Abstract
Cyanobacteria are vital photosynthetic prokaryotes, but some form harmful algal blooms (cyanoHABs) that disrupt ecosystems and produce toxins. The mechanisms by which these blooms form have yet to be fully understood, particularly the role of extracellular components. Here, we present a 2.4 Å cryo-EM structure of a pilus, termed the cyanobacterial tubular (CT) pilus, found in the cyanoHAB-forming Microcystis aeruginosa. The pilin exhibits a unique protein fold, forming a tubular pilus structure with tight, double-layer anti-parallel β-sheet interactions. We show that CT pili are essential for buoyancy by facilitating the formation of micro-colonies, which increases drag force and prevents sinking. The CT pilus surface is heavily glycosylated with ten monosaccharide modifications per pilin. Furthermore, CT pili can enrich microcystin, potentially enhancing cellular resilience, and co-localize with iron-enriched extracellular matrix components. Thus, we propose that this pilus plays an important role in the proliferation of cyanoHABs. This just discovered pilus family appears to be widely distributed across several cyanobacterial orders. Our structural and functional characterization of CT pili provide insights into cyanobacterial cell morphology, physiology, and toxin interactions, and identify potential targets for disrupting bloom formation.
Similar content being viewed by others
Introduction
Cyanobacteria are globally widespread photosynthetic prokaryotes that are crucial to biogeochemical cycling and offer a reservoir of novel compounds1 with potential applications in drug discovery2 and biofuels3. However, certain cyanobacterial species can form harmful algal blooms4 (cyanoHABs), disrupting aquatic ecosystems and posing a toxic threat to both wildlife and humans5. The first documented toxic cyanoHAB filled a chain of lakes in Australia in 18786, and cyanoHAB research has since become a highly active field of study7,8,9. One important goal is to describe the drivers of bloom formation, which allow a barely noticeable cyanobacterial growth to engulf an entire ecosystem in a matter of weeks. During this rapid growth period, numerous potent cyanotoxins can be produced, such as microcystins10, cylindrospermopsin11, and saxitoxin12.
Microcystins, the most common class of toxins found in cyanobacteria, were discovered in the 1950s and were referred to then as the “fast death factor”13. However, they are mysteriously beneficial secondary metabolites in Microcystis aeruginosa with various proposed functions, including regulation of oxidative stress14, defense against zooplankton predation15, allelopathic suppression of competing bacteria under nutrient limitation16, among others. Interestingly, during the life cycle of a cyanoHAB released toxins could rapidly diffuse into surrounding environments, thus implying that cyanobacteria might have mechanisms to maintain localized toxin concentrations near their production sites. Also, cyanotoxins have been implicated in the colony formation of Microcystis sp.17, suggesting they may be a factor which drives bloom-formation.
Beyond their role in toxin production, the remarkable morphological diversity and transition between different morphological states of cyanobacteria18 is believed to aid their dominance in a variety of ecological niches. Cyanobacteria range from unicellular forms to complex multicellular filaments found in orders such as Nostocales, Oscillatoriales, and Spirulinales. Filamentous cyanobacteria further vary in their branching patterns, the presence of sheaths, and the formation of coiled trichomes. Unicellular forms are quite common, including in the most frequently found freshwater cyanoHAB forming species M. aeruginosa19. These cells may form into micro-colonies and eventually into mat-like formations several inches thick and covering surface waters in matured blooms20,21. The transitions between these morphological states represent crucial checkpoints in cyanoHAB formation22,23,24. This raises an interesting question in fluid mechanics25. It is well known that cyanobacteria, including M. aeruginosa, can use gas vesicles as a floating device to rise in the water column for optimal access to light conditions. But after this positioning, vesicle production typically decreases due to increasing turgor pressure and accumulation of photosynthetic products26,27. During certain seasons, cyanobacteria may also sink to the sediments, possibly aided by glycogen inclusions and/or intracellular calcium carbonate inclusions28,29. Thus, how do cyanobacteria modulate their flow resistance to maintain optimal positioning within their environment? Such a process may be mediated by components of the extracellular matrix (ECM), such as polysaccharides30,31,32 and pili.
Another intriguing metabolic characteristic of cyanobacteria is their high demand for iron to carry out photosynthetic processes, compared to other non-photosynthetic bacteria33. Because of this demand, cyanoHAB growth is often limited by iron availability34,35. Under the neutral to slightly alkaline pH conditions of most freshwater habitats, iron predominantly exists in insoluble forms, such as ferric oxide and hydroxide, and the majority of soluble iron is in complex organic chelates36. While certain anaerobic respiring bacteria like Geobacter sulfurreducens can reduce insoluble iron through long-range electron transfer via extracellular cytochrome nanowires37,38,39, these bacteria typically inhabit sediments rather than surface waters and lack photosynthetic capabilities. Interestingly, extracellular pili in two cyanobacteria, the type IV pili of Synechocystis and the 11 nm unknown pili40 of M. aeruginosa, have been suggested to be conductive, as measured by conductive atomic force microscopy (AFM) on dried filament samples41. However, the claimed conductivity of type IV pili in a different bacterium, G. sulfurreducens, has recently been debunked42,43, leaving the conductivity of the 11 nm pili in M. aeruginosa and its other possible roles to be re-visited.
These interesting observations prompted us to examine the extracellular space of a model bloom-forming cyanobacterium, M. aeruginosa. Using cryo-EM, we report a 2.4 Å structure of its extracellular pilus. This pilus is the only filament type observed and appears as a rigid tube with an outer diameter of ~110 Å and a ~ 45 Å wide lumen. We named this pilus the cyanobacterial tubular (CT) pilus. The pilin, the building block of the pilus, has a unique protein fold. This sequence was annotated as a “dark cluster” in AFDB clusters44, and no significant structural homologs can be identified, except for similar sequences from other cyanobacteria. The CT pilus is primarily held together by tightly packed, double-layer anti-parallel β-sheet interactions. Such an interaction has not been observed in other microbial pili; the most similar filament packing may be the reported structure of a gas vesicle45, which has a single layer of tightly packed anti-parallel β-sheets. Furthermore, we show that the CT pilus likely has multiple functions, including colony formation, bridging cells to increase drag force, and binding microcystin toxins from the surrounding environment. We also propose that it may interact with ECM, which itself is enriched in iron. Sequence analysis shows that this pilus family is widely distributed and has conserved genes associated with several cyanobacteria orders, including Chroococcales, Nostocales, and Synechococcales, providing insights into cyanobacterial cell morphology, physiology, and toxin interactions.
Results
Cryo-EM structures of cyanobacterial tubular (CT) pilus
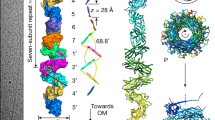
To investigate the questions raised above, we first examined the extracellular space of M. aeruginosa cultures (strain PCC 7806SL) using scanning electron microscopy (SEM) and negative staining under transmission electron microscopy (TEM). Most M. aeruginosa cells formed single-layer micro-colonies, displaying numerous pili protruding from cell surfaces (Fig. 1a and Supplementary Fig. 1). These pili were sheared from the cells by vortexing, and the soluble fraction was imaged using cryogenic electron microscopy (cryo-EM) to obtain an atomic pilus structure. Recent advancements in cryo-EM46 and deep learning modeling tools47 have enabled the routine identification of proteins directly from cryo-EM maps with resolutions of 4 Å or better48. Among over ten thousand collected micrographs, only one type of pilus was observed (Fig. 1b). This pilus, several microns in length, appeared straight and rigid under cryo-EM with a diameter of approximately 110 Å, an inner lumen diameter of 45–50 Å and a pitch of 89 Å based on 2D averages. The averaged power spectrum of aligned raw particles was indexed to estimate all possible helical symmetries. All possibilities were tested in helical reconstruction49, and the correct symmetry was determined to have a helical rise of 3.65 Å and a twist of −14.74°. Pilin subunits are tightly packed along this left-handed 1-start helix, forming a tubular pilus structure (Fig. 1c).
a Representative scanning electron microscopy (SEM) image of M. aeruginosa cells, taken from the 9 images recorded. Some pili bundles are indicated with yellow arrowheads. Scale bar, 10 µm. This low-magnification image shows the morphology of M. aeruginosa cells. Higher magnification images of M. aeruginosa cells under negative staining TEM, can be found in Supplementary Fig. 1. b Representative cryo-EM image of long and straight CT pili sheared from M. aeruginosa, taken from the 6935 images recorded. Scale bar, 50 nm. The upper right is a two-dimensional class average of the CT pilus, showing the pitch of 89 Å. c The 2.4 Å cryo-EM reconstruction (left) of CT-pilus. A left-handed 1-start helix is indicated. A zoom-in view (right) is shown to highlight map densities corresponding to a single pilin colored in light green. The diameters of outer surface and lumen are indicated. d The atomic model of one repeating subunit of CT pilus. Protein backbone is shown in green. N- and C-termini, along with secondary structure elements, are labeled. e Density (red) due to post-translational modifications on CT pilin, with fitted O-GlcNAc molecules shown to demonstrate the size of density. The map and model for the protein parts are colored green.
The final reconstruction, with helical symmetry applied, reached a resolution of 2.4 Å as judged by map-to-map Fourier shell correlation (FSC, Supplementary Fig. 2). The Cα tracing at such a resolution was straightforward, and the correct pilin was identified using ModelAngelo47 as pilin BH695_0841, as no other protein in the proteome could fully account for the cryo-EM density (Supplementary Fig. 3). Interestingly, the pilin has been reported in a very close strain (M. aeruginosa PCC 7806) as microcystin-related protein C50 (MrpC). It was suggested MrpC undergoes glycosylation, is secreted to the extracellular space (found largely at the cell surface), and has a complex relationship with microcystin and colony formation50. Notably, this pilin exhibits a unique protein fold: the N-terminal region includes two long β-hairpins, one located in the lumen and the other on the pilus surface, with the latter having a short turn in the middle mediated by proline and glycine residues. The C-terminal side of the pilin comprises a helix-turn-helix motif (Fig. 1d). To avoid confusion with well-established types of pili, we designate this tubular shape pilus as the cyanobacterial tubular (CT) pilus. Additional O-linked glycosylation densities, consistent with the size of monosaccharides, were observed on ten residues exclusively on the pilus surface, including five serines and five threonines (Fig. 1e). No glycosylation was detected on the lumen side.
CT pilin gene cluster and its distribution
To gain further insight into this glycosylation, we examined the gene cluster surrounding the CT pilin gene (Fig. 2a). Interestingly, adjacent to the pilin gene (BH695_0841), we identified a CT pilin-like protein (BH695_0840), an O-GlcNAc transferase (BH695_0839), an FkbM methyltransferase (BH695_0838), a putative β-sheet-rich adhesion protein (BH695_0837), and a cell surface protein (BH695_0836). These identifications were based on UniProt annotations, AlphaFold predictions51 (Fig. 2b and Supplementary Fig. 4), and Foldseek52 analysis of similar structures. Homologs of these genes are present (Fig. 2a) in other Microcystis species (order Chroococcales) and other cyanobacterial orders, such as Nostocales (Anabaena, Nodularia, Aphanizomenon, Sphaerospermopsis) and Synechococcales (Synechococcus).
a Gene structures of the CT pilus in M. aeruginosa and similar clusters identified in other cyanobacteria species. b AlphaFold predictions for proteins in the M. aeruginosa cluster. Putative annotations are based on sequence database and Foldseek searches. Predicted models are colored corresponding to gene colors. c Midpoint-rooted maximum likelihood phylogeny of CT pilin homologs. Tree branches are colored by organism taxonomy, with taxon names indicated. The position of M. aeruginosa CT pilin is marked with a blue star. Blue circles indicate approximate likelihood ratio test (aLRT) branch supports higher than 90%. The scale bar represents the number of substitutions per site. d Comparison of the CT pilin in M. aeruginosa with putative pilins in other cyanobacteria. Representative AlphaFold predictions are shown for the same strains used in (a). The order of these strains is indicated by the background color, which matches the order color scheme in (c).
In a very closely related M. aeruginosa strain, O-GlcNAc modification was identified on the immature pilin protein within the cell surface fraction, but this modification was subsequently processed to a lower molecular weight form after secretion into the extracellular space50. This finding was further supported in this study by two independent intact mass spectrometry measurements, which revealed a total mass of ~14,060 Da for a single pilin protein. This mass includes 12,348 Da from the protein component and an additional 1712 Da attributed to glycosylation (Supplementary Fig. 5). Moreover, using MS/MS analysis, we detected residue masses of 174 Da and 160 Da at the glycosylation sites (Supplementary Figs. 6–8), originating from Ser132 and Ser138, respectively. Two copies of the 160 Da residue plus eight copies of the 174 Da residue would precisely account for the observed 1712 Da mass of glycosylation. The intact mass spectrometry and MS/MS data suggest that these monosaccharide modifications at the serine and threonine sites are specific and likely processed from O-GlcNAc modifications during pilus assembly.
Pilin-like proteins have been implicated in various roles, including tip adhesion in chaperone-usher pathway (CUP) pili53, endo-pilus formation in the type II secretion system54, and potential gene duplication events55, however, like the function of most other pilin-like proteins, the function of the CT pilin-like protein remains unclear. To assess the broader distribution of the CT pilin, we performed a PSI-BLAST search, which revealed pilin homologs exclusively in cyanobacteria (Fig. 2c and Supp Data 1), primarily within the orders Chroococcales and Nostocales, with a few distant homologs in Synechococcales. AlphaFold predictions of these pilin homologs display an overall similar secondary structure, but the domain-domain orientations are quite different from experimentally determined structures (Fig. 2d). This discrepancy further demonstrates that this family of proteins has been sparsely sampled in the past, hindering accurate 3D structure prediction.
The lateral and axial interface of the CT pilus
The CT pilus exhibits a tubular architecture with extensive double-layered β-sheet interactions observed on both the surface and lumen (Fig. 3a and Supplementary Fig. 8). Each pilin subunit, containing long β-hairpins and a C-terminal helix-turn-helix motif, has a total surface area of ~ 9200 Å2. For pilin subunit S0, extensive lateral contacts with S+1 and S−1 are observed, each with a buried surface of ~2400 Å2, accounting for 52% of the total pilin surface (Fig. 3b). These substantial interfaces are primarily formed by two sets of anti-parallel β-sheets that extend continuously throughout the pilus, one on the outer surface and one in the lumen (Fig. 3c and Supplementary Fig. 8). While protein and peptide fibrils held together by β-sheets are common in amyloids56,57, the β-strands in such fibers are typically parallel and perpendicular to the helical axis. In contrast, the anti-parallel β-strands observed in the CT pilus are packed almost in parallel to the helical axis. In addition to the β-sheet interface, there is an interface between helices near the C-terminus, with hydrophobic residues present within the continuous interfaces between helices (Fig. 3d). The axial interface between pilins is smaller, with a buried surface of ~440 Å2. Although the axial interface has a smaller buried surface area compared to the lateral interface, the axial interface features notable hook-latch contacts. The helix-turn-helix motif and C-terminal loop of the pilin form a hook-like shape that continues along the 1-start helix. Loops from subunits on the adjacent upper level of the 1-start helix, including S+21 and S+22, insert into this hook, possibly enhancing the rigidity of the tube (Fig. 3e).
a Atomic model of the CT pilus. One pilin subunit (S0) is colored green. Two pilin subunits (S+1 and S−1) forming lateral contacts with S0 are colored blue. Pilin subunits forming axial contacts with S0 are colored lavender (S+22 and S−22) and pink (S+21 and S−21), respectively. b Zoomed-in view of all unique interfaces surrounding pilin S0. The lateral-1, lateral-2, and axial interfaces are further highlighted in (c–e), respectively. Ten glycans on the surface of pilin S0 are shown as red dots. c Surface β-sheets and lumen β-sheets at the lateral-1 interface. Pilin subunits are colored consistently with (a). Hydrogen bonds are labeled with dashed lines. d Lateral-2 interface between helices. Residues involved in the interface are displayed as sticks. Pilin subunits are colored consistently with (a). e Axial interface, with all involved residues shown as sticks. Pilin subunits are colored consistently with (a). f Cyclic voltammogram measurements of CT pili films on glassy carbon electrodes. Legend indicates the volume of CT pili solution used to deposit each film. Source data are provided as a Source Data file.
The pilin-pilin interface observed in the CT pilus structure does not support a molecular basis for conductivity. Current protein nanowires that transfer electrons are typically based on either heme-containing cytochrome filaments39 or iron-sulfur cluster containing proteins58. The CT pilus possesses neither of these features, raising some questions about its previously reported conductivity. Given the potential limitations of conductive AFM measurements on dried filaments48, we conducted bulk cyclic voltammetry (CV) measurements in solution. Upon measuring the electroactive surface area, we observed a decrease in current with the addition of more fibers (Fig. 3f). Additionally, the redox couple’s diffusion remained unimpeded, suggesting the absence of a charge transfer reaction and indicate the CT pili do not exhibit the expected signatures of conductivity. This finding was consistent with additional electrochemical impedance spectroscopy (EIS) measurements59, which demonstrated an increase in the semicircle size with the addition of more fibers (Supplementary Fig. 9), corresponding to an increase in resistance of the pilus films and corroborating the CT pili, in the CV and EIS setups used, do not exhibit behavior characteristic of a conductive filament film.
CT pilin belongs to a structurally uncharacterized protein cluster
We then investigated whether a similar structure, at either the single protein fold level or the filament architecture level, has been previously reported. Taking advantage of recent developments in protein structure prediction, we compared the CT pilin against the entire AlphaFold prediction database. Interestingly, the Foldseek-based AFDB clusters44 have annotated the CT pilin and related homologs as a “dark cluster”, defined as a structurally and functionally unknown protein cluster. All AlphaFold predictions within this cluster are from cyanobacteria with apparent sequence homology to the CT pilin, and Foldseek did not find sequences outside cyanobacteria with similar protein fold predictions. Next, we performed a similar analysis using DALI60, which yielded comparable results. The best hit had a Z-score of approximately 2.4 (not significant) and is a bursicon protein from fruit flies. While there are some similarities in secondary structure topology between the CT pilin and bursicon, and both have a signal peptide, their 3D structures do not align well (Fig. 4a, b). This suggests that possible species containing similar proteins have been very sparsely sampled in the past.
All models displayed use the following color scheme: α-helices are red, and β-sheets are green. Schematic representations are also used, with β-sheets shown as green arrows, α-helices as red cylinders, and loops as dark gray lines. a Cryo-EM structure and secondary structure of the CT pilin from M. aeruginosa. b AlphaFold prediction and predicted secondary structure of fruit fly bursicon protein (Q9VD83). This protein was one of the top hits in a DALI search against AFDB, with a non-significant Z-score of 2.4. c Cryo-EM structure and secondary structure of gas vesicle protein A (7R1C). d Protein interface between gas vesicle proteins, displaying a single layer anti-parallel β-sheet interaction.
At the filament packing level, a remotely similar architecture containing tightly packed anti-parallel β-sheets has only been observed in gas vesicles45. Gas vesicle protein A, the building block of gas versicles, is a shorter protein, with an N-terminal and a C-terminal short helix and a long β-hairpin in the middle (Fig. 4c). This long β-hairpin allows gas vesicle protein A to form single-layer, air-tight contacts with adjacent subunits to create the sealed vesicle (Fig. 4d). In contrast, the CT pilus has an additional layer of such β-sheets, potentially conferring greater rigidity. Since they are tightly packed as gas vessels, could this type of pilus contribute to gas storage? From a mathematical perspective, we think this is very unlikely because the lumen diameter of the pilus is too small. For example, the gas volume of a typical gas vesicle, as shown in Supp Fig. 1, is roughly equivalent to that of 200 5-µm long CT pili. While a single cyanobacterial cell may possess multiple gas vesicles, we have never seen a bacterium produce 200 pili under negative staining. Therefore, gas storage using CT pili would appear to be very energetically inefficient. We also considered whether these pili could function like swimming snorkels to release gas. However, because gases can diffuse readily across cell membranes61, it is also quite unlikely that pili are used for gas removal.
CT pili bridge cells together to avoid sinking
The structural characteristics of CT pili clearly indicate that they are long, rigid tubes. However, the reason M. aeruginosa produces so many of them remains unclear. Recent research on Synechococcus sp., a marine picocyanobacterium lacking both CT-pilin and gas vesicles, suggests that extracellular pili enhance drag force, allowing the organism to maintain an optimal position in the water column62. Specifically, genetic evidence, including knockout experiments, points to type IV pili as the filaments responsible for this phenomenon. We hypothesized that CT pili in M. aeruginosa serve a similar purpose, networking cells together to form colonies with increased drag force in fluid flow. This networking would enable M. aeruginosa cells to remain on the water surface for optimal light exposure, even if their gas vesicles are damaged.
There are limited genetic tools available for M. aeruginosa, but we were able to temporarily disrupt the function of CT pili through brief vortexing. This mechanical shearing removes extracellular pili, leaving them soluble but detached from the cells. We compared M. aeruginosa cells in cultures with and without a 2-min vortexing treatment, centrifuging both at 1000 × g for 0 to 5 min. Notably, pili-sheared cells rapidly pelleted within 2 min, while a significant portion of un-vortexed cells remained in suspension after 3–5 min (Fig. 5a and Supplementary Fig. 10). A similar phenomenon was also observed under natural gravity. Specifically, we compared M. aeruginosa cells in cultures with and without a 2-min vortexing treatment and allowed the cells to settle under natural gravity for 12 h. We observed that the un-vortexed cells remained in suspension, whereas the pili-sheared cells pelleted to the bottom of the tube (Supplementary Movie 1). This observation can be explained by the drag force in Stokes’ Law. The rigid CT pili create “rafts” between cells, effectively increasing their radius compared to individual cells lacking CT pili (Fig. 5b).
a M. aeruginosa cells in culture medium within Falcon tubes. The top tube was left untreated, while the bottom tube was vortexed for 2 min to shear off CT pili. Both tubes were centrifuged at 1000 × g for 1-min intervals over a 3-min period. b Schematic of drag force (Fd) acting on M. aeruginosa cells in solution, with or without CT pili. The radius of the particle increases for micro-colony forming cells mediated by CT pili. In contrast, the radius of pili-sheared cells is equivalent to the radius of an individual bacterium. c Cell morphology and pili production were monitored via negative staining transmission electron microscopy (TEM) during the process. Representative micrographs were shown, taken from the 5 images recorded per condition. The following observations were made (from left to right): 1. Before treatment: Micro-colonies with pili between cells were observed; 2. After mechanical shearing and spinning, Pili were present in the supernatant. 3. Pelleted cells: Cells lacked pili and did not form micro-colonies. 4. 12 h after treatment: Pili began to reappear on the cell surface. 5. Several days after treatment: Micro-colonies were observed again, with pili present between cells. Low-magnification micrographs were shown at the top, and zoomed-in views were shown at the bottom. The scale bar in low-magnification images is 1 µm. The scale bar in the zoomed-in images is 100 nm.
To further investigate this phenomenon, we tracked pili shearing and recovery using negative staining TEM. Before shearing, cells formed micro-colonies with pili observed between them (Fig. 5c). After mechanical shearing and centrifugation, the sheared pili remained soluble and were observed in the supernatant. The cell pellets at the bottom of the tube lacked CT pili on their surface (Fig. 5c). We then continued culturing the sheared M. aeruginosa cells. After 12 h, cells were still primarily unicellular with short pili observed on their surface. After additional days of culturing, M. aeruginosa cells have fully recovered their planktonic form. Under negative staining, micro-colonies with extensive CT pili between cells were observed (Fig. 5c).
Role of CT pili in iron and toxin accumulation
Examination of the outer surface of CT pili reveals an abundance of hydroxyl and other hydrophilic groups contributed by both glycans (ten per pilin) and surface amino acids (Supplementary Fig. 8). This pattern repeats along the filament due to the continuous β-sheet arrangement of pilin within the helical symmetry. We wondered whether this surface has been evolutionarily optimized for interaction with extracellular molecules, such as heteropolysaccharides within the ECM or essential minerals required by M. aeruginosa for various metabolic processes. To test this, we analyzed M. aeruginosa cultures by scanning TEM with energy-dispersive X-ray spectroscopy (STEM-EDS) and were able to locate micro-colonies, extracellular pili, and ECM between cells. As expected, STEM-EDS elemental imaging revealed peaks for carbon, nitrogen, oxygen, sodium, etc., primarily overlapping with cyanobacterial cells (Fig. 6a and Supplementary Fig. 11). Interestingly, the elemental images for metals including calcium, magnesium, molybdenum, cobalt, copper, zinc, and manganese showed no correlation with the extracellular pili or ECM. Previous reports indicate that calcium ions may promote the secretion of extracellular polysaccharides and cross-link with negatively charged EPS bound to the surface of Microcystis cells63,64. However, we did not observe a high calcium signal overlapping with ECM. This could be due to the calcium concentration in the medium not being sufficiently high to form the proposed extracellular cross-linking architecture. The iron level, however, was significantly higher in the ECM areas between cells compared to the bacterial cells themselves (Fig. 6a and Supplementary Figs. 11 and 12). The pili area outside the ECM did not co-localize with iron signals. We propose that the pili might be a scaffold for hosting ECM to enrich iron, and thus the pili may be used as a strategy to maintain iron concentration near the cell for bacterial growth. It is important to note that the BG-11 medium used is relatively iron-rich, which may not accurately reflect typical physiological conditions. Therefore, we cannot exclude the possibility that the observed signals are iron precipitates resulting from the air-dry imaging method. Using the same batch of M. aeruginosa cells employed for STEM-EDS imaging, we also examined them in aqueous settings via fluorescence microscopy. While we could not visualize pili or iron signals at such low-magnification, we did observe ECM with dense deposits that might contain iron. Furthermore, bacterial cells within the ECM tended to form colonies compared to those outside the ECM (Supplementary Fig. 13), suggesting colonization promoting CT pili are likely present within the ECM as observed in high resolution imaging methods. In summary, we propose CT pili co-localize with ECM, which may help enrich iron.
a Representative STEM-EDS of a group of cells with abundant CT pili. Additional set of images of other elements can be found in the Supplemental Fig. 6. Extracellular matrix (ECM) material is observed with the pili. The distribution of different elements is shown, including nitrogen, carbon, sodium, magnesium, iron, copper, manganese, and zinc. Scale bars, 5 µm. b–e Four sets of representative nanoSIMS images reveal the abundance of isotope-labeled MCLR toxin localized with filaments spanning between cells. For individual set of image, from left to right are secondary electrons (SE), carbon-12 anions (12C2-), and 13C/12C- ratios (13C12C/12C2). Isotope-labeled MCLR toxin was incubated with cells for 12 h before looking under the nanoSIMS. e is an example of filaments sheared from cells that still have signals of binding labeled MCLR. Scale bars, 5 µm.
Previous reports indicated that the abundance of MrpC, identified in this study as the CT pilin, is influenced by the presence or absence of microcystin. Microcystin-deficient mutants exhibit increased MrpC accumulation compared to the wild type, suggesting a complex regulatory relationship50. We hypothesized that CT pili might bind to microcystin, potentially enabling cyanobacterial cells to sense and maintain localized concentrations of released cyanotoxins. To investigate this, we prepared 13C,15N-labeled microcystin from isotope-labeled M. aeruginosa cultures as previously described65. M. aeruginosa cultures were then incubated with labeled microcystins for 12 h and analyzed using nanoscale secondary ion mass spectrometry (nanoSIMS). Strikingly, elevated isotope enrichment was consistently observed along CT pili filaments spanning between cells in nanoSIMS images (Fig. 6b–d). Additionally, some CT pili observed to be detached from cells also demonstrated clear isotope enrichment (Fig. 6e, bottom right). To rule out the possibility of remineralization and incorporation of isotope labels into CT pili, we also fixed some cultures prior to incubation with labeled microcystin. In these samples, isotope enrichment was again observed along CT pili. This finding suggests that the CT pilus itself has an affinity for secreted microcystins and thus possibly facilitates the maintenance of toxin concentrations near the cyanobacterial cells. Interestingly, microcystins were not visualized in the helical symmetry averaged cryo-EM maps of the CT pilus (Fig. 1). This suggests that the interaction between CT pilus and microcystin is either moderate affinity, which could be sheared off during the blotting process66, or non-specific, resulting in smeared density upon the application of helical symmetry during reconstruction.
Discussion
In this study, our research reveals a unique CT pilus structure in M. aeruginosa, distinguished by unique protein fold with extensive anti-parallel β-sheet interactions and glycosylation patterns. This pilus demonstrates diverse functionality, contributing not only to colony formation and positioning in the water column, but also to the accumulation of microcystins and ECM. While the precise mechanism of CT pilus biogenesis remains unclear, our findings underscore the intricate and multifaceted nature of these appendages in cyanobacterial biology and ecology.
From a bacterial “economics” perspective, M. aeruginosa invests substantial energy in producing abundant, long, and rigid extracellular CT pili. These protein polymers, comprising over 30,000 protein subunits per filament (estimated based on the observed length and helical rise), impose considerable metabolic demands on the bacteria, including protein synthesis, transport, assembly, and operation. Studies of bacterial flagella, for instance, have estimated that the energy cost for filament synthesis alone can range from 0.5% to 40% of a bacterium’s total energy budget, depending on the species67. With a likely similar energy requirement for CT pili, it is probable that the bacterium only produces these pili when they are essential. When are they needed? It is important to consider that M. aeruginosa also produces gas vesicles, which are flotation devices that help the bacteria reach an optimal position in the water column. If the cell utilizes both gas vesicles and CT pili simultaneously during movement, it will increase drag force and slow down its movement. This suggests that CT pili are likely produced once the cyanobacteria have already reached an optimal position for photosynthesis, and the pili act as a method to increase drag and help them remain stationary.
The relationship between extracellular pili and their role in increasing drag has also been reported in picocyanobacteria that lack gas vesicles62. Homologs of the CT pilin were not identified in the reported strain, and type IV pili were suggested to be responsible for this role based on genetic knockout evidence. This is an exciting discovery as type IV pili are typically flexible filaments with a persistence length (PL) in the order of ~10 μm. In contrast, CT pili are predominantly straight tubes under EM, rendering PL estimation impractical as it far exceeds the micrometer scale68. This difference in rigidity suggests that type IV pili are unlikely to form rigid rods like CT pili, but they could potentially connect bacterial cells through other methods, such as adhesion. However, it is also important to note that cytochrome nanowires, one family of extracellular appendages responsible for extracellular long-range electron transfer, have also been believed to be type IV pili for decades, due to genetic knockout evidence39,42. Therefore, future high-resolution cryo-EM studies are warranted in Synechococcales to validate the profile of observed extracellular pili.
Despite observing the CT pili’s interaction with ECM and microcystin, it remains unclear how CT pili are assembled and whether they can retract for cellular uptake of these compounds and recycling of pilins. The CT pilus may share some similarities with the CUP pili; but significant differences remain, especially its tubular morphology, which diverges significantly from that of typical bacterial flagella and pili (Supplementary Fig. 14). One notable similarity between CT and CUP pilus is the presence of an outer membrane porin protein, which might function as an assembly platform, analogous to the usher protein in the CUP pili. However, the adhesion protein fold observed here is not typically found in most CUP pili. Most CUP pilins possess an Ig-fold with a donor strand that inserts into the neighboring pilin53,69. The major pilins or tip pilins, often referred to as adhesin pilins in CUP systems, also have the same fold as major pilins. In this case, the putative adhesin in the CP pilus is beta-sheet-rich but exhibits a very different fold from CT pilin. More research is needed to better understand the biosynthesis of the CT pilus. Regarding pilus retraction, known retractile pili include type IV pili and conjugation pili70. Type IV pili are produced by the type II secretion system, and this process is driven by two ATPases rotating in opposite directions, one for assembly and the other for retraction71. While a putative type IV pilin has been detected in M. aeruginosa, we did not observe type IV pili in our cryo-EM imaging of all extracellular filaments. Notably, other iron uptake mechanisms, such as siderophore-mediated uptake72, have been reported in M. aeruginosa. Therefore, even if CT pili are incapable of retraction, bacterial cells could still acquire the enriched iron in the ECM near their surface through those mechanisms.
When a pilus has multiple functions, it raises the question of which function is primary. Another well-known example of a multi-functional pilus is the bacterial type IV pilus. The primary function of type IV pili is arguably twitching motility since the pilus can both extend and retract, and it has an adhesive tip. Other suggested functions of type IV pili, such as DNA uptake73, are then largely based on additional positively charged surface properties built upon the existing nature of type IV pili. Using type IV pili as an analogy, we propose that the primary function of CT pili is to serve as a structural scaffold which bridges cells together and hosts the ECM, forming micro-colonies to maintain their position in the water column. Other possible functions, such as enriching extracellular toxins and iron, likely emerged later and rely heavily on additional surface properties, such as O-linked glycosylation. Sequence alignment of CT pilin homologs in different cyanobacteria is consistent with this hypothesis. The observed glycosylation sites are partially conserved among other pilin homologs (Supplementary Fig. 15), where serine residues are sometimes replaced by threonine, and vice versa. However, only one site contains threonine in all pilins. For all other sites, there are putative pilins that possess neither serine nor threonine. This level of partial conservation suggests that those features might have appeared later than the primary function and, therefore, have become more diverse among different cyanobacteria.
In summary, we report a previous unknown family of bacterial pili with a unique protein fold. This discovery was greatly facilitated by recent developments in cryo-EM74,75 and deep-learning based methods47,51,76,77 that enable direct protein identification from near-atomic resolution maps, allowing the discovery of pili without prior knowledge. Future investigation into the CT pilus secretion system, potential retractile properties, and interplay with other cellular processes will undoubtedly deepen our understanding of cyanoHAB formation and its ecological repercussions.
Methods
M. aeruginosa PCC 7806SL growth condition and filament preparation
Cultures of M. aeruginosa PCC 7806SL were procured from Pasteur Culture Collection (Paris, France) and were grown/maintained in polyethylene culture flasks with vented caps and culture medium (BG-11 with 2 mM nitrate) at 22 °C under 25 µE of light (white fluorescent-lights, 6500 K) in an innova™ 4000 incubator shaker (60 rpm). From 100 mL of stationary-phase culture, cells were pelleted by centrifugation at 3200 × g for 10 min and the pellet was washed three times by gentle resuspension in phosphate-buffered saline buffer (PBS, pH 7.4) and centrifugation (3200 × g, 10 min). The washed pellet was resuspended in 2 mL of PBS and vortexed for 2 min to shear off the extracellular filaments. Cells were removed by centrifugation (3200 × g, 10 min) and the supernatant containing CT pili was collected and further centrifuged at 10,000 × g for 5 min. The resulting supernatant contained isolated CT pili, confirmed by SDS-PAGE.
Cryo-EM conditions and image processing
The cell appendage sample (ca. 3.5–4.5 μl) was applied to glow-discharged lacey carbon grids and then plunge frozen using an EM GP2 Plunge Freezer (Leica). The cryo-EM micrographs were collected on a 300 keV Titan Krios with a K3 camera at 1.07 Å/pixel and a total dose of ca. 50 e/Å2. First, patch motion correction and CTF estimation were done in cryoSPARC78,79,80. Next, particles were auto-picked by “Filament Tracer”. All auto-picked particles were subsequently 2D classified with multiple rounds, and all particles in bad 2D averages were removed. After this, the CT pilus dataset had 1,875,465 particles left with a shift of 19 pixels between adjacent boxes. Next, the possible helical symmetries were calculated from an averaged power spectrum generated from the raw particles within 2D classes (Supplementary Fig. 2). There are ~10 possibilities for possible helical symmetry, and all of them were tested in helical refinement. Reconstruction with the correct symmetry reached to 2.4 Å, with side chains of amino acids clearly resolved49. The hand of the 3D volume was determined by the hand of α-helices in the map81,82. The resolution of the final reconstruction was estimated by Map:Map FSC and Model:Map FSC. The final volume was then sharpened with a negative B-factor automatically estimated in cryoSPARC, and the statistics are listed in Supplementary Table 1.
Model building of M. aeruginosa CT pilus
The first step of model building is identifying the pilin protein from the experimental cryo-EM map. The pilin protein, produced by gene BH695_0841, was identified to be the best fit to the EM map among all proteins in this strain by the DeepTracer-ID77, and direct cryo-EM-based approaches such as ModelAngelo83 and DeepTracer76. AlphaFold prediction84 of the identified pilin was used as the starting model for the model building process. Pilin subunits within an asymmetric unit were first docked into the map using ChimeraX85. Pilin subunits were then manually refined in Coot86. Then a filament model was generated, and real space refined in PHENIX87. MolProbity was used to evaluate the quality of the filament model88. The refinement statistics of both mono-pili and tri-pili are shown in Supplemental Table 1. Data visualization was primarily done in ChimeraX85.
Homology searches and phylogenetic analysis
Homologs of M. aeruginosa CT pilin were collected by PSI-BLAST (5 iterations, inclusion E-value threshold of 1e-06) against the non-redundant protein sequence database at NCBI. All sequences longer than 400 amino acids were purged to avoid potential problems with sequence alignment. Multiple sequence alignment was built using MAFFT with the G-INS-1 option. Poorly conserved positions were removed using trimAl with the gap threshold of 0.289. Maximum likelihood phylogenetic analysis was performed using IQ-Tree90. The phylogeny was visualized using iTOL v691.
Electrophoresis and protein characterization
Isolated CT pili were analyzed by SDS-PAGE using a 4–12% polyacrylamide gel, 200 V potential, MES running buffer, SeeBlue™ Plus2 pre-stained protein standard (Invitrogen), and SimplyBlue™ SafeStain for visualization (Invitrogen). The only visible protein band of the isolated CT pili was excised and Trypsin-digested using an in-gel trypsin digestion kit (Thermo Scientific™). Trypsin-fragments were characterized by LC-MSMS using a Vanquish™ Neo UHPLC system (Thermo Scientific) and an Orbitrap Fusion™ Lumos™ Tribrid™ mass spectrometer (Thermo Scientific). Chromatographic separation was done with a Double nanoViper™ PepMap™ Neo UHPLC column (75 µm by 150 mm) and a solvent system of water (A) and 80/20 acetonitrile/water (B), both with 0.1 % formic acid. Elution was done with a flow rate of 0.35 µL/min, a column temperature of 35 °C, and a gradient that began at 3% B, increasing to 7% in 1 min, to 30% in 40 min, to 50% in 6 min, to 95% in 2 min, remaining 95% for 1 min, and returning to 3% immediately, held for 10 min to re-equilibrate the column. Mass spectra were collected with an Orbitrap resolution of 120 K (at m/z of 200) and a scan range of 200–2000 m/z, obtaining data-dependent higher energy collisional dissociation MS2 scans with a normalized collision energy (NCE) of 30 for confirmation of peptide sequences (i.e., b- and y-fragment ions). Apex detection and dynamic exclusion were set in the scan cycle. Using Proteome Discoverer Software (version 3.1, Thermo Scientific), a library trypsin-digest fragments was generated from the proteome of M. aeruginosa PCC7806SL and mass spectra were searched against this library using MSPep Search. Peptide spectral matches were also searched using Sequest HT and were used to determine protein identity. For intact mass and deconvolution analysis, mass spectra were collected with an Orbitrap Fusion Lumos Tribrid Mass Spectrometer coupled to a Vanquish Neo UHPLC System (Thermo Scientific). Samples of isolated CT pili were injected on a MAbPac™ Capillary Reversed Phase HPLC Column (150 µm × 150 mm, Thermo Scientific) and eluted using a mixture of 0.1% formic acid in water (solvent A) and 0.1% formic acid in 80% acetonitrile in water (solvent B). The solvent gradient began at a B:A ratio of 3:97 and was increased to 90:10 over 6 min at a flow rate of 2 µL/min with a column temperature of 60 °C. The EASY-Spray™ ion source spray voltage was set to 1800 V positive ion, the ion transfer tube temp to 305 °C, and the RF lens to 75%. The Orbitrap resolution was set to 7.5 K (at m/z of 200) with quad isolation, a scan range 400–2000 (m/z) in positive mode, an FT Full scan AGC target of 4 × 105, a normalized AGC target of 100%, and a cycle time of 3 s. The resolution was set to the lowest setting possible to shorten the transients and use only the high signal to noise beats before the protein dephases in the Orbitrap. Intact mass spectra were deconvoluted using BioPharma Finder (version 3.0, Thermo Scientific software). Pilin was loaded on a Waters nano-ease C4 column and eluted at 10 µl/min with a 20 min gradient of increasing acetonitrile. Spectral data was collected from 300–1500 m/z on a Waters Synapt G2-S(i) mass spectrometer operated in positive ion, resolution mode. The protein eluted in the post-run column wash at ~80% acetonitrile. The source spectra method was set to “Average Over Selected Retention Time” at 4.19–4.33 min, the deconvolution algorithm was set to “ReSpect” due to the lack of isotopically resolved source spectra, and the mass tolerance was set to 20 ppm. The output mass range for deconvolution was set from 8000 to 16,000 Da with peak model set to “Intact Protein”.
Scanning electron microscopy (SEM), negative-stain transmission electron microscopy (TEM), and scanning-TEM with energy-dispersive X-ray spectroscopy (STEM-EDS)
For SEM, 2 µL M. aeruginosa culture were pipetted onto a silicon wafer, then mounted onto an Al specimen holder and left to air-dry. Prior to SEM analysis, samples were sputter-coated with a 10–15 nm thin layer of Pt under an argon atmosphere at 8 mA for 30 s using a MicroNanoTools MNT-JS1600 plasma sputtering coater. Samples were imaged using a COXEM EM-30N scanning electron microscope (SEM). For TEM, samples of M. aeruginosa and CT pili were visualized by spotting 2 µL onto formvar-coated copper TEM-grids, allowing the drop to sit for 5–10 min on the grid, which was then wicked off with a Kimwipe® (the longer the sample was kept on the grid, the more cells/pili adhered to the formvar, but less than 5 min was insufficient). Samples were stained by spotting 2 µL of 1% uranyl acetate on the grid and wicking off after 30 s. The samples were analyzed using JEOL JEM-1400 series 120 kV Transmission Electron Microscope with an AMT-NanoSprint15L-MarkII camera. With this microscope, qualitative elemental analysis of M. aeruginosa samples was done by STEM-EDS, performed using a STEM unit with bright-field and dark-field imaging, and an Oxford AZtec energy TEM standard EDS detector and software. The goal of the EDS experiments was to obtain a qualitative understanding of the atomic composition of ECM colocalized with cell colonies, therefore spectra were deconvoluted and processed into the presented images directly by the Aztec software without any further processing.
Nanoscale secondary ion mass spectrometric (nanoSIMS) experiments
13C,15N-MCLR was purified from isotope-labeled M. aeruginosa cultures (grown on 13C bicarbonate and 15N nitrate for 3 generations) per the methods as previously described65. 5 mL aliquots of unlabeled M. aeruginosa cultures were centrifuged at 3200 × g for 10 min and cell pellets were resuspended in 5 mL of solutions containing approximately 100 µM 15N13C-MCLR. These were incubated for a week at 22 °C under 25 µE of white fluorescent-lights (6500 K). All were done in triplicate. After 1 week, 100 µL aliquots of the incubation mixtures were fixed with formalin (5% final concentration, 20 min, room temp) and centrifuged at 2000 × g for 10 min. The supernatants were discarded, and cell pellets were resuspended in 100 µL of sterile-filtered MilliQ water. These were centrifuged and decanted again, and the pellets were resuspended in 100 µL of 50/50 water/ethanol. These samples were stored at −20 °C at Florida Atlantic University (Boca Raton, Florida) and shipped overnight on dry-ice to Lawrence Livermore National Laboratory (LLNL) in Livermore, California. For nanoSIMS, cells were pipetted onto a Si wafer, air dried, and sputter coated with ~5 nm of gold. Isotope imaging was performed with a primary 133Cs+ ion beam set to 2 pA, corresponding to an approximately 150 nm diameter beam diameter at 16 keV. Rastering was performed over 20 × 20 microns2 analysis areas with a dwell time of 1 ms pixel−1 for 19–30 scans (cycles) and generated images containing 256 × 256 pixels. Sputtering equilibrium at each analysis area was achieved with an initial beam current of 90 pA to a depth of ~60 nm. After tuning the secondary ion mass spectrometer for mass resolving power of ~7000 (1.5× corrected), secondary electron images and quantitative secondary ion images were simultaneously collected for 12C2−, 13C12C−, 12C14N−, and 12C15N− on individual electron multipliers in pulse counting mode; note that 13C12C−/12C2− = 2 × 13C/12C and 12C15N−/12C14N− = 15N/14N. All nanoSIMS datasets were initially processed using L’Image (http://limagesoftware.net) to perform dead time and image shift correction of ion image data before creating 13C/12C and 15N/14N ratio images, which reflected the level of 13C and 15N incorporation into biomass.
Centrifuging experiments
Approximately 10 mL aliquots of M. aeruginosa culture were transferred to 50 mL conical centrifuge tubes and were either left undisturbed (Control) or were vortexed for 2 min to shear CT pili from cells (vortexed). These were then subjected to repeated rounds of centrifugation for 30-s intervals at 1000 × g. This was done using an IEC Centra CL2 benchtop centrifuge equipped with a 4 × 50 mL bucket rotor set to 1000 × g. For each 30-s interval documented, the centrifuge was run for 45 s, allotting an extra 15 s to account for the time it took the centrifuge to reach ~1000 × g. Cells were counted just prior to the first centrifugation, and cells in the supernatant were counted after each centrifuging interval using a Countess™ II FL automated cell counter. Bright-field and fluorescence images of cell cultures were obtained using this cell counter, equipped with an Invitrogen EVOS light cube with an excitation maximum of 445 nm.
Conductivity measurements
Electrochemical measurements were performed in a 3-electrode electrochemical cell with a glassy carbon working electrode, Pt wire counter electrode, and a Ag/AgCl reference electrode using a Gamry Ref600 potentiostat. The solution consisted of 10 mM potassium ferricyanide (K3Fe(CN)63−) in 0.1 M phosphate buffer at pH 7. Films of CT pili were dropcast onto glassy carbon electrodes and left to dry under vacuum for 1 h before electrochemical experiments. For CV, the potential versus reference was cycled between 0 and 600 mV at scan rates of 10, 50, 100, and 200 mV/s, consecutively. For EIS, the DC potential was set at 0 mV vs reference, and the AC oscillations were set to ±10 mV across a frequency range of 1–1,000,000 Hz.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The three-dimensional reconstruction of CT pilus has been deposited in the Electron Microscopy Data Bank with accession codes EMD-44895. The atomic model has been deposited in the Protein Data Bank with accession code 9BTQ. Unless otherwise stated, all data supporting the results of this study can be found in the article, supplementary, and source data files. The MS/MS mass spectrometry data have been deposited in PRIDE with accession code PXD066540. The intact mass spectrometry data have been deposited in PRIDE with accession code PXD066594. Source data are provided with this paper.
References
Sánchez-Baracaldo, P., Bianchini, G., Wilson, J. D. & Knoll, A. H. Cyanobacteria and biogeochemical cycles through Earth history. Trends Microbiol. 30, 143–157 (2022).
Tan, L. T. Bioactive natural products from marine cyanobacteria for drug discovery. Phytochemistry 68, 954–979 (2007).
Machado, I. M. & Atsumi, S. Cyanobacterial biofuel production. J. Biotechnol. 162, 50–56 (2012).
Plaas, H. E. & Paerl, H. W. Toxic cyanobacteria: a growing threat to water and air quality. Environ. Sci. Technol. 55, 44–64 (2021).
Paerl, H. W. & Huisman, J. Climate change: a catalyst for global expansion of harmful cyanobacterial blooms. Environ. Microbiol. Rep. 1, 27–37 (2009).
Francis, G. Poisonous Australian lake. Nature 18, 11–12 (1878).
Song, L. et al. Harmful cyanobacterial blooms: biological traits, mechanisms, risks, and control strategies. Annu. Rev. Environ. Resour. 48, 123–147 (2023).
Huisman, J. et al. Cyanobacterial blooms. Nat. Rev. Microbiol 16, 471–483 (2018).
Quiblier, C. et al. A review of current knowledge on toxic benthic freshwater cyanobacteria - Ecology, toxin production and risk management. Water Res. 47, 5464–5479 (2013).
Rastogi, R. P., Sinha, R. P. & Incharoensakdi, A. The cyanotoxin-microcystins: current overview. Rev. Environ. Sci. Bio 13, 215–249 (2014).
Yang, Y. M., Yu, G. L., Chen, Y. X., Jia, N. N. & Li, R. H. Four decades of progre124653ss in cylindrospermopsin research: the ins and outs of a potent cyanotoxin. J. Hazard. Mater. 406, 124653 (2021).
Llewellyn, L. E. Saxitoxin, a toxic marine natural product that targets a multitude of receptors. Nat. Prod. Rep. 23, 200–222 (2006).
Bishop, C. T., Anet, E. F. & Gorham, P. R. Isolation and identification of the fast-death factor in Microcystis aeruginosa NRC-1. Can. J. Biochem. Physiol. 37, 453–471 (1959).
Zillige, Y. et al. The cyanobacterial hepatotoxin microcystin binds to proteins and increases the fitness of under oxidative stress conditions. PLoS ONE 6, e17615 (2011).
Hansson, L. A., Gustafsson, S., Rengefors, K. & Bomark, L. Cyanobacterial chemical warfare affects zooplankton community composition. Freshw. Biol. 52, 1290–1301 (2007).
Babica, P., Bláha, L. & Marsálek, B. Exploring the natural role of microcystins—A review of effects on photoautotrophic organisms. J. Phycol. 42, 9–20 (2006).
Gan, N. Q. et al. The role of microcystins in maintaining colonies of bloom-forming spp. Environ. Microbiol. 14, 730–742 (2012).
Dvořák, P. et al. Diversity of the cyanobacteria. In Modern Topics in the Phototrophic Prokaryotes: Environmental and Applied Aspects (ed. Hallenbeck, P. C.) (Springer International Publishing, 2017).
Harke, M. J. et al. A review of the global ecology, genomics, and biogeography of the toxic cyanobacterium, spp. Harmful Algae 54, 4–20 (2016).
Wu, Z. X. & Song, L. R. Physiological comparison between colonial and unicellular forms of Kutz. (Cyanobacteria). Phycologia 47, 98–104 (2008).
Feng, G. Y., Cao, J., Chen, H. M., Meng, X. Z. & Duan, Z. P. Potential gap in understanding cyanoHABs: light-dependent morphological variations in colonial cyanobacterium. Harmful Algae 134, 102622 (2024).
Xu, G. et al. Effect of light-mediated variations of colony morphology on the buoyancy regulation of colonies. Water Res. 235, 119839 (2023).
Feng, G. Y., Zhu, W., Duan, Z. P. & Zhang, Y. The role of morphological changes in Microcystis adaptation to nutrient availability at the colonial level. Harmful Algae 115, 102235 (2022).
Zhang, Y., Zheng, B., Jiang, X. & Zheng, H. Role of gas vesicles and intra-colony spaces during the process of algal bloom formation. Water Environ. Res. 85, 539–548 (2013).
Purcell, E. M. Life at low Reynolds number. Am. J. Phys. 45, 3–11 (1977).
Oliver, R. L. & Walsby, A. E. Direct evidence for the role of light-mediated gas vesicle collapse in the buoyancy regulation of Anabaena flos-aquae (cyanobacteria). Limnol. Oceanogr. 29, 879–886 (1984).
Kinsman, R., Ibelings, B. W. & Walsby, A. E. Gas vesicle collapse by turgor pressure and its role in buoyancy regulation by Anabaena flos-aquae. Microbiology 137, 1171–1178 (1991).
Harke, M. J. et al. A review of the global ecology, genomics, and biogeography of the toxic cyanobacterium spp. Harmful Algae 54, 4–20 (2016).
Gaëtan, J. et al. Widespread formation of intracellular calcium carbonates by the bloom-forming cyanobacterium. Environ. Microbiol. 25, 751–765 (2023).
Duan, Z. P., Tan, X. & Zeng, Q. F. Key physiological traits and chemical properties of extracellular polymeric substances determining colony formation in a cyanobacterium. J. Oceano. Limnol. 40, 1720–1731 (2022).
Sato, M., Amano, Y., Machida, M. & Imazeki, F. Colony formation of highly dispersed by controlling extracellular polysaccharides and calcium ion concentrations in aquatic solution. Limnology 18, 111–119 (2017).
Liu, L. Z., Huang, Q. & Qin, B. Q. Characteristics and roles of extracellular polymeric substances (EPS) in cyanobacterial blooms: a short review. J. Freshw. Ecol. 33, 183–193 (2018).
Keren, N., Aurora, R. & Pakrasi, H. B. Critical roles of bacterioferritins in iron storage and proliferation of cyanobacteria. Plant Physiol. 135, 1666–1673 (2004).
Jiang, H.-B., Lu, X.-H., Deng, B., Liu, L.-M. & Qiu, B.-S. Adaptive mechanisms of the model photosynthetic organisms, cyanobacteria, to iron deficiency. In Microbial Photosynthesis (ed. Wang, Q.). (Springer Singapore, 2020).
Wilhelm, S. W. Ecology of iron-limited cyanobacteria: a review of physiological responses and implications for aquatic systems. Aquat. Microb. Ecol. 09, 295–303 (1995).
Wu, J. F. & Luther, G. W. Complexation of Fe(Iii) by natural organic-ligands in the Northwest Atlantic-Ocean by a competitive ligand equilibration method and a kinetic approach. Mar. Chem. 50, 159–177 (1995).
Wang, F. et al. Cryo-EM structure of an extracellular Geobacter OmcE cytochrome filament reveals tetrahaem packing. Nat. Microbiol 7, 1291–1300 (2022).
Wang, F. et al. Structure of Geobacter OmcZ filaments suggests extracellular cytochrome polymers evolved independently multiple times. Elife 11, e81551 (2022).
Wang, F. et al. Structure of Microbial Nanowires Reveals Stacked Hemes that Transport Electrons over Micrometers. Cell 177, 361–369.e310 (2019).
Nakasugi, K. & Neilan, B. A. Identification of pilus-like structures and genes in Microcystis aeruginosa PCC7806. Appl. Environ. Microbiol. 71, 7621–7625 (2005).
Sure, S. et al. Inquisition of Microcystis aeruginosa and Synechocystis nanowires: characterization and modelling. Anton. Leeuw. Int. J. G 108, 1213–1225 (2015).
Wang, F., Craig, L., Liu, X., Rensing, C. & Egelman, E. H. Microbial nanowires: type IV pili or cytochrome filaments?. Trends Microbiol. 31, 384–392 (2023).
Wang, F., Craig, L., Liu, X., Rensing, C. & Egelman, E. H. Models are useful until high-resolution structures are available. Trends Microbiol. 31, 550–551 (2023).
Barrio-Hernandez, I. et al. Clustering predicted structures at the scale of the known protein universe. Nature 622, 637–645 (2023).
Huber, S. T., Terwiel, D., Evers, W. H., Maresca, D. & Jakobi, A. J. Cryo-EM structure of gas vesicles for buoyancy-controlled motility. Cell 186, 975–986.e913 (2023).
Chua, E. Y. D. et al. Better, faster, cheaper: recent advances in cryo-electron microscopy. Annu. Rev. Biochem. 91, 1–32 (2022).
Jamali, K. et al. Automated model building and protein identification in cryo-EM maps. Nature 628, 450–457 (2024).
Baquero, D. P. et al. Extracellular cytochrome nanowires appear to be ubiquitous in prokaryotes. Cell 186, 2853–2864.e2858 (2023).
Wang, F., Gnewou, O., Solemanifar, A., Conticello, V. P. & Egelman, E. H. Cryo-EM of helical polymers. Chem. Rev. 122, 14055–14065 (2022).
Zilliges, Y. et al. An extracellular glycoprotein is implicated in cell-cell contacts in the toxic cyanobacterium PCC 7806. J. Bacteriol. 190, 2871–2879 (2008).
Abramson, J. et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 630, 493–500 (2024).
van Kempen, M. et al. Fast and accurate protein structure search with Foldseek. Nat Biotechnol 42, 243–246 (2024).
Busch, A. & Waksman, G. Chaperone-usher pathways: diversity and pilus assembly mechanism. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367, 1112–1122 (2012).
Guilvout, I. et al. Membrane platform protein PulF of the Klebsiella type II secretion system forms a trimeric ion channel essential for endopilus assembly and protein secretion. mBio 15, e0142323 (2024).
Liu, J. et al. Two distinct archaeal type IV pili structures formed by proteins with identical sequence. Nat. Commun. in press, (2024).
Yang, Y. et al. Structures of alpha-synuclein filaments from human brains with Lewy pathology. Nature 610, 791–795 (2022).
Wang, F. et al. Deterministic chaos in the self-assembly of beta sheet nanotubes from an amphipathic oligopeptide. Matter 4, 3217–3231 (2021).
Dietrich, H. M. et al. Membrane-anchored HDCR nanowires drive hydrogen-powered CO(2) fixation. Nature 607, 823–830 (2022).
Kumar, M. et al. Amino-acid-encoded biocatalytic self-assembly enables the formation of transient conducting nanostructures. Nat. Chem. 10, 696–703 (2018).
Holm, L. Dali server: structural unification of protein families. Nucleic Acids Res. 50, W210–W215 (2022).
Glitsch, M. Essentials of cell physiology. Surgery 34, 371–375 (2016).
Aguilo-Ferretjans, M. D. M. et al. Pili allow dominant marine cyanobacteria to avoid sinking and evade predation. Nat. Commun. 12, 1857 (2021).
Gu, P., Li, Q., Zhang, W., Zheng, Z. & Luo, X. Effects of different metal ions (Ca, Cu, Pb, Cd) on formation of cyanobacterial blooms. Ecotoxicol. Environ. Saf. 189, 109976 (2020).
Chen, H. & Lurling, M. Calcium promotes formation of large colonies of the cyanobacterium Microcystis by enhancing cell-adhesion. Harmful Algae 92, 101768 (2020).
Ricca, J. G. et al. Endogenous production and vibrational analysis of heavy-isotope-labeled peptides from cyanobacteria. Chembiochem 25, e202400019 (2024).
Weissenberger, G., Henderikx, R. J. M. & Peters, P. J. Understanding the invisible hands of sample preparation for cryo-EM. Nat. Methods 18, 463–471 (2021).
Schavemaker, P. E. & Lynch, M. Flagellar energy costs across the tree of life. Elife 11, e77266 (2022).
Treuner-Lange, A. et al. Tight-packing of large pilin subunits provides distinct structural and mechanical properties for the type IVa pilus. Proc. Natl. Acad. Sci. USA 121, e2321989121 (2024).
Allen, W. J., Phan, G. & Waksman, G. Pilus biogenesis at the outer membrane of Gram-negative bacterial pathogens. Curr. Opin. Struc. Biol. 22, 500–506 (2012).
Clarke, M., Maddera, L., Harris, R. L. & Silverman, P. M. F-pili dynamics by live-cell imaging. Proc. Natl. Acad. Sci. USA 105, 17978–17981 (2008).
Craig, L., Forest, K. T. & Maier, B. Type IV pili: dynamics, biophysics and functional consequences. Nat. Rev. Microbiol 17, 429–440 (2019).
Xing, W., Huang, W. M., Li, D. H. & Liu, Y. D. Effects of iron on growth, pigment content, photosystem II efficiency, and siderophores production of Microcystis aeruginosa and Microcystis wesenbergii. Curr. Microbiol. 55, 94–98 (2007).
Wang, F. et al. Cryoelectron microscopy reconstructions of the Pseudomonas aeruginosa and Neisseria gonorrhoeae type IV pili at sub-nanometer resolution. Structure 25, 1423–1435.e1424 (2017).
Egelman, E. H. The current revolution in Cryo-EM. Biophys. J. 110, 1008–1012 (2016).
Callaway, E. The revolution will not be crystallized: a new method sweeps through structural biology. Nature 525, 172–174 (2015).
Pfab, J., Phan, N. M. & Si, D. DeepTracer for fast de novo cryo-EM protein structure modeling and special studies on CoV-related complexes. Proc. Natl. Acad. Sci. USA 118, e2017525118 (2021).
Chang, L. et al. DeepTracer-ID: de novo protein identification from cryo-EM maps. Biophys. J. 121, 2840–2848 (2022).
Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Egelman, E. H. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy 85, 225–234 (2000).
Egelman, E. H. Reconstruction of helical filaments and tubes. Methods Enzymol. 482, 167–183 (2010).
Jamali, K. et al. Automated model building and protein identification in cryo-EM maps. Nature 628, 450–457 (2023).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 (2004).
Afonine, P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D. Struct. Biol. 74, 531–544 (2018).
Williams, C. J. et al. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018).
Capella-Gutierrez, S., Silla-Martinez, J. M. & Gabaldon, T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 (2009).
Minh, B. Q. et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534 (2020).
Letunic, I. & Bork, P. Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296 (2021).
Acknowledgements
This research was, in part, supported by the National Cancer Institute’s National Cryo-EM Facility at the Frederick National Laboratory for Cancer Research under contract 75N91019D00024. Electron microscopy screening was carried out in the UAB Cryo-EM Facility, supported by the Institutional Research Core Program and O’Neal Comprehensive Cancer Center (NIH grant P30 CA013148), with additional funding from NIH grant S10 OD024978. We are grateful to Dr. James Kizziah, and Dr. Tara Fox for assisting with the screening or data collection. We’d like to thank Dr. Ed Egelman for helpful discussions in the preparation of this manuscripts. The cryo-EM work in F.W. laboratory was supported by NIH grant GM138756. The conductivity measurements in A.I.H. and F.W. laboratories were supported by DOE grant DE-SC0020322 and DE-SC0024303. The work in J.W.L. laboratory was supported by contracts with the South Florida Water Management District (through J.W.L.) and the U.S. Geological Survey under Grant/Cooperative Agreement No. G18AC00311 (through J.G.R). Part of this work was funded by Lawrence Livermore National Laboratory’s (LLNL) Laboratory Directed Research Development grant # 20-ERD-061 and work at LLNL was performed under the auspices of the US Department of Energy under contract DE-AC52-07NA27344. The acquisition of a Transmission Electron Microscope was supported by a Department of Defense instrumentation/equipment grant (W911NF2110147) to V.M.
Author information
Authors and Affiliations
Contributions
J.G.R. and H.A.P. performed pili sample preparation. H.A.P. and F.W. performed cryo-EM and image analysis. S.H.N. performed SDS-PAGE and S.H.N., C.P.D, H.A.P., and P.E.P. performed mass spectrometry. J.G.R., R.R.S., and H.A.P. performed the negative staining analysis. J.G.R. and B.G.D., under the supervision of V.M., collected TEM and STEM-EDS images. X.M. and P.K.W. performed nanoSIMS analysis. A.G., under the supervision of A.I.H., performed conductivity measurements. F.W. and J.W.L. supervised the research, and J.G.R., J.W.L., and F.W. obtained funding. J.G.R. and F.W. wrote the manuscript with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Sandun Fernando, Pranab Goswami, Kevin Mace and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ricca, J.G., Petersen, H.A., Grosvirt-Dramen, A. et al. A family of tubular pili from harmful algal bloom forming cyanobacterium Microcystis aeruginosa. Nat Commun 16, 8082 (2025). https://doi.org/10.1038/s41467-025-63379-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-63379-1