Abstract
Marine heatwave (MHW) impacts on ecosystem functions and services remain poorly constrained due to limited time-resolved datasets integrating physical, chemical, and biological parameters at relevant scales. Here we show that combining over a decade of autonomous Biogeochemical (BGC)-Argo float measurements with water-column plankton community profiles reveals the impacts of MHWs on particulate organic carbon (POC) production, transformation, and transport in the northeastern subarctic Pacific Ocean. POC concentrations are exceptionally high during the 2015 and 2019 MHWs, linked to detritus enrichment and shifts in plankton community structure. Instead of being rapidly exported to depth, particles <100 µm accumulate in mesopelagic waters, where slow remineralization over the year reduces deep particle flux and carbon sequestration potential. This enhancement is absent in the 2014 and 2020 MHWs, underscoring variability in ecosystem responses to extreme events. These findings highlight the need for sustained, multi-platform observations to assess and predict carbon-cycle responses to thermal extremes.
Similar content being viewed by others
Introduction
Marine heatwaves manifest as prolonged warming events persisting from weeks to years. The resulting thermal stress can lead to increased mortality and biodiversity loss with concomitant changes in food web structure and biogeochemical cycling1,2. Such impacts can overshoot ecological resilience thresholds resulting in state changes that negatively impact ecosystem functions and services3. Ocean observations and models suggest that MHWs have been expanding in size and intensifying over the past few decades4. Even a reduction in anthropogenic carbon emissions is unlikely to decelerate spatial expansion and duration of these events for many years to come4.
From the standpoint of carbon cycling dynamics, elevated ocean temperature reduces CO2 uptake5 and promotes water column stratification; the latter decreases vertical mixing with a resulting impact on surface nutrient renewal and primary production6. These changes percolate through marine food webs with the potential to alter the size and composition of particulate organic carbon (POC) and the efficiency of the biological carbon pump (BCP), an integral global mechanism for carbon export and deep ocean sequestration7,8,9. Despite the magnitude of these effects, integrated analysis of plankton communities at the base of marine food webs and POC production and export during MHWs is limiting10.
Two successive and persistent MHWs impacted the Northeastern subarctic Pacific Ocean (NESAP) waters: the first started at the end of 2013 and persisted until 20156,11,12 and the second developed between 2019-2012,13. The first event called “The Blob” was catalyzed by suppressed wind stress during the winters of 2013-14. Air-sea teleconnections with the equatorial Pacific during the development of an extreme El-Niño added persistence to the warm event and increased upper ocean stratification11,12. Water column impacts included a decline in surface dissolved iron concentrations14, and changes in primary productivity6,15 and microbial community structure including heterotrophs 16 and phytoplankton17. The second MHW formed due to the weakening of the North Pacific high-pressure system and imprinted a low-density, low-salinity feature in the euphotic zone12,13. High productivity during the summer of 2019 was associated with a volcanic eruption and land fires15,18,19. Biogenic CaCO3 export was insignificant in comparison to non-MHW years20, indicating potential changes in phytoplankton community composition and the BCP. During both events higher trophic levels were also affected, leading to the collapse of Bering Sea snow crab populations21 and low birth rates among whales22.
Here, we combine high-resolution BGC-Argo profiling float data collected near the Line P transect in conjunction with the NASA EXPORTS program23 between 2010-22 with pigment-based measurements of phytoplankton functional groups and DNA metabarcoding of plankton community structure including microscopic prokaryotic (bacteria and archaea) and eukaryotic constituents to explore the impacts of MHWs on food webs and carbon transport processes in the NESAP. BGC-Argo profiling floats equipped with temperature, salinity, nitrate, optical backscatter (bbp, a proxy for POC) and fluorescence (a proxy for chlorophyll a concentration, hereafter referred as Chl) sensors profiled the area every 5–10 days, allowing high-resolution observations of net community production, particle production and export. Pigment and metabarcoding data collected near the float’s location provided biological context to link anomalous POC accumulation under thermal stress to plankton community structure. During both MHWs, the vertical transfer of small particles to a preferential mesopelagic depth indicated an intensification of zooplankton vertical migration, a well-known dominant export driver in the region24,25. The subsequent slow remineralization of non-sinking particles <100 µm over the following months decreased the prospects for deep carbon export.
Results
Physical and biogeochemical impacts
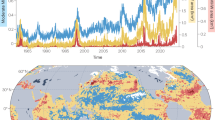
Both MHWs imprinted a warm and stratified water column effect (Fig. 1a). Indeed, stratification and stability at the base of the mixed layer was strongest during MHW years of 2014, 2015, 2019 and the non-MHW year of 2016, as observed in the Brunt-Väisälä frequency (Fig. 1a). Surface temperature anomalies >1°C during the first MHW persisted between 2014-15 (Fig. 1b), agreeing with previous reports6,11. Comparisons between three temperature products (BGC-Argo floats, the Argo-based climatology, and the gridded Armor3D reanalysis) confirmed strong agreement among datasets (R = 0.88–0.96), with minimal biases and low RMSE values (Fig. 1c, Table S1). The agreement across products suggests that the BGC-Argo data reliably capture regional temperature variability, supporting their use in tracking changes associated with marine heatwaves.
Temperature structure and variability in the NESAP from BGC-Argo floats and gridded products. a Brunt-Väisälä frequency (N2, s-2) over 0-100 m, highlighting stratification; the mixed layer depth (MLD) is overlaid in black (MLD defined by a 0.2 °C potential-temperature threshold). b Temperature anomaly (°C) from 0 to 1000 m relative to the 2004-2023 Argo gridded climatology; the diverging red–blue color scale is zero-centered (warm colors = positive anomalies, cool colors = negative anomalies). c Temperature at 50 m from three sources: BGC-Argo floats (blue triangles, monthly means with shaded ±1σ), Armor3D reanalysis (red line), and Argo gridded climatology (black line with black circles). d–f Time series of temperature anomaly (°C) at 50, 300, and 500 m from the Argo climatology; the red dashed line marks zero anomaly. Year ticks on the x-axes indicate calendar years. Statistical comparisons for panel (c) are reported in Table S1.
Elevated temperatures propagated to subsurface waters over subsequent years with temperature anomalies up to +0.2°C observed down to 300 m (Fig. 1d–f). During the second MHW, similar anomalies were observed between 2018-19 and propagated to deeper ocean layers down to 500 m, persisting until 2021.
BGC-Argo profiling float data collected in the vicinity of stations P20-P26 along the Line P transect (Fig. 2a) revealed peaks of small particles <100 µm, maxima in chlorophyll a (Chl), and net community production (NCP) during springs and late summers over the time series, signaling enhanced primary productivity during those seasons (Figs. S1–S4). Upper ocean stocks of the small POC fraction <100 µm increased during the productive season and were particularly elevated during the second MHW in the Spring of 2019 and Summer of 2020 (Fig. 2e). Baseline POC concentrations (i.e. POC during wintertime minima in January) were remarkably different between the two periods: the years preceding and during the first MHW (2010-15) had lower POC baselines in comparison to post-MHW (2018-22). For the first period, the minimum POC ranged between 248 mg m-2 in 2011 and 395 mg m-2 in 2012. For the second period, the POC baseline ranged between 596 mg m-2 in 2020 and 949 mg m-2 in 2021.
a Map of the Northeastern subarctic Pacific (NESAP) showing BGC-Argo float trajectories (colored dots; float IDs in legend), approximate float positions during the marine heatwave (MHW) peaks in 2015 and 2019 (black dots), and Line P stations sampled for pigments and genetics (magenta triangles, stations P20-P26). b, c Mean monthly chlorophyll-a to carbon ratio (Chl:C, dimensionless) integrated over 0-100 m. d, e Mean monthly particulate organic carbon (POC, mg m-2) for 0-100 m. f, g Mean monthly POC (mg m-2) for 100-300 m (upper mesopelagic). In each panel, lines are colored by year (see panel legends); MHW years (2014, 2015, 2019, 2020) are red. “Small particles” denotes the bbp-derived fraction <100 µm.
POC accumulation during each year’s productive season was estimated in relation to the baseline as ∆POC = POCmax - POCmin. For the first period (Fig. 2d), upper-ocean ∆POC had similar values regardless of the year corresponded to a MHW or non-MHW year: for example, ∆POC was 254 mg m-2 in 2015 (MHW) versus 252 mg m-2 in 2011 (non-MHW). During the second period (Fig. 2e), upper ocean POC was anomalously high in the spring of 2019 (MHW) and different from any other year on record. ∆POC increased to 2041 mg m-2 by April, a 3-fold increase from the baseline and 8x higher than in the MHW year of 2015. The late summer blooms promoted a peak in ∆POC at 760 mg m-2 in August of 2019, and 1016 mg m-2 in October of 2020 (Fig. 2c). The upper ocean ∆POC accumulations in 2019 and 2020 were not proportionally tracked by Chl, resulting in a significant drop in Chl:C ratios (Fig. 2c and e).
The productive seasons were followed by a subsurface increase in POC stocks within the upper mesopelagic layer between 100-300 m (Fig. 2f, g). This was a seasonal trend for the entire time series, resulting in higher POC concentrations at 200 m in relation to 100 m independent of the magnitude of the mixed layer shoaling during spring months (Fig. S5). Since this particle accumulation at a deeper depth horizon was unrelated to mixed-layer shoaling and given that the shoaling is minor in the region—only a few meters per month (Fig. S5a)—a mixed-layer pump could not have driven the preferential transfer of particles to deeper waters. Rather, the persistent accumulation at ~200 m likely reflects biologically mediated processes such as vertical migration or mesopelagic retention, which would explain the formation of a stable particle layer at that depth. Additionally, this accumulation pattern at a preferential depth does not correspond to particle export by gravitational sinking- when an exponential decrease of particle concentrations over depth follows a Martin curve6 due to active particle export coupled with microbial respiration.
MHW years experienced much higher accumulation during the spring of 2015 and 2019 than all other years (Fig. 2f, g). In 2015, there was a 3.2-fold change in mesopelagic POC stocks as ∆POC = 174 mg m-2. The mesopelagic accumulation did not follow upper-ocean accumulation. In 2019, mesopelagic ∆POC was 1155 mg m-2, marking a 3.7-fold increase from baseline values and the highest difference between deeper and shallower POC concentrations (Fig. S5b). This mesopelagic enhancement tracked a high upper ocean ∆POC (Fig. 2). During 2015 and 2019, there was a clear particle flux imbalance in the upper mesopelagic in comparison to non-MHW years during spring, when the system experienced lower ∆POC changes between 0.4- and 2.1-fold. The high ∆POC feature was also observed during the summer of 2020 (MHW year, ∆POC = 495 mg m-2), although smaller in comparison to the spring peak. The accumulated POC during those MHW years slowly decreased over the following months, and concentrations dropped down to near baseline values by winter.
The timing between POC accumulation in the mesopelagic and its subsequent removal during MHWs is imprinted as concentration anomalies in relation to non-MHW years: overall, stronger anomalies were observed in 2015 and 2019 (Fig. 3) for the upper ocean and the mesopelagic, especially around 200 m. The main source of particles to the deeper layer can be tracked to spring months, with a small contribution of summer months in 2015. In 2014 and 2020, positive anomalies were smaller, shorter in duration and driven by summer particles.
a–d Time series of particulate organic carbon (POC; mg m-3) anomalies in the upper 0-400 m for 2014, 2015, 2019 and 2020 MHW years. Anomaly = small-particle POC (<100 µm, bbp-derived) - monthly non-MHW baseline. Positive values (green) indicate POC accumulation relative to the non-warming baseline; negative values (purple) indicate deficits. Profiles were vertically smoothed (20 m bins) and gridded (5 m) before interpolation.
To link the anomalous POC accumulation with phytoplankton production as a potential source of carbon, we computed NCP using the mass balance model based on BGC-Argo profiling float nitrate data (Fig. S4)20. In 2015, the largest fraction of NCP happened during the spring bloom at about 2.8 mol C m-2 yr-1, the highest on record for the season. Elevated NCP did not result in elevated upper ocean ∆POC, but was closely followed by mesopelagic accumulation (Fig. 2d, f). In 2019, spring blooms resulted in NCP around 1.2 mol C m-2 yr-1, modest and comparable in magnitude to non-MHW years. There was a mismatch between modest spring NCP and high ∆POC (Fig. 2c, e), thus, POC accumulation with low Chl:C could not be attributed to increasing phytoplankton productivity and biomass alone. Summer NCP elevated annual production to about 3.8 mol C m-2 yr-1, but did not contribute significantly to the elevated ∆POC observed in the mesopelagic (Figs. 2g, 3c). Finally, the 2014 and 2020 summer blooms drove most of the NCP in those years at 2.4 mol C m-2 yr-1 and 0.4 mol C m-2 yr-1, respectively (Fig. S4). The 2020 low NCP did not track the elevated ∆POC that year (Fig. 2e, g).
Food web impacts
Phytoplankton pigment data (chlorophylls and carotenoids) from stations P20-P26 along the Line P transect were used to estimate seasonal phytoplankton composition (Fig. 4). Total chlorophyll a concentrations were generally low (mean = 0.395 ± 0.187 mg m-3), but were elevated in the spring of 2019 as compared to other years (0.829 mg m-3 vs. averaged 0.371 mg m-3, respectively). Phytoplankton communities were dominated by haptophytes, chlorophytes and pelagophytes. Spring communities had additional contributions from cyanobacteria and diatoms, while summer had contributions from diatoms and dinoflagellates (Fig. 4a, b). Increased chlorophyte abundances were observed during both MHWs (Figs. 4c, S6), while higher diatom abundances were observed only in the Spring of 2019, tracked by enhanced NCP (Fig. S4) and anomalous ∆POC (Fig. 2e, g).
Phytoplankton groups were estimated using CHEMTAX analysis of pigment concentrations measured using HPLC. A Average concentrations of major phytoplankton groups from hydrographic stations along the Line P transect (P20-P26). B Average relative concentrations of major phytoplankton groups across P20-P26. Wi winter, Sp spring, Su summer. C Principal components analysis (PCA) ordination biplot of phytoplankton group abundance, with points color-coded by year and shapes by season. Directions and length of vectors indicate how each phytoplankton group contributes to the annual and interannual variation in phytoplankton community composition in this region. Note that MHW years (2015 and 2019) are indicated by shades of red.
Prokaryotic and eukaryotic plankton community composition was determined using small subunit ribosomal RNA (16S and 18S rRNA) metabarcode sequencing on samples collected in the surface mixed layer (10−100 m) and upper mesopelagic waters (100−300 m) at stations P20 and P26, flanking the BGC-Argo float paths (Fig. 2) between 2010−2019 (2015−2019 only for eukaryotes). Community structure correlated with depth (Figs. S7–S9), which explained 70% of the variation in prokaryotic communities (PERMANOVA, p = 0.001) and 24% of the variation in eukaryotic communities (PERMANOVA, p = 0.001). Season only explained <10% of the community variation (PERMANOVA, p = 0.001), perhaps not surprising given the weak seasonal harmonic in biochemical parameters at P20 and P2626.
Across the time series, prokaryotic surface communities were dominated by cosmopolitan taxa like SAR11 within the Alphaproteobacteria, SAR86 and OM43 within the Gammaproteobacteria, Formosa within the Bacteroidia, and the cyanobacterial genus Synechococcus. In mesopelagic waters chemoautotrophic taxa were more abundant including Nitrosopumilus and Nitrosopelagicus, SAR324, Marinimicrobia, SUP05, and Nitrospina involved in coupled carbon, nitrogen, and sulfur cycling processes (Fig. S8). Dominant eukaryotic taxa included cyclopoid copepods (especially Oithona), and dinoflagellates distributed throughout the water column while other eukaryotes exhibited depth-specific trends (Fig. S9). For example, Ciliates affiliated with Spirotrichea and phototrophic taxa including the haptophytes Phaeocystis and Chrysochromulina, chlorophyte Bathycoccus, diatoms Bacillariophyceae and Mediophyceae, and Pelagophytes were more abundant in surface waters while radiolarians including Polycystinea, Acantharea, and RAD-B, and siphonophores were more abundant in upper mesopelagic waters. Dinoflagellate abundance shifted from Gymnodiniales in euphotic waters, to parasitic Syndinians in the upper mesopelagic (Fig. S9)—such heterogeneity was similar to observed in the Pacific, signaling a complex food-web26,27,28,29. Calanoid copepod sequences, especially the genus Paracalanus in the upper ocean, made up a larger fraction of copepod sequences in both heatwave years (2015 and 2019, Fig. S9) compared to non-heatwave years, however copepod sequences as a whole were notably lower in the 2019 upper mesopelagic layer compared to 2015 (Fig. S10). Typically found on the continental shelf in the Northeast Pacific30, the presence of sequences closely related to Paracalanus sp. warrants further investigation and validation.
The planktonic community inhabiting surface waters was relatively consistent between MHWs (Table S2) with only 8 prokaryotic and 6 eukaryotic taxa identified as significantly differentially abundant (Fig. 5). These included increased abundance of prokaryotic taxa Marinimicrobia, Pseudoalteromonas, Pirellulaceae and Cellvibrionaceae along with the eukaryotic diatoms Thalassiosiraceae and Fragilariopsis, MAST-3 stramenopile, and Syndiniales DG-I-Clade-5 in 2015 as well as various Gammaproteobacteria affiliated with Pseudomonas and Thiotricales and eukaryotic haptophytes in 2019. The magnitude of changes in mesopelagic community structure were more intense (Table S2), with numerous taxa coinciding with POC accumulation (Fig. 2f, g) identified as significantly differentially abundant (Fig. 5). For example, Synechococcus, SAR11 Clade I, SAR116, OCS116, SAR86, and Pseudohongiella were all more abundant in 2015 than in 2019 while HOC36 and UBA10353 were more abundant in 2019 (Fig. 5). Decreased SAR11 abundance in 2019 corresponded to a proportional increase in Nitrosopumilus, Nitrosopelagicus (though these were not identified as significant) and SAR324. Within the eukaryotic community, parasitic Syndiniales and heterotrophic dinoflagellates including Prorocentrum, Gyrodinium, and Gymnodinium taxa were identified as significantly differentially abundant in 2019. Increased abundance of these taxa corresponded to a decrease in Cyclopoid copepod abundance, typically the most abundant copepods in mesopelagic waters, and an increase in the Calanoid copepod Microcalanus (Fig. S9). Polycystinea and Acantharea radiolarians, especially Cladococcus and Acantharea F3, were significantly more abundant in 2015, while radiolarian groups RAD-A and RAD-B were elevated in 2019.
Heat trees showing changes in prokaryotic (A, B) and eukaryotic (C, D) taxon abundance during 2015 and 2019 MHWs in surface mixed layer (A, C) and mesopelagic (B, D) waters at P20 and P26. Significant differences were determined using a Wilcoxon Rank Sum Test. Only samples from spring and summer Line P cruises were considered in this comparison since these samples were from months when the BGC-Argo floats detected anomalies in POC concentrations. The log2 ratio of the median of proportions for each significantly different sample group is displayed by the color of the nodes and edges in the heat tree while the node size is scaled to the average relative abundance of that taxon in the associated depth range. Surface mixed layer and mesopelagic waters were considered separately for this statistical analysis, given clear differences in prokaryotic and eukaryotic community structure with depth (See Fig. S7).
Discussion
Integrated physical, chemical and biological parameter information collected over more than a decade of time-resolved observations shed light on marine food web structure, POC composition, and carbon export processes in NESAP waters during non-MHW years. A tightly coupled food web was observed in euphotic waters, with microzooplankton as the main consumers of heterotrophic bacteria and phytoplankton24,25,28,29. Elevated POC during the productive season was composed of detritus and fecal pellets of relatively small sizes ranging between 1–6 µm8,25,29 with the main drivers of POC export to mesopelagic waters linked to consumption of microzooplankton and picoeukaryotes9,24. Processed at different trophic levels in the euphotic zone, highly-worked particles <100 µm are typically the main carbon source for mesopelagic zooplankton dominated by heterotrophic and parasitic protists24,25,31 (Fig. S9).
In response to thermal stress during successive and persistent MHWs, the NESAP experienced changes in plankton community structure and POC production and export, as summarized in Table 1. At the start of the first MHW in 2014, production was nearly balanced by export with no anomalous ∆POC accumulation observed (Fig. 3a), similar to historical observations in the region25,29. As thermal stress persisted into 2015, POC export became imbalanced and resulted in mesopelagic particle accumulation (Fig. 3b) likely caused by changes in zooplankton community structure (Figs. 5, S9) and vertical migration patterns between sunlit and dark ocean waters. For instance, two radiolarian groups, Acantharea and especially Spumellarian genus Cladococcus, were elevated in summer 2015 (Fig. 5). These protozooplankton groups can be important carbon exporters out of the euphotic zone32,33,34,35,36 due to their strontium and silicate skeletons, respectively, and because they reside in epipelagic and mesopelagic waters31,36. Furthermore, radiolarians are thought to be a source of minipellets, 3-50 um in size and comprising a notable amount of particles in sediment traps32,37. In addition to the adult and juvenile organisms themselves34, these minipellets may have contributed to the accumulation of particles in the upper mesopelagic waters38 during the 2015 MHW. Similarly, the higher abundance of the small copepod Oithona in 2015 (Figs. 5, S9) could have further contributed to the production and continued accumulation of small slow to non-sinking fecal pellets26,39,40 in upper mesopelagic waters (Fig. 2e), decreasing the prospects for rapid, deep particle export.
The period following the 2015 MHW was marked by elevated background POC (Fig. 2e) in an increasingly stratified water column (Fig. 1a) promoting further particle accumulation. In the Spring of 2019 extreme POC accumulation in the upper ocean was not proportionally tracked by NCP (Fig. S4) and Chl (Figs. S2, 2b), leading to low Chl:C ratios. This anomaly in Chl:C, not caused by elevated NCP, indicates an intensification of food web carbon processing, resulting in excess carbon-rich detritus, which is already known to comprise most background POC in the NESAP29. The low Chl:C ratios also coincided with high diatom concentrations observed in the HPLC dataset which is not typical for the region (Fig. 4b), indicating that changes in phytoplankton composition could have played an additional role in changing particle composition and Chl:C ratios that season. Interestingly, elevated NCP (Fig. S4) and high Chl levels in 2019 (Fig. 4a) promoted by wildfires and volcanic eruptions15,18,19 did not result in mesopelagic POC accumulation (Fig. 3c), suggesting that phytoplankton production alone cannot account for anomalous ∆POC accumulation. Rather, thermal stress induced by MHWs could have modulated food web structure causing changes in carbon processing between trophic levels.
Increased abundance of parasitic Syndiniales in mesopelagic waters in 2019 compared to 2015 may have altered carbon processing between heatwave years41. Syndiniales have been implicated in particulate carbon flux attenuation by diverting carbon away from POC pools and into labile dissolved organic matter, fueling bacterial production42. This group of parasites were shown to be associated with dinoflagellates (Dinophyceae) and radiolarians, including RAD-B radiolarians31,33, both of which were elevated in 2019 compared to 2015 in upper mesopelagic waters and both important contributors to particle flux dynamics within and out of upper mesopelagic NESAP waters25. To that end, while small, calanoid copepods including Microcalanus and the warm-water, southern copepod Paracalanus were abundant in spring and summer 2019 respectively, consistent with observed positive anomalies of warm-water copepods during MHWs, there was an overall reduction in the abundance of copepod sequences in 2019 compared to prior years (Fig. S10). Without these key mesozooplankton grazers, control on microzooplankton (e.g. heterotrophic and mixotrophic dinoflagellates and ciliates) may have relaxed, allowing for their increased abundance in 2019 (Fig. 5) and further fueling associated parasite populations (Figs. S9, S10). To this end, with fewer copepods during the spring and summer, overall fecal pellet production likely declined, reducing export from the mesopelagic and contributing to small detrital particle accumulation.
The accumulated particles may have expanded the niche space for chemoautotrophic bacteria and archaea driving coupled carbon, nitrogen, and sulfur cycling processes across microscale oxyclines, with implications for nitrogen loss and climate-active trace gas production43. The decline in cosmopolitan SAR11 clades in the 2019 MHW relative to 2015 is noteworthy and could be linked to the heavy recycling and accumulation of organic matter. Experimental incubations in NESAP waters revealed SAR11 (and SAR86, also elevated in 2015) was more abundant when DOM was less degraded44. SAR11 and SAR116 (both more abundant in 2015) were more abundant during Phaeocystis (haptophyte) blooms in the Southern Ocean as compared to diatom blooms45. Thus, the 2019 MHW anomalous diatom bloom and resulting organic matter accumulation may have limited the success of SAR11.
Looking forward, more granular insight into the network topology of marine food webs is needed to resolve functional and activity-dependent changes in carbon processing pathways and the BCP in response to thermal stress and their associated impacts on deep particle export and carbon sequestration. This information can inform future Earth system models and provide key metrics for ocean health. It is therefore imperative to expand the time-series monitoring programs that provide essential parameter information and samples. At the same time, there is a need for new, environmentally sustainable methods and blue technologies that integrate sample collection with BGC-Argo float measurements to better resolve spatiotemporal changes in food web structure and carbon fluxes.
Methods
Temperature anomalies
The depth-resolved physical parameters shown in Fig. 1 were obtained from an Argo-based gridded product46- (RG09) Argo Climatology (https://sio-argo.ucsd.edu/RG_Climatology.html, last access: 06 January 2024). We selected data from Ocean Station Papa (50°N, 145°W), a core region used to define MHWs in NESAP11,12,13, to assess decadal-scale variability. Temperature anomalies at each depth were calculated as deviations from the climatological mean during the period 2004−2023, corresponding to the period matching BGC-Argo float and shipboard data. The mixed layer depth (MLD) was defined as the depth where the temperature is 0.2 °C lower than that at 10 meters47, which generally corresponds well with the depth at which the maximum Brunt-Väisälä frequency (N2) occurs (calculated using sw_bfrq Matlab® function).
To verify the consistency of temperature estimates used in anomaly calculations, and to assess potential spatial biases arising from the semi-lagrangian nature of the BGC-Argo floats, we compared monthly temperature records at 50 m from three sources: BGC-Argo floats, the gridded Roemmich and Gilson (2009) Argo climatology product, and the Armor3D Copernicus reanalysis48,49. The BGC-Argo data were averaged monthly within the study domain. Armor3D temperature fields were averaged over the spatial bounds of 47°N-53°N and 147°W-137°W, corresponding to the spatial extent sampled by the BGC-Argo floats, using monthly reprocessed data.
BGC-Argo float data analysis: POC and Chl estimates
Data from the three floats used in this study were freely acquired from the Argo Global Data Assembly Center (USGODAE; usgodae.org/ftp/outgoing/argo/). Only quality-controlled files (Sprof) and data flagged as “good” were considered. Variables used in this study include temperature, salinity, nitrate and bio-optics (i.e., fluorescence chlorophyll and backscatter). We focused the analysis in the upper ocean between the surface and 400 m, with data every 5–10 days and acquired between June 2010 and October 2022, with a gap between February 2016 and August 2018. The study region was constrained between 47˚-52.5˚N and 137.5-146˚W (Fig. 2a). Details on quality control and sensor performances have been published50,51. As previously described15, float-estimated chlorophyll a (Chl) in our study region is biased when compared to nearby discrete Chl measurement from repeat Line P cruises, requiring additional post-correction. We applied a correction factor to the adjusted Chl data for each float, estimated by taking the median of the ratio of float-adjusted Chl data to the Line P corrected float data15 in the upper 15 m. The following correction factors were divided by our adjusted Chl data to obtain corrected values extending to 2022.
Backscatter (bbp) data was separated into small and large particles52,53 (smaller or bigger than 100 µm, respectively), but only small particles were used in this study since large particles were a minor fraction of total bbp and showed no temporal variability (Figs. S1–S3). To estimate POC stocks (Fig. 2) at the different ocean layers, we first estimated the euphotic zone depth (Ez) assuming it comprises the primary production zone7,53 as:
Where Chl_Max is the depth of maxima Chl for each float profile. Mean euphotic zone depth was 98 ± 20 m for the entire time series, so we assumed Ez = 100 m for the subsequent calculations. We then estimated mean monthly particle stocks between surface-Ez and Ez-300 m for each year and converted bbp into total POC50.
Chl:C ratios were estimated using mean monthly integrated POC and Chl concentrations within the upper 100 m of depth.
POC anomalies (Fig. 3) derived from BGC-Argo floats were estimated by subtracting POC time series from each MHW year (2014, 2015, 2019 and 2020) from mean monthly anomalies during non-MHW years in a multi-step process. Since POC background signals were significatively different after the first MHW, we compiled non-MHW years into two datasets: the first was composed of float profiles from complete calendar years prior to 2013, and the second was composed of profiles from the years of 2018, 2021 and 2022. For each non-MHW dataset, we computed mean monthly [POC] and then linearly smoothed the data every 20 m (floats profiled approximately every 5 m for the top 100 m of depth, and every 10−20 m between 100−400 m). MHW time series were then created for each year by linearly interpolating the data every 20 m. Anomalies for each year were computed by subtracting their POC time series from non-MHW datasets (bsxfun function, Matlab®) and daily-interpolated for display. The first non-MHW dataset was used to estimate 2014 and 2015 anomalies, while the second was used to estimate the 2019 and 2020 anomalies.
To estimate the mixed layer rate of change, we first calculated mixed layer depths (MLDs)47, and then ∆MLD (m month-1) for each year as:
Where MLD_month is the mean monthly MLD, and MLD_month-1 is the mean monthly MLD of the previous month.
Net community production (carbon export) computed from BGC-Argo floats
The carbon export at the base of the euphotic zone was computed using float-measured nitrate minus a set of abiotic processes acting on the upper-layer nitrate budget, following the method outlined in Huang et al.20:
where subscripts of Bio, Obs, DIF, Adv, EP, and BG represent the biological activity, float measurements, diapycnal diffusion, wind-induced Ekman pumping, evaporation and precipitation, and background nitrate gradient change alongside the float trajectory, respectively. The biological term solved from the tracer budget represents the amount of photosynthetically-produced carbon exceeding the respiration consumption and can serve as a metric of carbon export under the assumption of steady-state when integrating over seasonal or annual scales54. The parameterizations of abiotic terms and associated uncertainty estimates are detailed in Huang et al.20. The nitrate-based biological term estimate was further converted to carbon unit via the Redfield ratio of 6.6. Due to limited knowledge of the seasonality of diapycnal diffusion coefficients, which may exponentially increase by several orders of magnitude in the fall and winter55, and considering that most biological production occurs in the spring and summer20, our analysis is limited to the relatively stratified spring and summer seasons to minimize error in the biological term estimate.
Spatiotemporal sensitivity analyses
We conducted a series of sensitivity analyses to assess whether methodological assumptions or float-specific variability introduced bias into the observed patterns of POC production and accumulation. First, to test the robustness of using a fixed 100 m integration depth for surface POC stocks, we compared estimates based on a variable Ez versus the fixed 100 m depth. Small-particle backscatter (bbp) was integrated from the surface to both depths and converted to POC using empirical coefficients (Eq. 2). The resulting regression analysis revealed a strong linear relationship (R² = 0.94) with a slope of 0.96 (Fig. S11), supporting the use of a fixed depth for consistent comparisons across the time series.
Next, we tested whether float-specific spatial variability could influence our results. We compared Chl concentrations from two floats deployed during overlapping periods but spatially apart (#5903274 and #5903714; Figs. 2a, S12a). Profiles collected within 3 days of each other showed a strong positive correlation in integrated Chl over the upper 100 m. We also calculated NCP from all floats equipped with nitrate sensors using the same method as in Fig. S4, and included floats lacking bio-optical sensors but profiling the region concurrently (Fig. S12b). A linear mixed-effects model with year and float ID as random intercepts showed that most variance in NCP was explained by year (σ² = 0.81), while float ID contributed negligible variance (σ² ≈ 0), indicating that floats operating during the same time periods yielded comparable NCP estimates.
Finally, we examined spatial variability in surface chlorophyll using monthly mean concentrations from the Aqua-MODIS satellite product56 (4 km resolution, 2008-2023) across four Line P stations located within the float trajectories. Seasonal chlorophyll patterns were highly consistent among stations (Pearson correlation coefficients = 0.85–0.98, Bonferroni-adjusted p-values < 0.05; Fig. S12c), and within-month differences across stations were not statistically significant (Repeated Measures ANOVA, F(4,50) = 0.213, p = 0.93). Discrete chlorophyll samples collected during Line P cruises further confirmed these seasonal trends (Fig. S12d). While individual profiles were strongly correlated regionally (P21 and P22, Spearman’s ρ = 0.77, adjusted p-value < 0.001; P24 and P25 Spearman’s ρ = 0.89, adjusted p-value < 0.001), there was weaker correspondence between more distant stations, reflecting the inherently patchy nature of chlorophyll distributions, these results collectively indicate that the observed trends in POC production and accumulation were not driven by float-specific or spatial biases.
Phytoplankton composition from pigment concentrations
Phytoplankton pigment concentrations (chlorophylls and carotenoids) were measured by high-performance liquid chromatography (HPLC) following the method detailed in Nemcek and Peña57 and analyzed to estimate the phytoplankton composition using CHEMTAX58, following the procedures detailed in Peña et al.17. Pigment samples have been routinely collected since 2011 on the Fisheries and Oceans Canada (DFO) Line P Hydrographic Surveys (waterproperties.ca/linep) which operate three times a year (generally February, June and August) since 1981 from the coast of British Columbia to station P26 (50°N, 145 W) 913 km offshore in the Northeast Subarctic Pacific. For this study, we focus on stations P20 to P26 (Ocean Station Papa), as they span the locations of the BGC-Argo floats (Fig. 2a). Pigment concentration data are from 5 m or mixed layer average when there was more than one depth sampled within the mixed layer. Post-CHEMTAX statistical analyses and data visualization were performed using R Statistical Software (v4.3.2)59. The observed patterns in phytoplankton pigment groups were generally consistent across these outermost stations of the Line P transect, with no significant difference by station (PerMANOVA p-value = 0.981, although phytoplankton community composition at P26 was somewhat distinct (Fig. S6). Therefore, average concentrations of major phytoplankton functional groups from hydrographic stations P20-P26 were used to examine trends in phytoplankton over the time series.
DNA-derived community composition
2-L seawater samples were collected and filtered onto 0.22 µm Sterivex filters for biomolecular analysis from Stations P20 and P26 during the Fisheries and Oceans Canada Line P seasonal occupation program from 2010 through 2019. While samples were collected throughout the entire water column at each station16, only samples from 10, 25, 50, 100, 150, 200, and 300 m were used in this study. Briefly, DNA was obtained using enzymatic lysis, followed by phenol:chloroform extraction, and purification and concentration using Amicon Ultra-4 Centrifugal Filter Units (described in greater detail Traving et al.). The V4-V5 region of the 16S rRNA gene was amplified from samples collected for the entirety of the biomolecular time series (2010−2019) using the primers 515F-Y(5′-GTGYCAGCMGCCGCGGTAA-3′) and 926 R (5′-CCGYCAATTYMTTTRAGTTT-3′), while the V4 region of the 18S rRNA gene was amplified from 2015−2019 samples using the primers V4F (5’-CCAGCASCYGCGGTAATTCC-3’)60 and V4RB (5’-ACTTTCGTTCTTGATYRR-3’)61. For 2010-Winter 2016 samples, 16S V4-V5 amplicon libraries were prepared by the Joint Genome Institute (California, USA) following the JGI iTag Library Preparation SOP (jgi.doe.gov/user-programs/pmo-overview/protocols-sample-preparation-information/), and sequenced on an Illumina MiSeq using a 600 cycle V3 kit. 16S V4-V5 libraries prepared for Spring 2016–2019 samples were prepared at the Hakai Institute (BC, Canada). Samples were amplified in duplicate using fusion primers (containing Nextera indices and Illumina adapters in addition to the primers; described in detail here https://doi.org/10.17504/protocols.io.3byl4bq1jvo5/v1), PCR replicates pooled, cleaned individually using SPRI beads at ratio of 0.8x, quantified using the Quant-iT PicoGreen dsDNA Assay (Invitrogen), and then all samples successfully amplified were pooled in equal amounts before sequencing on an Illumina MiSeq using a 600 cycle V3 kit (https://doi.org/10.17504/protocols.io.3byl49842go5/v1). The 16S-V4V5 library generated at the Hakai Institute was sequenced twice and data pooled to obtain a read depth similar to those libraries sequenced at the Joint Genome Institute. The 18S-V4 amplicon library was also prepared and sequenced at the Hakai Institute, using methods described in detail here https://doi.org/10.17504/protocols.io.rm7vzjjjxlx1/v1 and sequenced on an Illumina MiSeq using a 600 cycle V3 kit.
Illumina sequence data were processed in QIIME2 v. 2022.862. Briefly, primer sequences were removed from amplicon libraries using cutadapt (cutadapt trim-paired) and reads were denoised, amplicon sequence variants (ASVs) determined and chimeras removed using dada2 (dada2 denoise-paired, using the following settings for the 16S-V4V5 rRNA libraries run the Hakai Institute trunc-len-f 240, trunc-len-r 190, max-ee-f 4, max-ee-r 6; trunc-len-f 267, trunc-len-r 220, max-ee-f 3, max-ee-r 5 for the 16S-V4V5 libraries run at the Joint Genome Institute; trunc-len-f 220, trunc-len-r 200, max-ee-f 4, max-ee-r 6 for the 18S-V4 library). As multiple 16S-V4V5 libraries were sequenced, each MiSeq run was denoised separately, ASVs within a fixed length window were retained (rescript filter-seqs-length; global-min 300 and global-max 417), rare ASVs removed (feature-table filter-features; present a frequency of at least 0.001% of the average read depth for that run and in at least 2 samples), and then runs were merged at the ASV level (feature-table merge). Rare ASVs were similarly removed from the 18S-V4 library. ASVs were then classified using the QIIME2 Naive Bayes classifier trained to the SILVA database v.138.163 for the 16S rRNA genes. 18S rRNA ASVs were annotated with both the Protist Ribosomal References database (PR2 v5) and the MetaZooGene database (v2023-m07-15)64,65. MetaZooGene annotations were used for metazoan sequences while PR2 annotations were used for all other eukaryotes. A total of 3058 ASVs were retained and an average read depth of 119,853 sequences per sample were obtained across the five 16S-V4V5 MiSeq libraries. There was a large difference in the library richness between the sample libraries run at the Joint Genome Institute compared to that run at the Hakai Institute. Rarefying the dataset did not improve this richness difference. Therefore, for the 16S-V4V5 dataset we chose to filter the dataset to retain the top 80% of the ASVs using the function filterfun_sample(topf(0.8)) in the R package phyloseq, leaving 858 16S-V4V5 ASVs. For the 18S-V4 dataset, a total of 3068 ASVs were retained with an average read depth of 56,518 sequences per sample. Raw sequence data have been deposited in the NCBI SRA under the BioProjects PRJNA639229 and PRJNA640752.
All data visualization and statistical analysis of the resulting 16S and 18S rRNA ASV tables were performed using R Statistical Software (v4.3.2). Briefly, depth-based and interannual variation in prokaryotic, Opisthokonta (animal), and non-Opisthokonta (predominantly protistan) community composition was quantified using euclidean distance of CLR-transformed ASV counts agglomerated at the genus level. Principal components analysis (microViz R package)66 was used to display the variation in this data. PERMANOVA (adonis2 in the vegan R package)67 and pairwise PERMANOVAs (pairwiseAdonis R package, using the Benjamini-Hochberg correction)68 were used to determine the significance of any observed compositional differences between heatwave years within the surface and upper mesopelagic waters. A Wilcoxon Rank Sum Test (correcting for multiple comparisons using the Benjamini-Hochberg test) as implemented in the metarr R package69 was used to determine significant differences in the Spring and Summer prokaryotic and eukaryotic genera (and all higher taxonomic levels) between MHW years in both the surface mixed layer and upper mesopelagic waters. A genus was required to be present in at least 2% of the samples and have a mean relative abundance of 0.0005 to be included in the analysis. Results were visualized using a heat tree, where the log2 of ratio of the median of proportions for each significantly different sample group is displayed by the color of the nodes and edges while the node size is scaled to the average relative abundance of that taxon in the associated depth range.
Data availability
All raw and processed datasets, along with figure source data and code, are available in the public GitHub repository [https://github.com/hallamlab/NESAP_marine_heatwaves/tree/main/argo]. BGC-Argo float data are also freely available via the SOCCOM and GO-BGC data archives [https://doi.org/10.6075/J0ZG6SG1]70. The RG09 Argo climatology is provided by Scripps Institution of Oceanography (Roemmich & Gilson, 2009); Armor3D fields are from the Copernicus Marine Service (reprocessed monthly; https://doi.org/10.48670/moi-00052); and Line P pigment (HPLC) data are from the Fisheries and Oceans Canada Line P program. Amplicon sequence data used for community analyses are deposited in NCBI SRA under BioProjects PRJNA639229 and PRJNA640752.
References
Holbrook, N. J. et al. Keeping pace with marine heatwaves. Nat. Rev. Earth Environ. 1, 482–493 (2020).
Oliver, E. C. et al. Longer and more frequent marine heatwaves over the past century. Nat. Commun. 9, 1–12 (2018).
Smale, D. A. et al. Marine heatwaves threaten global biodiversity and the provision of ecosystem services. Nat. Clim. Chang. 9, 306–312 (2019).
Frölicher, T. L., Fischer, E. M. & Gruber, N. Marine heatwaves under global warming. Nature 560, 360–364 (2018).
Mignot, A. et al. Decrease in air-sea CO2 fluxes caused by persistent marine heatwaves. Nat. Commun. 13, 4300 (2022).
Bif, M. B., Siqueira, L. & Hansell, D. A. Warm events induce loss of resilience in organic carbon production in the northeast Pacific Ocean. Glob. Biogeochem. Cycles 33, 1174–1186 (2019).
Buesseler, K. O., Boyd, P. W., Black, E. E. & Siegel, D. A. Metrics that matter for assessing the ocean biological carbon pump. Proc. Natl. Acad. Sci. 117, 9679–9687 (2020).
Durkin, C. A. et al. Visual tour of carbon export by sinking particles. Glob. Biogeochem. Cycles 35, e2021GB006985 (2021).
McDonnell, A. M. P., Boyd, P. W. & Buesseler, K. O. Effects of sinking velocities and microbial respiration rates on the attenuation of particulate carbon fluxes through the mesopelagic zone. Glob. Biogeochem. Cycles 29, 175–193 (2015).
Henson, S. A. et al. Uncertain response of ocean biological carbon export in a changing world. Nat. Geosci. 15, 248–254 (2022).
Bond, N. A., Cronin, M. F., Freeland, H. & Mantua, N. Causes and impacts of the 2014 warm anomaly in the NE Pacific. Geophys. Res. Lett. 42, 3414–3420 (2015).
Scannell, H. A., Johnson, G. C., Thompson, L., Lyman, J. M. & Riser, S. C. Subsurface evolution and persistence of marine heatwaves in the Northeast Pacific. Geophys. Res. Lett. 47, e2020GL090548 (2020).
Amaya, D. J., Miller, A. J., Xie, S. P. & Kosaka, Y. Physical drivers of the summer 2019 North Pacific marine heatwave. Nat. Commun. 11, 1903 (2020).
Taves, R. C. et al. Relationship between surface dissolved iron inventories and net community production during a marine heatwave in the subarctic northeast Pacific. Environ. Sci. Process. Impacts 24, 1460–1473 (2022).
Long, J. S., Fassbender, A. J. & Estapa, M. L. Depth‐resolved net primary production in the Northeast Pacific Ocean: a comparison of satellite and profiling float estimates in the context of two marine heatwaves. Geophys. Res. Lett. 48, e2021GL093462 (2021).
Traving, S. J. et al. Prokaryotic responses to a warm temperature anomaly in northeast subarctic Pacific waters. Commun. Biol. 4, 1217 (2021).
Peña, M. A., Nemcek, N. & Robert, M. Phytoplankton responses to the 2014–2016 warming anomaly in the northeast subarctic Pacific Ocean. Limnol. Oceanogr. 64, 515–525 (2019).
Bhatt, U. S. et al. Emerging anthropogenic influences on the Southcentral Alaska temperature and precipitation extremes and related fires in 2019. Land 10, 82 (2021).
Theys, N. et al. Global monitoring of volcanic SO2 degassing with unprecedented resolution from TROPOMI onboard Sentinel-5. Precursor. Sci. Rep. 9, 2643 (2019).
Huang, Y., Fassbender, A. J., Long, J. S., Johannessen, S. & Bif, M. B. Partitioning the export of distinct biogenic carbon pools in the Northeast Pacific Ocean using a biogeochemical profiling float. Glob. Biogeochem. Cycles 36, e2021GB007178 (2022).
Szuwaski, C. S., Aydin, K., Fedewa, E. J., Garber-Yonts, B. & Litzow, M. A. The collapse of eastern Bering Sea snow crab. Science 382, 306–310 (2023).
Cartwright, R. et al. Fluctuating reproductive rates in Hawaii’s humpback whales, Megaptera novaeangliae, reflect recent climate anomalies in the North Pacific. R. Soc. Open. Sci. 6, 181463 (2019).
Siegel, D. A. et al. An operational overview of the EXport Processes in the Ocean from RemoTe Sensing (EXPORTS) Northeast Pacific field deployment. Elem. Sci. Anthr. 9, 00107 (2021).
McNair, H. M., Morison, F., Graff, J. R., Rynearson, T. A. & Menden‐Deuer, S. Microzooplankton grazing constrains pathways of carbon export in the subarctic North Pacific. Limnol. Oceanogr. 66, 2697–2711 (2021).
Shea, C. H. et al. Small particles and heterotrophic protists support the mesopelagic zooplankton food web in the subarctic northeast Pacific Ocean. Limnol. Oceanogr. 68, 1949–1963 (2023).
Kwong, L. E., Ross, T., Lüskow, F., Florko, K. R. & Pakhomov, E. A. Spatial, seasonal, and climatic variability in mesozooplankton size spectra along a coastal-to-open ocean transect in the subarctic Northeast Pacific. Prog. Oceanogr. 201, 102728 (2022).
Cohen, N. R. et al. Dinoflagellates alter their carbon and nutrient metabolic strategies across environmental gradients in the central Pacific Ocean. Nat. Microbiol. 6, 173–186 (2021).
Rivkin, R. B., Putland, J. N., Anderson, M. R. & Deibel, D. Microzooplankton bacterivory and herbivory in the NE subarctic Pacific. Deep Sea Res. Part. II Top. Stud. Oceanogr. 46, 2579–2618 (1999).
Stephens, B. M. et al. An upper-mesopelagic-zone carbon budget for the subarctic North Pacific. Biogeosciences 22, 3301–3328 (2025).
Pata, P. R. et al. Persistent zooplankton bioregions reflect long-term consistency of community composition and oceanographic drivers in the NE Pacific. Prog. Oceanogr. 206, 102849 (2022).
Anderson, S. R., Blanco-Bercial, L., Carlson, C. A. & Harvey, E. L. Role of syndiniales parasites in depth-specific networks and carbon flux in the oligotrophic ocean. ISME Commun 4, ycae014 (2024).
Lampitt, R. S. et al. Deep ocean particle flux in the Northeast Atlantic over the past 30 years: carbon sequestration is controlled by ecosystem structure in the upper ocean. Front. Earth Sci. 11, 1176196 (2023).
Steinberg, D. K. & Landry, M. R. Zooplankton and the ocean carbon cycle. Annu. Rev. Mar. Sci. 9, 413–444 (2017).
Li, L. & Endo, K. Phylogenetic positions of “pico-sized” radiolarians from middle layer waters of the tropical Pacific. Prog. Earth Planet. Sci. 7, 1–10 (2020).
Ishitani, Y., Ujiié, Y., de Vargas, C., Not, F. & Takahashi, K. Two distinct lineages in the radiolarian Order Spumellaria having different ecological preferences. Deep Sea Res. Part. II Top. Stud. Oceanogr. 61, 172–178 (2012).
Gutierrez-Rodriguez, A. et al. High contribution of Rhizaria (Radiolaria) to vertical export in the California Current Ecosystem revealed by DNA metabarcoding. ISME J 13, 964–976 (2019).
Gowing, Marcia M. and Mary W. Silver. Minipellets: a new and abundant size class of marine fecal pellets. J. Mar. Res. 43 (1985).
Wojtal, P. et al. Deconvolving mechanisms of particle flux attenuation using nitrogen isotope analyses of amino acids. Limnol. Oceanogr. 68, 1965–1981 (2023).
Pond, D. W. & Ward, P. Importance of diatoms for Oithona in Antarctic waters. J. Plankton Res. 33, 105–118 (2011).
Bode-Dalby, M. et al. Small is beautiful: the important role of small copepods in carbon budgets of the southern Benguela upwelling system. J. Plankton Res. 45, 110–128 (2023).
Coats, D. W. Parasitic life styles of marine dinoflagellates 1. J. Eukaryot. Microbiol. 46, 402–4 (1999).
Giering, S. L. et al. Reconciliation of the carbon budget in the ocean’s twilight zone. Nature 507, 480–483 (2014).
Wright, J. J., Konwar, K. M. & Hallam, S. J. Microbial ecology of expanding oxygen minimum zones. Nat. Rev. Microbiol. 10, 381–94 (2012).
Stephens, B. M. et al. Influence of amino acids on bacterioplankton production, biomass and community composition at Ocean Station Papa in the subarctic Pacific. Elem. Sci. Anthr. 11, 00095 (2023).
West, N. J., Landa, M. & Obernosterer, I. Differential association of key bacterial groups with diatoms and Phaeocystis spp. during spring blooms in the Southern Ocean. MicrobiologyOpen 13, e1428 (2024).
Roemmich, D. & Gilson, J. The 2004-2008 mean and annual cycle of temperature, salinity, and steric height in the global ocean from the Argo Program. Prog. Oceanogr. 82, 81–100 (2009).
de Boyer Montégut, C., Madec, G., Fischer, A. S., Lazar, L. & Iudicone, D. Mixed layer depth over the global ocean: an examination of profile data and a profile-based climatology. J. Geophys. Res. Ocean. 109, C12003 (2004).
Guinehut, S., Dhomps, A.-L., Larnicol, G. & Le Traon, P.-Y. High resolution 3D temperature and salinity fields derived from in situ and satellite observations. Ocean Sci 8, 845–857 (2012).
Mulet, S., Rio, M.-H., Mignot, A., Guinehut, S. & Morrow, R. A new estimate of the global 3D geostrophic ocean circulation based on satellite data and in-situ measurements. Deep Sea Res. Part. II Top. Stud. Oceanogr. 77, 70–81 (2012).
Johnson, K. S. et al. Biogeochemical sensor performance in the SOCCOM profiling float array. J. Geophys. Res. Oceans 122, 6416–6436 (2017).
Roesler, C. et al. Recommendations for obtaining unbiased chlorophyll estimates from in situ chlorophyll fluorometers: a global analysis of WET labs ECO sensors. Limnol. Oceanogr. Methods 15, 572–585 (2017).
Briggs, N., Dall’Olmo, G. & Claustre, H. Major role of particle fragmentation in regulating biological sequestration of CO2 by the oceans. Science 367, 791–793 (2020).
Bif, M. B., Long, J. S. & Johnson, K. S. Seasonality modulates particulate organic carbon dynamics in mid-latitudes of South Pacific and South Atlantic Oceans. J. Mar. Syst. 241, 103916 (2024).
Cronin, M. F., Pelland, N. A., Emerson, S. R. & Crawford, W. R. Estimating diffusivity from the mixed layer heat and salt balances in the North Pacific. J. Geophys. Res.: Ocean. 120, 7346–7362 (2015).
Emerson, S. Annual net community production and the biological carbon flux in the ocean. Glob. Biogeochem. Cycles 28, 14–28 (2014).
NASA Goddard Space Flight Center Ocean Biology Processing Group. MODIS-Aqua Ocean Color Data: Chlorophyll-a, Level-3 Monthly Climatology. NASA OB.DAAC. https://doi.org/10.5067/AQUA/MODIS_OC.2014.0 (2014).
Nemcek, N., & Peña, M. A. Institute of Ocean Sciences protocols for phytoplankton pigment analysis by HPLC. Canadian Technical Report of Fisheries and Aquatic Sciences, 3117: 80 p. (Fisheries & Oceans Canada, 2014).
Mackey, M. D., Mackey, D. J., Higgins, H. W. & Wright, S. W. CHEMTAX - a program for estimating class abundances from chemical markers: Application to HPLC measurements of phytoplankton. Mar. Ecol. Prog. Ser. 144, 265–283 (1996).
R Core Team. R: A Language and Environment for Statistical Computing. https://www.R-project.org/ (Foundation for Statistical Computing, 2023).
Stoeck, T. et al. Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Mol. Ecol. 19, 21–31 (2010).
Balzano, S., Abs, E. & Leterme, S. C. Protist diversity along a salinity gradient in a coastal lagoon. Aquat. Microb. Ecol. 74, 263–277 (2015).
Bolyen, E. et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857 (2019).
Quast, C. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucl. Acids Res. 41, D590–D596 (2012).
Guillou, L. et al. The Protist Ribosomal Reference database (PR2): a catalog of unicellular eukaryote small sub-unit rRNA sequences with curated taxonomy. Nucl. Acids Res. 41, D597–D604 (2012).
Bucklin, A. et al. Toward a global reference database of COI barcodes for marine zooplankton. Mar. Biol. 168, 78 (2021).
Barnett et al. microViz: an R package for microbiome data visualization and statistics. J. Open. Source Softw. 6, 3201 (2021).
Oksanen J, et al. Vegan: Community Ecology Package. R package version 2.6-8, https://CRAN.R-project.org/package=vegan (2024).
Martinez Arbizu P. pairwiseAdonis: Pairwise Multilevel Comparison using Adonis. R package version 01 (2017).
Foster, Z. S. L., Sharpton, T. J. & Grünwald, N. J. Metacoder: an R package for visualization and manipulation of community taxonomic diversity data. PLOS Comput. Biol. 13, e1005404 (2017).
Riser Stephen C. et al. SOCCOM and GO-BGC float data - Snapshot 2023-12-20. In Southern Ocean Carbon and Climate Observations and Modeling (SOCCOM) and Global Ocean Biogeochemistry (GO-BGC) Biogeochemical-Argo Float Data Archive. UC San Diego Library Digital Collections. https://doi.org/10.6075/J0ZG6SG1 (2024).
Acknowledgements
MBB and KSJ thank support from the David and Lucile Packard Foundation and the GO-BGC project under the NSF Award 1946578 with operational support from NSF Award 2110258. YH was supported by China NSF (grant number: 42406099) and Fundamental Research Funds for the Central Universities (grant number: 20720240105). SJT is supported by the Danish Center for Hadal Research (Grant No. DNRF145). Data was collected and made freely available by the Global Ocean Biogeochemistry Array (GO-BGC) Project funded by the National Science Foundation, Division of Ocean Sciences, and the International Argo Program and the national programs that contribute to it. The Argo Program is part of the Global Ocean Observing System. A heartfelt thanks to the scientists and crews of CCGS John P. Tully, especially Marie Robert for their world-class assistance during Line P cruises, and Fisheries and Oceans Canada for the financial support of Line P program. Additional support for sample collection and processing was provided by the Tula Foundation in support of CTEK and the G. Unger Vetlesen and Ambrose Monell Foundations, the Natural Sciences and Engineering Research Council of Canada, and the Canada Foundation for Innovation through grants awarded to SJH. We’d like to thank Jacki Long for their assistance with chlorophyll fluorescence corrections. CTEK would also like to thank Rose-Lynne Savage for preparing the 2016–2019 metabarcode libraries for sequencing, and Rute Carvalho for running these libraries. SJH would also like to thank the many undergraduate helpers in the Hallam lab and ocean-going technical support staff including Jade Shiller and Chris Payne for their support in sample collection and processing over the years.
Author information
Authors and Affiliations
Contributions
M.B.B. conceived the study, led data analysis and interpretation, integrated datasets, and wrote the manuscript with input from all co-authors. C.T.E.K. conducted genomics computing analyses, including all plankton community composition analyses, statistical analyses, and ecological interpretation. S.J.T. performed genomics computing analyses, including plankton community composition analyses, statistical analyses, and ecological interpretation. Y.H. conducted net community production (NCP) and net primary production (NPP) analyses and contributed to BGC-Argo data interpretation. J.A. performed molecular data processing and interpretation. M.A.P. provided phytoplankton pigment data and contributed to ecological interpretation. S.J.H. generated molecular datasets and contributed to ecological interpretation. K.S.J. contributed to study design and BGC-Argo data interpretation. All authors contributed to manuscript writing, editing, discussion of results, and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Bif, M.B., Kellogg, C.T.E., Huang, Y. et al. Marine heatwaves modulate food webs and carbon transport processes. Nat Commun 16, 8535 (2025). https://doi.org/10.1038/s41467-025-63605-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-63605-w