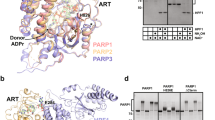
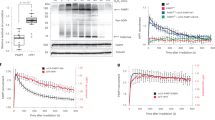
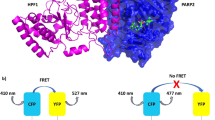
Abstract
PARP1 detection of DNA strand breaks allosterically leads to PARP1 synthesis of poly(ADP-ribose) modifications that signal DNA damage. HPF1 engages activated PARP1 to control modification site selection. Understanding of the mechanism of DNA break detection and catalytic activation is incomplete, due largely to limited structural information for full-length PARP1. Here, single-particle cryo-EM provides views of the full complement of PARP1 domains engaging a DNA single-strand break in the presence of HPF1 and a fragment of binding partner Timeless. Cryo-EM, single-molecule DNA dynamics, and small-angle X-ray scattering analysis indicate that PARP1 remains dynamic even when the multi-domain structure is organized on a DNA break, with the minimal catalytic region displaying high mobility relative to domains engaging damage. We propose that the organization of PARP1 domains on a DNA break releases a tethered, constitutively active catalytic region to modify molecules in a radius surrounding the DNA break site.
Similar content being viewed by others
Data availability
The raw cryo-EM data are deposited in the EM Public Image Archive under accession code EMPIAR-12081. Maps and atomic models are deposited in the EM Data Bank and Protein Data Bank (PDB) with accession codes EMD-48285, PDB: 9mi8 (PARP1 NTD/DNA) and EMD-48313, PDB: 9mja (PARP1 ART-HPF1-EB47). SEC-SAXS data of the PARP1-DNA-EB47-HPF1-Timeless complex is available at Simple Scattering (simplescattering.com; ID XS1A4OI7). The following structures from the PDB (www.rcsb.org) were used in this study:
• 4dqy (PARP1 Zn1, Zn3, WGR, HD, ART with a DNA double strand break)
• 2n8a (PARP1 Zn1, Zn2 with a DNA single strand break)
• 4xhu (PARP1 ART with a Timeless fragment)
• 6m3i (PARP1 ART with HPF1)
• 7aab (PARP1 HD-ART with EB47)
• 6tx2 (HPF1)
• 6tx3 (PARP2 ART with HPF1)
• 7s6m (PARP1 Zn1, Zn3, WGR, mutant HD, ART with a DNA double strand break)
• 7s81 (PARP1 Zn1, Zn3, WGR, HD with a DNA double strand break)
• 3odc (PARP1 Zn2 with a DNA double strand break)
• 3dsd (Archaeal Mre11 bound to DNA)
Source data are provided with this paper.
References
Lüscher, B. et al. ADP-ribosyltransferases, an update on function and nomenclature. FEBS J. https://doi.org/10.1111/febs.16142 (2021).
Suskiewicz, M. J., Prokhorova, E., Rack, J. G. M. & Ahel, I. ADP-ribosylation from molecular mechanisms to therapeutic implications. Cell 186, 4475–4495 (2023).
Rouleau-Turcotte, É & Pascal, J. M. ADP-ribose contributions to genome stability and PARP enzyme trapping on sites of DNA damage; paradigm shifts for a coming-of-age modification. J. Biol. Chem. 299, 105397 (2023).
Pandey, N. & Black, B. E. Rapid detection and signaling of DNA damage by PARP-1. Trends Biochem. Sci. 46, 744–757 (2021).
Duma, L. & Ahel, I. The function and regulation of ADP-ribosylation in the DNA damage response. Biochem. Soc. Trans. 51, 995–1008 (2023).
Buch-Larsen, S. C. et al. Mapping physiological ADP-ribosylation using activated ion electron transfer dissociation. Cell Rep. 32, 108176–108176 (2020).
Hanzlikova, H. et al. The importance of poly(ADP-ribose) polymerase as a sensor of unligated Okazaki fragments during DNA replication. Mol. Cell 71, 319–331 (2018).
Masson, M. et al. XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol. Cell. Biol. 18, 3563–3571 (1998).
Bonfiglio, J. J. et al. Serine ADP-ribosylation depends on HPF1. Mol. Cell 65, 932–940 (2017).
Suskiewicz, M. J. et al. HPF1 completes the PARP active site for DNA damage-induced ADP-ribosylation. Nature 579, 598–602 (2020).
Sun, F.-H. et al. HPF1 remodels the active site of PARP1 to enable the serine ADP-ribosylation of histones. Nat. Commun. 12, 1028 (2021).
Xie, S. et al. Timeless interacts with PARP-1 to promote homologous recombination repair. Mol. Cell 60, 163–176 (2015).
Young, L. M. et al. TIMELESS forms a complex with PARP1 distinct from its complex with TIPIN and plays a role in the DNA damage response. Cell Rep. 13, 451–459 (2015).
Petropoulos, M. et al. Transcription–replication conflicts underlie sensitivity to PARP inhibitors. Nature 628, 433–441 (2024).
Dawicki-McKenna, J. M. et al. PARP-1 activation requires local unfolding of an autoinhibitory domain. Mol. Cell 60, 755–768 (2015).
Langelier, M.-F., Planck, J. L., Roy, S. & Pascal, J. M. Structural basis for DNA damage–dependent poly(ADP-ribosyl)ation by human PARP-1. Science 336, 728–732 (2012).
Suskiewicz, M. J. et al. Updated protein domain annotation of the PARP protein family sheds new light on biological function. Nucleic Acids Res. https://doi.org/10.1093/nar/gkad514 (2023).
Desmarais, Y., Ménard, L., Lagueux, J. & Poirier, G. G. Enzymological properties of poly(ADP-ribose)polymerase: characterization of automodification sites and NADase activity. Biochim. Biophys. Acta Protein Struct. Mol. Enzymol. 1078, 179–186 (1991).
Gagné, J.-P. et al. Quantitative site-specific ADP-ribosylation profiling of DNA-dependent PARPs. DNA Repair 30, 68–79 (2015).
Prokhorova, E. et al. Serine-linked PARP1 auto-modification controls PARP inhibitor response. Nat. Commun. 12, 4055 (2021).
Bryant, H. E. et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434, 913–917 (2005).
Farmer, R., Gautam, B., Singh, S., Yadav, P. & Jain, P. Virtual screening of AmpC/β-lactamase as target for antimicrobial resistance in Pseudomonas aeruginosa. Bioinformation 4, 290–4 (2010).
Zandarashvili, L. et al. Structural basis for allosteric PARP-1 retention on DNA breaks. Science 368, eaax6367 (2020).
Langelier, M.-F., Zandarashvili, L., Aguiar, P. M., Black, B. E. & Pascal, J. M. NAD+ analog reveals PARP-1 substrate-blocking mechanism and allosteric communication from catalytic center to DNA-binding domains. Nat. Commun. 9, 844 (2018).
Jagtap, P. G. et al. The discovery and synthesis of novel adenosine substituted 2,3-dihydro-1H-isoindol-1-ones: potent inhibitors of poly(ADP-ribose) polymerase-1 (PARP-1). Bioorg. Med. Chem. Lett. 14, 81–85 (2004).
Langelier, M.-F., Servent, K. & Rogers, E. A third zinc-binding domain of human poly (ADP-ribose) polymerase-1 coordinates DNA-dependent enzyme activation https://doi.org/10.1074/jbc.m708558200 (2008).
Langelier, M.-F., Planck, J. L., Roy, S. & Pascal, J. M. Crystal structures of poly(ADP-ribose) polymerase-1 (PARP-1) zinc fingers bound to DNA: structural and functional insights into DNA-dependent PARP-1 activity. J. Biol. Chem. 286, 10690–701 (2011).
Eustermann, S. et al. Structural basis of detection and signaling of DNA single-strand breaks by human PARP-1. Mol. Cell 60, 742–754 (2015).
Le Cam, E. et al. Conformational analysis of a 139 base-pair DNA fragment containing a single-stranded break and its interaction with human poly(ADP-ribose) polymerase. J. Mol. Biol. 235, 1062–1071 (1994).
Xue, H. et al. A two-step mechanism governing PARP1-DNA retention by PARP inhibitors. Sci. Adv. 8, eabq0414 (2022).
Langelier, M.-F., Billur, R., Sverzhinsky, A., Black, B. E. & Pascal, J. M. HPF1 dynamically controls the PARP1/2 balance between initiating and elongating ADP-ribose modifications. Nat. Commun. 12, 6675 (2021).
Weissenberger, G., Henderikx, R. J. M. & Peters, P. J. Understanding the invisible hands of sample preparation for cryo-EM. Nat. Methods 18, 463–471 (2021).
Taylor, K. A. & Glaeser, R. M. Retrospective on the early development of cryoelectron microscopy of macromolecules and a prospective on opportunities for the future. J. Struct. Biol. 163, 214–223 (2008).
D’Imprima, E. et al. Protein denaturation at the air-water interface and how to prevent it. eLife 8, e42747 (2019).
Sefer, A. et al. Structural dynamics of DNA strand break sensing by PARP-1 at a single-molecule level. Nat. Commun. 13, 6569 (2022).
Langelier, M.-F., Ruhl, D. D., Planck, J. L., Kraus, W. L. & Pascal, J. M. The Zn3 domain of human poly (ADP-ribose) polymerase-1 (PARP-1) functions in both DNA-dependent poly (ADP-ribose) synthesis activity and chromatin compaction. J. Biol. Chem. 285, 18877–18887 (2010).
Armstrong, M. et al. Microscale fluid behavior during cryo-EM sample blotting. Biophys. J. 118, 708–719 (2020).
Ogden, T. E. H. et al. Dynamics of the HD regulatory subdomain of PARP-1; substrate access and allostery in PARP activation and inhibition. Nucleic Acids Res. 49, 2266–2288 (2021).
Rouleau-Turcotte, É, Krastev, D. B., Pettitt, S. J., Lord, C. J. & Pascal, J. M. Captured snapshots of PARP1 in the active state reveal the mechanics of PARP1 allostery. Mol. Cell 82, 2939–2951 (2022).
Abramson, J. et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 1–3, https://doi.org/10.1038/s41586-024-07487-w (2024).
Rosenberg, D. J., Hura, G. L. & Hammel, M. Size exclusion chromatography coupled small angle X-ray scattering with tandem multiangle light scattering at the SIBYLS beamline. Methods Enzymol. 677, 191–219 (2022).
Rambo, R. P. & Tainer, J. A. Characterizing flexible and intrinsically unstructured biological macromolecules by SAS using the Porod-Debye law. Biopolymers 95, 559–571 (2011).
Schneidman-Duhovny, D., Hammel, M., Tainer, J. A. & Sali, A. Accurate SAXS profile computation and its assessment by contrast variation experiments. Biophys. J. 105, 962–974 (2013).
Pelikan, M., Hura, G. & Hammel, M. Structure and flexibility within proteins as identified through small angle X-ray scattering. Gen. Physiol. Biophys. 28, 174–189 (2009).
Eustermann, S. et al. The DNA-binding domain of human PARP-1 interacts with DNA single-strand breaks as a monomer through its second zinc finger. J. Mol. Biol. 407, 149–170 (2011).
Mansoorabadi, S. O. et al. Conformational activation of poly(ADP-ribose) polymerase-1 upon DNA binding revealed by small-angle X-ray scattering. Biochemistry 53, 1779–1788 (2014).
Chappidi, N. et al. PARP1-DNA co-condensation drives DNA repair site assembly to prevent disjunction of broken DNA ends. Cell 187, 945–961.e18 (2024).
Wankowicz, S. A. & Fraser, J. S. Advances in uncovering the mechanisms of macromolecular conformational entropy. Nat. Chem. Biol. 21, 623–634 (2025).
Obaji, E., Maksimainen, M. M., Galera-Prat, A. & Lehtiö, L. Activation of PARP2/ARTD2 by DNA damage induces conformational changes relieving enzyme autoinhibition. Nat. Commun. 12, 3479 (2021).
Bilokapic, S., Suskiewicz, M. J., Ahel, I. & Halic, M. Bridging of DNA breaks activates PARP2–HPF1 to modify chromatin. Nature 585, 609–613 (2020).
Arnold, M. R. et al. Allosteric regulation of DNA binding and target residence time drive the cytotoxicity of phthalazinone-based PARP-1 inhibitors. Cell Chem. Biol. 29, 1694–1708.e10 (2022).
Langelier, M.-F., Steffen, J., Riccio, A.A., McCauley, M. & Pascal, J.M. Purification of DNA damage-dependent PARPs from E. coli for structural and biochemical analysis in Poly(ADP-Ribose) Polymerase. Methods in Molecular Biology 1608, 431–444 (Humana Press, 2017).
McKinney, S. A., Joo, C. & Ha, T. Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophys. J. 91, 1941–1951 (2006).
Bronson, J. E., Fei, J., Hofman, J. M., Gonzalez, R. L. & Wiggins, C. H. Learning rates and states from biophysical time series: a Bayesian approach to model selection and single-molecule FRET data. Biophys. J. 97, 3196–3205 (2009).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Scheres, S. H. W. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7, e42166 (2018).
Han, Y. et al. High-yield monolayer graphene grids for near-atomic resolution cryoelectron microscopy. Proc. Natl. Acad. Sci. USA 117, 1009–1014 (2020).
Wei, H. et al. Optimizing self-wicking nanowire grids. J. Struct. Biol. 202, 170–174 (2018).
Sanchez-Garcia, R. et al. DeepEMhancer: a deep learning solution for cryo-EM volume post-processing. Commun. Biol. 4, 874 (2021).
Frado, L.-L. & Craig, R. Electron microscopy of the actin-myosin head complex in the presence of ATP. J. Mol. Biol. 223, 391–397 (1992).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Suloway, C. et al. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 151, 41–60 (2005).
Punjani, A., Zhang, H. & Fleet, D. J. Non-uniform refinement: adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods 17, 1214–1221 (2020).
Punjani, A. & Fleet, D. J. 3DFlex: determining structure and motion of flexible proteins from cryo-EM. Nat. Methods 20, 860–870 (2023).
Goddard, T. D. et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018).
Meng, E. C. et al. UCSF ChimeraX: tools for structure building and analysis. Protein Sci. 32, e4792 (2023).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 66, 486–501 (2010).
Croll, T. I. ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr. Sect. D 74, 519–530 (2018).
Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. Sect. D 75, 861–877 (2019).
Lu, X.-J. & Olson, W. K. 3DNA: a versatile, integrated software system for the analysis, rebuilding and visualization of three-dimensional nucleic-acid structures. Nat. Protoc. 3, 1213–1227 (2008).
Mirdita, M. et al. ColabFold: making protein folding accessible to all. Nat. Methods 19, 679–682 (2022).
Hopkins, J. B. BioXTAS RAW 2: new developments for a free open-source program for small-angle scattering data reduction and analysis. J. Appl. Crystallogr. 57, 194–208 (2024).
Putnam, C. D., Hammel, M., Hura, G. L. & Tainer, J. A. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q. Rev. Biophys. 40, 191–285 (2007).
Svergun, D. I. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 25, 495–503 (1992).
Manalastas-Cantos, K. et al. ATSAS 3.0: expanded functionality and new tools for small-angle scattering data analysis. J. Appl. Crystallogr. 54, 343–355 (2021).
Rambo, R. P. & Tainer, J. A. Accurate assessment of mass, models and resolution by small-angle scattering. Nature 496, 477–481 (2013).
Schneidman-Duhovny, D., Hammel, M., Tainer, J. A. & Sali, A. FoXS, FoXSDock and MultiFoXS: single-state and multi-state structural modeling of proteins and their complexes based on SAXS profiles. Nucleic Acids Res. 44, W424–W429 (2016).
Langelier, M.-F., Servent, K. M., Rogers, E. E. & Pascal, J. M. A third zinc-binding domain of human poly(ADP-ribose) polymerase-1 coordinates DNA-dependent enzyme activation*. J. Biol. Chem. 283, 4105–4114 (2008).
Morin, A. et al. Collaboration gets the most out of software. eLife 2, e01456 (2013).
Acknowledgements
This work was supported in part by the Canadian Institutes of Health Research (CIHR: PJT374609 to J.M.P.) and a Cole Foundation doctoral award to M.V.M.C. Efforts to apply structural biology to characterize eukaryotic pathways related to human cancers are supported by the National Cancer Institute (CA92584). We thank staff and managers at the Facility for Electron Microscopy Research (FEMR) of McGill University and NYU Langone Health’s cryo-EM laboratory (RRID: SCR_019202) for assistance in data collection. A portion of this work was performed at the National Center for Cryo-EM Access and Training (NCCAT) and the Simons Electron Microscopy Center located at the New York Structural Biology Center, supported by the NIH Common Fund Transformative High Resolution Cryo-Electron Microscopy program (U24 GM129539) and by grants from the Simons Foundation (SF349247) and NY State Assembly. We thank the staff and managers at NCCAT for assistance and Hui (Alex) Wei for the preparation of graphene-coated grids. A portion of the work was conducted at the Advanced Light Source (ALS), a national user facility operated by Lawrence Berkeley National Laboratory on behalf of the Department of Energy, Office of Basic Energy Sciences, through the Integrated Diffraction Analysis Technologies (IDAT) program, supported by the DOE Office of Biological and Environmental Research. Structural biology applications used in this project were compiled and configured by SBGrid80. Molecular graphics and analyses performed with UCSF ChimeraX, developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, with support from National Institutes of Health R01-GM129325 and the Office of Cyber Infrastructure and Computational Biology, National Institute of Allergy and Infectious Diseases.
Author information
Authors and Affiliations
Contributions
A.S., M.F.L., M.V.M.C., and H.X. performed protein purifications. M.F.L. carried out PARP1 activity assays. A.S. and M.V.M.C. performed NS-EM of PARP1 complexes. H.X. performed single-molecule exchange and smFRET assays and analysis. J.D.M. and M.H. performed SEC-SAXS-MALS experiments. S.C. and M.H. aided A.S. with molecular dynamics analysis. A.S. carried out all cryo-EM grid preparation, image processing, and model building. A.S. and J.M.P. wrote the manuscript with input from all authors. J.M.P. and E.R. directed the study.
Corresponding author
Ethics declarations
Competing interests
J.M.P. is a co-founder of Hysplex with interests in PARP inhibitor development. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Sverzhinsky, A., Xue, H., Langelier, MF. et al. PARP1-HPF1 structure and dynamics on nicked DNA suggest a mechanism for acute and localized ADP-ribosylation. Nat Commun (2026). https://doi.org/10.1038/s41467-026-69375-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-026-69375-3