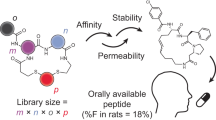
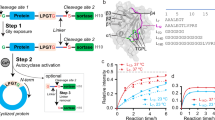
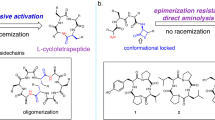
Abstract
Cyclic peptides exhibit remarkable stability, membrane permeability, and binding affinity, positioning them as promising therapeutics. However, their synthesis, particularly on-resin head-to-tail cyclization, remains challenging, with cyclization site selection critically influencing yield. Here, we introduce a machine learning (ML) approach to predict cyclization outcomes, leveraging CycloBot, our fully automated cyclic peptide synthesis platform. Using this system, we generate a standardized dataset of 306 cyclic peptides (2–14 residues) and develop an ML model achieving an average prediction accuracy of 84%. Experimental validation with 74 random and therapeutic peptides showed an 86% prediction consistency. To facilitate practical use, we built CycloPepper, a user-friendly platform available through both web and software interfaces, enabling rapid cyclization site assessment. This tool effectively identified potential cyclization sites for disease-targeting peptides, including cancer biomarkers. Our work illustrates the potential of ML-assisted synthesis to streamline cyclic peptide synthesis and accelerate therapeutic discovery.
Similar content being viewed by others
Data availability
All the relevant data generated in this study are provided in the Supplementary Information and Source Data. The training dataset of 306 cyclic peptides, along with all prediction results, has been deposited in the GitHub repository under https://github.com/Yongboxiao/Selection-of-Cyclization-Sites. Source data are provided with this paper.
Code availability
The Python code for the machine learning model described in this manuscript is available in a GitHub repository (https://github.com/Yongboxiao/Selection-of-Cyclization-Sites) and archived on https://doi.org/10.5281/zenodo.1815499434. Additionally, the CycloPepper platform can be accessed through its user-friendly website interface at http://www.cyclopepper.com/.
References
Thorstholm, L. & Craik, D. J. Discovery and applications of naturally occurring cyclic peptides. Drug Discov. Today Technol. 9, e13–e21 (2012).
Ji, X., Nielsen, A. L. & Heinis, C. Cyclic peptides for drug development. Angew. Chem. Int. Ed. 63, e202308251 (2024).
Muttenthaler, M., King, G. F., Adams, D. J. & Alewood, P. F. Trends in peptide drug discovery. Nat. Rev. Drug Discov. 20, 309–325 (2021).
Zorzi, A., Deyle, K. & Heinis, C. Cyclic peptide therapeutics: past, present and future. Curr. Opin. Chem. Biol. 38, 24–29 (2017).
Lai, S., Zhang, Q. & Jin, L. Natural and man-made cyclic peptide-based antibiotics. Antibiotics 12, 42 (2023).
Hill, T. A., Shepherd, N. E., Diness, F. & Fairlie, D. P. Constraining cyclic peptides to mimic protein structure motifs. Angew. Chem. Int. Ed. 53, 13020–13041 (2014).
Khatri, B., Nuthakki, V. R. & Chatterjee, J. Strategies to Enhance Metabolic Stabilities. In Cyclic Peptide Design (ed. Goetz, G.) 17–40 (Springer, https://doi.org/10.1007/978-1-4939-9504-2_2. 2019).
Bhat, A., Roberts, L. R. & Dwyer, J. J. Lead discovery and optimization strategies for peptide macrocycles. Eur. J. Med. Chem. 94, 471–479 (2015).
Bechtler, C. & Lamers, C. Macrocyclization strategies for cyclic peptides and peptidomimetics. RSC Med. Chem. 12, 1325–1351 (2021).
Chow, H. Y., Zhang, Y., Matheson, E. & Li, X. Ligation technologies for the synthesis of cyclic peptides. Chem. Rev. 119, 9971–10001 (2019).
Zhan, W., Duan, H. & Li, C. Recent advances in metal-free peptide stapling strategies. Chem. Bio. Eng. 1, 593–605 (2024).
Fang, P., Pang, W.-K., Xuan, S., Chan, W.-L. & Leung, K. C.-F. Recent advances in peptide macrocyclization strategies. Chem. Soc. Rev. 53, 11725–11771 (2024).
Zhang, C. et al. HighFold: accurately predicting structures of cyclic peptides and complexes with head-to-tail and disulfide bridge constraints. Brief. Bioinform. 25, bbae215 (2024).
Wang, X. et al. Towards efficient discovery of green synthetic pathways with Monte Carlo tree search and reinforcement learning. Chem. Sci. 11, 10959–10972 (2020).
Coley, C. W., Barzilay, R., Jaakkola, T. S., Green, W. H. & Jensen, K. F. Prediction of organic reaction outcomes using machine learning. ACS Cent. Sci. 3, 434–443 (2017).
Batra, R. et al. Machine learning overcomes human bias in the discovery of self-assembling peptides. Nat. Chem. 14, 1427–1435 (2022).
Li, J., Yanagisawa, K. & Akiyama, Y. CycPeptMP: enhancing membrane permeability prediction of cyclic peptides with multi-level molecular features and data augmentation. Brief. Bioinform. 25, bbae417 (2024).
Yoshida, M. et al. Using evolutionary algorithms and machine learning to explore sequence space for the discovery of antimicrobial peptides. Chem. 4, 533–543 (2018).
Huang, J. et al. Identification of potent antimicrobial peptides via a machine-learning pipeline that mines the entire space of peptide sequences. Nat. Biomed. Eng. 7, 797–810 (2023).
Lin, Y.-C. et al. Multidimensional design of anticancer peptides. Angew. Chem. Int. Ed. 54, 10370–10374 (2015).
Yuan, Q., Chen, K., Yu, Y., Le, N. Q. K. & Chua, M. C. H. Prediction of anticancer peptides based on an ensemble model of deep learning and machine learning using ordinal positional encoding. Brief. Bioinform. 24, bbac630 (2023).
Salveson, P. J. et al. Expansive discovery of chemically diverse structured macrocyclic oligoamides. Science 384, 420–428 (2024).
Watson, J. L. et al. De novo design of protein structure and function with RFdiffusion. Nature 620, 1089–1100 (2023).
Rettie, S. A. et al. Cyclic peptide structure prediction and design using AlphaFold2. Nat Commun 16, 4730 (2025).
Baker, M. 1,500 scientists lift the lid on reproducibility. Nature 533, 452–454 (2016).
Coley, C. W., Eyke, N. S. & Jensen, K. F. Autonomous discovery in the chemical sciences Part I: progress. Angew. Chem. Int. Ed. 59, 22858–22893 (2020).
Li, C. et al. Machine learning guides peptide nucleic acid flow synthesis and sequence design. Adv. Sci. 9, 2201988 (2022).
Mijalis, A. J. et al. A fully automated flow-based approach for accelerated peptide synthesis. Nat. Chem. Biol. 13, 464–466 (2017).
Hartrampf, N. et al. Synthesis of proteins by automated flow chemistry. Science 368, 980–987 (2020).
Wan, F. et al. Automated rapid synthesis of high-purity head-to-tail cyclic peptides via a diaminonicotinic acid scaffold. J. Am. Chem. Soc. 148, 986–996 (2026).
Hu, D., Shakerian, F., Zhao, J. & Li, S.-P. Chemistry, pharmacology and analysis of Pseudostellaria heterophylla: a mini-review. Chinese Med. 14, 21 (2019).
Dunbar, J. et al. SAbDab: the structural antibody database. Nucleic Acids Res. 42, D1140–D1146 (2014).
Walsh, I. et al. DOME: recommendations for supervised machine learning validation in biology. Nat Methods 18, 1122–1127 (2021).
Pan. Y. Yongboxiao/Selection-of-Cyclization-Sites: First release. Zenodo https://doi.org/10.5281/zenodo.18154994 (2026).
Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (Grant Nos. 22208290, C.L.; 22122109, X.H.; 22271253, X.H.; W2512004, X.H.). The China Postdoctoral Science Foundation (Grant No. 2023M743085, F.W.). National Key R&D Program of China (Grant No. 2022YFA1504301, X.H.). New Generation Artificial Intelligence-National Science and Technology Major Project (Grant No. 2025ZD0121905 X.H.).
Author information
Authors and Affiliations
Contributions
Y.P. contributed to the main idea, data collection, algorithm design, development of machine learning code, and manuscript writing. C.H. was involved in optimizing the machine learning algorithms, developing the software and website, and writing relevant sections of the manuscript. J.L. contributed to the algorithm design and provided valuable suggestions. F.W. assisted with data collection and experimental validation. X.H. and C.L. supervised the research and participated in the manuscript revision, and were responsible for the overall quality and direction of the work.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Denis Shields and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Pan, Y., Hu, C., Li, J. et al. CycloPepper: a machine learning platform for predicting cyclization outcomes and optimizing synthesis of therapeutic cyclopeptides. Nat Commun (2026). https://doi.org/10.1038/s41467-026-69441-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-026-69441-w