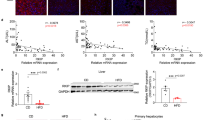
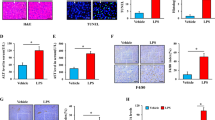
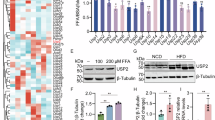
Abstract
Obesity and related metabolic disorders, including metabolic dysfunction-associated steatohepatitis (MASLD), have reached epidemic proportions worldwide. We unveil a previously unknown moonlighting role for arginase 1 (Arg1) in facilitating hepatic lipogenesis. Male mice lacking hepatic Arg1 exhibit diminished lipid accumulation in both liver and adipocytes, an effect mirrored in genetically- or diet-induced obesity models following Arg1 inhibitor treatment. Mechanistically, Arg1 competes with RSK2 and Elk1 for binding to the substrate-binding pocket of extracellular signal-regulated kinase 2 (ERK2) via its S-shaped motif, thereby enhancing ERK2 ubiquitination and degradation and upregulating the AKT/mTOR/PPARγ and Elk1/c-Fos/PPARγ cascades, ultimately augmenting lipogenesis. Peptides designed to mimic the ERK2 substrate-binding pocket disrupt the Arg1-ERK2 interaction and improve metabolic profiles in obesity and MASLD models. Our findings implicate Arg1 regulates hepatic lipid metabolism via its physical interaction with ERK2, highlighting the Arg1-ERK2 interaction as a promising therapeutic target for obesity and related metabolic disorders in male mice.
Similar content being viewed by others
Data availability
mRNA-seq data generated in this study have been deposited at the National Center for Biotechnology Information (NCBI) GenBank (PRJNA1186374 [https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1186374] and PRJNA1186081. Microscopy data reported in this paper will be shared by the lead contact upon request. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD059821. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request. Source data are provided in this paper.
References
Younossi, Z. M. Non-alcoholic fatty liver disease–a global public health perspective. J. Hepatol. 70, 531–544 (2019).
Kim, M. S. et al. Association between adiposity and cardiovascular outcomes: an umbrella review and meta-analysis of observational and Mendelian randomization studies. Eur. Heart J. 42, 3388–3403 (2021).
Samms, R. J. & Kusminski, C. M. A Mechanistic Rationale for Incretin-Based Therapeutics in the Management of Obesity. Annu. Rev. Physiol. 87, 279–299 (2025).
Xu, J. et al. Hepatic CDP-diacylglycerol synthase 2 deficiency causes mitochondrial dysfunction and promotes rapid progression of NASH and fibrosis. Sci. Bull. 67, 299–314 (2022).
Rouillard, N. A. et al. Bariatric surgery reduces long-term mortality in patients with metabolic dysfunction-associated steatotic liver disease and cirrhosis. Clin. Mol. Hepatol. 31, 227–239 (2025).
Anstee, Q. M., Reeves, H. L., Kotsiliti, E., Govaere, O. & Heikenwalder, M. From NASH to HCC: current concepts and future challenges. Nat. Rev. Gastroenterol. Hepatol. 16, 411–428 (2019).
Harrison, S. A. et al. Resmetirom (MGL-3196) for the treatment of non-alcoholic steatohepatitis: a multicentre, randomised, double-blind, placebo-controlled, phase 2 trial. Lancet 394, 2012–2024 (2019).
Mantovani, A., Byrne, C. D. & Targher, G. Efficacy of peroxisome proliferator-activated receptor agonists, glucagon-like peptide-1 receptor agonists, or sodium-glucose cotransporter-2 inhibitors for treatment of non-alcoholic fatty liver disease: a systematic review. Lancet Gastroenterol. Hepatol. 7, 367–378 (2022).
Brunner, K. T., Henneberg, C. J., Wilechansky, R. M., & Long, M. T. Nonalcoholic Fatty Liver Disease and Obesity Treatment. Curr. Obes. Rep. 8, 220–228 (2019).
Kim, O. Y. et al. Arginase I and the very low-density lipoprotein receptor are associated with phenotypic biomarkers for obesity. Nutrition 28, 635–639 (2012).
Therrell, B. L., Currier, R., Lapidus, D., Grimm, M. & Cederbaum, S. D. Newborn screening for hyperargininemia due to arginase 1 deficiency. Mol. Genet. Metab. 121, 308–313 (2017).
Jung, C., Figulla, H. R., Lichtenauer, M., Franz, M. & Pernow, J. Increased levels of circulating arginase I in overweight compared to normal weight adolescents. Pediatr. Diabetes 15, 51–56 (2014).
Bhatta, A. et al. Obesity-induced vascular dysfunction and arterial stiffening requires endothelial cell arginase 1. Cardiovasc. Res. 113, 1664–1676 (2017).
Ito, T. et al. Early obesity leads to increases in hepatic arginase I and related systemic changes in nitric oxide and L-arginine metabolism in mice. J. Physiol. Biochem. 74, 9–16 (2018).
Holbert, C. E., Cullen, M. T., Casero, R. A. Jr & Stewart, T. M. Polyamines in cancer: integrating organismal metabolism and antitumour immunity. Nat. Rev. Cancer 22, 467–480 (2022).
Xuan, M. et al. Polyamines: their significance for maintaining health and contributing to diseases. Cell Commun. Signal. 21, 348 (2023).
Monelli, E. et al. Angiocrine polyamine production regulates adiposity. Nat. Metab. 4, 327–343 (2022).
Sadasivan, S. K. et al. Exogenous administration of spermine improves glucose utilization and decreases bodyweight in mice. Eur. J. Pharmacol. 729, 94–99 (2014).
Hu, X., Li, Y., Cao, Y., Shi, F. & Shang, L. The role of nitric oxide synthase/ nitric oxide in infection-related cancers: Beyond antimicrobial activity. Biochim. Biophys. Acta Rev. Cancer 1879, 189156 (2024).
Clemente, S. et al. Arginase as a Potential Biomarker of Disease Progression: A Molecular Imaging Perspective. Int. J. Mol. Sci. 21, 5291 (2020).
Mabalirajan, U. et al. Beneficial effects of high dose of L-arginine on airway hyperresponsiveness and airway inflammation in a murine model of asthma. J. Allergy Clin. Immunol. 125, 626–635 (2010).
Kim, D. Y., Park, J. Y. & Gee, H. Y. Lactobacillus plantarum ameliorates NASH-related inflammation by upregulating L-arginine production. Exp. Mol. Med. 55, 2332–2345 (2023).
Savova, M. S., Mihaylova, L. V., Tews, D., Wabitsch, M. & Georgiev, M. I. Targeting PI3K/AKT signaling pathway in obesity. Biomed. Pharmacother. 159, 114244 (2023).
Kujiraoka, T. et al. Metabolic remodeling with hepatosteatosis induced vascular oxidative stress in hepatic ERK2 deficiency mice with high fat diets. Int. J. Mol. Sci. 23, 8521 (2022).
Wen, X. et al. Signaling pathways in obesity: mechanisms and therapeutic interventions. Signal Transduct. Target. Ther. 7, 298 (2022).
Allard, C., Miralpeix, C., López-Gambero, A. J. & Cota, D. mTORC1 in energy expenditure: consequences for obesity. Nat. Rev. Endocrinol. 20, 239–251 (2024).
Caron, A., Richard, D. & Laplante, M. The roles of mTOR complexes in lipid metabolism. Annu. Rev. Nutr. 35, 321–348 (2015).
Kassouf, T. & Sumara, G. Impact of conventional and Atypical MAPKs on the development of metabolic diseases. Biomolecules 10, 1256 (2020).
Bunch, H. et al. ERK2-topoisomerase II regulatory axis is important for gene activation in immediate early genes. Nat. Commun. 14, 8341 (2023).
Roskoski, R. Jr ERK1/2 MAP kinases: structure, function, and regulation. Pharmacol. Res. 66, 105–143 (2012).
Lavoie, H., Gagnon, J. & Therrien, M. ERK signalling: a master regulator of cell behaviour, life and fate. Nat. Rev. Mol. Cell Biol. 21, 607–632 (2020).
Artunc, F. et al. The impact of insulin resistance on the kidney and vasculature. Nat. Rev. Nephrol. 12, 721–737 (2016).
Cai, W. et al. STAT6/Arg1 promotes microglia/macrophage efferocytosis and inflammation resolution in stroke mice. JCI Insight 4, e131355 (2019).
Liu, L. et al. Sirt1 ameliorates monosodium urate crystal-induced inflammation by altering macrophage polarization via the PI3K/Akt/STAT6 pathway. Rheumatology 58, 1674–1683 (2019).
Wang, Y., Nakajima, T., Gonzalez, F. J. & Tanaka, N. PPARs as metabolic regulators in the liver: lessons from liver-specific PPAR-null mice. Int. J. Mol. Sci. 21, 2061 (2020).
Zhao, Y., Tan, H., Zhang, X. & Zhu, J. Roles of peroxisome proliferator-activated receptors in hepatocellular carcinoma. J. Cell. Mol. Med. 28, e18042 (2024).
Gallardo-Soler, A. et al. Arginase Iinduction by modified lipoproteins in macrophages: a peroxisome proliferator-activated receptor-gamma/delta-mediated effect that links lipid metabolism and immunity. Mol. Endocrinol. 22, 1394–1402 (2008).
Qiu, Y. et al. The global perspective on peroxisome proliferator-activated receptor γ (PPARγ) in ectopic fat deposition: a review. Int. J. Biol. Macromol. 253, 127042 (2023).
Chen, H. et al. PPAR-γ Signaling in nonalcoholic fatty liver disease: pathogenesis and therapeutic targets. Pharmacol. Ther. 245, 108391 (2023).
Kim, N. N. et al. Probing erectile function: S-(2-boronoethyl)-L-cysteine binds to arginase as a transition state analogue and enhances smooth muscle relaxation in human penile corpus cavernosum. Biochemistry 40, 2678–2688 (2001).
Khaidar, A., Marx, M., Lubec, B. & Lubec, G. L-Arginine reduces heart collagen accumulation in the diabetic db/db mouse. Circulation 90, 479–483 (1994).
Huang, X., Liu, G., Guo, J. & Su, Z. The PI3K/AKT pathway in obesity and type 2 diabetes. Int. J. Biol. Sci. 14, 1483–1496 (2018).
Zhang, H. et al. Molecular cloning and characterization of a putative mitogen-activated protein kinase (Erk1/2) gene: involvement in mantle immunity of Pinctada fucata. Fish Shellfish Immunol 80, 63–70 (2018).
Qu, F. et al. A molluscan extracellular signal-regulated kinase is involved in host response to immune challenges in vivo and in vitro. Fish Shellfish Immunol. 62, 311–319 (2017).
Zhang, J., Zhou, B., Zheng, C. F. & Zhang, Z. Y. A bipartite mechanism for ERK2 recognition by its cognate regulators and substrates. J. Biol. Chem. 278, 29901–29912 (2003).
Tanoue, T. & Nishida, E. Docking interactions in the mitogen-activated protein kinase cascades. Pharmacol. Ther. 93, 193–202 (2002).
Bardwell, A. J., Abdollahi, M. & Bardwell, L. Docking sites on mitogen-activated protein kinase (MAPK) kinases, MAPK phosphatases and the Elk-1 transcription factor compete for MAPK binding and are crucial for enzymic activity. Biochem. J. 370, 1077–1085 (2003).
Adams, M. R., Jessup, W., Hailstones, D. & Celermajer, D. S. L-arginine reduces human monocyte adhesion to vascular endothelium and endothelial expression of cell adhesion molecules. Circulation 95, 662–668 (1997).
Harhouri, K. et al. MG132-induced progerin clearance is mediated by autophagy activation and splicing regulation. EMBO Mol. Med. 9, 1294–1313 (2017).
Sala-Gaston, J. et al. Regulation of MAPK signaling pathways by the large HERC ubiquitin ligases. Int. J. Mol. Sci. 24, 4906 (2023).
Wang, K. et al. UBR5 regulates proliferation and radiosensitivity in human laryngeal carcinoma via the p38/MAPK signaling pathway. Oncol. Rep. 44, 685–697 (2020).
Liu, Y. et al. TRIM21-mediated ubiquitination and phosphorylation of ERK1/2 promotes cell proliferation and drug resistance in pituitary adenomas. Neuro Oncol. 27, 727–742 (2025).
Lu, Z., Xu, S., Joazeiro, C., Cobb, M. H. & Hunter, T. The PHD domain of MEKK1 acts as an E3 ubiquitin ligase and mediates ubiquitination and degradation of ERK1/2. Mol. Cell 9, 945–956 (2002).
Kanyo, Z. F., Scolnick, L. R., Ash, D. E. & Christianson, D. W. Structure of a unique binuclear manganese cluster in arginase. Nature 383, 554–557 (1996).
Khangulov, S. V., Sossong, T. M. Jr, Ash, D. E. & Dismukes, G. C. L. -arginine binding to liver arginase requires proton transfer to gateway residue His141 and coordination of the guanidinium group to the dimanganese(II,II) center. Biochemistry 37, 8539–8550 (1998).
García, D., Uribe, E., Lobos, M., Orellana, M. S. & Carvajal, N. Studies on the functional significance of a C-terminal S-shaped motif in human arginase type I: essentiality for cooperative effects. Arch. Biochem. Biophys. 481, 16–20 (2009).
Gao, K. et al. A synthetic peptide as an allosteric inhibitor of human arginase I and II. Mol. Biol. Rep. 48, 1959–1966 (2021).
Wang, X. et al. RARγ-C-Fos-PPARγ2 signaling rather than ROS generation is critical for all-trans retinoic acid-inhibited adipocyte differentiation. Biochimie 106, 121–130 (2014).
Jiao, S. et al. A peptide mimicking VGLL4 function acts as a YAP antagonist therapy against gastric cancer. Cancer Cell 25, 166–180 (2014).
Guan, T., Li, J., Chen, C. & Liu, Y. Self-assembling peptide-based hydrogels for wound tissue repair. Adv. Sci. 9, e2104165 (2022).
Linker, S. M. et al. Lessons for oral bioavailability: how conformationally flexible cyclic peptides enter and cross lipid membranes. J. Med. Chem. 66, 2773–2788 (2023).
Kovalainen, M. et al. Novel delivery systems for improving the clinical use of peptides. Pharmacol. Rev. 67, 541–561 (2015).
Kalezic, A., Korac, A., Korac, B. & Jankovic, A. L-Arginine induces white adipose tissue browning—a new pharmaceutical alternative to cold. Pharmaceutics 14, 1368 (2022).
Wu, Z., Satterfield, M. C., Bazer, F. W. & Wu, G. Regulation of brown adipose tissue development and white fat reduction by L-arginine. Curr. Opin. Clin. Nutr. Metab. Care 15, 529–538 (2012).
Oliva, L., Alemany, M., Remesar, X. & Fernández-López, J. A. The food energy/protein ratio regulates the rat urea cycle but not total nitrogen losses. Nutrients 11, 316 (2019).
Krieg, S., Fernandes, S. I., Kolliopoulos, C., Liu, M. & Fendt, S. M. Metabolic signaling in cancer metastasis. Cancer Discov. 14, 934–952 (2024).
Xu, D. et al. The evolving landscape of noncanonical functions of metabolic enzymes in cancer and other pathologies. Cell Metab. 33, 33–50 (2021).
Frödin, M., Jensen, C. J., Merienne, K. & Gammeltoft, S. A phosphoserine-regulated docking site in the protein kinase RSK2 that recruits and activates PDK1. EMBO J. 19, 2924–2934 (2000).
Dimitri, C. A., Dowdle, W., MacKeigan, J. P., Blenis, J. & Murphy, L. O. Spatially separate docking sites on ERK2 regulate distinct signaling events in vivo. Curr. Biol. 15, 1319–1324 (2005).
Davis, R. J. Signal transduction by the JNK group of MAP kinases. Cell 103, 239–252 (2000).
Rodríguez, J. & Crespo, P. Working without kinase activity: phosphotransfer-independent functions of extracellular signal-regulated kinases. Sci. Signal. 4, re3 (2011).
Smith, J. A., Poteet-Smith, C. E., Malarkey, K. & Sturgill, T. W. Identification of an extracellular signal-regulated kinase (ERK) docking site in ribosomal S6 kinase, a sequence critical for activation by ERK in vivo. J. Biol. Chem. 274, 2893–2898 (1999).
Arvind, R. et al. A mutation in the common docking domain of ERK2 in a human cancer cell line, which was associated with its constitutive phosphorylation. Int. J. Oncol. 27, 1499–1504 (2005).
Gonzalez, F. J., Jiang, C. & Patterson, A. D. An intestinal microbiota–farnesoid X receptor axis modulates metabolic disease. Gastroenterology 151, 845–859 (2016).
Dixon, E. D. et al. Inhibition of ATGL alleviates MASH via impaired PPARα signalling that favours hydrophilic bile acid composition in mice. J. Hepatol. 82, 658–675 (2025).
Wu, F., Zhang, Y. T., Teng, F., Li, H. H. & Guo, S. B. S100a8/a9 Contributes to sepsis-induced cardiomyopathy by activating ERK1/2-Drp1-mediated mitochondrial fission and respiratory dysfunction. Int. Immunopharmacol. 115, 109716 (2023).
Krepler et al. Personalized preclinical trials in BRAF inhibitor-resistant patient-derived xenograft models identify second-line combination therapies. Clin. Cancer Res. 22, 1592–1602 (2016).
Du, J. et al. CircNFIB Inhibits tumor growth and metastasis through suppressing MEK1/ERK signaling in intrahepatic cholangiocarcinoma. Mol. Cancer 21, 18 (2022).
Truong, B. et al. Lipid nanoparticle-targeted mRNA therapy as a treatment for the inherited metabolic liver disorder arginase Deficiency. Proc. Natl. Acad. Sci. USA 116, 21150–21159 (2019).
Acknowledgements
This research was supported by the Young Scientists Fund of the National Natural Science Foundation of China (Grant No. 82200649), the National Natural Science Foundation of China (Grant No. 82472241). We would also like to acknowledge the help of HUABIO and WZ Biosciences Inc. in providing peptides as well as virus strains. We are grateful to Shanghai Oe Biotech Co., Ltd., for providing sequencing services.
Author information
Authors and Affiliations
Contributions
M.S. and X.C. conceived and designed the project. Y.C. performed mRNA-seq experiments and analysis. M.S., Z.Z., Y.S., Q.T., Q.X., T.M., Z.W., M.C., Y.Z., and R.Y. bred mice, performed mouse experiments, and analyzed the data. M.S., J.G., and J.Y. provided liver disease model setup, study design, and joint discussions on the results. Y.S. and M.S. interpreted the data and drafted and revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Shao, M., Cao, X., Chen, Y. et al. Arginase 1 promotes hepatic lipogenesis by regulating ERK2/PPARγ signaling in a non-canonical manner. Nat Commun (2026). https://doi.org/10.1038/s41467-026-69731-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-026-69731-3