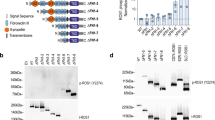
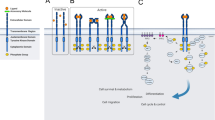
Abstract
Receptor tyrosine kinases (RTKs) are key regulators of cellular signaling and are often co-opted in cancer. ROS1 is an orphan RTK aberrantly expressed in multiple tumors, yet no approved biologic therapies target it, and its activation mechanism remains unknown. Here, we present Cryo-EM structures of mammalian ROS1 in ligand-free and NELL2-bound states, revealing how trimeric NELL2 induces both receptor clustering and a conformational switch that relieves receptor autoinhibition – both mechanisms are required for ROS1 activation. These structures, along with biochemical characterization, reflect a striking evolutionary divergence in regulatory logic compared to the invertebrate ortholog Sevenless (dROS1), highlighting how conserved RTKs can adopt fundamentally different activation strategies. Guided by these structural insights, we develop monoclonal antibodies that either block ligand binding or trap ROS1 in an inactive conformation. These agents potently suppress ROS1 signaling, representing distinct mechanistic classes of biologics that directly target ROS1 activity. Our findings elucidate a distinct mode of RTK regulation and establish a therapeutic framework for cancers driven by ROS1.
Similar content being viewed by others
Data availability
The refined structural models and corresponding density maps have been deposited in the Electron Microscopy Data Bank (EMDB) and Protein Data Bank (PDB) under accession codes: EMD-71895, 9PVP (ROS1), EMD-71938, 9PWQ (ROS1–NELL2), EMD-75142, 10FT (ROS1[N-term]–NELL2), EMD-75151, 10GH (ROS1–Fab-RX5 complex), EMD-47324, 9DZ4 (ROS1–Fab-CT4 complex). The source data are provided as a Source Data file. Source data are provided with this paper.
References
Lemmon, M. A. & Schlessinger, J. Cell signaling by receptor tyrosine kinases. Cell 141, 1117–1134 (2010).
Tomuleasa, C. et al. Therapeutic advances of targeting receptor tyrosine kinases in cancer. Signal Transduct. Target. Ther. 9, https://doi.org/10.1038/s41392-024-01899-w (2024).
Shibuya, M., Hanafusa, H. & Balduzzi, P. C. Cellular sequences related to 3 new Onc genes of avian-sarcoma virus (Fps, Yes, and Ros) and their expression in normal and transformed-cells. J. Virol. 42, 143–152 (1982).
Balduzzi, P. C., Notter, M. F. D., Morgan, H. R. & Shibuya, M. Some biological properties of 2 new avian-sarcoma viruses. J. Virol. 40, 268–275 (1981).
Kramer, H., Cagan, R. L. & Zipursky, S. L. Interaction of bride of sevenless membrane-bound ligand and the sevenless tyrosine-kinase receptor. Nature 352, 207–212 (1991).
Reinke, R. & Zipursky, S. L. Cell–cell interaction in the Drosophila retina: the bride of sevenless gene is required in photoreceptor cell R8 for R7 cell development. Cell 55, 321–330 (1988).
Cerutti, G. et al. Structures and pH-dependent dimerization of the sevenless receptor tyrosine kinase. Mol. Cell 84, 4677–4690 e4676 (2024).
Zhang, J. et al. Structural basis for the interaction between the Drosophila RTK Sevenless (dROS1) and the GPCR BOSS. Nat. Commun. 16, 808 (2025).
Acquaviva, J., Wong, R. & Charest, A. The multifaceted roles of the receptor tyrosine kinase ROS in development and cancer. Biochim. Biophys. Acta 1795, 37–52 (2009).
Springer, T. A. An extracellular β-propeller module predicted in lipoprotein and scavenger receptors, tyrosine kinases, epidermal growth factor precursor, and extracellular matrix components. J. Mol. Biol. 283, 837–862 (1998).
Sonnenberg, E., Godecke, A., Walter, B., Bladt, F. & Birchmeier, C. Transient and locally restricted expression of the ros1 protooncogene during mouse development. EMBO J. 10, 3693–3702 (1991).
Jun, H. J. et al. ROS1 signaling regulates epithelial differentiation in the epididymis. Endocrinology 155, 3661–3673 (2014).
Drilon, A. et al. ROS1-dependent cancers - biology, diagnostics and therapeutics. Nat. Rev. Clin. Oncol. 18, 35–55 (2021).
Birchmeier, C., Sharma, S. & Wigler, M. Expression and rearrangement of the ROS1 gene in human glioblastoma cells. Proc. Natl. Acad. Sci. USA 84, 9270–9274 (1987).
Charest, A. et al. Oncogenic targeting of an activated tyrosine kinase to the Golgi apparatus in a glioblastoma. Proc. Natl. Acad. Sci. USA 100, 916–921 (2003).
Rikova, K. et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 131, 1190–1203 (2007).
Takeuchi, K. et al. RET, ROS1 and ALK fusions in lung cancer. Nat. Med. 18, 378–381 (2012).
Nagasaka, M. et al. Pan-tumor survey of ROS1 fusions detected by next-generation RNA and whole transcriptome sequencing. BMC Cancer 23, 1000 (2023).
Watkins, D., Dion, F., Poisson, M., Delattre, J. Y. & Rouleau, G. A. Analysis of oncogene expression in primary human gliomas: evidence for increased expression of the ros oncogene. Cancer Genet. Cytogenet. 72, 130–136 (1994).
Mapstone, T., McMichael, M. & Goldthwait, D. Expression of platelet-derived growth factors, transforming growth factors, and the ros gene in a variety of primary human brain tumors. Neurosurgery 28, 216–222 (1991).
Grenier, K. et al. Routine clinically detected increased ROS1 transcripts are related with ROS1 expression by immunohistochemistry and associated with EGFR mutations in lung adenocarcinoma. JTO Clin. Res. Rep. 4, 100530 (2023).
Shih, C. H. et al. EZH2-mediated upregulation of ROS1 oncogene promotes oral cancer metastasis. Oncogene 36, 6542–6554 (2017).
Bajrami, I. et al. E-Cadherin/ROS1 inhibitor synthetic lethality in breast cancer. Cancer Discov. 8, 498–515 (2018).
Kiyozumi, D. et al. NELL2-mediated lumicrine signaling through OVCH2 is required for male fertility. Science 368, 1132–1135 (2020).
Watanabe, T. K. et al. Cloning and characterization of two novel human cDNAs (NELL1 and NELL2) encoding proteins with six EGF-like repeats. Genomics 38, 273–276 (1996).
Matsuhashi, S. et al. New gene, Nel, encoding a M(R) 93-K protein with Egf-Like repeats is strongly expressed in neural tissues of early-stage chick-embryos. Dev. Dynam. 203, 212–222 (1995).
Jaworski, A. et al. Operational redundancy in axon guidance through the multifunctional receptor Robo3 and its ligand NELL2. Science 350, 961–965 (2015).
Pak, J. S. et al. NELL2–Robo3 complex structure reveals mechanisms of receptor activation for axon guidance. Nat. Commun. 11, 1489 (2020).
Dawson, J. P. et al. Epidermal growth factor receptor dimerization and activation require ligand-induced conformational changes in the dimer interface. Mol. Cell. Biol. 25, 7734–7742 (2005).
Ou, S. H. I., Tan, J., Yen, Y. & Soo, R. A. ROS1 as a ‘druggable’ receptor tyrosine kinase: lessons learned from inhibiting the ALK pathway. Expert Rev. Anticancer 12, 447–456 (2012).
Trenker, R. & Jura, N. Receptor tyrosine kinase activation: from the ligand perspective. Curr. Opin. Cell Biol. 63, 174–185 (2020).
Feldman, R. A., Wang, L. H., Hanafusa, H. & Balduzzi, P. C. Avian-Sarcoma virus-Ur2 encodes a transforming protein which is associated with a unique protein-kinase activity. J. Virol. 42, 228–236 (1982).
Matsushime, H., Wang, L. H. & Shibuya, M. Human C-Ros-1 gene homologous to the V-Ros sequence of Ur2 sarcoma-virus encodes for a transmembrane receptor-like molecule. Mol. Cell. Biol. 6, 3000–3004 (1986).
Birchmeier, C., Oneill, K., Riggs, M. & Wigler, M. Characterization of Ros1 Cdna from a Human Glioblastoma Cell-Line. Proc. Natl. Acad. Sci. USA 87, 4799–4803 (1990).
Felix, J. et al. Structure and assembly mechanism of the signaling complex mediated by human CSF-1. Structure 23, 1621–1631 (2015).
Shim, A. H. et al. Structures of a platelet-derived growth factor/propeptide complex and a platelet-derived growth factor/receptor complex. Proc. Natl. Acad. Sci. USA 107, 11307–11312 (2010).
Wiesmann, C. et al. Crystal structure at 1.7 A resolution of VEGF in complex with domain 2 of the Flt-1 receptor. Cell 91, 695–704 (1997).
Li, T. et al. Structural basis for ligand reception by anaplastic lymphoma kinase. Nature 600, 148–152 (2021).
Uchikawa, E., Chen, Z., Xiao, G. Y., Zhang, X. & Bai, X. C. Structural basis of the activation of c-MET receptor. Nat. Commun. 12, 4074 (2021).
Jenni, S., Goyal, Y., von Grotthuss, M., Shvartsman, S. Y. & Klein, D. E. Structural basis of neurohormone perception by the receptor tyrosine kinase torso. Mol. Cell 60, 941–952 (2015).
Wiesmann, C., Ultsch, M. H., Bass, S. H. & de Vos, A. M. Crystal structure of nerve growth factor in complex with the ligand-binding domain of the TrkA receptor. Nature 401, 184–188 (1999).
Ferguson, K. M., Hu, C. & Lemmon, M. A. Insulin and epidermal growth factor receptor family members share parallel activation mechanisms. Protein Sci. 29, 1331–1344 (2020).
Lemmon, M. A. Ligand-induced ErbB receptor dimerization. Exp. Cell Res. 315, 638–648 (2009).
Jones, K. et al. Novel insight into mechanisms of ROS1 catalytic activation via loss of the extracellular domain. Sci. Rep. 14, 22191 (2024).
Lee, J. C. et al. Epidermal growth factor receptor activation in glioblastoma through novel missense mutations in the extracellular domain. PLoS Med. 3, 2264–2273 (2006).
Reinke, R. & Zipursky, S. L. Cell cell-interaction in the Drosophila retina—the bride of sevenless gene is required in photoreceptor cell-R8 for cell-R7 cell-development. Cell 55, 321–330 (1988).
Tomlinson, A., Mavromatakis, Y. E. & Arias, R. The role of Sevenless in Drosophila R7 photoreceptor specification. Dev. Biol. 454, 181–189 (2019).
Sevrioukov, E. A., Walenta, J. H., Sunio, A., Phistry, M. & Kramer, H. Oligomerization of the extracellular domain of Boss enhances its binding to the Sevenless receptor and its antagonistic effect on R7 induction. J. Cell Sci. 111, 737–747 (1998).
Hart, A. C., Kramer, H. & Zipursky, S. L. Extracellular domain of the boss transmembrane ligand acts as an antagonist of the Sev Receptor. Nature 361, 732–736 (1993).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Punjani, A., Zhang, H. & Fleet, D. J. Non-uniform refinement: adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods 17, 1214–1221 (2020).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Afonine, P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D Struct. Biol. 74, 531–544 (2018).
Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in. Acta Crystallogr. D 75, 861–877 (2019).
Pettersen, E. F. et al. UCSF chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Tonikian, R., Zhang, Y., Boone, C. & Sidhu, S. S. Identifying specificity profiles for peptide recognition modules from phage-displayed peptide libraries. Nat. Protoc. 2, 1368–1386 (2007).
Adams, J. J., Nelson, B. & Sidhu, S. S. Recombinant genetic libraries and human monoclonal antibodies. Methods Mol. Biol. 1060, 149–170 (2014).
Persson, H. et al. CDR-H3 diversity is not required for antigen recognition by synthetic antibodies. J. Mol. Biol. 425, 803–811 (2013).
Acknowledgements
We thank the Yale Cancer Biology Institute and its laboratories for valuable discussions. We thank Jianfeng Lin, Marc Llaguno at Yale Cryo-EM Resource Center for their assistance in Cryo-EM sample screening and data collection. Yale Cryo-EM Resource is funded in part by the NIH grant S10OD023603. We thank Guobin Hu and Jake Kaminsky at Brookhaven National Laboratory for their assistance in Cryo-EM data collection. The Laboratory for BioMolecular Structure (LBMS) is supported by the DOE Office of Biological and Environmental Research (KP1607011).
Author information
Authors and Affiliations
Contributions
D.E.K. designed the overall project with input from H.L. and C.R.A. H.L. generated all materials and performed all solution biophysical studies. H.L. performed cell assays (supported by J.Z., T.L., and Y.W.). H.L. performed cryo-EM grids preparation, sample screening, data collection, and processing. D.E.K. and H.L. analyzed the structures. D.E.K. and H.L. prepared the manuscript with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors have filed a patent application (Yale University, D.E.K., H.L. and C.R.A., PCT/US25/56600, Pending) related to the antibody sequences of RX5 and CT4 described in this manuscript. The patent application covers anti-ROS1 extracellular domain targeting antibodies (and derivatives) identified by phage display, including epitope-specific binders described in the manuscript, and their proposed use for modulating ROS1 signaling for potential therapeutic/diagnostic applications; the manuscript reports their discovery and use in structural and mechanistic studies of ROS1 activation. The other authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Zhe Zhang, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, H., Zhang, J., Li, T. et al. Clustering and a conformational switch drive activation of the mammalian receptor tyrosine kinase ROS1. Nat Commun (2026). https://doi.org/10.1038/s41467-026-69630-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-026-69630-7