Abstract
Long-range RNA-RNA pairing impacts the genome structure and function of SARS-CoV-2 variants. To understand the structure and function relationships of different SARS-CoV-2 variants that have emerged during the COVID-19 pandemic, we perform high-throughput structure probing and modelling of the genomic structures of the wildtype (WT), Alpha, Beta, Delta and Omicron variants of SARS-CoV-2. We observe that genomes of SARS-CoV-2 variants are generally structurally conserved, and that single-nucleotide variations and interactions with RNA binding proteins can impact RNA structures across the viruses. Importantly, using proximity ligation sequencing, we identify many conserved ultra-long-range RNA-RNA interactions, including one that spans more than 17 kb in both the WT virus and the Omicron variant. We show that mutations that disrupt this 17 kb long-range interaction reduce viral fitness at later stages of its infection cycle, while compensatory mutations partially restore virus fitness. Additionally, we show that this ultra-long-range RNA-RNA interaction structure binds directly to ADAR1 to alter the RNA editing levels on the viral genome. These studies deepen our understanding of RNA structures in the SARS-CoV-2 genome and their ability to interact with host factors to facilitate virus infectivity.
Similar content being viewed by others
Introduction
SARS-CoV-2 belongs to the family Coronaviridae characterized by large (26 – 32 kb) positive-strand RNA genomes1. Since its emergence in 2019, SARS-CoV-2 has caused millions of deaths world-wide2,3. In addition to the original WT strain, many SARS-CoV-2 variants including Alpha, Beta, Delta and Omicron variants of concern (VOCs) have emerged and circulated around the world3,4. Compared to the WT virus, VOCs contain different amounts of mutations along their genome which results in differences in their infectivity among humans. Among the VOCs, the Omicron variant contains 128 mutations as compared to the WT strain and is the most divergent VOC in terms of its genomic sequence (Supplementary Fig. 1a–c)5.
Numerous studies have shown that different RNA structures, including the frameshifting element (FSE), present within the SARS-CoV-2 genome are important for virus functions6,7,8,9. A number of studies devoted to mapping the secondary structure of the SARS-CoV-2 genome, both in infected cells and in virions, have been published since 202010,11,12,13,14,15,16,17,18. However, the locations of variable and conserved RNA structures across genomes of different VOCs, how single-nucleotide variations (SNVs) and RNA binding proteins (RBPs) impact RNA structures, and how ultra-long-range RNA-RNA pairing within the genome impacts virus functions are still under-explored.
Here, we probe the RNA secondary structures of WT, Alpha, Beta, Delta and Omicron variants to identify structurally variable and conserved regions along their genomes. We identify highly conserved regions along the SARS-CoV-2 genome, as well as structural changes due to mutations and RBPs binding to the RNA genome. We also identify conserved ultra-long-range RNA-RNA interactions that span distance more than 10 kb, across the variants, including a paired RNA structure element that spans 17 kb on the SARS-CoV-2 genome. Mutations impairing this interaction and compensatory mutations restoring this long-range interacting RNA structure reveal that it is important for virus fitness. We observe that this element can bind to ADAR1 and induce RNA editing along the virus genome. This study expands our understanding of the structure and function of the SARS-CoV-2 RNA genomes.
Results
Mapping of RNA structures across five different variants of SARS-CoV-2
To probe the RNA secondary structures along genomes of SARS-CoV-2 variants, we infected Vero-E6 TMPRSS2 cells using WT, Alpha, Beta, Delta or Omicron variants and treated the cells with a structure probing compound (NAI) at 48 hours post-infection (hpi) (Fig. 1a). We then extracted the RNAs, performed reverse transcription, library construction and deep sequencing, followed by mapping the reads to the Vero transcriptome as well as to the respective SARS-CoV-2 genomes.
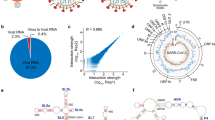
a Schematic showing the workflow for SHAPE-MaP probing and identifying potential RNA structural elements with regulatory functions in five virus variants. Virus genomes were probed inside infected cells using NAI, a SHAPE-like chemical, which modifies single-stranded RNA bases along the viral genome. The SHAPE reactivity profiling data was then used to constrain computational models to derive more accurate structural predictions for SARS-CoV-2. Image was created in BioRender. Wang, J. (2025) https://BioRender.com/69e2bov. b SHAPE-reactivity profiles for genomes of five SARS-CoV-2 variants. High reactivity indicates a higher probability of single-strandedness. c Structure models of 5’ UTR for five variants of SARS-CoV-2 created using the program RNAstructure with SHAPE-reactivities as constraints and visualized with VARNA. Source data are provided as a Source Data file.
We obtained a total of 689 million reads for at least 2 biological replicates (Supplementary Table 1), of which 256 million (37.1%) were aligned to the SARS-CoV-2 variants. As expected, NAI-treated samples showed an increase in mutation rates as compared to untreated samples for all variants (Supplementary Fig. 2a). We also observed highly consistent SHAPE-reactivities between two replicates (R = 0.86 – 0.93), indicating that our structure probing is robust (Supplementary Fig. 2b). Across the SARS-CoV-2 genomes, we were able to capture structural information for ~84% of the bases (Fig. 1b, Supplementary Fig. 2c), enabling us to identify RNA structures across the genomes of different variants. We confirmed that our SHAPE-reactivities recapitulate expected double- and single-stranded RNA signals on the 5’ and 3’UTRs of the SARS-CoV-2 genome (Fig. 1c, Supplementary Fig. 3a,b), and have high AUC-ROC against known SARS-CoV-2 structures (Supplementary Fig. 3c), indicating that our structure probing is accurate.
We had previously performed structure probing on a WT SARS-CoV-2 virus together with a 382-nucleotide deletion mutant (Δ382) to identify functional SARS-CoV-2 structures15. Now, probing RNA structures across five variants enabled us to identify a more conserved set of six low SHAPE reactivity and low Shannon entropy (LSLS) regions along their genomes (Fig. 2a, Supplementary Fig. 4a). Additionally, to identify structurally similar regions along the SARS-CoV-2 genomes, we performed Pearson correlation on sliding windows of SHAPE-reactivities along the different pairs of the genomes of SARS-CoV-2 variants (Supplementary Fig. 4b). We also used the reactivity along each virus genome to model local structures for the full genome and performed pair-wise local structure comparisons to identify the top 20 most structurally conserved elements across all the variants (Fig. 2a, b, Supplementary Fig. 5a, Supplementary Data 1). These structures include known conserved features such as the 5’UTR, the FSE, and conserved LSLS regions in the Orf3a described above (Fig. 2a, b). Using a set of 1581 sequences from the Coronaviridae family12, we observed that these 20 structures are supported by evolutionary covariation, indicating that they are likely to be functionally important (Fig. 2b). To test whether these conserved structures are important for virus fitness, we designed an antisense oligonucleotide (ASO) against a region in Orf7a (residues 27695-27714 in WT RNA genome) that is highly base-paired and structurally conserved across the analysed viruses. We observed that transfecting cells with this ASO before WT and Omicron infection reduced the replication of both viruses, as compared to cells transfected with an ASO that targets a randomly selected region of the SARS-CoV-2 genome, or with a non-targeting control ASO, indicating that this structure could be functionally important for virus replication (Supplementary Fig. 6a,b,c).
a Consensus regions (150 bases) with the lowest 20% SHAPE reactivity (black), Shannon entropy (blue), greatest difference (top 20%) between actual and shuffled energies (green), high SHAPE-MaP correlation (R ≥ 0.8, orange), and the top 20 structurally conserved regions (at least 85% identical base pairs, light blue) are shown along the SARS-CoV-2 genome map. b RNA structure models for the Omicron variant. The top 20 most conserved structures (light blue bars) were generated with the relevant RNAstructure program modules using SHAPE-MaP reactivity as a constraint and visualized with VARNA46,50,51. SHAPE-reactivities are mapped onto structure models as a low-to-high colour gradient, with red indicating bases with the highest accessibility. Base pair covariance was calculated via a protocol adapted from Sun, L. et al.12 and mapped to the corresponding pair with colour coding signifying strong (> 0.7), intermediate (0.5–0.7), and weak (0.4–0.5) covariance. The number of mutations (substitutions and indels) at a given genome position across the four VOCs (Alpha, Beta, Delta and Omicron) is displayed as purple gradients. The genome map shown for position reference is computed from a MSA of the WT SARS-CoV-2 and four VOCs. Source data are provided as a Source Data file.
SNVs and RBPs impact virus structure along the virus genome
As the VOCs have different mutations along their genome (Supplementary Fig. 1a–c), we examined the impact of the SNVs on RNA structure differences between these viruses. Across WT and four VOCs, we observed 720 pair-wise nucleotide differences, with most of the bases changing from C to U (Fig. 3a). Generally, RNA structures around SNVs were more variable than RNA structures around non-SNV bases (Fig. 3b, Supplementary Fig. 7a,b). Out of 720 pair-wise mutations across the different viruses, we observed that 310 ( ~ 43%) of them showed changes in RNA structures. Mutations that changed the original residue into purine bases (A/G) resulted in larger structural changes than SNVs that changed into pyrimidine bases (U/C, Supplementary Fig. 8a). As expected, bases that are mutated to A tend to become less paired, while bases that are mutated to G tend to become more paired (Supplementary Fig. 8a). As previously observed, RNA structure changes caused by SNVs are largely local and influence the reactivity of ~10 bases up- and downstream of the SNV (Fig. 3c). Scanning across the SARS-CoV-2 genomes, we observed that one of the most structurally heterogeneous regions is associated with an SNV (C- > U at base 7141 of the Delta variant, Supplementary Fig. 5a). Structure modelling of this region using SHAPE-reactivity as constraints showed that the region around position 7100 can form different structures in different variants (Fig. 3d). Additionally, many SNVs do not result in local structure changes, as seen by the relatively structurally stable regions in the RNA region encoding for the S-protein, despite the large number of mutations present in the gene (Fig. 3e). To determine whether other cellular factors such as RBPs can impact RNA structure changes in different SARS-CoV-2 variants, we identified 72 RBPs that are differentially expressed in Vero-E6 cells that are infected by different strains of SARS-CoV-2. These RBPs are experimentally identified to bind to SARS-CoV-212,19,20,21,22,23, and are known to have binding motifs (Fig. 3f–h, Supplementary Fig. 9a–c). While differences in RBP expression generally do not result in changes in RNA structures, we identified a few RBPs including HNRNPU and FXR1, whose expression was associated with the formation of different RNA structures in the SARS-CoV-2 genome (Fig. 3i, j, Supplementary Fig. 10a-c). In particular, we observed that a highly variable region around position 11000 along the SARS-CoV-2 genome folds into different predicted structures in the presence of different levels of HNRNPU (Fig. 3j, Supplementary Fig. 10a). To validate that HNRNPU binding indeed results in structure changes, we performed an in vitro assay by incubating a 500-base RNA fragment, centred around the HNRNPU binding site in the SARS-CoV-2 WT genome, in the presence or absence of the HNRNPU protein, and performing RNA structure probing and sequencing. Our SHAPE-MaP results show that adding HNRNPU protein results in an increased accessibility of a region upstream of the HNRNPU binding site (Fig. 3k), similar to what was observed in infected cells, whereby high HNRNPU expression (in WT infected cells) results in an increased accessibility of a region upstream of the HNRNPU binding site, as compared to cells with low HNRNPU levels (in Beta infected cells, Fig. 3k, Supplementary Fig. 10a,b). As HNRNPU is a novel RNA sensor involved in viral sensing, the different structures could be associated with differences in detecting different SARS-CoV-2 variants inside cells24,25.
a Bar-chart summarizing nucleotide substitutions across all genome pairs of WT and VOC. b Box-plot comparing ΔSHAPE reactivity at conserved versus variable sites (normal n = 168,921, substitutions n = 76,128). The box indicates the first and third quartiles, and the median is indicated as the line. The whiskers show the minimum and maximum values, and the circles are outliers. Significance was calculated by two-sided t-test. c Top: Line plots showing structure change as a base transitions to either double- or single-stranded. Bottom: Line plot indicates the degree of structure similarity at each base centered on the substitution. Light blue shaded bands indicate ± variance in structure conservation around a genome position (n = 720). d, e RNA secondary structure models are constrained by SHAPE reactivity and visualized with VARNA for the indicated SARS-CoV-2 variant. Nucleotide substitutions are circled in green. f Bar graph illustrating the number of differentially expressed RBPs in all VOC pairs. g Upset plot integrating six studies to define the SARS-CoV-2 RNA-bound proteome (RBPome). h Venn diagram depicting the intersection of several RBP sets from multiple databases. Confirmed SARS-CoV-2 RBPs are unique RBPs aggregated from literature sources, while Prospective - (DGE RBPs) are differentially expressed genes cross-referenced with Confirmed and ATtRACT Database RBPs lists. i Scatterplot correlating RBP expression with local structure similarity around RBP binding motifs. Experimentally confirmed RBPs are in light blue (Confirmed), while predicted RBPs from motif enrichments are coloured in black (Prospective). j Structure models of a region containing an HNRNPU motif (circled in purple) in WT, Beta, and Omicron show conserved stem-loops with reorganized long-range interactions. Structure changes are concordant with variant-specific HNRNPU expression. k Top, ΔSHAPE profile of HNRNPU binding region on SARS-CoV-2 genome comparing WT-infected with Beta-infected cells. Bottom, ΔSHAPE profile for an in vitro 500-nt fragment probed in the presence and absence of HNRNPU protein. Significantly less structured regions (positive change) are in light blue (two-tailed paired t-test, p < 0.01). HNRNPU motif is boxed in purple. Source data are provided as a Source Data file.
A long-range RNA-RNA interaction impacts virus fitness
As SARS-CoV-2 can form long-range RNA-RNA interactions within the genome that are hard to predict using SHAPE, we performed proximity ligation sequencing (SPLASH) to identify intra- and intermolecular RNA-RNA interactions in the cells infected with WT SARS-CoV-2 or the Omicron variant (Supplementary Table 2). This analysis identified 260 and 308 intramolecular interactions along the WT and Omicron genomes, respectively. We observed that 43−60% of the intramolecular RNA interactions in these two strains are long-range interactions that span more than 1 kb in distance (Fig. 4a, b, Supplementary Fig. 11a). Out of all 260 SPLASH interactions in the WT strain, 172 (66%) are present in Omicron, including 73 out of 155 (47%) longer-range interactions (> 1 kb, Fig. 4c, d). Both intra- and intermolecular RNA-RNA interactions are enriched in the structurally similar regions identified above (Fig. 4e, f). Additionally, we see an impact of SNV on intramolecular RNA interactions whereby only 54% of RNA-RNA interactions with SNVs (as compared to 66%) are conserved between WT and Omicron strains (Fig. 4g, Supplementary Fig. 11b). SNVs preferentially disrupt longer-range (> 1 kb, 39% conservation between WT and Omicron) versus shorter-range (≤ 1 kb, 85% conservation between WT and Omicron) RNA-RNA interactions (Fig. 4h, Supplementary Fig. 11b), suggesting that shorter-range RNA-RNA interactions are likely to be more conserved.
a Arc plots showing the top 40 pair-wise RNA-RNA interactions along the SARS-CoV-2 WT and Omicron genomes. Shared interactions between the WT and Omicron variants are highlighted in green. Unique interactions in WT (blue) and in Omicron (red) are plotted on the top and bottom panels, respectively. N = 2 independent experiments (total 4 replicates for each variant). b Bar-chart showing the abundance of SPLASH interactions that span different lengths along the WT and Omicron genomes. Interactions that span a distance longer than 1 kb are classified as long-range interactions and comprise 59.6% of all interactions in WT and 43.2% in Omicron, respectively. c Venn diagram showing the overlap of SPLASH interactions between SARS-CoV-2 WT (grey) and Omicron (pink) identified in infected Vero-E6/TMPRSS2 cells. d Multilevel doughnut chart showing unique or shared SPLASH interactions of SARS-CoV-2 WT and Omicron that span different distances along the genome. e Bar-chart showing interactions per base for short- (≤ 1 kb) and long-range (> 1 kb) interacting regions that fall within structurally conserved or non-conserved regions in WT and Omicron variants, respectively. f Bar-chart showing the number of RNA-RNA interactions between virus and host per base in structurally conserved and non-conserved regions. g Bar-chart showing the frequency of conserved interactions and conserved interactions with SNVs in WT and Omicron variants. h Bar-chart showing the frequency of conserved interactions with SNV that span short-range (≤ 1 kb) and long-range (> 1 kb) along the virus genome. P-values were calculated using a one-sided chi-square test. i Venn diagrams showing the intersection of RNA-RNA interactions that were experimentally identified from WT and Omicron-infected cells by SPLASH, as well as predicted interactions in all VOCs based on sequence conservation. Source data are provided as a Source Data file.
Out of a total of 260 RNA-RNA interactions detected, 78 interactions span longer than 10 kb in the WT strain. Sequence analysis revealed that from these ultra-long-range RNA-RNA interactions, 32 are predicted to be conserved across all VOCs, and 31 were experimentally confirmed to be conserved between WT SARS-CoV-2 and the Omicron variant (Fig. 4i, Supplementary Fig. 11c,d). One of these conserved long-range RNA-RNA interactions, named LR1, brings bases around position 2000 (in the nsp2 encoding region), to interact with bases around position 19760 (in the nsp15 encoding region, Fig. 5a). To explore the full ensemble of RNA structures that this interaction can form, we generated 1000 structures stochastically from the base-pairing probability of this region. We observed two main clusters, one corresponding to local structures (C1, C2, Fig. 5b) which account for a third of all structures in the ensemble, and the rest corresponding to long-range interaction structures (C3, C4, Fig. 5b).
a Top, Arc plot showing a conserved long-range RNA-RNA interaction (LR1) that spans 17 kb in distance along the SARS-CoV-2 genome. Bottom, structure model of the LR1 interaction. The mutations for LR1A and LR1B mutant substitutions are indicated in green and orange, next to the original bases, respectively. Compensatory mutant LR1AB contains all the mutations for LR1A and LR1B to restore the proposed RNA structure. b Clustering analysis of the LR1 secondary structure ensemble reveals two populations of distinct structural morphology among the four clusters identified in the structural landscape (n = 1000). Long-range interaction structures in clusters C1 & C2 (dark and light blue) and clusters C3 & C4 (dark and light orange) are highly homologous, differing by several base pair variations. Structure conformations (C3 & C4) that maximize long-range base pairing are more numerous and more stable than conformations that maximize local base pairing (C1 & C2). c Bar-chart showing the viral load of SARS-CoV-2 WT and its mutants in the supernatants of infected Vero-E6 cells. Each column represents the mean ± SD of three technical replicates from one representative experiment. The same trends were observed in n = 3 independent experiments. P-values are calculated by two-sided t-test. d Diagram showing that antisense oligonucleotide (ASO_LR1) disrupts the original long-range base-pairing of LR1. e Workflow showing transfecting Vero-E6 cells with ASO, followed by SARS-CoV-2 WT or Omicron infection. Image was created in BioRender. Wang, J. (2025) https://BioRender.com/xy543s1. f, g Box-plot showing the amount of SARS-CoV-2 WT and Omicron RNAs in cells that were transfected with different ASOs, at indicated timepoints. The box represents the 25–75th percentiles, the median is the line inside. The whiskers show the minimum and maximum values. N = 2 independent experiments (total 6 replicates). ASO_CTRL: ASO that does not target any region on the viral genome, ASO_1: ASO that targets a randomly selected region (genome position 1563-1582), ASO_LR1: ASO that targets LR1 to disrupt the long-range base-pairing structure. P-values were calculated by two-sided Mann-Whitney test. Source data are provided as a Source Data file.
To determine if the long-range base-pairing is indeed important for SARS-CoV-2 fitness, we introduced silent mutations affecting one side of the interacting regions (LR1A), another side of the interacting regions (LR1B), or both sides (LR1AB) into the genome of the SARS-CoV-2-Wuhan-Hu1 strain (Fig. 5a). Interestingly, we observed that mutating the A side resulted in lower copy numbers of the viruses being produced in infected cells at both 24 and 48 hpi, while mutating the B side resulted in a mild attenuation at 48 hpi. As the B side mutations introduced A to G substitutions, resulting in GU base-pairing, we expected the B mutations to be less disruptive, agreeing with our experimental data (Fig. 5a, c). Introducing compensatory mutations on both A and B sides partially rescues the low copy number of the LR1A mutant, indicating that the base-pairing is important for virus fitness (Fig. 5c). Interestingly, we observed that the mutation resulted in the reduction of virus titres only at late time points, 24 and 48 hpi. The mutant viruses generally displayed accelerated replication at earlier time points, suggesting that LR1 might influence later stages of the virus infection cycle, such as genome packaging (Supplementary Fig. 12a–d).
To confirm that LR1 base-pairing is indeed important for virus fitness, we designed an ASO that is complementary to bases 19758-19779 to inhibit the base-pairing of this region (Fig. 5d). Transfecting cells with the ASO, followed by SARS-CoV-2 infection, reduced the replication of both the WT SARS-CoV-2 and Omicron variants while transfecting control ASOs did not (Fig. 5e–g), supporting our hypothesis that this interaction is important for virus fitness across different variants. In vivo mouse experiments whereby mice that inhaled two different concentrations of LR1 ASO before being exposed to SARS-CoV-2 showed an attenuation of SARS-CoV-2 infection, including decreased weight loss and lower virus titres in mice (Supplementary Fig. 12e–h), suggesting that LR1 ASO could act as a prophylactic in mice.
To test which cellular factors could potentially bind to LR1 to regulate the growth of SARS-CoV-2, we generated a biotinylated version of a 158-base-long mini-construct that contains the LR1 base-pairing (mLR1) and incubated this RNA with cellular lysates (Fig. 6a). RNA pull-down followed by mass spectrometry was used to identify proteins that could bind to mLR1. We identified ADAR1 as an RNA binding protein that interacts with mLR1 across three biological replicates (Supplementary Fig. 13a, Supplementary Data 2), with ADAR1 binding strongest to mLR1-WT, and less so to the mutants mLR1A and mLR1B RNA (Supplementary Fig. 13b). To confirm this interaction, we performed a Western blot experiment using lysates from Vero-E6. We observed that ADAR1 was pulled-down with mLR1-WT and that this binding is significantly reduced in mutants mLR1A, mLR1B, and non-detectable for the highly structured Tetrahymena RNA control (Fig. 6b). We observed a similar result when the pull-down assays were performed using human lung cancer A549 cells (Supplementary Fig. 13c). To confirm that ADAR1 binds to this long-range LR1 interaction region inside cells, we also performed two biological replicates of enhanced crosslinking and immunoprecipitation (eCLIP)26 experiment using antibody against ADAR1 (Supplementary Fig. 13d). We observed highly reproducible ADAR1 binding across the SARS-CoV-2 genome (Supplementary Fig. 13e), and significant binding of ADAR1 to the 5’ arm of LR1 in the SARS-CoV-2 genome (Supplementary Fig. 13f). To test whether mLR1 indeed binds to ADAR1 directly, we performed an electrophoretic mobility shift assay (EMSA) using different amounts of ADAR1 protein. Again, we observed that WT mLR1 binds to ADAR1 the strongest, that this binding was partially disrupted by mutations introduced to the mLR1 (mLR1A or mLR1B), and is restored in the case of the mLR1AB compensatory mutant (Fig. 6c, d). We also observed that the binding between ADAR1 and the highly structured Tetrahymena RNA was weak, suggesting that ADAR1 indeed specifically binds to LR1 and not to structured RNAs in general (Fig. 6c, d).
a Structure model of a 158-base-long mini-construct that contains the long-range interaction LR1 base-pairing (mLR1). Mutations intended to disrupt the base-pairing from one side (mLR1A, green) or another side (mLR1B, orange) are indicated next to the original bases. Compensatory mutant mLR1AB contains both A and B mutations restoring the pairing of mLR1. b Top, PAGE showing proteins that were immunoprecipitated using biotinylated mLR1-WT RNA and negative control RNAs mLR1A (lane A), mLR1B (lane B) and Tetrahymena ribozyme (Teth). Input proteins from the whole cell lysate of Vero-E6 are also shown. Bottom, Western blot analysis of proteins from the pull-down assay using anti-ADAR1 antibody. c EMSA experiment showing the binding of different concentrations of ADAR1 with indicated RNAs. d Quantification of the bound RNA fraction shown in c. Data are presented as mean ± SD (n = 3 independent experiments). e A-to-I RNA editing level along SARS-CoV-2 WT, LR1A, LR1B and LR1AB mutant genomes in infected cells. N = 3 independent experiments (total 6 replicates). f Box-plot showing the distribution of editing level of each edited site. The total number of edited sites across three biological replicates is indicated. g Box-plot showing the distribution of the editing index (n = 6) in samples as described for panel (e). h A-to-I RNA editing level along SARS-CoV-2 WT genome in cells transfected using a random ASO (ASO_CTRL), an ASO that targets a different region (ASO_1, against 1563-1582), or an ASO designed against the LR1 region (ASO_LR1). N = 2 independent experiments (total 4 replicates). i Box-plot showing the distribution of the editing level of each edited site. Total number of edited sites across two biological replicates is indicated. j Box-plot showing the editing index in samples treated as described for panel h (n = 4). For box-plots, the box represents the 25–75th percentiles, median is indicated as the line. The whiskers show the minimum and maximum values. P-values were calculated using two-sided Wilcoxon Rank Sum Test. ns, not significant. Source data are provided as a Source Data file.
ADAR1 has been shown to edit the genome of SARS-CoV-2. Additionally, COVID-19 patients have also been observed to have increased A-to-I editing levels along host transcripts27,28. To confirm that ADAR1 binding results in changes in RNA editing on the SARS-CoV-2 genome, we performed RNA sequencing analysis on Vero-E6 cells infected by SARS-CoV-2-Wuhan-Hu1 (WT), LR1A mutant, LR1B mutant, and LR1AB compensatory mutant viruses. Across three biological replicates, we identified a total of 3022 A-to-I editing sites in the WT, 17 in the LR1A mutant, 1361 in the LR1B mutant, and 321 in the LR1AB mutant genomes (Fig. 6e). This suggests that A-to-I editing is severely repressed when LR1 is mutated and that restoring the base-pairing in the LR1AB mutant partially restores the number of editing sites on the genome. In addition to calculating the number of editing sites, we also calculated an editing index for each genome, which is the normalized number of editing sites that reflects the editing status of each sample. We observed that the LR1A mutant showed significantly lower editing index as compared to the WT, LR1B, and LR1AB mutants (Fig. 6g), indicating that it has lower editing levels. We confirmed our observation by calculating A-to-I editing upon ASO disruption of the LR1 pairing. Treating the infected Vero-E6 cells with a control ASO that does not target the SARS-CoV-2 genome (2071 A-to-I sites) and with an ASO that targets another region of the SARS-CoV-2 genome (1333 A-to-I sites) resulted in high levels of A-to-I editing along the virus genome as was also observed in untreated cells (1283 A-to-I sites). However, transfecting cells with ASO against LR1 prior to virus infection resulted in a ten-fold decrease of A-to-I editing sites (107 A-to-I sites), confirming that disrupting LR1 impacts A-to-I editing along the virus genome (Fig. 6h). Consistently, the editing index of ASO_LR1-treated cells was also significantly reduced as compared to control ASO-treated cells as well as untreated cells (Fig. 6j). In addition to quantifying the number of editing sites, we also quantified the percentage of editing at each edited site (editing level). We observed a decrease in editing levels upon mutating LR1 which was, again, partially restored by compensatory mutations (Fig. 6f). The same effect was observed when transfecting cells with the ASO designed against LR1, and not with the ASO against another part of the virus genome (Fig. 6i), confirming that LR1 binding to ADAR1 results in editing of the SARS-CoV-2 genome.
To determine whether this decrease in A-to-I editing in the LR1 mutants is due to decreased levels of ADAR1 expression in the infected cells, we performed RT-qPCR analysis to determine the amount of ADAR1 mRNA expression in SARS-CoV-2-Wuhan-Hu1 (WT), LR1A, LR1B, and LR1AB mutants infected cells. Interestingly, we observed a similar level of ADAR1 expression in all the infected cells (Supplementary Fig. 14a), indicating that the low RNA editing levels in LR1 mutants were not due to changes in ADAR1 levels, but likely due to differential recruitment of ADAR1. As ADAR1 has been shown to bind to SARS-CoV-2 nucleocapsid (N) protein to interact with the SARS-CoV-2 genome29, we also tested for the ability of mLR1 WT and mutant RNAs to bind to N protein by using the biotinylated mLR1 mini-constructs. Interestingly, we observed that WT mLR1 binds strongly to the N protein, the binding is disrupted in mLR1 mutants (mLR1A, mLR1B), and partially restored in mLR1AB compensatory mutant (Supplementary Fig. 14b,c), suggesting that both ADAR1 and N protein co-bind to the SARS-CoV-2 genome. We also observed that WT and mLR1 mutants do not bind to SARS-CoV-2 envelope (E) protein, suggesting that mLR1 binding to ADAR1 and N proteins is specific (Supplementary Fig. 14d). This is consistent with our observation above that the LR1 mutation impacts late stages of the virus infection and could be impacting genome encapsulation of the SARS-CoV-2.
Discussion
Numerous mutations have emerged in the SARS-CoV-2 genome during the years of the COVID-19 pandemic. These changes have increased transmission of SARS-CoV-2 as well as allowed escape from immune responses. However, how they impact RNA structures in the SARS-CoV-2 RNA is still understudied. Additionally, identifying ultra-long-range RNA-RNA base pairing and understanding their functions for RNA viruses remains a challenge in the field due to the extensive amount of search space that these interactions can form. Here, we utilized high-throughput structure mapping to map RNA structures across five different variants of SARS-CoV-2 to identify structurally similar RNA regions. We also used proximity ligation sequencing to identify shared long-range RNA-RNA interactions in the WT SARS-CoV-2 and Omicron variant and observed that SNVs preferentially impact long-range RNA structures. We identified 78 long-range RNA-RNA interactions that span more than 10 kb, out of which 31 were present in both the WT SARS-CoV-2 and Omicron variants. Interestingly, one of these interactions spans 17 kb in distance, can result in two alternative structures, and is important for virus fitness. Our experiments show that this long-range RNA element binds directly to ADAR1 and is associated with increased RNA editing along the SARS-CoV-2 genome.
A-to-I editing has been associated with SARS-CoV-2 infection27,30. However, how it impacts viral infection remains to be fully understood. Large-scale proteomic screens have identified ADAR1 as an interaction partner with the SARS-CoV-2 genome20,22. Previous literatures have also shown that the SARS-CoV-2 genome undergoes A-to-I editing, and that the transcriptome of SARS-CoV-2-infected cells shows elevated A-to-I editing28,30,31. Here, we identified an RNA element that could potentially recruit ADAR1 for RNA editing of the SARS-CoV-2 genome. As SARS-CoV-2 is sensitive to the interferon response and A-to-I editing of the virus genome could dampen the interferon response upon infection, we hypothesize that the editing of the SARS-CoV-2 genome could impact cellular sensing of the viral RNA, enabling the virus to replicate better inside cells. However, ADAR1 alone might not be sufficient for this effect to occur. ADAR1 is shown to work together with other proteins such as DHX9, which is another top hit in our LR1 immunoprecipitation-mass spectrometry data, to impact RNA sensing, complicating our understanding of how SARS-CoV-2 can be recognized inside cells32. Additionally, as disrupting the LR1 long-range interaction impacts virus fitness in the late stages of its lifecycle, and we observed binding of nucleocapsid protein with LR1, we further hypothesize that LR1 might be involved in packaging of the SARS-CoV-2 genomes through its interaction with the nucleocapsid protein and ADAR1. Our study highlights the complexity of SARS-CoV-2 RNA structures and demonstrates that ultra-long-range RNA-RNA interactions within the RNA genome can be functionally important by recruiting host proteins to impact its infectivity.
Methods
Cells and viruses
African green monkey Vero-E6 cells (ATCC CRL-1586) and Vero-E6 cells constitutively expressing human TMPRSS2 (Vero-E6/TMPRSS2; BPS Bioscience #78081) were used in this study. Vero-E6 cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM; Cytiva, Utah, USA) supplemented with 10% (v/v) heat-inactivated fetal bovine serum (FBS; Capricorn Scientific, Ebsdorfergrund, Germany). Vero-E6/TMPRSS2 cells were grown in DMEM supplemented with 10% (v/v) heat-inactivated FBS, 1% (v/v) antibiotic-antimycotic (Gibco, Carlsbad, CA, USA) and 3 µg/mL of puromycin (InvivoGen, San Diego, CA, USA). Both cell lines were maintained at 37 °C in a humidified 5% CO2 incubator.
The five SARS-CoV-2 variants used in this study were: SARS-CoV-2 wildtype (WT; Clade L, lineage B, hCoV-19/Singapore/1017/2020, EPI_ISL_574502), Alpha (lineage B.1.1.7, EPI_ISL_754083), Beta (lineage B.1.351.3, EPI_ISL_1173248), Delta (lineage B.1.617.2, EPI_ISL_2621925) and Omicron XBB (EPI_ISL_14917728). The viruses were isolated from the nasopharyngeal swabs of patients with COVID-19 confirmed by RT-qPCR. The virus isolates were propagated in Vero-E6 cells with no more than 3 passages prior to downstream experiments. All virus work was performed in the Biosafety Level 3 (BSL-3) laboratory and all protocols were approved by the BSL-3 Biosafety Committee and Institutional Biosafety Committee of the National University of Singapore.
SHAPE-MaP structure probing of five SARS-CoV-2 viruses in Vero-E6 cells
Vero-E6/TMPRSS2 cells were seeded in T75 flasks (Corning, NY, USA) at a seeding density of 2.5 × 106 cells/flask and incubated overnight. Cells were infected with SARS-CoV-2 WT, Alpha, Beta, Delta or Omicron XBB variants at a multiplicity of infection (MOI) of 0.01. At 1 hpi, inoculum was replaced with DMEM supplemented with 2% FBS, followed by incubation at 37 °C. At 48 hpi cells were washed once with PBS, trypsinized and collected by centrifugation at 300 × g for 5 min. The cell pellet was then resuspended in PBS and divided into three experimental samples: (i) 1:20 volume of 1 M 2-methylnicotinic acid imidazolide (NAI; final concentration of 50 mM) was added and cells were incubated at 37 °C for 15 min; (ii) 1:20 volume of dimethyl sulfoxide (DMSO) was added and cells were incubated at 37 °C for 15 min; and (iii) cells were left without any treatment. Total RNA was extracted using RNeasy Mini Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. The harvested RNA was heat-inactivated at 65 °C for 10 min before transferring it to BSL-2 laboratory for construction of cDNA libraries compatible for Illumina sequencing following the SHAPE-MaP protocol33.
Interactome mapping of SARS-CoV-2 viruses in Vero-E6 cells by SPLASH
Vero-E6/TMPRSS2 cells were seeded in 15 cm cell-culture dish (Corning, NY, USA) at a seeding density of 2.5 × 105 cells/mL and incubated overnight. Cells were then infected with SARS-CoV-2 WT or Omicron XBB at a MOI of 0.01. At 48 hpi, cells were washed once with PBS and incubated with biotinylated psoralen (BioPso; final concentration of 200 µM) and 0.01% digitonin in PBS at 37 °C for 5 min. The BioPso-treated cells were then irradiated at 365 nm on ice for 20 min using a CL-1000 ultraviolet crosslinker (UVP Inc.). Total cellular RNA was extracted using TRIzol LS reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s recommendations, followed by heat-inactivation at 65 °C for 10 min before transferring it to BSL-2 laboratory for preparation of SPLASH libraries following the published protocol34.
Construction of SARS-CoV-2 mutants
The SARS-CoV-2 mutants (LR1A, LR1B and LR1AB) were assembled using conventional restriction enzyme-based cloning, following previously described methods35. Briefly, sub- fragments containing mutations (Genscript) were introduced into pCCI-4K-SARS-CoV-2-Wuhan-Hu1 (MT926410) infectious cDNA (icDNA) plasmid using corresponding restriction enzymes. The correct mutations were confirmed with Sanger sequencing. Five microgram of plasmid DNA (SARS-CoV icDNA and its mutants) were transfected into BHK21-ACE2-N cells (a gift from Dr. Suzannah J. Rihn) grown in T25 flasks using lipofectamine LTX reagent (Thermo Fisher Scientific) following manufacturer’s instructions. After 4-5 days post-transfection, the supernatant was transferred to Vero-E6 cells in T25 flasks. Rescued viruses (P1 stocks) were collected 3-4 days post-infection and titred using focus-forming assay. All work with recombinant viruses was performed in the BSL-3 laboratory of University of Tartu.
Analysis of viral RNA kinetics by RT-qPCR
Vero-E6 cells were seeded in 6-well plates at a density of 4 × 105 cells per well one day before infection. The cells were infected with SARS-CoV-2-Wuhan-Hu1 (WT) or its mutants at a MOI of 0.001. Experiment was performed in triplicates and 0.5 mL inoculum (DMEM + 0.2% BSA) was used per well. At 1 hpi, the inoculum was replaced with 2 mL viral growth media and cells were cultured for 30 min to collect the 0 hpi sample. Samples for isolation of extracellular and intracellular viral RNAs were collected at 0, 6, 12, 24 and 48 hpi for RT-qPCR. For each timepoint, the media was collected, and cells were washed with PBS, trypsinized, and collected in 200 μL of 1x shield buffer supplemented with 10 μL protease K (R2132, Zymo Research Quick-RNA MagBead). The cell lysates were frozen at -80 °C for later RNA purification. Extracellular viral RNA was isolated using the MagMAX™ Viral/Pathogen Nucleic Acid Isolation Kit (A42352, Thermo Fisher Scientific) according to the manufacturer’s manual, utilizing the KingFisher™ Flex in a 96 deep well format. The purified extracellular viral RNA was then analysed using RT-qPCR by LightMix Modular SARS-CoV-2 (COVID-19) RdRp 530 (Roche), as described previously36. For analysis of intracellular viral RNAs, total cellular RNAs were purified using KingFisher Flex with the R2132 cells Quick RNA Magbead KingFisher Flex protocol. The cDNA of viral positive, negative and sub-genomic RNAs were synthesized using strand-specific primer according to the First strand cDNA synthesis kit manual (K1612, Thermo Fisher Scientific). PowerTrack SYBR Green Master Mix (A46109, Thermo Scientific) was used for qPCR analysis. The qPCR primers used for positive- and negative-strand RNA were SARS2-RdRp-F and SARS2-RdRp-R. The qPCR primers used for sub-genomic RNA were SARS2-N-F and SARS2-N-R, while the qPCR primers used to detect mRNA of GAPDH control were GAPDH-F and GAPDH-R. The obtained data was analysed using 2–∆∆Ct method. The sequences of primers used for cDNA synthesis and for qPCR are listed in Supplementary Table 3.
SARS-CoV-2 viral fitness assay
Vero-E6 cells seeded at a density of 2 × 105 cells per well in 24-well plates one day before transfection were grown in high glucose DMEM media supplemented with 5% FBS. The next day, cells were transfected with 100 nM ASO using Lipofectamine RNAiMAX reagent (Thermo Scientific). Six hours after ASO transfection, cells were infected with WT SARS-CoV-2 or Omicron XBB variant at a MOI of 0.01. Infected cells were collected at 16, 24, 48 and 72 hpi. Total RNAs were extracted by Qiagen Viral RNA Mini Kit. Viral RNA copy numbers were examined by RT-qPCR using Invitrogen SuperScript III Platinum One-Step RT-qPCR Kit according to the manufacturer’s instructions. The ASO sequences are listed in Supplementary Table 3.
RNA pull-down assay
In vitro transcribed RNAs (mLR1 mini-constructs and the highly structured Tetrahymena RNA) were biotinylated using Pierce RNA 3’ End Biotinylation Kit. RNAs were heated, put on ice for 2 min and RNA structure buffer (50 mM Tris pH 7.4, 10 mM MgCl2, 150 mM NaCl) was added. To allow proper folding the temperature was gradually increased from 0 °C to 37 °C. Vero-E6 or human lung cancer cells (A549) were trypsinized and collected by centrifugation at 300 × g for 5 min. Cells were lysed in Lysis/Binding buffer (50 mM Tris HCl, pH 7.5, 100 mM KCl, 5 mM MgCl2, 10% glycerol, 0.2% NP-40, 0.5 mM DTT, 1x protease inhibitor cocktail (Calbiochem), 50 U/mL SUPERase·In™ RNase Inhibitor) at 4 °C for 10 min. Lysates were centrifugated at 1300 × g for 5 min to remove cell debris and obtained supernatants were collected in new tubes. Twenty picomole of folded RNA was then added to the cell lysates and incubated at room temperature for 2 h with rotation. Next, twenty microliters washed Dynabeads MyOne Streptavidin C1 beads (Invitrogen) were added followed by further incubation at room temperature for 1 h. Beads were washed 5 times and enriched proteins were eluted in 1x protein loading buffer by heating at 95 °C for 5 min. Retrieved proteins were resolved using SDS-PAGE for mass spectrometry and Western blot analyses. For mass spectrometry analysis, pull-down proteins were resolved using SDS-PAGE followed by bands visualization using PageBlue protein staining solution (Thermo Scientific). Subsequently, areas of interest were excised from Coomassie-stained gel and subjected to in-gel digestion with trypsin, following general in-gel protocol37. Following peptide extraction, samples were vacuum dried, resuspended in solvent A (0.1% formic acid in water) and equal amount of sample was loaded on the C-18 Evotip (Evosep) according to manufacturer instructions. Samples were analyzed using Evosep liquid chromatography system (Evosep) coupled with a timsTOF-SCP (Bruker), with a 15 cm × 75 μm column Aurora Elite (IonOptics), in DIA-PASEF mode. Each sample was separated by SPD40 method using 0.1% formic acid in water and 99.9% acetonitrile with 0.1% formic acid. DIA-PASEF data was searched using DIA-NN 2.0 software38 using in-silico library created from Uniprot fasta for African green monkey (Chlorocebus sabaeus). Western blot analysis was performed according to standard procedures using a 1:1000 dilution of anti-ADAR1 (E6X9R) XP(R) rabbit monoclonal antibody (Cell Signalling Technology) and a 1:5000 dilution of secondary anti-rabbit antibody. The sequences of mLR1 mini-constructs and Tetrahymena are listed in Supplementary Table 3.
ADAR1 eCLIP experiment
The ADAR1 eCLIP experiment was performed as published protocol26. Vero-E6 cells were seeded in 10 cm cell-culture dish (Corning, NY, USA) at a seeding density of 2.5 × 105 cells/mL and incubated overnight. Cells were then infected with SARS-CoV-2 WT at a MOI of 0.01. At 48 hpi, cells were washed once with pre-chilled PBS, UV crosslinked at 400 mJ/cm2 on ice at 254 nm radiation. Crosslinked cells were then scraped and collected by centrifugation at 300 × g for 5 min at 4 °C. Cell pellets were lysed using iCLIP buffer (50 mM Tris-HCl pH 7.4, 100 mM NaCl, 1% NP-40, 0.1% SDS, 0.5% sodium deoxycholate), lysates were sonicated and digested with DNase and diluted RNase I. Cell lysates were immunoprecipitated using 10 µg of ADAR1(E6X9R) XP(R) rabbit monoclonal antibody (Cell Signalling Technology) with 125 µl of M-280 Sheep Anti-Rabbit IgG magnetic Dynabeads (Thermo Fisher Scientific, 11-203-D) by incubation with rotation overnight at 4 °C. Two percent volume of each lysate sample was collected for preparation of a parallel Input library. The remaining cell lysates were processed as the immunoprecipitated (IP) samples and washed by high salt buffer (50 mM Tris-HCl pH 7.4, 1 M NaCl, 1 mM EDTA, 1% NP-40, 0.1% SDS, 0.5% sodium deoxycholate) to remove unspecific protein-protein interactions. Bound RNA fragments were dephosphorylated and ligated to an RNA adaptor at 3’ end. Protein-RNA complexes from Input and ADAR1-IP samples were run on 4−12% Bis-Tris SDS-PAGE and transferred to nitrocellulose membrane. Membrane regions comprising the protein sizes to 110 kDa and above were excised, and RNA was released from the complexes with proteinase K. Input samples were dephosphorylated and 3’-end ligated to an RNA adaptor. All samples were reverse transcribed and cDNAs were 5’-end ligated to a DNA adaptor. Library was constructed and sequencing was performed using NovaSeq X Plus PE150.
Electrophoretic mobility shift assay (EMSA)
The assay was performed using agarose gel as previously described39 with some modifications. Twenty five nanomole of folded RNA was incubated with increasing concentration (0 to 1.25 μM) of ADAR1 protein (TP319761M, Origene), SARS-CoV-2 nucleocapsid (N) protein (Z03488, GenScript), or SARS-CoV-2 envelope (E) protein (RP-87682, Thermo Scientific) in binding buffer (20 mM Tris buffer pH 7.5, 2 mM DTT, 2 mM MgCl2, 15 mM NaCl) for 1 h at 4 °C. Sample was then mixed with 6x DNA loading buffer (R0611, Thermo Scientific) and loaded on 1% agarose gel with recommended concentration of GelRed in 0.5x Tris/Boric acid/EDTA (TBE) buffer. The electrophoresis was performed at 10 V/cm for 20 min at room temperature, gels were imaged using a ChemicDoc (Bio-rad).
Mice experiments
Animal experiments were conducted in a pathogen-free Animal Biosafety Level 2 (ABSL-2) facility in accordance with the National University of Singapore (NUS) Institutional Animal Care and Use Committee (IACUC) (Protocol No. R24i-0008) and the NUS Institutional Biosafety Committee (IBC) approved SOPs. Eight-week-old female K18-hACE2 transgenic mice (InVivos Ptd Ltd., Singapore) were used in this study. Mice were randomized and assigned to control and treatment groups. Studies were conducted in a blinded manner. The mice were acclimatized in the ABSL-2 facility for 72 h prior to the start of the experiment with 12 h light/dark cycles and controlled temperature (23 ± 2 °C) and humidity (50 ± 10%). K18-hACE2 transgenic mice were subjected to a four-day pre-treatment regimen with saline, low dose (400 µg) or high dose (1000 µg) of ASO LR1 via intranasal route once daily. The viral challenge was conducted through intranasal delivery in 25 µL (split across both nostrils) of approximately 1 × 103 PFU of ancestral SARS-CoV-2 WT variant. Body weight and physiological conditions were monitored daily throughout the experiment or until the humane endpoint was reached. Mice were sacrificed 4 days post-infection (dpi), with lung tissues harvested. Each organ was halved for the plaque assay (right lobes) and histopathological analysis (left lobe). To assess the viral load by plaque assay, lung tissues were homogenized with 500 µL of DMEM (Cytiva) supplemented with antibiotic and antimycotic (Gibco) and titrated in A549-hACE2 cells. The viral supernatants were serially diluted in 10-fold increments, and 100 µL of each serially diluted supernatant was added to the confluent A549-hACE2 cells into 24-well plates. After 1 h of incubation for viral adsorption, the inoculum was removed and washed once with PBS. About 1.2% microcrystalline cellulose-DMEM supplemented with antibiotic and antimycotic overlay media was added to each well and incubated at 37 °C, 5% CO2 for 72 h. The cells were then fixed in 10% formalin and counterstained with crystal violet. The number of plaques were determined, and the virus titre of individual sample was expressed in logarithm of plaque forming unit (PFU) per organ. For histopathological analysis, left lung lobes were fixed in 3.7% formaldehyde overnight. Tissues were routinely processed, embedded in paraffin blocks (Leica Surgipath Paraplast), sectioned at 4 µm thickness, and stained with haematoxylin and eosin (Thermo Scientific) following standard histological procedures. For immunohistochemistry (IHC), the lung sections were deparaffinized and rehydrated, followed by heat-mediated antigen retrieval, quenching of endogenous peroxidases and protein blocking. Sections were then covered with rabbit anti-SARS-CoV-2 nucleocapsid protein mAb (Abcam, 1:1000 dilution) for 1 h at room temperature. Subsequently, the sections were incubated with rabbit-specific HRP polymer secondary antibody (Abcam, no dilution), visualized using chromogenic substrate DAB solution, and counterstained with haematoxylin.
SHAPE-MaP analysis
Sequencing reads obtained from two replicates of SHAPE experiments were aligned with the respective sequences for the SARS-CoV-2 viruses (WT: EPI_ISL_574502, Alpha: EPI_ISL_754083, Beta: EPI_ISL_1173248, Delta: EPI_ISL_2621925, Omicron: EPI_ISL_14917728) and SHAPE values for each position were calculated using Shapemapper-2.1.5 with Bowtie-2.4.2 for reading alignment40. Read depths obtained in the sequencing experiments allowed for conclusive determination of SHAPE reactivities at approximately 74-99% of positions. A reference alignment of WT, Alpha, Beta, Delta and Omicron sequences was generated using mafft-7.520 with the L-INS-I strategy. Subsequently, the local correlation of SHAPE reactivity between replicates was calculated using Pearson correlation. As Pearson correlation between replicates for each data set was ≥0.86, replicates were pooled for subsequent analysis.
Modelling global local RNA structures
Global RNA structure models for each variant were derived from SHAPE reactivity profiles and reference sequences through Superfold (v1.0) with a 600 nt maximum base pairing distance constraint and default SHAPE slope (1.8) and intercept (-0.6) parameters. ScanFold41 was used to estimate local fold stability with step size and window size parameters of 10 and 120, respectively, and 50 randomizations per step. Regions were ranked by Z-score where the lowest 20% corresponds to the highest (top 20%) stabilization values (Fig. 2a, Supplementary Fig. 4a). Generation of specific local and long-range structures was achieved with RNAstructure 6.4 modules partition-smp, Fold-smp, and stochastic-smp and the ViennaRNA 2.6.4 module RNAcofold.
Identification of structurally conserved regions
Structure models were mapped to their respective aligned sequence to maintain consistency with multiple sequence alignment (MSA) numbering. Aligned structures were divided into 200 regions, each 150 bases long, to facilitate computing mean SHAPE-MaP reactivity, Shannon entropy, and a stability Z-score for each 150 nt block. Locations of the bottom 20th percentile SHAPE reactivity, Shannon entropy, and stability blocks were identified in all 5 variants. Consensus regions for each metric were calculated as any region present in at least 3 out of 5 variants. Pearson correlation was also calculated in 150 nt windows for all 10 combinations of variant pairs. Any region measured to have an r value ≥ 0.8 was considered highly correlated. Consensus regions of highly correlated reactivity were designated as those present in 7 out of 10 structure pairs. Structure conservation was quantified as the fraction of identical base paired and unpaired positions within a specified 150 nt window between any two of ten variant pair combinations. Highly conserved structures are defined as those 20 regions demonstrating a contiguous structure conservation ratio of 0.85 or higher in at least 7 out of the 10 compared variants.
Base pair covariance score calculation
The method for measuring base pair covariance was adapted from a procedure described by Sun et al.12and updated to utilize the Infernal 1.1.5 software suite42 and a data set of 1,581 sequences of viruses belonging to Coronaviridae family which were culled for redundancy with CD-HIT43 from a larger database of 10,000 entries obtained at https://www.bv-brc.org. The cmbuild, cmcalibrate, and cmsearch modules were called from Infernal 1.1.5 in succession on each conserved structure in all variants, up to a maximum of 3 iterations or until no further sequences could be added to the covariance model. A covariance score for each structure within the 20 groups of conserved structures was calculated from the resulting alignment in accordance with published study44.
Assessing the structural impact of SNVs
All SNV coordinates were gathered from the MSA and each was placed at the centre of a 100 nt window (±50 nt positions). Structure similarity at the corresponding genome coordinates of the two variants between which the SNV originates was then computed. The number of identical base (un)paired positions and mismatches were used to build a binary array of common (1) and distinct (0) identities. Arrays of a particular SNV type (A- > N, U- > N, G- > N, C- > N, N- > A, N- > U, N- > G, N- > C) were grouped into a 2D array and the mean value of each column was calculated as the overall structure conservation at that genome position.
Measuring the effect of RBP interaction on viral RNA structure
Position weight matrices (PWMs) for human RBP motifs were taken from the ATtRACT database (ver. 0.99β). Each RBP PWM was used to scan all variant genome sequences for possible motif matches. Any matching motif coordinates were recorded as the genome positions of the respective variant. Differential gene expression (DGE) analysis was performed on variant pairs with DESeq2 (R Bioconductor), and gene names were obtained from Ensembl IDs using biomaRt. We cross-referenced the list of differentially expressed genes with RBP names from the PWM database and literature sources to identify which RBPs are significantly regulated. Analogous to the SNV analysis, the genome coordinates of significantly differentially expressed RBP motifs were analysed within a 100-nt window to compute aggregate structure conservation across all structure comparisons of a given RBP. RBPs with the largest potential impact on local genome structure were defined as those whose motifs experience low structure conservation (≤ 0.5) between a given VOC pair and significant changes in gene expression (|log2FC | ≥ 1 and padj < 0.05). Pairs of SARS-CoV-2 structure models of RBP motifs at regions of low structure conservation and significant DGE were modelled and reported.
SPLASH analysis
Chimeric reads were divided into virus-virus and host-virus interactions for WT and Omicron XBB genomes. Virus-virus interactions were normalized to total virus-virus interactions and are shown in Fig. 4a (WT: blue, Omicron XBB: red). SPLASH hybrid structure models were generated with SHAPE-MaP reactivity as constrains using RNAcofold in the Vienna RNA package.
LR1 secondary structure clustering
Scaling and clustering procedures were adapted from previous study45. One thousand LR1 structures were sampled from the Boltzmann ensemble with the stochastic-smp module from RNAstructure 6.446 and folding free energy was calculated for all structures with efn2-smp. Duplicate structures were removed leaving N unique LR structures. Hamming base pair distances were calculated between all possible unique structure pairs to represent structural (dis)similarity as an N ⨯ N pairwise distance matrix. The scikit-learn multi-dimensional scaling (MDS) module was chosen to transform the distance matrix into two-dimensional embedding coordinates which are required for clustering analysis and plotting. The resulting embeddings were clustered with the HDBSCAN clustering algorithm. Optimal hyperparameter values were determined via grid search where the combination of values that maximized the Density Based Cluster Validity (DBCV) score were chosen for clustering.
RNA A-to-I editing analysis
The A-to-I editing analysis pipeline was referred to a previous study27 with some modifications. Raw reads were cleaned using FASTP47. Ten bases of 5’ and 3’ were trimmed and reads with more than 20% of unqualified bases were removed. The mean quality per base was fixed at a phred-score of 25. The reads that were shorter than 100-nt were discarded. The cleaned reads were mapped to the reference sequence using BWA48 aln (bwa aln -t 20) and mem (bwa mem -M -t 20 -k 50). The unmapped reads were extracted for editing site calling as described in the published study27. Editing site calling was performed by subsampled each library to the same reads’ depth. To identify edited reads, read was specially required to have a minimum number of edited sites ≥ 4 (with either A-to-G or T-to-C sites in a read). The putative misaligned reads matching any of the criteria as shown in the study27 were also excluded. Only reads with the number of mismatch and low-quality A-to-G/T-to-C sites ≤ 2 were considered as edited reads. For each sample, the editing index was defined as number of editing sites from edited viral reads divided by the total number of viral reads in our study. The editing level was quantified by calculating the percentage of editing counts out of total reads at each position.
Data analysis
Processing, analysis, and visualization of data were performed using python-3.11.7, R 4.3.3, and the associated modules numpy-1.26.4, scipy-1.12.0, scikit-learn-1.4.1, matplotlib-3.8.3, upsetplot-0.9.0, venn-0.1.3, seaborn 0.13.2, DESeq2-1.44.0, biomaRt-2.60.1, the stochastic-smp module from RNAstructure 6.4, and INFERNAL 1.1.5 modules cmbuild, cmcalibrate, and cmsearch. Calculation of statistical parameters of data sets (including means, medians, percentiles, and standard deviations) was performed using numpy. Statistical tests (Student’s t-tests) employed the appropriate functions in the scipy.stats module. Visualization was performed using matplotlib and seaborn for bar charts, pie charts, box-plots, line plots, scatter plots, upset plots, Venn diagrams, and violin plots unless otherwise stated. P-values that were < 0.05 were considered statistically significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data sets generated and analysed in this study have been deposited in the GEO at the NCBI under accession codes GSE279203, GSE285533 and GSE302215. The raw mass spectrometry spectra and search data were uploaded to the jPOST repository49 with the accession numbers JPST003909 and PXD065815 on ProteomeXchange. The authors declare that all other data supporting the findings of this study are available within the article and its Supplementary Information files. Source data are provided with this paper.
References
Gralinski, L. E. & Baric, R. S. Molecular pathology of emerging coronavirus infections. J. Pathol. 235, 185–195 (2015).
Cui, J., Li, F. & Shi, Z. L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 17, 181–192 (2019).
Markov, P. V. et al. The evolution of SARS-CoV-2. Nat. Rev. Microbiol. 21, 361–379 (2023).
Carabelli, A. M. et al. SARS-CoV-2 variant biology: immune escape, transmission and fitness. Nat. Rev. Microbiol. 21, 162–177 (2023).
Roemer, C. et al. SARS-CoV-2 evolution in the Omicron era. Nat. Microbiol. 8, 1952–1959 (2023).
Kelly, J. A. et al. Structural and functional conservation of the programmed -1 ribosomal frameshift signal of SARS coronavirus 2 (SARS-CoV-2). J. Biol. Chem. 295, 10741–10748 (2020).
Namy, O., Moran, S. J., Stuart, D. I., Gilbert, R. J. & Brierley, I. A mechanical explanation of RNA pseudoknot function in programmed ribosomal frameshifting. Nature 441, 244–247 (2006).
Vora, S. M. et al. Targeting stem-loop 1 of the SARS-CoV-2 5′ UTR to suppress viral translation and Nsp1 evasion. Proc. Natl Acad. Sci. 119, e2117198119 (2022).
Miao, Z., Tidu, A., Eriani, G. & Martin, F. Secondary structure of the SARS-CoV-2 5’-UTR. RNA Biol. 18, 447–456 (2021).
Manfredonia, I. et al. Genome-wide mapping of SARS-CoV-2 RNA structures identifies therapeutically-relevant elements. Nucleic acids Res. 48, 12436–12452 (2020).
Ziv, O. et al. The Short- and Long-Range RNA-RNA Interactome of SARS-CoV-2. Mol. cell 80, 1067–1077.e1065 (2020).
Sun, L. et al. In vivo structural characterization of the SARS-CoV-2 RNA genome identifies host proteins vulnerable to repurposed drugs. Cell 184, 1865–1883.e1820 (2021).
Huston, N. C. et al. Comprehensive in vivo secondary structure of the SARS-CoV-2 genome reveals novel regulatory motifs and mechanisms. Mol. cell 81, 584–598.e585 (2021).
Cao, C. et al. The architecture of the SARS-CoV-2 RNA genome inside virion. Nat. Commun. 12, 3917 (2021).
Yang, S. L. et al. Comprehensive mapping of SARS-CoV-2 interactions in vivo reveals functional virus-host interactions. Nat. Commun. 12, 5113 (2021).
Zhang, Y. et al. In vivo structure and dynamics of the SARS-CoV-2 RNA genome. Nat. Commun. 12, 5695 (2021).
Lan, T. C. T. et al. Secondary structural ensembles of the SARS-CoV-2 RNA genome in infected cells. Nat. Commun. 13, 1128 (2022).
Yang, S. L., Ponti, R. D., Wan, Y. & Huber, R. G. Computational and experimental approaches to study the RNA secondary structures of RNA viruses. viruses 14, 1795 (2022).
Flynn, R. A. et al. Discovery and functional interrogation of SARS-CoV-2 RNA-host protein interactions. Cell 184, 2394–2411.e2316 (2021).
Kamel, W. et al. Global analysis of protein-RNA interactions in SARS-CoV-2-infected cells reveals key regulators of infection. Mol. cell 81, 2851–2867 e2857 (2021).
Lee, S. et al. The SARS-CoV-2 RNA interactome. Mol. cell 81, 2838–2850 e2836 (2021).
Schmidt, N. et al. The SARS-CoV-2 RNA-protein interactome in infected human cells. Nat. Microbiol 6, 339–353 (2021).
Labeau, A. et al. Characterization and functional interrogation of the SARS-CoV-2 RNA interactome. Cell Rep. 39, 110744 (2022).
Bhattarai, K. & Holcik, M. Diverse roles of heterogeneous nuclear ribonucleoproteins in viral life cycle. Front. Virol. 2 https://doi.org/10.3389/fviro.2022.1044652 (2022).
Zhou, H. et al. Heterogeneous nuclear protein U degraded the m6A methylated TRAF3 transcript by YTHDF2 To promote porcine epidemic diarrhea virus replication. J. Virol. 97, e01751–01722 (2023).
Van Nostrand, E. L. et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat. Methods 13, 508–514 (2016).
Song, Y. et al. Virus-specific editing identification approach reveals the landscape of A-to-I editing and its impacts on SARS-CoV-2 characteristics and evolution. Nucleic acids Res. 50, 2509–2521 (2022).
Merdler-Rabinowicz, R. et al. Elevated A-to-I RNA editing in COVID-19 infected individuals. NAR Genomics Bioinform. 5, lqad092 (2023).
Kim, D.-K. et al. A proteome-scale map of the SARS-CoV-2–human contactome. Nat. Biotechnol. 41, 140–149 (2023).
Picardi, E., Mansi, L. & Pesole, G. Detection of A-to-I RNA editing in SARS-COV-2. Genes 13, 41 (2021).
Jin, Y.-Y. et al. Ocular A-to-I RNA editing signatures associated with SARS-CoV-2 infection. BMC Genomics 25, 431 (2024).
Cottrell, K. A. et al. Induction of viral mimicry upon loss of DHX9 and ADAR1 in breast cancer Cells. Cancer Res. Commun. 4, 986–1003 (2024).
Siegfried, N. A., Busan, S., Rice, G. M., Nelson, J. A. E. & Weeks, K. M. RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP). Nat. Methods 11, 959–965 (2014).
Aw, J. G. et al. In vivo mapping of eukaryotic RNA interactomes reveals principles of higher-order organization and regulation. Mol. cell 62, 603–617 (2016).
Rihn, S. J. et al. A plasmid DNA-launched SARS-CoV-2 reverse genetics system and coronavirus toolkit for COVID-19 research. PLOS Biol. 19, e3001091 (2021).
Corman, V. M. et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 25, 2000045 (2020).
Shevchenko, A., Tomas, H., Havli, J., Olsen, J. V. & Mann, M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856–2860 (2006).
Demichev, V., Messner, C. B., Vernardis, S. I., Lilley, K. S. & Ralser, M. DIA-NN: neural networks and interference correction enable deep proteome coverage in high throughput. Nat. Methods 17, 41–44 (2020).
Ream, J. A., Lewis, L. K. & Lewis, K. A. Rapid agarose gel electrophoretic mobility shift assay for quantitating protein: RNA interactions. Anal. Biochem 511, 36–41 (2016).
Busan, S. & Weeks, K. M. Accurate detection of chemical modifications in RNA by mutational profiling (MaP) with ShapeMapper 2. RNA (N. Y., N. Y.) 24, 143–148 (2018).
Andrews, R. J., Roche, J. & Moss, W. N. ScanFold: an approach for genome-wide discovery of local RNA structural elements-applications to Zika virus and HIV. PeerJ 6, e6136 (2018).
Nawrocki, E. P. & Eddy, S. R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinforma. (Oxf., Engl.) 29, 2933–2935 (2013).
Fu, L., Niu, B., Zhu, Z., Wu, S. & Li, W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinforma. (Oxf., Engl.) 28, 3150–3152 (2012).
Hofacker, I. L. RNA consensus structure prediction with RNAalifold. Methods Mol. Biol. 395, 527–544 (2007).
Ding, Y., Chan, C. Y. & Lawrence, C. E. Clustering of RNA secondary structures with application to messenger RNAs. J. Mol. Biol. 359, 554–571 (2006).
Reuter, J. S. & Mathews, D. H. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinforma. 11, 129 (2010).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinforma. (Oxf., Engl.) 34, i884–i890 (2018).
Li, H. & Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinforma. (Oxf., Engl.) 26, 589–595 (2010).
Okuda, S. et al. jPOST environment accelerates the reuse and reanalysis of public proteome mass spectrometry data. Nucleic acids Res. 53, D462–D467 (2024).
Busan, S. & Weeks, K. M. Visualization of RNA structure models within the Integrative Genomics Viewer. RNA (N. Y., N. Y.) 23, 1012–1018 (2017).
Darty, K., Denise, A. & Ponty, Y. VARNA: Interactive drawing and editing of the RNA secondary structure. Bioinforma. (Oxf., Engl.) 25, 1974–1975 (2009).
Acknowledgements
We thank Dr. Meng How Tan for productive discussions, members of the Wan laboratory, A*STAR, for their input and suggestions during the course of this work. YW is supported by funding from A*STAR, National Research Foundation of Singapore, and NRF grant NRF-CRP27-2021-0003. AM is the recipient of program grant (PRG1154) from Estonian Research Council.
Author information
Authors and Affiliations
Contributions
Y.W., R.G.H., A.M., S.L.Y. came up with the project. Y.W., R.G.H., A.M., S.L.Y. designed the experiments and the analysis together. S.L.Y., S.W. performed most of the experiments. L.D. performed most of the analysis. L.F.W., J.J.H.C., Y.H.W., C.K.M. and S.W. designed and performed the experiments that were carried out in BSL-3 laboratory. J.S.X. performed ADAR1 eCLIP experiment. R.S. designed and performed the mass spectrometry experiments. K.Y.T., S.Y.L., Z.Z. supported the experiments. J.H., Y.Z. and J.J.A.L. supported the analysis. Y.W., R.G.H. and S.L.Y. wrote the manuscript with everyone’s feedback.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Walter N. Moss, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yang, S.L., DeFalco, L., Wang, S. et al. A conserved long-range RNA interaction in SARS-CoV-2 recruits ADAR1 to enhance virus proliferation. Nat Commun 16, 8460 (2025). https://doi.org/10.1038/s41467-025-63297-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-63297-2