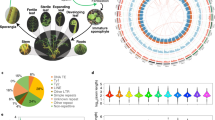
Abstract
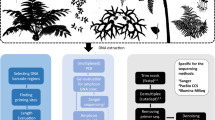
Ferns are essential for understanding plant evolution; however, their large and intricate genomes have kept their genetic landscape largely unexplored, with only a few genomes sequenced and limited transcriptomic data available. To bridge this gap, we generated extensive RNA-sequencing data across various organs from 22 representative fern species, resulting in high-quality transcriptome assemblies. These data enabled us to construct a time-calibrated phylogeny for ferns, encompassing all major clades, which revealed numerous instances of whole-genome duplication. We highlighted the distinctiveness of fern genetics, discovering that half of the identified gene families are unique to ferns. Our exploration of fern cell walls through biochemical and immunological analyses uncovered the presence of the lignin syringyl unit, along with evidence of its independent evolution in ferns. Additionally, the identification of an unusual sugar in fern cell walls suggests a divergent evolutionary trajectory in cell wall biochemistry, probably influenced by gene duplication and sub-functionalization. To facilitate further research, we have developed an online database that includes preloaded genomic and transcriptomic data for ferns and other land plants. We used this database to demonstrate the independent evolution of lignocellulosic gene modules in ferns. Our findings provide a comprehensive framework illustrating the unique evolutionary journey ferns have undertaken since diverging from the last common ancestor of euphyllophytes more than 360 million years ago.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The raw sequencing data are available at E-MTAB-13848, and the coding and protein sequences are available via Figshare at https://doi.org/10.6084/m9.figshare.26347330 (ref. 164). The co-expression networks are available at https://conekt.sbs.ntu.edu.sg/species/.
Code availability
The code used to perform the analyses is available on request.
References
Kenrick, P. & Crane, P. R. The origin and early evolution of plants on land. Nature 389, 33–39 (1997).
Lloyd, R. M. Mating systems and genetic load in pioneer and non-pioneer Hawaiian Pteridophyta. Bot. J. Linn. Soc. 69, 23–35 (1974).
Ellwood, M. D. F. & Foster, W. A. Doubling the estimate of invertebrate biomass in a rainforest canopy. Nature 429, 549–551 (2004).
González de León, S., Briones, O., Aguirre, A., Mehltreter, K. & Pérez-García, B. Germination of an invasive fern responds better than native ferns to water and light stress in a Mexican cloud forest. Biol. Invasions 23, 3187–3199 (2021).
Pryer, K. M. et al. Phylogeny and evolution of ferns (monilophytes) with a focus on the early leptosporangiate divergences. Am. J. Bot. 91, 1582–1598 (2004).
PPG I. A community-derived classification for extant lycophytes and ferns. J. Syst. Evol. 54, 563–603 (2016).
Mehltreter, K. et al. (eds) Fern Ecology (Cambridge Univ. Press, 2010); https://doi.org/10.1017/CBO9780511844898
Page, C. N. Ecological strategies in fern evolution: a neopteridological overview. Rev. Palaeobot. Palynol. 119, 1–33 (2002).
Cao, H. et al. Phytochemicals from fern species: potential for medicine applications. Phytochem. Rev. 16, 379–440 (2017).
Goswami, H. K., Sen, K. & Mukhopadhyay, R. Pteridophytes: evolutionary boon as medicinal plants. Plant Genet. Resour. 14, 328–355 (2016).
Shukla, A. K. et al. Expression of an insecticidal fern protein in cotton protects against whitefly. Nat. Biotechnol. 34, 1046–1051 (2016).
Shen, H. et al. Large-scale phylogenomic analysis resolves a backbone phylogeny in ferns. GigaScience 7, gix116 (2018).
Qi, X. et al. A well-resolved fern nuclear phylogeny reveals the evolution history of numerous transcription factor families. Mol. Phylogenet. Evol. 127, 961–977 (2018).
Nitta, J. H., Schuettpelz, E., Ramírez-Barahona, S. & Iwasaki, W. An open and continuously updated fern tree of life. Front. Plant Sci. 13, 909768 (2022).
Rensing, S. A. Why we need more non-seed plant models. New Phytol. 216, 355–360 (2017).
Christenhusz, M. J. M. & Chase, M. W. Trends and concepts in fern classification. Ann. Bot. 113, 571–594 (2014).
Testo, W. & Sundue, M. A 4000-species dataset provides new insight into the evolution of ferns. Mol. Phylogenet. Evol. 105, 200–211 (2016).
Clark, J. W. Genome evolution in plants and the origins of innovation. New Phytol. 240, 2204–2209 (2023).
Chomicki, G., Coiro, M. & Renner, S. S. Evolution and ecology of plant architecture: integrating insights from the fossil record, extant morphology, developmental genetics and phylogenies. Ann. Bot. 120, 855–891 (2017).
Weng, J.-K. & Chapple, C. The origin and evolution of lignin biosynthesis. New Phytol. 187, 273–285 (2010).
Rencoret, J. et al. New insights on structures forming the lignin-like fractions of ancestral plants. Front. Plant Sci. 12, 740923 (2021).
Renault, H., Werck-Reichhart, D. & Weng, J.-K. Harnessing lignin evolution for biotechnological applications. Curr. Opin. Biotechnol. 56, 105–111 (2019).
Cronk, Q. C. B. Plant evolution and development in a post-genomic context. Nat. Rev. Genet. 2, 607–619 (2001).
Fernández, P. et al. A 160 Gbp fork fern genome shatters size record for eukaryotes. iScience 27, 109889 (2024).
Khandelwal, S. Chromosome evolution in the genus Ophioglossum L. Bot. J. Linn. Soc. 102, 205–217 (1990).
Marchant, D. B. et al. Dynamic genome evolution in a model fern. Nat. Plants 8, 1038–1051 (2022).
Klekowski, E. J. & Baker, H. G. Evolutionary significance of polyploidy in the pteridophyta. Science 153, 305–307 (1966).
Haufler, C. H. Ever since Klekowski: testing a set of radical hypotheses revives the genetics of ferns and lycophytes. Am. J. Bot. 101, 2036–2042 (2014).
Clark, J. et al. Genome evolution of ferns: evidence for relative stasis of genome size across the fern phylogeny. New Phytol. 210, 1072–1082 (2016).
Wood, T. E. et al. The frequency of polyploid speciation in vascular plants. Proc. Natl Acad. Sci. USA 106, 13875–13879 (2009).
Nakazato, T., Barker, M. S., Rieseberg, L. H. & Gastony, G. J. in Biology and Evolution of Ferns and Lycophytes (eds Haufler, C. H. & Ranker, T. A.) 175–198 (Cambridge Univ. Press, 2008); https://doi.org/10.1017/CBO9780511541827.008
Barker, M. S. in Plant Genome Diversity Vol. 2: Physical Structure, Behaviour and Evolution of Plant Genomes (eds Greilhuber, J. et al.) 245–253 (Springer, 2013); https://doi.org/10.1007/978-3-7091-1160-4_15
Baniaga, A. E. & Barker, M. S. Nuclear genome size is positively correlated with median LTR-RT insertion time in fern and lycophyte genomes. Am. Fern J. 109, 248–266 (2019).
Huang, C.-H., Qi, X., Chen, D., Qi, J. & Ma, H. Recurrent genome duplication events likely contributed to both the ancient and recent rise of ferns. J. Integr. Plant Biol. 62, 433–455 (2020).
Soltis, D. E. & Soltis, P. S. Polyploidy and breeding systems in homosporous pteridophyta: a reevaluation. Am. Nat. 130, 219–232 (1987).
Nakazato, T., Jung, M.-K., Housworth, E. A., Rieseberg, L. H. & Gastony, G. J. Genetic map-based analysis of genome structure in the homosporous fern Ceratopteris richardii. Genetics 173, 1585–1597 (2006).
Marchant, D. B. et al. The C-Fern (Ceratopteris richardii) genome: insights into plant genome evolution with the first partial homosporous fern genome assembly. Sci. Rep. 9, 18181 (2019).
Huang, X. et al. The flying spider-monkey tree fern genome provides insights into fern evolution and arborescence. Nat. Plants 8, 500–512 (2022).
Li, F.-W. et al. Fern genomes elucidate land plant evolution and cyanobacterial symbioses. Nat. Plants 4, 460–472 (2018).
Fang, Y. et al. The genome of homosporous maidenhair fern sheds light on the euphyllophyte evolution and defences. Nat. Plants 8, 1024–1037 (2022).
Zhong, Y. et al. Genomic insights into genetic diploidization in the homosporous fern Adiantum nelumboides. Genome Biol. Evol. 14, evac127 (2022).
Leebens-Mack, J. H. et al. One thousand plant transcriptomes and the phylogenomics of green plants. Nature 574, 679–685 (2019).
Rahmatpour, N. et al. Analyses of Marsilea vestita genome and transcriptomes do not support widespread intron retention during spermatogenesis. New Phytol. 237, 1490–1494 (2023).
Vanneste, K., Sterck, L., Myburg, A. A., Van de Peer, Y. & Mizrachi, E. Horsetails are ancient polyploids: evidence from Equisetum giganteum. Plant Cell 27, 1567–1578 (2015).
Zhang, C. & Mirarab, S. ASTRAL-Pro 2: ultrafast species tree reconstruction from multi-copy gene family trees. Bioinformatics 38, 4949–4950 (2022).
Emms, D. M. & Kelly, S. STAG: species tree inference from all genes. Preprint at bioRxiv https://doi.org/10.1101/267914 (2018).
Le, S. Q. & Gascuel, O. An improved general amino acid replacement matrix. Mol. Biol. Evol. 25, 1307–1320 (2008).
Wang, F.-G. et al. Genome size evolution of the extant lycophytes and ferns. Plant Divers. 44, 141–152 (2022).
Chen, H. et al. Revisiting ancient polyploidy in leptosporangiate ferns. New Phytol. 237, 1405–1417 (2023).
Pelosi, J. A., Kim, E. H., Barbazuk, W. B. & Sessa, E. B. Phylotranscriptomics illuminates the placement of whole genome duplications and gene retention in ferns. Front. Plant Sci. 13, 882441 (2022).
Julca, I. et al. Comparative transcriptomic analysis reveals conserved programmes underpinning organogenesis and reproduction in land plants. Nat. Plants 7, 1143–1159 (2021).
Feng, X. et al. Genomes of multicellular algal sisters to land plants illuminate signaling network evolution. Nat. Genet. 56, 1018–1031 (2024).
Amorim-Silva, V. et al. TTL proteins scaffold brassinosteroid signaling components at the plasma membrane to optimize signal transduction in Arabidopsis. Plant Cell 31, 1807–1828 (2019).
Kazan, K. A new twist in SA signalling. Nat. Plants 4, 327–328 (2018).
Li, F.-W. et al. Anthoceros genomes illuminate the origin of land plants and the unique biology of hornworts. Nat. Plants 6, 259–272 (2020).
Monte, I. et al. Ligand-receptor co-evolution shaped the jasmonate pathway in land plants. Nat. Chem. Biol. 14, 480–488 (2018).
Piatkowski, B. T. et al. Phylogenomics reveals convergent evolution of red-violet coloration in land plants and the origins of the anthocyanin biosynthetic pathway. Mol. Phylogenet. Evol. 151, 106904 (2020).
Davies, K. M. et al. Evolution and function of red pigmentation in land plants. Ann. Bot. 130, 613–636 (2022).
Güngör, E. et al. Azolla ferns testify: seed plants and ferns share a common ancestor for leucoanthocyanidin reductase enzymes. New Phytol. 229, 1118–1132 (2021).
Hõrak, H., Kollist, H. & Merilo, E. Fern stomatal responses to ABA and CO2 depend on species and growth conditions. Plant Physiol. 174, 672–679 (2017).
Usadel, B. et al. Co-expression tools for plant biology: opportunities for hypothesis generation and caveats. Plant Cell Environ. 32, 1633–1651 (2009).
Pomar, F., Merino, F. & Barceló, A. R. O-4-Linked coniferyl and sinapyl aldehydes in lignifying cell walls are the main targets of the Wiesner (phloroglucinol–HCl) reaction. Protoplasma 220, 17–28 (2002).
Blaschek, L. et al. Cellular and genetic regulation of coniferaldehyde incorporation in lignin of herbaceous and woody plants by quantitative Wiesner staining. Front. Plant Sci. 11, 109 (2020).
Yamashita, D., Kimura, S., Wada, M. & Takabe, K. Improved Mäule color reaction provides more detailed information on syringyl lignin distribution in hardwood. J. Wood Sci. 62, 131–137 (2016).
Logan, K. J. & Thomas, B. A. Distribution of lignin derivatives in plants. New Phytol. 99, 571–585 (1985).
Lu, F., Wang, C., Chen, M., Yue, F. & Ralph, J. A facile spectroscopic method for measuring lignin content in lignocellulosic biomass. Green Chem. 23, 5106–5112 (2021).
Lapierre, C., Pollet, B. & Rolando, C. New insights into the molecular architecture of hardwood lignins by chemical degradative methods. Res. Chem. Intermed. 21, 397–412 (1995).
Kairouani, A. et al. Cell-type-specific control of secondary cell wall formation by Musashi-type translational regulators in Arabidopsis. eLife 12, RP88207 (2023).
Proost, S. & Mutwil, M. CoNekT: an open-source framework for comparative genomic and transcriptomic network analyses. Nucleic Acids Res. 46, W133–W140 (2018).
Weng, J. K., Li, X., Stout, J. & Chapple, C. Independent origins of syringyl lignin in vascular plants. Proc. Natl. Acad. Sci. USA 105, 7887–7892 (2008).
Weng, J. K. et al. Convergent evolution of syringyl lignin biosynthesis via distinct pathways in the lycophyte Selaginella and flowering plants. Plant Cell 22, 1033–1045 (2010).
Werck-Reichhart, D. & Feyereisen, R. Cytochromes P450: a success story. Genome Biol. 1, reviews3003.1 (2000).
Ferrari, C. et al. Expression atlas of Selaginella moellendorffii provides insights into the evolution of vasculature, secondary metabolism, and roots. Plant Cell https://doi.org/10.1105/tpc.19.00780 (2020).
Biswal, A. K. et al. Comparison of four glycosyl residue composition methods for effectiveness in detecting sugars from cell walls of dicot and grass tissues. Biotechnol. Biofuels 10, 182 (2017).
Mueller, K.-K. et al. Fern cell walls and the evolution of arabinogalactan proteins in streptophytes. Plant J. 114, 875–894 (2023).
Urbanowicz, B. R. et al. 4-O-Methylation of glucuronic acid in Arabidopsis glucuronoxylan is catalyzed by a domain of unknown function family 579 protein. Proc. Natl Acad. Sci. USA 109, 14253–14258 (2012).
Cantarel, B. L. et al. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37, D233–D238 (2009).
Sampedro, J. et al. AtBGAL10 is the main xyloglucan β-galactosidase in Arabidopsis, and its absence results in unusual xyloglucan subunits and growth defects. Plant Physiol. 158, 1146–1157 (2012).
Gille, S. et al. O-acetylation of Arabidopsis hemicellulose xyloglucan requires AXY4 or AXY4L, proteins with a TBL and DUF231 domain. Plant Cell 23, 4041–4053 (2011).
Rocha, J. et al. Structure of Arabidopsis thaliana FUT1 reveals a variant of the GT-B class fold and provides insight into xyloglucan fucosylation. Plant Cell 28, 2352–2364 (2016).
Mikkelsen, M. D. et al. Ancient origin of fucosylated xyloglucan in charophycean green algae. Commun. Biol. 4, 754 (2021).
Yuan, Y. et al. Mutations of Arabidopsis TBL32 and TBL33 affect xylan acetylation and secondary wall deposition. PLoS ONE 11, e0146460 (2016).
Jensen, J. K. et al. Identification of a xylogalacturonan xylosyltransferase involved in pectin biosynthesis in Arabidopsis. Plant Cell 20, 1289–1302 (2008).
Chiniquy, D. et al. PMR5, an acetylation protein at the intersection of pectin biosynthesis and defense against fungal pathogens. Plant J. Cell Mol. Biol. 100, 1022–1035 (2019).
Ishimaru, M., Smith, D. L., Mort, A. J. & Gross, K. C. Enzymatic activity and substrate specificity of recombinant tomato beta-galactosidases 4 and 5. Planta. 229, 447–456 (2009).
Temple, H. et al. Two members of the DUF579 family are responsible for arabinogalactan methylation in Arabidopsis. Plant Direct 3, e00117 (2019).
Zhong, R., Cui, D. & Ye, Z.-H. Members of the DUF231 family are O-acetyltransferases catalyzing 2-O- and 3-O-acetylation of mannan. Plant Cell Physiol. 59, 2339–2349 (2018).
Moller, I. et al. High-throughput mapping of cell-wall polymers within and between plants using novel microarrays. Plant J. Cell Mol. Biol. 50, 1118–1128 (2007).
Fry, S. C., Nesselrode, B. H. W. A., Miller, J. G. & Mewburn, B. R. Mixed-linkage (1→3,1→4)-beta-d-glucan is a major hemicellulose of Equisetum (horsetail) cell walls. New Phytol. 179, 104–115 (2008).
Sørensen, I. et al. Mixed-linkage (1→3),(1→4)-beta-d-glucan is not unique to the Poales and is an abundant component of Equisetum arvense cell walls. Plant J. Cell Mol. Biol. 54, 510–521 (2008).
Silva, G. B. et al. Cell wall polysaccharides from fern leaves: evidence for a mannan-rich Type III cell wall in Adiantum raddianum. Phytochemistry 72, 2352–2360 (2011).
Fry, S. C. Feruloylated pectins from the primary cell wall: their structure and possible functions. Planta 157, 111–123 (1983).
Lampugnani, E. R. et al. Cellulose synthesis—central components and their evolutionary relationships. Trends Plant Sci. 24, 402–412 (2019).
Tsekos, I. The sites of cellulose synthesis in algae: diversity and evolution of cellulose-synthesizing enzyme complexes. J. Phycol. 35, 635–655 (1999).
Brown, R. M. Jr The biosynthesis of cellulose. J. Macromol. Sci. A 33, 1345–1373 (1996).
Seifriz, W. The origin, composition, and structure of cellulose in the living plant. Protoplasma 21, 129–159 (1934).
Pear, J. R., Kawagoe, Y., Schreckengost, W. E., Delmer, D. P. & Stalker, D. M. Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc. Natl. Acad. Sci. USA 93, 12637–12642 (1996).
Harholt, J. et al. The glycosyltransferase repertoire of the spikemoss Selaginella moellendorffii and a comparative study of its cell wall. PLoS ONE 7, e35846 (2012).
Yin, Y., Huang, J. & Xu, Y. The cellulose synthase superfamily in fully sequenced plants and algae. BMC Plant Biol. 9, 99 (2009).
Goubet, F. et al. Cell wall glucomannan in Arabidopsis is synthesised by CSLA glycosyltransferases, and influences the progression of embryogenesis. Plant J. Cell Mol. Biol. 60, 527–538 (2009).
Cocuron, J.-C. et al. A gene from the cellulose synthase-like C family encodes a β-1,4 glucan synthase. Proc. Natl Acad. Sci. USA 104, 8550–8555 (2007).
Wang, W. et al. Arabidopsis CSLD1 and CSLD4 are required for cellulose deposition and normal growth of pollen tubes. J. Exp. Bot. 62, 5161–5177 (2011).
Bernal, A. J. et al. Functional analysis of the cellulose synthase-like genes CSLD1, CSLD2, and CSLD4 in tip-growing Arabidopsis cells. Plant Physiol. 148, 1238–1253 (2008).
Burton, R. A. et al. Cellulose synthase-like CslF genes mediate the synthesis of cell wall (1,3;1,4)-beta-d-glucans. Science 311, 1940–1942 (2006).
Liu, X. et al. Genome-wide bioinformatics analysis of cellulose synthase gene family in common bean (Phaseolus vulgaris L.) and the expression in the pod development. BMC Genom. Data 23, 9 (2022).
Bringmann, M. et al. POM-POM2/CELLULOSE SYNTHASE INTERACTING1 is essential for the functional association of cellulose synthase and microtubules in Arabidopsis. Plant Cell 24, 163–177 (2012).
Wang, Y. et al. LACCASE5 is required for lignification of the Brachypodium distachyon Culm. Plant Physiol. 168, 192–204 (2015).
Höfer, R. et al. Dual function of the cytochrome P450 CYP76 family from Arabidopsis thaliana in the metabolism of monoterpenols and phenylurea herbicides. Plant Physiol. 166, 1149–1161 (2014).
Rothfels, C. J. et al. The evolutionary history of ferns inferred from 25 low-copy nuclear genes. Am. J. Bot. 102, 1089–1107 (2015).
Morris, J. L. et al. The timescale of early land plant evolution. Proc. Natl Acad. Sci. USA 115, E2274–E2283 (2018).
Herendeen, P. S., Friis, E. M., Pedersen, K. R. & Crane, P. R. Palaeobotanical redux: revisiting the age of the angiosperms. Nat. Plants 3, 17015 (2017).
Condamine, F. L., Silvestro, D., Koppelhus, E. B. & Antonelli, A. The rise of angiosperms pushed conifers to decline during global cooling. Proc. Natl. Acad. Sci. USA 117, 28867–28875 (2020).
Bowman, J. L. et al. Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell 171, 287–304.e15 (2017).
Weng, J.-K., Banks, J. A. & Chapple, C. Parallels in lignin biosynthesis: a study in Selaginella moellendorffii reveals convergence across 400 million years of evolution. Commun. Integr. Biol. 1, 20–22 (2008).
Leroux, O. et al. Antibody-based screening of cell wall matrix glycans in ferns reveals taxon, tissue and cell-type specific distribution patterns. BMC Plant Biol. 15, 56 (2015).
Bartels, D. & Classen, B. Structural investigations on arabinogalactan-proteins from a lycophyte and different monilophytes (ferns) in the evolutionary context. Carbohydr. Polym. 172, 342–351 (2017).
Roberts, A. W. et al. Functional characterization of a glycosyltransferase from the moss Physcomitrella patens involved in the biosynthesis of a novel cell wall arabinoglucan. Plant Cell 30, 1293–1308 (2018).
Taketa, S. et al. Functional characterization of barley betaglucanless mutants demonstrates a unique role for CslF6 in (1,3;1,4)-β-d-glucan biosynthesis. J. Exp. Bot. 63, 381–392 (2012).
Sperry, J. S. Evolution of water transport and xylem structure. Int. J. Plant Sci. 164, S115–S127 (2003).
Baas, P. & Wheeler, E. A. Parallelism and reversibility in xylem evolution: a review. IAWA J. https://doi.org/10.1163/22941932-90000633 (1996).
Ruprecht, C. et al. Famnet: a framework to identify multiplied modules driving pathway expansion in plants. Plant Physiol. 170, 1878–1894 (2016).
Ruprecht, C. et al. Phylogenomic analysis of gene co-expression networks reveals the evolution of functional modules. Plant J. 90, 447–465 (2017).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018).
Luo, R. et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience 1, 18 (2012).
Grabherr, M. G. et al. Trinity: reconstructing a full-length transcriptome without a genome from RNA-seq data. Nat. Biotechnol. 29, 644–652 (2011).
Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V. & Zdobnov, E. M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212 (2015).
Bray, N. L., Pimentel, H., Melsted, P. & Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527 (2016).
Sensalari, C., Maere, S. & Lohaus, R. ksrates: positioning whole-genome duplications relative to speciation events in KS distributions. Bioinformatics 38, 530–532 (2022).
Camacho, C. et al. BLAST+: architecture and applications. BMC Bioinform. 10, 421 (2009).
Dongen, S. M. van. Graph Clustering by Flow Simulation. PhD dissertation, Utrecht Univ. (2000); https://dspace.library.uu.nl/handle/1874/848
Edgar, R. C. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 5, 113 (2004).
Yang, Z. PAML 4: Phylogenetic Analysis by Maximum Likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007).
Zwaenepoel, A. & Van de Peer, Y. wgd—simple command line tools for the analysis of ancient whole-genome duplications. Bioinformatics 35, 2153–2155 (2019).
Price, M. N., Dehal, P. S. & Arkin, A. P. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS ONE 5, e9490 (2010).
Proost, S. et al. i-ADHoRe 3.0—fast and sensitive detection of genomic homology in extremely large data sets. Nucleic Acids Res. 40, e11 (2012).
Fu, L., Niu, B., Zhu, Z., Wu, S. & Li, W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152 (2012).
Emms, D. M. & Kelly, S. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16, 157 (2015).
Steenwyk, J. L. et al. OrthoSNAP: a tree splitting and pruning algorithm for retrieving single-copy orthologs from gene family trees. PLoS Biol. 20, e3001827 (2022).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Capella-Gutiérrez, S., Silla-Martínez, J. M. & Gabaldón, T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 (2009).
Nguyen, L.-T., Schmidt, H. A., von Haeseler, A. & Minh, B. Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015).
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A. & Jermiin, L. S. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589 (2017).
Hoang, D. T., Chernomor, O., von Haeseler, A., Minh, B. Q. & Vinh, L. S. UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522 (2018).
Lehtonen, S. et al. Environmentally driven extinction and opportunistic origination explain fern diversification patterns. Sci. Rep. 7, 4831 (2017).
Zwaenepoel, A. & Van de Peer, Y. Inference of ancient whole-genome duplications and the evolution of gene duplication and loss rates. Mol. Biol. Evol. 36, 1384–1404 (2019).
Löytynoja, A. Phylogeny-aware alignment with PRANK. Methods Mol. Biol. 1079, 155–170 (2014).
Ronquist, F. et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542 (2012).
Szöllősi, G. J., Rosikiewicz, W., Boussau, B., Tannier, E. & Daubin, V. Efficient exploration of the space of reconciled gene trees. Syst. Biol. 62, 901–912 (2013).
Buchfink, B., Reuter, K. & Drost, H.-G. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat. Methods 18, 366–368 (2021).
Schwacke, R. et al. MapMan4: a refined protein classification and annotation framework applicable to multi-omics data analysis. Mol. Plant 12, 879–892 (2019).
Smith, A. R. et al. A classification for extant ferns. TAXON 55, 705–731 (2006).
Abramson, J. et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 630, 493–500 (2024).
Holm, L. Dali server: structural unification of protein families. Nucleic Acids Res. 50, W210–W215 (2022).
van Kempen, M. et al. Fast and accurate protein structure search with Foldseek. Nat. Biotechnol. 42, 243–246 (2024).
Gouy, M., Tannier, E., Comte, N. & Parsons, D. P. Seaview version 5: a multiplatform software for multiple sequence alignment, molecular phylogenetic analyses, and tree reconciliation. Methods Mol. Biol. 2231, 241–260 (2021).
Hoebler, C., Barry, J. L., David, A. & Delort-Laval, J. Rapid acid hydrolysis of plant cell wall polysaccharides and simplified quantitative determination of their neutral monosaccharides by gas–liquid chromatography. J. Agric. Food Chem. 37, 360–367 (1989).
Blakeney, A. B., Harris, P. J., Henry, R. J. & Stone, B. A. A simple and rapid preparation of alditol acetates for monosaccharide analysis. Carbohydr. Res. 113, 291–299 (1983).
Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549 (2018).
Mutwil, M. et al. Assembly of an interactive correlation network for the Arabidopsis genome using a novel heuristic clustering algorithm. Plant Physiol. 152, 29–43 (2010).
Zhang, H. et al. dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 46, W95–W101 (2018).
Letunic, I. & Bork, P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296 (2021).
Johnson, K. L. et al. Pipeline to identify hydroxyproline-rich glycoproteins. Plant Physiol. 174, 886–903 (2017).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B Methodol. 57, 289–300 (1995).
Ali, Z. CDS and PEP files. Figshare https://doi.org/10.6084/m9.figshare.26347330 (2024).
Acknowledgements
We acknowledge J. Fangel for contributions to Fig. 4e and S. Daniel for help with lignin biochemistry. We thank D. Maizels (http://www.scientific-art.com/) for the illustrations in Fig. 1. M.M. acknowledges funding from Singaporean Ministry of Education grant no. MOE-MOET32022-0002 ‘From tough pollen to soft matter’ and a Novo Nordisk Starting Grant. L.P. and B.C. (project no. 440046237) and J.d.V. (project no. 440231723; VR 132/4-2) acknowledge funding within the framework of MAdLand (http://madland.science), priority programme 2237 of the German Research Foundation (DFG). J.d.V. further thanks the European Research Council for funding under the European Union’s Horizon 2020 research and innovation programme (grant agreement no. 852725; ERC-StG ‘TerreStriAL’). S.d.V. acknowledges funding from the Lower Saxony Ministry of Science and Culture (Niedersachsen Vorab initiative) and DFG project no. 515101361. We thank L. Saulnier for discussion and help with identifying the unknown sugar. We also thank E. Haswell (https://elizabethhaswell.carrd.co) for her help with proofreading the manuscript. Finally, we thank J. C. Goh for his help in starting the project.
Author information
Authors and Affiliations
Contributions
Z.M.A. and B.C.H. were involved in the sampling of ferns. Z.M.A., Q.W.T., H.C., P.K.L., I.J., J.M.L., S.d.V., J.d.V., E.M. and Y.V.d.P. were involved in the bioinformatical analysis of the data. L.P. and B.C. were involved in the GC–MS and bioinformatical analyses. F.V., C.A., A.L. and R.S. were involved in tissue sectioning and microscopy and the lignin and sugar analyses. M.S.M. performed the sugar synthesis and analysis. B.J. and P.U. performed the CoMPP analysis. Z.M.A. and M.M. wrote the paper with help from all authors. M.M. conceptualized and supervised the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Plants thanks Thais Almeida and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–19.
Supplementary Information
Supplementary Methods.
Supplementary Data 1
Co-expression networks.
Supplementary Tables
Supplementary Tables 1–15.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ali, Z., Tan, Q.W., Lim, P.K. et al. Comparative transcriptomics in ferns reveals key innovations and divergent evolution of the secondary cell walls. Nat. Plants 11, 1028–1048 (2025). https://doi.org/10.1038/s41477-025-01978-y
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41477-025-01978-y
This article is cited by
-
Exploring fern pathosystems and immune receptors to bridge gaps in plant immunity
BMC Biology (2025)
-
Decoding the genome of Brainea insignis reveals insights into fern evolution and conservation
Nature Communications (2025)