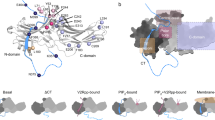
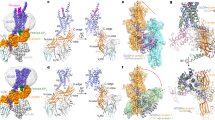
Abstract
Beta-arrestins (βarrs) are key regulators and transducers of G-protein coupled receptor signaling; however, little is known of how βarrs communicate with their downstream effectors. Here, we delineate structural mechanisms underlying βarr-mediated signal transduction. Using cryo-electron microscopy, we elucidate how βarr1 recruits and activates the non-receptor tyrosine kinase Src, a well-established signaling partner of βarrs. βarr1 engages Src SH3 through two distinct sites, each employing a different recognition mechanism: a polyproline motif in the N-domain and a non-proline-based interaction in the central crest region. At both sites βarr1 interacts with the aromatic surface of SH3, disrupting the autoinhibited conformation of Src and directly triggering its allosteric activation. This structural evidence establishes βarr1 as an active regulatory protein rather than a passive scaffold and suggests a potentially general mechanism for βarr-mediated signaling across diverse effectors.
Similar content being viewed by others
Data availability
The cryo-EM maps have been deposited in the EMDB under accession codes EMD-45977 (SH3–βarr1-CC complex), EMD-45982 (SH3–βarr1-N complex), and EMD-44881 (Src–βarr1-CC complex). The atomic coordinates have been deposited in the Protein Data Bank under accession codes 9CX3 (SH3–βarr1-CC complex); 9CX9 (SH3–βarr1-N complex); 9BT8 (Src–βarr1-CC complex). The HDX-MS data have been deposited to the ProteomeXchange Consortium [http://proteomecentral.proteomexchange.org] via the MassIVE repository [https://massive.ucsd.edu/] with the dataset identifier PXD073493. CXMS data have been deposited to the ProteomeXchange Consortium [http://proteomecentral.proteomexchange.org] via the PRIDE partner repository74 with the dataset identifier PXD073058. All other data generated or analyzed in this study are included in the article and its Supplementary Information. Source data are provided within the Source Data File. The manuscript refers to the following previously published PDB accession codes: 4JQI (βarr1–V2Rpp); 2PTK (Src); 1G4M (βarr1); 1FMK (Src); 6TKO (β1V2R–βarr1); 6U1N (M2V2R–βarr1); 6UP7 (NTSR1–βarr1); 4U5W (Hck–Nef); 2KNB (endophilin A1 SH3–parkin Ubl); 3A98 (DOCK2 SH3–ELMO 1); 1JT4 (Sla1 SH3-ubiquitin); 1Y57 (Src). Source data are provided with this paper.
References
Lefkowitz, R. J. Arrestins come of age: a personal historical perspective. Prog. Mol. Biol. Transl. Sci. 118, 3–18 (2013).
Peterson, Y. K. & Luttrell, L. M. The Diverse Roles of Arrestin Scaffolds in G Protein-Coupled Receptor Signaling. Pharm. Rev. 69, 256–297 (2017).
Yang, F. et al. Allosteric mechanisms underlie GPCR signaling to SH3-domain proteins through arrestin. Nat. Chem. Biol. 14, 876–886 (2018).
Pakharukova, N., Masoudi, A., Pani, B., Staus, D. P. & Lefkowitz, R. J. Allosteric activation of proto-oncogene kinase Src by GPCR-beta-arrestin complexes. J. Biol. Chem. 295, 16773–16784 (2020).
Zang, Y., Kahsai, A. W., Pakharukova, N., Huang, L. Y. & Lefkowitz, R. J. The GPCR-beta-arrestin complex allosterically activates C-Raf by binding its amino terminus. J. Biol. Chem. 297, 101369 (2021).
Kahsai, A. W. et al. Signal transduction at GPCRs: Allosteric activation of the ERK MAPK by β-arrestin. P Natl. Acad. Sci. USA. 120, (2023).
Karkkainen, S. et al. Identification of preferred protein interactions by phage-display of the human Src homology-3 proteome. EMBO Rep. 7, 186–191 (2006).
Li, S. S. Specificity and versatility of SH3 and other proline-recognition domains: structural basis and implications for cellular signal transduction. Biochem J. 390, 641–653 (2005).
Luttrell, L. M. et al. Beta-arrestin-dependent formation of beta2 adrenergic receptor-Src protein kinase complexes. Science 283, 655–661 (1999).
Xiao, K. et al. Revealing the architecture of protein complexes by an orthogonal approach combining HDXMS, CXMS, and disulfide trapping. Nat. Protoc. 13, 1403–1428 (2018).
Cahill, T. J. 3rd et al. Distinct conformations of GPCR-beta-arrestin complexes mediate desensitization, signaling, and endocytosis. Proc. Natl. Acad. Sci. USA 114, 2562–2567 (2017).
Shukla, A. K. et al. Structure of active beta-arrestin-1 bound to a G-protein-coupled receptor phosphopeptide. Nature 497, 137–141 (2013).
Lee, C. H., Saksela, K., Mirza, U. A., Chait, B. T. & Kuriyan, J. Crystal structure of the conserved core of HIV-1 Nef complexed with a Src family SH3 domain. Cell 85, 931–942 (1996).
Alvarado, J. J., Tarafdar, S., Yeh, J. I. & Smithgall, T. E. Interaction with the Src homology (SH3-SH2) region of the Src-family kinase Hck structures the HIV-1 Nef dimer for kinase activation and effector recruitment. J. Biol. Chem. 289, 28539–28553 (2014).
He, Y., Hicke, L. & Radhakrishnan, I. Structural basis for ubiquitin recognition by SH3 domains. J. Mol. Biol. 373, 190–196 (2007).
Trempe, J. F. et al. SH3 domains from a subset of BAR proteins define a Ubl-binding domain and implicate parkin in synaptic ubiquitination. Mol. Cell 36, 1034–1047 (2009).
Hanawa-Suetsugu, K. et al. Structural basis for mutual relief of the Rac guanine nucleotide exchange factor DOCK2 and its partner ELMO1 from their autoinhibited forms. P Natl. Acad. Sci. USA. 109, 3305–3310 (2012).
Teyra, J. et al. Comprehensive Analysis of the Human SH3 Domain Family Reveals a Wide Variety of Non-canonical Specificities. Structure 25, 1598–1610.e1593 (2017).
Saksela, K. & Permi, P. SH3 domain ligand binding: What’s the consensus and where’s the specificity? FEBS Lett. 586, 2609–2614 (2012).
Harrison, S. C. Variation on an Src-like theme. Cell 112, 737–740 (2003).
Xu, W., Harrison, S. C. & Eck, M. J. Three-dimensional structure of the tyrosine kinase c-Src. Nature 385, 595–602 (1997).
Porter, M., Schindler, T., Kuriyan, J. & Miller, W. T. Reciprocal regulation of Hck activity by phosphorylation of Tyr(527) and Tyr(416). Effect of introducing a high affinity intramolecular SH2 ligand. J. Biol. Chem. 275, 2721–2726 (2000).
Young, M. A., Gonfloni, S., Superti-Furga, G., Roux, B. & Kuriyan, J. Dynamic coupling between the SH2 and SH3 domains of c-Src and hck underlies their inactivation by C-terminal tyrosine phosphorylation. Cell 105, 115–126 (2001).
Bous, J. et al. Structure of the vasopressin hormone-V2 receptor-beta-arrestin1 ternary complex. Sci. Adv. 8, eabo7761 (2022).
Staus, D. P. et al. Structure of the M2 muscarinic receptor-beta-arrestin complex in a lipid nanodisc. Nature 579, 297–302 (2020).
Huang, W. et al. Structure of the neurotensin receptor 1 in complex with beta-arrestin 1. Nature 579, 303–308 (2020).
Lee, Y. et al. Molecular basis of beta-arrestin coupling to formoterol-bound beta(1)-adrenoceptor. Nature 583, 862–866 (2020).
O’Hayre, M. et al. Genetic evidence that β-arrestins are dispensable for the initiation of β-adrenergic receptor signaling to ERK. Sci Signal 10, (2017).
Stramiello, M. & Wagner, J. J. Dreceptor-mediated enhancement of LTP requires PKA, Src family kinases, and NR2B-containing NMDARs. Neuropharmacology 55, 871–877 (2008).
Kaya, A. I., Perry, N. A., Gurevich, V. V. & Iverson, T. M. Phosphorylation barcode-dependent signal bias of the dopamine D1 receptor. Proc. Natl. Acad. Sci. USA. 117, 14139–14149 (2020).
Violin, J. D. et al. J. β-adrenergic receptor signaling and desensitization elucidated by quantitative modeling of real time cAMP dynamics. J. Biol. Chem. 283, 2949–2961 (2008).
Qin, L. et al. Structural biology. Crystal structure of the chemokine receptor CXCR4 in complex with a viral chemokine. Science 347, 1117–1122 (2015).
Rosenbaum, D. M. et al. Structure and function of an irreversible agonist-beta(2) adrenoceptor complex. Nature 469, 236–240 (2011).
Chen, Q. Y. et al. Structures of rhodopsin in complex with G-protein-coupled receptor kinase 1. Nature 595, 600–605 (2021).
Scott, J. D. & Pawson, T. Cell Signaling in Space and Time: Where Proteins Come Together and When They’re Apart. Science 326, 1220–1224 (2009).
Moarefi, I. et al. Activation of the Src-family tyrosine kinase Hck by SH3 domain displacement. Nature 385, 650–653 (1997).
Alexandropoulos, K. & Baltimore, D. Coordinate activation of c-Src by SH3- and SH2-binding sites on a novel, p130(Cas)-related protein, Sin. Gene Dev. 10, 1341–1355 (1996).
Kypta, R. M., Goldberg, Y., Ulug, E. T. & Courtneidge, S. A. Association between the Pdgf Receptor and Members of the Src Family of Tyrosine Kinases. Cell 62, 481–492 (1990).
Cobb, B. S., Schaller, M. D., Leu, T. H. & Parsons, J. T. Stable Association of Pp60(Src) and Pp59(Fyn) with the Focal Adhesion-Associated Protein-Tyrosine Kinase, Pp125(Fak). Mol. Cell Biol. 14, 147–155 (1994).
Burnham, M. R. et al. Regulation of c-SRC activity and function by the adapter protein CAS. Mol. Cell Biol. 20, 5865–5878 (2000).
Bjorge, J. D., Jakymiw, A. & Fujita, D. J. Selected glimpses into the activation and function of Src kinase. Oncogene 19, 5620–5635 (2000).
Roskoski, R. Src protein-tyrosine kinase structure, mechanism, and small molecule inhibitors. Pharm. Res. 94, 9–25 (2015).
Thomsen, A. R. B. et al. GPCR-G Protein-beta-Arrestin Super-Complex Mediates Sustained G Protein Signaling. Cell 166, 907–919 (2016).
Nguyen, A. H. et al. Structure of an endosomal signaling GPCR-G protein-β-arrestin megacomplex. Nat. Struct. Mol. Biol. 26, 1123–1131 (2019).
Frame, M. C. Src in cancer: deregulation and consequences for cell behaviour. Biochim Biophys. Acta 1602, 114–130 (2002).
Benovic, J. L. et al. Functional desensitization of the isolated beta-adrenergic receptor by the beta-adrenergic receptor kinase: potential role of an analog of the retinal protein arrestin (48-kDa protein). Proc. Natl. Acad. Sci. USA. 84, 8879–8882 (1987).
Lohse, M. J., Benovic, J. L., Codina, J., Caron, M. G. & Lefkowitz, R. J. Beta-Arrestin - a Protein That Regulates Beta-Adrenergic-Receptor Function. Science 248, 1547–1550 (1990).
Gutkind, J. S. & Kostenis, E. Arrestins as rheostats of GPCR signalling. Nat. Rev. Mol. Cell Bio 19, 615–616 (2018).
Mitra, S. K. & Schlaepfer, D. D. Integrin-regulated FAK-Src signaling in normal and cancer cells. Curr. Opin. Cell Biol. 18, 516–523 (2006).
Staus, D. P. et al. Sortase ligation enables homogeneous GPCR phosphorylation to reveal diversity in beta-arrestin coupling. Proc. Natl. Acad. Sci. USA. 115, 3834–3839 (2018).
Paduch, M. et al. A. Generating conformation-specific synthetic antibodies to trap proteins in selected functional states. Methods 60, 3–14 (2013).
Shukla, A. K. et al. Visualization of arrestin recruitment by a G-protein-coupled receptor. Nature 512, 218–222 (2014).
Seeliger, M. A. et al. High yield bacterial expression of active c-Abl and c-Src tyrosine kinases. Protein Sci. 14, 3135–3139 (2005).
Nobles, K. N., Guan, Z., Xiao, K., Oas, T. G. & Lefkowitz, R. J. The active conformation of beta-arrestin1: direct evidence for the phosphate sensor in the N-domain and conformational differences in the active states of beta-arrestins1 and -2. J. Biol. Chem. 282, 21370–21381 (2007).
Rizk, S. S. et al. Allosteric control of ligand-binding affinity using engineered conformation-specific effector proteins. Nat. Struct. Mol. Biol. 18, 437–442 (2011).
Levinson, N. M., Seeliger, M. A., Cole, P. A. & Kuriyan, J. Structural basis for the recognition of c-Src by its inactivator Csk. Cell 134, 124–134 (2008).
Strachan, R. T. et al. Divergent Transducer-specific Molecular Efficacies Generate Biased Agonism at a G Protein-coupled Receptor (GPCR). J. Biol. Chem. 289, 14211–14224 (2014).
Attramadal, H. et al. Beta-Arrestin2, a Novel Member of the Arrestin Beta-Arrestin Gene Family. J. Biol. Chem. 267, 17882–17890 (1992).
Chen, S., Li, J., Vinothkumar, K. R. & Henderson, R. Interaction of human erythrocyte catalase with air-water interface in cryoEM. Microsc. (Oxf.) 71, i51–i59 (2022).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Bepler, T. et al. Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs. Nat. Methods 16, 1153–1160 (2019).
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Liu, Y. T., Fan, H., Hu, J. J. & Zhou, Z. H. Overcoming the preferred-orientation problem in cryo-EM with self-supervised deep learning. Nat. Methods 22, 113–123 (2025).
Sanchez-Garcia, R. et al. DeepEMhancer: a deep learning solution for cryo-EM volume post-processing. Commun. Biol. 4, 874 (2021).
Pettersen, E. F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput Chem. 25, 1605–1612 (2004).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr D. Biol. Crystallogr 66, 486–501 (2010).
Croll, T. I. ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr D. 74, 519–530 (2018).
Pettersen, E. F. et al. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Adams, P. D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D. Biol. Crystallogr 58, 1948–1954 (2002).
Kim, D. N. et al. Cryo_fit: Democratization of flexible fitting for cryo-EM. J. Struct. Biol. 208, 1–6 (2019).
Kidmose, R. T. et al. Namdinator - automatic molecular dynamics flexible fitting of structural models into cryo-EM and crystallography experimental maps. Iucrj 6, 526–531 (2019).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D. Biol. Crystallogr 66, 12–21 (2010).
Barker, S. C. et al. Characterization of pp60c-src tyrosine kinase activities using a continuous assay: autoactivation of the enzyme is an intermolecular autophosphorylation process. Biochemistry 34, 14843–14851 (1995).
Perez-Riverol, Y. et al. The PRIDE database at 20 years: 2025 update. Nucleic Acids Res. 53, D543–D553 (2024).
Acknowledgements
We acknowledge the use of cryo-EM microscopes at the Shared Materials Instrumentation Facility (Duke University) and thank Nilakshee Bhattacharya for assistance with microscope operation. We are grateful to Liyin Huang for their helpful discussions throughout this work and to Yangyang Li for administrative assistance. R.J.L. is an Investigator of the Howard Hughes Medical Institute. This work was supported, in part, by US National Institutes of Health (National Heart, Lung, and Blood Institute: R01 HL16037 to R.J.L.; National Institute of General Medical Sciences: R35GM133598 to A.G.). N.P. is supported by postdoctoral fellowships from Human Frontier Science Program (LT000174/2018) and European Molecular Biology Organization (ALTF 1071-2017). K.X. is supported by the Moonshot Biomarker Program of Allegheny Health Network Cancer Institute and Highmark Health, the Prostate Cancer Foundation Challenge Award (2023CHAL4223), the PA State Formula Grant (SAP #: 4100095527), and The Pittsburgh Foundation (cc#45126409).
Author information
Authors and Affiliations
Contributions
N.P. and R.J.L. conceived the study. N.P., R.R.A., J.K., A.W.K., B.P., K.X., and S.A. designed the experiments. N.P., B.N.T., L.L., D.K.B., A.W.K., B.P., K.X., R.O., and X.Z. performed biochemical experiments. N.P., B.N.T., J.K. and S.A. performed cellular functional assays. N.P., B.N.T., and D.K.B. performed structural biology experiments. N.P. and H.B. built and refined structural models. R.R.A. and S.L. performed HDX-MS experiments. N.P., B.N.T., H.B., D.K.B., R.R.A., J.K., A.W.K., B.P., and K.X. analyzed the data. N.P. wrote the manuscript with input from all authors. A.G. and R.J.L. supervised the work.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Pakharukova, N., Thomas, B.N., Bansia, H. et al. Mechanism of beta-arrestin 1 mediated Src activation via Src SH3 domain revealed by cryo-electron microscopy. Nat Commun (2026). https://doi.org/10.1038/s41467-026-69884-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-026-69884-1