Abstract
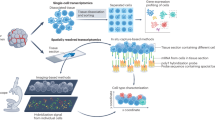
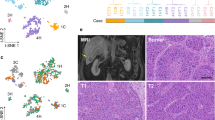
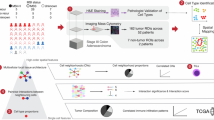
Spatial cellular context is crucial in shaping intratumor heterogeneity. However, understanding how each tumor establishes its unique spatial landscape and what factors drive the landscape for tumor fitness remains significantly challenging. Here, we analyze over 2 million cells from 50 tumor biospecimens using spatial single-cell imaging and single-cell RNA sequencing. We develop a deep learning-based strategy to spatially map tumor cell states and their surrounding environmental architecture, and find that different tumor cell states can be organized into distinct clusters, or “villages,” each supported by unique microenvironments. Notably, tumor cell villages exhibit village-specific molecular co-dependencies between tumor cells and their microenvironment and are associated with patient outcomes. Perturbation of molecular co-dependencies via random spatial shuffling of the microenvironment results in destabilization of the corresponding villages. We validate our findings using single-cell, spatial, and bulk transcriptome data from 740 liver cancer patients. This study provides insights into understanding tumor spatial landscape and its impact on tumor aggressiveness.
Similar content being viewed by others
Data availability
The single-cell spatial transcriptome data generated from this study have been deposited in Zenodo (https://zenodo.org/doi/10.5281/zenodo.13773977). The processed scRNA-seq data of the patients in this study are available through the Gene Expression Omnibus (accession number GSE189903). Raw sequencing data are considered protected information and are therefore available under restricted access through dbGaP under accession number phs003117.v1.p1. Access via the NCI’s dbGaP can be requested by qualified senior or principal investigators overseeing the research. The NCI’s Data Access Committee reviews such requests within 3 months and will make data available for up to 12 months. The publicly available 10X genomics visium datasets used in this study include samples from Liu et al.38 (Mendeley Data: skrx2fz79n, https://data.mendeley.com/datasets/skrx2fz79n), Wu et al.6 (http://lifeome.net/supp/livercancer-st/data.htm), Zhang et al.39 (GEO accession: GSE238264, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE238264), and Mo et al.42 (HTAN DCC Portal under the HTAN WUSTL Atlas, https://data.humantumoratlas.org/). Other publicly available data include bulk transcriptomic data of GSE14520 (LCI)43, GSE144269 (Mongolia, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE144269)44, and the TCGA database (TCGA-LIHC, https://portal.gdc.cancer.gov) and scRNA-seq data of GSE151530, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE1515301.
Code availability
Code used in this project have been deposited in github (https://github.com/MengLiu1/Tumor-cell-villages).
References
Ma, L. et al. Single-cell atlas of tumor cell evolution in response to therapy in hepatocellular carcinoma and intrahepatic cholangiocarcinoma. J. Hepatol. 75, 1397–1408 (2021.
Marjanovic, N. D. et al. Emergence of a high-plasticity cell state during lung cancer evolution. Cancer cell 38, 229–246.e213 (2020).
Peng, J. et al. Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res. 29, 725–738 (2019).
Gavish, A. et al. Hallmarks of transcriptional intratumour heterogeneity across a thousand tumours. Nature 618, 598–606 (2023).
Barkley, D. et al. Cancer cell states recur across tumor types and form specific interactions with the tumor microenvironment. Nat. Genet. 54, 1192–1201 (2022).
Wu, R. et al. Comprehensive analysis of spatial architecture in primary liver cancer. Sci. Adv. 7, eabg3750 (2021).
Denisenko, E. et al. Spatial transcriptomics reveals discrete tumour microenvironments and autocrine loops within ovarian cancer subclones. Nat. Commun. 15, 2860 (2024).
Ravi, V. M. et al. Spatially resolved multi-omics deciphers bidirectional tumor-host interdependence in glioblastoma. Cancer Cell 40, 639–655.e613 (2022).
Greenwald, A. C. et al. Integrative spatial analysis reveals a multi-layered organization of glioblastoma. Cell 187, 2485–2501.e2426 (2024).
Cui Zhou, D. et al. Spatially restricted drivers and transitional cell populations cooperate with the microenvironment in untreated and chemo-resistant pancreatic cancer. Nat. Genet. 54, 1390–1405 (2022).
De Zuani, M. et al. Single-cell and spatial transcriptomics analysis of non-small cell lung cancer. Nat. Commun. 15, 4388 (2024).
Kirschenbaum, D. et al. Time-resolved single-cell transcriptomics defines immune trajectories in glioblastoma. Cell 187, 149–165.e123 (2024).
Seferbekova, Z., Lomakin, A., Yates, L. R. & Gerstung, M. Spatial biology of cancer evolution. Nat. Rev. Genet. 24, 295–313 (2023).
Yeh, C. Y. et al. Mapping spatial organization and genetic cell-state regulators to target immune evasion in ovarian cancer. Nat. Immunol. 25, 1943–1958 (2024).
Ma, L. et al. Tumor cell biodiversity drives microenvironmental reprogramming in liver cancer. Cancer Cell 36, 418–430.e416 (2019).
Ma, L. & Wang, X. W. Dissecting liver tumor heterogeneity to improve health equity. Trends cancer 8, 286–290 (2022).
Ma, L., Khatib, S., Craig, A. J. & Wang, X. W. Toward a liver cell atlas: understanding liver biology in health and disease at single-cell resolution. Semin Liver Dis. 41, 321–330 (2021).
Zheng, H. et al. Single-cell analysis reveals cancer stem cell heterogeneity in hepatocellular carcinoma. Hepatology 68, 127–140 (2018).
He, S. S. et al. High-plex imaging of RNA and proteins at subcellular resolution in fixed tissue by spatial molecular imaging. Nat. Biotechnol. 40 https://doi.org/10.1038/s41587-022-01483-z (2022).
Ma, L. et al. Multiregional single-cell dissection of tumor and immune cells reveals stable lock-and-key features in liver cancer. Nat. Commun. 13, 7533 (2022).
Denisenko, E. et al. Systematic assessment of tissue dissociation and storage biases in single-cell and single-nucleus RNA-seq workflows. Genome Biol. 21, 1–25 (2020).
Axelrod, M. L., Cook, R. S., Johnson, D. B. & Balko, J. M. Biological consequences of MHC-II expression by tumor cells in cancer. Clin. cancer Res. 25, 2392–2402 (2019).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289-+ (2019).
Xu, C. et al. Arteries are formed by vein-derived endothelial tip cells. Nat. Commun. 5, 5758 (2014).
Siemerink, M. J. et al. CD34 marks angiogenic tip cells in human vascular endothelial cell cultures. Angiogenesis 15, 151–163 (2012).
Li, J. et al. Pan-cancer integrative analyses dissect the remodeling of endothelial cells in human cancers. Natl. Sci. Rev. 11, nwae231 (2024).
Goveia, J. et al. An integrated gene expression landscape profiling approach to identify lung tumor endothelial cell heterogeneity and angiogenic candidates. Cancer Cell 37, 21–36.e13 (2020).
Guo, B., Wen, X., Yu, S. & Yang, J. Single-cell sequencing reveals PHLDA1-positive smooth muscle cells promote local invasion in head and neck squamous cell carcinoma. Transl. Oncol. 55, 102301 (2025).
Ma, R. Y., Black, A. & Qian, B. Z. Macrophage diversity in cancer revisited in the era of single-cell omics. Trends Immunol. 43, 546–563 (2022).
Ma, L., Li, C. C. & Wang, X. W. Roles of cellular neighborhoods in hepatocellular carcinoma pathogenesis. Annu. Rev. Pathol. Mech. Dis. 20, 169–192 (2025).
Maestri, E. et al. Spatial proximity of tumor-immune interactions predicts patient outcome in hepatocellular carcinoma. Hepatology 10, 1097 (2023).
Pastushenko, I. et al. Identification of the tumour transition states occurring during EMT. Nature 556, 463–468 (2018).
Belfiore, A. et al. IGF2: a role in metastasis and tumor evasion from immune surveillance? Biomedicines 11, 229 (2023).
Pastushenko, I. & Blanpain, C. EMT transition states during tumor progression and metastasis. Trends Cell Biol. 29, 212–226 (2019).
Dunn, G. P., Koebel, C. M. & Schreiber, R. D. Interferons, immunity and cancer immunoediting. Nat. Rev. Immunol. 6, 836–848 (2006).
Pichlmair, A. et al. IFIT1 is an antiviral protein that recognizes 5′-triphosphate RNA. Nat. Immunol. 12, 624–630 (2011).
Zhou, Z. et al. Antiviral activities of ISG20 in positive-strand RNA virus infections. Virology 409, 175–188 (2011).
Liu, Y. et al. Identification of a tumour immune barrier in the HCC microenvironment that determines the efficacy of immunotherapy. J. Hepatol. 78, 770–782 (2023).
Zhang, S. et al. Spatial transcriptomics analysis of neoadjuvant cabozantinib and nivolumab in advanced hepatocellular carcinoma identifies independent mechanisms of resistance and recurrence. Genome Med. 15, 72 (2023).
Gargiulo, G., Serresi, M. & Marine, J.-C. Cell states in cancer: drivers, passengers, and trailers. Cancer Discov. 14, 610–614 (2024).
Velickovic, P. et al. Graph attention networks. Stat 1050, 10–48550 (2017).
Mo, C. K. et al. Tumour evolution and microenvironment interactions in 2D and 3D space. Nature 634 https://doi.org/10.1038/s41586-024-08087-4 (2024).
Roessler, S. et al. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res. 70, 10202–10212 (2010).
Candia, J. et al. The genomic landscape of Mongolian hepatocellular carcinoma. Nat. Commun. 11, 4383 (2020).
St Croix, B. et al. Genes expressed in human tumor endothelium. Science 289, 1197–1202 (2000).
Ateeq, B. et al. Therapeutic targeting of SPINK1-positive prostate cancer. Sci. Transl. Med. 3, 72ra17–72ra17 (2011).
Ozaki, N. et al. Serine protease inhibitor Kazal type 1 promotes proliferation of pancreatic cancer cells through the epidermal growth factor receptor. Mol. Cancer Res. 7, 1572–1581 (2009).
Hambardzumyan, D. & Bergers, G. Glioblastoma: defining tumor niches. Trends Cancer 1, 252–265 (2015).
Bray, F. et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA a Cancer J.clin.74, 229–263 (2024).
Toh, M. R. et al. Global epidemiology and genetics of hepatocellular carcinoma. Gastroenterology 164, 766–782 (2023).
Revsine, M. et al. Lineage and ecology define liver tumor evolution in response to treatment. Cell Rep. Med. 5, 101394 (2024).
Chaisaingmongkol, J. et al. Common molecular subtypes among Asian hepatocellular carcinoma and cholangiocarcinoma. Cancer cell 32, 57–70.e53 (2017).
Ohmuraya, M. et al. Autophagic cell death of pancreatic acinar cells in serine protease inhibitor Kazal type 3—deficient mice. Gastroenterology 129, 696–705 (2005).
Man, K.-F. et al. SPINK1-induced tumor plasticity provides a therapeutic window for chemotherapy in hepatocellular carcinoma. Nat. Commun. 14, 7863 (2023).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550 (2005).
Neftel, C. et al. An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell 178, 835–849.e821 (2019).
Puram, S. V. et al. Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell 171, 1611–1624.e1624 (2017).
Ji, A. L. et al. Multimodal analysis of composition and spatial architecture in human squamous cell carcinoma. Cell 182, 497–514.e422 (2020).
Cords, L. et al. Cancer-associated fibroblast classification in single-cell and spatial proteomics data. Nat. Commun. 14, 4294 (2023).
Zhang, M. et al. Single-cell transcriptomic architecture and intercellular crosstalk of human intrahepatic cholangiocarcinoma. J. Hepatol. 73, 1118–1130 (2020).
Morgan, D. & Tergaonkar, V. Unraveling B cell trajectories at single cell resolution. Trends Immunol. 43, 210–229 (2022).
Yang, Y. et al. Pan-cancer single-cell dissection reveals phenotypically distinct B cell subtypes. Cell 187, 4790–4811.e4722 (2024).
Zhang, Z. et al. Landscape of infiltrating B cells and their clinical significance in human hepatocellular carcinoma. Oncoimmunology 8, e1571388 (2019).
Guo, X. et al. Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat. Med. 24, 978–985 (2018).
Zhang, L. et al. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature 564, 268–272 (2018).
Zheng, C. H. et al. Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell 169, 1342 (2017).
Shiao, S. L. et al. Single-cell and spatial profiling identify three response trajectories to pembrolizumab and radiation therapy in triple negative breast cancer. Cancer Cell 42, 70–84.e78 (2024).
Cang, Z. et al. Screening cell-cell communication in spatial transcriptomics via collective optimal transport. Nat. Methods 20, 218–228 (2023).
Acknowledgements
We thank Drs. Xin Wei Wang, Eytan Ruppin, and Tom Misteli for helpful comments on the manuscript; Dr. Yuuki Ohara for assistance in interpreting the histology images; the patients, families, and nurses for contribution to this study. This work was supported by grants (ZIA BC 012079 [L.M.] and ZIA BC 012083 [L.M.]) from the intramural research program of the Center for Cancer Research, National Cancer Institute of the United States. J.U.M. is supported by grants from the Wilhelm Sander Foundation (2021.089.1). The contributions of the NIH authors were made as part of their official duties as NIH federal employees, are in compliance with agency policy requirements, and are considered Works of the United States Government. However, the findings and conclusions presented in this paper are those of the authors and do not necessarily reflect the views of the NIH or the U.S. Department of Health and Human Services.
Funding
Open access funding provided by the National Institutes of Health.
Author information
Authors and Affiliations
Contributions
L.M. developed study concept; J.U.M. directed clinical study; M.L. performed computational analysis; M.O.H. conducted experiments; D.C., H.P.L., W.W., L.W., M.F., and J.M.H. conducted additional experiments and data analysis; M.L. and L.M. interpreted data; L.M. and M.L. wrote the manuscript with help from M.O.H. and D.C. All authors read, edited, and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Ruidong Xue, Flavio Maina and Lei Chen for their contribution to the peer review of this work. [A peer review file is available].
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liu, M., Hernandez, M.O., Castven, D. et al. Tumor cell villages define the co-dependency of tumor and microenvironment in liver cancer. Nat Commun (2026). https://doi.org/10.1038/s41467-026-69797-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-026-69797-z