4, 155 mM NaCl, 2.96 mM Na2HPO4 [pH 7.4]) before trypsinization. Dislodged cells were pelleted at 1000 x g for 3 min, the supernatant removed, and the pellet resuspended in 1X PBS. The resuspended pellet was combined with 0.7 mm zirconia beads (Biospec Products; Bartlesville, OK), and the host cells were disrupted to release intracellular bacteria using an analog disruptor genie (Scientific Industries, Inc.; Bohemia, New York) for 2 min. Beads and host cell debris were pelleted at 1000 x g for 3 min. The supernatant containing host cell-free O. tsutsugamushi was collected and utilized for continuous cultivation in HeLa cells or for infection studies in EA.hy926 cells53. Synchronous infections were achieved by removing inocula and replacing with fresh complete Dulbecco’s modified Eagle’s medium (DMEM) with l-glutamine, 4.5 g d-glucose and 100 mg sodium pyruvate (Gibco, Gaithersburg, MD) supplemented with 10% (vol/vol) heat-inactivated fetal bovine serum (FBS) (Gemini Bioproducts, West Sacramento, CA), 1X minimal essential medium containing non-essential amino acids (Gibco) and 15 mM HEPES (Gibco) at 2–4 h p.i. Experiments were verified for achieving a multiplicity of infection (MOI) of 5-10 in at least 75% of the host cells by assessing coverslips of infected cells by immunofluorescence as described below. Samples in which less than 75% of the cells were infected were not used.
Genome equivalents (GE) quantification
Lysates for GE were collected by trypsinization followed by dilution in H2O and incubation at 100 °C for 15 min as described previously127. Briefly, at the time of analysis, samples were thawed, vortexed, and GE quantified by qPCR. O. tsutsugamushi str. Ikeda tsa56 gene (OTT_RS04590)53 or tsa47 (47-kDa type-specific antigen) gene (OTT_ RS06370) were detected using gene-specific primers (Supplementary Table 1) with PerfeCTa SYBR Green Fastmix (Quantbio, Beverly, MA), and a CFX384 Real Time PCR Detection System (Bio-Rad Laboratories). Thermal cycling conditions were 95 °C for 30 s followed by 40 cycles of 95 °C for 10 s to 54 °C for 10 s, and a 65 °C–95 °C melt curve. A standard curve was prepared using pCR4.0::Ot_tsa5650 or pCR4.0::Ot_tsa47. pCR4.0::Ot_tsa47 standard plasmid was generated by Azenta Genewiz from Azenta Life Sciences (https://www.genewiz.com/en/Public/Services/Gene-Synthesis/Gene-Synthesis) to include base pairs 354-844 of the tsa47 gene. O. tsutsugamushi GE mL-1 was calculated based on the standard curve as done previously53,127. O. tsutsugamushi yield was calculated by subtracting the GE mL-1 at 72 or 96 h p.i by the GE mL-1 of the O. tsutsugamushi inoculum for the corresponding experiment.
RNA-seq
EA.hy926 cells at 100% confluency were synchronously infected with O. tsutsugamushi. At 8, 48, and 72 h p.i., spent medium was removed and the cells washed with 1X PBS before trypsinization. Dislodged cells were pelleted at 1000 x g for 3 min, and total RNA was extracted using RNeasy mini kit (Qiagen [74104], Hilden, Germany). GE per sample was measured per timepoint. RNA concentration and purity were assessed using a NanoVue Plus Spectrophotometer (GE HealthCare, Chicago, IL) before storage at −80 °C. Three independent biological samples that met the requisite criteria of ≥20 ng/uL, OD260/280 ≥ 2.0, and OD260/230 ≥ 2.0 were submitted to Novogene (Sacramento, CA) where sample integrity was confirmed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA) prior to sequencing. Samples with an RNA integrity number of ≥ 4.0 and no gDNA contamination were sequenced on an Illumina high-throughput sequencing platform (Illumina, San Diego, CA). Spliced Transcripts Alignment to a Reference (STAR) program was employed to map raw reads to human reference genome (GRCh38/hg38)128. DESeq2 R software package 1.14.1 (Bioconducter, https://www.bioconductor.org/) was used to perform differential expression significant analysis between infected and uninfected samples per timepoint129. To control the false discovery rate (FDR) of the resulting P values, the Benjamini and Hochberg’s approach was employed and adjusted P values generated130,131. Genes with an adjusted P value of ≤ 0.05, as well as a log2(fold change) ≥ 1 or ≤ −1 of significantly differentially expressed genes in infected cells compared to the uninfected controls were considered differentially expressed genes (DEGs). DESeq2 R package was used to determine the Pearson’s correlation coefficient and PCAtools R package (Bioconducter) was employed to perform a principal components analysis.
STRING analysis
Interaction networks were defined using STRING132. Briefly, DEGs per timepoint were individually imported into STRING, and unconnected nodes were removed. MCL clustering was performed with a constricting inflation parameter of 2. GO terms sub-grouped into biological processes per cluster were defined and those with a FDR ≤ 0.05 were considered significantly enriched pathways.
qRT-PCR
Total RNA was isolated from uninfected or O. tsutsugamushi infected EA.hy926 cells, and concentration and purity were determined as described above. One µg of RNA was treated with amplification grade DNase I (Invitrogen) following the manufacturer’s protocol. Next, iScript Reverse Transcription Supermix (Bio-Rad) was used to generate cDNA. To ensure gDNA was successfully removed, reactions in the absence or presence of reverse transcriptase were subjected to PCR with MyTaq polymerase (Bioline, Taunton, MA) and primers for human GAPDH133 before being visualized by an agarose gel electrophoresis. gDNA-free cDNA was subjected to qPCR with PerfeCTa SYBR Green Fastmix and primers for TP53, HDM2, TP53 P1, TP53 P2, PMAIP1, CDKN1A, TP53I3, SESN1, ZMAT3 (Supplementary Table 1), GAPDH133, O. tsutsugamushi 16S rDNA (ott16s)133, or ank1349. Thermal cycling conditions used were 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s and 55 °C for 5 s, followed by a melt curve from 60 °C to 95 °C. 2-∆∆CT was calculated to determine the relative expression of each gene50.
Western blotting
Cells were washed with 1X PBS, harvested by trypsinization, pelleted at 1000 xg for 3 min, and resuspended in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 1 mM EDTA [pH 8]) containing Halt protease and phosphatase inhibitor cocktail (ThermoFisher Scientific, Waltham, MA). Suspensions were incubated on ice for 15-45 min and insoluble fractions were pelleted by centrifugation at 16,000 xg for 10 min. Supernatants were collected and stored at −20 °C. Protein concentration was quantified using the Bradford assay (Bio-Rad [#5000006], Hercules, CA). Equivalent amounts of protein were resolved on a 4 to 15% TGX polyacrylamide gels (Bio-Rad) at 110 V for 15 min followed by 200 V from 30 min. Proteins were transferred onto nitrocellulose membrane (Bio-rad; #1620115) in Towbin buffer (24 mM Tris, 192 mM glycine, 20% [vol/vol] methanol) at 100 V for 30 min. Blots were blocked with either 5% (vol/vol) nonfat dry milk or 5% (vol/vol) BSA in Tris-buffered saline plus Tween-20 (TBS-T; 25 mM Tris HCl, 137 mM NaCl, 2.7 mM KCl, 0.05% Tween-20, pH 7.4) for 30-60 min before incubation with primary antibody. Primary antibodies used were: mouse anti-p53 (Santa Cruz [sc-98]; 1:200), rabbit anti-p21 (Cell Signaling [#2947]; 1:1000), mouse anti-HDM2 (Invitrogen [#33-7100]; 1:500), mouse anti-Cyclin E (Santa Cruz, Dallas, TX [sc-377100]; 1:500), mouse anti-Cyclin A (Santa Cruz [sc-271682]; 1:250), mouse anti-Cyclin D3 (Cell Signaling, Danvers, MA [#2936]; 1:1000), rabbit anti-Cyclin D1 (Cell Signaling [#55506]; 1:1000), rabbit anti-Cyclin B1 (Cell Signaling [#4138]; 1:1000), rabbit anti-TCF11/NRF1 (Cell Signaling [#8052]; 1:1000), rabbit anti-GFP (Invitrogen [A11122]; 1:1000), rabbit anti-GMNN (Cell Signaling [#52508]; 1:1000), rabbit anti-cdc25a (Cell Signaling [#3652]; 1:1000), rabbit anti-phosphorylated-Histone H2A.X (ƔH2AX) (Cell Signaling [#2577]; 1:1,1000), rabbit anti-Caspase 7 (Cell Signaling [#9492]; 1:1000), rabbit anti-Caspase 3 (Cell Signaling [#9662]; 1:1000), rabbit anti-PARP1 (Invitrogen [MA5-15031]; 1:1000), rabbit anti-PKR (Cell signaling [#3072; 1:1000), rabbit anti-p-PKR (T446) (Abcam [ab32036]; 1:1000), rabbit anti-O. tsutsugamushi TSA56123 (1:1000), mouse anti-GAPDH (Santa Cruz [sc-365062]; 1:750). Horseradish peroxidase-conjugated horse anti-mouse or anti-rabbit (Cell Signaling Technology [mouse #7076, rabbit #7074]; 1:10,000) were used to detect bound primary antibodies. Blots were incubated with SuperSignal West Pico PLUS, SuperSignal West Dura, or SuperSignal West Femto chemiluminescent substrate (ThermoFisher Scientific) before visualization using the ChemiDoc Touch Imaging System (Bio-Rad). Densitometric values were determined using the Bio-Rad Image Lab 6.0 software.
Immunofluorescent microscopy
EA.hy926 cells were seeded in 24-well plates with 12 mm round glass coverslips (Electron Microscopy Sciences, Hatfield, PA) and synchronously infected with O. tsutsugamushi. The cells were fixed with methanol for 5 min followed by successive incubations with rabbit-anti TSA56123 (1:1,000) for 1 h in 1X PBS containing 5% (vol/vol) BSA and Alexa Fluor 488-conjugated goat anti-rabbit IgG (Invitrogen [A-11034]; 1:1000) at 1:1,000 with the addition of 0.1 mg mL-1 4′,6′-diamidino-2-phenylindole (DAPI; Invitrogen [D1306]). The coverslips were then mounted using ProLong Gold antifade reagent (Invitrogen) and imaged with a TCS SP8 microscope (Leica Microsystems, Germany).
Pharmacologic treatments
Chloramphenicol (CAM; 34 ug/mL) was added to uninfected and O. tsutsugamushi infected EA.hy926 cells at 2 h p.i. EA.hy926 cells were treated with Nutlin 3a (5 or 10 µM)6, C16 (100 nM), (hydroxyurea (HU; 80 µM)80, or RO-3306 (9 µM)134 18-24 h before infection with O. tsutsugamushi, and etoposide (1 µM)87,88,89 or staurosporine (1 µM)94,135 18-24 h prior to collection. Pharmacologic agents were diluted in complete DMEM and remained in culture unless otherwise stated. The effects of each pharmacologic inhibitor and serum starvation on host cell fitness was assessed by the MTT assay as described53. To determine the effects of hydroxyurea and RO-3306 on host cell-free O. tsutsugamushi, the bacteria were treated 30 min prior to being added to untreated EA.hy926 cells followed by GE quantification50,53,127.
Proliferation assay
EA.hy926 cells were seeded to achieve 60–70% confluency and serum starved in DMEM without FBS, HEPES, and non-essential amino acids for 24 h. After serum starvation, cells were labeled with the CellTrace Far Red Proliferation kit (ThermoFisher Scientific [C34564]) following the manufacturer’s protocol. Briefly, the medium was removed, 1 µM of CellTrace Far Red dye diluted in pre-warmed 1X PBS was added, and the cells were incubated at 35 °C in a humidified incuabor with 5% CO2 for 20 min. The dye solution was removed, and cells were washed twice with complete DMEM. Cells were incubated in complete DMEM at 35 °C with 5% CO2 for 10 min prior to O. tsutsugamushi infection. At the time of analysis, cells were dislodged with 0.526 mM EDTA (Versene) solution and the fluorescence intensity of CellTrace Far Red dye in 10,000 individual cells was quantified using a Becton Dickinson (BD) FACSMelody with BDFACSChorus 1.3.3 software (BD Biosciences, Franklin Lakes, NJ). Analysis was performed in FlowJo (version 10.8.1) software (BD Biosciences) with population gates isolating single cell populations. In parallel, immunofluorescence microscopy was performed with the adjustment of serum starvation prior to infection, and percentage of infected host cells at each time point was verified50.
RNAi
EA.hy926 cells grown to 100% confluency were transfected with ON-TARGETplus human Cyclin E (L-003213-00-0005, GE Healthcare Dharmacon, Inc., Lafayette, CO), CDC25A (L-003226-00-0005, GE Healthcare Dharmacon, Inc.), or GMNN (L-003270-00-0005, GE Healthcare Dharmacon, Inc.) SMARTpool siRNA, or non-targeting siRNA (D-001810-10-05, GE Healthcare Dharmacon, Inc.) at a final concentration of 30 pmol following the manufacturer’s protocol. After 48 h, siRNA was replenished, and cells were infected with O. tsutsugamushi. GE was measured for uninfected and O. tsutsugamushi infected samples.
Plasmid constructs
pGFP-Ank13 and GFP-Ank13I127R were generated previously49. In-Fusion Cloning Primer Design Tool v1.0 (www.takarabio.com) was used to designs primers to generate GFP-Ank13-F-boxAAAAA (Supplementary Table 1), which contained the subsequent point mutations, I451A, P452A, E454A, I459A, and D467A. GFP-Ank13-F-boxAAAAA was generated following the TaKaRa Bio USA (San Francisco, CA) In-Fusion Mutagenesis protocol with pGFP-Ank13 as a template49.
Transfections
EA.hy926 cells were grown to 100% confluency before transfection with GFP, GFP-Ank13, GFP-Ank13-F-boxAAAAA, or GFP-Ank13I127R plasmid DNA using Lipofectamine 2000 (Invitrogen) and incubation at 37 °C in a humidified incubator with 5% CO2. At 18-24 h post transfection, samples were washed with 1X PBS, trypsinized, and subjected to fluorescent-activated cell sorting (FACS) for collection of GFP positive cells as done previously50. Total RNA for RT-qPCR or lysates for immunoblot analysis were collected from enriched GFP populations.
Annexin V staining
Uninfected or O. tsutsugamushi-infected cells, or cells expressing GFP, GFP-Ank13, GFP-Ank13∆F-boxAAAAA, or GFP-Ank13I127R were treated with vehicle, etoposide, or staurosporine for 24 h. At the time of collection, uninfected and infected samples were incubated with FITC-Annexin V and propidium iodine (PI), while cells expressing GFP or GFP-Ank13 and mutants were incubated with PE-Annexin and 7-AAD and analyzed by flow cytometry on a BD FACSMelody with BDFACSChorus 1.3.3 software (BD Biosciences) as described51. Compensation and quadrant analysis were performed using FlowJo software (version 10.8.1; BD Biosciences). Cells in quadrant 1 (Q1; PI + /FITC- or 7-AAD + /PE-) and Q2 (PI + /FITC+ or 7-AAD + /PE + ) were considered late apoptotic/dead. Cells in Q3 (PI-/FITC+ or 7-AAD-/PE + ) were deemed early apoptotic while those in Q4 (PI-/FITC- or 7-AAD-/PE-) were considered live. The fold change in the percentage of late apoptotic/dead cells was calculated by subtracting the percentage of late apoptotic/dead cells per each vehicle control-treated sample from that of its etoposide- or staurosporine-treated counterpart and dividing the difference by its respective vehicle control.
Statistical analysis
Statistical analyses were performed using Prism 9.0 (GraphPad, San Diego, CA). Significance between two groups was determined by an unpaired student’s t-test. One-way analysis of variance followed by Tukey’s honest significance or Dunnett’s post hoc tests was used to test for significant differences between groups or each group compared to a control, respectively. Statistical significance was set at P values of ≤ 0.05.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data supporting the findings of this study are available within the paper and its Supplementary Information/Source Data files. Sequencing data has been deposited in GEO with accession number GSE303493. Source data are provided with this paper.
References
Gagnaire, A., Nadel, B., Raoult, D., Neefjes, J. & Gorvel, J. P. Collateral damage: insights into bacterial mechanisms that predispose host cells to cancer. Nat. Rev. Microbiol. 15, 109–128 (2017).
Siegl, C. & Rudel, T. Modulation of p53 during bacterial infections. Nat. Rev. Microbiol. 13, 741–748 (2015).
Lane, D. P. P53, the guardian of the genome. Nature 358, 15–6 (1992).
Miciak, J. & Bunz, F. Long story short: p53 mediates innate immunity. Biochim. Biophys. Acta 1865, 220–227 (2016).
Bergounioux, J. et al. Calpain activation by the Shigella flexneri effector VirA regulates key steps in the formation and life of the bacterium’s epithelial niche. Cell Host Microbe 11, 240–252 (2012).
Gonzalez, E. et al. Chlamydia infection depends on a functional MDM2-p53 axis. Nat. Commun. 5, 5201 (2014).
Groskreutz, D. J. et al. Respiratory syncytial virus decreases p53 protein to prolong survival of airway epithelial cells. J. Immunol. 179, 2741–2747 (2007).
Kaushansky, A. et al. Suppression of host p53 is critical for Plasmodium liver-stage infection. Cell Rep. 3, 630–637 (2013).
Querido, E. et al. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes Dev. 15, 3104–3117 (2001).
Sato, Y. et al. Degradation of phosphorylated p53 by viral protein-ECS E3 ligase complex. PLoS Pathog. 5, e1000530 (2009).
Scheffner, M., Werness, B. A., Huibregtse, J. M., Levine, A. J. & Howley, P. M. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 63, 1129–1136 (1990).
Siegl, C., Prusty, B. K., Karunakaran, K., Wischhusen, J. & Rudel, T. Tumor suppressor p53 alters host cell metabolism to limit Chlamydia trachomatis infection. Cell Rep. 9, 918–929 (2014).
Shin, Y. C. et al. Inhibition of the ATM/p53 signal transduction pathway by Kaposi’s sarcoma-associated herpesvirus interferon regulatory factor 1. J. Virol. 80, 2257–2266 (2006).
Diensthuber, D. et al. Clearing Chlamydia abortus infection in epithelial cells and primary human macrophages by use of antibiotics and the MDM2-p53-inhibitor nutlin-3. Diagn. Microbiol. Infect. Dis. 103, 115715 (2022).
Zou, Y. et al. Chlamydia trachomatis plasmid-encoded protein Pgp3 inhibits apoptosis via the PI3K-AKT-mediated MDM2-p53 axis. Mol. Cell Biochem 452, 167–176 (2019).
Cao, H. et al. The role of MDM2-p53 axis dysfunction in the hepatocellular carcinoma transformation. Cell Death Discov. 6, 53 (2020).
Pal, A., Tripathi, S. K., Rani, P., Rastogi, M. & Das, S. p53 and RNA viruses: the tug of war. Wiley Interdiscip. Rev. RNA 20, e1826 (2023).
Zaika, A. I., Wei, J., Noto, J. M. & Peek, R. M. Microbial regulation of p53 tumor suppressor. PLoS Pathog. 11, e1005099 (2015).
Zamaraev, A. V., Zhivotovsky, B. & Kopeina, G. S. Viral Infections: Negative Regulators of Apoptosis and Oncogenic Factors. Biochem. (Mosc. 85, 1191–1201 (2020).
Friborg, J. Jr., Kong, W., Hottiger, M. O. & Nabel, G. J. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature 402, 889–894 (1999).
Dobbelstein, M. & Roth, J. The large T antigen of simian virus 40 binds and inactivates p53 but not p73. J. Gen. Virol. 79, 3079–3083 (1998).
Ueda, H. et al. Functional inactivation but not structural mutation of p53 causes liver cancer. Nat. Genet. 9, 41–47 (1995).
Haller, D. et al. Cytoplasmic sequestration of p53 promotes survival in leukocytes transformed by Theileria. Oncogene 29, 3079–3086 (2010).
Pise-Masison, C. A. et al. Inactivation of p53 by human T-cell lymphotropic virus type 1 Tax requires activation of the NF-kappaB pathway and is dependent on p53 phosphorylation. Mol. Cell Biol. 20, 3377–3386 (2000).
Li, C. J., Wang, C., Friedman, D. J. & Pardee, A. B. Reciprocal modulations between p53 and Tat of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 92, 5461–5464 (1995).
Majumder, M., Ghosh, A. K., Steele, R., Ray, R. & Ray, R. B. Hepatitis C virus NS5A physically associates with p53 and regulates p21/waf1 gene expression in a p53-dependent manner. J. Virol. 75, 1401–1407 (2001).
Aschtgen, M. S. et al. Enterobacteria impair host p53 tumor suppressor activity through mRNA destabilization. Oncogene 41, 2173–2186 (2022).
Buti, L. et al. Helicobacter pylori cytotoxin-associated gene A (CagA) subverts the apoptosis-stimulating protein of p53 (ASPP2) tumor suppressor pathway of the host. Proc. Natl. Acad. Sci. USA 108, 9238–9243 (2011).
Wei, J. et al. Regulation of p53 tumor suppressor by Helicobacter pylori in gastric epithelial cells. Gastroenterology 139, 1333–1343 (2010).
Wei, J. et al. Pathogenic bacterium Helicobacter pylori alters the expression profile of p53 protein isoforms and p53 response to cellular stresses. Proc. Natl. Acad. Sci. USA 109, E2543–E2550 (2012).
Gunasekaran, K., Bal, D. & Varghese, G. M. Scrub Typhus and Other Rickettsial Infections. Indian J. Crit. Care Med. 25, S138–S143 (2021).
Richards, A. L. & Jiang, J. Scrub typhus: historic perspective and current status of the worldwide presence of orientia species. Trop. Med. Infect. Dis. 5, 49 (2020).
Blair, P. W. et al. Evidence of orientia spp. Endemicity among severe infectious disease cohorts, Uganda. Emerg. Infect. Dis. 30, 1442–1446 (2024).
Oakley, R. et al. Seroprevalence of Rickettsia Spp. and Orientia tsutsugamushi in Indigenous Populations from the Sierra Nevada de Santa Marta, Colombia. Vector Borne Zoonotic Dis. 24, 641–648 (2024).
Faccini-Martínez, Á et al. Serologic evidence of Orientia infection among rural population, Cauca Department, Colombia. Emerg. Infect. Dis. 29, 456–459 (2023).
Weitzel, T., Martínez-Valdebenito, C., Acosta-Jamett, G. & Abarca, K. Notes from the Field: scrub typhus outbreak - Los Lagos Region, Chile, January-February 2023. MMWR Morb. Mortal. Wkly Rep. 72, 753–754 (2023).
Weitzel, T. et al. Novel vector of scrub typhus in Sub-Antarctic Chile: evidence from human exposure. Clin. Infect. Dis. 74, 1862–1865 (2022).
Chen, K. et al. Detection of orientia spp. Bacteria in field-collected free-living eutrombicula chigger mites, United States. Emerg. Infect. Dis. 29, 1676–1679 (2023).
Richardson, E. A. et al. Orientia, Rickettsia, and the microbiome in rodent attached chiggers in North Carolina, USA. PLoS One 19, e0311698 (2024).
Saraswati, K., Day, N. P. J., Mukaka, M. & Blacksell, S. D. Scrub typhus point-of-care testing: a systematic review and meta-analysis. PLoS Negl. Trop. Dis. 12, e0006330 (2018).
Walker, D. H. & Mendell, N. L. A scrub typhus vaccine presents a challenging unmet need. NPJ Vaccines 8, 11 (2023).
Bhandari, M. et al. Revisiting scrub typhus: a neglected tropical disease. Comp. Immunol. Microbiol Infect. Dis. 90-91, 101888 (2022).
Shelite, T. R. et al. IL-33-dependent endothelial activation contributes to apoptosis and renal injury in orientia tsutsugamushi infected mice. PLoS Negl. Trop. Dis. 10, e0004467 (2016).
Shelite, T. R. et al. Hematogenously disseminated Orientia tsutsugamushi infected murine model of scrub typhus [corrected]. PLoS Negl. Trop. Dis. 8, e2966 (2014).
Sonthayanon, P. et al. Association of high Orientia tsutsugamushi DNA loads with disease of greater severity in adults with scrub typhus. J. Clin. Microbiol. 47, 430–434 (2009).
Soong, L. Dysregulated Th1 Immune And Vascular Responses In Scrub Typhus Pathogenesis. J. Immunol. 200, 1233–1240 (2018).
Trent, B. et al. Polarized lung inflammation and Tie2/angiopoietin-mediated endothelial dysfunction during severe Orientia tsutsugamushi infection. PLoS Negl. Trop. Dis. 14, e0007675 (2020).
Soong, L. et al. Strong type 1, but impaired type 2, immune responses contribute to Orientia tsutsugamushi-induced pathology in mice. PLoS Negl. Trop. Dis. 8, e3191 (2014).
Adcox, H. E. et al. Orientia tsutsugamushi nucleomodulin Ank13 Exploits the RaDAR nuclear import pathway to modulate host cell transcription. mBio 12, e0181621 (2021).
Adcox, H. E. et al. Orientia tsutsugamushi Ank5 promotes NLRC5 cytoplasmic retention and degradation to inhibit MHC class I expression. Nat. Commun. 15, 8069 (2024).
Allen, P. E. et al. Orientia tsutsugamushi alters the intranuclear balance of cullin-1 and c-MYC to inhibit apoptosis. Infect. Immun. 93, e0055924 (2025).
Kim, M. K. et al. Apoptosis of endothelial cell line ECV304 persistently infected with Orientia tsutsugamushi. Microbiol Immunol. 43, 751–757 (1999).
Sanchez, S. E., Chiarelli, T. J., Park, M. A. & Carlyon, J. A. Orientia tsutsugamushi infection reduces host gluconeogenic but not glycolytic substrates. Infect. Immun. 92, e0028424 (2024).
Atwal, S. et al. The obligate intracellular bacterium Orientia tsutsugamushi differentiates into a developmentally distinct extracellular state. Nat. Commun. 13, 3603 (2022).
Kee, S. H. et al. Disassembly of focal adhesions during apoptosis of endothelial cell line ECV304 infected with Orientia tsutsugamushi. Micro Pathog. 27, 265–271 (1999).
Nakayama, K. et al. The Whole-genome sequencing of the obligate intracellular bacterium Orientia tsutsugamushi revealed massive gene amplification during reductive genome evolution. DNA Res. 15, 185–199 (2008).
Tamura, A. et al. Isolation of Rickettsia tsutsugamushi antigenically different from Kato, Karp, and Gilliam strains from patients. Microbiol. Immunol. 28, 873–882 (1984).
Varghese, G. M. et al. Molecular epidemiology and genetic diversity of Orientia tsutsugamushi from patients with scrub typhus in 3 regions of India. Emerg. Infect. Dis. 21, 64–69 (2015).
Park, H. et al. Case report: fulminant myocarditis successfully treated with extracorporeal membrane oxygenation in ikeda strain orientia tsutsugamushi infection. Front. Cardiovasc. Med. 8, 795249 (2021).
Ohashi, N. et al. Demonstration of antigenic and genotypic variation in Orientia tsutsugamushi which were isolated in Japan, and their classification into type and subtype. Microbiol. Immunol. 40, 627–638 (1996).
Fromm, L., Mehl, J. & Keller, C. Orientia tsutsugamushi: a life between escapes. Microbiologyopen 12, e1380 (2023).
Cianci, F. & Verduci, I. Transmembrane chloride intracellular channel 1 (tmCLIC1) as a potential biomarker for personalized medicine. J. Pers. Med. 11, 635 (2021).
Mahauad-Fernandez, W. D. & Okeoma, C. M. The role of BST-2/Tetherin in host protection and disease manifestation. Immun. Inflamm. Dis. 4, 4–23 (2016).
Betancor, G. You shall not pass: MX2 proteins are versatile viral inhibitors. Vaccines 11, 930 (2023).
Wang, B., Niu, D., Lam, T. H., Xiao, Z. & Ren, E. C. Mapping the p53 transcriptome universe using p53 natural polymorphs. Cell Death Differ. 21, 521–532 (2014).
Kastenhuber, E. R. & Lowe, S. W. Putting p53 in Context. Cell 170, 1062–1078 (2017).
Laptenko, O. & Prives, C. Transcriptional regulation by p53: one protein, many possibilities. Cell Death Differ. 13, 951–961 (2006).
Patil, M. R. & Bihari, A. A comprehensive study of p53 protein. J. Cell Biochem. 123, 1891–1937 (2022).
Royds, J. A. & Iacopetta, B. p53 and disease: when the guardian angel fails. Cell Death Differ. 13, 1017–1026 (2006).
Gomez-Manzano, C. et al. Characterization of p53 and p21 functional interactions in glioma cells en route to apoptosis. J. Natl. Cancer Inst. 89, 1036–1044 (1997).
Shen, H. & Maki, C. G. Pharmacologic activation of p53 by small-molecule MDM2 antagonists. Curr. Pharm. Des. 17, 560–568 (2011).
Radhakrishnan, S. K. et al. Transcription factor Nrf1 mediates the proteasome recovery pathway after proteasome inhibition in mammalian cells. Mol. Cell 38, 17–28 (2010).
Marques, J. T. et al. Down-regulation of p53 by double-stranded RNA modulates the antiviral response. J. Virol. 79, 11105–11114 (2005).
Mitchell, J. K. et al. Hepatitis C virus indirectly disrupts DNA damage-Induced p53 responses by activating protein kinase R. mBio 8, e00121 (2017).
Cesaro, T. et al. PKR activity modulation by phosphomimetic mutations of serine residues located three aminoacids upstream of double-stranded RNA binding motifs. Sci. Rep. 11, 9188 (2021).
Hashimoto, Y. et al. C16, a PKR inhibitor, suppresses cell proliferation by regulating the cell cycle via p21 in colorectal cancer. Sci. Rep. 14, 9029 (2024).
Guo, Y., Wu, H., Wiesmüller, L. & Chen, M. Canonical and non-canonical functions of p53 isoforms: potentiating the complexity of tumor development and therapy resistance. Cell Death Dis. 15, 412 (2024).
Wee, P., Wang, R. C. & Wang, Z. Synchronization of HeLa cells to various interphases including G1, S, and G2 phases. Methods Mol. Biol. 2579, 87–97 (2022).
Musiałek, M. W. & Rybaczek, D. Hydroxyurea-the good, the bad and the ugly. Genes 12, 1096 (2021).
Ly, T., Endo, A. & Lamond, A. I. Proteomic analysis of the response to cell cycle arrests in human myeloid leukemia cells. Elife 4, e04534 (2015).
Ghasemi, M., Turnbull, T., Sebastian, S. & Kempson, I. The MTT assay: utility, limitations, pitfalls, and interpretation in bulk and single-cell analysis. Int. J. Mol. Sci. 22, 12827 (2021).
Montemurro, C. et al. Cell cycle-related metabolism and mitochondrial dynamics in a replication-competent pancreatic beta-cell line. Cell Cycle 16, 2086–2099 (2017).
de Jesús-Díaz, D. A., Murphy, C., Sol, A., Dorer, M. & Isberg, R. R. Host cell S phase restricts legionella pneumophila intracellular replication by destabilizing the membrane-bound replication compartment. mBio 8, e02345 (2017).
Meek, D. W. Tumour suppression by p53: a role for the DNA damage response? Nat. Rev. Cancer 9, 714–723 (2009).
Fragkos, M., Jurvansuu, J. & Beard, P. H2AX is required for cell cycle arrest via the p53/p21 pathway. Mol. Cell Biol. 29, 2828–2840 (2009).
Sharma, A., Singh, K. & Almasan, A. Histone H2AX phosphorylation: a marker for DNA damage. Methods Mol. Biol. 920, 613–626 (2012).
Montecucco, A., Zanetta, F. & Biamonti, G. Molecular mechanisms of etoposide. Excli J. 14, 95–108 (2015).
Jamil, S., Lam, I., Majd, M., Tsai, S. H. & Duronio, V. Etoposide induces cell death via mitochondrial-dependent actions of p53. Cancer Cell Int. 15, 79 (2015).
Karpinich, N. O., Tafani, M., Rothman, R. J., Russo, M. A. & Farber, J. L. The course of etoposide-induced apoptosis from damage to DNA and p53 activation to mitochondrial release of cytochrome c. J. Biol. Chem. 277, 16547–16552 (2002).
Mijit, M., Caracciolo, V., Melillo, A., Amicarelli, F. & Giordano, A. Role of p53 in the regulation of cellular senescence. Biomolecules 10, 420 (2020).
Reinhardt, H. C. & Schumacher, B. The p53 network: cellular and systemic DNA damage responses in aging and cancer. Trends Genet. 28, 128–136 (2012).
Rufini, A., Tucci, P., Celardo, I. & Melino, G. Senescence and aging: the critical roles of p53. Oncogene 32, 5129–5143 (2013).
Lakhani, S. A. et al. Caspases 3 and 7: key mediators of mitochondrial events of apoptosis. Science 311, 847–851 (2006).
Godard, T. et al. Early detection of staurosporine-induced apoptosis by comet and annexin V assays. Histochem. Cell Biol. 112, 155–161 (1999).
Wang, P., Yu, J. & Zhang, L. The nuclear function of p53 is required for PUMA-mediated apoptosis induced by DNA damage. Proc. Natl. Acad. Sci. USA 104, 4054–4059 (2007).
Nicolier, M., Decrion-Barthod, A. Z., Launay, S., Prétet, J. L. & Mougin, C. Spatiotemporal activation of caspase-dependent and -independent pathways in staurosporine-induced apoptosis of p53wt and p53mt human cervical carcinoma cells. Biol. Cell 101, 455–467 (2009).
Giengkam, S. et al. Orientia tsutsugamushi- comprehensive analysis of the mobilome of a highly fragmented and repetitive genome reveals the capacity for ongoing lateral gene transfer in an obligate intracellular bacterium. ASM mSphere 8, e0026823 (2023).
Gupta, S. et al. Functional charaterization of non-ankyrin repeat domains of orientia tsutsugamushi ank effectors reveals their importance for molecular pathogenesis. ASM Infect. Immun. 90, e0062821 (2022).
Jernigan, K. K. & Bordenstein, S. R. Ankyrin domains across the Tree of Life. PeerJ 2, e264 (2014).
VieBrock, L. et al. Orientia tsutsugamushi ankyrin repeat-containing protein family members are Type 1 secretion system substrates that traffic to the host cell endoplasmic reticulum. Front. Cell Infect. Microbiol. 4, 186 (2014).
Herbert, M. H., Squire, C. J. & Mercer, A. A. Poxviral ankyrin proteins. Viruses 7, 709–738 (2015).
Noroy, C. & Meyer, D. F. The super repertoire of type IV effectors in the pangenome of Ehrlichia spp. provides insights into host-specificity and pathogenesis. PLoS Comput. Biol. 17, e1008788 (2021).
Evans, S. M., Rodino, K. G., Adcox, H. E. & Carlyon, J. A. Orientia tsutsugamushi uses two Ank effectors to modulate NF-κB p65 nuclear transport and inhibit NF-κB transcriptional activation. PLoS Pathog. 14, e1007023 (2018).
McClure, E. E. et al. Engineering of obligate intracellular bacteria: progress, challenges and paradigms. Nat. Rev. Microbiol. 15, 544–558 (2017).
Beyer, A. R. et al. Orientia tsutsugamushi strain ikeda ankyrin repeat-containing proteins recruit SCF1 ubiquitin ligase machinery via poxvirus-like f-box motifs. J. Bacteriol. 197, 3097–3109 (2015).
Kim, M. K. et al. Orientia tsutsugamushi inhibits apoptosis of macrophages by retarding intracellular calcium release. Infect. Immun. 70, 4692–4696 (2002).
Kloc, M., Kubiak, J. Z., Li, X. C. & Ghobrial, R. M. The newly found functions of MTOC in immunological response. J. Leukoc. Biol. 95, 417–430 (2014).
Icard, P. & Simula, L. Metabolic oscillations during cell-cycle progression. Trends Endocrinol. Metab. 33, 447–450 (2022).
Inthawong, M. et al. A time-course comparative clinical and immune response evaluation study between the human pathogenic Orientia tsutsugamushi strains: Karp and Gilliam in a rhesus macaque (Macaca mulatta) model. PLoS Negl. Trop. Dis. 16, e0010611 (2022).
Koo, J. E., Hong, H. J., Dearth, A., Kobayashi, K. S. & Koh, Y. S. Intracellular invasion of Orientia tsutsugamushi activates inflammasome in asc-dependent manner. PLoS One 7, e39042 (2012).
Min, C. K. et al. A type I interferon and IL-10 induced by orientia tsutsugamushi infection suppresses antigen-specific T cells and their memory responses. Front. Immunol. 9, 2022 (2018).
Lin, C. et al. The ISG15-protease USP18 is a pleiotropic enhancer of HIV-1 replication. Viruses 16, 485 (2024).
Munch, C. C. et al. Multiple orientia clusters and Th1-skewed chemokine profile: a cross-sectional study in patients with scrub typhus from Nepal. Int J. Infect. Dis. 128, 78–87 (2023).
Xu, G. et al. CD8+ T cells provide immune protection against murine disseminated endotheliotropic Orientia tsutsugamushi infection. PLoS Negl. Trop. Dis. 11, e0005763 (2017).
Carra, G., Lingua, M. F., Maffeo, B., Taulli, R. & Morotti, A. P53 vs NF-kappaB: the role of nuclear factor-kappa B in the regulation of p53 activity and vice versa. Cell Mol. Life Sci. 77, 4449–4458 (2020).
Wangsanut, T., Brann, K. R., Adcox, H. E. & Carlyon, J. A. Orientia tsutsugamushi modulates cellular levels of NF-κB inhibitor p105. PLoS Negl. Trop. Dis. 15, e0009339 (2021).
Fontana, M. F. et al. Secreted bacterial effectors that inhibit host protein synthesis are critical for induction of the innate immune response to virulent Legionella pneumophila. PLoS Pathog. 7, e1001289 (2011).
Hempstead, A. D. & Isberg, R. R. Inhibition of host cell translation elongation by Legionella pneumophila blocks the host cell unfolded protein response. Proc. Natl. Acad. Sci. USA 112, E6790–E6797 (2015).
Lemaitre, B. & Girardin, S. E. Translation inhibition and metabolic stress pathways in the host response to bacterial pathogens. Nat. Rev. Microbiol 11, 365–369 (2013).
Mohr, I. & Sonenberg, N. Host translation at the nexus of infection and immunity. Cell Host Microbe 12, 470–483 (2012).
Bang, S. et al. Inhibition of eukaryotic translation by tetratricopeptide-repeat proteins of Orientia tsutsugamushi. J. Microbiol. 54, 136–144 (2016).
Rodino, K. G. et al. Orientia tsutsugamushi modulates endoplasmic reticulum-associated degradation to benefit its growth. Infect. Immun. 86, e00596 (2018).
Beyer, A. R. et al. Orientia tsutsugamushi Ank9 is a multifunctional effector that utilizes a novel GRIP-like Golgi localization domain for Golgi-to-endoplasmic reticulum trafficking and interacts with host COPB2. Cell Microbiol. 19, 12727 (2017).
Chung, M. H., Lee, J. S., Baek, J. H., Kim, M. & Kang, J. S. Persistence of Orientia tsutsugamushi in humans. J. Korean Med. Sci. 27, 231–235 (2012).
Im, J. H. et al. A case series of possibly recrudescent Orientia tsutsugamushi infection presenting as pneumonia. Jpn. J. Infect. Dis. 67, 122–126 (2014).
Chung, W. S., Lin, C. L., Hsu, W. H. & Kao, C. H. Scrub typhus increases the risk of developing acute coronary syndrome: a nationwide cohort study. Heart 100, 1844–1850 (2014).
Hart, T. M. et al. An atlas of human vector-borne microbe interactions reveals pathogenicity mechanisms. Cell 187, 4113–4127 e4113 (2024).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Anders, S. et al. Count-based differential expression analysis of RNA sequencing data using R and bioconductor. Nat. Protoc. 8, 1765–1786 (2013).
Benjamini, Y. & Hochberg, Y. On the adaptive control of the false discovery rate in multiple testing with independent statistics. J. Educ. Behav. Stat. 25, 60–83 (2000).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate- a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300 (1995).
Szklarczyk, D. et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 51, D638–D646 (2023).
Rodino, K. G. et al. The obligate intracellular bacterium Orientia tsutsugamushi targets NLRC5 to modulate the major histocompatibility complex class I pathway. Infect. Immun. 87, e00876 (2019).
Vassilev, L. T. et al. Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc. Natl. Acad. Sci. USA 103, 10660–10665 (2006).
Antonsson, A. & Persson, J. L. Induction of apoptosis by staurosporine involves the inhibition of expression of the major cell cycle proteins at the G(2)/m checkpoint accompanied by alterations in Erk and Akt kinase activities. Anticancer Res. 29, 2893–2898 (2009).
Acknowledgements
We thank Dr. Haley E. Adcox, Dr. Hanna J. Laukaitis-Yousey, Dr. E. Handly Mayton, and Jason R. Hunt for critical review of this manuscript. This work was supported by National Institutes of Health-National Institute of Allergy and Infectious Diseases (www.niaid.gov) grants 1R01 AI167857 to J.A.C. and 1F32 AI174656A1 to P.E.A.
Author information
Authors and Affiliations
Contributions
Conceptualization, P.E.A. and J.A.C.; investigation, P.E.A., D.L.A., S.B.; writing – original draft, P.E.A. and J.A.C.; writing – review and editing, P.E.A. and J.A.C.; supervision, J.A.C.; acquisition of grant support, J.A.C. and P.E.A.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Patrick Woida and the other, anonymous, reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Allen, P.E., Armistead, D.L., Blinova, S. et al. Orientia tsutsugamushi modulates p53, the cell cycle, and genotoxicity to maintain its intracellular niche. Nat Commun 16, 7728 (2025). https://doi.org/10.1038/s41467-025-63149-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-63149-z