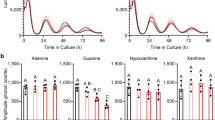
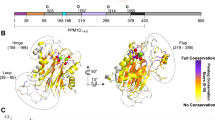
Abstract
Purine nucleotides are ubiquitous molecules essential for all life. The de novo biosynthesis of purines is a metabolic dependency that is frequently reprogrammed in cancers and is a well-established target for chemotherapies, immune modulation and antivirals. Here, we report cryo-electron microscopy structures of the multi-domain human phosphoribosylformylglycinamidine synthase, a central purine biosynthetic enzyme and foundational feature of the purinosome metabolon. These data capture, the proposed iminophosphate intermediate and provide the structural elucidation of an ammonia channel connecting the active sites of the glutaminase and synthase domains. Analysis of this series of structures and the accompanying biochemical data also reveal the molecular features and transient conformational changes that underlie allosteric regulation and catalytic coupling of the domains. This data resolves several longstanding mechanistic questions about this enzyme class and provides a strong foundation for therapeutic development.
Similar content being viewed by others
Data availability
The image files and processed data for the reported macromolecular structures are available in the RSCB PDB and EMDB under the accession codes: 9N9W and EMD-49179; 9NAL and EMD-49198; 9NB3 and EMD-49207. PDB codes of previously published structures used in this study are 3UGJ, 1T3T, 2HS4 and 3UMM. All data needed to evaluate the conclusions in the paper are present in the manuscript, Supplementary Information file, and/or the accompanying source data file. Additional data related to this paper may be requested from the authors. Source Data is provided as a Source Data file. Source data are provided with this paper.
References
Cronstein, B. N. & Angle, S. R. Purines and adenosine receptors in osteoarthritis. Biomolecules 13, 1760 (2023).
Wang, Y. et al. Cross talk between nucleotide synthesis pathways with cellular immunity in constraining hepatitis E virus replication. Antimicrob Agents Chemother. 60, 2834–2848 (2016).
Yamada, S., Mizukoshi, T., Sato, A. & Sakakibara, S. I. Purinosomes and purine metabolism in mammalian neural development: a review. Acta Histochem. Cytochem. 57, 89–100 (2024).
Zhang, Y., Morar, M. & Ealick, S. E. Structural biology of the purine biosynthetic pathway. Cell Mol. Life Sci. 65, 3699–3724 (2008).
Biscaglia, S., Ceconi, C., Malagu, M., Pavasini, R. & Ferrari, R. Uric acid and coronary artery disease: an elusive link deserving further attention. Int J. Cardiol. 213, 28–32 (2016).
Daniels, S. D. & Boison, D. Bipolar mania and epilepsy pathophysiology and treatment may converge in purine metabolism: a new perspective on available evidence. Neuropharmacology 241, 109756 (2023).
Agarwal, A., Banerjee, A. & Banerjee, U. C. Xanthine oxidoreductase: a journey from purine metabolism to cardiovascular excitation-contraction coupling. Crit. Rev. Biotechnol. 31, 264–280 (2011).
Moriwaki, Y., Yamamoto, T. & Higashino, K. Enzymes involved in purine metabolism-a review of histochemical localization and functional implications. Histol. Histopathol. 14, 1321–1340 (1999).
Cronstein, B. N. Low-dose methotrexate: a mainstay in the treatment of rheumatoid arthritis. Pharmacol. Rev. 57, 163–172 (2005).
Parker, W. B. Enzymology of purine and pyrimidine antimetabolites used in the treatment of cancer. Chem. Rev. 109, 2880–2893 (2009).
Carrera, C. J., Saven, A. & Piro, L. D. Purine metabolism of lymphocytes. Targets for chemotherapy drug development. Hematol. Oncol. Clin. North Am. 8, 357–381 (1994).
Cheviet, T., Lefebvre-Tournier, I., Wein, S. & Peyrottes, S. Plasmodium purine metabolism and its inhibition by nucleoside and nucleotide analogues. J. Med. Chem. 62, 8365–8391 (2019).
Hung, M. H. et al. Purine anabolism creates therapeutic vulnerability in hepatocellular carcinoma through m 6 A-mediated epitranscriptomic regulation. Hepatology 78, 1462–1477 (2023).
Mullen, N. J. & Singh, P. K. Nucleotide metabolism: a pan-cancer metabolic dependency. Nat. Rev. Cancer 23, 275–294 (2023).
Villa, E., Ali, E. S., Sahu, U. & Ben-Sahra, I. Cancer cells tune the signaling pathways to empower de novo synthesis of nucleotides. Cancers 11, 688 (2019).
Shi, D. D., Savani, M. R., Abdullah, K. G. & McBrayer, S. K. Emerging roles of nucleotide metabolism in cancer. Trends Cancer 9, 624–635 (2023).
Wishart, D. S. et al. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 34, D668–D672 (2006).
Ali, E. S. & Ben-Sahra, I. Regulation of nucleotide metabolism in cancers and immune disorders. Trends Cell Biol. 33, 950–966 (2023).
Biancur, D. E. et al. Functional genomics identifies metabolic vulnerabilities in pancreatic cancer. Cell Metab. 33, 199–210.e198 (2021).
Koundinya, M. et al. Dependence on the pyrimidine biosynthetic enzyme DHODH is a synthetic lethal vulnerability in mutant KRAS-driven cancers. Cell Chem. Biol. 25, 705–717.e711 (2018).
Li, L. et al. Identification of DHODH as a therapeutic target in small cell lung cancer. Sci. Transl. Med. 11, eaaw7852 (2019).
Mathur, D. et al. PTEN regulates glutamine flux to pyrimidine synthesis and sensitivity to dihydroorotate dehydrogenase inhibition. Cancer Discov. 7, 380–390 (2017).
Santana-Codina, N. et al. Oncogenic KRAS supports pancreatic cancer through regulation of nucleotide synthesis. Nat. Commun. 9, 4945 (2018).
Sykes, D. B. et al. Inhibition of dihydroorotate dehydrogenase overcomes differentiation blockade in acute myeloid leukemia. Cell 167, 171–186 e115 (2016).
Wang, X. et al (2019) Targeting pyrimidine synthesis accentuates molecular therapy response in glioblastoma stem cells. Sci. Transl. Med. 11, eaau4972 (2019).
Zhu, X. G. et al. Functional genomics in vivo reveal metabolic dependencies of pancreatic cancer cells. Cell Metab. 33, 211–221.e216 (2021).
Yamaoka, T. et al. Amidophosphoribosyltransferase limits the rate of cell growth-linked de novo purine biosynthesis in the presence of constant capacity of salvage purine biosynthesis. J. Biol. Chem. 272, 17719–17725 (1997).
Yamaoka, T. et al. Feedback inhibition of amidophosphoribosyltransferase regulates the rate of cell growth via purine nucleotide, DNA, and protein syntheses. J. Biol. Chem. 276, 21285–21291 (2001).
Goswami, M. T. et al. Role and regulation of coordinately expressed de novo purine biosynthetic enzymes PPAT and PAICS in lung cancer. Oncotarget 6, 23445–23461 (2015).
Lane, A. N. & Fan, T. W. Regulation of mammalian nucleotide metabolism and biosynthesis. Nucleic Acids Res. 43, 2466–2485 (2015).
Chitrakar, I., Kim-Holzapfel, D. M., Zhou, W. & French, J. B. Higher order structures in purine and pyrimidine metabolism. J. Struct. Biol. 197, 354–364 (2017).
Zhao, H., French, J. B., Fang, Y. & Benkovic, S. J. The purinosome, a multi-protein complex involved in the de novo biosynthesis of purines in humans. Chem. Commun. 49, 4444–4452 (2013).
Hardie, D. G., Ross, F. A. & Hawley, S. A. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat. Rev. Mol. Cell Biol. 13, 251–262 (2012).
Keller, K. E., Doctor, Z. M., Dwyer, Z. W. & Lee, Y. S. SAICAR induces protein kinase activity of PKM2 that is necessary for sustained proliferative signaling of cancer cells. Mol. Cell 53, 700–709 (2014).
Keller, K. E., Tan, I. S. & Lee, Y. S. SAICAR stimulates pyruvate kinase isoform M2 and promotes cancer cell survival in glucose-limited conditions. Science 338, 1069–1072 (2012).
Towler, M. C. & Hardie, D. G. AMP-activated protein kinase in metabolic control and insulin signaling. Circ. Res. 100, 328–341 (2007).
Soflaee, M. H. et al. Purine nucleotide depletion prompts cell migration by stimulating the serine synthesis pathway. Nat. Commun. 13, 2698 (2022).
An, S., Kumar, R., Sheets, E. D. & Benkovic, S. J. Reversible compartmentalization of de novo purine biosynthetic complexes in living cells. Science 320, 103–106 (2008).
An, S., Kyoung, M., Allen, J. J., Shokat, K. M. & Benkovic, S. J. Dynamic regulation of a metabolic multi-enzyme complex by protein kinase CK2. J. Biol. Chem. 285, 11093–11099 (2010).
French, J. B. et al. Spatial colocalization and functional link of purinosomes with mitochondria. Science 351, 733–737 (2016).
Zhao, H. et al. Quantitative analysis of purine nucleotides indicates that purinosomes increase de novo purine biosynthesis. J. Biol. Chem. 290, 6705–6713 (2015).
Sharma, N., Otsuka, Y., Scampavia, L., Spicer, T. P. & French, J. B. A high throughput assay for phosphoribosylformylglycinamidine synthase. SLAS Discov Adv. Life Sci. R. D. 31, 100212 (2025).
Ali, E. S. et al. ERK2 phosphorylates PFAS to mediate posttranslational control of de novo purine synthesis. Mol. Cell 78, 1178–1191.e1176 (2020).
Kim, J. H., Krahn, J. M., Tomchick, D. R., Smith, J. L. & Zalkin, H. Structure and function of the glutamine phosphoribosylpyrophosphate amidotransferase glutamine site and communication with the phosphoribosylpyrophosphate site. J. Biol. Chem. 271, 15549–15557 (1996).
Smith, J. L. et al. Structure of the allosteric regulatory enzyme of purine biosynthesis. Science 264, 1427–1433 (1994).
Zhou, G., Smith, J. L. & Zalkin, H. Binding of purine nucleotides to two regulatory sites results in synergistic feedback inhibition of glutamine 5-phosphoribosylpyrophosphate amidotransferase. J. Biol. Chem. 269, 6784–6789 (1994).
An, S., Deng, Y., Tomsho, J. W., Kyoung, M. & Benkovic, S. J. Microtubule-assisted mechanism for functional metabolic macromolecular complex formation. Proc. Natl. Acad. Sci. USA 107, 12872–12876 (2010).
Henzler-Wildman, K. & Kern, D. Dynamic personalities of proteins. Nature 450, 964–972 (2007).
Koshland, D. E. Jr Conformational changes: how small is big enough?. Nat. Med. 4, 1112–1114 (1998).
Motlagh, H. N., Wrabl, J. O., Li, J. & Hilser, V. J. The ensemble nature of allostery. Nature 508, 331–339 (2014).
He, J., Zou, L. N., Pareek, V. & Benkovic, S. J. Multienzyme interactions of the de novo purine biosynthetic protein PAICS facilitate purinosome formation and metabolic channeling. J. Biol. Chem. 298, 101853 (2022).
Shon, H. et al. Evidence supporting substrate channeling between domains of human PAICS: a time-course analysis of (13)c-bicarbonate incorporation. Biochemistry 61, 575–582 (2022).
Krahn, J. M. et al. Coupled formation of an amidotransferase interdomain ammonia channel and a phosphoribosyltransferase active site. Biochemistry 36, 11061–11068 (1997).
Sweetlove, L. J. & Fernie, A. R. The role of dynamic enzyme assemblies and substrate channelling in metabolic regulation. Nat. Commun. 9, 2136 (2018).
Morar, M., Anand, R., Hoskins, A. A., Stubbe, J. & Ealick, S. E. Complexed structures of formylglycinamide ribonucleotide amidotransferase from Thermotoga maritima describe a novel ATP binding protein superfamily. Biochemistry 45, 14880–14895 (2006).
Anand, R., Hoskins, A. A., Stubbe, J. & Ealick, S. E. Domain organization of Salmonella typhimurium formylglycinamide ribonucleotide amidotransferase revealed by X-ray crystallography. Biochemistry 43, 10328–10342 (2004).
Sharma, N. et al. Role of allosteric switches and adaptor domains in long-distance cross-talk and transient tunnel formation. Sci. Adv. 6, eaay7919 (2020).
Schendel, F. J., Mueller, E., Stubbe, J., Shiau, A. & Smith, J. M. Formylglycinamide ribonucleotide synthetase from Escherichia coli: cloning, sequencing, overproduction, isolation, and characterization. Biochemistry 28, 2459–2471 (1989).
Schendel, F. J. & Stubbe, J. Substrate specificity of formylglycinamidine synthetase. Biochemistry 25, 2256–2264 (1986).
Tanwar, A. S., Morar, M., Panjikar, S. & Anand, R. Formylglycinamide ribonucleotide amidotransferase from Salmonella typhimurium: role of ATP complexation and the glutaminase domain in catalytic coupling. Acta Crystallogr. D. Biol. Crystallogr. 68, 627–636 (2012).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Hoskins, A. A., Anand, R., Ealick, S. E. & Stubbe, J. The formylglycinamide ribonucleotide amidotransferase complex from Bacillus subtilis: metabolite-mediated complex formation. Biochemistry 43, 10314–10327 (2004).
Mouilleron, S. & Golinelli-Pimpaneau, B. Conformational changes in ammonia-channeling glutamine amidotransferases. Curr. Opin. Struct. Biol. 17, 653–664 (2007).
Sanchez-Garcia, R. et al. DeepEMhancer: a deep learning solution for cryo-EM volume post-processing. Commun. Biol. 4, 874 (2021).
Sharma, N., Singh, S., Tanwar, A. S., Mondal, J. & Anand, R. Mechanism of coordinated gating and signal transduction in purine biosynthetic enzyme formylglycinamidine synthetase. ACS Catal. 12, 1930–1944 (2022).
Li, C., Kappock, T. J., Stubbe, J., Weaver, T. M. & Ealick, S. E. X-ray crystal structure of aminoimidazole ribonucleotide synthetase (PurM), from the Escherichia coli purine biosynthetic pathway at 2.5 A resolution. Structure 7, 1155–1166 (1999).
Westheimer, F. H. Monomeric meta-phosphates. Chem. Rev. 81, 313–326 (1981).
Massiere, F. & Badet-Denisot, M. A. The mechanism of glutamine-dependent amidotransferases. Cell Mol. Life Sci. 54, 205–222 (1998).
Rivalta, I. et al. Allosteric pathways in imidazole glycerol phosphate synthase. Proc. Natl. Acad. Sci. USA 109, E1428–E1436 (2012).
Wurm, J. P. et al. Molecular basis for the allosteric activation mechanism of the heterodimeric imidazole glycerol phosphate synthase complex. Nat. Commun. 12, 2748 (2021).
Chovancova, E. et al. CAVER 3.0: a tool for the analysis of transport pathways in dynamic protein structures. PLoS Comput. Biol. 8, e1002708 (2012).
Tanwar, A. S., Sindhikara, D. J., Hirata, F. & Anand, R. Determination of the formylglycinamide ribonucleotide amidotransferase ammonia pathway by combining 3D-RISM theory with experiment. ACS Chem. Biol. 10, 698–704 (2015).
Mouilleron, S., Badet-Denisot, M. A. & Golinelli-Pimpaneau, B. Glutamine binding opens the ammonia channel and activates glucosamine-6P synthase. J. Biol. Chem. 281, 4404–4412 (2006).
Teplyakov, A., Obmolova, G., Badet, B. & Badet-Denisot, M. A. Channeling of ammonia in glucosamine-6-phosphate synthase. J. Mol. Biol. 313, 1093–1102 (2001).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Punjani, A. & Fleet, D. J. 3D variability analysis: Resolving continuous flexibility and discrete heterogeneity from single particle cryo-EM. J. Struct. Biol. 213, 107702 (2021).
Meng, E. C. et al. UCSF ChimeraX: tools for structure building and analysis. Protein Sci. 32, e4792 (2023).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 66, 486–501 (2010).
Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D. Biol. Crystallogr. 75, 861–877 (2019).
Williams, C. J. et al. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018).
Gibson, D. G. et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009).
Bernt, E. & Bergmeyer, H. U. l-Glutamate UV-assay with gutamate dehydrogenase and NAD. Methods of Enzymatic Analysis 2nd edn (Academic Press, 1974).
Bratton, A. C. & Marshall, E. K. A new coupling component for sulfanilamide determination. J. Biol. Chem. 128, 537–550 (1939).
Acknowledgements
The authors would like to thank the staff at The Hormel Institute Cryo-EM core facility, particularly Janarjan Bhandari, for their help with sample preparation and data collection, as well as all members of the J.B.F. lab for helpful discussions. This work was supported by the National Institute for General Medical Sciences and the National Cancer Institute, of the National Institutes of Health, under grant numbers R35GM124898 and R01CA299056. Partial support for N.S. was provided by an Eagles Fellowship and Paint the Town Pink grant from The Hormel Institute. The funding agencies played no role in the design or execution of this work.
Author information
Authors and Affiliations
Contributions
N.S. Methodology, investigation, formal analysis, validation, writing —preliminary draft, review and editing. W.Z. Methodology, investigation. J.B.F. Conceptualization and design, methodology, supervision, investigation, writing—original draft, review and editing, resources, project leadership, funding acquisition.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Sharma, N., Zhou, W. & French, J.B. Structural and molecular basis for allosteric regulation and catalytic coupling of human phosphoribosylformylglycinamidine synthase. Nat Commun (2026). https://doi.org/10.1038/s41467-026-69423-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-026-69423-y