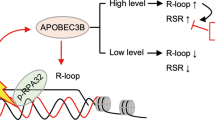
Abstract
Oestrogen receptor (ER) activation leads to the formation of DNA double strand breaks (DSB), promoting genomic instability and tumour heterogeneity. The single-stranded DNA cytosine deaminase APOBEC3B (A3B) serves as a co-activator of ER and is implicated in inducing DSBs at transcriptional enhancers regulated by ER. Using whole-genome sequencing in an engineered cell model lacking base excision repair (BER) function, we demonstrate that A3B preferentially targets transcriptionally active regulatory regions in an R-loop-dependent manner. Strand-specific DNA:RNA immunoprecipitation sequencing (ssDRIP-seq) and ssDNA-associated protein immunoprecipitation sequencing (SPI-seq) confirm that A3B binds to and deaminates ssDNA within R-loops, a process facilitated by ER transactivation. Furthermore, BER-mediated processing of A3B-induced uracil bases contributes to the formation of R-loop-associated DSBs, which are essential for ER-regulated gene activation. These findings establish a role for A3B in R-loop homeostasis and transcriptional regulation, with implications for understanding ER-driven genomic instability and potential therapeutic targeting of A3B.
Similar content being viewed by others
Data availability
The WGS, ChIP-seq, SPI-seq, ssDRIP-seq, DSBCapture-seq, and RNA-seq data generated in this study have been deposited in NCBI’s Gene Expression Omnibus (GEO) database under the GEO series accession code GSE193234. The processed RNA-seq data generated in this study are provided in Supplementary Data 1 Source data are provided in this paper.
References
Orzolek, I., Sobieraj, J. & Domagala-Kulawik, J. Estrogens, cancer and immunity. Cancers 14, https://doi.org/10.3390/cancers14092265 (2022).
Yang, J. A., Stires, H., Belden, W. J. & Roepke, T. A. The arcuate estrogen-regulated transcriptome: estrogen response element-dependent and -independent signaling of ERalpha in female mice. Endocrinology 158, 612–626 (2017).
Hewitt, S. C. & Korach, K. S. Estrogen receptors: new directions in the new millennium. Endocr. Rev. 39, 664–675 (2018).
Rajan, A. et al. Deregulated estrogen receptor signaling and DNA damage response in breast tumorigenesis. Biochim. Biophys. Acta Rev. Cancer 1875, 188482 (2021).
Periyasamy, M. et al. APOBEC3B-Mediated cytidine deamination is required for estrogen receptor action in breast cancer. Cell Rep. 13, 108–121 (2015).
Stork, C. T. et al. Co-transcriptional R-loops are the main cause of estrogen-induced DNA damage. Elife 5, https://doi.org/10.7554/eLife.17548 (2016).
Li, J. J. et al. Estrogen mediates Aurora-A overexpression, centrosome amplification, chromosomal instability, and breast cancer in female ACI rats. Proc. Natl. Acad. Sci. USA 101, 18123–18128 (2004).
Nik-Zainal, S. et al. Mutational processes molding the genomes of 21 breast cancers. Cell 149, 979–993 (2012).
Nik-Zainal, S. et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 534, 47–54 (2016).
Compe, E. & Egly, J. M. TFIIH: when transcription met DNA repair. Nat. Rev. Mol. Cell Biol. 13, 343–354 (2012).
Kamileri, I., Karakasilioti, I. & Garinis, G. A. Nucleotide excision repair: new tricks with old bricks. Trends Genet. 28, 566–573 (2012).
Fong, Y. W., Cattoglio, C. & Tjian, R. The intertwined roles of transcription and repair proteins. Mol. Cell 52, 291–302 (2013).
Schuermann, D., Weber, A. R. & Schar, P. Active DNA demethylation by DNA repair: Facts and uncertainties. DNA Repair 44, 92–102 (2016).
Perillo, B., Castoria, G. & Migliaccio, A. Exploiting the mechanism of estrogen-induced transcription to fight breast cancer. Exp. Mol. Med. 53, 1205–1206 (2021).
Puc, J. et al. Ligand-dependent enhancer activation regulated by topoisomerase-I activity. Cell 160, 367–380 (2015).
Ju, B. G. et al. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science 312, 1798–1802 (2006).
Green, A. M. & Weitzman, M. D. The spectrum of APOBEC3 activity: From anti-viral agents to anti-cancer opportunities. DNA Repair 83, 102700 (2019).
Harris, R. S. & Dudley, J. P. APOBECs and virus restriction. Virology 479-480, 131–145 (2015).
Henderson, S. & Fenton, T. APOBEC3 genes: retroviral restriction factors to cancer drivers. Trends Mol. Med. 21, 274–284 (2015).
Burns, M. B. et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature 494, 366–370 (2013).
Salamango, D. J. et al. APOBEC3B Nuclear localization requires two distinct N-terminal domain surfaces. J. Mol. Biol. 430, 2695–2708 (2018).
Mas-Ponte, D. & Supek, F. DNA mismatch repair promotes APOBEC3-mediated diffuse hypermutation in human cancers. Nat. Genet. 52, 958–968 (2020).
Taylor, B. J. et al. DNA deaminases induce break-associated mutation showers with implication of APOBEC3B and 3A in breast cancer kataegis. Elife 2, e00534 (2013).
Law, E. K. et al. The DNA cytosine deaminase APOBEC3B promotes tamoxifen resistance in ER-positive breast cancer. Sci. Adv. 2, e1601737 (2016).
Garcia-Muse, T. & Aguilera, A. R Loops: from physiological to pathological roles. Cell 179, 604–618 (2019).
Gu, S., Bodai, Z., Cowan, Q. T. & Komor, A. C. Base editors: expanding the types of DNA damage products harnessed for genome editing. Gene Genome Ed. 1, https://doi.org/10.1016/j.ggedit.2021.100005 (2021).
McCann, J. L. et al. APOBEC3B regulates R-loops and promotes transcription-associated mutagenesis in cancer. Nat. Genet. 55, 1721–1734 (2023).
Alexandrov, L. B. et al. The repertoire of mutational signatures in human cancer. Nature 578, 94–101 (2020).
Krokan, H. E. et al. Error-free versus mutagenic processing of genomic uracil-relevance to cancer. DNA Repair 19, 38–47 (2014).
Petljak, M. et al. Mechanisms of APOBEC3 mutagenesis in human cancer cells. Nature 607, 799–807 (2022).
Akre, M. K. et al. Mutation processes in 293-based clones overexpressing the DNA cytosine deaminase APOBEC3B. PLoS ONE 11, e0155391 (2016).
Hoopes, J. I. et al. APOBEC3A and APOBEC3B Preferentially deaminate the lagging strand template during DNA replication. Cell Rep. 14, 1273–1282 (2016).
Nikkila, J. et al. Elevated APOBEC3B expression drives a kataegic-like mutation signature and replication stress-related therapeutic vulnerabilities in p53-defective cells. Br. J. Cancer 117, 113–123 (2017).
Sui, Y. et al. Analysis of APOBEC-induced mutations in yeast strains with low levels of replicative DNA polymerases. Proc. Natl. Acad. Sci. USA 117, 9440–9450 (2020).
Serebrenik, A. A. et al. The deaminase APOBEC3B triggers the death of cells lacking uracil DNA glycosylase. Proc. Natl. Acad. Sci. USA 116, 22158–22163 (2019).
McLaren, W. et al. The ensembl variant effect predictor. Genome Biol. 17, 122 (2016).
Ernst, J. & Kellis, M. Chromatin-state discovery and genome annotation with ChromHMM. Nat. Protoc. 12, 2478–2492 (2017).
Lee, C. Y. et al. R-loop induced G-quadruplex in non-template promotes transcription by successive R-loop formation. Nat. Commun. 11, 3392 (2020).
Xu, W. et al. The R-loop is a common chromatin feature of the Arabidopsis genome. Nat. Plants 3, 704–714 (2017).
Cristini, A., Groh, M., Kristiansen, M. S. & Gromak, N. RNA/DNA Hybrid interactome identifies DXH9 as a molecular player in transcriptional termination and R-loop-associated DNA damage. Cell Rep. 23, 1891–1905 (2018).
Zhou, Z. X. et al. Mapping genomic hotspots of DNA damage by a single-strand-DNA-compatible and strand-specific ChIP-seq method. Genome Res. 23, 705–715 (2013).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Lensing, S. V. et al. DSBCapture: in situ capture and sequencing of DNA breaks. Nat. Methods 13, 855–857 (2016).
Differential Binding Analysis of ChIP-Seq peak data (https://github.com/aeron15/DiffBind (2015).
Williamson, L. M. & Lees-Miller, S. P. Estrogen receptor alpha-mediated transcription induces cell cycle-dependent DNA double-strand breaks. Carcinogenesis 32, 279–285 (2011).
Aguilera, A. & Garcia-Muse, T. R loops: from transcription byproducts to threats to genome stability. Mol. Cell 46, 115–124 (2012).
Sollier, J. et al. Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability. Mol. Cell 56, 777–785 (2014).
McLean, C. Y. et al. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28, 495–501 (2010).
Liberzon, A. et al. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015).
Cancer Genome Atlas Research, N. et al. The cancer genome atlas pan-cancer analysis project. Nat. Genet. 45, 1113–1120 (2013).
Hutter, C. & Zenklusen, J. C. The cancer genome atlas: creating lasting value beyond its data. Cell 173, 283–285 (2018).
Sollier, J. & Cimprich, K. A. Breaking bad: R-loops and genome integrity. Trends Cell Biol. 25, 514–522 (2015).
Cristini, A. et al. Dual processing of R-Loops and topoisomerase I induces transcription-dependent DNA double-strand breaks. Cell Rep. 28, 3167–3181 (2019).
Chatzinikolaou, G. et al. XPF interacts with TOP2B for R-loop processing and DNA looping on actively transcribed genes. Sci. Adv. 9, eadi2095 (2023).
Sartorelli, V. & Lauberth, S. M. Enhancer RNAs are an important regulatory layer of the epigenome. Nat. Struct. Mol. Biol. 27, 521–528 (2020).
Liu, J. et al. Endogenous DNA damage at sites of terminated transcripts. Nature 640, 240–248 (2025).
Long, J. et al. A common deletion in the APOBEC3 genes and breast cancer risk. J. Natl. Cancer Inst. 105, 573–579 (2013).
Wen, W. X. et al. Germline APOBEC3B deletion is associated with breast cancer risk in an Asian multi-ethnic cohort and with immune cell presentation. Breast Cancer Res. 18, 56 (2016).
Caval, V., Suspene, R., Shapira, M., Vartanian, J. P. & Wain-Hobson, S. A prevalent cancer susceptibility APOBEC3A hybrid allele bearing APOBEC3B 3’UTR enhances chromosomal DNA damage. Nat. Commun. 5, 5129 (2014).
Udquim, K. I., Zettelmeyer, C., Banday, A. R., Lin, S. H. & Prokunina-Olsson, L. APOBEC3B expression in breast cancer cell lines and tumors depends on the estrogen receptor status. Carcinogenesis 41, 1030–1037 (2020).
Zhang, C. et al. Characterisation of APOBEC3B-Mediated RNA editing in breast cancer cells reveals regulatory roles of NEAT1 and MALAT1 lncRNAs. Oncogene 43, 3366–3377 (2024).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
DePristo, M. A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011).
Karczewski, K. J. et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443 (2020).
Piskol, R., Ramaswami, G. & Li, J. B. Reliable identification of genomic variants from RNA-seq data. Am. J. Hum. Genet. 93, 641–651 (2013).
Wang, M. & Kong, L. pblat: a multithread blat algorithm speeding up aligning sequences to genomes. BMC Bioinformatics 20, 28 (2019).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Turner, S. D. qqman: an R package for visualizing GWAS results using Q-Q and manhattan plots. J. Open Source Softw. 3, https://doi.org/10.21105/joss.00731 (2018).
Kent, W. J. et al. The human genome browser at UCSC. Genome Res. 12, 996–1006 (2002).
Blokzijl, F., Janssen, R., van Boxtel, R. & Cuppen, E. MutationalPatterns: comprehensive genome-wide analysis of mutational processes. Genome Med. 10, 33 (2018).
Gaujoux, R. & Seoighe, C. A flexible R package for nonnegative matrix factorization. BMC Bioinform. 11, 367 (2010).
Huang, H. et al. Defining super-enhancer landscape in triple-negative breast cancer by multiomic profiling. Nat. Commun. 12, 2242 (2021).
Halasz, L. et al. RNA-DNA hybrid (R-loop) immunoprecipitation mapping: an analytical workflow to evaluate inherent biases. Genome Res. 27, 1063–1073 (2017).
Jenjaroenpun, P., Wongsurawat, T., Sutheeworapong, S. & Kuznetsov, V. A. R-loopDB: a database for R-loop forming sequences (RLFS) and R-loops. Nucleic Acids Res. 45, D119–D127 (2017).
Bedrat, A., Lacroix, L. & Mergny, J. L. Re-evaluation of G-quadruplex propensity with G4Hunter. Nucleic Acids Res. 44, 1746–1759 (2016).
Stempor, P. & Ahringer, J. SeqPlots - Interactive software for exploratory data analyses, pattern discovery and visualization in genomics. Wellcome Open Res. 1, 14 (2016).
Robinson, J. T. et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Feng, J., Liu, T., Qin, B., Zhang, Y. & Liu, X. S. Identifying ChIP-seq enrichment using MACS. Nat. Protoc. 7, 1728–1740 (2012).
Jalili, V., Matteucci, M., Masseroli, M. & Morelli, M. J. Using combined evidence from replicates to evaluate ChIP-seq peaks. Bioinformatics 31, 2761–2769 (2015).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Ross-Innes, C. S. et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 481, 389–393 (2012).
McLeay, R. C. & Bailey, T. L. Motif Enrichment Analysis: a unified framework and an evaluation on ChIP data. BMC Bioinform. 11, 165 (2010).
Bailey, T. L., Johnson, J., Grant, C. E. & Noble, W. S. The MEME suite. Nucleic Acids Res. 43, W39–W49 (2015).
Gu, Z., Eils, R. & Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849 (2016).
Harrow, J. et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 22, 1760–1774 (2012).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Liao, Y., Smyth, G. K. & Shi, W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 47, e47 (2019).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550 (2005).
Zhang, Y. et al. Discovery of APOBEC cytidine deaminases inhibitors using a BspH1 restriction enzyme-based biosensor. ChemistrySelect 7, e202201456 (2022).
Acknowledgements
This work is financially supported by Cancer Research UK (C309/A11566 and C2739/A22897) and ICR (London, United Kingdom). C.Z. was sponsored by the Science and Technology Commission of Shanghai Municipality (23S11901100). P.W. and C.Z. acknowledge additional grant support from the Wellcome Trust (212969/Z/18/Z and 094885/Z/10/Z), Cancer Research UK (C35696/A23187). P.W. is a CRUK Life Fellow and acknowledges support from CRUK Strategic Award C35696/A23187 and Infrastructure Award C309/A27413, and funding for the CRUK Children’s Brain Tumour Centre of Excellence (C9685/A26398/RG93685); Wellcome Trust (Biomedical Resource and Technology Development Grant 212969/Z/18/Z to support the Chemical Probes Portal), Chordoma Foundation, Marcus Foundation, Mark Foundation, Bone Cancer Research Trust, CRIS Cancer, and The Institute of Cancer Research. Z.B. was supported by the Science, Technology, and Innovation Commission of Shenzhen Municipality (ZDSYS20200811142605017). We thank Prof. Ping Yuan from Sun Yat-sen University, Dr. Mike Walton and Dr. Alexandra Vasile from ICR for helpful discussions.
Author information
Authors and Affiliations
Contributions
C.Z., O.W.R., P.A.C. and P.W. conceived and designed this study. C.Z., Y.L., Q.Z. and M.T. conducted the experiments and analysis unless otherwise noted. B.C., Z.B., K.M. and B.A.-L. conducted analysis of sequencing data and provided guidance on data analysis using sequencing data. C.Z., A.H., M.P.L., O.W.R., P.W. and P.A.C. prepared the manuscript.
Corresponding authors
Ethics declarations
Competing interests
C.Z., A.H., M.P.L., O.W.R., P.A.C., K.M., M.T., and P.W. are current or past employees of The Institute of Cancer Research, which has a commercial interest in a range of drug targets and operates a Rewards to Discoverers scheme, including A3B inhibitors, through which employees may receive financial benefits following the commercial licensing of a project. P.W. is an independent director at Storm Therapeutics, is a consultant/advisory board member at Astex Pharmaceuticals, CV6 Therapeutics, Black Diamond Therapeutics, Vividion Therapeutics and Nextechinvest; reports receiving a commercial research grant from Sixth Element Capital, Astex Pharmaceuticals, and Merck; has ownership interest in Storm Therapeutics, Chroma Therapeutics, and Nextechinvest; and has an unpaid consultant/advisory board relationship with the Chemical Probes Portal. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Tim Fenton and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhang, C., Lu, Yj., Chen, B. et al. R-loop editing by DNA cytosine deaminase APOBEC3B modulates the activity of oestrogen receptor enhancers. Nat Commun (2026). https://doi.org/10.1038/s41467-026-69679-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-026-69679-4