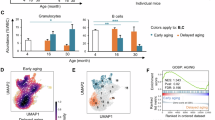
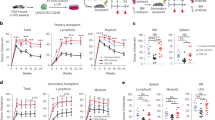
Abstract
Caloric restriction (CR) provides anti-aging benefits but has also been reported to be associated with reduced immune function, and how hematopoietic stem cells (HSCs) potentially contribute to this decline remains unclear. Using lifelong and short-term CR in male mice, we found reducing the energy supply decreases total white blood cell production and shifts hematopoiesis towards myeloid and thrombo-erythroid lineages, prioritizing cells essential for survival (red blood cells, platelets, innate immune cells) over adaptive immunity. HSCs under CR enter cell cycle to support myeloid differentiation rather than self-renewal. Lifelong CR inhibits age-associated transcriptome changes in HSCs, though age-associated profiles appear shortly after ad libitum feeding. Epigenetic profiling identified KDR as a key CR response regulator, and Kdr knockdown in aged HSCs recapitulated the youthful transcriptome of lifelong CR HSCs. Finally, we show PU.1 acts as an intracellular regulator of CR response, controlling HSC self-renewal and differentiation through increased target gene binding under CR conditions.
Similar content being viewed by others
Data availability
The raw data generated in this study have been deposited in the NCBI Gene Expression Omnibus database under accession code GSE284988. Source data are provided with this paper.
Code availability
All customized Python scripts used in the manuscript are available via GitHub repository URL (https://github.com/genomicspark/ESCA_Unit_Scripts) and has been archived in Zenodo for citation (https://doi.org/10.5281/zenodo.17857712)51. Source data are provided with this paper.
References
Fontana, L., Partridge, L. & Longo, V. D. Extending healthy life span—from yeast to humans. Science 328, 321–326 (2010).
Di Francesco, A., Di Germanio, C., Bernier, M. & de Cabo, R. A time to fast. Science 362, 770–775 (2018).
Mitchell, S. J. et al. Effects of sex, strain, and energy intake on hallmarks of aging in mice. Cell Metab. 23, 1093–1112 (2016).
Green, C. L. et al. The effects of graded levels of calorie restriction: IX. Global metabolomic screen reveals modulation of carnitines, sphingolipids and bile acids in the liver of C57BL/6 mice. Aging Cell 16, 529–540 (2017).
Mitchell, S. J. et al. Daily fasting improves health and survival in male mice independent of diet composition and calories. Cell Metab. 29, 221–228.e223 (2019).
Grossmann, A., Maggio-Price, L., Jinneman, J. C., Wolf, N. S. & Rabinovitch, P. S. The effect of long-term caloric restriction on function of T-cell subsets in old mice. Cell Immunol. 131, 191–204 (1990).
Kristan, D. M. Chronic calorie restriction increases susceptibility of laboratory mice (Mus musculus) to a primary intestinal parasite infection. Aging Cell 6, 817–825 (2007).
Gardner, E. M. Caloric restriction decreases survival of aged mice in response to primary influenza infection. J. Gerontol. A 60, 688–694 (2005).
Tang, D. et al. Dietary restriction improves repopulation but impairs lymphoid differentiation capacity of hematopoietic stem cells in early aging. J. Exp. Med. 213, 535–553 (2016).
Yilmaz, Ö.H. et al. mTORC1 in the Paneth cell niche couples intestinal stem-cell function to calorie intake. Nature 486, 490–495 (2012).
Yousefi, M. et al. Calorie restriction governs intestinal epithelial regeneration through cell-autonomous regulation of mTORC1 in reserve stem cells. Stem Cell Rep. 10, 703–711 (2018).
Cerletti, M. et al. Short-term calorie restriction enhances skeletal muscle stem cell function. Cell Stem Cell 10, 515–519 (2012).
Park, J.-H. et al. Calorie restriction alleviates the age-related decrease in neural progenitor cell division in the aging brain. Eur. J. Neurosci. 37, 1987–1993 (2013).
Ho, T. T. et al. Aged hematopoietic stem cells are refractory to bloodborne systemic rejuvenation interventions. J. Exp. Med. 218, e20210223 (2021).
Lazare, S. et al. Lifelong dietary intervention does not affect hematopoietic stem cell function. Exp. Hematol. 53, 26–30 (2017).
Beerman, I. et al. Functionally distinct hematopoietic stem cells modulate hematopoietic lineage potential during aging by a mechanism of clonal expansion. Proc. Natl. Acad. Sci. USA 107, 5465–5470 (2010).
Flohr Svendsen, A. et al. A comprehensive transcriptome signature of murine hematopoietic stem cell aging. Blood 138, 439–451 (2021).
Sun, N. et al. Clusterin drives myeloid bias in aged hematopoietic stem cells by regulating mitochondrial function. Nat. Aging 5, 1510–1527 (2025).
Tanaka, M. et al. Targeted disruption of oncostatin M receptor results in altered hematopoiesis. Blood 102, 3154–3162 (2003).
Sun, D. et al. Epigenomic profiling of young and aged hSCs reveals concerted changes during aging that reinforce self-renewal. Cell Stem Cell 14, 673–688 (2014).
Staber, P. H. et al. Sustained PU.1 levels balance cell-cycle regulators to prevent exhaustion of adult hematopoietic stem cells. Mol. Cell 49, 934–946 (2013).
Hosokawa, H. et al. Transcription factor PU.1 represses and activates gene expression in early T cells by redirecting partner transcription factor binding. Immunity 48, 1119–1134 (2018).
Brandhorst, S. et al. A periodic diet that mimics fasting promotes multi-system regeneration, enhanced cognitive performance, and healthspan. Cell Metab. 22, 86–99 (2015).
Warr, M. R. et al. FOXO3A directs a protective autophagy program in haematopoietic stem cells. Nature 494, 323–327 (2013).
Chinnapaka, S., Malekzadeh, H., Tirmizi, Z. & Ejaz, A. Caloric restriction mitigates age-associated senescence characteristics in subcutaneous adipose tissue-derived stem cells. Aging (Albany NY) 16, 7535–7552 (2024).
Yilmaz, O. H. et al. mTORC1 in the Paneth cell niche couples intestinal stem-cell function to calorie intake. Nature 486, 490–495 (2012).
Igarashi, M. & Guarente, L. mTORC1 and SIRT1 cooperate to foster expansion of gut adult stem cells during calorie restriction. Cell 166, 436–450 (2016).
Abreu, P., Serna, J. D. C., Munhoz, A. C. & Kowaltowski, A. J. Calorie restriction changes muscle satellite cell proliferation in a manner independent of metabolic modulation. Mech. Ageing Dev. 192, 111362 (2020).
Merchant, S. et al. Different effects of fatty acid oxidation on hematopoietic stem cells based on age and diet. Cell Stem Cell 32, 263–275.e265 (2025).
Mentch, S. J. et al. Histone methylation dynamics and gene regulation occur through the sensing of one-carbon metabolism. Cell Metab. 22, 861–873 (2015).
Gerber, H.-P. et al. VEGF regulates haematopoietic stem cell survival by an internal autocrine loop mechanism. Nature 417, 954–958 (2002).
Olsson, A.-K., Dimberg, A., Kreuger, J. & Claesson-Welsh, L. VEGF receptor signalling? in control of vascular function. Nat. Rev. Mol. Cell Biol. 7, 359–371 (2006).
Grunewald, M. et al. Counteracting age-related VEGF signaling insufficiency promotes healthy aging and extends life span. Science 373, eabc8479 (2021).
Huang, Y. et al. Distinct roles of VEGFR-1 and VEGFR-2 in the aberrant hematopoiesis associated with elevated levels of VEGF. Blood 110, 624–631 (2007).
Scott, E. W., Simon, M. C., Anastasi, J. & Singh, H. Requirement of transcription Factor PU.1 in the development of multiple hematopoietic lineages. Science 265, 1573–1577 (1994).
McKercher, S. R. et al. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J. 15, 5647–5658 (1996).
Chavez, J. S. et al. PU.1 enforces quiescence and limits hematopoietic stem cell expansion during inflammatory stress. J. Exp. Med. 218, e20201169 (2021).
Wilkinson, A. C. et al. Long-term ex vivo haematopoietic-stem-cell expansion allows nonconditioned transplantation. Nature 571, 117–121 (2019).
Corces, M. R. et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods 14, 959–962 (2017).
Buenrostro, J. D., Wu, B., Chang, H. Y. & Greenleaf, W. J. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 109, 21.29.1–21.29.9 (2015).
Zheng, X. et al. Low-cell-number epigenome profiling aids the study of lens aging and hematopoiesis. Cell Rep. 13, 1505–1518 (2015).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10–12 (2011).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2012).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2013).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Yang, W. R., Ardeljan, D., Pacyna, C. N., Payer, L. M. & Burns, K. H. SQuIRE reveals locus-specific regulation of interspersed repeat expression. Nucleic Acids Res 47, e27 (2019).
Mardis, E. R. et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N. Engl. J. Med. 361, 1058–1066 (2009).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Zong, L. et al. Epigenetic profiling of hematopoietic stem cells from male mice identifies KDR and PU.1 as regulators of aging transcriptome and caloric restriction response. Zenodo https://doi.org/10.5281/zenodo.17857712 (2025).
Robinson, J. T. et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011).
Acknowledgements
Many thanks to Drs. Rafael de Cabo, Myriam Gorospe and all members of TGB for invaluable support. We thank Dr. Fei Ma for assistance in preparing illustrations used in this manuscript, including generating and assembling figure elements created with BioRender. Kind support was shared by the Genomics Core and we’d like to specially acknowledge Jinshui Fan, William Wood, and Supriyo De for their assistance with data handling and sequencing advice. Thanks to Christopher Dunn at the NIA Flow Core for always being helpful and willing to share time and expertise. We would like to thank all the members of the NIA Comparative Medicine Section for their consistent efforts and high standards of animal care. Data analysis of this work utilized the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov). This research was supported entirely by the Intramural Research Program of the NIH, National Institute on Aging.
Funding
Open access funding provided by the National Institutes of Health.
Author information
Authors and Affiliations
Contributions
L.Z., F.T., W.K., M.T., and K.L. performed the experiments; B.P. performed bioinformatical analyses; L.Z. and I.B. designed the research, interpreted the results, and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Linda Resar, Gary Churchill and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zong, L., Park, B., Tekin-Turhan, F. et al. Epigenetic profiling of hematopoietic stem cells from male mice identifies KDR and PU.1 as regulators of aging transcriptome and caloric restriction response. Nat Commun (2026). https://doi.org/10.1038/s41467-026-69718-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-026-69718-0