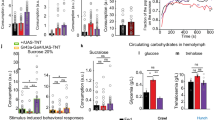
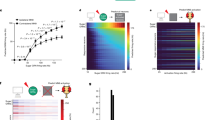
Abstract
Animals use gustatory information to decide whether to ingest nutritious substances or avoid toxic ones. Although certain neurons in the gustatory circuits respond to both aversive and appetitive signals, how these neurons resolve inputs with opposing valences is unknown. Here, we examine how the Drosophila melanogaster neuropeptide leucokinin (LK) affects gustatory information processing to elicit the appropriate feeding behaviors. We identify the subesophageal LK neurons (SELKs) as downstream synaptic partners of gustatory receptor neurons and show that these two groups are functionally connected. We then show that SELKs affect bitter avoidance through LK release and food intake in an acetylcholine-dependent manner. Our study uncovers a mechanism whereby strong activation of SELKs results in LK release, leading to feeding suppression, while weak activation results in acetylcholine-dependent feeding promotion. Thus, our results reveal that a single pair of neurons, SELKs, differentially controls opposing feeding behaviors via distinct neurotransmitters.
Similar content being viewed by others
Data availability
The behavioral and functional imaging data generated in this study have been deposited in the Figshare database under accession code https://doi.org/10.6084/m9.figshare.31049989. Source data are provided as a Source data file. Source data are provided with this paper.
Code availability
Custom codes used for data analyses are available at: https://github.com/Zandawala-lab/Savas-et-al-2025-Drosophila-SELK.
References
Rosenstein, D. & Oster, H. Differential facial responses to four basic tastes in newborns. Child Dev. 59, 1555–1568 (1988).
Clyne, P. J., Warr, C. G. & Carlson, J. R. Candidate taste receptors in Drosophila. Science 287, 1830–1834 (2000).
Scott, K. et al. A chemosensory gene family encoding candidate gustatory and olfactory receptors in Drosophila. Cell 104, 661–673 (2001).
Accolla, R., Bathellier, B., Petersen, C. C. & Carleton, A. Differential spatial representation of taste modalities in the rat gustatory cortex. J. Neurosci. 27, 1396–1404 (2007).
Marella, S. et al. Imaging taste responses in the fly brain reveals a functional map of taste category and behavior. Neuron 49, 285–295 (2006).
Dethier, V. G. The Hungry Fly: A Physiological Study of the Behavior Associated with Feeding (Harvard University Press, 1976).
Dahanukar, A., Lei, Y. T., Kwon, J. Y. & Carlson, J. R. Two Gr genes underlie sugar reception in Drosophila. Neuron 56, 503–516 (2007).
Weiss, L. A., Dahanukar, A., Kwon, J. Y., Banerjee, D. & Carlson, J. R. The molecular and cellular basis of bitter taste in Drosophila. Neuron 69, 258–272 (2011).
Meunier, N., Marion-Poll, F., Rospars, J. P. & Tanimura, T. Peripheral coding of bitter taste in Drosophila. J. Neurobiol. 56, 139–152 (2003).
Inagaki, H. K., Panse, K. M. & Anderson, D. J. Independent, reciprocal neuromodulatory control of sweet and bitter taste sensitivity during starvation in Drosophila. Neuron 84, 806–820 (2014).
Devineni, A. V., Sun, B., Zhukovskaya, A. & Axel, R. Acetic acid activates distinct taste pathways in Drosophila to elicit opposing, state-dependent feeding responses. Elife 8, https://doi.org/10.7554/eLife.47677 (2019).
Sterne, G. R., Otsuna, H., Dickson, B. J. & Scott, K. Classification and genetic targeting of cell types in the primary taste and premotor center of the adult Drosophila brain. Elife 10, https://doi.org/10.7554/eLife.71679 (2021).
Shiu, P. K., Sterne, G. R., Engert, S., Dickson, B. J. & Scott, K. Taste quality and hunger interactions in a feeding sensorimotor circuit. Elife 11, https://doi.org/10.7554/eLife.79887 (2022).
Scott, K. Gustatory processing in Drosophila melanogaster. Annu. Rev. Entomol. 63, 15–30 (2018).
Kendroud, S. et al. Structure and development of the subesophageal zone of the Drosophila brain. II. Sensory compartments. J. Comp. Neurol. 526, 33–58 (2018).
Snell, N. J. et al. Complex representation of taste quality by second-order gustatory neurons in Drosophila. Curr. Biol. 32, 3758–3772 (2022).
Chu, B., Chui, V., Mann, K. & Gordon, M. D. Presynaptic gain control drives sweet and bitter taste integration in Drosophila. Curr. Biol. 24, 1978–1984 (2014).
Reiter, S., Campillo Rodriguez, C., Sun, K. & Stopfer, M. Spatiotemporal coding of individual chemicals by the gustatory system. J. Neurosci. 35, 12309–12321 (2015).
Liu, H. & Fontanini, A. State dependency of chemosensory coding in the gustatory thalamus (VPMpc) of alert rats. J. Neurosci. 35, 15479–15491 (2015).
Sweazey, R. D. & Smith, D. V. Convergence onto hamster medullary taste neurons. Brain Res. 408, 173–184 (1987).
Nassel, D. R. & Zandawala, M. Recent advances in neuropeptide signaling in Drosophila, from genes to physiology and behavior. Prog. Neurobiol. 179, 101607 (2019).
Marder, E. Neuromodulation of neuronal circuits: back to the future. Neuron 76, 1–11 (2012).
Bargmann, C. I. & Marder, E. From the connectome to brain function. Nat. Methods 10, 483–490 (2013).
Cavey, M., Collins, B., Bertet, C. & Blau, J. Circadian rhythms in neuronal activity propagate through output circuits. Nat. Neurosci. 19, 587–595 (2016).
Yurgel, M. E. et al. A single pair of leucokinin neurons are modulated by feeding state and regulate sleep-metabolism interactions. PLoS Biol. 17, e2006409 (2019).
Zandawala, M. et al. Modulation of Drosophila post-feeding physiology and behavior by the neuropeptide leucokinin. PLoS Genet. 14, e1007767 (2018).
Nassel, D. R. Substrates for neuronal cotransmission with neuropeptides and small molecule neurotransmitters in Drosophila. Front. Cell. Neurosci. 12, 83 (2018).
Senapati, B. et al. A neural mechanism for deprivation state-specific expression of relevant memories in Drosophila. Nat. Neurosci. 22, 2029–2039 (2019).
Marder, E. & Weimann, J. Neurobiology of Motor Progamme Selection (Pergamon Press/Elsevier, 1992).
Huang, Y. C. et al. A single neuron in C. elegans orchestrates multiple motor outputs through parallel modes of transmission. Curr. Biol. 33, 4430–4445 (2023).
Vaaga, C. E., Borisovska, M. & Westbrook, G. L. Dual-transmitter neurons: functional implications of co-release and co-transmission. Curr. Opin. Neurobiol. 29, 25–32 (2014).
Kim, D.-I. et al. Encoding opposing valences through frequency-dependent transmitter switching in single peptidergic neurons. Preprint at bioRxiv https://doi.org/10.1101/2024.11.09.622790 (2024).
Talay, M. et al. Transsynaptic mapping of second-order taste neurons in flies by trans-Tango. Neuron 96, 783–795 (2017).
Snell, N. J., Fisher, J. D., Hartmann, G. G., Talay, M. & Barnea, G. Distributed representation of taste quality by second-order gustatory neurons in Drosophila. Preprint at bioRxiv https://doi.org/10.1101/2020.11.10.377382 (2020).
Nassel, D. R. Leucokinin and associated neuropeptides regulate multiple aspects of physiology and behavior in Drosophila. Int. J. Mol. Sci. 22, https://doi.org/10.3390/ijms22041940 (2021).
Liu, Y., Luo, J., Carlsson, M. A. & Nassel, D. R. Serotonin and insulin-like peptides modulate leucokinin-producing neurons that affect feeding and water homeostasis in Drosophila. J. Comp. Neurol. 523, 1840–1863 (2015).
Luo, J., Liu, Y. & Nassel, D. R. Insulin/IGF-regulated size scaling of neuroendocrine cells expressing the bHLH transcription factor dimmed in Drosophila. PLoS Genet. 9, e1004052 (2013).
Al-Anzi, B. et al. The leucokinin pathway and its neurons regulate meal size in Drosophila. Curr. Biol. 20, 969–978 (2010).
de Haro, M. et al. Detailed analysis of leucokinin-expressing neurons and their candidate functions in the Drosophila nervous system. Cell Tissue Res. 339, 321–336 (2010).
Zandawala, M., Marley, R., Davies, S. A. & Nassel, D. R. Characterization of a set of abdominal neuroendocrine cells that regulate stress physiology using colocalized diuretic peptides in Drosophila. Cell. Mol. Life Sci. 75, 1099–1115 (2018).
Galikova, M., Dircksen, H. & Nassel, D. R. The thirsty fly: ion transport peptide (ITP) is a novel endocrine regulator of water homeostasis in Drosophila. PLoS Genet. 14, e1007618 (2018).
Lopez-Arias, B., Dorado, B. & Herrero, P. Blockade of the release of the neuropeptide leucokinin to determine its possible functions in fly behavior: chemoreception assays. Peptides 32, 545–552 (2011).
Zheng, Z. et al. A complete electron microscopy volume of the brain of adult Drosophila melanogaster. Cell 174, 730–743 e722 (2018).
Isaacman-Beck, J. et al. SPARC enables genetic manipulation of precise proportions of cells. Nat. Neurosci. 23, 1168–1175 (2020).
Matsliah, A. et al. Codex: Connectome Data Explorer. https://doi.org/10.13140/RG.2.2.35928.67844 (2023).
Dorkenwald, S. et al. Neuronal wiring diagram of an adult brain. Nature 634, 124–138 (2024).
Schlegel, P. et al. Whole-brain annotation and multi-connectome cell typing of Drosophila. Nature 634, 139–152 (2024).
Klapoetke, N. C. et al. Independent optical excitation of distinct neural populations. Nat. Methods 11, 338–346 (2014).
Chen, T. W. et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300 (2013).
Inagaki, H. K. et al. Visualizing neuromodulation in vivo: TANGO-mapping of dopamine signaling reveals appetite control of sugar sensing. Cell 148, 583–595 (2012).
Marella, S., Mann, K. & Scott, K. Dopaminergic modulation of sucrose acceptance behavior in Drosophila. Neuron 73, 941–950 (2012).
Sareen, P. F., McCurdy, L. Y. & Nitabach, M. N. A neuronal ensemble encoding adaptive choice during sensory conflict in Drosophila. Nat. Commun. 12, 4131 (2021).
Moreira, J. M. et al. optoPAD, a closed-loop optogenetics system to study the circuit basis of feeding behaviors. Elife 8, https://doi.org/10.7554/eLife.43924 (2019).
Mohammad, F. et al. Optogenetic inhibition of behavior with anion channelrhodopsins. Nat. Methods 14, 271–274 (2017).
Simpson, J. H. Rationally subdividing the fly nervous system with versatile expression reagents. J. Neurogenet. 30, 185–194 (2016).
Davie, K. et al. A single-cell transcriptome atlas of the aging Drosophila brain. Cell 174, 982–998 (2018).
Kahsai, L., Kapan, N., Dircksen, H., Winther, A. M. & Nassel, D. R. Metabolic stress responses in Drosophila are modulated by brain neurosecretory cells that produce multiple neuropeptides. PLoS ONE 5, e11480 (2010).
Herrero, P., Magarinos, M., Torroja, L. & Canal, I. Neurosecretory identity conferred by the apterous gene: lateral horn leucokinin neurons in Drosophila. J. Comp. Neurol. 457, 123–132 (2003).
Kitamoto, T. & Salvaterra, P. M. A POU homeo domain protein related to dPOU-19/pdm-1 binds to the regulatory DNA necessary for vital expression of the Drosophila choline acetyltransferase gene. J. Neurosci. 15, 3509–3518 (1995).
Hamid, R. et al. Drosophila choline transporter non-canonically regulates pupal eclosion and NMJ integrity through a neuronal subset of mushroom body. Dev. Biol. 446, 80–93 (2019).
Dethier, V. G. Other tastes, other worlds. Science 201, 224–228 (1978).
Shiu, P. K. et al. A Drosophila computational brain model reveals sensorimotor processing. Nature 634, 210–219 (2024).
Sorkac, A. et al. retro-Tango enables versatile retrograde circuit tracing in Drosophila. Elife 12, https://doi.org/10.7554/eLife.85041 (2023).
Chanat, E. & Huttner, W. B. Milieu-induced, selective aggregation of regulated secretory proteins in the trans-Golgi network. J. Cell Biol. 115, 1505–1519 (1991).
Nordmann, J. J. & Morris, J. F. Method for quantitating the molecular content of a subcellular organelle: hormone and neurophysin content of newly formed and aged neurosecretory granules. Proc. Natl. Acad. Sci. USA 81, 180–184 (1984).
Karhunen, T., Vilim, F. S., Alexeeva, V., Weiss, K. R. & Church, P. J. Targeting of peptidergic vesicles in cotransmitting terminals. J. Neurosci. 21, RC127 (2001).
Molla-Albaladejo, R., Jimenez-Caballero, M. & Sanchez-Alcaniz, J. A. Molecular characterization of gustatory second-order neurons reveals integrative mechanisms of gustatory and metabolic information. Elife 13, https://doi.org/10.7554/eLife.100947 (2025).
Ohla, K. et al. Recognizing taste: coding patterns along the neural axis in mammals. Chem. Senses 44, 237–247 (2019).
Pfaffmann, C. The afferent code for sensory quality. Am. Psychol. 14, 226 (1959).
Kvello, P., Jorgensen, K. & Mustaparta, H. Central gustatory neurons integrate taste quality information from four appendages in the moth Heliothis virescens. J. Neurophysiol. 103, 2965–2981 (2010).
Wu, J. S. & Luo, L. A protocol for dissecting Drosophila melanogaster brains for live imaging or immunostaining. Nat. Protoc. 1, 2110–2115 (2006).
Groth, A. C., Fish, M., Nusse, R. & Calos, M. P. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics 166, 1775–1782 (2004).
Yu, S.-C. et al. New synapse detection in the whole-brain connectome of Drosophila. Preprint at bioRxiv https://doi.org/10.1101/2025.07.11.664377 (2025).
Iwabuchi, S., Kakazu, Y., Koh, J. Y. & Harata, N. C. Evaluation of the effectiveness of Gaussian filtering in distinguishing punctate synaptic signals from background noise during image analysis. J. Neurosci. Methods 223, 92–113 (2014).
The Open Lab Book. Measuring cell fluorescence using ImageJ https://theolb.readthedocs.io/en/latest/imaging/measuring-cell-fluorescence-using-imagej.html (2014).
Kwon, J. Y., Dahanukar, A., Weiss, L. A. & Carlson, J. R. A map of taste neuron projections in the Drosophila CNS. J. Biosci. 39, 565–574 (2014).
Perkins, L. A. et al. The transgenic RNAi project at Harvard Medical School: resources and validation. Genetics 201, 843–852 (2015).
Pfeiffer, B. D., Truman, J. W. & Rubin, G. M. Using translational enhancers to increase transgene expression in Drosophila. Proc. Natl. Acad. Sci. USA 109, 6626–6631 (2012).
Thistle, R., Cameron, P., Ghorayshi, A., Dennison, L. & Scott, K. Contact chemoreceptors mediate male-male repulsion and male-female attraction during Drosophila courtship. Cell 149, 1140–1151 (2012).
Ishimoto, H. & Kamikouchi, A. A feedforward circuit regulates action selection of pre-mating courtship behavior in female Drosophila. Curr. Biol. 30, 396–407 (2020).
Itskov, P. M. et al. Automated monitoring and quantitative analysis of feeding behaviour in Drosophila. Nat. Commun. 5, 4560 (2014).
Acknowledgements
This work was supported by NIH grant R01DC020703 (G.B.), Brown University Carney Institute for Brain Science, Suna Kıraç Fund for Brain Science (D.S.), Brown University Carney Institute for Brain Science, Graduate Award in Brain Science (D.S.) and NIH/NIDCD award F31DC019540 (A.M.C.). Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study. We thank Dr. Brian Kim for conducting preliminary behavioral studies. We thank Susan Brenner-Morton and Drs. Dick Nässel and Matthias Schlichting for sharing reagents. We would like to thank Drs. Karla Kaun, Alexander Fleischmann, Mustafa Talay, and members of the Barnea Laboratory for critical reading of the manuscript.
Author information
Authors and Affiliations
Contributions
D.S., M.Z., and G.B. conceptualized the study. D.S., A.O., R.A.M., A.M.C., Z.C., R.S., A.S., and M.Z. contributed to the acquisition and analysis of the data. D.S. and G.B. administered the project. D.S. and G.B. wrote the manuscript. The project was supervised by M.Z. and G.B.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Savaş, D., Okoro, A.M., Moșneanu, R.A. et al. Feeding decision-making by a single neuron via disparate neurotransmitters. Nat Commun (2026). https://doi.org/10.1038/s41467-026-69443-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-026-69443-8