Abstract
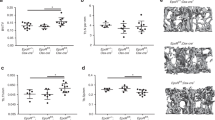
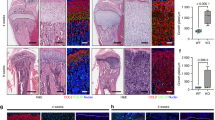
Osteoporotic fractures are a major global health burden. To uncover potential targets for fracture prevention, we use a proteome-wide Mendelian randomization (MR) approach combined with colocalization. Here we show that nine circulating proteins associate with forearm fracture risk, including sclerostin and osteoprotegerin targeted by existing osteoporosis treatments, and three other known bone-related proteins, providing proof of concept for our MRpipeline. Notably, we identify ephrin-A1 as a novel protective factor against fractures, a membrane-linked protein partly released into circulation that binds its high-affinity receptor EphA2 on osteoblasts. Experimental models and genetic analyses indicate that ephrin-A1 increases bone mineral density, supporting a mechanism by which this pathway may mediate fracture protection. Spatial expression analysis with the innovative 3D DeepBone technique suggests ephrin-A1 on endothelial cells interacts with EphA2 on adjacent osteoblasts at the bone surface. These findings position ephrin-A1–EphA2 signalling as a therapeutic target to strengthen bone and reduce fracture risk.
Similar content being viewed by others
Data availability
All GWAS summary statistics for the exposures and outcomes in the Mendelian randomisation analyses are available online: circulating proteins https://www.decode.com/summarydata/, eBMD and total body BMD http://www.gefos.org/, forearm fractures at the GWAS Catalogue under study accession number GCST90281273 (https://www.ebi.ac.uk/gwas). The human bone marrow single-cell RNA sequencing atlas can be explored at the CellxGene Portal:https://cellxgene.cziscience.com/collections/0391c84c-d57d-4741-9277-e4d58f9a3d0c. and primary data used in the present study are available at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE147287; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE147390; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE169396; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE190965; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE196678; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE202813. Source data are provided in this paper.
Code availability
All analyses were performed using publicly available software, tools, packages and databases. The MR analyses were conducted using R v4.4.3 (https://cran.r-project.org/) and the packages MendelianRandomization, LDlinkR, and dplyr. Colocalization analyses were performed using the pwcoco tool (https://github.com/jwr-git/pwcoco).
References
Baron, R. & Hesse, E. Update on bone anabolics in osteoporosis treatment: rationale, current status, and perspectives. J. Clin. Endocrinol. Metab. 97, 311–325 (2012).
Johnell, O. & Kanis, J. Epidemiology of osteoporotic fractures. Osteoporos. Int. 16, S3–S7 (2005).
Lorentzon, M. Treating osteoporosis to prevent fractures: current concepts and future developments. J. Intern. Med 285, 381–394 (2019).
Anastasilakis, A. D. et al. Osteonecrosis of the jaw and antiresorptive agents in benign and malignant diseases: a critical review organized by the ECTS. J. Clin. Endocrinol. Metab. 107, 1441–1460 (2022).
Black, D. M. et al. Atypical femur fracture risk versus fragility fracture prevention with bisphosphonates. N. Engl. J. Med. 383, 743–753 (2020).
Willers, C. et al. Osteoporosis in Europe: a compendium of country-specific reports. Arch. Osteoporos. 17, 23 (2022).
Eldjarn, G. H. et al. Large-scale plasma proteomics comparisons through genetics and disease associations. Nature 622, 348–358 (2023).
Sun, B. B. et al. Plasma proteomic associations with genetics and health in the UK Biobank. Nature 622, 329–338 (2023).
Austin, T. R. et al. A plasma protein-based risk score to predict hip fractures. Nat. Aging 4, 1064–1075 (2024).
Desai, T. A. et al. Identifying proteomic risk factors for overall, aggressive, and early onset prostate cancer using Mendelian Randomisation and tumour spatial transcriptomics. EBioMedicine 105, 105168 (2024).
Yuan, S. et al. Plasma proteins and onset of type 2 diabetes and diabetic complications: Proteome-wide Mendelian randomization and colocalization analyses. Cell Rep. Med. 4, 101174 (2023).
Nethander, M. et al. An atlas of genetic determinants of forearm fracture. Nat. Genet. 55, 1820–1830 (2023).
Cosman, F. et al. Romosozumab Treatment in Postmenopausal Women with Osteoporosis. N. Engl. J. Med. 375, 1532–1543 (2016).
Cummings, S. R. et al. Denosumab for prevention of fractures in postmenopausal women with osteoporosis. N. Engl. J. Med. 361, 756–765 (2009).
Saag, K. G. et al. Romosozumab or alendronate for fracture prevention in women with osteoporosis. N. Engl. J. Med. 377, 1417–1427 (2017).
Estrada, K. et al. Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat. Genet. 44, 491–501 (2012).
Nilsson, K. H. et al. RSPO3 is important for trabecular bone and fracture risk in mice and humans. Nat. Commun. 12, 4923 (2021).
Surakka, I. et al. MEPE loss-of-function variant associates with decreased bone mineral density and increased fracture risk. Nat. Commun. 11, 4093 (2020).
Zhou, S. et al. Converging evidence from exome sequencing and common variants implicates target genes for osteoporosis. Nat. Genet. 55, 1277–1287 (2023).
Morris, J. A. et al. An atlas of genetic influences on osteoporosis in humans and mice. Nat. Genet. 51, 258–266 (2019).
Nethander, M. et al. BMD-Related genetic risk scores predict site-specific fractures as well as trabecular and cortical bone microstructure. J. Clin. Endocrinol. Metab. 105, e1344–e1357 (2020).
Medina-Gomez, C. et al. Life-course genome-wide association study meta-analysis of total body BMD and assessment of agespecific effects. Am. J. Hum. Genet. 102, 88–102 (2018).
Pirozzi, F. et al. Proximal variants in CCND2 associated with microcephaly, short stature, and developmental delay: A case series and review of inverse brain growth phenotypes. Am. J. Med. Genet. A 185, 2719–2738 (2021).
Saleban, M., Harris, E. L. & Poulter, J. A. D-Type cyclins in development and disease. Genes 14, https://doi.org/10.3390/genes14071445 (2023).
Guan, Y., Ackert-Bicknell, C. L., Kell, B., Troyanskaya, O. G. & Hibbs, M. A. Functional genomics complements quantitative genetics in identifying disease-gene associations. PLoS Comput. Biol. 6, e1000991 (2010).
Chermside-Scabbo, C. J. et al. A proteomics approach to study mouse long bones: examining baseline differences and mechanical loading-induced bone formation in young-adult and old mice. Aging 16, 12726–12768 (2024).
Dew, G. et al. Localisation of matrix metalloproteinases and TIMP-2 in resorbing mouse bone. Cell Tissue Res. 299, 385–394 (2000).
Chu, N. T. L. et al. Quantitative multicolored deep imaging of human bones reveals a composite osteo-sinusoidal niche for mesenchymal stromal cells. Preprint at https://doi.org/10.1101/2025.10.07.680053 (2025).
Darling, T. K. & Lamb, T. J. Emerging Roles for Eph Receptors and Ephrin Ligands in Immunity. Front. Immunol. 10, 1473 (2019).
Irie, N. et al. Bidirectional signaling through ephrinA2-EphA2 enhances osteoclastogenesis and suppresses osteoblastogenesis. J. Biol. Chem. 284, 14637–14644 (2009).
Gomariz, A. et al. Quantitative spatial analysis of haematopoiesis-regulating stromal cells in the bone marrow microenvironment by 3D microscopy. Nat. Commun. 9, 2532 (2018).
Dimitrov, D. et al. Comparison of methods and resources for cell-cell communication inference from single-cell RNA-Seq data. Nat. Commun. 13, 3224 (2022).
Drake, M. T. et al. AGA2118, a Bispecific monoclonal antibody neutralizing both sclerostin and DKK1, increased bone formation, decreased bone resorption, and led to rapid BMD gains in a first in human, single- and multiple-dose, placebo-controlled, randomized study. in ASBMR Annual Meeting (Toronto, Canada, 2024).
Mendoza, R. et al. Ephrin-A1 and the sheddase ADAM12 are upregulated in COVID-19. Heliyon 7, e07200 (2021).
Wykosky, J. et al. Soluble monomeric EphrinA1 is released from tumor cells and is a functional ligand for the EphA2 receptor. Oncogene 27, 7260–7273 (2008).
Perepletchikova, D. & Malashicheva, A. Communication between endothelial cells and osteoblasts in regulation of bone homeostasis: Notch players. Stem Cell Res. Ther. 16, 56 (2025).
Tuckermann, J. & Adams, R. H. The endothelium-bone axis in development, homeostasis and bone and joint disease. Nat. Rev. Rheumatol. 17, 608–620 (2021).
Movérare-Skrtic, S. et al. B4GALNT3 regulates glycosylation of sclerostin and bone mass. EBioMedicine 91, 104546 (2023).
Matsuo, K. & Otaki, N. Bone cell interactions through Eph/ephrin: bone modeling, remodeling and associated diseases. Cell Adh. Migr. 6, 148–156 (2012).
Wang, Y. et al. Ephrin B2/EphB4 mediates the actions of IGF-I signaling in regulating endochondral bone formation. J. Bone Miner. Res. 29, 1900–1913 (2014).
Wang, Y. et al. Ablation of ephrin B2 in Col2 expressing cells delays fracture repair. Endocrinology 161, https://doi.org/10.1210/endocr/bqaa179 (2020).
Andrew, T., Antioniades, L., Scurrah, K. J., Macgregor, A. J. & Spector, T. D. Risk of wrist fracture in women is heritable and is influenced by genes that are largely independent of those influencing BMD. J. Bone Miner. Res. 20, 67–74 (2005).
Ralston, S. H. & Uitterlinden, A. G. Genetics of osteoporosis. Endocr. Rev. 31, 629–662 (2010).
Yang, J. et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet. 44, 369–375 (2012).
Yang, J., Lee, S. H., Goddard, M. E. & Visscher, P. M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88, 76–82 (2011).
Giambartolomei, C. et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 10, e1004383 (2014).
Dickinson, M. E. et al. High-throughput discovery of novel developmental phenotypes. Nature 537, 508–514 (2016).
Lawenius, L. et al. Transplantation of gut microbiota from old mice into young healthy mice reduces lean mass but not bone mass. Gut Microbes 15, 2236755 (2023).
Nilsson, K. H. et al. Estradiol and RSPO3 regulate vertebral trabecular bone mass independent of each other. Am. J. Physiol. Endocrinol. Metab. 322, E211–E218 (2022).
Lionikaite, V. et al. Vitamin A decreases the anabolic bone response to mechanical loading by suppressing bone formation. Faseb J. 33, 5237–5247 (2019).
Liu, Y. et al. Dissection of Cellular Communication between Human Primary Osteoblasts and Bone Marrow Mesenchymal Stem Cells in Osteoarthritis at Single-Cell Resolution. Int. J. Stem Cells 16, 342–355 (2023).
Qiu, X. et al. Single-cell RNA sequencing of human femoral head in vivo. Aging 13, 15595–15619 (2021).
Li, H. et al. Identification of phenotypically, functionally, and anatomically distinct stromal niche populations in human bone marrow based on single-cell RNA sequencing. Elife 12, https://doi.org/10.7554/elife.81656 (2023).
Mei, S. et al. Single-cell analysis of immune and stroma cell remodeling in clear cell renal cell carcinoma primary tumors and bone metastatic lesions. Genome Med. 16, 1 (2024).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019).
Virshup, I. et al. The scverse project provides a computational ecosystem for single-cell omics data analysis. Nat. Biotechnol. 41, 604–606 (2023).
Choi, H. M. T. et al. Third-generation in situ hybridization chain reaction: multiplexed, quantitative, sensitive, versatile, robust. Development 145, https://doi.org/10.1242/dev.165753 (2018).
Gorelashvili, M. G., Heinze, K. G. & Stegner, D. Platelets and Megakaryocytes: Volume 4, Advanced Protocols and Perspectives(Springer New York, New York, NY, 2018).
Jähn, K. & Stoddart, M. J. Mammalian Cell Viability: Methods and Protocols (Humana Press, Totowa, NJ, 2011).
Acknowledgements
This work was supported by the Swedish Research Council (2020-01392 and 2024-02412, C.O.; 2022-01156, S.M.-S.; 2020-02298, A.S.C.); the Swedish state under the agreement between the Swedish government and the county councils, the ALF-agreement (ALFGBG-720331, ALFGBG-965235, and ALFGBG-965744, C.O.; ALFGBG-1005555, S.M.-S.; ALFGBG-1006264, A.S.C.); the Novo Nordisk Foundation (NNF19OC0055250 and NNF22OC0078421, C.O.; NNF23OC0084522 and NNF25OC0105187, S.M.-S.; NNF21OC0070314, A.S.C.); the Lundberg Foundation (LU2024-0110, C.O.; LU2025-0079, A.S.C.); the Knut and Alice Wallenberg Foundation (KAW 2020.0230, C.O.); and the European Union (ERC Advanced Grant, HeMaFA, Project 101096347, C.O.). Views and opinions expressed are, however, those of the author(s) only and do not necessarily reflect those of the European Union or the European Research Council. Neither the European Union nor the granting authority can be held responsible for them. The funders had no role in study design, data collection, data analysis, data interpretation, or manuscript writing.
Funding
Open access funding provided by University of Gothenburg.
Author information
Authors and Affiliations
Contributions
The design of the study was performed in collaboration between S.M.-S., M.N. and C.O. Mendelian randomisation and colocalization were conducted by M.N. and C.O. Mechanistic studies were conducted by S.M.-S., K.H.N., P.H., U.H.L., and C.O. L.L. conducted the in situ hybridisation using mouse tissues, O.D. and A.S.C. conducted the human bone marrow single-cell RNA seq analyses, also including ligand-receptor interaction analyses. N.T.L.C., X.T. and A.S.C. conducted the analyses of human bone using 3D spatial expression. S.M.-S., M.N., A.S.C. and C.O. wrote the first draft of the manuscript. All authors contributed to subsequent drafts of the manuscript and made the decision to submit the manuscript for publication.
Corresponding authors
Ethics declarations
Competing interests
C.O. is an applicant on filed patent applications on the effect of probiotics on bone metabolism. All other authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Can Yang, Georg Duda and Shushan Zhao for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Movérare-Skrtic, S., Nethander, M., Li, L. et al. Identification of ephrin-A1–EphA2 signalling as a potential target for fracture prevention. Nat Commun (2026). https://doi.org/10.1038/s41467-026-69863-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-026-69863-6