Abstract
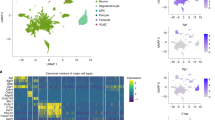
The arcuate nucleus of the hypothalamus plays a central role in sensing and integrating nutritional, hormonal, and neural signals that regulate feeding, energy homeostasis, growth, and reproduction, all of which show pronounced sex differences. However, the cellular mechanisms underlying these responses remain poorly understood. We performed snRNA-seq of the mediobasal hypothalamus, focusing on the arcuate nucleus, in female and male mice under different nutritional states. Analysis of 42 cell types revealed that Agrp neurons were most sensitive to nutritional changes, dopaminergic neurons showed strong sex-specific differences, and KNDy neurons were highly responsive to both sex and nutrition. Pomc neurons displayed moderate nutritional sensitivity. Most glial populations were stable, although microglia and oligodendrocytes showed moderate variation. Cell–cell communication analysis identified neurotrophic factor signaling as a key pathway regulated by sex and nutrition. This study represents a major effort to comprehensively characterize sex-specific differences in arcuate nucleus response across nutritional conditions.
Similar content being viewed by others
Data availability
The fully processed snRNA-seq dataset generated in this study have been deposited in the Gene Expression Omnibus (GEO) database under accession code GSE282955. Source data are provided as a Source Data file. Source data are provided with this paper.
Code availability
Detailed analysis codes used in this study are available on the Github repository 663 (https://github.com/jbeanphd/ARH_Sex_by_Nutr) https://doi.org/10.5281/zenodo.18276251.
References
World Health Organization. World Health Statistics 2023: Monitoring Health for the SDGs, Sustainable Development Goals. 1–119 https://www.who.int/publications-detail-redirect/9789240074323 (WHO, 2023).
Bryan, S. et al. NHSR 158. National Health and Nutrition Examination Survey 2017–March 2020 Pre-Pandemic Data Files. https://stacks.cdc.gov/view/cdc/106273 (NHSR, 2021) https://doi.org/10.15620/cdc:106273.
Collaboration, P. S Body-mass index and cause-specific mortality in 900 000 adults: collaborative analyses of 57 prospective studies. Lancet 373, 1083–1096 (2009).
Kannel, W. B. et al. Regional obesity and risk of cardiovascular disease; the Framingham study. J. Clin. Epidemiol. 44, 183–190 (1991).
Gesta, S., Tseng, Y.-H. & Kahn, C. R. Developmental origin of fat: tracking obesity to its source. Cell 131, 242–256 (2007).
Lee, C. G. et al. Adipokines, inflammation, and visceral adiposity across the menopausal transition: a prospective study. J. Clin. Endocrinol. Metab. 94, 1104–1110 (2009).
Xu, Y. et al. Distinct hypothalamic neurons mediate estrogenic effects on energy homeostasis and reproduction. Cell Metab. 14, 453–465 (2011).
Wislocki, G. B. & King, L. S. The permeability of the hypophysis and hypothalamus to vital dyes, with a study of the hypophyseal vascular supply. Am. J. Anat. 58, 421–472 (1936).
Gross, P. M. & Weindl, A. Peering through the windows of the brain. J. Cereb. Blood Flow. Metab. J. Int. Soc. Cereb. Blood Flow. Metab. 7, 663–672 (1987).
Zhang, Y. et al. Positional cloning of the mouse obese gene and its human homologue. Nature 372, 425–432 (1994).
Castro-Dufourny, I., Carrasco, R., Prieto, R. & Pascual, J. M. Jean Camus and Gustave Roussy: pioneering French researchers on the endocrine functions of the hypothalamus. Pituitary 20, 409–421 (2017).
Krashes, M. J. et al. Rapid, reversible activation of AgRP neurons drives feeding behavior in mice. J. Clin. Invest. 121, 1424–1428 (2011).
Zhang, X. & van den Pol, A. N. Hypothalamic arcuate nucleus tyrosine hydroxylase neurons play orexigenic role in energy homeostasis. Nat. Neurosci. 19, 1341–1347 (2016).
Fenselau, H. et al. A rapidly acting glutamatergic ARC→PVH satiety circuit postsynaptically regulated by α-MSH. Nat. Neurosci. 20, 42–51 (2017).
Zhan, C. et al. Acute and long-term suppression of feeding behavior by POMC neurons in the brainstem and hypothalamus, respectively. J. Neurosci. J. Soc. Neurosci. 33, 3624–3632 (2013).
Herbison, A. E. Control of puberty onset and fertility by gonadotropin-releasing hormone neurons. Nat. Rev. Endocrinol. 12, 452–466 (2016).
Thorner, M. O. et al. Physiological role of somatostatin on growth hormone regulation in humans. Metabolism 39, 40–42 (1990).
Campbell, J. N. et al. A molecular census of arcuate hypothalamus and median eminence cell types. Nat. Neurosci. 20, 484–496 (2017).
Deng, G. et al. Single-nucleus rna sequencing of the hypothalamic arcuate nucleus of C57BL/6J mice after prolonged diet-induced obesity. Hypertens. Dallas Tex. 76, 589–597 (2020).
Steuernagel, L. et al. HypoMap—a unified single-cell gene expression atlas of the murine hypothalamus. Nat. Metab. 4, 1402–1419 (2022).
Germain, P.-L., Lun, A., Garcia Meixide, C., Macnair, W. & Robinson, M. D. Doublet identification in single-cell sequencing data using scDblFinder. Research 10, 979 (2021).
Li, H. et al. Fly cell atlas: a single-nucleus transcriptomic atlas of the adult fruit fly. Science 375, eabk2432 (2022).
Lu, T.-C. et al. Aging fly cell atlas identifies exhaustive aging features at cellular resolution. Science 380, eadg0934 (2023).
Luecken, M. D. & Theis, F. J. Current best practices in single-cell RNA-seq analysis: a tutorial. Mol. Syst. Biol. 15, e8746 (2019).
Young, M. D. & Behjati, S. SoupX removes ambient RNA contamination from droplet-based single-cell RNA sequencing data. GigaScience 9, giaa151 (2020).
MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data | Genome Biology | Full Text. https://genomebiology.biomedcentral.com/articles/10.1186/s13059-015-0844-5in/hdWGCNA.R.
Luquet, S., Perez, F. A., Hnasko, T. S. & Palmiter, R. D. NPY/AgRP neurons are essential for feeding in adult mice but can be ablated in neonates. Science 310, 683–685 (2005).
van Maanen, J. H. & Smelik, P. G. Depletion of monoamines in the hypothalamus and prolactin secretion. Acta Physiol. Pharmacol. Neerl. 14, 519–520 (1967).
Tullai, J. W. et al. Immediate-early and delayed primary response genes are distinct in function and genomic architecture. J. Biol. Chem. 282, 23981–23995 (2007).
Jurgens, H. A. & Johnson, R. W. Dysregulated neuronal-microglial cross-talk during aging, stress and inflammation. Exp. Neurol. 233, 40–48 (2012).
Cai, Z. & Xiao, M. Oligodendrocytes and Alzheimer’s disease. Int. J. Neurosci. 126, 97–104 (2016).
Jin, S. et al. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 12, 1088 (2021).
Alhadeff, A. L. et al. A neural circuit for the suppression of pain by a competing need state. Cell 173, 140–152 (2018).
Burnett, C. J. et al. Hunger-driven motivational state competition. Neuron 92, 187–201 (2016).
Dietrich, M. O., Zimmer, M. R., Bober, J. & Horvath, T. L. Hypothalamic AGRP neurons drive stereotypic behaviors beyond feeding. Cell 160, 1222–1232 (2015).
Padilla, S. L. et al. Agouti-related peptide neural circuits mediate adaptive behaviors in the starved state. Nat. Neurosci. 19, 734–741 (2016).
Xu, Y., Elmquist, J. K. & Fukuda, M. Central nervous control of energy and glucose balance: focus on the central melanocortin system. Ann. N. Y. Acad. Sci. 1243, 1–14 (2011).
Castellano, J. M. et al. Early metabolic programming of puberty onset: impact of changes in postnatal feeding and rearing conditions on the timing of puberty and development of the hypothalamic kisspeptin system. Endocrinology 152, 3396–3408 (2011).
Navarro, V. M. Metabolic regulation of kisspeptin—the link between energy balance and reproduction. Nat. Rev. Endocrinol. 16, 407–420 (2020).
Padilla, S. L. et al. AgRP to Kiss1 neuron signaling links nutritional state and fertility. Proc. Natl. Acad. Sci. USA 114, 2413–2418 (2017).
Donato, J. et al. Leptin’s effect on puberty in mice is relayed by the ventral premammillary nucleus and does not require signaling in Kiss1 neurons. J. Clin. Invest. 121, 355–368 (2011).
Sun, J. et al. Caloric restriction in female reproduction: is it beneficial or detrimental? Reprod. Biol. Endocrinol. 19, 1 (2021).
Caron, M. G. et al. Dopaminergic receptors in the anterior pituitary gland. Correlation of [3H]dihydroergocryptine binding with the dopaminergic control of prolactin release. J. Biol. Chem. 253, 2244–2253 (1978).
Manjarrez-Gutiérrez, G., González-Ramírez, M., de Oca, A. B.-M., Herrera-Márquez, R. & Hernández-Rodríguez, J. Serotonin and dopamine in the hypothalamus of control and malnourished mother rats during pregnancy and lactation and body composition of their offspring. Nutr. Neurosci. 16, 225–232 (2013).
Lee, A. K. et al. Effect of high-fat feeding on expression of genes controlling availability of dopamine in mouse hypothalamus. Nutrition 26, 411–422 (2010).
Moore, S. W., Tessier-Lavigne, M. & Kennedy, T. E. Netrins and their receptors. Adv. Exp. Med. Biol. 621, 17–31 (2007).
Cavallaro, U. & Christofori, G. Cell adhesion and signalling by cadherins and Ig-CAMs in cancer. Nat. Rev. Cancer 4, 118–132 (2004).
Wu, Y., Dissing-Olesen, L., MacVicar, B. A. & Stevens, B. Microglia: dynamic mediators of synapse development and plasticity. Trends Immunol. 36, 605–613 (2015).
Huang, Y. et al. Maternal dietary fat during lactation shapes single nucleus transcriptomic profile of postnatal offspring hypothalamus in a sexually dimorphic manner in mice. Nat. Commun. 15, 2382 (2024).
Thaler, J. P. et al. Obesity is associated with hypothalamic injury in rodents and humans. J. Clin. Invest. 122, 153–162 (2012).
Lv, Z. et al. Clearance of β-amyloid and synapses by the optogenetic depolarization of microglia is complement selective. Neuron 112, 740–754 (2024).
Nave, K.-A. & Werner, H. B. Myelination of the nervous system: mechanisms and functions. Annu. Rev. Cell Dev. Biol. 30, 503–533 (2014).
Kohnke, S. et al. Nutritional regulation of oligodendrocyte differentiation regulates perineuronal net remodeling in the median eminence. Cell Rep. 36, 109362 (2021).
Saab, A. S. et al. Oligodendroglial NMDA receptors regulate glucose import and axonal energy metabolism. Neuron 91, 119–132 (2016).
Scarlett, J. M. et al. Peripheral mechanisms mediating the sustained antidiabetic action of FGF1 in the brain. Diabetes 68, 654–664 (2019).
Hwang, E. et al. Sustained inhibition of NPY/AgRP neuronal activity by FGF1. JCI Insight 7, e160891 (2022).
Bean, J. C. et al. Genetic labeling reveals novel cellular targets of schizophrenia susceptibility gene: distribution of GABA and non-GABA ErbB4-positive cells in adult mouse brain. J. Neurosci. 34, 13549–13566 (2014).
Santiago-Marrero, I. et al. Energy expenditure homeostasis requires ErbB4, an obesity risk gene, in the paraventricular nucleus. eNeuro 10, 0139 (2023).
Paxinos, G. & Franklin, K. B. J. The Mouse Brain in Stereotaxic Coordinates. (Academic Press, 2001).
Lein, E. S. et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176 (2007).
McLaughlin, C. N., Qi, Y., Quake, S. R., Luo, L. & Li, H. Isolation and RNA sequencing of single nuclei from Drosophila tissues. STAR Protoc. 3, 101417 (2022).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Browaeys, R., Saelens, W. & Saeys, Y. NicheNet: modeling intercellular communication by linking ligands to target genes. Nat. Methods 17, 159–162 (2020).
Morabito, S., Reese, F., Rahimzadeh, N., Miyoshi, E. & Swarup, V. hdWGCNA identifies co-expression networks in high-dimensional transcriptomics data. Cell Rep. Methods 3, 100498 (2023).
Acknowledgements
H.L. discloses support for the research of this work from the CPRIT Scholar in Cancer Research [RR200063], NIH/NIA [U01AG086143], NIH [DP2AT013275], the Longevity Impetus Grant, the Ted Nash Long Life Foundation, and the Welch Foundation. Y.X. discloses support for the research of this work from the Silver Endowment. X.F. discloses support for the research of this work from NIDDK [1F32DK138685-01A1]. M.W. discloses support for the research of this work from NIMHD [1F32HD112123-01A1].
Author information
Authors and Affiliations
Contributions
J.C.B., H.L., and Y.X. co-conceptualized the study. J.C.B. and J.J. performed the experiments and analyses, with assistance from T.C.L., H.L., K.M.M., D.A.T., S.V.J., M.E.B., J.C., Y.D., X.F., X.G., J.H., Y.L., H.L., Q.L., Y.L., Y.S., L.T., M.W., X.X., Y.Y., M.Y., X.L., M.S., F.W., O.Z.G., Y.Y., Y.H., C.W., and Y.Q., H.L., and Y.X. supervised the study.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Tune Pers and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Bean, J.C., Jian, J., Lu, TC. et al. Sex-specific differences in mediobasal hypothalamus in response to nutritional states. Nat Commun (2026). https://doi.org/10.1038/s41467-026-69239-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-026-69239-w