Abstract
TANK-Binding Kinase 1 (TBK1) is involved in autophagy and immune signaling. Dominant loss-of-function mutations in TBK1 have been linked to Amyotrophic Lateral Sclerosis (ALS), Fronto-temporal dementia (FTD), and ALS/FTD. However, pathogenic mechanisms remain unclear, particularly the cell-type specific disease contributions of TBK1 mutations. Here, we show that deleting Tbk1 from mouse motor neurons does not induce transcriptional stress, despite lifelong signs of autophagy deregulations. Conversely, Tbk1 deletion in microglia alters their homeostasis and reactive responses. In both spinal cord and brain, Tbk1 deletion leads to a pro-inflammatory, primed microglial signature with features of ageing and neurodegeneration. While it does not induce or modify ALS-like motor neuron damage, microglial Tbk1 deletion is sufficient to cause early FTD-like social recognition deficits. This phenotype is linked to focal microglial activation and T cell infiltration in the substantia nigra pars reticulata and pallidum. Our results reveal that part of TBK1-linked FTD disease originates from microglial dysfunction.
Similar content being viewed by others
Introduction
The TANK-Binding Kinase 1 (TBK1) is a ubiquitously expressed serine/threonine protein kinase, known for its role in autophagy, as it phosphorylates the autophagy receptors optineurin and p621, and for its key role in immune signaling, especially through the type-I interferon pathway1. Genetic mutations in TBK1 have been associated with many diseases. TBK1 duplications account for certain forms of normal tension glaucoma2. TBK1 homozygous loss-of-function mutations induce severe systemic autoinflammation3 driven by RIPK1-dependent cell death, as TBK1 is also a RIPK1 inhibitor4; while TBK1 heterozygous loss-of-function mutations cause Amyotrophic Lateral Sclerosis (ALS), Fronto-Temporal Dementia (FTD), and combined ALS/FTD5.
ALS and FTD represent extremes of a disease spectrum with shared pathological mechanisms, that share genetic causes including mutations in C9orf72, TARDBP, FUS, and TBK16. While ALS leads to progressive muscle paralysis and motor neuron (MN) death7, FTD is a major cause of dementia with deterioration of behavior and language6. The pathological hallmarks of most ALS cases, including TBK1-linked ALS/FTD, are neuronal cytoplasmic TDP-43 inclusions, which are also associated with p62 aggregates8, suggesting defects in protein homeostasis, including autophagy, to be part of cell-autonomous disease mechanisms in affected neurons. Microglial-driven neuroinflammation is also present around affected neurons9, and microglial cells were shown to contribute to neurodegeneration through non-cell-autonomous mechanisms, in studies mostly based on mutant SOD1 driven ALS mouse models10.
In mice, depending on the genetic background, systemic Tbk1 deletion leads to embryonic lethality due to necrotic liver failure11 or autoinflammation12. We have previously shown that heterozygous ubiquitous Tbk1 deletion is not sufficient to produce ALS/FTD-like neurodegeneration in mice13. Thus, to study the role of TBK1 in the CNS and its links to ALS/FTD, and to avoid deleterious systemic effects, cell-type specific deletion of Tbk1 is needed in mice.
With this study, we aimed to understand how loss of TBK1 leads to ALS and ALS/FTD by focusing on cell-specific disease contributions of Tbk1 loss in MN and in microglial cells. We focused on MN, as their demise is the cause for paralysis in ALS and TBK1 loss could contribute to neurodegeneration due to autophagy defects; and on microglial cells, as their central role in neuroinflammation makes them a cell-type important to the entire ALS/FTD disease spectrum. With the key role of TBK1 in immune cells, TBK1 loss could have major consequences in microglial cells and directly contribute to TBK1-linked ALS/FTD. While homozygous deletion of Tbk1 in MN has been reported as not sufficient to induce MN death in adult mice14, it was not assessed whether such MN already show signs of stress or develop them with ageing. Likewise, while the importance of TBK1 has been shown for different immune cell-types outside the CNS15,16, very little is known about the roles of TBK1 in microglial cells and deciphering the role of TBK1 in microglia could benefit the general understanding of ALS/FTD pathogenesis.
Thus, using mice, we assessed the consequences of deleting Tbk1 in either MN or microglia, and the relative importance of Tbk1 loss-of-function in the two cell-types with respect to ALS/FTD. We found that MN were resistant to Tbk1 deletion despite signs of autophagy deregulations. In contrast, Tbk1 deletion in microglia affected their biology in vitro and in vivo, and while it did not lead to ALS-like motor defects, it caused an FTD-like cognitive deficit. The phenotype was linked to an early shift of microglia to a pro-inflammatory, aged-like signature, and focal microglial activation in specific brain regions linked to behavior. We also found roles of TBK1 in defining homeostatic microglial density and restricting aged-like microglial priming. Our study provides evidence for an implication of microglia in TBK1-mediated FTD and suggests that part of the disease has its origin in microglial defects.
Results
Spinal cord motor neurons are resistant to Tbk1 loss despite lifelong p62 accumulations
To test if lifelong Tbk1 deletion in motor neurons (MN) only is sufficient to lead to ALS-like defects, we used Tbk1fl/fl and ChAT-Cre mice to delete both copies of Tbk1 in spinal cord MN (hereafter Tbk1-MN-KO mice) and compared to littermate control mice (Tbk1-MN-WT). Of note, the ChAT-Cre line used, targets >90% of spinal MN17 and we confirmed highly efficient Tbk1 deletion in MN (Supplementary Fig. 1a).
Ageing of Tbk1-MN-KO mice to 20 months did not result in an ALS-like motor phenotype (Supplementary Fig. 1b), and no MN loss was detected (Fig. 1a and Supplementary Fig. 1c), consistent with an earlier study that assessed this up to 10 months14.
a Quantification of motor neurons (MN) in the lumbar spinal cord of 20-month-old mice with MN specific full Tbk1 deletion (Chat-Cre/Tbk1fl/fl, hereafter Tbk1-MN-KO) compared to littermate control (Tbk1-MN-WT) mice. n = 4 mice per genotype (equally sex-mixed). Adjusted p value is indicated and was determined using a two-tailed unpaired t-test. b Representative confocal immunofluorescence images of the autophagy receptor p62 and the MN marker ChAT in lumbar spinal cord sections from 3-month-old Tbk1-MN-KO and Tbk1-MN-WT mice, showing MN with p62+ accumulations (puncta) only in Tbk1-MN-KO mice (arrowheads). Scale bar = 50 µm, scale bar in enlargements 10 µm. c Quantification of MN containing p62+ accumulations at 3 months (13.6%) and 20 months (26.1%). n = 4 mice per genotype and age (equally sex-mixed). Adjusted p values are indicated and were determined using a two-way ANOVA with uncorrected Fisher LSD. d Representative brightfield images of laser-microdissection of MN (for RNAseq), identified by Nissl staining, from mouse lumbar spinal cord sections. Scale bar = 100 µm. e Volcano plot and bar graph of significantly deregulated genes (DEGs) (DESeq2, log2FC ≥ 0.5, adjusted p values (FDR) < 0.05, see “Methods”) identified by RNAseq from laser-microdissected MN (pools of 200 MN per mouse), between 20-month-old Tbk1-MN-KO and Tbk1-MN-WT mice, showing beside expected Tbk1 downregulation only very few DEGs. n = 6 mice per genotype (females only). FC fold change, p.adj adjusted p value, UP upregulated, DOWN downregulated, nc not changed. f Acute MN injury, using the unilateral sciatic nerve crush/regeneration paradigm, was performed in 6-month-old Tbk1-MN-KO and Tbk1-MN-WT mice. Recovery of the sciatic functional index (SFI) was calculated based on toe-spread and paw print length, over 4 weeks after crush. n = 7–8 mice per genotype (females only). Adjusted p values were determined using repeated measures one-way ANOVA with Sidak’s multiple comparisons test. a, c Data are shown as single data points and (a, c, f) means ± SEM. ns not significant.
Next, we tested if Tbk1 deletion would lead to signs of autophagy deregulation by focusing on the autophagy receptor p62 (Sqstm1) as a TBK1 target. We found that already in young (3-month-old) Tbk1-MN-KO mice, 14% of MN exhibited round cytoplasmic p62+ accumulations (puncta), which further increased to 26% at 20 months of age, and almost none were present in MN of littermate control mice (Fig. 1b, c). While ChAT-Cre targets all cholinergic neurons17, in the spinal cord p62+ accumulations were only found in large ventral horn MN (Fig. 1b, c). Of note, p62+ accumulations have also been detected in iPSC-derived MN from TBK1-linked ALS patients18. To better define MN with p62+ accumulations, we used Osteopontin/SPP1 (Spp1) and MMP9 (Mmp9) to mark ALS resistant and vulnerable MN, respectively19. Compared to either SPP1+ or MMP9+ MN alone (representing 27% and 55% of all MN, respectively), p62+ accumulations were twofold enriched in the small subpopulation of SPP1+/MMP9+ MN (representing 18% of all MN, but comprising 36% of all MN with p62+ accumulations) (Supplementary Fig. 1d, e), which have been linked to remodeled MN during ageing and in ALS mice19.
We further confirmed our findings with a second MN specific Cre line (VAChT-cre). Consistent with this line only targeting 50% of all spinal MN20 (Supplementary Fig. 1f), we found p62+ accumulations in 8% of MN in 3-month-old Tbk1-MN-KO mice (VAChT-Cre × Tbk1fl/fl) (Supplementary Fig. 1g), but again without any MN death (Supplementary Fig. 1h).
Most of these p62+ accumulations (puncta) were also positive for the autophagy marker GABARAPL121, while half were ubiquitin positive (Supplementary Fig. 2a–c). STED (super-resolution) microscopy resolved the p62+ puncta into a spherical (1 μm wide) p62+ staining around a p62− vesicle-like core, consistent with large potentially stalled autophagosomes, as detected by EM (Supplementary Fig. 2d). We also found some p62+ puncta to resolve to p62+ staining around Cytochrome c+ mitochondria or to be associated with lysosomal LAMP1 (Supplementary Fig. 2e). Together, this suggests a deregulated autophagic flux.
However, on a histological level, this was not sufficient to induce overall signs of neuronal stress, and MN with p62+ accumulations had normal neurofilament (NF-L/NF-H) content and were not positive for phospho-cJUN, a stress-induced transcription factor (Supplementary Fig. 2f, g). Likewise, no overall signs of local microglial activation were present around MN with p62+ accumulations (Supplementary Fig. 2h–j).
To investigate long-term consequences (stress responses) of Tbk1 deletion in MN, we performed RNA sequencing (RNAseq) of laser-microdissected lumbar spinal cord MN from 20-month-old Tbk1-MN-KO mice (ChAT-Cre × Tbk1fl/fl) compared to control littermates (Fig. 1d, e). This only revealed 6 genes to be significantly changed besides the expected Tbk1 loss (Fig. 1d, e and Supplementary Data 1). Lastly, to test if an external stress would demask a vulnerability, we performed a sciatic nerve crush, but did not detect changes in overall speed of MN regeneration between Tbk1-MN-KO and littermate control mice (Fig. 1f).
Thus, despite lifelong signs of autophagy deregulations, spinal MN in mice are remarkably resistant to Tbk1 loss, without signs of cell-autonomous neurodegeneration or increased vulnerability.
Deletion of Tbk1 in microglia in vitro leads to stimulus-dependent altered responses
With the dual role of TBK1 in autophagy and immune signaling, we next tested if Tbk1 loss has a direct effect in microglial cells.
We used Tbk1fl/fl and Cx3cr1-Cre (MMRRC #036395-UCD) mice22,23 to delete 0, 1, or 2 Tbk1 copies selectively in microglia (Tbk1-µG-WT, Tbk1-µG-HET, Tbk1-µG-KO), and confirmed efficient Tbk1 deletion in primary microglial cells isolated from neonatal brains (Fig. 2a and Supplementary Fig. 3a–d). Of note, microglial yields and purity were unaffected by Tbk1 deletion (Supplementary Fig. 3e–h). Cells were stimulated for 24 h with pro-inflammatory lipopolysaccharides (LPS) or anti-inflammatory interleukin 4 (IL4), and RNAseq was performed (Fig. 2a and Supplementary Data 2).
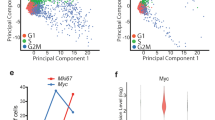
a Setup for transcriptional analysis of neonatal mouse microglia in vitro. Microglia are isolated from P0 mouse brains with microglial specific full Tbk1 deletion (BAC-Cx3cr1-CreMMRRC/Tbk1fl/fl, called Tbk1-µG-KO) and compared to microglia from littermates with only one Tbk1 copy deleted (Tbk1-µG-HET) or control littermates (Tbk1-µG-WT). Microglia are treated with pro- (LPS, poly I:C) or anti-inflammatory (IL4) stimulants, or not treated (unstimulated, Control), followed by RNAseq analysis. Note, cultures are >95% pure IBA1+ microglia (see Supplementary Fig. 3f, g). Created in BioRender. Lenoel, I. (2025) https://BioRender.com/ct8qnd6. b RNAseq results showing the number of genes significantly regulated (DESeq2, log2FC ≥ 1.0, adjusted p values (FDR) < 0.05, see “Methods”) in Tbk1-µG-KO vs Tbk1-µG-WT microglial cultures after no stimulation (Control), LPS, poly I:C, or IL4 stimulation. n = 5 independent experiments. c, e Principal component analysis (PCA) of RNAseq data of Tbk1-µG-KO, Tbk1-µG-HET, and Tbk1-µG-WT microglia after LPS (c) or poly I:C stimulation (e). d, f Gene set enrichment analysis (GSEA) showing regulation of immune and other (Hallmark) pathways between Tbk1-µG-KO and Tbk1-µG-WT microglial cells, after LPS (d) or poly I:C stimulation (f) (adjusted p values < 0.05, NES normalized enrichment scores specified for each pathway and ≥ ±1.5, see “Methods”). Note, for LPS/Tbk1-µG-KO, all inflammatory pathways found were downregulated (dampened), and the 4 shown are among the top downregulated pathways. For poly I:C/Tbk1-µG-KO, all inflammatory pathways found were upregulated (overactivated), and the 4 shown are among the top upregulated pathways. Effect of each stimulus on each pathway in the control condition is represented by a red up arrow (↑) for pathways activated in Tbk1-µG-WT LPS vs unstimulated microglia (see Supplementary Fig. 3k), and a black equal sign (=) for unaltered pathways.
LPS and IL4 led to the expected responses in WT microglia (Supplementary Fig. 3i–l). However, at baseline, no differences were detectable between untreated WT (Tbk1-µG-WT) and KO (Tbk1-µG-KO) microglia (Fig. 2b), and only very few in the IL4 response between KO and WT microglia (Fig. 2b).
In contrast, we detected robust differences in the LPS response between KO and WT microglia (Fig. 2b). Principal component analysis (PCA) also clearly separated KO from WT samples, with HET (Tbk1-µG-HET) samples showing a trend only (Fig. 2c). Unsupervised pathway analysis revealed that Tbk1 deletion led to an overall dampened inflammatory response after LPS stimulation (Fig. 2d). Among the genes with the strongest block of LPS-induction in KO microglia was the pro-inflammatory Nos2, and we confirmed complete lack of nitric oxide (NO) production (Supplementary Fig. 3m, n). In contrast, we did not measure a robust difference in overall phagocytic capacity between WT and KO microglia (Supplementary Fig. 3o–q), while we found large p62+ accumulations in LPS-stimulated KO but not WT microglia, suggesting autophagy defects (Supplementary Fig. 3r, s).
To test if this KO-linked dampened inflammatory response is present in other pathways involving TBK1, we also stimulated with polyinosinic-polycytidylic acid (poly I:C) to assess the TLR3 pathway (in contrast to TLR4 for LPS) (Fig. 2b, Supplementary Fig. 3i–l and Supplementary Data 2). PCA again clearly separated KO from WT microglia (Fig. 2e), but in contrast to LPS stimulation, pathway analysis revealed an over-activated inflammatory response in KO microglia after poly I:C stimulation (Fig. 2f).
With this in vitro approach, we found evidence that Tbk1 loss in microglia affects their responses to pro-inflammatory stimulation, but depending on the stimulus, it led to dampened or over-activated inflammatory responses.
Tbk1 loss in spinal cord microglia increases their density and induces a pro-inflammatory signature
To assess in vivo, if Tbk1 loss would affect microglia, we used again Tbk1fl/fl and Cx3cr1-Cre mice to delete Tbk1 selectively in microglia (Tbk1-µG-WT, Tbk1-µG-HET, Tbk1-µG-KO). We confirmed selective spinal cord and brain microglial CRE expression, without neuronal CRE expression or Tbk1 recombination, and highly efficient microglial Tbk1 deletion (Supplementary Fig. 4). Of note, our BAC-transgenic Cx3cr1-Cre line (MMRRC, #036395-UCD) is well characterized, and is within the CNS microglial specific, and also shows, in contrast to other constitutive Cx3cr1-Cre lines, no neuronal leakage (Supplementary Fig. 4)22,23.
We labelled spinal cord sections for microglia using IBA1 and PU.1, both at 4 and 22 months of age (Fig. 3a). Consistent with the known age-linked increased microglial reactivity24,25, more microglial cells were found in old compared to young control mice (1.6 times; 131 ± 8.2/mm2 vs 80 ± 19.3/mm2). Importantly, compared to Tbk1-µG-WT mice, microglial density was increased at 4 months in Tbk1-µG-KO mice, and this increase persisted in aged Tbk1-µG-KO mice (2.1 times at 4 months and 1.5 times at 22 months) (Fig. 3b). Of note, the observed effect was Tbk1 dose-dependent, with deletion of only one Tbk1 allele (Tbk1-µG-HET) resulting already in an intermediate increase in microglial density (Fig. 3b). Likewise, the increase in microglial density was sex-independent (Supplementary Fig. 5a), and the grey matter had similar area sizes in all the groups (Supplementary Fig. 5b).
a Representative immunofluorescence images of microglia identified by cytoplasmic IBA1 (green) and nuclear PU.1 (red) in the lumbar spinal cord of young (4-month-old) and aged (22-month-old) mice, with microglial specific deletion (Cx3cr1-CreMMRRC/Tbk1fl/fl) of one (Tbk1-µG-HET) or two (Tbk1-µG-KO) Tbk1 copies or control littermates (Tbk1-µG-WT). Scale bar = 100 µm. b Quantification of the number of microglial cells at 4 and 22 months in lumbar spinal cord ventral horn grey matter of male mice (for females see Supplementary Fig. 5a). n = 4 mice per genotype and age (except for Tbk1-µG-HET at 22 months, n = 3). Adjusted p values are indicated and were determined by ordinary two-way ANOVA with Tukey’s multiple comparisons test. c Setup for RNAseq analysis of microglia, isolated by mechanical dissociation and FACS from spinal cord of young (4-month-old) and aged (22-month-old) mice with microglial specific deletion of Tbk1 (Tbk1-µG-KO) and compared to control littermates (Tbk1-µG-WT). n = 4–5 samples per genotype and age, with each sample corresponding to microglia from 2 pooled spinal cords (females only). Created in BioRender. Lenoel, I. (2025) https://BioRender.com/ct8qnd6. d PCA reveals clustering of all the samples by age (n = 4–5 samples per genotype and age). e Number of significantly deregulated genes (DEGs) in microglia with Tbk1 deletion (Tbk1-µG-KO) at 4 and 22 months of age compared to control microglia (Tbk1-µG-WT), and the number of overlapping DEGs between the two age-groups. f Volcano plots of differentially regulated genes comparing Tbk1-µG-KO to Tbk1-µG-WT microglia at 4 and 22 months. Some relevant genes are highlighted. e, f DESeq2, log2FC ≥ 0.5, adjusted p values (FDR) < 0.05, see “Methods”. g GSEA showing the top Hallmark pathways upregulated in microglia upon microglial Tbk1 deletion (Tbk1-µG-KO) at 4 and 22 months compared to control microglia (Tbk1-µG-WT) (adjusted p values < 0.05, NES ≥ +2.1, see “Methods”). b Data are shown as single data points and means ± SEM.
We then assessed if these Tbk1-µG-KO microglial cells show signs of activation, although we did not detect any morphological changes (Supplementary Fig. 5c–e). Thus, we performed RNAseq on FACS-isolated CD45+/CD11b+ microglia from spinal cords of Tbk1-µG-WT (WT) and Tbk1-µG-KO (KO) mice at both 4 and 22 months of age (Fig. 3c–g and Supplementary Data 3). We found robust changes between KO and WT microglia and a significant overlap between the two age-groups (Fig. 3d–f). Gene Set Enrichment Analysis (GSEA) showed that the top (Hallmark) pathways were all linked to pro-inflammatory signaling and upregulated in KO microglia in both age-groups (Fig. 3g).
Thus, in the spinal cord, microglial Tbk1 loss leads to persistently increased microglial density, with a shift to a pro-inflammatory signature. This shows that in the spinal cord, microglial TBK1 is involved in setting homeostatic microglial density and state.
Deletion of Tbk1 in microglia in vivo dampens their response to an acute systemic inflammation
While we found a dampened pro-inflammatory response to LPS in vitro (Fig. 2d) and no differences between unstimulated Tbk1-KO and WT microglia (Fig. 2b), we found a shift to a pro-inflammatory microglial signature in vivo in the adult Tbk1-µG-KO spinal cord (Fig. 3c–g). To address this difference, we injected adult (4-month-old) Tbk1-µG-KO and -WT mice systemically with LPS, and 24 h later performed RNAseq on FACS-isolated spinal cord microglia. We found a similar dampened pro-inflammatory response to LPS in Tbk1-KO microglia in vivo (Fig. 4a–c, Supplementary Fig. 6a–d, and Supplementary Data 4) as in vitro (Fig. 2d). Therefore, despite the pro-inflammatory shift under homeostatic conditions, adult Tbk1-KO microglia were less capable to respond to LPS than WT cells. Indeed, certain genes induced by LPS in WT cells (e.g. Cxcl10) were not induced by LPS in KO microglia, despite being upregulated at baseline in homeostatic conditions in the KO cells (Supplementary Fig. 6a, d). This is also reflected by more genes down than upregulated between LPS KO vs WT cells, and most showing an inverted regulation compared to WT LPS vs unstimulated cells (Supplementary Fig. 6b, c).
a Setup for RNAseq of microglia 24 h after systemic (i.p.) LPS injection, isolated from spinal cords of 4-month-old mice with microglial specific Tbk1 deletion (Tbk1-µG-KO, KO), compared to control littermates (Tbk1-µG-WT, WT). b PCA reveals clustering of samples by treatment. Each point (sample) corresponds to microglia from 2 pooled spinal cords. n = 4 samples per genotype and age, females only. c GSEA showing top regulated immune (Hallmark) pathways between Tbk1-µG-KO and Tbk1-µG-WT microglia, after LPS treatment (adjusted p values < 0.05, NES ≥ ± 1.5, see “Methods”). LPS effect on each pathway in control condition is represented by a red up arrow if activated in WT-LPS vs unstimulated. d Setup to assess the effect of microglial Tbk1 deletion during chronic MN degeneration. Mice with microglial specific Tbk1 deletion were crossed to SOD1G93A ALS mice, to produce SOD1G93A mice with one (G93A/Tbk1-µG-HET) or two (G93A/Tbk1-µG-KO) Tbk1 copies deleted, compared to littermate SOD1G93A mice with control microglial Tbk1 (G93A/Tbk1-µG-WT). e–g Kaplan–Meier plots of ages reached at disease onset (e), early disease (f), and survival age reached at end-stage (g). n = 20-23 mice per genotype (equally sex-mixed). Mean ages in days (d) ± SEM. h Setup to assess the effect of microglial Tbk1 deletion during acute MN injury (unilateral sciatic nerve crush/regeneration) in 4-month-old Tbk1-µG-KO, Tbk1-µG-HET, and littermate control Tbk1-µG-WT mice. Shown are representative toe-spread images (Catwalk) before and after major recovery. i Recovery of the sciatic functional index (SFI) over 4 weeks after crush. n = 6-7 mice per genotype (males only). Adjusted p values are indicated and were determined by mixed effect models. j Recovery was also assessed long-term (over 17 weeks after crush) by measuring the compound muscle action potential (CMAP) using electromyography. n = 5 mice per genotype. Adjusted p values are indicated and were determined by repeated measure two-way ANOVA. a, d, h Created in BioRender. Lenoel, I. (2025) https://BioRender.com/ct8qnd6. i, j Data are shown as means ± SEM. ns not significant.
Thus, microglial Tbk1 deletion leads to different consequences depending on the physiological situation: a pro-inflammatory shift during homeostasis, but a dampened pro-inflammatory response during an acute inflammation (LPS).
Microglial Tbk1 deletion is not sufficient to induce or modify ALS-like motor neuron damage
We next asked if the increase in spinal cord microglial density and the pro-inflammatory shift induced by Tbk1 loss could have a deleterious effect on wildtype MN or influence ongoing ALS-like neurodegeneration.
First, we assessed whether Tbk1-µG-KO mice presented signs of ALS-like disease. Until up to 22 months of age, we did not detect any lumbar spinal cord MN loss, nor p62+ accumulations in MN, and no signs of a motor phenotype (Supplementary Fig. 6e–i). We also did not detect increased signs of MN stress as assessed by staining for phospho-cJUN (Supplementary Fig. 6j).
Secondly, as microglial responses to ongoing MN degeneration are able to influence disease progression in mutant SOD1 ALS mice10, we examined the consequences of microglial Tbk1 loss in SOD1G93A mice (Fig. 4d). However, deleting one or both microglial Tbk1 copies in SOD1G93A mice changed neither the disease onset nor the survival of the mice (Fig. 4e–g and Supplementary Fig. 7a, b). On a histological level, while we measured micro- and astro-gliosis in SOD1G93A/Tbk1-µG-WT mice, this was not changed by microglial Tbk1 deletion (Supplementary Fig. 7c–e).
Thirdly, as it is possible that the neurotoxic effect of SOD1G93A overexpression in microglia of SOD1G93A mice could override the effect of additional microglial Tbk1 deletion, we used as an alternative model of MN damage, the sciatic nerve crush paradigm, which is well known to be influenced by microglial responses26. However, we did not detect differences in the overall nerve regeneration speed between the 3 genotypes (Tbk1-µG-WT, Tbk1-µG-HET, Tbk1-µG-KO), as measured by pawprint analysis, and by recovery of the compound muscle action potential (Fig. 4h–j and Supplementary Fig. 7f, g).
Thus, while TBK1 can influence microglial responses to an acute inflammatory stimulus (LPS) in vitro and in vivo, and is involved in setting homeostatic spinal cord microglial density and state, microglial Tbk1 loss is not sufficient to induce or influence ALS-like spinal cord MN damage.
Microglial Tbk1 deletion is sufficient to induce an FTD-like social recognition deficit
Therefore, we assessed whether microglial Tbk1 loss and thus deregulations only in microglia are sufficient to lead to FTD-like symptoms.
We used the 3-chamber test to assess FTD-like social interaction deficits27 in mice with microglial-specific Tbk1 deletion (Fig. 5a). Young (4-month-old male) control (Tbk1-µG-WT) mice performed expectedly in the three trials of the first sociability phase, spending more time with stranger 1 than with the empty cylinder (Fig. 5b). Mice with one (Tbk1-µG-HET) or both (Tbk1-µG-KO) Tbk1 copies deleted in microglia, showed the same preference as littermate controls for a social target over an empty cage (Chi-square = 160.55, df = 1, p < 0.0001; n = 12 male mice per genotype), thus demonstrating no FTD-like sociability impairment (Fig. 5b). There was also no modification in the total distance moved and the mean velocity during the test, reflecting no motor deficits (Supplementary Fig. 8a, b). In addition, absence of sociability defects was confirmed by the resident-intruder test (Supplementary Fig. 8c, d).
a Workflow of the 3-chamber test to assess presence of sociability and social recognition (social novelty preference) defects in 4-month-old male mice with microglial specific deletion of one (Tbk1-µG-HET) or two (Tbk1-µG-KO) Tbk1 copies and compared to control littermates (Tbk1-µG-WT). Created in BioRender. Lenoel, I. (2025) https://BioRender.com/ct8qnd6. b Interaction times during the first three trials (trials 1–3) of the 3-chamber test (sociability phase). n = 12 mice per genotype (males only). Adjusted p values are indicated and were determined by a two-tailed type II Wald chi-square test with Tukey’s post hoc test. Results from trials 1–3 indicate no impairment of social interaction (sociability) due to microglial Tbk1 deletion. c Representative heatmaps of the mouse positions during the social recognition phase of the 3-chamber test (trial 4), shown for Tbk1-µG-KO and control Tbk1-µG-WT mice. d During trial 4, contact durations to stranger 1 (S1, known) and stranger 2 (S2, unknown) were measured. n = 12 mice per genotype. Adjusted p values are indicated and were determined by repeated measures two-way ANOVA with Sidak’s multiple comparisons test. e Calculation of the social recognition index (trial 4) per mouse. n = 12 mice per genotype. Adjusted p values are indicated and were determined by a two-tailed one sample t-test to the theoretical mean of 50%. Results from trial 4 (social recognition phase) indicate an impairment of social recognition due to microglial Tbk1 deletion. Of note, the observed social recognition (social novelty preference) deficit affects social memory, but spatial memory was unaffected (Supplementary Fig. 8e–g). d, e Data are shown as single data points and (b, d, e) means ± SEM. ns: not significant.
Then, we analyzed the second social recognition phase (which assesses social novelty preference) of the 3-chamber test, which consists of adding a new unfamiliar stranger 2 to the empty chamber. Importantly, while control Tbk1-µG-WT mice showed an expected preference for the new social target stranger 2 over the now familiar stranger 1, Tbk1-µG-KO mice had completely lost this preference between familiar and unfamiliar social targets. Respective recognition indexes were 73% for Tbk1-µG-WT and 53% for Tbk1-µG-KO mice (Fig. 5c–e). This social recognition deficit was Tbk1 dose-dependent, as mice with only one Tbk1 copy deleted in microglia (Tbk1-µG-HET) already showed an intermediate effect with a 65% recognition index. Social recognition can also be considered as social memory (reviewed in Cum et al.28). Thus, to assess if other forms of memory could be affected, we used the Morris water maze test and found Tbk1-µG-KO mice to perform well (Supplementary Fig. 8e–g), revealing that microglial Tbk1 deletion specifically affects social but not spatial memory.
Our results show that deleting Tbk1 solely from microglia is sufficient to impair social recognition, consistent with an FTD-like cognitive deficit, suggesting that part of the disease has its origins in microglial defects.
The behavioral deficit is associated with shifting microglia to a pro-inflammatory, aged-like signature
To understand how microglial Tbk1 loss leads to the observed social recognition deficit, we studied specifically the molecular changes induced in brain microglia.
Thus, we performed RNAseq on FACS-isolated CD45+/CD11b+ microglia from brains of young (4 months) and aged (22 months) Tbk1-µG-HET, Tbk1-µG-KO, and control Tbk1-µG-WT mice (Fig. 6a and Supplementary Data 5). Of note, microglial purity was unaffected by Tbk1 deletion (Supplementary Fig. 9a). PCA discriminated the samples mainly by age (Fig. 6b), consistent with the known age-linked microglial activation24,25. We found 1340/146 genes up/downregulated when comparing aged and young microglia (log2FC ≥ 0.5, adjusted p value < 0.05) that were enriched in the Mouse microglial ageing pathway of the HDSig database24 (adjusted p value = 8.16E-86, over-representation analysis).
a Setup to compare microglia from young and aged (female) mice with full or partial microglial specific Tbk1 deletion, to control littermates. Created in BioRender. Lenoel, I. (2025) https://BioRender.com/ct8qnd6. b PCA reveals clustering of all samples by age. c Significant DEGs in Tbk1-KO microglia at 4 and 22 months vs control microglia and overlapping DEGs between the two ages (DESeq2, log2FC ≥ 0.5, adjusted p values (FDR) < 0.05, see “Methods”) (for Tbk1-HET, see Supplementary Fig. 9j). d Venn diagram of DEGs comparing genotypes and ageing (with partial and total sums indicated). e GSEA showing the top 5 Hallmark pathways upregulated by Tbk1 deletion at 4 and 22 months compared to control microglia, and during ageing in control microglia (adjusted p values < 0.05, see “Methods”). f WGCNA revealed only one KO relevant module (see also Supplementary Fig. 9g, h). Data represented as single data points (corresponding to a single mouse sample, see Fig. 6a) with mean. Adjusted p values were determined by a two-tailed Wilcoxon test. g, h Upper panels: Red/blue squares indicate significant UP/DOWNregulations (red/blue DEGs). White squares indicate not significant (ns) changes. Lower panels: Gene heatmaps with unbiased clustering (centered VST expression values, see “Methods”). g Heatmap of the top 20 DEGs (ranked by adjusted p values; marked with a white cross in the upper panel) upregulated in microglia by Tbk1 deletion at either 4 or 22 months compared to control microglia, and their regulation during ageing in control microglia. Of note, as 10 DEGs were overlapping, this resulted in a total of 30 DEGs shown. h Heatmap of the top 30 DEGs (ranked as in (g)) upregulated during ageing in control microglia and compared to their regulation in microglia with Tbk1 deletion at 4 or 22 months (vs control microglia). Purple arrowheads highlight 3 disease-associated microglia (DAM) markers, Axl, Cst7, and Itgax. i, j GSEA to assess how full microglial Tbk1 deletion compares to partial. GSEA plots comparing KO to WT (i), and HET to WT (j), at 4 and 22 months, with the top 100 genes significantly upregulated by ageing in control microglia (genes ranked based on adjusted p values).
To reveal differences between the genotypes, we assessed differentially expressed genes (DEGs) and found robust changes between Tbk1-µG-KO (KO) and Tbk1-µG-WT (WT) microglia from both young and aged mice (Fig. 6c, Supplementary Fig. 9b, c, and Supplementary Data 5): at 4 months, 198 genes were up and 46 downregulated, between KO and WT samples, while at 22 months, 141 were up and 58 downregulated. There was also an overlap between the effects of Tbk1 deletion in young and aged KO microglia, with 46% (91/198, in young) and 65% (91/141, in aged) of upregulated DEGs in common (Fig. 6c).
When comparing the DEGs upregulated by Tbk1 loss at 4 and 22 months, to ageing of WT microglia, we found an overlap of common genes, with 66% at 4 months (131/198) and 46% at 22 months (65/141) (Fig. 6d and Supplementary Data 5). The shift was stronger in young than in aged KO microglia, since only part of the age-linked DEGs that increased in young, remained further increased in aged KO. This resulted in 56 genes commonly upregulated in both young and aged KO microglia as well as during WT microglial ageing (Fig. 6d). Thus, 62% (56/91) of DEGs upregulated in common in both young and aged KO microglia, were also upregulated during WT microglial ageing, revealing that the shift towards an aged microglial signature is an important component of the effect induced by microglial Tbk1 loss.
On a pathway level, GSEA showed that the top 5 (Hallmark) pathways upregulated in young and aged Tbk1-KO microglia, were identical to the top 5 pathways upregulated in control (WT) microglia during ageing (Fig. 6e and Supplementary Fig. 9d–f). All these pathways were related to inflammation, revealing that in vivo, microglial Tbk1 loss induces a pro-inflammatory phenotype. Thus, our results show that Tbk1 loss in microglia upregulates the same pro-inflammatory pathways that are also upregulated during control (WT) microglial ageing (Fig. 6e). Of note, the shared 56 genes common to the three groups were still linked to the same 5 pro-inflammatory pathways identified in the initial GSEA (Fig. 6d, e).
Weighted Gene Correlation Network Analysis (WGCNA) indeed revealed one module reflecting an effect of Tbk1 deletion (Fig. 6f and Supplementary Fig. 9g, h). This module’s eigengene profile showed an increase during ageing of WT microglia, thus, again consistent with a link between the effect of Tbk1 deletion and microglial ageing (Fig. 6f). Of note, the only other module with a clear profile reflected the expected age-linked microglial activation, independent of the genotype (Supplementary Fig. 9h).
The link between the effect of Tbk1 loss and microglial ageing was supported by intersecting the top DEGs from all the groups. We compared the top upregulated DEGs from KO microglia compared to control (WT), choosing the top 20 DEGs in young and the top 20 DEGs in aged, which resulted in 30 DEGs, as 10 were overlapping. We found that many of these 30 top DEGs were also upregulated in WT microglial ageing (90% at 4 and 60% at 22 months) (Fig. 6g and Supplementary Data 5). Reciprocally, comparing the top 30 upregulated DEGs during WT microglia ageing with the DEGs induced in KO microglia, revealed an overlap of 53% at 4 months and of 40% at 22 months (Fig. 6h). Thus, this shows that the main effect of Tbk1 loss in microglia is a shift of young KO microglia to a signature of aged WT microglia, and this partially persists during ageing.
Notably, among the top upregulated DEGs, we found 3 classic Disease-Associated Microglial (DAM) markers (Itgax, Cst7, Axl), which were all upregulated by both control (WT) ageing and by Tbk1 loss (Supplementary Fig. 9i), further supporting a disease-like shift of KO microglia due to Tbk1 loss. Of note, in our in vitro study, Itgax was a DEG consistently induced by both pro-inflammatory stimuli (LPS, poly I:C) in KO but not WT microglia (Supplementary Fig. 9k, l).
While our analysis was mostly based on full Tbk1 deletion (KO), we also tested if the same effect is already present in HET (Tbk1-µG-HET) microglia. Thus, we used the top 100 DEGs upregulated in WT microglia during ageing (log2FC ≥ 0.5, ranked on adjusted p value < 0.05) (Supplementary Data 5), to perform GSEA on differential expression results in KO and HET microglia compared to WT (Fig. 6i, j and Supplementary Fig. 9j). We found an enrichment for the ageing microglial gene signature in KO microglia at both 4 and 22 months, and also, albeit less so, in HET microglia at 4 and 22 months. This shows that already partial Tbk1 loss in microglia trends to shift them towards an aged microglial signature, which becomes fully apparent with complete Tbk1 deletion.
Finally, we compared the RNAseq analysis between Tbk1-KO brain and spinal cord microglia (Figs. 3c–g and 6a–e). By focusing on a link to ageing, we found a similar shift to an aged-like microglial signature in the spinal cord (Supplementary Fig. 10a, b) as in the brain (Fig. 6d, e). Thus, in both compartments microglial Tbk1 deletion induced an early shift to a pro-inflammatory, aged-like signature, with the same top 5 inflammatory pathways upregulated (Fig. 6e and Supplementary Fig. 10b) and robust overlap of DEGs between the two population of Tbk1-KO microglia (Supplementary Fig. 10c and Supplementary Data 5). However, the Tbk1 deletion induced gene deregulations were less strong in spinal cord than brain microglia (124 vs 198 upregulated DEGs, with 85 in common). As an example, the classic pro-inflammatory cytokine Tnf, and the senescence marker Cdkn1a (p21) were only upregulated in Tbk1-KO brain microglia (Supplementary Fig. 10d–f), indicating a stronger shift to a pro-inflammatory signature in Tbk1-KO brain than spinal cord microglia.
Additionally, we also complemented our RNAseq analysis of Tbk1 deletion on microglial responses to in vivo LPS stimulation, initially done in the spinal cord (Fig. 4a–c), with brain microglia and found a similar dampened inflammatory response of Tbk1-KO microglia to LPS stimulation for both compartments (Supplementary Fig. 10g–k and Supplementary Data 4).
Thus, these in vivo results show that Tbk1 loss in microglia induces an early shift to an aged-like state and indicate that TBK1 is implicated in restricting aged-like microglial activation. These molecular changes in microglia could be the cause for the social recognition deficits we detected in mice with microglial Tbk1 deletion.
Meta-analysis links Tbk1 deleted microglia to primed microglia from ageing and neurodegeneration
Next, we compared the Tbk1-KO microglial signature to actual neurodegenerative conditions and microglial priming, that has been described as the overlap between ageing and neurodegeneration24.
Thus, we performed a meta-analysis to compare our Tbk1-KO and control ageing microglia, with published transcriptomes of microglia from active neuroinflammatory stages of 3 different neurodegenerative models: Alzheimer’s disease (AD) APPNL-G-F/NL-G-F mice with amyloid pathology29, AD/FTD rTg(TauP301L)4510 mice with tauopathy29, and ALS SOD1G93A mice30 (Fig. 7a and Supplementary Data 6).
a UpSetPlot depicting the results of the meta-analysis comparing the present study’s transcriptome of young brain microglia with Tbk1 deletion (Tbk1-µG-KO 4 m), or microglial cells during control brain ageing (Tbk1-µG-WT, Ageing), to 3 published RNAseq datasets of microglia isolated at active neuroinflammatory stages of three neurodegenerative disease models, from brains of APPNL-G-F/NL-G-F and rTg(TauP301L)4510 mice and from spinal cord of SOD1G93A mice (Neurodegeneration), compared to intrinsic controls in each study. The 46 genes commonly upregulated in all 5 conditions are highlighted with a box, representing a core gene-set showing that microglia with Tbk1 deletion share features with primed microglia from ageing and neurodegeneration. b STRING-based analysis of the protein-protein interactions among the shared 46 genes of the core gene-set, with three markers of disease-associated microglia (DAM), Axl, Cst7, and Itgax, highlighted by purple arrowheads, and the central driver, Tnf, highlighted in red. c Heatmap of the shared 46 genes from the core gene-set showing the fold changes as found in the published datasets of microglial changes during neurodegeneration and from the present study of microglial changes during ageing and after Tbk1 deletion. Note the less strong upregulation of the 46 genes in ageing and after Tbk1 deletion, as compared to neurodegeneration models, indicating a priming. Purple arrowheads highlight the three markers of disease-associated microglia (DAM), Axl, Cst7, and Itgax, that are present throughout our analysis of Tbk1 deletion in microglial cells. d Cytoscape-based grouped network analysis of GO biological processes pathways related to the shared 46 genes of the core gene-set, with driver genes of each group highlighted (see “Methods”).
Focusing on our young Tbk1-KO (TBK1-µG-KO) microglia (when the social recognition deficit was detected), we found a common set of 46 DEGs that were upregulated in all 5 conditions (Fig. 7a). Most of these 46 genes encode for proteins known to interact together (STRING-based analysis), showing the biological relevance of this cluster (Fig. 7b). Thus, 1/3 of the DEGs in common between Tbk1-KO microglia and control microglial ageing (46/131), were also shared with reactive microglia during actual neurodegeneration (Supplementary Fig. 11a). This reveals that priming to an aged and neurodegenerative microglial signature is an important early consequence of microglial Tbk1 loss and could lead to the observed social recognition deficit. Of note, 22 of these 46 DEGs remained upregulated in our aged Tbk1-KO microglia, (Supplementary Fig. 11b, c), indicating that this primed state gets partially further increased with age.
Consistent with a primed state of Tbk1-KO microglia, comparable to aged microglia, the relative fold-changes of the 46 genes were substantially stronger in the 3 neurodegenerative disease models (Fig. 7c). This primed core-set of 46 DEGs contains mostly inflammatory genes (Fig. 7c), including the 3 DAM markers (Axl, Cst7, Itgax), we originally detected in young Tbk1-KO and aged control microglia (Fig. 6h and Supplementary Fig. 9i), suggesting a shift towards a neurodegenerative signature. Pathway analysis identified links to microglial activation, antigen presentation, response to IFN-I and chemotaxis, supporting the idea of a primed microglial state (Fig. 7d).
Together, this meta-analysis shows that microglial Tbk1 loss induced an aged-like, primed state that is associated with early behavioral deficits and gets amplified in microglia in different neurodegenerative disease conditions.
Microglial Tbk1 deletion leads to focal microglial activation in the brain and T cell infiltration
In a final approach, we asked if the molecular phenotype we found in whole brain isolated Tbk1-KO microglia has a defined spatial localization, which could reveal a cellular mechanism leading to the behavioral deficit.
Using sagittal brain sections, we found a focal increased IBA1 immunoreactivity in the substantia nigra pars reticulata (SNr) and (less strong) in the pallidum (Allen Brain Atlas; containing globus pallidus and substantia innominata) in 4-month-old Tbk1-µG-KO vs control mice (Fig. 8a, b and Supplementary Fig. 12a–d), with the SNr representing a region with one of the highest brain baseline microglial densities (Fig. 8b)31,32.
a Representative IBA1 immunofluorescence image of microglia in brains of 4-month-old Tbk1-µG-KO mice. Filled arrowhead: Pallidum (Pa), Empty: Substantia nigra pars reticulata (SNr). Enlargements show representative images of focal microglial activation in the SNr of Tbk1-µG-KO vs control Tbk1-µG-WT mice. Scale bar = 2 mm. b Quantification of IBA1 immunofluorescence intensity in different brain regions between Tbk1-µG-KO and control mice. n = 4 mice per genotype (males only). Adjusted p values determined by repeated measures two-way ANOVA with Sidak’s multiple comparisons test. c Representative IBA1 images of microglia (green) with nuclear PU.1 (red) in the SNr of Tbk1-µG-KO and Tbk1-µG-WT mice. Scale bar = 50 µm. Enlargement, microglial morphology. Scale bar = 10 µm. d–f Quantification of microglia characteristics in the SNr: d density, e processes, and f process length. n = 4 mice per genotype (males only). Adjusted p values determined by two-tailed unpaired t-tests. g Expression profile of a top DEG (Cxcl10) from the brain microglial RNAseq (Fig. 6g) comparing Tbk1-µG-KO to Tbk1-µG-WT mice. RNAseq TPM values are shown and fold changes indicated (DESeq2, log2FC ≥ 0.5, adjusted p values < 0.05). n = 5 (4-month-old, except Tbk1-µG-WT, n = 6) or n = 6 (22-month-old, except Tbk1-µG-WT, n = 7) mice per genotype (females only). h Representative Cxcl10 mRNA expression image (RNAscope) of 4-month-old Tbk1-µG-KO and Tbk1-µG-WT (male) mice brain sections. Scale bar = 2 mm. Enlargement showing Cxcl10 expression only in the SNr of Tbk1-µG-KO mice. Scale bar = 20 µm. i Representative images of microglia (IBA1+) and T cells (CD3+) in the SNr of 4-month-old Tbk1-µG-KO (KO) and Tbk1-µG-WT (WT) mice. Scale bar = 50 µm. j Quantification of CD3+ T cells density. n = 4 mice per genotype (males only). Adjusted p values determined by a two-tailed unpaired t-test. k Representative images of microglia (IBA1+), T cells (CD3+), and CD8+ subtype T cells in the SNr. Scale bar = 20 µm. h, k Full set of mice were stained (n = 4 mice per genotype, males only), with one representative image shown. b, d–g, j Data are shown as single data points and means ± SEM.
A detailed analysis of Tbk1-KO microglia in the SNr revealed increased numbers and robust morphological changes (Fig. 8c–f), suggesting partial activation and consistent with the pro-inflammatory transcriptional shift (Fig. 6d, e). Of note, anti-CD169 staining33 did not detect peripheral macrophages infiltrating the parenchyma of KO mice (Supplementary Fig. 12e). The microglial activation remained focal, without changes in numbers or morphology in other major brain regions (e.g., hippocampus, Supplementary Fig. 12f–j). Using RNAscope for Cxcl10, a top DEG from the whole brain microglial RNAseq (Figs. 6g and 8g), we found equally focal induction in the SNr and pallidum of Tbk1-µG-KO mice (Fig. 8h).
Next, we asked if this focal microglial activation in the SNr, could lead to increased phagocytic activity, signs of neuronal damage or synaptic changes (pruning). However, we did not detect changes in CD68 staining (for phagocytic activity) in Tbk1-KO microglia (Supplementary Fig. 12k, l), nor reduced dendritic (tyrosine-hydroxylase positive) staining of dopaminergic neurons, or overall reduced (GABAergic) neuronal numbers (Supplementary Fig. 13a–d). We also did not find overall changes in synaptic density and apposition of pre-/post-synaptic markers in the SNr (Supplementary Fig. 13e–j).
When looking for early neuronal stress, we found c-FOS+/Neurotrace+ neurons in the SNr only of Tbk1-µG-KO, but they were very rare (Supplementary Fig. 13k, l). However, we found in higher numbers, but still relatively rare, clusters of c-FOS+/Neurotrace+ and phospho-c-JUN+/Neurotrace+ structures, co-stained with nuclear Hoechst, in the cytoplasm of microglia, in the SNr exclusively of Tbk1-µG-KO mice (Supplementary Fig. 13k–n). This suggests phagocytosis by Tbk1-KO microglia of stressed cells, possibly SNr neurons, although the number is probably small, as undetected in the global neuronal count (Supplementary Fig. 13c, d).
Finally, but consistent with the microglial Tbk1-KO driven induction in the SNr of Cxcl10 (Fig. 8h), a major T lymphocyte-attracting chemokine, we found infiltrating T cells (mostly CD8+), exclusively in Tbk1-µG-KO mice (Fig. 8i–k). While the infiltration was less focal, it was still more concentrated in the SNr and pallidum than other brain regions.
Thus, we spatially localized the Tbk1-KO driven pro-inflammatory shift of the microglial signature, and found linked T cell infiltration. This could create a deregulated local cytokine environment at the base of the social recognition deficit of Tbk1-µG-KO mice.
Discussion
Cell-specific Tbk1 deletion in mice revealed that microglia were much more affected than MN. While we found increased microglial density and a pro-inflammatory signature in the spinal cord, this was not sufficient to induce or modify ALS-like MN degeneration. In contrast, microglial Tbk1 loss in the brain was sufficient to induce FTD-like social recognition deficits, linked to an early shift to an aged-like microglial signature sharing features with primed microglia from ageing and neurodegeneration. This phenotype was associated and likely driven by focal microglial activation and cytokine production in the substantia nigra pars reticulata and pallidum, together with T cell infiltration. Thus, in the context of FTD, this suggests that changes to microglia only, without initial activation by affected neurons, can lead to specific clinical symptoms.
Regarding ALS/FTD human iPSC-models, mutant C9orf72 microglia showed impaired phagocytosis and autophagy34, an overactivated response to LPS34,35, and toxicity towards MN35, although the effects could vary in strength36. Mutant PFN1 microglia showed autophagy dysregulation and deficient phagocytosis37. Together it shows that ALS/FTD iPSC derived microglia can show cell-autonomous deregulations.
Regarding ALS/FTD mouse models, for OPTN-linked ALS, microglial-specific Optn deletion was sufficient to induce an axonal pathology in mice38. Similarly, for C9orf72-linked and TARDBP-linked ALS/FTD, microglial-specific C9orf72-39 or Tardpb-deletions40, produced neuropathological signs in mice. But in these cases clinical phenotypes were not assessed40, or only assessed in systemic KO mice38,39. More directly, for GRN-linked FTD, microglial-specific Grn (progranulin) deletion was sufficient to induce (obsessive-compulsive) FTD-like behavioral defects41, which were also found in systemic Grn-KO mice42. In contrast, for SOD1-linked ALS, overexpressing mutant SOD1 in microglia, was not sufficient to induce an ALS-like phenotype43, while reducing mutant human SOD1 overexpression only in microglia, slowed disease progression, but not onset10. Together with our results, this suggests that while microglia are of prime importance for the ALS/FTD disease spectrum, depending on the specific disease form, microglial defects can either cause specific disease symptoms, or rather contribute to ongoing disease. Although mechanistically not yet fully resolved, our results support the emerging concept that microglial defects can directly cause neurological dysfunction.
To understand how microglial Tbk1 loss leads to social recognition defects, we performed RNAseq and revealed that Tbk1 deletion induced a pro-inflammatory, aged-like signature in young microglia. In mice, CNS ageing is associated with social recognition impairment, without changes to sociability44, similar to our young microglial Tbk1-KO mice, and consistent with a link between Tbk1 and microglial ageing. In support of this, we found the senescence marker Cdkn1a(p21) to be induced in Tbk1-KO brain microglia. By a meta-analysis we compared our Tbk1-KO microglia to microglia from different neurodegenerative disease models and during ageing and found a common core-set of upregulated genes. While the signature of Tbk1-KO microglia was also shifted in the spinal cord, this was less strong than in the brain, with Tnf (a central driver identified in the meta-analysis, Fig.7b) only induced in KO brain microglia. Thus, while Tbk1 loss affected microglia in both spinal cord and brain, this only led to a neurological phenotype in the brain, suggesting differences on how microglial defects per se contribute to ALS and FTD.
With respect to how Tbk1-KO microglia react under inflammatory conditions, we found that both in vitro and in vivo, Tbk1 deletion led to a dampened pro-inflammatory response to LPS. However, microglial Tbk1 deletion did not impact acute or chronic MN damage. In contrast, under homeostatic conditions, microglial Tbk1 deletion shifted microglia to a pro-inflammatory, aged-like signature and led to social recognition deficits. Thus, the effect of Tbk1 deletion is dependent on the physiological situation. With respect to TBK1-linked FTD, this could suggest that the effect on the disease is rather early, before microglia enter a chronic neuroinflammatory state later in the disease.
Regarding the focal microglial activation in the SNr and pallidum of microglial Tbk1-KO mice, the known high baseline microglial density of these regions31,32 could be a contributing factor. With respect to the circuits linked to social recognition (social memory/novelty preference), they involve diverse brain regions45. While the SNr and pallidum are not specifically known for social recognition, studies have linked the midbrain, including SN/VTA dopaminergic neurons and interpeduncular nucleus GABAergic neurons to social recognition46,47,48,49,50, and unrelated microglial-specific gene-deletions (Fmr1 and Cannabinoid receptor type 1) have also been linked to social recognition deficits51,52.
A possible downstream consequence of microglial defects could be deregulated synaptic pruning53. With respect to ALS/FTD, deregulated synaptic pruning was detected in FTD-linked systemic Grn-KO42 and ALS/FTD-linked systemic C9orf72-KO mice39, which both show behavioral defects. While deregulated synaptic pruning was not assessed in microglial-specific Grn-KO mice41, it was reported in microglial-specific C9orf72-KO mice, although if this leads to a behavioral phenotype was not assessed39. While we did not detect signs of synaptic changes in microglial Tbk1-KO mice, the observed cytokine induction could have more subtle effects on synapses. The classic view describes microglial cytokines as amplifiers of neuroinflammation54, but they can have physiological roles by acting on neuronal activity55. Thus, the microglial-produced cytokines in the affected SNr of microglial Tbk1-KO mice, together with the infiltrating T cells (also sources of cytokines), could act on neuronal activity, inducing the behavioral deficit we identified. Of note, during normal ageing in mice, increased CNS T cell infiltration has been reported and associated to the more pro-inflammatory state of aged microglia56, similar to what we found with our aged-like shifted young Tbk1-KO microglia.
Despite the increased spinal cord microglial density and pro-inflammatory signature, we did not detect overt signs of MN degeneration in microglial Tbk1-KO mice. Crossing systemic Tbk1+/− mice to mutant SOD1 ALS mice showed a dual-effect with earlier disease onset but extended survival14,57. This led to the hypothesis of a protective effect from reducing microglial Tbk1. However, we found no disease ameliorating effects of Tbk1-KO microglia in ALS mice, suggesting that the reported beneficial effects were not driven by Tbk1 deletion in microglia only. Furthermore, we recently showed that systemic knock-in mice with the rare human C-terminal TBK1E696K mutation, developed a mild motor phenotype58. In contrast to most other ALS-linked TBK1 mutations, this TBK1E696K mutation retains partial protein expression. However, we cannot rule-out a dominant-negative effect in addition to the loss-of-function of most other TBK1-mutations, and a direct contribution from microglia could not be assessed with this systemic model. Likewise, a recent study reported that systemic Tbk1+/− mice with myeloid deletion of one Tak1 copy (another RIPK1 inhibitor), produced signs of MN degeneration4. Together with another study of systemic TBK1 activity reduction, which exacerbated C9orf72 dipeptide-aggregate mediated neurodegeneration in mice59, these studies highlight the link of TBK1 loss to ALS/FTD. However, these approaches were not designed to assess the specific disease link of TBK1 in microglia.
Certain limitations apply to this study. The Cx3cr1-cre driver is also active in certain peripheral and CNS-associated macrophage populations (e.g., perivascular/meningeal macrophages). While we have not detected CNS infiltration of peripheral macrophages, it remains possible that part of the phenotype could have been produced by Tbk1 deletion in non-microglial macrophage populations, which could also have been part of our protocol to isolate microglia.
While we used complete microglial Tbk1 deletion, actual TBK1-linked ALS/FTD is due to haploinsufficiency5. However, central findings of our study (social recognition deficit, increased microglial density) showed already intermediate effects in Tbk1-µG-HET mice. Thus, in mice additional stress from full Tbk1 deletion is needed for disease-like defects.
A complex behavioral disorder like FTD is difficult to model in mice, however, certain behavioral defects in mice could be called FTD-like, including social interaction deficits identified in a 3-chamber test60,61,62. The inability of our microglial Tbk1-KO mice to distinguish between a known and a new social interactor can be considered a symptom comparable to the deteriorating social functions of FTD patients, which includes disinhibition behavior but also apathy (e.g., lack of interest to interact with novel stimuli). Memory defects could also be involved. While FTD is generally not linked to memory loss, early memory deficits are specifically associated with TBK1-linked FTD63. While we have found spatial memory to be normal in our mice, the social recognition deficit is linked to a social memory deficit, suggesting a possible link between microglial TBK1 loss and specific symptoms of TBK1-linked FTD.
In conclusion, we reveal that reducing Tbk1 in microglia only is sufficient to lead to an FTD-like social recognition defect. The phenotype is linked to an early shift towards a pro-inflammatory, aged-like microglial signature with focal microglial activation in the SNr and pallidum, together with T cell infiltration. This adds support to the importance of microglial disease contributions in the ALS/FTD spectrum and suggests that part of the TBK1-linked FTD disease has its origins in microglial defects.
Methods
Animals
All the mice were on a C57Bl6/J genetic background. The following transgenic mouse lines were used: Tbk1flox/flox mice (Model #11131, C57BL/6-Tbk1tm1.1mrl) were from Taconic Biosciences; VAChT-Cre mice (Vesicular Acetylcholine Transferase) were obtained from Dr. Hidemi Misawa (Keio University, Japan)20; ChAT-Cre mice (B6.129S-Chattm1(cre)Lowl/MwarJ, #031661) were from Jackson Laboratory (USA); BAC-transgenic Cx3CR1-Cre mice (Tg(Cx3cr1-Cre)MW126Gsat/Mmucd, MMRRC (Mutant Mouse Resource and Research Center), #036395-UCD)22,23 were obtained from Dr. Eric Huang (UCSF, USA), is not a knock-in line and does not affect the endogenous mouse Cx3cr1 locus; and SOD1G93A mice (B6.Cg-Tg(SOD1*G93A)1Gur/J, #004435) were from Jackson Laboratory. For all experiments, all the 3 different mouse genotypes per age were littermates. For mouse genotyping, primers used were: 5′-AAGAAGCCAAAAGATTGATTGC-3′, 5′-GATCATATCTCCAACAGTACAGAGG-3′ to detect Tbk1 and Tbk1flox alleles (179 vs 359 bp) and 5′-ATGCTTCTGTCCGTTTGCCG-3′, 5′-CCTGTTTTGCACGTTCACCG-3′ to detect the BAC-Cx3cr1-Cre transgene (271 bp). Mouse strains purchased from the Jackson Laboratory were genotyped according to the manufacturer’s instructions. Housing conditions were as follows: temperature 20 ± 2 °C, humidity: 55 ± 10%, light–dark cycle: 12–12 h, ventilation: approximately 12 cycles per hour of filtered/non-recycled air. All animal procedures (including end-stage definition) were performed in accordance with the guidelines for the care and use of experimental animals of the European Union and approved by the ethics committee for animal experimentation number 5 (CE5) of Ile-de-France (#36402/40885 at the CEF/UMS-28 and #47179 at the Paris Brain Institute).
In vitro analysis of primary mouse microglial cells
Microglial cell culture
Cultured microglial cells were derived from the brains of 0 to 1-day-old Tbk1-µG-WT, Tbk1-µG-HET, and Tbk1-µG-KO mice according to ref. 64. After genotyping, brains of 2–4 pups per genotype were recovered in cold dissection medium (1× PBS, 100 U/mL Penicillin and 100 µg/mL Streptomycin, Glucose 6 g/L). Olfactory bulbs and cerebellum were removed, hemispheres were split, and meninges removed. Hemispheres were cut in pieces using angled scissors and put in Dulbecco’s Modified Eagle Medium (DMEM, Gibco, 41965-039). Cells were dissociated, spun down (900 rpm, 9 min) and resuspended in culture medium (DMEM, 10% Fetal Calf Serum (FCS, Gibco, 16000-044), 5 U/mL Penicillin, 5 µg/mL streptomycin (Gibco, 15070-063), 50 μM ß-mercaptoethanol (Gibco, 31350-010)). Finally, cells were plated in dishes coated with poly-ornithine (1.5 μg/mL) and incubated (37 °C, 5% CO2) in 10 cm petri dishes with a density of 106 cells per plate. Medium was changed 7 days later. 14 days after plating, plates were shaken for 1 h and the floating cells recovered, spun down, and resuspended in cold DMEM, washed, resuspended in medium (DMEM, 10% FCS, 50 μM ß-mercaptoethanol) and incubated (37 °C, 5% CO2). Cells were plated in 24-well plates with 250,000 cells/well for RNA and supernatant collection, and 100,000 cells/well for immunofluorescence and phagocytosis analysis. 24 h later, medium was replaced with medium plus stimulus: Lipopolysaccharide (LPS, 100 ng/mL, Sigma, L6143), or Polyinosinic-polycytidylic acid (poly I:C, 50 µg/mL, Invivogen, Tlrl-pic) or Interleukin-4 (IL-4, 20 ng/mL, R&D systems, 404-ML) or regular medium. After 24 h, supernatant was collected and cells were either fixed for immunocytochemistry (4% PFA, 10 min) or RNA extracted (RNeasy Micro Kit, Qiagen).
Phagocytosis assay
After 22 h of stimulation, pHrodo E. coli bioparticles (Invitrogen, P35361) were added to the medium (1:10) and incubated for 2 h. Images were then taken and analyzed with the ArrayScan (ThermoFisher).
NO assay
Nitrite concentration in supernatants were determined using the Griess Reagent System (Promega, G2930) according to manufacturer’s instructions and the absorbance at 540 nm was measured with Spectramax i3× (Molecular Devices).
Immunocytochemistry
Fixed cells were incubated for 30 min in blocking buffer (PBS, 0.1% Triton ×-100 (PBST), 10% Normal Goat Serum (NGS)) and overnight at 4 °C with primary antibodies: mouse anti-CRE (1:500, Millipore MAB3120), rabbit anti-TBK1 (1:100, Abcam Ab40676), and biotin-Cd11c (1:50, Biolegend 117303). Cells were washed (PBST) and incubated with secondary antibodies (for 1 h diluted 1:500 in PBST): goat anti-rabbit (Alexa 488, A11008), goat anti-mouse (Alexa 594, A21125), and Hoechst (1:5000 in PBS, 5 min only, ThermoFisher 33342). Biotin-antibody was revealed using the Alexa Fluor-488 Tyramide SuperBoost Kit (ThermoFisher, B40932). Images were taken and analyzed using the ArrayScan (ThermoFisher) or AxioImager Z1 microscope (Zeiss) with ZenPro (v3.1) software.
RNAseq
RNA was purified using the RNeasy Micro Kit (Qiagen). 300 ng of RNA per sample were used to prepare RNAseq libraries, using the KAPA mRNA Hyperprep Kit (Roche). Final sample pooled library preps were sequenced on a NovaSeq 6000 Sequencing System (ILLUMINA) with the S1-200 cartridge corresponding to 2 × 26 millions of paired-end reads per sample, after demultiplexing. Quality of raw data has been evaluated with FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc). Poor quality sequences were trimmed or removed with Trimmomatic65 software to retain only good quality paired reads. The Illumina DRAGEN bio-IT platform (v3.8.4) was used to align reads on mm10 mouse reference genome and quantification with the gencode v25 annotation gtf file. Library orientation, library composition, and coverage along transcripts were checked with Picard tools. Following analyses were conducted with R software. Data were normalized with DESeq2 (v1.26.0) bioconductor packages, prior to differential analysis with glm framework likelihood ratio test from DESeq2 workflow. Multiple hypothesis adjusted p values were calculated with the Benjamini–Hochberg procedure to control FDR. Finally, enrichment analysis was conducted with clusterProfiler R package (v3.14.3) with Gene Set Enrichment Analysis on Hallmark database.
Tbk1 RT-qPCR in cultured microglial cells
300 ng of RNA was used to obtain cDNA with the SuperScript IV Kit using random hexamers (ThermoFisher), according to manufacturer’s instructions. cDNA was then used to perform qPCR with TaqMan universal PCR Master Mix (ThermoFisher), with Tbk1 primers (00451150, ThermoFisher) and Tbp primers (00446973, ThermoFisher) in 384-well-plates, using the LightCycler 480 (Roche). Data were analyzed using LightCycler 480 software (v1.5).
Disease-stage analysis in SOD1 G93A mice
A cohort of approximately 20 mice per genotype (10 males and 10 females) were followed weekly from the presymptomatic stage (around 50 days) until disease end-stage (around 165 days). The breeding strategy was designed to obtain all the genotypes in the same litter, and SOD1G93A/Tbk1-µG-WT mice were used as controls to detect genetic derivation (abnormal disease duration), which was a criterion for exclusion. Mice were followed for signs of paralysis and weighed weekly as an objective and unbiased measure to track the disease course. The time of disease onset was retrospectively determined as the time when mice reached either peak body weight or peak grip strength (Bioseb, grip test; average of three consecutive measures once a week). The time of early disease (symptomatic stage) was defined as the age at which the animals had lost 10% of their maximal weight or 35% of their maximal grip strength, which is accompanied by gait alterations and failure of the hindlimb splaying reflex, but without obvious signs of paralysis. The end stage is defined by the mouse’s incapacity to right itself up within 20 s when placed on its side. This is the endpoint used for euthanasia in mutant SOD1-expressing ALS model mice10.
Sciatic nerve crush in mice
Surgery
Mice received analgesia (Buprenorphine, Vetergesic, 0.1 mg/kg) prior to the surgery. Mice were then placed under isoflurane anesthesia, and the right sciatic nerve was exposed via an incision in the flank followed by the separation of underlying muscles. The sciatic nerve was crushed twice for 10 s, using a straight Castroviejo needle holder at the level of the obturator tendon. After surgery, mice received again analgesia (Buprenorphine, Vetergesic, 0.1 mg/kg) for 3 days, twice per day. Functional recovery was assessed by pawprint analysis (see below) and electrophysiology (see below).
Pawprint analysis
Pawprint analysis was started prior to the surgery, and was done daily, from 7 days after the surgery until 28 days. Mice were placed on the Catwalk XT (Noldus), with wet paws to obtain robust prints despite no weight on the injured limb. An optimum of 3 independent continuous runs were recorded, to get 3 independent pawprint measurements. Print length and toe-spread were measured using the Catwalk XT software (Noldus), and sciatic functional index was calculated according to (Inserra et al., 1998)66, using the pre-surgery measurements instead of the contralateral data.
Electromyography
Mice received analgesia (Buprenorphine, Vetergesic, 0.1 mg/kg) prior to the procedure. Mice were then placed under isoflurane anesthesia. Two stimulation needles were place subcutaneously near the lumbar region of the spinal cord, and two recording needles were placed in the gastrocnemius muscle. Signals were recorded using BioAmp (ADInstruments) and analyzed by LabChart8 (ADInstruments). Compound muscle action potential (CMAP) was measured with supra-maximal stimulations.
Mouse behavior
3-chamber test
4-month-old male mice were used to assess social behavior by a fully blinded experimenter (regarding the genotype). Sociability and social recognition (social memory, social novelty preference) were assessed by using the 3-chamber social task. Mice were first habituated to the 3-chamber cage (Noldus) and allowed to explore the three chambers with empty cylinders for 5 min. Trials 1–4 took place 24 h later. For trials 1–3 (sociability), the tested mouse was placed in the middle chamber, and an unfamiliar mouse of the same background, age, sex, and weight was placed in a cylinder in the left or right chamber. The other chamber contained an empty cylinder. The tested mouse was allowed to explore for 5 min and put in its home cage for 15 min before the next trial. All trials were recorded using a camera and MediaRecorder software (Noldus). Position of the stranger was alternated between trials, and the same stranger was used for all 3 trials of a tested mouse. For trial 4 (social recognition/memory), the tested mouse was placed in the middle chamber, the now-familiar mouse was placed in a cylinder in the left or right chamber, and a novel unfamiliar mouse was placed in the right or left chamber. The tested mouse was recorded and allowed to explore freely for 5 min. The recorded videos were analyzed offline with EthoVision XT software system (v17.0, Noldus). The time spent in the area around each cylinder was quantified to assess social interactions, while the total distance travelled, and average moving speed (mean velocity) was used to evaluate locomotor activity.
Resident-intruder test
Social interaction in the home cage between freely moving mice was assessed by the resident-intruder test. The mouse was placed in its home cage under a recording camera (Noldus), without lids, and environment stimuli were removed. An unknown mouse (different from the ones used for the 3-chamber test) of the same age, sex, weight, and genetic background was placed in the opposite side of the cage, and interactions were recorded for 5 min using MediaRecorder software (Noldus). The recorded videos were analyzed manually by a blinded experimenter and non-aggressive contact number and duration were quantified.
Morris water maze test
4-month-old male mice were used to assess spatial memory by a fully blinded experimenter (regarding the genotype). Mice were first habituated to swimming for 1 min in a circular pool of 1.5 m in diameter filled with opaque water (21 ± 1 °C) with non-toxic white paint. Mice were then trained for 7 days to locate a hidden platform using visual cues on the walls during 4 trials of maximum 1 min per day, each trial starting in a different corner of the pool. 48 h after the learning phase, a probe test was done by removing the platform and tracking mice for 1 min. During the whole test, mice were tracked using ANY-Maze software. Time to reach the platform was quantified during the training phase of the test, and time spent in the platform quadrant was quantified during the probe test. Animals that never learned to find the platform in less than 30 s were excluded from the analysis.
Mouse tissue collection
Mice were deeply anesthetized with xylazine (Rompun 0.2%) and ketamine (Imalgene 500) and euthanized by transcardial perfusion with 0.1 M PBS followed by 4% paraformaldehyde (liquid 32% PFA, Electron Microscopy Sciences, 15714) in phosphate buffer. Lumbar spinal cords and brains were collected and post-fixed in 4% PFA for respectively 4 and 24 h. Tissues were then cryoprotected in 30% sucrose in PBS for 72 h before freezing in isopentane at −40 °C.
For electron microscopy, mice were anesthetized as described and were transcardially perfused with 2% paraformaldehyde (liquid 32% PFA, Electron Microscopy Sciences, 15714) and 2% glutaraldehyde (liquid 10% glutaraldehyde, Electron Microscopy Sciences, 16010) in phosphate buffer.
Immunohistochemistry analyses
Antibody labeling
Lumbar spinal cord transverse serial cryosections of 30 μm thickness, and sagittal and coronal brain sections of 20 µm thickness were used to perform immunohistochemistry. Floating sections were incubated in a blocking solution containing 0.1 M PBS, 0.3% Triton ×-100 (or 1% Triton ×-100 for nuclear antigens) and 5% horse serum (Eurobio Scientific) for 1 h at room temperature, shaking. Sections were then incubated overnight at 4 °C with the following primary antibodies diluted in blocking solution: mouse anti-p62 (1:500, ab56416, Abcam), goat anti-ChAT (1:200, ab144p, Abcam), mouse anti-CRE (1:500, MAB3120, Merck Millipore), rabbit anti-IBA1 (1:500, W1W019-19741, Wako), goat anti-IBA1(1:1000, ab5076, Abcam), rabbit anti-PU.1 (1:100, 2258S, Cell Signaling Technology), rabbit anti-p-cJUN (1:200, 9261, Cell Signaling Technology), rabbit anti-GABARAPL1 (1:500, 11010-1-AP, Proteintech), mouse anti-Ubiquitin (1:300, FK2 BML-PW8810, Enzo Life Science), mouse anti-Cytochrome c (1:1000, 556432, BD sciences), rat anti-LAMP1 (1:500, 1D4B, DSHB), rabbit anti-NFL (1:500, ab9568, Abcam), mouse anti-NFH (1:500, RT97, MAB5262, Merck Millipore), mouse anti-GAD 67 (1:500, MAB5406, Merck Millipore), chicken anti-IBA1 (1:1000, 234 009, Synaptic Systems), rat anti-CD169 (1:200, MCA94G, Biorad), rat anti-CD3 (1:100, MCA500G, Biorad), rat anti-CD8 (1:100, MCA609G, Biorad), rat anti-CD68 (1:400, MCA1957, Biorad), mouse anti-Bassoon (1:200, ab82958, Abcam), rabbit anti-Homer1 (1:400, 160 003, Synaptic Systems), guinea pig anti-Synaptophysin1 (1:500, 101 004, Synaptic Systems), rabbit anti-PSD95 (1:500, 20665-1-AP, Proteintech), rabbit anti-VGAT (1:1000, 131 003, Synaptic Systems), rabbit anti-TH (1:1000, T9237-13, USBiological), rabbit anti-cFOS (1:1000, 226 008, Synaptic Systems, overnight at room temperature), rabbit anti-p21 (1:100, ab188224, Abcam). The different stainings were revealed using species-specific Alexa Fluor-488, -555, -594, or -647 secondary antibodies (1:500, Life Technologies), diluted in 0.1 M PBS, 0.3% Triton ×-100. Neurotrace dye (N21480, ThermoFisher, 20 min in PBS) was used to counterstain motor neurons. Nuclear stain was done using Hoechst (33342, ThermoFisher, 5 min in PBS). Fluorescence was preserved with Fluoromount G (00-4958-02, ThermoFisher). For high resolution STED microscopy, STAR secondary antibody (1:500, STORANGE-1001, Abberior) was used with mounting medium (solid antifade MM-2013, Abberior).
RNAScope
mRNA staining was performed on spinal cord sections for Tbk1 with the RNAscope Probe for Mm-Tbk1 (Biotechne, 418591) and on sagittal brain sections for Cxcl10 with the RNAscope Probe for Mm-Cxcl10 (Biotechne, 408921) according to manufacturer’s instructions and followed by immunohistochemistry to stain microglial cells (rabbit anti-IBA1, 1:500, Wako).
Image acquisition
Images were acquired using a AxioImager Z1 microscope (Zeiss) with ZenPro (v3.1) software, or an upright Spinning-Disk Confocal 3i microscope (Intelligent-Imaging) with a Hamamatsu Orca Flash 4.0 (sCMOS) camera, and Slidebook software (3i). Overview of brains were obtained using the Axioscan (Zeiss). For high resolution, images were acquired using Facility Line STED microscope (Abberior Instruments). Images were processed using Fiji software (ImageJ).
Image analysis
Morphology analysis of microglia was done using the skeleton plug-in in Fiji according to ref. 67 and at least 50 microglia were assessed per image. 3D analysis of images was done using IMARIS software (v10.1.1), to quantify synaptic density and phago-lysosomal volume. For analysis of MN in the spinal cord, the entire ventral spinal cord was analyzed per image. In the brain, images were taken only in the region of interest. For all quantifications, at least 4 images were taken and analyzed per condition/mouse.
Electron microscopy
Tissue processing
After tissue collection, spinal cords were sliced in 1 mm thick sections and post-fixed in 2% glutaraldehyde and 2% paraformaldehyde in sodium cacodylate buffer 0.1 M pH 7.4 (caco) for 2 h at 4 °C. Samples were washed 5 times with caco and then post-fixed for 1 h at RT in a caco solution containing 1% osmium tetroxide and 1.5% potassium ferricyanide. After extensive washes with distilled water, they were incubated for 1 h in 2% uranyl acetate in water. They were then dehydrated in a graded series of ethanol solutions. Final dehydration was performed twice in 100% acetone for 20 min. Samples were then progressively infiltrated with an epoxy resin, Epon 812® (EMS, Souffelweyersheim, France): 1 night in 50% resin/50% acetone at 4 °C in an airtight container, 2 × 2 h in pure fresh resin at room temperature. They were embedded in gelatin capsules (size 3, EMS, Souffelweyersheim, France) and the resin was polymerized at 56 °C for 48 h in a dry oven.
Sectioning
Blocks were cut with an UC7 ultramicrotome (Leica, Leica Microsystems SAS, Nanterre, France). Semi-thin sections (0.5 μm thick) were stained with 1% toluidine blue in 1% borax. Ultra-thin sections (70 nm thick) were recovered on copper grids and contrasted with Reynold’s lead citrate (Reynolds, ES (1963).
Imaging
Ultrathin sections were observed with a Hitachi HT7700 electron microscope (Milexia, Saint Aubin 91190, France) operating at 100 kV. Pictures (4096 × 4096 pixels) were taken with a Nanosprint12 camera.
RNAseq in vivo
Mouse motor neuron isolation
Motor neuron laser-microdissection was performed as in our earlier studies13,68. Briefly, mice were deeply anesthetized with xylazine (Rompun 0.2%) and ketamine (Imalgene 500) and euthanized by transcardial perfusion with ice-cold 0.1 M PBS. Lumbar spinal cords were rapidly recovered, rinsed, and frozen in −40 °C Isopentane in HistoPrep OCT compound (Fisher Scientific International), cryosectioned (16 μm) and sections mounted on RNase-free PEN-Membrane covered glass slides (Leica, 11505189). Sections were fixed for 60 s in 75% EtOH and stained for 30 s in 4% cresyl-violet acetate/0.3% acetic-acid (Sigma) to identify motor neurons, dehydrated in graded solutions of ethanol (70%, 95%, 100%) and dehydrated for 1 h before laser-microdissection (Leica LMD-7000). From one spinal cord, 200 large (diameter ≥ 25 μm) ventral horn motor neurons of the lumbar L4–L6 region were collected dryly, then immediately lysed in lysis buffer (NucleoSpin RNA XS Kit, Machery-Nagel) and the lysate stored at −80 °C. RNA was isolated with the NucleoSpin RNA XS Kit (with DNase treatment).
LPS injections
Mice received intra-peritoneal injections of lipopolysaccharide (LPS, Merck, L2654, 4 mg/kg) 24 h before microglial isolation. After 24 h, efficiency of the injection was confirmed by 10% of weight loss of the mice.
Mouse microglia isolation
Of note, we used a mechanical dissociation without enzymatic treatment to avoid any artefactual activation. Microglia isolation was performed as in an earlier study69. Briefly, mice were all females and were deeply anesthetized using xylazine (Rompun 0.2%) and ketamine (Imalgene 500) and euthanized by transcardial perfusion with ice-cold 0.1 M PBS followed by tissue collection. Brains were processed individually, and 2 spinal cords were pooled from 2 mice with the same genotype. For isolation after LPS, 2 spinal cords were harvested and two half brains were processed per individual datapoint. Tissues were Dounce homogenized in ice cold Hank’s balanced salt solution (HBSS) 10 times with the loose and 5 times with the tight pestles. Tissues were passed through a 70 μm cell strainer (Corning). Cells were pelleted by centrifugation (300 × g, 5 min), resuspended in 40% Percoll solution (GE Healthcare), HBSS, and centrifugated (30 min, 500 × g, 4 °C). Myelin was then removed by Percoll. The cell pellet was washed in HBSS and resuspended in PBS/2 mM EDTA (Life Technologies). Cells were stained with Zombie Green (1:1000, Biolegend) for 15 min at 4 °C. All samples were then washed and resuspended in ice cold FACS buffer (5% FCS, 2 mM EDTA, in 1× PBS) with TruStain FcX (Biolegend) for 10 min. Antibodies were then added, CD11b (PE, 1:100, Biolegend, 101207) and CD45 (SBB700, 1:20, Biorad, MCA1031), and incubated for 20 min at 4 °C. Finally, live CD45+, CD11b+ cells were sorted using a Biorad S3 cell sorter (see gating strategy as Supplementary Fig. 14). Approximately 30,000 cells were isolated per brain sample and 6000 per two spinal cords and directly lysed with RNA lysis buffer (RNeasy Micro Kit, Qiagen). Lysates were stored at −80 °C before RNA extraction according to manufacturer’s instructions (RNeasy micro kit, Qiagen, with DNase treatment). Quantity of RNA used for sequencing was adjusted to the number of cells sorted and equal amounts of cells per sample were sequenced.
Tbk1 RT-qPCR from isolated microglial cells
RNA from brain microglial cells of Tbk1-µG-WT and Tbk1-µG-KO mice was used to obtain cDNA with the SuperScript IV Kit (ThermoFisher), according to manufacturer’s instructions. Quantity of RNA used for cDNA was adjusted to the number of cells sorted and equal amounts per sample used. cDNA was then used to perform qPCR with TaqMan universal PCR Master Mix (ThermoFisher), and Tbk1 primers (00451150, ThermoFisher Scientific) and Tbp primers (00446973, ThermoFisher) in 96-well-plates, using the LightCycler 96 Real Time PCR System (Roche). Data were analyzed using LightCycler 96 software (v1.1).
Sample preparation and RNAseq
RNA obtained from isolated motor neurons and brain microglial cells were processed and sequenced together. RNA obtained from spinal cord microglial cells, and brain and spinal cord microglial cells after LPS injection were processed and sequenced together. Preparation of cDNA libraries for RNAseq was done using the SmartSeq2 method according to manufacturer’s instructions (SMART-Seq v4 Ultra Low Input RNA Kit for Sequencing, TaKaRa). For motor neurons, half (9.5 µL) of the total amount of eluted RNA (per sample) was used as starting material for reverse transcription. For brain microglia, the equivalent of 5000 cells was used as starting material for reverse transcription. For spinal cord and LPS-stimulated microglia, the equivalent of 3000 cells was used. For all, reverse transcription was followed by 12 cycles of pre-amplification. 150 pg of cDNA was used for RNAseq sequencing library preparation, according to manufacturer’s instructions (Nextera XT DNA Library Preparation, Illumina). Final sample pooled library preps were sequenced on a NovaSeq 6000 Sequencing System, ILLUMINA with S1-200 cartridge corresponding to 2 × 34 millions of (paired-end) reads per sample after demultiplexing. Quality of raw data was evaluated with FastQC. Poor quality sequences have been trimmed or removed with Trimmomatic software to retain only good quality paired reads. Illumina DRAGEN bio-IT Plateform (v3.8.4) was used to align reads on mm10 mouse reference genome (except for the in vivo LPS-stimulated microglial experiment that was aligned on mm39) and quantification with a gencode v25 annotation gtf file. Library orientation, library composition and coverage along transcripts were checked with Picard tools. Following analyses were conducted with R software. Data were normalized with DESeq2 (v1.26.0) bioconductor package, prior to differential analysis with glm framework likelihood ratio test from DESeq2 workflow. Multiple hypothesis adjusted p values were calculated with the Benjamini–Hochberg procedure to control FDR. Finally, enrichment analysis was conducted with clusterProfiler R package (v3.14.3) with Gene Set Enrichment Analysis, on Hallmark and custom Ageing database. Over-representation analysis was done using EnrichR.
Weighted correlation network analysis
WGCNA analysis was performed on 35 samples, corresponding to the three genotypes (Tbk1-µG-WT, Tbk1-µG-HET, and Tbk1-µG-KO; n = 5–7 per genotype and age) of microglial Tbk1 deletion in young and aged mice. Filtering was done first on the TPM value: only genes with a TPM = 1 in at least 5 samples (number of samples in the smallest group) were kept. We then excluded genes with the lowest variance (threshold set at 25%). At the end, the analysis was done on 11,534 genes. For the WGCNA analysis the authors recommend using VST-transformed counts (varianceStabilizingTransformation from DESeq2 package). Variance stabilizing transformation was done on the 35 samples. To build the signed network, we used a power = 12. Modules were identified using a minimum module size of 100 genes. Correlation was calculated between gene expressions (VST) and the module eigengene using R’s standard cor function (Pearson’s correlation). P values were calculated using WGCNA’s corPvalueStudent function. Eigengenes represent the weighted average expression profile in each module.
Meta-analysis
Upregulated genes in microglia in diseases were extracted from published works (log2FC > 0.5, adjusted p value < 0.05)29,30. Upset plot was generated using UpSetR70. STRING analysis was performed using Cytoscape (v3.10.1) and pathway analysis using the ClueGO application using the Gene Ontology biological process database.
Statistics and reproducibility
All manual counts were performed in a blinded manner. All data were presented as means with standard error of means as error bars. All statistics were performed using GraphPad Prism v10 (if not otherwise mentioned). Since sample sizes were too small, data was assumed to follow normal distribution, and statistical tests were chosen accordingly. All statistical tests were two-sided (if not otherwise mentioned) and the level of statistical significance was set at p or adjusted p < 0.05 (as mentioned). For all results that show representative images (micrographs), the experiment was done with at least n = 4 mice per genotype and condition, and if quantifications were done, at least 4 images were taken and analyzed per condition/mouse.
Brown–Forsythe tests were performed to test whether equal SDs could be assumed or not. Welch’s tests were used when SD were different. Two-tailed unpaired t-test, Welch’s t-test, one-way or two-way ANOVAs followed by Tukey’s multiple comparisons test or Dunnett’s multiple comparisons test were performed to compare different groups, or groups to a control group, respectively. Fisher LSD was performed to compare two groups and Sidak’s multiple comparisons test was performed to compare groups with repeated measures. Mixed effect models were used to compare repeated measures data when some data were missing. Chi-square test was used to compare the distribution of cells into subpopulations. For survival, or disease course analysis, Log-rank tests were performed.
For RNAseq data, the identification of DEGs was done using the DESeq2 method (with a two-tailed Wald test), and results were filtered by fold changes (log2FC ≥ 0.5 or log2FC ≥ 1.0, as mentioned) and (Benjamini–Hochberg) adjusted p values < 0.05 (FDR). Pathway enrichment GSEA for pathway enrichment is based on a Kolmogorov–Smirnov test according to Subramanian et al., 2005 (PMID: 16199517). Heatmaps show centered VST (Variance stabilizing transformation) expression values of the indicated genes (or as mentioned). Cytoscape gene/protein network analysis used a hypergeometric test, and group p values are indicated (p < 0.05) and were corrected with Bonferroni step down.
For the 3-chamber test sociability assessment, a linear mixed-effects model (LMM) was used to analyze the time of interaction with social elements across various genotypes. The fixed effects of the model included Social Element (S1 and E) and Genotype (Tbk1-µG-WT, Tbk1-µG-HET, and Tbk1-µG-KO), with Trial added as a covariate. A random intercept for each mouse was introduced to the model to control for intra-mouse variability due to the repeated measures design. To assess the significance of the main and interaction effects in the model, we conducted Type II Wald chi-square test using the ANOVA function in the car R package. The level of statistical significance was set at a p value < 0.05. These statistical analyses were conducted using R version 4.2.2 (R Development Core Team, 2022).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The RNAseq data generated in this study have been deposited in the GEO database under accession code GSE254229. This superseries contains: - GSE254228 for RNAseq of micro-dissected motor neurons (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE254228) - GSE254217 for RNAseq of in vitro microglia (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE254217) - GSE254227 for RNAseq of FACS-sorted brain microglia (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE254227) - GSE290810 for RNAseq of FACS-sorted spinal cord microglia, and LPS-stimulated brain and spinal cord microglia (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE290810) The full lists of DEGs and GSEA pathways from the RNAseq data are provided in Supplementary Data 1–6. Part of the RNAseq results (see Figs. 3c–g and 6) can also be visualized in our public web portal (https://boillee-rnaseq.icm-institute.org). Source Data are provided with this paper (Supplementary Data 7).
References
Ahmad, L., Zhang, S.-Y., Casanova, J.-L. & Sancho-Shimizu, V. Human TBK1: a gatekeeper of neuroinflammation. Trends Mol. Med. 22, 511–527 (2016).
Fingert, J. H. et al. Copy number variations on chromosome 12q14 in patients with normal tension glaucoma. Hum. Mol. Genet. 20, 2482–2494 (2011).
Taft, J. et al. Human TBK1 deficiency leads to autoinflammation driven by TNF-induced cell death. Cell 184, 4447–4463.e20 (2021).
Xu, D. et al. TBK1 suppresses RIPK1-driven apoptosis and inflammation during development and in aging. Cell 174, 1477–1491.e19 (2018).
Freischmidt, A. et al. Haploinsufficiency of TBK1 causes familial ALS and fronto-temporal dementia. Nat. Neurosci. 18, 631–636 (2015).
Couratier, P., Corcia, P., Lautrette, G., Nicol, M. & Marin, B. ALS and frontotemporal dementia belong to a common disease spectrum. Rev. Neurol. 173, 273–279 (2017).
Hardiman, O. et al. Amyotrophic lateral sclerosis. Nat. Rev. Dis. Primers 3, 17071 (2017).
Trist, B. G. et al. Co-deposition of SOD1, TDP-43 and p62 proteinopathies in ALS: evidence for multifaceted pathways underlying neurodegeneration. Acta Neuropathol. Commun. 10, 122 (2022).
Clarke, B. E. & Patani, R. The microglial component of amyotrophic lateral sclerosis. Brain 143, 3526–3539 (2020).
Boillee, S. et al. Onset and progression in inherited ALS determined by motor neurons and microglia. Science 312, 1389–1392 (2006).
Bonnard, M. Deficiency of T2K leads to apoptotic liver degeneration and impaired NF-kappaB-dependent gene transcription. EMBO J. 19, 4976–4985 (2000).
Marchlik, E. et al. Mice lacking Tbk1 activity exhibit immune cell infiltrates in multiple tissues and increased susceptibility to LPS-induced lethality. J. Leukoc. Biol. 88, 1171–1180 (2010).
Bruno, C. et al. Haploinsufficiency of TANK-binding kinase 1 prepones age-associated neuroinflammatory changes without causing motor neuron degeneration in aged mice. Brain Commun. 2, fcaa133 (2020).
Gerbino, V. et al. The loss of TBK1 kinase activity in motor neurons or in all cell types differentially impacts ALS disease progression in SOD1 mice. Neuron 106, 789–805.e5 (2020).
Gao, T. et al. Myeloid cell TBK1 restricts inflammatory responses. Proc. Natl. Acad. Sci. USA 119, e2107742119 (2022).
Xiao, Y. et al. The kinase TBK1 functions in dendritic cells to regulate T cell homeostasis, autoimmunity, and antitumor immunity. J. Exp. Med. 214, 1493–1507 (2017).
Rossi, J. et al. Melanocortin-4 receptors expressed by cholinergic neurons regulate energy balance and glucose homeostasis. Cell Metab. 13, 195–204 (2011).
Catanese, A. et al. Retinoic acid worsens ATG10-dependent autophagy impairment in TBK1-mutant hiPSC-derived motoneurons through SQSTM1/p62 accumulation. Autophagy 15, 1719–1737 (2019).
Morisaki, Y. et al. Selective expression of osteopontin in ALS-resistant motor neurons is a critical determinant of late phase neurodegeneration mediated by matrix metalloproteinase-9. Sci. Rep. 6, 27354 (2016).
Misawa, H. et al. Reappraisal of VAChT-Cre: preference in slow motor neurons innervating type I or IIa muscle fibers: MISAWA et al. Genesis 54, 568–572 (2016).
Chakrama, F. Z. et al. GABARAPL1 (GEC1) associates with autophagic vesicles. Autophagy 6, 495–505 (2010).
Zhao, X. et al. Noninflammatory changes of microglia are sufficient to cause epilepsy. Cell Rep. 22, 2080–2093 (2018).
Zhao, X.-F. et al. Targeting microglia using Cx3cr1-Cre lines: revisiting the specificity. eNeuro 6, ENEURO.0114-19.2019 (2019).
Holtman, I. R. et al. Induction of a common microglia gene expression signature by aging and neurodegenerative conditions: a co-expression meta-analysis. Acta Neuropathol. Commun. 3, 31 (2015).
Galatro, T. F. et al. Transcriptomic analysis of purified human cortical microglia reveals age-associated changes. Nat. Neurosci. 20, 1162–1171 (2017).
Salvany, S. et al. Microglial recruitment and mechanisms involved in the disruption of afferent synaptic terminals on spinal cord motor neurons after acute peripheral nerve injury. Glia 69, 1216–1240 (2021).
Moy, S. S. et al. Sociability and preference for social novelty in five inbred strains: an approach to assess autistic-like behavior in mice. Genes Brain Behav. 3, 287–302 (2004).
Cum, M. et al. A systematic review and meta-analysis of how social memory is studied. Sci. Rep. 14, 2221 (2024).
Sobue, A. et al. Microglial gene signature reveals loss of homeostatic microglia associated with neurodegeneration of Alzheimer’s disease. Acta Neuropathol. commun. 9, 1–17 (2021).
Chiot, A. et al. Modifying macrophages at the periphery has the capacity to change microglial reactivity and to extend ALS survival. Nat. Neurosci. 23, 1339–1351 (2020).
Lawson, L. J., Perry, V. H., Dri, P. & Gordon, S. Heterogeneity in the distribution and morphology of microglia in the normal adult mouse brain. Neuroscience 39, 151–170 (1990).
Brandi, E. et al. Brain region-specific microglial and astrocytic activation in response to systemic lipopolysaccharides exposure. Front. Aging Neurosci. 14, 910988 (2022).
Zondler, L. et al. Peripheral monocytes are functionally altered and invade the CNS in ALS patients. Acta Neuropathol. 132, 391–411 (2016).
Banerjee, P. et al. Cell-autonomous immune dysfunction driven by disrupted autophagy in C9orf72-ALS iPSC-derived microglia contributes to neurodegeneration. Sci. Adv. 9, eabq0651 (2023).
Vahsen, B. F. et al. C9orf72-ALS human iPSC microglia are pro-inflammatory and toxic to co-cultured motor neurons via MMP9. Nat. Commun. 14, 5898 (2023).
Lorenzini, I. et al. Moderate intrinsic phenotypic alterations in C9orf72 ALS/FTD iPSC-microglia despite the presence of C9orf72 pathological features. Front. Cell Neurosci. 17, 1179796 (2023).
Funes, S. et al. Expression of ALS-PFN1 impairs vesicular degradation in iPSC-derived microglia. Nat. Commun. 15, 2497 (2024).
Ito, Y. et al. RIPK1 mediates axonal degeneration by promoting inflammation and necroptosis in ALS. Science 353, 603–608 (2016).
Lall, D. et al. C9orf72 deficiency promotes microglial-mediated synaptic loss in aging and amyloid accumulation. Neuron 109, 2275–2291.e8 (2021).
Paolicelli, R. C. et al. TDP-43 depletion in microglia promotes amyloid clearance but also induces synapse loss. Neuron 95, 297–308.e6 (2017).
Krabbe, G. et al. Microglial NFκB-TNFα hyperactivation induces obsessive-compulsive behavior in mouse models of progranulin-deficient frontotemporal dementia. Proc. Natl. Acad. Sci. USA 114, 5029–5034 (2017).
Lui, H. et al. Progranulin deficiency promotes circuit-specific synaptic pruning by microglia via complement activation. Cell 165, 921–935 (2016).
Beers, D. R. et al. Wild-type microglia extend survival in PU.1 knockout mice with familial amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. USA 103, 16021–16026 (2006).
Shoji, H. & Miyakawa, T. Age-related behavioral changes from young to old age in male mice of a C57BL/6J strain maintained under a genetic stability program. Neuropsychopharmacol. Rep. 39, 100–118 (2019).
Wang, F. et al. The thalamic reticular nucleus orchestrates social memory. Neuron 112, 2368–2385.e11 (2024).
Molas, S. et al. A circuit-based mechanism underlying familiarity signaling and the preference for novelty. Nat. Neurosci. 20, 1260–1268 (2017).
Molas, S. et al. Dopamine control of social novelty preference is constrained by an interpeduncular-tegmentum circuit. Nat. Commun. 15, 1–14 (2024).
Tapper, A. R. & Molas, S. Midbrain circuits of novelty processing. Neurobiol. Learn Mem. 176, 107323 (2020).
Shan, Q., Tian, Y., Chen, H., Lin, X. & Tian, Y. Reduction in the activity of VTA/SNc dopaminergic neurons underlies aging-related decline in novelty seeking. Commun. Biol. 6, 1–16 (2023).
Bian, W.-J., Brewer, C. L., Kauer, J. A. & de Lecea, L. Adolescent sleep shapes social novelty preference in mice. Nat. Neurosci. 25, 912–923 (2022).
Hooshmandi, M. et al. Postnatal downregulation of Fmr1 in microglia promotes microglial reactivity and causes behavioural alterations in female mice. Mol. Autism 16, 17 (2025).
Hasegawa, Y. et al. Microglial cannabinoid receptor type 1 mediates social memory deficits in mice produced by adolescent THC exposure and 16p11.2 duplication. Nat. Commun. 14, 6559 (2023).
Hong, S. et al. Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science 352, 712–716 (2016).
Tastan, B. & Heneka, M. T. The impact of neuroinflammation on neuronal integrity. Immunol. Rev. 327, 8–32 (2024).
Zipp, F., Bittner, S. & Schafer, D. P. Cytokines as emerging regulators of central nervous system synapses. Immunity 56, 914–925 (2023).
Gemechu, J. M. & Bentivoglio, M. T cell recruitment in the brain during normal aging. Front Cell Neurosci. 6, 38 (2012).
Brenner, D. et al. Heterozygous Tbk1 loss has opposing effects in early and late stages of ALS in mice. J. Exp. Med. 216, 267–278 (2019).
Brenner, D. et al. A TBK1 variant causes autophagolysosomal and motoneuron pathology without neuroinflammation in mice. J. Exp. Med. 221, e20221190 (2024).
Shao, W. et al. Two FTD-ALS genes converge on the endosomal pathway to induce TDP-43 pathology and degeneration. Science 378, 94–99 (2022).
Scekic-Zahirovic, J. et al. Cytoplasmic FUS triggers early behavioral alterations linked to cortical neuronal hyperactivity and inhibitory synaptic defects. Nat. Commun. 12, 3028 (2021).
Yin, F. et al. Behavioral deficits and progressive neuropathology in progranulin-deficient mice: a mouse model of frontotemporal dementia. FASEB J. 24, 4639–4647 (2010).
Lopez-Herdoiza, M.-B. et al. C9ORF72 knockdown triggers FTD-like symptoms and cell pathology in mice. Front. Cell Neurosci. 17, 1155929 (2023).
Van Mossevelde, S. et al. Clinical features of TBK1 carriers compared with C9orf72, GRN and non-mutation carriers in a Belgian cohort. Brain 139, 452–467 (2016).
Thery, C., Chamak, B. & Mallat, M. Cytotoxic effect of brain macrophages on developing neurons. Eur. J. Neurosci. 3, 1155–1164 (1991).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Inserra, M. M., Bloch, D. A. & Terris, D. J. Functional indices for sciatic, peroneal, and posterior tibial nerve lesions in the mouse. Microsurgery 18, 119–124 (1998).
Young, K. & Morrison, H. Quantifying microglia morphology from photomicrographs of immunohistochemistry prepared tissue using ImageJ. J. Vis. Exp. 136, e57648 (2018).
Lobsiger, C. S., Boillée, S. & Cleveland, D. W. Toxicity from different SOD1 mutants dysregulates the complement system and the neuronal regenerative response in ALS motor neurons. Proc. Natl. Acad. Sci. USA 104, 7319–7326 (2007).
Hammond, T. R. et al. Single-cell RNA sequencing of microglia throughout the mouse lifespan and in the injured brain reveals complex cell-state changes. Immunity 50, 253–271.e6 (2019).
Lex, A., Gehlenborg, N., Strobelt, H., Vuillemot, R. & Pfister, H. UpSet: visualization of intersecting sets. IEEE Trans. Vis. Comput. Graph 20, 1983–1992 (2014).
Acknowledgements
We thank the following Paris Brain Institute core facilities (Paris, France): iGenSeq, Histomics, ICV-3C, DAC, DSI, ICM.Quant, ePHYS, and PhenoParc (which all received funding from the program Investissements d’avenir ANR-10-IAIHU-06). We thank D. Bouteiller and Y. Marie for RNAseq sample preparation (iGenSeq), J. Garrigue and B. Choisi for mouse genotyping (iGenSeq), A. Prigent and S. Ing for RNAscope analysis (Histomics), B. Gyorgy for RNAseq analysis (DAC), F.X. Lejeune for statistical analysis (DAC), S. Candelier for designing the RNAseq web portal (DSI), D. Akbar and G. Jimenez for help with image analysis (ICM.Quant), F. Deknuydt, A. Gestin, J. Van Wassenhove and S. Kerboua for assistance with cell sorting (ICV-3C), D. Langui for EM analysis (ICM.Quant), C. Dalle for electrophysiology analysis (ePHYS) and S. Louvegnies for help with behavioral analysis (PhenoParc). Part of this work was carried out in the AniFM animal core facility of the UMS28. We thank M. Lamaitre for advice and analysis of electromyology, and the technical staff from the animal housing facility, UMS28 CEF (Centre d’expérimentation fonctionnelle), M. Mihoc, D. Pintea, and O. Bregerie. We thank E. Huang (UCSF, USA) for providing the BAC-Cx3cr1-CreMMRRC #036395-UCD mice, and S. Berrard (Paris, France) for sharing the VAChT-Cre.FAST (VAChT-Cre.early) mice that were originally generated by H. Misawa (Tokyo, Japan). We thank B. Delatour, C. François, S. Hunot, J. Messeant, A. Bacci, E. Burguière, N. Renier, B. Lau, M. Dhenain, C. Mathis, C. Sephton, D. Seilhean, L. Dupuis, I. Munitic, E. Hedlund, F. René, and H. Hirbec for help and advice. Funding for this project came from: Association pour la Recherche sur la Sclérose Latérale Amyotrophique et autres Maladies du Motoneurone (ARSLA, France, to C.S.L.), Fondation pour la Recherche Medicale (FRM, Equipe FRM, EQU202103012581, France, to S.B.), NEURATRIS (Translational Research Infrastructure for Biotherapies in Neurosciences. France; to C.S.L., J.W. and D.B.), Aide à la Recherche des Maladies du Cerveau (ARMC, France), SLA Fondation Recherche (S.L.A.F.R., France), La longue route des malades de la SLA (France), Un pied devant l’autre (France) and Les invincibles all united associations, and private donors to ALS research at the Paris Brain Institute (France). I.L. received a PhD MESR (French Ministry for Higher Education and Research) fellowship, and a grant from ARSLA (4th year PhD). The research leading to these results has received funding from the national programs Investissements d’avenir ANR-10-IAIHU-0006 and ANR-11-INBS-0011/NeurATRIS (Translational Research Infrastructure for Biotherapies in Neurosciences).
Author information
Authors and Affiliations
Contributions
C.S.L., S.B. and I.L. designed the study. I.L., A.D., C.E.P., D.R., M.B., J.P., and N.S. performed the experiments. I.L., F.L., A.D., J.P., F.B., M.C., N.S., C.J.-M., D.Br. and M.R. analyzed the data. M.R., J.L., R.C., H.M., D.B., S.M., M.M., D.Br., J.H.W. and S.B. provided technical expertise and scientific input. C.S.L., S.B. and I.L. prepared the manuscript. All authors contributed to editing and reviewing the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Anja Capell and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Lenoel, I., Ribon, M., Lorenc, F. et al. ALS/FTD-linked TBK1 deficiency in microglia induces an aged-like microglial signature and drives social recognition deficits in mice. Nat Commun 16, 7951 (2025). https://doi.org/10.1038/s41467-025-63211-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-63211-w