Abstract
Research on chromosome organization and cell cycle progression in spherical bacteria, particularly Staphylococcus aureus, remains limited and fragmented. In this study, we established a working model to investigate chromosome dynamics in S. aureus using a Fluorescent Repressor-Operator System (FROS), which enabled precise localization of specific chromosomal loci. This approach revealed that the S. aureus cell cycle and chromosome replication cycle are not synchronized (i.e. they do not initiate simultaneously), with cells exhibiting two segregated origins of replication at the start of the cell cycle. The chromosome has a specific origin-terminus-origin conformation, with origins localizing near the membrane, towards the tip of each hemisphere, or the “cell poles”. We further used this system to assess the role of various proteins with a function in S. aureus chromosome biology, focusing on the ParB-parS and SMC-ScpAB systems. Our results demonstrate that ParB binds five parS chromosomal sequences and the resulting complexes are required for specific chromosomal inter-arm alignment, but play a minor role in chromosome segregation. In contrast, the SMC-ScpAB complex plays a key role in S. aureus chromosome biology, contributing to chromosome segregation and spatial organization. Additionally, we systematically assessed and compared the impact of proteins linking chromosome segregation to cell division—Noc, FtsK, SpoIIIE and XerC—on origin and terminus number and positioning. This work provides a comprehensive study of the factors governing chromosome dynamics and organization in S. aureus, contributing to our knowledge on chromosome biology of spherical bacteria.
Similar content being viewed by others
Introduction
Bacteria exhibit an exquisite spatiotemporal organization of cellular components. From generating their own shape, to dividing with precision and placing structures and organelles in specific locations, these microorganisms coordinate their cellular activities and machinery´s localization with remarkable accuracy. One key example is chromosome dynamics, which includes the spatial organization of the chromosome, its replication and proper segregation into daughter cells, all of which are essential for the faithful transmission of genetic information to the next generation1,2.
Studies on multiple bacterial model organisms have revealed specific arrangements for the chromosome during the processes of replication and segregation (Fig. S1). Some organisms like Caulobacter crescentus3, Myxococcus xanthus4, or the chromosome 1 of Vibrio cholerae5 display an organization known as longitudinal (Ori-Ter) chromosome arrangement, in which the replication origin (Ori) is in a polar (sub-polar in M. xanthus4) region and the replication terminus (Ter) sits on the opposite pole in newborn cells3,4,5. After the initiation of chromosome replication, one of the newly replicated Ori is segregated to the opposite cell pole, and the Ter region moves to the cell center, near the division site, creating an Ori-Ter-Ori arrangement. Such dynamics ensures that both daughter cells inherit a copy of the chromosome which is in the same orientation as in the mother cell.
Newborn cells of the model organism Bacillus subtilis, in slow-growing conditions, also present an Ori-Ter-Ori orientation of partially replicated chromosomes. However, after their complete replication, B. subtilis chromosomes adopt a Left-Ori-Right configuration, with the origins of each chromosome in the ¼ and ¾ positions of the cell6,7. As chromosome replication re-starts before the end of the cell cycle, the origins segregate to the poles and the septum regions (Fig. S1). The actinobacterium Corynebacterium glutamicum has a similar cycle, but starting with two completely replicated chromosomes and, after replication of the origins, two of them remain in the polar positions, while the other two are segregated towards the future division site8. On the other hand, Escherichia coli in slow-growing conditions adopts a Left-Ori-Right (or transverse) organization, with the origin in the cell center of newborn cells9,10,11,12. After the initiation of replication, the origins segregate to the ¼ and ¾ positions, where they remain until replication is finished and the cells divide (Fig. S1). Although phylogenetically distant to E. coli, a similar chromosomal arrangement can be found in both the ovococcoid firmicute Streptococcus pneumoniae13 and the actinobacterium Mycobacterium smegmatis14. Besides the diversity of chromosome arrangements, an additional factor that adds complexity to the process of chromosome segregation is the presence of multifork replication, that is, the initiation of new replication rounds in chromosomes that are still being replicated, which increases the number of origins per cell. This has been observed in some organisms, such as B. subtilis6,15,16 and E. coli17,18 in fast growth conditions, and in the slow-growing M. smegmatis19.
Chromosome organization is highly dependent on the correct segregation of the origin regions, which is mediated by multiple factors that vary among bacterial species. In B. subtilis20, C. crescentus21, V. cholerae22, M. xanthus23 or Pseudomonas aeruginosa24,25 among others, ParABS systems play a direct role in origin segregation. In brief, the ParB component of the system is a CTPase that interacts with itself and with parS sequences, generally located near the origin, forming a kinetochore-like structure that can be mobilized through cyclic dimerization and monomerization of the ParA component, an ATPase whose dimers interact both with ParB and the DNA26. The interaction of ParB with parS sequences stimulates ParB binding to CTP, which in turn promotes the formation of closed rings of ParB dimers that slide along the chromosome and spread to adjacent sequences27,28,29,30. Together with ParB´s ability to self-interact, these activities enable ParB to form highly concentrated nucleoprotein complexes31,32. There is evidence that these ParABS systems might be present in the majority of bacterial species33. A second function of ParB is to load SMC-ScpAB condensin-like complexes onto chromosomal parS sites34,35,36,37. These complexes contribute to the overall packaging of the nucleoid and to the segregation of sister chromosomes as they are being replicated38,39,40. SMC molecules dimerize spontaneously, forming a structure with long coiled-coil arms that connect a central hinge to two closely positioned ATPase domains (or heads) composed of the C- and N- terminus of each monomer41,42,43. ScpA interacts with SMC by bridging the two heads of the dimer, forming a ring-like structure that is able to entrap DNA, while ScpB dimers associate to ScpA and are required for the loading of SMC-ScpAB complexes onto the chromosome44,45. The current model proposes that ParB-parS nucleoprotein complexes directly interact with SMC and facilitate the loading of SMC-ScpAB complexes onto the chromosome37. These complexes then travel from the origin region to the terminus, juxtaposing and aligning the two chromosome arms and therefore contributing to the overall spatial organization of the chromosome36,39,40,46,47,48,49. Depending on the bacterial species, the role of SMC-ScpAB or ParABS systems can vary from being essential for survival (particularly in fast growing conditions) to their absence causing only mild phenotypes21,23,24,46,47,50,51,52,53,54,55,56,57. Besides specific proteins with a role in chromosome segregation, the physico-chemical properties of the chromosome are proposed to contribute to the spontaneous unmixing of sister chromatids58. In turn, chromosome organization plays a role in the regulation of other cellular processes, like cell division. In many organisms, nucleoid-associated proteins prevent the progression of septum-formation, coordinating both processes and preventing guillotining of the chromosome59,60,61,62,63.
Most knowledge on chromosome segregation comes from a selected group of rod-shaped model organisms. The geometry of the bacterium is a key player in its spatial organization, as the presence of topologically different regions allows to determine separate spaces in the cell64,65. For example, proteins sensing curvature are involved in chromosome positioning66, division site placement67,68 and cell morphogenesis69,70,71. Therefore, the existence of spherical bacteria which, during part of the cell cycle, appear to have a constant curvature in every direction of their surfaces, raises questions about how their chromosomes are spatially organized and how this organization is maintained.
The model organism used in this work, Staphylococcus aureus, is a firmicute with a nearly spherical morphology. S. aureus is a human opportunistic pathogen that generally inhabits the skin as a commensal but can cause a variety of infections. Moreover, many S. aureus strains have acquired resistance to beta-lactam antibiotics, and some pathogenic strains are resistant to many antibiotic classes72. Its widespread presence in the human population, combined with its drug resistance, makes S. aureus a significant threat to human health and a major cause of death from antibiotic-resistant infections73. Besides its clinical relevance, S. aureus is an important model organism in the bacterial cell biology field, as it is one of the few intensively studied coccoid organisms. Its cell cycle is divided into three stages74,75,76: it begins with Phase 1 (P1), characterized by a nearly spherical newborn cell. As the cell starts to build a septum, it transitions into Phase 2 (P2), during which the DNA must be segregated into each of the developing compartments or hemispheres. Previous data suggest that septum formation is determined by the orientation of the segregated chromosomes, linking nucleoid spatial organization with cell division61,77. Once the septum is completed, the cell enters Phase 3 (P3), characterized by two compartmentalized cytoplasmic spaces. At the end of P3 the septum splits rapidly, giving rise to two P1 cells.
Dynamics of chromosome segregation in S. aureus is currently understudied, with most information available deriving from studies of ParB localization as a proxy for Ori localization61,78,79. In the present study, we localized specific chromosomal loci in S. aureus and showed that newborn cells generally have a partially replicated chromosome with two segregated origins. We show that the origin has a preferential localization pattern towards the tip of each hemisphere, henceforth referred to as “cell poles”, while the terminus is restricted to the cell center, resulting in an Ori-Ter-Ori chromosomal organization. Furthermore, we systematically analyzed the role of proteins known to influence chromosome segregation/dynamics, offering a comprehensive understanding of the factors contributing to chromosome biology of S. aureus.
Results
S. aureus cell cycle and chromosome replication cycle are not coupled
The number, orientation and dynamics of the S. aureus chromosome have not been comprehensively investigated. Previous studies used ParB (also known as Spo0J) as a proxy for Ori localization, showing cells containing two to four origins61,77,78. However, none of these studies analyzed the number ParB foci per cell at the population level, limiting our ability to draw conclusions about S. aureus ploidy, replication cycle or Ori localization. Furthermore, fluorescently tagged versions of the native S. aureus ParB often produce diffuse foci, hindering precise quantitative analysis. Therefore, we adapted a Fluorescent Repressor-Operator System (FROS) initially developed for E. coli11 for the visualization of specific chromosomal loci in S. aureus. This system makes use of fluorescent derivatives of the Lac repressor (LacI) and the Tet repressor (TetR) that bind to, and allow visualization of, arrays containing multiple copies of lacO or tetO operator sequences, respectively, which are introduced at specific locations on the bacterial chromosome. For this purpose, we first introduced (lacO)48 and (tetO)48 arrays at chromosomal loci near the Ori or Ter regions. We then introduced the lacI and tetR genes fused to sequences encoding the fluorescent proteins eCFP and eYFP11, respectively, under the control of a cadmium-inducible promoter80 in the spa locus of S. aureus JE2 chromosome81, generating strains JE2_Ori_CFP_Ter_YFP and JE2_Ori_YFP_Ter_CFP which allowed us to simultaneously visualize two different loci in each strain. We tested two different cadmium induction times and two combinations of operator arrays, and observed the formation of fluorescent foci corresponding to Ori and Ter localization in the cells (Fig.S2A). Cells usually had two to four origins, similar to what had been reported using a ParB fluorescent derivative in S. aureus61,77,78. However, the exact number of foci per cell may be underestimated in this assay, as data displayed in Fig.S2 was acquired by imaging a single z-plane, potentially missing overlapping foci or foci out of the focal plane. Furthermore, we noticed that the average number of foci per cell varied slightly depending on the duration of cadmium induction and the type of operator sequence array (tetO/lacO) used to label each region (Fig. S2B). The observed variation could partially arise from different TetR-eYFP and LacI-eCFP cellular levels, fluorescent protein brightness, and background noise in the different channels (which could influence foci identification). Such variability could compromise quantitative studies comparing different strains. Fortuitously, we noticed that a fusion of mNeonGreen82 to TetR (TetR-mNG, strain JE2_FROSOri) was sufficiently expressed in the absence of inducer to allow clear foci identification. Thus, we decided to use this constitutive single-locus FROS system (Fig. 1A) when labelling only one chromosomal position was sufficient, eliminating potential variations due to induction time, to different properties of the fluorescent protein employed, and/or to cellular responses to the presence of cadmium.
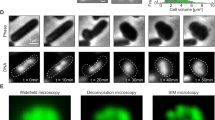
A Schematic of the single-locus FROS system used in this study. A sequence encoding the TetR repressor fused to the mNeonGreen (mNG) fluorescent protein, under the control of a cadmium-inducible promoter with leaky expression, was integrated in the spa locus (right). TetR-mNG binds to an array of 48 tetO operator sequences introduced at the locus of interest (left). B Bar chart representing the relative frequency of cells of the JE2_FROSOri_DnaN-Halo strain with 1-4 Ori foci in P1 (yellow, no septum) or P2/P3 (blue, incomplete and complete septum) cell cycle stages. Data are from three biological replicates (n = 100 each). C Bar chart showing the relative frequency of P1 and P2/P3 cells of the JE2_FROSOri_DnaN-Halo strain with Ori and replisome (visualized using DnaN as a proxy) either colocalizing (yellow) or not (blue). Data are from three biological replicates (n = 100 each). Examples of P1 cells with Ori-DnaN colocalization (yellow arrows) or no colocalization are shown on the right side; DnaN-Halo signal is shown in red and TetR-mNG (Ori) in green. Scale bar: 1 µm. D Classification of newborn cells by Ori number and Ori /replisome colocalization. Left, brightfield and fluorescence microscopy images of a representative cell from JE2_FROSOri_DnaN-Halo strain that underwent division between frame 0 and frame +1, showing two newborn cells with two Ori each in the latter frame (3 min interval between frames). JF549-labelled DnaN-Halo signal is shown in red and TetR-mNG (Ori) signal in green, scale bar: 1 µm. Right, bar chart showing the relative frequency of each class of newborn cells, categorized by Ori number, replisome assembly, and Ori/replisome colocalization (Y, yes; N, no). Data are from three biological replicates (n = 36, 36, 30). For panels B, C and D error bars indicate the standard deviation and are centered on the mean; individual data points are shown as colored circles; source data are provided as a Source Data file.
FROS labeling can potentially cause roadblocks in the chromosome, which could affect DNA replication83,84. To determine if the FROS system used in this work hinders DNA replication, we used marker frequency analysis (MFA) to quantify the relative abundance of DNA sequences across the entire genome85. This method has been used in bacteria, including in S. aureus and S. pneumoniae, to investigate defects in chromosome replication49,86, as a roadblock would cause a decrease in the frequency of downstream sequences. We found that S. aureus JE2_FROSOri and JE2_FROSTer strains had MFA profiles similar to that of the JE2 control strain (Fig. S3A), indicating that DNA replication is not impaired by the introduction of the FROS system.
In addition to the FROS system to localize different chromosomal loci, we constructed a HaloTag fusion87 to DnaN, a component of the replisome whose localization has been used as a proxy for replisome localization in studies of other organisms56,88. These tools allowed us to follow cellular localization of chromosome loci and of the replisome, enabling us to characterize the chromosome replication cycle in S. aureus.
In a first approach, we classified S. aureus cells according to the number of origin foci and correlated that information with the absence/presence of a visible septum. When quantifying the number of origins in a spherical cell, two factors can result in an underestimation: (i) two foci separated by a distance smaller than the resolution limit appear as a single focus; (ii) two foci with similar coordinates in the xy plane (parallel to the microscope slide) but different coordinate in the z axis (perpendicular to the microscope slide) appear as a single focus in a microscopy image. To overcome the latter limitation, cells can be imaged in three z-planes followed by manual analysis of each plane to count all detected foci. Using this approach, in the JE2_FROSOri_DnaN-Halo strain we did not observe cells without foci and rarely observed cells with a single Ori focus (<1%), with cells typically having two (~27%), three (~34%) or four (~38%) Ori foci (Fig. 1B). As expected, the number of origins increases as the cell cycle progresses, with P2 and P3 cells (i.e. cells with a division septum) having three or four Ori foci, indicating that four origins are the typical maximum.
We then asked when, during the cell cycle, is replication initiated. For that, we assessed colocalization of the origin and the replisome protein DnaN in strain JE2_FROSOri_DnaN-Halo. This colocalization was only observed in P1 cells (cells lacking any nascent or complete septa), suggesting that re-initiation of chromosome replication happens during that stage (Fig. 1C). Furthermore, the replisome was assembled, i.e. formed one or more foci (as opposed to having a diffuse cytoplasmic signal) in >95% of the cells (Fig. S4), indicating that DNA replication is ongoing during almost the entire cell cycle.
To directly show, in time-lapse movies, that cells start their cell cycle with two segregated origins of replication, JE2_FROSOri_DnaN-Halo cells were imaged every 3 min. By analyzing the frame immediately after the splitting of the mother cell in two daughter cells, we showed that more than 90% of newborn cells had two Ori foci (Fig. 1D). Additionally, around 70% of newborn cells had an assembled replisome that did not colocalize with the origins, indicating that cells are typically born with a partially replicated chromosome (Fig. 1D). We have also observed newborn cells (~20%) containing two origins either with a diffuse DnaN signal (no active replisome, indicating that the replication round is finished) or with DnaN colocalizing with the Ori foci (replisome initiating the next round of replication), indicating that in these cases cells are born with two completely replicated chromosomes (Fig. 1D). We then imaged strain JE2_FROSTer_DnaN-Halo, in which the FROS array is inserted near the Ter region. We observed that ~95% of newborn cells displayed a single Ter focus (Fig. S5) and only ~5% of cells exhibited two foci. Given that data obtained with Ori-labelled strain JE2_FROSOri_DnaN-Halo (Fig. 1D) indicate that ~20% of newborn cells are diploid, this discrepancy may be due to the Ter region being confined to a small region within the cell, causing two closely positioned foci to appear as one because of resolution limitations. Alternatively, the Ter regions of sister chromosomes could remain close due to an unknown cohesion mechanism. In any case, these data support our conclusion that the majority of cells are born with a partially replicated chromosome and a minority as diploids. When following the JE2_FROSOri_DnaN-Halo cells in longer time-lapse experiments, we could observe cells with two Ori that underwent the completion of one round of replication and initiated a new round, followed by origin segregation, resulting in cells with four Ori foci (Fig.S6). These observations further support that, during its cell cycle, S. aureus generally progresses from a partially replicated chromosome to two partially replicated chromosomes.
Collectively, the data show that the S. aureus cell cycle (from one cell division to the next) and chromosome replication cycle (a complete round of chromosome replication) are not necessarily coupled: during a single cell cycle, most cells complete one replication cycle and initiate another, resulting in cells with four origins and two partially replicated chromosomes that will be distributed to the two daughter cells. This chromosome replication cycle resembles what has been described for B. subtilis under slow-growing conditions6.
S. aureus chromosome adopts a longitudinal Ori-Ter-Ori conformation
Bacterial species generally have specific spatial arrangements of their chromosomes, particularly regarding the position of the Ori and Ter regions (Fig. S1). To study chromosome organization in S. aureus, we used the FROS system to label not only the Ori and Ter, but also the left and right arms of the chromosome (Fig. 2A, B). To systematically analyze the localization of labelled chromosome loci in thousands of cells grown at 37 °C in TSB, we developed a pipeline using the e-Hooke software version 1.189 for cell segmentation, and TrackMate90 for foci detection. This pipeline uses a maximum intensity projection of the TetR-mNG foci signals obtained from three z-planes. Foci detection data is then used to generate average maps of the locations of different chromosomal regions within the cell (Fig. 2C). To generate these maps, cells were first aligned to the slightly longer axis, which is perpendicular to the future division plane. The relative position of each focus center in individual cells was recorded and mapped onto a model cell with median dimensions for length and width specific for each dataset. Heat maps were then created by calculating the average localization of foci, which correlates with the probability of a focus being found at each location. The resulting data showed that origins are typically positioned at the cell periphery, on opposite ends of the longer cell axis (the cell poles), termini are confined to the cell center, and the left and right arms occupy intermediate locations (Fig. 2C).
A Schematic of the S. aureus chromosome showing the locations of the (tetO)48 array used to localize the chromosomal origin, terminus, left arm and right arm. B Fluorescence microscopy images of cells with indicated chromosomal regions labeled by TetR-mNG (green) and membrane labeled with FM 4-64 dye (red). Scale bar: 2 μm. Three independent colonies were imaged for each strain, showing similar results. C Heatmaps showing the average localization of detected fluorescent TetR-mNG foci in strains indicated in B. The color scale in each dataset ranges from red (maximum spot density) to dark blue (no spots detected), n number indicated in the lower right corner of each heatmap. D Bar charts showing the distribution of the number of origin (left, strain JE2_FROSOri) or terminus (right, strain JE2_FROSTer) foci for cells in each phase of the cell cycle. Data from three biological replicates, error bars indicate the standard deviation and are centered on the mean; individual data points are shown as colored circles. For the JE2_FROSOri n = 2309, 2001, 1097; for the JE2_FROSTer n = 2081, 2983, 2802. Source data are provided as a Source Data file. E Heatmaps showing the average localization of detected fluorescence spots for cells of strains JE2_FROSOri (Ori) and JE2_FROSTer (Ter) in each phase of the cell cycle. Color scale as in panel C. n number indicated in the lower right corner of each heatmap.
To further validate the results of Ori localization, we colocalized a ParB-YFP fusion (a well-established proxy for Ori localization20,21,33,79,91) with Ori labelled by TetR-mCherry, in the strain JE2_FROSOri-mCherry_ParB-YFP. We observed that ParB and TetR foci colocalized and that their average localization overlapped, with both proteins most often localizing at the cell poles (Fig. S7A). A similar polar localization for ParB was also observed in strain JE2_ParB-YFP, which lacks the FROS system, confirming that the presence of the FROS array does not interfere with Ori localization (Fig. S7B). Altogether, these findings indicate that S. aureus cells adopt an Ori-Ter-Ori organization, with origins located in close proximity to the cell periphery.
The analyzed cells of the JE2_FROSOri and JE2_FROSTer strains were also manually classified according to their cell cycle phase, as described in ref. 74, allowing us to quantify the number of origins and termini per cell at each cell cycle phase (Fig. 2D). Similarly to the data shown in Fig. 1B, we observed that P1 cells had two (~40%), three (~25%) or four (~25%) origins, with the number of Ori per cell increasing as the cell cycle progresses (Fig. 2D). As for the termini, the majority of P1 cells had a single focus (~60%), while most P2 and P3 cells had 2 foci (~80%) (Fig. 2D). We also generated heatmaps of cells automatically classified according to their cell cycle phase (Fig. 2E), which showed that the Ori´s polar localization and the Ter´s central positioning were maintained throughout the cell cycle.
The observation that cells typically have a maximum of four origins and two termini suggests that S. aureus cells do not undergo multifork replication. This is supported by MFA analysis (Fig. S3) which yielded a calculated Ori/Ter ratio of 2.49 for S. aureus JE2 strain, in line to what has been previously reported (ratio of 2.16 for S. aureus strain HG00386, and of 2.64 in S. aureus strain SH100092). Cells that undergo a single replication cycle per cell cycle exhibit Ori/Ter ratios between 1 (corresponding to cells that have not initiated replication) and 2 (corresponding to cells that have replicated the origin but not yet the terminus) as observed in C. crescentus93 and Agrobacterium tumefaciens94. In contrast, species that consistently perform multifork replication exhibit significantly higher Ori/Ter ratios, for example ratios of 4-6 in E. coli95,96, approximately 5 in B. subtilis16,97 and above 5 in M. smegmatis98. Therefore, our data indicate that S. aureus appears capable of very limited multifork replication compared to these organisms, in line with less than 5% of cells having more than four Ori foci (Fig. 2D).
To investigate whether the number of Ori/cell was dependent on growth rate, we imaged the JE2_FROSOri strain grown at 25 °C and compared with cells grown at 37 °C. Ori localization pattern was polar at both temperatures (Fig. S8A) but the number of Ori foci/cell was lower 25 °C, across all cell cycle phases (Fig. S8B). This likely reflects the slower pace of DNA replication at 25 °C, which is also supported by lower Ori/Ter ratio (1.99 at 25 °C vs. 2.49 at 37 °C) obtained by MFA (Fig. S3B).
Additionally, we imaged the JE2_FROSOri strain in stationary phase (12 h of growth at 37 °C) and observed 1-2 Ori foci per cell (Fig. S9). Together with the MFA data (Fig. S3B), that yields a calculated Ori/Ter ratio of 1.22, these results indicate that most stationary phase cells have completed chromosome replication. This finding is consistent with previous observations in B. subtilis, where the number of replication origins decreases in stationary phase16.
Collectively, these results confirm that S. aureus cells in active growth generally initiate the cell cycle with two origins, indicating that origin segregation usually occurs before septum synthesis is initiated. As the cell cycle progresses, a new round of chromosome replication produces cells with four origins and two termini.
SMC complex, but not ParB, plays a major role in chromosome segregation in S. aureus
Having established the spatial organization and dynamics of the staphylococcal chromosome, we aimed to investigate the role of proteins reported to be involved in bacterial chromosome segregation and/or organization, specifically ParB and SMC. ParB is a key component of the ParABS system in various bacterial species26. However, no ParA homolog has been identified in S. aureus, meaning that its ParABS system is incomplete, which makes the role of ParB in this bacterium particularly intriguing. It has been previously assumed that, similar to other ParABS systems, S. aureus ParB binds to Ori-proximal parS sequences, with one study predicting four such sequences in its genome33. To experimentally determine the parS sites, we performed Chromatin ImmunoPrecipitation Sequencing (ChIP-Seq) analysis using JE2_ParB-3xFLAG strain. We identified ParB enrichment at five loci, all near the Ori region (Fig. S10A). At each of these five locations, we found a parS motif, three of which (parS1, parS3 and parS5) coincide with those previously predicted33 (Fig. S10B). Moreover, each ParB enrichment peak spanned 8–10 kb encompassing the parS sequence, indicating that, like in other organisms25,28,46,99,100,101, S. aureus ParB nucleates around origin-proximal parS sequences and spreads to neighboring regions.
To understand the role of ParB and the SMC complex on the global organization of the chromosome, we used High-throughput Chromosome Conformation Capture (Hi-C) assay102, a technique that involves crosslinking nearby DNA regions to capture the chromosome conformation by detecting the frequency of interaction between DNA loci across the whole genome. The S. aureus chromosome contact map for JE2 wild type strain (Fig. 3A) displays a primary diagonal and a secondary diagonal. The primary diagonal has stronger signal, resulting from a high frequency of contacts between adjacent sequences in the chromosome. Similar to what was previously reported in other bacteria, chromosome interaction domains which have the strongest domain boundaries, generally correlated with highly transcribed genes such as ribosomal operons, were present along the primary diagonal (Fig. 3A, black arrows)46,102,103,104. Moreover, we were able to observe an origin domain with an increased number of internal contacts, similar to what has been described in B. subtilis105 as well as an intra-right arm hairpin near the origin (evidenced by a bow-like pattern, blue arrows in Fig. 3A). Finally, the secondary diagonal has weaker signal, resulting from inter-arm, long distance DNA contacts. This interaction pattern supports that the chromosome has an Ori-Ter longitudinal organization.
A–E normalized Hi-C maps showing contact frequencies for pairs of genomic regions (5 kb bins) which are represented by the Hi-C score (lower right color scale) in the indicated strains. Each map is generated using data from a representative replicate for each strain. The chromosomal origin is at the center of the axes. The primary diagonal, running from lower-left to upper-right, represents local, short-range interactions, while the secondary diagonal, running from upper-left to lower-right, represents interactions between loci equidistant from the Ori and on opposite chromosomal arms. A green dotted line crosses the maps horizontally and vertically through the center (Ori), where the two diagonals intersect in JE2 (A), but not in JE2_Δ4parS (D). In A, black arrows indicate strong boundaries between chromosomal interaction domains, blue arrows point towards a bow-shaped pattern that indicates the presence of an intra-right arm hairpin. Below each map, a scheme shows the proposed chromosome arrangement, with the Ori in green, the parS sites in red and the Ter in yellow. These schematics show the overall folding of the chromosome and do not include local domains. Pairs of loci that, in JE2, would be on opposite arms and in physical proximity (long-range contacts), are represented as colored line segments. Note that the specific pattern of long-range contacts that results in the secondary diagonal in JE2, is lost in the JE2_ΔparB and JE2_Δ5parS strains. In these strains SMC is no longer loaded at specific parS sites, leading to different possible chromosome arrangements, three examples of which are represented.
Upon deletion of parB, the secondary diagonal disappeared (Fig. 3B) and the contacts of the origin domain were reduced, consistent with previous findings37,46,47,105,106. This can be explained by the loss of DNA interactions mediated by ParB nucleoprotein complexes near the Ori and the failure to load SMC at specific parS loci, which is required for proper chromosomal arm alignment. Furthermore, deletion or mutation of the five identified parS sites also abolished the secondary diagonal, confirming that ParB and parS form a functional unit for chromosome organization (Fig. 3C). Importantly, we constructed the strain JE2_Δ4parS, in which four parS sites were deleted or mutated and parS2 (47 kb away from the Ori) remained as the sole parS on the chromosome. This strain showed a more defined secondary diagonal compared to the wild type JE2, shifted towards the right arm, where parS2 is located (Fig. 3D). Presumably, this occurs because SMC is loaded onto the chromosome from the single parS2 locus, resulting in the alignment of the chromosomal arms. This is consistent with previous studies showing that a single parS site can determine the chromosomal arm alignment in other bacteria34,39,47,105,107,108,109.
To assess the role of the SMC complex in chromosome organization, we made a clean deletion of the genes scpA and scpB (JE2_ΔscpAB), whose inactivation in B. subtilis leads to the same phenotype as that of SMC110. Similarly to deletion of parB, deletion of scpAB also abolished the secondary diagonal in the HiC map (Fig. 3E), consistent with the idea that ParB loading SMC complexes to parS sites generates arm alignments. In addition, deletion of scpAB caused a reduction in the Hi-C signal outside of the primary diagonal (i.e. long-range DNA interactions) and an increase in the signal of the primary diagonal (i.e. short-range DNA interactions), in comparison to both JE2_ΔparB and JE2 wild type (Fig. 3). Indeed, when we analyzed the global contact probability curve, Pc(s) curve, which shows the averaged contact probability for all loci separated by a set distance and indicates the compaction status of the chromosome111, we found that the JE2 and JE2_∆parB had almost identical curves, but JE2_∆scpAB showed reduced long-range DNA interaction (Fig. S10C), similar to previous findings in B. subtilis37. These results indicate that the SMC complex is responsible for promoting DNA interactions between regions that are more than 400 kb apart, but this activity does not require specific loading of the SMC complex at parS sites. In the absence of ParB or parS sites, SMC presumably loads randomly all over the chromosome, providing a level of chromosome contacts that are overall similar to those observed in the wild type JE2 strain. However, the specific chromosomal regions involved in these contacts differed from those observed in JE2, as indicated by the loss of the secondary diagonal in the Hi-C maps of JE2_ΔparB or JE2_Δ5parS (Fig. 3). Consistent with this, studies in B. subtilis have shown that SMC can load non-specifically onto the chromosome (albeit with low efficiency) in the absence of ParB, through an unknown mechanism37.
To assess the role of ParB and the SMC complex in chromosome segregation, we examined the localization and number of Ori and Ter foci, as well as the occurrence of anucleate cells, in mutants lacking these proteins. Deletion of parB in the background of strains JE2_FROSOri and JE2_FROSTer did not alter the localization of Ori and Ter foci in comparison with the parental control strains (Fig. 4A), but led to a reduction in cells with four Ori foci (Fig. 4B). Importantly, the JE2_ΔparB strain produced only 0.14 ± 0.04% of anucleate cells (vs. 0.03 ± 0.05% in JE2 wild type, Fig. S11A), consistent with previous reports suggesting that parB deletion causes only a very mild chromosome segregation defect in S. aureus78,86.
A Fluorescence microscopy images of the indicated strains showing localization of Ori (top) or Ter (bottom) labeled with the FROS system (green) and membrane labeled with FM 4–64 dye (red). Scale bar: 2 μm. Heatmaps of the average localization of detected fluorescence spots of each strain are shown below each microscopy image. The color scale in each dataset ranges from red (maximum spot density) to dark blue (no spots detected), n number indicated in the lower right corner of each heatmap. B Bar charts showing the relative distribution of the number of Ori (left) and Ter (right) foci in cells of strains shown in A. Data from three biological replicates, error bars indicate the standard deviation and are centered on the mean; individual data points are shown as colored circles. n number for each replicate is shown under each strain’s name. Source data are provided as a Source Data file.
The function of the SMC complex in chromosome segregation was evaluated using two different mutants: the JE2_ΔscpAB strain described above and a second mutant in which we introduced an array of premature STOP codons near the start of the smc gene (JE2_smcSTOP strain), preventing potential polar effects on the two essential genes (ftsY and ffh) downstream of smc112. Both mutants exhibited ~17% of anucleate cells (Fig. S11A). Importantly, ΔscpAB and smcSTOP mutants phenotypes could be complemented with ectopic expression of scpAB and 3xflag-smc, respectively (Fig. S11A), confirming that anucleate cells arise from the lack of ScpAB or SMC. We note that previous reports on the effect of deletion of smc had conflicting results, with anucleate cells frequencies varying from 1-2%78,113 to 10%114, while deletion of scpB led to the production of ~14% of anucleate cells115. Given that we observed 17% of anucleate cells in two different SMC-ScpAB mutants, and that we were able to complement their phenotype, we hypothesize that the previously reported mutants with low frequency of anucleate cells might contain suppressor mutations.
Consistent with the increased number of anucleate cells, mutants lacking a functional SMC complex showed a higher number of cells with no Ori foci and a decrease in cells with 3 or 4 Ori foci when compared with the parental strain (Fig. 4B). In agreement, MFA analysis (Fig. S11B) indicated that both the JE2_ smcSTOP and JE2_ ΔscpAB strains have reduced Ori/Ter ratios (1.92 and 2.05 respectively, compared to 2.49 for the wildtype). However, this reduction in Ori/Ter ratio alone cannot fully account for the decreased number of cells with 3 or 4 Ori foci because JE2 strain grown at 25 °C also has an Ori/Ter of 1.99 (Fig. S3B), yet 47% of its cells exhibited 3 or 4 Ori foci, whereas strains lacking SMC or ScpAB showed less than 20% of these cells (Fig. 4B). Therefore, the SMC-ScpAB complex has a role in segregating the origins.
Not only the number, but also the localization of origins in both ΔscpAB or smcSTOP backgrounds was altered, with a distribution no longer restricted to the cell poles, though still in close proximity to the membrane (Fig. 4A). The termini also showed a more diffuse pattern, although still positioned around the cell center. Altogether, our data indicate that the SMC complex plays a key role in the spatial organization and segregation of the S. aureus chromosome.
Finally, we constructed a double mutant lacking both parB and scpAB (JE2_∆parB_∆scpAB) and found that it was very similar to JE2_∆scpAB in terms of anucleate cells frequency (Fig. S11). Together with the Hi-C data from the JE2_ΔparB and JE2_ΔscpAB strains (Fig. 3B, E), this result suggests that in S. aureus, which is missing ParA, the main role of ParB is to load SMC complexes at the parS sites. Since ∆parB alone had little effect on chromosome segregation but smcSTOP and ∆scpAB each had a strong defect in chromosome segregation, we conclude that the SMC complex does not need to be specifically loaded onto the parS sites to segregate chromosomes. In the absence of ParB/parS, SMC-ScpAB molecules are likely still loaded onto the chromosome (in a random manner or via unidentified loading sites) as long-range contacts are maintained (Fig. S10C). These molecules contribute to chromosome segregation (Fig. S11A), even though specific inter-arm alignment is lost (Fig. 3B).
Role of factors connecting the divisome with the chromosome
One important link between the cell division machinery and chromosome segregation is the FtsK protein family of DNA pumps that ensure segregation of the chromosomes before the completion of the division septum116,117. In E. coli, FtsK works together with the recombinase XerCD complex to resolve chromosome dimers that would otherwise prevent proper chromosome segregation118. S. aureus has two FtsK family proteins, FtsK and SpoIIIE. Each protein is individually dispensable, but the presence of at least one is required for correct chromosome segregation119. Both the Ori-labelled and the Ter-labelled FtsK mutants strains JE2_FROSOri_ΔftsK and JE2_FROSTer_ΔftsK, respectively, showed the previously reported phenotype for an S. aureus ΔftsK mutant, characterized by delayed cell splitting at the end of division120, which can lead to the synthesis of new septa in the hemispheric compartments (Fig. 5A, white arrows). The localization pattern of Ori and Ter foci was altered, with both regions occupying larger cell areas on average (Fig. 5A), probably due, at least in part, to this cell division defect. Moreover, the FtsK mutants had increased numbers of Ori and Ter foci per cell, as JE2_FROSOri_ΔftsK displayed a higher proportion of cells with three, four or more Ori foci compared to the parental strain, while JE2_FROSTer_ΔftsK mutant produced about 10% of cells with more than two termini, compared to only 1% in the JE2_FROSTer parental strain (Fig. 5B, C). MFA analysis did not show differences in the replication profile of the JE2_FROSTer_ΔftsK strain compared to its parental strain (Fig. S12A) indicating that the cells are not over-replicating their chromosomes. Therefore, the cause of the increased number of chromosomes per cell is likely a consequence of the delayed cell separation at the end of division caused by the deletion of ΔftsK, which leads to a longer cell cycle Phase 3120 allowing chromosome replication to be completed and re-initiated before cells physically divide.
A Fluorescence microscopy images of the indicated strains showing localization of Ori (top) or Ter (bottom) labeled with the FROS system (green) and membrane labeled with FM 4–64 dye (red). Scale bar: 2 μm. Heatmaps of the average localization of detected fluorescence spots of each strain are shown below each microscopy image. The color scale in each dataset ranges from red (maximum spot density) to dark blue (no spots detected), n number indicated in the lower right corner of each heatmap. White arrows point to cells with a second septum, dividing a hemisphere into two compartments, a phenotype described for S. aureus ΔftsK mutants. Bar charts showing the relative distribution of number of origin (B) and terminus (C) foci in cells of the strains shown in A. Data from three biological replicates, error bars indicate the standard deviation and are centered on the mean; individual data points are shown as colored circles. n number for each replicate is shown under each strain’s name. Source data are provided as a Source Data file.
To evaluate the role of SpoIIIE in chromosome segregation, we first imaged the Ori-labelled JE2_FROSOri_ΔspoIIIE strain, which showed a reduction in cells with 4 Ori foci (15% compared to 25% in the parental strain, Fig. 5B) but no major change in Ori localization (Fig. 5A). We then imaged the Ter-labelled JE2_FROSTer_ΔspoIIIE strain which, strikingly, produced around 17% of cells with no terminus foci (compared to 4.5% in the JE2_FROSTer parental strain). A similar, but more pronounced, phenotype was observed in the JE2_FROSTer_ΔxerC strain, with 35% of cells lacking a terminus focus. MFA revealed a decreased DNA copy number at the terminus region in the population of ΔxerC mutant cells (Fig. S12B, blue arrows). Thus, we hypothesize that the absence of XerC can cause severe alterations, or even DNA degradation, in the terminus region (where the tetO array is located), preventing the formation TetR-mNG foci in the affected cells. Interestingly, MFA did not show an obvious decrease in the terminus region in the JE2_FROSTer_ΔspoIIIE strain, indicating that perhaps another process may be interfering with the TetR association to the tetO arrays in this strain (and potentially also in the JE2_FROSTer_ΔxerC strain). Furthermore, in ΔxerC and ΔspoIIIE mutants, we observed two peaks, one in each replication arm, indicating increased DNA amplification in these genomic regions (Fig. S12B, red arrows). These regions encompass genes that are annotated as encoding phage proteins, including capsid proteins, phage terminases and phage DNA primases (genes SAUSA300_1921-1940 in the left arm peak and SAUSA300_0809-0815 in the right arm peak). Therefore, it is likely that the observed DNA amplification in these regions is due to the activation of prophages, triggered by the deletion of xerC or spoIIIE. Despite the changes in the number of origins and termini in mutants lacking spoIIIE or xerC, the cellular localization of Ori and Ter foci remained largely unchanged (Fig. 5A). However, Ter positioning appeared less confined to the cell center, suggesting that these proteins may play a role in restricting the spatial localization of the Ter region.
We also investigated the role of the nucleoid occlusion protein Noc, which in both B. subtilis and S. aureus prevents the assembly of the divisome over the nucleoid to avoid its guillotining61,63. Additionally, in S. aureus, Noc is a negative regulator of the initiation of DNA replication86. In agreement with published data, the JE2_FROSOri_Δnoc strain exhibited over 30% of cells with more than four origin foci (Fig. 5A, B), and showed an increased number of cells with more than two terminus foci (Fig. 5C). Despite their increased number, Ori foci remained spaced from each other, although they were more dispersed around the cell periphery compared to the JE2_FROSOri parental strain (Fig. 5A). Overall, our data supports the role of Noc as a key regulator of chromosome replication in S. aureus.
Discussion
The spatial and temporal organization of the bacterial chromosome may seem particularly challenging for a nearly spherical bacterium, as there are fewer geometric cues available compared to rod-shaped or asymmetric cells. Yet, S. aureus elegantly solves this problem for chromosome segregation, by not synchronizing its chromosome replication cycle (one complete round of chromosome replication) with its cell division cycle (from completion of one cell division to the next). S. aureus newborn cells, in fast growing conditions, typically have two origins of replication and one active replisome (Fig. 6A), similar to B. subtilis in slow growing media6. The origins tend to localize at the cell periphery, near the membrane, positioned opposite to each other, at the cell´s poles. This arrangement establishes an axis of chromosome segregation, breaking the internal spherical symmetry of the cell. As chromosomes segregate along this axis, the space between them creates a region for the division site, where a septum can form without guillotining the DNA (Fig. 6B). Mechanistically, this is mediated by the nucleoid occlusion protein Noc, which binds the origin-proximal region of the chromosome and prevents spurious FtsZ assembly in those regions61. A second round of replication can begin in Phase 1 cells, i.e., even before septum synthesis starts. This is supported by the observation that ~35% of Phase 1 cells have three to four segregated origins (Fig. 1B), indicating that the presence of a septum is not required for origin segregation. As the cell cycle progresses and the septum begins to be synthesized (Fig. 6B), the cell becomes increasingly divided in two hemispherical compartments. These compartments now have a long axis (parallel to the nascent septum) and a short axis (perpendicular to the nascent septum). Our data show that, within each hemisphere, the two origins generally segregate away from each other along a long axis parallel to the septum (Fig. 6C). Therefore, when P3 cells split and give rise to newborn P1 cells, the future division plane is already defined within the spherical cytoplasmic compartment, located between the segregated chromosomes, where the septum will form. In turn, the septum demarcates the possible directions for the next round of chromosome segregation. Interestingly, some organisms that rely on a ParABS system to segregate the duplicated origins to opposite cell poles show a faint horizontal line on the Hi-C map46,103. This Hi-C signature represents specific contacts of the origin region with DNA along the entire chromosome, which is consistent with the idea that ParA uses the chromosome as a track to direct the Ori to the opposite cell pole121,122,123,124. The absence of such a signal in the JE2 Hi-C map (Fig. 3A) is consistent with S. aureus lacking ParA and supports the idea that the S. aureus Ori is not actively transported across the chromosome.
The cell envelope (brown), origins (green circles), active replisomes (magenta circles), termini (yellow circles) and the chromosome (orange and brown) are illustrated. A Typical newborn cell (P1), with two segregated origins and a hemi-replicated chromosome. B P2 cell, after the new round of replication has begun, and origins segregation has initiated. C P3 cell, which generally has four segregated origins and each hemi-replicated chromosome occupying one of the hemispheres, which become spherical again after cell division. Consequently, the future division septum is already defined in newborn cells, located between segregated chromosomes. Notice that the cell cycle and chromosome replication cycle are not coupled, and Ori segregation can occur as early as P1, leading to ~35% of P1 cells having three or four origins.
Some fast-growing organisms, such as B. subtilis15 and E. coli17, as well as slow-growing M. smegmatis19 can undergo multifork replication, which occurs when multiple rounds of replication take place during one cell cycle, usually under rich media conditions. For S. aureus, we did not detect cells with assembled replisomes colocalizing with the origins while a second set of replisomes was located further away from the origins. However, MFA results (Fig. S3) suggest that S. aureus may engage in limited multifork replication, as the JE2 strain had an Ori/Ter ratio of approximately 2.49, higher than expected if a single round of replication occurred per cell cycle, which would result in a Ori/Ter ratio lower than 2. Nevertheless, this ratio is considerably lower than in bacteria known to undergo extensive multifork replication, which have Ori/Ter ratios above 416,95,96,97,98. Interestingly, the ovococcal S. pneumoniae is thought not to engage in multifork replication13,92. It is possible that the small size and the geometry of S. aureus (and perhaps other coccoid bacteria) make it difficult to extensively use multiple rounds of replication in a single cell cycle. The S. aureus Ori/Ter ratio decreased to 1.99 when the growth rate was slower (25 °C, TSB) and to 1.22 in stationary phase. Collectively this suggests that during active growth S. aureus tends to maintain the 2 to 4 Ori cycle, but in a reduced nutrient environment it can lower its ploidy by slowing down chromosome replication with respect to the cell division cycle.
Regarding the spatial arrangement of S. aureus chromosome, the Ori is primarily localized at the cell periphery, near the membrane, throughout the cell cycle (Fig. 2C, E), even in mutants affecting its number (Δnoc, Fig. 5A) or segregation (ΔscpAB, smcSTOP, Fig. 4A). This localization is not exclusive of S. aureus, as other bacteria also position their origins in close proximity to the membrane, such as B. subtilis during sporulation or S. pneumoniae125,126, or in polar or sub-polar regions, like C. crescentus, or M. xanthus3,4. These bacteria employ molecular mechanisms to restrict the movement of the origins, leading us to hypothesize that S. aureus likely has a similar mechanism, perhaps resembling those in other firmicutes, such as the RacA protein in B. subtilis126,127,128 or RocS in S. pneumoniae125.
The forces driving chromosomal segregation in S. aureus are not yet fully understood, but our data from the ΔscpAB and smcSTOP mutants strongly suggest that the SMC complex plays a crucial role in the process, likely by promoting the unmixing of the sister chromosomes as they are replicated39. Furthermore, the steep increase in cells with a single Ori focus in the ΔscpAB and smcSTOP mutants (Fig. 4B) suggests that the SMC complex has an important role in the segregation of the Ori regions, similar to what has been described for S. pneumoniae13 although in this bacterium the absence of SMC results in only a small percentage (~2%) of anucleate cells57,129. This role of SMC in Ori segregation in S. aureus would explain the drastic decrease in cells with three or four origins in these mutants, compatible with the possibility that chromosomes are replicated but origins remain together and are thus indistinguishable using the FROS system. Such a failure in origin segregation would compromise the overall chromosome segregation process, resulting in the production of anucleate cells, as observed (Fig. S11A). Importantly, ParB is not essential for overall chromosome segregation in S. aureus, although its presence organizes the loading of the SMC complexes (Fig. 3B). In the ΔparB background, we measured a reduction in cells with four Ori foci, suggesting that ParB-mediated loading of the SMC complexes slightly increases the efficiency or speed of Ori segregation. Overall, our findings indicate that ParB bound to parS loads SMC, which globally organizes the chromosome, creating a defined inter-arm alignment. This is consistent with previous work showing that in S. aureus, deletion of parB reduces the number of SMC foci78 and in line with data from S. pneumoniae49,57, B. subtilis37,103,105, P. aeruginosa47, or C. glutamicum46. However, this specific alignment by itself is not a major contributor to chromosome segregation. Rather, SMC loading onto the chromosome (not necessarily at parS sites) and its translocation away from the loading position, would generate DNA loops (long-range DNA contacts), contributing to chromosome compaction and segregation. Interestingly, the possibility that B. subtilis SMC can also load onto the chromosome, presumably at random positions, in the absence of ParB, albeit at a much reduced rate, has been recently reported37. In addition to ParB and the SMC complex, other factors like nucleoid-associated proteins (NAPs) may also contribute to maintaining S. aureus chromosome conformation. For example, the NAP protein HU, which is essential in B. subtilis130, S. pneumoniae131 and S. aureus132, has been shown to contribute to DNA supercoiling in S. aureus133 and to maintain chromosome structure in S. pneumoniae49. Future research will be needed to assess the contribution of NAPs to S. aureus chromosome positioning and organization.
Two additional proteins involved in chromosome segregation in S. aureus are the DNA pumps FtsK and SpoIIIE. Each protein is individually dispensable, but at least one must be present for correct chromosome segregation, suggesting partial redundancy119. However, FtsK and SpoIIIE do not colocalize, and while ftsK deletion causes cell morphology defects such as multi-septated cells and cell size heterogenicity, deleting spoIIIE leads to an increase of cells with condensed chromosomes, altogether indicating that they have partially independent functions119. In this study, deletion of ftsK led to an increase in the number of Ori and Ter foci (Fig. 5B, C). Previous research has shown that FtsK mutants have a delay in P3, resulting in cells remaining for longer in a pre-divisional stage. This delay is due to an additional role of S. aureus FtsK in promoting the export of the autolysin Sle1, a peptidoglycan hydrolase that plays an important role in splitting the septum at the end of the cell cycle120. It is plausible that during this delay, chromosome replication has time to finish, explaining the observed increase in the number of Ori and Ter foci. This increase in the number of Ter foci could also indicate that FtsK is involved in keeping replicated Ter together, which would explain the less restricted Ter positioning observed in the heatmaps in the ΔftsK background. Interestingly, in sporulating B. subtilis the two Ter regions of sister chromosomes are held together by an unknown mechanism134,135.
In contrast, deletion of spoIIIE results in approximately 17% of cells lacking a terminus focus, a phenotype similar to that observed in the ΔxerC mutant. In E. coli, the XerC recombinase works together with FtsK to resolve chromosome dimers during the final stages of chromosome replication and segregation118,136. In S. aureus, previous studies have shown that deleting either spoIIIE or xerC increases the number of cells with condensed nucleoids, with this effect being more pronounced in the ΔxerC mutant119. However, we found no correlation between cells with condensed nucleoids and cells lacking Ter foci. Furthermore, our data suggest that the deletion of xerC could give rise to alterations (or even in DNA degradation) in the Ter region, potentially explaining why a subset of cells lack a Ter focus (Fig. S12B). However, the literature presents conflicting evidence regarding Ter degradation in E. coli xerC mutants. While one study reports Ter degradation137, another finds minimal differences compared to the wild-type strain138. Collectively our findings serve as a starting point for further investigation into possible functional connections between SpoIIIE and XerC, a link suggested by a previous study119.
Finally, our data supports the proposed role of Noc as a key regulator of initiation of DNA replication86, given that its absence led to a sharp increase in the number of cells with more than four origins.
This study provides the first comprehensive characterization of chromosome positioning and dynamics in a small, spherical bacterium, highlighting the role of chromosome segregation in division site positioning. When comparing to other firmicutes, S. aureus chromosome organization and replication cycle resembles that of slow-growing B. subtilis, where newborn cells typically start with one hemi-replicated chromosome and origins positioned at opposite poles6. However, key differences were observed, as in S. aureus origins are consistently associated with the cell periphery, with segregation occurring along an axis parallel to the septum. On the other hand, similar to S. pneumoniae, S. aureus has a ParABS system lacking ParA, and has a mild phenotype for parB deletions57,129. However, while in S. pneumoniae the newly replicated origins immediately mark the future cell division sites13,92, in S. aureus the Ori tends to localize in the polar regions and does not colocalize with the divisome. Another key difference is the importance of the SMC complex, whose absence causes a severe segregation defect in S. aureus, but a mild one in S. pneumoniae57,129. Future research will determine whether other spherical coccoid organisms follow a similar pattern.
Methods
Bacterial growth conditions
Strains of E. coli were grown in lysogeny broth (LB, VWR) or on lysogeny broth agar (LA, VWR) at 37 °C. S. aureus was grown in tryptic soy broth (TSB, Difco) and in M9 minimal medium (KH2PO4 3.4 g L−1, VWR; K2HPO4 2.9 g L−1, VWR; di-ammonium citrate 0.7 g L−1, Sigma-Aldrich; sodium acetate 0.26 g L−1, Merck; glucose 1% (w/v), Merck; MgSO4 0.7 mg L−1, Sigma-Aldrich; CaCl2 7 mg L−1, Sigma-Aldrich; casamino acids 1% (w/v), Difco; minimum essential medium amino acids 1×, Thermo Fisher Scientific; and minimum essential medium vitamins 1×, Thermo Fisher Scientific) with agitation (200 rpm) or on tryptic soy broth agar (TSA, VWR). When required, media were supplemented with antibiotics (100 μg mL−1 ampicillin, Sigma-Aldrich; 10 μg mL−1 erythromycin, Apollo Scientific; 10 μg mL−1 chloramphenicol, Sigma-Aldrich). For blue/white colony screening, TSA plates were supplemented with 5-bromo-4-chloro-3-indolyl β-d-galactopyranoside (X-Gal, Apollo Scientific) at 100 μg mL−1. When required, cadmium chloride (Fluka) was added to liquid cultures at 1 µM and Isopropyl β-D-1-thiogalactopyranoside (IPTG, NZYtech) added at 100 µM. For the JE2_smcSTOP+ conditional mutant, the strain was always grown in the presence of 50 ng mL−1 of anhydrotetracycline (ATC, Sigma-Aldrich), except for imaging under depletion conditions, in which case both the overnight precultures and the diluted exponential cultures were not supplemented with ATC.
Plasmid and strain construction
The complete lists of strains, plasmids and oligonucleotides are in Supplementary Tables 1, 2, and 3, respectively. Plasmids were assembled as described in Supplementary Table 2, propagated in E. coli DC10B and purified using the QIAprep Spin miniprep kit (Qiagen) and verified by sequencing. Purified plasmids were used to transform by electroporation S. aureus RN4220 cells as previously described139 and subsequently transduced into other S. aureus strains using the bacteriophage 80α140. S. aureus strain construction was done as indicated in Supplementary Table 1, using derivatives of the temperature-sensitive vector pMAD141, by performing allelic replacement through double homologous recombination, creating marker-less strains. Allelic replacement was confirmed by colony polymerase chain reaction (PCR).
Molecular biology methods
Amplification of DNA fragments for plasmid construction was carried out using a Phusion high-fidelity polymerase kit (Thermo Scientific) following the manufacturer instructions. For PCR using as a template a S. aureus bacterial colony, a small portion of the colony was resuspended in phosphate buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4) and cells were disrupted mechanically (by adding glass sand and three cycles of shaking for 45 s at a speed of 6.5 m s−1 in a FastPrep-24, MP Biomedicals) or enzymatically (by incubation at 37 °C for 1 h in the presence of 10 µg mL−1 of lysostaphin, Sigma-Aldrich, L9043) and the lysate was used as PCR template. For the PCR reaction, the Phire Hot Start II PCR Master Mix (Thermo Scientific) was employed following the manufacturer instructions.
Cloning was performed using restriction enzymes (FastDigest, Thermo Scientific) indicated, for each construct, in Supplementary Table 2. Fragments were ligated using T4 DNA ligase (Thermo Scientific). For Gibson Assembly, the Gibson Assembly Master Mix (NEB) was employed.
Microscopy
S. aureus strains were streaked from cryo-stocks onto TSA plates. Single colonies were used to inoculate independent cultures in TSB that were grown overnight at 37 °C with agitation, or for 12 h for experiments requiring stationary phase cells. The next day, the cultures were diluted 1:200 in TSB or M9 minimal medium and grown at 37 °C (or 25 °C for Fig. S8) with agitation until they reached mid-exponential phase (OD600 0.6–0.8). Fluorescent dyes for membrane labelling (FM4-64, 5 µg mL−1, Invitrogen; CellBrite Fix 640 3.3 nM, Biotium), DNA labelling (Hoechst 33342, 1 µg mL−1, Invitrogen) or HaloTag (HT) labelling (Janelia Fluor 549 HT ligand, 500 nM, Janelia Research Campus) were added when required to 1 mL of the exponential growth culture, which was then incubated for 20 minutes at 37 °C with agitation. Afterwards, the culture was centrifuged at 10000 ×g for one minute, the pellet was resuspended in 50 µL of PBS and 1 µL of the suspension was spotted on a pad of 1.2% Topvision Agarose (Thermo Fisher) in PBS.
For time-lapse microscopy, the aforementioned procedure was followed, but cells were spotted on pads of 1.2% Topvision Agarose (Thermo Fisher Scientific) in M9 minimal medium. The cells were kept at 37 °C during imaging and were imaged every three minutes.
Imaging was performed in a DeltaVision OMX SR microscope equipped with a hardware-based focus stability (HW UltimateFocus) and an environmental control module (set to 37° for time-lapses). Z-stacks of three epifluorescence images with a step size of 500 nm were acquired using a 405 nm laser (100 mW, at 10% maximal power; for the Hoechst 33342 DNA dye), a 488 nm laser (100 mW, at 15% maximal power for the mNeonGreen fusions), a 568 nm laser (100 mW, at 30% maximal power; for JF549-labelled DnaN-Halo and FM4-64 membrane dye) or a 640 nm laser (100 mW, at 40% maximal power; for the CellBrite Fix 640 dye), each with an exposure time of 100 ms. When required, a maximum intensity projection of the three images from each z-stack, fluorescence channel alignment and Structured Imaging Microscopy (SIM) image reconstruction was performed using SoftWoRx v7.2.1.
For cell cycle automated classification (Fig. 2E), quantification of anucleate cells (Fig. S11A), ParB imaging (Fig. S7) and the dual labeling FROS system (Fig. S2A), cells were imaged in a Zeiss Axio Observer microscope equipped with a Plan-Apochromat 100×/1.4 oil Ph3 objective, a Retiga R1 CCD camera (QImaging), a white-light source HXP 120 V (Zeiss) and the software ZEN blue v2.0.0.0 (Zeiss). For image acquisition, the filters (Semrock USA) Brightline TXRED-4040B (FM4-64), Brightline GFP-3035B (mNeonGreen), Brightline YFP-2427B (YFP), Brightline CFP-2432C (CFP) and Brightline DAPI-1160A (Hoechst 33342) were used.
Image processing and automated analysis
Images were examined using ImageJ Fiji142, which was also used to produce crops of illustrative regions. Lateral drift in time-lapse datasets was corrected with the ImageJ plugin NanoJ143.
For cell cycle automated classification and generation of foci average heatmaps from images obtained using the Zeiss Axio Observer microscope (Fig. 2E and S7), crops of single cells and automated cell cycle phase analysis were generated using eHooke software version 1.1, as previously described in ref. 89.
For foci quantification and generation of foci average heatmaps from images acquired in the OMX microscope (Figs. 2C, 2D, 4A, 4B, 5A, 5B, 5C, S8, S9), cell segmentation was performed using an in-house fine-tuned StarDist model144 applied on images with fluorescence signal from membrane labelling. When mentioned, the cell cycle phase analysis was performed manually (Fig. 2D, S8B).
After cell segmentation (and cell cycle classification if required) we used a PCA transform applied to the coordinates of the pixels that constitute the cell outline to calculate the orientation of the major axis of each cell. Then cell crops were aligned by their major axes as previously described120. Foci localization was determined using TrackMate 7.11.190 using the Laplacian of Gaussian filter with subpixel localization. The blob diameter was set to 0.24 μm, the quality threshold was manually adjusted for each field of view, and the results were exported as an .xml file. In each cell crop, foci were represented as a circle of 1 pixel radius, intensity 1, and the same relative coordinates as the foci, in a rectangle with the same dimensions as the cell crop (model image), with background set to 0. All model images were then resized to a common width and height equal to the median of the width and height of all cell crops. Heatmaps were generated by averaging all model images, and colored using the coolwarm colormap provided by matplotlib145.
High-throughput Chromosome Conformation Capture (Hi-C)
The Hi-C procedure used for S. aureus was adapted from a previously described protocol used for B. subtilis103,146. Briefly, S. aureus strains were streaked from cryo-stocks onto TSA plates. Single colonies were used to inoculate independent cultures in TSB that were grown overnight at 37 °C with agitation, in duplicate. The next day, the cultures were diluted 1:1000 in TSB and grown at 37 °C with agitation until they reached early-exponential phase (OD600 0.3-0.4). Cells were crosslinked by adding formaldehyde (Sigma-Aldrich) to a final concentration of 7% at room temperature (RT) for 30 min and quenched with 125 mM glycine (Sigma-Aldrich). Cells were lysed using Ready-Lyse Lysozyme (Epicentre, R1802M) and 200 µg mL−1 lysostaphin (Sigma-Aldrich, L9043) at RT for 1 h, followed by the treatment with 1% SDS (v/v) at RT for 30 min. Solubilized chromatin was digested with DpnII for 2 h at 37 °C. The digested ends were filled in with Klenow and Biotin-14-dATP, dGTP, dCTP, dTTP. The products were ligated with T4 DNA ligase at 16 °C for about 20 h. Crosslinks were reversed at 65 ˚C for 17–20 h in the presence of EDTA, proteinase K and 0.5% SDS. The DNA was then extracted twice with phenol/chloroform/isoamylalcohol (25:24:1) (PCI), precipitated with ethanol, and resuspended in 20 µL of 0.1X TE buffer (10 mM Tris-HCl, 1 mM EDTA). Biotin from non-ligated ends was removed using T4 polymerase (4 h at 20 °C) followed by extraction with PCI. The DNA was then sheared by sonication for 12 min with 20% amplitude using a Qsonica Q800R2 water bath sonicator. The sheared DNA was used for library preparation with the NEBNext UltraII kit (E7645). Biotinylated DNA fragments were purified using 5 µL streptavidin beads. DNA-bound beads were used for PCR in a 50 µL reaction for 14 cycles. PCR products were purified using Ampure beads (Beckman, A63881) and sequenced at the Indiana University Center for Genomics and Bioinformatics using NextSeq500. Paired-end sequencing reads were mapped to the genome of S. aureus JE2 (NCBI Reference Sequence GCF_002085525.1) using the same pipeline described previously103. The genome was divided into 5-kb bins. Subsequent analysis and visualization were done using R scripts. Hi-C scores, which quantify the interaction between loci and correct for biases in the abundance of the different bins in each experiment, were calculated as described in ref.103.
Whole Genome Sequencing (WGS) and Marker Frequency Analysis (MFA)
For genomic DNA extraction, cells from the relevant strains were grown overnight in TSB at 37 °C or 25 °C with agitation, in duplicate. The next day the cultures were diluted 1:1000 in 50 mL of TSB and grown at 37 °C or 25 °C with agitation until the early-exponential phase (OD600 0.3–0.4). Then cultures were centrifuged at 6000 × g for 10 min and the supernatants were discarded. Cells were resuspended in 180 µL of Enzymatic lysis buffer (20 mM TRIS, VWR; 2 mM sodium EDTA; 1.2% (v/v) Triton X-100, Sigma, adjusted to pH 8 using HCl, Sigma-Aldrich), supplemented with 100 µg mL−1 of lysostaphin (Sigma-Aldrich, L9043) and were incubated at 37 °C for 15 min. Afterwards, the samples were processed using the DNeasy Blood & Tissue Kit (Quiagen) following the indications from the manufacturer. The extracted DNA was sonicated using a Qsonica Q800R2 water bath sonicator, prepared using the NEBNext UltraII kit (E7645), and sequenced at the Indiana University Center for Genomics and Bioinformatics using NextSeq2000. The reads were mapped to the genome of S. aureus JE2 (NCBI Reference Sequence GCF_002085525.1) using CLC Genomics Workbench (CLC Bio, QIAGEN). The mapped reads were normalized by the total number of reads. Plotting and analysis were performed using R scripts. MFA plots show the genome-wide DNA replication profile and examine the relative abundance of specific DNA sequences across the genome85.
Chromatin ImmunoPrecipitation sequencing (ChIP-seq)
The S. aureus strains JE2_ParB-3xFLAG and JE2_3xFLAG-mNG were grown overnight in TSB at 37 °C with agitation. The next day the cultures were diluted 1:200 in 50 mL of TSB and incubated at 37 °C with agitation until they reached the mid-exponential phase (OD600 0.6–0.8). Cultures of the strain JE2_3xFLAG-mNG were supplemented with 100 µM IPTG to induce expression of 3xflag-mng. Formaldehyde (Sigma-Aldrich) was added to a final concentration of 1% (v/v) and the mixture was incubated at room-temperature with shaking for 30 min. Afterwards, glycine was added to a final concentration of 125 mM and cultures were further incubated at room-temperature with shaking for 10 min. The mixture was cooled down on ice and centrifuged at 7000 × g for 10 min at 4 °C. The pellet was resuspended in ice-cold PBS and centrifuged again as in the previous step. This was repeated three times, before snap-freezing the pellet in liquid nitrogen and storing it at −80 °C. When required, samples were thawed, resuspended in 300 µl IP buffer (50 mM Tris/HCl pH 7.5, 5 mM EDTA, 150 mM NaCl, 25 mM sucrose, 1 µg mL−1 lysostaphin (Sigma-Aldrich, L9043), 0.3 µg mL−1 RNase A (Sigma-Aldrich) and 1 tablet of cOmplete protease inhibitor cocktail, EDTA free, Roche, per 10 mL of buffer) and incubated at 37 °C for 1 h with shaking. Afterwards the samples were cooled on ice, followed by addition of TritonX-100 to a final concentration of 1% (v/v). Samples were then sonicated using a Bioruptor Plus bath sonicator at 4 °C using 50 cycles of alternating 30 s on and 90 s off in the high-power mode, followed by centrifugation at 20000 × g for 10 min at 4 °C. The supernatants were mixed with 50 µL of anti-Flag M2 agarose beads (Sigma, pre-washed in 1 mL of IP buffer supplemented with 1% Triton X-100), and the mixture was incubated overnight at 4 °C with tumbling. Afterwards, the IP-samples were centrifuged at 800 × g for 2 min at 4 °C, the supernatant was discarded, and the beads were resuspended in 1 mL of IP buffer with 1% Triton X-100. The IP-samples were centrifuged as in the previous step and resuspended in 1 mL of High-Salt Buffer (50 mM Tris/HCl pH 7.5, 5 mM EDTA, 700 mM NaCl, 0.1% Na-deoxycholate, Calbiochem, 1% Triton X-100). IP-samples were centrifuged as in the previous step and resuspended in 1 mM of TE buffer (10 mM Tris/HCl pH 8, 1 mM EDTA), this step was done twice. Then, the IP-samples were centrifuged and resuspended in 300 µL of Reversal Buffer (RB, 10 mM Tris/HCl pH 8, 1 mM EDTA, 300 mM NaCl), followed by addition of SDS to a final concentration of 1% (w/v). All samples were then incubated at 65 °C with 1500 rpm shaking for 14–16 h. Afterwards, the IP-samples were centrifuged at 800 × g for 2 min and the supernatant was transferred to a new tube. Then 300 µL of phenol-chloroform-isoamyl alcohol mix (Roth) were added to each sample, the mixture was vigorously mixed by vortexing for 10 s and centrifuged at 20000 × g for 5 min at RT. 250 µL were taken from the aqueous phase and transferred to a new tube where they were combined with 25 μL of 3 M sodium-acetate (pH 5.2), 1.5 μL of 20 mg mL−1 glycogen and 690 μl of absolute ethanol. The samples were placed at −80 °C for 1 h and then centrifuged at 20000 × g for 15 min at RT. The supernatant was discarded and the pellet was washed with 1 mL of ice-cold 70% (v/v) ethanol. Samples were centrifuged at 20000 × g for 1 min at RT and the supernatant was discarded. The pellet was left to air-dry. Afterwards the pellet was resuspended in 25 μl of Nuclease-free water and incubated at 55 °C for 10 min with gentle shaking. Samples were sent to Lausanne Genomic Technologies Facility for next-generation sequencing. Sequencing results were assembled to the S. aureus JE2 reference genome (NCBI Reference Sequence GCF_002085525.1) using the CLC workbench (Qiagen) and plotted using Microsoft Excel.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Hi-C and WGS data were deposited to the NCBI Gene Expression Omnibus (accession no. GSE286260). ChIP-seq data was also deposited in the NCBI Gene Expression Omnibus (accession no. GSE286023). Source data are provided with this paper.
Code availability
The codes used to create average localization heatmaps were deposited to github (https://github.com/BacterialCellBiologyLab/AverageCellLoc/releases/tag/1.0.0).
References
Gogou, C., Japaridze, A. & Dekker, C. Mechanisms for chromosome segregation in bacteria. Front. Microbiol. 12, 685687 (2021).
X. Wang, P. M. Llopis, D. Z. Rudner, Organization and segregation of bacterial chromosomes. Nat. Rev. Genet.14, https://doi.org/10.1038/nrg3375 (2013).
Viollier, P. H. et al. Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication. Proc. Natl. Acad. Sci. 101, 9257–9262 (2004).
Harms, A., Treuner-Lange, A., Schumacher, D. & Søgaard-Andersen, L. Tracking of chromosome and replisome dynamics in Myxococcus xanthus reveals a novel chromosome arrangement. PLOS Genet. 9, e1003802 (2013).
David, A. et al. The two cis-acting sites, parS1 and oriC1, contribute to the longitudinal organisation of Vibrio cholerae chromosome I. PLOS Genet. 10, e1004448 (2014).
Wang, X., Montero Llopis, P. & Rudner, D. Z. Bacillus subtilis chromosome organization oscillates between two distinct patterns. Proc. Natl Acad. Sci. 111, 12877–12882 (2014).
Webb, C. D. et al. Bipolar localization of the replication origin regions of chromosomes in vegetative and sporulating cells of Bacillus subtilis. Cell 88, 667–674 (1997).
Böhm K. et al. Novel chromosome organization pattern in Actinomycetales-overlapping replication cycles combined with diploidy. mBio 8, e00511-17 (2017).
Nielsen, H. J., Ottesen, J. R., Youngren, B., Austin, S. J. & Hansen, F. G. The Escherichia coli chromosome is organized with the left and right chromosome arms in separate cell halves. Mol. Microbiol. 62, 331–338 (2006).
Wang, X., Liu, X., Possoz, C. & Sherratt, D. J. The two Escherichia coli chromosome arms locate to separate cell halves. Genes Dev. 20, 1727–1731 (2006).
Lau, I. F. et al. Spatial and temporal organization of replicating Escherichia coli chromosomes. Mol. Microbiol. 49, 731–743 (2003).
Gras, K., Fange, D. & Elf, J. The Escherichia coli chromosome moves to the replisome. Nat. Commun. 15, 6018 (2024).
van Raaphorst, R., Kjos, M. & Veening, J.-W. Chromosome segregation drives division site selection in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. 114, E5959–E5968 (2017).
Santi, I. & McKinney, J. D. Chromosome organization and replisome dynamics in Mycobacterium smegmatis. mBio 6, e01999–14 (2015).
Yoshikawa, H., O’Sullivan, A. & Sueoka, N. Sequential replication of the Bacillus subtilis chromosome, III. regulation of initiation. Proc. Natl Acad. Sci. 52, 973–980 (1964).
Böttinger, B., Semmler, F., Zerulla, K., Ludt, K. & Soppa, J. Regulated ploidy of Bacillus subtilis and three new isolates of Bacillus and Paenibacillus. FEMS Microbiol. Lett. 365, https://doi.org/10.1093/femsle/fnx282 (2018).
Cooper, S. & Helmstetter, C. E. Chromosome replication and the division cycle of Escherichia coli B/r. J. Mol. Biol. 31, 519–540 (1968).
Nielsen, H. J., Youngren, B., Hansen, F. G. & Austin, S. Dynamics of Escherichia coli chromosome segregation during multifork replication. J. Bacteriol. 189, 8660–8666 (2007).
Trojanowski, D., Hołówka, J., Ginda, K., Jakimowicz, D. & Zakrzewska-Czerwińska, J. Multifork chromosome replication in slow-growing bacteria. Sci. Rep. 7, 43836 (2017).
Lin, D. C. & Grossman, A. D. Identification and characterization of a bacterial chromosome partitioning site. Cell 92, 675–685 (1998).
Toro, E., Hong, S.-H., McAdams, H. H. & Shapiro, L. Caulobacter requires a dedicated mechanism to initiate chromosome segregation. Proc. Natl. Acad. Sci. 105, 15435–15440 (2008).
Kadoya, R., Baek, J. H., Sarker, A. & Chattoraj, D. K. Participation of chromosome segregation protein ParAI of Vibrio cholerae in chromosome replication. J. Bacteriol. 193, 1504–1514 (2011).
Iniesta, A. A. ParABS system in chromosome partitioning in the bacterium Myxococcus xanthus. PLOS ONE 9, e86897 (2014).
Vallet-Gely, I. & Boccard, F. Chromosomal organization and segregation in Pseudomonas aeruginosa. PLoS Genet. 9, e1003492 (2013).
Lagage, V., Boccard, F. & Vallet-Gely, I. Regional control of chromosome segregation in Pseudomonas aeruginosa. PLOS Genet. 12, e1006428 (2016).
Jalal, A. S. B. & Le, T. B. K. Bacterial chromosome segregation by the ParABS system. Open Biol. 10, 200097 (2020).
Soh, Y.-M. et al. Self-organization of parS centromeres by the ParB CTP hydrolase. Science 366, 1129–1133 (2019).
Osorio-Valeriano, M. et al. The CTPase activity of ParB determines the size and dynamics of prokaryotic DNA partition complexes. Mol. Cell 81, 3992–4007.e10 (2021).
Osorio-Valeriano, M. et al. ParB-type DNA segregation proteins are CTP-dependent molecular switches. Cell 179, 1512–1524.e15 (2019).
Jalal, A. S., Tran, N. T. & Le, T. B. ParB spreading on DNA requires cytidine triphosphate in vitro. eLife 9, e53515 (2020).
Guilhas, B. et al. ATP-driven separation of liquid phase condensates in bacteria. Mol. Cell 79, 293–303.e4 (2020).
Tišma, M. et al. ParB proteins can bypass DNA-bound roadblocks via dimer-dimer recruitment. Sci. Adv. 8, eabn3299 (2022).
Livny, J., Yamaichi, Y. & Waldor, M. K. Distribution of centromere-like parS sites in Bacteria: insights from comparative genomics. J. Bacteriol. 189, 8693–8703 (2007).
Gruber, S. & Errington, J. Recruitment of condensin to replication origin regions by ParB/SpoOJ promotes chromosome segregation in B. subtilis. Cell 137, 685–696 (2009).
Sullivan, N. L., Marquis, K. A. & Rudner, D. Z. Recruitment of SMC by ParB-parS organizes the origin region and promotes efficient chromosome segregation. Cell 137, 697–707 (2009).
Tran, N. T., Laub, M. T. & Le, T. B. K. SMC progressively aligns chromosomal arms in Caulobacter crescentus but is antagonized by convergent transcription. Cell Rep. 20, 2057–2071 (2017).
Bock, F. P., Liu, H. W., Anchimiuk, A., Diebold-Durand, M.-L. & Gruber, S. A joint-ParB interface promotes SMC DNA recruitment. Cell Rep. 40, 111273 (2022).
Gruber, S. et al. Interlinked sister chromosomes arise in the absence of condensin during fast replication in B. subtilis. Curr. Biol. 24, 293–298 (2014).
Wang, X., Brandão, H. B., Le, T. B. K., Laub, M. T. & Rudner, D. Z. Bacillus subtilis SMC complexes juxtapose chromosome arms as they travel from origin to terminus. Science 355, 524–527 (2017).
Wang, X., Tang, O. W., Riley, E. P. & Rudner, D. Z. The SMC condensin complex is required for origin segregation in Bacillus subtilis. Curr. Biol. 24, 287–292 (2014).
Nunez, R. V., Polyhach, Y., Soh, Y.-M., Jeschke, G. & Gruber, S. Gradual opening of SMC arms in prokaryotic condensin. Cell Rep. 35, 109051 (2021).
Lee, B.-G. et al. Cryo-EM structures of holo condensin reveal a subunit flip-flop mechanism. Nat. Struct. Mol. Biol. 27, 743–751 (2020).
Bürmann, F., Funke, L. F. H., Chin, J. W. & Löwe, J. Cryo-EM structure of MukBEF reveals DNA loop entrapment at chromosomal unloading sites. Mol. Cell 81, 4891–4906.e8 (2021).
Krepel, D., Cheng, R. R., Di Pierro, M. & Onuchic, J. N. Deciphering the structure of the condensin protein complex. Proc. Natl. Acad. Sci. 115, 11911–11916 (2018).
Wilhelm, L. et al. SMC condensin entraps chromosomal DNA by an ATP hydrolysis dependent loading mechanism in Bacillus subtilis. eLife 4, e06659 (2015).
Böhm, K. et al. Chromosome organization by a conserved condensin-ParB system in the actinobacterium Corynebacterium glutamicum. Nat. Commun. 11, 1485 (2020).
Lioy, V. S., Junier, I., Lagage, V., Vallet, I. & Boccard, F. Distinct activities of bacterial condensins for chromosome management in Pseudomonas aeruginosa. Cell Rep. 33, 108344 (2020).
Schwartz, M. A. & Shapiro, L. An SMC ATPase mutant disrupts chromosome segregation in Caulobacter. Mol. Microbiol. 82, 1359–1374 (2011).
Mazzuoli, M.-V. et al. HU promotes higher order chromosome organization and influences DNA replication rates in Streptococcus pneumoniae. Nucleic Acids Res. 53, gkaf312 (2025).
Anand, D., Schumacher, D. & Søgaard-Andersen, L. SMC and the bactofilin/PadC scaffold have distinct yet redundant functions in chromosome segregation and organization in Myxococcus xanthus. Mol. Microbiol. 114, 839–856 (2020).
Donovan, C., Schwaiger, A., Krämer, R. & Bramkamp, M. Subcellular localization and characterization of the ParAB system from Corynebacterium glutamicum. J. Bacteriol. 192, 3441–3451 (2010).
Güthlein, C., Wanner, R. M., Sander, P., Böttger, E. C. & Springer, B. A mycobacterial smc null mutant is proficient in DNA repair and long-term survival. J. Bacteriol. 190, 452–456 (2008).
Jakimowicz, D. et al. Characterization of the mycobacterial chromosome segregation protein ParB and identification of its target in Mycobacterium smegmatis. Microbiology 153, 4050–4060 (2007).
Jecz, P., Bartosik, A. A., Glabski, K. & Jagura-Burdzy, G. A single parS sequence from the cluster of four sites closest to oriC is necessary and sufficient for proper chromosome segregation in Pseudomonas aeruginosa. PloS One 10, e0120867 (2015).
Jensen, R. B. & Shapiro, L. The Caulobacter crescentus smc gene is required for cell cycle progression and chromosome segregation. Proc. Natl. Acad. Sci. 96, 10661–10666 (1999).
Jung, A. et al. Two-step chromosome segregation in the stalked budding bacterium Hyphomonas neptunium. Nat. Commun. 10, 3290 (2019).
Minnen, A., Attaiech, L., Thon, M., Gruber, S. & Veening, J.-W. SMC is recruited to oriC by ParB and promotes chromosome segregation in Streptococcus pneumoniae. Mol. Microbiol. 81, 676–688 (2011).
Jun, S. & Wright, A. Entropy as the driver of chromosome segregation. Nat. Rev. Microbiol. 8, 600–607 (2010).
Bernhardt, T. G. & de Boer, P. A. J. SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over chromosomes in E. coli. Mol. Cell 18, 555–564 (2005).
Thanbichler, M. & Shapiro, L. MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell 126, 147–162 (2006).
Veiga, H., Jorge, A. M. & Pinho, M. G. Absence of nucleoid occlusion effector Noc impairs formation of orthogonal FtsZ rings during Staphylococcus aureus cell division. Mol. Microbiol. 80, 1366–1380 (2011).
Wu, L. J. et al. Noc protein binds to specific DNA sequences to coordinate cell division with chromosome segregation. EMBO J. 28, 1940–1952 (2009).
Wu, L. J. & Errington, J. Coordination of cell division and chromosome segregation by a nucleoid occlusion protein in Bacillus subtilis. Cell 117, 915–925 (2004).
Ramamurthi, K. S., Lecuyer, S., Stone, H. A. & Losick, R. Geometric cue for protein localization in a bacterium. Science 323, 1354–1357 (2009).
Shapiro, L., McAdams, H. H. & Losick, R. Why and how bacteria localize proteins. Science 326, 1225–1228 (2009).
Ramamurthi, K. S. & Losick, R. Negative membrane curvature as a cue for subcellular localization of a bacterial protein. Proc. Natl Acad. Sci. 106, 13541–13545 (2009).
Varma, A., Huang, K. C. & Young, K. D. The Min system as a general cell geometry detection mechanism: branch lengths in Y-shaped Escherichia coli cells affect Min oscillation patterns and division dynamics. J. Bacteriol. 190, 2106–2117 (2008).
de Boer, P. A., Crossley, R. E. & Rothfield, L. I. A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell 56, 641–649 (1989).
Hussain, S. et al. MreB filaments align along greatest principal membrane curvature to orient cell wall synthesis. eLife 7, e32471 (2018).
Taylor, J. A. et al. Distinct cytoskeletal proteins define zones of enhanced cell wall synthesis in Helicobacter pylori. eLife 9, e52482 (2020).
Ursell, T. S. et al. Rod-like bacterial shape is maintained by feedback between cell curvature and cytoskeletal localization. Proc. Natl. Acad. Sci. 111, E1025–E1034 (2014).
Lee, A. S. et al. Methicillin-resistant Staphylococcus aureus. Nat. Rev. Dis. Prim. 4, 1–23 (2018).
Antimicrobial Resistance Collaborators Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399, 629–655 (2022).
Monteiro, J. M. et al. Cell shape dynamics during the staphylococcal cell cycle. Nat. Commun. 6, 8055 (2015).
Pinho, M. G. & Foster, S. J. Cell growth and division of Staphylococcus aureus. Annu. Rev. Microbiol. 78, 293–310 (2024).
Barbuti, M. D., Myrbråten, I. S., Morales Angeles, D. & Kjos, M. The cell cycle of Staphylococcus aureus: an updated review. MicrobiologyOpen 12, e1338 (2023).
Saraiva, B. M. et al. Reassessment of the distinctive geometry of Staphylococcus aureus cell division. Nat. Commun. 11, 4097 (2020).
Chan, H., Söderström, B. & Skoglund, U. Spo0J and SMC are required for normal chromosome segregation in Staphylococcus aureus. MicrobiologyOpen 9, e999 (2020).
Pinho, M. G. & Errington, J. A divIVA null mutant of Staphylococcus aureus undergoes normal cell division. FEMS Microbiol. Lett. 240, 145–149 (2004).
Charpentier, E. et al. Novel cassette-based shuttle vector system for Gram-positive bacteria. Appl. Environ. Microbiol. 70, 6076–6085 (2004).
Fey, P. D. et al. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. mBio 4, https://doi.org/10.1128/mbio.00537-12 (2013).
Shaner, N. C. et al. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat. Methods 10, 407–409 (2013).
Payne, B. T. I. et al. Replication fork blockage by transcription factor-DNA complexes in Escherichia coli. Nucleic Acids Res. 34, 5194–5202 (2006).
Possoz, C., Filipe, S. R., Grainge, I. & Sherratt, D. J. Tracking of controlled Escherichia coli replication fork stalling and restart at repressor-bound DNA in vivo. EMBO J. 25, 2596–2604 (2006).
Huang, D. et al. The in vivo measurement of replication fork velocity and pausing by lag-time analysis. Nat. Commun. 14, 1762 (2023).
Pang, T., Wang, X., Lim, H. C., Bernhardt, T. G. & Rudner, D. Z. The nucleoid occlusion factor Noc controls DNA replication initiation in Staphylococcus aureus. PLOS Genet. 13, e1006908 (2017).
Los, G. V. et al. HaloTag: a novel protein labeling technology for cell imaging and protein analysis. ACS Chem. Biol. 3, 373–382 (2008).
Chen, P. J. et al. Interdependent progression of bidirectional sister replisomes in E. coli. eLife 12, e82241 (2023).
Saraiva, B. M., Krippahl, L., Filipe, S. R., Henriques, R. & Pinho, M. G. eHooke: A tool for automated image analysis of spherical bacteria based on cell cycle progression. Biol. Imaging 1, e3 (2021).
Tinevez, J.-Y. et al. TrackMate: An open and extensible platform for single-particle tracking. Methods 115, 80–90 (2017).
Lewis, P. J. & Errington, J. Direct evidence for active segregation of oriC regions of the Bacillus subtilis chromosome and co-localization with the SpoOJ partitioning protein. Mol. Microbiol. 25, 945–954 (1997).
Gallay, C. et al. CcrZ is a pneumococcal spatiotemporal cell cycle regulator that interacts with FtsZ and controls DNA replication by modulating the activity of DnaA. Nat. Microbiol 6, 1175–1187 (2021).
Guo, M. S., Haakonsen, D. L., Zeng, W., Schumacher, M. A. & Laub, M. T. A bacterial chromosome structuring protein binds overtwisted DNA to stimulate type II topoisomerases and enable DNA replication. Cell 175, 583–597.e23 (2018).
Ren, Z. et al. Conformation and dynamic interactions of the multipartite genome in Agrobacterium tumefaciens. Proc. Natl Acad. Sci. USA 119, e2115854119 (2022).
L. Fernández-Coll, et al. The absence of (p)ppGpp renders initiation of Escherichia coli chromosomal DNA synthesis independent of growth rates. mBio 11, https://doi.org/10.1128/mbio.03223-19 (2020).
Fan, J. et al. RNA polymerase redistribution supports growth in E. coli strains with a minimal number of rRNA operons. Nucleic Acids Res. 51, 8085–8101 (2023).
Murray, H. & Koh, A. Multiple regulatory systems coordinate DNA replication with cell growth in Bacillus subtilis. PLOS Genet. 10, e1004731 (2014).
Ghosh, S., Samaddar, S., Kirtania, P. & Das Gupta, S. K. A DinB ortholog enables mycobacterial growth under dTTP-limiting conditions induced by the expression of a mycobacteriophage-derived ribonucleotide reductase gene. J. Bacteriol. 198, 352–362 (2015).
Breier, A. M. & Grossman, A. D. Whole-genome analysis of the chromosome partitioning and sporulation protein Spo0J (ParB) reveals spreading and origin-distal sites on the Bacillus subtilis chromosome. Mol. Microbiol. 64, 703–718 (2007).
Tran, N. T. et al. Permissive zones for the centromere-binding protein ParB on the Caulobacter crescentus chromosome. Nucleic Acids Res. 46, 1196–1209 (2018).
de Balaguer, F. A. et al. CTP promotes efficient ParB-dependent DNA condensation by facilitating one-dimensional diffusion from parS. Elife 10, e67554 (2021).
Le, T. B. K., Imakaev, M. V., Mirny, L. A. & Laub, M. T. High-resolution mapping of the spatial organization of a bacterial chromosome. Science 342, 731–734 (2013).
Wang, X. et al. Condensin promotes the juxtaposition of DNA flanking its loading site in Bacillus subtilis. Genes Dev. 29, 1661–1675 (2015).
Lioy, V. S. et al. Multiscale structuring of the E. coli chromosome by nucleoid-associated and condensin proteins. Cell 172, 771–783.e18 (2018).
Marbouty, M. et al. Condensin- and replication-mediated bacterial chromosome folding and origin condensation revealed by Hi-C and super-resolution imaging. Mol. Cell 59, 588–602 (2015).
Karaboja, X. et al. XerD unloads bacterial SMC complexes at the replication terminus. Mol. Cell 81, 756–766.e8 (2021).
Umbarger, M. A. et al. The three-dimensional architecture of a bacterial genome and its alteration by genetic perturbation. Mol. Cell 44, 252–264 (2011).
Anchimiuk, A. et al. A low SMC flux avoids collisions and facilitates chromosome organization in Bacillus subtilis. eLife 10, e65467 (2021).
Brandão, H. B., Ren, Z., Karaboja, X., Mirny, L. A. & Wang, X. DNA-loop-extruding SMC complexes can traverse one another in vivo. Nat. Struct. Mol. Biol. 28, 642–651 (2021).
Soppa, J. et al. Discovery of two novel families of proteins that are proposed to interact with prokaryotic SMC proteins, and characterization of the Bacillus subtilis family members ScpA and ScpB. Mol. Microbiol. 45, 59–71 (2002).
Naumova, N. et al. Organization of the mitotic chromosome. Science 342, 948–953 (2013).
Chaudhuri, R. R. et al. Comprehensive identification of essential Staphylococcus aureus genes using Transposon-Mediated Differential Hybridisation (TMDH). BMC Genomics 10, 291 (2009).
Bottomley, A. L. et al. Coordination of chromosome segregation and cell division in Staphylococcus aureus. Front. Microbiol. 8, 1575 (2017).
Yu, W., Herbert, S., Graumann, P. L. & Götz, F. Contribution of SMC (Structural Maintenance of Chromosomes) and SpoIIIE to chromosome segregation in Staphylococci. J. Bacteriol. 192, 4067–4073 (2010).
Ramos-León, F. et al. PcdA promotes orthogonal division plane selection in Staphylococcus aureus. Nat. Microbiol. 9, 2997–3012 (2024).
Bigot, S., Sivanathan, V., Possoz, C., Barre, F.-X. & Cornet, F. FtsK, a literate chromosome segregation machine. Mol. Microbiol. 64, 1434–1441 (2007).
Chan, H., Mohamed, A. M. T., Grainge, I. & Rodrigues, C. D. A. FtsK and SpoIIIE, coordinators of chromosome segregation and envelope remodeling in bacteria. Trends Microbiol. 30, 480–494 (2022).
Aussel, L. et al. FtsK Is a DNA motor protein that activates chromosome dimer resolution by switching the catalytic state of the XerC and XerD recombinases. Cell 108, 195–205 (2002).
Veiga, H. & Pinho, M. G. Staphylococcus aureus requires at least one FtsK/SpoIIIE protein for correct chromosome segregation. Mol. Microbiol. 103, 504–517 (2017).
Veiga, H. et al. Cell division protein FtsK coordinates bacterial chromosome segregation and daughter cell separation in Staphylococcus aureus. EMBO J. 42, e112140 (2023).
Lim, H. C. et al. Evidence for a DNA-relay mechanism in ParABS-mediated chromosome segregation. eLife 3, e02758 (2014).
Hu, L., Vecchiarelli, A. G., Mizuuchi, K., Neuman, K. C. & Liu, J. Brownian ratchet mechanism for faithful segregation of low-copy-number plasmids. Biophys. J. 112, 1489–1502 (2017).
Surovtsev, I. V., Campos, M. & Jacobs-Wagner, C. DNA-relay mechanism is sufficient to explain ParA-dependent intracellular transport and patterning of single and multiple cargos. Proc. Natl Acad. Sci. 113, E7268–E7276 (2016).
Surovtsev, I. V., Lim, H. C. & Jacobs-Wagner, C. The slow mobility of the ParA partitioning protein underlies its steady-state patterning in Caulobacter. Biophys. J. 110, 2790–2799 (2016).
Mercy, C. et al. RocS drives chromosome segregation and nucleoid protection in Streptococcus pneumoniae. Nat. Microbiol. 4, 1661–1670 (2019).
Schumacher, M. A., Lee, J. & Zeng, W. Molecular insights into DNA binding and anchoring by the Bacillus subtilis sporulation kinetochore-like RacA protein. Nucleic Acids Res. 44, 5438–5449 (2016).
Ben-Yehuda, S. et al. Defining a centromere-like element in Bacillus subtilis by identifying the binding sites for the chromosome-anchoring protein RacA. Mol. Cell 17, 773–782 (2005).
Ben-Yehuda, S., Rudner, D. Z. & Losick, R. RacA, a bacterial protein that anchors chromosomes to the cell poles. Science 299, 532–536 (2003).
Kjos, M. & Veening, J.-W. Tracking of chromosome dynamics in live Streptococcus pneumoniae reveals that transcription promotes chromosome segregation. Mol. Microbiol. 91, 1088–1105 (2014).
Micka, B., Groch, N., Heinemann, U. & Marahiel, M. A. Molecular cloning, nucleotide sequence, and characterization of the Bacillus subtilis gene encoding the DNA-binding protein HBsu. J. Bacteriol. 173, 3191–3198 (1991).
Ferrándiz, M.-J., Carreño, D., Ayora, S. & de la Campa, A. G. HU of Streptococcus pneumoniae is essential for the preservation of DNA supercoiling. Front. Microbiol. 9, 493 (2018).
Yao, J. et al. Use of targetrons to disrupt essential and nonessential genes in Staphylococcus aureus reveals temperature sensitivity of Ll.LtrB group II intron splicing. RNA 12, 1271–1281 (2006).
Sharkey, L. K. R. et al. The two-component system WalKR provides an essential link between cell wall homeostasis and DNA replication in Staphylococcus aureus. mBio 14, e0226223 (2023).
Willis, C., Errington, J. & Wu, L. J. Cohesion of sister chromosome termini during the early stages of sporulation in Bacillus subtilis. J. Bacteriol. 202, e00296–20 (2020).
Bogush, M., Xenopoulos, P. & Piggot, P. J. Separation of chromosome termini during sporulation of Bacillus subtilis depends on SpoIIIE. J. Bacteriol. 189, 3564–3572 (2007).
Keller, A. N. et al. Activation of Xer-recombination at dif: structural basis of the FtsKγ–XerD interaction. Sci. Rep. 6, 33357 (2016).
Prikryl, J., Hendricks, E. C. & Kuempel, P. L. DNA degradation in the terminus region of resolvase mutants of Escherichia coli, and suppression of this degradation and the Dif phenotype by recD. Biochimie 83, 171–176 (2001).
Sinha, A. K. et al. Division-induced DNA double strand breaks in the chromosome terminus region of Escherichia coli lacking RecBCD DNA repair enzyme. PLOS Genet. 13, e1006895 (2017).
Veiga, H. & Pinho, M. G. Inactivation of the SauI type I restriction-modification system is not sufficient to generate Staphylococcus aureus strains capable of efficiently accepting foreign DNA. Appl. Environ. Microbiol. 75, 3034–3038 (2009).
Oshida, T. & Tomasz, A. Isolation and characterization of a Tn551-autolysis mutant of Staphylococcus aureus. J. Bacteriol. 174, 4952–4959 (1992).
Arnaud, M., Chastanet, A. & Débarbouillé, M. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl. Environ. Microbiol. 70, 6887–6891 (2004).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Laine, R. F. et al. NanoJ: a high-performance open-source super-resolution microscopy toolbox. J. Phys. D. 52, 163001 (2019).
U. Schmidt, M. Weigert, C. Broaddus, G. Myers, “Cell detection with star-convex polygons” in Medical Image Computing and Computer Assisted Intervention – MICCAI 2018, A. F. Frangi, J. A. Schnabel, C. Davatzikos, C. Alberola-López, G. Fichtinger, Eds. (Springer International Publishing, Cham, 2018), pp. 265–273.
Hunter, J. D. Matplotlib: a 2D graphics environment. Comput. Sci. Eng. 9, 90–95 (2007).
Liao, Q. & Wang, X. Using chromosome conformation capture combined with deep sequencing (Hi-C) to study genome organization in bacteria. Methods Mol. Biol. 2866, 231–243 (2025).
Acknowledgements
We thank members of the Pinho lab, P. Pereira (ITQB-NOVA) and S. Filipe (FCT-NOVA) for stimulating discussions and support, H. Veiga (ITQB-NOVA) for reviewing the manuscript, N. Reichmann (ITQB-NOVA) for constructing plasmid pBCB33, N. Meiresonne and T. den Blaauwen (Univ of Amsterdam) for plasmid psav-mSc-I, L. Lavis (Janelia Research Campus, Ashburn) for the generous gift of JF549-HTL, and the Indiana University Center for Genomics and Bioinformatics for high throughput sequencing. This study was funded by the European Research Council through ERC Advanced Grant 101096393 (to MGP), by La Caixa Foundation grant HR23-00221 (to MGP), by Fundação para a Ciência e a Tecnologia (FCT) through grant 2022.01678.PTDC (to SS) and contract 2022.03033.CEECIND (to SS), MOSTMICRO-ITQB R&D Unit (UIDB/04612/2020, UIDP/04612/2020 to ITQB-NOVA) and LS4FUTURE Associated Laboratory (LA/P/0087/2020 to ITQB-NOVA), and by National Institutes of Health R01GM141242, R01GM143182, and R01AI172822 (to X.W.). Research is a contribution of the GEMS Biology Integration Institute, funded by the National Science Foundation DBI Biology Integration Institutes Program, Award #2022049 (X.W.).
Author information
Authors and Affiliations
Contributions
A.I.M., S.S. and M.G.P. designed the research. S.S. generated the FROS system and CT performed initial tests. S.S., A.I.M., D.S.B. and M.S. constructed the plasmids and strains for the study. A.I.M. performed the microscopy experiments. A.D.B developed the software to automate the analysis of microscopy data with input from A.I.M., S.S. and M.G.P. S.S. performed the ChIP-Seq experiment. Q.L. performed the Hi-C and WGS experiments and analyzed the data together with X.W. The data was overall analyzed by A.I.M., S.S., X.W. and M.G.P. A.I.M., X.W. and M.G.P wrote the manuscript with input from other authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Izquierdo-Martinez, A., Schäper, S., Brito, A.D. et al. Chromosome segregation dynamics during the cell cycle of Staphylococcus aureus. Nat Commun 16, 8626 (2025). https://doi.org/10.1038/s41467-025-63634-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-63634-5