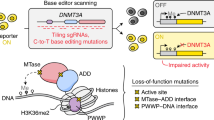
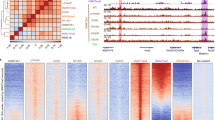
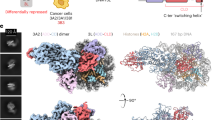
Abstract
DNA methyltransferase DNMT3A-mediated DNA methylation is important for genomic imprinting and transcriptional regulation. However, how the regulatory domains of DNMT3A cooperate with its methyltransferase domain and histone marks to orchestrate genomic methylation remains unclear. Here we report the cryo-EM structure of DNMT3A2 with regulatory factor DNMT3L, revealing an intricate domain interaction underlying multilayered autoinhibition. The PWWP domain interacts with the ADD and methyltransferase domains to block the target recognition domain and the H3K36me2-binding pocket, thereby coupling the H3K36me2 binding with DNMT3A activation, adding a layer of allosteric regulation distinct from the previously characterized ADD-H3K4me0 regulation. Molecular dynamics simulations of the DNMT3A-DNMT3L complex further reveals that relief of DNMT3A autoinhibition involves disengagement of the CpG-recognition loop of the target recognition domain from autoinhibitory interaction, leading to enhanced accessibility of the target recognition domain loop for DNA binding and DNMT3A activation. Importantly, our combined structural, biochemical and genomic methylation analysis demonstrates that disrupting the PWWP-ADD interaction by disease-associated DNMT3A mutations leads to impaired DNMT3A autoinhibition and substrate specificity, providing a potential explanation to aberrant DNA methylation in disease.
Similar content being viewed by others
Data availability
The atomic model for the DNMT3A2-DNMT3L complex has been deposited in the Protein Data Bank under accession code 9PRW. The cryo-EM density map has been deposited in EMDB under the accession number of EMD-71814. The PDB accession codes 2PVC2PV03LLR, 4QBQ, 4U7P, 5CIU, 5YX2 and 8EIH were used in this study. The Cut&Tag and DNA methylation profiling data deposited in NCBI Gene Expression Omnibus under accession number GSE24701913 were used in this study. Source Data are provided in the Source Data file. Source data are provided with this paper.
References
Bird, A. DNA methylation patterns and epigenetic memory. Genes Dev. 16, 6–21 (2002).
Jones, P. A. & Gonzalgo, M. L. Altered DNA methylation and genome instability: a new pathway to cancer? Proc. Natl. Acad. Sci. USA 94, 2103–2105 (1997).
Moore, L. D., Le, T. & Fan, G. DNA methylation and its basic function. Neuropsychopharmacology 38, 23–38 (2013).
Okano, M., Bell, D. W., Haber, D. A. & Li, E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99, 247–257 (1999).
Lister, R. et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462, 315–322 (2009).
Jeltsch, A. & Jurkowska, R. Z. Allosteric control of mammalian DNA methyltransferases - a new regulatory paradigm. Nucleic Acids Res. 44, 8556–8575 (2016).
Liang, G. et al. Cooperativity between DNA methyltransferases in the maintenance methylation of repetitive elements. Mol. Cell Biol. 22, 480–491 (2002).
Goll, M. G. & Bestor, T. H. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 74, 481–514 (2005).
Jones, P. A. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 13, 484–492 (2012).
Gu, T. et al. The disordered N-terminal domain of DNMT3A recognizes H2AK119ub and is required for postnatal development. Nat. Genet. 54, 625–636 (2022).
Weinberg, D. N. et al. Two competing mechanisms of DNMT3A recruitment regulate the dynamics of de novo DNA methylation at PRC1-targeted CpG islands. Nat. Genet. 53, 794–800 (2021).
Chen, X. et al. Structural basis for the H2AK119ub1-specific DNMT3A-nucleosome interaction. Nat. Commun. 15, 6217 (2024).
Gretarsson, K. H. et al. Cancer-associated DNA hypermethylation of Polycomb targets requires DNMT3A dual recognition of histone H2AK119 ubiquitination and the nucleosome acidic patch. Sci. Adv. 10, eadp0975 (2024).
Wapenaar, H. et al. The N-terminal region of DNMT3A engages the nucleosome surface to aid chromatin recruitment. EMBO Rep. 25, 5743–5779 (2024).
Tajima, S. et al. Domain structure of the Dnmt1, Dnmt3a, and Dnmt3b DNA methyltransferases. Adv. Exp. Med. Biol. 1389, 45–68 (2022).
Dhayalan, A. et al. The Dnmt3a PWWP domain reads histone 3 lysine 36 trimethylation and guides DNA methylation. J. Biol. Chem. 285, 26114–26120 (2010).
Dukatz, M. et al. H3K36me2/3 binding and DNA binding of the DNA methyltransferase DNMT3A PWWP domain both contribute to its chromatin interaction. J. Mol. Biol. 431, 5063–5074 (2019).
Weinberg, D. N. et al. The histone mark H3K36me2 recruits DNMT3A and shapes the intergenic DNA methylation landscape. Nature 573, 281–286 (2019).
Otani, J. et al. Structural basis for recognition of H3K4 methylation status by the DNA methyltransferase 3A ATRX-DNMT3-DNMT3L domain. EMBO Rep. 10, 1235–1241 (2009).
Zhang, Y. et al. Chromatin methylation activity of Dnmt3a and Dnmt3a/3L is guided by interaction of the ADD domain with the histone H3 tail. Nucleic Acids Res. 38, 4246–4253 (2010).
Chen, T., Tsujimoto, N. & Li, E. The PWWP domain of Dnmt3a and Dnmt3b is required for directing DNA methylation to the major satellite repeats at pericentric heterochromatin. Mol. Cell Biol. 24, 9048–9058 (2004).
Guo, X. et al. Structural insight into autoinhibition and histone H3-induced activation of DNMT3A. Nature 517, 640–644 (2015).
Li, B. Z. et al. Histone tails regulate DNA methylation by allosterically activating de novo methyltransferase. Cell Res. 21, 1172–1181 (2011).
Xu, W. et al. DNMT3A reads and connects histone H3K36me2 to DNA methylation. Protein Cell 11, 150–154 (2020).
Bourc’his, D., Xu, G. L., Lin, C. S., Bollman, B. & Bestor, T. H. Dnmt3L and the establishment of maternal genomic imprints. Science 294, 2536–2539 (2001).
Chedin, F., Lieber, M. R. & Hsieh, C. L. The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by Dnmt3a. Proc. Natl. Acad. Sci. USA 99, 16916–16921 (2002).
Hata, K., Okano, M., Lei, H. & Li, E. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development 129, 1983–1993 (2002).
Veland, N. et al. DNMT3L facilitates DNA methylation partly by maintaining DNMT3A stability in mouse embryonic stem cells. Nucleic Acids Res. 47, 152–167 (2019).
Ooi, S. K. et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448, 714–717 (2007).
Gowher, H., Liebert, K., Hermann, A., Xu, G. & Jeltsch, A. Mechanism of stimulation of catalytic activity of Dnmt3A and Dnmt3B DNA-(cytosine-C5)-methyltransferases by Dnmt3L. J. Biol. Chem. 280, 13341–13348 (2005).
Jia, D., Jurkowska, R. Z., Zhang, X., Jeltsch, A. & Cheng, X. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature 449, 248–251 (2007).
Anteneh, H., Fang, J. & Song, J. Structural basis for impairment of DNA methylation by the DNMT3A R882H mutation. Nat. Commun. 11, 2294 (2020).
Zhang, Z. M. et al. Structural basis for DNMT3A-mediated de novo DNA methylation. Nature 554, 387–391 (2018).
Heyn, P. et al. Gain-of-function DNMT3A mutations cause microcephalic dwarfism and hypermethylation of Polycomb-regulated regions. Nat. Genet. 51, 96–105 (2019).
Ley, T. J. et al. DNMT3A mutations in acute myeloid leukemia. N. Engl. J. Med. 363, 2424–2433 (2010).
Remacha, L. et al. Gain-of-function mutations in DNMT3A in patients with paraganglioma. Genet. Med. 20, 1644–1651 (2018).
Tatton-Brown, K. et al. Mutations in the DNA methyltransferase gene DNMT3A cause an overgrowth syndrome with intellectual disability. Nat. Genet. 46, 385–388 (2014).
Yang, L., Rau, R. & Goodell, M. A. DNMT3A in haematological malignancies. Nat. Rev. Cancer 15, 152–165 (2015).
Huang, Y. H. et al. Systematic profiling of DNMT3A variants reveals protein instability mediated by the DCAF8 E3 ubiquitin ligase adaptor. Cancer Discov. 12, 220–235 (2022).
Tatton-Brown, K. et al. The Tatton-Brown-Rahman syndrome: a clinical study of 55 individuals with de novo constitutive DNMT3A variants. Wellcome Open Res. 3, 46 (2018).
Lu, J. et al. Structure-guided functional suppression of AML-associated DNMT3A hotspot mutations. Nat. Commun. 15, 3111 (2024).
Lu, R. et al. A model system for studying the DNMT3A hotspot mutation (DNMT3A(R882)) demonstrates a causal relationship between its dominant-negative effect and leukemogenesis. Cancer Res. 79, 3583–3594 (2019).
Nguyen, T. V. et al. The R882H DNMT3A hot spot mutation stabilizes the formation of large DNMT3A oligomers with low DNA methyltransferase activity. J. Biol. Chem. 294, 16966–16977 (2019).
Russler-Germain, D. A. et al. The R882H DNMT3A mutation associated with AML dominantly inhibits wild-type DNMT3A by blocking its ability to form active tetramers. Cancer Cell 25, 442–454 (2014).
Cho, C. C. et al. Histone modification-driven structural remodeling unleashes DNMT3B in DNA methylation. Sci. Adv. 11, eadu8116 (2025).
Lu, J. et al. Structural basis for the allosteric regulation and dynamic assembly of DNMT3B. Nucleic Acids Res. 51, 12476–12491 (2023).
Rondelet, G., Dal Maso, T., Willems, L. & Wouters, J. Structural basis for recognition of histone H3K36me3 nucleosome by human de novo DNA methyltransferases 3A and 3B. J. Struct. Biol. 194, 357–367 (2016).
Simon, M. D. et al. The site-specific installation of methyl-lysine analogs into recombinant histones. Cell 128, 1003–1012 (2007).
Hu, J. L. et al. The N-terminus of histone H3 is required for de novo DNA methylation in chromatin. Proc. Natl. Acad. Sci. USA 106, 22187–22192 (2009).
Brohm, A. et al. Methylation of recombinant mononucleosomes by DNMT3A demonstrates efficient linker DNA methylation and a role of H3K36me3. Commun. Biol. 5, 192 (2022).
Gao, L. et al. Comprehensive structure-function characterization of DNMT3B and DNMT3A reveals distinctive de novo DNA methylation mechanisms. Nat. Commun. 11, 3355 (2020).
Yan, X. J. et al. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat. Genet. 43, 309–315 (2011).
Kikuchi, A. et al. Structural basis for activation of DNMT1. Nat. Commun. 13, 7130 (2022).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Bepler, T. et al. Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs. Nat. Methods 16, 1153–1160 (2019).
Wang, N. et al. Structural basis of human monocarboxylate transporter 1 inhibition by anti-cancer drug candidates. Cell 184, 370–383.e313 (2021).
Sanchez-Garcia, R. et al. DeepEMhancer: a deep learning solution for cryo-EM volume post-processing. Commun. Biol. 4, 874 (2021).
Pettersen, E. F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 (2004).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 66, 213–221 (2010).
Case, D. A. et al. AmberTools. J. Chem. Inf. Model 63, 6183–6191 (2023).
Peters, M. B. et al. Structural survey of zinc containing proteins and the development of the zinc AMBER force field (ZAFF). J. Chem. Theory Comput. 6, 2935–2947 (2010).
Hyman, A. A., Weber, C. A. & Julicher, F. Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 30, 39–58 (2014).
Ramirez, F. et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016).
Meers, M. P., Tenenbaum, D. & Henikoff, S. Peak calling by Sparse Enrichment Analysis for CUT&RUN chromatin profiling. Epigenet. Chromatin 12, 42 (2019).
Akalin, A. et al. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 13, R87 (2012).
Acknowledgements
This work was supported by NIH grants (R35GM119721 to J.S., R35GM138181 and R01CA266978 to C.L., and R01GM109045 to C.A.C.), NSF grant MCB-2437134 to C.A.C. and the Academic Senate Award of UC Riverside to J.S. E.V. and N.K. were supported by GAANN fellowship of Department of Education (P200A210136). N.K. was also supported by the administrative supplement of NIH R35GM119721. A portion of this research was supported by NIH grant U24GM129547 and performed at the PNCC at OHSU and accessed through EMSL (grid.436923.9), a DOE Office of Science User Facility sponsored by the Office of Biological and Environmental Research. Collection of the Cryo-EM data was assisted by Dr. Rose Marie Haynes. We thank Dr. Lingchao Zhu in ACIF NMR facility of UC Riverside for assistance in NMR data collection and UC Riverside High Performance Computer Cluster for providing computation resources.
Author information
Authors and Affiliations
Contributions
J.L., E.V., J.C., K.H.G., N.K. and Z.S. performed the experiments, C.L., C.C. and J.S. supervised the study. J.L. and J.S. wrote the manuscript and all authors approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Hanna Yuan and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. [A peer review file is available.]
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lu, J., Vig, E., Chen, J. et al. Structural insight into hierarchical DNMT3A autoinhibition and its dysregulation in disease. Nat Commun (2026). https://doi.org/10.1038/s41467-026-69563-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-026-69563-1