Abstract
The β-barrel assembly machinery (BAM) inserts β-barrel proteins into the outer membrane of Gram-negative bacteria, forming an essential permeability barrier. The core BAM component, BamA, is a β-barrel protein with an N-terminal periplasmic extension comprising five polypeptide transport-associated (POTRA) domains. Whilst BamA’s structure is well characterised, it remains unclear how β-barrel and POTRA domain motions are coordinated. Using BamA variants with mutations in the hinge region between these two domains, we demonstrate that hinge flexibility is required for BAM function. Cryo-electron microscopy suggests that hinge rigidity impairs function by structurally decoupling these domains. A screen for spontaneous suppressors identified a mutation at position T434 in an extracellular loop of BamA, which has been previously shown to suppress BAM defects. Studying this variant provides insights into its function as a general rescue mechanism. Our findings underscore how BamA’s sequence has been evolutionarily optimised for efficient function.
Similar content being viewed by others
Introduction
The cell envelope of diderm bacteria consists of two membranes that enclose the periplasm, an aqueous, oxidising compartment containing the peptidoglycan cell wall. The inner membrane is a typical phospholipid bilayer, while the outer membrane exhibits a distinctive composition, with its inner leaflet composed of phospholipids and its outer leaflet consisting of lipopolysaccharides (LPS)1,2. The outer membrane plays a major role in maintaining the integrity of the cell, serving both as a permeability barrier against toxic compounds3 and a load-bearing structure that enables the buildup of periplasmic turgor4,5. Two groups of proteins are present in the outer membrane. Lipoproteins, which are globular proteins anchored to the membrane by a lipid moiety6, and outer membrane proteins (OMPs) that are integral membrane proteins adopting a β-barrel conformation7,8. OMPs are synthesised in the cytoplasm and are secreted, unfolded, into the periplasm via the Sec pathway. They are escorted, unfolded, to the outer membrane by periplasmic chaperones, which deliver them to the β-Barrel Assembly Machinery (BAM) for insertion into the outer membrane2,9.
BamA, the central component of the BAM complex, is a 16-stranded β-barrel that belongs to the highly conserved OMP85 superfamily10,11,12,13. At its N-terminus, BamA features five polypeptide transport-associated (POTRA) domains located in the periplasm. In the bacterium Escherichia coli, the BAM holocomplex includes four accessory lipoproteins (BamB, BamC, BamD, and BamE), which interact with the POTRA domains14,15,16. BamA and BamD are essential, but all BAM components are necessary for optimal function10,17,18.
Several cryo-electron microscopy (cryoEM) and crystal structures have provided detailed insights into the mechanism(s) used by BAM to insert β-barrels into the outer membrane. Central to this mechanism is the ability of BamA to adopt multiple conformations, which can be grouped into two distinct families: an inward-open conformation19,20, where the BamA β-barrel is sealed at its lateral gate involving β-strands 1 and 16, and an outward-open conformation19,21,22, in which the lateral gate in BamA opens between β-strands 1 and 16. Cycling between these conformations is essential for the insertion and folding of OMPs19,23. Currently available data suggest that OMP folding is initiated by binding of a sequence in the C-terminal β-strand of the substrate (known as the β-signal) to the first β-strand (β1) of BamA24,25. Thereafter, sequential addition of β-strands towards the N-terminus of the substrate ultimately results in the formation of an asymmetrical hybrid BamA-substrate barrel in which BamA is found in its outward-open conformation26,27. Promoted by outer membrane tension26 and BamA-induced membrane destabilization11,28,29,30,31, the client OMP is finally released into the membrane. Conformational changes also occur in the POTRA domains during the OMP folding and assembly cycle: they are located under the barrel, extending into the periplasm, and occlude access to the lumen of the BamA β-barrel when the lateral gate is open, but move away when the lateral gate is closed, opening a periplasmic entry pore to the barrel lumen17,31.
Despite extensive knowledge about BamA structure and function, many questions remain regarding its mechanical properties and structural transitions and how these factors relate to the regulation of BAM’s foldase activity. For instance, while the function of the lateral gate and β1 in initiating OMP folding has been well documented23,25,32, and demonstrated by the bactericidal natural products, darobactin and dynobactin, that bind at this site33,34, the precise contribution of the POTRA domains in this process remains poorly understood. Additionally, how the movements between the POTRA and β-barrel domains of BamA are coordinated remains unknown. The rotation of the POTRA domains and associated lipoproteins appears to be coupled with the opening and closing of the lateral gate19, but it is still unclear which movement induces the other.
Here, using a BamA variant with mutations in the hinge region between its β-barrel and POTRA domains, in combination with in vivo, structural, biochemical and proteomics studies, we obtained mechanistic insights into how motions in the β-barrel domain are coupled to the movement of the POTRA domains throughout the BamA functional cycle. First, we find that introducing an LVPR tetrapeptide in the hinge, which acts to rigidify the linker region between the BamA β-barrel and the POTRA domains, impairs BAM function in vitro and in bacteria by causing conformational decoupling between the BamA barrel and its POTRA domains. In contrast, inserting a flexible GSGS sequence had no impact on bacterial growth and only minor effects on BAM function, confirming that maintaining flexibility in the hinge region is necessary for optimal BAM activity. The growth defects we observed with the LVPR insertion are thus mainly due to sequence-specific effects on hinge flexibility.
A search for spontaneous suppressors able to rescue the defects caused by the LVPR insertion led to the identification of a BamA mutant in which threonine 434, a residue located on an extracellular loop of the barrel, is replaced by an alanine (BamAT434A). Interestingly, mutations affecting T434 have been found previously to suppress various BAM defects, including inhibition by darobactin, a BAM-specific antibiotic, and the absence of BAM accessory lipoproteins35,36. Intrigued by the fact that mutating T434 appears to function as a general reset mechanism to rescue compromised BAM function, we characterised BamAT434A. Our data suggest that while the T434A mutation does not severely affect BAM folding activity, it results in altered BamA conformational dynamics and suboptimal adaptation to stress, leading to outer membrane defects. Together, our results demonstrate that the motions of the BamA β-barrel and POTRA domains are intricately coupled, highlighting how evolution has precisely tuned the native BamA sequence to ensure optimal OMP assembly and maintain outer membrane integrity.
Results
We previously reported the identification of a BamA variant (BamALVPR) with decreased assembly function37. In this variant, a short peptide (lysine–valine–proline–arginine—LVPR) is introduced into the hinge region between the β-barrel and POTRA domains of BamA (Fig. 1a). This insertion, originally intended to introduce a thrombin cleavage site between the two domains of BamA, was found to be toxic. Cells expressing BamALVPR displayed a growth defect in rich medium (LB) at 37 °C. Interestingly, however, these cells grow well in minimal medium at 30 °C, where a lower rate of OMP folding into the outer membrane is required, suggesting that the BamALVPR variant cannot cope with a higher flux of substrates37. Consequently, we decided to use BamALVPR as a molecular tool to explore the functional importance of the hinge region between the β-barrel and periplasmic domains of BamA and gain mechanistic insights into the conformational dynamics of the BAM complex.
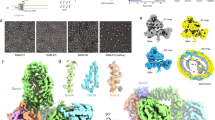
a The hinge region (V418-G424) between the β-barrel and the POTRA domains of BamA is depicted in pink on a BAM structure (PDB 5D0O19). LVPR is inserted before residue G424. b, c Cells expressing BamALVPR have growth defects and a more permeable outer membrane due to decreased OMP levels. bamA::kan cells expressing variants of BamA from a plasmid were grown in LB at 30 °C until mid-log phase. b (top) Ten-fold serial dilutions were made in LB, plated onto LB or MacConkey agar plates and incubated at 37 °C. b (bottom) Sensi-Disc Susceptibility Tests were used to test strain sensitivity to antibiotics (see “Methods” section). The diameter of growth inhibition around each disk was recorded in millimetres. Values correspond to means ± standard deviations (SD) (n = 4 independent experiments). ND not detectable (i.e. no growth inhibition). c Western blot of trichloroacetic acid (TCA) precipitates of annotated cells, using primary antibodies against different OMPs (representative of n = 3, biological replicates). d, e Cells harbouring the BamALVPR mutant have an altered envelope proteome and a SigmaE signature. d Volcano plot showing the difference in protein abundance between cells expressing WT BamA or BamALVPR. P-values were evaluated by a two-sample t-test. Detected OMPs are labelled in red, and periplasmic chaperones and stress-related proteins are in blue. Proteins above the black line, have statistically significantly different expression levels. e SigmaE activity measured by β-galactosidase assay was reported using the transcriptional rpoHP3’-lacZ fusion on the chromosome. Error bars represent the SEM (n = 5 biological replicates, cells were grown in LB medium at 30 °C). P-values were evaluated by a mixed model, with random plate effect, and multiple comparisons with adjustment by the Dunnett method (****P < 0.0001). f The hinge region of BamA is conserved to be disordered. Weblogo sequence and secondary structure conservation plots for the BamA hinge region (E “extended strand in parallel and/or anti-parallel β-barrel”, C “coil”) (see “Methods” section). g The LVPR tetrapeptide has a restricted C-alpha dihedral angle distribution compared to NTGS (WT) and GSGS. Data extracted from 1 μs molecular dynamics simulations in explicit solvent (n = 3) (see “Methods” section).
Cells expressing BamALVPR have an impaired outer membrane with decreased OMP levels
To start our investigation, we assessed the outer membrane integrity of cells expressing BamALVPR in a range of non-permissive conditions. We found that these cells are more sensitive to large antibiotics (vancomycin, bacitracin, novobiocin, rifampin) and bile salts than bacteria expressing wild-type BamA (Fig. 1b). Given that proper assembly of the outer membrane is crucial for its function as a permeability barrier, increased sensitivity to antibiotics and detergents is indicative of outer membrane assembly defects38. Second, we probed the expression levels of LptD, a protein required for LPS insertion into the outer membrane39,40,41,42; LptD is challenging to fold and is particularly sensitive to BAM malfunction43,44,45,46. Notably, LptD levels were found to be decreased dramatically in cells expressing BamALVPR in rich media at 30 °C (Fig. 1c). Next, we used label-free quantitative proteomics to analyse the impact of BamALVPR expression on the entire E. coli proteome (Fig. 1d). To circumvent the severe growth defect in rich medium, cells were grown in minimal medium at 30 °C. In the quantitative proteomics experiment, 16 OMPs were detected, and the expression levels of 11 of these decreased in cells expressing BamALVPR, including the major OMPs OmpA and OmpC, which was confirmed by western blot (Fig. 1c). Together, these results indicate that introducing the LVPR peptide into the hinge region between the β-barrel and POTRA domains of BamA affects the function of BAM, resulting in outer membrane defects and decreased OMP folding. Accumulation of unfolded OMPs occurs when BAM activity is compromised, triggering the SigmaE stress response35,47,48. Consistent with this, SigmaE regulon members such as SurA and BepA were found to be more abundant in cells expressing BamALVPR (Fig. 1d) and activation of SigmaE was confirmed under both growth conditions using a reporter strain (Fig. 1e and Supplementary Fig. 1b). Together, the results show that the insertion of LVPR into the hinge region between the β-barrel and POTRA domains of BamA results in outer membrane assembly defects by compromising BAM function.
Flexibility is required in the hinge region of BamA for optimal activity
We next asked whether the impairment in BamA’s function observed in BamALVPR results from intrinsic features of the LVPR sequence or simply from the addition of a tetrapeptide between the β-barrel domain and periplasmic POTRA domain 5 (POTRA5). Proline residues are known to restrict protein conformational space49,50; and thus, the impact of the insertion on BamA function might have been due to restricted flexibility of the hinge region. Interestingly, analysis of the secondary structure of the hinge region in >2000 BamA homologues revealed that, despite variable amino acid composition, this region is consistently predicted to be disordered, underscoring the critical importance of maintaining its flexibility (Fig. 1f). To test the hinge region’s capacity to accommodate the insertion of a tetrapeptide while preserving its flexibility, we replaced LVPR with the Glycine–Serine–Glycine–Serine (GSGS) sequence, generating BamAGSGS. Molecular dynamics simulations of NTGS (wild-type), LVPR, and GSGS sequences, as isolated peptide sequences, followed by a dihedral angle distribution analysis of the Cα atoms, showed that for the LVPR sequence, the dihedral angle distribution is highly restricted, indicating low levels of conformational flexibility. In contrast, NTGS and GSGS exhibit broad dihedral angle distributions, confirming that the presence of these sequences in the hinge are compatible with maintaining flexibility in this region (Fig. 1g). Remarkably, cells expressing BamAGSGS exhibit a phenotype similar to that of bacteria expressing wild-type BAM: they grow on rich medium at 37 °C, are resistant to bile salts, and only moderately sensitive to large antibiotics (reduced zone of inhibition in the presence of bacitracin compared to BamALVPR, and no growth inhibition by novobiocin unlike BamALVPR) (Fig. 1b). Furthermore, levels of OmpA, OmpC, and LptD are similar to those observed in wild-type cells, both in rich and minimal media (Fig. 1c and Supplementary Fig. 1a). Finally, the SigmaE response in bacteria expressing BamAGSGS is only moderately increased when cells are grown in LB, and does not turn on in minimal media (Fig. 1e and Supplementary Fig. 1b). These findings indicate that inserting a tetrapeptide between the two domains of BamA has only a slight effect on BAM function when the tetrapeptide is flexible, but significantly impairs function when access to conformational space is limited, as in LVPR.
The activity of BamALVPR is decreased in an in vitro folding assay
Next, we compared the folding activity of BAM containing either BamALVPR or BamAGSGS to complexes containing wild-type BamA using an in vitro OMP folding assay with a reconstituted BAM complex31. In these experiments, we analysed the folding kinetics of two 8-stranded OMP β-barrels, OmpX and a truncated version of OmpA lacking its periplasmic domain (tOmpA). These experiments showed that inserting the LVPR peptide markedly affects BAM activity: after a 3-h folding period, only about 54% of tOmpA and 30% of OmpX achieved their folded state, compared to 96% and 84% folding efficiency, respectively, with wild-type BamA (Fig. 2a, b and Supplementary Table 1). The GSGS insertion in BamA does not substantially affect BAM’s ability to fold tOmpA (75% of tOmpA was folded in the presence of BAMGSGS), consistent with the in vivo results presented above that show wild-type levels of growth for cells expressing BamAGSGS. However, no benefits from BAMGSGS compared to BAMLVPR were observed for the OmpX substrate (Fig. 2b), which could reflect some OMP specificity on BAM’s ability to fold its substrates, at least under the conditions used for our in vitro folding assay (note that wild-type BAM folds OmpX more slowly than tOmpA, despite both OMPs being 8-stranded (Supplementary Table 2)). Incubation of these OMP substrates with the different BAM complexes for longer periods (overnight) yielded high yields (70–90%) for both proteins, indicating that the mutations introduced into BamA reduce its catalytic efficiency, but do not prevent folding (Supplementary Tables 1 and 2). Overall, the in vitro data support the in vivo observations presented above and suggest that although lengthening the hinge of BamA has an impact on the function of BAM, rigidifying this region is even more deleterious, leading to the lethality of the LVPR insertion in rich media.
Rigidifying the hinge region of BamA (LVPR insertion) has a greater impact on the ability of BAM to fold tOmpA than when the intrinsic flexibility of this region is preserved (GSGS insertion), but the LVPR and GSGS insertions equally perturb the folding of OmpX. a, b Quantification of folded and unfolded bands from SDS-PAGE band-shift assay plotted as fraction folded against time for tOmpA or OmpX, respectively. The folded fraction values calculated from at least two technical replicate measurements are shown, and dashed lines represent a single exponential fit to the data. Folding experiments were performed at 25 °C. Average initial rates, normalised data and mean/ranges for the replicate data presented are listed in Supplementary Tables 1 and 2.
The β-barrel domain is decoupled from the POTRA domains in BamALVPR
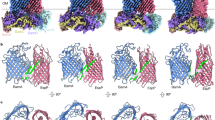
To better understand the impact of the hinge insertions on BAM function, we next used cryoEM to solve the structures of BAMLVPR and BAMGSGS in dodecyl maltoside (DDM) micelles to 4.3 Å and 4.2 Å resolution, respectively (Fig. 3a, b and Supplementary Fig. 2). The structures are similar, including the subunit interaction interface (fitted model RMSD: 2.2 Å, Fig. 3c), although the inserted hinge region and the base of the barrel around the insert (β1–3) are more poorly resolved (Supplementary Fig. 3). Strikingly, while the periplasmic lipoproteins and BamA’s POTRA domains are arranged in a BAM outward-open conformation with POTRA5 obscuring the base of the β-barrel (Fig. 3d and Supplementary Fig. 2), the BamA transmembrane barrel resembles the inward-open conformation with a closed lateral gate (Fig. 3d, e). Although the barrel’s lateral gate is unambiguously closed, the base of the barrel is narrower than in the canonical inward-open conformation, with BAMLVPR being more constricted than BAMGSGS (Fig. 3e and Supplementary Fig. 2). This is likely due to constraints imposed by the relative rigidity of the LVPR insert compared to the GSGS insert that prevents a full relaxation to the lateral gate closed conformation. At low contour levels, there is also evidence of a subpopulation of particles with a narrower barrel in the BAMGSGS density, although this was too weak to separate via classification (Supplementary Fig. 3), further indicating the highly flexible nature of the GSGS linker. The results are strikingly different to the structure of wild-type BAM in DDM micelles obtained by cryoEM, which adopts an outward-open conformation21,51, and of BAM in DDM micelles in the presence of the BAM inhibitor, darobactin B, in which BamA adopts a lateral closed conformation in which the POTRA domains are rotated to block the entrance to the BamA barrel in a canonical inward-open conformation51. Hence, our results show that increasing the length of the linker between the BamA barrel and POTRA5 leads to structural uncoupling between the BamA barrel and its POTRA domains, facilitating their independent motion. While a previously published cryoEM structure of wild-type BAM also displayed an outward-open conformation of the periplasmic domains but with a closed lateral gate52, in this structure the base of the barrel and extracellular loop 3 remain in their outward-open conformation, with only the highly flexible extracellular loops 1 and 2 shifting to the inward-open state (Supplementary Fig. 2). This observation indicates that the hinge insertion is necessary to fully decouple the motion of the two domains. It is important to note that we have chosen to focus our discussion here on the clear differences observed between the structures determined for BAMLVPR and BAMGSGS and their relationship to other available BAM structures, especially with respect to the status of the lateral gate and the orientation of the periplasmic domains. There may be additional small differences in these structures, for example, as a result of subtle variations in lipoprotein orientation, that could be functionally important, but these differences are at the resolution limit of the cryoEM maps and hence are not highlighted or discussed further. Higher resolution maps would be needed to resolve the significance of any such smaller changes between the different structures.
CryoEM structure of a BAMLVPR and b BAMGSGS in DDM micelles; in both structures, the hinge region and the initial β-strands of the barrel are of lower resolution and hence presumably destabilised relative to the back of the barrel (c.f. insets). c BAMLVPR and BAMGSGS are globally similar, structures aligned on BamA transmembrane β8-11 (BamA POTRA1 was poorly resolved and not modelled). d Comparison between BamA of canonical inward-open and outward-open conformations and BAMLVPR, only the transmembrane domain and POTRA5 are shown for clarity. BAMGSGS is similar to BAMLVPR (Supplementary Fig. 2). e Comparison between the transmembrane domains of BAMLVPR, BAMGSGS and BAM-WT inward-open, showing lateral-gate closure and incomplete expansion at the base of the barrel. Inset: Inward-open like orientation of extracellular loop 3 in BAMLVPR and BAMGSGS, which is disordered and not resolved in BAM-WT outward-open. PDBs: 8BWC51 (outward-open), 5D0O19 (inward-open).
A mutation in the extracellular loop 1 suppresses the effect of BamALVPR
We next embarked on a search for spontaneous suppressors that could restore the growth defect of cells expressing BamALVPR under non-permissive conditions (LB, 42 °C). One such mutant (BamALVPR*) mapped to bamA, and, upon sequencing, we discovered the replacement of threonine 434 by an alanine (Fig. 4a). T434 is located in extracellular loop 1 (eL1) between β1 and β2 of the BamA β-barrel, near the seam of the BamA β-barrel involving β-strands β1 and β16. When the lateral gate is closed19,20, but not when it is open19,21,22, the hydroxyl side chain of T434 forms a hydrogen bond with a backbone hydrogen of F804, a residue that lies in the C-terminal β-strand (β16) of the BamA barrel. Replacement of T434 by an alanine will prevent the formation of this hydrogen bond.
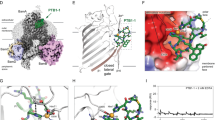
a Zoom of the β-barrel of BamA (PDB 4C4V12) with T434 and F804 represented in dark blue (side chains shown). β-strands 1 and 16 are coloured in light grey, and eL1 is represented in pink. The boxed area is enlarged and depicted below. Possible hydrogen bonds between the hydroxyl side chain of T434 and the backbone of F804 are represented by dashed blue lines. b, c The T434A mutation in BamA partially restores the phenotypical defects induced by the LVPR insertion in the hinge region of BamA. bamA::kan cells expressing variants of BamA from a plasmid were grown in LB medium at 30 °C until mid-log phase. Results for BamAWT and BamALVPR are from Fig. 1b, e, shown for comparison. b (top) Ten-fold serial dilutions were made in LB media, and the bacteria were plated onto LB or MacConkey agar plates and incubated at 37 °C. b (bottom) Sensi-Disc Susceptibility Test Discs were used to test strain sensitivity to compounds (see “Methods” section). Plates were incubated overnight at 30 °C. The diameter of the zone of inhibition of growth around each disk was recorded in millimetres. Values correspond to means ± SD (n = 4 independent experiments). ND not detectable. c SigmaE activity measured by β-galactosidase assay as reported using the transcriptional rpoHP3’-lacZ fusion on the chromosome. Error bars represent the SEM (n = 5 biological replicates). P-values were evaluated by a mixed model, with random plate effect, and multiple comparisons with adjustment by the Dunnett method (**P = 0.004). d BAMLVPR and BAMLVPR* are structurally identical, structures aligned on BamA transmembrane β-strands 8–11 (POTRA1 was poorly resolved and not modelled).
Cells expressing BamALVPR* have no growth defects in rich medium at 37 °C (Fig. 4b) and partially restore levels of OmpA, OmpC, and LptD (Fig. 1c). They exhibit only modest outer membrane permeability to antibiotics and detergents (Fig. 4b). However, SigmaE activity is still upregulated and only weakly decreased compared with bacteria expressing BamALVPR (Fig. 4c). Resolving the cryoEM structure of BAMLVPR* confirmed that the T434A substitution had no major impact on the overall structure of BAMLVPR (model RMSD: 1.0 Å, Fig. 4d). It is remarkable that a single amino acid substitution in eL1 can restore the functional defects caused by an insertion in the hinge region of BamA, located ~3 nm away in the periplasm. This suggests a communication between this loop and the hinge region, where alterations in one region can compensate for the disruptions in the other.
Mutation of threonine 434 results in an altered outer membrane and affects the conformational plasticity of BAM
Interestingly, two previous studies have reported mutations that involve T434, which also act as suppressors of loss of BAM function. First, the T434A mutation was identified in several different triple mutants that rescue BAM from darobactin inhibition35. Darobactin is a novel class of antibiotic that exerts its mechanism of action by mimicking the β-signal of OMP β-barrels, hydrogen bonding to the β1 strand of BamA33. This interaction stabilises the inward-open conformation of BamA, and inhibits the essential docking of the OMP β-signal to the BamA β-barrel required to initiate OMP folding at the BAM complex35,51. Second, a BamA mutant harbouring a T434I mutation was identified in a search for suppressors of the ΔbamBΔbamE synthetic lethal phenotype36. Intrigued by the ability of mutations affecting T434 to rescue various BAM defects, we introduced the T434A mutation into wild-type BamA (named BamAT434A) to investigate its impact on BAM activity and gain insights into how it can compensate for the effects of the LVPR insertion in the hinge region.
The introduction of the T434A substitution into wild-type BamA was found to have no impact on BAM activity in vitro, as BAMT434A did not exhibit any folding defects in our in vitro folding assay (Fig. 5a and Supplementary Fig. 7). The point mutation is also well tolerated in vivo, as cells expressing BamAT434A did not exhibit any growth defects under the tested conditions (Fig. 4b, top) and were as sensitive to large antibiotics as cells expressing wild-type BamA (Fig. 4b, lower). Additionally, there was no significant change in the abundance of LptD, OmpA or OmpC as judged by western blotting (Fig. 1c) nor of most OMPs (apart from small changes in OmpX and MipA levels) as detected by proteomic analysis (Supplementary Fig. 8). Thus, BAM appears to tolerate the T434A substitution in BamA without major effects on its function. Interestingly, we found that BamAT434A was more efficient than the wild type at folding tOmpA and OmpX in the presence of darobactin B, both in terms of initial rate of OMP folding (Fig. 5a and Supplementary Fig. 7) and in the yield of folded OMP after overnight incubation (63% to 80% and 5% to 37%, respectively (Supplementary Table 3)). These findings align with results obtained in vivo35 and suggest that the T434A substitution is a major contributor to the resistance phenotype observed with darobactin.
a The T434A mutation on BamA decreases the effect of darobactin B on the folding activity of BAM in an in vitro folding assay. Quantification of the folded fraction of tOmpA against time, from the SDS-PAGE band-shift assay. The folded fraction values calculated from two technical repeats are shown, and dashed lines represent a single exponential fit to the data. b, c Cells expressing a BamA variant with the T434A point-mutation have outer membrane defects. bamA::kan cells expressing variants of BamA from a plasmid were grown in LB medium at 37 °C until mid-log phase. b Cells were spotted on a gradient plate containing a fixed concentration of SDS (0.5% (w/v)) and a gradient of EDTA (0–0.4 mM) (top) and incubated overnight at 37 °C. For the deoxycholate (DOC) 1% (w/v) plate (bottom), ten-fold serial dilutions were made in LB and spotted onto the plate, and the plates were incubated at 30 °C. c Ten-fold serial dilutions were made in LB medium and plated on LB agar plates (top) or plates containing 15 μg/mL of vancomycin (bottom). Plates were incubated for 5 days at 15 °C.
However, while the T434A substitution did not severely impact BAM function, it did affect outer membrane integrity. Cells expressing BamAT434A were more sensitive to SDS-EDTA at 37 °C and deoxycholate (DOC) at 30 °C compared to cells expressing wild-type BamA, indicating membrane defects (Fig. 5b). Interestingly, these cells were more resistant to vancomycin at 15 °C compared to the wild type (Fig. 5c). Sensitivity to vancomycin at this temperature is thought to result from transient cracks in the brittle LPS layer53,54. Thus, increased resistance to vancomycin also suggests altered outer membrane properties due to the T434A mutation. Together, these observations suggest that although the T434A mutation has no impact on the function of BAM in normal growth conditions, it impairs the adaptability of the complex to some stresses.
Preventing the formation of the hydrogen bond between T434 and F804 likely changes the conformational dynamics of BamA, allowing this mutation to mitigate toxic mechanisms such as those caused by hinge insertion and inhibition with darobactin while altering BAM’s optimal functioning. To investigate the conformational dynamics of BAM and BAM variants, we utilised differential hydrogen-deuterium exchange mass spectrometry (HDX-MS) (see “Methods” section). The rate and extent of deuterium exchange in an HDX-MS experiment are determined by the solvent accessibility and intra-protein hydrogen bonding of each residue. Consequently, HDX-MS can reveal subtle changes in the conformational dynamics of proteins and protein assemblies55. Significant differences were observed in the extent of deuterium incorporation in BAMT434A and wild-type BAM, suggesting that this single residue substitution results in altered conformational dynamics across the complex (Fig. 6a, b). Notably, BamB, the POTRA domains (POTRA2/3), and eL6 of BamA were significantly more protected from hydrogen-deuterium exchange in BAMT434A compared with wild-type BAM complexes, suggesting reduced conformational flexibility or reduced solvent accessibility in the mutant complex. The alterations in the dynamics of eL6 are of particular interest, considering the high conservation of residues 675–700 in this loop among members of the OMP85 superfamily56,57 and its critical role in BAM function58. The large distance separating residue 434 and those affected by its substitution adds further weight to the observation that residue 434 must be part of an allosteric network connecting the status of the BamA β-barrel domain not only to the location of its POTRA domains (Fig. 6a, b), but also to the associated BamB lipoprotein. Further to this, comparative HDX-MS of BAMLVPR with wild-type BAM demonstrates that the insertion of LVPR also perturbs BAM dynamics (Fig. 6c), with the LVPR insertion resulting in increased protection from hydrogen exchange in the periplasmic region of the protein (specifically POTRA2, BamB and BamD). Combined, this analysis suggests that BAM conformational dynamics are precisely controlled, and that mutations/insertions in the BamA sequence in eL1 and in the β-barrel-POTRA5 linker can perturb the precise molecular motions of, and intermolecular interactions within, BAM that are critical for its foldase function.
a Structure of BAM in a lateral-open conformation (PDB 5LJO21) with each subunit coloured. b, c BAM structures showing regions of HDX protection and deprotection in the BAM complex in the presence of the b T434A mutation or c LVPR insertion. Regions protected or deprotected from hydrogen exchange in the mutant complexes are highlighted in blue or red, respectively. Regions in white show no change in deuterium uptake, while those in black denote sequences for which peptides were not detected. Sequence coverage maps are shown in Supplementary Fig. 9, and Wood’s plots are shown in Supplementary Figs. 10–14.
Discussion
Understanding the molecular details of how BAM inserts OMPs into the bacterial outer membrane has been the subject of intensive research since the discovery of the complex in the early 2000s10,14,59. Significant insights into the mechanism of BAM have been gained through structural resolution of the complex in crystals and using cryoEM, revealing the complex to adopt varied conformations which can be grouped into two distinct categories: inward-open, in which the BamA lateral gate is closed and the BamA β-barrel is accessible from the periplasm, and outward-open, wherein the lateral gate is open and access to the BamA barrel from the periplasm is blocked by its POTRA5 domain19,20,21,22. In parallel, biochemical studies designed to capture OMP folding intermediates on the BAM complex25,26,27,32, and other approaches31,51,60,61 have revealed details of how BAM facilitates OMP folding into the outer membrane via growth of the OMP barrel, templated from the BamA β1 strand in the lateral gate. However, many questions remain unresolved, particularly concerning how the conformational changes of the BamA β-barrel are linked to, and coordinated with, the movement of its POTRA domains and associated BamB-E lipoproteins that reside in the periplasm. Recently, BAM lipoproteins were found to play a crucial role in membrane destabilisation, presumably facilitating OMP insertion into the outer membrane when released from BAM31. While the dynamics and flexibility of BamA’s POTRA domains have been studied in detail and shown to be critical for BAM activity17,28,62, how POTRA domain movements are coordinated with motions at the BamA β-barrel to achieve BAM function remains elusive.
Here, we show that the flexibility between the POTRA domains and the β-barrel domain of BamA is crucial for the optimal functionality of the complex. We show that inserting a rigid tetrapeptide (LVPR) into the hinge region between POTRA5 and the BamA β-barrel impairs BAM activity in vitro and in bacteria, whereas adding a flexible GSGS insertion at the same site has little effect on BAM activity. This suggests that the rigidity imposed by the LVPR insertion is responsible for the observed severe phenotype, but the molecular mechanism underlying this observation remains unclear. Notably, our findings with BAMLVPR align with previous studies showing that tethering POTRA5 to the base of the BamA β-barrel—thereby reducing flexibility between the POTRA and β-barrel domains—is lethal in vivo19 and results in substantially decreased BAM-assisted OMP folding activity in vitro31. Furthermore, our structural data show that maintaining a flexible hinge region between POTRA5 and the BamA β-barrel aids in coordinating the motions between the barrel and POTRA domains in BamA, as the insertion of the rigid LVPR sequence leads to new states in which coordination of the movement of the lateral gate and rotation of POTRA5 are uncoupled. Given the resolution of our cryoEM maps (4.3 Å and 4.2 Å), we cannot rule out that subtle structural differences that are at, or beyond, the resolution limit of our cryoEM maps may be involved in the structural coupling of the BamA lateral gate domain structure and the organisation of the POTRA domains and the BamB, C, D, E lipoproteins. Additional mechanistic studies are needed to determine precisely how signals are transduced between the POTRA domains and the BamA β-barrel and to clarify the exact role of the hinge sequence in this coordination. For example, it remains unclear whether insertion in the hinge region could impact the binding of BAM to SurA, the chaperone that delivers OMPs to the BAM complex63, or BepA, a chaperone/protease that plays a key role in OM quality control64. Indeed, recent cryoEM structures have shown that SurA forms extensive interactions with POTRA1, POTRA2 and BamB63,65, whereas evidence from crosslinking suggests that BepA engages in interactions with POTRA3, BamC and BamD64. All of the interprotein contacts mediating recruitment of these key folding factors to BAM could be affected by altering BAM dynamics and the coupling of the motions of the BamA lateral gate to those of the POTRA domains and their associated lipoproteins. Whilst here we show that the LVPR insertion results in a reduction in folding efficiency in an in vitro BAM folding assay, future studies investigating how altering BAM dynamics impacts engagement with these folding factors are needed to answer these questions. Such studies could include, for example, a comprehensive biophysical characterisation of the affinities and nature of the interactions between BAMLVPR with SurA/BepA, as alterations in these interactions could be determinants of the growth defects observed in the BamALVPR strain.
Another key finding of our work is the identification of the T434A suppressor mutation, located on eL1 of the BamA β-barrel, which can partially restore defects caused by the distal insertion of the sequence LVPR in the hinge region. Remarkably, mutations affecting T434 have previously been shown to rescue BAM from two distinct stresses. First, the T434I mutation was identified as a suppressor of the conditionally lethal ΔbamBΔbamE double mutant, whose lethality arises from BamA being jammed by the lipoprotein stress sensor RcsF, likely locking BamA in the inward-open conformation36,37. Second, a T434A mutation was found in two different sets of three mutations capable of rescuing BAM from the inhibitory effects of darobactin A, a β-strand mimetic that binds to β1 at the BamA lateral gate, preventing its opening and shifting the equilibrium of the BAM complex toward the inward-open conformation33,35,51. Confirming these results and the direct role of T434 in the allosteric control network of BamA, we found through an in vitro folding assay that BAMT434A can fold the OMPs, tOmpA and OmpX, in the presence of darobactin B, albeit with reduced efficiency compared to that in the absence of darobactin. Thus, in both cases—the ΔbamBΔbamE mutant and darobactin inhibition—the dynamic, coordinated cycling of BAM is disrupted, but can be restored by mutating T434, similar to the observations presented here for BAMLVPR. Overall, therefore, the T434 mutation seems to act as a reset button, restoring BAM activity in the face of various defects. A hydrogen bond between the sidechain of T434 and the mainchain of F804 in β16 of the BamA β-barrel, is disrupted by the T434A mutation. We propose that this facilitates the opening of the lateral gate, thereby helping to rescue compromised BAM function.
Further supporting the idea that the T434A mutation modifies the dynamics of the BAM complex, HDX-MS results revealed that eL6 of the BamA β-barrel is less accessible to solvent and hence stabilised in BAMT434A compared to the wild type. This impact on eL6 is interesting given the conservation of this loop in all members of the OMP85 superfamily and the physiological importance of eL6 in BamA, which was shown to be important for OMP substrate insertion56,58. Finally, the observation of increased protection from hydrogen exchange of residues that reside distal to T434, including residues in POTRA2/3 and BamB that lie more than 4 nm from T434, starts to reveal an allosteric coupling mechanism that spans the length of the entire BAM complex. Our data show that both the T434A mutation and LVPR insertion modulate BAM dynamics in disparate regions of the complex distal to the sites of modification, suggesting different allosteric impacts on BAM dynamics. Combined with the impacts of the T434A and LVPR variants on BAM function, this suggests a functional role for such an allosteric network, although the precise molecular details of such allosteric coupling remain to be discovered.
If the T434A mutation provides a functional advantage by rescuing BAM from different blockages, why has it not been selected by evolution? The answer may lie in the results of our phenotypic assays, where we compared the behaviour of cells expressing BamAT434A to that of wild-type cells. Notably, cells expressing BamAT434A show increased sensitivity to SDS-EDTA and DOC, indicating altered outer membrane properties, likely due to suboptimal activity of BamAT434A under these conditions. Additionally, these cells exhibit increased resistance to vancomycin at 15 °C, further supporting the idea of modified BamAT434A function. At lower temperatures, cells are generally more sensitive to vancomycin due to cold-induced transient cracks in the brittle LPS layer53,54. They also adapt by modifying their phospholipid composition, decreasing the length and saturation of acyl chains to maintain membrane fluidity66. Thus, the increased resistance to vancomycin likely results from altered BamAT434A activity compared to the wild type. Together, these observations suggest that in the wide variety of conditions that Gram-negative bacteria encounter, the fitness cost of expressing BamAT434A likely outweighs any potential benefits of heightened ability to survive under different growth conditions. These findings are reminiscent of previous findings on the essential E. coli chaperone, GroEL, which, akin to BAM, has also evolved to fold a diversity of substrates. Specialisation of the GroEL sequence towards enhanced folding of a single substrate was also found to compromise the folding of others67,68. This underscores the delicate balance required to fine-tune the activity of essential proteins—BamA and GroEL—for bacterial growth and survival and illustrates the compromises inherent in evolutionary adaptations.
Methods
Media
M9 minimal media was prepared with 1× solution of M9 salts (MP Biomedicals) and supplemented with 0.2% (w/v) glucose, 1 mM MgSO4, and 0.1 mM CaCl2.
Bacterial strains, plasmids, and primers
Details of the bacterial strains, plasmids and primers used in this study are listed in Supplementary Tables 4–6, respectively. MG1655 was used as the wild-type strain throughout this study. To delete bamA from the chromosome, we followed the same workflow as previously described in ref. 37. The plasmid pBamAGSGS was generated by site-directed mutagenesis using primers “RRA93-bamA GSGS-F” and “RRA94-bamA GSGS-R”, and pBamA as template. The plasmid pNCA162 was generated by mutagenesis using primers “BamA hinge F” and BamA hinge R” and pJH114 as a template. The T434A point mutation was introduced by site-directed mutagenesis using primers “RRA97-BamAT434A-F” and “RRA98-BamAT434A-R”.
Suppressor selection
To select for BamALVPR suppressor mutations, 0.1 mL of an overnight culture of SEN1772 grown in M9 minimal medium was plated onto LB agar. Bacterial colonies with spontaneous BamALVPR mutations appeared after overnight incubation at 42 °C. The BamALVPR containing plasmid (with spontaneous mutations) was isolated from the bacteria using the Monarch Plasmid Miniprep Kit (New England BioLabs). DNA sequencing was performed using primers against the gene of interest, bamA (Eurofins). Plasmids carrying the suppressor mutation in bamALVPR were transformed into a DH300 strain where the chromosomal bamA was deleted as described above.
Spotting assay for growth
Cells were grown in LB medium at 30 °C until they reached an OD600 of 0.5. Ten-fold serial dilutions were made in LB, and the cells were plated onto LB or MacConkey agar, and incubated at 30 °C or 37 °C. For sensitivity to DOC or vancomycin, cells were grown in LB medium at 37 °C until they reached an OD600 of 0.5. Ten-fold serial dilutions were made in LB, and 2 μL of culture was plated onto LB agar plates containing 15 μg mL−1 vancomycin or 1% DOC, and plates were incubated at 15 °C or 30 °C, respectively.
SDS-EDTA gradient plates were prepared by pouring 25 mL of LB agar containing 0.5% (w/v) SDS in a tilted square petri dish. Once the layer was dry, the plate was laid flat and 25 mL of LB agar containing the maximum concentration of the gradient was poured (0.5% (w/v) SDS and 0.4 mM EDTA). Cells were grown in LB medium at 37 °C until they reached an OD600 of 0.5. Two microliters of culture was directly spotted onto the gradient plate. Plates were incubated at 37 °C.
Uncropped images of the plates are available in the Source Data file.
Sensidisc assays
BBL Sensi-Disc Antimicrobial Susceptibility Test Discs (BBL) were used to test strain sensitivity to bacitracin (10 IU), rifampin (25 μg), vancomycin (30 μg) or novobiocin (5 μg). Discs containing the compounds at the quantities indicated were used in disk diffusion assays as follows. A 0.1 mL inoculum taken from overnight cultures grown in LB was mixed with 3 mL of LB top agar (LB with 0.75% (w/v) agar) and poured over an LB agar plate. The discs containing antibiotics were placed on top of the LB top agar, and the plates were incubated overnight at 30 °C. The diameter of the zone of inhibition of growth around each disk was recorded in millimetres. Data from four biologically independent replicates were acquired.
Western blot analysis
Cells were grown in LB media at 30 °C to an OD600 of 0.5, and the proteins were precipitated by adding 10% (w/v) trichloroacetic acid (TCA). The pellets were washed with acetone and finally solubilised in 50 μL of SDS-sample buffer. Samples were boiled and loaded onto a 4-12% bis-Tris gel (Nupage, Invitrogen), and MOPS was used as a running buffer. After transfer of the proteins from the SDS-PAGE gel onto a nitrocellulose membrane, the membrane was blocked with 5% (w/v) skim milk in TBS (25 mM Tris, 3 mM KCl, 140 mM NaCl, pH 7.4) supplemented with 0.1% (v/v) Tween 20 (TBS-T). Membranes were then incubated with primary antibodies diluted in TBS-T, 1% (w/v) skim milk: anti-LptD (Shin-ichi Matsuyama) (1/5000 dilution), anti-BamA (homemade)37 (1/10,000 dilution), anti-OmpA (L4, Bernstein)69 (1/5000 dilution) or anti-OmpC (Gentaur A64454) (1/5000 dilution). After incubation with primary antibody and washes with TBS-T, the membrane was incubated with horseradish peroxidase (HRP) conjugated donkey anti-rabbit IgG (Cytiva NA934) (1/10,000 dilution) in TBST-T 5% (w/v) skimmed milk. The membranes were washed, and the labelled protein was detected using a chemiluminescent HRP substrate (Immobilon, Millipore). Western blots were visualised using a CCD camera (ImageQuant 800, Cytiva). Uncropped and unprocessed images of the blots, as well as replicates, are available in the Source Data file.
Proteomics
Cells were either grown in 100 mL M9 glucose minimal medium at 30 °C (SEN1603 and SEN1722) or in 100 mL LB medium at 37 °C (NCAs580 and NCAs584) until an OD600 of 0.8. Cells were harvested by centrifugation at 5000×g for 20 min at 4 °C. OM extracts were prepared by suspending the cell pellet in 1 mL TSE buffer (0.2 M Tris (pH 8), 0.5 M sucrose, 1 mM EDTA). Cells were incubated in TSE buffer on ice for 30 min, transferred to a 2 mL microcentrifuge tube, and centrifuged at 18,000×g for 30 min at 4 °C. Supernatants were removed to a new microcentrifuge tube. The outer membrane fraction was collected by ultracentrifugation at 100,000×g for 1 h. The pellet contained the OM extract, and the proteins were resuspended in 100 μL of 1% (v/v) DDM. The protein concentration was measured by Bicinchoninic acid (BCA) assay. The proteins were precipitated with ice-cold ethanol, and the protein pellet was suspended in Rapigest (Waters) for tryptic digestion. Digested material was analysed in data-dependent acquisition mode on an Orbitrap Velos (comparing BamA and BamALVPR strains) or Orbitrap Eclipse (comparing BamA and BamAT434A strains). For experiments using the Orbitrap Velos, peptides were analysed using an EASY-nLC 1000 Liquid Chromatograph (Thermo Fisher Scientific) connected to a custom-made 30-cm capillary emitter column (75 μm inner diameter, 3 μm Reprosil-Pur 120 C18 media). Mass spectrometry analysis was performed on a linear quadrupole ion trap—orbitrap (LTQ-Orbitrap) Velos mass spectrometer (Thermo). The total acquisition time was set to 90 min, with a gradient of 3–22% (v/v) acetonitrile in 0.1% formic acid. For the survey scan, the resolving power was set at 60,000 with a scan range of 305–1350 amu. Data acquisition was performed in data-dependent analysis mode. MS/MS data were obtained by fragmenting up to the twenty most intense ions in the linear ion trap. For experiments using the Orbitrap Eclipse, peptides were separated using a Vanquish Neo LC (Thermo Fisher Scientific) system. The peptides were trapped on a PepMap Neo C18 trap cartridge (Thermo Fisher Scientific, 5 μm particle size, 300 μm × 0.5 cm) before separation using an Easy-spray reverse-phase column (Thermo Fisher Scientific, 2 μm particle size, 75 μm × 500 mm). Peptides were separated by gradient elution of 2–40% (v/v) solvent B (0.1% (v/v) formic acid in acetonitrile) in solvent A (0.1% (v/v) formic acid in water) over 80 min at 250 nL min−1. The eluate was infused into an Orbitrap Eclipse mass spectrometer (Thermo Fisher Scientific) operating in positive-ion mode. Data acquisition was performed in data-dependent analysis mode. Each high-resolution full scan (m/z 380–1400, R = 60,000) was followed by high-resolution product ion scans (R = 30,000), with a stepped normalised higher energy collision induced dissociation energies of 21%, 26% and 31%. A cycle time of 3 s was used. Dynamic exclusion of the 60 s was used.
Data were searched against the E. coli K12 proteome (MaxQuant v2.4.2.0) and protein abundance changes quantified by label-free quantification (statistical analyses performed using Perseus v2.0.10.0), employing a false discovery rate (FDR) of 0.05 and s0 value of 0.1. Replicate measurements from three independent grows were analysed. Raw data have been deposited to the ProteomeXchange Consortium via the PRIDE70 partner repository with the dataset identifier PXD057225.
Reporting SigmaE stress response activation
Stress responses were monitored by measuring β-galactosidase activity as previously described in ref. 71. To monitor the impact of the LVPR, GSGS, LVPR* or T434A insertions in BamA on the SigmaE stress response, cells were grown overnight in LB or in M9 minimal media at 30 °C. Overnight cultures were diluted 1:250 (in M9) or 1:500 (in LB) and harvested once an OD600 of 0.5 was reached. Twenty microliters of cell suspension was transferred to a 48-well plate and this was frozen at −20 °C until processing. For β-galactosidase measurement, the plates were thawed, and 80 μL of permeabilisation solution (100 mM Na2HPO4, 20 mM KCl, 2 mM MgSO4, 5.4 μL/mL β-mercaptoethanol, 0.8 mg/mL hexadecyltrimethylammonium bromide, 0.4 mg/mL sodium DOC) was added. Plates were incubated for 60 min at 30 °C. Then, 600 μL of substrate solution (60 mM Na2HPO4, 40 mM NaH2PO4, 1 mg/mL o-nitrophenyl-β-D-galactoside, 2.7 μL/mL β-mercaptoethanol) was added. The reaction was stopped by the addition of 700 μL of 1 M Na2CO3. Samples were centrifuged to remove cell debris, and 500 μL of supernatant was moved to a new 48-well plate. The optical density was measured at 420 nm using a BioTek Synergy 1 plate reader (software: Gen5 2.06). The standardised amount of β-galactosidase activity was reported in Miller units71. 5 biological replicates (for LB) or 3 biological replicates (for M9) were performed, and data are reported as mean values ± standard error of the mean (SEM). P-values were evaluated by a mixed model, with random plate effect, and multiple comparisons with adjustment by the Dunnett method. Graphs were generated with GraphPad Prism 9 software, and statistical analyses were done on JMP Pro v.17.2.0.
AlphaFold models of BamA homologues and analysis of secondary structure in the hinge region
Our initial dataset included 3992 complete RefSeq bacterial genomes (one for each species) covering 34 phyla, each one being the RefSeq reference or representative genome for the corresponding species (Supplementary Data 1). Based on the gene annotation in the genomes, 2179 complete bamA genes were identified (one copy in 2085 genomes, two copies in 92 genomes and three copies in 2 genomes). There were 23 duplicate accessions, which were removed. The remaining 2156 accessions were queried against UniProt, the corresponding accessions being retrieved for 765 of them (Supplementary Data 2). Among these, AlphaFold2 models were available in the Alphafold Protein Structure Database at the EMBL-EBI [https://alphafold.ebi.ac.uk/] for only 761 entries. We computed locally the remaining 1395 AlphaFold2 models, using a local implementation of AlphaFold72 based on MMseqs273 and ColabFold74. The secondary structure of all models was analysed using DSSP version 2.2.175. The region corresponding to residues 413–432 in BamA from E. coli was extracted and used for the generation of sequence and secondary structure conservation plots using a local implementation of WebLogo version 3.7.1276. All files were deposited in the repository [https://github.com/IorgaLab/BamAset] [https://doi.org/10.5281/zenodo.13986908]77.
Molecular dynamics simulations
Three-dimensional models of BamALVPR and BamAGSGS were built with MODELLER 10.078 using the structure of WT BamA (PDB 5LJO21) as a template. The tetrapeptides NTGS, LVPR and GSGS were extracted from WT BamA, BamALVPR and BamAGSGS, respectively. Molecular dynamics simulations were performed using GROMACS 2020.379 and the AMBER99SB-ILDN force field80. Each tetrapeptide was placed in a cubic periodic box of TIP3P waters, with a minimum distance of 1.0 nm on each side. The simulation box, obtained after hydration and neutralisation with Na+ and Cl– ions to physiological ionic strength (150 mM), was energy-minimised until convergence using a steepest descents algorithm to remove close contacts of water molecules with the solute. Molecular dynamics simulations with position restraints on the solute atoms were then performed for 100 ps (50 ps NVT and 50 ps NPT) to relax the water molecules around the solute, followed by the production run of 1 μs. During the position restraints and production runs, the Parrinello–Rahman and V-rescale methods were used for pressure and temperature coupling, respectively. Electrostatics were calculated with the particle mesh Ewald method. The P-LINCS algorithm was used to constrain bond lengths, and a time step of 2 fs was used throughout. All production simulations were performed using constant temperature, pressure, and number of particles (NPT) and periodic boundary conditions. The cut-off distance for Lennard-Jones interactions and electrostatic interactions was set at 1.0 nm. The dihedral angle distribution was extracted using the gmx angle module implemented in GROMACS 2020.3 and plotted with Xmgrace. All files were deposited in the repository [https://github.com/IorgaLab/BamAset] [https://doi.org/10.5281/zenodo.13986908]77. The molecular dynamics checklist can be found in Supplementary Table 7.
OMP purification
OMP substrates tOmpA, and OmpX were expressed as inclusion bodies in E. coli BL21 (DE3) cells as previously described in ref. 81. Inclusion bodies were solubilised in 25 mM Tris.HCl pH 8.0, 6 M guanidine HCl, and insoluble material removed by centrifugation (20,000×g, 20 min, 4 °C). Solubilised inclusion bodies were further purified by size exclusion chromatography using a Superdex 75 HiLoad 26/600 column (GE Healthcare).
SurA purification
The plasmid encoding SurA with an N-terminal 6x His-tag followed by a TEV cleavage site81 was transformed into BL21 (DE3) cells (Stratagene). Cells were grown in LB supplemented with 40 μg/mL kanamycin at 37 °C at 200 rpm until the OD600 reached ~0.6. The temperature was reduced to 20 °C, and induction of expression was performed by the addition of 0.4 mM IPTG. After incubation for ~18 h, the cells were harvested by centrifugation and resuspended in 20 mM Tris.HCl, pH 7.2, 150 mM NaCl, 20 mM imidazole, containing EDTA-free protease inhibitor tablets (Roche). Cells were lysed with a cell disruptor (Constant Cell Disruption Systems), the lysate was centrifuged (20 min, 4 °C, 39,000×g), and then applied to a HisTrap column (5 mL, Cytiva). The column was washed (20 mM Tris.HCl, pH 7.2, 150 mM NaCl and 20 mM imidazole), and elution of SurA was performed with 20 mM Tris.HCl, 150 mM NaCl, 500 mM imidazole, pH 7.2. The eluate was dialysed overnight against 20 mM Tris.HCl, 150 mM NaCl, pH 8.0, and then TEV protease was added along with 0.1% (v/v) β-mercaptoethanol. TEV cleavage was allowed to proceed overnight at 4 °C. The cleavage reaction was applied to a HisTrap column (5 mL, Cytiva) and the eluate, which contained the cleaved SurA, was dialysed against 20 mM Tris.HCl, 150 mM NaCl, pH 8.0, concentrated to ~200 μM with Vivaspin 20 concentrators (Sartorius; 5 kDa MWCO), aliquoted, snap-frozen in liquid nitrogen and stored at −80 °C.
BAM variant purification
pTrc99a vectors containing the respective BAM variants were used to transform BL21 (DE3) E. coli. Expression and purification followed the same protocol as described in ref. 31.
Reconstitution of BAM variants into E. coli polar lipid proteoliposomes
E. coli polar lipid extract, purchased as powder from Avanti Polar Lipids, was dissolved in 80:20 (v/v) chloroform/methanol at 20 mg/mL. Appropriate volumes were dried to thin films in clean Pyrex tubes at 42 °C under N2 gas, and were further dried by vacuum desiccation for ≥3 h. WT BAM, BAMLVPR, BAMT434A in TBS pH 8.0, 0.05% (w/v) DDM were mixed with E. coli polar lipid extract films solubilised in TBS pH 8.0, 0.05% (w/v) DDM in a 1:2 (w/w) ratio. To remove detergent and promote liposome formation, the mixtures were dialysed against 2 L of 20 mM Tris.HCl pH 8.0, 150 mM KCl using 12–14 kDa MWCO D-Tube™ Maxi Dialysers (Merck) at room temperature for 48 h with a total of six buffer changes. Following dialysis, proteoliposomes were pelleted twice by ultracentrifugation at 135,000×g for 30 min at 4 °C and were resuspended in TBS pH 8.0. Protein concentration was determined using a BCA assay (ThermoScientific), and successful reconstitution was determined by SDS–PAGE.
In vitro folding assays
Denatured OMP substrates were prepared in TBS pH 8.0, 8 M urea by buffer exchanging using Zeba™ Spin Desalting Columns (0.5 mL, 7 k MWCO, Thermo Scientific) and diluted to a concentration of 20 μM. A pre-fold solution containing 4 μM denatured OMP, 20 μM SurA, TBS pH 8.0, 1.6 M urea was immediately diluted 2-fold into a solution of 2 μM BAM-proteoliposomes to give final concentrations 1 μM BAM variant, 2 μM OMP, 0.8 M urea, 10 μM SurA. Samples were taken periodically, and the reaction was quenched by dilution into 4× SDS loading buffer (final concentration of 50 mM glycine-NaOH, pH 6.8, 10% (v/v) glycerol, 1.5% (w/v) SDS, 0.0015% (w/v) bromophenol blue), and a sample taken at the 3-h timepoint was boiled (>95 °C, 10 min). Reaction samples were loaded onto a 15% (w/v) acrylamide/bis-acrylamide (37.5:1) Tris-tricine SDS-PAGE gels and run at 60 mA until the dye-front reached the end of the gel. Gels were stained in InstantBlue™ (Experion) and were imaged using an Alliance Q9 Advanced gel doc (UVITEC, Cambridge, UK). Substrate folded and unfolded band intensities were quantified using ImageJ software (V1.53) (Fiji) and plotted as a fraction folded (IF/(IF + IUF)) against time. Data were fitted to a single exponential function in Igor Pro (V8.04), and initial rates were calculated by fitting the first 5% of the time course to a linear function. Uncropped and unprocessed gels used for quantification are available in the Source Data file.
CryoEM grid preparation and data collection
Purified BAM (both wild-type and variants) (0.05% DDM, TBS, pH 8.0) was diluted to ~3.3 mg/mL. The 1.2/1.3 Quantifoil copper 300 mesh grids were glow-discharged in air-vacuum for 20 s at 60 mA in a GlowQube Plus (Electron Microscopy Sciences). Three microliters of sample were applied to each grid, then blotted for 6 s with Whatman #1 filter paper at 4 °C and >80% humidity, before being plunge frozen into liquid ethane (Vitrobot Mark IV, Thermofisher). Data were collected on a 300 kV Titan Krios (ThermoFisher) in transmission electron microscopy mode using EPU (ThermoFisher) equipped with a Falcon4i detector without (BAMLVPR) or with a Selectris energy filter (10 eV slit width; BAMGSGS, BAMLVPR*). A 100 μm objective aperture was used. Additional data collection parameters for each sample are shown in Supplementary Table 8.
CryoEM image processing
Image processing was carried out in RELION 3.182/4.083 unless otherwise stated. All map sharpening, masking, and Fourier shell correlation (FSC) calculations were done in RELION, while angular distribution plots were generated in cryoSPARC. CryoEM processing workflows are shown in Supplementary Figs. 4–6.
BAMLVPR: 1850 micrographs were collected and motion-corrected (1.1 e−/Å2/frame). Following contrast transfer function (CTF) estimation (CtfFind484), particles were initially picked with crYOLO’s general model85, yielding 421,707 particles extracted into a 240-pixel box with 4× binning, which were reduced to 54,325 particles with high-resolution features with two rounds of 2D classification. These particles were used to train a new crYOLO model, which picked 765,341 particles with a low confidence threshold of 0.1, extracted into a 240-pixel box with 4× binning. Following two rounds of 2D classification, 45,498 particles were used to generate an initial model, and the best 177,882 particles were unbinned and re-extracted into 320-pixel boxes. Following two rounds of 3D classification (into three classes), 127,056 particles remained in a high-resolution class, which gave a 6.9 Å structure in an initial 3D refinement. The particles were then subjected to four cycles of polishing/CTF-refinement and masked 3D refinement (T = 8), yielding a final model at 4.3 Å resolution.
BAMGSGS: 7813 micrographs were collected and motion-corrected (0.9 e−/Å2/frame). Following CTF estimation, particles were picked from 2313 micrographs with the crYOLOs general model, yielding 342,172 particles, which were extracted with 4x binning in a 240-pixel box. Following a single round of 2D classification, 31,400 BAM-like particles were used to generate an initial model, followed by one round of 3D classification, resulting in 17,810 best particles. These were then used to train a new crYOLO model, which picked 975,473 with a low confidence threshold of 0.1. Two rounds of 2D classification yielded 589,795 particles, and a round of 3D classification resulted in 269,212 good particles. The particle stack was then unbinned and extracted into a 320-pixel box. An additional two rounds of 3D classification in RELION yielded 123,985 particles, which were subjected to ab initio modelling in cryoSPARC to further classify them to a final dataset of 75,556 particles. These particles gave a 5.2 Å model in cryoSPARC’s Non-Uniform refinement86, and were subsequently subjected to three cycles of polishing/CTF-refinement in RELION and cryoSPARC Non-Uniform refinement, resulting in a final model at 4.2 Å resolution.
BAMLVPR-T434A (LVPR*): 1504 micrographs were collected and motion-corrected (1.0 e−/Å2/frame). Following CTF estimation, 159,488 particles were picked with crYOLO's general model and extracted in a 240-pixel box with 4x binning. A single round of 2D classification yielded 49,764 BAM-like particles, which were subsequently used to generate an initial model and to train a new crYOLO model, which picked 393,859 particles at a low confidence threshold of 0.1. Two rounds of 2D classification cleaned the particle stack to 161,586 particles, followed by a single round of 3D classification to 79,991 particles, which were then extracted unbinned into 400-pixel boxes. The particles were subject to a final round of 2D classification, yielding a final stack of 62,266 particles. An additional 3D classification did not improve reconstructions. The particles were then reconstructed using cryoSPARCs Non-Uniform refinement86, resulting in a model of 6.2 Å, and subsequently subjected to two cycles of polishing/CTF-refinement in RELION and cryoSPARC non-uniform refinement86, resulting in a final model of 4.0 Å.
CryoEM model building
For BAMLVPR, an initial model was constructed using the BamA barrel from PDB 5D0O19 and the POTRA/periplasmic domains from PDB 8BWC51, with the LVPR insert. The model was then optimised in ISOLDE87 and subsequently passed through real-space refinement in PHENIX88 (v1.20) with secondary structure restraints, and manually optimised in COOT89. Geometry was assessed using molprobity90. BAMGSGS and BAMLVPR-T434A followed the same process, but using BAMLVPR (with appropriate mutations) as the initial model. Model statistics are shown in Supplementary Table 8. Images were generated in ChimeraX 1.7 or Pymol 2.5.5. Structure images in Figs. 1a and 4a were generated in ChimeraX-1.8 and 1.10.
HDX-MS
An automated HDX robot (LEAP Technologies) was coupled to an Acquity M-Class LC fitted with an HDX manager (Waters). Samples comprised BAM, BAMLVPR or BAMT434A (10 μM) in 10 mM potassium phosphate, pH 8.0, 0.02% n-dodecyl-β-D-maltoside (DDM). To initiate the HDX experiment, 95 μL of deuterated buffer (10 mM potassium phosphate, pH 8.0, 0.02% DDM) was added to 5 μL of protein-containing solution, and the mixture was incubated at 4 °C for 0.5 min, 2 min, or 30 min, as outlined previously in ref. 91.
For each time point and condition, three replicate measurements were performed. The HDX reaction was quenched by adding 100 μL of quench buffer (10 mM potassium phosphate, 4 M guanidine hydrochloride, pH 2.2) to 50 μL of the labelling reaction. The quenched sample (50 μL) was proteolysed by flowing through an immobilised pepsin column (Enzymate, Waters). The produced peptides were trapped on a VanGuard Pre-column [Acquity UPLC BEH C18 (1.7 μm, 2.1 mm × 5 mm, Waters)] for 3 min and the peptides were separated using a C18 column (75 μm × 150 mm, Waters, UK) by gradient elution of 0–40% (v/v) acetonitrile (0.1% v/v formic acid) in H2O (0.3% v/v formic acid) over 7 min at 40 μL min−1.
Peptides were detected using a Synapt G2Si mass spectrometer (Waters) operating in HDMSE mode, with dynamic range extension enabled. Ion mobility separation was used to separate peptides prior to fragmentation by collision-induced dissociation (CID) in the transfer cell. CID data were used for peptide identification, and uptake quantification was performed at the peptide level. Data were analysed using PLGS (v3.0.2) and DynamX (v3.0.0) software (Waters). Search parameters in PLGS were as follows: peptide and fragment tolerances = automatic, min fragment ion matches = 1, digest reagent = non-specific, false discovery rate = 4. Restrictions for peptides in DynamX were as follows: minimum intensity = 1000, minimum products per amino acid = 0.3, max sequence length = 25, max ppm error = 10, and file threshold = 3. The software Deuteros 2.0 was used to identify peptides with statistically significant increases/decreases in deuterium uptake (hybrid significance test with a 95 % confidence interval, P < 0.05) and to prepare Wood’s plots92. The raw HDX-MS data have been deposited to the ProteomeXchange Consortium via the PRIDE70 partner repository with the dataset identifier PXD057055. A summary of the HDX-MS data, as recommended by reported guidelines, is shown in Supplementary Table 9. Sequence coverage maps of BAM subunits are shown in Supplementary Fig. 9, and Wood’s plots are shown in Supplementary Figs. 10–14.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
CryoEM reconstructions and corresponding coordinates have been deposited in the Electron Microscopy Data Bank (EMBD) and the Protein Data Bank (PDB), respectively: BAM-LVPR (EMD-51930 [https://www.ebi.ac.uk/pdbe/entry/emdb/EMD-51930], 9H84), BAM-GSGS (EMD-51931 [https://www.ebi.ac.uk/pdbe/entry/emdb/EMD-51931], 9H85) and BAM-LVPR* (EMD-51933 [https://www.ebi.ac.uk/pdbe/entry/emdb/EMD-51933], 9H89). Previously published structures referenced in this study are available in the PDB under accession codes: 5D0O; 4C4V; 8BWC; 6LYU. The raw proteomics data have been deposited to the ProteomeXchange Consortium via PRIDE70 partner repository with the dataset identifier PXD057225. The raw HDX-MS data have been deposited to the ProteomeXchange Consortium via the PRIDE70 partner repository with the dataset identifier PXD057055. All files for the AlphaFold models of BamA homologues, the analysis of the secondary structure in the hinge region, and molecular dynamics simulations files (input.tpr files, final coordinates in.gro format for the three replicas, and raw data for the dihedral angle distribution plots) were deposited in the repository [https://github.com/IorgaLab/BamAset] [https://doi.org/10.5281/zenodo.13986908]77. The source data underlying Figs. 1b, c, e, 2, 4b, c, and 5, Supplementary Tables 1–3, and Supplementary Figs 1 and 7 are provided as a Source Data file. Source data are provided with this paper.
References
Silhavy, T. J., Kahne, D. & Walker, S. The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2, a000414 (2010).
Guest, R. L. & Silhavy, T. J. Cracking outer membrane biogenesis. Biochim Biophys. Acta Mol. Cell Res 1870, 119405 (2023).
Konovalova, A., Kahne, D. E. & Silhavy, T. J. Outer membrane biogenesis. Annu Rev. Microbiol 71, 539–556 (2017).
Rojas, E. R. et al. The outer membrane is an essential load-bearing element in Gram-negative bacteria. Nature 559, 617–621 (2018).
Deghelt, M. et al. Peptidoglycan-outer membrane attachment generates periplasmic pressure to prevent lysis in Gram-negative bacteria. Nat. Microbiol. 10, 1963–1974 (2025).
El Rayes, J., Rodriguez-Alonso, R. & Collet, J. F. Lipoproteins in Gram-negative bacteria: new insights into their biogenesis, subcellular targeting and functional roles. Curr. Opin. Microbiol. 61, 25–34 (2021).
Rollauer, S. E., Sooreshjani, M. A., Noinaj, N. & Buchanan, S. K. Outer membrane protein biogenesis in Gram-negative bacteria. Philos. Trans. R. Soc. Lond. B Biol. Sci. https://doi.org/10.1098/rstb.2015.0023 (2015).
Horne, J. E., Brockwell, D. J. & Radford, S. E. Role of the lipid bilayer in outer membrane protein folding in Gram-negative bacteria. J. Biol. Chem. https://doi.org/10.1074/jbc.REV120.011473 (2020).
Schiffrin, B., Brockwell, D. J. & Radford, S. E. Outer membrane protein folding from an energy landscape perspective. BMC Biol. 15, 123 (2017).
Voulhoux, R., Bos, M. P., Geurtsen, J., Mols, M. & Tommassen, J. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science 299, 262–265 (2003).
Noinaj, N. et al. Structural insight into the biogenesis of beta-barrel membrane proteins. Nature 501, 385–390 (2013).
Albrecht, R. et al. Structure of BamA, an essential factor in outer membrane protein biogenesis. Acta Crystallogr. D. Biol. Crystallogr. 70, 1779–1789 (2014).
Ni, D. et al. Structural and functional analysis of the beta-barrel domain of BamA from Escherichia coli. FASEB J. 28, 2677–2685 (2014).
Wu, T. et al. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell 121, 235–245 (2005).
Sklar, J. G. et al. Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc. Natl. Acad. Sci. USA 104, 6400–6405 (2007).
Kim, S. et al. Structure and function of an essential component of the outer membrane protein assembly machine. Science 317, 961–964 (2007).
Wu, R., Stephenson, R., Gichaba, A. & Noinaj, N. The big BAM theory: an open and closed case?. Biochim. Biophys. Acta Biomembr. 1862, 183062 (2020).
Malinverni, J. C. et al. YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli. Mol. Microbiol. 61, 151–164 (2006).
Gu, Y. et al. Structural basis of outer membrane protein insertion by the BAM complex. Nature 531, 64–69 (2016).
Han, L. et al. Structure of the BAM complex and its implications for biogenesis of outer-membrane proteins. Nat. Struct. Mol. Biol. 23, 192–196 (2016).
Iadanza, M. G. et al. Lateral opening in the intact beta-barrel assembly machinery captured by cryo-EM. Nat. Commun. 7, 12865 (2016).
Bakelar, J., Buchanan, S. K. & Noinaj, N. The structure of the beta-barrel assembly machinery complex. Science 351, 180–186 (2016).
Noinaj, N., Kuszak, A. J., Balusek, C., Gumbart, J. C. & Buchanan, S. K. Lateral opening and exit pore formation are required for BamA function. Structure 22, 1055–1062 (2014).
Robert, V. et al. Assembly factor Omp85 recognizes its outer membrane protein substrates by a species-specific C-terminal motif. PLoS Biol. 4, e377 (2006).
Doyle, M. T. & Bernstein, H. D. Bacterial outer membrane proteins assemble via asymmetric interactions with the BamA beta-barrel. Nat. Commun. 10, 3358 (2019).
Doyle, M. T. et al. Cryo-EM structures reveal multiple stages of bacterial outer membrane protein folding. Cell 185, 1143–1156 e1113 (2022).
Tomasek, D. et al. Structure of a nascent membrane protein as it folds on the BAM complex. Nature 583, 473–478 (2020).
Iadanza, M. G. et al. Distortion of the bilayer and dynamics of the BAM complex in lipid nanodiscs. Commun. Biol. 3, 766 (2020).
Schiffrin, B. et al. Effects of periplasmic chaperones and membrane thickness on BamA-catalyzed outer-membrane protein folding. J. Mol. Biol. 429, 3776–3792 (2017).
Gessmann, D. et al. Outer membrane beta-barrel protein folding is physically controlled by periplasmic lipid head groups and BamA. Proc. Natl. Acad. Sci. USA 111, 5878–5883 (2014).
White, P. et al. The role of membrane destabilisation and protein dynamics in BAM catalysed OMP folding. Nat. Commun. 12, 4174 (2021).
Lee, J. et al. Formation of a beta-barrel membrane protein is catalyzed by the interior surface of the assembly machine protein BamA. Elife. https://doi.org/10.7554/eLife.49787 (2019).
Kaur, H. et al. The antibiotic darobactin mimics a beta-strand to inhibit outer membrane insertase. Nature 593, 125–129 (2021).
Miller, R. D. et al. Computational identification of a systemic antibiotic for gram-negative bacteria. Nat. Microbiol 7, 1661–1672 (2022).
Imai, Y. et al. A new antibiotic selectively kills Gram-negative pathogens. Nature 576, 459–464 (2019).
Tata, M. et al. High-throughput suppressor screen demonstrates that RcsF monitors outer membrane integrity and not Bam complex function. Proc. Natl. Acad. Sci. USA. https://doi.org/10.1073/pnas.2100369118 (2021).
Rodriguez-Alonso, R. et al. Structural insight into the formation of lipoprotein-beta-barrel complexes. Nat. Chem. Biol. 16, 1019–1025 (2020).
Ruiz, N., Falcone, B., Kahne, D. & Silhavy, T. J. Chemical conditionality: a genetic strategy to probe organelle assembly. Cell 121, 307–317 (2005).
Braun, M. & Silhavy, T. J. Imp/OstA is required for cell envelope biogenesis in Escherichia coli. Mol. Microbiol. 45, 1289–1302 (2002).
Bos, M. P., Tefsen, B., Geurtsen, J. & Tommassen, J. Identification of an outer membrane protein required for the transport of lipopolysaccharide to the bacterial cell surface. Proc. Natl. Acad. Sci. USA 101, 9417–9422 (2004).
Wu, T. et al. Identification of a protein complex that assembles lipopolysaccharide in the outer membrane of Escherichia coli. Proc. Natl. Acad. Sci. USA 103, 11754–11759 (2006).
Chng, S. S., Ruiz, N., Chimalakonda, G., Silhavy, T. J. & Kahne, D. Characterization of the two-protein complex in Escherichia coli responsible for lipopolysaccharide assembly at the outer membrane. Proc. Natl. Acad. Sci. USA 107, 5363–5368 (2010).
Chimalakonda, G. et al. Lipoprotein LptE is required for the assembly of LptD by the beta-barrel assembly machine in the outer membrane of Escherichia coli. Proc. Natl. Acad. Sci. USA 108, 2492–2497 (2011).
Chng, S. S. et al. Disulfide rearrangement triggered by translocon assembly controls lipopolysaccharide export. Science 337, 1665–1668 (2012).
Ruiz, N., Chng, S. S., Hiniker, A., Kahne, D. & Silhavy, T. J. Nonconsecutive disulfide bond formation in an essential integral outer membrane protein. Proc. Natl. Acad. Sci. USA 107, 12245–12250 (2010).
Mikheyeva, I. V., Sun, J., Huang, K. C. & Silhavy, T. J. Mechanism of outer membrane destabilization by global reduction of protein content. Nat. Commun. 14, 5715 (2023).
Mecsas, J., Rouviere, P. E., Erickson, J. W., Donohue, T. J. & Gross, C. A. The activity of sigma E, an Escherichia coli heat-inducible sigma-factor, is modulated by expression of outer membrane proteins. Genes Dev. 7, 2618–2628 (1993).
Hart, E. M. et al. A small-molecule inhibitor of BamA impervious to efflux and the outer membrane permeability barrier. Proc. Natl. Acad. Sci. USA 116, 21748–21757 (2019).
MacArthur, M. W. & Thornton, J. M. Influence of proline residues on protein conformation. J. Mol. Biol. 218, 397–412 (1991).
Schimmel, P. R. & Flory, P. J. Conformational energies and configurational statistics of copolypeptides containing L-proline. J. Mol. Biol. 34, 105–120 (1968).
Haysom, S. F. et al. Darobactin B stabilises a lateral-closed conformation of the BAM complex in E. coli cells. Angew. Chem. Int. Ed. Engl. https://doi.org/10.1002/anie.202218783 (2023).
Xiao, L. et al. Structures of the beta-barrel assembly machine recognizing outer membrane protein substrates. FASEB J. 35, e21207 (2021).
Stokes, J. M. et al. Cold stress makes Escherichia coli susceptible to glycopeptide antibiotics by altering outer membrane integrity. Cell Chem. Biol. 23, 267–277 (2016).
Nikaido, H. Restoring permeability barrier function to outer membrane. Chem. Biol. 12, 507–509 (2005).
Calabrese, A. N. & Radford, S. E. Mass spectrometry-enabled structural biology of membrane proteins. Methods 147, 187–205 (2018).
Noinaj, N., Rollauer, S. E. & Buchanan, S. K. The beta-barrel membrane protein insertase machinery from Gram-negative bacteria. Curr. Opin. Struct. Biol. 31, 35–42 (2015).
Diederichs, K. A., Buchanan, S. K. & Botos, I. Building better barrels—beta-barrel biogenesis and insertion in bacteria and mitochondria. J. Mol. Biol. 433, 166894 (2021).
Leonard-Rivera, M. & Misra, R. Conserved residues of the putative L6 loop of Escherichia coli BamA play a critical role in the assembly of beta-barrel outer membrane proteins, including that of BamA itself. J. Bacteriol. 194, 4662–4668 (2012).
Doyle, M. T. & Bernstein, H. D. Function of the Omp85 superfamily of outer membrane protein assembly factors and polypeptide transporters. Annu Rev. Microbiol. 76, 259–279 (2022).
Mamou, G. et al. Peptidoglycan maturation controls outer membrane protein assembly. Nature 606, 953–959 (2022).
Thewasano, N., Germany, E. M., Maruno, Y., Nakajima, Y. & Shiota, T. Categorization of Escherichia coli outer membrane proteins by dependence on accessory proteins of the beta-barrel assembly machinery complex. J. Biol. Chem. 299, 104821 (2023).
Warner, L. R., Gatzeva-Topalova, P. Z., Doerner, P. A., Pardi, A. & Sousa, M. C. Flexibility in the periplasmic domain of BamA is important for function. Structure 25, 94–106 (2017).
Fenn, K. L. et al. Outer membrane protein assembly mediated by BAM-SurA complexes. Nat. Commun. 15, 7612 (2024).
Daimon, Y. et al. The TPR domain of BepA is required for productive interaction with substrate proteins and the beta-barrel assembly machinery complex. Mol. Microbiol. 106, 760–776 (2017).
Lehner, P. A. et al. Architecture and conformational dynamics of the BAM-SurA holo insertase complex. Sci. Adv. 11, eads6094 (2025).
Sinensky, M. Homeoviscous adaptation-a homeostatic process that regulates the viscosity of membrane lipids in Escherichia coli. Proc. Natl. Acad. Sci. USA 71, 522–525 (1974).
Wang, J. D., Herman, C., Tipton, K. A., Gross, C. A. & Weissman, J. S. Directed evolution of substrate-optimized GroEL/S chaperonins. Cell 111, 1027–1039 (2002).
Horwich, A. L. & Fenton, W. A. Chaperonin-assisted protein folding: a chronologue. Q Rev. Biophys. 53, e4 (2020).
Hussain, S. & Bernstein, H. D. The Bam complex catalyzes efficient insertion of bacterial outer membrane proteins into membrane vesicles of variable lipid composition. J. Biol. Chem. 293, 2959–2973 (2018).
Perez-Riverol, Y. et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 50, D543–D552 (2022).
Miller, J. Experiments in Molecular Genetics (Cold Spring Harbor, 1972).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Mirdita, M., Steinegger, M. & Soding, J. MMseqs2 desktop and local web server app for fast, interactive sequence searches. Bioinformatics 35, 2856–2858 (2019).
Mirdita, M. et al. ColabFold: making protein folding accessible to all. Nat. Methods 19, 679–682 (2022).
Kabsch, W. & Sander, C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22, 2577–2637 (1983).
Crooks, G. E., Hon, G., Chandonia, J. M. & Brenner, S. E. WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 (2004).
Csoma, N., Cho, S.-H. & Collet, J.-F. & Bogdan I. BamAset: v0.2. Zenodo https://doi.org/10.5281/zenodo.13986908 (2025).
Sali, A. & Blundell, T. L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 (1993).
Van Der Spoel, D. et al. GROMACS: fast, flexible, and free. J. Comput Chem. 26, 1701–1718 (2005).
Lindorff-Larsen, K. et al. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins 78, 1950–1958 (2010).
Calabrese, A. N. et al. Inter-domain dynamics in the chaperone SurA and multi-site binding to its outer membrane protein clients. Nat. Commun. 11, 2155 (2020).
Zivanov, J., Nakane, T. & Scheres, S. H. W. Estimation of high-order aberrations and anisotropic magnification from cryo-EM data sets in RELION-3.1. IUCrJ 7, 253–267 (2020).
Kimanius, D., Dong, L., Sharov, G., Nakane, T. & Scheres, S. H. W. New tools for automated cryo-EM single-particle analysis in RELION-4.0. Biochem J. 478, 4169–4185 (2021).
Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Wagner, T. et al. SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun. Biol. 2, 218 (2019).
Punjani, A., Zhang, H. & Fleet, D. J. Non-uniform refinement: adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods 17, 1214–1221 (2020).
Croll, T. I. ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr. D. Struct. Biol. 74, 519–530 (2018).
Afonine, P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D. Struct. Biol. 74, 531–544 (2018).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 66, 486–501 (2010).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D. Biol. Crystallogr. 66, 12–21 (2010).
Schiffrin, B. et al. Dynamic interplay between the periplasmic chaperone SurA and the BAM complex in outer membrane protein folding. Commun. Biol. 5, 560 (2022).
Lau, A. M., Claesen, J., Hansen, K. & Politis, A. Deuteroments in molecular gs 2.0: peptide-level significance testing of data from hydrogen deuterium exchange mass spectrometry. Bioinformatics 37, 270–272 (2021).
Acknowledgements
We are indebted to the members of the Collet, Radford, Ranson and Calabrese laboratories for many helpful suggestions and discussions. We thank S-I Matsuyama (Rikkyo University, Japan) and H. Bernstein (NIH, Bethesda, USA) for providing antibodies, H. Bernstein for providing plasmids and C.A. Gross (UCSF, San Francisco, USA) for providing strains. J.-F.C. acknowledge support of the Fonds de la Recherche Scientifique (FNRS) grant agreements WELBIO-CR-2019C-03, the Fédération Wallonie-Bruxelles (ARC 17/22-087) awarded to J.-F.C. N.C. was a research fellow of FNRS. R.R.-A. was funded by the European Commission via the International Training Network Train2Target (no. 721484). A.N.C. and M.O. acknowledge support of a Sir Henry Dale Fellowship (awarded to A.N.C.) jointly funded by Wellcome and the Royal Society (220628/Z/20/Z). A.K.C. acknowledges the award of a studentship funded by EPSRC (EP/W524372/1, project reference 2879844). A.N.C. and S.E.R. acknowledge funding from the BBSRC (BB/Y00034X/1). Funding from BBSRC (BB/M012573/1) and Wellcome (208385/Z/17/Z) enabled the purchase of mass spectrometry equipment. S.E.R. is funded by a Royal Society Professorial Fellowship (RSRP\R1\211057). J.M.M. was funded by Wellcome (222373/Z/21/Z). J.M.M., S.E.R. and N.A.R. also thank MRC for funding (MR/Y012453/1). We thank E.O.S. (G0G0818N/EOS, ID 30550343) and the University of Leeds for funding J.M.W. via an International Institutional Award (BB/Y514160/1). CryoEM data were collected at the Astbury Biostructure Laboratory, funded by the University of Leeds and Wellcome (108466/Z/15/Z; 221524/Z/20/Z), and we thank facility staff for their technical input. B.I.I. is supported by a grant from the French National Research Agency (ANR) through the PPR Antibioresistance programme (ANR-20-PAMR-0010). T.F.S. was funded by the German Federal Ministry of Education and Research (BMBF, via grant GBi2S) and German Centre for Infection Research (DZIF) 09.918.
Author information
Authors and Affiliations
Contributions
Writing: J.-F.C., S.E.R., A.N.C., N.C., J.M.M. and N.A.R. Conceptualisation: N.C., R.R.A., S.-H.C., J.-F.C., S.E.R., and A.N.C. Investigation, strain construction, construct cloning, and protein expression and purification: N.C., J.M.W. and R.R.A. Proteomics: A.N.C., M.O., and A.K.C. Phylogenetic analysis and molecular dynamics simulations: B.I.I. Structure determination and analysis: J.M.M., N.A.R. HDX: A.N.C. and J.M.W. Provided darobactin B for the study: T.F.S. Data analysis and interpretation: N.C., J.M.M, J.M.W., R.R.A., A.N.C., S.E.R., and J.-F.C. All authors discussed the results and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Csoma, N., Machin, J.M., Whitehouse, J.M. et al. Molecular insights into how the motions of the β-barrel and POTRA domains of BamA are coupled for efficient function. Nat Commun 16, 8832 (2025). https://doi.org/10.1038/s41467-025-63897-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-63897-y