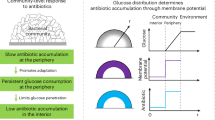
Abstract
Antibiotic resistance is a growing global health threat. Although antibiotic activity is well studied in homogeneous liquid cultures, many infections are caused by spatially structured multicellular populations where consumption of scarce nutrients establishes strong spatial variations in their abundance. These nutrient variations have long been hypothesized to help bacterial populations tolerate antibiotics, since liquid culture studies link antibiotic tolerance to metabolic activity, and thus, local nutrient availability. Here, we test this hypothesis by visualizing cell death in structured Escherichia coli populations exposed to select nutrients and antibiotics. We find that nutrient availability acts as a bottleneck to antibiotic killing, causing death to propagate through the population as a traveling front. By integrating our measurements with biophysical theory and simulations, we establish quantitative principles that explain how collective nutrient consumption can limit the progression of this “death front,” protecting a population from a nominally deadly antibiotic dose. While increasing nutrient supply can overcome this bottleneck, in some cases, excess nutrient unexpectedly promotes the regrowth of resistant cells. Altogether, this work provides a key step toward predicting and controlling antibiotic treatment of spatially structured bacterial populations, yielding biophysical insights into collective behavior and guiding strategies for effective antibiotic stewardship.
Similar content being viewed by others
Data availability
The compressed imaging data generated in this study are provided in the Supplementary Movie files on Zenodo https://zenodo.org/records/14990206. Additional experimental and simulation data generated in this study are provided in the Source Data file. Source data are provided with this paper.
Code availability
The code generated during the current study are available at Code Ocean with DOI: 10.24433/CO.4205040.v1.
References
Murray, C. J. L. et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399, 629–655 (2022).
Iskandar, K. et al. Antibiotic discovery and resistance: the chase and the race. Antibiotics 11, 182 (2022).
Meylan, S., Andrews, I. W. & Collins, J. J. Targeting antibiotic tolerance, pathogen by pathogen. Cell 172, 1228–1238 (2018).
Yang, J. H., Bening, S. C. & Collins, J. J. Antibiotic efficacy – context matters. Curr. Opin. Microbiol. 39, 73–80 (2017).
Van Acker, H. & Coenye, T. The role of reactive oxygen species in antibiotic-mediated killing of bacteria. Trends Microbiol. 25, 456–466 (2017).
Bren, A., Glass, D. S., Kohanim, Y. K., Mayo, A. & Alon, U. Tradeoffs in bacterial physiology determine the efficiency of antibiotic killing. Proc. Natl. Acad. Sci. 120, e2312651120 (2023).
Lobritz, M. A. et al. Antibiotic efficacy is linked to bacterial cellular respiration. Proc. Natl. Acad. Sci. 112, 8173–8180 (2015).
Harms, A., Maisonneuve, E. & Gerdes, K. Mechanisms of bacterial persistence during stress and antibiotic exposure. Science 354, aaf4268 (2016).
Ojkic, N., Serbanescu, D. & Banerjee, S. Antibiotic resistance via bacterial cell shape-shifting. mBio 13, e00659–22 (2022).
Jo, J., Price-Whelan, A. & Dietrich, L. E. P. Gradients and consequences of heterogeneity in biofilms. Nat. Rev. Microbiol. 20, 593–607 (2022).
Brauner, A., Fridman, O., Gefen, O. & Balaban, N. Q. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat. Rev. Microbiol. 14, 320–330 (2016).
Ceri, H. et al. The Calgary biofilm device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J. Clin. Microbiol. 37, 1771–1776 (1999).
Jacqueline, C. & Caillon, J. Impact of bacterial biofilm on the treatment of prosthetic joint infections. J. Antimicrob. Chemother. 69, i37–i40 (2014).
Kirby, A. E., Garner, K. & Levin, B. R. The relative contributions of physical structure and cell density to the antibiotic susceptibility of bacteria in biofilms. Antimicrob. Agents Chemother. 56, 2967–2975 (2012).
Sharma, D., Misba, L. & Khan, A. U. Antibiotics versus biofilm: an emerging battleground in microbial communities. Antimicrob. Resistance Infect. Control 8, 76 (2019).
Kolpen, M. et al. Bacterial biofilms predominate in both acute and chronic human lung infections. Thorax 77, 1015–1022 (2022).
Flemming, H.-C. et al. Who put the film in biofilm? The migration of a term from wastewater engineering to medicine and beyond. npj Biofilms Microbiomes 7, 10 (2021).
Cámara, M. et al. Economic significance of biofilms: a multidisciplinary and cross-sectoral challenge. npj Biofilms Microbiomes 8, 42 (2022).
Stewart, P. S. & Franklin, M. J. Physiological heterogeneity in biofilms. Nat. Rev. Microbiol. 6, 199–210 (2008).
Eng, R. H., Padberg, F. T., Smith, S. M., Tan, E. N. & Cherubin, C. E. Bactericidal effects of antibiotics on slowly growing and nongrowing bacteria. Antimicrob. Agents Chemother. 35, 1824–1828 (1991).
Greulich, P., Scott, M., Evans, M. R. & Allen, R. J. Growth-dependent bacterial susceptibility to ribosome-targeting antibiotics. Mol. Syst. Biol. 11, 796 (2015).
Lee, A. J. et al. Robust, linear correlations between growth rates and β-lactam mediated lysis rates. Proc. Natl. Acad. Sci. 115, 4069–4074 (2018).
Levin, B. R. & Rozen, D. E. Non-inherited antibiotic resistance. Nat. Rev. Microbiol. 4, 556–562 (2006).
Roberts, M. E. & Stewart, P. S. Modeling antibiotic tolerance in biofilms by accounting for nutrient limitation. Antimicrob. Agents Chemother. 48, 48–52 (2004).
Anderl, J. N., Zahller, J., Roe, F. & Stewart, P. S. Role of nutrient limitation and stationary-phase existence in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob. Agents Chemother. 47, 1251–1256 (2003).
Walters, M. C., Roe, F., Bugnicourt, A., Franklin, M. J. & Stewart, P. S. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob. Agents Chemother. 47, 317–323 (2003).
Fux, C., Costerton, J., Stewart, P. & Stoodley, P. Survival strategies of infectious biofilms. Trends Microbiol. 13, 34–40 (2005).
Highmore, C. J. et al. Translational challenges and opportunities in biofilm science: a BRIEF for the future. npj Biofilms Microbiomes 8, 68 (2022).
Quan, K. et al. Water in bacterial biofilms: pores and channels, storage and transport functions. Crit. Rev. Microbiol. 48, 283–302 (2022).
Sauer, K. et al. The biofilm life cycle: expanding the conceptual model of biofilm formation. Nat. Rev. Microbiol. 20, 608–620 (2022).
Wimpenny, J., Manz, W. & Szewzyk, U. Heterogeneity in biofilms: Table 1. FEMS Microbiol. Rev. 24, 661–671 (2000).
Eigentler, L., Davidson, F. A. & Stanley-Wall, N. R. Mechanisms driving spatial distribution of residents in colony biofilms: an interdisciplinary perspective. Open Biol. 12, 220194 (2022).
Relucenti, M. et al. Microscopy methods for biofilm imaging: focus on SEM and VP-SEM Pros and Cons. Biology 10, 51 (2021).
Stevanovic, M. et al. Nutrient gradients mediate complex colony-level antibiotic responses in structured microbial populations. Front. Microbiol. 13, 740259 (2022).
Nagy, K. et al. Emergence of resistant Escherichia coli mutants in microfluidic on-chip antibiotic gradients. Front. Microbiol. 13, 820738 (2022).
Zhang, Q. et al. Acceleration of emergence of bacterial antibiotic resistance in connected microenvironments. Science 333, 1764–1767 (2011).
Hol, F. J. H., Hubert, B., Dekker, C. & Keymer, J. E. Density-dependent adaptive resistance allows swimming bacteria to colonize an antibiotic gradient. ISME J. 10, 30–38 (2016).
Kim, S., Masum, F., Kim, J.-K., Chung, H. J. & Jeon, J. S. On-chip phenotypic investigation of combinatory antibiotic effects by generating orthogonal concentration gradients. Lab Chip 19, 959–973 (2019).
Falagas, M. E., Vouloumanou, E. K., Samonis, G. & Vardakas, K. Z. Fosfomycin. Clin. Microbiol. Rev. 29, 321–347 (2016).
Sojo-Dorado, J. et al. Effectiveness of fosfomycin for the treatment of multidrug-resistant Escherichia coli bacteremic urinary tract infections: a randomized clinical trial. JAMA Netw. Open 5, e2137277 (2022).
Grabein, B., Graninger, W., Rodríguez Baño, J., Dinh, A. & Liesenfeld, D. Intravenous fosfomycin–back to the future. Systematic review and meta-analysis of the clinical literature. Clin. Microbiol. Infect. 23, 363–372 (2017).
Silver, L. L. Fosfomycin: mechanism and resistance. Cold Spring Harb. Perspect. Med. 7, a025262 (2017).
Castañeda-García, A., Blázquez, J. & Rodríguez-Rojas, A. Molecular mechanisms and clinical impact of acquired and intrinsic fosfomycin resistance. Antibiotics 2, 217–236 (2013).
Martín-Gutiérrez, G. et al. Urinary tract conditions affect fosfomycin activity against Escherichia coli strains harboring chromosomal mutations involved in fosfomycin uptake. Antimicrob. Agents Chemother. 62, e01899–17 (2018).
Detter, G., Knothe, H., Schönenbach, B. & Plage, G. Comparative study of fosfomycin activity in Mueller-Hinton media and in tissues. J. Antimicrob. Chemother. 11, 517–524 (1983).
Ishihama, Y. et al. Protein abundance profiling of the Escherichia coli cytosol. BMC Genomics 9, 102 (2008).
Gil-Gil, T. & Martínez, J. L. Fosfomycin resistance evolutionary pathways of Stenotrophomonas maltophilia in different growing conditions. Int. J. Mol. Sci. 23, 1132 (2022).
Carreón-Rodríguez, O. E., Gosset, G., Escalante, A. & Bolívar, F. Glucose transport in Escherichia coli: from basics to transport engineering. Microorganisms 11, 1588 (2023).
Stewart, P. S. Biofilm accumulation model that predicts antibiotic resistance of Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 38, 1052–1058 (1994).
Bhattacharjee, T. & Datta, S. S. Bacterial hopping and trapping in porous media. Nat. Commun. 10, 2075 (2019).
Bhattacharjee, T. & Datta, S. S. Confinement and activity regulate bacterial motion in porous media. Soft Matter 15, 9920–9930 (2019).
Bhattacharjee, T., Amchin, D. B., Ott, J. A., Kratz, F. & Datta, S. S. Chemotactic migration of bacteria in porous media. Biophys. J. 120, 3483–3497 (2021).
Bay, R. K., Hancock, A. M., Dill-Macky, A. S., Luu, H. N. & Datta, S. S. 3D printing bacteria to study motility and growth in complex 3D porous media. J. Vis. Exp. 66166, https://app.jove.com/t/66166 (2004).
Hancock, A. M. & Datta, S. S. Interplay between environmental yielding and dynamic forcing modulates bacterial growth. Biophys. J. 123, 957–967 (2024).
Martínez-Calvo, A. et al. Morphological instability and roughening of growing 3D bacterial colonies. Proc. Natl. Acad. Sci. 119, e2208019119 (2022).
Liu, H. Y., Prentice, E. L. & Webber, M. A. Mechanisms of antimicrobial resistance in biofilms. npj Antimicrob. Resist. 2, 27 (2024).
Olsen, I. Biofilm-specific antibiotic tolerance and resistance. Eur. J. Clin. Microbiol. Infect. Dis. 34, 877–886 (2015).
Cowart, S. L. & Stachura, M. E. Glucosuria. In Clinical Methods: The History, Physical, and Laboratory Examinations, 3rd edn. (eds, Walker, H. K., Hall, W. D. & Hurst, J. W.) (Butterworths, Boston, 1990) http://www.ncbi.nlm.nih.gov/books/NBK245/.
Stewart, P. S. Antimicrobial tolerance in biofilms. Microbiol. Spectr. 3, 3.3.07 (2015).
Mattei, M. R. et al. Continuum and discrete approach in modeling biofilm development and structure: a review. J. Math. Biol. 76, 945–1003 (2018).
Moore-Ott, J. A., Chiu, S., Amchin, D. B., Bhattacharjee, T. & Datta, S. S. A biophysical threshold for biofilm formation. eLife 11, e76380 (2022).
Zhang, Y. et al. Persistent glucose consumption under antibiotic treatment protects bacterial community. Nat. Chem. Biol. 21, 238–246 (2025).
Monod, J. The growth of bacterial cultures. Annu. Rev. Microbiol. 3, 371–394 (1949).
Scott, M., Klumpp, S., Mateescu, E. M. & Hwa, T. Emergence of robust growth laws from optimal regulation of ribosome synthesis. Mol. Syst. Biol. 10, 747 (2014).
Kovárová-Kovar, K. & Egli, T. Growth kinetics of suspended microbial cells: from single-substrate-controlled growth to mixed-substrate kinetics. Microbiol. Mol. Biol. Rev. 62, 646–666 (1998).
Kim, K. et al. Mapping single-cell responses to population-level dynamics during antibiotic treatment. Mol. Syst. Biol. 19, e11475 (2023).
El-Halfawy, O. M. & Valvano, M. A. Antimicrobial heteroresistance: an emerging field in need of clarity. Clin. Microbiol. Rev. 28, 191–207 (2015).
Andersson, D. I., Nicoloff, H. & Hjort, K. Mechanisms and clinical relevance of bacterial heteroresistance. Nat. Rev. Microbiol. 17, 479–496 (2019).
Xu, L. et al. Epidemiology, mechanisms, and clinical impact of bacterial heteroresistance. npj Antimicrob. Resist. 3, 7 (2025).
Pereira, C., Larsson, J., Hjort, K., Elf, J. & Andersson, D. I. The highly dynamic nature of bacterial heteroresistance impairs its clinical detection. Commun. Biol. 4, 521 (2021).
Nicoloff, H., Hjort, K., Levin, B. R. & Andersson, D. I. The high prevalence of antibiotic heteroresistance in pathogenic bacteria is mainly caused by gene amplification. Nat. Microbiol. 4, 504–514 (2019).
Abbott, I. J. et al. Oral fosfomycin efficacy with variable urinary exposures following single and multiple doses against Enterobacterales : the importance of heteroresistance for growth outcome. Antimicrob. Agents Chemother. 64, e01982–19 (2020).
Balaban, N. Q. et al. Definitions and guidelines for research on antibiotic persistence. Nat. Rev. Microbiol. 17, 441–448 (2019).
Wang, Y. et al. Heteroresistance is associated with in vitro regrowth during colistin treatment in carbapenem-resistant Klebsiella pneumoniae. Front. Microbiol. 13, 868991 (2022).
Band, V. I. et al. Carbapenem-resistant Klebsiella pneumoniae exhibiting clinically undetected colistin heteroresistance leads to treatment failure in a murine model of infection. mBio 9, e02448–17 (2018).
Hughes, D. & Andersson, D. I. Evolutionary consequences of drug resistance: shared principles across diverse targets and organisms. Nat. Rev. Genet. 16, 459–471 (2015).
Barrick, J. E. & Lenski, R. E. Genome dynamics during experimental evolution. Nat. Rev. Genet. 14, 827–839 (2013).
Spratt, M. R. & Lane, K. Navigating environmental transitions: the role of phenotypic variation in bacterial responses. mBio 13, e02212–22 (2022).
Verdon, N., Popescu, O., Titmuss, S. & Allen, R. J. Habitat fragmentation enhances microbial collective defence. J. R. Soc. Interface 22, 20240611 (2025).
Baym, M. et al. Spatiotemporal microbial evolution on antibiotic landscapes. Science 353, 1147–1151 (2016).
Geyrhofer, L. & Brenner, N. Coexistence and cooperation in structured habitats. BMC Ecol. 20, 14 (2020).
Grimbergen, A. J., Siebring, J., Solopova, A. & Kuipers, O. P. Microbial bet-hedging: the power of being different. Curr. Opin. Microbiol. 25, 67–72 (2015).
Villa Martín, P., Muñoz, M. A. & Pigolotti, S. Bet-hedging strategies in expanding populations. PLOS Comput. Biol. 15, e1006529 (2019).
Morawska, L. P., Hernandez-Valdes, J. A. & Kuipers, O. P. Diversity of bet-hedging strategies in microbial communities–recent cases and insights. WIREs Mechanisms Dis. 14, e1544 (2022).
Lowery, N. V., McNally, L., Ratcliff, W. C. & Brown, S. P. Division of labor, bet hedging, and the evolution of mixed biofilm investment strategies. mBio 8, e00672–17 (2017).
Kowalski, C. H., Morelli, K. A., Schultz, D., Nadell, C. D. & Cramer, R. A. Fungal biofilm architecture produces hypoxic microenvironments that drive antifungal resistance. Proc. Natl. Acad. Sci. 117, 22473–22483 (2020).
Beebout, C. J., Sominsky, L. A., Eberly, A. R., Van Horn, G. T. & Hadjifrangiskou, M. Cytochrome bd promotes Escherichia coli biofilm antibiotic tolerance by regulating accumulation of noxious chemicals. npj Biofilms Microbiomes 7, 35 (2021).
Berryhill, B. A. et al. What’s the matter with MICs: bacterial nutrition, limiting resources, and antibiotic pharmacodynamics. Microbiol. Spectr. 11, e04091–22 (2023).
Crabbé, A., Jensen, P. O., Bjarnsholt, T. & Coenye, T. Antimicrobial tolerance and metabolic adaptations in microbial biofilms. Trends Microbiol. 27, 850–863 (2019).
De Vos, M. G. J., Zagorski, M., McNally, A. & Bollenbach, T. Interaction networks, ecological stability, and collective antibiotic tolerance in polymicrobial infections. Proc. Natl. Acad. Sci. 114, 10666–10671 (2017).
Townsend, E. M. et al. Development and characterisation of a novel three-dimensional inter-kingdom wound biofilm model. Biofouling 32, 1259–1270 (2016).
Stewart, P. S. Diffusion in biofilms. J. Bacteriol. 185, 1485–1491 (2003).
Wilking, J. N., Angelini, T. E., Seminara, A., Brenner, M. P. & Weitz, D. A. Biofilms as complex fluids. MRS Bull. 36, 385–391 (2011).
Paone, P. & Cani, P. D. Mucus barrier, mucins and gut microbiota: the expected slimy partners? Gut 69, 2232–2243 (2020).
Stewart, P. S. Biophysics of biofilm infection. Pathog. Dis. 70, 212–218 (2014).
Patange, O. et al. Escherichia coli can survive stress by noisy growth modulation. Nat. Commun. 9, 5333 (2018).
Nadezhdin, E., Murphy, N., Dalchau, N., Phillips, A. & Locke, J. C. W. Stochastic pulsing of gene expression enables the generation of spatial patterns in Bacillus subtilis biofilms. Nat. Commun. 11, 950 (2020).
Moreno-Gámez, S. et al. Wide lag time distributions break a trade-off between reproduction and survival in bacteria. Proc. Natl. Acad. Sci. 117, 18729–18736 (2020).
Oliveira, N. M. et al. Suicidal chemotaxis in bacteria. Nat. Commun. 13, 7608 (2022).
Schink, S. J., Biselli, E., Ammar, C. & Gerland, U. Death rate of E. coli during starvation Is set by maintenance cost and biomass recycling. Cell Syst. 9, 64–73.e3 (2019).
Fagerlind, M. G. et al. Dynamic modelling of cell death during biofilm development. J. Theor. Biol. 295, 23–36 (2012).
Webb, J. S. et al. Cell Death in Pseudomonas aeruginosa Biofilm Development. J. Bacteriol. 185, 4585–4592 (2003).
Le Quellec, L. et al. Measuring single-cell susceptibility to antibiotics within monoclonal bacterial populations. PLOS One 19, e0303630 (2024).
Coates, J. et al. Antibiotic-induced population fluctuations and stochastic clearance of bacteria. eLife 7, e32976 (2018).
Allison, K. R., Brynildsen, M. P. & Collins, J. J. Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature 473, 216–220 (2011).
Gil-Gil, T., Laborda, P., Martínez, J. L. & Hernando-Amado, S. Use of adjuvants to improve antibiotic efficacy and reduce the burden of antimicrobial resistance. Expert Rev. Anti Infective Ther. 23, 31–47 (2025).
Song, S. & Wood, T. K. Combatting persister cells with substituted indoles. Front. Microbiol. 11, 1565 (2020).
Dhanda, G., Acharya, Y. & Haldar, J. Antibiotic adjuvants: a versatile approach to combat antibiotic resistance. ACS Omega 8, 10757–10783 (2023).
Cohen, N., Lobritz, M. & Collins, J. Microbial persistence and the road to drug resistance. Cell Host Microbe 13, 632–642 (2013).
Levin-Reisman, I. et al. Antibiotic tolerance facilitates the evolution of resistance. Science 355, 826–830 (2017).
Levin-Reisman, I., Brauner, A., Ronin, I. & Balaban, N. Q. Epistasis between antibiotic tolerance, persistence, and resistance mutations. Proc. Natl. Acad. Sci. 116, 14734–14739 (2019).
Andersson, D. I. & Hughes, D. Microbiological effects of sublethal levels of antibiotics. Nat. Rev. Microbiol. 12, 465–478 (2014).
Linares, J. F., Gustafsson, I., Baquero, F. & Martinez, J. L. Antibiotics as intermicrobial signaling agents instead of weapons. Proc. Natl. Acad. Sci. 103, 19484–19489 (2006).
Cordisco, E. & Serra, D. O. Moonlighting antibiotics: the extra job of modulating biofilm formation. Trends Microbiol. 33, 459–471 (2025).
Bottery, M. J., Pitchford, J. W. & Friman, V.-P. Ecology and evolution of antimicrobial resistance in bacterial communities. ISME J. 15, 939–948 (2021).
Meredith, H. R., Srimani, J. K., Lee, A. J., Lopatkin, A. J. & You, L. Collective antibiotic tolerance: mechanisms, dynamics and intervention. Nat. Chem. Biol. 11, 182–188 (2015).
Vega, N. M. & Gore, J. Collective antibiotic resistance: mechanisms and implications. Curr. Opin. Microbiol. 21, 28–34 (2014).
Sorg, R. A. et al. Collective resistance in microbial communities by intracellular antibiotic deactivation. PLOS Biol. 14, e2000631 (2016).
Orazi, G. & O’Toole, G. A. It takes a village: mechanisms underlying antimicrobial recalcitrance of polymicrobial biofilms. J. Bacteriol. 202, https://journals.asm.org/doi/10.1128/JB.00530-19 (2019).
Acknowledgements
We acknowledge support from National Science Foundation (NSF) grants CBET-1941716, DMR-2011750, and EF-2124863 as well as the Camille Dreyfus Teacher-Scholar and Pew Biomedical Scholars Programs, the Eric and Wendy Schmidt Transformative Technology Fund, and the Princeton Catalysis Initiative. This work was supported in part by the NSF Graduate Research Fellowship Program (to A.M.H.) under grant no. DGE-2039656; any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the NSF. We thank Katherine Sniezek for constructing the strain used in these experiments and for instructions on the population analysis profiling assay, as well as Mark Brynildsen, Ned Wingreen, Bruce Levin, the late Kevin Wood, and members of the Datta Lab for stimulating discussions and useful feedback.
Author information
Authors and Affiliations
Contributions
A.M.H. and S.S.D. conceptualized and designed the overall research project; A.M.H., A.S.D-M., and C.D. performed all experiments and experimental analyses; A.M.H. developed the theoretical model and performed all calculations and analyses with help from J.A.M.; A.M.H., M.S.D., and S.S.D. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hancock, A.M., Dill-Macky, A.S., Moore, J.A. et al. A nutrient bottleneck controls antibiotic efficacy in structured bacterial populations. Nat Commun (2026). https://doi.org/10.1038/s41467-026-69625-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-026-69625-4