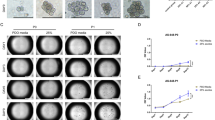
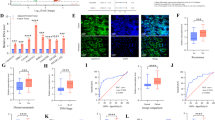
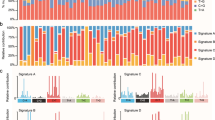
Abstract
The majority of gastric cancer cells proliferate in a Wnt ligand-dependent manner. To investigate this, we generated mice harboring Kras, Tgfbr2, and Trp53 (KTP) as well as with the same mutations plus Wnt1 expression (WKTP) in gastric mucosa. While KTP mice develop gastric metaplasia, WKTP mice develop dysplastic tumors, highlighting the role of ligand-dependent Wnt signaling in primary tumorigenesis. Organoids derived from WKTP mice form liver metastases following splenic transplantation, whereas KTP organoids do not. Notably, Apc disruption fails to induce metastasis of KTP cells, suggesting that stromal Wnt signaling promotes metastasis. Mechanistically, tumor-derived Wnt ligands cooperate with TGFβ signaling to induce Has2 expression in cancer-associated fibroblasts (CAFs), leading to hyaluronan accumulation in the metastatic microenvironment. Strikingly, hyaluronidase expression in WKTP cells significantly suppresses liver metastasis. Here we show the critical role of ligand-dependent Wnt signaling and Has2-mediated hyaluronan deposition in metastasis, offering potential therapeutic strategy against gastric cancer metastasis.
Similar content being viewed by others
Data availability
The spatial transcriptomics (Visium) data generated in this study have been deposited in the DDBJ under the BioProject accession number PRJDB20422 and Run accession number DRR656668. Source data underlying Figs. 1c, e, g, 2c, f, 3c, f, i, 4c, d, g, 5c, e, 6b, c, d, f, and 7a, b, d, g, as well as Supplementary Figs. 1, 2d, 6b, 7c, 8a, b, and 9a are provided in the Source Data file with this paper. All other data supporting the findings of this study are available within the Article and its Supplementary Information. Any additional materials, including mouse models and organoids, are available from the corresponding authors upon completion of a material transfer agreement. Source data are provided with this paper.
References
Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249 (2021).
Smyth, E. C., Nilsson, M., Grabsch, H. I., van Grieken, N. C. & Lordick, F. Gastric cancer. Lancet 396, 635–648 (2020).
Thrift, A. P., Wenker, T. N. & El-Serag, H. B. Global burden of gastric cancer: epidemiological trends, risk factors, screening and prevention. Nat. Rev. Clin. Oncol. 20, 338–349 (2023).
Riihimaki, M., Hemminki, A., Sundquist, K., Sundquist, J. & Hemminki, K. Metastatic spread in patients with gastric cancer. Oncotarget 7, 52307–52316 (2016).
Bass, A. J. et al. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 513, 202–209 (2014).
Muzny, D. M. et al. Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337 (2012).
Totoki, Y. et al. Multiancestry genomic and transcriptomic analysis of gastric cancer. Nat. Genet. 55, 581–594 (2023).
Nusse, R. & Clevers, H. Wnt/β-catenin signaling, disease, and emerging therapeutic modalities. Cell 169, 985–999 (2017).
Koushyar, S., Powell, A. G., Vincan, E. & Phesse, T. J. Targeting Wnt signaling for the treatment of gastric cancer. Int. J. Mol. Sci. 21, 3927 (2020).
Nanki, K. et al. Divergent routes toward Wnt and R-spondin niche independency during human gastric carcinogenesis. Cell 174, 856–869 (2018).
Han, R., Yang, J., Zhu, Y. & Gan, R. Wnt signaling in gastric cancer: current progress and future prospects. Front. Oncol. 14, 1410513 (2024).
Li, Y., Wang, Y., Zou, Q., Li, S. & Zhang, F. KLF3 transcription activates WNT1 and promotes the growth and metastasis of gastric cancer via activation of the WNT/β-catenin signaling pathway. Lab. Invest. 103, 100078 (2023).
Xu, H. et al. Liquid tumor microenvironment enhances WNT signaling pathway of peritoneal metastasis of gastric cancer. Sci. Rep. 13, 11125 (2023).
Oshima, H. et al. Carcinogenesis in mouse stomach by simultaneous activation of the Wnt signaling and prostaglandin E2 pathway. Gastroenterology 131, 1086–1095 (2006).
Oshima, H., Oguma, K., Du, Y. C. & Oshima, M. Prostaglandin E2, Wnt, and BMP in gastric tumor mouse models. Cancer Sci. 100, 1779–1785 (2009).
Seidlitz, T. et al. Mouse models of human gastric cancer subtypes with stomach-specific CreERT2-mediated pathway alterations. Gastroenterology 157, 1599–1614 (2019).
Fatehullah, A. et al. A tumor-resident Lgr5+ stem-cell-like pool drives the establishment and progression of advanced gastric cancers. Nat. Cell Biol. 23, 1299–1313 (2021).
Mao, J. et al. Role of Wnt/β-catenin signaling in the gastric cancer stem cells proliferation and salinomycin treatment. Cell Death Dis. 5, e1039 (2014).
Kok, S. Y. et al. Malignant subclone drives metastasis of genetically and phenotypically heterogeneous cell clusters through fibrotic niche generation. Nat. Commun. 12, 863 (2021).
Costa-Silva, B. et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 17, 816–826 (2015).
Jiang, H. et al. Revealing the transcriptional heterogeneity of organ-specific metastasis in human gastric cancer using single-cell RNA sequencing. Clin. Transl. Med. 12, e730 (2022).
Caligiuri, G. & Tuveson, D. A. Activated fibroblasts in cancer: perspectives and challenges. Cancer Cell 41, 434–449 (2023).
Geng, Y., and Schwabe, R.F. Hepatic stellate cell heterogeneity: Functional aspect and therapeutic implications. Hepatology https://doi.org/10.1097/hep.0000000000001386 (2025).
Yamamoto, Y. et al. The heterogeneity of cancer-associated fibroblast subpopulations: their origins, biomarkers, and roles in the tumor microenvironment. Cancer Sci. 114, 16–24 (2023).
Mihara, E. et al. Active and water-soluble form of lipidated Wnt protein is maintained by a serum glycoprotein afamin/α-albumin. eLife 5, e11621 (2016).
Martinez-Ordonez, A. et al. Hyaluronan driven by epithelial aPKC deficiency remodels the microenvironment and creates a vulnerability in mesenchymal colorectal cancer. Cancer Cell 41, 252–271 (2023).
Bourguignon, V. & Flamion, B. Respective roles of hyaluronidases 1 and 2 in endogenous hyaluronan turnover. FASEB J. 30, 2108–2114 (2016).
Leushacke, M. et al. Lgr5-expressing chief cells drive epithelial regeneration and cancer in the oxyntic stomach. Nat. Cell Biol. 19, 774–786 (2017).
Kinoshita, H. et al. Three types of metaplasia model through Kras activation, Pten deletion, or Cdh1 deletion in the gastric epithelium. J. Pathol. 247, 35–47 (2019).
Oshima, H. et al. Suppressing TGFβ signaling in regenerating epithelia in an inflammatory microenvironment is sufficient to cause invasive intestinal cancer. Cancer Res. 75, 766–776 (2015).
Huh, W. J. et al. Tamoxifen induces rapid, reversible atrophy, and metaplasia in mouse stomach. Gastroenterology 142, 21–24 (2012).
Tigue, M. L. et al. Wnt signaling in the phenotype and function of tumor-associated macrophage. Cancer Res. 83, 3–11 (2023).
Akhmetshina, A. et al. Activation of canonical Wnt signaling is required for TGF-β-mediated fibrosis. Nat. Commun. 3, 735 (2012).
Sun, Y. et al. FGF9 promotes expression of HAS2 in palatal elevation via the Wnt/β-catenin/TCF7L2 pathway. Biomolecules 12, 1639 (2022).
Liu, M., Tolg, C. & Turley, E. Dissecting the dual nature of hyaluronan in the tumor microenvironment. Front. Immunol. 10, 947 (2019).
McGuire, J. et al. Hyaluronidase inhibitor delphinidin inhibits cancer metastasis. Sci. Rep. 14, 14958 (2024).
Sakai, E. et al. Combined mutation of Apc, Kras, and Tgfbr2 effectively drives metastasis of intestinal cancer. Cancer Res. 78, 1334–1346 (2018).
Khurana, S. S. et al. The hyaluronic acid receptor CD44 coordinates normal and metaplastic gastric epithelial progenitor cell proliferation. J. Biol. Chem. 288, 16085–16097 (2013).
Meyer, A. R. et al. Cystine/glutamate antiporter (xCT) is required for chief cell plasticity after gastric injury. Cell. Mol. Gastroenterol. Hepatol. 8, 379–405 (2019).
Chen, Y. et al. Type I collagen deletion in αSMA+ myofibroblasts augments immune suppression and accelerates progression of pancreatic cancer. Cancer Cell 39, 548–565 (2021).
Bhattacharjee, S. et al. Tumor restriction by type I collagen oppose tumor-promoting effects of cancer-associated fibroblasts. J. Clin. Invest. 131, e146987 (2021).
Chytil, A., Magnunson, M. A., Wright, C. V. & Moses, H. L. Conditional inactivation of the TGF-β type II receptor using Cre:Lox. Genesis 32, 73–75 (2002).
Olive, K. P. et al. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell 119, 847–860 (2004).
Jackson, E. L. et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 15, 3243–3248 (2001).
Jameson, J. M., Cauvi, G., Sharp, L. L., Witherden, D. A. & Havran, W. L. T cell-induced hyaluronan production by epithelial cells regulates inflammation. J. Exp. Med. 201, 1269–1279 (2005).
Tan, S. H. et al. AQP5 enriches for stem cells and cancer origins in the distal stomach. Nature 578, 437–443 (2020).
Miyoshi, H., Ajima, R., Luo, C. T., Yamaguchi, T. & Stappenbeck, T. Wnt5a potentiates TGF-β signaling to promote colonic crypt regeneration after tissue injury. Science 338, 108–113 (2012).
Morita, A., Nakayama, M., Wang, D., Murakami, K. & Oshima, M. Frequent los of metastatic ability in subclones of Apc, Kras, Tgfbr2, and Trp53 mutant intestinal tumor organoids. Cancer Sci. 114, 1437–1450 (2023).
Madan, B. et al. Wnt addiction of genetically defined cancers reversed by PORCN inhibition. Oncogene 35, 2197–2207 (2016).
Cheng, D. et al. Discovery of pyridinyl acetamide derivatives as potent, selective and orally bioavailable porcupine inhibitors. ACS Med. Chem. Lett. 7, 676–680 (2016).
Riccalton-Banks, L., Bhandari, R., Fry, J. & Shakesheff, K. M. A simple method for the simultaneous isolation of stellate cells and hepatocytes from rat liver tissue. Mol. Cell. Biochem. 248, 97–102 (2003).
Franzen, O., Gan L-M., & Bjorkegren J.L.M. PanglaoDB: a web server for exploration of mouse and human single-cell RNA sequencing data. Database https://doi.org/10.1093/database/baz046 (2019).
Acknowledgements
We thank Manami Watanabe and Ayako Tsuda for their technical assistance. This work was supported by Grants-in-Aid for Scientific Research (A) (22H00454 to M.O.) and (B) (23K02899 to H.O.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan; AMED (24ama22152h0002 to H.O.) from the Japan Agency for Medical Research and Development, Japan.
Author information
Authors and Affiliations
Contributions
Y.F., H.O., R.M., M.N., and K.M. performed experiments; C.P.H. and SJ.C. performed bioinformatics analysis; S.Y., Y.I., and D.M. analyzed patient materials; H.O. and N.B. generated mouse models; H.O., N.I, and M.O. discussed results; M.O. supervised research; H.O. and M.O. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Jason Mills, Jae-Il Park, and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Furutani, Y., Oshima, H., Hong, C.P. et al. Ligand-dependent Wnt signaling promotes gastric cancer metastasis through hyaluronan expression in microenvironment. Nat Commun (2026). https://doi.org/10.1038/s41467-026-69470-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-026-69470-5